Abstract
Ess1 is a prolyl isomerase that regulates the structure and function of eukaryotic RNA polymerase II. Ess1 works by catalyzing the cis/trans conversion of pSer5–Pro6 bonds, and to a lesser extent pSer2–Pro3 bonds, within the carboxy-terminal domain (CTD) of Rpb1, the largest subunit of RNA pol II. Ess1 is conserved in organisms ranging from yeast to humans. In budding yeast, Ess1 is essential for growth and is required for efficient transcription initiation and termination, RNA processing, and suppression of cryptic transcription. In mammals, Ess1 (called Pin1) functions in a variety of pathways, including transcription, but it is not essential. Recent work has shown that Ess1 coordinates the binding and release of CTD-binding proteins that function as co-factors in the RNA pol II complex. In this way, Ess1 plays an integral role in writing (and reading) the so-called CTD code to promote production of mature RNA pol II transcripts including non-coding RNAs and mRNAs.
Keywords: Ess1/Pin1, Peptidyl prolyl cis/trans isomerase, Transcription regulation, CTD code, Saccharomyces cerevisiae
1. Introduction
1.1. Scope of this review
This review will focus on the yeast enzyme Ess1 (Essential 1), originally discovered in the 1980s, and later shown to play a key role in RNA polymerase II (pol II) transcription. The human ortholog of Ess1, called Pin1, has been extensively studied, with thousands of publications appearing since its isolation in 1996 [1]. Pin1 targets a wide range of substrates and is proposed to play important roles in cell growth, development, signal transduction, apoptosis, DNA replication and repair, stress and immune responses, cancer, inflammatory and neurodegenerative disease, viral latency, and stem cell pluripotency. A large number of reviews are available that cover these topics [2–9]. With respect to the role of Ess1/Pin1 on regulation of RNA pol II, there are two excellent although somewhat dated reviews [10,11], and another that covers the role of Pin1 on transcription during the cell cycle [12]. The present review will not include studies on transcription factors or signaling molecules reportedly regulated by Pin1, e.g. NFκB, p53, and β-catenin [13–16]. Instead, the goal is to introduce the basic structure and biochemistry of the Ess1 (and Pin1) enzyme and to discuss how Ess1 controls the RNA pol II machinery.
1.2. Organization of this review
First, a timeline of discoveries will be presented to provide context and to clarify the relationship between Ess1 family members. Second, the structures and enzymatic activities of Pin1 and Ess1 will be described. Third, work that linked Ess1 (and Pin1) to transcription by RNA pol II and current models for how prolyl isomerization regulates transcription-coupled events will be described. Along the way, the nature of the carboxy-terminal domain (CTD) of Rpb1, the largest subunit of RNA pol II will be introduced. Understanding the “CTD-code” hypothesis is essential to appreciate the role that Ess1 plays in RNA pol II transcription. Finally, a few transcription-related functions of Ess1 will be described, and some commentary given on current limitations to research in the field and new directions we expect to see in the future.
2. Discovery of Ess1 and family members
2.1. Yeast Ess1 was first
Ess1 was discovered by serendipity in the early 1980s during the quest to discover oncogenes in organisms other than their retroviral hosts — remarkably, even in yeast cells. Working in the laboratory of Peter Shank, the author carried out low stringency hybridization to identify a gene that cross-hybridized with the v-sis oncogene, but which turned out to be unrelated [17]. This gene was named ESS1, on account of it being essential as shown using gene disruption by homologous recombination [18]. This was a new method, and at the time it was a surprise to learn that most genes in yeast were not essential [19 –21]. Using elutriated cells it was shown that ESS1 is expressed constitutively throughout the cell cycle, but only in actively growing yeast. ESS1 transcript levels diminish as cells enter stationary phase. Although ESS1 is essential in most (but not all) strains of Saccharomyces cerevisiae, the amount of Ess1 protein is in excess, as pedigree analysis showed that cells in which the ESS1 gene is removed grow up to seven generations prior to arrest [17]. In rich media there are ∼200,000 molecules of Ess1 per cell, whereas only ∼400 appear to be sufficient for growth [22]. Early mutational analysis of ESS1 using a conditional tRNA suppressor indicated a defect late in mitosis or cell wall separation [18] a finding more clearly demonstrated using shut-off and temperature-sensitive (ts) mutant experiments [1,23].
2.2. Its an isomerase!
At the time of its discovery (1984) the sequence of Ess1 did not reveal similarity to any known protein (unpublished). In that same year, 1984, Gunter Fischer and colleagues [24] reported the discovery of an enzymatic activity capable of interconverting the cis and trans forms of a peptide substrate at the normally restricted prolyl bond (Fig. 1). These “foldases” as they were known were presumed to help fold nascent peptides into proteins as they exited the ribosome. Their activity was shown to be distinct from that of chaperones in that they targeted a single type of bond, those that precede the amino acid proline. The enzymes, called peptidyl prolyl cis/trans isomerases (prolyl isomerases or PPIases) catalyze the reaction in both directions [25 –28]. The cis/trans interconversion is non-covalent and does not require ATP, but instead uses energy derived from conformational changes in the protein substrates.
Fig. 1.
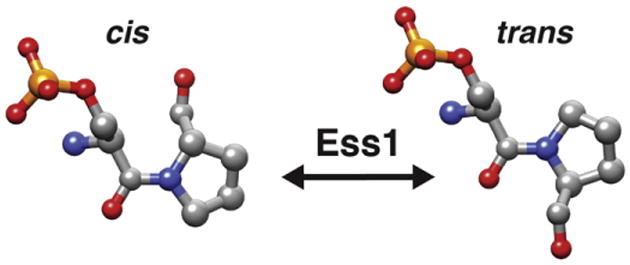
Model for phospho-Ser-Pro peptidyl bond isomerization. cis and trans isomers are shown. Note the 180° difference in the position of the proline's carbonyl group. Oxygens are shown in red, nitrogens in blue, carbons in gray, the phosphate in orange.
It was soon revealed that cyclophilin and FK506-binding protein, which are the targets of immunosuppressive drugs, are in fact prolyl isomerases [29–31]. Finally, in 1994 Rahfeld et al. [32], described a new class of PPIases in Escherichia coli called parvulins (from parvulus, Latin for very small) that are not sensitive to immunosuppressive drugs. One year later, Hani et al. [33] recovered ESS1 in a yeast screen (that will be discussed later) and aptly noted the similarity between Ess1 (called Ptf1 in their paper) and the newly-described parvulin class of PPIases. This was a breakthrough, as it revealed a likely bio chemical activity for Ess1 and showed that the parvulin class of PPIase extended to eukaryotic organisms.
2.3. Ess1 is highly conserved
A distinguishing feature of Ess1 is the presence of an amino-terminal WW domain. WW domains are eukaryotic protein-interaction modules about 40 residues in length characterized by two signature tryptophan residues spaced 20–22 aa apart [34 –36]. WW domains bind proline-rich sequences and are not found in prokaryotic (or archaeal) parvulins. The presence of the distinctive WW domain combined with the parvulin-type PPIase catalytic domain facilitated the identification of Ess1 orthologs (Fig. 2). Ess1 orthologs have been found in all fungi and animals that have been examined. The first was found in Drosophila melanogaster encoded by a gene called dodo, so named because it is located within the flightless region of the X-chromosome that also contains penguin, flightless and tweety [37]. Remarkably, the dodo gene driven from a yeast promoter completely rescued (complemented) yeast cells in which ESS1 was deleted.
Fig. 2.
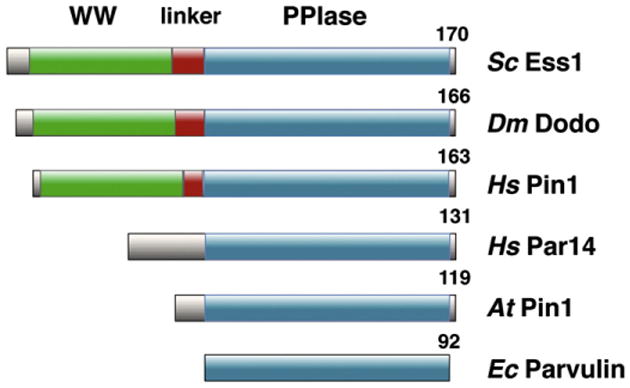
Domain organization of eukaryotic parvulins. The WW proline-binding domain, the catalytic domain (PPIase), and the linker region are highlighted. Sc, Saccharomyces cerevisiae; Dm, Drosophila melanogaster; Hs, Homo sapiens; At, Arabidopsis thaliana. A bacterial parvulin (Ec, Escherichia coli) is shown for comparison.
The next ortholog discovered was human Pin1, so named because of its discovery in a two-hybrid assay as a Protein Interacting with NIMA, a cell cycle kinase from Aspergillus nidulans [1]. Indeed, human PIN1 fully rescued yeast cells lacking ESS1 [1]. The notion that Pin1 is a mitotic regulator [38, 39] originally derived from the fact that (1) it was isolated by interaction with NIMA (Never In Mitosis), and (2) the observation that following shut off of a GAL1–PIN1 construct in ess1Δ mutants, cells accumulated in mitosis as large-budded cells and (3) in mammalian cells, Pin1 antisense constructs showed increased chromosome condensation consistent with a mitotic defect [1]. It is still not clear in yeast or mammalian cells, whether Ess1/Pin1 is a bona fide mitotic regulator, or whether the cell cycle arrest in yeast (which takes nearly 12 h to occur) is an indirect consequence of other defects (such as in transcription, see below). Pin1 knockouts in mice showed that Pin1 is not essential although it may help promote the G0 to G1 transition [40].
The ability to complement yeast ess1 mutants has been used as a litmus test for homology. The simplest method is to complement an ess1 ts-mutant rather than use a deletion mutant, since most non-yeast labs prefer not to carry out the requisite tetrad dissection of a diploid ESS1/ess1Δ strain and the subsequent genetic analysis. Orthologs that complement Ess1 in budding yeast include Xenopus lavis Pin1 (unpublished data), Trypanosoma cruzi Pin1 [41], Schizosaccharomyces pombe Pin1 [42], Candida albicans Ess1 [43], and Crytococcus deformans Ess1 [44]. The yeast complementation assay has also been extremely useful to monitor the function and enzymatic activity of Ess1-related enzymes in vivo [23, 45, 46]. As discussed below, some plant parvulins, which lack WW domains can complement Ess1 in yeast. Finally, it is worth noting that in contrast to other PPIases such as the cyclophilins and FKBPs, which are encoded by multigene families, eukaryotes seem to contain only one bona fide Ess1 ortholog (containing both a WW and PPIase domain), making their analysis more straightforward.
2.4. Other eukaryotic parvulins
Eukaryotes also contain a small number of parvulin-class PPIases that lack a N-terminal WW domain (Fig. 2). Humans contain at least two Ess1/Pin1-related proteins, hPar14 (parvulin 14 kDa) and hPar17 (parvulin 17 kDa), both encoded by the same gene. hPar14 is expressed in many tissues, and has PPIase activity, albeit with a 1000-fold lower catalytic rate and a different target specificity (Arg-Pro) compared to Ess1/Pin1 [47,48]. Instead of a WW domain, hPar14 has a basic N-terminal extension that promotes nuclear localization and DNA-binding activity [49]. hPar14 is thought to function in pre-ribosomal RNA processing [50,51] and does not complement yeast ess1 mutants (unpublished data; [52]. Par17 is a longer isoform of Par14 that is found only in primates and is targeted to mitochondria [53]. Although Par14 is not found in S. cerevisiae or C. albicans, it is present in filamentous fungi like A. nidulans and Neurospora crassa [54]. The function of Par14 in those organisms is not clear. There is another PIN1-like gene in humans, hPIN1L, which is 89% identical to PIN1 over its length, but it contains a frameshift that would result in a truncated protein [55]. The murine PIN1L is not expressed in any tissue suggesting that PIN1L is simply a processed pseudogene [56]. Finally, human Gas7b protein, while not a parvulin (no PPIase activity) contains a WW domain similar to that of hPin1, and like hPin1 it binds (and may compete for) phosphorylated Tau protein and may be linked to Alzheimer's disease [57].
Plants too, contain parvulin-class PPIases. However, none are strict orthologs of Ess1/Pin1 (i.e. contain both a WW and PPIase domain). The first discovered was Arabidopsis thaliana Pin1 (PIN1At), which lacks an N-terminal WW domain yet catalyzes the isomerization of phospho-Ser-Pro substrates in vitro similar to Ess1/Pin1 [58]. There are two other parvulins in A. thaliana, AtPIN2 and AtPIN3, but their sequences are more similar to hPar14 and E. coli Par10, respectively, and they likely do not share the target specificity of Ess1/Pin1 [59]. Surprisingly, PIN1At and other plant parvulins, MdPin1 from apple (Malus domestica) and DlPar13 (Digitalis lanata), despite lacking WW domains, all rescue yeast ess1 ts-mutants when overexpressed [52,60]. MdPin1 and DlPar13 also show specificity for phospho-Ser-Pro peptides [52,60]. In plants, parvulins may function in auxin production [61], and as shown for A. thaliana, Pin1At isomerizes transcription factors that regulate the developmental timing of flowering pathways [62].
In summary, all eukaryotes appear to have parvulin-class PPIases, with fungi and animals containing both Ess1/Pin1-type isomerases (WW + PPIase domains) and other parvulins (lacking WW domains). By contrast, all plant parvulins lack WW domains. Nonetheless, a subset of plant parvulins shows Ess1/Pin1-like activities toward peptide substrates. Lastly, an archaeal parvulin has been discovered, and it is most similar to hPar14 in that it lacks a WW domain and has a substrate preference distinct from Ess1/Pin1 [63].
3. Structure and specificity of Ess1/Pin1
3.1. Overall features of Ess1 and Pin1 enzymes
Ess1 and its fungal and metazoan orthologs range from about 163–178 residues in length (Fig. 1). While the sequence identities between them are relatively modest (typically 38%–44%), the relative position of the WW and PPIase domains is the same, and several key residues are essentially invariant. These include the signature tryptophan residues of the Ess1 WW domain, W15, W38 (W11, W34 in Pin1), and several residues that map to the Ess1 catalytic site including H64, S118, C120, H164 (H59, S111, C113, H157 in Pin1). These and other highly conserved residues were shown to be functionally important for growth in vivo [23,46]. For sequence alignments see Arevalo-Rodriguez et al. [64]. The most comprehensive and illustrative mutational analysis was carried out by Berhsin et al. [45] who used a plasmid shuffle assay to introduce a library of 5000 Pin1 mutants (35,000 substitutions) into a yeast ess1Δ mutant background. By screening for functionality (rather than loss of function) they identified both tolerated and invariant substitutions. There were many surprises, including the fact that several in variant residues (e.g. C113, H157 in Pin1) could, in fact, be substituted. The results are important for understanding catalytic function (see below). An interesting and somewhat unique feature of these enzymes is that the two domains, despite being completely dissimilar, bind the same target sequence–pSer–Pro or pThr–Pro. As discussed in more detail below, the WW domain binds with ∼10-fold higher affinity than does the PPIase catalytic domain.
3.2. Structures of Pin1 and CaEss1
The first structure of a eukaryotic parvulin, Pin1, was solved by the Noel laboratory [65] (Fig. 3A). The Pin1 structure revealed the N- terminal WW domain and C-terminal catalytic domain are tethered by a short flexible linker, not all of which was visible in the crystal. The Pin1 WW domain assumes a compact β-strand structure similar to WW domains in other proteins. The WW domain in Pin1 (and Ess1) is a Type IV domain [66,67] with a strong preference for substrate peptides containing phosphorylated serine or threonine preceding proline (pSer–Pro, pThr–Pro) [68]. The PPIase catalytic site is distal to the hydrophobic cleft formed between the two domains (Fig. 3A). The loop that forms the entrance to the active site contains a series of conserved basic residues (K63, R68 and R69in Pin1), explaining the enzyme's preference for phosphorylated substrates (69). A second Pin1 structure from the Noel group [69] shows a phosphorylated CTD peptide bound to the WW domain and positioned in the hydrophobic cleft (Fig. 3B). A number of direct contacts (not shown) are made between residues in the WW domain and Pro3 and Pro5 residues in the peptide substrate (both in the trans configuration) as well as to phospho-Ser5 (but not to phospho-Ser2). Notably, while there may be contacts from the long α-helix (K101) and another residue in the PPIase domain (P153), the majority of the contacts with substrate are made by the WW domain. This structure also shows an alternate conformation in which the loop at the entrance to the active site that contains the three basic residues is extended away from the body of the protein (Fig. 3B). This suggests that a mouse-trap type mechanism might occur in the catalytic domain that could be linked to substrate binding to the WW domain.
Fig. 3.
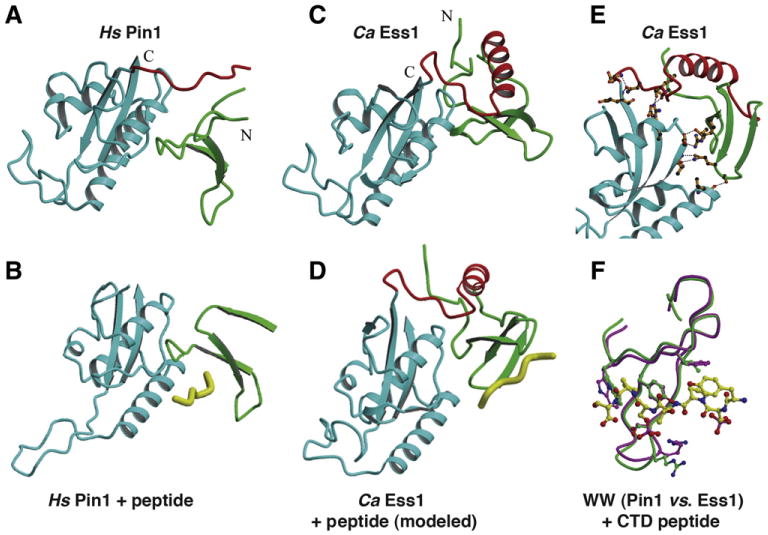
Structures of the human Pin1 and Candida albicans Ess1. In figures (A–E) the PPIase domains are shown in blue, the WW domains in green, and the linker regions in red. (A) Crystal structure of human Pin1 at 1.4 Å resolution (PDB ID: 1PIN), with the N and C termini labeled [65]. Not all of the linker is resolved in this structure indicating disorder. (B) Co-crystal structure of human Pin1 bound to a doubly-phosphorylated (pSer2/pSer5) CTD peptide at1.8 Å resolution (PDB ID: 1F8A) [69]. All contacts to the peptide are from theWW domain, none from the PPIase domain. Note the open conformation of the active-site loop. (C) Crystal structure of C. albicans Ess1 at 1.6 Å resolution (PDB ID: 1YW5) [75]. Note the fully structured linker with prominent α-helix indicated in red, and the WW domaini n a different juxtaposition than in Pin1. (D) Structure of the CaEss1 with a CTD peptide modeled on the WW domain, using the same interactions seen in the Pin1 structure of (B). (E) Detailed view of CaEss1 showing multiple contacts between residues in the linker region and the WW domain to the main body of the protein (PPIase domain). None of these contacts occur in Pin1. (F) Superposition of the WW domains of Pin1 (purple) and CaEss1 (green) showing an essentially identical position for binding of a CTD peptide.
Panels A–F are courtesy of P. Van Roey, and (E, F) are from Li et al. [75], reprinted with permission from the American Chemical Society © 2005.
In solution, the WW and PPIase domains of Pin1 are mobile relative to one another, and coalesce upon binding of substrate peptide to the WW domain [70,71]. Recent studies indicate there may be an intermediate state in which transient contacts between the WW and PPIase do mains may stimulate substrate binding [72]. These and other studies suggest a very dynamic Pin1 enzyme that undergoes dramatic conformational changes both with and without added substrate [73]. The use of bivalent peptides capable of binding both domains has been useful for understanding the flexibility of Pin1 as well as for developing high-affinity (nM) inhibitors [74].
There is currently no structure available for S. cerevisiae Ess1. How ever, the structure of the C. albicans Ess1 is known [75] and provides an informative comparison to that of human Pin1 (Fig. 3C). The individual WW and PPIase domains of CaEss1 are virtually superimposable on those of Pin1, however, there is a striking difference in the linker region that joins the two domains (shown in red). In CaEss1, this linker is 12 residues longer, is highly structured, contains a four-turn α-helix, and makes multiple contacts to the PPIase domain (Fig. 3E). Direct contacts are also observed between the WW domain and the PPIase domain in CaEss1 that do not occur in Pin1 (Fig. 3E). As a consequence, the WW is displaced upward, away from the long α-helix in the PPIase domain. While these differences may not have direct implications for the catalytic mechanism, they are likely to have profound implications for high-affinity WW-binding to pSer-Pro substrates.
First, the WW and PPIase domains are juxtaposed differently in CaEss1 effectively eliminating the hydrophobic pocket seen in Pin1 (where substrate peptide binds). Modeling studies indicate the interaction between the CaEss1 WW domain and a phospho-CTD peptide substrate would occur on a different surface of the protein from that seen for Pin1 (Fig. 3D), even though the individual contacts would likely be nearly identical (Fig. 3E) [75]. Second, the high degree of flexibility measured for Pin1 in solution [70,71] is not likely to exist for CaEss1 due to the multiple contacts between the linker region and the WW and PPIase domains (Fig. 4E), which essentially locks them in place. This was confirmed by NMR solution studies that indicate CaEss1 is highly-structured throughout its length and tumbles as a unit, even in the absence of substrate [76]. Third, the path for a substrate to take from the initial binding site on the WW domain to the PPIase catalytic domain in each enzyme is likely to be different. This would be especially important for long substrates like the CTD of RNA pol II. For a further discussion of the functional implications of the Pin1/Ess1 structures see Li et al. [75] and Lippens et al. [77].
Fig. 4.
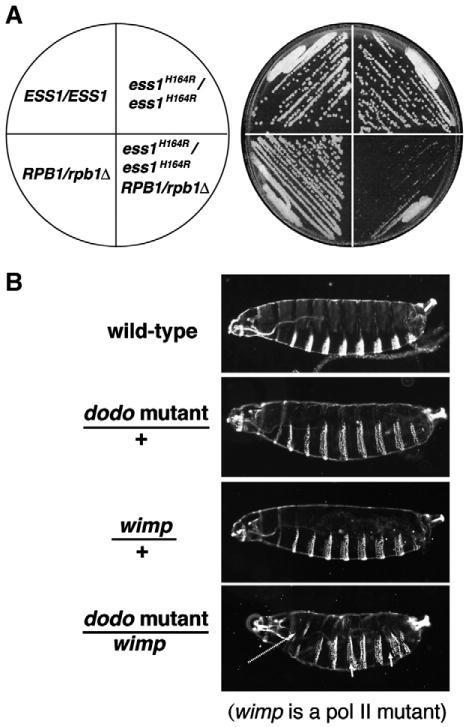
Genetic interactions between ESS1 and RNA polymerase II are conserved from yeast to metazoans. (A) Diploid yeast cells of the indicated genotype were grown at permissive temperature (30 °C) on rich medium (YEPD). Reducing the dosage of the largest subunit of RNA pol II (RBP1/rpb1Δ) combined with an ess1H164R ts-mutant is synthetic lethal. (B) Reduced maternal dosage of the fruit fly Ess1 (Dodo) enhances the larval defects in the reduced activity RNA polymerase II mutant, wimp. wimp is a dominant negative maternal-effect mutant in Rpb2 [121]. Cuticle preparations of first instar larvae of the indicated genotype are shown with the anterior to the left and dorsal up. Thoracic and abdominal defects occur in the double mutant (arrows). Bristle patterns reveal segmental defects including fusions and a general disorganization of anterior–posterior axial patterning. Viability is also reduced (N. Singh and S.D. Hanes, unpublished results).
Data in (A) are from Wu et al. [23], and are reprinted with permission from the Nature Publishing Company.
What is the evolutionary significance of the differences between Pin1 CaEss1 structures? Although speculative, it is tempting to suggest that the fungal enzyme, whose structure is less flexible would have a more restricted substrate specificity, while the human enzyme, with its high degree of conformational flexibility between domains would be able to target a greater variety of substrates. Sequence alignments re veal that the fungal enzymes have longer linker sequences than the metazoan enzymes, and 2° structure predictions show that these longer linkers could form α-helices, whereas the metazoan enzymes have short linkers rich in prolines and glycines that would prevent α-helix formation [75]. While metazoan enzymes have been shown to fully complement in yeast, the fungal enzymes have never been tested for complementing ability in animal cells. If the fungal enzymes have narrower specificities, then they would not be expected to fully rescue in animals.
3.3. Substrate specificity and catalytic mechanism
Prolyl isomerases accelerate the cis–trans isomerization of the peptide bond within peptide substrates by a factor of 103–106 [27,78,79]. For example, the rate of isomerization of CTD peptides is increased by S. cerevisiae Ess1 by 103 from a spontaneous rate of ∼1 turnover/min to a catalyzed rate of 47/;17/s [22]. The rate of spontaneous and catalyzed isomerization within intact proteins may be lower. An in vivo expression titration system was used to measure the total number of Ess1 molecules per cell required for yeast viability, and by extension, the number of turnovers required for viability under different growth conditions [22]. In rich medium, ∼6000 Ess1-catalyzed turnovers/s are required per cell for robust growth. However, as few as 20–300 turnovers/s will support minimal growth. Under stress conditions (e.g. hygromycin B, caffeine), much higher levels of Ess1 are required for viability, since ∼6000 turnovers/s was not sufficient for growth [22].
3.3.1. Measuring binding and isomerization by Ess1/Pin1-type PPIases
Despite their small size (∼19.5 kDa) the Ess1/Pin1-type parvulins area challenge to study biochemically. This derives from the fact that (1) both the PPIase and WW domains bind the same phospho-Ser/ Thr-Pro substrates, (2) in isolation, the catalytic domain has a very low affinity for substrates, (3) the reaction is reversible – the products are not easily “captured” and are in turn used as substrates, and (4) it is difficult to mea sure isomerization using physiological targets (intact proteins). It is a testament to the ingenuity of researchers in the field, including Kurt Wuthrich, Harold Scheraga, Franz Schmid, Gunter Fischer and others [24,80–83], that a number of useful methods have been developed to study isomerization of peptide and protein substrates by these enzymes, reviewed in [27].
Binding to proline-rich peptides or proteins can be measured using a variety of general techniques including filter immunoblots [84], two-hybrid analysis [85], GST-pulldown [86], fluorescence anisotropy [68], circular dichroism (CD) [87], NMR [88], and most recently by biolayer interferometry (BLI) [89]. Only apparent dissociation constants (Kapp), however, can be determined for intact Ess1/Pin1 proteins because of the dual-binding mode. Expression of individual WW or PPIase domains or mutants thereof is necessary to resolve individual binding constants. For full-length Pin1, Kapp was measured by fluorescence anisotropy to be 10,30 or 60 μM for CTD peptide doubly phosphorylated or phosphorylated on Ser5 or Ser2, respectively [69]. The bulk of the binding affinity was contributed by the WW domain. For example, on the doubly-phosphorylated CTD peptide, the Kd for the isolated WW domain was 34 μM, while the Kd for the PPIase domain was 390 μM.
For intact Ess1, fluorescence anisotropy was used to estimate Kapp to be 60 μM and 240 μM for CTD peptides phosphorylated on Ser5 and Ser2, respectively [22]. Using a different method (BLI), the Kapp of intact Ess1 for a Ser5 phosphorylated peptide was estimated at 2.1–2.6 μM [89]. Binding affinities of the isolated WW-domain of Ess1 were measured using CD to be ∼70 μM for pSer2 peptides, 80–100 μM for pSer5 peptides and ∼20 μM for doubly-phosphorylated peptides [87]. For both Pin1 and Ess1, binding to unphosphorylated Ser/Thr-Pro peptides is es sentially undetectable. Despite the quantitatively different results report ed by different laboratories using different methods and substrates, a few common themes emerge. First, Ser/Thr-Pro substrates must be phosphorylated to bind. Second, longer doubly-phosphorylated substrates tend to bind better. And third, even with the best substrates, the binding affinity is relatively weak, typically in the micromolar range. A summary is pro vided in Table 1.
Table 1.
Binding affinities and turnover rates of eukaryotic parvulins.
| Protein | Substrate | Kda (μM) | Method | Refs. | Turnover | Method | Refs. |
|---|---|---|---|---|---|---|---|
| Sc Ess1 | Unphosp. CTD | >3000 | FA; BLI | [22,89] | 0 s−1 | NMR | [22] |
| >300 | |||||||
| pSer2 CTD | 241 ± 23 | FA | [22] | 2.8 s−1 | NMR | [22] | |
| pSer5 CTD | 61 ± 4.9 | FA; BLI | [22,89] | 17.7 s−1 | NMR | [22] | |
| 2.6 ± 0.7 | |||||||
| Sc Ess1 WW | Unphosp. CTD | n.b. | CD | [87] | n.a. | n.a. | n.a. |
| pSer2 CTDb | 67 ± 11 | CD | [87] | n.a. | n.a. | n.a. | |
| pSer5 CTDb | 79 ± 13 | CD | [87] | n.a. | n.a. | n.a. | |
| pSer2,5 CTDb | 21 ± 3 | CD | [87] | n.a. | n.a. | n.a. | |
| Sc Ess1 | Ac-ApSPY-pNA | n.d. | n.a. | n.a. | 17,200 mM−1 s−1 | Protease-coupled | [114] |
| Hs Pin1 | Ac-ApSPY-pNA | n.d. | n.a. | n.a. | 3370 mM−1 s−1 | Protease-coupled | [94] |
| Sc Ess1 | Ac-AApSPR-NH-Np | n.d. | n.a. | n.a. | 12,000 mM−1 s−1 | Protease-coupled | [90] |
| Hs Pin1 | Ac-AApSPR-NH-Np | n.d. | n.a. | n.a. | 9500 mM−1 s−1 | Protease-coupled | [90] |
| Hs Pin1 | pSer2 CTD | 61 ± 6.3 | FA | [69] | n.d. | n.a. | n.a. |
| pSer5 CTD | 30 ± 0.39 | FA | [69] | n.d. | n.a. | n.a. | |
| pSer2,5 CTD | 10 ± 0.8 | FA | [69] | n.d. | n.a. | n.a. | |
| Hs Pin1 WW | pSer2 CTD | 110 ± 23 | FA | [69] | n.a. | n.a. | n.a. |
| pSer5 CTD | 34 ± 5.9 | FA | [69] | n.a. | n.a. | n.a. | |
| pSer2,5 CTD | 34 ± 6.2 | FA | [69] | n.a. | n.a. | n.a. | |
| Hs Pin1 PPIase | pSer2 CTD | n.b. | FA | [69] | n.d. | n.a. | n.a. |
| pSer5 CTD | >500 | FA | [69] | n.d. | n.a. | n.a. | |
| pSer2,5 CTD | 390 ± 6.82 | FA | [69] | n.d. | n.a. | n.a. | |
| Ca Ess1 | Suc-AEPF-pNA | n.d. | n.a. | n.a. | 19 s−1 | Protease-coupled | [75] |
| Hs Par14 | Suc-ARPF-pNA | n.d. | n.a. | n.a. | 4.0 mM−1 s−1 | Protease-coupled | [47] |
| At Pin1 | Ac-WFYpSPRLR-NH | n.d. | n.a. | n.a. | 1s−1 | NMR | [58] |
n.b. = no binding detected; n.a. = not applicable; n.d. = not determined; FA = fluorescence anisotropy; BLI = biolayer interferometry; NMR = nuclear magnetic resonance chemical shift; CD = circular dichroism thermal denaturation; caution is advised when comparing values obtained by different groups using different methods; the concentrations of protein and substrates used as well as the temperatures and buffer conditions may differ. Turnover rates are given as turnovers per second (s−1), or as Kcat/Km (mM−1 s−1).
For full-length proteins apparent Kd.
Circularly permutated relative to other studies.
Accurate measurement of isomerization rates in vitro is possible, but has its limitations because the simplest assays use unnatural substrates, whereas natural substrates are difficult or impossible to assay. Methods include the standard protease-coupled assay that uses non-physiological substrates [24], a set of fluorescence-based assays that rely on solvent or pH jumps but offer more flexibility in allowable peptide sequences [90], and dynamic NMR methods (chemical exchange) that can monitor individual isomerizations within natural peptide or protein substrates [91,92]. For a review that describes these methods with additional refer ences see [27]. As expected, measuring isomerization in intact proteins is the most difficult but has been done with well-behaved proteins such as RNaseA and bovine pancreatic trypsin inhibitor (BPTI) [93]. Unfortunately, at this time there is no reliable way to monitor isomerization in vivo.
For Pin1, isomerization rates have been measured by multiple methods on a wide variety of peptide substrates, with Tyr-pSer-Pro-Arg motif-containing peptides showing the highest rates [90,94]. Ess1-catalyzed isomerization of CTD peptides (AS[YS2PTS5PS]YS) was measured using chemical exchange and revealed a 6-fold higher rate on a peptide phosphorylated on Ser5 (17.7 turnovers/s) than on Ser2 (2.8 turnovers/s) [22] (Table 1). This finding correlated with genetic suppression experiments in yeast that suggested that Ser5-Pro6 in the CTD was the more relevant in vivo target [95]. Based on these and other findings discussed below (Section 5.3), it appears that Ess1 may bind doubly-phosphorylated CTD substrates best, but prefers to catalyze isomerization of the pSer5-Pro6 bond within the CTD repeats.
3.3.2. A specificity problem
Similar to the situation for many DNA-binding proteins [96,97], the Ess1/Pin1-type isomerases appear to have a “specificity problem”. Ser-Pro and Thr-Pro sites are ubiquitous in proteins and are phosphorylated by cyclin-dependent and mitogen-activated kinases, CDKs and MAPKs [98 –100]. How do Ess1/Pin1-type isomerases recognize bona fide substrates given the widespread occurrence of pSer-Pro and pThr-Pro motifs in the proteome? While there are some minor preferences for flanking residues [86], the totality of studies show promiscuous binding to pSer/Thr-Pro-containing targets of in vitro. As a consequence, either these PPIases do in fact target multitudes of proteins in the cell, or their interactions are restricted in some unknown way. Several potential mechanisms come to mind. First, the pSer/Thr-Pro motifs within mature proteins might be buried within their three-dimensional architecture and therefore might not be accessible. Second, these sites might be masked by the binding of competing proteins. Third, these PPIases might require cooperative or combinatorial binding with other proteins to help target them to physiological substrates. There is currently little evidence to distinguish among these possibilities. We suspect that in yeast, Ess1 may simply require that substrates have multiple repeats (or proximally-located copies within their 3D-structures) of the pSer/ Thr-Pro motif, as found in the CTD of RNA pol II discussed in Section 5.2. While the issue of specificity is largely ignored in the literature, it is likely to be important particularly in higher organisms where there are so many substrates of Pin1 reported (see also Lippens for discussion) [77].
3.3.3. Catalytic mechanism and inhibitors
The mechanism of the prolyl isomerization by Pin1 was originally proposed to involve a covalent intermediate formed by nucleophilic attack by the active site cysteine (C113) [65]. However, the aforementioned mutagenesis study suggested otherwise, since a C113D substitution was functional in vivo and retained about 30% catalytic activity in vitro [45]. Indeed, C. albicans Ess1, which has activity comparable to that of Pin1 [75], normally carries an aspartate at this position. Detailed studies with inhibitors and substrate analogs have revealed more likely catalytic mechanisms for Pin1 [73,101 –104]. Evidence supporting a twisted-amide mechanism was described by Etzkorn and colleagues [101]. They suggested that the phosphate group of the pSer-Pro motif and the proline carbonyl are locked in place by hydrogen bonds from K63, R68 and R69, and Q131 of Pin1, respectively. The rotation involves a “jump-rope” type motion resulting in rotation of the serine carbonyl group via a transition state that bears an intramolecular hydrogen bond within the substrate from the Pro-X amide NH group to the pro line nitrogen. This bond is proposed to stabilize the transition-state intermediate.
The basis for the twisted-amide bond model derives from older studies with cyclophilins and FKBPs [105,106], as well as the structures of Pin1-inhibitor complexes from the Noel laboratory [102]. In support of this model, substrates with a proline at the +1 position (pSer/pThr-Pro-Pro), which lack the amino hydrogen required for the hydrogen bond that stabilizes the twisted state (β-turn), were found to bind the PPIase domain less well than other substrates, although this could instead be due to loss of a potential hydrogen bond to the carbonyl oxygen of the pSer/pThr [86].
Chemical inhibitors have been helpful in understanding not only the catalytic mechanism but also the basis for binding and selectivity of both the PPIase and WW domains. These studies are driven by the potential clinical significance of Pin1 in a number of human diseases. The first reported inhibitor, juglone [107], has been widely used in biological studies. This is unfortunate, as it is a relatively non-specific inhibitor. While it inhibits the parvulin-class Ess1/Pin1 isomerases but not cyclophilins or FKBPs, it works by covalent modification of Cys residues. Since active-site Cys residues are common in many enzymes such as pyruvate decarboxylase, glutathione-S-transferase and RNA polymerase II, results obtained from the use of juglone are of questionable value [90].
Other potent and specific inhibitors have been identified. These include peptidomimetics such as D-isomer and cyclic peptides, and conformationally-locked isosteres [84,90,102,108–111]. Selective WW-domain inhibitors have also been identified [112]. Studies using cis-locked or trans-locked inhibitors revealed, among other things, that the PPIase domain prefers the cis-isomer, while the WW domain is rather non-selective, and that there is intramolecular signaling between the WW and PPIase domains [113]. On substrates with multiple pSer/pThr-Pro bonds such as the CTD, these findings imply that targeting would occur via the WW domain if the substrate bonds are initially in trans, and that binding to the WW domain could potentially alter catalytic activity of the PPIase domain. Flanking sequences may also influence WW vs. PPIase preferences, for example, as mentioned, proline at the +1 position (pSer/pThr-Pro-Pro) favors WW-domain binding over PPIase binding [86].
In summary, although certain aspects of the Ess1/Pin1 family of enzymes are reasonably well-understood from a structural and biochemical standpoint, further studies will be needed to fully understand the catalytic mechanism and to identify potential binding differences with distinct substrates, as well as to determine how the sequences of amino acid residues flanking the X-Pro target affect catalytic rates. An other future goal is to understand how longer, multi-site substrates bind and to determine the path taken from binding site on the WW do main to the active site of the PPIase domain. Finally, understanding the structural and functional differences between metazoan and fungal enzymes may provide clues to their evolutionary divergence, crosstalk between the two domains, and highlight differences in their respective substrate spectrum.
4. Ess1 plays a role in RNA polymerase II transcription
4.1. Early studies linking Ess1 to transcription
The first hint that Ess1 was involved in transcription came in 1995 [33]. At the time, this link to transcription was overlooked probably be cause the relevant experiments, a genetic screen for rescue of a 3′-end processing defect that identified ESS1 (called PTF1 in that study), were simply cited as unpublished results. Instead, the paper emphasized the similarity of Ess1 (Ptf1) to bacterial parvulin-class PPIases, which of course was also very important. A follow-up study, published in early 1999 described the screen and showed that ess1 mutants read through poly(A) termination sites embedded in reporter constructs [114]. This prescient study also demonstrated Ess1/Ptf1 PPIase activity for the first time. Biochemical studies showed that Pin1 and Ess1 interacted in vitro with the phosphorylated form of RNA polymerase II or phospho-CTD peptides, respectively [115,116], although these studies did not provide evidence that the interaction was functional.
During this time, conditional ess1 mutants were generated and used to carry out an unbiased high-copy suppressor screen to look for clues about Ess1 function [23]. The expectation, based on the supposed mitotic function of Ess1 and Pin1 was that cell cycle regulators would be identified. Instead, all but one suppressor was transcription-related and remaining suppressor was cyclophilin A, another PPIase. Among the suppressors, YKL005C, now known as BYE1 (bypass of Ess1) encodes a likely elongation factor that interacts directly with RNA polymerase II [117,118]. Another, FCP1, encodes a CTD-phosphatase [119] and a third, SAP30, encodes a component of a histone deacetylase complex [120].
In the same study, Ess1 was shown to interact biochemically and in vivo (using two hybrid and genetics) with RNA polymerase II, and ess1 mutations showed defects in transcription of individual genes [23]. A powerful genetic experiment that functionally linked Ess1 to RNA polymerase II function in yeast cells is reproduced in Fig. 4A. It shows the synthetic-lethal effect of reducing both Ess1 and the largest subunit of RNA pol II (Rpb1), which bears the pSer-Pro containing CTD. Remarkably, a similar result is seen in whole animals, where a dodo mutation combined with a reduced activity RNA pol II allele (called wimp) [121] results in cuticular defects in embryos, whereas mutation of either one alone does not (unpublished data; Fig. 4B). Ess1 mutants were also shown to be synthetic lethal with CTD-truncation alleles and interact genetically with SRB2 (synthetic lethal) a gene originally isolated as a suppressor of CTD truncations and part of the mediator complex [23,95].
Based on these studies, a model for Ess1 function was proposed in which Ess1 binds the phosphorylated form of the RNA pol II CTD, and induces conformational changes that regulate the binding of protein co-factors required for the transcription cycle [23]. A number of genetic studies supported the idea that Ess1 function is important during multiple stages of the transcription cycle including initiation, elongation and termination [85,95,118,122]. In particular, these and other studies suggested that Ess1 promotes the activity of CTD phosphatases and op poses the action of CTD kinases [123], reviewed in [64]. The model was expanded to include Ess1 and isomerization of the CTD as an integral part of the CTD code [124] (described in Section 5).
Following the initial studies in yeast, work from the Manley laboratory linked human Pin1 to transcription. In these studies, Xu et al. [125] used in vitro assays and in vivo approaches with the inhibitor juglone, as well as pin1-/- knockout mouse embryo fibroblasts [40] and Pin1 overexpressing HeLa cells. They showed that Pin1 inhibits the CTD-phosphatase activity of Fcp1, which was also shown by Palancade et al. [126] who found that inhibition was likely due to steric hindrance (at least in vitro), and not necessarily requiring PPIase activity. In addition, Pin1 stimulated CTD phosphorylation by Cdc2/CyclinB and promoted hyperphosphorylation of RNA pol II [125]. Finally, they showed that Pin1 inhibited in vitro transcription (and splicing) and in later work pro posed that Pin1 acts to shut down transcription during the mitotic phase of the cell cycle [127]. While these studies confirmed a conserved role for Pin1 in mammalian transcription via regulation of RNA pol II, the effects differed in “direction” with those observed in yeast. As described below, in yeast, Ess1 seems to promote de-phosphorylation of the CTD, while in mammalian cells, Pin1 seemed to promote phosphorylation of the CTD. It is possible that the difference is due to distinct cell cycle mechanics, for example the lack of nuclear envelope breakdown or transcription shut down during mitosis in yeast. To date, however, there is no adequate ex perimental explanation for the observed differences. Perhaps careful measurements of cell cycle-dependent CTD modification (along specific loci and genome-wide) might help resolve the differences.
4.2. Loss of Ess1 has effects on transcription genome-wide
4.2.1. Use of temperature-sensitive (ts) alleles
Ess1 mutants show a variety of transcription-related defects. Since ESS1 is essential, almost all functional studies have been done using conditional (ts) mutants. The most commonly used allele is ess1H164R, which has a mutation in the catalytic site (H164R) that reduces PPIase activity ∼10,000 fold [22] and renders the cells temperature sensitive [23]. Cells bearing this allele grow normally at 23–30 °C, but fail to grow at 37 °C. The defect is likely due to the catalytic deficiency since the Ess1 (H164R) protein binds CTD peptides about as well as the wild-type protein [22]. Other ts-alleles commonly used include A144T [23], which also has a stop codon substitution leading to a longer protein (33 additional residues) (unpublished data), and G127D, and another PPIase domain mutation independently isolated by two groups [23,114]. Both A144T and G127D mutant proteins are less stable than the H164R mutant protein at 37 °C [23].
To better understand the yeast Ess1 literature, two important points need to be kept in mind. First, defects in transcription in the ts-mutants are apparent even at permissive temperature, especially for the ess1H164R allele [128,129]. This is probably why genetic interactions can be observed at semi-permissive (34 °C) and permissive temperatures (25°, 30 °C) [85,95,118,122]. The ability to detect defects at these temperatures allows the experimenter to avoid potential complicating factors associated with prolonged incubation at (37 °C) including heat shock response, and transcriptional reprogramming and the onset of cell death. The second point is related, but more subtle. The fact that ess1H164R mutant, whose catalytic activity is dramatically reduced, is even viable at permissive temperature indicates that very little Ess1 activity is required for growth. Or put another way, normally (in rich media) Ess1 activity is present in great excess. This was demonstrated by Gemmill et al. [22]. In addition, we and others have found that the catalytic activity of Ess1 (H164R) protein does not appear to be thermo-labile [22], and the protein is not degraded in cells shifted to 37 °C (although its level is somewhat reduced) [22,23]. These findings indicate that the temperature-sensitivity is likely due to a heightened requirement for Ess1 activity at the elevated temperature (i.e. a stress condition), and not strictly due to diminished Ess1 activity at the elevated temperature. This conclusion is consistent with findings that high levels of Ess1 are required for viability under other stress conditions, such as addition of hygromycin B or caffeine [22]. Thus, genetic and bio chemical effects observed at restrictive temperatures must take into ac count the increased requirement for Ess1 activity and the reason for the temperature-sensitivity.
4.2.2. Ess1 is important for efficient termination of mRNAs and small non-coding RNAs
Hani et al. [114] observed a decrease in total poly(A)-plus RNA following a shift of a ts-mutant (ess1G127D) to restrictive temperature. They also observed read through of an ACT1 promoter-lacZ construct into which was inserted an ADH1 terminator-poly(A) sequence, suggesting a defect in 3′-end processing. This defect was confirmed using a different allele (ess1H164R) and a different terminator (ADH2) [118,122], and readthrough was measured to be nearly 20% in ess1H164R mutants relative to control cells [118]. These were early indications that Ess1 might help coordinate recruitment of the termination and/or 3′-end mRNA processing machinery to the RNA pol II complex. Details are discussed in Section 5.
Efforts to gauge the global importance of Ess1 for mRNA 3′-end for mation used genome-wide approaches that included high-density tiling arrays. Surprisingly, these efforts did not uncover broad readthrough of mRNAs in ess1H164R mutants [129]. While some examples of mRNAs readthrough transcription were observed, the majority of effects were on small non-coding RNAs (discussed below). It was not until the use of genetic backgrounds in which mRNA decay pathways were inactivated that transcription readthrough in ess1 mutants was revealed to be wide-spread [128]. About half of the fourteen genes examined in ess1H164R cells that also carried a deletion of the UPF1 gene showed significant readthrough transcription. UPF1 (also knownasNAM7)encodes an RNA helicase required for nonsense-mediated decay (NMD) [130]. Indeed ess1H164R upf1Δ double mutants showed synthetic growth defects. This was interpreted [128] to indicate that in double mutant cells the accumulation of readthrough transcripts might contribute to their demise, whereas in ess1H164R UPF1+ cells, where the NMD decay pathway is operational, aberrant transcripts are rapidly degraded [131,132].
Mutations in other RNA surveillance/decay pathway genes, XRN1 and RRP6 were also synthetic lethal with ess1H164R. XRN1 encodes a cytoplasmic 5′ → 3′ exonuclease located in P-bodies and RRP6 encodes a 3′ → 5′ exonuclease that is part of the nuclear exosome complex. Both function in RNA processing and in destruction of aberrant RNAs [133,134]. While no genome-wide analysis of ess1H164R mutants has been done in mRNA decay-deficient backgrounds, based on experiments done thus far it is likely that 47/;50% of all mRNA genes will re quire Ess1 for efficient transcription termination/3′-end processing [128].
Using an in vitro 3′ mRNA processing assay, Krishnamurthy et al. [122] showed that Ess1 was dispensable for efficient cleavage and polyadenylation. If this is also true in vivo, it would suggest that the readthrough defects in ess1 mutants are due to termination defects and not to the requisite 3′ cleavage and poly(A) addition events that occur prior to actual termination and RNA pol II disengagement from the DNA template. However, lack of activity in vitro must be interpreted with caution. In vitro transcription also does not require Ess1 [135]. It is possible for example, that in vitro, the processing factors are present in such excess that Ess1's effects would not be required.
Ess1 is also critically important for the termination and 3′-end processing of small non-coding RNAs. This was discovered somewhat indirectly. Using standard ORF microarrays, it was shown that the expression of ∼10% of all protein-coding genes was affected in ess1H164R mutants. What was striking, however, was that the small set of genes whose expression increased was nearly identical to those ob served in microarray experiments with ssu72 mutants [136]. As will be described in more detail below, SSU72 encodes a pSer5-specific CTD phosphatase. Further analysis revealed that the reason these protein-coding genes showed increased microarray signals in ess1H164R mutant cells was because of transcriptional readthrough from adjacent small nucleolar RNA (snoRNA) genes, the same results found in the ssu72 study. snoRNA genes are transcribed by RNA pol II but their transcripts are not polyadenylated or exported and serve as guides in rRNA processing events [137]. In ess1H164R mutants, nearly all independently-transcribed snoRNA genes (∼30) show transcriptional readthrough [129]. In some cases readthrough transcription led to decreases in the expression of downstream genes. No effects were observed for small nuclear RNAs, e.g. U1–U5, which are part of the nuclear spliceosome. Preliminary work in human cells indicates that Pin1 siRNA knockdown cells show similar readthrough of at least some independently-transcribed snoRNAs (unpublished data).
Ess1 is also required to keep cryptic unstable transcripts (CUTs) [138] under control [129]. Tiling array analysis revealed that a large numbers of CUTs are stabilized in ess1H164R mutant cells. This includes hundreds of CUTs already been identified in other mutant backgrounds (e.g. rrp6) and a similar number that appear to be unique to ess1 mutants [129]. The CUTs were in 5′, 3′ and intergenic regions of protein-coding genes in both sense and antisense directions. Their ubiquity throughout the genome suggests that there is a global defect in transcription repression in the ess1H164R mutant, perhaps chromatin-mediated. Many CUTs seemed to be stabilized due to failure to terminate at their normal sites, which is required to initiate their degradation [139,140]. Instead the elongated transcripts extended into neighboring genes or promoters, oriented in the same or opposite direction, often resulting in their increased or decreased expression [141–144]. Analogous results were observed for ess1 mutants in C. albicans using high-throughput RNA-seq analysis [145]. In summary, ess1 mutants show genomic chaos due to faulty mRNA and small non-coding RNA termination/3′-end formation and to high levels of cryptic transcription.
4.2.3. Ess1 affects initiation and elongation
In several studies, ess1 mutants failed to activate reporter genes under inducing conditions [23,122,146]. Since these reporters (LexA-lacZ, PHO5-lacZ, GAL10-lacZ, INO1-lacZ) are driven by different activator proteins (LexA-GAL4, Pho4, Gal4, Spt23), the loss of expression was not likely due to an activator-specific defect, but more likely to a general defect in initiation. As expected if Ess1 has a role in initiation, chromatin immunoprecipitation (ChIP) data showed that Ess1 is present at the 5′ end of several highly-expressed genes [23,122].
The idea that Ess1 plays a role in initiation is supported by genetic interactions observed between mutations in ESS1 and SUA7 (TFIIB), KIN28 (TFIIH kinase), and SRB10 (a kinase component of the mediator complex), all of which have roles in initiation or preinitiation. Overexpression of SUA7 suppresses ess1 growth defects, suggesting a positive role for Ess1 in initiation. That is, loss of Ess1 function is overcome by high levels of the initiation factor TFIIB [122]. KIN28 and ESS1 have an antagonistic relationship [95] suggesting that Ess1 may reverse the action of the Kin28 kinase, which generates Ser5-phosphorylated CTD at the time of promoter escape and promotes 5′ capping [147]. Finally and most remarkably, an srb10Δ mutation fully restores viability to an ess1Δ mutant [95]. The Srb10 (Cdk8) kinase, part of the mediator complex, phosphorylates the CTD and was thought to have an inhibitory role prior to pre-initiation complex formation (PIC) [148]. Ess1 might promote dephosphorylation of the CTD after its phosphorylation by Srb10. If true, then the genetic results can be explained as follows: When Srb10's negative effect on PIC is removed, Ess1 is no longer needed because initiation occurs unperturbed. The mechanism is likely more complex, as Srb10/Cdk8 also targets other proteins in the initiation complex and has been shown to promote initiation and elongation [149,150].
Several lines of evidence suggest that Ess1 may also control elongation, probably by slowing it. First, there are strong genetic interactions between ESS1 and CTK1 (elongation-related CTD kinase), DST1 (elongation factor TFIIS), and SPT4/SPT5 (elongation factor complex, DSIF) [23,95]. For example, ESS1 and DST1 (which promotes elongation) oppose one another genetically. Second, ess1 mutants show increased readthrough of an ARTAR-artificial pause/arrest site reporter. Third, ess1 mutants are resistant to the elongation inhibitor 6-azauracil (6-AU) suggesting Ess1 normally slows elongation, and in ess1 mutants, elongation rates would increase [23]. This result, by itself would not be significant since changes in 6-AU sensitivity can result from a number mechanisms unrelated to elongation rates. For example, 6-AU resistance could be due to increased transcription of IMD2, a gene that allows cells to become 6-AU tolerant [151], and indeed, IMD2 expression is in creased in ess1 mutants [129]. Finally, using a well-defined in vitro elongation system, it was shown that extracts from ess1 mutant cells are up to 40% more efficient at elongating a purified template than extracts from the control wild-type cells (unpublished data). Adding back purified Ess1 protein to a mutant extract reduced efficiency elongation. The mechanism is currently unknown.
In summary, the use of conditional mutants has revealed a number of transcriptional defects that occur when Ess1 activity is compromised. The findings raise an important question: How does one protein play so many different roles in transcription and RNA processing? The answer seems to be that Ess1 targets the RNA polymerase enzyme itself and by doing so is able to influence multiple steps in the process.
5. Mechanism(s) of Pol II regulation by Ess1
5.1. Overview
The major target of Ess1 in yeast is RNA pol II. To understand how Ess1 targets RNA pol II and controls its functions, one must appreciate the structure of the RNA pol II CTD and how it functions in RNA synthesis and processing. In a nutshell, the CTD functions as molecular Velcro to bind proteins required for nearly all aspects of RNA pol II function, and the role of Ess1 is to modulate the stickiness of that Velcro. Like the hook portion of Velcro, the CTD is flexible and can attract a variety of different binding proteins, and these proteins can have distinct types of fasteners (CTD-binding domains). Ess1 can change the shape of the “hooks” in the CTD thus favoring the binding of one protein over another. In the absence of Ess1, there is a loss of coordination of protein ex change on the CTD that leads to defects in RNA production and in some organisms, cell death or disease.
5.2. The CTD and the CTD code hypothesis
5.2.1. CTD basics
Eukaryotic RNA polymerase II is a large 12 subunit enzyme, the largest of which is Rpb1, which in yeast is encoded by the RP021 (RPB1) gene. Rpb1 contains a C-terminal domain (CTD) unique to eukaryotic pol II that consists of the repeats of the heptad sequence (Y1S2P3T4S5P6S7)n. There are 26 nearly identical repeats of this sequence in the budding yeast CTD and 52 not quite so identical repeats in the human CTD. As a general rule the more complex the organism the more repeats, and the more that some repeats diverge from the consensus. Before proceeding, it should be pointed out that the function of the CTD is a fascinating topic and has been the subject of many studies. Treatment of the CTD in the present review is limited to issues important for understanding Ess1 function. For more detailed information and collections of references please refer to the outstanding reviews that are available [152–164].
Several important points need to be emphasized at this juncture. First, the CTD is essential. Partial truncations are tolerated, but complete deletions are inviable [165,166]. Second, not all of the repeats function identically or have the same importance. Mutational analysis has demonstrated that proximal (more N-terminal) vs. distal (more C-terminal) show different genetic interactions and behave differently [165,167]. This is especially interesting to consider given that the beginning of the CTD is located near the RNA exit channel on the polymerase holoenzyme, and helps recruit RNA processing enzymes. Third, the CTD is highly flexible. It is unstructured in solution, and can take on a variety of conformations (how many is not known) depending on what protein it is bound to. Fourth, the CTD repeat unit shown above is not the functional unit although it is typically written this way for convenience. In stead the functional unit seems to be the better part of a di-heptad repeat, and the spacing between repeats is critical [168]. Structure studies of a C albicans capping enzyme shows that protein binding occurs in a manner in which an individual repeat is looped out and the protein makes contacts with non-adjacent repeats [169]. In all likelihood, the functional repeat unit will depend upon which CTD-binding protein is being considered. Fifth, the CTD is covalently and non-covalently modified and these modifications help determine what proteins can bind (elaborated below).
In the context of this review it is interesting to note the evolutionary correlation between WW domain-containing PPIases and CTD-containing polymerases. Organisms that have WW-domain containing PPIases (Ess1/Pin1-type) have CTD-containing RNA polymerases. In contrast, archaea, which do not have WW domain-PPIases, do not have a CTD repeat. The reverse is not always true, however. Plants, which do not have WW domain-PPIases do have a CTD-containing Rpb1 subunit. So, while there are other ways to target PPIases to the CTD [58,60], a WW-domain on an isomerase almost certainly means that the enzyme will target a CTD-containing RNA pol II.
5.2.2. The CTD code: covalent and non-covalent modification
The sequence and composition of the CTD are not likely to have occurred by accident. Evolutionary pressure likely resulted in the selection of residues with maximum versatility. This short motif can be covalently modified by phosphorylation, primarily at serines 2,5, and 7, but also at Tyr1 and Thr4 [170]. In mammals, the Ser and Thr residues can also be glycosylated [171,172], while degenerate Arg7 and Lys7 residues can be methylated [173], or potentially acetylated, methylated, sumoylated or ubiquitylated [157]. Moreover, the two Ser-Pro bonds can be non-covalently modified by cis/trans isomerization [22,69].
The potential for different combinations of modifications along a sequence of CTD repeats is vast. For interesting discussions about evolution of the CTD and its potential information content, see the following references [152,164,168,174,175]. Importantly, the Ser phosphorylations show characteristic patterns on the CTD as the RNA pol II complex travels down the length of a gene [176–179]. This led to the idea that the modifications constitute a “CTD code” that signals the recruitment and eviction of protein co-factors to the polymerase to promote its transitions (e.g. initiation → elongation) and/or to help recruit the RNA processing machinery [124,154,155,180]. Proline isomerization by Ess1 would be an integral part of this code (Fig. 5).
Fig. 5.
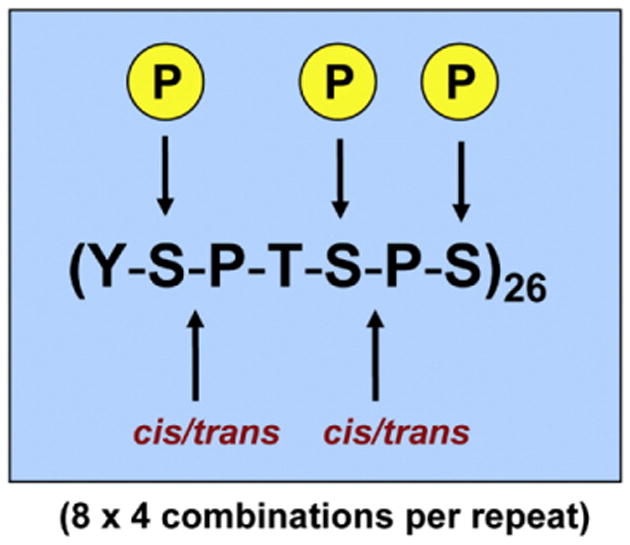
Simplified version of the CTD code. One repeat of the heptad sequence found at the carboxy-terminus of the Rpb1 subunit is shown. There are 26 repeats in yeast and 52 in humans. The Ser2, Ser5 and Ser7 residues can be phosphorylated in any combination (8 total), and the Ser2-Pro3 and Ser5-Pro6 bonds can exist in the cis or trans conformation (4 different combinations). Thus, there are at least 8 × 4 = 32 (×26 repeats) for a total of 832 potential configurations. Not shown are the potential phosphorylations at Tyr1, Thr4, as well as glycosylation at Thr4, and in humans, acetylation and methylation at degenerate Arg and Lys residues at position 7. See Egloff et al. [157] and other reviews cited in the text for details.
The patterns of CTD phosphorylation have been studied genome-wide in yeast [181–183]. The general pattern observed for serine phosphorylations (S2, S5, S7) across protein coding genes is shown in Fig. 6A. Ser5 and Ser7 phosphorylation peak early at the 5′ end of the gene near the transcription start site, while Ser2 increases over the body of the gene reaching peaking at the 3′ end near the transcription termination site. The most recent of these studies confirms this pattern is uniform across nearly all protein-coding genes [184]. Similar patterns are observed in mammalian cells [152]. Most of the kinases and phosphatases that control the phosphorylation state of serines within the CTD in yeast have been identified and have mammalian counterparts [180,185,186]. In higher organisms, there are additional CTD-kinases that respond to cell cycle or extracellular signals. The pattern of phosphorylation along the CTD does not seem to be critical for RNA pol II activity per se, and in deed the CTD is dispensable for transcription in vitro [187–190]. Instead the CTD and its modifications are important for recruitment of protein co-factors needed for RNA processing steps such as 5′-capping, splicing, 3′ cleavage, polyadenylation and mRNA packaging for nuclear export (reviewed in) [154,155,158]. CTD phosphorylation is also likely to be important for RNA pol II to elongate efficiently through chromatin and for it to recruit histone-modifying enzymes [163].
Fig. 6.
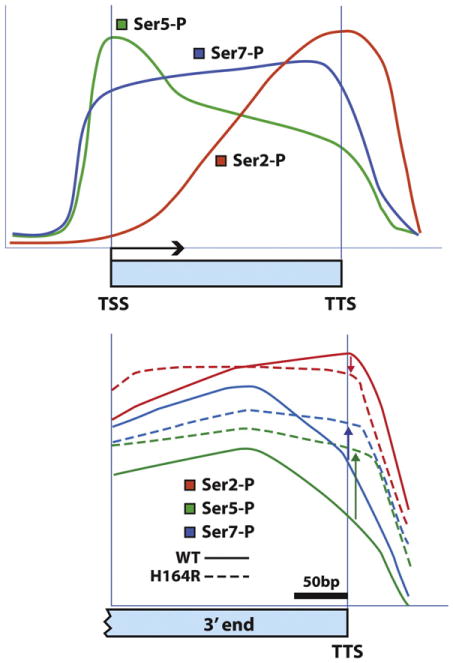
Phosphorylation pattern of the CTD as RNA pol II travels down a gene. (A) Patterns of Ser2, Ser5, and Ser7 phosphorylation are averaged across the length of protein-coding genes in yeast. TSS, transcription start site; TTS, transcription termination site. The updated pattern is derived from genome-wide ChIP-chip using newer, more specific monoclonal antibodies [152,184,191]. pSer5 peaks early and diminishes as RNA pol II proceeds from 5′ → 3′. pSer7 starts high and stays high until after 3′ cleavage and termination. pSer2 increases and reaches a maximum at the termination site. (B) Comparison of Ser phosphorylation levels at the 3′ termini of genes in wild-type vs. ess1H164R mutant cells. Note the increase in both pSer5 and pSer7 as polymerase nears the TTS in ess1 mutants. pSer2 levels are slightly diminished. These results indicate that in normal cells, Ess1 helps reduce phosphorylation levels of Ser5 and Ser7, probably by making the CTD a better substrate for the cis-specific Ssu72 phosphatase (see text for details).
Figure B is adapted from Bataille et al. [184], and is reprinted with permission from Elsevier © 2012.
5.3. Ess1 isomerizes the CTD preferentially targeting pSer5-Pro6
Using two-dimensional NMR (NOESY) to measure chemical ex change of proline γ-carbon protons, it was shown that Ess1 isomerizes a CTD peptide, AS(YSPTpSPS)YS, which contains serine phosphorylated at position 5, at a rate of 17.7 turnovers/s [22]. This was about six-fold faster than a comparable peptide, AS(YpSPTSPS)YS, phosphorylated at position 2 (2.8 turnovers/s). Ess1 also appears to target the Ser5 phosphorylated form of RNA pol II in vivo, as co-immunoprecipitation experiments using Ess1 antibodies retrieves more pSer5 than pSer2 modified enzyme [128]. An important caveat is that the monoclonal antibodies used to detect the pSer5-specific form of RNA pol II (H14), also recognize the pSer2/pSer5 doubly-phosphorylated forms [191]. Indeed, genome-wide ChIP-chip data using additional antibodies suggest that Ess1 preferentially associates with the doubly-phosphorylated form of RNA pol II CTD (unpublished).
The preference of Ess1 for targeting pSer5-Pro6 was anticipated by genetic experiments in which CTD serines 2 and 5 were mutated to glutamic acid or alanine [165] and tested for growth in ess1 mutant backgrounds [95]. Both S2E and S5E “partial” mutations (some repeats are wild-type) in the CTD were synthetic lethal with the ess1H164R mutant, indicating that they function in the same or converging pathways. More informative however, was the finding that an S5A mutation sup pressed ess1H164R mutant, allowing it to grow at elevated temperature, whereas a comparable S2A mutation did not. The interpretation is that Ess1's in vivo function is to promote dephosphorylation of Ser5, and if that residue is converted to alanine (de-phospho mimic), Ess1 function becomes redundant. Consistent with this idea, when Ser5 is converted to glutamic acid (phospho-mimic), Ess1 is unable to promote dephosphorylation, hence the synthetic lethality. Ess1 probably works in the same pathway as Ser2 but at a distinct step, consistent with the failure of S2A to suppress (and S2E mutations being synthetic lethal). See Wilcox et al. for additional details [95]. Finally, it is worth noting that the S5A substitutions only suppressed ess1H164R when located proximally in the CTD (more N-terminal), not when they were distal (more C-terminal), suggesting that Ess1 has the most impact on the CTD positioned near the RNA exit channel of the RNA polymerase. Taken together, the biochemical and genetic data support the idea that Ess1 is recruited by both the pSer5 and pSer2/pSer5 doubly-phosphorylated CTD, and it shows a preference for isomerizing the pSer5-Pro6 bond over the pSer2-Pro3 bond.
5.4. Ess1 controls binding and activity of proteins required for transcription and co-transcriptional RNA processing
5.4.1. Ess1 regulates the competition for CTD-binding
Ess1-catalyzed cis/trans isomerization of the CTD is likely to be critical for regulation of co-factor binding to RNA pol II during the transcription cycle. The best example for the role of Ess1 is in snoRNA termination. While it is not yet possible to monitor the isomer state of the CTD in vivo, chromatin IP can be used to measure the recruitment of proteins along individual genes in wild-type vs. ess1 mutant cells. Using this approach it was shown that ess1 coordinates the recruitment of Nrd1 and Pcf11 to the 3′-end of snoRNAs [129]. Nrd1, along with Nab3 and Sen1 functions in 3′-end cleavage by recruiting the nuclear exosome to process the transcripts of short non-coding RNAs [139,140,192–195]. Pcf11, which is important for mRNA cleavage and termination is also required for snoRNA termination [196–199]. In ess1 mutant cells, Nrd1 levels were increased at the 3′ ends of snoRNA genes, while Pcf11 levels were decreased. Both proteins bind to the nascent RNA, as well as to the phosphorylated CTD, Nrd1 preferring the Ser5-phosphorylated form and Pcf11 preferring the Ser2-phosphorylated form [197–199]. ChIP data suggest that Ess1 can referee the competition between CTD-binding proteins [129]. For example, in normal cells Ess1 promotes Nrd1 eviction from snoRNA 3′ ends to allow Pcf11 binding and termination. In ess1 mutants, Nrd1 stays bound blocking Pcf11 binding. Consistent with this competition model, overexpression of Pcf11 rescues the growth defect of ess1 mutant cells and reduces Nrd1 binding to snoRNA loci [129].
There is other evidence that Ess1 controls co-factor recruitment to the RNA pol II complex. Compared to wild-type cells, ess1 mutants show aberrantly high recruitment of TBP, TFIIB and Ceg1 (capping enzyme) to the initiation regions of cryptic transcripts [128]. This helps ex plain why CUT expression is increased in the ess1 mutants. These examples suggest that faulty recruitment of RNA pol II co-factors and a “mis-coordination” of a normally ordered process could explain many, if not all of the transcriptional defects observed in ess1 mutant cells (and Pin1 knockout cells).
How might Ess1 control recruitment of proteins to the CTD? Several distinct mechanisms are possible. First, an indirect mechanism would be to control the phosphorylation state of the CTD, thereby controlling protein binding (Section 5.4.2). A second mechanism, would be to directly control binding by stimulating isomerization of the CTD so that a preferred isomer is made more available (Section 5.4.3). Other potential mechanisms would be for Ess1 to control accessibility via chromatin structure modification, or to control the activity or intracellular localization of the co-factors themselves (Sections 6.1, 6.2).
5.4.2. Ess1 controls phosphorylation state of the CTD
Evidence in yeast indicates that Ess1 helps lower CTD phosphorylation levels by assisting CTD-specific phosphatases. Overexpression of either CTD phosphatase, Fcp1 or Ssu72 suppresses the growth defect of ess1 mutants [23,122,123], while overexpression of at least one CTD kinase (Ctk1) exacerbates the growth defect [95]. These genetic experiments predicted that CTD phosphorylation in ess1 mutants will be abnormally high. Indeed, levels of CTD phosphorylation on Ser5 and Ser7 levels are increased significantly in extracts from ess1 mutant cells vs. wild type [122,128,129]. Moreover, overexpression of wild-type Ess1 but not a catalytically-deficient mutant (C120R) reduces phosphorylation of Ser5 below wild-type levels [129]. pSer2 levels are largely un affected. That loss of Ess1 activity causes CTD hyperphosphorylation was confirmed using ChIP to monitor Ser2, Ser5 and Ser7 levels on individual genes or genome-wide in ess1 mutants [128,184]. Interestingly, in the genome-wide study [184], the most pronounced increase in pSer5 (and Ser7) levels occurred at the 3′ ends of protein-coding genes (Fig. 6B), consistent with the observed defects in termination/3′-end formation noted previously. Changes in the CTD phosphorylation pattern would help explain, for example why Nrd1, which favors binding to pSer5-CTD, increased abnormally at the 3′ end of genes in ess1 mutants.
Ssu72 is a Ser5/Ser7-specific phosphatase [136,184,200,201] so it is easy to understand why its overexpression rescues ess1 mutants, where Ser5 and Ser7 are elevated. Less clear is why overexpression of Fcp1, whichisthoughttobeSer2-specific, would suppressess1 mutants, since pSer2 levels are not significantly affected. One explanation is that at high concentration, its substrate-specificity might be compromised so that it dephosphorylates Ser5 and Ser7. Alternatively, it might be that Ser2 dephosphorylation by Fcp1 stimulates Ssu72 to dephosphorylate neighboring Ser5 residues (and vice versa). Crosstalk between these two phosphatases has been shown to occur [184].
In summary, Ess1 promotes dephosphorylation of the CTD at Ser5 and Ser7, and this activity is likely to be an essential part of controlling the so-called CTD code. Human Pin1 also seems to regulate CTD phosphorylation levels, but the one study in which this was examined, the direction was opposite; Pin1 seemed to increase CTD phosphorylation (on Ser5) [125]. Below, a mechanism by which Ess1 controls CTD phos-phatase activity is considered.
5.4.3. CTD-binding proteins show cis/trans stereoselectivity
A major prediction of the CTD code is that prolyl isomerization will affect the binding of proteins to the CTD. While ChIP experiments clearly show that Ess1 affects the binding of proteins to individual genes in vivo, the results cannot directly demonstrate that binding is affected by isomer status per se. However, over the last decade or so, nearly a dozen structures of CTD-binding proteins bound to CTD peptides have been solved (Table 2), and a number of themes have emerged. First, CTD-binding proteins use a variety of distinct structural motifs to bind the CTD. Second, the CTD takes on a different conformation depending on the protein to which it is bound. And third, CTD-binding proteins do, in fact, display a distinct preference for cis or trans isomers of the CTD (as well as different phosphorylation states). Biochemical evidence has also indicated that the activity as well as the binding of some proteins may be affected by the cis or trans state of the substrate peptide. A number of excellent reviews are available that summarize the results of the structural studies [202–204]. Below I will highlight a few key findings for Ssu72, Nrd1 and Pcf11, three proteins whose function has already been linked to Ess1.
Table 2.
Summary of structural studies for CTD-binding proteins.
| Protein (binding motif) | Phosphor. | CTD peptide | Conformation | Structure | Refs. |
|---|---|---|---|---|---|
| Pin1 (WW) (human PPIase) | Ser2-P, Ser5-P | YpSPTpSPS | trans | Extended coil | [69] |
| Cgt1 (C. albicans capping enzyme) | Ser5-P | 4 repeat | trans | β-Like, looped | [169] |
| Pcf11 (CID) (yeast termination factor) | Ser2-P | PTSPSYpSPTSPS | trans | β-Spiral | [210,211] |
| Scp1 (BRCT) (human phosphatase) | Ser5-P | PSYSPTpSPS | trans | Extended coil | [207] |
| SCAF8 (CID) (human splicing factor) | Ser2-P or Ser5-P | Various | trans | β-Turn | [230] |
| MCE (human capping enzyme guanyltransferase subunit) | Ser5-P | [(pSPSYSP)T]3 | trans | Extended β-like | [204] |
| Rtt103 (CID) (yeast exo termination factor) | Ser2-P | 4 repeat (binds w/Pcf11) | trans | β-Turn | [212] |
| Ssu72 (Drosophila CTD phosphatase) | Ser5-P | PTpSPSYS | cis | β-Turn | [205] |
| Ssu72 (human CTD phosphatase) | Ser5-P | SYSPTpSPSYS | cis | β-Turn | [206] |
| Nrd1 (CID) (yeast termination factor) | Ser5-P | 2 repeat | cis | β-Turn | [213] |
| Set2 (H3K36 methyltransferase) | Ser2-P, Ser5-P | SPSYpSPTpSPSYpSPTpSPS | ? | n.d. | [231,232] |
CID = CTD interacting domain; WW = WW-domain; BRCT = BRCA1 C-terminal domain.
Structures of the Ssu72 orthologs from human and Drosophila have been solved [205,206]. Both were co-crystallized with CTD peptides phosphorylated on Ser5, although the sequences were in a different register relative to the consensus heptad repeat (Table 2). These structures were the first to show the CTD peptide bound to a protein in the cis-isomeric state. This cis configuration of the pSer5 CTD peptide with Ssu72 contrasts with that found in the co-crystal of the Scp1 phosphatase, which binds a trans-isomer of a pSer5 CTD peptide [207]. The human Ssu72 structure was solved using a full-length protein (a C12S catalytic mutant) in a ternary complex with a scaffolding protein Symplekin and a decamer CTD peptide. The functional equivalent of Symplekin in yeast, the Pta1 3′-processing factor, interacts genetically with Ess1 [122]. In the ternary structure, the CTD peptide makes a nearly 180° turn at the cis-proline and fits within a narrow groove of the Ssu72 active site. Only a cis peptide can be accommodated in this highly-constrained environment. Interactions are seen with CTD residues Thr4, pSer5, Pro6 and Tyr1 of the next repeat. Another structure of the ternary complex between Symplekin, Ssu72 and a CTD peptide was solved, but in this case the peptide was phosphorylated on Ser7 and bound in the opposite orientation as the pSer5 peptide, with all proline bonds in the trans conformation [208].
In the Drosophila Ssu72 structure (a C13D/D144N catalytic mutant), a number of interactions can be seen between residues in the Ssu72 active site and the substrate that help explain the isomer-specificity [205]. In addition, the cis-configuration of the heptamer peptide seems to be stabilized by an intramolecular hydrogen bond between hydroxyl group of the Thr4 residue (located at the – 1 position relative pSer5-Pro6) and the proline backbone carbonyl. This stabilizing interaction by Thr4 may explain Ssu72's preference for pSer5 over pSer2. In the CTD, pSer2 is pre ceded by Tyr2, which would be unable to make this hydrogen bond, and lead to steric clashes destabilizing the cis-conformation. While the intra molecular bond is likely to be important, it is probably not required, be cause while phosphorylation of Thr4 prevents bond formation and lowers Ssu72 activity by about 4-fold, the co-crystal structure indicates the overall configuration of a CTD peptide within the active site remains nearly identical [209].
Most relevant to Ess1 were the biochemical findings of Werner-Allen et al. [205], which showed that Ess1 stimulated the phosphatase activity of Ssu72 on CTD substrates, consistent with prior genetic and molecular studies [129,184]. They found that only about 12% of the phosphorylated CTD peptide in solution was present in the cis conformation. This low percentage of cis-isomers was rate-limiting for Ssu72 phosphatase activity, and isomerization by Ess1 provided a kinetic ad vantage. Not only did Ess1 stimulate the phosphatase activity of Ssu72 on a small CTD peptide, it also did so on a “full-length” GST-CTD fusion protein (26 repeats) and as expected, the stimulation was saturable and required isomerization, since catalytic mutants (C120S, S122P, H164R) did not have any effect [205]. These findings are critical in that they pro vide a mechanism by which Ess1 can control the phosphorylation state of the CTD and thus participate in writing the CTD-code. While it is not known what percentage of CTD proline bonds are in the cis or trans configuration in vivo, these studies show that cis-trans interconversion by Ess1 would provide a kinetic advantage to isomer-specific enzymes like Ssu72.
Previous work showed that Ess1 controls the competition for binding to the CTD by the Pcf11 and Nrd1 3′-processing factors [129]. The mechanism was suggested to be indirect, through changes in CTD phosphorylation, as Pcf11 and Nrd1 have preferences for pSer2 and pSer5, respectively. While this may be part of the mechanism, it is also possible that Ess1 acts more directly because Pcf11 and Nrd1 also show strong and distinct isomer-specific preferences.
The solution and crystal structures of the CTD-interacting domain (CID) of Pcf11 bound to CTD peptides phosphorylated on Ser2 were deter mined [210,211]. CID domains are found in many proteins and are characterized by a right-handed bundle of eight α-helices that creates a groove for CTD binding. The solution study shows that CTD peptides phosphorylated at Ser2 exist in a disordered state, with the proline bonds in both the cis (<30%) and trans (47/; 70%) conformations [211]. Spontaneous intraconversion was measured from the millisecond to second time scale or slower. The Pcf11 CID selected the all trans form of a CTD peptide using an induced-fit binding mechanism to impart a (β-spiral structure on the CTD. Indeed, in the crystal structure, the central Ser2 is phosphorylated within the CTD peptide and both pSer2-Pro3 and pSer5-Pro6 bonds are in trans. In this structure cis bonds would not be ac commodated. Interestingly, Pcf11 binds the CTD cooperatively along with another CID-containing 3′-end processing protein, Rtt103, which also recognizes a pSer-CTD peptide with all trans prolyl bonds [212].
Nrd1 stands in sharp contrast with respect to its isomer preference. The solution structure of the CTD-interacting domain (CID) of Nrd1 bound to a two-repeat CTD peptide phosphorylated on Ser5 was solved by NMR [213]. The 14-residue CTD motif adopts a β-turn configuration in which both pSer5-Pro6 bonds are in the cis conformation. The cis conformation of the peptidyl-prolyl bonds is required to maximize the specific contacts required for CTD recognition. This is the only known CID that prefers the cis conformation. Given this specificity, it is possible that Ess1 plays a direct role in Nrd1 binding (i.e. independent of pSer status) that has not previously been appreciated.
In summary, proline isomerization of the CTD by Ess1 is likely to play an integral role in regulating the binding and/or activity of transcriptional co-factors. For both Ssu72 and Nrd1, the low abundance of cis-conformers would be made more kinetically accessible by increased rates of isomerization by Ess1. For Pcf11, which binds cooperatively with Rtt109, and requires a substrate with multiple repeats with sequential trans-trans configurations, Ess1 could also assist in a kinetic manner. It is worth noting that the spontaneous interconversion rates measured in peptides in vitro are probably much higher than what occurs in vivo in intact proteins, even the relatively unstructured CTD. Thus, Ess1 isomerization of the CTD would keep it in a constant state of flux, enabling the rapid exchange of proteins at a rate sufficient for transcription and RNA processing. In the case of coupled reactions (e.g. isomerization/dephosphorylation), Ess1 would influence the equilibrium state of the CTD.
5.5. Ess1 is the traffic cop of the RNA pol II transcription cycle
The genetic, biochemical and structural data suggest that Ess1 functions at multiple steps of the transcription cycle in a “Traffic Cop” type mechanism (Fig. 7). That is, by changing the cis/trans state of pSer2-Pro3 and pSer5-Pro6, Ess1 coordinates the flow of traffic of proteins binding to and being released from the CTD, relieving congestion and helping to make each step more efficient. There is probably not an absolute requirement for Ess1 at any given step, as proteins would eventually bind and release, perhaps in response to spontaneous cis/ trans interconversion. For example, termination of snoRNAs and mRNAs still occurs correctly ∼ 80% or more of the time, and transcription in vitro occurs without Ess1/Pin1 [118,128,129,135]. However, without Ess1, the inefficiencies in the transcription cycle, the aberrant RNAs produced, and cryptic transcription that occurs, probably combine to lead to cell death, at least in yeast. In organisms where Ess1/Pin1 is not essential, the defects may be better tolerated or there may exist compensatory mechanisms. However, these inefficiencies may insidiously contribute to the developmental and disease phenotypes observed in Pin1 under- or overexpressing tissues. A simple example was shown in embryos with reduced Ess1 (dodo) in a pol II-sensitized background (Fig. 4B). Also, as noted above, the requirement for Ess1/Pin1 is probably higher during times of stress, when the efficiency of the transcription cycle is probably more critical, especially at stress-responsive loci.
Fig. 7.
Ess1 is the traffic cop of the transcription cycle. Diagram shows the transcription cycle from preinitiation complex formation, to initiation, elongation, RNA splicing, 3′-end processing, termination and re-initiation. During these steps, the CTD of Rpb1 undergoes a complex regime of dynamic covalent (phosphorylation) and non-covalent (isomerization) modifications. These changes in the CTD effect the recruitment and eviction of transcriptional co-factors that are required for of RNA synthesis and maturation. Only a single CTD heptad repeat (Y1S2P3T4S5P6S7) plus a few flanking residues is shown. In this model, Ess1 plays the role of a traffic cop: it helps direct the heavy traffic of protein co-factors so that they can bind and be released from the CTD in a coordinated and efficient manner. Ess1 isomerization is probably not required for transcription and RNA processing per se, but increases its efficiency, just as you can take a traffic cop away from a busy intersection, cars would still get through, just not as efficiently and with a lot more crashes. Arrows from Ess1 show steps that Ess1 promotes, and blocked arrows show steps that Ess1 attenuates.
The backbone of this figure was adapted from Zhang [162] © Creative Commons 2012.
The fact that Ess1's role in transcription is conserved from yeast to humans is also indicative of its importance. And, in some plants that lack Ess1, its role seems to have been subsumed by other prolyl isomerases (e.g. cyclophilins) [214]. Finally, with regard to the mechanism of action, Ess1 on its own cannot change the equilibrium between cis and trans isomers, but instead it provides a kinetic effect that accelerates the availability of the “correct” isomer for a given CTD-binding protein. In combination with a protein(s) that binds one isoform over the other (i.e. a coupled reaction) Ess1 can change the local equilibrium (the CTD-bound protein removes one isomer from the equation). This effect may be necessary to boost the abundance of the cis isomer, which is probably severely underrepresented in the CTD.
6. Other transcription-related roles for Ess1
6.1. An Ess 1-chromatin connection
Ess1 may play a role in organizing chromatin structure by controlling recruitment of histone-modifying enzymes. This would not be entirely surprising given that the pol II CTD is important for recruitment of histone modifiers [163,215–217]. Genetic interactions between ESS1 and genes encoding histone acetylase-deacetylase components, GCN5, RPD3, and SAP30 infer that Ess1 inhibits histone deacetylase (HDAC) activity [23,218]. Combined with biochemical and pharmacologic experiments, the overall effect of Ess1 is likely to be an increase in histone acetylation, thus leading to gene activation [218]. The mechanism by which Ess1 may control histone acetylation state is not known.
ESS1 also shows genetic interactions with SET1, SET2 and JHD2, enzymes responsible for the methylation state of histone H3 lysine 4 (H3K4) and H3 lysine 36 (H3K36). For example, set1Δ is synthetically lethal with ess1H164R and jhd2Δ suppresses ess1H164R [128]. As the genetics predicts, levels of H3K4 trimethylation are sharply reduced in ess1 mutant cells. These results suggest that Ess1 might recruit Set1 (H3K4 methylase), which is known to prefer the pSer5 CTD and is associated with active transcription, or that Ess1 inhibits recruitment of Jhd2 (H3K4 demethylase). The reduction in H3K4 trimethylation levels in ess1 mutants might explain why cryptic transcripts in these cells are elevated. Loss of Ess1 would result in reduced H3K4me3 at cryptic promoters, which can result in a failure to recruit the histone deacetylase complex (Rpd3L) leading to derepression [219].
Despite the strong genetic suppression of ess1H164R mutants by set2Δ, the levels of H3K36 trimethylation, predicted to increase, did not change detectably in ess1 mutants. Changes in H3K36 trimethylation are typically more difficult to detect as the modification is relatively stable (B. Strahl, U.N.C., pers. comm.). Set2 contains a WW-domain and binds the doubly-phosphorylated CTD (pSer2/pSer5) and is associated with elongating polymerase [220,221]. In summary, there is a strong link between Ess1 and chromatin modification, but the mechanisms by which Ess1 affects the enzymes involved in histone acetylation and methylation are unknown.
6.2. Transcription factor localization; Ess1 targets NLS and NES motifs
In a large-scale synthetic genetic array screen, Ess1 was linked to a number of transcription regulators including the cell cycle transcription factors Swi6 and Whi5 [89]. Surprisingly, the expression of the genes encoding these proteins was not affected. Instead their nuclear-cytoplasmic shuttling was defective in ess1 mutant cells. Using GFP-tagged proteins and in vitro binding assays, it was shown that Ess1 is required for nuclear localization of Swi6 and Whi5, and that Ess1 binds peptides corresponding to the nuclear localization sequence (NLS) of Swi6, and the NLS and nuclear export sequences (NES) of Whi5 [89]. The NLS and NES sequences each contain between one and three Ser-Pro motifs, and Ess1 binding as assayed using biolayer interferometry, was dependent upon phosphorylation of these serines. These results suggest a model in which Ess1 regulates localization of Swi6 and Whi5 either indirectly by stimulating their dephosphorylation (a prerequisite for nuclear entry), or directly by causing conformational changes that effect interactions with nuclear pore complexes (both importins and exportins). The exact mechanism remains to be determined. The Cdc14 phosphatase is known to dephosphorylate the NLS and NES sites in Swi6 and Whi5 [222,223], but it is not known whether it is isomer-specific. Swi6 and Whi5 are the only targets of Ess1 other than the CTD identified so far in yeast. Control of nuclear localization of transcription regulators has been previously shown for Pin1 in mammalian cells [16,224], but the de tailed mechanisms have not been worked out.
7. More questions than answers
While much is known about Ess1 and its role in transcription, more remains to be learned. The tools of genetics, genomics, and the bio chemistry of binding and isomerization have been informative. We know of many steps in transcription that require Ess1 for efficiency and have a good idea of how Ess1 might affect the enzymes required for co-transcriptional RNA processing. However, more remains to be discovered. For example, RNA splicing, which is relatively rare in yeast, is likely to depend upon Ess1 for efficiency (more so for Pin1 in humans) as it is a co-transcriptional process and is influenced by elongation rates [154,155,158,225]. mRNA export is also likely to be dependent upon Ess1. Genes encoding a number of mRNA export factors show interactions with ESS1 (unpublished data). Essentially all CTD-related activities in the cell, including those not directly related to RNA biogenesis, such as transcription-coupled repair, recombination, and gene silencing will likely be influenced by Ess1. There are already examples linking the CTD and Pin1 to DNA repair [226–229]. And, if Swi6 and Whi5 serve as examples, then it is likely that other Ess1 targets will be discovered in yeast.
Perhaps more pressing, however, is to determine the exact mechanisms by which Ess1 functions in transcription, such as during elongation and mRNA 3′-end formation. Studies thus far reveal a gap between the existing in vivo work, mostly genetic/genomic and in vitro work with purified isomerases and peptides. A major impediment is that it is not currently possible to monitor the isomerization state of the CTD or any other protein in vivo. New tools need to be developed, such as isomer-specific monoclonal antibodies, or FRET-based or other dynamic in vivo assays to detect changes in the isomerization state of substrate proteins. In addition, better in vitro systems are likely to be required to detect eve fects of Ess1, which may be subtle and require that multiple factors (whose binding Ess1 influences in vivo) are present in the appropriate rate-limiting amounts. Having a large excess of a particular enzyme, for example a CTD phosphatase, will likely obscure the effect of isomer-specificity on substrate preferences. Moreover, in the case of competing proteins, excessive amounts of one protein (or an absence of it) may obscure the effects of Ess1 on binding by another protein. These are difficult challenges that will require the efforts of many.
In the short term, determining the structures of additional CTD-binding proteins with peptide substrates will add important information to the repertoire of cis- or trans-specific enzymes. Genome-wide approaches, such as ChIP-chip and ChIP-seq will reveal the global effects of Ess1 on recruitment of transcriptional co-factors and chromatin modifiers. Also, detailed examination of individual genes by genetic approaches combined with ChIP and perhaps ChIP/reChIP-type methods and detailed RNA analysis will help elucidate the effects of Ess1 on individual steps in transcription.
Finally, one of the most interesting challenges is to figure out how Ess1, and indeed all CTD-binding proteins, operate on the long and repeated heptapeptide motifs in the CTD. Even in yeast, with only 26 repeats, there are 52 potential sites for Ess1 binding and isomerization. Does Ess1 target all these sites, and which sites get isomerized and when? Which if any processes, such as isomerization and phosphorylation/dephosphorylation are processive and what is the mechanism of processivity? And does Ess1 isomerization help direct different proteins to different regions along the length of the CTD? In short, we have finally begun to recognize the importance of Ess1-dependent isomerization of the CTD for transcription, but understand very little of how it works. It is essentially an unfinished book, waiting to be completed by creative and dedicated investigators.
Acknowledgments
The author wishes to thank David Atencio, Patrick Van Roey, and Tom Duncan for help with the figures, and David Atencio and Tom Duncan for comments on the manuscript. Thanks go out to former members of the Hanes laboratory who worked on Ess1, including Xiaoyun Wu, Navjot Singh, Cathy Wilcox, Gina Devasahayam, Ping Ren, Trent Gemmill, Dhanushki Samaranayake, and collaborators Vishnu Chaturvedi, Joe Heitman, Randy Morse, Mike Palumbo, Francois Robert, Jesper Svejstrup, Patrick Van Roey, and Joe Wade. This paper is dedicated to Peter Shank, in whose laboratory ESS1 was discovered, on the occasion of his retirement from Brown University and to Keith A. Bostian, in whose laboratory ESS1 was first characterized. Ess1 work in the Hanes laboratory was supported by the National Science Foundation (MCB-0613001) and the National Institutes of Health (R01-GM55108).
References
- 1.Lu KP, Hanes SD, Hunter T. A human peptidyl-prolyl isomerase essential for regulation of mitosis. Nature. 1996;380:544–547. doi: 10.1038/380544a0. [DOI] [PubMed] [Google Scholar]
- 2.Joseph JD, Yeh ES, Swenson KI, Means AR, Winkler KE. The peptidyl-prolyl isomerase Pin1. Prog Cell Cycle Res. 2003;5:477–487. [PubMed] [Google Scholar]
- 3.Wulf G, Ryo A, Liou YC, Lu KP. The prolyl isomerase Pin1 in breast development and cancer. Breast Cancer Res. 2003;5:76–82. doi: 10.1186/bcr572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Etzkorn FA. Pin1 flips Alzheimer's switch. ACS Chem Biol. 2006;1:214–216. doi: 10.1021/cb600171g. [DOI] [PubMed] [Google Scholar]
- 5.Yeh ES, Means AR. PIN1, the cell cycle and cancer. Nat Rev Cancer. 2007;7:381–388. doi: 10.1038/nrc2107. [DOI] [PubMed] [Google Scholar]
- 6.Takahashi K, Uchida C, Shin RW, Shimazaki K, Uchida T. Prolyl isomerase, Pin1: new findings of post-translational modifications and physiological substrates in cancer, asthma and Alzheimer's disease. Cell Mol Life Sci. 2008;65:359–375. doi: 10.1007/s00018-007-7270-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee TH, Pastorino L, Lu KP. Peptidyl-prolyl cis–trans isomerase Pin1 in ageing, cancer and Alzheimer disease. Expert Rev Mol Med. 2011;13:e21. doi: 10.1017/S1462399411001906. [DOI] [PubMed] [Google Scholar]
- 8.Liou YC, Zhou XZ, Lu KP. Prolyl isomerase Pin1 as a molecular switch to determine the fate of phosphoproteins. Trends Biochem Sci. 2011;36:501–514. doi: 10.1016/j.tibs.2011.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Esnault S, Shen ZJ, Malter JS. Pinning down signaling in the immune system: the role of the peptidyl-prolyl isomerase Pin1 in immune cell function. Crit Rev Immunol. 2008;28:45–60. doi: 10.1615/critrevimmunol.v28.i1.30. [DOI] [PubMed] [Google Scholar]
- 10.Shaw PE. Peptidyl-prolyl isomerases: a new twist to transcription. EMBO J Rep. 2002;3:521–526. doi: 10.1093/embo-reports/kvf118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shaw PE. Peptidyl-prolyl cis/trans isomerases and transcription: is there a twist inthe tail? EMBO J Rep. 2007;8:40–45. doi: 10.1038/sj.embor.7400873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu YX, Manley JL. Pinning down transcription: regulation of RNA polymerase IIactivity during the cell cycle. Cell Cycle. 2004;3:432–435. [PubMed] [Google Scholar]
- 13.Ryo A, Suizu F, Yoshida Y, Perrem K, Liou YC, Wulf G, Rottapel R, Yamaoka S, Lu KP. Regulation of NF-kappaB signaling by Pin1-dependent prolyl isomerization and ubiquitin-mediated proteolysis of p65/RelA. Mol Cell. 2003;12:1413–1426. doi: 10.1016/s1097-2765(03)00490-8. [DOI] [PubMed] [Google Scholar]
- 14.Mantovani F, Piazza S, Gostissa M, Strano S, Zacchi P, Mantovani R, Blandino G, Del Sal G. Pin1 links the activities of c-Abl and p300 in regulating p73 function. Mol Cell. 2004;14:625–636. doi: 10.1016/j.molcel.2004.05.007. [DOI] [PubMed] [Google Scholar]
- 15.Zacchi P, Gostissa M, Uchida T, Salvagno C, Avolio F, Volinia S, Ronai Z, Blandino G, Schneider C, Del Sal G. The prolyl isomerase Pin1 reveals a mechanism to control p53 functions after genotoxic insults. Nature. 2002;419:853–857. doi: 10.1038/nature01120. [DOI] [PubMed] [Google Scholar]
- 16.Ryo A, Nakamura M, Wulf G, Liou YC, Lu KP. Pin1 regulates turnover and sub-cellular localization of beta-catenin by inhibiting its interaction with APC. Nat Cell Biol. 2001;3:793–801. doi: 10.1038/ncb0901-793. [DOI] [PubMed] [Google Scholar]
- 17.Hanes SD. PhD Thesis Division of Biology and Medicine. Brown University; Providence, RI: 1988. Isolation, Sequence and Mutational Analysis of ESS1, a Gene Essential for Growth in Saccharomyces cerevisiase. [Google Scholar]
- 18.Hanes SD, Shank PR, Bostian KA. Sequence and mutational analysis of ESS1, a gene essential for growth in Saccharomyces cerevisiae. Yeast. 1989;5:55–72. doi: 10.1002/yea.320050108. [DOI] [PubMed] [Google Scholar]
- 19.Goebl MG, Petes TD. Most of the yeast genomic sequences are not essential for cell growth and division. Cell. 1986;46:983–992. doi: 10.1016/0092-8674(86)90697-5. [DOI] [PubMed] [Google Scholar]
- 20.Shortle D, Haber JE, Botstein D. Lethal disruption of the yeast actin gene by integrative DNA transformation. Science. 1982;217:371–373. doi: 10.1126/science.7046050. [DOI] [PubMed] [Google Scholar]
- 21.Rothstein RJ. One-step gene disruption in yeast. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- 22.Gemmill TR, Wu X, Hanes SD. Vanishingly low levels of Ess1 prolyl-isomerase activity are sufficient for growth in Saccharomyces cerevisiae. J Biol Chem. 2005;280:15510–15517. doi: 10.1074/jbc.M412172200. [DOI] [PubMed] [Google Scholar]
- 23.Wu X, Wilcox CB, Devasahayam G, Hackett RL, Arevalo-Rodriguez M, Cardenas ME, Heitman J, Hanes SD. The Ess1 prolyl isomerase is linked to chromatin remodeling complexes and the general transcription machinery. EMBO J. 2000;19:3727–3738. doi: 10.1093/emboj/19.14.3727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fischer G, Bang H, Mech C. Determination of enzymatic catalysis for the cis–trans-isomerization of peptide binding in proline-containing peptides. Biomed Biochim Acta. 1984;43:1101–1111. [PubMed] [Google Scholar]
- 25.Schiene C, Fischer G. Enzymes that catalyse the restructuring of proteins. Curr Opin Struct Biol. 2000;10:40–45. doi: 10.1016/s0959-440x(99)00046-9. [DOI] [PubMed] [Google Scholar]
- 26.Fanghanel J, Fischer G. Insights into the catalytic mechanism of peptidyl prolyl cis/trans isomerases. Front Biosci. 2004;9:3453–3478. doi: 10.2741/1494. [DOI] [PubMed] [Google Scholar]
- 27.Schiene-Fischer C, Aumuller T, Fischer G. Peptide bond cis/trans isomerases: a biocatalysis perspective of conformational dynamics in proteins. Top Curr Chem. 2013;328:35–67. doi: 10.1007/128_2011_151. [DOI] [PubMed] [Google Scholar]
- 28.Schmid FX. Prolyl isomerase: enzymatic catalysis of slow protein-folding reactions. Annu Rev Biophys Biomol Struct. 1993;22:123–142. doi: 10.1146/annurev.bb.22.060193.001011. [DOI] [PubMed] [Google Scholar]
- 29.Handschumacher RE, Harding MW, Rice J, Drugge RJ, Speicher DW. Cyclophilin: a specific cytosolic binding protein for cyclosporin A. Science. 1984;226:544–547. doi: 10.1126/science.6238408. [DOI] [PubMed] [Google Scholar]
- 30.Harding MW, Galat A, Uehling DE, Schreiber SL. A receptor for the immunosuppressant FK506 is a cis–trans peptidyl-prolyl isomerase. Nature. 1989;341:758–760. doi: 10.1038/341758a0. [DOI] [PubMed] [Google Scholar]
- 31.Siekierka JJ, Hung SH, Poe M, Lin CS, Sigal NH. A cytosolic binding protein for the immunosuppressant FK506 has peptidyl-prolyl isomerase activity but is distinct from cyclophilin. Nature. 1989;341:755–757. doi: 10.1038/341755a0. [DOI] [PubMed] [Google Scholar]
- 32.Rahfeld JU, Rucknagel KP, Schelbert B, Ludwig B, Hacker J, Mann K, Fischer G. Confirmation of the existence of a third family among peptidyl-prolyl cis/trans isomerases. Amino acid sequence and recombinant production of parvulin. FEBS Lett. 1994;352:180–184. doi: 10.1016/0014-5793(94)00932-5. [DOI] [PubMed] [Google Scholar]
- 33.Hani J, Stumpf G, Domdey H. PTF1 encodes an essential protein in Saccharomyces cerevisiae, which shows strong homology with a new putative family of PPIases. FEBS Lett. 1995;365:198–202. doi: 10.1016/0014-5793(95)00471-k. [DOI] [PubMed] [Google Scholar]
- 34.Sudol M, Chen HI, Bougeret C, Einbond A, Bork P. Characterization of a novel protein-binding module—the WW domain. FEBS Lett. 1995;369:67–71. doi: 10.1016/0014-5793(95)00550-s. [DOI] [PubMed] [Google Scholar]
- 35.Macias MJ, Wiesner S, Sudol M. WW and SH3 domains, two different scaffolds to recognize proline-rich ligands. FEBS Lett. 2002;513:30–37. doi: 10.1016/s0014-5793(01)03290-2. [DOI] [PubMed] [Google Scholar]
- 36.Bork P, Sudol M. The WW domain: a signalling site in dystrophin? Trends Biochem Sci. 1994;19:531–533. doi: 10.1016/0968-0004(94)90053-1. [DOI] [PubMed] [Google Scholar]
- 37.Maleszka R, Hanes SD, Hackett RL, de Couet HG, Miklos GL. The Drosophila melanogaster dodo (dod) gene, conserved in humans, is functionally interchange able with the ESS1 cell division gene of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1996;93:447–451. doi: 10.1073/pnas.93.1.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Crenshaw DG, Yang J, Means AR, Kornbluth S. The mitotic peptidyl-prolyl isomerase, Pin1, interacts with Cdc25 and Plx1. EMBO J. 1998;17:1315–1327. doi: 10.1093/emboj/17.5.1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shen M, Stukenberg PT, Kirschner MW, Lu KP. The essential mitotic peptidyl-prolyl isomerase Pin1 binds and regulates mitosis-specific phosphoproteins. Genes Dev. 1998;12:706–720. doi: 10.1101/gad.12.5.706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fujimori F, Takahashi K, Uchida C, Uchida T. Mice lacking Pin1 develop normally, but are defective in entering cell cycle from G (0) arrest. Biochem Biophys Res Commun. 1999;265:658–663. doi: 10.1006/bbrc.1999.1736. [DOI] [PubMed] [Google Scholar]
- 41.Erben ED, Daum S, Tellez-Inon MT. The Trypanosoma cruzi PIN1 gene encodes a parvulin peptidyl-prolyl cis/trans isomerase able to replace the essential ESS1 in Saccharomyces cerevisiae. Mol Biochem Parasitol. 2007;153:186–193. doi: 10.1016/j.molbiopara.2007.03.004. [DOI] [PubMed] [Google Scholar]
- 42.Huang HK, Forsburg SL, John UP, O'Connell MJ, Hunter T. Isolation and characterization of the Pin1/Ess1p homologue in Schizosaccharomyces pombe. J Cell Sci. 2001;114:3779–3788. doi: 10.1242/jcs.114.20.3779. [DOI] [PubMed] [Google Scholar]
- 43.Devasahayam G, Chaturvedi V, Hanes SD. The Ess1 prolyl isomerase is required for growth and morphogenetic switching in Candida albicans. Genetics. 2002;160:37–48. doi: 10.1093/genetics/160.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ren P, Rossettini A, Chaturvedi V, Hanes SD. The Ess1 prolyl isomerase is dispensable for growth but required for virulence in Cryptococcus neoformans. Microbiology. 2005;151:1593–1605. doi: 10.1099/mic.0.27786-0. [DOI] [PubMed] [Google Scholar]
- 45.Behrsin CD, Bailey ML, Bateman KS, Hamilton KS, Wahl LM, Brandl CJ, Shilton BH, Litchfield DW. Functionally important residues in the peptidyl-prolyl isomerase Pin1 revealed by unigenic evolution. J Mol Biol. 2007;365:1143–1162. doi: 10.1016/j.jmb.2006.10.078. [DOI] [PubMed] [Google Scholar]
- 46.Zhou XZ, Kops O, Werner A, Lu PJ, Shen M, Stoller G, Kullertz G, Stark M, Fischer G, Lu KP. Pin1-dependent prolyl isomerization regulates dephosphorylation of Cdc25C and tau proteins. Mol Cell. 2000;6:873–883. doi: 10.1016/s1097-2765(05)00083-3. [DOI] [PubMed] [Google Scholar]
- 47.Uchida T, Fujimori F, Tradler T, Fischer G, Rahfeld JU. Identification and characterization of a 14 kDa human protein as a novel parvulin-like peptidyl prolyl cis/trans isomerase. FEBS Lett. 1999;446:278–282. doi: 10.1016/s0014-5793(99)00239-2. [DOI] [PubMed] [Google Scholar]
- 48.Rulten S, Thorpe J, Kay J. Identification of eukaryotic parvulin homologues: a new subfamily of peptidylprolyl cis–trans isomerases. Biochem Biophys Res Commun. 1999;259:557–562. doi: 10.1006/bbrc.1999.0828. [DOI] [PubMed] [Google Scholar]
- 49.Surmacz TA, Bayer E, Rahfeld JU, Fischer G, Bayer P. The N-terminal basic domain of human parvulin hPar14 is responsible for the entry to the nucleus and high-affinity DNA-binding. J Mol Biol. 2002;321:235–247. doi: 10.1016/s0022-2836(02)00615-0. [DOI] [PubMed] [Google Scholar]
- 50.Fujiyama S, Yanagida M, Hayano T, Miura Y, Isobe T, Fujimori F, Uchida T, Takahashi N. Isolation and proteomic characterization of human Parvulin-associating preribosomal ribonucleoprotein complexes. J Biol Chem. 2002;277:23773–23780. doi: 10.1074/jbc.M201181200. [DOI] [PubMed] [Google Scholar]
- 51.Fujiyama-Nakamura S, Yoshikawa H, Homma K, Hayano T, Tsujimura-Takahashi T, Izumikawa K, Ishikawa H, Miyazawa N, Yanagida M, Miura Y, Shinkawa T, Yamauchi Y, Isobe T, Takahashi N. Parvulin (Par14), a peptidyl-prolyl cis–trans isomerase, is a novel rRNA processing factor that evolved in the metazoan lineage. Mol Cell Proteomics. 2009;8:1552–1565. doi: 10.1074/mcp.M900147-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Metzner M, Stoller G, Rucknagel KP, Lu KP, Fischer G, Luckner M, Kullertz G. Functional replacement of the essential ESS1 in yeast by the plant parvulin DlPar13. J Biol Chem. 2001;276:13524–13529. doi: 10.1074/jbc.M007005200. [DOI] [PubMed] [Google Scholar]
- 53.Kessler D, Papatheodorou P, Stratmann T, Dian EA, Hartmann-Fatu C, Rassow J, Bayer P, Mueller JW. The DNA binding parvulin Par17 is targeted to the mitochondrial matrix by a recently evolved prepeptide uniquely present in Hominidae. BMC Biol. 2007;5:37. doi: 10.1186/1741-7007-5-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mueller JW, Bayer P. Small family with key contacts: par14 and par17 parvulin proteins, relatives of Pin1, now emerge in biomedical research. Perspect Med Chem. 2008;2:11–20. doi: 10.4137/pmc.s496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Campbell HD, Webb GC, Fountain S, Young IG. The human PIN1 peptidyl-prolyl cis/trans isomerase gene maps to human chromosome 19p13 and the closely related PIN1L gene to 1p31. Genomics. 1997;44:157–162. doi: 10.1006/geno.1997.4854. [DOI] [PubMed] [Google Scholar]
- 56.Fanghanel J, Akiyama H, Uchida C, Uchida T. Comparative analysis of enzyme activities and mRNA levels of peptidyl prolyl cis/trans isomerases in various organs of wild type and Pin1-/- mice. FEBS Lett. 2006;580:3237–3245. doi: 10.1016/j.febslet.2006.04.087. [DOI] [PubMed] [Google Scholar]
- 57.Akiyama H, Gotoh A, Shin RW, Koga T, Ohashi T, Sakamoto W, Harada A, Arai H, Sawa A, Uchida C, Uchida T. A novel role for hGas7b in microtubular maintenance: possible implication in tau-associated pathology in Alzheimer disease. J Biol Chem. 2009;284:32695–32699. doi: 10.1074/jbc.M109.035998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Landrieu I, De Veylder L, Fruchart JS, Odaert B, Casteels P, Portetelle D, Van Montagu M, Inze D, Lippens G. The Arabidopsis thaliana PIN1At gene encodes a single-domain phosphorylation-dependent peptidyl prolyl cis/trans isomerase. J Biol Chem. 2000;275:10577–10581. doi: 10.1074/jbc.275.14.10577. [DOI] [PubMed] [Google Scholar]
- 59.He Z, Li L, Luan S. Immunophilins and parvulins. Superfamily of peptidyl prolyl isomerases in Arabidopsis. Plant Physiol. 2004;134:1248–1267. doi: 10.1104/pp.103.031005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yao JL, Kops O, Lu PJ, Lu KP. Functional conservation of phosphorylation-specific prolyl isomerases in plants. J Biol Chem. 2001;276:13517–13523. doi: 10.1074/jbc.M007006200. [DOI] [PubMed] [Google Scholar]
- 61.Dharmasiri N, Dharmasiri S, Jones AM, Estelle M. Auxin action in a cell-free system. Curr Biol. 2003;13:1418–1422. doi: 10.1016/s0960-9822(03)00536-0. [DOI] [PubMed] [Google Scholar]
- 62.Wang Y, Liu C, Yang D, Yu H, Liou YC. Pin1At encoding a peptidyl-prolyl cis/trans isomerase regulates flowering time in Arabidopsis. Mol Cell. 2010;37:112–122. doi: 10.1016/j.molcel.2009.12.020. [DOI] [PubMed] [Google Scholar]
- 63.Jaremko L, Jaremko M, Elfaki I, Mueller JW, Ejchart A, Bayer P, Zhukov I. Structure and dynamics of the first archaeal parvulin reveal a new functionally important loop in parvulin-type prolyl isomerases. J Biol Chem. 2011;286:6554–6565. doi: 10.1074/jbc.M110.160713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Arevalo-Rodriguez M, Wu X, Hanes SD, Heitman J. Prolyl isomerases in yeast. Front Biosci. 2004;9:2420–2446. doi: 10.2741/1405. [DOI] [PubMed] [Google Scholar]
- 65.Ranganathan R, Lu KP, Hunter T, Noel JP. Structural and functional analysis of the mitotic rotamase Pin1 suggests substrate recognition is phosphorylation dependent. Cell. 1997;89:875–886. doi: 10.1016/s0092-8674(00)80273-1. [DOI] [PubMed] [Google Scholar]
- 66.Ng CA, Kato Y, Tanokura M, Brownlee RT. Structural characterisation of PinA WW domain and a comparison with other group IV WW domains, Pin1 and Ess1. Biochim Biophys Acta. 2008;1784:1208–1214. doi: 10.1016/j.bbapap.2008.04.026. [DOI] [PubMed] [Google Scholar]
- 67.Sudol M, Hunter T. NeW wrinkles for an old domain. Cell. 2000;103:1001–1004. doi: 10.1016/s0092-8674(00)00203-8. [DOI] [PubMed] [Google Scholar]
- 68.Lu PJ, Zhou XZ, Shen M, Lu KP. Function of WW domains as phosphoserine- or phosphothreonine-binding modules. Science. 1999;283:1325–1328. doi: 10.1126/science.283.5406.1325. [DOI] [PubMed] [Google Scholar]
- 69.Verdecia MA, Bowman ME, Lu KP, Hunter T, Noel JP. Structural basis for phosphoserine–proline recognition by group IV WW domains. Nat Struct Biol. 2000;7:639–643. doi: 10.1038/77929. [DOI] [PubMed] [Google Scholar]
- 70.Bayer E, Goettsch S, Mueller JW, Griewel B, Guiberman E, Mayr LM, Bayer P. Structural analysis of the mitotic regulator hPin1 in solution: insights into domain architecture and substrate binding. J Biol Chem. 2003;278:26183–26193. doi: 10.1074/jbc.M300721200. [DOI] [PubMed] [Google Scholar]
- 71.Jacobs DM, Saxena K, Vogtherr M, Bernado P, Pons M, Fiebig KM. Peptide binding induces large scale changes in inter-domain mobility in human Pin1. J Biol Chem. 2003;278:26174–26182. doi: 10.1074/jbc.M300796200. [DOI] [PubMed] [Google Scholar]
- 72.Matena A, Sinnen C, van den Boom J, Wilms C, Dybowski JN, Maltaner R, Mueller JW, Link NM, Hoffmann D, Bayer P. Transient domain interactions enhance the affinity of the mitotic regulator Pin1 toward phosphorylated peptide ligands. Structure. 2013;21:1769–1777. doi: 10.1016/j.str.2013.07.016. [DOI] [PubMed] [Google Scholar]
- 73.Labeikovsky W, Eisenmesser EZ, Bosco DA, Kern D. Structure and dynamics of Pin1 during catalysis by NMR. J Mol Biol. 2007;367:1370–1381. doi: 10.1016/j.jmb.2007.01.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Daum S, Lucke C, Wildemann D, Schiene-Fischer C. On the benefit of bivalency in peptide ligand/pin1 interactions. J Mol Biol. 2007;374:147–161. doi: 10.1016/j.jmb.2007.09.019. [DOI] [PubMed] [Google Scholar]
- 75.Li Z, Li H, Devasahayam G, Gemmill T, Chaturvedi V, Hanes SD, Van Roey P. The structure of the Candida albicans Ess1 prolyl isomerase reveals a well-ordered linker that restricts domain mobility. Biochemistry. 2005;44:6180–6189. doi: 10.1021/bi050115l. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.McNaughton L, Li Z, Van Roey P, Hanes SD, LeMaster DM. Restricted domain mobility in the Candida albicans Ess1 prolyl isomerase. Biochim Biophys Acta. 2010;1804:1537–1541. doi: 10.1016/j.bbapap.2010.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lippens G, Landrieu I, Smet C. Molecular mechanisms of the phospho-dependent prolyl cis/trans isomerase Pin1. FEBS J. 2007;274:5211–5222. doi: 10.1111/j.1742-4658.2007.06057.x. [DOI] [PubMed] [Google Scholar]
- 78.Park ST, Aldape RA, Futer O, DeCenzo MT, Livingston DJ. PPIase catalysis by human FK506-binding protein proceeds through a conformational twist mechanism. J Biol Chem. 1992;267:3316–3324. [PubMed] [Google Scholar]
- 79.Kofron JL, Kuzmic P, Kishore V, Colon-Bonilla E, Rich DH. Determination of kinetic constants for peptidyl prolyl cis–trans isomerases by an improved spectro-photometric assay. Biochemistry. 1991;30:6127–6134. doi: 10.1021/bi00239a007. [DOI] [PubMed] [Google Scholar]
- 80.Grathwohl C, Wuthrich K. Nmr studies of the molecular conformations in the linear oligopeptides H-(L-Ala)n-L-Pro-OH. Biopolymers. 1976;15:2043–2057. doi: 10.1002/bip.1976.360151013. [DOI] [PubMed] [Google Scholar]
- 81.Cheng HN, Bovey FA. cis–trans equilibrium and kinetic studies of acetyl-l-proline and glycyl-l-proline. Biopolymers. 1977;16:1465–1472. doi: 10.1002/bip.1977.360160707. [DOI] [PubMed] [Google Scholar]
- 82.Stimson ER, Montelione GT, Meinwald YC, Rudolph RK, Scheraga HA. Equilibrium rium ratios of cis- and trans-proline conformers in fragments of ribonuclease A from nuclear magnetic resonance spectra of adjacent tyrosine ring resonances. Bio chemistry. 1982;21:5252–5262. doi: 10.1021/bi00264a021. [DOI] [PubMed] [Google Scholar]
- 83.Schmid FX, Baldwin RL. Acid catalysis of the formation of the slow-folding species of RNase A: evidence that the reaction is proline isomerization. Proc Natl Acad Sci U S A. 1978;75:4764–4768. doi: 10.1073/pnas.75.10.4764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wildemann D, Erdmann F, Alvarez BH, Stoller G, Zhou XZ, Fanghanel J, Schutkowski M, Lu KP, Fischer G. Nanomolar inhibitors of the peptidyl prolyl cis/trans isomerase Pin1 from combinatorial peptide libraries. J Med Chem. 2006;49:2147–2150. doi: 10.1021/jm060036n. [DOI] [PubMed] [Google Scholar]
- 85.Wu X, Chang A, Sudol M, Hanes SD. Genetic interactions between the ESS1 prolyl-isomerase and the RSP5 ubiquitin ligase reveal opposing effects on RNA polymerase II function. Curr Genet. 2001;40:234–242. doi: 10.1007/s00294-001-0257-8. [DOI] [PubMed] [Google Scholar]
- 86.Innes BT, Bailey ML, Brandl CJ, Shilton BH, Litchfield DW. Non-catalytic participation of the Pin1 peptidyl-prolyl isomerase domain in target binding. Front Physiol. 2013;4:18. doi: 10.3389/fphys.2013.00018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Myers JK, Morris DP, Greenleaf AL, Oas TG. Phosphorylation of RNA polymerase II CTD fragments results in tight binding to the WW domain from the yeast prolyl isomerase Ess1. Biochemistry. 2001;40:8479–8486. doi: 10.1021/bi0027884. [DOI] [PubMed] [Google Scholar]
- 88.Kern D, Drakenberg T, Wikstrom M, Forsen S, Bang H, Fischer G. The cis/trans interconversion of the calcium regulating hormone calcitonin is catalyzed by cyclophilin. FEBS Lett. 1993;323:198–202. doi: 10.1016/0014-5793(93)81338-z. [DOI] [PubMed] [Google Scholar]
- 89.Atencio D, Barnes C, Duncan TM, Willis IM, Hanes SD. The yeast Ess1 prolyl isomerase controls Swi6 and Whi5 nuclear localization, Genes, Genome, Gen. 2014 doi: 10.1534/g3.113.008763. http://dx.doi.org/10.1534/g3.113.008763 (Epub ahead of print) (January 27) [DOI] [PMC free article] [PubMed]
- 90.Zhang Y, Fussel S, Reimer U, Schutkowski M, Fischer G. Substrate-based design of reversible Pin1 inhibitors. Biochemistry. 2002;41:11868–11877. doi: 10.1021/bi0262395. [DOI] [PubMed] [Google Scholar]
- 91.Landrieu I, Smet C, Wieruszeski JM, Sambo AV, Wintjens R, Buee L, Lippens G. Exploring the molecular function of PIN1 by nuclear magnetic resonance. Curr Protein Pept Sci. 2006;7:179–194. doi: 10.2174/138920306777452303. [DOI] [PubMed] [Google Scholar]
- 92.Schutkowski M, Bernhardt A, Zhou XZ, Shen M, Reimer U, Rahfeld JU, Lu KP, Fischer G. Role of phosphorylation in determining the backbone dynamics of the serine/threonine–proline motif and Pin1 substrate recognition. Biochemistry. 1998;37:5566–5575. doi: 10.1021/bi973060z. [DOI] [PubMed] [Google Scholar]
- 93.Wedemeyer WJ, Welker E, Scheraga HA. Proline cis–trans isomerization and protein folding. Biochemistry. 2002;41:14637–14644. doi: 10.1021/bi020574b. [DOI] [PubMed] [Google Scholar]
- 94.Yaffe MB, Schutkowski M, Shen M, Zhou XZ, Stukenberg PT, Rahfeld JU, Xu J, Kuang J, Kirschner MW, Fischer G, Cantley LC, Lu KP. Sequence-specific and phosphorylation-dependent proline isomerization: a potential mitotic regulatory mechanism. Science. 1997;278:1957–1960. doi: 10.1126/science.278.5345.1957. [DOI] [PubMed] [Google Scholar]
- 95.Wilcox CB, Rossettini A, Hanes SD. Genetic interactions with C-terminal domain (CTD) kinases and the CTD of RNA Pol II suggest a role for ESS1 in transcription initiation and elongation in Saccharomyces cerevisiae. Genetics. 2004;167:93–105. doi: 10.1534/genetics.167.1.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Mann RS. The specificity of homeotic gene function. Bioessays. 1995;17:855–863. doi: 10.1002/bies.950171007. [DOI] [PubMed] [Google Scholar]
- 97.Biggin MD, McGinnis W. Regulation of segmentation and segmental identity by Drosophila homeoproteins: the role of DNA binding in functional activity and specificity. Development. 1997;124:4425–4433. doi: 10.1242/dev.124.22.4425. [DOI] [PubMed] [Google Scholar]
- 98.Hall FL, Vulliet PR. Proline-directed protein phosphorylation and cell cycle regulation. Curr Opin Cell Biol. 1991;3:176–184. doi: 10.1016/0955-0674(91)90136-m. [DOI] [PubMed] [Google Scholar]
- 99.Lim S, Kaldis P. Cdks, cyclins and CKIs: roles beyond cell cycle regulation. Development. 2013;140:3079–3093. doi: 10.1242/dev.091744. [DOI] [PubMed] [Google Scholar]
- 100.Pelech SL. Networking with proline-directed protein kinases implicated in tau phosphorylation. Neurobiol Aging. 1995;16:247–256. doi: 10.1016/0197-4580(94)00187-6. discussion 257–261. [DOI] [PubMed] [Google Scholar]
- 101.Mercedes-Camacho AY, Mullins AB, Mason MD, Xu GG, Mahoney BJ, Wang X, Peng JW, Etzkorn FA. Kinetic isotope effects support the twisted amide mechanism of Pin1 peptidyl-prolyl isomerase. Biochemistry. 2013;52:7707–7713. doi: 10.1021/bi400700b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zhang Y, Daum S, Wildemann D, Zhou XZ, Verdecia MA, Bowman ME, Lucke C, Hunter T, Lu KP, Fischer G, Noel JP. Structural basis for high-affinity peptide inhibition of human Pin1. ACS Chem Biol. 2007;2:320–328. doi: 10.1021/cb7000044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Vohringer-Martinez E, Duarte F, Toro-Labbe A. How does Pin1 catalyze the cis–trans prolyl peptide bond isomerization? A QM/MM and mean reaction force study. J Phys Chem B. 2012;116:12972–12979. doi: 10.1021/jp307946h. [DOI] [PubMed] [Google Scholar]
- 104.Bailey ML, Shilton BH, Brandl CJ, Litchfield DW. The dual histidine motif in the active site of Pin1 has a structural rather than catalytic role. Biochemistry. 2008;47:11481–11489. doi: 10.1021/bi800964q. [DOI] [PubMed] [Google Scholar]
- 105.Harrison RK, Stein RL. Mechanistic studies of peptidyl prolyl cis–trans isomerase: evidence for catalysis by distortion. Biochemistry. 1990;29:1684–1689. doi: 10.1021/bi00459a003. [DOI] [PubMed] [Google Scholar]
- 106.Fischer S, Michnick S, Karplus M. A mechanism for rotamase catalysis by the FK506 binding protein (FKBP) Biochemistry. 1993;32:13830–13837. doi: 10.1021/bi00213a011. [DOI] [PubMed] [Google Scholar]
- 107.Hennig L, Christner C, Kipping M, Schelbert B, Rucknagel KP, Grabley S, Kullertz G, Fischer G. Selective inactivation of parvulin-like peptidyl-prolyl cis/trans isomerases by juglone. Biochemistry. 1998;37:5953–5960. doi: 10.1021/bi973162p. [DOI] [PubMed] [Google Scholar]
- 108.Duncan KE, Dempsey BR, Killip LE, Adams J, Bailey ML, Lajoie GA, Litchfield DW, Brandl CJ, Shaw GS, Shilton BH. Discovery and characterization of a nonphosphorylated cyclic peptide inhibitor of the peptidylprolyl isomerase, Pin1. J Med Chem. 2011;54:3854–3865. doi: 10.1021/jm200156c. [DOI] [PubMed] [Google Scholar]
- 109.Liu T, Liu Y, Kao HY, Pei D. Membrane permeable cyclic peptidyl inhibitors against human peptidylprolyl isomerase Pin1. J Med Chem. 2010;53:2494–2501. doi: 10.1021/jm901778v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wang XJ, Xu B, Mullins AB, Neiler FK, Etzkorn FA. Conformationally locked isostere of phosphoSer-cis-Pro inhibits Pin1 23-fold better than phosphoSer-trans-Pro isostere. J Am Chem Soc. 2004;126:15533–15542. doi: 10.1021/ja046396m. [DOI] [PubMed] [Google Scholar]
- 111.Zhao S, Etzkorn FA. A phosphorylated prodrug for the inhibition of Pin1. Bioorg Med Chem Lett. 2007;17:6615–6618. doi: 10.1016/j.bmcl.2007.09.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Smet C, Wieruszeski JM, Buee L, Landrieu I, Lippens G. Regulation of Pin1 peptidyl-prolyl cis/trans isomerase activity by its WW binding module on a multi-phosphorylated peptide of Tau protein. FEBS Lett. 2005;579:4159–4164. doi: 10.1016/j.febslet.2005.06.048. [DOI] [PubMed] [Google Scholar]
- 113.Namanja AT, Wang XJ, Xu B, Mercedes-Camacho AY, Wilson KA, Etzkorn FA, Peng JW. Stereospecific gating of functional motions in Pin1. Proc Natl Acad Sci U S A. 2011;108:12289–12294. doi: 10.1073/pnas.1019382108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Hani J, Schelbert B, Bernhardt A, Domdey H, Fischer G, Wiebauer K, Rahfeld JU. Mutations in a peptidylprolyl-cis/trans-isomerase gene lead to a defect in 3′-end formation of a pre-mRNA in Saccharomyces cerevisiae. J Biol Chem. 1999;274:108–116. doi: 10.1074/jbc.274.1.108. [DOI] [PubMed] [Google Scholar]
- 115.Albert A, Lavoie S, Vincent M. A hyperphosphorylated form of RNA polymerase II is the major interphase antigen of the phosphoprotein antibody MPM-2 and interacts with the peptidyl-prolyl isomerase Pin1. J Cell Sci. 1999;112(Pt 15):2493–2500. doi: 10.1242/jcs.112.15.2493. [DOI] [PubMed] [Google Scholar]
- 116.Morris DP, Phatnani HP, Greenleaf AL. Phospho-carboxyl-terminal domain binding and the role of a prolyl isomerase in pre-mRNA 3′-end formation. J Biol Chem. 1999;274:31583–31587. doi: 10.1074/jbc.274.44.31583. [DOI] [PubMed] [Google Scholar]
- 117.Kinkelin K, Wozniak GG, Rothbart SB, Lidschreiber M, Strahl BD, Cramer P. Structures of RNA polymerase II complexes with Bye1, a chromatin-binding PHF3/DIDO homologue. Proc Natl Acad Sci U S A. 2013;110:15277–15282. doi: 10.1073/pnas.1311010110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Wu X, Rossettini A, Hanes SD. The ESS1 prolyl isomerase and its suppressor BYE1 interact with RNA pol II to inhibit transcription elongation in Saccharomyces cerevisiae. Genetics. 2003;165:1687–1702. doi: 10.1093/genetics/165.4.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Kobor MS, Archambault J, Lester W, Holstege FC, Gileadi O, Jansma DB, Jennings EG, Kouyoumdjian F, Davidson AR, Young RA, Greenblatt J. An unusual eukaryotic protein phosphatase required for transcription by RNA polymerase II and CTD dephosphorylation in S. cerevisiae. Mol Cell. 1999;4:55–62. doi: 10.1016/s1097-2765(00)80187-2. [DOI] [PubMed] [Google Scholar]
- 120.Zhang Y, Sun ZW, Iratni R, Erdjument-Bromage H, Tempst P, Hampsey M, Reinberg D. SAP30, a novel protein conserved between human and yeast, is a component of a histone deacetylase complex. Mol Cell. 1998;1:1021–1031. doi: 10.1016/s1097-2765(00)80102-1. [DOI] [PubMed] [Google Scholar]
- 121.Parkhurst SM, Ish-Horowicz D. Wimp, a dominant maternal-effect mutation, reduces transcription of a specific subset of segmentation genes in Drosophila. Genes Dev. 1991;5:341–357. doi: 10.1101/gad.5.3.341. [DOI] [PubMed] [Google Scholar]
- 122.Krishnamurthy S, Ghazy MA, Moore C, Hampsey M. Functional interaction of the Ess1 prolyl isomerase with components of the RNA polymerase II initiation and termination machineries. Mol Cell Biol. 2009;29:2925–2934. doi: 10.1128/MCB.01655-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Kops O, Zhou XZ, Lu KP. Pin1 modulates the dephosphorylation of the RNA polymerase II C-terminal domain by yeast Fcp1. FEBS Lett. 2002;513:305–311. doi: 10.1016/s0014-5793(02)02288-3. [DOI] [PubMed] [Google Scholar]
- 124.Buratowski S. The CTD code. Nat Struct Biol. 2003;10:679–680. doi: 10.1038/nsb0903-679. [DOI] [PubMed] [Google Scholar]
- 125.Xu YX, Hirose Y, Zhou XZ, Lu KP, Manley JL. Pin1 modulates the structure and function of human RNA polymerase II. Genes Dev. 2003;17:2765–2776. doi: 10.1101/gad.1135503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Palancade B, Marshall NF, Tremeau-Bravard A, Bensaude O, Dahmus ME, Dubois MF. Dephosphorylation of RNA polymerase II by CTD-phosphatase FCP1 is inhibited by phospho-CTD associating proteins. J Mol Biol. 2004;335:415–424. doi: 10.1016/j.jmb.2003.10.036. [DOI] [PubMed] [Google Scholar]
- 127.Xu YX, Manley JL. Pin1 modulates RNA polymerase II activity during the transcription cycle. Genes Dev. 2007;21:2950–2962. doi: 10.1101/gad.1592807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Ma Z, Atencio D, Barnes C, DeFiglio H, Hanes SD. Multiple roles for the Ess1 prolyl isomerase in the RNA polymerase II transcription cycle. Mol Cell Biol. 2012;32:3594–3607. doi: 10.1128/MCB.00672-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Singh N, Ma Z, Gemmill T, Wu X, Rossettini A, Rabeler C, Beane O, DeFiglio H, Palumbo M, Morse R, Hanes SD. The Ess1 prolyl isomerase is required for transcription termination of small non-coding regulatory RNAs via the Nrd1 pathway. Mol Cell. 2009;36:255–266. doi: 10.1016/j.molcel.2009.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Nazarenus T, Cedarberg R, Bell R, Cheatle J, Forch A, Haifley A, Hou A, Wanja Kebaara B, Shields C, Stoysich K, Taylor R, Atkin AL. Upf1p, a highly conserved protein required for nonsense-mediated mRNA decay, interacts with the nuclear pore proteins Nup100p and Nup116p. Gene. 2005;345:199–212. doi: 10.1016/j.gene.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 131.Amrani N, Dong S, He F, Ganesan R, Ghosh S, Kervestin S, Li C, Mangus DA, Spatrick P, Jacobson A. Aberrant termination triggers nonsense-mediated mRNA decay. Biochem Soc Trans. 2006;34:39–42. doi: 10.1042/BST20060039. [DOI] [PubMed] [Google Scholar]
- 132.Muhlemann O, Eberle AB, Stalder L, Zamudio Orozco R. Recognition and elimination of nonsense mRNA. Biochim Biophys Acta. 2008;1779:538–549. doi: 10.1016/j.bbagrm.2008.06.012. [DOI] [PubMed] [Google Scholar]
- 133.Porrua O, Libri D. RNA quality control in the nucleus: the Angels' share of RNA. Biochim Biophys Acta. 2013;1829:604–611. doi: 10.1016/j.bbagrm.2013.02.012. [DOI] [PubMed] [Google Scholar]
- 134.Parker R. RNA degradation in Saccharomyces cerevisae. Genetics. 2012;191:671–702. doi: 10.1534/genetics.111.137265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Chao SH, Greenleaf AL, Price DH. Juglone, an inhibitor of the peptidyl-prolyl isomerase Pin1, also directly blocks transcription. Nucleic Acids Res. 2001;29:767–773. doi: 10.1093/nar/29.3.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Ganem C, Devaux F, Torchet C, Jacq C, Quevillon-Cheruel S, Labesse G, Facca C, Faye G. Ssu72 is a phosphatase essential for transcription termination of snoRNAs and specific mRNAs in yeast. EMBOJ. 2003;22:1588–1598. doi: 10.1093/emboj/cdg141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Reichow SL, Hamma T, Ferre-D'Amare AR, Varani G. The structure and function of small nucleolar ribonucleoproteins. Nucleic Acids Res. 2007;35:1452–1464. doi: 10.1093/nar/gkl1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Wyers F, Rougemaille M, Badis G, Rousselle JC, Dufour ME, Boulay J, Regnault B, Devaux F, Namane A, Seraphin B, Libri D, Jacquier A. Cryptic pol II transcripts are degraded by a nuclear quality control pathway involving a new poly(A) polymerase. Cell. 2005;121:725–737. doi: 10.1016/j.cell.2005.04.030. [DOI] [PubMed] [Google Scholar]
- 139.Arigo JT, Eyler DE, Carroll KL, Corden JL. Termination of cryptic unstable tran scripts is directed by yeast RNA-binding proteins Nrd1 and Nab3. Mol Cell. 2006;23:841–851. doi: 10.1016/j.molcel.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 140.Thiebaut M, Kisseleva-Romanova E, Rougemaille M, Boulay J, Libri D. Transcription termination and nuclear degradation of cryptic unstable transcripts: a role for the Nrd1-Nab3 pathway in genome surveillance. Mol Cell. 2006;23:853–864. doi: 10.1016/j.molcel.2006.07.029. [DOI] [PubMed] [Google Scholar]
- 141.Dichtl B. Transcriptional shortCUTs. Mol Cell. 2008;31:617–618. doi: 10.1016/j.molcel.2008.08.014. [DOI] [PubMed] [Google Scholar]
- 142.Rougemaille M, Libri D. Control of cryptic transcription in eukaryotes. Adv Exp Med Biol. 2011;702:122–131. doi: 10.1007/978-1-4419-7841-7_10. [DOI] [PubMed] [Google Scholar]
- 143.Colin J, Libri D, Porrua O. Cryptic transcription and early termination in the control of gene expression. Genet Res Int. 2011;2011:653494. doi: 10.4061/2011/653494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Rougemaille M, Libri D. Control of cryptic transcription in eukaryotes. Adv Exp Med Biol. 2010;702:122–131. [PubMed] [Google Scholar]
- 145.Samaranayake D, Atencio D, Morse R, Wade JT, Chaturvedi V, Hanes SD. Role of Ess1 in growth, morphogenetic switching, and RNA polymerase II transcription in Candida albicans. PLoS One. 2013;8:e59094. doi: 10.1371/journal.pone.0059094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Siepe D, Jentsch S. Prolyl isomerase Pin1 acts as a switch to control the degree of substrate ubiquitylation. Nat Cell Biol. 2009;11:967–972. doi: 10.1038/ncb1908. [DOI] [PubMed] [Google Scholar]
- 147.Hong SW, Hong SM, Yoo JW, Lee YC, Kim S, Lis JT, Lee DK. Phosphorylation of the RNA polymerase II C-terminal domain by TFIIH kinase is not essential for transcription of Saccharomyces cerevisiae genome. Proc Natl Acad Sci U S A. 2009;106:14276–14280. doi: 10.1073/pnas.0903642106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Hengartner CJ, Myer VE, Liao SM, Wilson CJ, Koh SS, Young RA. Temporal regulation of RNA polymerase II by Srb10 and Kin28 cyclin-dependent kinases. Mol Cell. 1998;2:43–53. doi: 10.1016/s1097-2765(00)80112-4. [DOI] [PubMed] [Google Scholar]
- 149.Liu Y, Kung C, Fishburn J, Ansari AZ, Shokat KM, Hahn S. Two cyclin-dependent kinases promote RNA polymerase II transcription and formation of the scaffold complex. Mol Cell Biol. 2004;24:1721–1735. doi: 10.1128/MCB.24.4.1721-1735.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Aristizabal MJ, Negri GL, Benschop JJ, Holstege FC, Krogan NJ, Kobor MS. High-throughput genetic and gene expression analysis of the RNAPII-CTD reveals unexpected connections to SRB10/CDK8. PLoS Genet. 2013;9:e1003758. doi: 10.1371/journal.pgen.1003758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Riles L, Shaw RJ, Johnston M, Reines D. Large-scale screening of yeast mutants for sensitivity to the IMP dehydrogenase inhibitor 6-azauracil. Yeast. 2004;21:241–248. doi: 10.1002/yea.1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Eick D, Geyer M. The RNA polymerase II carboxy-terminal domain (CTD) code. Chem Rev. 2013;113:8456–8490. doi: 10.1021/cr400071f. [DOI] [PubMed] [Google Scholar]
- 153.Heidemann M, Hintermair C, Voss K, Eick D. Dynamic phosphorylation patterns of RNA polymerase II CTD during transcription. Biochim Biophys Acta. 2013;1829:55–62. doi: 10.1016/j.bbagrm.2012.08.013. [DOI] [PubMed] [Google Scholar]
- 154.Buratowski S. Progression through the RNA polymerase II CTD cycle. Mol Cell. 2009;36:541–546. doi: 10.1016/j.molcel.2009.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Perales R, Bentley D. “Cotranscriptionality”: the transcription elongation complex as a nexus for nuclear transactions. Mol Cell. 2009;36:178–191. doi: 10.1016/j.molcel.2009.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Egloff S, Murphy S. Cracking the RNA polymerase II CTD code. Trends Genet. 2008;24:280–288. doi: 10.1016/j.tig.2008.03.008. [DOI] [PubMed] [Google Scholar]
- 157.Egloff S, Dienstbier M, Murphy S. Updating the RNA polymerase CTD code: adding gene-specific layers. Trends Genet. 2012;28:333–341. doi: 10.1016/j.tig.2012.03.007. [DOI] [PubMed] [Google Scholar]
- 158.Hsin JP, Manley JL. The RNA polymerase II CTD coordinates transcription and RNA processing. Genes Dev. 2012;26:2119–2137. doi: 10.1101/gad.200303.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Drogat J, Hermand D. Gene-specific requirement of RNA polymerase II CTD phosphorylation. Mol Microbiol. 2012;84:995–1004. doi: 10.1111/j.1365-2958.2012.08071.x. [DOI] [PubMed] [Google Scholar]
- 160.Bartkowiak B, Mackellar AL, Greenleaf AL. Updating the CTD story: from tail to epic. Genet Res Int. 2011;2011:623718. doi: 10.4061/2011/623718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Schwer B, Shuman S. Deciphering the RNA polymerase II CTD code in fission yeast. Mol Cell. 2011;43:311–318. doi: 10.1016/j.molcel.2011.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Zhang DW, Rodriguez-Molina JB, Tietjen JR, Nemec CM, Ansari AZ. Emerging views on the CTD code. Genet Res Int. 2012;2012:347214. doi: 10.1155/2012/347214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Jeronimo C, Bataille AR, Robert F. The writers, readers, and functions of the RNA polymerase II C-terminal domain code. Chem Rev. 2013;113:8491–8522. doi: 10.1021/cr4001397. [DOI] [PubMed] [Google Scholar]
- 164.Corden JL. RNA polymerase II C-terminal domain: tethering transcription to transcript and template. Chem Rev. 2013;113:8423–8455. doi: 10.1021/cr400158h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.West ML, Corden JL. Construction and analysis of yeast RNA polymerase II CTD deletion and substitution mutations. Genetics. 1995;140:1223–1233. doi: 10.1093/genetics/140.4.1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Nonet ML, Young RA. Intragenic and extragenic suppressors of mutations in the heptapeptide repeat domain of Saccharomyces cerevisiae RNA polymerase II. Genetics. 1989;123:715–724. doi: 10.1093/genetics/123.4.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Fong N, Bentley DL. Capping, splicing, and 3′ processing are independently stimulated by RNA polymerase II: different functions for different segments of the CTD. Genes Dev. 2001;15:1783–1795. doi: 10.1101/gad.889101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Stiller JW, Cook MS. Functional unit of the RNA polymerase II C-terminal domain lies within heptapeptide pairs. Eukaryot Cell. 2004;3:735–740. doi: 10.1128/EC.3.3.735-740.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Fabrega C, Shen V, Shuman S, Lima CD. Structure of an mRNA capping enzyme bound to the phosphorylated carboxy-terminal domain of RNA polymerase II. Mol Cell. 2003;11:1549–1561. doi: 10.1016/s1097-2765(03)00187-4. [DOI] [PubMed] [Google Scholar]
- 170.Heidemann M, Hintermair C, Voss K, Eick D. Dynamic phosphorylation pat terns of RNA polymerase II CTD during transcription. BBA-Gene Regulatory Mechanisms. 2013;1829:55–62. doi: 10.1016/j.bbagrm.2012.08.013. [DOI] [PubMed] [Google Scholar]
- 171.Kelly WG, Dahmus ME, Hart GW. RNA polymerase II is a glycoprotein. Modification of the COOH-terminal domain by O-GlcNAc. J Biol Chem. 1993;268:10416–10424. [PubMed] [Google Scholar]
- 172.Ranuncolo SM, Ghosh S, Hanover JA, Hart GW, Lewis BA. Evidence of the involvement of O-GlcNAc-modified human RNA polymerase II CTD in transcription in vitro and in vivo. J Biol Chem. 2012;287:23549–23561. doi: 10.1074/jbc.M111.330910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Sims RJ, 3rd, Rojas LA, Beck D, Bonasio R, Schuller R, Drury WJ, 3rd, Eick D, Reinberg D. The C-terminal domain of RNA polymerase II is modified by site-specific methylation. Science. 2011;332:99–103. doi: 10.1126/science.1202663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Stiller JW, McConaughy BL, Hall BD. Evolutionary complementation for polymerase II CTD function. Yeast. 2000;16:57–64. doi: 10.1002/(SICI)1097-0061(20000115)16:1<57::AID-YEA509>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 175.Karagiannis J. Decoding the informational properties of the RNA polymerase II carboxy terminal domain. BMC Res Notes. 2012;5:241. doi: 10.1186/1756-0500-5-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Komarnitsky P, Cho EJ, Buratowski S. Different phosphorylated forms of RNA po-lymerase II and associated mRNA processing factors during transcription. Genes Dev. 2000;14:2452–2460. doi: 10.1101/gad.824700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Schroeder SC, Schwer B, Shuman S, Bentley D. Dynamic association of capping enzymes with transcribing RNA polymerase II. Genes Dev. 2000;14:2435–2440. doi: 10.1101/gad.836300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.O'Brien T, Hardin S, Greenleaf A, Lis JT. Phosphorylation of RNA polymerase II C-terminal domain and transcriptional elongation. Nature. 1994;370:75–77. doi: 10.1038/370075a0. [DOI] [PubMed] [Google Scholar]
- 179.Lee JM, Greenleaf AL. A protein kinase that phosphorylates the C-terminal repeat domain of the largest subunit of RNA polymerase II. Proc Natl Acad Sci U S A. 1989;86:3624–3628. doi: 10.1073/pnas.86.10.3624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Lin PS, Marshall NF, Dahmus ME. CTD phosphatase: role in RNA polymerase II cycling and the regulation of transcript elongation. Prog Nucleic Acid Res Mol Biol. 2002;72:333–365. doi: 10.1016/s0079-6603(02)72074-6. [DOI] [PubMed] [Google Scholar]
- 181.Kim H, Erickson B, Luo W, Seward D, Graber JH, Pollock DD, Megee PC, Bentley DL. Gene-specific RNA polymerase II phosphorylation and the CTD code. Nat Struct Mol Biol. 2010;17:1279–1286. doi: 10.1038/nsmb.1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Tietjen JR, Zhang DW, Rodriguez-Molina JB, White BE, Akhtar MS, Heidemann M, Li X, Chapman RD, Shokat K, Keles S, Eick D, Ansari AZ. Chemical-genomic dissection of the CTD code. Nat Struct Mol Biol. 2010;17:1154–1161. doi: 10.1038/nsmb.1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Mayer A, Lidschreiber M, Siebert M, Leike K, Soding J, Cramer P. Uniform transitions of the general RNA polymerase II transcription complex. Nat Struct Mol Biol. 2010;17:1272–1278. doi: 10.1038/nsmb.1903. [DOI] [PubMed] [Google Scholar]
- 184.Bataille AR, Jeronimo C, Jacques PE, Laramée L, Fortin ME, Forest A, Bergeron M, Hanes SD, Robert F. A universal RNA polymerase II CTD cycle is orchestrated by complex interplays between kinase, phosphatase and isomerase enzymes along genes. Mol Cell. 2012;45:158–170. doi: 10.1016/j.molcel.2011.11.024. [DOI] [PubMed] [Google Scholar]
- 185.Hirose Y, Ohkuma Y. Phosphorylation of the C-terminal domain of RNA polymerase II plays central roles in the integrated events of eucaryotic gene expression. J Biochem. 2007;141:601–608. doi: 10.1093/jb/mvm090. [DOI] [PubMed] [Google Scholar]
- 186.Majello B, Napolitano G. Control of RNA polymerase II activity by dedicated CTD kinases and phosphatases. Front Biosci. 2001;6:D1358–D1368. doi: 10.2741/majello. [DOI] [PubMed] [Google Scholar]
- 187.Kang ME, Dahmus ME. RNA polymerases IIA and IIO have distinct roles during transcription from the TATA-less murine dihydrofolate reductase promoter. J Biol Chem. 1993;268:25033–25040. [PubMed] [Google Scholar]
- 188.Zehring WA, Lee JM, Weeks JR, Jokerst RS, Greenleaf AL. The C-terminal repeat domain of RNA polymerase II largest subunit is essential in vivo but is not required for accurate transcription initiation in vitro. Proc Natl Acad Sci U S A. 1988;85:3698–3702. doi: 10.1073/pnas.85.11.3698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Buratowski S, Sharp PA. Transcription initiation complexes and upstream activation with RNA polymerase II lacking the C-terminal domain of the largest subunit. Mol Cell Biol. 1990;10:5562–5564. doi: 10.1128/mcb.10.10.5562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Kim WY, Dahmus ME. The major late promoter of adenovirus-2 is accurately tran scribed by RNA polymerases IIO, IIA, and IIB. J Biol Chem. 1989;264:3169–3176. [PubMed] [Google Scholar]
- 191.Chapman RD, Heidemann M, Albert TK, Mailhammer R, Flatley A, Meisterernst M, Kremmer E, Eick D. Transcribing RNA polymerase II is phosphorylated at CTD residue serine-7. Science. 2007;318:1780–1782. doi: 10.1126/science.1145977. [DOI] [PubMed] [Google Scholar]
- 192.Steinmetz EJ, Conrad NK, Brow DA, Corden JL. RNA-binding protein Nrd1 directs poly(A)-independent 3′-end formation of RNA polymerase II transcripts. Nature. 2001;413:327–331. doi: 10.1038/35095090. [DOI] [PubMed] [Google Scholar]
- 193.Carroll KL, Ghirlando R, Ames JM, Corden JL. Interaction of yeast RNA-binding proteins Nrd1 and Nab3 with RNA polymerase II terminator elements. RNA. 2007;13:361–373. doi: 10.1261/rna.338407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Vasiljeva L, Buratowski S. Nrd1 interacts with the nuclear exosome for 3′ processing of RNA polymerase II transcripts. Mol Cell. 2006;21:239–248. doi: 10.1016/j.molcel.2005.11.028. [DOI] [PubMed] [Google Scholar]
- 195.Steinmetz EJ, Warren CL, Kuehner JN, Panbehi B, Ansari AZ, Brow DA. Genome-wide distribution of yeast RNA polymerase II and its control by Sen1 helicase. Mol Cell. 2006;24:735–746. doi: 10.1016/j.molcel.2006.10.023. [DOI] [PubMed] [Google Scholar]
- 196.Amrani N, Minet M, Wyers F, Dufour ME, Aggerbeck LP, Lacroute F. PCF11 encodes a third protein component of yeast cleavage and polyadenylation factor I. Mol Cell Biol. 1997;17:1102–1109. doi: 10.1128/mcb.17.3.1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Zhang Z, Fu J, Gilmour DS. CTD-dependent dismantling of the RNA polymerase II elongation complex by the pre-mRNA 3′-end processing factor, Pcf11. Genes Dev. 2005;19:1572–1580. doi: 10.1101/gad.1296305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Kim M, Vasiljeva L, Rando OJ, Zhelkovsky A, Moore C, Buratowski S. Distinct pathways for snoRNA and mRNA termination. Mol Cell. 2006;24:723–734. doi: 10.1016/j.molcel.2006.11.011. [DOI] [PubMed] [Google Scholar]
- 199.Licatalosi DD, Geiger G, Minet M, Schroeder S, Cilli K, McNeil JB, Bentley DL. Functional interaction of yeast pre-mRNA 3′ end processing factors with RNA polymerase II. Mol Cell. 2002;9:1101–1111. doi: 10.1016/s1097-2765(02)00518-x. [DOI] [PubMed] [Google Scholar]
- 200.Krishnamurthy S, He X, Reyes-Reyes M, Moore C, Hampsey M. Ssu72 Is an RNA polymerase II CTD phosphatase. Mol Cell. 2004;14:387–394. doi: 10.1016/s1097-2765(04)00235-7. [DOI] [PubMed] [Google Scholar]
- 201.Zhang DW, Mosley AL, Ramisetty SR, Rodriguez-Molina JB, Washburn MP, Ansari AZ. Ssu72 phosphatase-dependent erasure of phospho-Ser7 marks on the RNA polymerase II C-terminal domain is essential for viability and transcription termination. J Biol Chem. 2012;287:8541–8551. doi: 10.1074/jbc.M111.335687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Jasnovidova O, Stefl R. The CTD code of RNA polymerase II: a structural view. Wiley Interdiscip Rev RNA. 2013;4:1–16. doi: 10.1002/wrna.1138. [DOI] [PubMed] [Google Scholar]
- 203.Meinhart A, Kamenski T, Hoeppner S, Baumli S, Cramer P. A structural perspective of CTD function. Genes Dev. 2005;19:1401–1415. doi: 10.1101/gad.1318105. [DOI] [PubMed] [Google Scholar]
- 204.Ghosh A, Shuman S, Lima CD. Structural insights to how mammalian capping enzyme reads the CTD code. Mol Cell. 2011;43:299–310. doi: 10.1016/j.molcel.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Werner-Allen JW, Lee CJ, Liu P, Nicely NI, Wang S, Greenleaf AL, Zhou P. cis-Proline-mediated Ser(P)5 dephosphorylation by the RNA polymerase II C-terminal domain phosphatase Ssu72. J Biol Chem. 2011;286:5717–5726. doi: 10.1074/jbc.M110.197129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Xiang K, Nagaike T, Xiang S, Kilic T, Beh MM, Manley JL, Tong L. Crystal structure of the human symplekin-Ssu72-CTD phosphopeptide complex. Nature. 2010;467:729–733. doi: 10.1038/nature09391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Zhang Y, Kim Y, Genoud N, Gao J, Kelly JW, Pfaff SL, Gill GN, Dixon JE, Noel JP. Determinants for dephosphorylation of the RNA polymerase II C-terminal domain by Scp1. Mol Cell. 2006;24:759–770. doi: 10.1016/j.molcel.2006.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Xiang K, Manley JL, Tong L. An unexpected binding mode for a Pol II CTD peptide phosphorylated at Ser7 in the active site of the CTD phosphatase Ssu72. Genes Dev. 2012;26:2265–2270. doi: 10.1101/gad.198853.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Luo Y, Yogesha SD, Cannon JR, Yan W, Ellington AD, Brodbelt JS, Zhang Y. Novel modifications on C-terminal domain of RNA polymerase II can fine-tune the phosphatase activity of Ssu72. ACS Chem Biol. 2013;8:2042–2052. doi: 10.1021/cb400229c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Meinhart A, Cramer P. Recognition of RNA polymerase II carboxy-terminal domain by 3′-RNA-processing factors. Nature. 2004;430:223–226. doi: 10.1038/nature02679. [DOI] [PubMed] [Google Scholar]
- 211.Noble CG, Hollingworth D, Martin SR, Ennis-Adeniran V, Smerdon SJ, Kelly G, Taylor IA, Ramos A. Key features of the interaction between Pcf11 CID and RNA polymerase II CTD. Nat Struct Mol Biol. 2005;12:144–151. doi: 10.1038/nsmb887. [DOI] [PubMed] [Google Scholar]
- 212.Lunde BM, Reichow SL, Kim M, Suh H, Leeper TC, Yang F, Mutschler H, Buratowski S, Meinhart A, Varani G. Cooperative interaction of transcription termination factors with the RNA polymerase II C-terminal domain. Nat Struct Mol Biol. 2010;17:1195–1201. doi: 10.1038/nsmb.1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Kubicek K, Cerna H, Holub P, Pasulka J, Hrossova D, Loehr F, Hofr C, Vanacova S, Stefl R. Serine phosphorylation and proline isomerization in RNAP II CTD control recruitment of Nrd1. Genes Dev. 2012;26:1891–1896. doi: 10.1101/gad.192781.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Domingues MN, de Campos BM, de Oliveira ML, de Mello UQ, Benedetti CE. TAL effectors target the C-terminal domain of RNA polymerase II (CTD) by inhibiting the prolyl-isomerase activity of a CTD-associated cyclophilin. PLoS One. 2012;7:e41553. doi: 10.1371/journal.pone.0041553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Govind CK, Qiu H, Ginsburg DS, Ruan C, Hofmeyer K, Hu C, Swaminathan V, Workman JL, Li B, Hinnebusch AG. Phosphorylated Pol II CTD recruits multiple HDACs, including Rpd3C(S), for methylation-dependent deacetylation of ORF nucleosomes. Mol Cell. 2010;39:234–246. doi: 10.1016/j.molcel.2010.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Ng HH, Robert F, Young RA, Struhl K. Targeted recruitment of Set1 histone methylase by elongating Pol II provides a localized mark and memory of recent transcriptional activity. Mol Cell. 2003;11:709–719. doi: 10.1016/s1097-2765(03)00092-3. [DOI] [PubMed] [Google Scholar]
- 217.Smolle M, Workman JL. Transcription-associated histone modifications and cryptic transcription. BBA-Gene Regulatory Mechanisms. 2013;1829:84–97. doi: 10.1016/j.bbagrm.2012.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Arevalo-Rodriguez M, Cardenas ME, Wu X, Hanes SD, Heitman J. Cyclophilin A and Ess1 interact with and regulate silencing by the Sin3-Rpd3 histone deacetylase. EMBO J. 2000;19:3739–3749. doi: 10.1093/emboj/19.14.3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Wang SS, Zhou BO, Zhou JQ. Histone H3 lysine 4 hypermethylation prevents aberrant nucleosome remodeling at the PHO5 promoter. Mol Cell Biol. 2011;31:3171–3181. doi: 10.1128/MCB.05017-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Li J, Moazed D, Gygi SP. Association of the histone methyltransferase Set2 with RNA polymerase II plays a role in transcription elongation. J Biol Chem. 2002;277:49383–49388. doi: 10.1074/jbc.M209294200. [DOI] [PubMed] [Google Scholar]
- 221.Phatnani HP, Jones JC, Greenleaf AL. Expanding the functional repertoire of CTD kinase I and RNA polymerase II: novel phosphoCTD-associating proteins in the yeast proteome. Biochemistry. 2004;43:15702–15719. doi: 10.1021/bi048364h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Geymonat M, Spanos A, Wells GP, Smerdon SJ, Sedgwick SG. Clb6/Cdc28 and Cdc14 regulate phosphorylation status and cellular localization of Swi6. Mol Cell Biol. 2004;24:2277–2285. doi: 10.1128/MCB.24.6.2277-2285.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Taberner FJ, Quilis I, Igual JC. Spatial regulation of the start repressor Whi5. Cell Cycle. 2009;8:3010–3018. [PubMed] [Google Scholar]
- 224.Nakatsu Y, Sakoda H, Kushiyama A, Ono H, Fujishiro M, Horike N, Yoneda M, Ohno H, Tsuchiya Y, Kamata H, Tahara H, Isobe T, Nishimura F, Katagiri H, Oka Y, Fukushima T, Takahashi S, Kurihara H, Uchida T, Asano T. Pin1 associates with and induces translocation of CRTC2 to the cytosol, thereby suppressing cAMP-responsive element transcriptional activity. J Biol Chem. 2010;285:33018–33027. doi: 10.1074/jbc.M110.137836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Pandit S, Wang D, Fu XD. Functional integration of transcriptional and RNA processing machineries. Curr Opin Cell Biol. 2008;20:260–265. doi: 10.1016/j.ceb.2008.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Winsor TS, Bartkowiak B, Bennett CB, Greenleaf AL. A DNA damage response system associated with the phosphoCTD of elongating RNA polymerase II. PLoS One. 2013;8:e60909. doi: 10.1371/journal.pone.0060909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Steger M, Murina O, Huhn D, Ferretti LP, Walser R, Hanggi K, Lafranchi L, Neugebauer C, Paliwal S, Janscak P, Gerrits B, Del Sal G, Zerbe O, Sartori AA. Prolyl isomerase PIN1 regulates DNA double-strand break repair by counteracting DNA end resection. Mol Cell. 2013;50:333–343. doi: 10.1016/j.molcel.2013.03.023. [DOI] [PubMed] [Google Scholar]
- 228.Jung SJ, Kim HJ, Yang YJ, Seal JH, Jung BY, Han JW, Lee HW, Cho EJ. Role of RNA polymerase II carboxy terminal domain phosphorylation in DNA damage response. J Microbial. 2005;43:516–522. [PubMed] [Google Scholar]
- 229.Sartor AA, Steger M. Prolyl isomerization: a new PIN code for DSB repair. Cell Cycle. 2013;12:2717–2718. doi: 10.4161/cc.26077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Becker R, Loll B, Meinhart A. Snapshots of the RNA processing factor SCAF8 bound to different phosphorylated forms of the carboxyl-terminal domain of RNA polymerase II. J Biol Chem. 2008;283:22659–22669. doi: 10.1074/jbc.M803540200. [DOI] [PubMed] [Google Scholar]
- 231.Li M, Phatnani HP, Guan Z, Sage H, Greenleaf AL, Zhou P. Solution structure of the Set2-Rpb1 interacting domain of human Set2 and its interaction with the hyperphosphorylated C-terminal domain of Rpb1. Proc Natl Acad Sci U S A. 2005;102:17636–17641. doi: 10.1073/pnas.0506350102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Vojnic E, Simon B, Strahl BD, Sattler M, Cramer P. Structure and carboxyl-terminal domain (CTD) binding of the Set2 SRI domain that couples histone H3 Lys36 methylation to transcription. J Biol Chem. 2006;281:13–15. doi: 10.1074/jbc.C500423200. [DOI] [PubMed] [Google Scholar]


