Abstract

The N-terminus of the B-chain of insulin may adopt two alternative conformations designated as the T- and R-states. Despite the recent structural insight into insulin–insulin receptor (IR) complexes, the physiological relevance of the T/R transition is still unclear. Hence, this study focused on the rational design, synthesis, and characterization of human insulin analogues structurally locked in expected R- or T-states. Sites B3, B5, and B8, capable of affecting the conformation of the N-terminus of the B-chain, were subjects of rational substitutions with amino acids with specific allowed and disallowed dihedral φ and ψ main-chain angles. α-Aminoisobutyric acid was systematically incorporated into positions B3, B5, and B8 for stabilization of the R-state, and N-methylalanine and d-proline amino acids were introduced at position B8 for stabilization of the T-state. IR affinities of the analogues were compared and correlated with their T/R transition ability and analyzed against their crystal and nuclear magnetic resonance structures. Our data revealed that (i) the T-like state is indeed important for the folding efficiency of (pro)insulin, (ii) the R-state is most probably incompatible with an active form of insulin, (iii) the R-state cannot be induced or stabilized by a single substitution at a specific site, and (iv) the B1–B8 segment is capable of folding into a variety of low-affinity T-like states. Therefore, we conclude that the active conformation of the N-terminus of the B-chain must be different from the “classical” T-state and that a substantial flexibility of the B1–B8 segment, where GlyB8 plays a key role, is a crucial prerequisite for an efficient insulin–IR interaction.
Insulin is a 51-amino acids polypeptide hormone that is a primary regulator of carbohydrate homeostasis, and insufficient or defective insulin secretion, or impaired insulin signaling, leads to a metabolic disorder, known as diabetes mellitus. Insulin is the post-translational product of a single-chain precursor proinsulin, and its conversion to two-chain insulin, and the subsequent storage of the mature hormone, occurs in pancreatic β-cells.1
Processed human insulin (HI) consists of the 21-amino acid A-chain and 30-amino acid B-chain that are interconnected by two (A7–B7 and A20–B19) disulfide bridges and stabilized further by one intrachain (A6–A11) disulfide bridge. Insulin is stored as a Zn2+-stabilized hexamer, which dissociates in portal circulation to the functional Zn2+-free monomer that binds to the insulin receptor (IR).2 Extensive mutagenesis and functional studies of this hormone have revealed side chains responsible for insulin–IR interactions.3−7 High-affinity binding of insulin to so-called site 1 of the IR involves mainly GlyA1, IleA2, ValA3, GlnA5, TyrA19, and AsnA21 on the A-chain and GlyB8, LeuB11, ValB12, LeuB15, and the B23–B26 segment on the B-chain.8,9 The importance of some of these residues in the formation of the insulin–IR interface was confirmed by the first crystal structures of insulin complexes with two different constructs of the IR ectodomain.10 The formation of the insulin–IR complex is accompanied by extensive conformational changes in the hormone, particularly in the N- and C-termini of its B-chain, that constitute the most flexible and accessible parts of the molecule. The C-terminus of the B-chain must be detached from its central B9–B19 β-helix11−14 upon IR binding to expose hidden A1–A3 and A19 residues, which are some of the hot spots of IR interactions.8,10 The importance of the B-chain C-terminal residues in IR binding has also been demonstrated by their critical role in the negative cooperativity character of this interaction.15
However, the continuing ambiguity with regard to the conformational behavior of the B22–B30 segment at the hormone–IR interface is paralleled by the lack of clarity about the physiological (i.e., in hexamer/storage hormone form and in its IR complex) fold of the B-chain N-terminal B1–B8 residues. The observed large, spatially diverged (nearly 30 Å) structural changes at the N-terminus of the B-chain span two alternative insulin conformational forms designated as the T- and R-states.16,17 In the T-state, residues B1–B8 are in an extended conformation, which is followed by the central, structurally conserved α-helix of residues B9–B19. In contrast, this helix if further fully extended by residues B1–B8 in the R-state of the hormone. Extensive structural and spectroscopic studies indicated that the T- and R-states can lead to three main allosteric insulin oligomers designated as T6, T3R3/T3Rf3, and R6.16−22 Whereas the conformations of the two independent monomers in three dimers forming the T6 or R6 hexamers are nearly identical, the dimeric unit in the T3Rf3 hexamer consists of one T-state and one Rf-state17,23,24 insulin, in which B1–B3 segments of the Rf3 molecules are “frayed” into a nonhelical and extended conformation.24 These hexamer structural states are related in solution by dynamic equilibria (T6 ↔ T3Rf3 ↔ R6),21 with the T6 state being dominant.25 Spectroscopic and structural studies of the allosteric behavior of insulin revealed that anions from the Hofmeister lyotropic series, such as Cl– or SCN–, can induce only the T6 → T3Rf3 conformational change,21,26 and that the full R6 hexamer in HI requires the presence of phenol derivatives or other cyclic alcohols (e.g., phenol, cresol, resorcinol, etc.).16,24,27,28 Moreover, the full R-state has never been observed in any dimeric or monomeric forms of insulin. The physiological relevance of the T- and R-states is still unclear, but the importance of the T-state for proper proinsulin and/or insulin folding has been suggested.22,29 Although the R-state was linked with insulin biological activity,29,30 a high activity of an insulin analogue that cannot adopt an R-like fold31 might indicate that the R-state is not involved in IR binding. Therefore, an unambiguous assignment of the biological relevance of T- and R-states is still required. This not only is necessary for a full understanding of the insulin–IR interaction and the nature of the insulin storage oligomers but also may have a positive impact on the rational design of the new, more effective insulin analogues.
The GlyB8 site is known to be the key, pivotlike, structural residue implicated in the interconversion of the T- and R-states. Even if B8 substitutions are accompanied by a loss of binding affinity,29,32 the study of the structure–function relationship of B8-modified analogues in a rationally stabilized T- or R-state may shed some light on the physiological relevance of these conformations. In the T-state, GlyB8 lies at the right half of the Ramachandran plot that is typical for d-amino acids with positive φ angles [approximately 59°, as in Protein Data Bank (PDB) entry 1mso]. In contrast, GlyB8 in the R-state has negative φ angles (approximately −67°, as in PDB entry 1znj) and occupies the left half-segment of the Ramachandran plot characteristic of a right-handed α-helix (Figure S1 of the Supporting Information).
Here, we present the synthesis and detailed characterization of analogues that were rationally designed to be structurally locked in “a predicted” R- or T-state. The allowed and disallowed dihedral φ and ψ main-chain angles of the specific, non-natural amino acids have been considered in this process (Figure S1 of the Supporting Information). Three amino acids [α-aminoisobutyric acid (Aib), d-proline, and N-methylalanine (NMeAla)] with the desired conformational properties have been systematically introduced into positions B3, B5, and B8 at the N-terminus of the B-chain, as they are known to affect its conformation.21,22,33 Aib was selected for the rational induction of the R-like helical conformation of the B1–B8 segment, as the amino acid with a high helical propensity that often facilitates folding into the right- or left-handed α-helices.34,35 The enforcement of the T-state was designed by the use of d-Pro and NMeAla as these amino acids cannot adopt the right-handed α-helix φ and ψ angles. The structure–function relationship of these insulin analogues is discussed on the basis of their functional and structural [X-ray, nuclear magnetic resonance (NMR), and circular dichroism (CD)] properties.
Experimental Procedures
Solid-Phase Peptide Synthesis of Insulin and Analogue Chains
The individual wild-type A-chain and modified B-chains (AibB3; AibB5; AibB8; AibB8, LysB28, ProB29; and d-ProB8 or NMeAlaB8) were synthesized by stepwise coupling of the corresponding Fmoc amino acid on Fmoc-Asn(Trt)-Wang LL resin and Fmoc-Thr(OtBu)-Wang LL resin (Nova Biochem, San Diego, CA), respectively, using an automatic solid-phase synthesizer (ABI 433A, Applied Biosystems, Foster City, CA). HBTU/HOBt in DMF was used as a coupling reagent. Fully protected peptides were cleaved from the resins by a TFA/H2O/TIS/EDT/phenol/thioanisol mixture (90:3:1:1:2:3) and were precipitated with cold diethyl ether. The LysB28 ↔ ProB29 swap was introduced into [AibB8,LysB28,ProB29]-insulin to promote its monomeric behavior36 for solution NMR studies.
Sulfitolysis
Insulin crude A- or B-chains (100 μmol) in reduced (SH) forms were dissolved and stirred in 25 mL of freshly prepared sulfiltolysis buffer [100 mM Tris, 250 mM Na2SO3, 80 mM Na2S4O6, and 7 M GuaHCl (pH 8.6)] for 3 h at room temperature to convert SH groups to S-sulfonates. The chains were then desalted on a Sephadex G10 column (4 cm × 85 cm) in 50 mM NH4HCO3 and purified using reversed-phase high-performance liquid chromatography (RP-HPLC) (Nucleosil C18 column, 250 mm × 21 mm, 5 μm).
Disulfide Bridge Combination
Insulin A-chain (30 mg) and B-chain (15 mg) S-sulfonate derivatives were dissolved in 2 and 1 mL of degassed 0.1 M Gly/NaOH buffer (pH 10.5), respectively. The exact molar concentrations of individual chains were determined by UV spectrophotometry at 280 nm using molar extinction coefficients of 3480 and 3230 M–1 cm–1 for the A-chain and the B-chain, respectively. The solutions of chains were combined, and dithiothreitol (DTT, aliquoted from Pierce catalog no. 20291) in a minimal volume of a degassed 0.1 M Gly/NaOH buffer (pH 10.5) was rapidly added to the peptide solution to give a SH:SSO3– molar ratio of 1.2. This solution was stirred for 30 min in a capped vessel at room temperature. After the reduction of SSO3– to SH, 3 mL of aerated 0.1 M Gly/NaOH buffer (pH 10.5) was added, and the resulting solution was stirred for 48 h at 4 °C in an open vessel to permit air oxidation. Glacial acetic acid (3 mL) was added to the mixture to terminate the reaction. The resulting mixture was applied to a low-pressure column (Sephadex G-50 in 1 M acetic acid, 2 cm × 75 cm). The fractions containing the respective analogues were purified using RP-HPLC (Nucleosil C18 column, 250 mm × 8 mm, 5 μm). The molecular weight of resulted analogues was confirmed by HR mass spectroscopy (LTQ Orbitrap XL, Thermo Fisher Scientific, Waltham, MA),37,38 and the purity of analogues was controlled by RP-HPLC (Nucleosil C18 column, 250 mm × 4 mm, 5 μm).
IR Isoform A Binding Affinity Assays
The binding affinity for isoform A of the human IR (IR-A) was determined using the method of Morcavallo et al.39 using IM-9 lymphocytes (ATCC), which are rich in IR-A expression. The IM-9 cell line was cultured in RPMI 1640 containing 10% fetal bovine serum. For the assay, 2.0 × 106 cells per milliliter were incubated with increasing concentrations of insulin or an analogue and human [125I]monoiodotyrosyl-A14-insulin (PerkinElmer Life Sciences, 2200 Ci/mmol, 20000 cpm, ∼0.01 nM) for 2.5 h at 15 °C in HEPES binding buffer [100 mM HEPES, 100 mM NaCl, 5 mM KCl, 1.3 mM MgSO4, 1 mM EDTA, 10 mM glucose, 15 mM NaOAc, and 1% BSA (w/v) (pH 7.6)] (500 μL). After incubation, 2 × 200 μL was centrifuged at 13000g for 10 min. Radioactive pellets were counted using a Wizard 1470 Automatic γ Counter (PerkinElmer Life Sciences). Binding data were analyzed with Excel using a one-site fitting program developed in the laboratory of P. De Meyts (A. V. Groth and R. M. Shymko, Hagedorn Research Institute, Gentofte, Denmark, a kind gift of P. De Meyts) and GraphPad Prism 5, which take the potential ligand depletion into account using equations developed by Swillens.40 The dissociation constant of human [125I]insulin was set to 0.3 nM.
IR Isoform B and IGF-1 Receptor Binding Affinity Assays
The methodology for the determination of receptor binding affinities for isoform B of the IR (IR-B) and for the receptor of IGF-1 (IGF-1R) is described in the Supporting Information.
Circular Dichroism
The CD spectra were recorded in a quartz cuvette with an optical path length of 1 cm or 0.5 mm (Starna Cells) using a J-815 spectropolarimeter (Jasco) at room temperature. The far- and near-UV CD spectra were used to measure changes in protein secondary and tertiary structures. The spectral regions were 200–260 and 245–300 nm, with a scanning speed of 10 nm/min, a response time of 8 s, a resolution of 0.5 nm, a bandwidth of 1 nm, and a sensitivity of 100 mdeg. The final spectrum was obtained as an average of five accumulations. The spectra were corrected for the baseline by subtracting the spectra of the corresponding polypeptide-free solution. Analogues or HI (0.25 mg/mL, 43 μM) were measured in 25 mM Tris-HCl buffer (pH 7.8) with 0.33 mol of Zn2+ (zinc acetate) per mole of analogue or HI with or without 25 mol of phenol per mole of analogue or HI.
NMR Spectroscopy and Structure Calculations
NMR spectra were acquired from a 0.35 mL sample of 0.5 mM [AibB8,LysB28,ProB29]-insulin in 20% d4-acetic acid (pH 1.9). All the NMR data were collected at 25 °C on a 600 MHz Bruker Avance spectrometer equipped with a triple-resonance (15N, 13C, 1H) cryoprobe. A series of homonuclear spectra were recorded to determine sequence-specific resonance assignments for [AibB8,LysB28,ProB29]-insulin, in particular, two-dimensional (2D) TOCSY spectra with a mixing time of 55 ms and 2D DQF-COSY and 2D NOESY spectra, which were acquired with a nuclear Overhauser effect (NOE) mixing time of 200 ms. Residues involved in forming stable backbone hydrogen bonds were identified by monitoring the rate of backbone amide exchange in 2D TOCSY spectra of [AibB8,LysB28,ProB29]-insulin dissolved in a 20% d4-acetic acid/80% D2O mixture. The family of converged structures for [AibB8]-insulin was initially calculated using Cyana version 2.1.41 The combined automated NOE assignment and structure determination protocol was used to automatically assign the NOE cross-peaks identified in 2D NOESY spectrum and to produce preliminary structures. Subsequently, five cycles of simulated annealing combined with redundant dihedral angle constraints (Redac)42 were used to produce a set of converged structures with no significant restraint violations (distance and van der Waals violations of <0.2 Å), which were further refined in explicit solvent using the YASARA software with the YASARA force field.43 The 35 structures with the lowest total energy were selected. Analysis of the family of structures obtained was conducted using Molmol, iCING,44,45 and PyMol (http://www.pymol.org).
X-ray Studies
Crystallizations of all insulin analogues were performed with the in-house insulin crystallization screens that cover most of the parameters of the reported insulin crystallogenesis. Crystallization conditions, data collection, refinement and models statistics, and PDB entries are listed in Table S1 of the Supporting Information. X-ray data were processed with xia2,46 and model building and refinement were performed with the CCP4 suite of programs47 and COOT.48 Crystal structures were determined with Molrep49 with B1–B6 and B23–B30 truncated hexamer-derived insulin monomers as a model (PDB entry 1mso)50 and refined with Refmac version 5.8.51 Figures were made with CCP4mg.47 For structural comparisons, the relevant structures were superimposed on the Cα atoms of residues B9–B19 with the LSQ fit option in COOT.48
Results
Synthesis of the Analogues
Six new insulin analogues modified at the N-terminus of the B-chain have been prepared by the total synthesis and chain combination. The yields of chain recombination reactions were different depending on the amino acid substitution (the average yield from several recombination reactions was calculated to a starting amount of B-chain S-sulfonate as a limiting factor of the reaction). Aib-containing analogues gave generally low yields. AibB5-insulin was obtained with the lowest (0.5%) yield of all the analogues prepared here. The average yields of [AibB3]-insulin, [AibB8]-insulin, and [AibB8,LysB28,ProB29]-insulin were 1.7, 5, and 2%, respectively. In contrast, the analogues with the expected T-state conformation were produced with increased yields; [NMeAlaB8]-insulin and [d-ProB8]-insulin gave yields of 10 and 25%, respectively. For comparison, the average recombination yields of HI in our laboratory are typically in the range of 8–12% (data not shown).
IR Binding Affinities of the Analogues
IR binding affinities for IR-A in membranes of human IM-9 lymphocytes of all the prepared analogues are listed in Table 1, with the corresponding binding curves provided in Figure S2 of the Supporting Information. Both [AibB8]-insulin and [AibB8,LysB28,ProB29]-insulin exhibit very low receptor binding affinities of 0.6 and 0.25%, respectively. The substitution of the B8 site with d-ProB8 and NMeAlaB8 amino acids (adopting mostly dihedral angles from the right half of the Ramachandran plot) also had a deleterious effect on the binding potencies of the resulting analogues. [d-ProB8]-insulin retained only 0.08% of the affinity of HI, and the binding affinity of [NMeAlaB8]-insulin was under the range of our measurement (<0.01%). [AibB5]-insulin had a binding affinity that was 4.4% of that of HI. The α-Aib substitution at position B3 has the least impact on IR affinity as [AibB3]-insulin retained 28.4% of the binding potency of HI.
Table 1. Values of Kd and Relative Binding Affinities of Human Insulin and Insulin Analogues for IR-A in Membranes of Human IM-9 Lymphocytes.
| analogue | Kda (nM) | potencyb (%) |
|---|---|---|
| human insulin | 0.27 ± 0.04 (5) | 100 ± 15 |
| [AibB3]-insulin | 0.94 ± 0.02 (3) | 28.4 ± 0.6 |
| [AibB5]-insulin | 6.09 ± 1.04 (3) | 4.4 ± 0.8 |
| [AibB8]-insulin | 48.4 ± 0.54 (4) | 0.6 ± 0.007 |
| [AibB8,LysB28,ProB29]-insulin | 106 ± 22.0 (4) | 0.25 ± 0.05 |
| [d-ProB8]-insulin | 345 ± 20.5 (3) | 0.08 ± 0.005 |
| [NMeAlaB8]-insulin | >1500 (3) | <0.01 |
Each value represents the mean ± the standard deviation of multiple determinations (n).
Relative receptor binding affinity (potency) defined as [(Kd of human insulin)/(Kd of analogue)] × 100.
Additionally, two analogues, [AibB8]-insulin and [NMe-AlaB8]-insulin, were tested for their binding affinities for IR-B and the IGF-1 receptor (IGF-1R) in membranes of mouse embryonal fibroblasts. The binding curves are shown in Figure S3 of the Supporting Information, and the data are listed in Table S2 of the Supporting Information. Like those for IR-A, both analogues displayed low binding affinities for IR-B (0.27% for [AibB8]-insulin and <0.05% for [NMeAlaB8]-insulin), and their binding affinities for IGF-1R were below the measurable range (<0.01%).
Circular Dichroism
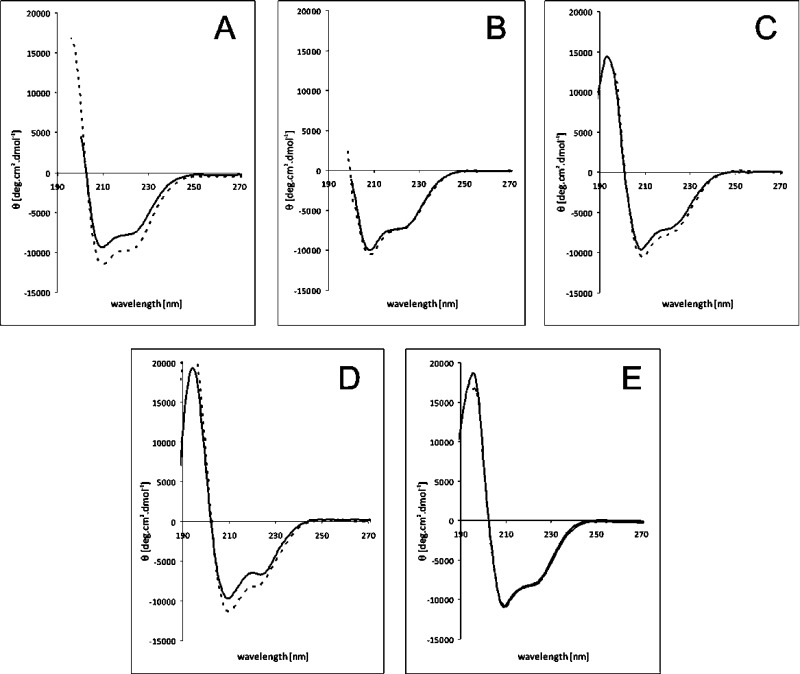
The T → R transition abilities of HI and studied analogues were inferred from the CD spectra. The far-UV (200–260 nm) and near-UV (245–300 nm) CD spectra were acquired in the presence and absence of phenol, an inducer of the T → R transition (Figure 1 and Figure S4 of the Supporting Information). Typically, the addition of phenol results in an increase in the magnitude of the CD signals at 208 and 222 nm, characteristic of the helix-associated mean residue, and in an increase in the magnitude of the negative CD signal around 250 nm.21,28 The presence of individual secondary structure elements within the molecule (Table S3 of the Supporting Information) was calculated from far-UV spectra using CD Spectra Deconvolution version 2.11 developed in 2001 by G. Böhm (Institut für Biotechnologie Martin-Luther-Universität Halle-Wittenberg, Halle-Wittenberg, Germany).
Figure 1.
Far-UV CD spectra of insulin analogues in the absence (—) and presence (---) of phenol: (A) human insulin, (B) [AibB3]-insulin, (C) [AibB8]-insulin, (D) [d-ProB8]-insulin, and (E) [NMeAlaB8]-insulin.
Representative curves for the T- and R-states of HI (Figure 1A) demonstrate that the presence of phenol increases the negative ellipticity in insulin spectra at 208 and 222 nm. In contrast to those of HI, the spectra of [AibB3]-insulin and [AibB8]-insulin analogues exhibit only small differences in the absence or presence of phenol (Figure 1B,C), which may be indicative of a much smaller increase in the α-helical content (Table S3 of the Supporting Information). The low yield of the [AibB5]-insulin analogue prevented its spectroscopic analysis. The [d-ProB8]-insulin analogue showed a significant increase in the magnitude of the negative CD signal in the presence of phenol, especially at 208 nm (Figure 1D), indicating a possible, and unexpected, T → R transition ability of this analogue. In contrast, the spectra of [NMeAlaB8]-insulin showed only a minimal change after addition of phenol in the near- and far-UV regions, confirming the conservation of the T-state conformation regardless of the presence or absence of phenol (Figure 1E and Figure S4D of the Supporting Information).
Solution Structure of the [AibB8,LysB28,ProB29]-Insulin Analogue
The NMR structure of [AibB8,LysB28,ProB29]-insulin has been determined as none of the Aib analogues yielded any suitable crystals. This analogue provided good-quality NMR spectra under acidic conditions [20% d4-acetic acid (pH 1.9)], which is indicative of the expected monomeric behavior for this modified insulin. However, the apparent line broadening of amide proton signals, in comparison with that of the previously studied insulin analogues, may suggest some degree of chemical exchange between two, or more, similar conformational states. Essentially complete sequence-specific assignment of 1H NMR resonances has been achieved using the combination of homonuclear 2D TOCSY, NOESY, and DQF-COSY experiments. In particular, 96.8% of all proton resonances were assigned with the exception of the poorly resolved Hξ signals of all three phenylalanine side chains (PheB1, PheB24, and PheB25), Hα and Hβ of CysA11, Hα of CysB7, and HN and Hβ3 of LeuB11. The 1H resonance assignments were used for automated assignment of the NOEs identified in the 2D NOESY spectrum using the CANDID protocol implemented in Cyana.41 This yielded unique assignments for 94.1% (606 of 644) of the NOE peaks observed, providing 488 nonredundant 1H–1H distance constraints. The 35 satisfactorily converged [AibB8,LysB28,ProB29]-insulin structures [obtained from 100 random starting conformations using 570 NMR-derived structural constraints, including distance restraints for hydrogen and disulfide bonds (>11 constraints per residue)] were further refined in explicit water using YASARA.43 The numbers of observed NOE peaks, distance constraints, and structural statistics for obtained structures are listed in Table S4 of the Supporting Information.
The solution structure calculated from the NMR data for [AibB8,LysB28,ProB29]-insulin is shown in Figure 2. The GlyB8Aib mutation did not have a significant impact on the conformation of the B8 site of this analogue or on the conformation of the B1–B8 segment. Unexpectedly, the N-terminus of the B-chain of the analogue adopts a conformation similar to that of the T-state of HI (Figure 2B). The main difference between the structures of this analogue and HI is at the C-termini of their B-chains. The C-terminus of the B-chain of the analogue is disordered from PheB24 upward, likely because of the LysB28 ↔ ProB29 swap, as we observed the same phenomenon in the NMR structure of [LysB28,ProB29]-insulin under the same experimental conditions (data not shown).
Figure 2.

Solution structure of the [AibB8,LysB28,ProB29]-insulin analogue. (A) Best-fit superposition of the protein backbone for 35 converged structures. The A-chain is colored red, AibB8 blue, and the B-chain gray. (B) Overlay of the representative (closest to mean) structure of the ensemble and HI (light yellow). The side chain of AibB8 is colored blue.
Crystal Structure of [d-ProB8]-Insulin
[d-ProB8]-insulin crystallized as a dimer with one molecule in the asymmetric unit. The overall conformation of each single molecule of this analogue is similar to the T-like state. However, the structure of the B1–B3 region is different. PheB1 is relocated by ∼7.5 Å from its T-state position, and the hydrogen-bonded (2.8 Å) side-chain GluB4OE and main-chain COPheB1 are engaged in the B1–B4 pseudo-β-turn. This is different from the previously observed “open” state, i.e., O-state,52 or an I-state (intermediate state),13 and it is here termed the Ic-state (I-compact state). The compactness of the Ic-state is a consequence of the tethering of the B1 phenyl ring into a hydrophobic cavity lined by LeuB17, ValB2, ValB18, AlaB14, and LeuA16 (Figure 3A).
Figure 3.

(A) Conformation of the N-terminus of the B-chain of [d-ProB8]-insulin (backbone colored green) and [NMeAlaB8]-insulinII (backbone colored coral). The wild-type T-state insulin (PDB entry 1mso) backbone is colored white; all nitrogen atoms are colored blue and oxygen atoms red. Hydrogen bonds are shown as dashed lines (see the text for distances). Only the relevant side chains discussed in the text are shown. (B) Impact of B8 site substitutions on the hydrogen bond network in the B9–B19 helix in [d-ProB8]-insulin and [NMeAlaB8]-insulinII. The coloring scheme in analogues is the same as in panel A. The R-state insulin (PDB entry 1g7a) backbone is colored gold. The NMe group is marked with a red asterisk, and labels of positions of B-helix hydrogen bond donor and acceptor groups are colored black (see the text for distances).
The GlyB8 → d-Pro mutation did not significantly impact the conformation of the B8 site that is overall similar to that found in O- and I-state insulins. It is characterized by the occurrence of the B8GlyCO–HNB12Val hydrogen bond (3.1 Å) that is also present in the R-state, which is broken in the “typical” T-state insulins. Additionally, the second feature of this analogue is the preservation of the T-state typical B7CysCO–NHHisB10 hydrogen bond-like contact (∼3 Å, with imperfect helical hydrogen bond geometry), which switches to a B7CysCO–NHLeuB11 hydrogen bond pattern in the R-state B-helix (Figure 3B).
Crystal Structure of [NMeAlaB8]-Insulin
This analogue crystallized in two crystal forms. Crystal form I ([NMeAlaB8]-insulinI) was isomorphous with the [d-ProB8]-insulin; hence, the [NMeAlaB8]-insulinI structure is identical to that of the d-ProB8Gly mutant. The N-methylated amide in [NMeAlaB8]-insulinI mimics the N–Cδ bond of d-ProB8 (Figure 3B). Interestingly, this analogue yielded another crystal form (crystal form II, [NMeAlaB8]-insulinII) that contains two molecules in the asymmetric unit, which are engaged in two typical insulin dimers by crystallographic 2-fold symmetry. The dimeric nature of [NMeAlaB8]-insulinII may be surprising as it was crystallized under “typical” monomeric conditions.
The [NMeAlaB8]-insulinII structure follows the typical I-state insulin conformation in which the B1–B4 region moves even farther from the core of the insulin, resulting in an ∼16 Å distance between B1 Cα atoms in this analogue and in T-state insulin molecules (∼12.6 Å between B1 Cα atoms in the I- and Ic-states) (Figure 3A). Also, the Ic-like B1CO–B4 side-chain pseudo-β-turn is replaced by a typical I-state B3 side-chain–B5 main-chain pseudo-β-turn stabilized by AsnB3OD1–HNHisB5 (2.8 Å) and AsnB3OD1–ND1HisB5 (3.2 Å) hydrogen bonds. Further “opening” of the N-terminus of B1–B4 in [NMeAlaB8]-insulinII does not impact the other part of the N-terminus of B5–B8; hence, the conformation of the B8 site, and its surrounding regions (i.e., its hydrogen bond pattern), is identical in both [NMeAlaB8]-insulin crystal forms and [d-ProB8]-insulin. All ψ and φ angles for d-Pro-/NMeAla- and AibB8-substituted insulins occupy the −/+ quarter of the Ramachandran plot.
Structural Context of Crystal Packing
The crystallographic dimers in [d-ProB8]- and [NMeAlaB8]-insulinI form a trimer of dimers around the crystallographic 3-fold axis. This brings three B1 side chains within 3.8 Å of each other and locates them over a hydrophobic surface formed by these three dimers. From the relevant residues of the N-termini, only the OD1 atom of AsnB3 forms an intermolecular hydrogen bond to OE2 of GlnB13.
The crystal packing in [NMeAlaB8]-insulinII is different. PheB1 is in the proximity of the hydrophobic B19–A20 disulfide bond from a neighboring molecule, and its phenyl ring may also be involved in a π-cation interaction with the guanidinium group of symmetry-related ArgB22. The B-chain N-terminal ValB2NH forms a hydrogen bond with symmetry-related OE2GluA17, and OE1 of GlnB4 interacts also via crystal symmetry with ND2 of AsnA18.
Despite all these interactions, N-termini of [d-ProB8]- and [NMeAlaB8]-insulinI/II are not especially prohibited from other conformations as they are exposed to relatively open voids of the crystal space.
The [AibB8]-insulin and [AibB3]-insulin analogues have not been crystallized despite the use of a wide range of crystallization conditions.
Discussion
The significance of the T → R transition at the N-terminus of the B-chain of insulin, and the relevance of individual states, has been a subject of vigorous debate. This study of analogues with modifications at the N-terminus of the B-chain, designed to selectively stabilize the conformation of the T/R-related structural states, aimed to provide further insight into this rather complex issue. Here, we modified HI at three N-terminal positions: B3, B5, and B8. The reasons behind the selection of each of these positions were different. Site B8 is a crucial structural pivot for the T → R interconversion of the N-terminus of the B-chain. Sites B3 and B5 also seem to affect the T → R transition; however, their substitutions are not deleterious with respect to insulin–IR binding affinity.21,33,53−55 To probe the role of enforced B1–B8 R-state-like helix formation, α-aminoisobutyric acid (Aib)34,35 was incorporated at positions B3, B5, and B8. In contrast, the assurance of the T-state of insulin was intended by the introduction of a d-Pro or NMeAla amino acids at position B8, as they cannot adopt ψ and φ angles that are typical for the right-handed α-helices (Figure S1 of the Supporting Information).
Structural and Functional Features of Site B3
The lack of a detrimental effect of the deletion of the B1–B4 segment on insulin activity indicates that AsnB3 is not essential for IR binding.39 However, the loss of the AsnB3 side chain caused by its deamidation can affect the physicochemical properties of the hormone.56,57
[AibB3]-insulin retained nearly 30% of HI binding affinity, being the most potent analogue of the series reported here. The lack of a detrimental effect of the Asn → AibB3 replacement on the IR binding affinity is not surprising because of the general, relatively high tolerance for substitutions at the B3 site, which has been documented for [SerB3]-insulin (97% binding affinity), [ProB3]-insulin (54%),58 [AlaB3]-insulin (134%),59 and the [LysB3,GluB29]-insulin analogue (insulin glulisine) with a wild-type-like affinity33 (see Table S5 of the Supporting Information). Changes in the structure and reduced flexibility of [AibB3]-insulin were observed also in the CD spectra of this analogue, although the low yield of chain combination (only 1.7%) did not allow measurements of the near-UV CD spectra (245–300 nm), where the structural effect of phenol would be more evident. Also, the far-UV spectra (200–260 nm) showed a small phenol-related effect on the structure of [AibB3]-insulin, likely an indication of a decreased rate of the T → R transition (Figure 1B) and small differences in the helical content of [AibB3]-insulin in the absence or presence of phenol (Table S3 of the Supporting Information). Moreover, AsnB3 in HI is not involved in any significant interaction in the T/R hexamers. Therefore, the rigidity of the local main chain due to the presence of two methyl groups at B3 Cα atoms in [AibB3]-insulin may be responsible for its reduced affinity and decreased T → R transition ability as the one-methyl side-chain [AlaB3]-insulin analogue has an affinity of 134%.
Structural and Functional Features of Site B5
HisB5 is conserved among eutherian mammals and plays an important role in insulin aggregation, binding of phenolic ligands, folding, structure, and stability but, like the B3 site, does not seem to be crucial for the biological activity of this hormone.22,53−55 However, HisB5 seems to be co-responsible for the low binding affinity of insulin for IGF-1R.58 Some importance of HisB5 is also reflected in its ability to coordinate zinc ions,16,25 its contribution to the stability of the R6 insulin hexamers by bonding to phenol ligand,16 and the impact of insulin folding.
The hexamer-stabilizing role of HisB5 is highlighted by the retention of 40% of the affinity by a naturally occurring ArgB5 mutation (hystricomorph mammals, fish, and birds) that inhibits hexamer formation and the T → R transition.60,61 Many B5 site analogues, e.g., [AlaB5]-insulin,62,63 [ThrB5]-insulin,54 or [AlaB5]-DKP insulin55 (and [ArgB5]-insulin as well22), were obtained only in very low yields, and the synthesis of [MetB5]-insulin was unsuccessful.22 Such detrimental effects of the HisB5 substitutions on (pro)insulin self-assembly are also confirmed in vivo(55,64,65) by the naturally occurring HisB5Asp mutation (H29D), which causes permanent neonatal diabetes mellitus due to the disruption of proinsulin folding.64,65 Therefore, the infinitesimal yield (0.5%) of our [AibB5]-insulin analogue reflects this trend; the amount of this analogue was sufficient only for binding assays and did not allow structural studies.
Interestingly, the decrease in the number of methyl side-chain groups at the B5 site increases the analogue’s IR affinity from 4.4% in [AibB5]-insulin to 31% in [AlaB5]-insulin.59 It seems that the IR affinity of the B5 analogues is quite tolerant to mutations [affinities in the range of 20–50% (see Table S5 of the Supporting Information)], with an exception of acidic amino acids (e.g., [AspB5]-insulin with 0.4% binding affinity66). Thus, the low affinity of the [AibB5]-insulin may result mainly from the increased rigidity of the α-AibB5 main chain. The already reported structures of [ArgB5]- and [TyrB5]-insulin confirmed the importance of the B5 site for the T → R transition.22,67 [ArgB5]-insulin crystallized only in the T-state, confirming the blocked T → R transition for this naturally occurring insulin.22 In contrast, [TyrB5]-insulin formed T3Rf3 hexamer even without an excess of chloride ions or cyclic alcohols.67 Therefore, it seems that the replacement of HisB5 with an appropriate or specific amino acid can indeed significantly affect the conformation of the N-terminus of the B-chain.
Structural and Functional Features of Site B8
In contrast to the substitutions of sites B3 and B5, mutations of GlyB8 usually have a detrimental effect on IR binding. GlyB8 is evolutionarily strictly conserved in all insulins61 and in other members of the insulin superfamily, such as IGFs, relaxin, or bombyxin.68 Glycine has the largest allowed chiral angle space, reflected in its large area in the Ramachandran plot (Figure S1 of the Supporting Information). To narrow the B8 conformational spectrum, we prepared [AibB8]- and [AibB8,LysB28,ProB29]-insulin with the aim of “locking” them in the R-state. Subsequently, [d-ProB8]- and [NMeAlaB8]-insulin were prepared to yield the opposite, structurally “stabilized” T-state hormones. Interestingly, all these analogues exhibited <1% binding affinity for IR-A (Table 1), with the affinity of [NMeAlaB8]-insulin being below the detection limit of the measurements (see Figure S2C of the Supporting Information). We also determined similar low, or nondetectable, binding affinities of [AibB8]- and [NMeAlaB8]-insulin for IR-B and IGF-1R (Table S2 of the Supporting Information). These low binding affinities are in agreement with the weak affinities of all (except SerB8, with 23% of the affinity of HI) the B8 analogues published to date (Table S5 of the Supporting Information).
The crucial role of GlyB8 is also underlined by the two naturally occurring (pro)insulin gene mutations, GlyB8Ser (G32S) and GlyB8Arg (G32R), that cause permanent neonatal diabetes mellitus.64,65,71,72 Also, the in vitro data for the GlyB8Arg (G32R) mutant suggest that the negative effect of this mutation results from the disruptive disulfide pairing during proinsulin folding in the endoplasmic reticulum.64 Impaired folding of [AibB8,LysB28,ProB29]-insulin and [AibB8]-insulin was also observed in this work, although the yields were not as negligible as previously reported for l-amino acid B8 analogues (or the analogues’ precursors).29,32,59,69,70,73,74
The d-amino acids substitutions of site B8 had more deleterious effects on analogues’ affinities than substitutions with l-amino acids (Table S5 of the Supporting Information).29,32,69,70 However, d-amino acid substitutions at site B8 were usually associated with an increase in the efficiency of chain combination,29,32,69,74 also confirmed here by good synthetic yields of [d-ProB8]-insulin and [NMeAlaB8]-insulin analogues. While the [NMeAlaB8]-insulin analogue had a yield comparable to that of HI, the yield of the [d-ProB8]-insulin analogue was nearly 3-fold higher than the yield of native insulin. It indicates that the conformational space of the d-Pro φ angle at the B8 site has a positive impact on insulin folding. These high synthetic yields of the R-state “prohibited” ([d-ProB8]- and [NMeAlaB8]-insulin) analogues support previous hypotheses about the importance of the T-state (or T-like state) of insulin for an efficient folding of the hormone and its precursors29,74 and indicate T-like solution states of these analogues.
Higher yields of the B8 analogues allowed their extensive structural characterization by CD, NMR, and X-ray crystallography. The solution properties of the B1–B8 N-termini of these analogues, especially their response and interactions with R-state-stabilizing phenol, were monitored by CD spectroscopy. The far- and near-UV CD spectra of [AibB8]-insulin showed a small effect of addition of phenol compared to those of HI (Figure 1C and Figure S4B of the Supporting Information), and [NMeAlaB8]-insulin responded with a negligible change in the CD spectra to the presence of this ligand (Figure 1E and Figure S4D of the Supporting Information). The structural resilience of [NMeAlaB8]-insulin, its locked T-state and lack of any R-like structure in the presence of phenol, likely resulted from the inability of this analogue to contribute to the α-helix hydrogen bond pattern due to replacement of the B8 amide hydrogen with a methyl group.
Surprisingly, the CD spectra of the T-state locked [d-ProB8]-insulin showed some phenol-induced changes (Figure 1D and Figure S4C of the Supporting Information). However, it is possible that these spectral variations may result from certain subtle changes in local interactions or structural arrangements in this analogue, and not from spectral effects accompanying typical T → R conformational changes. However, their exact nature cannot be explained by its crystal structure.
Although crystallization screening is not an ultimate proof of a protein’s solution behavior, it is symptomatic that [d-ProB8]- and [NMeAlaB8]-insulin crystallized only in the Ic- and I-states, even despite the presence of phenol and zinc in some crystallization media. This, together with the superior folding abilities of these analogues, indicates their strong preference for the T-state-like conformation. Moreover, [d-ProB8]- and [NMeAlaB8]-insulin also showed a predisposition to dimerization as all their crystal structures are exclusively dimeric. Interestingly, all differently packed [d-ProB8]- and [NMeAlaB8]-insulinII crystal structures, and the solution structure of [AibB8,LysB28,ProB29]-insulin, adopted a range of I-state-like conformations. The variations of these I-state-type conformers (intermediates between the T- and R-states) have already been observed in crystal structures of the highly active insulin analogues,13 [GluB9]-insulin,52 despenta-75 and deshepta-insulins,76 and an extensively engineered insulin monomer.77 The Ic-state of [d-ProB8]- and [NMeAlaB8]-insulinI resembles the T-like state, but with B1–B8 packed more closely against the insulin core, i.e., toward a conformation observed in [Cys(B4–B10),desB30]-insulin,31 in which B1–B8 formed a tight β-sheet with the adjacent A-chain. However, the N-terminal β-sheet-like fold in these B8 site analogues is loosened up because of the lack of a B1CO–HNA13 hydrogen bond.
Although the T-like conformations of the N-termini in [d-ProB8]- and [NMeAlaB8]-insulin were expected, the T-like state in the NMR structure of [AibB8,LysB28,ProB29]-insulin was surprising. The presence of the helix-inducing Aib amino acid at site B8 was not sufficient to force B1–B8 into R-like, or Rf-like, states. Interestingly, all N-termini in [d-ProB8]-, [NMeAlaB8]-, and [AibB8,LysB28,ProB29]-insulin (and some other T-like-state analogues) feature a B7CO–NHB10 hydrogen bond at the beginning of the B9–B19 helix (Figure 3). In the R-state, the register of this hydrogen bond must change into a B7CO–NHB11 contact to follow the proper B1(B3)–B19 α-helix hydrogen bond pattern. Interestingly, this R-state NHB10 → NHB11 intrahelical shift of a hydrogen bond acceptor combines B1(B3)–B19 helix formation with the observed isomerization of the B7–A7, and closely associated A6–A11, disulfide bonds. The isomerization of B7–A7 and A6–A11 disulfide bridges is frequently observed in T → R transitions but cannot be easily quantified because of different crystal packing, mutations, allosteric ligands, etc., found in these structures. However, it is plausible that these two structural rearrangements, a switch of the NHB10 → NHB11 B-helix hydrogen bond pattern and combined B7–A7 and A6–A11 disulfide isomerization, present substantial T-state conformational locks. Hence, the conformational freedom of the glycine at site B8 not only is required for efficient insulin folding but also provides minimal steric hindrance and maximal chiral flexibility, if these site B8-associated conformational changes are indeed involved in effective insulin–IR complex formation.
The higher (130%) affinity of [Cys(B4–B10),desB30]-insulin,31 with an N-terminal β-sheet that follows the “tight” T-state-like fold restrained by an additional SS link, is not surprising in the context of this study and previous studies,13 and the emergence of a wide spectrum of active and inactive T-like (e.g., I to Ic) states. Moreover, an additional B4–B10 disulfide bond in [Cys(B4–B10),desB30]-insulin does not exclude the possibility of a modification of this structure toward, for example, I-like states. This could be facilitated, for example, by B4–B10 disulfide bond isomerization, synchronized with similar structural responses in B7–A7 and A6–A11 disulfides. Interestingly, the active, compact conformation of [Cys(B4–B10),desB30]-insulin is observed only in its hexamer crystal structure; therefore, its alternative conformation may occur in monomeric and dimeric forms of this analogue.
As the R- and Rf-states were never observed in insulin monomers and dimers, it seems that the full T → R/Rf transition requires substantial cooperative, entropic contributions from zinc coordination, including various ligands (e.g., halogens, SCN–, phenols, etc.) and cannot be achieved by a single, helix-preferring, even non-natural, amino acid substitution. However, B1–B8 structural rearrangements toward the R-state involving a significant chaperone-like contribution from IR cannot be excluded.
In conclusion, this study (i) provided further proof of the importance of the T-like state for the folding efficiency of (pro)insulin, (ii) underlined the incompatibility of the R-state with an active form of insulin, (iii) indicated the complex character of the T → R transition that cannot be induced or stabilized by a single substitution at site B8, and (iv) provided structural evidence of the ability of the B1–B8 segment to form a variety of structurally T- or I-like states, correlated with low hormone affinities. Moreover, this study revealed that the substantial flexibility of the B1–B8 segment, especially site B8, is necessary for the effective binding to IR. Undoubtedly, further cocrystallization and structural studies of insulin with site 2-containing IR constructs will be necessary to shed light on the insulin B-chain N-terminal “active” conformation and to delineate the physiological roles of individual components of the B1–B8 segment.
Acknowledgments
We thank Prof. Axel Wollmer for helpful discussions and advice in planning the CD experiments, Dr. John P. Mayer for useful advice about the recombination of insulin chains, and Miroslava Blechová (Institute of Organic Chemistry and Biochemistry, Academy of Sciences of the Czech Republic) for the synthesis of peptide chains.
Glossary
Abbreviations
- DIPEA
N,N-diisopropylethylamine
- DQF-COSY
double-quantum-filtered correlation spectroscopy
- EDT
1,2-ethanedithiol
- HEPES
N-(2-hydroxyethyl)piperazine-N′-2-ethanesulfonic acid
- HI
human insulin
- IR
insulin receptor
- NOESY
nuclear Overhauser effect spectroscopy
- TIS
triisopropylsilane
- TOCSY
total correlation spectroscopy.
Supporting Information Available
Ramachandran plots for selected amino acids (Figure S1), data collection and refinement statistics for crystal structures of insulin analogues (Table S1), IR-A assay binding curves for human insulin and analogues (Figure S2), methodology for the determination of binding affinities for IR-B and IGF-1R, binding curves of analogues for IR-B and IGF-1R (Figure S3), values of Kd of analogues for IR-B and IGF-R (Table S2), near-UV (245–305 nm) CD spectra for insulin and analogues (Figure S4), secondary structure content of HI or insulin analogues with or without phenol calculated from CD spectra (Table S3), NMR constraints and structural statistics for the NMR structure of [AibB8,LysB28,ProB29]-insulin (Table S4), literature values of insulin receptor binding affinities of previously published insulin analogues (Table S5), and supplemental references. This material is available free of charge via the Internet at http://pubs.acs.org.
This work was supported by Grant Agency of the Czech Republic Grant P207/11/P430 (to L.Ž.) and Grant P208/11/0105 (to M.U.), the Ministry of Education of the Czech Republic (Program “NAVRAT” LK11205, to V.V.), the Medical Research Council (Grant MR/K000179/1, to A.M.B.), the Specific University Research (MSMT No. 20/2013, A1_FCHI_2014_003), and Research Project RVO:61388963 of the Academy of Sciences of the Czech Republic (to the Institute of Organic Chemistry and Biochemistry, Academy of Sciences of the Czech Republic).
The authors declare no competing financial interest.
Supplementary Material
References
- Steiner D. F.; Chan S. J.; Welsh J. M.; Kwok S. C. (1985) Structure and evolution of the insulin gene. Annu. Rev. Genet. 19, 463–484. [DOI] [PubMed] [Google Scholar]
- Baker E. N.; Blundell T. L.; Cutfield J. F.; Cutfield S. M.; Dodson E. J.; Dodson G. G.; Hodgkin D. M.; Hubbard R. E.; Isaacs N. W.; Reynolds C. D. (1988) The structure of 2Zn pig insulin crystals at 1.5 Å resolution. Philos. Trans. R. Soc. London, Ser. B 319, 369–456. [DOI] [PubMed] [Google Scholar]
- Mirmira R. G.; Nakagawa S. H.; Tager H. S. (1991) Importance of the character and configuration of residues B24, B25, and B26 in insulin-receptor interactions. J. Biol. Chem. 266, 1428–1436. [PubMed] [Google Scholar]
- Nakagawa S. H.; Tager H. S. (1987) Role of the COOH-terminal B-chain domain in insulin-receptor interactions. Identification of perturbations involving the insulin mainchain. J. Biol. Chem. 262, 12054–12058. [PubMed] [Google Scholar]
- Zakova L.; Kazdova L.; Hanclova I.; Protivinska E.; Sanda M.; Budesinsky M.; Jiracek J. (2008) Insulin analogues with modifications at position B26. Divergence of binding affinity and biological activity. Biochemistry 47, 5858–5868. [DOI] [PubMed] [Google Scholar]
- Zakova L.; Barth T.; Jiracek J.; Barthova J.; Zorad S. (2004) Shortened insulin analogues: Marked changes in biological activity resulting from replacement of TyrB26 and N-methylation of peptide bonds in the C-terminus of the B-chain. Biochemistry 43, 2323–2331. [DOI] [PubMed] [Google Scholar]
- Antolikova E.; Zakova L.; Turkenburg J. P.; Watson C. J.; Hanclova I.; Sanda M.; Cooper A.; Kraus T.; Brzozowski A. M.; Jiracek J. (2011) Non-equivalent Role of Inter- and Intramolecular Hydrogen Bonds in the Insulin Dimer Interface. J. Biol. Chem. 286, 36968–36977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Meyts P.; Whittaker J. (2002) Structural biology of insulin and IGF1 receptors: Implications for drug design. Nat. Rev. Drug Discovery 1, 769–783. [DOI] [PubMed] [Google Scholar]
- Pullen R. A.; Lindsay D. G.; Wood S. P.; Tickle I. J.; Blundell T. L.; Wollmer A.; Krail G.; Brandenburg D.; Zahn H.; Gliemann J.; Gammeltoft S. (1976) Receptor-binding region of insulin. Nature 259, 369–373. [DOI] [PubMed] [Google Scholar]
- Menting J. G.; Whittaker J.; Margetts M. B.; Whittaker L. J.; Kong G. K. W.; Smith B. J.; Watson C. J.; Zakova L.; Kletvikova E.; Jiracek J.; Chan S. J.; Steiner D. F.; Dodson G. G.; Brzozowski A. M.; Weiss M. A.; Ward C. W.; Lawrence M. C. (2013) How insulin engages its primary binding site on the insulin receptor. Nature 493, 241–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu B.; Huang K.; Chu Y. C.; Hu S. Q.; Nakagawa S.; Wang S. H.; Wang R. Y.; Whittaker J.; Katsoyannis P. G.; Weiss M. A. (2009) Decoding the Cryptic Active Conformation of a Protein by Synthetic Photoscanning. J. Biol. Chem. 284, 14597–14608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodson E. J.; Dodson G. G.; Hubbard R. E.; Reynolds C. D. (1983) Insulin’s structural behavior and its relation to activity. Biopolymers 22, 281–291. [DOI] [PubMed] [Google Scholar]
- Jiracek J.; Zakova L.; Antolikova E.; Watson C. J.; Turkenburg J. P.; Dodson G. G.; Brzozowski A. M. (2010) Implications for the active form of human insulin based on the structural convergence of highly active hormone analogues. Proc. Natl. Acad. Sci. U.S.A. 107, 1966–1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakova L.; Kletvikova E.; Veverka V.; Lepsik M.; Watson C. J.; Turkenburg J. P.; Jiracek J.; Brzozowski A. M. (2013) Structural integrity of the B24 site in human insulin is important for hormone functionality. J. Biol. Chem. 288, 10230–10240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Meyts P.; Van Obberghen E.; Roth J. (1978) Mapping of the residues responsible for the negative cooperativity of the receptor-binding region of insulin. Nature 273, 504–509. [DOI] [PubMed] [Google Scholar]
- Derewenda U.; Derewenda Z.; Dodson E. J.; Dodson G. G.; Reynolds C. D.; Smith G. D.; Sparks C.; Swenson D. (1989) Phenol stabilizes more helix in a new symmetrical zinc insulin hexamer. Nature 338, 594–596. [DOI] [PubMed] [Google Scholar]
- Kaarsholm N. C.; Ko H. C.; Dunn M. F. (1989) Comparison of solution structural flexibility and zinc binding domains for insulin, proinsulin, and miniproinsulin. Biochemistry 28, 4427–4435. [DOI] [PubMed] [Google Scholar]
- Smith G. D.; Dodson G. G. (1992) The Structure of a Rhombohedral R6 Insulin Hexamer That Binds Phenol. Biopolymers 32, 441–445. [DOI] [PubMed] [Google Scholar]
- Choi W. E.; Borchardt D.; Kaarsholm N. C.; Brzovic P. S.; Dunn M. F. (1996) Spectroscopic evidence for preexisting T- and R-state insulin hexamer conformations. Proteins: Struct., Funct., Genet. 26, 377–390. [DOI] [PubMed] [Google Scholar]
- Roy M.; Brader M. L.; Lee R. W.; Kaarsholm N. C.; Hansen J. F.; Dunn M. F. (1989) Spectroscopic signatures of the T to R conformational transition in the insulin hexamer. J. Biol. Chem. 264, 19081–19085. [PubMed] [Google Scholar]
- Shneine J.; Voswinkel M.; Federwisch M.; Wollmer A. (2000) Enhancing the T → R transition of insulin by helix-promoting sequence modifications at the N-terminal B-chain. Biol. Chem. 381, 127–133. [DOI] [PubMed] [Google Scholar]
- Wan Z. L.; Huang K.; Hu S. Q.; Whittaker J.; Weiss M. A. (2008) The structure of a mutant insulin uncouples receptor binding from protein allostery: An electrostatic block to the TR transition. J. Biol. Chem. 283, 21198–21210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley G.; Dodson E.; Dodson G.; Hodgkin D.; Mercola D. (1976) Structure of insulin in 4-zinc insulin. Nature 261, 166–168. [DOI] [PubMed] [Google Scholar]
- Ciszak E.; Smith G. D. (1994) Crystallographic evidence for dual coordination around zinc in the T3R3 human insulin hexamer. Biochemistry 33, 1512–1517. [DOI] [PubMed] [Google Scholar]
- Jacoby E.; Kruger P.; Karatas Y.; Wollmer A. (1993) Distinction of Structural Reorganization and Ligand-Binding in the T→ R Transition of Insulin on the Basis of Allosteric Models. Biol. Chem. Hoppe-Seyler 374, 877–885. [DOI] [PubMed] [Google Scholar]
- Brzovic P. S.; Choi W. E.; Borchardt D.; Kaarsholm N. C.; Dunn M. F. (1994) Structural asymmetry and half-site reactivity in the T to R allosteric transition of the insulin hexamer. Biochemistry 33, 13057–13069. [DOI] [PubMed] [Google Scholar]
- Smith G. D.; Dodson G. G. (1992) Structure of a Rhombohedral R6 Insulin Phenol Complex. Proteins: Struct., Funct., Genet. 14, 401–408. [DOI] [PubMed] [Google Scholar]
- Wollmer A.; Rannefeld B.; Johansen B. R.; Hejnaes K. R.; Balschmidt P.; Hansen F. B. (1987) Phenol-Promoted Structural Transformation of Insulin in Solution. Biol. Chem. Hoppe-Seyler 368, 903–911. [DOI] [PubMed] [Google Scholar]
- Nakagawa S. H.; Zhao M.; Hua Q. X.; Hu S. Q.; Wan Z. L.; Jia W.; Weiss M. A. (2005) Chiral mutagenesis of insulin. Foldability and function are inversely regulated by a stereospecific switch in the B chain. Biochemistry 44, 4984–4999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss M. A. (2009) The Structure and Function of Insulin: Decoding the TR Transition. In Insulin and IGFs (Litwack G., Ed.) pp 33–49, Academic Press, New York. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinther T. N.; Norrman M.; Ribel U.; Huus K.; Schlein M.; Steensgaard D. B.; Pedersen T. A.; Pettersson I.; Ludvigsen S.; Kjeldsen T.; Jensen K. J.; Hubalek F. (2013) Insulin analog with additional disulfide bond has increased stability and preserved activity. Protein Sci. 22, 296–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua Q. X.; Nakagawa S.; Hu S. Q.; Jia W.; Wang S.; Weiss M. A. (2006) Toward the active conformation of insulin: Stereospecific modulation of a structural switch in the B chain. J. Biol. Chem. 281, 24900–24909. [DOI] [PubMed] [Google Scholar]
- Becker R. H. A. (2007) Insulin glulisine complementing basal insulins: A review of structure and activity. Diabetes Technol. Ther. 9, 109–121. [DOI] [PubMed] [Google Scholar]
- Marshall G. R.; Hodgkin E. E.; Langs D. A.; Smith G. D.; Zabrocki J.; Leplawy M. T. (1990) Factors Governing Helical Preference of Peptides Containing Multiple α,α-Dialkyl Amino-Acids. Proc. Natl. Acad. Sci. U.S.A. 87, 487–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karle I. L.; Balaram P. (1990) Structural Characteristics of α-Helical Peptide Molecules Containing Aib Residues. Biochemistry 29, 6747–6756. [DOI] [PubMed] [Google Scholar]
- Ciszak E.; Beals J. M.; Frank B. H.; Baker J. C.; Carter N. D.; Smith G. D. (1995) Role of C-terminal B-chain residues in insulin assembly: The structure of hexameric LysB28ProB29-human insulin. Structure 3, 615–622. [DOI] [PubMed] [Google Scholar]
- Wang S. H.; Hu S. Q.; Burke G. T.; Katsoyannis P. G. (1991) Insulin analogues with modifications in the β-turn of the B-chain. J. Protein Chem. 10, 313–324. [DOI] [PubMed] [Google Scholar]
- Chance R. E., Hoffmann J. A., Kroeff E. P., Johnson M. G., Schirmer E. W., and Bromer W. W. (1981) The production of human insulin using recombinant DNA technology and a new chain combination procedure. In Proceedings of the 7th American Peptide Symposium (Rich D. H., and Gross E., Eds.) pp 721–728, Pierce Chemical Co., Rockford, IL. [Google Scholar]
- Morcavallo A.; Genua M.; Palummo A.; Kletvikova E.; Jiracek J.; Brzozowski A. M.; Iozzo R. V.; Belfiore A.; Morrione A. (2012) Insulin and Insulin-like Growth Factor II Differentially Regulate Endocytic Sorting and Stability of Insulin Receptor Isoform A. J. Biol. Chem. 287, 11422–11436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swillens S. (1995) Interpretation of Binding Curves Obtained with High Receptor Concentrations: Practical Aid for Computer-Analysis. Mol. Pharmacol. 47, 1197–1203. [PubMed] [Google Scholar]
- Herrmann T.; Guntert P.; Wuthrich K. (2002) Protein NMR structure determination with automated NOE assignment using the new software CANDID and the torsion angle dynamics algorithm DYANA. J. Mol. Biol. 319, 209–227. [DOI] [PubMed] [Google Scholar]
- Guntert P.; Wuthrich K. (1991) Improved efficiency of protein structure calculations from NMR data using the program DIANA with redundant dihedral angle constraints. J. Biomol. NMR 1, 447–456. [DOI] [PubMed] [Google Scholar]
- Harjes E.; Harjes S.; Wohlgemuth S.; Muller K. H.; Krieger E.; Herrmann C.; Bayer P. (2006) GTP-Ras disrupts the intramolecular complex of C1 and RA domains of Nore1. Structure 14, 881–888. [DOI] [PubMed] [Google Scholar]
- Koradi R.; Billeter M.; Wuthrich K. (1996) MOLMOL: A program for display and analysis of macromolecular structures. J. Mol. Graphics 14, 51–55. [DOI] [PubMed] [Google Scholar]
- Doreleijers J. F.; da Silva A. W. S.; Krieger E.; Nabuurs S. B.; Spronk C. A. E. M.; Stevens T. J.; Vranken W. F.; Vriend G.; Vuister G. W. (2012) CING: An integrated residue-based structure validation program suite. J. Biomol. NMR 54, 267–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter G. (2010) xia2: An expert system for macromolecular crystallography data reduction. J. Appl. Crystallogr. 43, 186–190. [Google Scholar]
- Bailey S. (1994) The Ccp4 Suite: Programs for Protein Crystallography. Acta Crystallogr. D50, 760–763. [DOI] [PubMed] [Google Scholar]
- Emsley P.; Cowtan K. (2004) Coot: Model-building tools for molecular graphics. Acta Crystallogr. D60, 2126–2132. [DOI] [PubMed] [Google Scholar]
- Vagin A.; Teplyakov A. (1997) MOLREP: An automated program for molecular replacement. J. Appl. Crystallogr. 30, 1022–1025. [Google Scholar]
- Smith G. D.; Pangborn W. A.; Blessing R. H. (2003) The structure of T-6 human insulin at 1.0 Å resolution. Acta Crystallogr. D59, 474–482. [DOI] [PubMed] [Google Scholar]
- Murshudov G. N.; Vagin A. A.; Dodson E. J. (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D53, 240–255. [DOI] [PubMed] [Google Scholar]
- Yao Z. P.; Zeng Z. H.; Li H. M.; Zhang Y.; Feng Y. M.; Wang D. C. (1999) Structure of an insulin dimer in an orthorhombic crystal: The structure analysis of a human insulin mutant (B9 Ser → Glu). Acta Crystallogr. D55, 1524–1532. [DOI] [PubMed] [Google Scholar]
- Nakagawa S. H.; Tager H. S. (1991) Implications of invariant residue LeuB6 in insulin-receptor interactions. J. Biol. Chem. 266, 11502–11509. [PubMed] [Google Scholar]
- Sohma Y.; Hua Q. X.; Liu M.; Phillips N. B.; Hu S. Q.; Whittaker J.; Whittaker L. J.; Ng A.; Roberts C. T.; Arvan P.; Kent S. B. H.; Weiss M. A. (2010) Contribution of Residue B5 to the Folding and Function of Insulin and IGF-I. J. Biol. Chem. 285, 5040–5055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua Q. X.; Liu M.; Hu S. Q.; Jia W. H.; Arvan P.; Weiss M. A. (2006) A conserved histidine in insulin is required for the foldability of human proinsulin: Structure and function of an Ala(B5) analog. J. Biol. Chem. 281, 24889–24899. [DOI] [PubMed] [Google Scholar]
- Brange J.; Langkjaer L.; Havelund S.; Volund A. (1992) Chemical-Stability of Insulin. 1. Hydrolytic Degradation during Storage of Pharmaceutical Preparations. Pharmacol. Res. 9, 715–726. [DOI] [PubMed] [Google Scholar]
- Huus K.; Havelund S.; Olsen H. B.; de Weert M. V.; Frokjaer S. (2006) Chemical and thermal stability of insulin: Effects of zinc and ligand binding to the insulin zinc-hexamer. Pharmacol. Res. 23, 2611–2620. [DOI] [PubMed] [Google Scholar]
- Gauguin L.; Klaproth B.; Sajid W.; Andersen A. S.; McNeil K. A.; Forbes B. E.; De Meyts P. (2008) Structural basis for the lower affinity of the insulin-like growth factors for the insulin receptor. J. Biol. Chem. 283, 2604–2613. [DOI] [PubMed] [Google Scholar]
- Kristensen C.; Kjeldsen T.; Wiberg F. C.; Schaffer L.; Hach M.; Havelund S.; Bass J.; Steiner D. F.; Andersen A. S. (1997) Alanine scanning mutagenesis of insulin. J. Biol. Chem. 272, 12978–12983. [DOI] [PubMed] [Google Scholar]
- Bajaj M.; Blundell T. L.; Horuk R.; Pitts J. E.; Wood S. P.; Gowan L. K.; Schwabe C.; Wollmer A.; Gliemann J.; Gammeltoft S. (1986) Coypu Insulin: Primary Structure, Conformation and Biological Properties of a Hystricomorph Rodent Insulin. Biochem. J. 238, 345–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conlon J. M. (2001) Evolution of the insulin molecule: Insights into structure-activity and phylogenetic relationships. Peptides 22, 1183–1193. [DOI] [PubMed] [Google Scholar]
- Marki F.; de Gasparo M.; Eisler K.; Kamber B.; Riniker B.; Rittel W.; Sieber P. (1979) Synthesis and biological activity of seventeen analogues of human insulin. Hoppe-Seyler’s Z. Physiol. Chem. 360, 1619–1632. [DOI] [PubMed] [Google Scholar]
- Cao Q. P.; Geiger R.; Langner D.; Geisen K. (1986) Biological-Activity In vivo of Insulin Analogs Modified in the N-Terminal Region of the B-Chain. Biol. Chem. Hoppe-Seyler 367, 135–140. [DOI] [PubMed] [Google Scholar]
- Liu M.; Haataja L.; Wright J.; Wickramasinghe N. P.; Hua Q. X.; Phillips N. F.; Barbetti F.; Weiss M. A.; Arvan P. (2010) Mutant INS-Gene Induced Diabetes of Youth: Proinsulin Cysteine Residues Impose Dominant-Negative Inhibition on Wild-Type Proinsulin Transport. PLoS One 5, e13333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edghill E. L.; Flanagan S. E.; Patch A. M.; Boustred C.; Parrish A.; Shields B.; Shepherd M. H.; Hussain K.; Kapoor R. R.; Malecki M.; MacDonald M. J.; Stoy J.; Steiner D. F.; Philipson L. H.; Bell G. I.; Hattersley A. T.; Ellard S.; Collaborative N. D. I. (2008) Insulin mutation screening in 1,044 patients with diabetes: Mutations in the INS gene are a common cause of neonatal diabetes but a rare cause of diabetes diagnosed in childhood or adulthood. Diabetes 57, 1034–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke G. T.; Hu S. Q.; Ohta N.; Schwartz G. P.; Zong L.; Katsoyannis P. G. (1990) Superactive insulins. Biochem. Biophys. Res. Commun. 173, 982–987. [DOI] [PubMed] [Google Scholar]
- Tang L.; Whittingham J. L.; Verma C. S.; Caves L. S.; Dodson G. G. (1999) Structural consequences of the B5 histidine → tyrosine mutation in human insulin characterized by X-ray crystallography and conformational analysis. Biochemistry 38, 12041–12051. [DOI] [PubMed] [Google Scholar]
- Chen H.; Feng Y. M. (2001) Contribution of the residue Glu9, Glu46, and Phe49 to the biological activity of insulin-like growth factor-1. IUBMB Life 51, 33–37. [DOI] [PubMed] [Google Scholar]
- Nakagawa S. H., Zhao M., Hua Q. X., and Weiss M. A.. The importance of residue B8 in insulin activity, structure and folding. In Proceedings of the 15th American Peptide Symposium (Tam J. P., and Kaumaya P. T. P., Eds.) pp 471–472, Pierce Chemical Co., Rockford, IL. [Google Scholar]
- Guo Z. Y.; Zhang Z.; Jia X. Y.; Tang Y. H.; Feng Y. M. (2005) Mutational analysis of the ahsolutely conserved B8Gly: Consequence on foldability and activity of insulin. Acta Biochim. Biophys. Sin. 37, 673–679. [DOI] [PubMed] [Google Scholar]
- Stoy J.; Steiner D. F.; Park S. Y.; Ye H. G.; Philipson L. H.; Bell G. I. (2010) Clinical and molecular genetics of neonatal diabetes due to mutations in the insulin gene. Rev. Endocr. Metab. Disord. 11, 205–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoy J.; Edghill E. L.; Flanagan S. E.; Ye H. G.; Paz V. P.; Pluzhnikov A.; Below J. E.; Hayes M. G.; Cox N. J.; Lipkind G. M.; Lipton R. B.; Greeley S. A. W.; Patch A. M.; Ellard S.; Steiner D. F.; Hattersley A. T.; Philipson L. H.; Bell G. I. (2007) Insulin gene mutations as a cause of permanent neonatal diabetes. Proc. Natl. Acad. Sci. U.S.A. 104, 15040–15044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markussen J.; Diers I.; Engesgaard A.; Hansen M. T.; Hougaard P.; Langkjaer L.; Norris K.; Ribel U.; Sorensen A. R.; Sorensen E. (1987) Soluble, prolonged-acting insulin derivatives. II. Degree of protraction and crystallizability of insulins substituted in positions A17, B8, B13, B27 and B30. Protein Eng. 1, 215–223. [DOI] [PubMed] [Google Scholar]
- Zhao M., Nakagawa S. H., Hua Q. X., and Weiss M. A. (1997) Exploring the foldability and function of insulin by combinatorial peptide chemistry. In Proceedings of the 15th American Peptide Symposium (Tam J. P., and Kaumaya P. T. P., Eds.) pp 369–371, Pierce Chemical Co., Rockford, IL. [Google Scholar]
- Brange J. (1997) The new era of biotech insulin analogues. Diabetologia 40, S48–S53. [DOI] [PubMed] [Google Scholar]
- Bao S. J.; Xie D. L.; Zhang J. P.; Chang W. R.; Liang D. C. (1997) Crystal structure of desheptapeptide(B24-B30)insulin at 1.6 Å resolution: Implications for receptor binding. Proc. Natl. Acad. Sci. U.S.A. 94, 2975–2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen H. B.; Ludvigsen S.; Kaarsholm N. C. (1996) Solution structure of an engineered insulin monomer at neutral pH. Biochemistry 35, 8836–8845. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.