Abstract
To better understand the roles of Sm proteins in forming the cores of many RNA-processing ribonucleoproteins, we determined the crystal structure of an atypical Sm-like archaeal protein (SmAP3) in which the conserved Sm domain is augmented by a previously uncharacterized, mixed α/β C-terminal domain. The structure reveals an unexpected SmAP3 14-mer that is perforated by a cylindrical pore and is bound to 14 cadmium (Cd2+) ions. Individual heptamers adopt either “apical” or “equatorial” conformations that chelate Cd2+ differently. SmAP3 forms supraheptameric oligomers (SmAP3)n = 7,14,28 in solution, and assembly of the asymmetric 14-mer is modulated by differential divalent cation-binding in apical and equatorial subunits. Phylogenetic and sequence analyses substantiate SmAP3s as a unique subset of SmAPs. These results distinguish SmAP3s from other Sm proteins and provide a model for the structure and properties of Sm proteins >100 residues in length, e.g., several human Sm proteins.
Sm proteins are key components of the ribonucleoprotein (RNP) assemblies that are required for high-fidelity cellular RNA processing, including rRNA and tRNA processing, mRNA decapping and decay, and intron splicing in pre-mRNA (1). Although the primary functions of Sm⋅⋅⋅RNA interactions in many RNPs are unclear, the importance of Sm proteins is illustrated by their roles in forming the cores of the uridine-rich small nuclear RNPs (U snRNPs) that further assemble into a large, transiently stable spliceosome that catalyzes the final step of eukaryotic pre-mRNA processing (intron excision/exon ligation; ref. 2). Each spliceosomal snRNP consists of a unique snRNA and up to dozens of snRNP-specific proteins (3). The only subset of proteins common to different snRNPs is the archetypal Sm heteroheptamer (e.g., human Sm D1⋅D2⋅F⋅E⋅G⋅D3⋅B/B′; ref. 4), which assembles snRNP cores by binding to uridine-rich, single-stranded regions of snRNA via discrete intermediates (e.g., D3⋅B, D1⋅D2, and E⋅F⋅G heteromers; ref. 5). Sm proteins also may function as nuclear localization signals and in hypermethylation of snRNA caps (6). Functional complexes of RNA and homologous Sm protein septets, such as the Sm-like (Lsm) 1 → 7 and Lsm 2 → 8 paralogs of yeast, are thought to assemble in a similar manner as canonical Sm proteins (7).
The Sm domain is highly conserved across many species, and its widespread phylogenetic distribution suggests its importance in the early evolution of RNA metabolism. Homologs of snRNP core Sm proteins occur in eukaryotes ranging from yeast to human. The existence of Sm-like archaeal proteins (SmAPs) implies an ancient origin of Sm proteins (8–10). Most recently, the Escherichia coli Hfq protein (a host factor required for bacteriophage replication) and its orthologs were shown to be Sm-like proteins in several eubacterial lineages (11), thus expanding the scope of possible Sm-like functions to include nonsplicing roles (12). Organisms may contain from ≈1–3 Sm-like proteins (as in archaea) to multiple sets of seven Sm and Lsm proteins (as in humans).
Crystal structures of Sm, Lsm, and SmAP domains reveal a highly conserved fold, capable of forming a variety of cyclic quaternary structures. Sm proteins exhibit a strong tendency to assemble into circular heptamers (13), and cryo-electron microscopic reconstructions confirm the ring-shaped Sm core of the U1 snRNP (14). However, Sm proteins are not restricted to heptamers: the eubacterial Lsm protein Hfq forms a cyclic homohexamer, and Archaeoglobus fulgidus (Afu) SmAP2 may form cyclic hexamers (15) and heptamers (16). The structures of two human Sm heterodimers (17), the Hfq protein (18), and several homoheptameric SmAPs (Fig. 1, boldface; refs. 8–10 and 15) show that Sm monomers fold as a highly bent, five-stranded antiparallel β-sheet capped by an N-terminal α-helix, and form oligomers that surround a conserved cationic pore. The inner surface of this pore seems to be the oligouridine-binding site (9, 18, 19). We now report the crystal structure and metal-dependent assembly properties of an unexpected SmAP3 14-mer from the hyperthermophilic crenarchaeote Pyrobaculum aerophilum (Pae).
Fig 1.
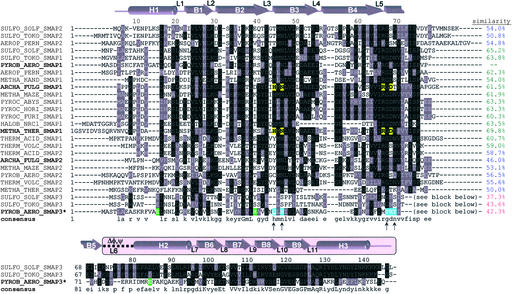
Sequence analysis and secondary structure of SmAPs. A composite multiple sequence alignment of SmAPs shows that the conserved Sm domain of ≈70 residues is augmented by a 60-residue, mixed α/β CTD (pink-shaded background) that is highly conserved in the SmAP3 subfamily. This CTD is linked to the Sm domain by an ≈8-residue variable loop (L6) whose conformation differs between apical and equatorial monomers (Δφ,ψ). Residue numbers and secondary structure elements (top line) are depicted for the Pae SmAP3 monomer. Pairwise sequence similarity scores to Pae SmAP1 discriminate the SmAP1 (green), SmAP2 (blue), and SmAP3 (red) subfamilies. Highlighted residues are likely involved in binding RNA (yellow, blue) or divalent cation (green), as discussed in the text; arrows indicate the positions of uridine-binding residues in the consensus sequence.
Methods
Protein Preparation and Crystallization.
Recombinant Pae SmAP3 (14.6 kDa) containing a protease-sensitive C-terminal His6-tag was produced by standard PCR cloning and overexpression methods in E. coli, and was purified by heat treatment of cell lysate, Ni2+-affinity chromatography, tryptic removal of the tag, and cation exchange chromatography. SeMet-substituted SmAP3 was prepared by overexpression in M9 minimal media containing SeMet (20). Diffraction-quality crystals of SeMet-Pae SmAP3 (≈85 mg/ml) were grown by vapor diffusion only after addition of 20 mM uridine-5′-monophosphate (UMP) and 10 mM CdCl2 to hanging drops (see text; ref. 21), and a monoclinic crystal (P21; a = 83.32 Å, b = 172.43 Å, c = 148.11 Å, β = 89.99°) was used to determine the structure reported here by multiwavelength anomalous dispersion (MAD) phasing. Evaluation of noncrystallographic symmetry (NCS) via native Patterson and self-rotation functions, along with crystal packing considerations, revealed an asymmetric unit (a.u.) composed of 28 monomers (VM = 2.60 Å3/Da), arranged as two asymmetric 14-mers that are related by a pseudo 2-fold rotational symmetry axis.
Data Collection, MAD Phasing, and Refinement.
Diffraction data at three wavelengths were collected at 105 K (Table 1) and processed with denzo/scalepack (22). MAD phasing proceeded by usual methods for heavy atom (Se) location (shelxd; http://shelx.uni-ac.gwdg.de/SHELX/), phase refinement (mlphare; ref. 23), and density modification (24). All three MAD datasets were required for shelxd to find any Se sites (the single-wavelength anomalous scattering approach failed), and only 50 of the 84 Se sites per a.u. were used for initial phasing. Phase extension to 2.0 Å and further density modification in resolve (25) permitted automatic secondary structure fragment matching for roughly half of the 28-mer. Fortuitously, autobuilt residues were distributed randomly across the structure, so the NCS that relates the 28 monomers was taken advantage of to build a relatively complete composite model for the Sm domain of SmAP3. C-terminal domains (CTDs) in either apical or equatorial conformations were manually built into the improved maps. No NCS restraints were applied during refinement of the final 28-mer in CNS (26). Refinement rounds ended with inspection of the model and experimental electron density (Fo, φMAD), as well as σA-weighted 2Fo − Fc, Fo − Fc, and simulated annealing omit maps. The final model was validated by Ramachandran analysis and programs such as errat (27), and contains 3,566 of 3,584 residues in the SmAP3 28-mer. Several heteroatoms and solvent molecules also were observed (Table 1), including the 28 Cd2+ ions that were first noticed as peaks >+4σ in anomalous difference electron density.
Table 1.
Data collection, phasing, and atomic refinement of Pae SmAP3
| Data set | Inflection | Peak | High-energy remote |
|---|---|---|---|
| Data collection & MAD phasing | |||
| Wavelength, Å | 0.97954 | 0.97933 | 0.97186 |
| Resolution range, Å | 100.0–2.0 | 100.0–2.0 | 100.0–2.0 |
| No. of reflections (total/unique) | 1,697,271/270,390 | 1,705,654/269,604 | 1,693,038/270,937 |
| Completeness, %* | 95.6 [73.3] | 95.8 [73.9] | 96.3 [76.3] |
| I/σ(I) | 13.4 [2.2] | 14.0 [2.7] | 14.5 [2.9] |
| Rmerge, %† | 12.5 [51.6] | 13.0 [50.4] | 11.0 [39.8] |
| No. of Se sites per a.u. (used/expected)‡ | — | 50/84 | — |
| Phasing resolution range, Å | 50.0–2.5 | 50.0–2.5 | 50.0–2.5 |
| Rcullis§ | |||
| acentric | — | 0.93/0.76 | 0.91/0.80 |
| centric | — | 0.91 | 0.88 |
| Figure of merit¶ | 0.48/0.82 | — | — |
| Model refinement | |||
| Resolution range (Å) | 20.0–2.0 | ||
| No. of reflections (total/test set) | 262,801/13,049 | ||
| No. of protein residues∥ | 3,566/3,584 | ||
| 〈B〉 (protein atoms, Å2) | 31.3 | ||
| No. of solvent molecules (〈B〉, Å2) | |||
| Water | 1,888 (35.6) | ||
| Acetate | 15 (32.0) | ||
| Glycerol | 12 (48.7) | ||
| Cd2+ | 28 (40.0) | ||
| Na+ | 5 (70.3) | ||
| rmsd (bonds/angles) | 0.018 Å/1.74° | ||
| Rcryst/Rfree, %** | 19.1/23.6 | ||
| PDB submission code |
Statistics for the highest resolution shell are given in square brackets.
Rmerge(I) = ∑hkl((∑i |Ihkl,i − 〈Ihkl〉|)/∑i Ihkl,i).
Number of Se sites calculated by shelxd and used for phasing (out of 84 sites expected per a.u.).
Rcullis = (∑hkl ∥FPH ± FP| − FH,calc|)/∑hkl |FPH ± FP|. Statistics for acentric reflections are given as isomorphous/anomalous.
Values are given before/after density modification and phase extension to 2.0 Å.
Number of SmAP3 residues built, out of 128 residues/monomer × 28 monomers = 3,584 expected residues.
Rcryst = ∑hkl ∥Fobs| − |Fcalc∥/∑hkl |Fobs|. Rfree was computed identically, except that 4.6% of the reflections were omitted as a test set.
Sequence and Structure Analyses.
A database of all 26 known SmAPs was compiled via iterative psi-blast (28) searches of a current, nonredundant database of protein sequences at the National Center for Biotechnology Information. Pae SmAP1 was used as a query, because searches with full-length Pae SmAP3 retrieved only the other two SmAP3s (Sulfolobus tokodaii and Sulfolobus solfataricus, Fig. 1). A composite multiple sequence alignment was constructed by separately aligning residues from the N-terminal Sm domains and C-terminal domains (using clustalw; ref. 29), because inclusion of SmAP3 CTD sequences results in inaccurate alignments (see text). Pairwise alignments were computed by the Smith–Waterman algorithm. Similar protein 3D structures [rms deviation (rmsd) < ≈2.5 Å] were superimposed by the Kabsch least squares method, whereas more dissimilar structures (rmsd > 2.5 Å by align) were optimally aligned by combinatorial extension (30). Ensemble structural analyses of SmAP3 monomers were performed via error-scaled difference distance matrices in the program escet (31). grasp (32) and cns (26) were used for electrostatic and surface area calculations, respectively.
Analytical Ultracentrifugation.
Sedimentation velocity runs were used to examine SeMet-Pae SmAP3 (1.35 mg/ml) in 10 mM Tris⋅Cl pH 7.81, 150 mM NaCl, supplemented with either (i) no divalent metal, (ii) 10 mM CdCl2, or (iii) 0.01, 0.10, 1.0, 3.0, 10, and 20 mM ZnCl2. Experiments were performed in a Beckman Optima XL-A analytical ultracentrifuge at 50,000 rpm and 20°C by using absorption optics at 280 nm and a 12-mm pathlength double sector cell. Sedimentation coefficients were determined from g(s) distribution plots by using the Beckman origin-based software (version 3.01). The peak sedimentation coefficient was corrected for density and viscosity to an S20,wat value by using a value for the partial specific volume at 20°C of 0.743 (calculated from the amino acid composition and corrected to 20°C). Values were linearly scaled to a maximum g(S20,wat) = 1.0 for illustrative purposes (see Fig. 3d).
Fig 3.
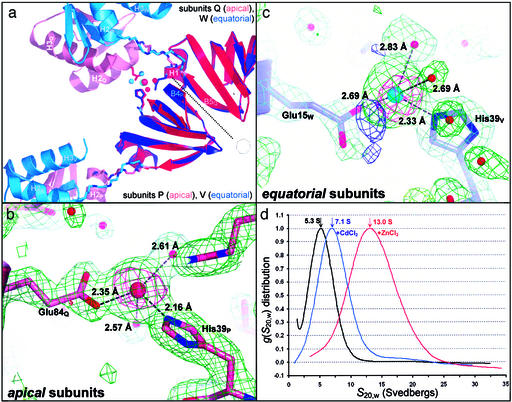
SmAP3 14-mer assembly is mediated by differential divalent cation-binding in apical and equatorial subunits. (a) The SmAP3 14-mer binds Cd2+ with a 1:1 stoichiometry, but binding sites differ in apical vs. equatorial heptamers. Ribbon diagrams are shown for a rigid-body superimposition of two adjacent SmAP3 apical subunits (P, Q) and two adjacent equatorial subunits (V, W), by using just the Sm core domain for least squares alignment (same coloring as Fig. 2). The Sm–Sm interface (β4P/V⋅⋅⋅β5Q/W, dotted line) and the location of the 7-fold axis/pore are indicated (heptagon). Stick models are shown for the backbones of the conformationally variant linker (Δφ,ψ in Fig. 1) and the following Cd2+-chelating residues: His-39V (equa), Glu-15W (equa), His-39P (apic), and Glu-84Q (apic). Cd2+ and waters are drawn as space-filling (apical, red; equatorial, blue). The distorted tetrahedral Cd2+-binding sites in apical (b) and equatorial (c) subunits are shown, along with 2Fo − Fc (+1.3σ, green), Fo − Fc (−3.5σ, red; +3.5σ, blue), and experimentally phased anomalous difference electron densities (ΔFano, +4.2σ, magenta). (d) The distribution of sedimentation coefficients for SmAP3 in the absence of divalent metal ion (5.3S, black) shifts to greater values on addition of 10 mM CdCl2 (7.1S, blue) or ZnCl2 (13.0S, red). As discussed in the text, these increases are incompatible with shape-only changes in SmAP3 heptamers and are likely due to divalent cation-mediated assembly of 14- and 28-mers.
Coordinates.
Final model coordinates and diffraction intensity data have been submitted to the Protein Data Bank (PDB ID code ).
Results
Structure of the SmAP3 14-mer.
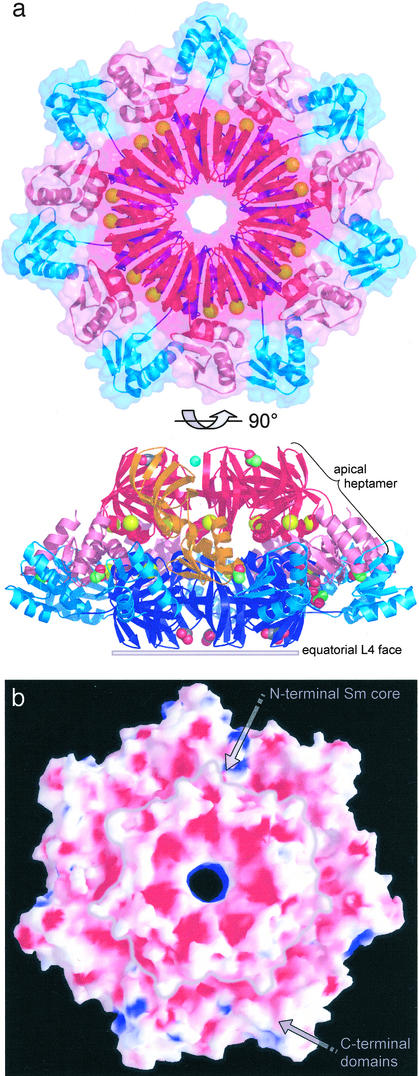
The Pae SmAP3 crystal structure reveals that SmAP3 forms homoheptamers that adopt one of two conformations (apical or equatorial) and assemble into 14-mers with a 1:1 stoichiometry. After using sequence analysis to identify the SmAP3 open reading frame as a potentially novel, extended Sm-like protein from Pae (Fig. 1), the structure of a SeMet-substituted version of Pae SmAP3 was determined by MAD phasing, and refined to 2.0-Å resolution (Table 1). The final model contains 28 SmAP3 monomers per a.u. and several types of solvent molecules and heteroatoms, including 28 Cd2+ ions (Fig. 2a). The 28 SmAP3 monomers are arranged as distinct 14-mers that scarcely contact one another and that are related by pseudo 2-fold rotational symmetry. The two 14-mers are nearly identical to within model coordinate error, and their overall shape can be described as a conical frustum ≈65 Å in height, ≈68 Å across the apical L4 face (top), and ≈120 Å in outer diameter (Fig. 2a, 90° view). The occluded surface area in the heptamerapic⋅⋅⋅heptamerequa interface (21,486 ± 44 Å2) supports the significance of the SmAP3 14-mer; this extensive interface is largely formed by apolar contacts and hydrogen bonds between the interdigitated apical and equatorial C-terminal domains (Fig. 2a).
Fig 2.
Overall structure of the SmAP3 14-mer. (a) A ribbon diagram of the Pae SmAP3 14-mer is shown, in which the Sm and C-terminal domains of the apical and equatorial heptamers are distinguished by various hues of red (apic) and blue (equa): Smapic (red), C-terminalapic (pink), Smequa (blue), and C-terminalequa (aqua). One apical SmAP3 monomer is highlighted (orange), and non-protein atoms (Table 1) are displayed as space-filling models: acetate (green carbons), glycerol (gray carbons), Na+ (blue), and Cd2+ (yellow, delimiting the Sm and C-terminal domains). Massive deviation of the 14-mer from D7 point group symmetry is clear from the view orthogonal to the 7-fold axis, and from the differing Cd2+-binding sites in the apic and equa heptamers. A transparent molecular surface of the 14-mer, viewed down the apical face, illustrates that the C-terminal domains are largely responsible for the extensive heptamerapic–heptamerequa interface. (b) The equatorial face of the 14-mer is shown, colored by the electrostatic potential at the molecular surface (red = −9.5 kT, blue = +10.5 kT); a gray line indicates the border of the Sm core of the equatorial L4 face (as in a). The intense negative potential across most of this face is found in other SmAPs and is likely to be important in controlling SmAP⋅⋅⋅RNA interactions.
Pae SmAP3 monomers exhibit remarkable structural plasticity, as indicated by the assembly of a 14-mer from two heptamers that are in different conformations. The bipartite SmAP3 monomer consists of an N-terminal Sm domain (80-residues) augmented by a 60-residue C-terminal domain (Fig. 1). The backbone of the ≈8-residue segment that links the Sm and C-terminal domains (Fig. 1, Δφ,ψ) adopts one of two conformations (Fig. 3a): in the equatorial conformer, the Δφ,ψ linker (centered on Asp74-Ser75-Thr76) positions the CTDs nearly coplanar with the disk-shaped Sm core heptamer whereas, in the more compact apical conformer, the CTDs are angled by ≈30° toward the equatorial domain (with respect to a plane drawn through the apical Sm core heptamer; Fig. 2a). The apical loop-L4 face effectively caps the 14-mer and breaks the dihedral point group symmetry seen in 14-mers of other Sm proteins (33) and 22-mers of trp RNA-binding attenuation protein (TRAP), another single-stranded RNA-binding protein (34); the 14-mer retains C7 symmetry. Beyond the two Δφ,ψ conformations, the hierarchic assembly mechanism of SmAP3 14-mers is unclear. For example, do apical heptamers assemble from preexisting apical SmAP3 monomers, or do mixed-conformation states exist, and undergo a cooperative transition to all-apical or all-equatorial heptamers? Heptamerapic ↔ heptamerequa transitions may be modulated by differential divalent cation-binding in the Sm and C-terminal domains, as discussed below.
The Sm portion of Pae SmAP3 is nearly identical to the oligonucleotide/oligosaccharide-binding fold of known Sm structures, whereas the CTD is atypical. Root mean square deviations (rmsd) for Cα positions between the Sm domain of SmAP3 and other Sm domains vary from 0.9 Å (against Pae SmAP1 and human Sm D3) to 1.5 Å [against Methanobacterium thermautotrophicum (Mth) SmAP1], with most of the variation found in loop L4 (Fig. 1). The core Sm heptamer of SmAP3 and other Sm heptamers superimpose with similarly small rmsd. Aside from large-scale differences due to the variable Δφ,ψ segment, minor structural variation within the ensemble of 28 independently refined SmAP3 monomers was analyzed via error-scaled difference distance matrices (31); notably, monomers can be grouped into heptamers correctly (i.e., as they occur in the structure), solely on the basis of pairwise rmsd. The mean rmsd for all pairwise alignments is slightly higher for the C-terminal domains (0.4 Å on all atoms) than the Sm domains (0.2 Å), consistent with their location on the periphery of the 14-mer and with crystallographic temperature factors. Overall structural features that are conserved between SmAP3 and other Sm proteins are: (i) the orientation of heptamers in the 14-mer (exposing the loop L4 faces) and (ii) the calculated electrostatic potential at the SmAP3 surface, which reveals a highly acidic L4 face (Fig. 2b), as found for Mth (8) and Pae (10) SmAP1, and Afu SmAP2 (15). The pronounced electrostatic asymmetries of SmAP surfaces may modulate putative SmAP⋅⋅⋅RNA interactions, e.g., by minimizing unproductive RNA-binding near the acidic loop L4 face.
A Partially Conserved Ligand-Binding Site.
The SmAP3 14-mer surrounds a cylindrical pore that may form a single-stranded RNA (ssRNA)-binding site, similar to that found in other SmAPs. However, the geometric and chemical features of this partially conserved ligand-binding site distinguish SmAP3 from known Sm structures. In addition to being slightly more acidic than the highly cationic pores found in other SmAPs and Hfq (Fig. 2b), the SmAP3 pore is slightly wider. The hourglass-shaped pores that coincide with the 6- or 7-fold axis of NCS in other SmAPs vary in diameter from ≈6 Å (Afu SmAP2 hexamer) to ≈13 Å (Afu, Mth SmAP1 heptamers), whereas the SmAP3 pore is ≈15 Å wide at its narrowest point. Structures of Afu (9) and Mth (33) SmAP1 bound to oligouridine or UMP show that the uracil bases make several specific contacts to residues around the circumference of the pore, the primary interaction being intercalation between a conserved histidine/arginine pair (Fig. 1). Although the SmAP3 pore does not contain this motif, other pore-lining residues are likely to form an ssRNA-binding site: the SmAP3 Tyr67-Arg68-Tyr69 sequence lies in a similar place as the Afu/Mth His46/Arg72 pair, and its structure seems ideal for uracil intercalation (unpublished results). Such a model is supported by observed stacking of uracil on a conserved tyrosine in the structure of Hfq bound to AUUUUUG ssRNA (18), and by the suspected intercalation of uracil between Tyr/Arg in Afu SmAP2 (15). Apparently, the conserved His46(Mth) residue of the SmAP1 subfamily is replaced by tyrosine in many SmAP2 proteins (Fig. 1). No electron density was seen for uracil through the course of refinement, even though SmAP3 was crystallized in the presence of 10 mM UMP. But, this may be a result of the 14-mer assembly: the likely UMP-binding site lies on the opposite face of the heptamer from the highly acidic L4 face, and this face is occluded in the heptamerapic-heptamerequa interface.
SmAP3 Assembly and Cd2+-Binding Sites.
An unexplored feature of Sm proteins is their cofactor- or metal-binding properties. Crystallographic and biophysical characterization of Pae SmAP3 reveals that it binds to the divalent metal ions cadmium and zinc (Fig. 2a), and that the mode of binding differs in the apical and equatorial conformers (Fig. 3a). We discovered cadmium-binding fortuitously, as CdCl2 was a required additive for the formation of high quality crystals (21), and Cd2+ sites were readily identified as prominent peaks in anomalous difference electron density. In terms of local structure, the binding sites are nearly identical in the equatorial and apical subunits (Fig. 3 b and c): Cd2+ is chelated in a distorted tetrahedral geometry by an imidazole nitrogen of histidine (Nɛ2), a glutamate oxygen, and two waters (consistent with the large enthalpy of dehydration of Cd2+). However, apical and equatorial binding sites clearly differ in terms of global structure (Fig. 3a): in the equatorial conformer, the glutamate is contributed by the Sm domain of an adjacent subunit (giving the Glu15/His39 pair) whereas, in the apical binding site, glutamate is supplied by the C-terminal domain of an adjacent subunit (Glu84/His39 pair). Both the apical and equatorial glutamates are conserved in the SmAP3 subfamily (Fig. 1). As inferred from bond lengths, Cd2+ may be bound more tightly in apical than in equatorial subunits (Fig. 3 b vs. c). However, the role of metal binding in causing (or resulting from) the conformational difference between apical and equatorial subunits is unclear, as is the relationship between metal binding and oligomerization.
The oligomeric state of Pae SmAP3 increases on binding to Cd2+ and Zn2+ in solution, as assayed by sedimentation velocity ultracentrifugation (Fig. 3d). The sedimentation coefficient of apo-(SmAP3)n corresponds most closely to a heptamer (S20,w = 5.3S, n = 7, 102.2-kDa heptamer), but is shifted to larger values in samples containing Cd2+ (blue curve, Fig. 3d) or Zn2+ (red curve). The shifts in S20,w on metal binding agree with predictable changes in the stoichiometry of the SmAP3 oligomer. Given the dependence of sedimentation coefficient on molecular weight (S ≈ Mr2/3), the SmAP3+Zn2+ data are consistent with a SmAP3 28-mer (S20,w = 13.0S, possibly the species observed in the crystallographic a.u.). An interesting continuum of S20,w values is produced by titration of SmAP3 with ZnCl2 concentrations varying over several orders of magnitude: the sigmoidal S20,w vs. log([ZnCl2]) curve has an inflection point near 1 mM ZnCl2, and saturates near the 13.0S particle at 10–20 mM ZnCl2 (unpublished results). An S20,w value of 8.3S is calculated for a 14-mer, based on the 5.3S apo-SmAP3 heptamer. Thus, the SmAP3+Cd2+ data cannot be as easily interpreted in terms of a (SmAP3)7n oligomer, but may correspond to a distorted 14-mer or a long-lived assembly intermediate. As with Zn2+ binding, the increased S20,w on Cd2+ binding is likely attributable to a fast heptamer ↔ 28-mer equilibrium, but with a lower binding affinity of SmAP3 for Cd2+ than Zn2+. All increases in SmAP3 sedimentation coefficients on metal-binding are too large to be accounted for by shape changes alone (e.g., a metal-induced shift from equatorial heptamers to the more compact apical conformer), suggesting that differential metal-binding modulates SmAP3 oligomerization in an ion-dependent manner.
Our results show that the following three aspects of SmAP3 assembly are interdependent: (i) divalent cation binding, (ii) conformational variation between apical and equatorial subunits, and (iii) distribution of oligomeric states. Along with recent data that suggest that the Hfq hexamer forms 12-mers (35) and that Pae and Mth SmAP1s form 14-mers (33), demonstration of higher-order Pae SmAP3 oligomers in solution supports the significance of the crystallographic SmAP 14-mer, although its biological function is unknown. Head-to-head stacking of heptamers may prevent their polymerization into fibers (33). More likely, modulation of SmAP3 14-mer assembly via divalent metal binding may regulate RNA binding, because the putative ssRNA-binding site is distal to the exposed L4 face, and is occluded in the heptamer–heptamer interface.
Discussion
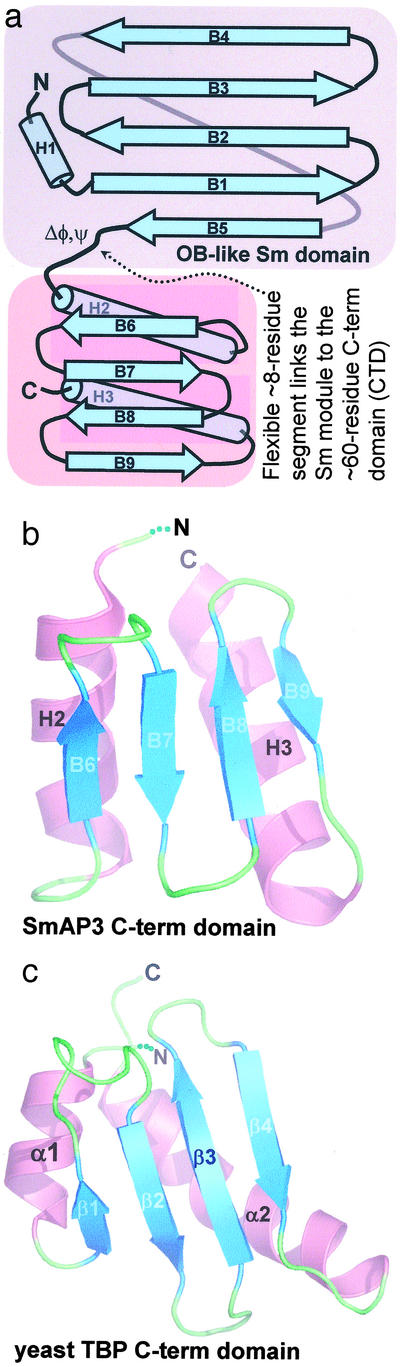
An unexpected feature of SmAP3 is the presence of a novel C-terminal domain. Compared with other SmAPs, the SmAP3 sequences are extended by nearly 70 C-terminal residues (Fig. 1). Multiple sequence alignments based solely on sequence similarity suggest that half of these extra residues are in the Sm domain (in variable loop L4, Fig. 1), whereas the remainder form an unstructured C-terminal tail (as is thought for the longer human Sm B and D3 proteins). Instead, the Pae SmAP3 structure shows that these additional residues form a compact CTD in which two α-helices lie on one side of an antiparallel four-stranded β-sheet (Fig. 4). Although other protein domains have similar αβ4α topologies (e.g., residues 160–224 of ribosomal elongation factor G), the following two criteria lead us to suggest that the SmAP3 CTD is a novel fold: (i) searches against protein structure databases (dali; ref. 36) produce only weak hits, with a maximal z-score of 4.8 (2.5 Å rmsd) for a DNA-contacting domain from the yeast TATA-box binding protein (ref. 37; yTBPc, Fig. 4c); (ii) the helical packing differs between SmAP3 CTD and yTBPc. The antiparallel α-helices of the SmAP3 CTD (Fig. 4b) form more extensive tertiary contacts than the nearly perpendicular helices of the yTBPc domain (Fig. 4c). Although the sequences of the Sm domains of SmAP3s are highly variable, strong conservation of the three SmAP3 CTD sequences suggests that other SmAP3s are structurally similar to Pae SmAP3 (Fig. 1). Similarly, the SmAP3 CTD may serve as a model for the 3D structure of Sm and Lsm proteins that are >100 residues, such as human SmB.
Fig 4.
Topology of SmAP3 and the novel fold of its C-terminal domain. (a) A schematic of the SmAP3 topology shows the oligonucleotide/oligosaccharide-binding-like fold of the conserved N-terminal Sm domain (gray background) and the mixed α/β fold of the C-terminal domain (CTD, pink background). Comparison of the 3D structures of the SmAP3 CTD (b) and the C-term domain of yeast TATA-box binding protein (c) suggests that the SmAP3 CTD is a novel α/β fold (see text).
The SmAP3 structure shows that the highly conserved Sm module may be fused to other domains to obtain proteins with the potential for novel nucleic acid binding properties. Recent evidence suggests roles for SmAP1 in pre-tRNA processing (9) and DNA-binding (33), but it is unclear how the CTD of SmAP3 would modify any such function. The primary function of this auxiliary domain may be biochemical (e.g., contacting nucleic acid) or structural (e.g., extending the acidic L4 face and stabilizing the 14-mer by adding >15,000 Å2 of buried surface area to the 4,300-Å2 portion of the heptamer–heptamer interface formed by just the Sm domains; Fig. 2). Several proteins with RNA- or DNA-binding functions contain mixed α/β domains that either (i) have similar 3D structures as the SmAP3 CTD, but differ topologically, or (ii) have similar topologies, but differ in 3D structure (unpublished results). The recent structure of a heptameric voltage-gated mechanosensitive channel [E. coli MscS (38)] shows that Sm-like domains may be used for physiological functions entirely unrelated to RNA metabolism.
By showing that it is an Sm protein, the Pae SmAP3 structure supports a gene duplication model for the evolution of modern, eukaryotic Sm proteins. Consistent with such a model, we find archaeal genomes encoding intermediate numbers of SmAPs (between 1 and 7 Sm ORFs): several genomes contain two SmAPs, and Pae, S. tokodaii, and S. solfataricus are the first examples of genomes with three SmAPs (Fig. 1). Patterns of sequence similarity scores between SmAPs and details of their clustering into phylogenetic clades (Fig. 1 and unpublished data) suggest that SmAP2 and SmAP3 paralogs arose by SmAP1 duplications, rather than horizontal gene transfer. The recently discovered eubacterial Lsm protein Hfq is thought to have evolved similarly (11), although the evolutionary relationship between SmAPs and eubacterial Sm proteins is not obvious. Also unclear is the generality of the structural and metal-binding features of SmAP3: whether or not other Sm proteins bind to divalent metals and assemble into 14-mers. These results raise intriguing questions regarding the role of conformational flexibility and the novel C-terminal domain of SmAP3, the existence of similar domains in other (extended) Sm proteins, and possible cellular roles of divalent metal binding in mediating Sm⋅⋅⋅RNA interactions.
Acknowledgments
We thank Dr. Sorel Fitz-Gibbon (University of California, Los Angeles) for the phosmid vector containing the Pae SmAP3 gene; Drs. Duilio Cascio and Michael Sawaya (University of California, Los Angeles) for data collection at beamline X8C of the National Synchrotron Light Source (Brookhaven National Laboratory); Dr. Thomas Schneider (University of Göttingen) for advice on the escet program; and Drs. James Bowie and Linda Columbus for discussions. This work was supported by the U.S. Department of Energy and the National Institutes of Health. D.E. is an Investigator of the Howard Hughes Medical Institute.
Abbreviations
RNP, ribonucleoprotein
snRNP, small nuclear RNP
SmAP, Sm-like archaeal protein
NCS, noncrystallographic symmetry
a.u., asymmetric unit
MAD, multiwavelength anomalous dispersion
rmsd, rms deviation
Afu, Archaeoglobus fulgidus
Pae, Pyrobaculum aerophilum
Mth, Methanobacterium thermautotrophicum
CTD, C-terminal domain
ssRNA, single-stranded RNA
UMP, uridine-5′-monophosphate
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID code ).
References
- 1.Yu Y.-T., Scharl, E. C., Smith, C. M. & Steitz, J. A. (1999) in The RNA World, eds. Gesteland, R. F., Cech, T. R. & Atkins, J. F. (Cold Spring Harbor Lab. Press, Plainview, NY), pp. 487–524.
- 2.Collins C. A. & Guthrie, C. (2000) Nat. Struct. Biol 7, 850-854. [DOI] [PubMed] [Google Scholar]
- 3.Stevens S. W., Ryan, D. E., Ge, H. Y., Moore, R. E., Young, M. K., Lee, T. D. & Abelson, J. (2002) Mol. Cell 9, 31-44. [DOI] [PubMed] [Google Scholar]
- 4.Walke S., Bragado-Nilsson, E., Seraphin, B. & Nagai, K. (2001) J. Mol. Biol. 308, 49-58. [DOI] [PubMed] [Google Scholar]
- 5.Raker V. A., Plessel, G. & Luhrmann, R. (1996) EMBO J. 15, 2256-2269. [PMC free article] [PubMed] [Google Scholar]
- 6.Will C. L. & Luhrmann, R. (2001) Curr. Opin. Cell Biol. 13, 290-301. [DOI] [PubMed] [Google Scholar]
- 7.Achsel T., Brahms, H., Kastner, B., Bachi, A., Wilm, M. & Luhrmann, R. (1999) EMBO J. 18, 5789-5802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Collins B. M., Harrop, S. J., Kornfeld, G. D., Dawes, I. W., Curmi, P. M. & Mabbutt, B.C. (2001) J. Mol. Biol. 309, 915-923. [DOI] [PubMed] [Google Scholar]
- 9.Toro I., Thore, S., Mayer, C., Basquin, J., Seraphin, B. & Suck, D. (2001) EMBO J. 20, 2293-2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mura C., Cascio, D., Sawaya, M. R. & Eisenberg, D. S. (2001) Proc. Natl. Acad. Sci. USA 98, 5532-5537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sun X., Zhulin, I. & Wartell, R. M. (2002) Nucleic Acids Res. 30, 3662-3671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moller T., Franch, T., Hojrup, P., Keene, D. R., Bachinger, H. P., Brennan, R. G. & Valentin-Hansen, P. (2002) Mol. Cell 9, 23-30. [DOI] [PubMed] [Google Scholar]
- 13.Plessel G., Luhrmann, R. & Kastner, B. (1997) J. Mol. Biol. 265, 87-94. [DOI] [PubMed] [Google Scholar]
- 14.Stark H., Dube, P., Luhrmann, R. & Kastner, B. (2001) Nature 409, 539-542. [DOI] [PubMed] [Google Scholar]
- 15.Toro I., Basquin, J., Teo-Dreher, H. & Suck, D. (2002) J. Mol. Biol. 320, 129-142. [DOI] [PubMed] [Google Scholar]
- 16.Achsel T., Stark, H. & Luhrmann, R. (2001) Proc. Natl. Acad. Sci. USA 98, 3685-3689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kambach C., Walke, S., Young, R., Avis, J. M., de la Fortelle, E., Raker, V. A., Luhrmann, R., Li, J. & Nagai, K. (1999) Cell 96, 375-387. [DOI] [PubMed] [Google Scholar]
- 18.Schumacher M. A., Pearson, R. F., Moller, T., Valentin-Hansen, P. & Brennan, R. G. (2002) EMBO J. 21, 3546-3556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Urlaub H., Raker, V. A., Kostka, S. & Luhrmann, R. (2001) EMBO J. 20, 187-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Van Duyne G. D., Standaert, R. F., Karplus, P. A., Schreiber, S. L. & Clardy, J. (1993) J. Mol. Biol. 229, 105-124. [DOI] [PubMed] [Google Scholar]
- 21.Trakhanov S., Kreimer, D. I., Parkin, S., Ames, G. F. & Rupp, B. (1998) Protein Sci. 7, 600-604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Otwinowski Z. & Minor, W. (1997) in Methods in Enzymology, eds. Sweet, R. & Carter, C. W., Jr. (Academic, San Diego), Vol. 276A, pp. 307–326. [DOI] [PubMed] [Google Scholar]
- 23.Dodson E. J., Winn, M. & Ralph, A. (1997) Methods Enzymol. 276, 620-633. [DOI] [PubMed] [Google Scholar]
- 24.Cowtan K. & Main, P. (1998) Acta Crystallogr. D Biol. Crystallogr. 54, 487-493. [DOI] [PubMed] [Google Scholar]
- 25.Terwilliger T. C. (2001) Acta Crystallogr. D Biol. Crystallogr. 57, 1755-1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brunger A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J. S., Kuszewski, J., Nilges, M., Pannu, N. S., et al. (1998) Acta Crystallogr. D Biol. Crystallogr. 54, 905-921. [DOI] [PubMed] [Google Scholar]
- 27.Colovos C. & Yeates, T. O. (1993) Protein Sci. 2, 1511-1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Altschul S. F., Madden, T. L., Schaffer, A. A., Zhang, J., Zhang, Z., Miller, W. & Lipman, D. J. (1997) Nucleic Acids Res. 25, 3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Higgins D. G., Thompson, J. D. & Gibson, T. J. (1996) Methods Enzymol. 266, 383-402. [DOI] [PubMed] [Google Scholar]
- 30.Shindyalov I. N. & Bourne, P. E. (1998) Protein Eng. 11, 739-747. [DOI] [PubMed] [Google Scholar]
- 31.Schneider T. R. (2002) Acta Crystallogr. D Biol. Crystallogr. 58, 195-208. [DOI] [PubMed] [Google Scholar]
- 32.Nicholls A., Sharp, K. A. & Honig, B. (1991) Proteins 11, 281-296. [DOI] [PubMed] [Google Scholar]
- 33.Mura C., Kozhukhovsky, A., Gingery, M., Phillips, M. & Eisenberg, D. (2003) Protein Sci. 12, 832-847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Antson A. A., Dodson, E. J., Dodson, G., Greaves, R. B., Chen, X. & Gollnick, P. (1999) Nature 401, 235-242. [DOI] [PubMed] [Google Scholar]
- 35.Arluison V., Derreumaux, P., Allemand, F., Folichon, M., Hajnsdorf, E. & Regnier, P. (2002) J. Mol. Biol. 320, 705-712. [DOI] [PubMed] [Google Scholar]
- 36.Holm L. & Sander, C. (1993) J. Mol. Biol. 233, 123-138. [DOI] [PubMed] [Google Scholar]
- 37.Kim Y., Geiger, J. H., Hahn, S. & Sigler, P. B. (1993) Nature 365, 512-520. [DOI] [PubMed] [Google Scholar]
- 38.Bass R. B., Strop, P., Barclay, M. & Rees, D. C. (2002) Science 298, 1582-1587. [DOI] [PubMed] [Google Scholar]