Abstract
Small non-coding RNAs (sRNAs) are important players of gene expression regulation in bacterial pathogens. MtvR is a 136-nucleotide long sRNA previously identified in the human pathogen Burkholderia cenocepacia J2315 and with homologues restricted to bacteria of the Burkholderia cepacia complex. In this work we have investigated the effects of expressing MtvR in Escherichia coli and Pseudomonas aeruginosa. Results are presented showing that MtvR negatively regulates the hfq mRNA levels in both bacterial species. In the case of E. coli, this negative regulation is shown to involve binding of MtvR to the 5′-UTR region of the hfq Ec mRNA. Results presented also show that expression of MtvR in E. coli and P. aeruginosa originates multiple phenotypes, including reduced resistance to selected stresses, biofilm formation ability, and increased susceptibility to various antibiotics.
Introduction
Small non-coding RNAs (sRNAs) are increasingly recognized as important players in gene expression regulation in bacteria, and more recently, in the regulation of virulence determinants in bacterial pathogens [1]. Many of the sRNAs characterized so far exert their action by base-pairing with their target mRNAs in the region of the ribosome-binding site, thus affecting their stability and/or translation [2]. This mode of regulation has been shown to be advantageous for bacteria when a fast response to external stimuli is required, providing a fine tuning of gene expression [3]. This also enables the bacterial cell with a precise gene expression regulation and the means for a rapid adaptation in response to specific environmental changes [4]. These advantages are consistent with the involvement of many described bacterial sRNAs in response to stress [5]. In addition, sRNAs and the associated complex regulatory circuits are also used by bacterial pathogens to adapt to the host environment, and to coordinately express specific virulence determinants during different stages of infection [6].
Trans-encoded sRNAs are arguably the most extensively characterized class of bacterial sRNAs [6]. These sRNAs can target one or more mRNAs, sharing with them a limited complementarity. In order to interact with their targets, most of the trans-encoded sRNAs require the aid of the RNA chaperone Hfq, which plays a role as a facilitator of sRNA-mRNA interactions [7].
Most of the sRNAs recently described are only conserved among closely related bacterial species [8]. This is the case of MtvR, a 136 nucleotide-long sRNA identified in Burkholderia cenocepacia J2315, with homologues restricted to bacteria of the Burkholderia genus [9]. B. cenocepacia is a member of the so called Burkholderia cepacia complex (Bcc), a group of closely related species that emerged in the 1980s as important opportunistic pathogens among patients afflicted with the genetic disease cystic fibrosis [10], and more recently, among hospitalized non-cystic fibrosis patients, especially cancer patients (reviewed in [11]).
Previous work from our research group has shown that the sRNA MtvR targets multiple genes in the clinical isolate B. cenocepacia J2315, affecting cell size, growth and survival under nutrient deprivation, biofilm formation, antibiotics resistance, and virulence to the nematode Caenorhabditis elegans [9]. Due to the pleiotropic phenotypes observed, MtvR was proposed as a global regulatory sRNA in B. cenocepacia [9]. hfq was previously shown as one of the MtvR multiple targets in B. cenocepacia, with the sRNA negatively regulating hfq translation by specifically binding to its 5′ leader region [9]. In the present study, we investigated the possible roles played by MtvR in the non-Bcc organisms Escherichia coli and Pseudomonas aeruginosa, two bacterial species with no MtvR homologues, but harboring Hfq-encoding genes. Despite being an exogenous sRNA in both bacterial species, results here presented indicate that MtvR modulates the levels of the Hfq mRNA and protein in both species, affecting their resistance to selected stresses, including to antibiotics.
Materials and Methods
Bacterial Strains, Culture Conditions, Plasmids and Primers
Bacterial strains and plasmids are described in Table 1, and oligonucleotides are listed in Table 2. P. aeruginosa strains were maintained on PIA (Pseudomonas Isolation Agar, Becton Dickinson) plates, supplemented with 650 µg.ml−1 trimethoprim, when appropriate. E. coli strains were maintained on Lennox Broth (LB) plates supplemented with 100 µg.ml−1 trimethoprim or 50 µg.ml−1 kanamycin or chloramphenicol, when appropriate. Unless otherwise stated, liquid cultures were carried out using LB liquid medium, supplemented with antibiotics when appropriate, with orbital agitation (250 rev min−1) at 37°C. Expression of MtvR in E. coli or P. aeruginosa was achieved by induction of plasmid pCGR12 (Table 1) with 0.1% (v/v) L-arabinose. Plasmid pCGR34 was derived from pCR II by cloning into the XbaI-HindIII sites the E. coli MC4100 hfq (hfq Ec) coding sequence, including the complete 5′ leader region and a 6×histidine tag encoded at the C-terminus. Plasmid pCGR35, containing the genetic fusion composed of the hfq Ec leader -β-galactosidase, was engineered as follows: the LacZα fragment was amplified by PCR from pBBR1MCS and blunt-end ligated to the EcoRV site of pMLBAD. A 380 bp fragment containing the 5′-UTR region of the hfq Ec gene was obtained by XbaI-BamHI restriction of a PCR fragment obtained using as template E. coli MC4100 chromosomal DNA. After restriction, the fragment was filled-in with the Klenow large fragment of DNA polymerase I (Invitrogen), and then blunt-ligated to the filled-in XbaI site of the cloned LacZα fragment. Plasmid pCGR36 derives from pCR II, and contains the hfq Ec 5′ leader region together with the coding sequence in the XbaI-HindIII sites, allowing T7-driven transcription. Plasmid pCGR37 derives from pMLBAD, and contains the 255 nt fragment corresponding to the P. aeruginosa PA14 hfq (hfq Pa, obtained by PCR using as template P. aeruginosa PA14 chromosomal DNA), cloned in the PstI-SalI sites (3′ to 5′ direction of the coding sequence). Plasmid pCGR38 derives from pMLBAD and contains the 140 bp cDNA fragment corresponding to the MtvR sRNA cloned in the EcoRI/XbaI sites and the 255 bp cDNA fragment corresponding to the P. aeruginosa PA14 hfq cloned in the SalI/PstI sites (tandem cloning, pBAD promoter control). All plasmid constructions were verified by DNA sequencing.
Table 1. Bacterial strains and plasmids used in this work.
| Strain or plasmid | Description | Reference or source |
| P. aeruginosa WT | P. aeruginosa PA14, clinical isolate | [37] |
| P. aeruginosa WT-hfqsil | P. aeruginosa PA14 with hfq Pa silenced after transformationwith pCGR37 | This study |
| P. aeruginosa WT/phfq | P. aeruginosa PA14 expressing heterologous hfq Bc aftertransformation with pSAS3 | This study |
| P. aeruginosa WT/phfq+MtvR | P. aeruginosa PA14 expressing heterologous hfq Bc andMtvR after transformation with pSAS3 and pCGR12 | This study |
| P. aeruginosa WT/pMLBAD | P. aeruginosa PA14 after transformation with pMLBAD | This study |
| P. aeruginosa WT/MtvR | P. aeruginosa PA14 expressing MtvR after transformationwith pCGR12 | This study |
| P. aeruginosa WT-hfqsil/MtvR | P. aeruginosa PA14 with hfq Pa silenced and expressing MtvR,after transformation with pCGR38 | This study |
| E. coli WT | E. coli MC4100, laboratory strain | [38] |
| E. coli Δhfq | E. coli GS081, CmR | [39] |
| E. coli WT/phfq | E. coli MC4100 after transformation with pCGR34, expressingtagged hfq Ec | This study |
| E. coli WT/phfq+MtvR | E. coli MC4100 after transformation with pCGR34 and pCGR12,expressing tagged hfq Ec and MtvR | This study |
| E. coli WT/pMLBAD | E. coli MC4100 after transformation with pMLBAD | This study |
| E. coli WT/MtvR | E. coli MC4100 after transformation with pCGR12,expressing MtvR | This study |
| E. coli Δhfq/pMLBAD | E. coli GS081 transformed with pMLBAD | This study |
| E. coli Δhfq/phfq | E. coli GS081 transformed with pSAS3, expressingheterologous hfq Bc | This study |
| E. coli Δhfq/MtvR | E. coli GS081 transformed with pCGR12, expressing MtvR | This study |
| E. coli Δhfq/phfq+MtvR | E. coli GS081 transformed with pCGR34 and pCGR12,expressing tagged hfq Ec and MtvR | This study |
| Plasmids | ||
| pCR II | AmpR; KmR; used for generating in vitro transcription templates | Invitrogen |
| pMLBAD | TmpR; used for inducible gene expression | [40] |
| pCGR4 | pET23a+ with the hfq Bc encoding sequence cloned | [14] |
| pSAS3 | pMLBAD with the hfq Bc encoding sequence cloned | [14] |
| pCGR12 | pMLBAD with the 140 bp cDNA fragment corresponding tothe MtvR sRNA cloned in the EcoRI/XbaI sites(pBAD promoter control) | [9] |
| pCGR34 | pCR II with the 609 bp cDNA fragment corresponding tothe E. coli MC4100 hfq full mRNA (5′-UTR and CDS) with6 histidines at the C-terminus, cloned in the XbaI/HindIIIsites (T7 promoter control) | This study |
| pCGR35 | pMLBAD with the hfq Ec 5′-UTR-LacZ DNA fragment (pBABpromoter disrupted, only replicative) | This study |
| pCGR36 | pCR II with the DNA fragment corresponding to E. coli MC4100hfq cloned in the XbaI/HindIII sites | This study |
| pCGR37 | pMLBAD with the 255 bp cDNA fragment corresponding to theP. aeruginosa PA14 hfq cloned in the PstI/SalI sites | This study |
| pCGR38 | pMLBAD with the 140 bp cDNA fragment corresponding tothe MtvR sRNA cloned in the EcoRI/XbaI sites and the255 bp cDNA fragment corresponding to the P. aeruginosaPA14 hfq cloned in the SalI/PstI sites(tandem cloning, pBAD promoter control) | This study |
Table 2. Oligonucleotides and primers used in this work.
| Name | Purpose | Sequence 5′ –3′ | Source or reference |
| UF | Cloning mtvR | TTTCTAGATATTGACGGCGGCGGGT | [9] |
| LF | Cloning mtvR | TTAAGCTTAAATTATAGCGCCCCAATTA | [9] |
| NP | Northern analysis of MtvR | CTATCACCCGCCTGTGTCGCCA | [9] |
| HFQ | Northern blot probe for hfq | AAAGGGCAATTGTTACAAG | [12] |
| CGRO117 | Amplification of P. aeruginosa hfq | TTCTGCAGACCGGACGGCTCGGTACCAC | This study |
| CGRO118 | Amplification of P. aeruginosa hfq | TCTGTCGACTTCCGGAGCGAGACCGGAGT | This study |
| CGRO119 | Amplification of E. coli hfq | CCTCTAGACCAGAACAGGCGCGTGACGA | This study |
| CGRO120 | Amplification of E. coli hfq | TTAAGCTTGAAACCGGGCGAGACGGGAC | This study |
| CGRO123 | Fwd primer hfq his-tag | TTTCTAGAGCACGTCCCGCAAGGGCTAG | This study |
| CGRO124 | Rev primer hfq his-tag | TTGGATCCATTGTGGTGGTGGTGGTGGGACGAGGCTTCCGC | This study |
| M13FWD | LacZ amplification | GTAAAACGACGGCCAGT | Invitrogen |
| M13REV | LacZ amplification | AGCGGATAACAATTTCACACAGGA | Invitrogen |
| CGRO125 | Fwd primer uhpA | CTGGGGCTGGAACCTGATTT | This study |
| CGRO126 | Rev primer uhpA | CGCAGCAATGAGTTCATCCG | This study |
| CGRO127 | Fwd primer uhpT | TTCCTGCCGTTCATGCTGAT | This study |
| CGRO128 | Rev primer uhpT | GAGGCCATAAGATTCCGGGG | This study |
| 5S | Northern blot probe for 5S rRNA | TTCGGGATGGGAAGGGGTGGGA | [12] |
DNA Manipulation Techniques
Total DNA was obtained from the indicated cell cultures using the High pure PCR template preparation kit (Roche). PCR amplification was performed using TaqPlatinum (Invitrogen) DNA polimerase and adequate primers and DNA templates (Table 2). PCR products were purified using the NucleoExtract II kit (Nagel-Machery) as previously described [9]. After nuclease restriction, fragments were directionally cloned into the indicated plasmids. All plasmid constructions were confirmed by DNA sequencing.
RNA Manipulation Techniques
Total RNA isolation, quantification of RNA concentration and visual quality control, and RNA labeling with biotin were performed as previously described [12].
Northern Blot Analysis
The expression levels of MtvR, hfq Ec or hfq Pa were assessed by Northern blot analysis using 2 µg of total RNA and adequate oligonucleotide probes (Table 2), previously labeled with biotin, based on previously described methods [13]. Briefly, RNA samples were separated in 8% TBE-urea pre-cast gels (BioRad), using constant current of 200 mA. RNA was then transferred to a BrightStar plus membrane (Ambion) using the wet transfer system (BioRad), at 100 V for 1h. Samples were probed with adequate biotinylated oligonucleotides at 40°C, for 16 h. Hybridization signals were detected using the BrightStar Biodetect kit (Ambion) and Kodak MX X-ray films. The 5S RNA was used as loading control in all Northern blot experiments. Relative expression was estimated with the ImageJ software suite, using the band intensities of the 5S RNA for normalization.
RNA Decay Experiments
The RNA decay rate was assessed by Northern blot analysis using 2 µg of total RNA, purified from cells of E. coli (Ec) or P. aeruginosa (Pa), harvested from 24 h-cultures, immediately before (t0) or after the addition of rifampicin to induce transcriptional arrest. This antibiotic was used at final concentrations of 250 µg ml−1. Aliquots taken after 5, 10, 15 and 20 min (for hfq), or 2, 5, 10 and 15 min (for MtvR) of transcription arrest, were processed and analyzed by Northern blot, as described above. RNA decay rates were calculated based on the exponential fit expression: t1/2 (min): ae−bt, and using the slope of semi-log plots.
RNA in vitro Transcription and Labeling
The DNA templates for in vitro transcription of the hfq Ec mRNA full transcript, the 5′-UTR of hfq Ec, the hfq Ec coding region (CDS), and the MtvR sRNA were obtained by endonuclease restriction of the appropriate plasmids (Table 1). All RNA transcripts were generated from the T7 promoter, using the MEGAshortscript kit (Ambion). The transcripts MtvR (136 nt), 5′-UTR of hfq Ec (155 nt), and the CDS of hfq Ec (307 nt) were purified from 8%–7 M urea polyacrylamide gels, while the hfq Ec full transcript (462 nt) was purified from a 1% TBE/agarose gel. RNA transcripts were processed and labeled based on previously described methods [13]. Signal intensity was tested using the dot-blot procedure, and detected using the Bright Star Biodetect Kit (Ambion).
Electrophoretic Mobility Shift Assays
EMSA experiments to assess the binding affinity of MtvR to the hfq Ec 5′-UTR, the hfq Ec coding region (CDS), and the hfq Ec full mRNA, were performed as previously described [12]. Briefly, 25 nM of the MtvR, together with 0, 0.5, 1, 5, 10, 50 or 100 nM of the hfq Ec full transcript, or with 0, 5, 10, 25, 50, 75, 100, 150, 200, 250 or 500 nM of the hfq Ec CDS, or with 0, 0.5, 1, 5, 10, 50 or 100 nM the 5′-UTR of hfq Ec, were incubated in 25 µl of RNA binding buffer (20 mM Tris.Cl, pH 8.0, 1 mM DTT, 1 mM MgCl2, 10 mM KCl and 10 mM KH2PO4) [14] for 30 min at 25°C. The ability of 25 nM of the 5′-end labeled (with 11-UTP-biotin) hfq Ec 5′-UTR to bind to 50, 100 or 250 nM of B. cenocepacia J2315 Hfq6 (HfqBc), of 100 nM of hfq Ec full RNA to bind to HfqBc (concentrations ranging from 10 to 1000 nM of HfqBc), or to HfqBc (concentrations ranging from 0.5 to 250 nM) in the presence of MtvR (concentrations ranging from 0.1 to 100 nM), were also evaluated by EMSA experiments using native 8% polyacrylamide gel containing 10% glycerol.
Non-labeled yeast tRNA (Ambion) was added in excess to each sample to minimize non-specific binding. Incubation, resolution of RNA-RNA and RNA-protein complexes, and detection of band-shifts, were performed as previously described [14]. The curves generated from the data plots were fitted by non-linear least squares regression assuming a bimolecular model in which the K d values represent the protein concentration at half the maximal RNA binding.
Reverse Transcription-PCR Experiments
Total RNA was extracted from cells of exponentially growing cultures of E. coli WT strain, E. coli WT expressing MtvR, or E. coli Δhfq mutant, using the already described methods. Reverse transcription reactions were performed using the First Strand cDNA synthesis kit (Fermentas), with an incubation of 75 min at 45°C, in a total volume of 50 µL, and 500 ng of total RNA. The cDNA samples were used in PCR experiments with TaqMed (Citomed) DNA polymerase, and the oligonucleotide pairs CGRO125 and CGRO126 (uhpA), or CGRO127 and CGRO128 (uhpT) (Table 2). Cycling conditions were as follows: 45 cycles of denaturation at 95°C for 1 min, annealing at 55°C for 1 min, extension at 72°C for 0.5 min, and a final extension at 72°C for 10 min. cDNA pools were resolved in 2%-TBE agarose gels. Control reactions using DNA were included. 500 ng of total RNA from each strain were also loaded on the same gel, as loading controls.
Western Blot Analysis
The effect of MtvR expression on the levels of the HfqEc protein was assessed by Western blotting. For this purpose, plasmid pCGR34 was introduced into the E. coli Δhfq mutant strain. Plasmid pCGR34 is able to drive the transcription of hfq Ec as a derivative containing a 6×His-tag at the C-terminus (HfqEc-His), from its own promoter. This strain was further transformed with plasmid pCGR12, which allows MtvR expression. The cultures were grown for 16 h in LB liquid medium, supplemented with 150 µg ml−1 trimethoprim, 50 µg ml−1 kanamycin and 0.1% L-arabinose. Total proteins from culture aliquots of 1 ml were purified with Illustra TriplePrep kit (GE Healthcare), based on previously described methods [13]. Aliquots containing 10 µg of total protein were used for Western blot detection of the HfqEc-His protein, using the polyclonal pentaHis-IgG-HRP antibody (Invitrogen). The α-GroEL, used as loading control in Western-blot experiments, was detected with a goat anti-GroEL antibody (SicGen, Portugal). Signals were detected using a ECL system (GE Healthcare). Fold-change values were estimated using as unitary value the number of pixels counted for the reference, normalized with the internal standard.
β-galactosidase Assays
β-galactosidase assays were performed based on previously described methods [15] using a SpectraMax 250 microtiter plate reader (Molecular Devices). Briefly, E. coli strains WT, WT transformed with the plasmid that allows MtvR expression, or transformed with the control vector pMLBAD, were grown at 37°C in LB medium for 24 h with antibiotics (when appropriate) and diluted 1000-fold into 50 mL of fresh medium at 37°C. Cultures were grown with agitation to an OD640 of 0.1 before inducing MtvR expression by the addition of L-arabinose (0.1% final concentration). Specific β-galactosidase activities (OD640 ∼1.0) were calculated using the formula Vmax/OD600. The reported results represent data from at least four independent experiments.
Antibiotic Susceptibility and Biofilm Formation Experiments
The susceptibility of E. coli WT, P. aeruginosa WT, and of the respective derivatives to the antibiotics chloramphenicol, ciprofloxacin, tetracycline, tobramycin, gentamycin and ampicillin, was assessed by the broth micro-dilution method, in Mueller-Hinton medium (Gibco) supplemented with 0.1% arabinose, based on previously described methods [16], and following the CLSI guidelines [17]. The ability of E. coli WT, P. aeruginosa WT and the respective derivatives to form biofilms was assessed after 24, 48 and 72-h of growth on LB liquid medium at 28°C, based on previously published methods [18]. Briefly, appropriate volumes of overnight liquid cultures of bacterial strains were used to inoculate LB liquid medium and grown at 28°C with orbital agitation, until the mid-exponential phase was reached. The cultures were subsequently diluted to a standardized culture OD640 of 0.5, and 20 µl of these cell suspensions were used to inoculate the wells of a 96-well polystyrene microtiter plate (Greiner Bio-One) containing 180 µl of LB medium. Plates were incubated at 28°C without agitation for 24, 48 and 72 h. The biofilm formed was quantified by measuring the absorbance at 590 nm using a VERSAmax microplate reader (Molecular Devices). All measurements were performed in triplicate, using biological duplicates.
Growth and Nutrient Deprivation Kinetics
Cultures of E. coli WT, P. aeruginosa WT and the respective derivatives were carried out at 37°C in liquid LB supplemented with 0.1% L-arabinose. Growth was followed spectrophotometrically at 640 nm. The ability of E. coli WT, P. aeruginosa WT and respective derivatives to survive to nutrient limiting conditions was assayed in M9 minimal medium supplemented with 0.1% L-arabinose instead of glucose, based on previously described methods [12]. Determination of total CFU’s was performed by plating aliquots of each culture in solid LB supplemented with 0.1% L-arabinose. All measurements were performed in quadruplicate, using biological triplicates.
Stress Susceptibility Experiments
The susceptibility of E. coli WT or P. aeruginosa WT and derivative strains to the stresses imposed by growth in LB solid medium containing 0.1% (w/v) L-arabinose, and supplemented with 0.05% SDS, 2.5% (w/v) NaCl, 2.5% (v/v) ethanol, or 25 µM methyl viologen, were performed as previously described [12]. Results are the means of at least 5 independent experiments.
Statistics Analysis
An unpaired two-tailed chi-square test was used to calculate the P values for β-galactosidase assays. Analysis of data from Northern and Western blotting was performed using a paired one-tailed t test to calculate the P values (P<0.01 [*]; p<0.005 [**]). Error bars represent the means of the standard deviation. Images shown are representative of the experiments performed. All experiments were repeated independently at least 4 times, using biological triplicates, with a minimum n value of 12.
Bioinformatics
BLAST searches were performed using the Integrated Microbial Genomes (IMG) webserver [19], using an E-value ≤1e−50 as cut-off. MtvR putative targets were predicted using the sRNATarget [20], the TargetRNA [21], and the RNAPredator [22] programs, with a cut-off of 1, a minimum seed of 7 nucleotides, and an hybridization target region window size of −100 to +30 around the translation start site. The MtvR sRNA sequence was aligned with the RNA sequence of the putative mRNA targets, using the sequences retrieved from NCBI for the Escherichia coli str. K12 substr. DH10B and the Pseudomonas aeruginosa UCBPP-PA14 genome sequences, using the RNAHybrid web tool [23], using a minimum seed of 7 nucleotides [24] in the region around the start codon.
Results
MtvR Regulates the mRNA Levels of E. coli and P. aeruginosa hfq Genes by Promoting their Accelerated Decay
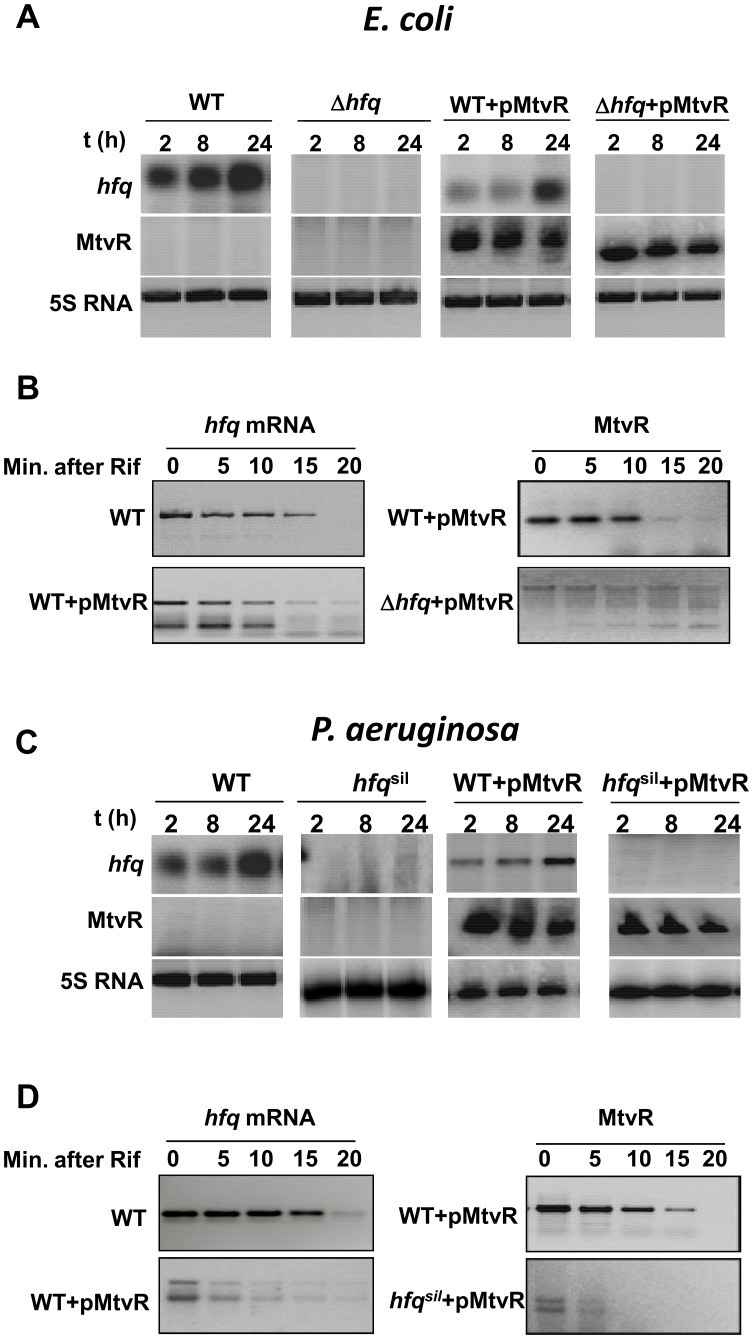
We have investigated if MtvR affects hfq expression in E. coli and P. aeruginosa. For this purpose, we transformed E. coli and P. aeruginosa WT strains with plasmid pCGR12, which allows MtvR expression. Results in Fig. 1A and 1C confirm that the sRNA is expressed, respectively, in E. coli and P. aeruginosa. The ectopic expression of MtvR for 2, 8 or 24 h led, respectively, to a reduction of 1.4±0.09, 1.29±0.08 or 1.21±0.03 fold of the hfq Ec mRNA levels (Fig. 1A), and to a reduction of the hfq Pa mRNA levels of 2.60±0.07, 2.30±0.13 or 1.8±0.3 fold (Fig. 1C). Expression of MtvR also affected the stability of the E. coli and P. aeruginosa hfq mRNAs, leading to the reduction of the hfq Ec mRNA half-life time (t1/2) from 13.4±1.0 to 8.6±0.5 min (Fig. 1B), and of the hfq Pa mRNA t1/2 from 18.6±1 to 9.9±0.4 min (Fig. 1D). Unlike the single band corresponding to hfq Ec or hfq Pa mRNAs observed for the respective controls, the expression of MtvR resulted in the detection of two bands corresponding to hfq Ec or hfq Pa mRNA decay products, suggesting that the processing of both mRNAs in the presence of MtvR involves pathways distinct from those used by the E. coli or P. aeruginosa WT strains. Interestingly, our results also indicate that MtvR requires the Hfq proteins from E. coli or P. aeruginosa for stability, as suggested by the decrease of the sRNA t1/2 from 17.3±0.7 min in the E. coli WT strain to 8.5±0.4 min in the E. coli hfq mutant (Fig. 1B), and from 5.2±0.7 min in the P. aeruginosa WT strain to 2.6±0.4 min in the P. aeruginosa strain with the hfq Pa gene silenced (Fig. 1D). Together, these results indicate that MtvR is stabilized by HfqEc and HfqPa, while the mRNAs hfq Ec and hfq Pa are targets of this sRNA.
Figure 1. MtvR regulates the mRNA levels of the E. coli and P. aeruginosa hfq genes.
(A) Levels the hfqEc mRNA and the MtvR transcript in cells of the E. coli WT or the E. coli Δhfq mutant, expressing (+pMtvR) or not the sRNA. (B) Stability of the hfq Ec mRNA (left panel) in cells expressing (+pMtvR) or not MtvR, and of the MtvR transcript (right panel) in cells of the WT or Δhfq mutant, expressing MtvR (+pMtvR). (C) Levels of the hfqPa mRNA and the MtvR transcript in cells of the P. aeruginosa WT or WT with the hfq gene silenced (hfq sil), expressing (+pMtvR) or not MtvR. (D) Analysis of the stability of the P. aeruginosa hfqPa mRNA and of the MtvR transcript, in cells of the P. aeruginosa strains WT or the WT with the hfq gene silenced (hfqsil), expressing (+pMtvR) or not MtvR. The levels of the 5S rRNA were used as loading control.
The MtvR sRNA Binds to the 5′ Leader Region of the hfq mRNA
Based on both experimental and bioinformatics analyses indicating that hfq Ec mRNA is a target of MtvR, we have conducted further experiments to gain additional insights into the molecular details of the observed negative regulatory effects exerted by MtvR on the hfq Ec mRNA.
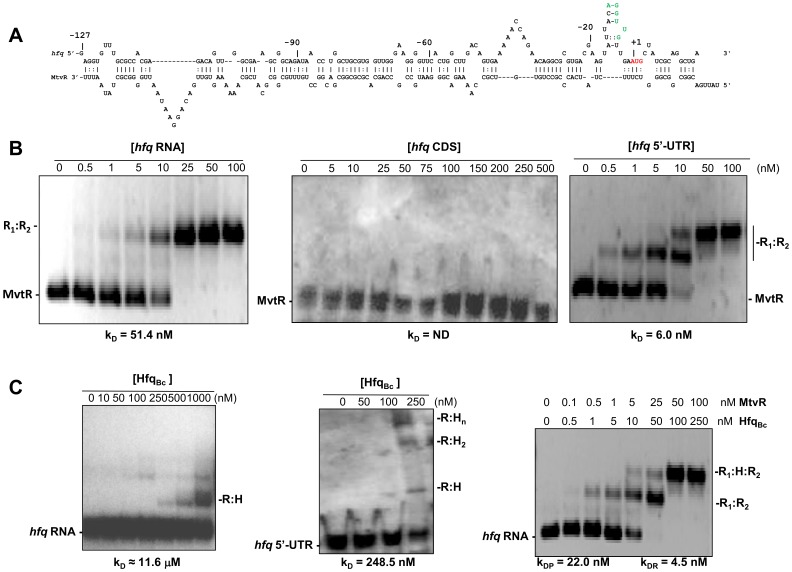
The interaction between MtvR and the hfq Ec 5′ leader region was investigated using the RNAHybrid software, which predicted a strong interaction between the two RNA molecules, suggesting that these two RNAs are able to form extended RNA duplexes, occluding the Ribosome Binding Site (green letters in Fig. 2A) and the start codon (Red letters in Fig. 2A).
Figure 2. MtvR regulates hfq Ec expression through interaction with its 5′-leader region.
(A) Schematic representation of the nucleotide interaction between the 5′-UTR of hfq Ec and the MtvR sRNA, highlighting in green and red lettering, respectively, the RBS and the AUG translation start site in the hfq Ec 5′-UTR. (B) EMSA experiments using 25 nM of biotin-labeled MtvR transcript and the indicated concentrations of: left panel, the hfq Ec full mRNA; center panel, the hfq Ec coding region (CDS); right panel, the 5′-UTR of hfq Ec. C) EMSA experiments performed to assess the ability of HfqBc to bind to: left panel, 25 nM of the biotin-labeled hfq Ec full mRNA; center panel, 25 nM of the biotin-labeled hfq Ec 5′-UTR. The ability of mixtures containing HfqBc together with the tested concentrations of the MtvR transcript to bind to 25 nM of the biotin-labeled hfq Ec full mRNA is shown in the right panel. H(n), HfqBc(n); R1, MtvR; R2, hfq Ec RNA; kDP, affinity constant for the protein; kDR, affinity constant for the RNA species.
This RNA-RNA interaction was experimentally confirmed by EMSA experiments. For this purpose, MtvR was transcribed in vitro and biotin-labeled, and the hfq Ec RNA was transcribed from a T7 promoter using as template plasmid pCGR36, yielding the complete transcriptional unit of hfq Ec, composed of 475 nt corresponding to the 5′ leader region of hfq Ec, and 306 nt corresponding to the hfq Ec coding sequence (CDS). Results presented in Fig. 2B (left panel) show that MtvR binds to the hfqEc RNA, with an apparent KD of 51.4 nM. Similar band-shift assays performed with the hfqEc CDS instead of the full RNA, revealed that MtvR requires the hfqEc 5′-UTR for binding (Fig. 2B, center panel), since no MtvR displacement could be detected, at least for the concentrations used of the hfq Ec CDS. An additional experiment was performed using the sRNA together with the 5′-UTR of hfqEc. Results shown in Fig. 2B (right panel) indicate that MtvR binds to the 5′-UTR of hfqEc, with an apparent KD of 6.0 nM.
The E. coli Hfq protein was previously shown to be auto-regulated, through the interaction of the protein with two distinct binding sites within the 5′ leader of the mRNA, resulting in the inhibition of the formation of the translation initiation complex [25]. Since the action of sRNAs is often mediated by the Hfq RNA chaperone and the Hfq protein is involved in auto-regulation in E. coli, apparently without the requirement for sRNAs, we have conducted EMSA experiments with the hfqEc RNA (composed of the 5′-UTR and the coding sequence), or the hfqEc 5′-UTR together with the Hfq of B. cenocepacia J2315 (HfqBc, which lacks amino acid residues beyond position 79). Results obtained indicate that HfqBc can form complexes with the hfqEc RNA only in relatively high concentrations (500 nM or higher) (Fig. 2C, left panel), with an apparent KD of 11.6 µM. The HfqBc protein also needs to be present in concentrations as high as 250 nM in order to bind to the hfqEc 5′-UTR RNA (Fig. 2C, center panel). Interestingly, in the presence of the MtvR sRNA, the HfqBc protein is able to bind to the two RNAs at a concentration 500-fold lower compared to the hfqEc RNA alone (Fig. 2C, left panel).
These results suggest that MtvR together with HfqBc can synergistically bind to the hfq Ec mRNA, more efficiently than HfqBc alone.
MtvR Expression Affects E. coli Hfq Translation
Our data strongly suggests that MtvR might also impact the HfqEc levels. To further investigate the impact of MtvR on the levels of HfqEc, we have transformed the E. coli WT strain and the strain expressing MtvR with plasmid pCGR34, which allows expression of HfqEc-His from its own promoter. This was performed since the polyclonal anti-Hfq IgG that was previously raised (unpublished data) is specific for the HfqBc, and no signals on Western blots could be detected for the HfqEc using this antibody (data not shown).
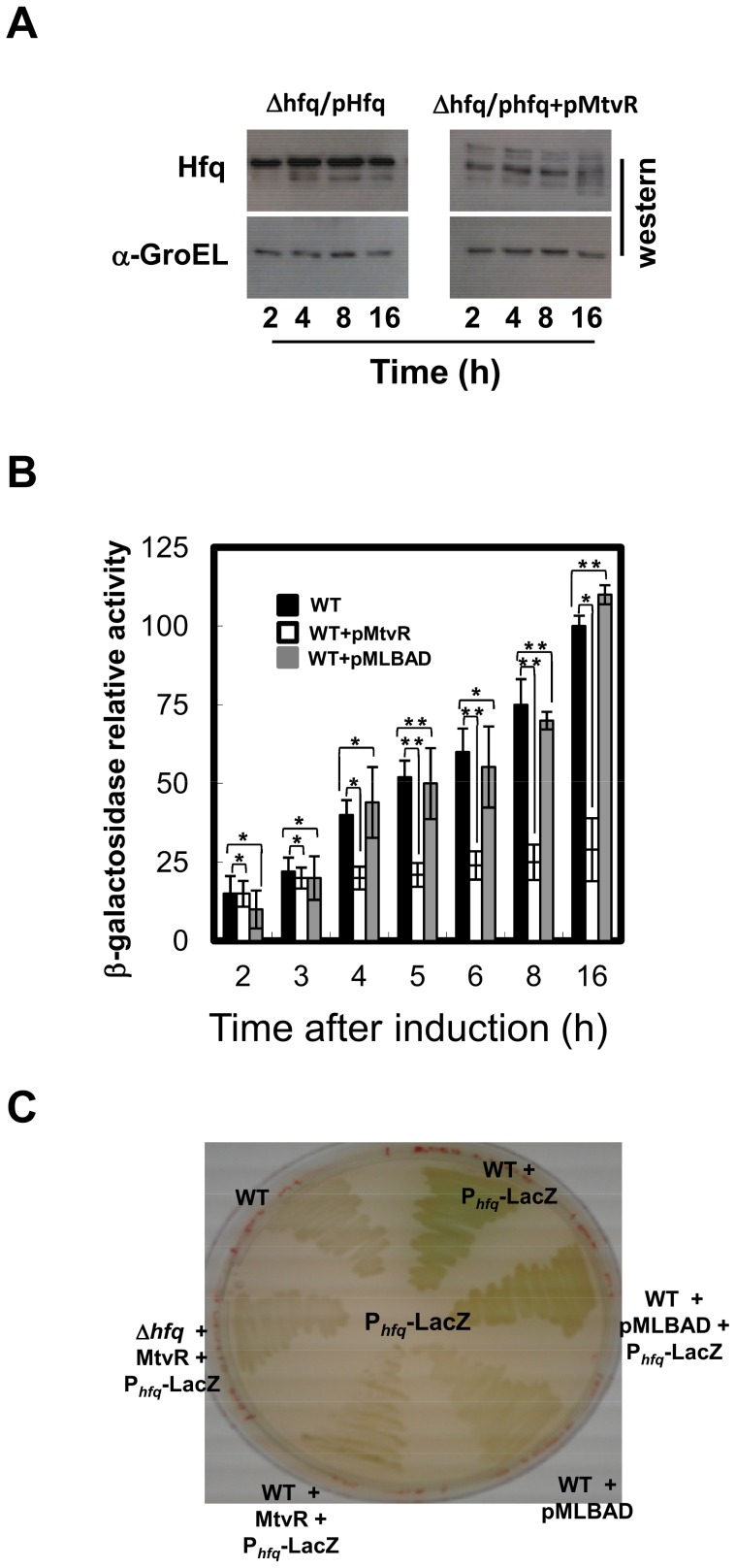
Results from Western blot analysis showed that MtvR expression reduced hfq Ec translation by 1.87-, 2.09-, 2.30- or 2.77-fold, at 2, 4, 8 or 16 h, respectively (Fig. 3A).
Figure 3. MtvR affects E. coli Hfq translation.
(A) Western blot analysis of the levels of the 6×His-tagged HfqEc protein in cells of the E. coli Δhfq mutant expressing only the tagged protein (left panel, Δhfq/phfq), or also expressing the MtvR transcript (Right panel, Δhfq/phfq+pMtvR). The GroEL levels were used as loading control. (B) β-galactosidase relative activity in cells of the E. coli strains WT (black bars), WT expressing MtvR (+pMtvR, white bars), or WT with the empty vector (pMLBAD, grey bars), harboring the hfq Ec 5′-UTR-LacZ fusion (phfq-LacZ). C) β-galactosidase activity of the indicated E. coli strains, grown in solid LB media, supplemented with X-Gal and 0.1% L-arabinose. The WT strain transformed with pMLBAD was used as control.
The in vivo assessment of MtvR interaction with the 5′ leader region of hfq Ec was achieved by measuring the β-galactosidase activity of the 5′-UTR-LacZ fusion. With this purpose, E. coli derivatives expressing MtvR were transformed with plasmid pCGR35. The WT strain was transformed with pMLBAD and used as control. Results obtained (Fig. 3B and 3C) show that the lowest values of β-galactosidase activity were registered when MtvR was expressed. The inability of MtvR to induce a complete translational blocking might derive from its dependence on a RNA chaperone for increased stability. These results are consistent with a negative regulatory action exerted by MtvR on hfq Ec mRNA, occurring by binding to its 5′ leader region, which most probably prevents an efficient translation of the messenger RNA, by coupling enhanced mRNA decay with impaired ribosome loading.
MtvR Expression Affects E. coli and P. aeruginosa Growth Abilities
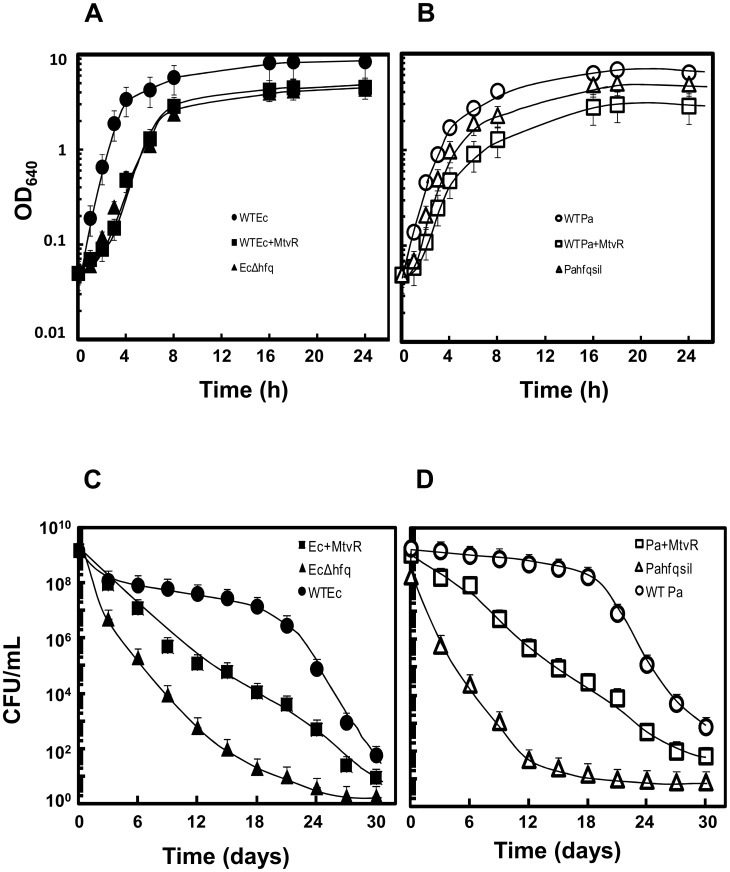
Data presented so far indicates that MtvR is able to regulate hfq EC and hfq Pa, and, at least, HfqEc. This led us to investigate the roles of MtvR on the growth abilities of E. coli and P. aeruginosa. The specific growth rate and biomass yield reached after 24 h of batch growth of the E. coli expressing MtvR were reduced to approximately the same level as that of the Δhfq mutant strain (Fig. 4A). In the case of P. aeruginosa, the expression of MtvR reduced the specific growth rate and biomass yield reached after 24 h of batch growth to levels even lower than those observed for the strain with the hfq Pa silenced (Fig. 4B). When challenged with long-term carbon starvation, E. coli and P. aeruginosa strains expressing MtvR exhibited a reduction of their survival ability more pronounced than the reduction observed for the E. coli Δhfq mutant and the P. aeruginosa with the hfq gene silenced (hfq sil), respectively.
Figure 4. MtvR expression affects E. coli and P. aeruginosa growth kinetics and survival to prolonged nutrient deprivation.
Growth curves in liquid LB medium supplemented with 0.1% L-arabinose (A, B) and survival in M9 minimal medium supplemented with 0.1% L-arabinose, at 37°C for 30 days (C, D), of (A, C) E. coli strains WT (circles), WT expressing MtvR (squares) or the Δhfq Ec mutant (triangles), or (B,D) P. aeruginosa strains WT (circles), WT expressing MtvR (squares) or the WT strain with the hfq Pa gene silenced (triangles).
Since the growth rate and biomass yield or resistance to nutrient deprivation registered for the WT strains of E. coli or P. aeruginosa and the respective transformants harboring pMLBAD were similar, these results were not included in Fig. 4 to keep it simpler.
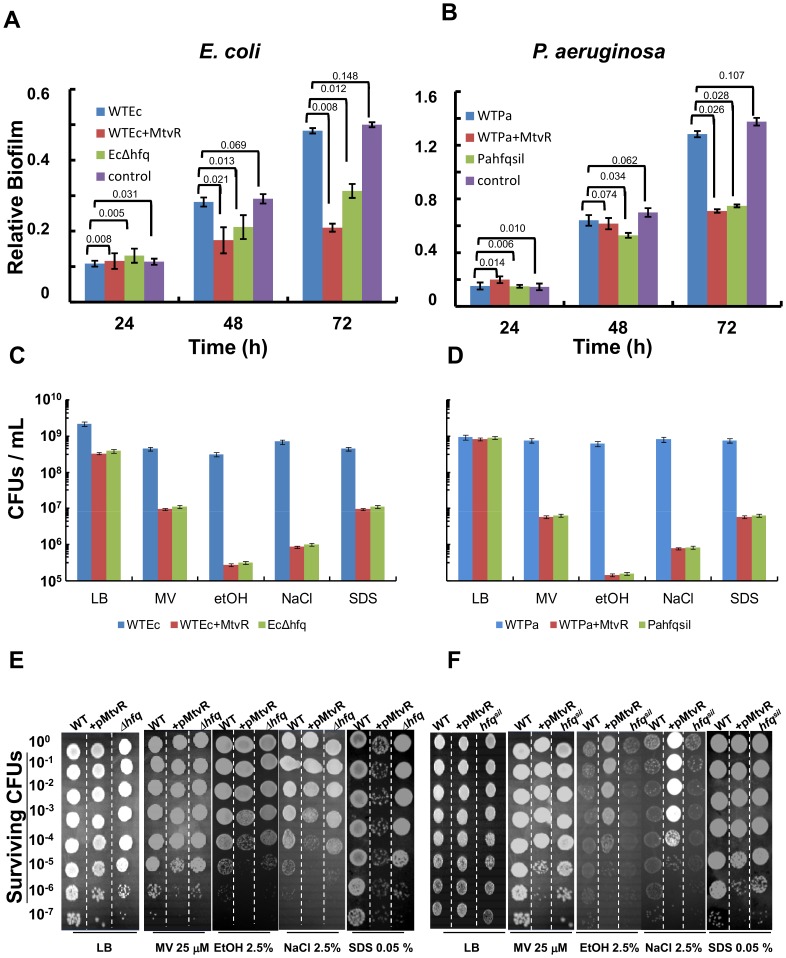
MtvR Affects the Biofilm Formation Ability of E. coli and P. aeruginosa
In a previous study MtvR was shown to play a role on biofilm formation ability in Bcc bacteria [9]. Therefore, we decided to investigate the effect of expressing MtvR on the biofilm formation ability of E. coli and P. aeruginosa. Since for the different strains under study differences in the biomass yield were registered, results were expressed as relative biofilm amount, i.e., the ratio of biofilm amount estimated using the crystal violet and the biomass concentration assessed by the OD640 of cultures. Results obtained are presented in Figs. 5A and 5B and indicate that MtvR expression affected negatively the ability of both E. coli and P. aeruginosa to form biofilms in vitro, particularly evident at 72 h. The observed effects might result from the down-regulation of hfq Ec and hfq Pa, since the strains E. coli Δhfq and P. aeruginosa hfq sil formed relative biofilm amounts comparable to those formed by the respective WT strains expressing MtvR.
Figure 5. MtvR expression in E. coli and P. aeruginosa reduces biofilm formation ability and increases susceptibility to stresses.
Relative biofilm formation ability (panels A, B) and susceptibility to the stress imposed by growth on the surface of LB solid medium supplemented or not (LB) with the indicated concentrations of methyl viologen (MV), ethanol (etOH), NaCl or SDS (SDS) (panels C,D), of strains of (panels A, C) E. coli WT (WTEc), WT expressing MtvR (WTEc+MtvR), or the Δhfq Ec mutant (EcΔhfq), and (panels B, D) P. aeruginosa WT (WTPa), WT expressing MtvR (WTPa+MtvR), or the WT strain with the hfq Pa gene silenced (Pahfqsil). Relative biofilm formation was estimated by dividing the total amount of biofilm formed by the total amount of biomass (see Materials and Methods section). Panels E and F show photographs illustrative of results from a single representative susceptibility experiment with the E. coli (panel E) and P. aeruginosa (panel F) strains WT (WT), WT expressing MtvR (+MtvR), and the E. coli Δhfq Ec mutant (Δhfq), or the P. aeruginosa WT with the hfq Pa gene silenced (hfq sil). Susceptibilities were assessed by spot inoculation of serially diluted bacterial suspensions with an initial OD640 of 1.0. Error bars represent standard deviation of the means. Numbers above bars in panels (A) and (B) are the estimated P-values.
E. coli and P. aeruginosa Exhibit Enhanced Susceptibility to Stresses When Expressing MtvR
The effects of MtvR expression on the susceptibility of E. coli and P. aeruginosa strains to oxidative, osmotic and membrane stresses were investigated by spot-inoculation of bacterial culture aliquots on LB solid media supplemented with methyl viologen, NaCl, ethanol or SDS. Illustrative photographs of a set of results obtained for E. coli and P. aeruginosa are shown, respectively, in Figs. 5E and 5F. Methyl viologen is a charged quaternary ammonium compound that generates reactive oxygen species under aerobic conditions [26], used to generate oxidative stress conditions. Ethanol and NaCl were used to increase the medium osmolarity, while SDS was used as a membrane integrity-perturbing agent. These stressors were chosen to mimic the environmental conditions oxidative stress, high osmolarity, and extracytoplasmic stress, conditions that are faced, for instance, when P. aeruginosa colonizes/infects the cystic fibrosis lung [10].
Results presented in Fig. 5C show that when compared to the WT strain, the numbers of total CFUs of the E. coli strains expressing MtvR or the Δhfq Ec mutant, exposed to methyl viologen or SDS, were reduced by more than 1 log. This reduction was higher, 2 to 3 logs, for cells of the strains expressing MtvR or Δhfq Ec when exposed to stressors NaCl or ethanol, respectively. Under non-stress conditions, about one log reduction in the total CFU were registered for the E. coli strains expressing MtvR and the mutant Δhfq Ec when compared to the WT strain (Fig. 5C). It is worth to note that the reductions in total CFUs registered for the E. coli strains expressing MtvR or the Δhfq mutant were quite similar, suggesting that the observed increased susceptibility to the tested stressors might result from the down-regulation of hfq Ec.
In the case of P. aeruginosa, no differences on the total CFUs were registered for the strains WT, WT expressing MtvR, and the WT with the hfq Pa gene silenced under non-stress conditions (Fig. 5D). However, when compared to the P. aeruginosa WT strain, approximately 2, 3, or 4-log reduction in the total CFU were registered for the strains P. aeruginosa expressing MtvR and P aeruginosa hfq sil when exposed, respectively, to methyl viologen or SDS, NaCl, or ethanol (Fig. 5D). Since no significant differences in the total CFUs were observed for the E. coli strains WT and WT transformed with pMLBAD, and P. aeruginosa WT and P. aeruginosa transformed with pMLBAD, only the results obtained for the E. coli and P. aeruginosa WT strains are shown, respectively, in Figs. 5C to 5F.
MtvR Expression Enhances Antibiotic Susceptibility in E. coli and P. aeruginosa
The sRNA MtvR was recently shown to play a role on resistance to antibiotics in Bcc [9]. Therefore, we investigated the effects of MtvR expression on the E. coli and P. aeruginosa susceptibility to the antibiotics chloramphenicol, ciprofloxacin, tetracycline, tobramycin, gentamycin and ampicillin. Results presented in Table 3 show that MtvR expression reduced E. coli MIC values by 8-fold for tetracycline, 4-fold for chloramphenicol and ciprofloxacin, and 2-fold for tobramycin, gentamycin and ampicillin. The observed increased susceptibility to these antibiotics in the Δhfq Ec mutant strain was identical to the observed due to MtvR expression, except for chloramphenicol and tetracycline. For these two antibiotics, the susceptibility only increased by 2- and 4-fold, respectively. The effect of MtvR expression in the Δhfq Ec led to an even more drastic effect in susceptibility, especially to tobramycin and gentamycin, with MIC values lowering 16- and 128-fold, respectively (Table 3).
Table 3. Antibiotic susceptibility of E. coli WT and derivative strains.
| Strain | CHL | CIP | TET | NM | GNT | AMP |
| E. coli WT | 4 | 1 | 2 | 8 | 16 | 4 |
| E. coli Δhfq | 2* | 0.25 | 0.25 | 2 | 8 | 2 |
| E. coli WT/phfq | 16 | 8 | 32 | 32 | 128 | 64 |
| E. coli WT/phfq+MtvR | 2 | 0.25 | 0.5 | 2 | 1 | 0.5 |
| E. coli WT/pMLBAD | 4 | 1 | 2 | 8 | 16 | 4 |
| E. coli WT/MtvR | 1 | 0.125 | 0.125 | 4 | 8 | 2 |
| E. coli Δhfq/pMLBAD | 64* | ≤0.125 | 0.125 | 1 | 8 | 2 |
| E. coli Δhfq/phfq | >512* | 1 | 2 | 8 | 16 | 4 |
| E. coli Δhfq/MtvR | 8* | ≤0.125 | ≤0.125 | 0.5 | 0.125 | 1 |
| E. coli Δhfq/phfq+MtvR | 64* | 0.125 | 0.125 | 4 | 8 | 2 |
CHL, chloramphenicol; CIP, ciprofloxacin; TET, tetracycline; NM, tobramycin; GNT, gentamycin; AMP, ampicillin. Numbers with asterisks represent strains that have a chromosomal chloramphenicol resistance cassette.
We have also investigated the role of MtvR on the P. aeruginosa susceptibility to chloramphenicol, ciprofloxacin, tetracycline, tobramycin, gentamycin and ampicillin. Results in Table 4 indicate that the antibiotic susceptibility to the tested antibiotics increased in cells expressing MtvR. MtvR expression induced a 4-fold reduction in most of the MIC values, with the exception of gentamycin, for which a MIC value 2-fold lower than the WT strain was registered (Table 4). In the strain with the hfq Pa gene silenced, the antibiotic susceptibility was also affected, although to a lesser extent. The MIC values for chloramphenicol and ampicillin remained unchanged in the strain with the hfq silenced (Table 4). A more drastic effect on antibiotic susceptibility was observed in the strain with the hfq gene silenced and expressing MtvR. In fact, an impressive 64-fold reduction in the MIC value for chloramphenicol, 32-fold for tobramycin and gentamycin, and 16-fold for ampicillin were registered for this strain (Table 4).
Table 4. Antibiotic susceptibility of P. aeruginosa WT and derivative strains.
| Strain | CHL | CIP | TET | NM | GNT | AMP |
| P. aeruginosa WT | 64 | 1 | 16 | 16 | 128 | >512 |
| P. aeruginosa hfqsil | 64 | 0.5 | 8 | 4 | 64 | 512 |
| P. aeruginosa WT/phfq | 128 | 1 | 64 | 128 | 256 | >512 |
| P. aeruginosa WT/phfq+MtvR | 16 | 0.125 | 2 | 4 | 16 | 64 |
| P. aeruginosa WT/pMLBAD | 64 | 1 | 16 | 16 | 128 | >512 |
| P. aeruginosa WT/MtvR | 16 | 0.125 | 2 | 4 | 16 | 64 |
| P. aeruginosa hfqsil/MtvR | 1 | 0.125 | 1 | 0.5 | 8 | 32 |
CHL, chloramphenicol; CIP, ciprofloxacin; TET, tetracycline; NM, tobramycin; GNT, gentamycin; AMP, ampicillin.
MtvR has Additional Putative Targets in E. coli
Some of the phenotypes here reported for the E. coli strain expressing MtvR differ from those observed for the E. coli Δhfq Ec mutant, suggesting that MtvR might regulate additional mRNAs in this bacterium. Therefore, we have used the programs TargetRNA [21], RNAPredator [22] and sRNATarget [20] to predict possible MtvR additional mRNA targets within the genome of E. coli K12 strain MG1655. A total of 11 (Table S1 in File S1) and 3 (Table S2 in File S1) putative mRNA targets were predicted by TargetRNA and RNAPredator, respectively, assuming only putative hybridizations in the −20 to +20 nt region around the start codon. Using the sRNATarget with the same restrictions, and excluding genes of unknown functions, 52 distinct mRNA targets were predicted (Table S3 in File S1). Interestingly, several targets were predicted to interact with MtvR in more than one region (Table S4 in File S1).
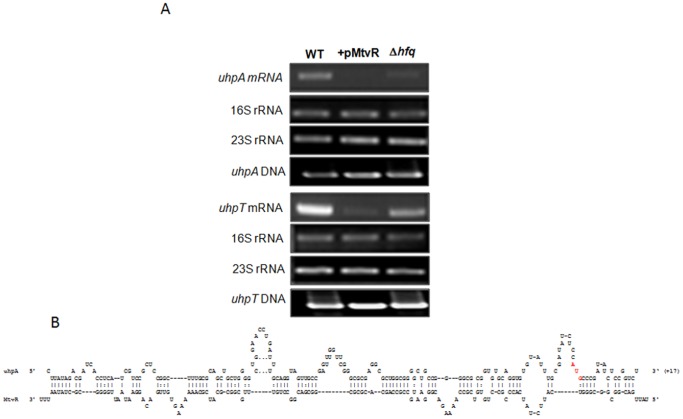
The gene uhpA of E. coli was the only common target of MtvR predicted by TargetRNA and sRNATarget. The uhpA encodes the response regulator of a two-component regulatory system where UhpB is a histidine kinase that controls the synthesis of the sugar-phosphate transporter UhpT [27]. These findings prompted us to investigate the effects of MtvR expression on the transcript levels corresponding to the uhpA mRNA in E. coli using RT-PCR. We have also investigated the levels of the uhpT mRNA, which is regulated by the UhpA-UhpB two-component regulatory system [27]. Results presented in Fig. 5 show that MtvR expression led to undetectable uhpA-derived cDNA, opposed to the observed for the WT strain (Fig. 6A). However, cDNA corresponding to uhpA mRNA could be detected in the Δhfq Ec mutant strain, although with a reduced intensity relative to the WT strain. These observations suggest that Hfq might be involved in uhpA regulation.
Figure 6. The MtvR sRNA also targets uhpA gene in E. coli.
(A) Reverse-transcription analysis of the effect of MtvR on the mRNA levels of uhpA. Total RNA was obtained from late-exponentially growing cells of the E. coli strains WT and WT expressing MtvR (+pMtvR), or the Δhfq mutant (Δhfq). Reverse-transcription experiments were also performed for uhpT, induced by UhpA. The 16S and 23S rRNA bands were used as loading controls. PCR experiments were also performed using DNA, for reference. Images shown are representative of 3 independent experiments. B) Schematic representation of the nucleotide interaction between the 5′-UTR of uhpA and MtvR, highlighting in red lettering the AUG translation start site in the 5′-UTR of uhpA.
The levels of cDNA corresponding to uhpT mRNA were highly reduced in E. coli cells expressing MtvR when compared to those observed for the WT strain (Fig. 6A). This observation is consistent with the requirement of UhpA (in its phosphorylated form) for the transcriptional activation of uhpT [27].
Since the levels of cDNA corresponding to uhpT mRNA were higher in the Δhfq Ec mutant than in the strain expressing MtvR, we concluded that HfqEc is unlikely to be involved in the direct regulation of uhpT mRNA. The regulation exerted by MtvR on uhpA mRNA might involve partial base-pairing of the two molecules, predicted to occur within the 5′ region of uhpA mRNA, leading to the formation of a RNA duplex (with a predicted energy of −117.0 kcal/mol) that occludes part of the RBS and start codon (Fig. 6B).
We also have used the TargetRNA [21], the RNAPredator [22] and the sRNATarget [20] programs to predict possible MtvR mRNA targets within the genome of P. aeruginosa UCBPP-PA14 (Tables S5 and S6 in File S1). No common targets were predicted.
Discussion
The sRNA MtvR was recently identified as a trans-encoded sRNA that occurs exclusively among members of the Burkholderia genus [9]. In these bacteria, MtvR acts as a global regulatory RNA, and strains with the sRNA silenced or overexpressed exhibited pleiotropic phenotypes related to growth and survival when challenged with stress, motility, biofilm formation, resistance to antibiotics and virulence [9]. In addition, MtvR was shown to regulate the levels of at least 17 mRNA targets in the cystic fibrosis isolate B. cenocepacia J2315 [9]. Results presented in this work also show that, at least in E. coli, MtvR targets other genes besides hfq Ec, as is the case of uhpT.
Several trans-encoded sRNAs have been described as regulating multiple targets, most probably due to the limited complementarities shared with their mRNA targets [6]. This limited base-pairing is thought to justify the need of most of the trans-encoded sRNAs to bind to the RNA chaperone Hfq to effectively interact with their targets [6].
Despite the absence of homologues to MtvR in E. coli or P. aeruginosa, here we present evidence that this sRNA is able to regulate the levels of the hfq mRNA in both species.
The Hfq protein of Burkholderia is 83% identical to the E. coli [14]. The E. coli Hfq is composed of 102 amino acid residues, while the P. aeruginosa and B. cenocepacia Hfq proteins are composed of 82 and 79 amino acid residues, respectively. All 3 proteins contain the conserved Sm1 and Sm2 motifs, and have a secondary structure compose by a N-terminal α-helix, followed by 5 β-strands. The amino acid residues 8–68 correspond to the conserved core of the 3 proteins.
Earlier studies on the E. coli Hfq have shown that the protein relative abundance is growth-phase dependent [28], being hfq Ec transcription regulated by multiple mechanisms [29]. More recently, Vecerek et al. (2005) presented evidence indicating that in E. coli the synthesis of HfqEc is auto-regulated at the translational level [25]. These authors have shown that HfqEc binds to the two HfqEc-binding sites in the 5′-UTR region of hfq Ec mRNA, inhibiting the formation of the translation initiation complex [30], in an interaction that involves the C-terminal region of the protein. This interaction occurs in vitro at protein concentrations ranging 50–200 nM [30]. In the case of Bcc bacteria, the HfqBc protein lacks the C-terminal region [12] suggesting a lack of auto-regulation. In fact, the B. cenocepacia hfq Bc mRNA was recently shown to be regulated through a mechanism involving sequestration of the RBS by MtvR, leading to accelerated mRNA decay and reduced protein translation [9].
Interestingly, the 5′-UTR region of the Bcc hfq Bc is 55% identical to the E. coli hfq Ec 5′-UTR (data not shown). Our results show that the B. cenocepacia shorter protein is also able to bind to the E. coli hfq Ec 5′ leader region, but with a ∼100-fold lower affinity. We also show that the sRNA can act synergistically with HfqBc, increasing its affinity to hfq Ec mRNA by more than 500-fold.
A few examples of functional analysis of sRNAs on heterologous systems have been reported. For instance, AbdelRahman et al. (2010) have used a heterologous co-expression system to demonstrate that the Chlamydia trachomatis non-coding RNA CTIG270 regulates the expression of FtsI by inducing ftsI mRNA degradation [31].
The data presented here show that despite being absent in E. coli, MtvR regulates at least, the levels of the hfq, uhpA and uhpT mRNAs. In B. cenocepacia J2315, MtvR affects the mRNA levels of at least 17 genes, among 309 predicted targets [9]. It is therefore possible that MtvR also interacts with other mRNAs, as suggested by the observed phenotypes of cells expressing the sRNA.
Perhaps the most interesting phenotypes observed due to MtvR expression in E. coli and P. aeruginosa are those related to increased antibiotic susceptibility. Bacterial resistance to antibiotics is a problem of increasingly concern, since data from the Centers for Disease Control and Prevention evidence a rapidly increasing rate of infections due to fluoroquinolone-resistant P. aeruginosa [32].
A recent study revealed that the involvement of MtvR in the regulation of an hfq-like gene also impacted the bacterium resistance to several antibiotics, leading to a phenotype conversion from resistant to susceptible [9]. A study by Yamada et al. (2010) revealed that mutations in the hfq gene from E. coli resulted in susceptibility to acriflavine, benzalkonium, cefamandole, chloramphenicol, crystal violet, nalidixic acid, novobiocin, oxacillin and rhodamine 6G [33]. In addition, Moon & Gottesman (2009) reported on the requirement of Hfq for resistance to polymyxin B through a mechanism involving the sRNA MgrR [34]. In Stenotrophomonas maltophilia, Hfq was also shown to play a role in resistance to tobramycin and amikacin, most likely due to the regulation of efflux pumps [35].
Anti-sense acting oligonucleotides are being used as components of peptide-morpholino oligonucleotide conjugates (PMO) that can act as bactericidal agents. For instance, a PMO targeting the highly conserved region of the E. coli gyrA was recently shown to effectively inactivate several species of Gram-positive and Gram-negative bacteria, when used in the micro-molar range [36].
Results presented in this work show that when expressing MtvR, the MIC values for the studied antibiotics were reduced by 2 to 128 fold in E. coli and P. aeruginosa, pointing out this sRNA as an interesting molecule, with potential to be exploited as an adjuvant in antimicrobial therapies. In fact, multi-target sRNAs, like MtvR, are potential candidates for the development of PMOs that can be used as antimicrobials, or in combination with already available antibiotics, to fight infections by multi-resistant bacteria, as is the case of P. aeruginosa.
Supporting Information
Supporting Information File combining Supplementary Methods, Supplementary Results, Table S1 (Putative MtvR targets in the genome of E. coli predicted by TargetRNA), Table S2 (Putative MtvR targets in the genome of E. coli predicted by RNAPredator), Table S3 (Putative MtvR targets in the genome of E. coli predicted by sRNATarget), Table S4 (Regions with homology to within the genome of E. coli), Table S5 (Putative MtvR targets in the genome of P. aeruginosa UCBPP-PA14 predicted by TargetRNA), and Table S6 (Putative MtvR targets in the genome of P. aeruginosa UCBPP-PA14 predicted by RNAPredator), and Supplementary References.
(PDF)
Acknowledgments
The authors acknowledge Dr. Gisela Storz for the kind gift of E. coli strains MC4100 and GS081.
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. All data are included within the manuscript.
Funding Statement
This work was partially supported by Fundação Ciência e Tecnologia, Portugal (contracts PTDC/BIA-MIC/119091/2010 and PTDC/BBB-BIO/1958/2012), a post-Doc grant to CGR, and doctoral grants to AMG and JRF, respectively. SAS and PJPDC acknowledge research grants from projects PTDC/BBB-BIO/1958/2012 and PTDC/BIA-MIC/119091/2010, respectively. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Chao Y, Vogel J (2010) The role of Hfq in bacterial pathogens Curr Opin Microbiol. 13: 24–33. [DOI] [PubMed] [Google Scholar]
- 2. Gottesman S, Storz G (2011) Bacterial small RNA regulators: Versatile roles and rapidly evolving variations. Cold Spring Harb Perspect Biol 3: a003798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Shimoni Y, Friedlander G, Hetzroni G, Niv G, Altuvia S, et al. (2007) Regulation of gene expression by small non-coding RNAs: a quantitative view. Mol Syst Biol 3: 138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Harris JF, Micheva-Viteva S, Li N, Hong-Geller E (2013) Small RNA-mediated regulation of host–pathogen interactions. Virulence 4: 785–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bobrovskyy M, Vanderpool CK (2013) Regulation of bacterial metabolism by small RNAs using diverse mechanisms. Ann Rev Genet 47: 209–232. [DOI] [PubMed] [Google Scholar]
- 6. Han Y, Liu L, Fang N, Yang R, Zhou D (2013) Regulation of pathogenicity by noncoding RNAs in bacteria. Future Microbiol 8: 579–591. [DOI] [PubMed] [Google Scholar]
- 7. Vogel J, Luisi BF (2011) Hfq and its constellation of RNA. Nat Rev Microbiol 9: 578–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Pichon C, Felden B (2008) Small RNA gene identification and mRNA target predictions in bacteria. Bioinformatics 24: 2807–2813. [DOI] [PubMed] [Google Scholar]
- 9. Ramos CG, Grilo AM, Feliciano JR, da Costa PJP, Leitão JH (2013) MtvR is a global regulatory sRNA in Burkholderia cenocepacia . J Bacteriol 195: 3514–3523. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 10. Govan JR, Deretic V (1996) Microbial pathogenesis in cystic fibrosis: mucoid Pseudomonas aeruginosa and Burkholderia cepacia . Microbiol Rev 60: 539–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Leitão JH, Sousa SA, Ferreira AS, Ramos CG, Silva IN, et al. (2010) Pathogenicity, virulence factors, and strategies to fight against Burkholderia cepacia complex pathogens and related species. Appl Microbiol Biotechnol 87: 31–40. [DOI] [PubMed] [Google Scholar]
- 12. Ramos CG, Sousa SA, Grilo AM, Feliciano JR, Leitão JH (2011) The second RNA chaperone Hfq2, is also required for survival to stress and the full virulence of Burkholderia cenocepacia J2315. J Bacteriol 193: 1515–1526. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 13. Ramos CG, Da Costa PJP, Döring G, Leitão JH (2012) The novel cis-encoded small RNA h2cR is a negative regulator of hfq2 in Burkholderia cenocepacia . PLoS ONE 7: e47896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sousa SA, Ramos CG, Moreira LM, Leitão JH (2010) The hfq gene is required for stress resistance and full virulence of Burkholderia cepacia to the nematode Caenorhabditis elegans . Microbiology 156: 896–908. [DOI] [PubMed] [Google Scholar]
- 15. Zhou Y, Gottesman S (1998) Regulation of proteolysis of the stationary-phase sigma factor RpoS. J Bacteriol 180: 1154–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Leitão JH, Sousa SA, Cunha MV, Salgado MJ, Melo-Cristino J, et al. (2008) Variation of the antimicrobial susceptibility profiles of Burkholderia cepacia complex clonal isolates obtained from chronically infected cystic fibrosis patients: a five-year survey in the major Portuguese treatment center. Eur J Clin Microbiol Infect Dis 27: 1101–1111. [DOI] [PubMed] [Google Scholar]
- 17.CLSI (2013) Performance Standards for Antimicrobial Susceptibility Testing; Twenty-Third Informational Supplement. Clinical and Laboratory Standards Institute Document M100–S23.
- 18. Ferreira AS, Leitão JH, Sousa SA, Cosme AM, Sá-Correia I, et al. (2007) Functional analysis of Burkholderia cepacia genes bceD and bceF, encoding a phosphotyrosine phosphatase and a tyrosine autokinase, respectively: Role in exopolysaccharide biosynthesis and biofilm formation. Appl Environ Microbiol 73: 524–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Markowitz VM, Chen IMA, Palaniappan K, Chu K, Szeto E, et al. (2010) The integrated microbial genomes system: an expanding comparative analysis resource. Nucleic Acids Res 30: D382–D390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhao Y, Li H, Hou Y, Cha L, Cao Y, et al. (2008) Construction of two mathematical models for prediction of bacterial sRNA targets. Biochem Biophys Res Commun 372: 346–350. [DOI] [PubMed] [Google Scholar]
- 21. Tjaden B, Goodwin SS, Opdyke JA, Guillier M, Fu DX, et al. (2006) Target prediction for small, noncoding RNAs in bacteria. Nucleic Acids Res 34: 2791–2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Eggenhofer F, Tafer H, Stadler PF, Hofacker IL (2011) RNApredator: fast accessibility-based prediction of sRNA targets. Nucleic Acids Res 39: W149–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Rehmsmeier M, Steffen P, Höchsmann M, Giegerich R (2004) Fast and effective prediction of microRNA/target duplexes. RNA 10: 1507–1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Fröhlich KS, Vogel J (2009) Activation of gene expression by small RNA. Curr Opin Microbiol 12: 674–682. [DOI] [PubMed] [Google Scholar]
- 25. Vecerek B, Moll I, Blasi U (2005) Translational autocontrol of the Escherichia coli hfq RNA chaperone gene. RNA 11: 976–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hassan HM, Fridovich I (1979) Paraquat and Escherichia coli. Mechanism of production of extracellular superoxide radical. J Biol Chem 254: 10846–10852. [PubMed] [Google Scholar]
- 27. Wright JS, Kadner RJ (2001) The phosphoryl transfer domain of UhpB interacts with the response regulator UhpA. J Bacteriol 183: 3149–3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kajitani M, Kato A, Wada A, Inokuchi Y, Ishihama A (1994) Regulation of the Escherichia coli hfq gene encoding the host factor for phage Q beta. J Bacteriol 176: 531–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Tsui HCT, Leung HCE, Winkler ME (1994) Characterization of broadly pleiotropic phenotypes caused by an hfq insertion mutation in Escherichia coli K 12. Mol Microbiol 13: 35–49. [DOI] [PubMed] [Google Scholar]
- 30. Vecerek B, Rajkowitsch L, Sonnleitner E, Schroeder R, Blasi U (2008) The C-terminal domain of Escherichia coli Hfq is required for regulation. Nucleic Acids Res 36: 133–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. AbdelRahman YM, Rose LA, Belland RJ (2011) Developmental expression of non-coding RNAs in Chlamydia trachomatis during normal and persistent growth. Nucleic Acids Res 39: 1843–1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. National Nosocomial Infections Surveillance (NNIS) System (2004) Report, data summary from January 1992 through June 2004, issued October 2004. American Journal of Infection Control 32: 470–485. [DOI] [PubMed] [Google Scholar]
- 33. Yamada J, Yamasaki S, Hirakawa H, Hayashi-Nishino M, Yamaguchi A, et al. (2010) Impact of the RNA chaperone Hfq on multidrug resistance in Escherichia coli . J Antimicrob Chemother 65: 853–858. [DOI] [PubMed] [Google Scholar]
- 34. Moon K, Gottesman S (2009) A PhoQ/P-regulated small RNA regulates sensitivity of Escherichia coli to antimicrobial peptides. Mol Microbiol 74: 1314–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Roscetto E, Angrisano T, Costa V, Casalino M, Förstner KU, et al. (2012) Functional characterization of the RNA chaperone Hfq in the opportunistic human pathogen Stenotrophomonas maltophilia . J Bacteriol 194: 5864–5874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wesolowski D, Alonso D, Altman S (2013) Combined effect of a peptide–morpholino oligonucleotide conjugate and a cell-penetrating peptide as an antibiotic. Proc Natl Acad Sci U S A 110: 8686–8689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Rahme LG, Stevens EJ, Wolfort SF, Shao J, Tompkins RG, et al. (1995) Common virulence factors for bacterial pathogenicity in plants and animals. Science 268: 1899–1902. [DOI] [PubMed] [Google Scholar]
- 38. Casadaban MJ (1976) Transposition and fusion of the lac genes to selected promoters in Escherichia coli using bacteriophage lambda and Mu. J Mol Biol 104: 541–555. [DOI] [PubMed] [Google Scholar]
- 39. Zhang A, Wassarman KM, Ortega J, Steven AC, Storz G (2002) The Sm-like Hfq protein increases OxyS RNA interaction with target mRNAs. Mol Cell 9: 11–22. [DOI] [PubMed] [Google Scholar]
- 40. Lefebre MD, Valvano MA (2002) Construction and evaluation of plasmid vectors optimized for constitutive and regulated gene expression in Burkholderia cepacia complex isolates. Appl Environ Microbiol 68: 5956–5964. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information File combining Supplementary Methods, Supplementary Results, Table S1 (Putative MtvR targets in the genome of E. coli predicted by TargetRNA), Table S2 (Putative MtvR targets in the genome of E. coli predicted by RNAPredator), Table S3 (Putative MtvR targets in the genome of E. coli predicted by sRNATarget), Table S4 (Regions with homology to within the genome of E. coli), Table S5 (Putative MtvR targets in the genome of P. aeruginosa UCBPP-PA14 predicted by TargetRNA), and Table S6 (Putative MtvR targets in the genome of P. aeruginosa UCBPP-PA14 predicted by RNAPredator), and Supplementary References.
(PDF)
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. All data are included within the manuscript.