Abstract
The tumor suppressor gene p53 regulates apoptosis in response to DNA damage. Promoter selectivity of p53 depends on mainly its phosphorylation. Particularly, the phosphorylation at serine-46 of p53 is indispensable in promoting pro-apoptotic genes that are, however, poorly determined. In the current study, we identified palmdelphin as a pro-apoptotic gene induced by p53 in a phosphorylated serine-46-specific manner. Upregulation of palmdelphin was observed in wild-type p53-transfected cells, but not in serine-46-mutated cells. Expression of palmdelphin was induced by p53 in response to DNA damage. In turn, palmdelphin induced apoptosis. Intriguingly, downregulation of palmdelphin resulted in necroptosis-like cell death via ATP depletion. Upon DNA damage, palmdelphin dominantly accumulated in the nucleus to induce apoptosis. These findings define palmdelphin as a target of serine-46-phosphorylated p53 that controls cell death in response to DNA damage.
Keywords: palmdelphin, p53, DNA damage, apoptosis
Mutations in the tumor suppressor gene p53 are frequently identified in human cancers. p53 functions in the cellular responses to the variety of stresses such as radiation, oxidative stress, and DNA-damaging agents by inducing cell cycle arrest, DNA repair, or apoptosis.1, 2, 3, 4, 5, 6 Various functions of p53 are regulated by its post-translational modifications.7 The apoptotic function of p53 requires phosphorylation at serine-46 (ser46).6, 8 Available evidence suggested that protein kinases, HIPK2,4, 5, 9, 10 ATM kinase,11 and DYRK24 phosphorylate p53 at ser46 under certain conditions. For instance, HIPK2 phosphorylates ser46 in response to UV exposure and DNA single-strand break to induce apoptosis.4, 10 Likewise, in response to a DNA double-strand break, DYRK2 targets p53 to phosphorylate ser46 to induce pro-apoptotic genes.4 At present, many pro-apoptotic genes induced by p53 are identified while the downstream targets of the ser46 phosphorylation are very limited with only p53AIP1.8 In order to elucidate the molecular mechanisms underlying apoptosis, it is important to uncover which proteins are regulated by p53 when ser46 is phosphorylated.
p53 is also involved in the necrotic cell death.12 Necrosis was long considered as a non-programmed form of cell death, whereas recent evidence suggested programmed necrosis called necroptosis. Necroptosis is the result of interplay among several signaling cascades. In a transcription-independent manner, p53 forms complexes with PTP regulator cyclophilin D (CypD) in the mitochondria and triggers mitochondrial permeability transition pore opening that leads to necrosis.12 However, a transcription-dependent pathway of the p53-dependent necrosis remains unclear.
Abnormal genetic alterations in palmdelphin (PALMD) are observed in the malignant tumors.13, 14 PALMD is an isoform of the paralemmin families that are lipid-anchored proteins having a role in the cell shape control and cell dynamics. PALMD is harbored on the human chromosomal region at 1p22-p21 and within this region, human disease genes such as atrioventricular canal defect 1, venous malformations with glomus cells, autosomal dominant osteopetrosis type II, and Waardenburg syndrome type 2B have been mapped.15, 16 The molecular and functional nature of PALMD is obscure.
Current study demonstrates PALMD as a target of phosphorylated p53 at ser46. PALMD is targeted to the nucleus to induce apoptosis in response to DNA damage.
Results
p53 induces PALMD expression when ser46 is phosphorylated
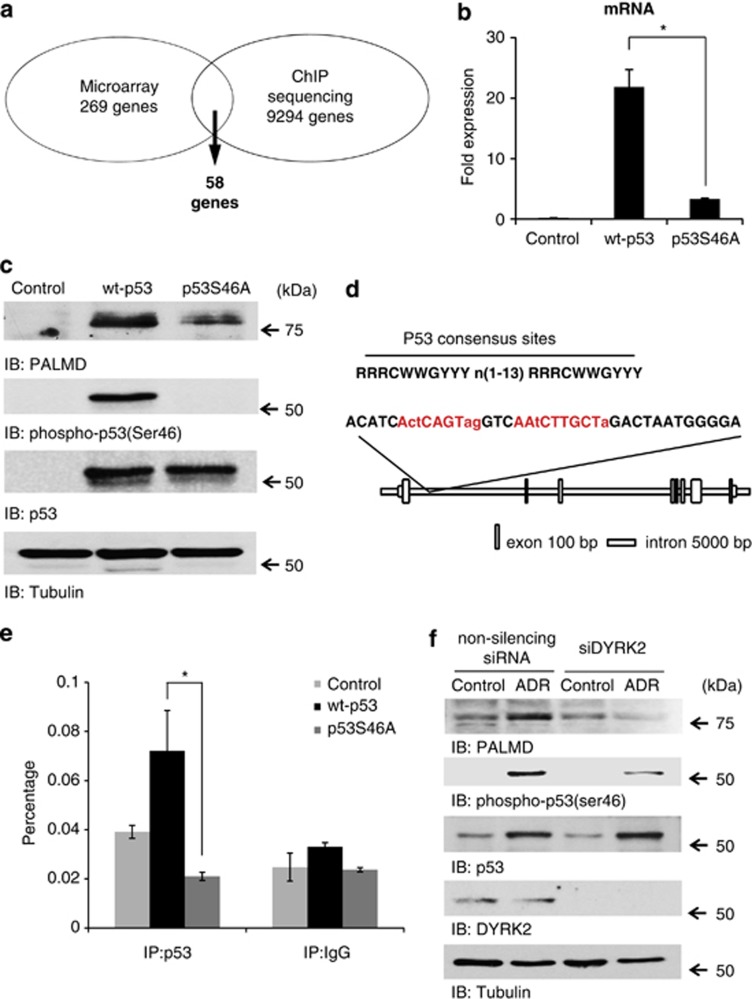
To investigate target genes of the ser46 phosphorylation, microarray and chromatin immunoprecipitation (ChIP) sequencing were performed. The results of microarray revealed 269 genes that are specific to the ser46 phosphorylation of p53 (Figure 1a). In the ChIP sequencing, wild-type p53 (wt-p53) bound onto 27 694 regions of 9294 genes (Figure 1a). Comparison of two assays showed 58 genes as candidates of p53-target genes when ser46 is phosphorylated (Figure 1a). From them, PALMD showed the highest specificity to wt-p53 as well as to phospho-ser46 (Table 1). mRNA expression of PALMD was elucidated by real-time PCR (Figure 1b), and protein expression level was determined by western blotting (Figure 1c). The expression of PALMD was higher in the wt-p53-transfected fraction (Figures 1b and c). In addition, the induction of PALMD expression in ser15-mutated p53 (p53S15A)-transfected cells was similar to that in wt-p53-transfected cells; however, it was decreased in p53S46A-transfected cells (Supplementary Figure 1a). To clarify PALMD as a p53-target gene when ser46 is phosphorylated, the ChIP assay was performed. The p53 consensus site was included in the highest peak region of ChIP sequencing (Figure 1d) and this region was amplified for the ChIP assay. As expected, only wt-p53 had an ability to bind onto PALMD but neither control nor p53S46A (Figure 1e). Given that ser46 is phosphorylated by DYRK2 after genotoxic stimuli,4 PALMD expression in DYRK2-silenced cells was examined. Importantly, DYRK2-depleted cells failed to induce PALMD expression in response to DNA damage (Figure 1f). Together, these results indicate that PALMD is a p53-target gene when ser46 is phosphorylated.
Figure 1.
PALMD is induced by p53 when ser46 is phosphorylated. (a) A genome-wide search of the pro-apoptotic genes promoted by phosphorylated p53 at ser46. Results of microarray and ChIP sequencing were overlapped to identify novel targets of phospho-ser46 of p53. (b) Control vector (control), wild-type p53 (wt-p53), and mutant p53 at serine-46 (p53S46A) were transfected into SaOS2 cells. Real-time PCR was performed and data represents fold change in PALMD relative to the GAPDH. *P<0.01. (c) Samples described in b were analyzed by western blotting. Protein-specific bands were detected with anti-PALMD (top panel), anti-phospho-p53(Ser46) (second panel), anti-p53 (third panel), or anti-tubulin (bottom panel). (d) The highest peak region of PALMD by ChIP sequencing that includes p53 consensus sites (red color). This region was amplified in the ChIP assay. (e) The binding of p53 onto PALMD was identified by the ChIP assay. Cell lysates from samples described in b were immunoprecipitated with anti-p53 or anti-immunoglobulin (Ig)G. ChIP assay and subsequent real-time PCR were performed. The data were normalized with the level of input control. *P<0.01. (f) DYRK2-depleted cells failed to induce PALMD expression. U2OS cells were transfected with non-silencing siRNA or siDYRK2. Cells were treated with ADR or left untreated. Cell lysates were immune-stained with anti-PALMD, anti-phospho-p53(Ser46), anti-p53, anti-DYRK2, or anti-tubulin
Table 1. Candidate genes induced by phsopho-p53 at ser46.
| Gene | wt-p53/control | wt-p53/p53S46A |
|---|---|---|
| PALMD | 8 | 1.49 |
| SFN | 5 | 1.48 |
| COL13A1 | 4 | 1.62 |
| NT5E | 3 | 1.51 |
| PAK6 | 3 | 1.65 |
Abbreviation: wt, wild type
PALMD is induced by p53 in response to DNA damage
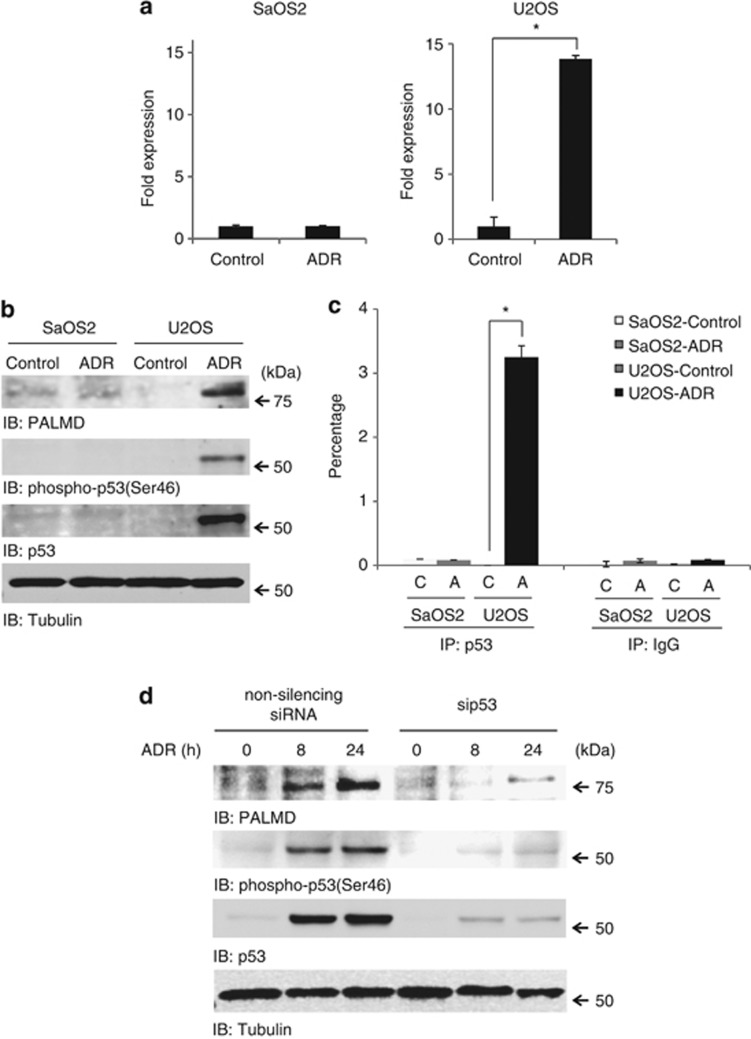
Since PALMD was regulated by exogenously transfected p53 in a phospho-ser46-specific manner in SaOS2 cells, we aimed to analyze its expression under control of endogenously expressing p53 using U2OS cells. p53 was stabilized and phosphorylated at ser46 (Figures 2b and d) that follows induction of PALMD in both mRNA (Figure 2a) and in protein levels (Figure 2b) in response to ADR exposure. However, the expression of PALMD was not changed after DNA damage in SaOS2 (Figures 2a and 2b). PALMD expression in other cell lines, H1299 and A549, showed the same result (Supplementary Figure 1b). In addition, PALMD expression was not induced under mild DNA damage, which is sufficient to phosphorylate serine-15 but not serine-46 (Supplementary Figure 1c, lane 3). Moreover, p53 targets PALMD upon ADR exposure (Figure 2c). Importantly, silencing of p53 did not promote PALMD expression (Figure 2d). Therefore, these findings suggest that PALMD is induced by p53 in response to DNA damage.
Figure 2.
PALMD is promoted by p53 in response to DNA damage. SaOS2 (p53-deficient) and U2OS (p53-proficient) cells were treated with ADR (ADR or A) or left untreated (control or C). (a) mRNA expression of PALMD in response to DNA damage. Data represent fold change of PALMD in ADR-treated cells relative to the control. *P<0.01. (b) Protein expression level of PALMD in response to ADR exposure. Cell lysates were immunoblotted against anti-PALMD (top panel), anti-phospho-p53(Ser46) (second panel), anti-p53 (third panel), or anti-tubulin (bottom panel). (c) The binding of endogenously expressing p53 onto PALMD under stressed condition. Cell lysates were immunoprecipitated with anti-p53 or anti-immunoglobulin (Ig)G. ChIP assay and subsequent real-time PCR were performed. The data were normalized with the level of input control. *P<0.01. (d) PALMD level in p53-depleted cells. U2OS cells were transfected with non-silencing siRNA or p53 siRNA (sip53). Cells were treated or left untreated with ADR and harvested at indicated times. Cell lysates were immunoblotted with anti-PALMD (top panel), anti-phospho-p53(Ser46) (second panel), anti-p53 (third panel), or anti-tubulin (bottom panel)
PALMD induces apoptosis in response to DNA damage
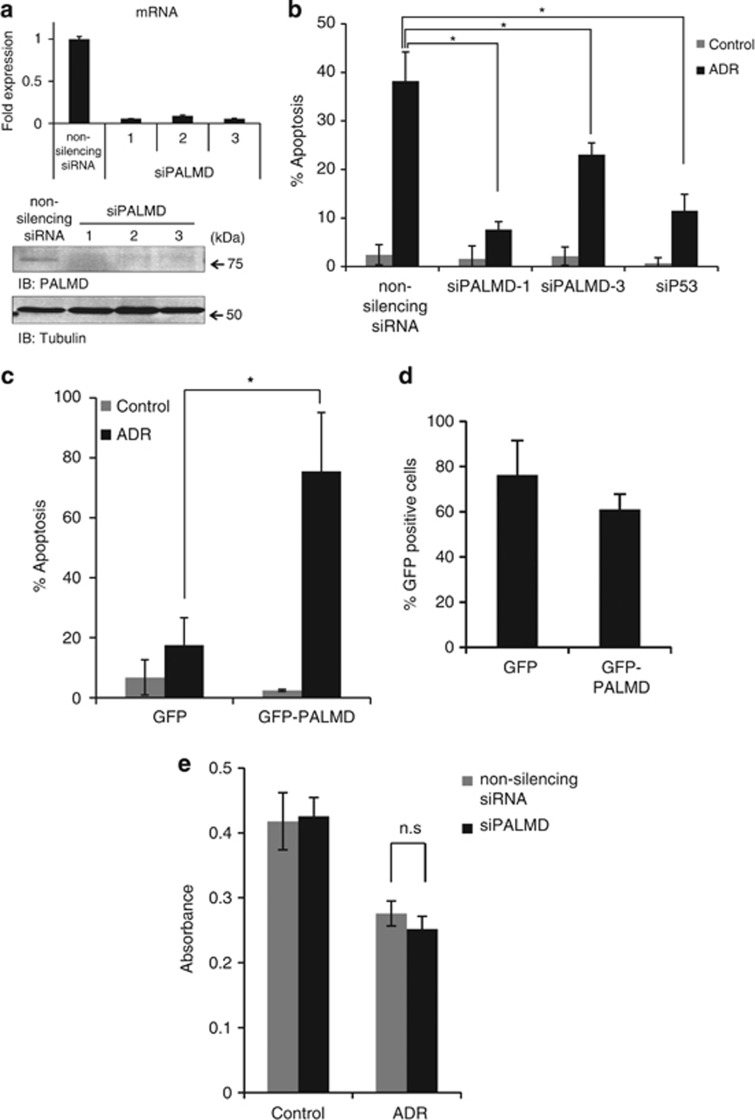
As phosphorylation of p53 at ser46 is essential for induction of apoptosis, we investigated whether PALMD is involved in the p53-dependent apoptosis. To prove this possibility, the TUNEL assay was performed to elucidate whether apoptosis is suppressed by the knockdown of PALMD (Figure 3b) or induced by overexpression of PALMD (Figure 3c). Three different short interfering RNAs (siRNAs), siPALMD-1, siPALMD-2, and siPALMD-3, were examined to silence PALMD expression. All siRNAs suppressed PALMD (Figure 3a). The suppression of PALMD substantially attenuated apoptosis in response to DNA damage (Figure 3b). As a control, knockdown of p53 also decreased apoptosis under ADR exposure (Figure 3b). Overexpressing PALMD with GFP-PALMD presented higher apoptotic cells than control (Figure 3c). Of note, transfection quality of GFP plasmids was examined (Figure 3d). As we have showed the apoptotic function of PALMD by the TUNEL assay, the cell viability assay was carried out to support these results. Unexpectedly, the cell viability of U2OS cells after silencing of PALMD in response to DNA damage remained unchanged (Figure 3e).
Figure 3.
PALMD induces apoptosis in response to DNA damage. (a) The specificity of the siRNAs of PALMD. Three different siRNAs, siPALMD-1, siPALMD-2, and siPALMD-3 or non-silencing siRNA were transfected into U2OS cells. After 24 h incubation, cells were harvested and total RNA was isolated. mRNA (upper panel, data represent fold expression of PALMD relative to the non-silencing siRNA) and protein expression (lower panel) were examined. (b) Apoptotic assay in PALMD-depleted cells. U2OS cells were seeded into four-well chamber slides and transfected with non-silencing siRNA, siPALMD-1, siPALMD-3, or sip53. Cells were treated with ADR or left untreated and incubated for 24 h. The results represent the percentage of the TUNEL (terminal deoxynucleotidyl transferase dUTP nick end labeling)-positive cells. *P<0.01. (c) Apoptotic assay in PALMD-overexpressing cells. Empty vector as a control (green fluorescent protein (GFP)) and GFP-tagged PALMD (GFP-PALMD) were transfected into U2OS cells. Cells were left untreated or treated with ADR and incubated for 6 h. Apoptotic cells were analyzed with TUNEL-positive cells. *P<0.01. (d) The transfection efficiency of GFP and GFP-PALMD into U2OS cells. (e) Cell viability in PALMD-depleted cells. U2OS cells were seeded in 96-well plates and transfected with non-silencing siRNA or siPALMD-1 and treated with ADR. The MTS assay was performed after 24 h incubation. The data represent the mean±S.D. from three independent experiments, each performed in triplicates
Downregulation of PALMD leads to necroptosis-like cell death
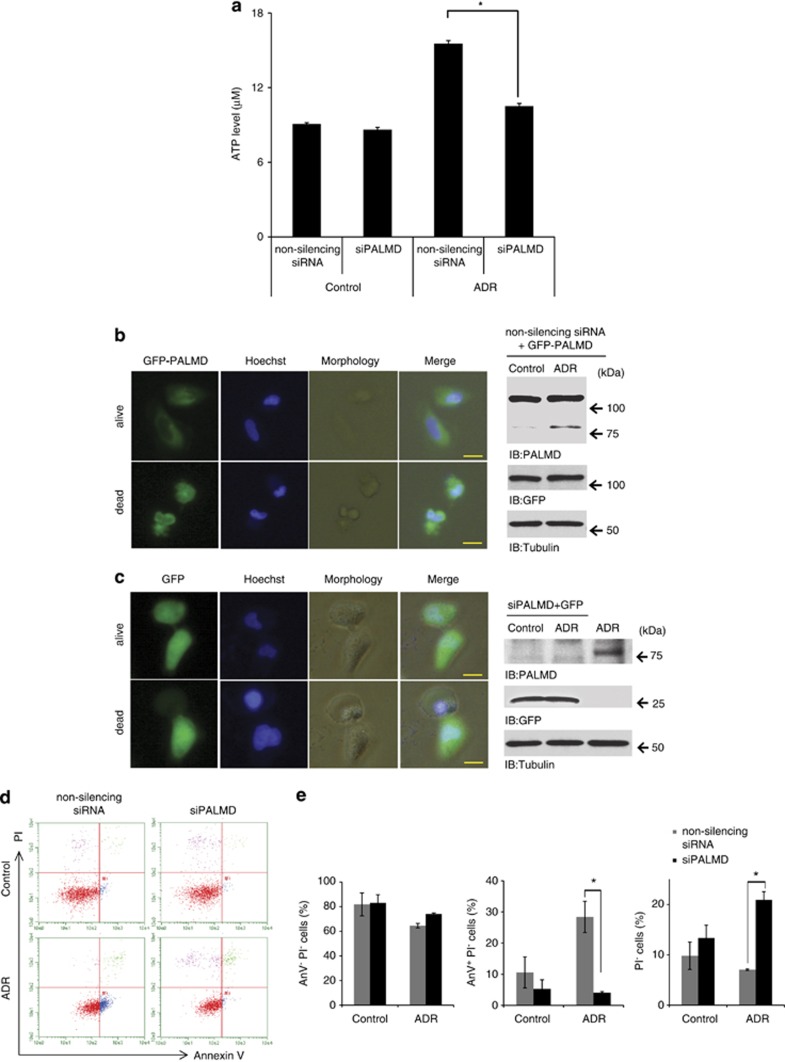
We found that PALMD induces apoptosis, however, knockdown of PALMD remained unchanged in cell viability after DNA damage. Therefore, PALMD loss was expected to induce other type of cell death. Given that both the apoptotic cell death and the necrotic cell death suppress each other17, 18, 19 and those were distinguished by the intracellular ATP level,17, 18 we measured the ATP concentration in PALMD-downregulated cells. After DNA damage, the concentration of ATP was increased in control cells. However, in PALMD-downregulated cells, the concentration of ATP significantly decreased (Figure 4a). The morphology of the cell death was examined by time-lapse imaging. When PALMD was overexpressed, cells exhibited apoptotic features such as cell shrinkage, membrane blebbing, and chromatin condensation (Figure 4b). Moreover, cells showed secondary necrosis after 13–14 h (data not shown). Interestingly, in the PALMD-downregulated cells, cells ruptured into the environment and chromatin was condensed (Figure 4c). To monitor transfection efficiency, western blotting (right panels in Figures 4b and c) and the immunoflourencence assay (Supplementary Figure 2) were carried out. In addition, double staining with propodium iodide (PI) and annexin V (AnV) analysis showed significant decrease of PI−AnV+ population (apoptotic20) and increase of PI-positive population (necrotic21, 22) in PALMD-depleted cells (Figures 4d and e).
Figure 4.
Loss of PALMD causes necroptosis-like cell death in response to DNA damage through ATP depletion. (a) ATP concentration in PALMD-depleted cells. U2OS cells were transfected with non-silencing siRNA or siPALMD-1 (siPALMD) and treated or left untreated with ADR. The data represent the mean±S.D. from three independent experiments, each performed in triplicates. *P<0.01. (b, left) Cell death morphology in PALMD-overexpressing cells. U2OS cells were co-transfected with green fluorescent protein (GFP)-tagged PALMD and non-silencing siRNA and treated with ADR. The morphologies of the cell death were illustrated by time-lapse imaging and cell image was captured at 5-min intervals from treatment. Upper panels present illustration of the cells just after treatment (alive) and lower panels show cell death (dead). The nuclei were stained with Hoechst. The scale bars indicate 20 μm. (b, right) The transfection efficiency of GFP-PALMD and non-silencing siRNA was monitored. Cell lysates were immunoblotted with anti-PALMD, anti-GFP, or anti-tubulin. (c, left) Cell death morphology in PALMD-depleted cells. U2OS cells were co-transfected with GFP vector and siPALMD-1. Further procedure was performed as described in b. (d) PALMD-involved cell death was confirmed by fluorescence-activated cell sorting analysis. U2OS cells were transfected with non-silencing siRNA or siPALMD-1 (siPALMD), and treated or left untreated with ADR. Cells were double stained with PI and AnV. PI−AnV− populations were defined as viable cells, PI−AnV+ populations were identified as apoptotic and PI-positive populations indicated as necrotic/necroptotic cells. (e) Percentage of viable (left panel), apoptotic (middle panel), or necrotic/necroptotic (right panel) cells from the data described in d was calculated. The data represent the mean±S.D. from three independent experiments. *P<0.01
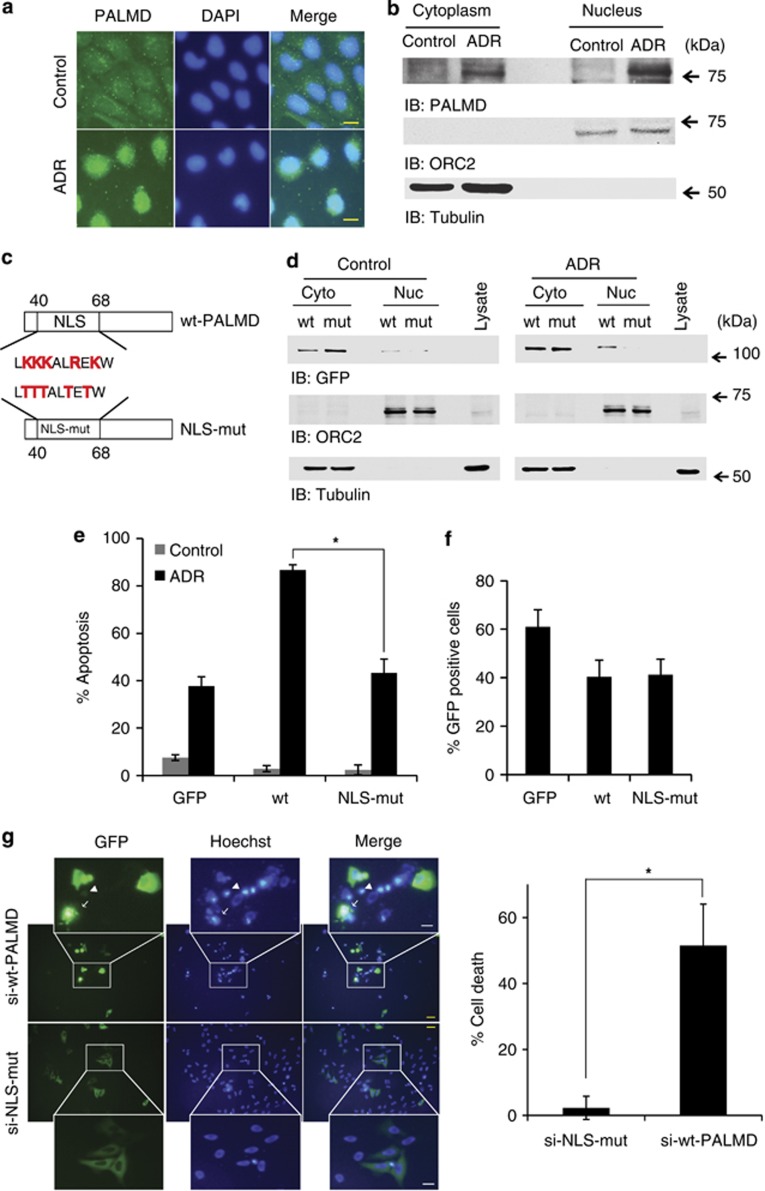
PALMD dominantly accumulates in the nucleus in response to DNA damage
PALMD is involved in the p53-dependent apoptosis. However, a mechanism by which PALMD promotes apoptosis has remained to be clarified. Although previous evidence suggested that PALMD localizes predominantly in the cytosol,23 a current study aimed to know whether PALMD changes location in response to DNA damage. Remarkably, the expression of PALMD increased in both cytoplasm and nucleus, although it was dominantly accumulated in the nucleus after ADR exposure (Figures 5a and b). Control cells showed low expression of PALMD that was mainly located in the cytoplasm (Figures 5a and b). The putative nuclear localization signal (NLS) was found within PALMD (Figure 5c). To determine whether this NLS is functional, GFP-tagged wild-type PALMD (wt-PALMD or wt) or NLS mutant (NLS-mut) was transfected into U2OS cells. NLS-mut remained in the cytoplasm, suggesting that the signal is required to move into the nucleus (Figure 5d; Supplementary Figure 3a). The transfection efficiency of wt-PALMD and NLS-mut into U2OS cells is shown in Supplementary Figure 3b. To further investigate whether NLS-mut induces apoptosis, the TUNEL assay was performed. Predictably, the expression of NLS-mut reduced induction of apoptosis compared with that of intact PALMD (Figure 5e). The transfection efficiency of plasmids is shown in Figure 5f. Furthermore, ectopic expression of PALMD showed ∼50% of the cells underwent death, which was attenuated in NLS-mut-transfected cells (Figure 5g). Consequently, these findings suggest the possibility that nuclear localization of PALMD is necessary for induction of apoptosis.
Figure 5.
Accumulation of PALMD is required to induce apoptosis. (a) PALMD localization by immunoflourescence in U2OS cells in normal condition (upper panels) and in stressed condition by ADR (lower panels). U2OS cells were seeded in four-well chamber slides and treated or left untreated with ADR. The nuclei were stained with DAPI. The scale bars indicate 20 μm. (b) PALMD localization by the subcellular fractionation assay. U2OS cells were treated or left untreated with ADR and fractionated as cytoplasmic and nucleic fractions. Both fractions were immunoblotted with anti-PALMD (top panel), anti-ORC2 (second panel), or anti-tubulin (bottom panel). (c) The map of the NLS construct. To design mutant NLS (NLS-mut), arginine and lysine indicated in red within NLS region of wild-type PALMD (wt-PALMD) were substituted with threonine and tagged to the green fluorescent protein (GFP) vector. (d) Subcellular fractionation assay of wt-PALMD and NLS-mut-transfected-cells. U2OS cells were transfected with wt-PALMD (wt) or NLS-mut. Cells were treated (ADR, right panels) or left untreated with ADR (control, left panels) and incubated for 24 h. Cell lysates were fractionated into cytoplasmic (Cyto) and nucleic (Nuc) fractions. Immunoblotting was performed using anti-GFP (top panel), anti-ORC2 (middle panel), or anti-tubulin (bottom panel). (e) Apoptotic assay of wt-PALMD or NLS-mut-transfected cells. U2OS cells were transfected with empty GFP vector (GFP), wt-PALMD (wt) or NLS-mut. Cells were treated with ADR and incubated for 6 h. The percentage of apoptotic cells was quantified with TUNEL (terminal deoxynucleotidyl transferase dUTP nick end labeling)-positive cells. (f) Transfection efficiency of GFP, wt-PALMD, or NLS-mut. (g) Cells undergo death with wt-PALMD or NLS-mut transfection when endogenous PALMD is depleted. The siRNA resistant-PALMD (si-wt-PALMD) or NLS-mut (si-NLS-mut) were co-transfected with siPALMD-1 and incubated for 24 h. The nuclei were illustrated with Hoechst. Cell death was defined with cell blebbings and apoptotic body (arrow), and chromatin condensation (arrow head). The scale bars indicate 40 μm (yellow) or 10 μm (white). The percentage of the cell death in GFP-positive cells is shown in the right panel. The data represent the mean±S.D. from three independent experiments, each performed in triplicates. *P<0.01
Discussion
In the current study, we isolated and identified PALMD as a p53-target pro-apoptotic gene in the phospho-ser46-specific manner. Upon exposure to the genotoxic stress, p53 is stabilized and activated by the phosphorylation to promote its function in the cell cycle arrest, DNA repair, and apoptosis. Previous evidence have suggested that the phosphorylation of ser46 is essential to promote apoptosis.8, 24 Various protein kinases such as HIPK2,9, 10 ATM,11 and DYRK24 activate p53 by phosphorylating ser46 to induce apoptosis. However, the pro-apoptotic gene promoted by p53 in a phospho-ser46-specific manner is largely unknown. Recently, microarray has become the basic tool to search gene expression differences between two samples such as normal cells and cancer cells. In addition to the microarray, ChIP sequencing is a powerful tool used widely to uncover transcription targets.25 Current study has used both analyses to demonstrate target genes of phospho-ser46 of p53. We found PALMD as a p53-target pro-apoptotic gene when ser46 is phosphorylated. Although, we showed that PALMD induces apoptosis in response to DNA damage, interestingly, we observed that loss of PALMD resulted in necroptosis-like cell death. For decades, necrosis was known as accidental cell death. However, growing evidence have demonstrated that certain types of necrosis are tightly controlled by orchestrated cellular pathways that are called necroptosis.17, 18, 19, 26, 27, 28 In the necroptotic cell death, poly-ADP-ribose polymerase becomes hyperactivated by reactive oxygen species that consumes cellular NAD and leads a cell to the ATP depletion.26, 29 Moreover, during necrosis, the cellular membrane is broken and cellular contents rupture into the environment.19 Our results showed ATP depletion in PALMD-downregulated cells in response to DNA damage. Those cells showed cell rupture into the environment, suggesting that PALMD is required for the apoptosis, otherwise cells die with necroptosis-like cell death. p53 regulates necrosis in a transcription-independent way through interacting with CypD by opening the mitochondrial permeability transition pore.12 Thus, the transcription-dependent way of p53 to induce necrosis is supposed to be existing. Therefore, we propose that p53 may decide cell death fate, apoptosis, or necrosis, through PALMD regulation.
PALMD is a newly identified gene and neither its functional analysis nor molecular mechanism is established. Paralemmin proteins anchor to the cytoplasm through its C-terminal CAAX motif by palmytoylation and prenylation.15, 16, 23, 30 Previously, two splicing variants of PALMD were found, one contains the CAAX motif as other paralemmin proteins and another consists of the KVVI motif at the C terminus.16 Unlike other paralemmin proteins, PALMD is found predominantly in the cytosol,23 which is because of the KVVI motif.16 In the current study, the splicing variant containing the CAAX motif was not amplified. Therefore, the regulation of p53 on PALMD to induce apoptosis depends upon KVVI domain. Moreover, our findings also suggested that the movement of PALMD into the nucleus is important to induce apoptosis; however, sequestering PALMD in the cytoplasm remained alive. Together, these results suggest that the localization of PALMD decides whether cells die with apoptosis or live. Our data showed that in the normal condition, PALMD expression was very low but not absent in the cytoplasm, which may be required to keep the cell alive. However, in response to DNA damage, p53 induces PALMD expression in a phospho-ser46-specific manner, and accumulated PALMD in the cytoplasm moves into the nucleus and triggers apoptosis. This function is required for the cell to die with apoptosis, and if this function is blocked the cell dies with nectoptosis. However, the molecular mechanisms of PALMD to induce those functions are currently under investigation.
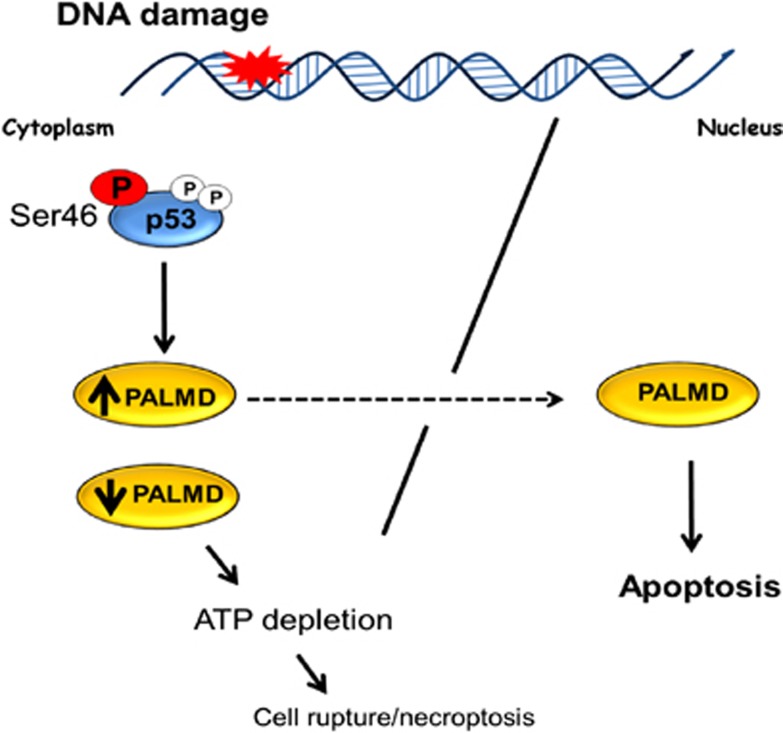
In conclusion, PALMD is demonstrated as the target of ser46 phosphorylation of p53. PALMD targets to the nucleus and leads to apoptosis in response to DNA damage. The loss of PALMD results in necroptosis-like cell death through ATP depletion (Figure 6).
Figure 6.
Proposed model of the molecular function of PALMD. In response to DNA damage, p53 is stabilized and activated by phosphorylation at ser46. Activated p53 promotes PALMD expression. Accumulated PALMD in the cytoplasm localizes into the nucleus in response to DNA damage to induce apoptosis. In the other side, if PALMD is not induced, cells die with necroptosis-like cell death through ATP depletion
Materials and Methods
Cell culture
SaOS2 cells (p53-deficient human osteosarcoma cell line) and U2OS cells (p53-proficient human osteosarcoma cell line) were cultured in Dulbecco's modified Eagle's medium and RPMI 1640 medium, respectively. Both media were supplemented with 10% (v/v) heat-inactivated fetal bovine serum, 100 μg/ml penicillin, 100 μg/ml streptomycin, and 2 mM L-glutamine. Cells were treated with 0.2 μg/ml adriamycin (ADR; Sigma-Aldrich, St. Louis, MO, USA).
Construction of the plasmids and siRNAs
Flag-tagged p53 (wt-p53) and mutant p53 in which the residue ser46 is substituted with alanine (p53S46A) were constructed as previously described.4 The cDNA of PALMD was amplified by PCR using pfuUltra High-Fidelity DNA polymerase (Stratagene, La Jolla, CA, USA) from SaOS2 cells using primers; forward primer—5′-GCCCTCGAGGAATGGAAGAAGCTGAGCTG-3′, reverse primer—5′-CGGGGTACCGGTGGTACAACTCTTAGATC-3′. The NLS was searched using cNLS mapper (http://nls-mapper.iab.keio.ac.jp). To construct mutant NLS in which the residue arginine and lysine were substituted with threonine, forward primer—5′-ACTACCACAGCCTTGACTGAGAAATGGCTTCTAGATGG-3′ and reverse primer—5′-AGTCAAGGCTGTGGTAGTCAAATGCTGGTGCTTTAG-3′ were used. cDNAs were cloned into the pEGFP-C1 vector (GFP vector) or pcDNA3-flag vector (Flag vector). Gene-specific siRNAs of PALMD (siPALMD-1; NM_017734.4_stealth_838, siPALMD-2; NM_017734.4_stealth_1149, and siPALMD-3; NM_017734.4_stealth_1759) (Invitrogen, Carlsbad, CA, USA), non-silencing siRNA (Qiagen, Hilden, Germany), and siDYRK2 (Qiagen)4 were used. siRNA mutant wt-PALMD and NLS-mut were designed against siPALMD-1 using the following primers: forward primers—5′-GAGGAGTCAATCGAGGATATTTATGCTAATATCCCTGACC-3′, reverse primer—5′-TAAATATCCTCGATTGACTCCTCTGCTCTTTCTTCTCTTTCC-3′. The mutated sites are shown in Supplementary Figure 4a. siPALMD-1-resistant plasmids were examined and the results are presented in Supplementary Figure 4b.
Cell transfection
p53-plasmid DNAs were transfected using FuGENE 9 transfection reagent (Roche, Hilden, Germany) following the manufacturer's protocol. The GFP-tagged and flag-tagged PALMD-plasmids were transfected using Lipofectamine 2000 (Invitrogen) following the supported protocol. Transfections of validated gene-specific siRNAs were performed using Lipofectamine RNAi MAX (Invitrogen) according to the supplied protocol.
ChIP and ChIP sequencing assay
ChIP and ChIP sequencing were performed as described previously.3 Briefly, empty flag vector and flag-tagged p53 were transfected into SaOS2 cells to perform the ChIP sequencing assay. In the ChIP assay, SaOS2 and U2OS cells were treated with ADR or left untreated and incubated for 24 h. Cell lysates were immunoprecipitated with mouse monoclonal anti-p53 (Santa Cruz Biotechnology, Santa Cruz, CA, USA) or mouse monoclonal anti-IgG antibodies (Santa Cruz Biotechnology). Immunoprecipitated DNA fragments were amplified and subjected to semi-quantitative RT-PCR. The data were normalized for the level of input control. To amplify the PALMD promoter region, forward primer—5′-TCACGATGTTTCCTCACACC-3′ and reverse primer—5′-CCTTTTCCTTCTTCCCCATT-3′ were utilized. ChIP sequencing was performed by Takara Bio Inc. (Shiga, Japan). The data of ChIP sequencing can be accessed through NCBI's Gene Expression Omnibus under NCB accession number GSE51268.31
Microarray analysis
Microarray was performed as described previously.32 Briefly, the plasmids, empty flag vector, wt-p53, or p53S46A were transfected into SaOS2 cells and incubated for 48 h. Total RNA was isolated from cells using the RNaesy kit (Qiagen) and reverse transcribed into cDNA and synthesized biotin-labeled cRNA following the manufacturer's protocol (Affymetrix, Santa Clara, CA, USA). The Human Genome U133 Plus 2.0 array (Affimetrix, comprised 1 300 000 distinct oligonucleotides, features >47 000 transcripts and variants, including ∼39 000 of the best-characterized human genes) was used. Arrays were stained using Fluidic station 450 according to the protocol, EukGE-WS2v5_450, and scanned using the Affimetrix GeneChip Scanner 3000. The complete expression data sets are available in online data repository ArrayExpress, under accession number E-MEXP-2643.32
Screening of the results of microarray and ChIP sequencing
To screen microarray, whole-gene expression was normalized with results of flag vector-transfected cells (control). The genes higher expressed than control sample were filtered for p53 responsible genes, and to identify p53S46A-related genes, the genes expressed in mutant fraction were excluded. To screen ChIP sequencing, candidate genes indicated by the downstream region were removed. Overlapping of both results revealed p53-target genes in a phospho-ser46-specific manner.
Real-time RT-PCR analysis
Isolation of total RNA was performed using TRIsure (Nippon Gene, Toyama, Japan) according to the manufacturer's protocol after 48 h of the transfection. Total RNA (2 μg) was reverse transcribed into cDNA using Superscript III First-Strand Synthesis System for RT-PCR (Takara Bio Inc.) following the supplied protocol. The PCR reaction was performed using the KAPA SYBR Green Master Mix (5 μl KAPA SYBR Green Master Mix, 0.3 μl each of 10 μM primers, and 4.7 μl cDNA). PCR was performed using 7900 Fast Real-Time RT-PCR System (Applied Biosystems, Foster City, CA, USA) under the following condition: 42 cycles of two-step PCR (95 °C for 2 sec, 60 °C for 1 min). Forward primer—5′-CAATTGAGCGGACAACAGAA-3′ and reverse primer—5′-TTGGAAGGTCAGGGATATTAGC-3′ were used to amplify cDNA of the PALMD. Gene expressions were normalized with GAPDH.
Immunoblotting and antibodies
Cultured cells were washed twice with PBS and resuspended in lysis buffer (50 mM Tris-HCl, pH 7.6, 150 mM NaCl, 10 mM NaF, 1 mM Na3VO4, 1 mM phenylmethylsulfonyl fluoride, 1 mM dithiothretiol, 10 mg/ml aprotinin, 1 mg/ml leupeptin, 1 mg/ml pepstatin A, and 1% NP-40). Cell lysates were centrifuged for 5 min at 4 °C. The supernatants were separated by sodium dodecyl sulfate-PAGE and transferred to nitrocellulose membranes. The membranes were incubated with anti-PALMD (Proteintech, Chicago, IL, USA), anti-p53 (Santa Cruz Biotechnology), anti-phospho-Ser46 (BioAcademia, Osaka, Japan), or anti-tubulin (Sigma-Aldrich). Immune complexes were incubated with secondary antibodies and visualized by chemiluminescense (PerkinElmer, Waltham, MA, USA) and Light capture II (Cooled CCD Camera System; ATTO, Tokyo, Japan) using software, CS Analyzer (ATTO).
TUNEL assay
U2OS cells were cultured in poly-D-lysine-coated four-well chamber slides. Cells were transfected with non-silencing siRNA, gene-specific siRNAs (siPALMD-1, siPALMD-3, and sip53), or GFP-tagged plasmids. Then cells were treated with 0.2 μg/ml ADR and incubated for 24 or 6 h, respectively. The apoptotic effect was measured using the Dead End Fluorometric TUNEL system (Promega, Madison, WI, USA). Morphological changes in the nuclear condensation of the cells undergoing apoptosis were detected by staining with 4′, 6′-diamidino-2-phenylindole (DAPI).
Cell viability assay
The number of viable cells was counted by trypan blue exclusion using a hemocytometer. Cells (1 × 104) were seeded in 96-well plates and transfected with non-silencing siRNA and siPALMD-1. In the following day, cells were treated with 0.2 μg/ml ADR. MTS assays were performed using the CellTiter 96 AQ One Solution Cell Proliferation Assay Kit (Promega) following the manufacture's protocol. The absorbance was measured at 490 nm with use of a multilabel counter (PerkinElmer).
Immunoflourencence assay
U2OS cells (1.5 × 105) were cultured in four-well chamber slides. In the following day, cells were treated with 0.2 μg/ml ADR and incubated for 24 h. Cells were fixed with 3% paraformaldehyde for 25 min at 4 °C and rinsed with PBS and incubated with 0.1% Triton X-100 for 15 min at room temperature. After washing with PBS and subsequent blocking with 10% goat serum for 1 h, cells were incubated with anti-PALMD (1 : 100 dilution) overnight at 4 °C. Secondary antibodies were then applied for 1 h at room temperature. Nuclei were stained with DAPI.
Subcellular fractionation assay
Subcellular fractionation assay was performed as described elsewhere.3, 4 Briefly, cell lysates were fractionated as nuclear and cytoplasmic fractions. Purity of the fractions was monitored by immunoblot analysis with anti-tubulin and anti-ORC2 (MBL, Woburn, MA, USA). Immune complexes were incubated with secondary antibodies and visualized by chemiluminescense (PerkinElmer) or Light capture II (Cooled CCD Camera System; ATTO) using software, CS Analyzer (ATTO).
Imaging of the cell death morphology
U2OS cells were cultured in a 35-mm cell culture dish (Corning). Cells were co-transfected with non-silencing siRNA and GFP-tagged PALMD or siPALMD-1 and GFP vector. In the following day, cells were treated with 0.2 μg/ml ADR and the nucleus was visualized with Hoechst (Bisbenzimide H33342; Sigma, Kanagawa, Japan). Then, cells were incubated in an Incubation system for microscopes (TOKAI HIT, Shizuoka, Japan) under 5% CO2 at 37 °C combined with a fluorescence microscope (Bio-Zero BZ-8000; Keyence, Osaka, Japan) equipped with a phase-difference lens (Nikon, Tokyo, Japan). The cells were captured at 5-min intervals. For the flourescence imaging, a halogen lamp was used with excitation (BP520-540HQ) and emission (BP555-600HQ) filters. Image acquisition and analysis were performed using analyzer software (BZ-H1TL; Keyence).
Intracellular ATP concentration
U2OS cells were transfected with non-silencing siRNA or siPALMD-1. In the following day, cells were treated with 0.2 μg/ml ADR or left untreated and incubated for 24 h. ATP concentration was determined using the ATP detection kit (Invitrogen) according to the supplied protocol, and the luminescence was measured using Infinite M200 PRO (TECAN, Männedorf, Switzerland).
Flow cytometry (FACS) analysis
U2OS cells were transfected with non-silencing siRNA or siRNA of PALMD and incubated for 24 h. Cells were treated with ADR (0.2 μg/ml) or left untreated. Cells (1 × 105) were stained with PI (Life Technology, Carlsbad, CA, USA) and FITC-AnV (BD Biosciences, San Jose, CA, USA) for 15 min at 37 °C. FACS analysis was performed using Guava EasyCyte Mini, Guava Technology.
Statistics
Statistical analysis was performed using two-tailed t-tests. Data represent the mean±S.D. P<0.05 was considered as statistically significant.
Acknowledgments
This work was supported by grants from the Ministry of Education, Science and Culture of Japan (to KY), the Mochida Memorial Foundation for Medical and Pharmaceutical Research (to KY), Uehara Memorial Foundation (to KY), Japan Foundation for Applied Enzymology (to KY), Takeda Science Foundation (to KY), the Naito Foundation (to KY), Suzuken Memorial Foundation (to KY), the NOVARTIS Foundation for the Promotion of Science (to KY), the Sumitomo Foundation (to KY), the Astellas Foundation for Research on Medical Resources (to KY), Project Mirai Cancer Research Grants (to KY), the Jikei University Graduate Research Fund (to KY), and Global COE Program, Tokyo Medical and Dental University (to ND).
Glossary
- PALMD
palmdelphin
- ChIP
chromatin immunoprecipitation
- ATM
ataxia telangiectasia mutated
- DYRK2
dual-specificity tyrosine phosphorylation regulated kinase 2
- HIPK2
homodomain interacting protein kinase 2
- ADR
adriamycin
- PI
propidium iodide
- AnV
annexin V
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Cell Death and Disease website (http://www.nature.com/cddis)
Edited by G Melino
Supplementary Material
References
- Levine AJ. p53, the cellular gatekeeper for growth and division. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- Nakamura Y. Isolation of p53-target genes and their functional analysis. Cancer Sci. 2004;95:7–11. doi: 10.1111/j.1349-7006.2004.tb03163.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K, Dashzeveg N, Lu ZG, Taira N, Miki Y, Yoshida K. Programmed cell death 6, a novel p53-responsive gene, targets to the nucleus in the apoptotic response to DNA damage. Cancer Sci. 2012;103:1788–1794. doi: 10.1111/j.1349-7006.2012.02362.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taira N, Nihira K, Yamaguchi T, Miki Y, Yoshida K. DYRK2 is targeted to the nucleus and controls p53 via Ser46 phosphorylation in the apoptotic response to DNA damage. Mol Cell. 2007;25:725–738. doi: 10.1016/j.molcel.2007.02.007. [DOI] [PubMed] [Google Scholar]
- Taira N, Yoshida K. Post-translational modifications of p53 tumor suppressor: determinants of its functional targets. Histol Histopathol. 2012;27:437–443. doi: 10.14670/HH-27.437. [DOI] [PubMed] [Google Scholar]
- Yoshida K, Miki Y. The cell death machinery governed by the p53 tumor suppressor in response to DNA damage. Cancer Sci. 2010;101:831–835. doi: 10.1111/j.1349-7006.2010.01488.x. [DOI] [PubMed] [Google Scholar]
- Bode AM, Dong Z. Post-translational modification of p53 in tumorigenesis. Nat Rev Cancer. 2004;4:793–805. doi: 10.1038/nrc1455. [DOI] [PubMed] [Google Scholar]
- Oda K, Arakawa H, Tanaka T, Matsuda K, Tanikawa C, Mori T, et al. p53AIP1, a potential mediator of p53-dependent apoptosis, and its regulation by Ser-46-phosphorylated p53. Cell. 2000;102:849–862. doi: 10.1016/s0092-8674(00)00073-8. [DOI] [PubMed] [Google Scholar]
- Hofmann TG, Moller A, Sirma H, Zentgraf H, Taya Y, Droge W, et al. Regulation of p53 activity by its interaction with homeodomain-interacting protein kinase-2. Nat Cell Biol. 2002;4:1–10. doi: 10.1038/ncb715. [DOI] [PubMed] [Google Scholar]
- D'Orazi G, Cecchinelli B, Bruno T, Manni I, Higashimoto Y, Saito S, et al. Homeodomain-interacting protein kinase-2 phosphorylates p53 at Ser 46 and mediates apoptosis. Nat Cell Biol. 2002;4:11–19. doi: 10.1038/ncb714. [DOI] [PubMed] [Google Scholar]
- Saito S, Goodarzi AA, Higashimoto Y, Noda Y, Lees-Miller SP, Appella E, et al. ATM mediates phosphorylation at multiple p53 sites, including Ser(46), in response to ionizing radiation. J Biol Chem. 2002;277:12491–12494. doi: 10.1074/jbc.C200093200. [DOI] [PubMed] [Google Scholar]
- Vaseva AV, Marchenko ND, Ji K, Tsirka SE, Holzmann S, Moll UM. p53 opens the mitochondrial permeability transition pore to trigger necrosis. Cell. 2012;149:1536–1548. doi: 10.1016/j.cell.2012.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreu N, Escarceller M, Feather S, Devriendt K, Wolf AS, Estivill X, et al. PALML, a novel paralemmin-related gene mapping on human chromosome 1p21. Gene. 2001;278:33–40. doi: 10.1016/s0378-1119(01)00719-3. [DOI] [PubMed] [Google Scholar]
- Hu B, Copeland NG, Gilbert DJ, Jenkins NA, Kilimann MW. The paralemmin protein family: identification of paralemmin-2, an isoform differentially spliced to AKAP2/AKAP-KL, and of palmdelphin, a more distant cytosolic relative. Biochem Biophys Res Commun. 2001;285:1369–1376. doi: 10.1006/bbrc.2001.5329. [DOI] [PubMed] [Google Scholar]
- Eguchi Y, Shimizu S, Tsujimoto Y. Intracellular ATP levels determine cell death fate by apoptosis or necrosis. Cancer Res. 1997;57:1835–1840. [PubMed] [Google Scholar]
- Zamaraeva MV, Sabirov RZ, Maeno E, Ando-Akatsuka Y, Bessonova SV, Okada Y. Cells die with increased cytosolic ATP during apoptosis: a bioluminescence study with intracellular luciferase. Cell Death Differ. 2005;12:1390–1397. doi: 10.1038/sj.cdd.4401661. [DOI] [PubMed] [Google Scholar]
- Han J, Zhong CQ, Zhang DW. Programmed necrosis: backup to and competitor with apoptosis in the immune system. Nat Immunol. 2011;12:1143–1149. doi: 10.1038/ni.2159. [DOI] [PubMed] [Google Scholar]
- Brown V, Elborn JS, Bradley J, Ennis M. Dysregulated apoptosis and NFkappaB expression in COPD subjects. Respir Res. 2009;10:24. doi: 10.1186/1465-9921-10-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabon L, Galan-Malo P, Bouharrour A, Delavallee L, Brunelle-Navas MN, Lorenzo HK, et al. BID regulates AIF-mediated caspase-independent necroptosis by promoting BAX activation. Cell Death Differ. 2012;19:245–256. doi: 10.1038/cdd.2011.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinhart L, Belz K, Fulda S. Smac mimetic and demethylating agents synergistically trigger cell death in acute myeloid leukemia cells and overcome apoptosis resistance by inducing necroptosis. Cell Death Dis. 2013;4:e802. doi: 10.1038/cddis.2013.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu B, Petrasch-Parwez E, Laue MM, Kilimann MW. Molecular characterization and immunohistochemical localization of palmdelphin, a cytosolic isoform of the paralemmin protein family implicated in membrane dynamics. Eur J Cell Biol. 2005;84:853–866. doi: 10.1016/j.ejcb.2005.07.002. [DOI] [PubMed] [Google Scholar]
- Okamura S, Arakawa H, Tanaka T, Nakanishi H, Ng CC, Taya Y, et al. p53DINP1, a p53-inducible gene, regulates p53-dependent apoptosis. Mol Cell. 2001;8:85–94. doi: 10.1016/s1097-2765(01)00284-2. [DOI] [PubMed] [Google Scholar]
- Collas P. The current state of chromatin immunoprecipitation. Mol Biotechnol. 2010;45:87–100. doi: 10.1007/s12033-009-9239-8. [DOI] [PubMed] [Google Scholar]
- Fu D, Jordan JJ, Samson LD. Human ALKBH7 is required for alkylation and oxidation-induced programmed necrosis. Genes Dev. 2013;27:1089–1100. doi: 10.1101/gad.215533.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verrax J, Dejeans N, Sid B, Glorieux C, Calderon PB. Intracellular ATP levels determine cell death fate of cancer cells exposed to both standard and redox chemotherapeutic agents. Biochem Pharmacol. 2011;82:1540–1548. doi: 10.1016/j.bcp.2011.07.102. [DOI] [PubMed] [Google Scholar]
- Yang X, Wang J, Liu C, Grizzle WE, Yu S, Zhang S, et al. Cleavage of p53-vimentin complex enhances tumor necrosis factor-related apoptosis-inducing ligand-mediated apoptosis of rheumatoid arthritis synovial fibroblasts. Am J Pathol. 2005;167:705–719. doi: 10.1016/S0002-9440(10)62045-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elkholi R, Chipuk JE. How do I kill thee? Let me count the ways: p53 regulates PARP-1 dependent necrosis. Bioessays. 2014;36:46–51. doi: 10.1002/bies.201300117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutzleb C, Sanders G, Yamamoto R, Wang X, Lichte B, Petrasch-Parwez E, et al. Paralemmin, a prenyl-palmitoyl-anchored phosphoprotein abundant in neurons and implicated in plasma membrane dynamics and cell process formation. J Cell Biol. 1998;143:795–813. doi: 10.1083/jcb.143.3.795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R, Domrachev M, Lash AE. Gene expression omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura J, Kudoh T, Miki Y, Yoshida K. Identification of dihydropyrimidinase-related protein 4 as a novel target of the p53 tumor suppressor in the apoptotic response to DNA damage. Int J Cancer. 2011;128:1524–1531. doi: 10.1002/ijc.25475. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.