Abstract
Background
The bulk of human genes undergo alternative splicing (AS) upon response to physiological stimuli. AS is a great source of protein diversity and biological processes and is associated with the development of many diseases. Pheochromocytoma is a neuroendocrine tumor, characterized by an excessive Ca2+-dependent secretion of catecholamines. This underlines the importance of balanced control of calcium transport via regulation of gene expression pattern, including different calcium transport systems, such as plasma membrane Ca2+-ATPases (PMCAs), abundantly expressed in pheochromocytoma chromaffin cells (PC12 cells). PMCAs are encoded by four genes (Atp2b1, Atp2b2, Atp2b3, Atp2b4), whose transcript products undergo alternative splicing giving almost 30 variants.
Results
In this scientific report, we propose a novel mechanism of regulation of PMCA alternative splicing in PC12 cells through cooperation of the nuclear factor of activated T-cells (NFAT) and histone deacetylases (HDACs). Luciferase assays showed increased activity of NFAT in PC12 cells, which was associated with altered expression of PMCA. RT-PCR experiments suggested that inhibition of the transcriptional activity of NFAT might result in the rearrangement of PMCA splicing variants in PC12 cells. NFAT inhibition led to dominant expression of 2x/c, 3x/a and 4x/a PMCA variants, while in untreated cells the 2w,z/b, 3z,x/b,c,e,f, and 4x/b variants were found as well. Furthermore, chromatin immunoprecipitation experiments showed that NFAT1-HDAC4 or NFAT3-HDAC4 complexes might be involved in regulation of PMCA2x splicing variant generation.
Conclusions
We suggest that the influence of NFAT/HDAC on PMCA isoform composition might be important for altered dopamine secretion by PC12 cells.
Introduction
Alternative splicing of pre-mRNA is a major post-transcriptional source of protein diversity, which is essential for a variety of biological processes, both under physiological and pathological conditions [1]. Recent genome-wide association studies have shown that 94% of human multi-exon genes undergo alternative splicing [2]. In the nervous system, alternative splicing is switched on and off during different processes, including learning, memory, synaptogenesis or neurotransmission, by modulation of neurotransmitter release, ion channel functions, and receptor specificity [3]–[5]. In the nervous system, alternative splicing of genes encoding the neural cell adhesion molecule (NCAM), NMDA receptors, and calcium pumps, for example the plasma membrane Ca2+-ATPase (PMCA), undergoes cell activity-induced changes [6]–[10]. Instabilities in alternative splicing regulatory sequences and disturbances in the binding of regulatory proteins to these sequences are important causes of numerous human diseases [11]. This is especially true for neurodegenerative diseases, neurological tumors and mental disorders [12], [13]. One of the commonly known neuropathologies is the pheochromocytoma neuroendocrine tumor, which causes widespread consequences such as hypertension or cardiac arrhythmia, as well as psychiatric disturbances [14]. Pheochromocytoma is localized in the adrenal medulla and is characterized by an excessive secretion of catecholamines, i.e. epinephrine, norepinephrine and dopamine.
Pheochromocytoma chromaffin cells (PC12 cells) release neurotransmitters in the process of Ca2+-regulated exocytosis [15]. Thus, PC12 cells are equipped with the neuronal type of secretory machinery, demanding tight Ca2+-dependent genetic control over alternative splicing of mRNAs encoding proteins involved in the maintenance of calcium homeostasis and secretory response [16]. Accordingly, alterative splicing has been found to influence the expression profile of various mRNAs encoding secretory proteins [17], including elements of membrane fusion complex: SNAP25, syntaxin 1 and synaptobrevin 1 [18]–[21] and mRNAs encoding calcium transporters (calcium pumps, ions exchangers, calcium channels). A great number of examples have been given on the alternative splicing of mRNAs for voltage gated calcium channels [22], [23], sodium calcium exchangers [24], [25], and plasma membrane Ca2+-ATPases (PMCAs). The latter proteins are the main subject of the several important studies [8], [26]. PMCAs are responsible for pumping Ca2+ ions out of the cell and maintenance of low cytosolic calcium ions concentration ([Ca2+]c). PMCAs are encoded by four genes (Atp2b1, Atp2b2, Atp2b3, Atp2b4) and some exons of these genes might be excluded from or included into the final mRNA/transcript by the process of alternative splicing generating almost 30 mRNA transcript variants [8], [10]. PC12 cells express all PMCA isoforms, and most of the splicing variants [26]. Alternative splicing of PMCAs affects two strategic regions of the pump: the acidic phospholipid-binding domain (splice site A) and the Ca2+-calmodulin binding domain (splice site C) [8]. Thus, alternative splicing generates PMCA variants of different structure and biochemical properties, such as affinity for calcium ions, velocity of calcium ion transport or ability to interact with a different signaling proteins (e.g. calcineurin, nitric oxide synthase, calmodulin or 14-3-3 protein) [9], [27]–[29]. Expression profile of the alternatively spliced variants of PMCAs has been well established in various tissues [8], [27], [30], [31]. However, the molecular basis of generation of alternative transcripts of PMCAs, including molecular mechanisms and regulatory proteins that may induce or arrest this process, remains unclear. Alternative splicing has been recently described as a co-transcriptional process requiring activity of transcription factors, histone modifying proteins and other regulatory proteins involved in chromatin rearrangement [32]–[39]. Transcriptional factors may influence alternative splicing by interaction with RNA polymerase II, which is responsible for targeting of the splicing machinery to the site of transcription [40]. One of the transcription factors whose activity has been linked with alternative splicing is nuclear factor of activated T cells (NFAT). NFAT was found to influence the alternative splicing of mRNAs of allograft inflammatory factor-1 (AIF-1) [41], of interferon responsive transcript-1 (IRT-1) [20], and of synaptotagmin-like 2 protein [42]. Interestingly, NFAT has been proposed to be responsible for the control of the expression of the PMCA1 and PMCA4 isoforms [43]–[48]. As already mentioned, histone modification could be another important tool for the control and moderation of the alternative splicing process. Several histone-binding proteins were found to interact with splicing factors [49]–[53]. Among the proteins that modify histones, histone deacetylases (HDACs) play an extremely important role, both in the context of the regulation of gene expression, by influencing the availability of DNA as well as in the context of alternative splicing of mRNA [54]. Furthermore, HDACs were found to interact with NFATs and to repress their activity [55]. More precisely, the class IIa of HDACs have been shown to repress cardiac hypertrophy by inhibiting cardiac-specific transcription factors such as myocyte enhancer factor 2 (MEF2), GATA4, and NFAT in the heart [56]. On the other hand, it was shown that NFATc1 favored the binding of HDAC3 to the proximal region of the osteocalcin gene promoter, enhancing the expression of the gene [57]. Finally, our recent studies have suggested that overactive NFAT signaling is responsible for the repression of genes Vamp1 and Vamp2 in PC12 cells, stressing the importance of NFAT activity in these cell types [58]. The interdependence between HDAC and NFAT suggests that these proteins could counteract or cooperate during the regulation of alternative splicing of mRNAs of PMCAs. In this report we would like to propose putative regulatory mechanisms of Ca2+-dependent alternative splicing of plasma membrane Ca2+-ATPases (PMCAs), that might be switched on during excessive secretion of catecholamines in pheochromocytoma. Thus, this study may shed a new light on the role of the genetic diversity of PMCA in a number of different neuropathologies as well as during cognitive processes based on neurotransmission.
Materials and Methods
Cell Culture and Cell Lines
PC12 cell line were obtained from the Department of Molecular Neurochemistry, Medical University of Lodz (purchased from American Type Culture Collection, ATCC No.: CRL-1722). PC12 cells were cultured in RPMI-1640 medium (Sigma Aldrich, USA) with 10% heat-inactivated horse serum and 5% heat-inactivated fetal bovine serum (Gibco, Invitrogen), in an atmosphere of 5% CO2/95% air at 37°C. In some experiments the inhibitor of NFAT (1 µM 11R-VIVIT) (Calbiochem Merck Chemicals, Germany) was added for 48 h to the cell culture. Cell lines deficient in PMCA2 (_2) or PMCA3 (_3) were established as described previously [59], [60]. Control cells (C) were transfected with empty pcDNA3.1 (+) plasmid. Stable cell lines were obtained by a 4-week selection with 1 mg/ml G418 and used for the experiments at passages 15–30. Using flow cytometry, cells were routinely tested propidium iodide staining in terms of cell cycle and apoptosis, and none of used conditions (5 mM KCl, 59 mM KCl or 1 µM 11R-VIVIT) did affected the measured parameters.
Quantitative PCR
Total RNA was isolated using TRIzol Reagent (Invitrogen). RNA aliquots of 5 µg were subjected to reverse transcription using MuMLV reverse transcriptase (Promega, USA), according to the manufacturer’s protocol. The obtained cDNA (3 µl) was used in quantitative PCR (qPCR) with the SYBR Green reagent (Applied Biosystem) according to the manufacturer’s protocol. qPCR data were normalized to Gapdh and 18SrRNA expression and calculated according to the ΔΔCT method [61]. The calculations for 11R-VIVIT treated cells were carried out according to a modified ΔΔCT method as follows: ΔΔCT = ΔC11R-VIVIT-treated−ΔCnon-treated. PMCA isoform expression was verified by RT-PCR. The primers were designed for the R. norvegicus genome using the GenScript Primer Design Tool (USA) (Table 1) and used at 1 µM concentration.
Table 1. Primers for PMCA isoforms expression analysis.
| gene name | sequence (5′ → 3′) | strand | Tm °C | amplicon size(bp) | NCBI number |
| Atp2b1 | AGAAGTTCACCGTCATCAGG | F | 57.71 | 92 | NM_053311.1 |
| ATCACCGTACTTCACTTGGG | R | 57.51 | |||
| Atp2b2 | TTGCTGTCAGGAACTCATGT | F | 56.76 | 79 | NM_012508.5 |
| TGCCAGTTTGAGAGTTGACA | R | 57.95 | |||
| Atp2b3 | GAAAGCAGGATTGGTGATGT | F | 57.58 | 164 | NM_133288.1 |
| CAACCAACACAGTGACTCCA | R | 58.06 | |||
| Atp2b4a | GAAATCCAGCCACTCAACAG | F | 58.29 | 148 | NM_001005871.1 |
| ACATGATCAGACCTGCCTTC | R | 57.64 | |||
| Atp2b4b | GAATAACAATGCCGTGGACT | F | 57.54 | 189 | NM_001005871.1 |
| GGAGCAGCTGATGAAACAAT | R | 57.89 |
RT-PCR for Alternative Splicing of PMCAs
To estimate the alternative splicing of PMCA isoforms the RT-PCR was performed. RNA aliquots of 15 µg or 10 µg were subjected to reverse transcription (RT) as described above. The obtained cDNA (10 µl) was used to assess PMCA alternative splicing at site A and at site C. PCR was performed using recombinant Taq DNA polymerase (Invitrogen, USA) and 2 µM primers (Table 2) designed as described [62]. The PCR product bands were visualized by electrophoresis in 1.5% agarose gel with 0.5 µg/ml ethidium bromide by UV transillumination.
Table 2. Primers for PMCA alternative splicing analysis.
| gene(accession number) | sequence (5′−3′) | Tm (°C) | strand | splice site | splice variant | PCR product(bp) |
| Atpb2b1 (NM_053311.1) | CTTACCTTACTTGGAGCTG | 55.00 | F | A | w | 504 |
| GTTGTTATCCTTCATCATTTTCTT | 57.00 | R | ||||
| ATCTTCTGCACAATTGTCTTAG | 57.00 | F | C | b | 358 | |
| GAGCTACGAATGCATTCACC | 58.00 | R | ||||
| Atpb2b2(NM_012508.5) | CTGTGGGTGTCAACTCTCAA | 58.00 | F | A | w, y, x, z | 348, 306, 255, 213 |
| GTGAGCTTGCCCTGAAGCA | 59.00 | R | ||||
| CATCTTCTGCACCATCGTTC | 58.00 | F | C | b, a, c | 417, 644, 472 | |
| AGCCATGAAGTTATGGATGGA | 57.00 | R | ||||
| Atpb2b3(NM_133288.1) | CATGTCATGGAAGGTTCTGG | 58.00 | F | A | y, x, z | 200, 521, 479 |
| GTTATTGTCCTTCATCATTTTCTT | 57.00 | R | ||||
| ATCTTCTGTACCATTGTCCTG | 57.00 | F | C | b, a, c, e, g | 203, 357, 290, 444, 424 | |
| GAGCTACGGAATGCTTTCAC | 58.00 | R | ||||
| Atpb2b4(NM_001005871.1) | GTGACTGCTGTGGGAATCAA | 58.00 | F | A | x, z | 524, 488 |
| GTTGTTGTCCTTCATCATTTTCTT | 57.00 | R | ||||
| TCTGCTCTGTTGTTTAGGCA | 56.00 | F | C | b, a | 343, 518 | |
| ATGAAATACTTTGACCACTCTG | 57.00 | R |
Total Cell Lysate Preparation and Immunoblotting
Cells were harvested and washed with PBS at room temperature. Cells were incubated for 40 min in an ice-cold lysis buffer (10 mM Tris-HCl, pH 7.5, 80 mM NaCl, 1% Triton X-100, 1 mM dithiothreitol, 1 mM PMSF, 10 mM NaF, 2 mM Na3VO4 and 10 µg/ml protein inhibitor cocktail, Sigma Aldrich). The lysates were centrifuged at 800×g for 5 min at 4°C and stored at −20°C. The protein content was determined by the Bradford assay. Total cell lysates were analyzed by SDS-PAGE and immunoblotting as described [60] using the antibodies described in table 2 (Table 2 and Table S1).
Luciferase Reporter Assay
Luciferase reporter plasmids with NFAT-dependent promoter (pGL3-NFAT-luc), Renilla luciferase control plasmids (pRL-SV40), promoter-less plasmid pGL3-luc and plasmid overexpressing NFAT (pNFAT+/+) were gifts from Dr. Wieslawa Lesniak from the Nencki Institute of Experimental Biology. PC12 cells (2×105) were transfected with X-tremeGENE Transfections Reagent (Roche Applied Science, Germany) with the following plasmidcombination: pGL3-NFAT-luc with pRL-SV40, pGL3-luc with pRL-SV40 (negative control), pNFAT+/+, with pGL3-NFAT-luc and with pRL-SV40 (positive control). Cells were harvested 48 h after transfection and lysed in lysis buffer (Thermo Scientific Pierce). Firefly and Renila luciferase activities were assayed with Pierce Renilla-Firefly Luciferase Dual Assay Kit (Thermo Scientific Pierce). The luminescent signal from Renilla luciferase was measured at λmax = 535 nm and that from firefly luciferase at λmax = 613 nm. The working solution contained substrates for both luciferases (coelenterazine and D-luciferin), and the reactions occurred simultaneously with flash-type kinetics. The luminescent signals were spectrally resolvable using filters. The activity of NFAT was determined based on the luminescence signal from firefly luciferase and standardized to the signal from Renilla luciferase. The luminescence emission was determined by SpectraMax M5e Microplate Reader (Molecular Devices, Sunnyvale, California, United States). The efficiency of transfection was verified by transfections with plasmids overexpressing EGFP and assessed by means of fluorescence microscopy to be 20%.
Immunoprecipitation
Cells were lysed in RIPA buffer and samples for immunoprecipitation were obtained with agarose beads with protein A/G (Santa Cruz, USA) as described [19]. Samples were incubated for 2 h at 4°C with mouse monoclonal anti-NFAT1 antibody (Abcam, USA) or with rabbit polyclonal anti-NFAT3 antibody (Cell Signaling, USA). The obtained immunoprecipitates were subjected to SDS-PAGE and immunoblotting with the following antibodies: mouse IgG1 anti-HDAC1 (10E2), mouse IgG1 anti-HDAC2 (3F3), mouse IgG1 anti-HDAC3 (7G6C5), rabbit polyclonal anti-HDAC4 (D15C3), rabbit polyclonal anti-HDAC5, rabbit polyclonal anti-HDAC6 (D2E5) (Cell Signaling, USA).
Chromatin Immunoprecipitation (ChIP)
PC12 cells (2×107) were cross-linked with 0.5% formaldehyde for 10 min at room temperature. Cross-linking was stopped by adding 125 mM glycine on ice. Cells were solubilized in a buffer containing 10 mM Tris-HCl (pH 8.0), 1% Triton X-100, 1% sodium deoxycholate, 1 mM PMSF and PIC for 10 min at 4°C. Pellets obtained by centrifugation at 1000×g for 5 min were suspended in RIPA buffer and sonicated using a Bioruptor Sonicator (Diagenode, Belgium) to shear chromatin into 500 bp fragments. Sonicated chromatin was subjected to immunoprecipitation using ChIP-grade agarose beads with protein G (Cell Signaling), blocked with 1% bovine albumin and 1% salmon sperm DNA. Then anti-NFAT1 and anti-NFAT3 antibodies were added and the obtained DNA-protein complexes were further complexed with anti-HDAC4 antibody (Cell Signaling). The obtained protein-DNA complexes were eluted with 100 mM sodium acetate and 1% SDS for 30 min and treated with RNase for 6 h at 65°C and proteinase K o/n at 45°C. DNA was isolated using the phenol/chloroform/isoamyl reagent (Sigma Aldrich, USA) and subjected to RT-PCR with primers used for analysis of the alternative splicing pattern of PMCA isoforms.
Statistical Analysis
All data are presented as means ± SEM of n observations. All data were analyzed by the two-sample paired Student’s t test at 95% or 99% confidence. Two-way ANOVA test was used for internal comparison between all experimental groups (3 cell lines) in terms of treatment with 1 µM 11R-VIVIT. For qPCR the nonparametric paired Wilcoxon signed rank test was used at 95% or 99% confidence.
Results
Altered Composition of PMCA Isoforms Influence Calcium Homeostasis
The goal of our study was to present changes exerted by stable suppression of PMCA2 or PMCA3 isoform. Thus, we used an in vitro cellular model with permanently downregulated PMCA2 and PMCA3 expression, which was validated in our several other studies [58]–[60]. At this point a comment should be added why the RNAi method was not appropriate regarding assumptions of this research. RNAi involves short-lived molecules and induces transient changes, thus the decay rate of any observed changes may vary considerably. In case of stable transfection we could control the level of PMCA isoforms and monitor long term effects of their suppression.
We have already shown that experimental reduction in the PMCA2 or PMCA3 content in PC12 cells caused a significant drop in the efficiency of calcium removal [59]. This data suggested that the control of [Ca2+]c in PC12 cells, especially the calcium clearance, was tightly dependent on the composition of PMCA isoforms, especially PMCA2 and PMCA3 isoforms, exhibiting the highest Ca2+ transport velocity and the highest affinity for Ca2+ [29]. The expression profile of PMCAs depends on a fine-tuned regulation by Ca2+-dependent molecular tools such as transcription factors or alternative splicing factors. Experimental downregulation of PMCA2 or PMCA3 created specific local calcium environment which was apparently important for the expression patter of the remained isoforms. Since we have observed an activation of Ca2+/calcineurin-dependent transcription factor NFAT, in the next stage of studies we have examined whether NFAT could be involved in determination of PMCA composition.
Overactivation of NFAT in the Cells with Reduced Content of PMCA2 or PMCA3
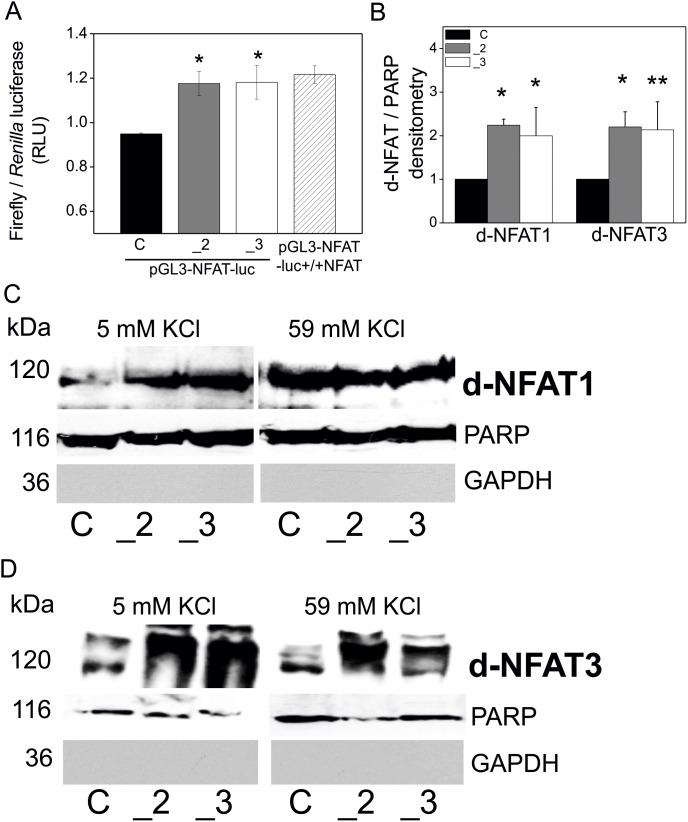
Luciferase reporter assays performed using constructs containing NFAT-dependent promoter revealed that NFAT transcriptional activity was significantly higher in the cells with a reduced content of PMCA2 or PMCA3. Moreover, this increase in NFAT activation was comparable to the cells overexpressing NFAT (transfected with constructs pGL3-NFAT-luc-+/+NFAT) (Fig. 1A). In addition, a statistically significant increase in the protein content of dephosphorylated NFAT1 and NFAT3 in the nuclei has been detected in PMCA2- and PMCA3-deficient cells, especially under resting conditions. This was demonstrated by densitometrical measurements of the immunoblots of NFAT1 and NFAT3, standardized to the content of nuclear poly (ADP-ribose) polymerase (PARP) and normalized to control cells (y = 1) (Fig. 1B). Increased level of dephosphorylated NFAT1 and NFAT3 in the nuclei in PMCA-deficient cells under resting conditions and in all cell types upon plasma membrane depolarization was shown by representative immunoblots (Fig. 1C and Fig. 1D). These findings concerning NFAT activity reinforce the hypothesis on the existence of a NFAT-PMCA regulatory loop.
Figure 1. NFAT activation in PC12 cells with reduce PMCAs content.
PC12 cells were transfected with plasmids encoding firefly luciferase under NFAT-dependent promoter (pGL3-NFAT-luc) and reference plasmids with Renilla luciferase (pRL-SV40). Negative controls were wild type PC12 cells transfected with promoterless pGL3-luc plasmids and positive controls were wild type PC12 transfected with plasmids overexpressing NFAT together with the pGL3-NFAT-luc (pGL3-NFAT-luc-NFAT+/+). NFAT activity was determined with a luciferase reporter dual assay (Thermo Scientific Pierce) and showed as the ratio of the luminescence signals derived from Firefly and Renilla luciferases. Bars represent mean values ± SEM. Symbols: control cells (C), PMCA2-deficient cells (_2), PMCA3-deficient cells (_3) and pGL3-NFAT-luc+/+NFAT – wild type cells overexpressing NFAT. Student’s t-test was used for comparison of control cells with PMCA2- or PMCA3-deficient cells. *P≤0.05, n = 5 (A). Nuclear content of dephosphorylated NFAT1 and NFAT3 was analyzed by immunoblotting. Protein bands were quantified densitometrically, standardized to PARP (nuclear marker) and normalized to control cells, expressed as y = 1. Bars represent mean values ± SEM. Student’s t-test was used for comparison of control cells with PMCA2- or PMCA3-deficient cells. *P≤0.05, n = 6 (B). Representative immunoblots of nuclear content of dephosphorylated NFAT1 (C) and NFAT3 (D) are demonstrated. Symbols correspond to: control cells (C), PMCA2-deficient cells (_2), PMCA3-deficient cells (_3).
PMCA Isoforms Expression Pattern upon Altered Transcriptional Control by NFAT
Following the above, the expression level of genes encoding PMCA isoforms: Atp21b1 (PMCA1), Atp21b2 (PMCA2), Atp21b3 (PMCA3), Atp21b4a (PMCA4a), Atp21b4b (PMCA4b) was determined by qPCR for PC12 cells non-treated (Fig. 2A), and compared with the cells treated with NFAT inhibitor (1 µM 11R-VIVIT) (Fig. 2B). On one hand, these experiments confirmed downregulation of PMCA2 or PMCA3 in respective cell lines. On the other hand, these experiments showed that inhibition of NFAT influenced significantly the expression pattern of PMCA4b as well as PMCA2 and PMCA3.
Figure 2. PMCA isoforms expression pattern versus transcriptional control by NFAT in PC12 cells.
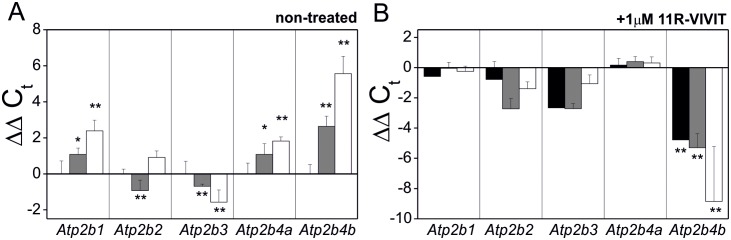
Expression of genes encoding PMCA isoforms: Atp21b1 (PMCA1), Atp21b2 (PMCA2), Atp21b3 (PMCA3), Atp21b4a (PMCA4a), Atp21b4b (PMCA4b) was determined by qPCR for PC12 cells non-treated (A), and treated with NFAT inhibitor (1 µM 11R-VIVIT) (B). Bars represent mean values ± SEM. Wilcoxon test was used for ΔCt from qPCR data (n = 3) for comparison between control cells (standardized to y = 0) and PMCA2- or PMCA3-deficient cells non-treated with 11R-VIVIT (n = 3). Two-way ANOVA test was used for comparison between non-treated and 11R-VIVIT treated cells. *P≤0.05, **P≤0.01.
Alternative Splicing of PMCAs Depends on Activity of NFAT in Splicing Regulatory Regions
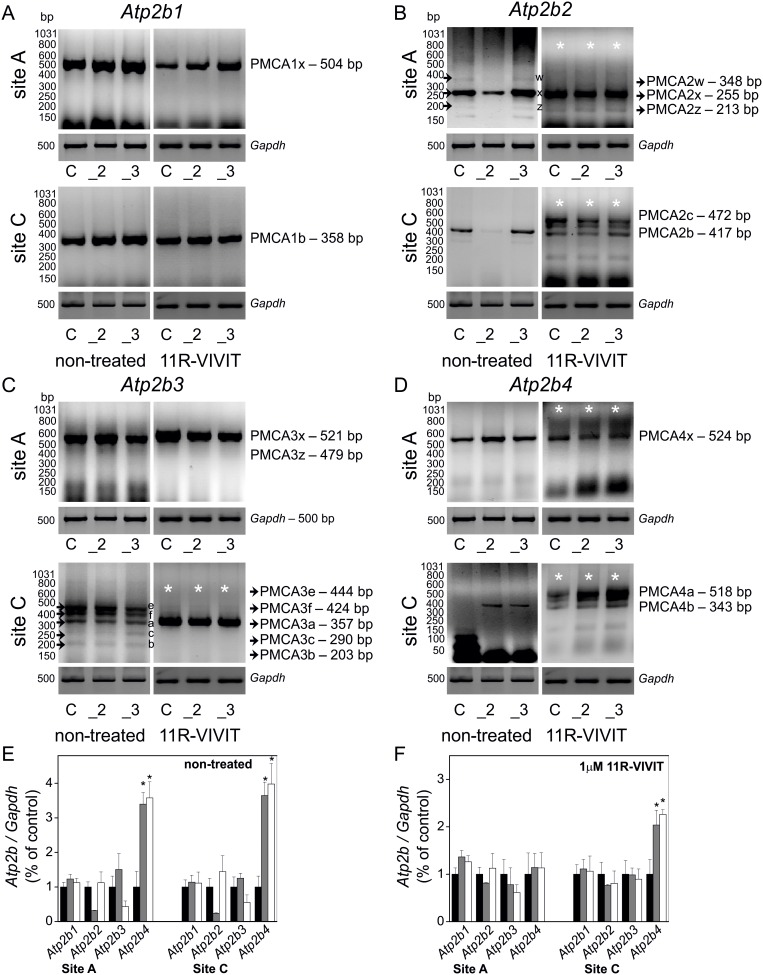
Diversity of PMCAs is not only due to the fact that these calcium pumps are encoded by four separate genes but mostly due to alternative splicing of mRNA. Thus, following the above findings suggesting increased NFAT activity and contribution to PMCAs expression profiling, a detailed analysis of the alternative splicing pattern of PMCA transcripts was performed. In order to obtain full information on the composition of PMCA splice variants the PCR method with primers flanking the appropriate splicing sites was applied. PMCA splicing variant composition was tested in control cells and in cells with a reduced content of neurospecific isoforms PMCA2 or PMCA3. This study was performed in order to verify whether a reduction in the content of neurospecific PMCAs might be compensated by other PMCA splicing variants. It revealed that downregulation of PMCA2 or PMCA3 did not statistically significantly influence the expression of PMCA1 (Atp2b1), and that the PMCA1x/b variant was abundantly and predominately expressed in PC12 cells (Fig. 3A, left). These outcomes were in accordance with the known data on PC12 cells [62], [63]. The expression of Atp2b1 transcript was in agreement with the protein level of PMCA1, detected with a specific antibody recognizing the PMCA1b form (Fig. S1A). In the case of Atp2b2 expression pattern, the PMCA2x/b variant predominated in all cell lines and its level was elevated in a compensatory mechanism in cells with a reduced amount of PMCA3 (Fig. 3B, left). This finding was verified at a protein level with a specific antibody recognizing the PMCA2b form (Fig. S1A). Regarding the expression pattern of Atp2b3, the most abundant variant was PMCA3x/a and in cells with a reduced amount of PMCA2 a compensatory increase in PMCA3x/a level was observed (Fig. 3C, left). PMCA3x/a transcript expression also correlated with the content of a protein recognized by specific anti-PMCA3a antibody (Fig. S1A). PMCA4x/b expression increased significantly in both PMCA2- and PMCA3-deficient cells (Fig. 3D, left). This was confirmed at a protein level by (Fig. S1A).
Figure 3. Alternative splicing of PMCA in PMCA2- or PMCA3-deficient PC12 cells upon NFAT inhibition.
Alternative splicing pattern at sites A and C of mRNA transcripts of PMCAs: Atp21b1 (PMCA1) (A), Atp21b2 (PMCA2) (B), Atp21b3 (PMCA3) (C), Atp21b4 (PMCA4) (D) was determined by RT-PCR in non-treated and 1 µM 11R-VIVIT-treated PC12 cells. RT-PCR product bands were quantified densitometrically, standardized to Gapdh and normalized to control cells, expressed as y = 1, both for non-treated (E) and 11R-VIVIT-treated cells (F). Student’s t-test was used for comparison of control cells with PMCA2- or PMCA3-reduced cells (n = 3). Bars represent mean values ± SEM. *P≤0.05. Symbols: control cells (C), PMCA2-deficient cells (_2), PMCA3-deficient cells (_3). Black arrows indicate the PCR product bands for PMCA2 site A and PMCA3 site C. White asterisks on the images of gels indicate the PCR product bands generated by alternative splicing that underwent a significant change upon NFAT inhibition with 11R-VIVIT.
Putative contribution of NFAT to the generation of PMCA splice variants was tested in PC12 cells incubated in the presence of NFAT inhibitor, 1 µM 11R-VIVIT for 48 hours. Upon this treatment the expression level of the PMCA1x/b variant did not significantly alter (Fig. 3A, right). In the case of PMCA2 splicing pattern, NFAT inhibition led to predominant expression of PMCA2x/c and PMCA2x/b, while the PMCA2z/b and PMCA2w/b variants, as well as PMCA2z/c and PMCA2w/c, were almost unchanged (Fig. 3B, right). Regarding the profile of PMCA3 splicing, a brain-specific variant PMCA3x/a predominated in all cell lines upon NFAT inhibition, while the expression of other splicing forms at site A (PMCA3e,f,c,b) was completely abolished (Fig. 3C, right). Finally, NFAT inhibition led to predominant expression of PMCA4x/a over the PMCA4x/b variant, which is a brain-specific variant exhibiting higher affinity for Ca2+ ions and better efficiency in Ca2+ removal (Fig. 3D, right). All RT-PCR data on alternative splicing pattern of PMCAs were quantified densitometrically. RT-PCR product bands were measure densitometrically, standardized to Gapdh and normalized to control cells, expressed as y = 1, both for non-treated (Fig. 3E) and 11R-VIVIT-treated cells (Fig. 3F). Taking into account the above results, it is very likely that the activity of NFAT is necessary during alternative splicing of PMCA. In particular, NFAT might be involved in the formation of PMCA2w,z, and PMCA3e,f,c,b splice variants. Bioinformatic analysis of the spliced regions in introns and UTR of genes coding for PMCAs revealed the presence of target motifs for NFAT (5′-TTTCCC-3′, and 5′-GGGAAA-3′). Based on the bioinformatic analysis and distribution of these motifs, it can be assumed that NFAT might bind to the regulatory splicing sequences alone or in complexes with other regulatory proteins.
Interaction of NFATs with HDACs in PC12 Cells
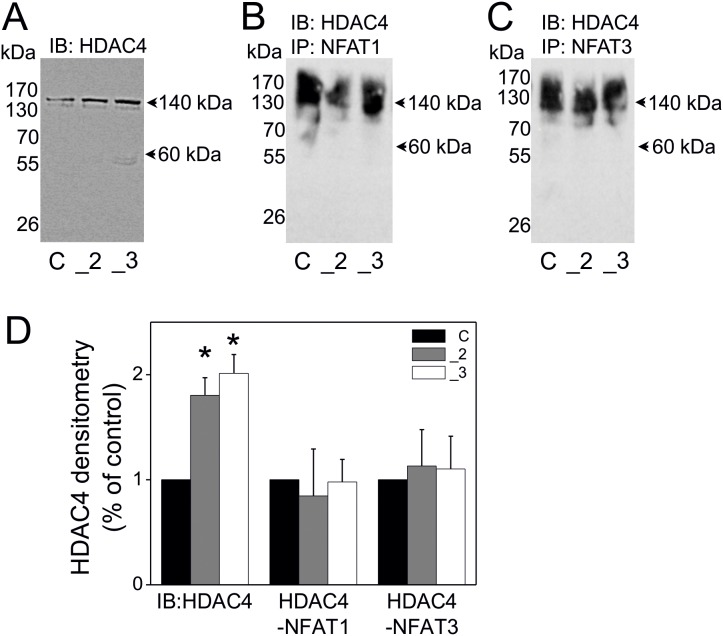
As suggested above NFAT might work alone or in complexes with other proteins [64]. NFATs were found to cooperate with HDACs, where NFAT1c mediated HDAC-dependent transcriptional repression [57]. Moreover, both NFATs and HDACs were found to be involved in regulation of alternative splicing [41], [42], [49]. To check whether NFAT cooperates with HDACs in PC12 cells with different PMCA status we first analyzed the presence of various HDACs in total cellular lysates obtained from these cells. This analysis revealed that HDAC4 was predominantly expressed in all examined PC12 cell lines (Fig. 4A). Densitometry analysis showed that in the PMCA2- and PMCA3-reduced cell lines the amount of HDAC4 was significantly higher than in control cells (Fig. 4D). We have tested as well HDAC1, HDAC2, HDAC3, HDAC5 and HDAC6 isoforms, however due to weak signal and very low or residual protein level of these isoforms, and thus, due to low importance these data are not shown in this paper. To study the putative interaction between NFAT1 or NFAT3 and HDAC4 the co-immunoprecipitation assays were performed. These experiments suggested that NFAT might interact with the HDAC4 isoform, both in the case of NFAT1 (ubiquitous) (Fig. 4B) and NFAT3 (neurospecific) (Fig. 4C). The content of immuneprecipitates was similar in all cell lines, as verified densitometrically and expressed as percentage of control cells (Fig. 4D).
Figure 4. The interaction of NFAT1 and NFAT3 with HDAC4 isoform in PMCA2- or PMCA3-deficient PC12 cells.
RIPA-total cellular extracts were subjected to immunoblotting to verify HDAC4 protein content and served as inputs of immunoprecipitation (A). The cellular extracts (inputs) were incubated with protein A/G agarose beads and with anti-NFAT1 antibody (B) or with anti-NFAT3 antibody (C) and the obtained immunoprecipitates were subjected to immunoblotting for HDAC4. All immunoblots and immunoprecipitates were measured densitometrically and expressed as % of control cells (D). Student’s t-test was used for comparison of control cells with PMCA2- or PMCA3- deficient cells. *P≤0.05, n = 3. Symbols: control cells (C), PMCA2-deficient cells (_2), PMCA3-deficient cells (_3).
Cooperation of NFAT with HDAC4 in Regulation of Alternative Splicing of PMCAs
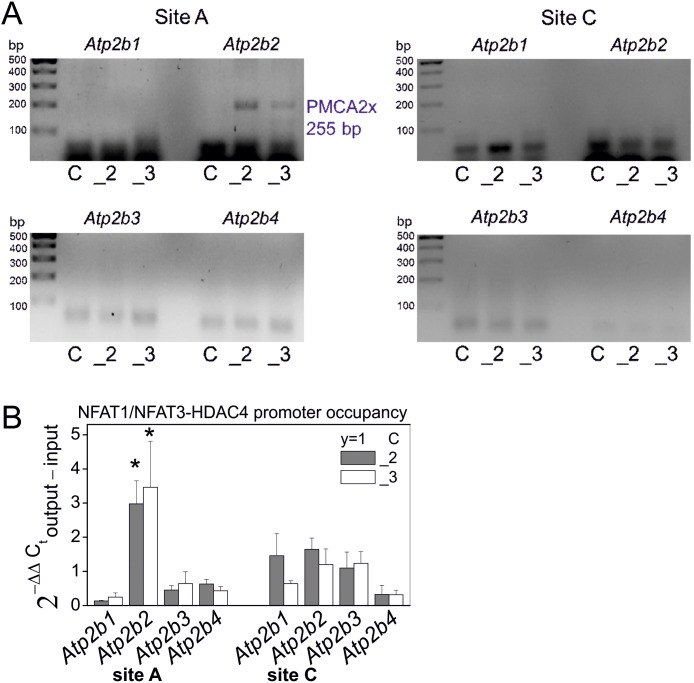
Regarding the protein interactions suggested above and formation of protein complexes consisting of NFAT1 and HDAC4 or NFAT3 and HDAC4, in the next step we examined whether these protein complexes may play a role in the regulation of alternative splicing of PMCAs. For that, binding of the NFAT1/NFAT3-HDAC4 complex to the splicing regulatory sites of PMCA isoforms has been studied with the use of chromatin immunoprecipitation technique with some modifications. Cross-linked chromatin and proteins were incubated with anti-NFAT1 and anti-NFAT3 antibodies. The DNA-protein complexes were further immunoprecipitated with anti-HDAC4 antibody. According to this protocol, generation of the PCR products of PMCA alternative splicing variants should be linked with the NFAT1/NFAT3-HDAC4 complex. Among all possible combinations of PMCA splicing variants only the PMCA2x splicing variant was detected under these conditions, suggesting that its generation might be related with HDAC4-NFAT1/NFAT3 binding to the splicing regulatory sites of the gene encoding PMCA2 (Fig. 5A), as shown in the supplementary material 2 (Fig. S2). The qPCR data, were expressed as fold of change (2−ΔΔC) calculated from the difference: ΔCT of output (immunoprecipitated DNA with HDAC/NFATs) – ΔCT of input (total DNA) and revealed that PMCA2x splicing variant generation was statistically significantly related to the NFAT1/NFAT3-HDAC4 complex activity, according to nonparametric paired Wilcoxon signed rank test at 95% confidence (Fig. 5B).
Figure 5. HDAC4-NFAT1/NFAT3 complex contribution to regulation of PMCA alternative splicing in PC12 cells.
HDAC/NFAT involvement in regulation of alternative splicing of PMCA transcripts was analyzed by qPCR-chromatin immunoprecipitation. The analysis was performed for the splicing at site A and at site for four PMCA isoforms and the PCR products were migrated in 1% agarose gels (A). The qPCR data, were expressed as fold of change (2−ΔΔC) calculated from the difference: ΔCT of output (immunoprecipitated DNA with HDAC/NFATs) – ΔCT of input (total DNA) and statistics were calculated according to nonparametric paired Wilcoxon signed rank test at 95% confidence, where PMCA2-deficient cells (_2) or PMCA3-deficient cells (_3) were compared to control cells assigned to y = 1 value (B). Symbols: control cells (C), PMCA2-deficient cells (_2), PMCA3-deficient cells (_3), n = 3.
Discussion
Our understanding of the regulation of the expression pattern of PMCA isoforms and their splicing variants remains incomplete. Only a few transcription factors involved in the regulation of the formation PMCA transcripts have been identified. It has been found that c-myc was able to repress PMCA4 expression during activation of B lymphocytes, which additionally resulted in decreased Ca2+ clearance, and interestingly, in an augmented NFAT level [65]. A similar research has been conducted in pancreatic β cells showing that c-myc expression has led to downregulation of PMCA1 and PMCA2 [66], [67]. Important data, showing that PMCA4 expression could be induced by NFAT1c in osteoclasts during the growth of bone mass, have been provided recently [43]. Furthermore, Kim et al have shown that PMCA-mediated increase in Ca2+ efflux prevented NFATc1 activation, forming a negative regulatory loop [43]. In our study we propose another novel role of NFATs. Namely, our results suggest that NFAT1 and NFAT3 activation may repress the expression of PMCA2, PMCA3 and PMCA4 isoforms, especially the PMCA4b variant. Moreover, we showed in our latest studies that overactivation of NFAT might be responsible for the repression of genes Vamp1 and Vamp2 in PC12 cells, thus indicating its role in regulation of the primary function of PC12, i.e. Ca2+-dependent secretory response [58]. Importantly, we showed that this coincided with a decrease in Ca2+ efflux, activating NFAT, and was accompanied by a dramatic arrest of the secretory machinery and a significant decrease in catecholaminesecretion. Summarizing, our data and the results of Kim et al suggest opposite NFAT functions and opposite interrelation between NFAT activity and PMCA expression. These contradictory scenarios are equally possible, especially during different physiological processes, such as growth of bone mass during osteoclast differentiation or catecholamine secretion by chromaffin tumor cells. Finally, it is worth to mention that NFAT has also been involved in regulation of the expression of other calcium transporters, such as voltage-dependent calcium channels (VDCC) [68], [69] or sodium-calcium exchanger (NCX) [70]. In general, downregulation or upregulation of PMCAs may be a common component of activating triggers in a wide variety of different cell types during various physiological process.
The biodiversity of PMCAs is based not only on the transcription of four independent genes but also relies on alternative splicing of the nascent pre-mRNA, transcribed from the DNA template wrapped on histones. Chromatin organization creates several points at which alternative splicing might be regulated, both at mRNA and chromatin level. Since our results suggested that NFAT was a repressor of PMCA transcription, we subsequently examined the possible role of NFAT in the regulation of alternative splicing of PMCAs in pheochromocytoma cells. We have undertaken these studies because of the recent evidence showing that alternative splicing occurs and is regulated co-transcriptionally [46]–[51] and because of the physiological importance of alternative splicing for the catecholamine secretion process. It remains undisputed that the functional variety of secretory proteins and calcium transporters depends largely on alternative splicing. For example, alternative splicing affects the formation of the exocytotic membrane fusion complex SNARE (SNAP (Soluble NSF Attachment Protein) Receptor) by altering the expression profile the following elements of this complex: synaptosomal-associated protein 25 (SNAP25), syntaxin 1 and synaptobrevin 1 [18]–[21]. Furthermore, there are several examples of alternative splicing of mRNA transcripts of several calcium transportersincluding PMCAs, responsible for calcium removal, as well as channels and ions exchangers responsible for calcium influx, i.e. VDCC [22], [23], [71], [72] and NCX [24], [25], [73]. The above examples underline the importance of the alternative splicing process in catecholamine secretion. Since in this report we propose NFAT as a transcriptional factor involved in splicing regulation, it is essential to discuss the relationship between transcription and alternative splicing. For instance, it has been shown that the process of alternative splicing depends on the gene promoter driving the transcription [74]–[77]. Moreover, alternative splicing might rely on the recruitment of transcription factors or co-activators to gene promoters [38], [39]. It is very likely that NFAT binding to the promoter regions of NFAT-regulated genes may alter alternative splicing mechanisms by trans-activation or trans-repression. Some RNA sequences that are called splicing recognition sites and are located in exons or introns, might stimulate or block splicing by binding specific splicing regulatory proteins such as serine/arginine rich proteins, snRNP or transcriptional factors [36], [78]–[80]. Based on our bioinformatic analysis and the relationship between inhibition of NFAT transcriptional activity and PMCA expression pattern, it is very likely that NFAT might bind to some splicing regulatory sequences. Moreover, NFAT has been found to bind not only DNA motifs [81], [82], but also to interact with some RNA sequences [83], [84], usually acting as NFAT repressors, like the non-coding RNA repressor of NFAT (NRON) [85]–[87]. It remains unclear which of the putative sequences in the PMCA coding genes that were showed in our in sillico analysis inhibit and which stimulate NFAT activity. Our study demonstrated the relationship between NFAT and PMCA expression pattern, based on experiments with inhibitory peptide 11R-VIVIT at physiological level. Distortion of this relationship results in disturbed calcium homeostasis and disturbed catecholamine secretion in pheochromocytoma. More studies with the use of the selected sequences are needed in the future to establish the exact regulatory sites for NFAT in PMCAs genes, regarding both regulation of expression and alternative splicing pattern.
Furthermore, in this report we found that NFAT might cooperate with histone deacetylases; more precisely we suggest an interaction of NFAT1 and NFAT3 with HDAC4. Our results are supported by many other data. For instance, it has been found that NFAT1 may cooperate with HDAC4, HDAC5 and HDAC7 in chromatin remodeling [57], [88], [89]. More precisely, it has been shown that NFAT1 might act with HDAC4 in a repressive complex during osteoblast differentiation [90]. Interestingly, both NFATs and HDACs have been found to be involved in the regulation of alternative splicing. NFAT1 has been suggested to participate in the regulation of alternative splicing of the allograft inflammatory factor-1 gene [41], and of synaptotagmin-like 2 gene during activation of murine T-cells [42]. HDACs have been proposed to contribute to the process of alternative splicing by regulating co-transcriptional spliceosome assembly [51]. Regarding the data provided in the literature on the functional interaction between NFATs and HDACs and given that both NFATs and HDACs are involved in the alternative splicing process, the next question addressed in this study concerned the role of this cooperation during alternative splicing of PMCAs. Our data suggest the contribution of the presumed complexes, NFAT1-HDAC4 and NFAT3-HDAC4 to alternative splicing of the mRNA transcript of PMCA2. More precisely, these complexes are supposed to enhance the generation of the 2x variant. Based on our bioinformatic analysis we suggest that the mentioned protein complexes might occupy some parts of the promoter region of the PMCA2 gene, and modulate the availability of some exons to alternative splicing. This is in accordance with the influence of some transcription factors on the formation of the nearby spliceosome. On the other hand, this might also suggest binding of the NFAT-HDAC complexes to the intronic splicing regulatory sites, preceding the excluded exon 7 and exon 8 but not to the intronic fragments of the gene preceding the included exon 9, resulting in the removal of exons 7 and 8 of the PMCA2 transcript leading to the formation of PMCA2x variant. The involvement of NFAT in this kind of splicing regulation is highly probable regarding our bioinformatic analysis showing numerous NFAT-specific motifs before exons 7 and 8, but not before exon 9 of the PMCA2 transcript. Generation of PMCA2x transcript with the contribution of NFAT1/NFAT3-HDAC4 complex could be a compensatory mechanism in response to experimental downregulation of PMCA2 or PMCA3 isoform.
It is worth to underline that the splicing recognition sites in the intronic/exonic regions of PMCAs genes were enriched in NFAT binding motifs (5′-GGAAA-3′ or 5′-TTTCCC-3′). PMCA2 and PMCA3 coding sequences contain relatively the highest number of NFAT-specific motifs, while those of PMCA1 contain a lower number of these motifs. Interestingly, NFAT binding elements were found in the proximity of the TATA box motifs (5′-TATAAA-3′), especially in the case of PMCA2 splicing regulatory regions before the spliced exons 7 and 8. This is similar to the interleukin-2 (IL-2) gene promoter [91] or COX-3 gene promoter [92]. Another example of NFAT connection with TATA box-proximal region has been given for the regulation of the promoter of the gene coding for p21 [93]. This data further reinforce the possible involvement of NFAT in regulation of the expression of PMCAs, especially PMCA2x.
Finally, an issue worth to be stressed is that an increased content of the endocrine-specific PMCA2x splicing form has been particularly observed upon deficiency of neurospecific variant PMCA3x. PMCA2x and PMCA3x variants exhibit the highest efficiency in calcium removal and the highest affinity for calcium ions among PMCA isoforms [64], thus it is obvious that a decrease in the content of one of these forms might be compensated by an up-regulation of the other. Such modification in PMCA composition probably serves to retrieve the affected calcium signaling and to rescue a disturbed secretory response in PC12 cells. Several examples of PMCA compensatory expression has been provided, including the compensatory expression of PMCA4 upon reduction of PMCA2 [94]. These data further support our results, because we have similarly observed an up-regulation of PMCA4b in cells with a reduced PMCA2 content, as well as in cells with a reduction in PMCA3 content. Interestingly, the expression of the PMCA4a variant was increased upon NFAT inhibition. This suggested a putative involvement of NFAT in maintaining the balance between expression of PMCA4b (upon NFAT activation) and PMCA4a (upon NFAT inhibition). This is in accordance with the results showing that NFAT might occupy the promoters and might be responsible for the control of the expression of PMCA1 and PMCA4 isoforms in osteoclasts [43]. Summarizing, all changes in PMCA composition were accompanied by activation of endogenous NFAT1 and NFAT3 suggesting their involvement in the control of PMCA expression pattern and, via interaction with HDAC4, in the control of alternative splicing of the PMCA2x variant. Thus, NFATs-HDACs complexes may play a role in the fine tuning of the process of catecholamine secretion via determination of the optimal expression profile of calcium transporters such as PMCAs.
Conclusions
The aim of this study was to verify whether the alternative splicing pattern of PMCAs in PC12 cells might be controlled by a protein complex composed of NFATs and HDACs. In this report we suggest that HDAC4 possibly operates in a complex with NFAT1 or with NFAT3 that might be involved in regulation of alternative splicing of PMCAs. This complex probably contributes to the generation of the splicing variant PMCA2x. Moreover, this was probably a compensatory response to altered composition of PMCAs, namely to experimental reduction of PMCA2 or PMCA3 isoform. Furthermore, this was linked with an altered calcium homeostasis. We propose that NFAT1 or NFAT3, in complexes with HDAC4, might occupy regions in the putative splicing regulatory sites in pre-mRNA of genes coding for selected PMCA isoforms.
Supporting Information
Protein level of PMCA1, PMCA2, PMCA3, PMCA4 in control, PMCA2- or PMCA3-deficient PC12 cells. Protein level was analyzed by immunoblotting as described in Materials and methos. 25 ug of cell lysates was used for the presented immunoblots for PMCA1, PMCA2, PMCA3, and 10 ug for PMCA4. Protein bands were quantified by densitometric analysis, i.e. the content of PMCAs was standardized to Δ-actin level in all cell lines and then normalized to control cells, expressed as y = 1. Student’s t-test was used for comparison of control cells with PMCA2- or PMCA3-reduced cells. *P≤0.05, **P≤0.01; n = 6. Symbols: control cells (C), PMCA2-deficient cells (_2), PMCA3-deficient cells (_3) (A). The antibodies used are presented in the table and the predicted PMCA forms, recognized by the antibodies, are marked as ‘expected gel band size’ (B).
(TIF)
Detection of NFAT-target sequences in spliced regions of exon/intron junctions of genes encoding PMCA1, PMCA2, PMCA3, PMCA4. Selected gene regions or selected single nucleotides in exon/intron frames of genes encoding PMCA1, PMCA2, PMCA3, PMCA4 were assigned as sites of recognition during alternative splicing according to ensemble database (http://www.ensembl.org). These regions were next analyzed in terms of the presence of NFAT-target sequences (TTTCCC and GGGAAA), as well as TATA box motifs (TATAAA).
(PDF)
Antibodies used in this study.
(DOCX)
Acknowledgments
The authors wish to thank to dr Wieslawa Lesniak from Laboratory of Calcium Binding Proteins of Department Of Molecular and Cellular Neurobiology at the Nencki Institute for fruitful discussion and suggestions.
Funding Statement
This work is supported by Grant No. 401 533340 from the Polish National Science Center. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Kim E, Goren A, Ast G (2008) Alternative splicing: current perspectives. Bioessays 30: 38–47. [DOI] [PubMed] [Google Scholar]
- 2.Chen L, Tovar-Corona JM, Urrutia AO (2012) Alternative splicing: a potential source of functional innovation in the eukaryotic genome. Int J Evol Biol doi:10.1155/2012/596274. [DOI] [PMC free article] [PubMed]
- 3. Lee CJ, Irizarry K (2003) Alternative splicing in the nervous system: an emerging source of diversity and regulation. Biol Psychiatry 54: 771–776. [DOI] [PubMed] [Google Scholar]
- 4. Calarco JA, Superina S, O’Hanlon D, Gabut M, Raj B, et al. (2009) Regulation of vertebrate nervous system alternative splicing and development by an SR-related protein. Cell 138: 898–910. [DOI] [PubMed] [Google Scholar]
- 5. Grabowski PJ, Black DL (2001) Alternative RNA splicing in the nervous system. Prog Neurobiol 65: 289–308. [DOI] [PubMed] [Google Scholar]
- 6. Xie J (2008) Control of alternative pre-mRNA splicing by Ca2+ signals. Biochim Biophys Acta 1779: 438–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Li Q, Lee JA, Black DL (2007) Neuronal regulation of alternative pre-mRNA splicing. Nat Rev Neurosci 8: 819–831. [DOI] [PubMed] [Google Scholar]
- 8. Strehler EE, Zacharias DA (2001) Role of alternative splicing in generating isoform diversity among plasma membrane calcium pumps. Physiol Rev 81: 21–50. [DOI] [PubMed] [Google Scholar]
- 9. Domi T, Di Leva F, Fedrizzi L, Rimessi A, Brini M (2007) Functional specificity of PMCA isoforms? Ann N Y Acad Sci 1099: 237–246. [DOI] [PubMed] [Google Scholar]
- 10. Brini M, Carafoli E (2009) Calcium pumps in health and disease. Physiol Rev 89: 1341–1378. [DOI] [PubMed] [Google Scholar]
- 11. Kornblihtt AR (2005) Promoter usage and alternative splicing. Curr Opin Cell Biol 17: 262–268. [DOI] [PubMed] [Google Scholar]
- 12. Licatalosi DD, Darnell RB (2006) Splicing regulation in neurologic disease. Neuron 52: 93–101. [DOI] [PubMed] [Google Scholar]
- 13. Grabowski P (2011) Alternative splicing takes shape during neuronal development. Curr Opin Genet Dev 21: 388–394. [DOI] [PubMed] [Google Scholar]
- 14. Waguespack SG, Rich T, Grubbs E, Ying AK, Perrier ND, et al. (2010) A current review of the etiology, diagnosis, and treatment of pediatric pheochromocytoma and paraganglioma. J Clin Endocrinol Metab 95: 2023–2037. [DOI] [PubMed] [Google Scholar]
- 15. García AG, García-De-Diego AM, Gandía L, Borges R, García-Sancho J (2006) Calcium signaling and exocytosis in adrenal chromaffin cells. Physiol Rev 86: 1093–1131. [DOI] [PubMed] [Google Scholar]
- 16. Unsicker K (1993) The chromaffin cell: paradigm in cell, developmental and growth factor biology. J Anat 183: 207–221. [PMC free article] [PubMed] [Google Scholar]
- 17.Südhof TC, Rizo J (2011) Synaptic vesicle exocytosis. Cold Spring Harb Perspect Biol 3: pii: a005637. doi:10.1101/cshperspect.a005637. [DOI] [PMC free article] [PubMed]
- 18. Nagy G, Milosevic I, Fasshauer D, Müller EM, de Groot BL, et al. (2005) Alternative splicing of SNAP-25 regulates secretion through nonconservative substitutions in SNARE domain. Mol Biol Cell 16: 5675–5685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Nakayama T, Kamiguchi H, Akagawa K (2012) Syntaxin 1C, a soluble form of syntaxin, attenuates membrane recycling by destabilizing microtubules. J Cell Sci 125: 817–830. [DOI] [PubMed] [Google Scholar]
- 20. Berglund L, Hoffmann HJ, Dahl R, Petersen TE (1996) VAMP-1 has a highly variable C-terminus generated by alternative splicing, Biochem. Biophys Res Commun 264: 777–780. [DOI] [PubMed] [Google Scholar]
- 21. Isenmann S, Khew-Goodall Z, Gamble J, Vadas M, Wattenberg BW (1998) A splice-isoform of vesicle-associated membrane protein-1 (VAMP-1) contains a mitochondrial targeting signal. Mol Biol Cell 9: 1649–1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Asadi S, Javan M, Ahmadiani A, Sanati MH (2009) Alternative splicing in the synaptic protein interaction site of rat Ca(v)2.2 (alpha (1B)) calcium channels: changes induced by chronic inflammatory pain. J Mol Neurosci 39: 40–48. [DOI] [PubMed] [Google Scholar]
- 23. Lipscombe D, Andrade A, Allen SE (2013) Alternative splicing: functional diversity among voltage-gated calcium channels and behavioral consequences. Biochim Biophys Acta 1828: 1522–1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Van Eylen F, Kamagate A, Herchuelz A (2001) A new Na/Ca exchanger splicing pattern identified in situ leads to a functionally active 70 kDa NH(2)-terminal protein, Cell Calcium. 30: 191–198. [DOI] [PubMed] [Google Scholar]
- 25. Quednau BD, Nicoll DA, Philipson KD (1997) Tissue specificity and alternative splicing of the Na+/Ca2+ exchanger isoforms NCX1, NCX2, and NCX3 in rat. Am J Physiol 272: C1250–C1261. [DOI] [PubMed] [Google Scholar]
- 26. Duman JG, Chen L, Hille B (2008) Calcium transport mechanisms of PC12 cells. J Gen Physiol 131: 307–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Talarico EF, Mangini NJ (2007) Alternative splice variants of plasma membrane calcium-ATPases in human corneal epithelium. Exp Eye Res 85: 869–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Oceandy D, Stanley JP, Cartwright EJ, Neyses L (2007) The regulatory function of plasma-membrane Ca2+-ATPase (PMCA) in the heart. Biochem Soc Trans 35: 927–930. [DOI] [PubMed] [Google Scholar]
- 29. Zaidi A (2010) Plasma membrane Ca2+-ATPases: Targets of oxidative stress in brain aging and neurodegeneration. World J Biol Chem 1: 271–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Abramowitz J, Aydemir-Koksoy A, Helgason T, Jemelka S, Odebunmi T, et al. (2000) Expression of plasma membrane calcium ATPases in phenotypically distinct canine vascular smooth muscle cells. J Mol Cell Cardiol 32: 777–789. [DOI] [PubMed] [Google Scholar]
- 31. Enyedi A, Strehler EE (2011) Regulation of apical membrane enrichment and retention of plasma membrane Ca2+-ATPase splice variants by the PDZ-domain protein NHERF2. Commun Integr Biol 4: 340–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Waks Z, Klein AM, Silver PA (2000) Cell-to-cell variability of alternative RNA splicing. Mol Syst Biol 5: 506 doi:10.1038/msb.2011.32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kelemen O, Convertini P, Zhang Z, Wen Y, Shen M, et al. (2013) Function of alternative splicing. Gene 514: 1–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Garg K, Green P (2007) Differing patterns of selection in alternative and constitutive splice sites. Genome Res 17: 1015–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Will CL, Lührmann R (2011) Spliceosome structure and function, Cold Spring Harb Perspect Biol 3: pii: a003707. doi:10.1101/cshperspect.a003707. [DOI] [PMC free article] [PubMed]
- 36. Chasin LA (2007) Searching for splicing motifs. Adv Exp Med Biol 623: 85–106. [DOI] [PubMed] [Google Scholar]
- 37. Ling JQ, Hoffman AR (2007) Epigenetics of long-range chromatin interactions. Pediatr Res 61: 11R–16R. [DOI] [PubMed] [Google Scholar]
- 38. Auboeuf D, Dowhan DH, Li X, Larkin K, Ko L, et al. (2004) CoAA, a nuclear receptor coactivator protein at the interface of transcriptional coactivation and RNA splicing. Mol Cell Biol 24: 442–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Nogues G, Kadener S, Cramer P, Bentley D, Kornblihtt AR (2002) Transcriptional activators differ in their abilities to control alternative splicing. J Biol Chem 277: 43110–43114. [DOI] [PubMed] [Google Scholar]
- 40. Misteli T, Spector DL (1999) RNA polymerase II targets pre-mRNA splicing factors to transcription sites in vivo. Mol Cell 3: 697–705. [DOI] [PubMed] [Google Scholar]
- 41. Nilsson LM, Xing C, Bengtsson JME, Nilsson-Öhman J, Lydrup ML, et al. (2008) NFAT regulates the alternative splicing of Allograft Inflammatory Factor-1 gene: Role in neointima formation. FASEB J 22: 49. [Google Scholar]
- 42. Mascarell L, Auger R, Kanellopoulos JM, Truffa-Bachi P (2006) The usage of alternative splice sites in Mus musculus synaptotagmin-like 2 gene is modulated by cyclosporin A and FK506 in T-lymphocytes. Mol Immunol 43: 1846–1854. [DOI] [PubMed] [Google Scholar]
- 43. Kim HJ, Prasad V, Hyung SW, Lee ZH, Lee SW, et al. (2012) Plasma membrane calcium ATPase regulates bone mass by fine-tuning osteoclast differentiation and survival. J Cell Biol 199: 1145–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Venables JP, Tazi J, Juge F (2012) Regulated functional alternative splicing in Drosophila. Nucleic Acids Res 40: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Allen SE, Darnell RB, Lipscombe D (2010) The neuronal splicing factor Nova controls alternative splicing in N-type and P-type CaV2 calcium channels, Channels. 4: 483–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Pandya-Jones A, Black DL (2009) Co-transcriptional splicing of constitutive and alternative exons, RNA. 15: 1896–1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kornblihtt AR, de la Mata M, Fededa JP, Munoz MJ, Nogues G (2004) Multiple links between transcription and splicing. RNA 10: 1489–1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Girard C, Will CL, Peng J, Makarov EM, Kastner B, et al. (2012) Post-transcriptional spliceosomes are retained in nuclear speckles until splicing completion. Nat Commun 3: 994. [DOI] [PubMed] [Google Scholar]
- 49. Luco RF, Pan Q, Tominaga K, Blencowe BJ, Pereira-Smith OM, et al. (2010) Regulation of Alternative Splicing by Histone Modifications. Science 327: 996–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Sims RJ, Millhouse S, Chen CF, Lewis BA, Erdjument-Bromage H, et al. (2007) Recognition of trimethylated histone h3 lysine 4 facilitates the recruitment of transcription postinitiation factors and pre-mRNA splicing. Mol Cell 28: 665–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Gunderson FQ, Johnson TL (2009) Acetylation by the Transcriptional Coactivator Gcn5 Plays a Novel Role in Co-Transcriptional Spliceosome Assembly. Plos Genet 5: e1000682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Santoro M, Piacentini R, Masciullo M, Bianchi ML, Modoni A, et al.. (2013) Alternative splicing alterations of Ca2+ handling genes are associated with Ca2+ signal dysregulation in DM1 and DM2 myotubes. Neuropathol Appl Neurobiol 29: doi:10.1111/nan.12076. [DOI] [PubMed]
- 53. Loomis RJ, Naoe Y, Parker JB, Savic V, Bozovsky MR, et al. (2009) Chromatin binding of SRp20 and ASF/SF2 and dissociation from mitotic chromosomes is modulated by histone H3 serine 10 phosphorylation. Mol Cell 33: 450–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Delcuve GP, Khan DH, Davie JR (2012) Roles of histone deacetylases in epigenetic regulation: emerging paradigms from studies with inhibitors. Clin Epigen 12: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Shen T, Liu Y, Randall WR, Schneider MF (2006) Parallel mechanisms for resting nucleo-cytoplasmic shuttling and activity dependent translocation provide dual control of transcriptional regulators HDAC and NFAT in skeletal muscle fiber type plasticity. J Muscle Res Cell Motil 27: 405–411. [DOI] [PubMed] [Google Scholar]
- 56.Kee HJ, Kook H (2011) Roles and targets of class I and IIa histone deacetylases in cardiac hypertrophy. J Biomed Biotechnol 928326: doi:10.1155/2011/928326. [DOI] [PMC free article] [PubMed]
- 57. Choo MK, Yeo H, Zayzafoon M (2009) NFATc1 mediates HDAC-dependent transcriptional repression of osteocalcin expression during osteoblast differentiation, Bone. 45: 579–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Kosiorek M, Zylinska L, Zablocki K, Pikula S (2014) Calcineurin/NFAT Signaling Represses Genes Vamp1 and Vamp2 via PMCA-Dependent Mechanism during Dopamine Secretion by Pheochromocytoma Cells. PloSOne 9: e92176 doi:10.1371/journal.pone.0092176. eCollection 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Zylinska L, Kozaczuk A, Szemraj J, Kargas C, Kowalska I (2007) Functional importance of PMCA isoforms in growth and development of PC12 cells. Ann N Y Acad Sci 1099: 254–269. [DOI] [PubMed] [Google Scholar]
- 60. Kosiorek M, Podszywalow-Bartnicka P, Zylinska L, Zablocki Z, Pikula S (2011) Interaction of plasma membrane Ca2+-ATPase isoform 4 with calcineurin A: implications for catecholamine secretion by PC12 cells. Biochem Biophys Res Commun 411: 235–240. [DOI] [PubMed] [Google Scholar]
- 61. Yuan JS, Reed A, Chen F, Stewart CN (2006) Statistical analysis of real-time PCR data, BMC Bioinformatics. 7: 85–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Kamagate A, Herchuelz A, Bollen A, Van Eylen F (2000) Expression of multiple plasma membrane Ca2+-ATPases in rat pancreatic islet cells. Cell Calcium 27: 231–246. [DOI] [PubMed] [Google Scholar]
- 63. Carafoli E, Brini M (2000) Calcium pumps: structural basis for and mechanism of calcium transmembrane transport. Curr Opin Chem Biol 4: 1520–1561. [DOI] [PubMed] [Google Scholar]
- 64. Holton ML, Wang W, Emerson M, Neyses L, Armesilla AL (2010) Plasma membrane calcium ATPase proteins as novel regulators of signal transduction pathways. World J Biol Chem 1: 201–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Park H, Tsang M, de Alboran IM, Nicks A, Wilson L, et al. (2007) Myc stimulates B lymphocyte differentiation and amplifies calcium signaling. J Cell Biol 179: 717–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Jonas JC, Laybutt DR, Steil GM, Trivedi N, Pertusa JG, et al. (2001) High glucose stimulates early response gene c-Myc expression in rat pancreatic beta cells. J Biol Chem 276: 35375–35381. [DOI] [PubMed] [Google Scholar]
- 67. Ximenes HM, Kamagate A, Van Eylen F, Carpinelli A, Herchuelz A (2003) Opposite effects of glucose on plasma membrane Ca2+-ATPase and Na/Ca exchanger transcription, expression, and activity in rat pancreatic beta-cells. J Biol Chem 278: 22956–22963. [DOI] [PubMed] [Google Scholar]
- 68. Chiang CS, Huang CH, Chieng H, Chang YT, Chang D, et al. (2009) The Ca(v)3.2 T-type Ca2+ channel is required for pressure overload-induced cardiac hypertrophy in mice. Circ Res 104: 522–530. [DOI] [PubMed] [Google Scholar]
- 69. Cano E, Canellada A, Minami T, Iglesias T, Redondo JM (2005) Depolarization of neural cells induces transcription of the Down syndrome critical region 1 isoform 4 via a calcineurin/nuclear factor of activated T cells-dependent pathway. J Biol Chem 280: 29435–29443. [DOI] [PubMed] [Google Scholar]
- 70. Desai-Shah M, Cooper RL (2009) Different mechanisms of Ca2+ regulation that influence synaptic transmission: comparison between crayfish and Drosophila neuromuscular junctions. Synapse 63: 1100–1121. [DOI] [PubMed] [Google Scholar]
- 71. Santoro M, Masciullo M, Bonvissuto D, Bianchi ML, Michetti F, et al. (2013) Alternative splicing of human insulin receptor gene (INSR) in type I and type II skeletal muscle fibers of patients with myotonic dystrophy type 1 and type 2. Mol Cell Biochem 380: 259–265. [DOI] [PubMed] [Google Scholar]
- 72. Liao P, Zhang HY, Soong TW (2009) Alternative splicing of voltage-gated calcium channels: from molecular biology to disease. Pflugers Arch 458: 481–487. [DOI] [PubMed] [Google Scholar]
- 73. Shubair M, Oriowo MA, Khan I (2012) Expression of alternatively spliced variants of Na-Ca-exchanger-1 in experimental colitis: role in reduced colonic contractility. Mol Cell Biochem 370: 15–21. [DOI] [PubMed] [Google Scholar]
- 74. Cramer P, Caceres JF, Cazalla D, Kadener S, Muro AF, et al. (1999) Coupling of transcription with alternative splicing: RNA pol II promoters modulate SF2/ASF and 9G8 effects on an exonic splicing enhancer. Mol Cell 4: 251–258. [DOI] [PubMed] [Google Scholar]
- 75. Cramer P, Pesce CG, Baralle FE, Kornblihtt AR (1997) Functional association between promoter structure and transcript alternative splicing. Proc Natl Acad Sci U S A 94: 11456–11460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Pagani F, Stuani C, Zuccato E, Kornblihtt AR, Baralle FE (2008) Promoter architecture modulates CFTR exon 9 skipping. J Biol Chem 278: 1511–1517. [DOI] [PubMed] [Google Scholar]
- 77. Chasin LA (2007) Searching for splicing motifs. Adv Exp Med Biol 623: 85–106. [DOI] [PubMed] [Google Scholar]
- 78. Long JC, Caceres JF (2009) The SR protein family of splicing factors: master regulators of gene expression. Biochem J 417: 15–27. [DOI] [PubMed] [Google Scholar]
- 79. Han SP, Tang YH, Smith R (2010) Functional diversity of the hnRNPs: past, present and perspectives. Biochem J 430: 379–392. [DOI] [PubMed] [Google Scholar]
- 80. Bonnal S, Vigevani L, Valcárcel J (2012) The spliceosome as a target of novel antitumour drugs. Nat Rev Drug Discov 11: 847–859. [DOI] [PubMed] [Google Scholar]
- 81. Reddy AS (2007) Alternative splicing of pre-messenger RNAs in plants in the genomic era. Annu Rev Plant Biol 58: 267–294. [DOI] [PubMed] [Google Scholar]
- 82. Badran BM, Wolinsky SM, Burny A, Willard-Gallo KE (2002) Identification of three NFAT binding motifs in the 5′-upstream region of the human CD3gamma gene that differentially bind NFATc1, NFATc2, and NF-kappa B p50. J Biol Chem 277: 47136–47148. [DOI] [PubMed] [Google Scholar]
- 83. Barrandon C, Spiluttini B, Bensaude O (2008) Non-coding RNAs regulating the transcriptional machinery. Biol Cell 100: 83–95. [DOI] [PubMed] [Google Scholar]
- 84. Willingham AT, Orth AP, Batalov S, Peters EC, Wen BG, et al. (2005) A strategy for probing the function of noncoding RNAs finds a repressor of NFAT. Science 309: 1570–1573. [DOI] [PubMed] [Google Scholar]
- 85. Langland JO, Kao PN, Jacobs BL (1999) Nuclear factor-90 of activated T-cells: A double-stranded RNA-binding protein and substrate for the double-stranded RNA-dependent protein kinase, PKR. Biochemistry 38: 6361–6368. [DOI] [PubMed] [Google Scholar]
- 86. Mercer TR, Mattick JS (2013) Structure and function of long noncoding RNAs in epigenetic regulation. Nat Struct Mol Biol 20: 300–307. [DOI] [PubMed] [Google Scholar]
- 87. Im SH, Rao A (2004) Activation and deactivation of gene expression by Ca2+/calcineurin-NFAT-mediated signaling. Mol Cell 18: 1–9. [PubMed] [Google Scholar]
- 88.Kee HJ, Kook H (2011) Roles and targets of class I and IIa histone deacetylases in cardiac hypertrophy. J Biomed Biotechnol e928326. [DOI] [PMC free article] [PubMed]
- 89. Ranger AM, Gerstenfeld LC, Wang J, Kon T, Bae H, et al. (2000) The nuclear factor of activated T cells (NFAT) transcription factor NFATp (NFATc2) is a repressor of chondrogenesis. J Exp Med 191: 9–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Walters RD, Drullinger LF, Kugel JF, Goodrich JA (2013) NFATc2 recruits cJun homodimers to an NFAT site to synergistically activate interleukin-2 transcription. Mol Immunol 56: 48–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Abraham F, Sacerdoti F, De León R, Gentile T, Canellada A (2012) Angiotensin II activates the calcineurin/NFAT signaling pathway and induces cyclooxygenase-2 expression in rat endometrial stromal cells. PLoS One 7: e37750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Mammucari C, Tommasi di Vignano A, Sharov AA, Neilson J, Havrda MC, et al. (2005) Integration of Notch 1 and calcineurin/NFAT signaling pathways in keratinocyte growth and differentiation control. Dev Cell 8: 665–676. [DOI] [PubMed] [Google Scholar]
- 93. Di Leva F, Domi T, Fedrizzi L, Lim D, Carafoli E T (2008) he plasma membrane Ca2+ ATPase of animal cells: structure, function and regulation. Arch Biochem Biophys 476: 65–74. [DOI] [PubMed] [Google Scholar]
- 94. Hogan PG, Chen L, Nardone J, Rao A (2003) Transcriptional regulation by calcium, calcineurin, and NFAT. Genes Dev 17: 2205–2232. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Protein level of PMCA1, PMCA2, PMCA3, PMCA4 in control, PMCA2- or PMCA3-deficient PC12 cells. Protein level was analyzed by immunoblotting as described in Materials and methos. 25 ug of cell lysates was used for the presented immunoblots for PMCA1, PMCA2, PMCA3, and 10 ug for PMCA4. Protein bands were quantified by densitometric analysis, i.e. the content of PMCAs was standardized to Δ-actin level in all cell lines and then normalized to control cells, expressed as y = 1. Student’s t-test was used for comparison of control cells with PMCA2- or PMCA3-reduced cells. *P≤0.05, **P≤0.01; n = 6. Symbols: control cells (C), PMCA2-deficient cells (_2), PMCA3-deficient cells (_3) (A). The antibodies used are presented in the table and the predicted PMCA forms, recognized by the antibodies, are marked as ‘expected gel band size’ (B).
(TIF)
Detection of NFAT-target sequences in spliced regions of exon/intron junctions of genes encoding PMCA1, PMCA2, PMCA3, PMCA4. Selected gene regions or selected single nucleotides in exon/intron frames of genes encoding PMCA1, PMCA2, PMCA3, PMCA4 were assigned as sites of recognition during alternative splicing according to ensemble database (http://www.ensembl.org). These regions were next analyzed in terms of the presence of NFAT-target sequences (TTTCCC and GGGAAA), as well as TATA box motifs (TATAAA).
(PDF)
Antibodies used in this study.
(DOCX)