Abstract
Osteogenesis Imperfecta (OI) is a heritable disorder of connective tissue characterized by brittle bones, fractures and extraskeletal manifestations1. How structural mutations of type I collagen (dominant OI) or of its post-translational modification machinery (recessive OI) can cause abnormal quality and quantity of bone is poorly understood. Notably, the clinical overlap between dominant and recessive forms of OI suggests common molecular pathomechanisms2. Here, we show that excessive transforming growth factor-beta (TGFβ) signaling is a mechanism of OI in both recessive (Crtap−/−) and dominant (Col1a2tm1.1Mcbr) OI mouse models. In the skeleton, we find higher expression of TGFβ target genes, ratio of pSmad2/Smad2 protein, and in vivo Smad2 reporter activity. Anti-TGFβ treatment using the neutralizing antibody 1D11 corrects the bone phenotype in both forms of OI, and improves the lung abnormalities in Crtap−/− mice. Moreover, type I collagen of Crtap−/− mice shows reduced binding to the small leucine rich proteoglycan decorin, a known regulator of TGFβ activity3–4. Hence, altered TGFβ matrix-cell signaling is a primary mechanism in the pathogenesis of OI, and could be a promising target for the treatment of OI.
Most cases of OI are caused by autosomal dominant mutations in the genes encoding type I collagen (COL1A1 and COL1A2)1. In recent years, mutations in additional genes encoding the proteins involved in post-translational collagen modification have been identified as causing recessive forms. The first described was in cartilage associated protein (CRTAP), a member of the prolyl-3-hydroxylase complex that is responsible for 3-hydroxylation of proline residue 986 of α1(I) in type I collagen2,5. Hypomorphic CRTAP mutations lead to partial loss of 3-hydroxyproline (3Hyp) in fibrillar collagen, overmodification of other residues and result in recessive OI type VII, which clinically overlaps with dominant forms2. The physiological function of 3Hyp is incompletely understood, but biochemical and genetic studies suggest that it is involved in collagen-protein interactions and required for normal bone mineralization6–7.
The extracellular matrix (ECM) is an important reservoir for signaling molecules and their regulators. In bone, TGFβ acts as a central coordinator of bone remodeling by coupling the activity of bone resorbing osteoclasts and bone forming osteoblasts8. TGFβ is produced by osteoblasts9, secreted predominantly as inactive latent forms10, and deposited into the bone matrix11. Here, it can be released and activated during bone resorption by osteoclasts12. As an additional level of regulation, active TGFβ can be bound by proteoglycans13, which modulate its bioactivity4 in association with collagen fibrils3. Because type I collagen is the most abundant component of the ECM in bone, we hypothesized that alterations of collagen observed in OI can affect the signaling modulating function of the bone matrix. Consistent with this, Crtap−/− mice show phenotypic overlap with animal models of upregulated TGFβ signaling14–16. For example, TGFβ overexpression results in low bone mass14. In addition, Crtap−/− mice exhibit an enlargement in alveolar airway space in lungs, similar to that observed in a model of Marfan syndrome, where dysregulated TGFβ signaling contributes to the lung pathology15. Therefore, we first studied the status of TGFβ signaling in the Crtap−/− mouse model of recessive OI.
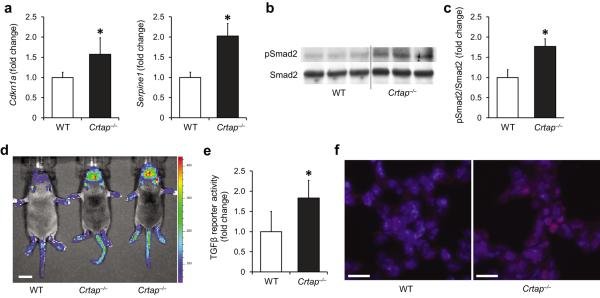
Compared with wild type (WT) samples, calvarial bone of Crtap−/− mice showed a higher expression of the TGFβ targets Cdkn1a (cyclin-dependent kinase inhibitor 1a, P21) and Serpine1 (plasminogen activator inhibitor-1), consistent with elevated TGFβ activity (Fig. 1a). To confirm activation of the intracellular TGFβ signaling pathway, we evaluated the status of Smad2, a second messenger protein, which becomes phosphorylated after activation of TGFβ receptors. Consistently, immunoblot analyses demonstrated a greater ratio of phosphorylated Smad2 (pSmad2) to total Smad2 in calvarial bone samples of Crtap−/− compared with WT mice, indicating higher TGFβ signaling (Fig. 1b,c). To confirm higher TGFβ activity in vivo, we intercrossed Crtap−/− mice with reporter mice expressing luciferase in response to TGFβ (SBE-luc mice). Compared with WT/SBE-luc littermates, Crtap−/−/SBE-luc mice showed a more intense bioluminescence of areas over skeletal structures, indicating higher TGFβ activity in vivo (Fig. 1d; in 3 litters Crtap−/− mice show a mean 2.86 fold (SD±0.34) bioluminescence signal at the head/calvaria compared with WT mice). To test whether the higher TGFβ signaling is intrinsic to bone, i.e. tissue autonomous, we cultured bone marrow stromal cells (BMSCs) under osteogenic conditions in vitro. By using a TGFβ reporter cell line, we found that conditioned medium from Crtap−/− BMSCs exhibited greater TGFβ activity compared with medium from WT BMSCs (Fig. 1e). Together, these findings indicate that loss of Crtap enhances TGFβ signaling in bone in a tissue autonomous fashion.
Figure 1.
Excessive TGFβ signaling in Crtap−/− mice. (a) Quantitative RT-PCR of TGFβ target genes Cdkn1a and Serpine1 in calvarial bone of P3 WT and Crtap−/− mice. Results are shown as fold change of the mean of WT group±SD; n=5 per group. (b) Western blot analysis of P3 calvarial protein extracts showing amounts of activated Smad2 (pSmad2) relative to total Smad2 protein in Crtap−/− versus WT mice; n=3 per group. (c) Quantification of the Western blot shown in b. Results are shown as fold change of the mean of WT group±SD. (d) Bioluminescence in regions that overlap with skeletal structures in Crtap−/− compared with WT mice that were intercrossed to TGFβ-reporter mice (SBE-Luc mice). Representative image of 3 litters at P10 is shown (scale bar=1cm). (e) TGFβ activity in conditioned medium from WT and Crtap−/− bone marrow stromal cells cultured under osteogenic conditions. Results are shown as fold change of the mean of WT group±SD, n=6 per group. (f) Immunostaining of lungs (P10) for pSmad2 (red) in WT and Crtap−/− mice, DAPI (blue) staining of nuclei (40X magnification). Representative images of n=3 mice per group are shown (scale bar=20μm). *P<0.05 WT versus Crtap−/−.
Patients with severe OI can exhibit lung abnormalities, and respiratory failure is one of the leading causes of death in these individuals17–18. Crtap−/− mice show an enlarged alveolar airway space compared with WT mice, a feature associated with higher TGFβ signaling in other models15–16. Accordingly, lungs of Crtap−/− mice showed more intense staining for pSmad2 in alveolar cells compared with lungs of WT mice, indicating that higher TGFβ activity is also present in extraskeletal tissues (Fig. 1f).
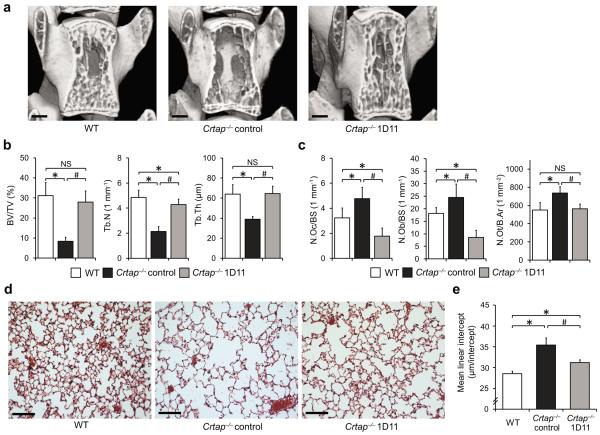
To understand whether upregulated TGFβ signaling represents a causal mechanism contributing to the bone and lung phenotypes in Crtap−/− mice, we performed a rescue experiment with a pan-TGFβ neutralizing antibody (1D11). Eight week old Crtap−/− mice were treated with 1D11 for eight weeks; control Crtap−/− and WT mice received a non-specific control antibody (13C4). 1D11 did not change body weight of the treated Crtap−/− mice, indicating that TGFβ inhibition did not affect their general nutritional status (Supplementary Fig. 1). Mass spectrometric and collagen cross-links analyses showed that 1D11 did not considerably change the status of P986 3-hydroxylation or collagen crosslinks in Crtap−/− mice, suggesting that dysregulated TGFβ signaling is a consequence of the altered collagen, and not directly involved in intracellular collagen processing or extracellular fibril assembly (Supplementary Fig. 2). As previously reported, Crtap−/− mice exhibit reduced bone mass and abnormal trabecular bone parameters (Fig. 2a,b)5. MicroCT imaging analysis of vertebrae and femurs demonstrated that compared with control Crtap−/− mice, TGFβ inhibition significantly improved trabecular bone parameters to near WT levels (Fig. 2a,b and Supplementary Table 1,2). The effects of TGFβ inhibition on the skeleton with 1D11 have been reported previously in WT and E-selectin ligand-1 knockout (Esl-1−/−) mice9, a model with higher TGFβ activity due to a defect in TGFβ production. While 1D11 moderately elevated bone volume/tissue volume (BV/TV) by 33% in WT mice, Esl-1−/− mice exhibited a 106% improvement. This suggests that targeting TGFβ in a pathophysiological situation where it is upregulated, could lead to a relatively more pronounced positive effect. In the present study, 1D11 improved the BV/TV at the spine by 235% in Crtap−/− mice, supporting that the dysregulated TGFβ signaling is a major contributor to the low bone mass.
Figure 2.
Phenotypic correction of Crtap−/− mice after treatment with the TGFβ neutralizing antibody 1D11. (a) MicroCT images of L4 vertebral bodies of 16-week-old wildtype (WT), control antibody-treated Crtap−/−, and 1D11-treated Crtap−/− mice after treatment for 8 weeks (scale bar=500μm). (b) MicroCT analysis results of L4 vertebral bodies for bone volume/total volume (BV/TV), trabecular number (Tb.N) and trabecular thickness (Tb.Th) in WT, control Crtap−/− and 1D11 treated Crtap−/− mice. Results are shown as means±SDs, n=8 per group. (c) Histomorphometric analysis of L4 for osteoclast (N.Oc/BS) and osteoblast (N.Ob/BS) numbers per bone surface, and numbers of osteocytes per bone area (N.Ot/B.Ar) in WT, control Crtap−/− and 1D11 treated Crtap−/− mice. Results are shown as means±SDs, n=6 per group. (d) Hematoxylin/eosin staining of inflated lungs of 16-week-old wildtype (WT), control Crtap−/−, and 1D11-treated Crtap−/− mice after treatment for 8 weeks. Representative images of n=8 mice per group are shown (scale bar=100 μm). (e) Quantification of the distance between alveolar structures by the mean-linear-intercept (MLI) method in lungs of WT, control Crtap−/− and 1D11-treated Crtap−/− mice. Results are shown as means±SDs, n=8 mice per group, 10 images analyzed per mouse. *P<0.05 for Crtap−/− vs. WT, #P<0.05 for Crtap−/− 1D11 vs. Crtap−/− control. NS, not significant.
At the femur midshaft, the parameters of cortical architecture in Crtap−/− mice were reduced compared to WT mice. Following 1D11 treatment these parameters were no longer significantly different from WT mice (Supplementary Table 3). To test if 1D11 treatment resulted in improved bone strength, we performed biomechanical testing of the femurs. TGFβ inhibition resulted in higher maximum load and ultimate strength in treated compared with control Crtap−/− mice. This indicates improved whole bone and tissue strength and suggests improved resistance to fracture. However, 1D11 had no effects on the higher brittleness of the OI bone as post-yield displacement in both control and 1D11 treated Crtap−/− mice remained reduced (Supplementary Table 4). This likely reflects the abnormal mineralization associated with altered collagen structure and crosslinking. Together, these findings indicate that upregulated TGFβ signaling is a major contributor to the bone phenotype in OI resulting from Crtap deficiency and that TGFβ inhibition restores bone mass, microstructural parameters and improves whole bone strength.
To understand the effects of TGFβ inhibition at the cellular level, we performed histomorphometric analyses. In sections of vertebrae in this study we found higher osteoclast (Oc) and osteoblast (Ob) numbers per bone surface in control Crtap−/− compared to WT mice, indicating higher bone remodeling (Fig. 2c and Supplementary Table 5). Consistently, the serum bone turnover markers osteocalcin (OCN) and C-terminal cross-linked telopeptide of bone collagen (CTX) were elevated in eight week old (OCN and CTX) and 16 week old (CTX only) control Crtap−/− compared with WT mice (Supplementary Fig. 3). Similar changes have been described in patients with OI, showing higher Oc and Ob numbers consistent with elevated bone turnover19–20. Notably, mouse models of upregulated TGFβ signaling also show higher bone resorption and abnormal bone remodeling8,14. Most reports of the effects of TGFβ on bone cells are consistent with a model where TGFβ stimulates the recruitment and initial differentiation of Oc and Ob precursors at the site of bone repair8,21–22, followed by insulin-like growth factor 1 mediated Ob differentiation23. However, at persistently high doses TGFβ can inhibit Ob differentiation by repressing Runx224. Therefore, fine tuning of TGFβ availability is crucial for the local coupling of bone resorption with formation during bone remodeling and its imbalance can lead to bone pathology8,21.
In contrast to control Crtap−/− mice, bone sections of 1D11 treated Crtap−/− mice revealed reduced Oc and Ob numbers, which were even lower than in WT mice, indicating a supraphysiologic suppression of bone remodeling resulting from TGFβ inhibition at the dose of 1D11 used (Fig. 2c). Our findings are different from previous studies in WT mice, where 1D11 treatment resulted in lower Oc but higher Ob numbers25. This may reflect distinct effects of TGFβ inhibition in a pathological situation with higher TGFβ signaling and bone remodeling compared with normal bone. TGFβ inhibits later differentiation of osteoblast precursors24, and upregulated TGFβ signaling could thereby lead to a higher proportion of immature osteoblastic cells. Vice versa, a higher number or proportion of immature Ob's could result in higher TGFβ secretion. The finding that TGFβ inhibition reduces the elevated Ob numbers in Crtap−/− mice suggests that the upregulated TGFβ signaling causally contributes to the higher number of Ob's.
We also observed higher osteocyte (Ot) numbers per bone area in control Crtap−/− compared with WT mice, which were reduced to WT levels in 1D11 treated Crtap−/− mice (Fig. 2c and Supplementary Table 5). In OI patients, a higher Ot density has been observed in individuals with more severe forms of the disease, likely reflecting the presence of immature primary bone due to a defect in physiological bone maturation26. Similarly, overexpression of TGFβ in WT mice results in higher Ot density14. As a possible explanation, TGFβ can inhibit Ob apoptosis during the transition of Ob's to Ot's, and thereby lead to a higher Ot density27. Collectively, these findings indicate that excessive TGFβ signaling contributes to a high bone turnover status and impaired bone maturation in Crtap−/− mice and that inhibition of TGFβ signaling reverses these alterations.
We were also interested in whether TGFβ inhibition affected the lung phenotype of Crtap−/− mice and found that lungs of Crtap−/− mice treated with 1D11 showed a 60% improvement in the distance between alveolar structures compared with control Crtap−/− mice (Fig 2d,e). This finding indicates that excessive TGFβ signaling is also a contributor to the lung abnormalities in Crtap−/− mice. Upregulated TGFβ signaling has been linked to developmental pulmonary abnormalities and disease in mature lungs. For example, TGFβ overexpression in lungs results in impaired lung development and enlarged airway space28 and upregulated TGFβ signaling is a pathomechanism of emphysema and bronchial asthma29–30. Given the partial rescue of the lung phenotype with 1D11 in Crtap−/− mice, it is possible that dysregulated TGFβ signaling affects lung development when anatomic structures are established, in addition to maintaining pulmonary tissue at later stages.
We next asked how collagen alterations due to loss of Crtap result in dysregulated TGFβ signaling. Biochemical analyses indicate that collagen prolyl-3-hydroxylation does not fundamentally affect the stability of collagen molecules, but instead may affect collagen-protein interactions6. An attractive hypothesis is that loss of 3Hyp and/or collagen overmodification could affect collagen interaction with small leucine-rich proteoglycans (SLRPs). SLRPs bind to both type I collagen and TGFβ, and thereby modulate TGFβ activity3–4. For example, the SLRP decorin inhibits distinct effects of TGFβ in osteosarcoma cells31 whereas it enhances TGFβ activity in preosteoblastic cells4. The binding region of decorin on type I collagen is suggested to center at residues 961/962 of the triple helical domain32, which is located in close proximity to the P986 residue. It is possible, that the P986 3Hyp position marks an interacting site for decorin-type I collagen binding, thereby mediating the sequestration of TGFβ to collagen.
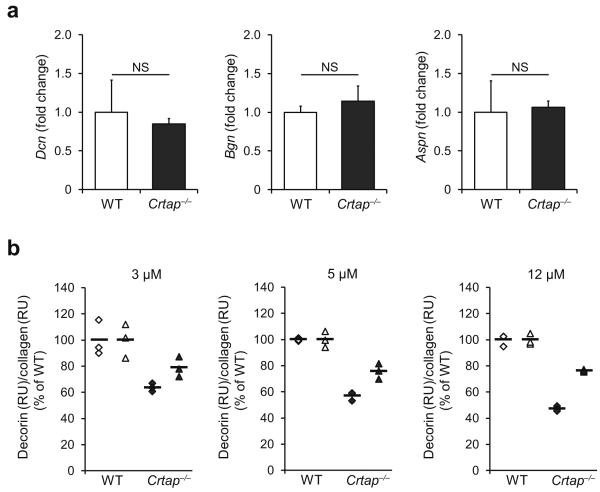
Hence, we hypothesize that decorin binding to collagen is critical for TGFβ regulation and that this binding is disrupted with altered collagen structure in OI. We found that while loss of Crtap did not alter the expression of decorin and other SLRPs in calvarial bone (Fig. 3a) nor the qualitative abundance of decorin in trabecular bone (Supplementary Fig.4), it did reduce binding of decorin to type I collagen (Fig. 3b; mean reductions of decorin binding to Crtap−/− vs. WT type I collagen at 3, 5 and 12 μM of decorin were 28.5%, 33.5% and 38.1%, respectively). Based on the requirement of collagen binding for decorin to effectively reduce TGFβ bioactivity3, it is possible that this altered binding in OI affects decorin's ability to sequester mature TGFβ in the matrix and modulate TGFβ function. This could contribute to dysregulated TGFβ signaling, even if no major changes in absolute TGFβ levels are present (Supplementary Fig.5 and 6). This notion is supported by the finding that COL1A1 and COL1A2 mutations in severe forms of dominant OI cluster in regions that are known to bind proteoglycans33, further supporting the relevance of proteoglycan-collagen interactions for normal bone homeostasis. This implies that other proteoglycans that are competing with decorin for the collagen binding site34 may also contribute to dysregulated TGFβ activity, and that additional signaling pathways could be altered35.
Figure 3.
Reduced decorin binding to type I collagen of Crtap−/− mice. (a) Quantitative RTPCR of calvarial bone of P3 mice for the small leucine-rich proteoglycans decorin (Dcn), biglycan (Bgn), and asporin (Aspn) in WT and Crtap−/− mice. Results given as fold change of the mean of WT group±SD, n=5 per group. NS, not significant. (b) Surface plasmon resonance analysis measuring the binding of recombinant decorin core protein to type I collagen of WT and Crtap−/− mice. Three technical replicates at each of the indicated concentrations of decorin were performed in two independent biological replicates (◆ replicate 1, ▲ replicate 2). Results are shown as the percentage of the mean of WT (bars indicate mean per group).
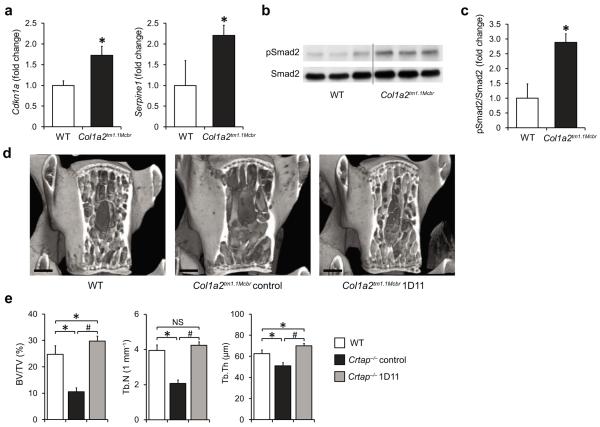
Because of the clinical overlap of some recessive and dominant forms of OI, it is possible that dysregulation of TGFβ signaling is a common disease mechanism. To address this hypothesis, we investigated the status of TGFβ signaling in a mouse model of dominant OI. Knock-in mice carrying a G610C mutation in the Col1a2 gene (Col1a2tm1.1Mcbr) phenocopy a dominantly inherited, moderate form of OI that was identified in an Amish population. Compared with bone samples of WT mice, we found higher expression of the TGFβ target genes Cdkn1a and Serpine1 in Col1a2tm1.1Mcbr mice, indicating upregulation of TGFβ signaling (Fig. 4a). Consistently, immunoblot analyses showed a greater ratio of pSmad2/total Smad2 in bone of Col1a2tm1.1Mcb compared with WT mice, similar to our observation in Crtap−/− mice (Fig. 4b,c).
Figure 4.
Inhibition of upregulated TGFβ signaling improves the bone phenotype in a mouse model of dominant OI (Col1a2tm1.1Mcbr). (a) Quantitative RT-PCR of TGFβ target genes Cdkn1a and Serpine1 in calvarial bone of P3 WT and Col1a2tm1.1Mcbr mice. Results are shown as fold change of the mean of WT group±SD; n=3 per group. (b) Western blot analysis showing activated Smad2 (pSmad2) relative to total Smad2 protein in P3 calvaria of WT and Col1a2tm1.1Mcbr mice; n=3 per group. (c) Quantification of the Western blot seen in b. Results are shown as fold change of the mean of WT group±SD. (d) MicroCT images of L4 vertebral bodies of 16-week-old wildtype (WT), control antibody-treated Col1a2tm1.1Mcbr and 1D11-treated Col1a2tm1.1Mcbr mice after treatment for 8 weeks (scale bar=500 μm). (e) MicroCT analysis results of L4 vertebral bodies for bone volume/total volume (BV/TV), trabecular number (Tb.N) and thickness (Tb.Th) in WT, control Col1a2tm1.1Mcbr and 1D11 treated Col1a2tm1.1Mcbr mice. Results are shown as means±SDs, n=6 per group. *P<0.05 for Crtap−/− vs. WT, #P<0.05 for Crtap−/− 1D11 vs. Crtap−/− control. NS, not significant.
To test if higher TGFβ signaling also represents a causal mechanism in dominant OI, eight week old Col1a2tm1.1Mcbr mice were treated with the TGFβ-neutralizing antibody 1D11 for eight weeks; control Col1a2tm1.1Mcbr and WT mice were treated with the control antibody 13C4. Similar to Crtap−/− mice, 1D11-treatment restored the trabecular bone parameters at the spine to WT levels (Fig. 4d,e and Supplementary Table 6). Together, these findings indicate that dysregulated TGFβ signaling is also an important contributor to the pathogenesis of dominant OI, and that anti-TGFβ therapy corrects the bone phenotype in dominant OI.
From a clinical-translational perspective, potential negative effects of systemic TGFβ inhibition in OI patients have to be considered. While Tgfβ1−/− mice develop a severe dysregulation of the immune system with inflammatory disease within the first weeks of life36, we did not observe obvious negative effects on general health, behavior or growth in both 1D11 treated Crtap−/− and Col1a2tm1.1Mcbr mice, suggesting that the effects of a partial pharmacological inhibition of TGFβ in adult mice are different from a complete loss of TGFβ1 during development. In humans, Fresolimumab (GC1008, Genzyme; similar to 1D11 in its affinity and specificity to the three isoforms of TGFβ) has been tested in phase I clinical studies in patients with primary focal segmental glomerulosclerosis37, idiopathic pulmonary fibrosis38 and cancer39. In these studies, Fresolimumab was in general well-tolerated, with possible dose-related adverse events including skin rashes or lesions, epistaxis, gingival bleeding and fatigue.
The molecular mechanisms of OI are incompletely understood. As a result, current treatment options for OI patients are mainly limited to anti-osteoporosis therapies with anti-resorptive drugs. Of note, a recent randomized controlled trial of the anabolic agent teriparatide showed that adult patients with severe OI responded differently than those with mild OI40. This suggests genotypic differences in response to therapies targeted at modifying cell signaling and that TGFβ-inhibition may be a promising target in severe OI due to collagen and collagen post-translational modification gene mutations. Overall, our data support the concept of dysregulated matrix-cell signaling as a mechanism in the pathogenesis of different forms of brittle bone disease and point to a disease-specific mechanism-based strategy for the treatment of OI by neutralizing overactive TGFβ activity.
Online Methods
Animals, anti-TGFβ treatment and tissue collection
We generated Crtap−/− mice as previously described5 and maintained them on a mixed C57Black/6J /129Sv genetic background. Mice harboring a G610C mutation in the Col1a2 gene (Col1a2tm1.1Mcbr)41 were a gift from Matthew Warman (Children's Hospital Boston, Harvard Medical School), and bred to wildtype C57Bl/6J mice. We used mice that were heterozygous for the Col1a2tm1.1Mcbr allele for experiments. TGFβ-reporter mice that express luciferase in response to the Smad2/3 dependent TGFβ signaling pathway (SBE-luc mice), were obtained from Jackson Laboratory (B6.Cg-Tg(SBE/TK-luc)7Twc/J)42 and bred to Crtap+/− mice for 2 generations to generate Crtap−/− mice and wildtype littermates expressing the reporter transgene. All mice were housed in the Baylor College of Medicine Animal Vivarium and animal experiments were performed following the approved protocol of the Animal Care and Use Committee (IACUC) at the Baylor College of Medicine.
For protein and RNA analyses, we isolated calvaria of P3 mice, cleaned them of extraskeletal tissue and snap froze them in liquid nitrogen. For immunostaining of Crtap−/− P10 mice lungs, we equally inflated the lungs of each mouse immediately after sacrifice by gravity with 4% paraformaldehyde at a constant pressure of 25 cm H2O and then suture closed them at the trachea. Lungs were then gently dissected from the thorax and fixed in 4% paraformaldehyde overnight.
We treated eight week old female Crtap−/− and Col1a2tm1.1Mcbr mice with the pan-TGFβ neutralizing antibody 1D11 (provided by Genzyme/Sanofi) for 8 weeks (10 mg kg−1 body weight, I.P. injections 3 times each week). Control Crtap−/−, Col1a2tm1.1Mcbr and WT mice received a control antibody (13C4) of the same IgG1 isotype. Mutant female mice (Crtap−/− and Col1a2tm1.1Mcbr) of each litter were randomly assigned to treatment (1D11) or control group. After treatment, mice were sacrificed and we collected lumbar spines and femurs and fixed them in 10% formalin for microCT and bone histomorphometry. We stored contralateral femurs of Crtap−/− mice in −20°C wrapped in saline soaked gauze until biomechanical testing was performed. We equally inflated lungs of Crtap−/− mice, collected and fixed them as described for P10 mice. No blinding was possible during treatment, because 1D11 or control antibody were injected according to group allocation. In all subsequent analyses the investigators were blinded to genotype and treatment group.
Immunoblotting
We extracted protein from snap frozen P3 calvaria samples by transferring them to 300 μl lysis buffer (0.0625 M Tris-HCl pH 7.5, 2% SDS, 5 mM NaF, 2 mM Na3VO4 and RocheComplete proteinase inhibitor) and homogenized for 1 minute, followed by incubation at 95°C for 40 minutes. We transferred the supernatant to Centrifugal Filter Units/Amicon Ultra 3K (Millipore) and centrifuged to concentrate the protein. We measured the total protein concentration of the lysate using the Micro BCA reagent (Pierce) following the manufacturer's directions. 40 μg of calvaria protein extracts were suspended in laemmeli buffer containing 5% β-mercaptoethanol and separated on Mini Protean TGX SDS-PAGE gels (gradient 4–20%; Bio-Rad) and transferred onto PVDF membranes for western blot analyses. We incubated PVDF membranes with pSmad2 monoclonal antibody (Cell Signaling #3108, 1:750 in TBST containing 5% BSA overnight), followed by secondary HRP-linked anti-rabbit antibody (GE, 1:5000 in TBST containing 5% BSA for 2 hours), treatment with ECL Plus Western Blotting Detection System (GE) and exposed them to X-ray films. Subsequently, we stripped antibodies from membranes using ReBlot Plus reagent (Millipore), and incubated them with Smad2 monoclonal antibody (Cell Signaling #5339, 1:2000 in TBST containing 5% BSA overnight), followed by similar secondary antibody incubation and ECL mediated visualization. X-ray films were scanned and the density of each band was quantified using ImageJ software (National Institutes of Health).
Quantitative realtime PCR
We extracted total RNA from snap frozen P3 mouse calvaria using Trizol reagent (Invitrogen). The Superscript III RT system (Invitrogen) was used to synthesize cDNA from total RNA according to the manufacturer's protocol. We performed quantitative RT-PCR on a LightCycler.v 1.5 (Roche) using gene-specific primers and SYBR Green I reagent (Roche). β2-Microglobulin was used as the reference gene for normalizing cDNA concentrations.
In vivo bioluminescence imaging
We injected P10 Ctrap−/− mice and wildtype littermates that expressed the TGFβ-reporter transgene (SBE-Luc mice), with D-luciferin (Goldbio, 150 mg kg−1, IP), anaesthetized them with isoflurane and performed imageing 10 minutes after injection using a bioluminescence imaging system (Xenogen).
Primary osteoblast culture, TGFβ-reporter cells
We isolated bone marrow cells from tibias and femurs of approximately 2 month old Crtap−/− and wildtype mice and cultured them in α-MEM supplied with 10% FBS, 100 U mL−1 penicillin and 100 ug mL−1 streptomycin. We changed media every second day and discarded unattached cells . After 7 days we reseeded the attached cells, defined as bone marrow stromal cells (BMSCs), to 24-well plates at 2.5×104 cells per cm2 and cultured them in osteogenic medium (α-MEM, 10% FBS, 500 μM ascorbic acid, and 10 mM β-glycerophosphate) for 3 days. We collected conditioned medium and incubated this with PAI-luciferase reporter mink lung epithelial cells43 (These cells were obtained from the laboratory of Dr. Daniel Rifkin, Department of Cell Biology, New York University Medical Center, New York. These cells were not recently further profiled or tested for mycoplasma contamination). After 24 hours we collected the cell lysates for luciferase activity assays, which were measured using the Dual-Luciferase Reporter System (Promega). The results were normalized to the total protein amount quantified using the Micro BCA reagent (Pierce).
MicroCT, bone histomorphometry
We scanned lumbar vertebrae and femurs using a Scanco μCT-40 microCT for quantification of trabecular and cortical bone parameters. We analyzed vertebral and femoral trabecular bone parameters using the Scanco analysis software by manually contouring the trabecular bone of vertebral body L4 as well as the distal metaphyseal section of the femur. The cortical bone parameters at the center of the femoral midshaft were quantified using the automated thresholding algorithm included in the software.
Scanned undecalcified Crtap−/− mouse spine samples were then embedded in plastic for sectioning. We performed toluidine blue staining and TRAP staining using standard protocols for visualization and quantification of Ob's and Oc's, respectively, using the Bioquant Osteo Image Analysis System.
Immunostaining and histology
For immunohistochemistry, we collected hind limbs of P5 mice , fixed them overnight in 4% paraformaldehyde and embedded them in paraffin. After deparaffinization and rehydration, we performed heat-induced antigen retrieval (Dako, S1700) followed by treatment with hyaluronidase for 30 min (2 mg ml−1; Sigma). Endogenous peroxidase was blocked using 3% hydrogen peroxide for 10 min. After incubation with blocking solution (3% normal goat serum, 0.1% BSA, 0.1% Triton X-100 in PBS), we incubated sections in antibodies for TGFβ1 (G1221, Promega) and decorin (LF-113, kindly provided from Larry Fisher, National Institute of Dental and Craniofacial Research, Bethesda, MD, USA) for 60 min (1:25 dilution each in PBS, control samples were incubated in PBS only) at 37°C, and subsequently incubated them with secondary antibody (SuperPicTure Ploymer Detection kit, Invitrogen). We added substrate DAB according to the manufacturer's recommendations and dehydrated and mounted samples using Cytoseal XYL xylene based mounting medium (Thermo Scientific). We processed sections of WT and mutant littermates at the same time. Images of the trabecular bone were taken with a light microscope (Axioplan 2, Zeiss) using identical exposure times for WT and mutant littermates.
We equally inflated lungs of P10 and 16 week old Crtap−/− mice during tissue collection, fixed them in 4% paraformaldehyde and performed paraffin embedding. We used lungs of P10 Crtap−/− and wildtype mice for immunostaining for pSmad2. Briefly, we treated paraffin sections with xylene, rehydrated and heated them for 20 minutes for antigen retrieval (pH 6; Dako). We then incubated sections in blocking solution (3% normal Donkey serum, 0.1% BSA, 0.1% Triton X-100 in PBS), and subsequently incubated them with rabbit anti-pSmad2 antibody (1:500) (Cell signaling, #3101), donkey anti-rabbit secondary antibody conjugated to Alexa flour 594 (1:600) (Invitrogen), and mounted them with Prolong Gold anti-fade reagent with DAPI (Invitrogen). Fluorescent images from these sections were taken using a Zeiss microscope (Axiovision Software) using identical exposure times.
For lung histology and morphometry of 16 week old mice, we stained parasagittal sections using a standard protocol for Hematoxylin and Eosin staining. We used the mean linear intercept (MLI) method to quantify the distance between alveolar structures as previously described44. Briefly, we captured 10 histological fields per mouse at 20X magnification from all lobes of both lungs using a light microscope (Axioplan 2, Zeiss) and measured the MLI using modified ImageJ software (National Institutes of Health, modified by Paul Thompson). After manual removal of blood vessels, large airways and other nonalveolar structures, the software automatically thresholds the alveolar tissue in each image and overlays a line grid comprised of 1,353 lines with each line measuring 21 pixels over the image. The number of lines that intercepted alveolar structures was used to calculate the MLI.
Biomechanical testing by 3-point bending
We tested Crtap−/− and WT femurs by three point bending using a span of 6mm with an Instron 5848 device (Instron Inc., Norwood MA). All the femurs were tested wet at room temperature. We preloaded femurs to 1N at a rate of 0.05 N s−1 for 5 seconds. Following the pre-loading, we loaded the femurs to failure at a rate of 0.1 mm sec−1. Load and displacement data was captured at rate of 40 Hz by using BLUEHILL Software (Instron 5848). To determine the Yield Point, a region was identified after the preload and before the maximum load on the load-displacement curve. We separated this region was into 5 segments from which the fitted line of the segment with greatest slope was taken. Next, a 0.012 mm offset was implemented on the line. The point of intersection between the offset line and the load-displacement curve was the 0.012 Offset Yield Point. This yield point corresponded more closely to a 0.2% offset strain, which is commonly chosen in the literature. The elastic region was identified as the region from the completion of the preload to the Yield Point. Post-Yield region was identified as the region from the Yield Point until the point at which the change in load exceeded −1 N, indicating failure. Elastic Displacement was the displacement during which specimen remained in the elastic region. Post-Yield Displacement was the displacement during which specimen remained in the Post-Yield region. Total Displacement was calculated as the sum of Elastic Displacement and Post-Yield Displacement. Using the trapezoidal numerical integration method, we calculated Energy to failure as the area under the load-displacement curve. We determined Maximum Load by finding the highest load value recorded by BLUEHILL, before the specimen failed. To calculate Stiffness, we applied the Least Square fit method to the steepest segment of the elastic region of the Load-Displacement curve. Stiffness was the slope of least square fit line. Geometric data (diameter and moment of inertia) obtained from microCT analysis of the femoral midshaft were utilized to calculate the intrinsic material properties: ultimate strength, toughness to failure and elastic modulus.
Serum bone turnover markers
We quantified serum osteocalcin (OCN) using the Mouse Osteocalcin EIA Kit from Biomedical Technologies Inc. and C-terminal cross-linked telopeptide of bone collagen (CTX) using the RatLapsTM EIA Kit from Immunodiagnostic Systems Ltd. Both analyses were performed according to the manufacturer`s protocols.
Collagen SDS-PAGE, mass spectrometry and crosslinks analyses
For mass spectrometry, we prepared type I collagen from Crtap−/− and wildtype tibiae. We defatted bone with chloroform/methanol (3:1 v/v) and demineralized it in 0.5 M EDTA, 0.05 M Tris-HCl, pH 7.5, all steps at 4°C. We finely minced the bone samples and solubilized collagen by heat denaturation (90°C) in SDS-PAGE sample buffer. Collagen α-chains were cut from SDS-PAGE gels and subjected to in-gel trypsin digestion. We performed electrospray MS on the tryptic peptides using an LCQ Deca XP ion-trap mass spectrometer equipped with in-line liquid chromatography (LC) (ThermoFinnigan) using a C8 capillary column (300 μm × 150 mm; Grace Vydac 208 MS5.315) eluted at 4.5 μl min. Sequest search software (ThermoFinnigan) was used for peptide identification using the NCBI protein database.
We quantified pyridinoline cross-links (HP and LP) by HPLC after hydrolyzing demineralized bone in 6 N HCl as described45.
Surface plasmon resonance analysis
We performed surface plasmon resonance experiments using a BIACore X instrument (GE Healthcare Bio-Science Corp.). We immobilized purified native mouse tendon type I collagen from three wild type and three Crtap−/− mice (pooled for each genotype) on CM5 sensor chips by amide coupling at a concentration of about 0.05 ng mm−2 (500 RU) and 0.08 ng mm−2 (800 RU), respectively. The experiments were conducted at a flow rate of 10 μl min−1 and 20°C in HBS-P buffer (10 mM Hepes buffer, pH 7.4, containing 150 mM NaCl and 0.005% Surfactant P20). We then injected recombinant human decorin core protein (R&D systems) onto both type I CM5 chips. We determined the concentration of the stock solution of human decorin by amino acid analysis. The binding response of decorin to wild type and Crtap−/− mouse type I collagen was normalized by the amounts of immobilized type I collagen on the CM5 sensor chips. We used three concentrations of decorin (3, 5 and 12 μM), for each concentration the analysis was repeated three times. We performed this experiment twice with collagen isolated from different mice each time.
Statistical methods
For comparisons between 2 groups we used unpaired, two-tailed Student's t-tests. For comparisons between 3 groups we performed One Way Analysis of Variance (ANOVA) if equal variance and normal distribution of groups was confirmed, followed by all pairwise multiple comparison using the Holm-Sidak method. If the equal variance or normal distribution test failed, we performed Kruskal-Wallis One Way ANOVA on Ranks , followed by all pairwise multiple comparison using the Tukey Test. A P value less than 0.05 was considered statistically significant for Student's t-test, ANOVA and Kruskal-Wallis One Way ANOVA on Ranks. For posthoc pairwise multiple comparisons, each P value was compared to a critical level depending on the rank of the P value and the total number of comparisons made to determine if differences between groups are significant. We used Sigma Plot V11.0 (Systat Software Inc.) for statistical analyses. The effects of 1D11 on bone and lungs of OI mice were unknown at study start. To determine the initial sample size per group of mice we calculated that to detect a minimal difference of 20% in bone mass (BV/TV) by MicroCT between 1D11 and control treated OI mice with a 90% power, a group size of 8 mice is required.
Supplementary Material
Acknowledgements
We thank D. Spencer and his lab (Baylor College of Medicine) for providing the IVIS camera system and training, M. Starbuck (MD Anderson Cancer Center) and F. Gannon (Baylor College of Medicine) for consultation and advice in bone histomorphometry, M. Warman and C. Jacobsen (Boston Children's Hospital) for kindly providing the Col1a2tm1.1Mcbr mice and helpful information. We also want to thank L. Fisher (US National Institute of Dental and Craniofacial Research, Bethesda, Marryland) for providing the decorin antibody LF-113. We also want to thank M. Bagos for help with microCT analyses, A. Abraham (University of Texas Health Science Center Medical School at Houston) for help with the biomechanical testing, W. Song and S. Liu (Genzyme/Sanofi) for their help with serum bone turnover analyses, the US National Institute of Health for providing the Image J software, and A. Choi and H. Lam (Harvard Medical School) for providing the Image J modification for lung morphometry, which was generated by P. Thompson. In addition, we want to thank R. Morello, G. Sule, D. Baldridge, and G. Ghosal for their helpful discussions, and P. Fonseca for editorial assistance. T.K. Sampath is an employee of Genzyme/Sanofi. None of the other authors have any conflicts of interest to declare. This work was supported by a research grant from the German Research Foundation/Deutsche Forschungsgemeinschaft (I.G.), a Michael Geisman Fellowship from the Osteogenesis Imperfecta Foundation (I.G.), grant support from Shriners Hospitals for Children (H.P.B.), US National Institute of Health Grants 5F31DE020954 (E.H.), 5F31DE022483 (C.L.), P01 HD70394 (B.L. and D.E.), and the Howard Hughes Medical Institute Foundation (B.L.). This work was also supported by the BCM Intellectual and Developmental Disabilities Research Center (HD024064) from the Eunice Kennedy Shriver US National Institute Of Child Health & Human Development and the Rolanette and Berdon Lawrence Bone Disease Program of Texas.
Footnotes
Author contributions: I.G. and B.L. conceptualized the study. T.Y., T.K.S., C.A., D.E. and H.P.B. contributed to the design of the study and experiments. I.G., T.Y., S.A., E.H., C.L., MM.J., T.B., E.M., Y.C., B.D., Y.I., MA.W. and C.A. performed and analyzed experiments. I.G., T.Y. and B.L. wrote the manuscript with contributions from all authors. B.L. supervised the project.
References
- 1.Rauch F, Glorieux FH. Osteogenesis imperfecta. Lancet. 2004;363:1377–1385. doi: 10.1016/S0140-6736(04)16051-0. [DOI] [PubMed] [Google Scholar]
- 2.Baldridge D, et al. CRTAP and LEPRE1 mutations in recessive osteogenesis imperfecta. Hum Mutat. 2008;29:1435–1442. doi: 10.1002/humu.20799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Markmann A, Hausser H, Schonherr E, Kresse H. Influence of decorin expression on transforming growth factor-beta-mediated collagen gel retraction and biglycan induction. Matrix Biol. 2000;19:631–636. doi: 10.1016/s0945-053x(00)00097-4. [DOI] [PubMed] [Google Scholar]
- 4.Takeuchi Y, Kodama Y, Matsumoto T. Bone matrix decorin binds transforming growth factor-beta and enhances its bioactivity. J Biol Chem. 1994;269:32634–32638. [PubMed] [Google Scholar]
- 5.Morello R, et al. CRTAP is required for prolyl 3- hydroxylation and mutations cause recessive osteogenesis imperfecta. Cell. 2006;127:291–304. doi: 10.1016/j.cell.2006.08.039. [DOI] [PubMed] [Google Scholar]
- 6.Mizuno K, Peyton DH, Hayashi T, Engel J, Bachinger HP. Effect of the -Gly-3(S)-hydroxyprolyl-4(R)-hydroxyprolyl- tripeptide unit on the stability of collagen model peptides. FEBS J. 2008;275:5830–5840. doi: 10.1111/j.1742-4658.2008.06704.x. [DOI] [PubMed] [Google Scholar]
- 7.Homan EP, et al. Differential effects of collagen prolyl 3-hydroxylation on skeletal tissues. PLoS Genet. 2014;10:e1004121. doi: 10.1371/journal.pgen.1004121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tang Y, et al. TGF-beta1-induced migration of bone mesenchymal stem cells couples bone resorption with formation. Nat Med. 2009;15:757–765. doi: 10.1038/nm.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang T, et al. E-selectin ligand 1 regulates bone remodeling by limiting bioactive TGF-beta in the bone microenvironment. Proc Natl Acad Sci U S A. 2013;110:7336–7341. doi: 10.1073/pnas.1219748110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dallas SL, et al. Characterization and autoregulation of latent transforming growth factor beta (TGF beta) complexes in osteoblast-like cell lines. Production of a latent complex lacking the latent TGF beta-binding protein. J Biol Chem. 1994;269:6815–6821. [PubMed] [Google Scholar]
- 11.Hering S, et al. TGFbeta1 and TGFbeta2 mRNA and protein expression in human bone samples. Exp Clin Endocrinol Diabetes. 2001;109:217–226. doi: 10.1055/s-2001-15109. [DOI] [PubMed] [Google Scholar]
- 12.Oreffo RO, Mundy GR, Seyedin SM, Bonewald LF. Activation of the bone-derived latent TGF beta complex by isolated osteoclasts. Biochem Biophys Res Commun. 1989;158:817–823. doi: 10.1016/0006-291x(89)92795-2. [DOI] [PubMed] [Google Scholar]
- 13.Hildebrand A, et al. Interaction of the small interstitial proteoglycans biglycan, decorin and fibromodulin with transforming growth factor beta. Biochem J. 1994;302(Pt 2):527–534. doi: 10.1042/bj3020527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Erlebacher A, Derynck R. Increased expression of TGF-beta 2 in osteoblasts results in an osteoporosis-like phenotype. J Cell Biol. 1996;132:195–210. doi: 10.1083/jcb.132.1.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Neptune ER, et al. Dysregulation of TGF-beta activation contributes to pathogenesis in Marfan syndrome. Nat Genet. 2003;33:407–411. doi: 10.1038/ng1116. [DOI] [PubMed] [Google Scholar]
- 16.Baldridge D, et al. Generalized connective tissue disease in Crtap−/− mouse. PLoS One. 2010;5:e10560. doi: 10.1371/journal.pone.0010560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thiele F, et al. Cardiopulmonary dysfunction in the Osteogenesis imperfecta mouse model Aga2 and human patients are caused by bone-independent mechanisms. Hum Mol Genet. 2012;21:3535–3545. doi: 10.1093/hmg/dds183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McAllion SJ, Paterson CR. Causes of death in osteogenesis imperfecta. J Clin Pathol. 1996;49:627–630. doi: 10.1136/jcp.49.8.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rauch F, Travers R, Parfitt AM, Glorieux FH. Static and dynamic bone histomorphometry in children with osteogenesis imperfecta. Bone. 2000;26:581–589. doi: 10.1016/s8756-3282(00)00269-6. [DOI] [PubMed] [Google Scholar]
- 20.Ward LM, et al. Osteogenesis imperfecta type VII: an autosomal recessive form of brittle bone disease. Bone. 2002;31:12–18. doi: 10.1016/s8756-3282(02)00790-1. [DOI] [PubMed] [Google Scholar]
- 21.Janssens K, ten Dijke P, Janssens S, Van Hul W. Transforming growth factor-beta1 to the bone. Endocr Rev. 2005;26:743–774. doi: 10.1210/er.2004-0001. [DOI] [PubMed] [Google Scholar]
- 22.Fuller K, Lean JM, Bayley KE, Wani MR, Chambers TJ. A role for TGFbeta(1) in osteoclast differentiation and survival. J Cell Sci. 2000;113(Pt 13):2445–2453. doi: 10.1242/jcs.113.13.2445. [DOI] [PubMed] [Google Scholar]
- 23.Xian L, et al. Matrix IGF-1 maintains bone mass by activation of mTOR in mesenchymal stem cells. Nat Med. 2012;18:1095–1101. doi: 10.1038/nm.2793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alliston T, Choy L, Ducy P, Karsenty G, Derynck R. TGF-beta-induced repression of CBFA1 by Smad3 decreases cbfa1 and osteocalcin expression and inhibits osteoblast differentiation. EMBO J. 2001;20:2254–2272. doi: 10.1093/emboj/20.9.2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Edwards JR, et al. Inhibition of TGF-beta signaling by 1D11 antibody treatment increases bone mass and quality in vivo. J Bone Miner Res. 2010;25:2419–2426. doi: 10.1002/jbmr.139. [DOI] [PubMed] [Google Scholar]
- 26.Sarathchandra P, Pope FM, Kayser MV, Ali SY. A light and electron microscopic study of osteogenesis imperfecta bone samples, with reference to collagen chemistry and clinical phenotype. J Pathol. 2000;192:385–395. doi: 10.1002/1096-9896(2000)9999:9999<::AID-PATH704>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 27.Karsdal MA, et al. Matrix metalloproteinase-dependent activation of latent transforming growth factor-beta controls the conversion of osteoblasts into osteocytes by blocking osteoblast apoptosis. J Biol Chem. 2002;277:44061–44067. doi: 10.1074/jbc.M207205200. [DOI] [PubMed] [Google Scholar]
- 28.Gauldie J, et al. Transfer of the active form of transforming growth factor-beta 1 gene to newborn rat lung induces changes consistent with bronchopulmonary dysplasia. Am J Pathol. 2003;163:2575–2584. doi: 10.1016/s0002-9440(10)63612-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Morty RE, Konigshoff M, Eickelberg O. Transforming growth factor-beta signaling across ages: from distorted lung development to chronic obstructive pulmonary disease. Proc Am Thorac Soc. 2009;6:607–613. doi: 10.1513/pats.200908-087RM. [DOI] [PubMed] [Google Scholar]
- 30.Marwick JA, et al. Cigarette smoke-induced oxidative stress and TGF-beta1 increase p21waf1/cip1 expression in alveolar epithelial cells. Ann N Y Acad Sci. 2002;973:278–283. doi: 10.1111/j.1749-6632.2002.tb04649.x. [DOI] [PubMed] [Google Scholar]
- 31.Hausser H, Groning A, Hasilik A, Schonherr E, Kresse H. Selective inactivity of TGF-beta/decorin complexes. FEBS Lett. 1994;353:243–245. doi: 10.1016/0014-5793(94)01044-7. [DOI] [PubMed] [Google Scholar]
- 32.Keene DR, et al. Decorin binds near the C terminus of type I collagen. J Biol Chem. 2000;275:21801–21804. doi: 10.1074/jbc.C000278200. [DOI] [PubMed] [Google Scholar]
- 33.Marini JC, et al. Consortium for osteogenesis imperfecta mutations in the helical domain of type I collagen: regions rich in lethal mutations align with collagen binding sites for integrins and proteoglycans. Hum Mutat. 2007;28:209–221. doi: 10.1002/humu.20429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schonherr E, et al. Interaction of biglycan with type I collagen. J Biol Chem. 1995;270:2776–2783. doi: 10.1074/jbc.270.6.2776. [DOI] [PubMed] [Google Scholar]
- 35.Nikitovic D, et al. The biology of small leucine-rich proteoglycans in bone pathophysiology. J Biol Chem. 2012;287:33926–33933. doi: 10.1074/jbc.R112.379602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Christ M, et al. Immune dysregulation in TGF-beta 1-deficient mice. J Immunol. 1994;153:1936–1946. [PubMed] [Google Scholar]
- 37.Trachtman H, et al. A phase 1, single-dose study of fresolimumab, an anti-TGF-beta antibody, in treatment-resistant primary focal segmental glomerulosclerosis. Kidney Int. 2011;79:1236–1243. doi: 10.1038/ki.2011.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lonning S, Mannick J, McPherson JM. Antibody targeting of TGF-beta in cancer patients. Curr Pharm Biotechnol. 2011;12:2176–2189. doi: 10.2174/138920111798808392. [DOI] [PubMed] [Google Scholar]
- 39.Morris JCS, G.I., Tan AR, Lawrence DP, Olencki TE, Dezube BJ, Hsu FJ, Reiss M, Berzofsky JA. Phase I/II study of GC1008: A human anti-transforming growth factor-beta (TGFb) monoclonal antibody (MAb) in patients with advanced malignant melanoma (MM) or renal cell carcinoma (RCC) J. Clin. Oncol. 2008;26(Suppl.) doi: 10.1371/journal.pone.0090353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Orwoll ES, et al. Evaluation of teriparatide treatment in adults with osteogenesis imperfecta. J Clin Invest. 2014;124:491–498. doi: 10.1172/JCI71101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Daley E, et al. Variable bone fragility associated with an Amish COL1A2 variant and a knock-in mouse model. J Bone Miner Res. 2010;25:247–261. doi: 10.1359/jbmr.090720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lin AH, et al. Global analysis of Smad2/3-dependent TGF-beta signaling in living mice reveals prominent tissue-specific responses to injury. J Immunol. 2005;175:547–554. doi: 10.4049/jimmunol.175.1.547. [DOI] [PubMed] [Google Scholar]
- 43.Abe M, et al. An assay for transforming growth factor-beta using cells transfected with a plasminogen activator inhibitor-1 promoter-luciferase construct. Anal Biochem. 1994;216:276–284. doi: 10.1006/abio.1994.1042. [DOI] [PubMed] [Google Scholar]
- 44.Chen ZH, et al. Autophagy protein microtubule-associated protein 1 light chain-3B (LC3B) activates extrinsic apoptosis during cigarette smoke-induced emphysema. Proc Natl Acad Sci U S A. 2010;107:18880–18885. doi: 10.1073/pnas.1005574107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Eyre D. Collagen cross-linking amino acids. Methods Enzymol. 1987;144:115–139. doi: 10.1016/0076-6879(87)44176-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.