Abstract
The open XML format mzML, used for representation of MS data, is pivotal for the development of platform-independent MS analysis software. Although conversion from vendor formats to mzML must take place on a platform on which the vendor libraries are available (i.e. Windows), once mzML files have been generated, they can be used on any platform. However, the mzML format has turned out to be less efficient than vendor formats. In many cases, the naïve mzML representation is fourfold or even up to 18-fold larger compared with the original vendor file. In disk I/O limited setups, a larger data file also leads to longer processing times, which is a problem given the data production rates of modern mass spectrometers. In an attempt to reduce this problem, we here present a family of numerical compression algorithms called MS-Numpress, intended for efficient compression of MS data. To facilitate ease of adoption, the algorithms target the binary data in the mzML standard, and support in main proteomics tools is already available. Using a test set of 10 representative MS data files we demonstrate typical file size decreases of 90% when combined with traditional compression, as well as read time decreases of up to 50%. It is envisaged that these improvements will be beneficial for data handling within the MS community.
Open XML formats for representation of MS data have been developed by the proteomics community to facilitate exchange and vendor neutral analysis of mass spectrometry data. Initially two formats, mzXML (1) and mzData (http://psidev.info/), existed in parallel, until these formats were merged into the single standard format mzML (2). The mzML format has been adopted widely by the proteomics community and is supported by many data processing tools. However, although successfully used in many pipelines, the mzML format has not reached its full usage potential, mainly because of large file sizes in comparison to the raw vendor formats. The file size problem has become more marked with the introduction of recent high-resolution high-frequency mass spectrometers. As an example, a raw data file from a data-independent acquisition experiment using an AB SCIEX TripleTOF resulted in a vendor format data file of 2.5 GB. Conversion of this file to standard mzML resulted in a 46.7 GB file, with a conversion time of about 12 min on a desktop computer (later called high-end I, Fig. 1A) dedicated to the conversion process. If the file is compressed using gzip to lower the storage footprint, the size drops to 21.6 GB, but the conversion now takes 2 h instead. This (extreme) example pinpoints the need for increased efficiency in the standardized representation of MS data.
Fig. 1.
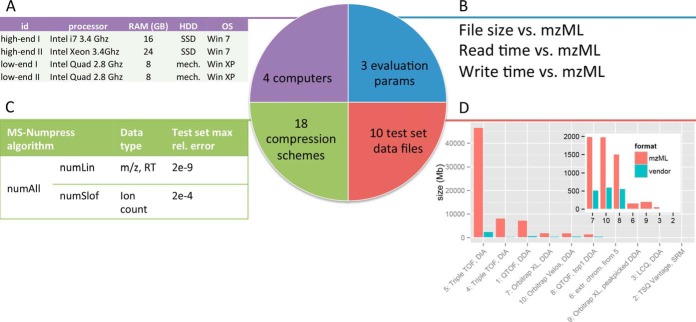
Overview of the compression scheme evaluation experiment. A, Evaluation was performed on four desktop computers of varying capacity, the most notable parameter being the use of solid state drive (SSD) hard drives compared with traditional mechanical hard drives. B, File size, write time and read time were used to evaluate compression. C, Main results include two new numerical compression methods optimized for the three different MS data types m/z, ion count and retention time (RT), with relative errors far below instrument precision. D, File sizes of vendor and mzML versions of the 10 used MS data files. The inset shows a magnification of the seven smallest files.
Across the community, there is little previous work done on compression of MS data. In two technical papers (3, 4), Miguel et al. describe lossless and near-lossless compression methods for QTOF data, achieving compression factors above 10. No measurements are provided on the compression time however, and the algorithms are benchmarked on a very small set of data files. Blanckenburg et al. describe a lossy compression technique for Fourier transform ion cyclotron resonance data (5), where known nonmetabolite data points are discarded. Outside the MS community, potential benefits could come from recent work in the numerical computation field (6, 7), were many data types are similar to MS data in terms of precision and smoothness. Nevertheless, perhaps the most relevant recent advance is the emergence of the mz5 format (8), which yields performance increases via a binary representation and optimized libraries, as well as some regular data compression. Although this format, based on the open HDF5 standard (The HDF Group, Champaign, IL, USA), is an efficient representation of mzML files, it suffers from the fact that the files are not readable without native libraries or specialized software, which, to some extent, has hampered its uptake. Also, while mz5 can be “lossless,” default compression implies removal of zero intensity scans, which means the original data cannot be reconstructed, and some algorithms require zero intensity scans for correct functioning.
The standard XML representation used in mzML can be easily viewed as text on any operating system, and it is relatively easy to write a parser in any programming language. We thus sought to overcome the mzML efficiency shortcomings by introducing better compression of the binary data found in mzML files while still leaving the metadata in XML format, and propose such an extension to the format here. Furthermore, we envisage that decompression of this binary data should be easy to incorporate into software tools via permissively licensed stand-alone source code files for C++ and Java, which do not require any external dependences. We here also exemplify the facility of usage by implementing support in several popular tools for proteomics data analysis.
Experimental Procedures and Results
To efficiently compress the three main types of binary data present in mzML files: (1) mass to charge ratios, (2) ion counts, and (3) retentions times, we have developed three new near-lossless compression algorithms, while ensuring for each data type that precision losses are well below the precision of the most advanced mass spectrometers of today. The Numpress Linear Prediction Compression algorithm (hereafter called numLin, relative error < 2e-9) takes advantage of the linearly increasing values in m/z and retention time data, and is optimized for high-resolution m/z data. Ion count data does not linearly increase but requires less stored precision because of the lower instrument precision, and Numpress Short Logged Float (numSlof1, relative error < 2e-4) is optimized for this data type. We also developed a second ion count compression (Numpress Positive Integer Count, numPic) and a lossless transformation (Numpress Linear Prediction Transformation, numSafe), which are not used further here, but are presented in the supplementary materials. Although the least significant of the 16 double-precision decimals are lost in the first conversion to the compressed format for all the algorithms, compression and decompression after this does not incur further losses. To maximize speed, the algorithms are highly local in memory and only need a single traversal of the data. For a complete description of the algorithms we refer to Supplemental Methods, and to the reference implementations in Java and C++, found at https://github.com/ms-numpress/ms-numpress under the Apache 2.0 license.
To compare MS-Numpress to current alternatives for storing mzML data, we extensively evaluated size, write time, and read time of available compression schemes on a varied set of data files using different computers (Fig. 1). For this we constructed a test set of 10 MS data files from different vendors, instruments, and experiment types (Fig. 1D and supplemental Table S1). The test set files included data-dependent acquisition (DDA), selected reaction monitoring (SRM) and data-independent (DIA/SWATH) acquisition modes, and both simple and complex samples, giving a heterogeneous set of distributions of MS1 and MS2 spectrum data and chromatogram binary data arrays of different lengths (supplemental Fig. S1). These files were converted to mzML, imzML (9) and mz5 (8), both without compression, using zlib compression, and using gzip of the entire file. Files were also compressed in multiple different setups using MS-Numpress compressions, resulting in a total of 18 tested compression schemes (Fig. 1C and supplemental Table S2). To avoid clutter, minor results are left out here, and readers interested in imzML-data or individual Numpress results are referred to the supplementary material. The different compression schemes were compared based on file size, read time and write time (Fig. 1B). Benchmarking was performed on four dedicated desktop computers of varying capacity (Fig. 1A), using a custom msconvert (10) build, and timed using a script written in Python. Write time was measured as the total time for an msconvert conversion from the vendor raw format. Because this includes the vendor read time it gives a constant offset, but this constant is in general small compared with the write time. For read benchmarking a custom program was made, that reads files using the ProteoWizard (10) API. To ensure that all data is read, this program explicitly reads all binary values in the spectra and chromatograms found in the file. Test files, results, and program binaries are available at the Swestore repository (http://webdav.swegrid.se/snic/bils/lu_proteomics/pub).
The use of near-lossless compression introduces the question of whether one can be sure that no analytically relevant data is lost. We measured the relative errors for the compressed versions of all the files in the test set (supplemental Tables S3 to S6), and found relative errors to be smaller than 2e-9 (0.002 ppm) for numLin compressed m/z data and smaller than 2e-4 (0.02%) for numSlof compressed ion counts. To validate that the small errors introduced by Numpress compression do not have adverse effect on common proteomics analyses, we converted two Orbitrap DDA LC-MS/MS mzML files to compressed (combination of numLin and numSlof) versions and back to uncompressed mzML again, and compared analysis results obtained from the doubly converted files to those obtained using the original files. MS/MS identification using Mascot after extraction of MGF (Mascot Generic Format) peak lists yielded identical lists of identified peptides at a 1% peptide to spectrum match (PSM) false discovery rate (FDR, Supplementary data). Extraction of features from the MS1 data using msInspect (11) yielded the same lists of features, with differences only in the least significant decimals of some reported m/z values and intensities (Supplementary Data). When comparing lists of integrated precursor intensities for the peptides that were identified using MS/MS, the lists contained the same entries with a 0.004% maximum observed relative intensity difference introduced by the double conversion, confirming that the Numpress compression schemes could be safely used for proteomics analyses.
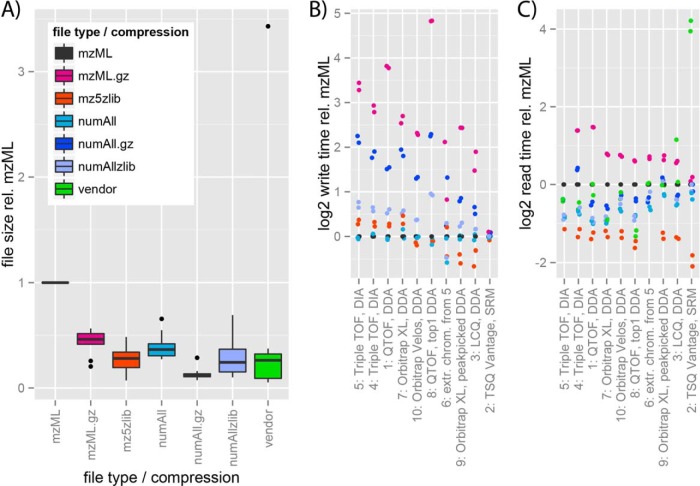
We found that to achieve minimal file size, a combination of numLin for m/z or retention time data and numSlof for ion count data (numAll), with subsequent gzipping of the entire file, was the most optimal in terms of file size. This yielded an average file size reduction of 87% compared with standard mzML across all 10 test set files (Fig. 2A), with 138% longer write times (Fig. 2B) but 21% shorter read times (Fig. 2C) on average across all tested computers and files (supplemental Table S7). This format is also half the size of the binary mz5 with zlib compression (mz5zlib), and also smaller than all vendor formats except for AB SCIEX's .wiff files (supplemental Fig. S2 and Table S7). In our read speed tests, the text based mzML formats cannot quite compete with the binary mz5, although the difference is small (20%) in the largest files (Fig. 2C). The effects of minimizing disk I/O through compression are the most visible in the largest files on the lower performance computers (supplemental Figs. S3 and S4), where the numAll alternatives catch up to mz5zlib. Overall, read times ranged over 3 orders of magnitude (supplemental Fig. S5), and write times over four orders of magnitude (supplemental Fig. S6).
Fig. 2.
File size, read and write time compared with standard mzML. A, Standard box plot of file size relative to mzML for 7 data formats. B, C, log2 write time B, and read time C, subtracted by log2 mzML write and read time, respectively, for the 10 test files and seven data formats. Test files are sorted in descending mzML size, and dots represent measurements on individual data files on one of two SSD-hard drive equipped computers. Writing of vendor formats could not be tested with this setup and is thus missing in B. Missing read times are because of errors in execution (mzML.gz for file 5 and mz5zlib for file 6) as discussed in the supplementary material, where also global statistics for all compression schemes (supplemental Table S7) and absolute timing figures (supplemental Figs. S5, S6) are provided.
Two of the test computers were equipped with SSD hard drives, which are fast enough to open up the disk I/O bottleneck, and reveal the next bottleneck: processor speed. On these machines the expensiveness of gzipping becomes apparent, with cost increasing with file size (Fig. 2B), and the largest gains from the fast disk I/O are seen for the processor-light schemes mzML, mz5zlib, and numAll (supplemental Figs. S7 and S8). Zlib compression of individual data arrays also shows minor slow-down of the write operation, whereas the numAll scheme does not affect write times at all (Fig. 2C).
Even though numAll decreased read times by on average 36% compared with standard mzML (supplemental Table S7), mz5zlib further decreased read times by 25% (total 61% from mzML), and this might have a number of reasons. Some hypotheses for explaining this are (1) the aggressive file caching of mz5 provides block read benefits, (2) large amount of string-string comparisons while building the mzML object model is costly, or (3) Base64 decoding is costly. Whereas (3) is not solvable while keeping a one-file, text-based format, both (1) and (2) could be improved in optimized reader implementations.
As the MS-Numpress compression techniques showed high degrees of compression, which was our primary goal, we set out to implement the technique as part of several proteomics pipelines in order to ensure easy adoption by the proteomics community. The initial implementation in ProteoWizard (10) enables conversion to the format from all major mass spectrometer raw data formats, and provides read access to tools that use the ProteoWizard API for reading files, for example MyriMatch (12) and Skyline (13).
Compression should be especially effective for workflows that use high-resolution profile data for quantitation, because of the large data files this implies, and we therefore implemented support for reading of mzML files with numpress compressed binaries in OpenMS (14), msInspect (11), and the Proteios Software Environment (15, 16). The implementation in OpenMS also implies that the complete MS-Numpress compression and decompression algorithms are directly available in the Python scripting language because of the recent Python-wrapping of the complete OpenMS library (17).
The jmzML (18) library has also been extended with MS-Numpress read and write support for numLin, numSlof, and numPic. The jmzML library is embedded in numerous Java-based mass spectrometry software solutions, including PRIDE Converter (19) and Proteosuite, an open source framework for the analysis of quantitative proteomics data (http://www.proteosuite.org/). Such solutions will now implicitly support MS-Numpress compressed data when utilizing the latest jmzML library.
Support for mzML with MS-Numpress compression was implemented in the tools of the Trans-Proteomics Pipeline (20, 21). Reading was also implemented in the X!Tandem search engine (22). Finally, we implemented support for MS-Numpress in the Anubis (23) tool for SRM.
The main advantages of using the new MS-Numpress-compressed mzML format is probably seen where small file sizes are of highest importance, for example in data sharing over the Internet. Minimal file size is also of utmost importance in distributed or cloud computing, for example shown by Dowsey et al. for two-dimensional gel electrophoresis alignment (24). Because of the file size importance, simple peak lists formats are currently still used extensively for MS/MS database searches, and the more complete file representations provided by mzML have mainly been used for data sharing and quantitative workflows. However, with the increased number of MS/MS spectra acquired with modern mass spectrometers it is envisaged that compressed data formats could become more widely used also for MS/MS database searches. As an example, we downloaded a raw data file from a single-shot LC-MS/MS analysis of a yeast lysate, performed on an Orbitrap Fusion acquired for a recent publication from the Coon lab (25). Conversion of the file to Mascot Generic Format (MGF, http://www.matrixscience.com) using ProteoWizard and no filtering yielded a file size of 1508 MB (715 MB gzipped), with only the centroid MS/MS data retained. ProteoWizard conversion of the original file using identical parameters to mzML with numAll compression resulted in a 944 MB (409 MB gzipped) file, while still retaining the MS1. The file size shrinks to 821 MB (343 MB gzipped) if only the MS/MS data is retained in the mzML file. Interestingly, an MS/MS search in X!Tandem resulted in slightly more peptide identifications at a 1% PSM FDR using the mzML file than using the MGF with a precursor mass tolerance of 7 ppm (Supplementary Data), probably because of a higher precision in the precursor mass in the mzML file, showing that there is no apparent drawback in using the compressed format for MS/MS database searches.
Our results demonstrate the power of some very simple techniques to improve the mzML format with respect to disk space and handling time. There are undoubtedly other algorithms that could further improve on the degree of compression or handling times, but for standard formats we believe it is crucial to provide simple and robust solutions, to minimize both the cost of implementation in tools, and the risk of mistakes in the algorithm. We further provide implemented support for the new algorithms in several tools, and thus give immediate access to the proteomics community. MS-Numpress will also be evaluated through the Proteomics Standards Initiative (PSI) process for formal inclusion in the next mzML release. We hope that this work may also stimulate additional data compression algorithm ideas, which, in the end, will lead to an amendment to the mzML standard to improve data handling for all mzML users.
Supplementary Material
Acknowledgments
We thank Jari Häkkinen for reviewing the C++ MS-Numpress library and to Giorgio Arrigoni for providing the Agilent QTOF data file.
Footnotes
Author contributions: J.T., A.R.J., E.W.D., and F.L. designed research; J.T. and F.F.G. performed research; J.T., A.W.D., F.F.G., S.P., B.P., H.R., L.M., and A.R.J. analyzed data; J.T., A.W.D., F.F.G., S.P., L.M., J.M., A.R.J., E.W.D., and F.L. wrote the paper.
* J.M. and J.T. were supported by the Swedish Research Council (projects 2008:3356 and 621–2012-3559), the Swedish Foundation for Strategic Research (grant FFL4), the Crafoord Foundation (grant 20100892), the Wallenberg Academy Fellow KAW (2012.0178) and European research council starting grant (ERC-2012-StG-309831). A.W.D.'s contribution was supported by UK Biotechnology and Biological Sciences Research Council (BBSRC) grant BB/K016733/1, and facilitated by the Manchester Biomedical Research Centre. F.G.G.G, S.P. and A.R.J. gratefully acknowledge support for this work from BBSRC (BB/I00095X/1, BB/K01997X/1). E.W.D. was supported by the National Institute of General Medical Sciences grant No. R01 GM087221, the National Science Foundation MRI grant No. 0923536, the National Human Genome Research Institute grant No. RC2 HG005805, and EU FP7 grant ‘ProteomeXchange’ (grant number 260558). F.L. was supported by the Swedish Foundation for Strategic Research (RBb08-0006), the Swedish Research Council (BILS, project 829-2009-6257) and the Mistra Biotech program.
1 The abbreviations used are:
- numSlof
- numpress short logged float
- XML
- extensible markup language
- DDA
- data-dependent acquisition
- SRM
- selected reaction monitoring
- DIA
- data-independent acquisition
- numLin
- numpress linear prediction
- numPic
- numpress positive integer count
- numSafe
- numpress linear prediction transformation (lossless)
- numAll
- combination of numLin and numSlof compression
- mz5zlib
- mz5 with zlib compression
- SSD
- solid-state drive
- MGF
- mascot generic format.
REFERENCES
- 1. Pedrioli P. G. A., Eng J. K., Hubley R., Vogelzang M., Deutsch E. W., Raught B., Pratt B., Nilsson E., Angeletti R. H., Apweiler R., Cheung K., Costello C. E., Hermjakob H., Huang S., Julian R. K., Kapp E., McComb M. E., Oliver S. G., Omenn G., Paton N. W., Simpson R., Smith R., Taylor C. F., Zhu W., Aebersold R. (2004) A common open representation of mass spectrometry data and its application to proteomics research. Nat. Biotechnol. 22, 1459–1466 [DOI] [PubMed] [Google Scholar]
- 2. Martens L., Chambers M., Sturm M., Kessner D., Levander F., Shofstahl J., Tang W. H., Römpp A., Neumann S., Pizarro A. D., Montecchi-Palazzi L., Tasman N., Coleman M., Reisinger F., Souda P., Hermjakob H., Binz P.-A., Deutsch E. W. (2011) mzML–a community standard for mass spectrometry data. Mol. Cell. Proteomics 10, R110.000133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Miguel A. C., Keane J. F., Whiteaker J., Zhang H., Paulovich A. G. (2006) Compression of LC/MS Proteomic Data. Proc. 19th IEEE Symp. Comput.-Based Med. Syst. CBMS06, [Google Scholar]
- 4. Miguel A. C., Kearney-Fischer M., Keane J. F., Whiteaker J., Feng L.-C., Paulovich A. G. (2007) Near-lossless compression of mass spectra for proteomics. Acoust. Speech Signal Process. 2007 ICASSP 2007 IEEE Int. Conf. 1, I-369–I-372 [Google Scholar]
- 5. Blanckenburg B., Burgt Y. E. M., Deelder A. M., Palmblad M. (2010) “Lossless” compression of high resolution mass spectra of small molecules. Metabolomics 6, 335–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Engelson V., Fritzson D., Fritzson P. (2000) Lossless Compression of High-volume Numerical Data from Simulations. Data Compression Conf. 574–586 [Google Scholar]
- 7. Ratanaworabhan P., Ke J., Burtscher M. (2006) Fast lossless compression of scientific floating-point data. Data Compression Conf. 2006 DCC 2006 Proc. 1, 133–142 [Google Scholar]
- 8. Wilhelm M., Kirchner M., Steen J. A. J., Steen H. (2012) mz5: space- and time-efficient storage of mass spectrometry data sets. Mol. Cell. Proteomics 11, O111.011379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Römpp A., Schramm T., Hester A., Klinkert I., Both J.-P., Heeren R. M. A., Stöckli M., Spengler B. (2011) imzML: Imaging Mass Spectrometry Markup Language: A common data format for mass spectrometry imaging. Methods Mol. Biol. 696, 205–224 [DOI] [PubMed] [Google Scholar]
- 10. Chambers M. C., Maclean B., Burke R., Amodei D., Ruderman D. L., Neumann S., Gatto L., Fischer B., Pratt B., Egertson J., Hoff K., Kessner D., Tasman N., Shulman N., Frewen B., Baker T. A., Brusniak M.-Y., Paulse C., Creasy D., Flashner L., Kani K., Moulding C., Seymour S. L., Nuwaysir L. M., Lefebvre B., Kuhlmann F., Roark J., Rainer P., Detlev S., Hemenway T., Huhmer A., Langridge J., Connolly B., Chadick T., Holly K., Eckels J., Deutsch E. W., Moritz R. L., Katz J. E., Agus D. B., MacCoss M., Tabb D. L., Mallick P. (2012) A cross-platform toolkit for mass spectrometry and proteomics. Nat. Biotechnol. 30, 918–920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bellew M., Coram M., Fitzgibbon M., Igra M., Randolph T., Wang P., May D., Eng J., Fang R., Lin C., Chen J., Goodlett D., Whiteaker J., Paulovich A., McIntosh M. (2006) A suite of algorithms for the comprehensive analysis of complex protein mixtures using high-resolution LC-MS. Bioinforma. Oxf. Engl. 22, 1902–1909 [DOI] [PubMed] [Google Scholar]
- 12. Tabb D. L., Fernando C. G., Chambers M. C. (2007) MyriMatch: highly accurate tandem mass spectral peptide identification by multivariate hypergeometric analysis. J. Proteome Res. 6, 654–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. MacLean B., Tomazela D. M., Shulman N., Chambers M., Finney G. L., Frewen B., Kern R., Tabb D. L., Liebler D. C., MacCoss M. J. (2010) Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinforma. Oxf. Engl. 26, 966–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kohlbacher O., Reinert K., Gröpl C., Lange E., Pfeifer N., Schulz-Trieglaff O., Sturm M. (2007) TOPP–the OpenMS proteomics pipeline. Bioinforma. Oxf. Engl. 23, e191–e197 [DOI] [PubMed] [Google Scholar]
- 15. Häkkinen J., Vincic G., Månsson O., Wårell K., Levander F. (2009) The proteios software environment: an extensible multiuser platform for management and analysis of proteomics data. J. Proteome Res. 8, 3037–3043 [DOI] [PubMed] [Google Scholar]
- 16. Sandin M., Ali A., Hansson K., Månsson O., Andreasson E., Resjö S., Levander F. (2013) An adaptive alignment algorithm for quality-controlled label-free LC-MS. Mol. Cell. Proteomics MCP 12, 1407–1420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Röst H. L., Schmitt U., Aebersold R., Malmström L. (2014) pyOpenMS: A Python-based interface to the OpenMS mass-spectrometry algorithm library. Proteomics 14, 74–77 [DOI] [PubMed] [Google Scholar]
- 18. Côté R. G., Reisinger F., Martens L. (2010) jmzML, an open-source Java API for mzML, the PSI standard for MS data. Proteomics 10, 1332–1335 [DOI] [PubMed] [Google Scholar]
- 19. Côté R. G., Griss J., Dianes J. A., Wang R., Wright J. C., van den Toorn H. W. P., van Breukelen B., Heck A. J. R., Hulstaert N., Martens L., Reisinger F., Csordas A., Ovelleiro D., Perez-Rivevol Y., Barsnes H., Hermjakob H., Vizcaíno J. A. (2012) The PRoteomics IDEntification (PRIDE) Converter 2 framework: an improved suite of tools to facilitate data submission to the PRIDE database and the ProteomeXchange consortium. Mol. Cell. Proteomics 11, 1682–1689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Keller A., Eng J., Zhang N., Li X., Aebersold R. (2005) A uniform proteomics MS/MS analysis platform utilizing open XML file formats. Mol. Syst. Biol. 1, 2005.0017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Deutsch E. W., Mendoza L., Shteynberg D., Farrah T., Lam H., Tasman N., Sun Z., Nilsson E., Pratt B., Prazen B., Eng J. K., Martin D. B., Nesvizhskii A. I., Aebersold R. (2010) A guided tour of the Trans-Proteomic Pipeline. Proteomics 10, 1150–1159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Craig R., Beavis R. C. (2004) TANDEM: matching proteins with tandem mass spectra. Bioinforma. Oxf. Engl. 20, 1466–1467 [DOI] [PubMed] [Google Scholar]
- 23. Teleman J., Karlsson C., Waldemarson S., Hansson K., James P., Malmström J., Levander F. (2012) Automated selected reaction monitoring software for accurate label-free protein quantification. J. Proteome Res. 11, 3766–3773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Dowsey A. W., Dunn M. J., Yang G.-Z. (2004) ProteomeGRID: towards a high-throughput proteomics pipeline through opportunistic cluster image computing for two-dimensional gel electrophoresis. Proteomics 4, 3800–3812 [DOI] [PubMed] [Google Scholar]
- 25. Hebert A. S., Richards A. L., Bailey D. J., Ulbrich A., Coughlin E. E., Westphall M. S., Coon J. J. (2014) The One Hour Yeast Proteome. Mol. Cell. Proteomics 13, 339–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.