Abstract
The development of affinity purification technologies combined with mass spectrometric analysis of purified protein mixtures has been used both to identify new protein–protein interactions and to define the subunit composition of protein complexes. Transcription factor protein interactions, however, have not been systematically analyzed using these approaches. Here, we investigated whether ectopic expression of an affinity tagged transcription factor as bait in affinity purification mass spectrometry experiments perturbs gene expression in cells, resulting in the false positive identification of bait-associated proteins when typical experimental controls are used. Using quantitative proteomics and RNA sequencing, we determined that the increase in the abundance of a set of proteins caused by overexpression of the transcription factor RelA is not sufficient for these proteins to then co-purify non-specifically and be misidentified as bait-associated proteins. Therefore, typical controls should be sufficient, and a number of different baits can be compared with a common set of controls. This is of practical interest when identifying bait interactors from a large number of different baits. As expected, we found several known RelA interactors enriched in our RelA purifications (NFκB1, NFκB2, Rel, RelB, IκBα, IκBβ, and IκBε). We also found several proteins not previously described in association with RelA, including the small mitochondrial chaperone Tim13. Using a variety of biochemical approaches, we further investigated the nature of the association between Tim13 and NFκB family transcription factors. This work therefore provides a conceptual and experimental framework for analyzing transcription factor protein interactions.
Mapping the complex network of interactions between proteins provides insight into how a cell's protein machinery functions (1–4). Much progress has been made in uncovering these networks, broadening our understanding of many proteins' biological functions and elucidating the architecture of many multi-subunit protein complexes (5). Despite this, there have been few studies focused on investigating proteins associated with transcription factors (3, 6). There are technical challenges to address when trying to identify transcription-factor-associated proteins. The endogenous transcription factors might be expressed at relatively low levels, and their interactions might vary according to the state of the cell or in response to signals. If affinity-tagged transcription factors are expressed ectopically at higher levels, it might be possible to uncover bona fide interactions, but this can result in gene expression changes in transcription-factor-expressing cells. Such changes need to be considered when analyzing experimental data. Here we focused on identifying transcription-factor-associated proteins using affinity-tagged, constitutively expressed transcription factors as bait in affinity purification mass spectrometry experiments.
Multidimensional protein identification technology (MudPIT)1 has been widely used to identify “prey” proteins that co-purify with an affinity-tagged “bait” protein (7). Although genetically tractable model systems such as yeast allow affinity tagging of the endogenous protein under the control of its own promoter, this is not as easy with higher eukaryotic systems, and a constitutively overexpressed recombinant bait is often used (8). There are advantages and drawbacks to this approach. First, having high amounts of bait might be propitious for the identification of bona fide bait interactors that are present in very low amounts in cells. Second, DNA constructs overexpressing tagged recombinant baits are often commercially available; it is straightforward to screen a significant number of bait proteins for new interactors in medium-throughput studies using these constructs. However, this strategy does not reflect the typical expression pattern of the endogenous counterpart of the bait in vivo, and so some interactions identified might be artifacts. Alternative strategies for affinity purification mass spectrometry include using promoters that maintain constitutive expression but express baits at lower levels or that use inducible promoters so that expression of the bait can be controlled as a function of time. Whichever system is used, it is important to identify bait-associated proteins that physically associate with the bait during purification, and not simply contaminants that do not result from bait enrichment.
Control experiments are commonly used to evaluate the subset of proteins that are enriched specifically by the presence of the bait protein during purification and filter out contaminants that do not depend directly on the bait enrichment (for example, proteins that interact nonspecifically with the affinity resin used for purification) (9). Previously, the set of proteins co-purifying with the bait has been compared either to the set of proteins purified from cells expressing the affinity tag alone (10) or to proteins purified from an untransfected parental cell line (11). A concern is that such control cells are not exposed to the bait protein during growth and therefore do not have the opportunity to respond to the presence of the bait as the experimental cells do. If the bait is a protein that might have significant effects on the relative abundance of cellular proteins (such as a transcription factor), it is possible that these effects could alter the pool of proteins present in the cellular extracts used for purification. As a consequence, the population of proteins purifying nonspecifically with the bait might be altered. This is of particular concern when using sensitive techniques such as MudPIT for identifying protein–protein interactions and when using baits, such as affinity-tagged transcription factors, that might lead to an increase in the expression of many cellular proteins.
To begin to address this concern, we used the well-characterized transcription factor RelA as a bait for identifying RelA-associated proteins (12). First, we developed a label-free quantitative proteomics workflow to generate lists of putative RelA interactors using typical experimental controls. Next we confirmed that the nonspecific contaminants co-purifying in the controls were largely the more abundant cellular proteins. Having observed by means of RNA sequencing that overexpression of a tagged transcription factor bait caused significant changes in global patterns of gene expression, we used an alternative set of control experiments to ask whether these altered patterns of gene expression were sufficient to cause false positive identifications of RelA-associated proteins. Finally, we focused on one of the novel RelA-associated proteins that we had identified, Tim13 (13), and investigated the nature of its association with RelA and the NFκB family member p105 (NFκB1) (14).
EXPERIMENTAL PROCEDURES
Materials
Anti-α-tubulin mouse monoclonal antibody (T9026), anti-FLAG (M2) agarose, and anti-FLAG peptide were purchased from Sigma. Anti-p65 rabbit polyclonal antibody was from Abcam® (Cambridge, MA). Magne™ HaloTag® magnetic affinity beads were purchased from Promega (Madison, WI). The following full-length human clones, used for subcloning, were from the Kazusa DNA Research Institute (Kisarazu, Chiba Japan): RELA (AB464351), NFKB1 (AB489154), NFKB2 (AB384801), and JUN (AB385120).
Construction of Vectors for Expression of Affinity-tagged Bait Proteins
The vectors FLAG pcDNA5/FRT PacI PmeI and Halo pcDNA5/FRT PacI PmeI were constructed by inserting a DNA fragment encoding either a FLAG or a Halo tag between the NheI and KpnI restriction sites of the vector pcDNA5/FRT (Invitrogen). DNA fragments were synthesized in PCR reactions using the following pairs of primers: NheI FLAG fwd (5′-CAGGCTAGCATCATGCCAGACTACAAGGACGATGATGACAAG-3′) and KpnI PacI FLAG rev (5′-CAGGGTACCTTAATTAACTTGTCATCATCGTCCTTGTAGTCTGGCAT-3′), or NheI Halo fwd (5′-CAGGCTAGCATCATGGCAGAAATCGGTACTGGCTTTCC-3′) and KpnI PacI Halo rev (5′-CAGGGTACCTTAATTAAGTTATCGCTCTGAAAGTACAGATCCTCAGTGG-3′). Sequences containing the ORFs of the bait proteins flanked by SgfI and PmeI restriction sites were then subcloned between the PacI and PmeI restriction sites in the FLAG pcDNA5/FRT PacI PmeI and Halo pcDNA5/FRT PacI PmeI vectors.
Preparation of Whole Cell Extracts
Approximately 2 × 107 HEK293T cells were transfected with 7.5 μg of plasmid DNA encoding the proteins indicated in the figure legends. Cells were harvested 48 h post-transfection and rinsed twice in ice-cold PBS. Cell pellets were frozen at −80 °C and later thawed, and the cells were resuspended in 300 μl of ice-cold buffer containing 50 mm Tris·HCl (pH 7.5), 150 mm NaCl, 1% Triton® X-100, 0.1% sodium deoxycholate, 0.1 mm benzamidine HCl, 55 μm phenanthroline, 10 μm bestatin, 20 μm leupeptin, 5 μm pepstatin A, and 1 mm PMSF. We then homogenized the lysates by passing the cells through a 26-gauge needle five times, and the lysates were then centrifuged at 21,000 × g for 30 min at 4 °C to remove insoluble material.
Purification of Protein Complexes
300 μl of whole cell extract was diluted with 700 μl of TBS (50 mm Tris·HCl, pH 7.4, 137 mm NaCl, 2.7 mm KCl) and centrifuged at 21,000 × g for 10 min at 4 °C. FLAG-tagged bait complexes were purified via anti-FLAG agarose immunoaffinity chromatography. The diluted lysates were incubated with 50 μl of anti-FLAG (M2) agarose beads for 2 h at 4 °C. The beads were washed four times with wash buffer containing 50 mm Tris·HCl, pH 7.4, 137 mm NaCl, 2.7 mm KCl, and 0.05% Nonidet® P40, and bound proteins were eluted from the beads with 100 μl of TBS containing 0.3 mg/ml FLAG peptide. Eluates were centrifuged through Micro Bio-Spin columns (Bio-Rad) to remove any traces of affinity resin. Halo-tagged bait complexes were purified using Magne™ HaloTag® magnetic affinity beads (Promega). Diluted lysates were incubated for 2 h at 4 °C with beads prepared from 100 μl of Magne™ HaloTag® bead slurry according to the manufacturer's instructions. The beads were washed four times with wash buffer, and bound proteins were eluted from the beads via incubation with 2 units of AcTEV™ Protease (Invitrogen) in 100 μl of buffer containing 50 mm Tris·HCl, pH 8.0, 0.5 mm EDTA, and 0.005 mm DTT for 2 h at 25 °C. Eluates were then centrifuged through Micro Bio-Spin columns (Bio-Rad). Where indicated, anti-FLAG agarose eluates prepared from cells expressing FLAG-Tim13 and Halo-p105 were further analyzed by means of anion exchange chromatography using an ÄKTA fast protein liquid chromatography system (Amersham Biosciences). Eluates were dialyzed in buffer A (25 mm HEPES·NaOH, pH 7.5, 1 mm DTT) containing 0.1 m NaCl and applied to a HiTrap DEAE Sepharose column (GE Healthcare) equilibrated in buffer A (0.1 m NaCl). The column was eluted with a 19-ml linear gradient from 0.1 to 1.0 m NaCl in buffer A, and 0.5-ml fractions were collected.
Expression of Recombinant Proteins in Insect Cells
Sequences coding for either Halo-p105434–968 or FLAG-Tim13 were subcloned into pBacPAK8, and recombinant baculoviruses were generated using the BacPAK expression system (Clontech, Mountain View, CA). Sf21 cells were cultured at 27 °C in Sf-900III SFM (Life Technologies™, Grand Island, NY). Dishes containing ∼1 × 107 Sf21 cells were infected with the baculoviruses indicated. Forty-eight hours after infection, cells were harvested and lysed in 1 ml of ice-cold buffer containing 50 mm HEPES-NaOH, pH 7.9, 0.15 m NaCl, 5 mm MgCl2, 0.2% Triton X-100, 20% (v/v) glycerol, 0.1% sodium deoxycholate, 0.1 mm benzamidine HCl, 55 μm phenanthroline, 10 μm bestatin, 20 μm leupeptin, 5 μm pepstatin A, and 1 mm PMSF. Lysates were centrifuged at 20,000 × g for 30 min at 4 °C.
Analysis of Global mRNA Levels
Total RNA was isolated from ∼1 × 108 HEK293T cells expressing Halo-RelA, Halo-NFkB1(p105), or the Halo tag alone using RNeasy Midi kits (Qiagen, Valencia, CA). During purification, samples were treated with DNase to remove any contaminating DNA. TruSeq RNA sample preparation kits (Illumina, San Diego, CA) were used to prepare individually barcoded libraries for Illumina sequencing according to the manufacturer's instructions. Libraries were pooled for multiplex sequencing with 50 nt read lengths using two lanes of an Illumina Genome Analyzer IIx. The resulting sequences were mapped to the human genome using the TopHat aligner (15). Gene expression was quantified, and RNA levels from samples overexpressing either Halo-RelA or Halo-NFkB1 were then compared with RNA levels from control cells to identify differentially expressed genes using Cuffdiff version 1.0.3 (16, 17).
Analysis of Protein Complexes via MudPIT Mass Spectrometry
Affinity purified proteins were precipitated with trichloroacetic acid, washed twice with acetone, and resuspended in buffer containing 100 mm Tris·HCl, pH 8.5, and 8 m urea (18). Disulfide bonds were reduced with Tris(2-carboxylethyl)-phosphine hydrochloride, and the samples were then treated with chloroacetamide to prevent disulfide bond reformation. Proteins were then digested with endoproteinase Lys-C and trypsin as described previously. The resulting peptides were loaded onto fused silica microcapillary columns packed with three phases of chromatography resin (reverse phase, strong cation exchange, reverse phase). Peptides were gradually eluted from the columns directly into the mass spectrometer for analysis using 10 ∼2-h chromatography steps (7). The resulting .raw files describing the MS/MS spectra were processed by the in-house software package RAWDistiller v. 1.0 to generate .ms2 files from which MS/MS spectra were matched to 29,375 human protein sequences (from the National Center of Biotechnology Information November 2010 release) using the SEQUEST algorithm (version 27, rev. 9) (19). Enzyme specificity was not imposed during searching. The mass tolerance was set at 3 amu for precursor ions and 0 amu for fragment ions. A static modification of +57 Da was added to cysteine residues to account for carboxamidomethylation; no variable modifications were searched. Matches that were not sufficiently accurate were filtered out using DTASelect prior to the assembly of a protein list for each sample (20). Parameters for filtering included a minimum DeltCn of 0.08; minimum XCorr values of 1.8 (singly charged spectra), 2.0 (doubly charged spectra), and 3.0 (triply charged spectra); a maximum Sp rank of 10; and a minimum peptide length of 7 amino acids. Only fully tryptic peptides were considered. Using these selection criteria, we obtained an average spectral false discovery rate (FDR) for the 60 MudPIT runs of 0.27% ± 0.17% (standard deviation), and the average protein FDR was 2.95% ± 2.41% (standard deviation). A list of proteins present only in the majority of replicate experimental samples was generated using Contrast and NSAF7 software (20). The parsimony option in Contrast was used to remove proteins that were subsets of others. If proteins were identified by the same set of peptides (including at least one peptide unique to the set to distinguish between isoforms), they were grouped together; one representative accession number was used to describe the set. Finally the abundance of these proteins in experimental and control samples was assessed using spectral counting to calculate dNSAF values (21); proteins with a high probability of being enriched in experimental but not control samples were determined using the PLGEM signal-to-noise method (22, 23). Proteins were then sorted according to PLGEM signal-to-noise values, and FDRs were calculated from PLGEM determined p values (23, 24). Identified proteins were then categorized according to their associated FDRs as category I (FDR = <0.1%), category II (FDR = 0.1% to 1%), or category III (FDR = 1% to 5%).
RESULTS
Identifying a Set of Bait-associated Proteins via MudPIT Mass Spectrometry
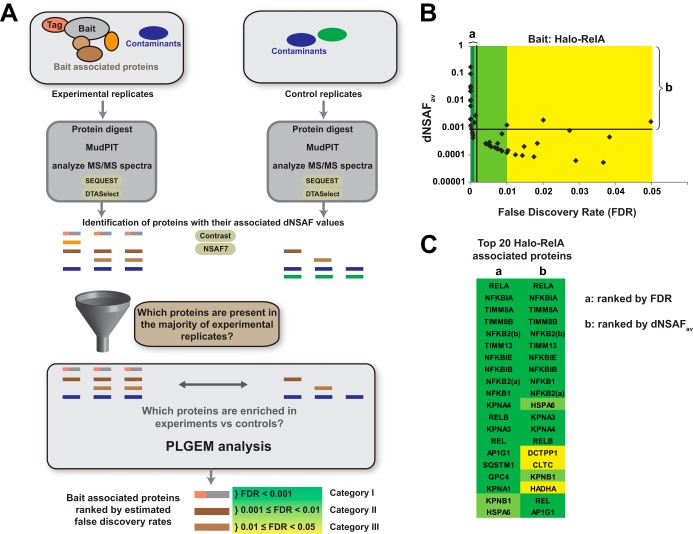
To identify proteins associated with the transcription factor RelA, we expressed either the Halo tag alone (control) or Halo-RelA (experiment) in HEK293 cells (control: nine replicates; experiment: six replicates). After subjecting the resulting whole cell extracts to Halo affinity chromatography, we identified proteins present in each purification using MudPIT (25) (reviewed in Ref. 7) (Fig. 1A). To analyze MudPIT data, we used the SEQUEST algorithm (19) and DTASelect (20) to assemble a list of proteins identified in each sample (26). We next determined which proteins were enriched in the Halo-RelA samples but not in control samples expressing the Halo tag alone (Halo-RelA-associated proteins). These Halo-RelA-associated proteins (prey) might co-purify as a result of direct physical interactions between the Halo-RelA bait and the prey, or through interactions mediated by other molecules. To create identifications that would be useful in follow-up studies, we removed proteins that were detected in ≤50% of the experimental Halo-RelA samples, limiting the number of false positive identifications at the expense of rejecting a proportion of genuine bait interactors (false negatives) (Fig. 1A). Having defined this shortlist of possible Halo-RelA-associated proteins, we next asked which of these were enriched in experimental relative to control samples. To make this quantitative comparison, we first assessed the relative amounts of each protein in each sample by calculating dNSAF values for each protein (21), and we then used the dNSAF values as input for the PLGEM (22). Finally, we used the estimated p values calculated by PLGEM to estimate FDRs (23, 24) and defined three categories of likely Rel-A-associated proteins based on the calculated FDRs; these were 0 ≤ FDR < 0.001 (Category I), 0.001 ≤ FDR < 0.01 (Category II), and 0.01 ≤ FDR < 0.05 (Category III) (Fig. 1A). Our analysis identified 50 Halo-RelA-associated proteins (18 Category I, 18 Category II, and 14 Category III). Category I identifications with low FDRs tended to be proteins that were more abundant in the purifications (with greater dNSAF values) (Fig. 1B). Lastly, to confirm the robustness of this approach, we repeated our analyses using tagged transcription factors not in the NFκB family as controls (supplemental Data S3).
Fig. 1.
A workflow for identifying bait interacting proteins using MudPIT mass spectrometry and PLGEM statistical analysis. A, affinity-tagged bait-associated proteins purified from a minimum of three replicate experiments are digested into peptide fragments and analyzed via MudPIT as described in the text. The resulting MS/MS spectra are matched to peptides generated from an in silico digest of the human proteome database and a list of putative bait interacting proteins assembled using the SEQUEST and DTASelect algorithms. In order to reduce the likelihood of false positive identifications, replicate experiments were compared using in-house-developed software (NSAF7); any proteins not found in a majority of the experimental samples were excluded from further analysis. For each protein identified in >50% of the experimental samples, dNSAF values for experimental samples were compared with dNSAF values for control samples; the PLGEM algorithm was used to assess the likelihood of a protein being enriched in experimental samples and calculate a p value for each comparison. The resulting p values were used to calculate false discovery rates (FDRs) using the method of Benjamini and Hochberg. Finally, three categories of bait-associated proteins were defined using the FDR values. B, the workflow applied to the identification of 50 Halo-RelA-associated proteins using nine control and six experimental purifications (supplemental Data S1 and S2), with the average experimental dNSAF and FDR values indicated for each identification. The top 20 RelA-associated proteins (including RelA) are indicated, ranked either by FDR (a) or by average dNSAF with an FDR cutoff of 0.05 (b). C, the identities of the top 20 RelA-associated proteins shown in B, indicated with the respective Human Genome Gene Nomenclature Committee official gene symbol. The two isoforms of the NFκB2 protein are indicated as NFKB2(a) and NFKB2(b).
Are Halo-RelA-associated Proteins Interacting Physically with the Bait?
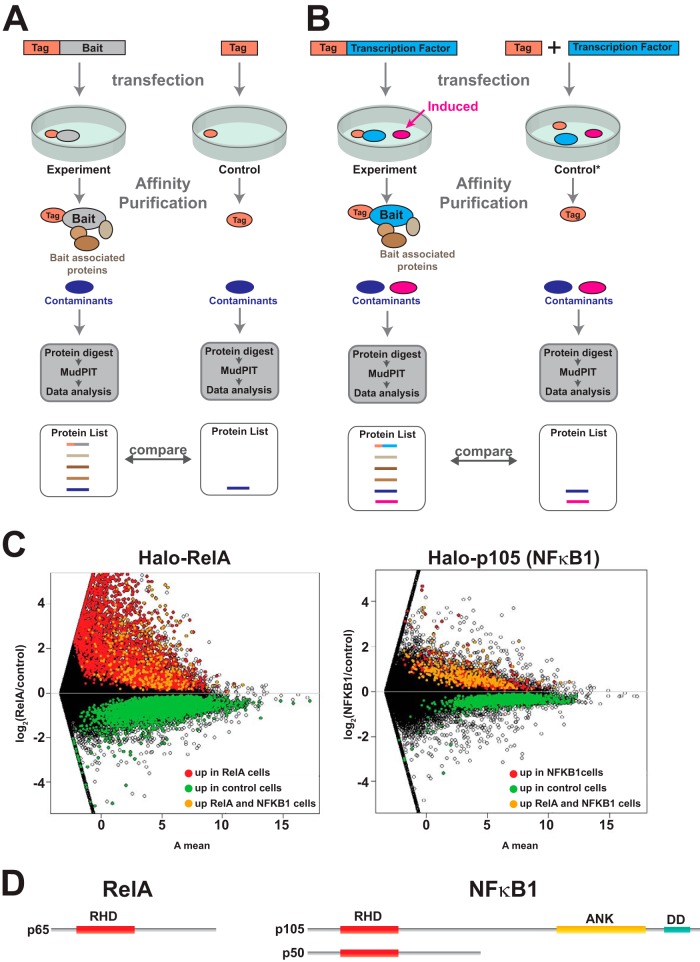
The RelA-associated proteins might have been present in our samples as the result of either direct or indirect interactions with the Halo-RelA bait. An alternative possibility is that a protein might co-purify in detectable quantities for reasons not involving physical interactions between bait and prey; for example, proteins might co-purify simply as a result of their high abundance in the cellular lysate. We thought it was important to address this possibility, particularly in the case where expression of the tagged bait might cause significant changes in gene expression between experimental and control samples—for example, when the bait is a transcription factor (compare Figs. 2A and 2B). If this is the case, then alternative control experiments are needed (Fig. 2B).
Fig. 2.
Ectopically expressed bait affects global gene expression. A, comparing proteins purified from experimental, affinity-tagged bait transfected cells with affinity-tag-only control transfected cells assuming that the transfections do not differ in their effects on global patterns of gene expression. In this model, the same contaminants (shown in dark blue) co-purify nonspecifically in both bait and control experiments and are accounted for during data processing. B, Alternatively transfecting cells with DNA encoding the bait might cause significant changes in global gene expression (for example, if the bait is a transcription factor). It is possible that a protein induced or up-regulated following transfection with such bait might co-purify nonspecifically as a contaminant (pink). In this case a different control might be needed (control denoted by asterisk). C, changes in global gene expression patterns among three biological replicates of cells transfected with the Halo-control plasmid and three replicates of cells transfected with either the transcription factor Halo-RelA or the NFκB family member Halo-p105 (NFκB1). Changes in gene expression are represented using an MA plot (intensity ratio versus average intensity). Differentially expressed genes are shown in color (genes called by the Cuffdiff algorithm with an adjusted p value < 0.05 and fold change (FC) > 1.4) (16). D, the DNA binding transcription factor RelA contains a Rel homology domain (RHD). In addition to the RHD, p105 (NFκB1) contains ankyrin repeats (ANK) and a death domain (DD). The p105 protein is partially processed into the NFκB1 transcription factor p50.
Overexpressing Transcription Factors Can Cause Significant Global Changes in Gene Expression
To confirm that a bait transcription factor might alter the gene expression profile of experimental cells relative to controls, we used RNA sequencing to compare the global patterns of gene expression in cells overexpressing the RelA transcription factor with that of control cells expressing only the Halo tag (Fig. 2C). We found 6007 differentially expressed genes in cells overexpressing the Halo-RelA transcription factor. We compared this with gene expression changes caused by overexpression of another NFkB family member, NFκB1(p105). Although the p105 protein is partially processed into the transcription factor NFκB (p50) in cells (27, 28), the full-length p105 protein contains ankyrin repeat domains (Fig. 2D) and can function as an inhibitor of NFκB-activated transcription (29). In contrast to the cells overexpressing Halo-RelA, we found only 732 differentially expressed genes in cells overexpressing p105, 456 of which were differentially expressed in both Halo-RelA- and Halo-p105-expressing cells.
Abundant Proteins Can Co-purify Nonspecifically during Affinity Purification
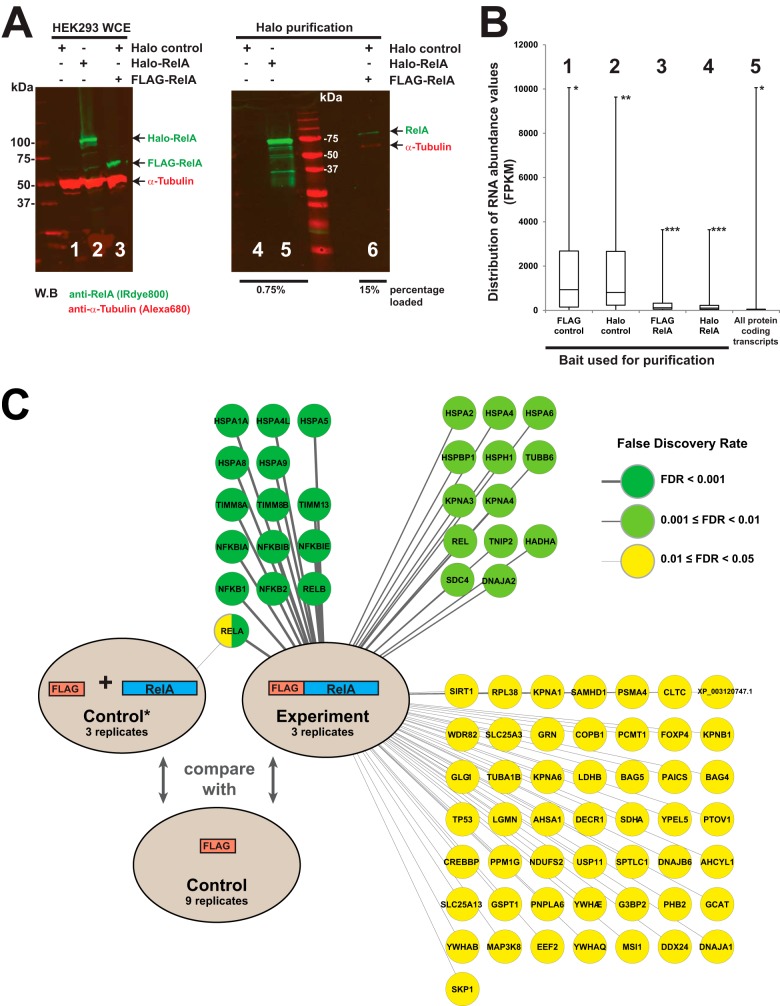
Having confirmed that Halo Rel-A transfected cells had a significantly altered gene expression profile, we next asked whether highly expressed proteins might co-purify nonspecifically as contaminants during affinity purification. We transfected cells with Halo-control (expressing the 33-kDa Halo tag), Halo-RelA, or both Halo-control and FLAG-RelA and subjected the resulting cell lysates to Halo affinity purification (Fig. 3A). We analyzed the eluates from these purifications via fractionation with SDS-PAGE followed by Western blotting and detected proteins using the LiCor Odyssey infrared imaging system, which has been shown to be particularly sensitive for detecting small amounts of protein (30). The Halo-RelA and smaller FLAG-RelA proteins were expressed at similar levels in the whole cell extracts (Fig. 3A, lanes 1–3). After purification, as expected, we were able to detect RelA in cells transfected with Halo-RelA but not in cells transfected with the Halo control plasmid when a modest quantity (0.75%) of the eluate was analyzed via Western blotting (lanes 4 and 5). Interestingly, we were also able to detect a small quantity of nonspecifically enriched FLAG-RelA, together with what we presumed was contaminating α-tubulin, when we analyzed a larger sample of the eluate (15%) from cells overexpressing FLAG-RelA (lane 6). These results suggest that small amounts of proteins present in large quantities in whole cell extract (FLAG-RelA and α-tubulin) can be retained in the solid phase during a typical purification procedure at levels that allow detection via the relatively sensitive techniques we used.
Fig. 3.
Small amounts of highly expressed proteins are purified nonspecifically during affinity purification. A, small but detectable amounts of recombinant overexpressed FLAG-RelA are observed after enrichment of proteins by Halo purification. HEK293T cells were transfected with DNA plasmids expressing the Halo tag alone, Halo-RelA, or FLAG-RelA, and the resulting whole cell extracts were subjected to Halo affinity purification as described in the text. Proteins present in the eluates were fractionated via SDS-PAGE and visualized through Western blotting. RelA protein was detected using anti-RelA rabbit polyclonal primary antibodies and IRDye™-800-labeled goat anti-rabbit IgG secondary antibodies (green); α-tubulin was detected using anti-α-tubulin mouse monoclonal primary antibodies and Alexa Fluor 680–labeled anti-mouse IgG secondary antibodies (red). A Li-Cor Odyssey infrared imaging system was used to detect the fluorescently labeled secondary antibodies. B, the distribution of RNA transcript abundances for different groups of proteins identified via mass spectrometry (groups 1–4, supplemental Data S4) or for all cellular proteins (group 5). FKPM values used to assess mRNA abundance were derived from RNA purified from cells transfected with either Halo-control (groups 1, 2, and 5) or Halo-RelA plasmids (groups 3 and 4). Box plots of the distribution of FKPM values indicate sample minimum, lower quartile, median, upper quartile, and sample maximum value. Groups of proteins analyzed are the top 50 detected in the control purifications via MudPIT (groups 1 and 2), the top 50 enriched in the experimental RelA purifications (groups 3 and 4), and all protein-coding transcripts in Halo-control transfected cells (group 5). FKPM values for the overexpressed RelA baits were not calculated for groups 3 and 4. Proteins in each group with the highest FKPM values were *the ribosomal protein RPL14 (groups 1 and 5), **the ribosomal protein RPS3 (group 2), and ***the NFκB inhibitor NFKBIA (groups 3 and 4). C, small amounts of contaminating, overexpressed Halo-RelA, but no other proteins, were identified via MudPIT/PLGEM analysis (FDR cutoff of 0.05) in FLAG purified Halo-RelA-expressing cells. In contrast, 77 bait-associated proteins were identified in FLAG-RelA-expressing cells subject to FLAG immunoaffinity purification (FDR cutoff of 0.05) (supplemental Data S1 and S5).
To gain additional evidence that nonspecific purification of a protein might result from its high abundance, we wanted to ask whether the proteins co-purifying in our control samples tended to be proteins that were the more abundant proteins in cells. To test this, we used our RNA sequencing and MudPIT data to compare the distribution of the FKPM values of RNA transcripts corresponding to groups of proteins present in our FLAG-control and Halo-control purifications with the distribution of the FKPM values of the group of all cellular proteins (compare groups 1 and 2 with group 5 in Fig. 3B). The FKPM values calculated by Cufflinks reflect the relative abundance of RNA transcripts (16, 31). We found that groups of proteins identified in the control purifications had transcript FKPM values distributed much higher than the group of all cellular proteins. This was perhaps not surprising, as many cellular proteins either are not expressed in cells or are expressed at very low levels, and identification of proteins via MudPIT would naturally be limited to proteins present in detectable amounts. We therefore decided to also compare the RNA abundance of proteins in our control purifications with the RNA abundance of proteins identified in experimental purifications (compare groups 1 and 2 with groups 3 and 4 in Fig. 3B). We observed that the abundance of RNA transcripts of proteins consistently purified from control cells tended to be much greater than the abundance of transcripts associated with proteins enriched in experimental purifications. Taken together, the results presented in Figs. 3A and 3B suggest that high abundance of a protein may result in its purification as a nonspecific contaminant.
Do the Changes in Gene Expression Caused by Overexpression of the Transcription Factor RelA Result in False Positive Identification of RelA-associated Proteins?
We next wanted to see whether the changing patterns of gene expression resulting from transfecting cells with RelA were sufficient to cause false positive identification of up-regulated proteins in purifications. We analyzed anti-FLAG immunoaffinity purifications from cells co-transfected with FLAG-control and Halo-RelA. When we compared proteins identified in three replicates of these samples with proteins identified from cells transfected with only the FLAG-control DNA, we detected RelA as a (contaminating) protein enriched in the cells overexpressing Halo-RelA (FDR < 0.05). This was consistent with the results of the reciprocal experiment outlined in Fig. 3A. Our analysis did not identify any other proteins as significantly enriched from the extracts of Halo-RelA overexpressing cells that had been subjected to anti-FLAG agarose immunoaffinity chromatography. This suggests that the changes in gene expression caused by overexpressing Halo-RelA were insufficient to cause spurious purification and aberrant identification of up-regulated proteins. In contrast, when we analyzed three replicates of FLAG purified extracts of cells expressing FLAG-RelA and compared these with controls transfected with the FLAG-control DNA alone, we identified 77 FLAG-RelA-associated proteins at a modest FDR of 0.05, including 14 FLAG-RelA-associated proteins with FDR values less than 0.001 (Fig. 3C). The results from a number of follow-up experiments using different baits and/or different purification techniques with similar analyses also suggested that spurious purification of up-regulated proteins was unlikely to result in false protein identifications (supplemental Data S5).
Investigating the Nature of the Association between the Mitochondrial Protein Tim13 and the RelA and NFκB1 (p105) Proteins
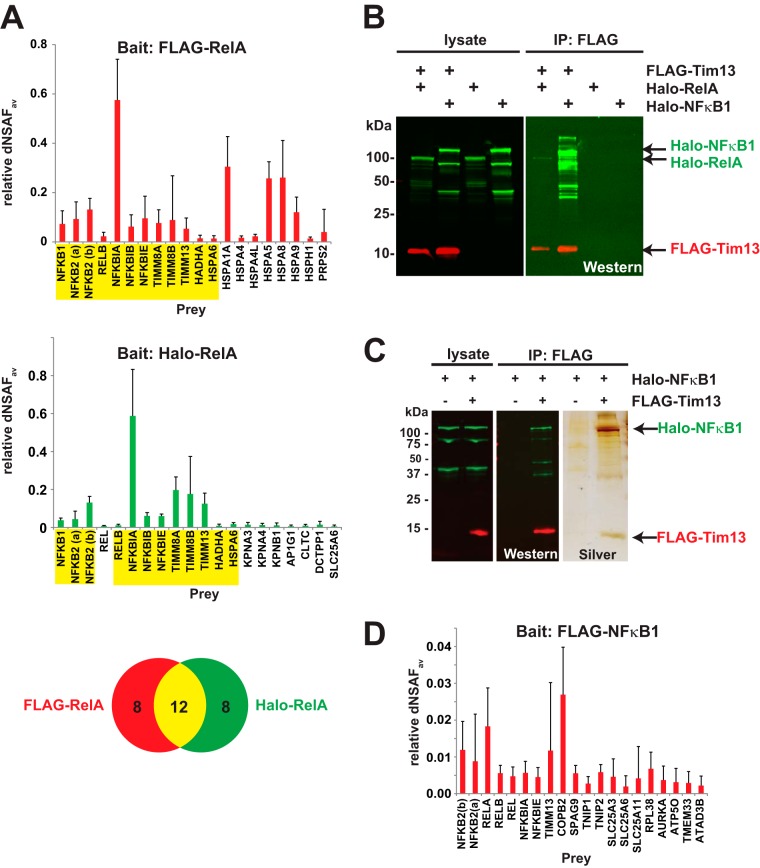
Having identified a set of proteins that consistently co-purified with overexpressed FLAG-RelA, we investigated whether any of these proteins were potential binding partners of NFκB family proteins. When we compared the 20 most abundant proteins enriched in FLAG-RelA or Halo-RelA purifications (FDR < 0.05) (Fig. 4A), we found 12 prey proteins that co-purified with both Halo-RelA and FLAG-RelA. These included four of the NFκB family transcription factors (NFκB1, NFκB2, RelA, and RelB) that can bind in different combinations to form dimers (32); also included were members of the IκB family (IκBα, IκBβ, and IkBε), known to bind to NFκB dimers (33). Thus our strategy works for identifying biologically relevant interactions. Novel associations included the mitochondrial trifunctional protein α subunit encoded by the HADHA gene and several components of the mitochondrial Tim8–Tim13 complex encoded by the genes TIMM8A, TIMM8B, and TIMM13. As the Tim13 protein had not been previously described in association with NFκB transcription factors, we wanted to investigate the nature of its association in more depth. First, we asked whether the Tim13 protein associates with other NFκB family members. We co-expressed FLAG-Tim13 with either Halo-RelA or Halo-NFκB1 (p105) and purified the resulting complexes using anti-FLAG immunoaffinity chromatography (Fig. 4B). FLAG-Tim13 co-immunoprecipitated significant amounts of Halo-NFκB1 and modest amounts of Halo-RelA (Fig. 4B). We further analyzed FLAG purified FLAG-Tim13/Halo-NFκB1 eluates by means of SDS-PAGE followed by silver staining (Fig. 4C). We detected Tim13 and NFκΒ1 but only relatively modest amounts of other proteins in these eluates. One possible explanation for this might be that FLAG-Tim13 interacts directly with Halo-NFkB1. Finally, we used MudPIT to examine endogenous proteins co-purifying with FLAG purified FLAG-NFκB1 and found Tim13 to be among the top 20 most abundant proteins enriched in these samples (FDR < 0.05) (Fig. 4D). Taken together, these experiments are consistent with a physical interaction between NFκB1 and Tim13.
Fig. 4.
The mitochondrial proteins Tim8A, Tim8B, and Tim13 associate with NFκB family members. A, proteins purified from cells expressing FLAG-RelA (six replicates) or Halo-RelA (six replicates) were analyzed via MudPIT and compared with proteins detected in either FLAG-control transfected cells (eight replicates) or Halo-control transfected cells (nine replicates) (supplemental Data S1). The bait normalized dNSAF values of the 20 most abundant prey identifications are shown for either FLAG-RelA (red) or Halo-RelA (green) baits, with those common to both highlighted in yellow (FDR < 0.05 and not including the RelA bait). Error bars show standard deviation. B, C, co-purification of Halo-RelA or Halo-NFκB1 with FLAG-Tim13. Lysates from HEK293 cells expressing Halo-RelA, Halo-NFκB1, or FLAG-Tim13, as indicated in the figure, were used for FLAG affinity purifications as described in the text. Bound proteins were eluted with 0.3 mg/ml FLAG peptide, analyzed via SDS-PAGE, and visualized by means of either Western blotting or silver staining. FLAG-Tim13 was detected using mouse anti-FLAG(M2) monoclonal antibodies and Alexa Fluor 680-labeled anti-mouse IgG secondary antibodies (red); Halo-RelA or Halo-NFκB1 was detected with rabbit anti-Halo antibodies and IRDye™-800 labeled goat anti-rabbit IgG secondary antibodies (green). D, proteins purified from cells expressing FLAG-NFκB1 (five replicates) or transfected with FLAG-control DNA (eight replicates) were analyzed via MudPIT. Bait normalized dNSAF values for the 20 most abundant identifications are shown (FDR < 0.05 and not including the NFκB1 bait).
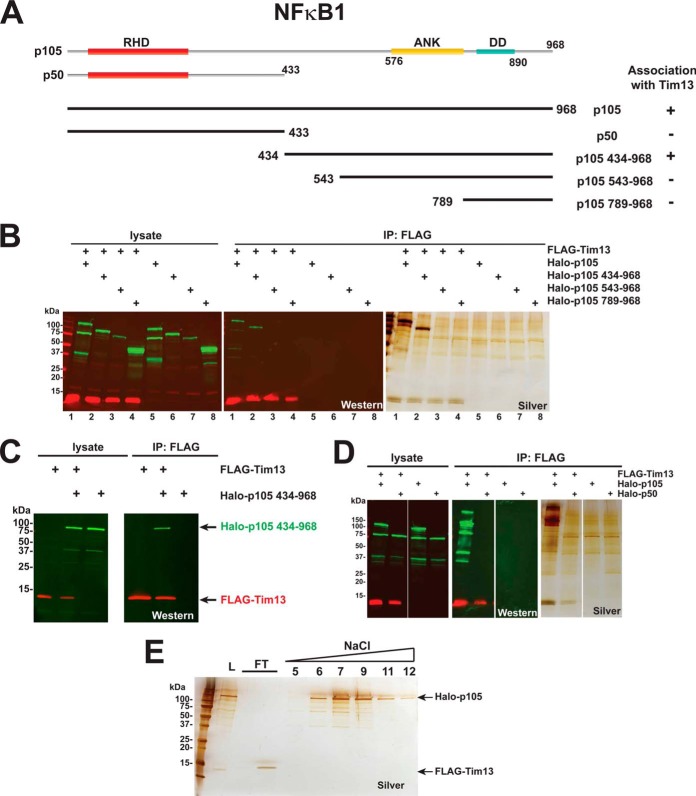
Having identified NFκB1 (p105) as a possible Tim13 interaction partner, we wanted to identify regions of p105 that might be important for their association. We co-expressed FLAG-Tim13 with each of the truncated Halo-p105 proteins described in Fig. 5A and subjected the resulting lysates to anti-FLAG chromatography (Figs. 5B and 5D). FLAG-Tim13 co-immunoprecipitated Halo-p105434–968 (Fig. 5B, lane 2) but not Halo-p50 (Fig. 5D), suggesting that the Tim13-p105 association might depend on a region within the C terminus of p105. In addition, FLAG-Tim13 overexpressed in Sf21 insect cells also co-immunoprecipitated Halo-p105434–968 (Fig. 5C). Shorter regions of the C terminus of p105, which lacked residues 435–542, did not co-immunoprecipitate with Tim13 (Fig. 5B, lanes 3 and 4). This region of p105 (residues 435–542) substantially overlaps a region previously identified as the p105 processing inhibitory domain (residues 474–544) (34). Protein interactions are diverse in their strength and permanence (35, 36). To test whether we could disrupt the association between Tim13 and NFκB1 (p105), we subjected our FLAG purified Tim13/p105 eluates to chromatographic separation on a HiTrap DEAE column. The Halo-p105 protein bound the column and was eluted at ∼410 mm NaCl, whereas the FLAG-Tim13 flowed through the column (Fig. 5E). This would be consistent with a weak interaction between Tim13 and NFκB1 that was not stable under the conditions of ion exchange chromatography.
Fig. 5.
Investigating the association between Tim13 and NFκB1 (p105). A, diagram of recombinant Halo-NFκB1 mutants. The Halo-p105 (NFkB1) synthetic construct (GenBank: AB489154.1) encoding the 968 amino acid p105 protein (GenBank: BAH24114.1) was used to construct plasmids encoding N-terminal Halo-tagged versions of the mutants shown. The positions of the Rel homology domain (RHD), ankyrin repeats (ANK), and the death domain (DD) are indicated. B, D, the recombinant affinity-tagged proteins indicated were expressed in HEK293 cells, and proteins were purified from the resulting lysates using anti-FLAG agarose chromatography. Samples were resolved via SDS-PAGE, FLAG-tagged (red) or Halo-tagged (green) proteins were detected via Western blotting, and total protein was detected via silver staining. C, co-purification of insect cell expressed Tim13 and p105 434–968. Lysates from Sf21 cells co-infected with FLAG-Tim13, Halo-p105 434–968, or both were subjected to anti-FLAG agarose chromatography as described in “Experimental Procedures.” Eluates were analyzed via SDS-PAGE, and proteins were detected via Western blotting. FLAG-tagged proteins were detected with mouse anti-FLAG (M2) monoclonal antibodies and IRDye™ 680-labeled goat anti-mouse IgG secondary antibodies; Halo-tagged proteins were detected with rabbit anti-Halo antibodies and IRDye™ 800-labeled goat anti-rabbit secondary antibodies. Fluorescently labeled secondary antibodies were detected with a Li-Cor Odyssey infrared imaging system. E, ion exchange chromatography of FLAG purified p105/Tim13. FLAG-Tim13 and Halo-p105 were co-expressed in HEK293 cells, and protein complexes were then purified by means of anti-FLAG affinity chromatography. The resulting eluates were adjusted to a conductivity equivalent to 100 mm NaCl (load L). Proteins were then resolved on a 1-ml HiTrap DEAE fast protein liquid chromatography column; aliquots of the indicated fractions were analyzed via SDS-PAGE and silver stained.
DISCUSSION
MudPIT has been used widely in affinity purification mass spectrometry experiments to define sets of proteins that co-purify specifically with affinity-tagged bait proteins (5). Because of the sensitivity achieved, there is an increased probability of detecting small amounts of protein contaminants that co-purify nonspecifically and would normally be beyond the limits of detection. Small but detectable amounts of contaminating proteins might be retained during purification, either simply as a result of their high abundance or due to an unusually high affinity for the resin used for the purification. Typically, such contaminants are also retained during control purifications using cells lacking the affinity-tagged bait and eliminated as putative bait interactors during analysis (Fig. 1). Previous studies have compared extracts from cells expressing the bait protein with cells lacking a tagged bait or with cells transfected with DNA expressing the epitope tag alone (10, 11). This approach has the advantage that one set of controls can be compared with results from many different types of bait, which is particularly useful when mass spectrometer time is at a premium. However, this also assumes that the cells expressing the affinity-tagged bait and the cells used for controls only differ in the presence or absence of the bait during purification. In fact, there is also the potential for differences in the populations of cells due to the presence or absence of the bait during cell growth. This might be of particular concern when investigating an affinity-tagged transcription factor, which might have profound effects on global patterns of gene expression (compare Fig. 2A with Fig. 2B).
We used the well-characterized NFκB DNA binding transcription factor RelA to begin to address whether this concern might lead to false positive identifications of transcription-factor-associated proteins. Endogenous RelA is thought to be sequestered in the cytoplasm in unstimulated cells by inhibitor proteins (IκBs) (32, 37). In response to a stimulus—for example, the binding of the cytokine TNFα to its receptor—IκBs are targeted for degradation, allowing NFκB transcription factor dimers to translocate to the nucleus and activate transcription of their target genes (32, 38). In order to identify RelA-associated proteins, we first established a workflow for processing data from MudPIT analyses of Halo purified samples. We used either Halo-RelA or Halo-control transfected cells to enable us to define groups of bait-associated proteins that were enriched in experimental samples relative to control samples (Fig. 1). We identified many well-characterized components of the NFκB pathway using this method (Fig. 1C). For example, among the 18 high-confidence RelA-associated proteins that we identified (FDR < 0.001) were the five NFκB transcription factors (RelA, RelB, Rel, NFκB1, and NFκB2), as well as three members of the IκB family (IκBα, IκBβ, and IκBε). Both the biological function of and the physical interactions between these proteins have been well documented (32). We next determined that using this approach might be problematic, as the control and experimental populations of cells in these purifications differ markedly in their global patterns of gene expression (Fig. 2C). Consequently, we asked whether a protein's abundance might influence its purification as a contaminant (Figs. 3A and 3B). This seems possible for at least some contaminants. Exogenous overexpressed FLAG-RelA was detectable in Western blots of Halo purified samples (Fig. 3A); in addition, FLAG purified overexpressed Halo-RelA was detectable via MudPIT (Fig. 3C). Common contaminants in typical control purifications also tend to be highly expressed in cells (Fig. 3B).
Having determined both that the bait transcription factor protein significantly affects patterns of gene expression and that contaminants might co-purify as a result of their cellular abundance, we asked whether these effects were enough to cause false positive identifications of RelA-associated proteins. The results presented in Fig. 3C and those of several follow-up studies (supplemental Data S5) suggest that in a number of cases where overexpression of the bait might be expected to have profound effects on gene expression, this does not lead to false positive identifications of bait-associated proteins.
In addition to a number of known RelA interacting proteins co-purifying with RelA, we observed that the mitochondrial protein Tim13 consistently co-purified with both FLAG-RelA and Halo-RelA (Fig. 4A). This was surprising, as neither Tim13 nor its binding partners Tim8A and Tim8B have been previously reported as NFκB-associated proteins. However, some evidence supports the possible biological role of an association between NFκB proteins and these small mitochondrial chaperones located in the intermembrane space. There have been reports that RelA and p50 (NFκB1) can localize to mitochondria and influence mitochondrial gene expression (39). In addition, a pool of RelA and its inhibitor IκBα has been localized to the mitochondrial intermembrane space (40). As the co-purification of the Tim8 and Tim13 proteins with RelA might have biological relevance, we decided to further investigate the nature of this association. We found that only modest amounts of RelA co-immunoprecipitated with recombinant Tim13 (Fig. 4B), so we tested the possibility that Tim13 might be associating with RelA via one of the other RelA binding proteins. Indeed, Tim13 co-immunoprecipitated significant amounts of NFκB1 (Figs. 4B and 4C), and endogenous Tim13 also co-purified with FLAG-NFκB1 (Fig. 4D). Taken together, these results are consistent with a physical interaction between Tim13 and NFκB1. To gain more support for a model involving such an interaction, we sought to define a region of NFκB1 important for its association with Tim13 (Fig. 5A). We found that Tim13 associated with NFκB mutants that included amino acids 435 to 542 (Figs. 5B to 5D). This region overlaps the processing inhibitory domain, which resides between residues 474 and 544 of p105. The processing inhibitory domain has previously been reported to be involved in inhibiting the processing of p105 into the transcription factor p50 (34). Next, we sought to test the stability of the Tim13–NFκB1 association. Protein–protein interactions in cells are diverse, and many important interactions involved with signaling pathways have modest affinity (41). In support of a model involving a transient interaction between Tim13 and NFκB1, we found that these proteins separated when subjected to the conditions of ion exchange chromatography (Fig. 5E). Taken together, the evidence presented in Figs. 4 and 5 helps to explain why we identified the Tim8/Tim13 proteins enriched in our RelA purifications in addition to the other previously characterized NFκB interacting proteins.
The mapping of interactions between transcription factors and their associated proteins has the potential to illuminate the complex mechanisms governing transcription factor function. Several previous studies have focused on defining transcription factor interaction partners using a variety of technologies. Recently Lambert and coworkers used a yeast two-hybrid approach to identify 59 proteins associated with the transcription factor Hoxa1 (42); these include proteins involved in diverse cellular processes including cell signaling, cell adhesion, and vesicular trafficking. Other studies have sought to identify transcription factor interactors by expressing recombinant affinity-tagged factors under the control of foreign promoters and analyzing purified complexes via affinity purification mass spectrometry; these include investigations into c-Myc associated proteins (43, 44) and NFκB associated proteins (3). Such approaches have used a variety of controls to determine the set of proteins co-purifying specifically with the affinity-tagged bait; these include proteins co-purifying with an unrelated tagged bait (3), proteins co-purifying with overexpressed bait lacking the affinity tag (43), and proteins co-purifying with cells expressing only the affinity tag (44). The expression of an affinity-tagged transcription factor as bait can lead to a marked change in gene expression in the bait transfected cells relative to controls not transfected with the transcription factor. We have examined whether control purifications expressing the tag alone are sufficient for identifying a set of bait-associated proteins that are enriched in affinity-tagged bait purifications, or whether using such controls likely results in false positive identifications of bait-associated proteins. In the examples we have examined, we have established that “tag only” controls are sufficient and do not result in false positive identifications. This is important for studies using techniques such as MudPIT in which mass spectrometer time is limited and where using a common set of controls when examining a variety of different tagged baits may be necessary.
Supplementary Material
Acknowledgments
We thank Drs. Mihaela Sardiu and Stephanie Kong for their critical reading of the manuscript and Dr. Danette Daniels of Promega Corporation for technical support during the course of this project.
Footnotes
Author contributions: C.A.B., Z.L., and M.P.W. designed research; C.A.B., Z.L., G.B., B.G., G.H., and C.S. performed research; C.A.B., M.L., Z.W., and L.F. contributed new reagents or analytic tools; C.A.B., Z.L., G.B., B.G., G.H., C.S., and M.P.W. analyzed data; C.A.B. and M.P.W. wrote the paper.
* This work was supported by the Stowers Institute for Medical Research.
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- MudPIT
- multidimensional protein identification technology
- dNSAF
- distributed normalized spectral abundance factor
- FDR
- false discovery rate
- FKPM
- fragments per kilobase per million fragments measured
- HEK293
- human embryonic kidney 293 cell line
- NFκB
- nuclear factor κ-light-chain-enhancer of activated B cells
- PLGEM
- Power Law Global Error Model.
REFERENCES
- 1. Sowa M. E., Bennett E. J., Gygi S. P., Harper J. W. (2009) Defining the human deubiquitinating enzyme interaction landscape. Cell 138, 389–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Behrends C., Sowa M. E., Gygi S. P., Harper J. W. (2010) Network organization of the human autophagy system. Nature 466, 68–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bouwmeester T., Bauch A., Ruffner H., Angrand P. O., Bergamini G., Croughton K., Cruciat C., Eberhard D., Gagneur J., Ghidelli S., Hopf C., Huhse B., Mangano R., Michon A. M., Schirle M., Schlegl J., Schwab M., Stein M. A., Bauer A., Casari G., Drewes G., Gavin A. C., Jackson D. B., Joberty G., Neubauer G., Rick J., Kuster B., Superti-Furga G. (2004) A physical and functional map of the human TNF-alpha/NF-kappa B signal transduction pathway. Nat. Cell Biol. 6, 97–105 [DOI] [PubMed] [Google Scholar]
- 4. Breitkreutz A., Choi H., Sharom J. R., Boucher L., Neduva V., Larsen B., Lin Z.-Y., Breitkreutz B.-J., Stark C., Liu G., Ahn J., Dewar-Darch D., Reguly T., Tang X., Almeida R., Qin Z. S., Pawson T., Gingras A.-C., Nesvizhskii A. I., Tyers M. (2010) A global protein kinase and phosphatase interaction network in yeast. Science 328, 1043–1046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sardiu M. E., Cai Y., Jin J., Swanson S. K., Conaway R. C., Conaway J. W., Florens L., Washburn M. P. (2008) Probabilistic assembly of human protein interaction networks from label-free quantitative proteomics. Proc. Natl. Acad. Sci. U.S.A. 105, 1454–1459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Giambruno R., Grebien F., Stukalov A., Knoll C., Planyavsky M., Rudashevskaya E. L., Colinge J., Superti-Furga G., Bennett K. L. (2013) Affinity purification strategies for proteomic analysis of transcription factor complexes. J. Proteome Res. 12, 4018–4027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Banks C., Kong S., Washburn M. (2012) Affinity purification of protein complexes for analysis by multidimensional protein identification technology. Protein Expr. Purif. 86, 105–119 [DOI] [PubMed] [Google Scholar]
- 8. Junttila M. R., Saarinen S., Schmidt T., Kast J., Westermarck J. (2005) Single-step Strep-tag purification for the isolation and identification of protein complexes from mammalian cells. Proteomics 5, 1199–1203 [DOI] [PubMed] [Google Scholar]
- 9. Mellacheruvu D., Wright Z., Couzens A. L., Lambert J. P., St-Denis N., Li T., Mitev Y. V., Hauri S., Sardiu M. E., Low T. Y., Halim V. A., Bagshaw R. D., Hubner N. C., Al-Hakim A., Bouchard A., Faubert D., Fermin D., Dunham W. H., Goudreault M., Lin Z. Y., Badillo B. G., Pawson T., Durocher D., Coulombe B., Aebersold R., Superti-Furga G., Colinge J., Heck A. J. R., Choi H., Gstaiger M., Mohammed S., Cristea I. M., Bennett K. L., Washburn M. P., Raught B., Ewing R. M., Gingras A. C., Nesvizhskii A. (2013) The CRAPome: a contaminant repository for affinity purification mass spectrometry data. Nat. Methods 24, S50–S51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Daniels D. L., Méndez J., Mosley A. L., Ramisetty S. R., Murphy N., Benink H., Wood K. V., Urh M., Washburn M. P. (2012) Examining the complexity of human RNA polymerase complexes using HaloTag technology coupled to label free quantitative proteomics. J. Proteome Res. 11, 564–575 [DOI] [PubMed] [Google Scholar]
- 11. Sato S., Tomomori-Sato C., Parmely T. J., Florens L., Zybailov B., Swanson S. K., Banks C. A., Jin J., Cai Y., Washburn M. P., Conaway J. W., Conaway R. C. (2004) A set of consensus mammalian mediator subunits identified by multidimensional protein identification technology. Mol. Cell 14, 685–691 [DOI] [PubMed] [Google Scholar]
- 12. Nolan G. P., Ghosh S., Liou H. C., Tempst P., Baltimore D. (1991) DNA binding and I kappa B inhibition of the cloned p65 subunit of NF-kappa B, a rel-related polypeptide. Cell 64, 961–969 [DOI] [PubMed] [Google Scholar]
- 13. Jin H., Kendall E., Freeman T. C., Roberts R. G., Vetrie D. L. (1999) The human family of Deafness/Dystonia peptide (DDP) related mitochondrial import proteins. Genomics 61, 259–267 [DOI] [PubMed] [Google Scholar]
- 14. Meyer R., Hatada E. N., Hohmann H. P., Haiker M., Bartsch C., Rothlisberger U., Lahm H. W., Schlaeger E. J., van Loon A. P., Scheidereit C. (1991) Cloning of the DNA-binding subunit of human nuclear factor kappa B: the level of its mRNA is strongly regulated by phorbol ester or tumor necrosis factor alpha. Proc. Natl. Acad. Sci. 88, 966–970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Trapnell C., Pachter L., Salzberg S. L. (2009) TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25, 1105–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Trapnell C., Williams B. A., Pertea G., Mortazavi A., Kwan G., van Baren M. J., Salzberg S. L., Wold B. J., Pachter L. (2010) Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28, 511–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Roberts A., Pimentel H., Trapnell C., Pachter L. (2011) Identification of novel transcripts in annotated genomes using RNA-Seq. Bioinformatics 27, 2325–2329 [DOI] [PubMed] [Google Scholar]
- 18. Swanson S., Florens L., Washburn M. P. (2009) Generation and analysis of multidimensional protein identification technology datasets. Methods Mol. Biol. 492, 1–20 [DOI] [PubMed] [Google Scholar]
- 19. Eng J. K., McCormack A. L., Yates J. R. (1994) An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom. 5, 976–989 [DOI] [PubMed] [Google Scholar]
- 20. Tabb D. L., McDonald W. H., Yates J. R. (2002) DTASelect and Contrast: tools for assembling and comparing protein identifications from shotgun proteomics. J. Proteome Res. 1, 21–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhang Y., Wen Z., Washburn M. P., Florens L. (2010) Refinements to label free proteome quantitation: how to deal with peptides shared by multiple proteins. Anal. Chem. 82, 2272–2281 [DOI] [PubMed] [Google Scholar]
- 22. Pavelka N., Fournier M. L., Swanson S. K., Pelizzola M., Ricciardi-Castagnoli P., Florens L., Washburn M. P. (2008) Statistical similarities between transcriptomics and quantitative shotgun proteomics data. Mol. Cell. Proteomics 7, 631–644 [DOI] [PubMed] [Google Scholar]
- 23. Shao C., Liu Y., Ruan H., Li Y., Wang H. (2010) Shotgun proteomics analysis of hibernating arctic ground squirrels. Mol. Cell. Proteomics 9, 313–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Benjamini Y., Hochberg Y. (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B 57, 289–300 [Google Scholar]
- 25. Washburn M. P., Wolters D., Yates J. R. (2001) Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat. Biotechnol. 19, 242–247 [DOI] [PubMed] [Google Scholar]
- 26. Florens L., Washburn M. P. (2006) Proteomic analysis by multidimensional protein identification technology. Methods Mol. Biol. 328, 159–175 [DOI] [PubMed] [Google Scholar]
- 27. Lin L., DeMartino G. N., Greene W. C. (1998) Cotranslational biogenesis of NF-kappaB p50 by the 26S proteasome. Cell 92, 819–828 [DOI] [PubMed] [Google Scholar]
- 28. Moorthy A. K., Savinova O. V., Ho J. Q., Wang V. Y.-F., Vu D., Ghosh G. (2006) The 20S proteasome processes NF-kappaB1 p105 into p50 in a translation-independent manner. EMBO J. 25, 1945–1956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Liou H., Nolan G., Ghosh S. (1992) The NF-kappa B p50 precursor, p105, contains an internal I kappa B-like inhibitor that preferentially inhibits p50. EMBO J. 1, 3003–3009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Schutz-Geschwender A., Zhang Y., Holt T., McDermitt D., Olive D. M. (2004) Quantitative, Two-color Western Blot Detection with Infrared Fluorescence, LI-COR Biosciences, Lincoln, NE [Google Scholar]
- 31. Mortazavi A., Williams B., McCue K. (2008) Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 5, 1–8 [DOI] [PubMed] [Google Scholar]
- 32. Oeckinghaus A., Ghosh S. (2009)The NF-kappaB family of transcription factors and its regulation. Cold Spring Harb. Perspect. Biol. 1, a000034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Jacobs M. D., Harrison S. C. (1998) Structure of an IκBα/NF-κB complex. Cell 95, 749–758 [DOI] [PubMed] [Google Scholar]
- 34. Cohen S., Lahav-Baratz S., Ciechanover A. (2006) Two distinct ubiquitin-dependent mechanisms are involved in NF-kappaB p105 proteolysis. Biochem. Biophys. Res. Commun. 345, 7–13 [DOI] [PubMed] [Google Scholar]
- 35. Nooren I., Thornton J. (2003) Diversity of protein–protein interactions. EMBO J. 22, 3486–3492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Perkins J. R., Diboun I., Dessailly B. H., Lees J. G., Orengo C. (2010) Transient protein-protein interactions: structural, functional, and network properties. Structure 18, 1233–1243 [DOI] [PubMed] [Google Scholar]
- 37. Baeuerle P. A., Baltimoire D. (1988) IκB: specific inhibitor of the NF-κB transcription factor. Science 242, 540–546 [DOI] [PubMed] [Google Scholar]
- 38. Sen R., Baltimore D. (1986) Inducibility of κ immunoglobulin enhancer-binding protein NF-κB by a posttranslational mechanism. Cell 47, 921–928 [DOI] [PubMed] [Google Scholar]
- 39. Cogswell P. C., Kashatus D. F., Keifer J. A., Guttridge D. C., Reuther J. Y., Bristow C., Roy S., Nicholson D. W., Baldwin A. S. (2003) NF-kappa B and I kappa B alpha are found in the mitochondria. Evidence for regulation of mitochondrial gene expression by NF-kappa B. J. Biol. Chem. 278, 2963–2968 [DOI] [PubMed] [Google Scholar]
- 40. Bottero V., Rossi F., Samson M., Mari M., Hofman P., Peyron J. F. (2001) Ikappa b-alpha, the NF-kappa B inhibitory subunit, interacts with ANT, the mitochondrial ATP/ADP translocator. J. Biol. Chem. 276, 21317–21324 [DOI] [PubMed] [Google Scholar]
- 41. Haslam N. J., Shields D. C. (2012) Peptide-binding domains: are limp handshakes safest? Sci. Signal. 5, pe40. [DOI] [PubMed] [Google Scholar]
- 42. Lambert B., Vandeputte J., Remacle S., Bergiers I., Simonis N., Twizere J.-C., Vidal M., Rezsohazy R. (2012) Protein interactions of the transcription factor Hoxa1. BMC Dev. Biol. 12, 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Agrawal P., Yu K., Salomon A. R., Sedivy J. M. (2010) Proteomic profiling of Myc-associated proteins. Cell Cycle 9, 4908–4921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Koch H. H. B., Zhang R., Verdoodt B., Bailey A., Zhang C.-D., Yates J. R., 3rd, Menssen A., Hermeking H., Yates J. R. (2007) Large-scale identification of c-MYC-associated proteins using a combined TAP/MudPIT approach. Cell Cycle 6, 205–217 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.