Abstract
Saccharomyces cerevisiae shows high growth activity under low pH conditions and can be used for producing acidic chemicals such as organic acids as well as fuel ethanol. However, ethanol can also be a problematic by-product in the production of chemicals except for ethanol. We have reported that a stable low-ethanol production phenotype was achieved by disrupting 6 NADH-dependent alcohol dehydrogenase genes of S. cerevisiae. Moreover, the genes encoding the NADH-dependent glycerol biosynthesis enzymes were further disrupted because the ADH-disrupted recombinant strain showed high glycerol production to maintain intracellular redox balance. The recombinant strain incapable producing ethanol and glycerol could have the potential to be a host for producing metabolite(s) whose biosynthesis is coupled with NADH oxidation. Indeed, we successfully achieved almost 100% yield for L-lactate production using this recombinant strain as a host. In addition, the potential of our constructed recombinant strain for efficient bioproduction, particularly under anaerobic conditions, is also discussed.
Keywords: alcohol dehydrogenase, bioproduction, ethanol, glycerol, L-lactate, Saccharomyces cerevisiae
Introduction
The yeast Saccharomyces cerevisiae has been widely used in the production of alcoholic beverages and the fuel ethanol because of its high ethanol production ability. Moreover, since S. cerevisiae exhibits high growth activity under low pH conditions, it has been utilized for the production of acidic chemicals such as lactate and succinate. When producing useful chemicals, the production of ethanol as a by-product by S. cerevisiae can be problematic. Indeed, metabolic engineering approaches of S. cerevisiae have been reported for reducing ethanol production and enhancing the productivity of target metabolite(s).1-4
In cellular metabolism, redox balance (i.e., intracellular balance between NAD(P)H and NAD(P)+) is maintained. Under aerobic conditions, NADH produced via glycolysis and the TCA cycle is oxidized in the respiratory chain. On the other hand, under anaerobic conditions, NADH produced via glycolysis is oxidized in fermentation pathways, such as ethanol and lactate production pathways. For S. cerevisiae, NADH produced via glyceraldehyde-3-phosphate dehydrogenase is mainly oxidized via the ethanol biosynthesis catalyzing alcohol dehydrogenases (ADHs).5 In addition, other metabolic reactions such as glycerol biosynthesis pathways are also involved in NADH oxidation. Since the intracellular redox imbalance can be problematic for bioproduction, it is very important to consider the intracellular redox balance in metabolic engineering.
Recently, we examined the characteristics of an ADH1-disrupted S. cerevisiae strain in continuous culture under anaerobic conditions.6 However, complete loss of ethanol production ability in this recombinant strain could not be achieved, and glycerol production was enhanced under anaerobic conditions. Therefore, all NADH-dependent ethanol biosynthesis pathways and glycerol biosynthesis pathways were disrupted in S. cerevisiae.7 The utility of this S. cerevisiae recombinant strain that was incapable of both ethanol and glycerol biosynthesis for bioproduction under anaerobic conditions was further examined.
In this paper, maintenance of the intracellular redox balance between NADH and NADPH in S. cerevisiae is summarized. In addition, characteristics of S. cerevisiae recombinant strains that do not produce ethanol and glycerol and the potential of such recombinant strains for bioproduction under anaerobic conditions are discussed.
Maintenance of Intracellular Redox Balance in S. cerevisiae
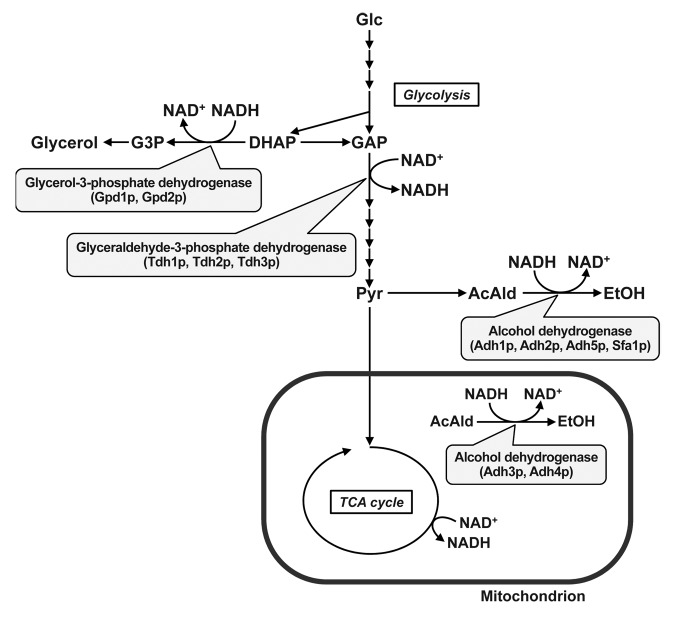
In metabolic networks, many oxidation and reduction reactions are included. For oxidation and reduction reactions, reducing powers such as NADH and NADPH and the reduced forms NAD+ and NADP+, respectively, are used, and the intracellular balances for such reducing powers (i.e., NADH/NAD+ and NADPH/NADP+ ratios) have to be maintained. In most microorganisms, NADH is produced in the glycolysis and the TCA cycle and is oxidized to NAD+ by the respiratory chain under aerobic conditions. For S. cerevisiae, NADH produced in glycolysis is dominantly oxidized by ethanol fermentation pathways catalyzed by NADH-dependent ADHs to maintain the redox balance (Fig. 1).5 Metabolic flux analysis revealed that 74% of carbons incorporated in the cells as glucose flowed to ethanol biosynthesis.8 S. cerevisiae possesses 6 NADH-dependent ADHs encoding ADH1, ADH2, ADH3, ADH4, ADH5 and SFA1.9 Among these, Adh1p is a 150-kDa protein and is the major ADH for ethanol production. Adh2p is thought to be utilized for ethanol assimilation. Adh1p, Adh2p and Adh5p are localized to the cytoplasm, and Adh3p and Adh4p are localized to mitochondria. The Sfa1p protein is annotated as a bifunctional enzyme ADH/S-(hydroxymethyl)glutathione dehydrogenase.
Figure 1. Central carbon metabolism of S. cerevisiae. The oxidation and reduction of NADH in glycolysis and ethanol and glycerol biosynthesis reactions are shown. Glc, glucose; GAP, glyceraldehyde-3-phosphate; DHAP, dihydroxyacetone phosphate; Pyr, pyruvate; AcAld, acetaldehyde; G3P, glycerol-3-phosphate.
In S. cerevisiae, the metabolic flux of glyceraldehyde-3-phosphate dehydrogenase in glycolysis, wherein NAD+ is reduced, is larger than that of ADHs coupled with NADH oxidation, because of the high carbon requirement for anabolic metabolism (i.e., biomass production) such as amino acid and fatty acid biosynthesis. Therefore, other NADH oxidation reactions are necessary to maintain the NADH/NAD+ balance. It has been reported that glycerol production was enhanced by disrupting the ADH1 gene.1 In addition, disrupting glycerol biosynthesis reactions enhanced ethanol production.10 Therefore, glycerol biosynthesis reactions also contribute to maintaining the NADH/NAD+ balance. In glycerol biosynthesis reactions, glycerol-3-phosphate dehydrogenases encoding GPD1 and GPD2 convert dihydroxyacetone phosphate to glycerol-3-phosphate and require NADH as a reducing power.11 Metabolic flux analysis revealed that 8.5% of incorporated carbons flowed into glycerol production.8
Other NADH oxidation mechanisms include respiration of cytosolic NADH by external mitochondrial NADH dehydrogenase and the glycerol-3-phosphate shunt and NADH oxidation by mitochondrial NADH dehydrogenase in the respiratory chain.5 Moreover, shuttle mechanisms such as the malate-oxaloacetate shuttle, malate-aspartate shuttle and malate-pyruvate shuttle have also been proposed.5
Characteristics of S. cerevisiae Recombinant Strains Incapable of Ethanol Biosynthesis under Anaerobic Conditions
As described above, since ethanol becomes a by-product in bioproduction processes other than ethanol production, reduction of ethanol production is important for such bioproduction, particularly anaerobic bioproduction. To that end, we analyzed the ADH1-disrupted strain of S. cerevisiae and examined the culture characteristics of the constructed strain.6 As expected, ethanol yield in the ADH1-disrupted strain was lower than that in the parental strain. Moreover, glycerol yield in the ADH1-disrupted strain was higher than that in the parent strain, suggesting that oxidation of NADH produced by glycolysis was oxidized by glycerol biosynthesis reactions, as a substitute to ethanol biosynthesis pathways.
It is noteworthy that the ADH1-disrupted strain showed unstable phenotypes in continuous culture under oxygen-limited conditions.6 The ethanol production levels became low at the initial steady-state by 100 h. However, ethanol production levels gradually increased after reaching the initial steady-state. The ethanol production yield was increased, while the glycerol production yield was decreased, indicating that NADH produced via glycolysis was oxidized by the remaining ethanol biosynthesis reactions in the ADH1-disrupted strain under oxygen-limited conditions rather than by glycerol biosynthesis reactions. Moreover, expression of the genes encoding ADH isozymes in the ADH1-disrupted strain, particularly ADH2 and ADH4, were upregulated after long-term continuous culture, probably to compensate for the requirement of oxidation of NADH produced via glycolysis. Therefore, the recombinant strain in which the 6 NADH-dependent ADH isozyme genes (i.e., ADH1, ADH2, ADH3, ADH4, ADH5 and SFA1) were disrupted was constructed and examined under continuous culture. Ethanol production levels became low throughout the cultivation, but glycerol production levels became high, suggesting that glycerol biosynthesis reactions were utilized for NADH oxidation in the recombinant strain in which the 6 NADH-dependent ADH genes were disrupted. The low ethanol production phenotype is stably shown when all the NADH-dependent ADH genes in S. cerevisiae are disrupted.
L-lactate production by the S. cerevisiae Recombinant Strain Incapable of Ethanol and Glycerol Biosynthesis under Anaerobic Conditions
Ethanol production decreased clearly and stably following disruption of all the NADH-dependent ADH genes, but glycerol production was increased to oxidize NADH produced via glycolysis under oxygen-limited conditions. Since glycerol is considered a by-product for producing the target product, reduction of glycerol production was also required. Therefore, the GPD1 and GPD2 genes encoding NADH-dependent glycerol-3-phosphate dehydrogenases were disrupted further.6 In addition, the PDC1 gene encoding the major pyruvate dehydrogenase that converts pyruvate to acetaldehyde, which is a substrate for ADHs, was also disrupted to ensure the reduction of ethanol production. As a result, the recombinant strain incapable of ethanol and glycerol biosynthesis was constructed.
Since the recombinant strain incapable of ethanol and glycerol biosynthesis does not possess the 2 major NADH oxidation pathways, it can be expected that production of the target product whose biosynthesis is coupled with NADH oxidation can be achieved by a metabolic engineering approach. We introduced the gene for human L-lactate dehydrogenase (LDH) into the recombinant strain incapable of ethanol and glycerol biosynthesis and examined L-lactate production under anaerobic conditions, which was achieved by N2 gas flow into the jar bioreactor.7 LDH converts 1 mol pyruvate to 1 mol L-lactate; this conversion is coupled with oxidation of 1 mol NADH. If the theoretical maximum yield is achieved, 2 mol L-lactate is produced from 1 mol glucose, and 2 mol NADH produced by the glyceraldehyde-3-phosphate dehydrogenase reaction are oxidized, which is coupled with the LDH reaction, to maintain redox balance. In addition, since the Michaelis-Menten constant for human LDH is lower than those of some lactic acid bacteria,12-14 we selected human LDH for constructing L-lactate-producing recombinant strain.
The recombinant strain incapable of both ethanol and glycerol biosynthesis could not grow under anaerobic conditions probably due to the NADH accumulation. Therefore, cells of the recombinant strain cultivated aerobically were suspended in fresh medium at a high density and packed in the jar bioreactor. Then, L-lactate production was examined by incubating the packed cells with gentle agitation and a N2 gas purge (Fig. 2). Although no cell growth was observed as expected, glucose was consumed and L-lactate was produced. Moreover, almost all glucose incorporated into the cell was efficiently converted to L-lactate; i.e., the mol-based L-lactate yield was around 200%. The efficient L-lactate production process under anaerobic conditions could be established by using the S. cerevisiae recombinant strain incapable of ethanol and glycerol biosynthesis.
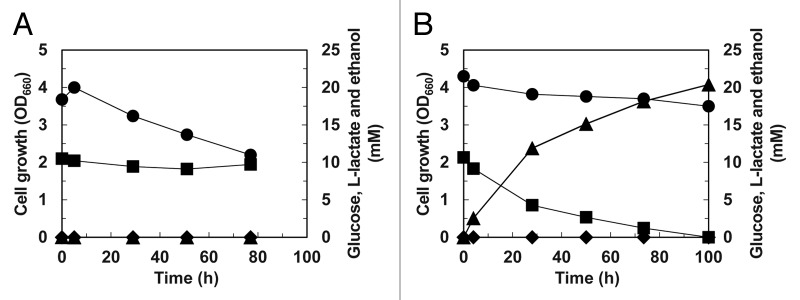
Figure 2. L-Lactate production by the human LDH-carrying S. cerevisiae recombinant strain with the 6 NADH-dependent ADH isozyme genes. Cells of the recombinant strains carrying empty plasmid (A) and the human-LDH-expression plasmid (B) were incubated in the jar bioreactor with a N2 gas purge and slow agitation. Cell growth (OD660; circles) and concentrations of glucose (squares), ethanol (diamonds) and L-lactate (triangles) are shown.
Potential of the S. cerevisiae Recombinant Strain Incapable of Ethanol and Glycerol Biosynthesis
The S. cerevisiae recombinant strain incapable of ethanol and glycerol biosynthesis could be used for anaerobic bioproduction. In particular, the metabolite(s) whose biosynthesis is coupled with NADH oxidation such as L-lactate could be suitable targets for anaerobic bioproduction using this recombinant strain.
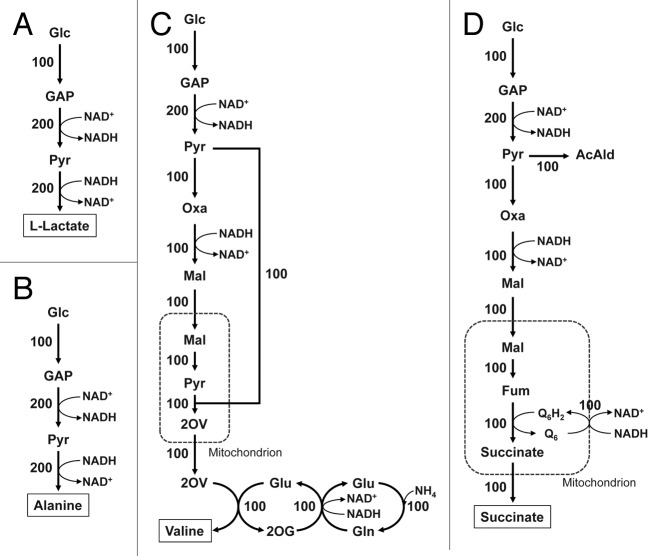
Recently, in silico metabolic simulation using genome-scale models have been utilized in metabolic engineering.15-17 In this simulation, metabolic flux distribution at a steady-state can be simulated as the biomass formation is maximized. This simulation platform can predict metabolic state changes against culture environments and gene knockouts for improvement of target metabolite production. Therefore, in silico metabolic simulation using a genome-scale metabolic model of S. cerevisiae iND75018 was applied to search for target metabolites for anaerobic bioproduction by the recombinant strain incapable of ethanol and glycerol biosynthesis. The feasibility of production of selected metabolite(s) whose biosynthesis is alternative to that of ethanol and glycerol in terms of NADH oxidation under anaerobic conditions can be surveyed by our in silico simulation studies. As a result, we found 20 native S. cerevisiae metabolites whose biosynthesis reactions involved oxidization of NADH as an alternative to ethanol and glycerol biosynthesis reactions based on the simulation: alanine, asparagine, aspartate, cysteine, D-glucosamine-6-phosphate, D-sorbitol, dTTP, fumarate, isoleucine, malate, methionine, ornithine, (R)-pantothenate, phenylalanine, proline, succinate, threonine, tryptophan, tyrosine and valine.7 We further simulated whether each metabolite could be produced under anaerobic conditions, and the only the solution where the target could be produced without biomass formation was feasible. In this simulation, the fluxes for biosynthesis of the identified 20 metabolites, except for the target metabolite, as well as ethanol and glycerol biosynthesis were set to zero in advance. Among the 20 metabolites, alanine, valine and succinate were produced without biomass formation under anaerobic conditions; feasible solutions on the production of target metabolites without biomass formation could be estimated. Figure 3 shows the simulated flux distributions when producing alanine, valine and succinate as well as L-lactate, whose production requires heterologous metabolic pathway, under completely anaerobic conditions. In all cases, glucose incorporated into the cells was completely converted to pyruvate, and the converted pyruvate was further completely converted to the target metabolites. Moreover, there was no biomass formation; i.e., achieving the theoretical maximum yield and maintenance of intracellular redox balance was simulated.
Figure 3. Simulated metabolic flux distributions during production of L-lactate (A), alanine (B), valine (C) and succinate (D) under anaerobic conditions in the recombinant strain incapable of both ethanol and glycerol biosynthesis. Metabolic simulation was performed using a genome-scale metabolic model of S. cerevisiae iND750 based on the concept of flux balance analysis; glucose and oxygen uptake rates assumed 1 and 0 mmol g dry cell−1 h−1, respectively, and the metabolic flux distributions at the metabolic steady-state were estimated as the biomass formation was maximized using linear programming. In these simulations, the fluxes for biosynthesis of the identified 20 metabolites, except for the target metabolite, as well as ethanol and glycerol biosynthesis were set to zero in advance. For simulation of L-lactate production (A), the heterologous reaction corresponding to LDH was added. In all cases, no biomass formation was achieved. The metabolic flux distributions normalized by the glucose uptake rate as 100 are shown. Glc, glucose; GAP, glyceraldehyde-3-phosphate; Pyr, pyruvate; Oxa, oxaloacetate; Mal, malate, Fum, fumarate; 2OV, 2-oxoisovalerate; AcAld, acetoaldehyde; Q6H2, ubiquinone (reduced form); Q6, ubiquinone (oxidized form).
Based on the concepts of synthetic biology, introduction of heterologous metabolic pathway(s) into the host species is effective for producing non-native metabolite(s).19,20 If the biosynthesis pathways for the target metabolite involve NADH-coupled reactions, utilization of our S. cerevisiae recombinant strain with elimination of both ethanol and glycerol biosynthesis as a host would be effective as shown above for the case of L-lactate production (Fig. 2). We recently established a platform to design heterologous metabolic pathways to produce metabolite(s) that cannot be produced by the host.21 By using our simulation platform, the design of the metabolic pathways and bioproduction are facilitated. Moreover, similar to the L-lactate production described above, by constructing the recombinant strain producing the target metabolite(s) based on the strain incapable of ethanol and glycerol biosynthesis, efficient anaerobic production processes with nearly the theoretical maximum yield might be able to be established. However, further studies are required.
We constructed the S. cerevisiae recombinant strain incapable of both ethanol and glycerol biosynthesis for bioproduction. As shown in our report on L-lactate production, high yields of target product can be achieved using this recombinant strain as a host. However, glucose consumption and target product production rates were very low. Moreover, the cell growth rate under aerobic conditions was also low in comparison with the parent strain. In our recombinant strain, glycerol biosynthesis is impaired due to the disruption of GPD1 and GPD2 genes. In the GPD1 and GPD2 knockout strain of S. cerevisiae, one of the important metabolites for phospholipid biosynthesis, glycerol-3-phoshate, which is a precursor for phosphatidic acid biosynthesis, cannot be produced from dihydroxyacetone phosphate directly. However, S. cerevisiae possess the bypass reactions for glycerol-3-phoshate biosynthesis; glycerol-3-phoshate is produced from 1-acyl-dihydroxyacetone phosphate which is produced by acylation of dihydroxyacetone phosphate.22-25 Thus, the GPD1 and GPD2 knockout strain is viable without nutrient supplementation. Additionally, the PDC1 gene encoding pyruvate decarboxylase involved in acetaldehyde biosynthesis, which is necessary for cytosolic acetyl-CoA biosynthesis, is also disrupted in our recombinant strain. However, since S. cerevisiae possesses 3 genes encoding pyruvate decarboxylase isozymes (PDC1, PDC5 and PDC6),26-28 our recombinant strain can produce acetaldehyde by the Pdc5p and Pdc6p proteins. Therefore, the reason why our recombinant strain shows low cell growth, glucose consumption and target product production rates of our recombinant strain is caused by disruption of the major metabolic reactions involved in NADH oxidation (i.e., ethanol and glycerol biosynthesis pathways) rather than the insufficiency of nutrients.
For efficient bioproduction, it is necessary to improve these parameters for the recombinant strain. We will continue to study our constructed recombinant strain for efficient bioproduction by S. cerevisiae.
Disclosure of Potential Conflicts of Interest
No potential conflict of interest was disclosed.
References
- 1.Cordier H, Mendes F, Vasconcelos I, François JM. A metabolic and genomic study of engineered Saccharomyces cerevisiae strains for high glycerol production. Metab Eng. 2007;9:364–78. doi: 10.1016/j.ymben.2007.03.002. [DOI] [PubMed] [Google Scholar]
- 2.van Maris AJ, Winkler AA, Porro D, van Dijken JP, Pronk JT. Homofermentative lactate production cannot sustain anaerobic growth of engineered Saccharomyces cerevisiae: possible consequence of energy-dependent lactate export. Appl Environ Microbiol. 2004;70:2898–905. doi: 10.1128/AEM.70.5.2898-2905.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tokuhiro K, Ishida N, Nagamori E, Saitoh S, Onishi T, Kondo A, Takahashi H. Double mutation of the PDC1 and ADH1 genes improves lactate production in the yeast Saccharomyces cerevisiae expressing the bovine lactate dehydrogenase gene. Appl Microbiol Biotechnol. 2009;82:883–90. doi: 10.1007/s00253-008-1831-5. [DOI] [PubMed] [Google Scholar]
- 4.Okano K, Tanaka T, Ogino C, Fukuda H, Kondo A. Biotechnological production of enantiomeric pure lactic acid from renewable resources: recent achievements, perspectives, and limits. Appl Microbiol Biotechnol. 2010;85:413–23. doi: 10.1007/s00253-009-2280-5. [DOI] [PubMed] [Google Scholar]
- 5.Bakker BM, Overkamp KM, Kötter P, Luttik MA, Pronk JT, van Dijken JP, et al. van Maris AJ. van Dijken JP Stoichiometry and compartmentation of NADH metabolism in Saccharomyces cerevisiae. FEMS Microbiol Rev. 2001;25:15–37. doi: 10.1111/j.1574-6976.2001.tb00570.x. [DOI] [PubMed] [Google Scholar]
- 6.Ida Y, Furusawa C, Hirasawa T, Shimizu H. Stable disruption of ethanol production by deletion of the genes encoding alcohol dehydrogenase isozymes in Saccharomyces cerevisiae. J Biosci Bioeng. 2012;113:192–5. doi: 10.1016/j.jbiosc.2011.09.019. [DOI] [PubMed] [Google Scholar]
- 7.Ida Y, Hirasawa T, Furusawa C, Shimizu H. Utilization of Saccharomyces cerevisiae recombinant strain incapable of both ethanol and glycerol biosynthesis for anaerobic bioproduction. Appl Microbiol Biotechnol. 2013;97:4811–9. doi: 10.1007/s00253-013-4760-x. [DOI] [PubMed] [Google Scholar]
- 8.Jouhten P, Rintala E, Huuskonen A, Tamminen A, Toivari M, Wiebe M, Ruohonen L, Penttilä M, Maaheimo H. Oxygen dependence of metabolic fluxes and energy generation of Saccharomyces cerevisiae CEN.PK113-1A. BMC Syst Biol. 2008;2:60. doi: 10.1186/1752-0509-2-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Smidt O, du Preez JC, Albertyn J. The alcohol dehydrogenases of Saccharomyces cerevisiae: a comprehensive review. FEMS Yeast Res. 2008;8:967–78. doi: 10.1111/j.1567-1364.2008.00387.x. [DOI] [PubMed] [Google Scholar]
- 10.Guadalupe Medina V, Almering MJ, van Maris AJ, Pronk JT. Elimination of glycerol production in anaerobic cultures of a Saccharomyces cerevisiae strain engineered to use acetic acid as an electron acceptor. Appl Environ Microbiol. 2010;76:190–5. doi: 10.1128/AEM.01772-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ansell R, Granath K, Hohmann S, Thevelein JM, Adler L. The two isoenzymes for yeast NAD+-dependent glycerol 3-phosphate dehydrogenase encoded by GPD1 and GPD2 have distinct roles in osmoadaptation and redox regulation. EMBO J. 1997;16:2179–87. doi: 10.1093/emboj/16.9.2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pettit SM, Nealon DA, Henderson AR. Purification of lactate dehydrogenase isoenzyme-5 from human liver. Clin Chem. 1981;27:88–93. [PubMed] [Google Scholar]
- 13.Garvie EI. Bacterial lactate dehydrogenases. Microbiol Rev. 1980;44:106–39. doi: 10.1128/mr.44.1.106-139.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.LeVan KM, Goldberg E. Properties of human testis-specific lactate dehydrogenase expressed from Escherichia coli. Biochem J. 1991;273:587–92. doi: 10.1042/bj2730587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blazeck J, Alper H. Systems metabolic engineering: genome-scale models and beyond. Biotechnol J. 2010;5:647–59. doi: 10.1002/biot.200900247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Oberhardt MA, Palsson BO, Papin JA. Applications of genome-scale metabolic reconstructions. Mol Syst Biol. 2009;5:320. doi: 10.1038/msb.2009.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Park JM, Kim TY, Lee SY. Constraints-based genome-scale metabolic simulation for systems metabolic engineering. Biotechnol Adv. 2009;27:979–88. doi: 10.1016/j.biotechadv.2009.05.019. [DOI] [PubMed] [Google Scholar]
- 18.Duarte NC, Herrgård MJ, Palsson BO. Reconstruction and validation of Saccharomyces cerevisiae iND750, a fully compartmentalized genome-scale metabolic model. Genome Res. 2004;14:1298–309. doi: 10.1101/gr.2250904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee JW, Na D, Park JM, Lee J, Choi S, Lee SY. Systems metabolic engineering of microorganisms for natural and non-natural chemicals. Nat Chem Biol. 2012;8:536–46. doi: 10.1038/nchembio.970. [DOI] [PubMed] [Google Scholar]
- 20.Yadav VG, De Mey M, Lim CG, Ajikumar PK, Stephanopoulos G. The future of metabolic engineering and synthetic biology: towards a systematic practice. Metab Eng. 2012;14:233–41. doi: 10.1016/j.ymben.2012.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chatsurachai S, Furusawa C, Shimizu H. ArtPathDesign: Rational heterologous pathway design system for the production of nonnative metabolites. J Biosci Bioeng. 2013;116:524–7. doi: 10.1016/j.jbiosc.2013.04.002. [DOI] [PubMed] [Google Scholar]
- 22.Athenstaedt K, Weys S, Paltauf F, Daum G. Redundant systems of phosphatidic acid biosynthesis via acylation of glycerol-3-phosphate or dihydroxyacetone phosphate in the yeast Saccharomyces cerevisiae. J Bacteriol. 1999;181:1458–63. doi: 10.1128/jb.181.5.1458-1463.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Racenis PV, Lai JL, Das AK, Mullick PC, Hajra AK, Greenberg ML. The acyl dihydroxyacetone phosphate pathway enzymes for glycerolipid biosynthesis are present in the yeast Saccharomyces cerevisiae. J Bacteriol. 1992;174:5702–10. doi: 10.1128/jb.174.17.5702-5710.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tillman TS, Bell RM. Mutants of Saccharomyces cerevisiae defective in sn-glycerol-3-phosphate acyltransferase. Simultaneous loss of dihydroxyacetone phosphate acyltransferase indicates a common gene. J Biol Chem. 1986;261:9144–9. [PubMed] [Google Scholar]
- 25.Zheng Z, Zou J. The initial step of the glycerolipid pathway: identification of glycerol 3-phosphate/dihydroxyacetone phosphate dual substrate acyltransferases in Saccharomyces cerevisiae. J Biol Chem. 2001;276:41710–6. doi: 10.1074/jbc.M104749200. [DOI] [PubMed] [Google Scholar]
- 26.Seeboth PG, Bohnsack K, Hollenberg CP. pdc1(0) mutants of Saccharomyces cerevisiae give evidence for an additional structural PDC gene: cloning of PDC5, a gene homologous to PDC1. J Bacteriol. 1990;172:678–85. doi: 10.1128/jb.172.2.678-685.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kellermann E, Seeboth PG, Hollenberg CP. Analysis of the primary structure and promoter function of a pyruvate decarboxylase gene (PDC1) from Saccharomyces cerevisiae. Nucleic Acids Res. 1986;14:8963–77. doi: 10.1093/nar/14.22.8963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hohmann S. Characterization of PDC6, a third structural gene for pyruvate decarboxylase in Saccharomyces cerevisiae. J Bacteriol. 1991;173:7963–9. doi: 10.1128/jb.173.24.7963-7969.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]