Abstract
Until recently, knowledge of the impact of abuse drugs on gene and protein expression in the brain was limited to less than 100 targets. With the advent of high-throughput genomic and proteomic techniques investigators are now able to evaluate changes across the entire genome and across thousands of proteins in defined brain regions and generate expression profiles of vulnerable neuroanatomical substrates in rodent and non-human primate drug abuse models and in human post-mortem brain tissue from drug abuse victims. The availability of gene and protein expression profiles will continue to expand our understanding of the short- and long-term consequences of drug addiction and other addictive disorders and may provide new approaches or new targets for pharmacotherapeutic intervention. This chapter will review gene expression data from rodent, non-human primate and human post-mortem studies of cocaine abuse and will provide a preliminary proteomic profile of human cocaine abuse and explore how these studies have advanced our understanding of addiction.
Keywords: microarray, RNA amplification, gene expression, molecular fingerprint, qPCR, transcriptome, proteome, brain, post-mortem, monkey
Introduction
The efforts to complete sequencing of the human genome have enabled new endeavors into the function of these genes in human disorders and have provided a wealth of knowledge about the molecular underpinnings of behavior. The next challenge in addiction biology is the utilization of this information to determine the function of the genes and proteins in the context of human disease. The advent of high-throughput screening technologies has produced a paradigm shift in the manner in which scientists are able to detect and identify molecular mechanisms related to disease. Microarray and proteomic analysis strategies allow the simultaneous assessment of thousands of genes and proteins of known and unknown function — thereby enabling a global biological view of addictive disorders. Broad-scale evaluations of gene and protein expression are well suited to the study of drug abuse, particularly in light of the complexity of the brain compared with other tissues, the multigenic nature of drug addiction, the vast representation of expressed genes in the brain, and our relatively limited knowledge of the molecular pathology of this illness.
The content of this chapter will include recent studies employing genomic and proteomic strategies to develop a comprehensive understanding of the changes induced by cocaine, a commonly abused stimulant. Furthermore, the chapter will focus on studies employing rodent and non-human primate models as well as studies examining the neuropathology identified in post-mortem human tissue of individuals with chronic histories of illicit substance abuse. The chapter is limited to studies on cocaine due to the fact that this is the most-studied abused drug with respect to genomic and proteomic strategies and thus may provide an investigative template for studying other abuse substances.
The use and abuse of illicit drugs has continued to increase and poses one of the most significant public health care concerns in American society. A recent report indicates that approximately 13.6 million Americans are current users of illicit drugs (e.g. marijuana, 11 million; cocaine, 1.8 million; heroin 130,000) and over 4 million Americans meet the diagnostic criteria for dependence on illicit drugs (SAMHSA, 2002). Despite intense behavioral and biological research, few effective pharmacotherapeutic strategies exist, with the arguable exception of methadone and LAAM treatment programs for opiate dependence. In order to devise effective treatment strategies, it is necessary to understand the interactions of behavioral, pharmacological and biochemical factors that underlie use and abuse. Substance abuse is the culmination of a number of contributing factors spanning scientific disciplines from behavior to molecular biology. As such, to understand the biology of addiction requires a multidisciplinary approach to identify the contributing factors, synthesize the information in the appropriate biological context and eventually relate this context to the behavioral abnormality. The development of new and innovative medications for drug addiction requires multidisciplinary research approaches examining the spectrum of drug-induced effects from behavior to the biological and biochemical effects in discrete neuronal populations.
A generally accepted tenet in drug abuse research is that drugs can function as reinforcing stimuli. Hence, with respect to drug abuse, the reinforcing effects of certain drugs contribute largely to their abuse liability. A significant amount of research investigating the neurobiology of drug abuse is conducted in animal models which closely resemble characteristics of human drug intake l. Criteria should include, but not be limited to the following: (1) behaviors are contingent upon drug delivery, (2) behaviors are engendered and maintained by drug delivery, and (3) drug delivery increases the frequency of those behaviors. The self-administration paradigm meets these criteria, unlike the other procedures, and is widely accepted as an appropriate model for studying the reinforcing effects of drugs. Generally, the self-administration paradigm involves the emission of specific behavior(s) (e.g. lever-press; nose-poke) that is maintained by drug administration (e.g. intravenous, oral, or intracranial). Advantages of self-administration include the following: (1) substances abused by humans can function as positive reinforcing stimuli under laboratory conditions, (2) general concordance between substances abused by humans and those self-administered by laboratory animals, (3) a variety of species readily acquire and maintain self-administration under a number of operant schedules and (4) the ability to generate clear dose-effect curves using this procedure (Hemby et al., 1997b; Hemby, 1999). Procedures such as place conditioning are hindered by the lack of objectively quantifiable behaviors, lack of dose dependency and most importantly by the fact that drug administration is not contingent on the behavior of the animal.
The concept of the contingency is critical for researchers attempting to draw conclusions regarding the involvement of specific neural substrates in drug reinforcement. The majority of studies investigating the neurobiological basis of drug administration have used experimenter-controlled drug administration and extrapolated the relevance of those findings to reinforcement mechanisms (Di Chiara and Imperato, 1988). However, a growing body of literature has demonstrated pronounced neurochemical differences resulting from the context and contingency of drug administration (self-administered versus experimenter delivered) (Wilson et al., 1994; Hemby et al., 1995, 1997a,b). Neurobiological differences between rats self-administering drugs and rats receiving experimenter-administered infusions are based on the context of drug presentation and suggest inferences of reinforcement mechanisms drawn from studies using experimenter-drug administration protocols may be misleading. These studies clearly indicate a need for reliance on accepted behavioral models when asserting relevance of biological findings to behavioral phenomena such as reinforcement. While reinforcement does not solely explain drug abuse, it allows for the quantification of the initiation and maintenance of drug self-administration.
Neuroanatomy of cocaine addiction
Similar to other psychiatric illnesses, drug abuse is a heterogeneous disorder with multiple causes all of which can lead to the same functional endpoint — namely addiction. While the regulation of individual transcripts and proteins have been suggested as mediators of the addictive process, a more probable scenario is that the coordinate regulation of multiple genes and proteins in defined neuroanatomical loci are either the mediators of addictive behaviors or are modulated by chronic drug use. Over the past 20 years, the driving theoretical construct in drug abuse research has been the psychomotor-stimulant theory of addiction which attempts to provide a unifying theory for the neurobiological basis of all abused drugs (Wise and Bozarth, 1987). The theory indicates that both the stimulant and the reinforcing effects of all abused drugs are mediated by a common neural mechanism, the mesolimbic dopamine system. The pathway originates in the mesencephalon, ventral tegmental area (VTA) and projects to several basal forebrain regions including the nucleus accumbens (NAc), ventral caudate-putamen, bed nucleus of the stria terminalis, diagonal band of Broca, olfactory tubercles, prefrontal and anterior cingulate cortices. Administration of drugs that are abused by humans lead to activation of this pathway in humans, non-human primates and rodents (Porrino, 1993; Lyons et al., 1996; Volkow et al., 1997). Activation of this circuit has been correlated with subjective reports of craving and euphoria in cocaine addicts (Volkow et al., 1997; Childress et al., 1999).
Dopaminergic projections from the VTA to the NAc have been implicated in the reinforcing effects of psychomotor stimulants (cocaine and amphetamine) and alcohol, whereas the role of this pathway in opiate reinforcement remains controversial (Hemby et al., 1997b). Previous studies have shown that rats will self-administer cocaine, amphetamine, opiates, and alcohol directly into regions of this pathway. Altering the functional integrity of the mesolimbic pathway by dopamine-selective neurotoxic lesions and dopamine D1 and D2 receptor blockade attenuate psychomotor stimulant self-administration. Similar manipulations of the other monoamines serotonin and norepinephrine fail to significantly influence drug intake. Thirdly, microdialysis studies indicate that extracellular dopamine concentrations are elevated during cocaine and amphetamine self-administration sessions (Hemby et al., 1997b). Taken together, the most recent research indicates that the neurobiological substrates of drug abuse are not the same across all dug classes and probably involve a myriad of neurotransmitter and receptor systems.
Functional genomics
Over the past 10 years, approximately 20 studies have employed various high-throughput gene expression strategies to examine stimulant-induced changes in various brain regions of animal models and humans. Several obstacles prevent the assimilation of the results from these studies into an overarching understanding of stimulant-induced transcriptional regulation such as species, brain regions, route and contingency of administration, dose and duration of drug administration, length of time since the final drug administration, experimental variables in microarray analysis, validation of findings with alternative techniques, etc. Although several studies have examined the effects of stimulants on gene expression, there is minimal literature on stimulant-induced proteomic analysis on a broad scale; however, preliminary data will be presented on proteomic analysis of human cocaine overdose victims.
Rodent studies: non-contingent administration
Several studies have examined the effects of cocaine administration on the coordinate expression of genes in rodent brain regions associated with the mesocorticolimbic pathway, including the NAc (Toda et al., 2002), prefrontal cortex (PFC) (Freeman et al., 2002; Toda et al., 2002), hippocampus (Freeman et al., 2001a), lateral hypothalamus (Ahmed et al., 2005) and VTA (Backes and Hemby, 2003). In the one study, rats were administered cocaine three times per day (15 mg/kg; intraperitoneal) for 14 days (Freeman et al., 2002) as an analogous “binge” paradigm, and gene expression was evaluated in the hippocampus using RNA pools. Using stringent inclusion criteria of 50% induction or 33% reduction, the authors noted only five transcripts were differentially regulated — all were upregulated in the cocaine-treated rats: protein kinase A alpha (PKAcα), metabotropic glutamate receptor 5 (mGluR5) and voltage-gated potassium channel 1.1 (Kv1.1), survival of motor neuron (SMN) and protein phosphatase 2A alpha subunit (PP2Aα). From this set, only mGluR5, PKCα, and Kv1.1 showed analogous changes in protein levels in this region. Interestingly, the authors note that protein tyrosine kinase 2 (PYK2), protein kinase C epsilon (PKCε) and β catenin, proteins found to be elevated in the NAc of cynomolgus monkeys, were also elevated in the hippocampus of cocaine-treated rats suggesting these changes are not region or treatment-specific regimen.
In a separate study, changes in gene expression in the PFC of the same subjects (Freeman et al., 2002) were examined by screening 588 rat genes (BD Bioscience Clonetech Atlas cNDA Expression Array). Cocaine administration induced the expression of activity-regulated cytoskeletal protein (ARC), NGFI-B and HMG-CoA synthase I and decreased the expression of casein kinase II alpha (CKIIa), glycogen synthase 3 alpha (GSK3α), and fos-related antigen (FRA1). The upregulation of NGFI-B was confirmed by quantitative PCR; however the remaining encoded proteins of the differentially expressed transcripts were assessed by Western blot analysis. Interestingly, only ARC protein levels were increased in the PFC similar to the mRNA levels — which may be due in part to the somatodendritic localization of ARC in neurons. The authors also examined proteins that had been shown to be upregulated in the hippocampus of rats and NAc of monkeys administered cocaine including PYK2, mitogen-activated kinase I (MEK), β-catenin, PKCα, PKCε, – of which only PYK2 was found to be upregulated in the frontal cortex of cocaine-reated rats. The study provides confirmatory data from previous studies showing increased ARC mRNA expression following cocaine administration (Fosnaugh et al., 1995; Tan et al., 2000; Ujike et al., 2002) as well as extending current knowledge on the ability of cocaine to induce genes and protein involved in neuroplasticity.
Additional insight into prefrontal and striatal synaptic dysfunction came from a cDNA micro-array study which screened 1176 rat genes (BD Bioscience Clontech Atlas cNDA Expression Array) in samples of NAc core, NAc shell, striatum and dorsal PFC of rats following 3 weeks of withdrawal from 7 days of cocaine administration (intraperitoneal; 15 mg/kg on days 1 and 7, 30 mg/ kg on days 2–6) (Toda et al., 2002). Nine genes were identified with at least 40% increase or 29% decrease relative to controls in one of the four brain regions studied. In the PFC, the authors noted a significant downregulation of the neurotrophic tyrosine kinase receptor type 2 (Ntrk2) in the PFC of cocaine-treated rats. Ntrk2 is the receptor for brain-derived neurotrophic factor (BDNF) previously shown to be involved in the behavioral effects of cocaine in the VTA and NAc (Berhow et al., 1996; Horger et al., 1999; Pierce and Bari, 2001; Freeman and Pierce, 2002). Though not significantly different at the protein level in the PFC, protein levels of the Ntrk2 truncated isoforms p95 and p145 were upregulated in the core of the NAc — a region receiving inputs from the distal regions such as the VTA, hippocampus, etc. Interestingly, the NAc core region exhibited changes in the expression of five transcripts: mitochondrial ATP synthase subunit D (ATP5H), adenosine receptor 1 (ADORA1/A1), leukocyte common antigen-related tyrosine phosphatase (LAR), RET ligand 2 (Retl2) (also known as glial cell line-derived neurotrophic factor family receptor alpha 2; Gfra2). The authors also identified a cocaine-induced downregulation of gastric inhibitory peptide (GIP) mRNA (also known as glucose-dependent insulinotropic polypeptide) — recently shown to be upregulated by chronic clozapine administration in the striatum (Sondhi et al., 2006) suggesting mediation of this transcript by dopamine given the reciprocal regulation by cocaine and clozapine. More recently, Gip was shown to be expressed in rat hippocampus and involved in a regulatory function in progenitor cell proliferation in the dentate gyrus (Nyberg et al., 2005). Examination of transcript-encoded transcripts showed significantly elevated levels of adenosine 1 receptor protein in the NAc core which may represent a compensatory response to the cocaine-induced upregulation of the D1/Gs signaling cascade documented previously (Nestler, 2001; Scheggi et al., 2004; Zhang et al., 2005), a decreased Gi/Go function (Nestler et al., 1990), elevated adenosine levels (Manzoni et al., 1998), or some combination thereof.
Kreek and colleagues further examined cocaine-induced gene expression in the striatum following acute (3 hourly injection of 15 mg/kg for 1 day) and chronic (3 hourly injections of 15 mg/kg for 3 days) “binge” administration using the Affymetrix rat genome U34A containing approximately 8000 gene/EST clusters (Yuferov et al., 2003). The authors noted 117 upregulated and 22 downregulated transcripts as a result of cocaine administration. Upregulated transcripts included immediate-early genes, “effector” and scaffolding proteins and receptors and signal transduction proteins, while downregulated transcripts was comprised primarily of transcripts related to mitochondrial function along with transcripts encoding signal transduction proteins. RNAse protection assays were used to confirm differential expression as noted by array analysis. In addition to expanding our understanding of cocaine-induced regulation of several gene families and pathways, the authors revealed upregulation of the Per2 clock gene and the somatostatin receptor 2 following “binge” cocaine administration. Previously, disruption of Per genes have been shown to block cocaine-induced sensitization in Drosophila (Andretic et al., 1999) and mice (Abarca et al., 2002); however, the localization to the striatum is interesting in that previous studies have found expression limited to the suprachiasmatic nucleus (Masubuchi et al., 2000). The elevated expression of SSTR2 may possibly reflect a less-studied mechanism of cocaine-regulated dopamine release in the striatum as noted by the authors. Additional studies that examine the cellular origin and localization of the Per 2 transcript and protein and the role of SSTR2 in the behavioral effects of cocaine are warranted.
Rodent studies: self-administration
The previous studies have expanded the knowledge base of the cocaine’s effects in the brain and provided novel insights into the pharmacological effects of cocaine in various brain regions; however, all used the non-contingent administration of cocaine and thus may have limited applicability to understanding the abuse liability/reinforcing effects of cocaine. As discussed in the Introduction, inferences of reinforcement mechanisms drawn from studies using experimenter drug administration protocols may be misleading as several studies have shown significant differences between experimenter- and self-administered drugs of abuse (Wilson et al., 1994; Hemby et al., 1995, 1997a, b; Hemby, 1999). To date, two studies have combined rodent intravenous self-administration procedures with functional genomics procedures. Ahmed and colleagues examined gene expression profiles in samples of NAc, lateral hypothalamus, septum, VTA, medial PFC and amygdala from rats self-administering cocaine or serving as controls using pooled samples of RNA on the Affymetrix Neurobiology RNU434 chips (Ahmed et al., 2005). The cocaine self-administration group was divided into two subgroups: short access (ShA; 1 h/day; 250 mg/infusion) and long access (LhA; 6 h/day; 250 mg/infusion access) in which one press of a level resulted in the delivery of the dose of cocaine through the intravenous catheter. This procedure results in a marked escalation of cocaine intake within the first hour of access and has been proposed as a model of compulsive drug intake (Ahmed and Koob, 1998, 1999; Ahmed et al., 2002). Interestingly, the lateral hypothalamus exhibited the greatest number of genes that were regulated by cocaine self-administration access (ShA and LhA) and by the escalation paradigm (LhA versus ShA) when compared to the other brain regions studied and differential expression of select transcripts were confirmed by qPCR. Transcripts altered by the escalation paradigm were members of several functional classes including functional and structural plasticity, receptors, synthetic and metabolic enzymes, neurotransmitter release, and proteins coding for neuronal growth and survival.
The aforementioned studies utilized dissected brain regions from rats to generate molecular profiles of cocaine administration. As noted in the previous section on the neuroanatomical basis of reinforcement, the circuitry that mediates the reinforcing effects of cocaine and others drugs of abuse is well-defined and includes dopaminergic cell bodies in the VTA that projects to several forebrain and cortical regions. The advent of discrete cell microdissection and laser capture microdissection (LCM) combined with RNA amplification strategies makes it possible to evaluate expression patterns in defined cell populations in the brain (Ginsberg et al., 1999, 2000, 2004; Hemby et al., 2002; Fasulo and Hemby, 2003). Whereas previous studies have examined regional gene expression profiles in the VTA as a function of cocaine administration, the effects of cocaine self-administration on VTA dopamine neurons remain largely unknown even though these cells are a critical substrate of drug reinforcement. To this end, the expression profile of 95 transcripts following 1 or 20 days of intravenous cocaine self-administration was assessed in dopamine neurons of the VTA in rats (Backes and Hemby, 2003). Tyrosine hydroxylase immunopositive cells were microdissected from the VTA using LCM microdissection and aRNA amplification was used to provide a linear amplification of the mRNA from each rat (Van Gelder et al., 1990; Eberwine et al., 1992; Eberwine, 2001; Hemby et al., 2002). Five GABA-A receptor subunit mRNAs (α4, α6, β2, γ2, and δ) were downregulated at both 1 and 20 days of cocaine self-administration. In contrast, the catalytic subunit of protein phosphatase 2A (PP2α), GABA-A α1 and Gαi2 were significantly increased at both time points. Additionally, calcium/calmodulin-dependent protein kinase IIα (CaMKIIα) mRNA levels were increased initially followed by a slight decrease after 20 days, whereas neuronal nitric oxide synthase (nNOS) mRNA levels were initially decreased but returned to near control levels by day 20. These results indicate that alterations of specific GABA-A receptor subtypes and other signal transduction transcripts appear to be specific neuroadaptations associated with cocaine self-administration. Moreover, as subunit composition determines the functional properties of GABA-A receptors, the observed changes may indicate alterations in the excitability of dopamine transmission underlying long-term biochemical and behavioral effects of cocaine.
Transgenic mouse studies
In an elegant series of experiments, Nestler and colleagues utilized ΔFosB and CREB-inducible transgenic mice with targets know to be involved in the behavioral effects of cocaine to ascertain their effects on the down-stream regulation of gene expression. Previous studies have shown that repeated cocaine administration leads to sustained elevation of ΔFosB levels in brain regions associated with the behavioral effects of cocaine (Hope et al., 1994; Moratalla et al., 1996; Nestler, 2001; Nestler et al., 2001; McClung and Nestler, 2003; Perrotti et al., 2005; Brenhouse and Stellar, 2006). Using the ΔFosB-inducible transgenic mouse model, the investigators were able to demonstrate increased levels of cyclin-dependent kinase 5 (cdk5) mRNA following induction and similarly increased following chronic cocaine administration (Bibb et al., 2001) using a 588 cDNA mouse array (BD Bioscience Clontech Atlas cNDA Expression Array). More importantly, a functional role of cdk5 in cocaine-mediated behaviors was shown by antagonism of cdk5 in the striatum and attenuation of kainate peak currents in the striatum following cocaine administration (Bibb et al., 2001). In a separate study using the ΔFosB-inducible transgenic mouse model, the authors employed the higher density Affymetrix DNA mouse array and found significantly higher levels of NFκB mRNA and protein in the transgenic mice and similar elevations in NFκB protein levels in wild-type mice administered cocaine (20 mg/kg; 14 days) (Ang et al., 2001).
Comparison of the effects of ΔFosB- and CREB-inducible transgenic mouse models on transcription in the NAc revealed that the majority of transcripts induced by CREB occurred after 2 weeks of expression and were sustained at 8 weeks of expression (McClung and Nestler, 2003). Conversely, ΔFosB expression generated dichotomous patterns of gene expression at 2 and 8 weeks with the 2-week expression pattern for ΔFosB similar to CREB expression. The longer ΔFosB expression was similar to effects observed following expression of the dominant-negative CREB. Interestingly, acute cocaine administration (5 days; 10 mg/kg) induced 21% of the genes induced by CREB expression alone whereas chronic cocaine administration (15 mg/g; 20 days) induced 27% of the genes induce by ΔFosB expression alone, leading the authors to conclude that the effects of short-term cocaine administration are more dependent on CREB, whereas chronic administration is dependent on ΔFosB. The list of genes attributable to the induction of CREB and ΔFosB is lengthy and will not be reviewed in here entirely for the sake of brevity; however it is important to note that these studies have significantly expanded the knowledge of transcriptional regulation by these transcription factors and the understanding of the neuroadaptive effects of cocaine administration.
Using a similar approach, Caron and colleagues examined the striatal transcriptomes of three transgenic mouse models, dopamine, norepinephrine, and vesicular monoamine 2 transporter knockouts and a cocaine-treated mouse model using the Affymetrix mouse Genechips (MG U74v2 Set) containing approximately 36,000 gene clusters (Yao et al., 2004). Twenty-six transcripts were altered in all three knockouts and six genes were also found to be altered following chronic cocaine administration (20 mg/kg per day for 5 days followed by 14 days of withdrawal) — adenylate cyclase 1 (signal transduction and plasticity), Pin/Dic-2 (involved in NOS activity and signaling) and post-synaptic density protein 95 kDa (PSD-95; involved in scaffolding of NMDA receptors and plasticity). In situ hybridization indicated a significant decrease in PSD-95 levels in the NAc and striatum of all knockdowns and the cocaine-treated groups, and qPCR confirmed similar decreases in the whole striatum — separate qPCR assessments in NAc and caudate-putamen were not performed. Similarly PSD-95 protein levels were decreased in the NAc, caudate-putamen and in whole striatum of all three knockouts and the cocaine-treated mice. In addition, all four groups exhibited altered synaptic plasticity of cortical accumbal plasticity.
Non-human primates
One of the first published studies to utilize array technology examined the effects of chronic intramuscular injections of cocaine in cynomolgus monkeys on gene expression in the NAc using a low-density human macroarray from Clonetech consisting of 588 probes (Freeman et al., 2001b). Pools of mRNA from each group were hybridized to two separate arrays leading to the identification of 18 transcripts designated as differentially expressed and included. Unfortunately, the complete list of differentially expressed transcripts is not provided in the manuscript and the website containing the complete dataset is no longer functional. Of the 18 differentially expressed transcripts, eight were selected for post-hoc analysis using Western blot procedures. Four of the eight selected encoded proteins exhibited significant increases in abundance (as hypothesized from the array data) and included PKAα subunit (catalytic; PKAα), the beta subunit of cell adhesion tyrosine kinase, MEK1 and β-catenin. Differences in the protein expression of the remaining four targets did not agree with the array data, which could be due to several factors including post-transcriptional degradation, differences in spatial trafficking of mRNA and protein in neurons, or more practical factors such as the extrapolation of data from pooled RNA samples. An additional limitation of this study is the cross-species hybridization of monkey cDNA (generated using human PCR primers) with human extended oligo probes. The generation of targets for the Clontech assay is a PCR-based method in which primers are used which correspond to the human cDNA sequence. In this case, the overriding assumption is that the Macaca fascicularis cDNA is identical to the human cDNA sequence for the transcripts of interest such that the primers would readily anneal to the monkey cDNA and prime the PCR reaction. The lack of specificity of the human primers for cynomolgus cDNA may lead to an underestimation of the abundance of target transcripts and/or may represent the amplification of multiple transcripts in the cynomolgus monkeys.
Nonetheless, the authors aptly point out that the confirmed targets are members of a common biochemical pathway that interact with CREB and AP-1 proteins shown previously to be regulated in rodent models following cocaine administration.
More recently, Hemby and colleagues have used a non-human primate cocaine self-administration model to validate protein and mRNA changes observed in human post-mortem tissue of cocaine-overdose victims (Hemby et al., 2005b). Unfortunately, attempts to recapitulate changes observed in cocaine overdose victims and non-human primate models in rodent self-administration models have not succeeded (Tang et al., 2004; Hemby et al., 2005a). Additional studies are needed to specifically address the ability of the rodent model to recapitulate biochemical changes observed in the primate brain. Whereas rodent models have provided significant information on drug-induced alterations, non-human primate models more closely approximate the anatomy and biochemical milieu of the human brain. For instance, differences between rodents and primates in frontal lobe anatomy (Preuss, 1995) are likely to be reflected in prefrontal–accumbal glutamatergic neurotransmission. In addition, mid-brain dopamine projections in rodents have been ascribed to different midbrain nuclei; however, studies in primates suggest a more complex pattern (Lynd-Balta and Haber, 1994; Williams and Goldman-Rakic, 1998). The use of non-human primates may allow the development of a more clear and clinically relevant characterization of the biochemical changes associated with cocaine use.
Human post-mortem studies
Understanding the consequences of long-term cocaine abuse on post-mortem brain tissues requires vigorous investigation with the benefit of revealing whether the adaptations observed in rodent and non-human primates are applicable to human brain, and which changes are state or trait markers in human drug abusers. Findings in postmortem brains often provide the first leads that can be investigated in living brain, for example the loss of dopamine in Parkinson’s disease (Kish et al., 1988), changes in the levels of the dopamine transporter (Little et al., 1993a, b; Staley et al., 1994a, b; ) or opiate system (Hurd and Herkenham, 1993; Staley et al., 1997) with chronic cocaine exposure, and the downregulation of the nicotinic ACh receptor after chronic nicotine (Breese et al., 1997). Although there are many difficulties with post-mortem brain studies, this approach is one of the most promising ways to view biochemical changes relevant to human drug abusers and to educate the public about the consequences of cocaine abuse. Whereas animal studies have advanced our understanding of the neurobiological basis of drug addiction, the evaluation of similar questions in human tissue are few, yet are essential. By assessing changes in defined biochemical pathways in human post-mortem tissue, the fundamental molecular and biochemical processes associated with long-term cocaine use can be ascertained.
Bannon and colleagues examined gene expression in the NAc of post-mortem brain tissue of human cocaine abusers and controls using Affymetrix Human U133A and U133B arrays with represent over 39,000 transcripts (Albertson et al., 2004). Forty-nine transcripts were present in all pairs (n = 10) of cocaine and control cases and were differentially expressed in the NAc of cocaine abusers. Transcripts were members of several functional classes including signal transduction, transcriptional and translational processing, neurotransmission and synaptic function, glia, structural and cell adhesion, receptors/transporters/ion channels, cell cycle and growth, and lipid and protein processing. The authors noted a significant upregulation of cocaine and amphetamine-related transcript (CART), a transcript previously discovered following cocaine administration in rats (Douglass et al., 1995; Douglass and Daoud, 1996). In addition, several myelin-associated transcripts were significantly decreased in the NAc of cocaine abusers including myelin basic protein (MBP), proteolipid protein 1 (PLP) and myelin-associated oligodendrocyte basic protein (MOBP) and a significant increase in T-cell differentiation protein (MAL2) — which were confirmed by qPCR. Immunohistochemistry revealed a similar decrease in MBP immunoreactivity in the NAc of these subjects as well. These data provide molecular basis of previous studies which suggested altered white-matter density and myelin expression in cocaine abusers (Volkow et al., 1988; Wiggins and Ruiz, 1990; Lim et al., 2002).
In a separate cohort, Hemby and colleagues used targeted macroarrays consisting of 96 cDNAs to compare gene and protein expression patterns between cocaine overdose victims and age-matched controls in the VTA and lateral substantia nigra (l-SN) (Tang et al., 2003). Evaluated transcripts included ionotropic glutamate receptor (iGluR) subunits, GABAA receptor subunits, dopamine receptors, G-protein subunits, regulators of G-protein signaling and other GTPases, transcriptional regulation, cell growth and death, and others (CART, cannabinoid receptor 1, and serotonin receptors 2A, 2C, and 3). Array analysis revealed significant upregulation of numerous transcripts in the VTA, but not l-SN, of cocaine overdose victims including NMDAR1, GluR2, GluR5, and KA2 receptor mRNAs. Corresponding Western blot analysis revealed VTA-selective upregulation of CREB, NR1, GluR2, GluR5, and KA2 protein levels in cocaine overdose victims. These results indicate that selective alterations of CREB and certain iGluR subunits appear to be associated with chronic cocaine use in humans in a region-specific manner. Extending these studies, we recently examined the extent of altered iGluR subunit expression in the NAc and putamen in cocaine overdose victims (Hemby et al., 2005b). Results revealed statistically significant increases in the NAc, but not in the putamen, of NR1 and GluR2/3 with trends in GluR1 and GluR5 in cocaine-overdose victims (COD). In order to determine that changes were related to cocaine intake and not to other factors in the COD victims, the effects of cocaine intravenous self-administration in rhesus monkeys for 18 months (unit dose of 0.1 mg/kg/injection and daily drug intake of 0.5 mg/kg/session) were examined. Statistically significant elevations were observed for NR1, GluR1, GluR2/3, and GluR5 (P < 0.05) and a trend toward increased NR1 phosphorylated at Serine 896 (p < 0.07) in the NAc but not putamen of monkeys self-administering cocaine compared to controls (Hemby et al., 2005b). These results extend previous results by demonstrating an upregulation of NR1, GluR2/3, and GluR5 in the NAc and suggest these alterations are pathway specific and likely mediate in part the persistent drug intake and craving in the human cocaine abuser.
Proteomics
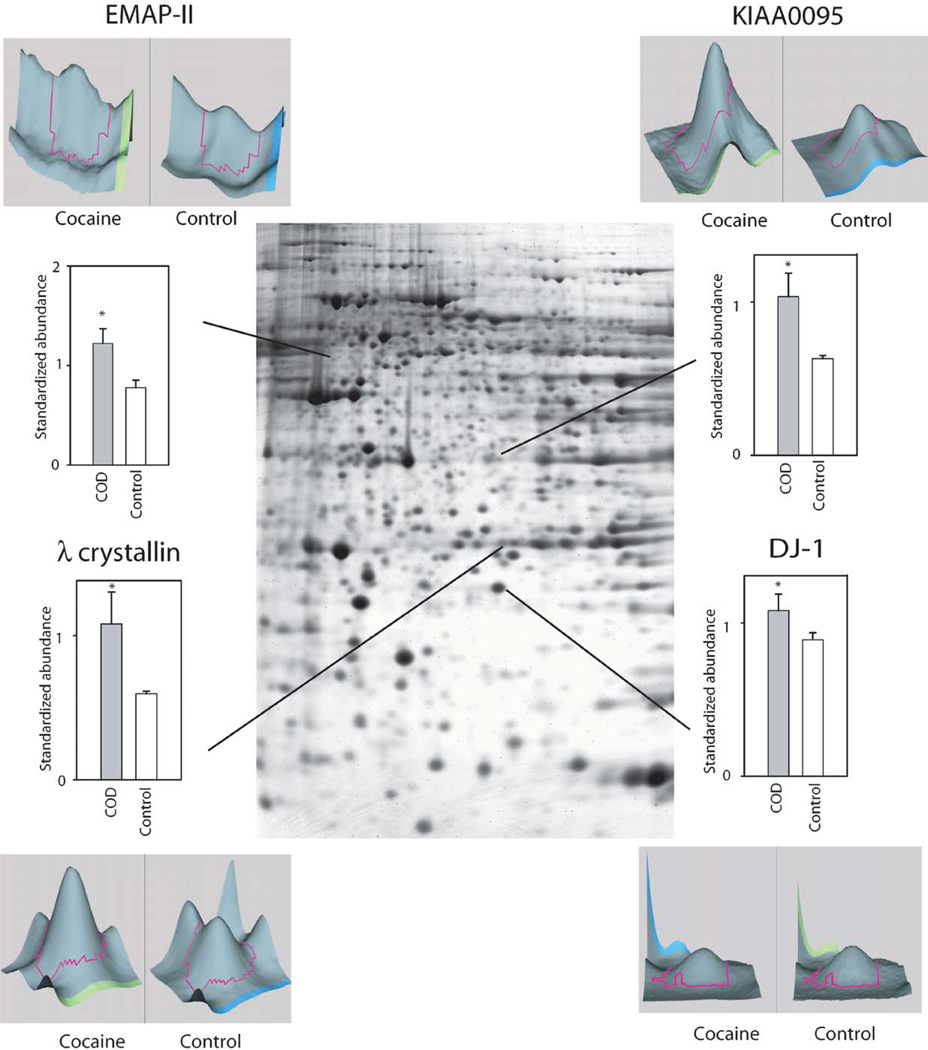
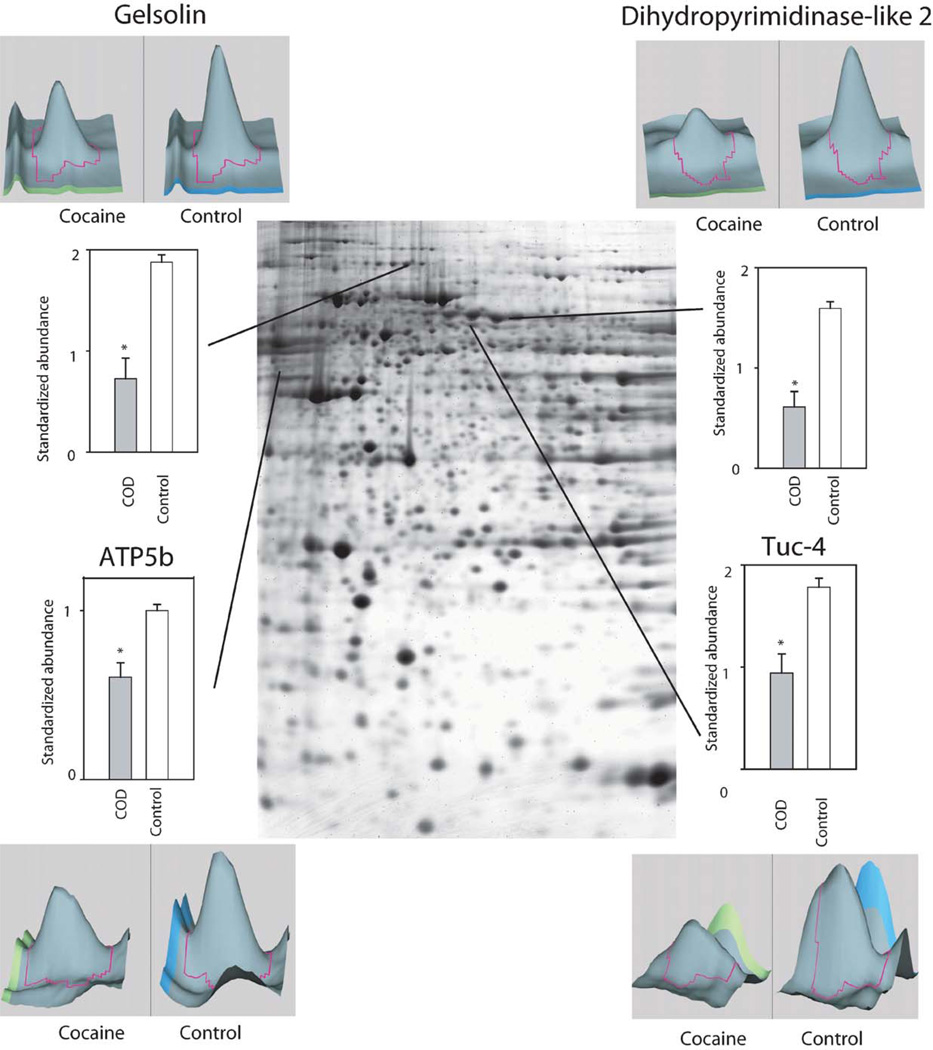
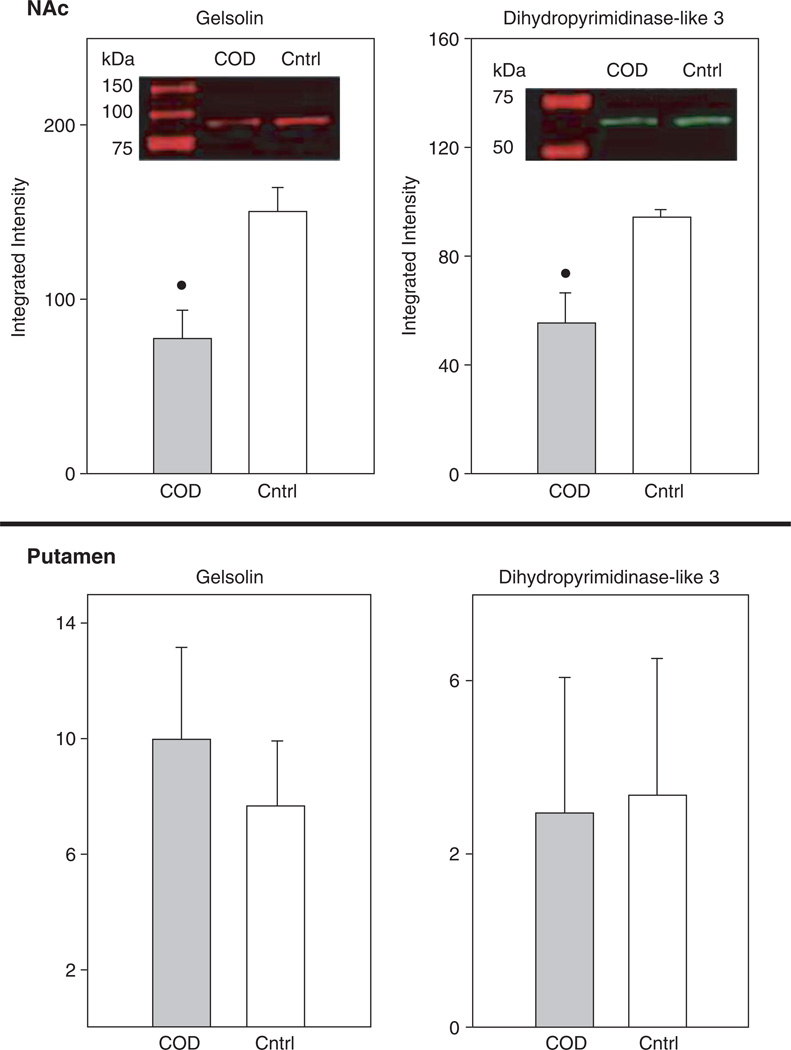
Whereas several studies have assessed gene and subsequent protein expression as a function of cocaine administration in humans and animal models, to date there are few studies using high-throughput proteomic technologies to examine drug-induced global protein expression patterns in brain regions (Freeman and Hemby, 2004; Freeman et al., 2005; Kim et al., 2005). In order to begin to fill this void in the field of the neurobiology of cocaine addiction, our lab has embarked on several studies in rhesus monkey cocaine self-administration models and in human post-mortem tissue from COD victims. Initial efforts have focused on changes in the NAc given the role of this brain region in the addictive processes of cocaine and the growing gene expression databases. In a preliminary study, cytosolic fractions of NAc proteins from human COD and controls (n = 5/group) were separated and quantified by two-dimensional difference gel electrophoresis (2-DIGE) and identified by matrix-assisted laser desorption/ionization time-of-flight (MALDI ToF/ToF) mass spectroscopy (see Chapter 4 for detailed explanation of procedures). Greater than 1000 spots were detected across the five pairs (COD and controls) of which 340 spots were excised, digested in-gel with trypsin, and subsequently analyzed by MALDI ToF/ToF (see Supplemental Table I). Fifty-two percent of the spots were positively identified including 11 upregulated proteins including DJ1 (Parkinson’s disease 7 (PARK7; autosomal recessive, early onset)), ubiquitin carboxyl-terminal esterase L1 (UCHL1; PARK5), lamda crystallin, endothelial monocyte-activating polypeptide 2 (EMAP-II) and others (Fig. 1). DJ1, a redox-sensitive chaperone that protects neurons against oxidative stress and cell death, and UCHL1, a neuronal de-ubiquitinating enzyme, are both associated with Parkinson Disease (Abou-Sleiman et al., 2006). Combined with elevated a-synuclein levels in human COD victims (Qin et al., 2005), these data support the suggestion by Deborah Mash and colleagues that chronic cocaine use induce Parkinson-like pathology in striatal regions. Eighteen-positively identified proteins were found to be downregulated in the NAc of COD victims including gelsolin, ATP5b, dihydropyrimidinase-like 3 (DRP3/TUC-4) and dihydropyrimidinase-like 2 (DRP2) (Fig. 2). Decreased expression of gelsolin and DRP3/TUC-4 and gelsolin was confirmed by immunoblotting (Fig. 3) Gelsolin has been reported to exhibit antiapoptotic properties in neurons (Harms et al., 2004) and fibroblasts (Ahn et al., 2003a, b) such that decreased gelsolin expression may render NAc cells more susceptible to apopotosis and oxidative stress due to cocaine exposure. DRP2 and 3 are generally associated with nerve terminal activity, more specifically axonal restructuring and decreased expression may imply decreased plasticity of NAc cells with chronic cocaine exposure. Efforts are underway to assimilate genomic and proteomic databases in a more systematic manner. The application of proteomics holds great promise to understanding the biology of psychiatric diseases, including substance abuse disorders. Further investigation of the changes found and a more comprehensive examination of the human proteome, which may provide the biological understanding and identification of novel therapeutic targets for treatment of cocaine dependence.
Fig. 1.
Preliminary data of representative proteins exhibiting increased abundance in COD victims. Signal intensities for specific gel spots from COD victims and control subjects were compared. Included in the figure are the proteins quantified by the 2DIGE technique using the normalization by Cy2-labeled pool sample and have statistical significance difference in expression profiles between the two groups (*p<0.05, t-test). Examples of proteins are provided with representative 3-D plots of individual COD and control spots.
Fig. 2.
Preliminary data of representative proteins exhibiting decreased abundance in COD victims. Signal intensities for specific gel spots from COD victims and control subjects were compared. Included in the figure are the proteins quantified by the 2DIGE technique using the normalization by Cy2-labeled pool sample and have statistical significant difference in expression profiles between the two groups (*p<0.05, t-test). Examples of proteins are provided with representative 3-D plots of individual COD and control spots.
Fig. 3.
Western blot analysis of gelsolin and DRP3. Assessment of protein levels from the samples used for 2DIGE revealed significant decreases in gelsolin and DRP3 in agreement with the 2DIGE analyis. Moreover, these changes were specific to the NAc and not observed in the putamen. (*p<0.05, t-test). β tubulin was used as a loading control and no differences were observed for this protein.
Conclusion
In conclusion, relevant gene expression profiles for cocaine abuse and other substance abuse disorders are being generated expanding our knowledge of drug-induced changes in the brain that may underlie persistent drug taking and relapse. Results from rodent, non-human primate and human post-mortem studies indicate significant impairments in neuronal function and plasticity in several brain regions. To date the majority of studies have utilized rodents to model human cocaine intake, however growing evidence indicates the need to refine rodent and non-human primate models to better recapitulate human drug intake and associated neuropathologies. As in other psychiatric and neurological illnesses, researchers should identify the molecular pathologies associated with cocaine addiction in humans and attempt to recapitulate such biological alterations in animal models.
The neurobiological and molecular characteristics of cocaine addiction, although specific to cocaine, may generalize to other drug dependencies. Understanding the coordinated involvement of multiple proteins with chronic cocaine abuse provides insight into the molecular basis of drug dependence and may offer novel targets for pharmacotherapeutic intervention. Although significant advances have been made in the identification of neurochemical and neurobiological substrates involved in the behavioral effects of abused drugs, the relationship between these effects and resultant alterations in gene and protein expression remains in its infancy. The relationship between altered gene and protein expression and the addictive effects of specific drugs remains understudied. The application of this information to the development of treatment strategies has not been fruitful for several reasons. One explanation is that research in the areas of neurobehavioral pharmacology and molecular biology has proceeded in relative isolation of each other. To date, there have been few published studies combining models of self-administration with genomic and proteomic approaches. Other possible explanations include (1) the inappropriate use of experimental models, (2) reliance on non-neuronal systems or neuronal tissue not directly involved in the reinforcing effects of the drug, and (3) the lack of definable neural substrates at the cellular or biochemical level. The combination of appropriate behavioral models of drug reinforcement, specific neurobiological systems and state-of-the-art molecular techniques will provide the most pertinent data for understanding the molecular basis of drug reinforcement and for potentially establishing novel targets for pharmacotherapeutic intervention.
A more detailed understanding of the molecular and biochemical cascades in specific neuronal populations and the interactions between well-defined neuronal populations within discrete brain regions could lead to a greater knowledge of the basic neurobiological processes involved in drug reinforcement. Future efforts investigating the biological basis of drug reinforcement should be directed at specific cellular targets in brain regions considered to be involved in drug reinforcement. The integration of basic neuroscience and behavior offers the most productive avenue for delineating the complexity of the neurobiological underpinnings of drug reinforcement and the subsequent development of effective pharmacotherapies to treat addiction.
Table 1.
Identified and matched proteins
| Spot # | Protein GI # |
Protein name | Theoretical MW |
Theoretical pI |
Peptide count |
Mascot score |
Confidence interval |
t-test value |
Average ratio |
Protein homolgues/other protein names |
|---|---|---|---|---|---|---|---|---|---|---|
| 526 | 28595 | Aldolase A; fructose-bisphosphate aldolase | 39851.5 | 8.3 | 15 | 196 | 100 | −1.1286 | ALDOA | |
| 769 | 136066 | Triosephosphate isomerase | 26909.8 | 6.45 | 4 | 74 | 99.944 | 0.05221 | −1.3549 | TIM |
| 772 | 136066 | Triosephosphate isomerase | 26909.8 | 6.45 | 4 | 74 | 99.944 | 0.1059 | 1.3564 | TIM |
| 775 | 136066 | Triosephosphate isomerase | 26909.8 | 6.45 | 4 | 74 | 99.944 | 0.111 | −1.1886 | TIM |
| 459 | 180570 | Creatine kinase | 42876.4 | 5.3 | 17 | 571 | 100 | 0.5089 | −1.0726 | CKB |
| 401 | 285975 | Rab GDI | 51088 | 5.94 | 13 | 200 | 100 | 0.654 | −1.0426 | GDP dissociation inhibitor 2, GDI2 |
| 259 | 334284 | GP120 | 58060.7 | 5.35 | 7 | 87 | 99.997 | 0.2132 | −1.3476 | |
| 660 | 387016 | Phosphoglycerate mutase | 28867.8 | 8.77 | 2 | 62 | 99.176 | 0.09345 | −1.1645 | PGAM2, phosphoglycerate mutase 2 (muscle) |
| 450 | 423123 | Tpr protein | 238769.7 | 5.05 | 20 | 63 | 99.315 | 0.1194 | −1.2695 | |
| 951 | 494781 | Fatty acid-binding protein | 14774.7 | 6.34 | 6 | 236 | 100 | 0.4423 | 1.0726 | |
| 162 | 763431 | Albumin | 52047.8 | 5.69 | 7 | 80 | 99.986 | 0.06642 | −1.8004 | |
| 639 | 999892 | Chain A, triosephosphate isomerase | 26806.8 | 6.51 | 5 | 274 | 100 | 2.9063 | ||
| 221 | 1465733 | Cytosolic NADP(+)-dependent malic enzyme | 63858.9 | 5.88 | 4 | 59 | 98.318 | 0.4025 | − 1.3355 | ME1 |
| 531 | 2118269 | Zebrin II | 39797.4 | 6.67 | 8 | 198 | 100 | 1.4454 | Similar to human Aldolase C | |
| 322 | 2183299 | Aldehyde dehydrogenase 1 | 55427.2 | 6.3 | 12 | 219 | 100 | 0.6272 | 1.0482 | ALDH1A1 |
| 741 | 2737906 | Plasminogen-related protein A | 7983.9 | 8.44 | 5 | 61 | 99.01 | 0.2901 | 1.8309 | LOC285189 |
| 705 | 2914390 | Chain B, hemoglobin mutant | 15834.2 | 6.76 | 4 | 84 | 99.995 | 0.4753 | −1.1197 | |
| 953 | 2981643 | Chain B, hemoglobin | 15980.2 | 6.75 | 4 | 93 | 99.999 | 0.6047 | −1.0872 | |
| 864 | 2982080 | Familial als mutant G37r, chain A | 16122 | 5.87 | 2 | 133 | 100 | 0.158 | 1.7454 | |
| 861 | 2982080 | Familial als mutant G37r, chain A | 16122 | 5.87 | 2 | 133 | 100 | 0.9453 | −1.0013 | |
| 797 | 3205211 | Non-muscle myosin heavy chain | 72555.1 | 5.18 | 9 | 62 | 99.117 | 0.05802 | 1.4379 | |
| 413 | 3766197 | ATP-specific succinyl-CoA synthetase beta subunit | 46732.3 | 5.84 | 4 | 62 | 99.117 | 0.3475 | 1.5502 | Succinate-CoA ligase, ADP-forming, beta subunit; SUCLA2 |
| 112 | 3811317 | Tryptophan hydroxylase isoform 1 | 6476.5 | 9.7 | 4 | 56 | 96.486 | 0.8722 | 1.3864 | TPH |
| 150 | 4389275 | Serum albumin | 67988.5 | 5.69 | 25 | 679 | 100 | 0.1761 | 2.0119 | |
| 161 | 4389275 | Serum albumin | 68424.7 | 5.67 | 35 | 975 | 100 | 0.4513 | 1.597 | |
| 523 | 4502561 | Capping protein (actin filament), gelsolin-like | 38778.6 | 5.88 | 3 | 67 | 99.727 | 0.2784 | −1.2687 | CAPG |
| 280 | 4503377 | Hydropyrimidinase-like 2; collapsin response mediator | 62710.7 | 5.95 | 14 | 325 | 100 | 0.3267 | −1.3266 | CRMP2; DRP2; DPYSL2 |
| 282 | 4503377 | Hydropyrimidinase-like 2; collapsin response mediator | 62710.7 | 5.95 | 14 | 380 | 100 | 0.6647 | −1.0535 | CRMP2; DRP2; DPYSL2 |
| 272 | 4503971 | Rab GDI-alpha | 51177.4 | 5 | 8 | 131 | 100 | 0.2966 | 1.5102 | GDP Dissociation Inhibitor 1; GDI1; oligophrenin 2; OPHN2; RHOGDI |
| 261 | 4503971 | GDP Dissociation inhibitor 1 | 51177.4 | 5 | 9 | 112 | 100 | 0.3715 | 1.3108 | GDI1 |
| 58 | 4504165 | Gelsolin | 86043.3 | 5.9 | 15 | 258 | 100 | 0.2562 | −1.6123 | GSN |
| 385 | 4504169 | Glutathione synthetase | 52523.3 | 5.67 | 17 | 346 | 100 | 0.06022 | −2.0733 | GSS; GSHS; MGC14098 |
| 391 | 4504169 | Glutathione synthetase | 52523.3 | 5.67 | 13 | 227 | 100 | 0.4307 | −1.1292 | GSS; GSHS; MGC14098 |
| 667 | 4505585 | Platelet-activating factor acetylhydrolase | 25724.2 | 5.57 | 2 | 60 | 98.724 | 0.8345 | 1.0289 | PAFAH1B2; platelet-activating factor acetylhydrolase, isoform Ib, beta subunit 30 kDa |
| 366 | 4506019 | Protein phosphatase 3, catalytic subunit, alpha isoform | 52172.7 | 5.82 | 7 | 72 | 99.912 | 0.9398 | −1.0099 | Calcineurin A alpha, PPP3CA, PP2BCA |
| 275 | 4506089 | Mitogen-activated protein kinase 4 | 63039.9 | 6.05 | 8 | 64 | 99.417 | 0.06896 | −1.9117 | MAPK4, p63MAPK |
| 314 | 4506089 | Mitogen-activated protein kinase 4 | 63039.9 | 6.05 | 11 | 72 | 99.916 | 0.3501 | −1.2684 | MAPK4, p63MAPK |
| 920 | 4507793 | Ubiquitin-conjugating enzyme E2N | 17184 | 6.13 | 2 | 66 | 99.694 | 0.5976 | −1.0522 | Uniquitin-conjugating enzyme 13, UBC13; bendless, ubchen |
| 633 | 4557032 | Lactate dehydrogenase B | 36900.2 | 5.71 | 13 | 559 | 100 | 0.1127 | −1.4306 | LDHB |
| 783 | 4557032 | Lactate dehydrogenase B | 36900.2 | 5.71 | 13 | 559 | 100 | 0.5403 | −1.1423 | LDHB |
| 636 | 4557032 | Lactate dehydrogenase B | 36900.2 | 5.71 | 8 | 219 | 100 | 0.8655 | 1.0184 | LDHB |
| 862 | 4557797 | Nucleoside-diphosphate kinase 1 isoform b | 17308.7 | 5.83 | 8 | 274 | 100 | 1.1544 | Non-metastatic cells 1; NME1; NM23A | |
| 95 | 4557871 | Transferrin | 79280.5 | 6.81 | 9 | 105 | 100 | 0.1594 | 2.0797 | TF |
| 97 | 4557871 | Transferrin | 79280.5 | 6.81 | 9 | 105 | 100 | 0.8981 | 1.443 | TF |
| 418 | 4758426 | Guanine deaminase | 51483.8 | 5.44 | 11 | 323 | 100 | 0.2742 | −1.1109 | GDA |
| 684 | 4758484 | Glutathione S-transferase omega 1 | 27833.1 | 6.23 | 7 | 132 | 100 | 0.434 | −1.0991 | GSTO1 |
| 708 | 4758484 | Glutathione S-transferase omega 1 | 27833.1 | 6.23 | 7 | 132 | 100 | 0.4908 | 1.1479 | |
| 647 | 4758638 | Peroxiredoxin 6 | 25133.2 | 6 | 9 | 326 | 100 | 0.1888 | 1.2268 | PRDX6 |
| 764 | 4758638 | Peroxiredoxin 6 | 25133.2 | 6 | 9 | 326 | 100 | 0.6421 | 1.0663 | |
| 567 | 4759036 | Regucalcin;senescence marker protein-30 | 33801.7 | 5.89 | 5 | 64 | 99.48 | 0.3902 | 1.2342 | RGN, SMP30 |
| 168 | 4827056 | WD repeat-containing protein 1 isoform 2 | 58593.2 | 6.41 | 3 | 59 | 98.279 | 0.09829 | −1.8217 | WDR1 |
| 514 | 4885063 | Aldolase C, fructose-bisphosphate; | 39830.4 | 6.41 | 15 | 422 | 100 | 0.3166 | −1.1063 | ALDOC |
| 557 | 5031777 | Isocitrate dehydrogenase 3 (NAD+) alpha precursor | 40022.2 | 6.47 | 6 | 95 | 100 | 0.7043 | −1.0422 | IDH3A |
| 873 | 5031851 | Stathmin 1; metablastin; | 17291.9 | 5.76 | 5 | 147 | 100 | 0.547 | 1.0666 | Leukemia-associated phosphoprotein p18; LAP18 |
| 549 | 5174391 | Aldo-keto reductase family 1, member A1 | 36892 | 6.32 | 12 | 263 | 100 | 0.08951 | −1.492 | ALDR1 |
| 644 | 5174539 | Cytosolic malate dehydrogenase | 36631.1 | 6.91 | 8 | 206 | 100 | 0.06213 | −1.639 | MDH1 |
| 768 | 5174539 | Cytosolic malate dehydrogenase | 36631.1 | 6.91 | 5 | 159 | 100 | 0.6754 | 1.1273 | MDH1 |
| 762 | 5174539 | Cytosolic malate dehydrogenase | 36631.1 | 6.91 | 5 | 159 | 100 | 0.8077 | 1.0359 | MDH1 |
| 170 | 5729877 | HSP70 protein 8 isoform 1 | 71082.3 | 5.37 | 23 | 437 | 100 | 0.1846 | −1.8623 | HSPA8 |
| 561 | 5803187 | Transaldolase 1; dihydroxyacetone transferase; glycerone transferase | 37687.5 | 6.36 | 11 | 178 | 100 | 0.4273 | 1.3777 | TALDO1 |
| 189 | 6005938 | Utrophin, dystrophin-related protein | 396472.1 | 5.21 | 22 | 60 | 98.809 | 0.07498 | −1.7499 | Dystrophin-like protein, DMDL, DRP1 |
| 354 | 6137677 | Mitochondrial aldehyde dehydrogenase | 54394.4 | 5.7 | 7 | 111 | 100 | 0.2883 | −1.1245 | |
| 121 | 6470150 | BiP protein | 71001.6 | 5.23 | 15 | 90 | 99.999 | 0.06848 | −1.4204 | HSPA5; heat shock 70 kDa protein 5 (glucose-regulated protein, 78kDa) |
| 576 | 6688197 | PAP-inositol-1,4-phosphatase | 33743.3 | 5.46 | 9 | 243 | 100 | 0.1324 | 1.1991 | 3′(2′), 5′-bisphosphate nucleotidase 1; BPNT1 |
| 892 | 6806898 | Synuclein, alpha | 11365 | 7.88 | 3 | 67 | 99.727 | 0.9546 | 1.0552 | SNCA |
| 99 | 6912526 | Nasopharyngeal epithelium-specific protein 1 | 46224 | 9.99 | 9 | 64 | 99.526 | 0.1088 | 1.6362 | NESG1 |
| 441 | 7670399 | Unnamed protein product | 43689.4 | 6.1 | 10 | 149 | 100 | 0.09942 | 1.6662 | MEK1 |
| 637 | 7677074 | Lamda crystallin | 33793.2 | 5.68 | 6 | 171 | 100 | 0.7142 | −1.0275 | CRYL1 |
| 755 | 8393948 | Phosphoglycerate mutase 2 | 28907.9 | 8.85 | 2 | 70 | 99.857 | 0.1175 | 1.3015 | Pgam2; Pgmut; PGAM-M; D14Mgh1 |
| 402 | 9966913 | Actin-related protein 3-beta | 40185.1 | 5 | 6 | 126 | 100 | 0.5354 | 1.0727 | ARP11 |
| 638 | 10092677 | Hypothetical protein | 32077.4 | 6.12 | 6 | 81 | 99.99 | 0.06068 | 1.5844 | |
| 724 | 10092677 | Hypothetical protein dJ37E16.5 | 32077.4 | 6.12 | 8 | 261 | 100 | 0.897 | 1.1513 | Pyridoxal phosphate phosphatase, PLP |
| 725 | 10092677 | Hypothetical protein dJ37E16.5 | 32077.4 | 6.12 | 6 | 81 | 99.99 | 1.7072 | Pyridoxal phosphate phosphatase, PLP | |
| 556 | 10241724 | Hypothetical protein | 31816.9 | 5.84 | 7 | 14 | 100 | 0.09672 | 1.3355 | Isocitrate/isopropylmalate dehydrogenase |
| 116 | 10433666 | Unnamed protein product | 88418.5 | 6.68 | 9 | 56 | 96.868 | 0.3911 | 1.2484 | Ring finger protein 20, RNF20 |
| 337 | 10434221 | Unnamed protein product | 63177.7 | 8.73 | 9 | 57 | 97.144 | 0.1442 | −1.4787 | Hypothetical protein FLJ10498, FLJ10498 |
| 439 | 11374664 | Isocitrate dehydrogenase (NADP) (EC 1.1.1.42), cytosolic | 46596.5 | 6.19 | 6 | 67 | 99.757 | 0.7066 | −1.1504 | |
| 242 | 12804225 | CCT5, chaperonin-containing TCP1, subunit 5 (epsilon) | 59886.9 | 5.45 | 11 | 159 | 100 | 0.1577 | −1.611 | |
| 680 | 12860410 | Unnamed protein product | 15612.5 | 10.08 | 5 | 51 | 88.887 | 0.6782 | 1.1754 | AU RNA binding protein/ enoyl-coenzyme A hydratase |
| 516 | 13279173 | Similar to COP9 | 46524.8 | 5.5 | 15 | 346 | 100 | 0.1907 | 1.1899 | COP9 constitutive photomorphogenic homolog subunit 4, COPS4 |
| 728 | 13435960 | Similar to hypothetical protein FLJ23571 | 41024.6 | 9.4 | 10 | 60 | 98.664 | 0.3842 | −1.1525 | Hypothetical protein DKFZp434B227 |
| 534 | 13435960 | Similar to hypothetical protein FLJ23571 | 41024.6 | 9.4 | 7 | 50 | 87.531 | 0.6963 | −1.0929 | Hypothetical protein DKFZp434B227 |
| 317 | 13623415 | Fascin 1 | 55151.3 | 6.84 | 18 | 311 | 100 | 0.1424 | −1.3277 | FSCN1 |
| 149 | 13676857 | HSP70 protein 2 | 69977.9 | 5.56 | 20 | 467 | 100 | 0.885 | 1.0331 | HSPA2 |
| 332 | 13938355 | Unknown | 55708.4 | 5.4 | 15 | 345 | 100 | 0.07185 | −1.6655 | ATPase, H+ transporting, lysosomal 56/58 kDa, V1 subunit B, isoform 2, ATP6V1B2 |
| 359 | 15099973 | Thrombospondin immunoglobulin heavy chain variable region | 12778.3 | 8.67 | 4 | 54 | 94.557 | 0.3817 | 1.2396 | |
| 888 | 15680064 | Similar to stathmin 1/ oncoprotein 18 | 17325.9 | 5.76 | 4 | 58 | 98.069 | 0.3664 | 1.0782 | STMN1 |
| 880 | 15824412 | Neuronal protein 22 | 22629.2 | 6.84 | 6 | 143 | 100 | 0.3978 | 1.2247 | NP22, NP25 |
| 669 | 15930083 | Calbindin 2 | 31663.6 | 5.06 | 7 | 83 | 99.993 | 1.1627 | Calretinin, calbindin 29kDa | |
| 760 | 16198390 | Unknown (protein for MGC:27286) | 33535.7 | 5.4 | 6 | 238 | 100 | 0.1244 | 1.2993 | CGI-150 protein |
| 659 | 16198390 | Unknown (protein for MGC:27286) | 33535.7 | 5.4 | 4 | 78 | 99.98 | 0.1916 | 1.1935 | CGI-150 protein |
| 581 | 16307182 | Similar to transaldolase 1 | 35534.5 | 9.07 | 9 | 124 | 100 | 0.8523 | −1.0085 | TALDO1 |
| 473 | 16924319 | Unknown (protein for IMAGE:3538275) | 40819.4 | 5.78 | 16 | 644 | 100 | 0.913 | −1.0861 | Actin |
| 643 | 17389815 | Triosephosphate isomerase 1 | 26909.8 | 6.45 | 4 | 74 | 99.944 | 0.09687 | 1.7767 | TPI |
| 348 | 18202063 | Endothelial-monocyte-activating polypeptide II (EMAP-II) | 39975.2 | 9.37 | 7 | 60 | 98.809 | 0.1527 | −1.3207 | Small inducible cytokine subfamily E, memeber 1 |
| 950 | 18202063 | Endothelial-monocyte-activating polypeptide II (EMAP-II) | 39975.2 | 9.37 | 5 | 53 | 93.893 | 0.499 | 1.1811 | Small inducible cytokine subfamily E, memeber 1 |
| 117 | 18256043 | Glycyl-tRNA synthetase | 81798.7 | 6.24 | 8 | 98 | 100 | 0.1313 | −1.7071 | Gars |
| 911 | 18307562 | Unnamed protein product | 69825.2 | 9.55 | 7 | 62 | 99.249 | 0.1397 | 7.2612 | |
| 295 | 18307562 | Unnamed protein product | 69825.2 | 9.55 | 5 | 49 | 85.009 | 0.3296 | −1.1903 | |
| 224 | 18307562 | Unnamed protein product | 69825.2 | 9.55 | 8 | 58 | 97.678 | 0.4037 | −1.1735 | |
| 301 | 19705447 | CDC-ike kinase 3 | 59262.5 | 9.53 | 8 | 61 | 99.01 | 0.3224 | −1.1692 | Clk3 |
| 896 | 19716076 | Myeloid cell nuclear-differentiation factor | 46244.3 | 9.72 | 8 | 64 | 99.492 | 0.1551 | 1.41 | |
| 325 | 19913428 | ATPase, H+ transporting, lysosomal 56/58 kD, V1 subunit B, isoform 2 | 56807 | 5.57 | 11 | 216 | 100 | 0.1551 | −1.3285 | ATP6V1B2 |
| 443 | 19923206 | Glutamate–ammonia ligase | 42664.5 | 6.43 | 9 | 173 | 100 | 0.1807 | −1.4274 | GLUL |
| 71 | 20072188 | Aconitase 2 | 86252.3 | 7.62 | 21 | 472 | 100 | 0.1309 | −1.8516 | |
| 934 | 20385874 | Beta-tropomyosin | 17808.9 | 4.6 | 6 | 63 | 99.315 | 0.4222 | 1.5841 | |
| 541 | 20563689 | Mannose phosphate isomerase isoform | 29908.3 | 5.99 | 4 | 80 | 99.988 | 0.8909 | 1.135 | MPI |
| 585 | 20862467 | Hypothetical protein XP_164064 | 14450.5 | 9.57 | 5 | 56 | 96.566 | 0.5297 | 1.0918 | |
| 400 | 20864657 | Similar to Retrovirus-related POL polyprotein | 21374.9 | 9.35 | 6 | 65 | 99.547 | 0.6174 | −1.1668 | Similar to Cas-Br-M ectropic retroviral-transforming sequence b |
| 712 | 20865698 | Similar to protein phosphatase 1, regulatory (inhibitor) subunit 12A | 22293.5 | 9.19 | 6 | 51 | 90.541 | 0.07508 | 1.1249 | PPP1R12A |
| 186 | 20868874 | Hypothetical protein XP_160082 | 25606.8 | 6.41 | 6 | 54 | 94.43 | 0.09973 | −1.5525 | |
| 800 | 20887601 | Hypothetical protein XP_157898 | 21094.5 | 8.58 | 6 | 58 | 98.069 | 0.1626 | 1.3765 | |
| 981 | 20892463 | RIKEN cDNA 1300010H20 | 13156.8 | 9.79 | 4 | 54 | 95.036 | 0.9819 | 1.0705 | Similar to NADH:ubiquinone oxidoreductase B15 subunit |
| 455 | 20892491 | Similar to creatine kinase, brain | 19243.9 | 7.82 | 4 | 59 | 98.279 | 0.9706 | 1.0978 | |
| 55 | 20901108 | Hypothetical protein XP_157013 | 13206.1 | 4.7 | 4 | 53 | 93.893 | 0.4558 | −1.169 | |
| 616 | 20978314 | GTP-ase ran | 24606.6 | 6.6 | 2 | 52 | 90.757 | 0.1235 | −1.5667 | RAN, member RAS oncogene family |
| 795 | 20978314 | GTP-ase ran | 24606.6 | 6.6 | 2 | 52 | 90.757 | 0.9104 | 1.1077 | RAN, member RAS oncogene family |
| 48 | 20984919 | Similar to interferon-inducible protein 10 (IP-10) receptor | 89422.8 | 5.14 | 24 | 429 | 100 | 0.09089 | −1.9122 | |
| 311 | 21313234 | RIKEN cDNA 1300006M19 | 57494.2 | 8.87 | 7 | 58 | 98.024 | 0.05004 | −1.6899 | |
| 570 | 22041696 | Similar to ribosomal protein L7a, cytosolic | 13035.2 | 10.46 | 5 | 55 | 95.677 | 0.1715 | −1.1965 | |
| 658 | 22748619 | Tropomyosin 3 | 28262.3 | 4.72 | 7 | 58 | 97.833 | 0.2513 | −1.1211 | TPM3, alpha-tropomyosin 3 |
| 615 | 22748619 | Tropomyosin 3 | 28262.3 | 4.72 | 8 | 69 | 99.839 | 0.6958 | −1.0386 | TPM3, alpha-tropomyosin 3 |
| 361 | 23208520 | DNA polymerase kappa | 11938 | 9.04 | 5 | 56 | 97.009 | 0.5065 | −1.0928 | |
| 284 | 23308577 | PHGDH, phosphoglycerate dehydrogenase | 57355.7 | 6.29 | 5 | 124 | 100 | 0.06459 | −1.6773 | 3-Phosphoglycerate dehydrogenase |
| 796 | 23395758 | TPA: aflatoxin B1-aldehyde reductase | 40019.9 | 6.7 | 7 | 63 | 99.403 | 0.7007 | −1.0226 | Aldo–keto reductase family 7, member A2 (aflatoxin aldehyde reductase), AKR7A2 |
| 255 | 24987750 | Protein phosphatase 3, catalytic subunit, alpha isoform | 43482.6 | 5.9 | 7 | 78 | 99.98 | 0.07135 | −1.3094 | Calcineurin A alpha, PPP3CA, PP2BCA |
| 252 | 24987750 | Protein phosphatase 3, catalytic subunit, alpha isoform | 42696 | 5.26 | 6 | 61 | 98.836 | 0.1063 | −1.4861 | Calcineurin A alpha, PPP3CA, PP2BCA |
| 399 | 24987750 | Protein phosphatase 3, catalytic subunit, alpha isoform | 3380.3 | 5.26 | 8 | 152 | 100 | 0.1853 | −1.1264 | Calcineurin A alpha, PPP3CA, PP2BCA |
| 723 | 24987750 | Protein phosphatase 3, catalytic subunit, alpha isoform | 43380.3 | 5.26 | 5 | 89 | 99.998 | 0.8823 | 1.3475 | Calcineurin A alpha, PPP3CA, PP2BCA |
| 106 | 25020592 | Hypothetical protein XP_206488 | 11818.2 | 10.44 | 4 | 49 | 84.66 | 0.7306 | 1.6247 | |
| 375 | 25777739 | Aldehyde dehydrogenase 9A1 | 54679.3 | 5.69 | 13 | 299 | 100 | 0.07027 | −1.6495 | ALDH9A1 |
| 376 | 25777739 | Aldehyde dehydrogenase 9A1 | 54679.3 | 5.69 | 13 | 299 | 100 | 0.453 | −1.1089 | ALDH9A1 |
| 378 | 26330804 | Unnamed protein product | 14753.8 | 10.76 | 4 | 52 | 91.949 | 0.1878 | 1.2013 | RIKEN cDNA 5730406M06 gene, 5730406M06Rik |
| 687 | 26336324 | Unnamed protein product | 47225 | 8.58 | 6 | 56 | 96.32 | 3.1461 | RIKEN cDNA 1500032A09 gene, 1500032A09Rik | |
| 409 | 27480797 | Similar to hypothetical protein DKFZp434D0917.1 | 26958.3 | 8.97 | 4 | 54 | 94.43 | 0.949 | 1.0125 | |
| 453 | 27503783 | Similar to mitochondrial translational release factor 1 | 52843 | 8.75 | 5 | 54 | 94.168 | 0.4426 | 1.4356 | RF1; MTTRF1; MGC47721 |
| 964 | 27574235 | Chain B deoxyhemoglobin | 16090.3 | 6.75 | 3 | 108 | 100 | 0.2269 | 1.6911 | |
| 949 | 27574235 | Chain B deoxyhemoglobin | 16090.3 | 6.75 | 3 | 108 | 100 | 0.2754 | 1.3964 | |
| 642 | 27658930 | Similar to ATP-dependent chromatin remodeling protein SNF2H | 42705.1 | 9.1 | 6 | 55 | 95.775 | 0.2464 | −1.1293 | |
| 789 | 27658930 | Similar to ATP-dependent chromatin remodeling protein SNF2H | 42705.1 | 9.15 | 6 | 51 | 90.095 | 0.265 | −1.1694 | |
| 624 | 27677648 | Similar to 60S ribosomal protein L7 | 17571.1 | 9.43 | 5 | 50 | 87.241 | 0.5553 | −1.0843 | |
| 278 | 27707686 | Similar to ribosomal protein L19 | 14078 | 9.87 | 4 | 52 | 91.949 | 0.2165 | −1.3871 | |
| 296 | 27714549 | Similar to ribosomal protein L24 | 12066.3 | 11.2 | 4 | 50 | 86.943 | 0.1397 | −1.5458 | |
| 351 | 27717139 | Similar to 60S ribosomal protein L29 | 13101.2 | 10.94 | 4 | 52 | 90.757 | 0.8939 | −1.0125 | |
| 411 | 27960434 | Colon cancer autoantigen protein | 83725.6 | 6.11 | 10 | 61 | 98.986 | 0.5941 | 1.153 | Serologically defined colon cancer antigen 8; Sdccag8 |
| 702 | 28376635 | Rab37 | 24268.2 | 5.97 | 7 | 64 | 99.417 | 0.9899 | 1.0827 | |
| 335 | 28422545 | UDP glucose pyrophosphorylase 2 | 57075.8 | 8.16 | 11 | 101 | 100 | 0.1491 | −1.3007 | UGPP2, UDPG |
| 265 | 28552838 | Hypotheical protein XP_289117 | 12857.5 | 9.25 | 4 | 57 | 97.569 | 0.06322 | −1.6146 |
Acknowledgments
Support for this project comes from NIH grants DA013234 and DA DA013772. Special appreciation to Nilesh Yannu, Kaitlin Duschene, Wenxue Tang, and Willard Freeman for technical assistance. I would like to express my appreciation for the altruism and support of families of the patients studied.
References
- Abarca C, Albrecht U, Spanagel R. Cocaine sensitization and reward are under the influence of circadian genes and rhythm. Proc. Natl. Acad. Sci. USA. 2002;99:9026–9030. doi: 10.1073/pnas.142039099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abou-Sleiman PM, Muqit MMK, Wood NW. Expanding insights of mitochondrial dysfunction in Parkinson’s disease. 2006;7:207–219. doi: 10.1038/nrn1868. [DOI] [PubMed] [Google Scholar]
- Ahmed SH, Kenny PJ, Koob GF, Markou A. Neurobiological evidence for hedonic allostasis associated with escalating cocaine use. Nat. Neurosci. 2002;5:625–626. doi: 10.1038/nn872. [DOI] [PubMed] [Google Scholar]
- Ahmed SH, Koob GF. Transition from moderate to excessive drug intake: change in hedonic set point. Science. 1998;282:298–300. doi: 10.1126/science.282.5387.298. [DOI] [PubMed] [Google Scholar]
- Ahmed SH, Koob GF. Long-lasting increase in the set point for cocaine self-administration after escalation in rats. Psychopharmacology. 1999;146:303–312. doi: 10.1007/s002130051121. [DOI] [PubMed] [Google Scholar]
- Ahmed SH, Lutjens R, van der Stap LD, Lekic D, Romano-Spica V, Morales M, Koob GF, Repunte-Canonigo V, Sanna PP. Gene expression evidence for remodeling of lateral hypothalamic circuitry in cocaine addiction. Proc. Natl. Acad. Sci. USA. 2005;102:11533–11538. doi: 10.1073/pnas.0504438102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn JS, Jang IS, Kim DI, Cho KA, Park YH, Kim K, Kwak CS, Chul Park S. Aging-associated increase of gelsolin for apoptosis resistance. Biochem. Biophys. Res. Commun. 2003a;312:1335–1341. doi: 10.1016/j.bbrc.2003.11.061. [DOI] [PubMed] [Google Scholar]
- Ahn JS, Jang IS, Rhim JH, Kim K, Yeo EJ, Park SC. Gelsolin for senescence-associated resistance to apoptosis. Ann. N. Y. Acad. Sci. 2003b;1010:493–495. doi: 10.1196/annals.1299.090. [DOI] [PubMed] [Google Scholar]
- Albertson DN, Pruetz B, Schmidt CJ, Kuhn DM, Kapatos G, Bannon MJ. Gene expression profile of the nucleus accumbens of human cocaine abusers: evidence for dysregulation of myelin. J. Neurochem. 2004;88:1211–1219. doi: 10.1046/j.1471-4159.2003.02247.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andretic R, Chaney S, Hirsh J. Requirement of circadian genes for cocaine sensitization in Drosophila. Science. 1999;285:1066–1068. doi: 10.1126/science.285.5430.1066. [DOI] [PubMed] [Google Scholar]
- Ang E, Chen J, Zagouras P, Magna H, Holland J, Schaeffer E, Nestler EJ. Induction of nuclear factor-kappaB in nucleus accumbens by chronic cocaine administration. J. Neurochem. 2001;79:221–224. doi: 10.1046/j.1471-4159.2001.00563.x. [DOI] [PubMed] [Google Scholar]
- Backes E, Hemby SE. Discrete cell gene profiling of ventral tegmental dopamine neurons after acute and chronic cocaine self-administration. J. Pharmacol. Exp. Ther. 2003;307:450–459. doi: 10.1124/jpet.103.054965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berhow MT, Hiroi N, Nestler EJ. Regulation of ERK (extracellular signal-regulated kinase), part of the neurotrophin signal transduction cascade, in the rat mesolimbic dopamine system by chronic exposure to morphine or cocaine. J. Neurosci. 1996;16:4707–4715. doi: 10.1523/JNEUROSCI.16-15-04707.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibb JA, Chen J, Taylor JR, Svenningsson P, Nishi A, Snyder GL, Yan Z, Sagawa ZK, Ouimet CC, Nairn AC, Nestler EJ, Greengard P. Effects of chronic exposure to cocaine are regulated by the neuronal protein Cdk5. Nature. 2001;410:376–380. doi: 10.1038/35066591. [DOI] [PubMed] [Google Scholar]
- Breese CR, Marks MJ, Logel J, Adams CE, Sullivan B, Collins AC, Leonard S. Effect of smoking history on [3 H]nicotine binding in human postmortem brain. J. Pharmacol. Exp. Ther. 1997;282:7–13. [PubMed] [Google Scholar]
- Brenhouse HC, Stellar JR. c-Fos and deltaFosB expression are differentially altered in distinct subregions of the nucleus accumbens shell in cocaine-sensitized rats. Neuroscience. 2006;137:773–780. doi: 10.1016/j.neuroscience.2005.09.039. [DOI] [PubMed] [Google Scholar]
- Childress AR, Mozley PD, McElgin W, Fitzgerald J, Reivich M, O’Brien CP. Limbic activation during cue-induced cocaine craving. Am. J. Psychiatry. 1999;156:11–18. doi: 10.1176/ajp.156.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Chiara G, Imperato A. Drugs abused by humans preferentially increase synaptic dopamine concentration in the mesolimbic system of freely moving rats. Proc. Natl. Acad. Sci. USA. 1988;85:5274–5278. doi: 10.1073/pnas.85.14.5274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglass J, Daoud S. Characterization of the human cDNA and genomic DNA encoding CART: a cocaine-and amphetamine-regulated transcript. Gene. 1996;169:241–245. doi: 10.1016/0378-1119(96)88651-3. [DOI] [PubMed] [Google Scholar]
- Douglass J, McKinzie AA, Couceyro P. PCR differential display identifies a rat brain mRNA that is transcriptionally regulated by cocaine and amphetamine. J. Neurosci. 1995;15:2471–2481. doi: 10.1523/JNEUROSCI.15-03-02471.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberwine J. Single-cell molecular biology. Nat. Neurosci. 2001;4(Suppl):1155–1156. doi: 10.1038/nn1101-1155. [DOI] [PubMed] [Google Scholar]
- Eberwine J, Yeh H, Miyashiro K, Cao Y, Nair S, Finnell R, Zettel M, Coleman P. Analysis of gene expression in single live neurons. Proc. Natl. Acad. Sci. USA. 1992;89:3010–3014. doi: 10.1073/pnas.89.7.3010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fasulo WH, Hemby SE. Time-dependent changes in gene expression profiles of midbrain dopamine neurons following haloperidol administration. J. Neurochem. 2003;87:205–219. doi: 10.1046/j.1471-4159.2003.01986.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fosnaugh JS, Bhat RV, Yamagata K, Worley PF, Baraban JM. Activation of arc, a putative “effector” immediate early gene, by cocaine in rat brain. J. Neurochem. 1995;64:2377–2380. doi: 10.1046/j.1471-4159.1995.64052377.x. [DOI] [PubMed] [Google Scholar]
- Freeman AY, Pierce RC. Neutralization of neutrophin-3 in the ventral tegmental area or nucleus accumbens differentially modulates cocaine-induced behavioral plasticity in rats. Synapse. 2002;46:57–65. doi: 10.1002/syn.10123. [DOI] [PubMed] [Google Scholar]
- Freeman WM, Brebner K, Amara SG, Reed MS, Pohl J, Phillips AG. Distinct proteomic profiles of amphetamine self-administration transitional states. Pharmacogenom. J. 2005;5:203–214. doi: 10.1038/sj.tpj.6500309. [DOI] [PubMed] [Google Scholar]
- Freeman WM, Brebner K, Lynch WJ, Robertson DJ, Roberts DC, Vrana KE. Cocaine-responsive gene expression changes in rat hippocampus. Neuroscience. 2001a;108:371–380. doi: 10.1016/s0306-4522(01)00432-8. [DOI] [PubMed] [Google Scholar]
- Freeman WM, Brebner K, Patel KM, Lynch WJ, Roberts DC, Vrana KE. Repeated cocaine self-administration causes multiple changes in rat frontal cortex gene expression. Neurochem. Res. 2002;27:1181–1192. doi: 10.1023/a:1020929526688. [DOI] [PubMed] [Google Scholar]
- Freeman WM, Hemby SE. Proteomics for protein expression profiling in neuroscience. Neurochem. Res. 2004;29:1065–1081. doi: 10.1023/b:nere.0000023594.21352.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman WM, Nader MA, Nader SH, Robertson DJ, Gioia L, Mitchell SM, Daunais JB, Porrino LJ, Friedman DP, Vrana KE. Chronic cocaine-mediated changes in non-human primate nucleus accumbens gene expression. J. Neurochem. 2001b;77:542–549. doi: 10.1046/j.1471-4159.2001.00252.x. [DOI] [PubMed] [Google Scholar]
- Ginsberg SD, Crino PB, Hemby SE, Weingarten JA, Lee VM, Eberwine JH, Trojanowski JQ. Predominance of neuronal mRNAs in individual Alzheimer’s disease senile plaques. Ann. Neurol. 1999;45:174–181. [PubMed] [Google Scholar]
- Ginsberg SD, Elarova I, Ruben M, Tan F, Counts SE, Eberwine JH, Trojanowski JQ, Hemby SE, Mufson EJ, Che S. Single-cell gene expression analysis: implications for neurodegenerative and neuropsychiatric disorders. Neurochem. Res. 2004;29:1053–1064. doi: 10.1023/b:nere.0000023593.77052.f7. [DOI] [PubMed] [Google Scholar]
- Ginsberg SD, Hemby SE, Lee VM, Eberwine JH, Trojanowski JQ. Expression profile of transcripts in Alzheimer’s disease tangle-bearing CA1 neurons. Ann. Neurol. 2000;48:77–87. [PubMed] [Google Scholar]
- Harms C, Bosel J, Lautenschlager M, Harms U, Braun JS, Hortnagl H, Dirnagl U, Kwiatkowski DJ, Fink K, Endres M. Neuronal gelsolin prevents apoptosis by enhancing actin depolymerization. Mol. Cell Neurosci. 2004;25:69–82. doi: 10.1016/j.mcn.2003.09.012. [DOI] [PubMed] [Google Scholar]
- Hemby SE. Recent advances in the biology of addiction. Curr. Psychiatry Rep. 1999;1:159–165. doi: 10.1007/s11920-999-0026-9. [DOI] [PubMed] [Google Scholar]
- Hemby SE, Co C, Koves TR, Smith JE, Dworkin SI. Differences in extracellular dopamine concentrations in the nucleus accumbens during response-dependent and response-independent cocaine administration in the rat. Psychopharmacology (Berl.) 1997a;133:7–16. doi: 10.1007/s002130050365. [DOI] [PubMed] [Google Scholar]
- Hemby SE, Ginsberg SD, Brunk B, Trojanowski JQ, Eberwine JH. Gene expression profile for schizophrenia: discrete neuron transcription patterns in the entorhinal cortex. Arch. Gen. Psychiatry. 2002;59:631–640. doi: 10.1001/archpsyc.59.7.631. [DOI] [PubMed] [Google Scholar]
- Hemby SE, Horman B, Tang W. Differential regulation of ionotropic glutamate receptor subunits following cocaine self-administration. Brain Res. 2005a;1064:75–82. doi: 10.1016/j.brainres.2005.09.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemby SE, Johnson BA, Dworkin SI. Neurobiological basis of drug reinforcement. In: Johnson BA, Roache JD, editors. Drug Addiction and Its Treatment: Nexus of Neuroscience and Behavior. Philadelphia: Lippincott-Raven Publishers; 1997b. pp. 137–169. [Google Scholar]
- Hemby SE, Martin TJ, Co C, Dworkin SI, Smith JE. The effects of intravenous heroin administration on extracellular nucleus accumbens dopamine concentrations as determined by in vivo microdialysis. J. Pharmacol. Exp. Ther. 1995;273:591–598. [PubMed] [Google Scholar]
- Hemby SE, Tang W, Muly EC, Kuhar MJ, Howell L, Mash DC. Cocaine-induced alterations in nucleus accumbens ionotropic glutamate receptor subunits in human and non-human primates. J. Neurochem. 2005b;95:1785–1793. doi: 10.1111/j.1471-4159.2005.03517.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hope BT, Nye HE, Kelz MB, Self DW, Iadarola MJ, Nakabeppu Y, Duman RS, Nestler EJ. Induction of a long-lasting AP-1 complex composed of altered Fos-like proteins in brain by chronic cocaine and other chronic treatments. Neuron. 1994;13:1235–1244. doi: 10.1016/0896-6273(94)90061-2. [DOI] [PubMed] [Google Scholar]
- Horger B, Iyasere C, Berhow M, Messer CJ, Nestler EJ, Taylor JR. Enhancement of locomotor activity and conditioned reward to cocaine by brain-derived neurotrophic factor. J. Neurosci. 1999;19:4110–4122. doi: 10.1523/JNEUROSCI.19-10-04110.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurd YL, Herkenham M. Molecular alterations in the neostriatum of human cocaine addicts. Synapse. 1993;13:357–369. doi: 10.1002/syn.890130408. [DOI] [PubMed] [Google Scholar]
- Kim SY, Chudapongse N, Lee SM, Levin MC, Oh JT, Park HJ, Ho IK. Proteomic analysis of phosphotyrosyl proteins in morphine-dependent rat brains. Brain Res. Mol. Brain Res. 2005;133:58–70. doi: 10.1016/j.molbrainres.2004.09.018. [DOI] [PubMed] [Google Scholar]
- Kish SJ, Shannak K, Hornykiewicz O. Uneven pattern of dopamine loss in the striatum of patients with idiopathic Parkinson’s disease. Pathophysiologic and clinical implications. N. Engl. J. Med. 1988;318:876–880. doi: 10.1056/NEJM198804073181402. [DOI] [PubMed] [Google Scholar]
- Lim KO, Choi SJ, Pomara N, Wolkin A, Rotrosen JP. Reduced frontal white matter integrity in cocaine dependence: a controlled diffusion tensor imaging study. Biol. Psychiatry. 2002;51:890–895. doi: 10.1016/s0006-3223(01)01355-5. [DOI] [PubMed] [Google Scholar]
- Little KY, Kirkman JA, Carroll FI, Breese GR, Duncan GE. [125I]RTI-55 binding to cocaine-sensitive dopaminergic and serotonergic uptake sites in the human brain. J. Neurochem. 1993a;61:1996–2006. doi: 10.1111/j.1471-4159.1993.tb07435.x. [DOI] [PubMed] [Google Scholar]
- Little KY, Kirkman JA, Carroll FI, Clark TB, Duncan GE. Cocaine use increases [3H]WIN 35428 binding sites in human striatum. Brain Res. 1993b;628:17–25. doi: 10.1016/0006-8993(93)90932-d. [DOI] [PubMed] [Google Scholar]
- Lynd-Balta E, Haber SN. The organization of midbrain projections to the ventral striatum in the primate. Neuroscience. 1994;59:609–623. doi: 10.1016/0306-4522(94)90181-3. [DOI] [PubMed] [Google Scholar]
- Lyons D, Friedman DP, Nader MA, Porrino LJ. Cocaine alters cerebral metabolism within the ventral striatum and limbic cortex of monkeys. J. Neurosci. 1996;16:1230–1238. doi: 10.1523/JNEUROSCI.16-03-01230.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzoni O, Pujalte D, Williams J, Bockaert J. Decreased presynaptic sensitivity to adenosine after cocaine withdrawal. J. Neurosci. 1998;18:7996–8002. doi: 10.1523/JNEUROSCI.18-19-07996.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masubuchi S, Honma S, Abe H, Ishizaki K, Namihira M, Ikeda M, Honma K. Clock genes outside the suprachiasmatic nucleus involved in manifestation of locomotor activity rhythm in rats. Eur. J. Neurosci. 2000;12:4206–4214. [PubMed] [Google Scholar]
- McClung CA, Nestler EJ. Regulation of gene expression and cocaine reward by CREB and DeltaFosB. Nat. Neurosci. 2003;6:1208–1215. doi: 10.1038/nn1143. [DOI] [PubMed] [Google Scholar]
- Moratalla R, Elibol B, Vallejo M, Graybiel AM. Network level changes in expression of inducible Fos-Jun proteins in the striatum during chronic cocaine treatment and withdrawal. Neuron. 1996;17:147–156. doi: 10.1016/s0896-6273(00)80288-3. [DOI] [PubMed] [Google Scholar]
- Nestler EJ. Molecular basis of long-term plasticity underlying addiction. Nat. Neurosci. Rev. 2001;2:119–128. doi: 10.1038/35053570. [DOI] [PubMed] [Google Scholar]
- Nestler EJ, Barrot M, Self DW. DeltaFosB: a sustained molecular switch for addiction. Proc. Natl. Acad. Sci. USA. 2001;98:11042–11046. doi: 10.1073/pnas.191352698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nestler EJ, Terwilliger RZ, Walker JR, Sevarino KA, Duman RS. Chronic cocaine treatment decreases levels of the G protein subunits Gi alpha and Go alpha in discrete regions of rat brain. J. Neurochem. 1990;55:1079–1082. doi: 10.1111/j.1471-4159.1990.tb04602.x. [DOI] [PubMed] [Google Scholar]
- Nyberg J, Anderson MF, Meister B, Alborn AM, Strom AK, Brederlau A, Illerskog AC, Nilsson O, Kieffer TJ, Hietala MA, Ricksten A, Eriksson PS. Glucose-dependent insulinotropic polypeptide is expressed in adult hippocampus and induces progenitor cell proliferation. J. Neurosci. 2005;25:1816–1825. doi: 10.1523/JNEUROSCI.4920-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrotti LI, Bolanos CA, Choi KH, Russo SJ, Edwards S, Ulery PG, Wallace DL, Self DW, Nestler EJ, Barrot M. DeltaFosB accumulates in a GABAergic cell population in the posterior tail of the ventral tegmental area after psychostimulant treatment. Eur. J. Neurosci. 2005;21:2817–2824. doi: 10.1111/j.1460-9568.2005.04110.x. [DOI] [PubMed] [Google Scholar]
- Pierce RC, Bari AA. The role of neurotrophic factors in psychostimulant-induced behavioral and neuronal plasticity. Rev. Neurosci. 2001;12:95–110. doi: 10.1515/revneuro.2001.12.2.95. [DOI] [PubMed] [Google Scholar]
- Porrino LJ. Functional consequences of acute cocaine treatment depend on route of administration. Psychopharmacology (Berl.) 1993;112:343–351. doi: 10.1007/BF02244931. [DOI] [PubMed] [Google Scholar]
- Preuss TM. Do rats have prefrontal cortex? The Rose–Woolsey–Akert Program reconsidered. J. Cogn. Neurosci. 1995;7:1–24. doi: 10.1162/jocn.1995.7.1.1. [DOI] [PubMed] [Google Scholar]
- Qin Y, Ouyang Q, Pablo J, Mash DC. Cocaine abuse elevates alpha-synuclein and dopamine transporter levels in the human striatum. Neuroreport. 2005;16:1489–1493. doi: 10.1097/01.wnr.0000175617.39054.ba. [DOI] [PubMed] [Google Scholar]
- SAMHSA. Results from the 2001 national household survey on drug abuse: Volume I. Summary of National Findings. Rockville, MD: Substance Abuse and Mental Health Services Administration; 2002. [Google Scholar]
- Scheggi S, Rauggi R, Gambarana C, Tagliamonte A, De Montis MG. Dopamine and cyclic AMP-regulated phosphoprotein-32 phosphorylation pattern in cocaine and morphine-sensitized rats. J. Neurochem. 2004;90:792–799. doi: 10.1111/j.1471-4159.2004.02510.x. [DOI] [PubMed] [Google Scholar]
- Sondhi S, Castellano JM, Chong VZ, Rogoza RM, Skoblenick KJ, Dyck BA, Gabriele J, Thomas N, Ki K, Pristupa ZB, Singh AN, MacCrimmon D, Voruganti P, Foster J, Mishra RK. cDNA array reveals increased expression of glucose-dependent insulinotropic polypeptide following chronic clozapine treatment: role in atypical antipsychotic drug-induced adverse metabolic effects. Pharmacogenom. J. 2006;6:131–140. doi: 10.1038/sj.tpj.6500346. [DOI] [PubMed] [Google Scholar]
- Staley JK, Basile M, Flynn DD, Mash DC. Visualizing dopamine and serotonin transporters in the human brain with the potent cocaine analogue [125I]RTI-55: in vitro binding and autoradiographic characterization. J. Neurochem. 1994a;62:549–556. doi: 10.1046/j.1471-4159.1994.62020549.x. [DOI] [PubMed] [Google Scholar]
- Staley JK, Hearn WL, Ruttenber AJ, Wetli CV, Mash DC. High affinity cocaine recognition sites on the dopamine transporter are elevated in fatal cocaine overdose victims. J. Pharmacol. Exp. Ther. 1994b;271:1678–1685. [PubMed] [Google Scholar]
- Staley JK, Rothman RB, Rice KC, Partilla J, Mash DC. Kappa2 opioid receptors in limbic areas of the human brain are upregulated by cocaine in fatal overdose victims. J. Neurosci. 1997;17:8225–8233. doi: 10.1523/JNEUROSCI.17-21-08225.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan A, Moratalla R, Lyford GL, Worley P, Graybiel AM. The activity-regulated cytoskeletal-associated protein arc is expressed in different striosome-matrix patterns following exposure to amphetamine and cocaine. J. Neurochem. 2000;74:2074–2078. doi: 10.1046/j.1471-4159.2000.0742074.x. [DOI] [PubMed] [Google Scholar]
- Tang W, Wesley M, Freeman WM, Liang B, Hemby SE. Alterations in ionotropic glutamate receptor subunits during binge cocaine self-administration and withdrawal in rats. J. Neurochem. 2004;89:1021–1033. doi: 10.1111/j.1471-4159.2004.02392.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W-X, Fasulo WH, Mash DC, Hemby SE. Molecular profiling of midbrain dopamine regions in cocaine overdose victims. J. Neurochem. 2003;85:911–924. doi: 10.1046/j.1471-4159.2003.01740.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toda S, McGinty JF, Kalivas PW. Repeated cocaine administration alters the expression of genes in corticolimbic circuitry after a 3-week withdrawal: a DNA macroarray study. J. Neurochem. 2002;82:1290–1299. doi: 10.1046/j.1471-4159.2002.01083.x. [DOI] [PubMed] [Google Scholar]
- Ujike H, Takaki M, Kodama M, Kuroda S. Gene expression related to synaptogenesis, neuritogenesis, and MAP kinase in behavioral sensitization to psychostimulants. Ann. N. Y. Acad. Sci. 2002;965:55–67. doi: 10.1111/j.1749-6632.2002.tb04151.x. [DOI] [PubMed] [Google Scholar]
- Van Gelder RN, von Zastrow ME, Yool A, Dement WC, Barchas JD, Eberwine JH. Amplified RNA synthesized from limited quantities of heterogeneous cDNA. Proc. Natl. Acad. Sci. USA. 1990;87:1663–1667. doi: 10.1073/pnas.87.5.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkow ND, Valentine A, Kulkarni M. Radiological and neurological changes in the drug abuse patient: a study with MRI. J. Neuroradiol. 1988;15:288–293. [PubMed] [Google Scholar]
- Volkow ND, Wang GJ, Fischman MW, Foltin RW, Fowler JS, Abumrad NN, Vitkun S, Logan J, Gatley SJ, Pappas N, Hitzemann R, Shea CE. Relationship between subjective effects of cocaine and dopamine transporter occupancy. Nature. 1997;386:827–830. doi: 10.1038/386827a0. [DOI] [PubMed] [Google Scholar]
- Wiggins RC, Ruiz B. Development under the influence of cocaine. I. A comparison of the effects of daily cocaine treatment and resultant undernutrition on pregnancy and early growth in a large population of rats. Metab. Brain Dis. 1990;5:85–99. doi: 10.1007/BF01001049. [DOI] [PubMed] [Google Scholar]
- Williams SM, Goldman-Rakic PS. Widespread origin of the primate mesofrontal dopamine system. Cereb. Cortex. 1998;8:321–345. doi: 10.1093/cercor/8.4.321. [DOI] [PubMed] [Google Scholar]
- Wilson JM, Nobrega JN, Corrigall WA, Coen KM, Shannak K, Kish SJ. Amygdala dopamine levels are markedly elevated after self-but not passive-administration of cocaine. Brain Res. 1994;668:39–45. doi: 10.1016/0006-8993(94)90508-8. [DOI] [PubMed] [Google Scholar]
- Wise RA, Bozarth MA. A psychomotor stimulant theory of addiction. Psychol. Rev. 1987;94:469–492. [PubMed] [Google Scholar]
- Yao WD, Gainetdinov RR, Arbuckle MI, Sotnikova TD, Cyr M, Beaulieu JM, Torres GE, Grant SG, Caron MG. Identification of PSD-95 as a regulator of dopamine-mediated synaptic and behavioral plasticity. Neuron. 2004;41:625–638. doi: 10.1016/s0896-6273(04)00048-0. [DOI] [PubMed] [Google Scholar]
- Yuferov V, Kroslak T, Laforge KS, Zhou Y, Ho A, Kreek MJ. Differential gene expression in the rat caudate putamen after “binge” cocaine administration: advantage of triplicate microarray analysis. Synapse. 2003;48:157–169. doi: 10.1002/syn.10198. [DOI] [PubMed] [Google Scholar]
- Zhang D, Zhang L, Tang Y, Zhang Q, Lou D, Sharp FR, Zhang J, Xu M. Repeated cocaine administration induces gene expression changes through the dopamine D1 receptors. Neuropsychopharmacology. 2005;30:1443–1454. doi: 10.1038/sj.npp.1300680. [DOI] [PubMed] [Google Scholar]