Abstract
A molecular screening of wild-caught rodents was conducted in French Guiana, South America to identify hosts of the hantavirus Maripa described in 2008 in a hantavirus pulmonary syndrome (HPS) case. Over a 9-year period, 418 echimyids and murids were captured. Viral RNA was detected in two sigmodontine rodents, Oligoryzomys fulvescens and Zygodontomys brevicauda, trapped close to the house of a second HPS case that occurred in 2009 and an O. fulvescens close to the fourth HPS case identified in 2013. Sequences from the rodents had 96% and 97% nucleotide identity (fragment of S and M segments, respectively) with the sequence of the first human HPS case. Phylogenetic reconstructions based on the complete sequence of the S segment show that Maripa virus is closely related to Rio Mamore hantavirus. Using environmental descriptors of trapping sites, including vegetation, landscape units, rain, and human disturbance, a maximal entropy-based species distribution model allowed for identification of areas of higher predicted occurrence of the two rodents, where emergence risks of Maripa virus are expected to be higher.
Hantaviruses (Bunyaviridae) are distributed worldwide.1 Although certain types are non-pathogenic in humans, others are responsible for hemorrhagic fever with renal syndrome in the Old World and pulmonary syndrome in the New World. In Europe and North America, epidemiology of hantavirus is closely related to the population dynamics of their rodent hosts.2,3 In South America, the fine-scale relationships between hantaviruses and their hosts are far less well-understood4,5 in relation to the huge diversity of rodent taxa.6 In addition, the ecology of most species of rodents is almost unknown, and for many of them, their taxonomy continues to be debated. To date, knowledge on hantavirus diversity in the Amazonian region is sparse, with only a few reports on the Amazonas, Mato Grosso, and Para states of Brazil.7–10 In French Guiana, a French department located on the northeast coast of South America, the circulation of hantavirus was first suggested after a serological survey performed on a selected population of patients suffering from atypical pneumonia.11 In 2008, the occurrence of the first biological confirmed human case led to the identification of a new strain, Maripa virus, which was closely related to the Rio Mamoré species (GenBank accession number KC876041).11 This hantavirus was then responsible for three fatal human cases in 2009, 2010, and 2013.
Since 2005, an ecoepidemiological survey has been implemented in French Guiana to characterize rodent communities and putative hantavirus hosts. The objectives were to (1) investigate the presence of hantavirus in wild species without a priori targeting capture efforts on supposed areas of virus occurrence, (2) clarify the phylogenetic position of Maripa virus, and (3) identify more suitable habitats, with higher predicted occurrences for species identified as potential hosts and expected to be associated with higher emergence risks, by applying species distribution models (SDMs).
Rodents were sampled in 14 sites located in primary forests, edge and secondary forest habitats, and savannas for a total effort of 28,670 traps/night. The animals were identified using external and/or skull morphology and confirmed with the cytochrome oxydase I sequence.12 In total, 418 rodents (18 species) were captured and analyzed (Table 1).
Table 1.
Serological and molecular evidence of hantavirus infection in wild-caught rodents in French Guiana
| Species | SNV-positive IgG | RT-PCR results* |
|---|---|---|
| Holochilus sciureus | 0/3 | 0/2 |
| Makalata didelphoides | 0/17 | 0/5 |
| Mesomys hispidus | 0/9 | 0/7 |
| Mus musculus | 0/23 | 0/5 |
| Neacomys paracou | 0/1 | nd |
| Nectomys rattus | 0/6 | nd |
| Oecomys auyantepui | 0/13 | 0/5 |
| Oecomys bicolor | 0/12 | 0/8 |
| Oecomys rex | 0/2 | nd |
| Oecomys rutilus | 0/1 | 0/1 |
| Oligoryzomys fulvescens | 0/5 | 2/5† |
| Oryzomys megacephalus | 0/7 | 0/5 |
| Oryzomys yunganus | 0/9 | 0/8 |
| Proechimys cayennensis | 0/201 | 0/112 |
| Proechimys cuvieri | 0/56 | 0/33 |
| Rattus rattus | 0/10 | 0/9 |
| Rhipidomys nitela | 0/7 | 0/6 |
| Z. brevicauda | 0/36 | 2/25† |
| Total | 0/418 | 4/236 |
Taxonomy follows ref. 33. nd = not done.
Based on the amplification of the 434-bp fragment of the S segment.
Number of positive rodents/number of tested rodents.
Sera from 418 rodents were tested for immunoglobulin G (IgG) antibodies directed against Sin Nombre Virus (SNV) using an indirect enzyme-linked immunosorbent assay (ELISA). Briefly, microtiter plates were coated with an SNV recombinant nucleocapsid antigen (positive antigen, lot SPR 569; Centers for Disease Control and Prevention [CDC], Atlanta, GA) or a recombinant non-viral antigen (negative antigen, lot SPR 568; CDC) diluted 1:2,000 in phosphate-buffered saline (PBS; 1×). Each tested serum was diluted 1:100 in 5% skim milk in PBS 1× containing 0.5% Tween 20. The presence of bound IgG from a wide variety of rodents was detected using an equal volume of two conjugates: anti-Rattus norvegicus and anti-Peromyscus leucopus (Kirkegaard and Perry Laboratories Inc., Gaithersburg, MD). Positive control consisted of serum from deer mice known to be infected by SNV (lot 703110; CDC), whereas negative sera were collected from mice that previously tested negative for SNV. Serum specimens were considered SNV-positive if the net absorbance values (after subtraction of absorbance values with and without antigen) were greater than 0.2 for a 1:100 dilution. Among 418 rodents tested, none were found positive for the detection of IgG antibodies against SNV (Table 1).
Total RNA was extracted from lung and/or kidney tissues of 236 rodents using the NucliSENS easyMAG bio-robot (bioMérieux, Marcy l'Etoile, France) (Table 1). Complementary DNA (cDNA) was prepared with the SuperScript III Reverse Transcriptase (Invitrogen; Life Technologies, Inc., Life Technologies, Carlsbad, NM) and random hexamers (Roche, Mannheim, Germany). Presence of the virus was investigated by nested reverse transcription polymerase chain reaction (RT-PCR) using primers previously designed for amplification of fragments of the S and M segments of New World hantaviruses.13 Amplification products were cloned using the TOPO-TA cloning system (Invitrogen, Life Technologies, Carlsbad, NM). Plasmids were sent for sequencing to Beckman Coulter Genomics (Takeley, United Kingdom). Sequences were aligned using MEGA 5.14 The phylogenetic tree was inferred from alignment of nucleotide sequences. The global time reversible (GTR) model, with γ-distribution and (I) proportion of invariable sites, was identified as the optimal model of nucleotide evolution (MrModeltest 2.2).15 Phylogenetic relationships were inferred with Bayesian approach performed with Mr. Bayes 3.1.b.16 Markov Chain Monte Carlo simulations were run for 1,000,000 generations with four simultaneous chains using a sample frequency of 1,000 and a burn-in of 25%. Validation of the inference was assessed based on the SD of split frequencies, which was less than the expected threshold value of 0.01; 4 of 236 animals analyzed molecularly were positive: two Zygodontomys brevicauda and two Oligoryzomys fulvescens (Cricetidae and Sigmodontinae) (Table 1). Three of those animals (2 Z. brevicauda and 1 O. fulvescens) were collected around the second human case of hantavirus pulmonary syndrome (HPS). Hantavirus sequences obtained on the S segment from the three rodents trapped in 2010 (GenBank accession numbers JN982967, JN982968, and JN982969) were identical on the 345 bp analyzed and are closely related (95.7% and 100% identity at the nucleotide level) to Maripa hantavirus detected from the first and second HPS cases from French Guiana, respectively.11,17 Partial sequences of the M segment from these three animals were also identical on the 292 bp analyzed (both at nucleotide and amino acid levels) and had 97% identity at the nucleotide level with the first HPS strain (GenBank accession numbers KJ209653, KJ209654, and KJ209655). The fourth rodent (an O. fulvescens) found positive was captured around the fourth human case that occurred in 2013. The 345 bp of the S segment (GenBank accession number KJ206656) showed 95.7% identity at the nucleotide level and 99.1% identity at the amino acid level with the sequences identified from the three first rodents. This sequence is closely related to the first and second human cases (99.1% and 95.7% in nucleotides, respectively).
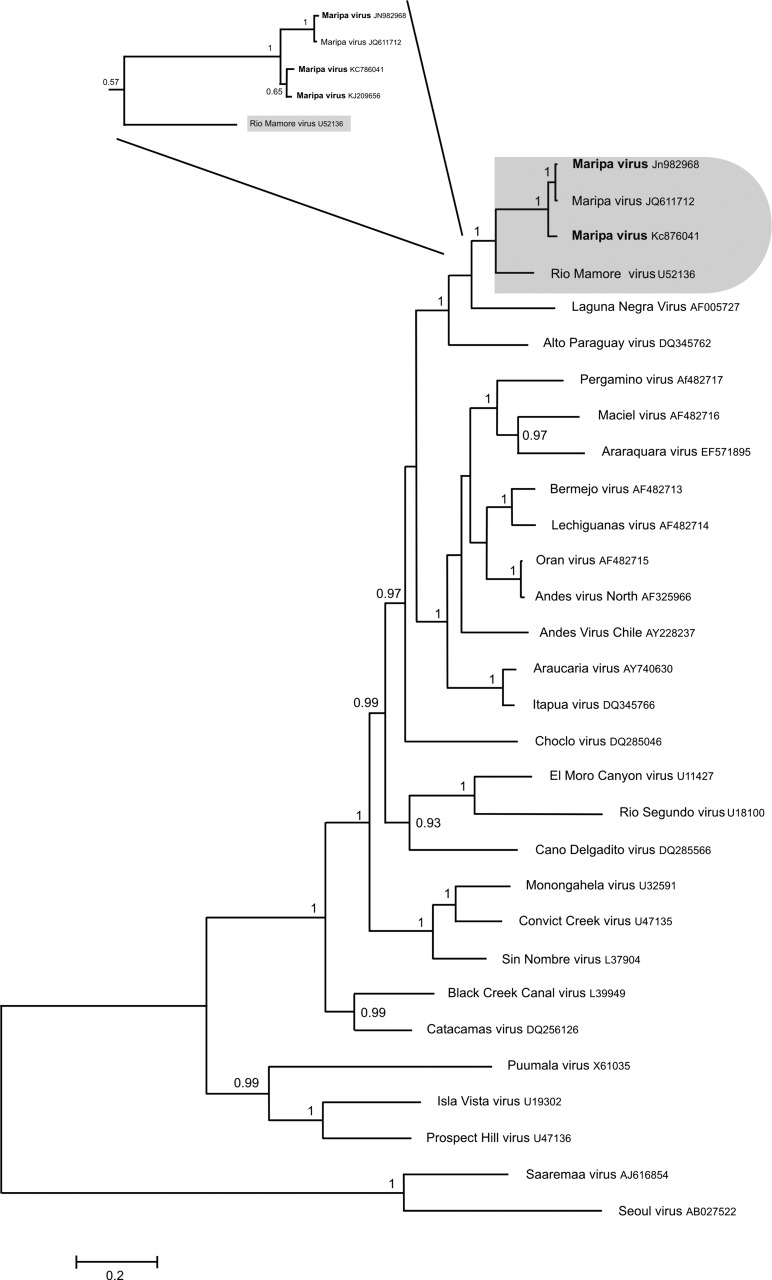
For phylogenetic purposes, we determined the complete sequence (length of 1,904 bp) of the S segment for the hantavirus detected in O. fulvescens captured in 2010 (GenBank accession number JN982968). The coding sequence of Maripa virus isolated from O. fulvescens presented 96.3% and 100% nucleotide identity and 99.8% and 100% amino acid identity with the two human strains of Maripa virus KC876041 and JQ611712, respectively. It presented 85.7% nucleotide identity (1,284 bp, S coding region) and 97.7% amino acid identity (428 amino acids analyzed) with the Bolivian Rio Mamoré strain. Phylogenetic analysis based on the S coding region showed that Maripa viruses are closely related to the Bolivian Rio Mamoré strain (posterior probability value [pp] = 1) (Figure 1). These two strains are closely associated with Laguna Negra and Alto Paraguay viruses (pp = 1).
Figure 1.
Phylogenetic tree constructed using Bayesian methods (1,000,000 replicates; Mr. Bayes, version 3.1b) based on 1,308 bp of the S segment of 30 hantaviruses. The tree is based on the GTR + I + G model of nucleotide evolution. The virus names are associated with their accession numbers. The Maripa virus sequences generated in this study are in bold. The association between Maripa viruses and Rio Mamoré virus is labeled. Support for nodes is provided by the posterior probabilities of the corresponding clades. All resolved nodes have posterior probability greater than 0.8. Scale bar indicates nucleotide sequence divergence among hantavirus sequences. Focus corresponds to the phylogenetic tree based on 345 bp of the S segment of the hantavirus detected in Oligoryzomys captured around the four human cases.
Viral identification in O. fulvescens and Z. brevicauda in the immediate vicinity of fatal human cases suggests that these species could be potential reservoirs of the hantavirus Maripa strain. Additionally, capture data show the presence of O. fulvescens and Z. brevicauda around three of four human cases. Nevertheless, only O. fulvescens was found positive for hantavirus near two different cases. We can, thus, hypothesize a spillover from O. fulvescens to Z. brevicauda. The absence of SNV antibodies suggests either a very recent infection or a very low infecting dose unable to stimulate a humoral IgG response.18,19 Spillover and host switching could explain the infection of two different species: the ecology and respective reservoir performance of Z. brevicauda and O. fulvescens should be scrutinized. Considering that Z. brevicauda and O. fulvescens have already been identified as competent reservoirs of different hantaviruses20,21 and that it is now accepted that a single hantavirus strain can infect different rodent species,22–24 our results suggest that Maripa may be present in a rodent's community of diverse sympatric and/or syntopic competence at the same collection site.
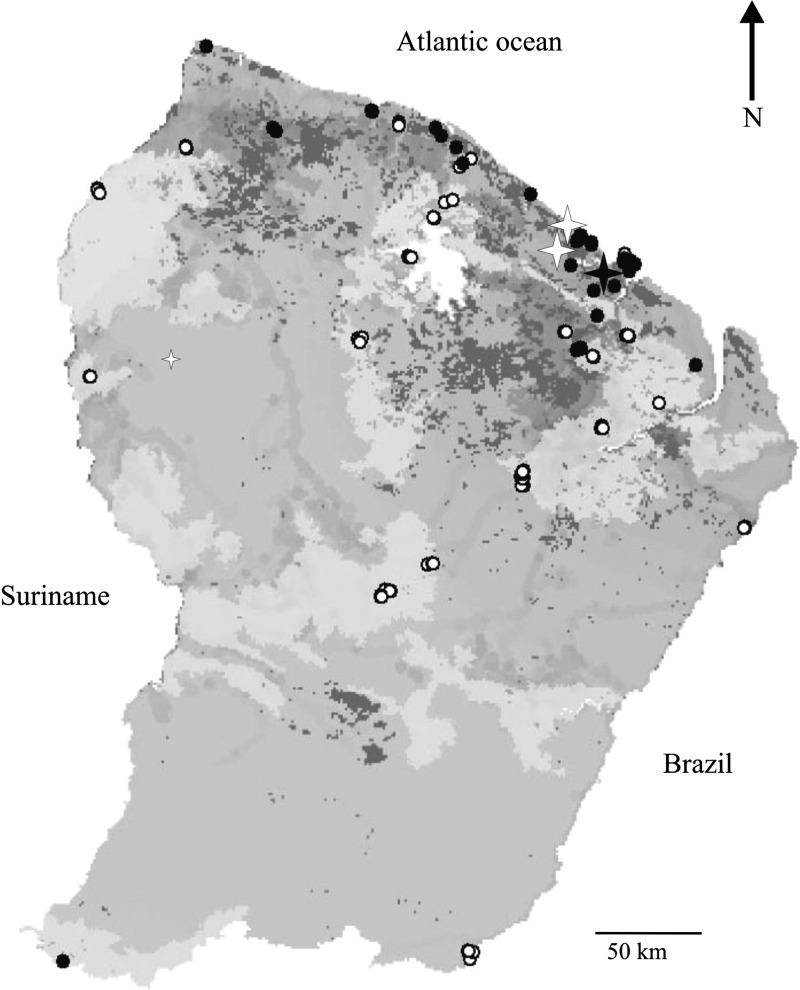
Among SDMs, maximum entropy analysis has a recognized efficiency in processing presence-only data and small datasets.25 MaxEnt3.3.324 estimates the probability distribution of the maximum entropy of each environmental variable across the study area calculated with the constraint that the expected value of each environmental variable under this estimated distribution matches the empirical average generated from environmental values associated with species occurrence data.26 Based on the rodent trapping sessions carried out in French Guiana for 15 years, including published27 and unpublished vouchered specimens (Paris National Museum of Natural History and our own field collections), both positive species seem to be rather rare and unevenly distributed, with 28 records of Z. brevicauda and 14 records of O. fulvescens. The SDM was applied to all the cumulated records of the two species using the 116 sampling locations corresponding to all rodent trapping sessions. Four environmental layers were used; vegetation types were defined with high-definition remote sensing,28 landscape units,29 rainfall, and human footprint (representing the distribution and strength of pressures on natural habitats).30 To control the likely geographic bias of sightings distribution (i.e., trapping effort), the model was forced to use environmental layers restricted to the areas of sampling during the learning stage.31 Predicted areas of occurrence were then projected at the country scale. The model was run using 75% of records for training and the remaining 25% of records for testing, and 50,000 iterations with a bootstrap replicate strategy were used. The null model hypothesis32 was used to test the reliability of the model, generating 99 random distributions. Low-relief hydromorphic and lowland coastal plain, open forests and savannas, and areas with a higher disturbance index have an increased occurrence probability. The resulting predicted distribution map (null hypothesis test: P < 0.01) is shown in Figure 2. We identified northern habitats of French Guiana, characterized by mosaics of more open vegetation types, as more favorable areas for Z. brevicauda and O. fulvescens; ecological closeness of the species, rather than their phylogenetic relatedness, may explain their status as Maripa hosts and enforces the spillover hypothesis. In various natural habitats, SDMs also introduced the human footprint as a dynamic variable favoring an increase in species occurrence, with a likely associated risk of emergence. The role of the anthropogenic disturbance, thus, requires further attention, and lessons for prevention policy are clear: some natural habitats more likely host rodent populations, and increased human pressures on these habitats will increase the likelihood of population expansion.
Figure 2.
Predicted locations of the most appropriate environmental conditions for Z. brevicauda and O. fulvescens. The gradient of colors from light to dark grey illustrates the gradient of the most appropriate environmental conditions. The filled dots indicate all French Guiana localities where one of these two species was documented (the literature, museum specimens, and our own unpublished materials), whereas the empty dots indicate all other rodent trapping sites where these two species have not been recorded. The filled star indicates the locality of capture of the rodents found positive for hantavirus and the associated HPS; the empty stars indicate the two other human HPS.
This report is one of the first to predict more suitable habitats for the emergence of hantavirus in the Amazonian region dominated by forest habitats but including numerous patches of open habitats with substantial and increasing human-induced disturbance. The closest previous records of HPS were located more than 1,000 km away.8,10 The geographical relationship between the predicted distribution of the two host species reservoirs, Z. brevicauda and O. fulvescens, and the emergence risks must nevertheless be taken with caution. Indeed, hantavirus emergence is related to locally and temporally restricted rodent proliferation phenomena: these proliferations are caused by small-scale ecological events that the model may fail to detect.
ACKNOWLEDGMENTS
This study was conducted within the Virus-Reservoirs-Urbanisation-Surveillance de l#x0027;Emergence en Amérique du Sud (ViRUSES) program. F.C. acknowledges the technical expertise of Michel Gillioz at the Natural History Museum of Geneva, who curated our specimens with care.
Footnotes
Financial support: This study was supported by Fonds Européen de Développement Régional (FEDER) and assistance from Région Guyane and Direction Régionale pour la Recherche et la Technologie. This study benefited from European Commission Grant REGPOT-CT-2011-285837-STRonGer in the frame of FP7 and an Investissement d'Avenir Grant managed by Agence Nationale de la Recherche (Centre d’Etude de la Biodiversité Amazonienne [CEBA]; ANR-10-LABX-25-01).
Authors' addresses: Benoît de Thoisy, Amandine Guidez, Damien Donato, Vincent Lacoste, and Anne Lavergne, Laboratoire des Interactions Virus-Hôtes, Institut Pasteur de la Guyane, Cayenne, French Guiana, E-mails: bdethoisy@pasteur-cayenne.fr, aguidez@hotmail.fr, ddonato@pasteur-cayenne.fr, vlacoste@pasteur-cayenne.fr, and alavergne@pasteur-cayenne.fr. Séverine Matheus, Laboratoire de Virologie, Centre National de Reference des Arbovirus, Virus Influenza et Hantavirus, Laboratoires Associés–Pôle Antilles Guyane, Institut Pasteur de la Guyane, Cayenne, French Guiana, E-mail: smatheus@pasteur-cayenne.fr. François Catzeflis, Laboratoire de Paléontologie, Paléobiologie et Phylogénie, Institut des Sciences de l'Evolution, UMR5554, Montpellier, France, E-mail: francois.catzeflis@univ-montp2.fr. Luc Clément and Sébastien Barrioz, Association Kwata, Cayenne, French Guiana, E-mails: lucclement@hotmail.fr and seb.kwata@orange.fr. Jean-François Cornu, Unité Mixte de Recherche (UMR) Biologie des ORganismes et des Ecosystèmes (BOREA), Département Milieux et Peuplements Aquatiques, Museum National d’Histoire Naturelle (MNHN), Centre National d’Etude Scientifique (CNRS) 7208, Institut de Recherche pour le Développement (IRD) 207, Université Pierre et Marie Curie (UPMC), Muséum National d'Histoire Naturelle, Cayenne, French Guiana, and Office National des Forêts, Cayenne, French Guiana, E-mail: jfrancoiscornu@wanadoo.fr. Olivier Brunaux and Stéphane Guitet, Office National des Forêts, Cayenne, French Guiana, E-mails: olivier.brunaux@onf.fr and stephane.guitet@cirad.fr.
References
- 1.Jonsson CB, Figueiredo LT, Vapalahti O. A global perspective on hantavirus ecology, epidemiology, and disease. Clin Microbiol Rev. 2010;23:412–441. doi: 10.1128/CMR.00062-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allen LJS, McCormack RK, Jonsson CB. Mathematical models for hantavirus infection in rodents. Bull Math Biol. 2006;68:511–524. doi: 10.1007/s11538-005-9034-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sauvage F, Langlais M, Pontier D. Predicting the emergence of human hantavirus disease using a combination of viral dynamics and rodent demographic patterns. Epidemiol Infect. 2007;135:46–56. doi: 10.1017/S0950268806006595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carroll DS, Mills JN, Montgomery JM, Bausch DG, Blair PJ, Burans JP, Felices V, Gianella A, Iihoshi N, Nichol ST, Olson JG, Rogers DS, Salazar M, Ksiazek TG. Hantavirus pulmonary syndrome in central Bolivia: relationships between reservoir hosts, habitats, and viral genotypes. Am J Trop Med Hyg. 2005;72:42–46. [PubMed] [Google Scholar]
- 5.Mills JN, Schmidt K, Ellis BA, Calderón G, Enria D, Ksiazek TG. A longitudinal study of hantavirus infection in three sympatric reservoir species in agroecosystems on the Argentine Pampa. Vector Borne Zoonotic Dis. 2007;7:229–240. doi: 10.1089/vbz.2006.0614. [DOI] [PubMed] [Google Scholar]
- 6.Patterson BD. Accumulating knowledge on the dimensions of biodiversity: systematic perspectives on Neotropical mammals. Biodivers Lett. 1994;2:79–86. [Google Scholar]
- 7.Rosa ES, Mills JN, Padula PJ, Elkhoury MR, Ksiazek TG, Mendes WS, Santos ED, Araújo GC, Martinez VP, Rosa JF, Edelstein A, Vasconcelos PF. Newly recognized hantaviruses associated with hantavirus pulmonary syndrome in northern Brazil: partial genetic characterization of viruses and serologic implication of likely reservoirs. Vector Borne Zoonotic Dis. 2005;5:11–19. doi: 10.1089/vbz.2005.5.11. [DOI] [PubMed] [Google Scholar]
- 8.Medeiros DB, da Rosa ES, Marques AA, Simith DB, Carnero AR, Chiang JO, Prazeres ITE, Vasconcelos PFC, Nunes MRT. Circulation of hantaviruses in the influence area of the Cuiabá-Santarém Highway. Mem Inst Oswaldo Cruz. 2010;105:665–671. doi: 10.1590/s0074-02762010000500011. [DOI] [PubMed] [Google Scholar]
- 9.Travassos da Rosa ES, Medeiros DB, Nunes MR, Simith DB, de Souza Pereira A, Elkhoury MR, Lavocat M, Marques AAR, Via AV, D'Andrea P, Bonvincino CR, Lemos ERS, Vasconcelos PFC. Pygmy rice rat as potential host of Castelo dos Sonhos Hantavirus. Emerg Infect Dis. 2011;17:1527–1530. doi: 10.3201/eid1708.101547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Firth C, Tokarz A, Simith DB, Nunes MRT, Bhat M, Rosa EST, Medeiros DBA, Palacio G, Vasconcelos PFC, Lipkin WI. Diversity and distribution of hantaviruses in South America. J Virol. 2012;84:13756–13766. doi: 10.1128/JVI.02341-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Matheus S, Djossou F, Moua D, Bourbigot AM, Hommel D, Lacoste V, Dussart P, Lavergne A. Hantavirus pulmonary syndrome, French Guiana. Emerg Infect Dis. 2010;16:739–741. doi: 10.3201/eid1604.090831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lim BK. Preliminary assessment of neotropical mammal DNA barcodes: an underestimation of biodiversity. Open Zool J. 2012;5:10–17. [Google Scholar]
- 13.Johnson AM, Bowen MD, Ksiazek TG, Williams RJ, Bryan RT, Mills JN, Peters CJ, Nichol ST. Laguna Negra virus associated with HPS in western Paraguay and Bolivia. Virology. 1997;238:115–127. doi: 10.1006/viro.1997.8840. [DOI] [PubMed] [Google Scholar]
- 14.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nylander JAA. MrModeltest v2. Uppsala, Sweden: Evolutionary Biology Centre, Uppsala University; 2004. [Google Scholar]
- 16.Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 17.Matheus S, Lavergne A, de Thoisy B, Dussart P, Lacoste V. Complete genome sequence of a novel hantavirus variant of Rio Mamoré virus, Maripa virus, from French Guiana. J Virol. 2012;86:5399. doi: 10.1128/JVI.00337-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Billings AN, Rollin PE, Milazzo ML, Molina CP, Eyzaguirre EJ, Livingstone W, Ksiazek TG, Fulhorst CF. Pathology of Black Creek Canal virus infection in juvenile hispid cotton rats (Sigmodon hispidus) Vector Borne Zoonotic Dis. 2010;10:621–628. doi: 10.1089/vbz.2009.0156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Boone JD, Otteson EW, McGwire KC, Villard P, Rowe JE, St Jeor SC. Ecology and demographics of hantavirus infections in rodent populations in the Walker River Basin of Nevada and California. Am J Trop Med Hyg. 1998;59:445–451. doi: 10.4269/ajtmh.1998.59.445. [DOI] [PubMed] [Google Scholar]
- 20.Vincent MJ, Quiroz E, Gracia F, Sanchez AJ, Ksiazek TG, Kitsutani PT, Ruedas LA, Tinnin DS, Caceres L, Garcia A, Rollin PE, Mills JN, Peters CJ, Nichol ST. Hantavirus pulmonary syndrome in Panama: identification of novel hantaviruses and their likely reservoirs. Virology. 2000;277:14–19. doi: 10.1006/viro.2000.0563. [DOI] [PubMed] [Google Scholar]
- 21.Suzan G, Giermakowski JT, Marce E, Suzan-Azpiri H, Armién B, Yates TL. Modeling hantavirus reservoir species dominance in high seroprevalence areas on the Azuero Peninsula of Panama. Am J Trop Med Hyg. 2006;74:1103–1110. [PubMed] [Google Scholar]
- 22.Chu YK, Goodin D, Owen RD, Koch D, Jonsson CB. Sympatry of 2 hantavirus strains, Paraguay, 2003–2007. Emerg Infect Dis. 2009;15:1977–1980. doi: 10.3201/eid1512.090338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Milazzo ML, Cajimat MN, Hanson JD, Bradley RD, Quintana M, Sherman C, Velásquez RT, Fulhorst CF. Catacamas virus, a hantaviral species naturally associated with Oryzomys couesi (Coues' oryzomys) in Honduras. Am J Trop Med Hyg. 2006;75:1003–1010. [PMC free article] [PubMed] [Google Scholar]
- 24.Raboni SM, Hoffmann FG, Oliveira RC, Teixeira BR, Bonvincino CR, Stella RC, Carstensen S, Bordignon J, D'Andrea PS, Lemos ERS, do Santos CND. Phylogenetic characterization of hantaviruses from wild rodents and hantavirus pulmonary syndrome cases in the state of Parana (southern Brazil) J Gen Virol. 2009;90:2166–2171. doi: 10.1099/vir.0.011585-0. [DOI] [PubMed] [Google Scholar]
- 25.Wisz MS, Hijmans RJ, Li R, Peterson AT, Graham CH, Guisan A. Effects of sample size on the performance of species distribution models. Divers Distrib. 2008;14:763–773. [Google Scholar]
- 26.Phillips SJ, Anderson RP, Schapire RE. Maximum entropy modeling of species geographic distributions. Ecol Modell. 2006;190:231–259. [Google Scholar]
- 27.Voss RS. An introduction to the Neotropical muroid rodent genus Zygodontomys. B Am Mus Nat Hist. 1991;210:1–113. [Google Scholar]
- 28.Gond V, Freycon V, Molino JF, Brunaux O, Ingrassia F, Joubert P, Pekel JF, Prévost MF, Thierron V, Trombe PJ, Sabatier D. Broad-scale spatial pattern of forest landscapes types in the Guiana shield. Int J Appl Earth Obs Geoinf. 2011;13:357–367. [Google Scholar]
- 29.Guitet S, Cornu JF, Brunaux O, Betbeder J, Carozza JM, Richard-Hansen C. Landform and landscape mapping, French Guiana (South America) J Maps. 2013 doi:10.1080/17445647.2013.785371. [Google Scholar]
- 30.de Thoisy B, Richard-Hansen C, Goguillon B, Joubert P, Obstancias J, Winterton P, Brosse S. Rapid evaluation of threats to biodiversity: human footprint score and large vertebrate species responses in French Guiana. Biodivers Conserv. 2010;19:1567–1584. [Google Scholar]
- 31.Dudík M, Schapire RE, Phillips SJ. Correcting sample selection bias in maximum entropy density estimation. Adv Neural Inf Process Syst. 2005;18:323–330. [Google Scholar]
- 32.Raes N, ter Steege H. A null-model for significance testing of presence-only species distribution models. Ecography (Cop.) 2007;30:727–736. [Google Scholar]
- 33.Wilson DE, Reeder DM. Mammal Species of the World. A Taxonomic and Geographic Reference. Baltimore, MD: The Johns Hopkins University Press; 2005. [Google Scholar]