Abstract
Cells respond to genotoxic stress by activating the DNA damage response (DDR). When injury is severe or irreparable, cells induce apoptosis or cellular senescence to prevent transmission of the lesions to the daughter cells upon cell division. Resistance to apoptosis is a hallmark of cancer that challenges the efficacy of cancer therapy. In this work, the effects of ionizing radiation on apoptosis-resistant E1A + E1B transformed cells were investigated to ascertain whether the activation of cellular senescence could provide an alternative tumor suppressor mechanism. We show that irradiated cells arrest cell cycle at G2/M phase and resume DNA replication in the absence of cell division followed by formation of giant polyploid cells. Permanent activation of DDR signaling due to impaired DNA repair results in the induction of cellular senescence in E1A + E1B cells. However, irradiated cells bypass senescence and restore the population by dividing cells, which have near normal size and ploidy and do not express senescence markers. Reversion of senescence and appearance of proliferating cells were associated with downregulation of mTOR, activation of autophagy, mitigation of DDR signaling, and expression of stem cell markers.
Keywords: apoptosis resistance, DNA damage response, DNA repair, polyploidy, mTOR, autophagy, stem cells markers, senescence reversion
Cellular senescence is a tumor suppressor program that is activated in response to various stimuli, including DNA damage, chromatin reorganization, and elevated oncogene signaling.1-7 Senescent cells are characterized by arrest of proliferation while maintaining metabolic activity and viability. They display a number of features including cell hypertrophy and flattening,8 expression of senescence-associated β-galactosidase (SA-β-Gal),9 activation of negative cell cycle regulators,2,10 development of senescence-associated secretory phenotype (SASP),11,12 and chromatin reorganization13 including senescence-associated heterochromatic foci (SAHF)14 and DNA segments with chromatin alterations reinforcing senescence (DNA-SCARS).15 DNA-SCARS represent persistent foci that contain DNA damage response factors (DDR foci) such as phosphorylated histone H2AXSer139 (termed γH2AX), p53-binding protein (53BP1), ataxia-telangiectasia mutated (ATM), and Rad3-related (ATR) kinases,15 as well as some others.
Mammalian target of rapamycin (mTOR) is a member of the phosphoinositide-3-kinase-related kinases (PIKK) family, which integrates multiple signaling pathways and serves as a central regulator of cellular senescence. mTOR forms 2 distinct complexes, mTORC1 and mTORC2,16,17 that negatively regulate autophagy.18-20 Autophagy is an evolutionarily conserved mechanism that provides cell survival in response to a variety of stresses, including exposure to IR. Activation of autophagy is required for development and maintenance of senescent phenotype.18
Ionizing radiation (IR) is among the factors that induce cellular senescence. Exposure to IR generates various DNA lesions, among which DNA double-strand breaks (DSBs) are the most harmful, as they can lead to mutations, genomic instability, and apoptosis when unrepaired. Irradiated cells initiate a complex of events resulting in the activation of DDR, checkpoint controls, and DNA repair. The initial steps of DDR include activation of PIKK family kinases ATM, ATR, and DNA-PK followed by phosphorylation and activation of multiple downstream targets, among which are histone H2AX and 53BP1.21-27 Two major mechanisms of DSBs repair in mammals are homologous recombination (HR) and non-homologous end joining (NHEJ).24 When DNA lesions are severe or irreparable, the DDR signaling remains activated, leading to apoptosis or cellular senescence.1,11,28-31
Tumor cells often acquire resistance to apoptosis that results in the selection of the most malignant cells.32 However, apoptosis-resistant cells retain the ability to undergo cellular senescence.33 Although senescence is canonically defined as a terminal arrest of cell division, recent works demonstrate that various types of senescence can be reversed.34-37
This work aimed to study the effects of IR on apoptosis-resistant E1A + E1B-transformed cells with special emphasis on determining whether an alternative to apoptosis tumor suppressor program, such as cellular senescence, can be activated. We revealed that in response to IR, E1A + E1B cells undergo G2/M cell cycle arrest followed by restart of DNA replication, which culminates in the formation of polyploid giant mono- and multinuclear cells. Irradiated E1A + E1B cells demonstrate a delayed DNA repair that leads to a sustained activation of DDR signaling and results in the induction of reversible cellular senescence. Finally, we show that the giant polyploid cells were eventually replaced by a population of proliferating cells that did not express SA-β-Gal. Reversion of IR-induced senescence in E1A + E1B cells was associated with suppression of mTOR activity, induction of autophagy, mitigation of DDR signaling, and expression of stem-cell markers Nanog and Oct3/4.
Results
Irradiated E1A + E1B cells arrest cell cycle progression in G2/M phase and resume DNA replication without cell division resulting in the formation of giant polyploid cells
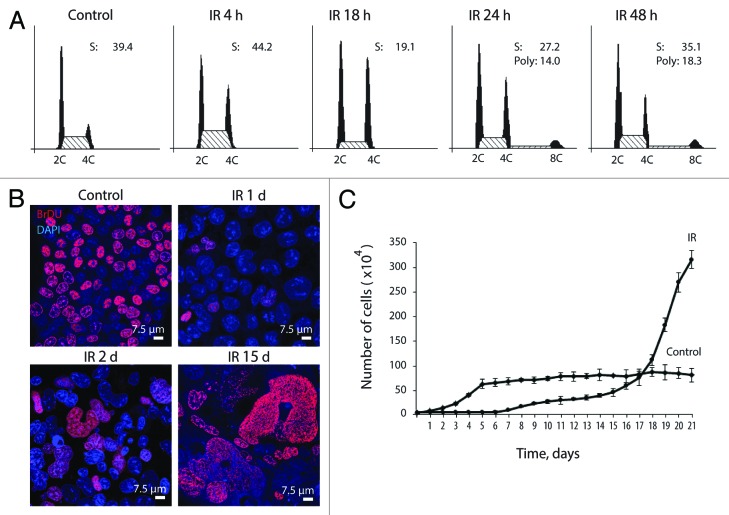
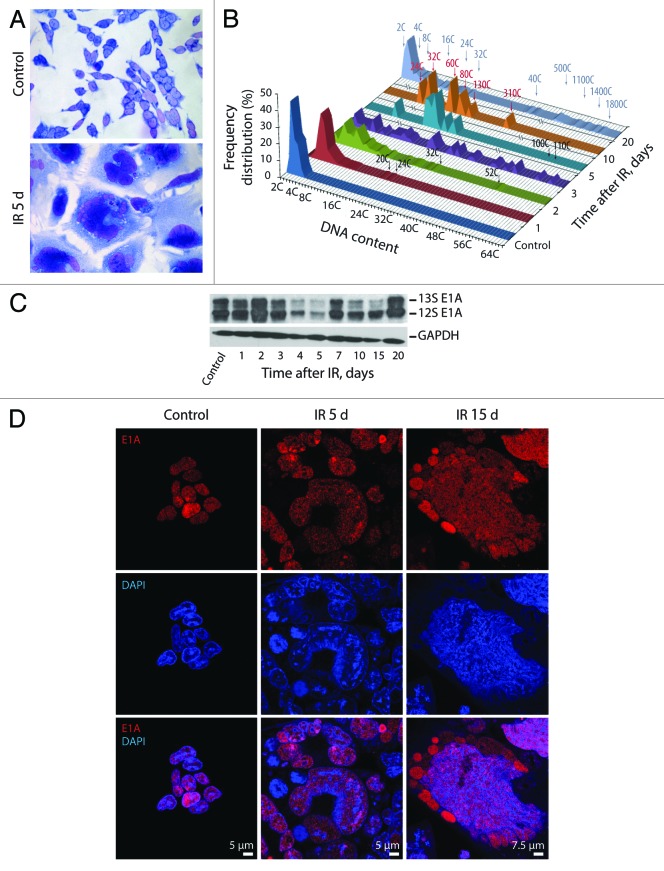
Irreversible arrest of cell cycle progression and proliferation is a hallmark of cellular senescence. To evaluate antiproliferative effect of IR on apoptosis-resistant cells, the ability of cells to arrest cell cycle progression, DNA replication, and proliferation was analyzed. The experimental data demonstrate that E1A + E1B cells undergo the G2/M cell cycle arrest followed by restart of DNA replication 24 h after irradiation that leads to the accumulation of polyploid cells (Fig. 1A). BrdU incorporation assay shows that DNA replication in E1A + E1B cells decreased dramatically 1 d post-exposure to IR but resumed already on the second day after irradiation and remained active in the following days (Fig. 1B). At the same time, the proliferation of irradiated cells was completely suppressed until day 7 post-exposure to IR (Fig. 1C). Importantly, replication of DNA in proliferation-arrested cells resulted in the formation of giant multi- and mononuclear cells, which often contained micronuclei (Fig. 2A). We analyzed the ploidy of giant cells by mean of Feulgen DNA staining with the subsequent DNA cytometry. Cells with DNA content over 16C were revealed already on the first and second days after exposure to IR, while 3 d after irradiation, over 60% of cells in the population reached a highly polyploid state, with the DNA content up to 60C (Fig. 2B). Moreover, giant cells continued DNA replication in the following days and reached the ploidy over 1500C (Fig. 2B), demonstrating loss of control on the coordination of DNA replication and cell division.
Figure 1. Irradiated E1A + E1B cells arrest cell cycle progression in G2/M phase and suppress proliferation while continue to replicate DNA. (A) Cell cycle distribution analyzed by flow cytometry of propidium iodide-stained cells. Percentage of cells in S phase and percent of polyploid cells are shown. (B) Analysis of DNA-replication in cells according to BrdU incorporation. Non-irradiated and IR-treated cells were pulse-labeled with BrdU for 1 h, followed by immunofluorescent staining. (C) Growth curves of irradiated and untreated E1A + E1B cells. Cells were seeded in initial density of 3 × 104 cells per 30-mm dish and counted daily. Mean data with standard deviation are shown.
Figure 2. Exposure of E1A + E1B cells to IR results in the formation of giant polyploid cells, which are characterized by high level of E1A protein expression. (A) Microphotographs of cells stained with hematoxylin and eosin. Images were acquired in transmitted light, magnification 10 × 40. (B) Frequency distribution of cells according to DNA content was calculated by DNA cytometry of Feulgen-stained samples. Analysis of E1A expression in non-irradiated and IR-exposed cells by western blot (C) and immunofluorescent staining (D).
Uncontrolled DNA replication in E1A + E1B cells may depend on the expression of E1A protein, which can bind to and inactivate negative regulators of the cell cycle such as pRb,38,39 leading to the release of E2F transcription factors and, therefore, transcription of S-phase genes.40-42 According to our data, the expression of E1A protein in E1A + E1B cells remained high throughout the period of observation also in giant cells (Fig. 2C and D); therefore, it may provide replicative activity in irradiated cells.
Impaired DDR in E1A + E1B cells results in the persistence of DDR foci
Ionizing radiation induces rapid accumulation of DDR factors, including γH2AX and 53BP1 at the sites of DNA damage, resulting in the formation of DDR foci. Typically, DDR foci can already be detected 3 min after irradiation, reaching a maximum size and number 30 min after exposure to IR and dissociating within 24 h.43 However, the persistence of DDR foci leads to apoptosis or cellular senescence.29,44
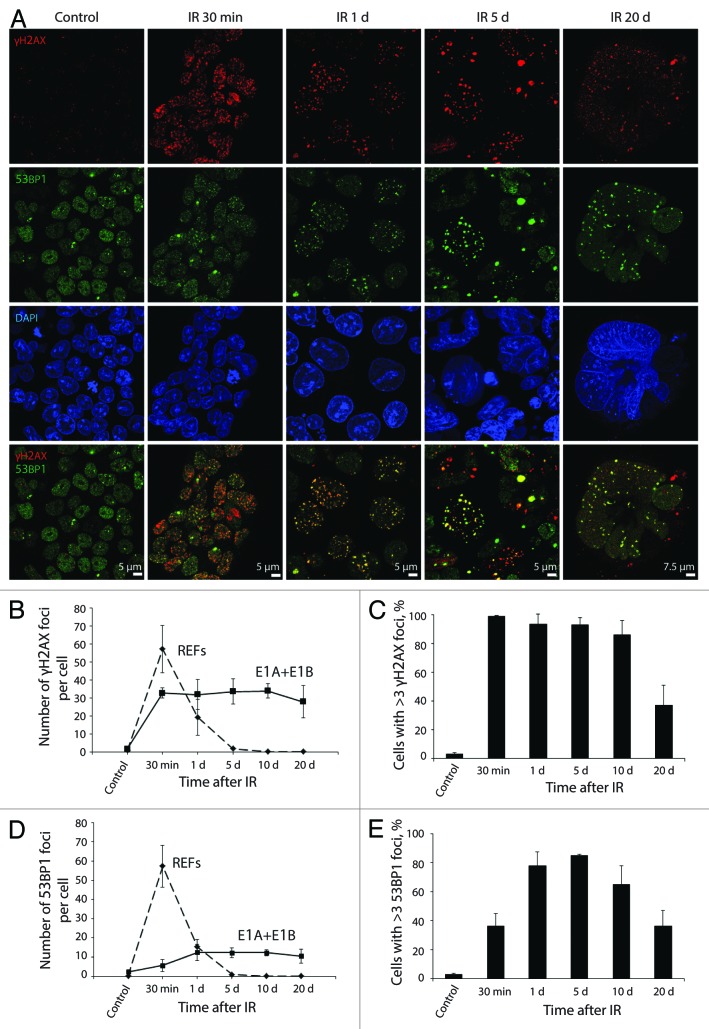
Therefore we studied the kinetics of γH2AX and 53BP1 foci formation and dissociation in E1A + E1B cells. The number of γH2AX foci reached the maximum 30 min after irradiation, whereas the maximal level of 53BP1 foci was detected only 1 d post-exposure to IR (Fig. 3A, B, and D). Notably, the translocation of 53BP1 to the sites of lesions was delayed, as it retained uniform distribution in the nuclei 30 min after irradiation (Fig. 3A). Furthermore, less than 40% of E1A + E1B cells showed 53BP1 foci formation 30 min post-IR treatment followed by a 2-fold increase on day 1 after irradiation (Fig. 3E). The kinetics of γH2AX and 53BP1 foci resolution in E1A + E1B cells was impaired as they persisted in most of the cells until day 20 post-exposure to IR (Fig. 3). γH2AX and 53BP1 foci remained colocolized until day 20 after IR treatment and increased in size (Fig. 3A). We compared the kinetics of γH2AX and 53BP1 foci formation and dissociation in E1A + E1B cell and rat embryonic fibroblasts (REFs). Unlike E1A + E1B cells, the maximal number of both γH2AX and 53BP1 foci in REFs was detected 30 min after irradiation and was two- and 10-fold higher respectively (Fig. 3B and D). Besides, the DDR foci did not persist in REFs and were completely resolved already 1 d post IR treatment (Fig. 3B and D; Fig. S1). Consequently, the kinetics of 53BP1 foci formation, and kinetics of both γH2AX and 53BP1 foci dissociation were impaired in E1A + E1B cells and resulted in the persistence of DDR foci.
Figure 3. Kinetics of γH2AX and 53BP1 foci formation and resolution in E1A + E1B cells. (A) Colocalization and persistence of γH2AX and 53BP1 foci in E1A + E1B cells after exposure to IR. Cells were irradiated or left untreated and stained with antibodies against γH2AX and 53BP1. Confocal images are shown. (B) Number of γH2AX foci per cell in E1A + E1B cells and REFs. (C) The percentage of cells with γH2AX foci. (D) Number of 53BP1 foci per cell in E1A + E1B cells and REFs. (E) The percentage of cells with 53BP1 foci. Note for (B) and (D): only cells with foci were included in the analysis. Note for (C) and (E): untreated cells contain 0–3 DDR foci per cell; therefore, cells with more than 3 foci were counted. (B–E) Mean data with the standard deviation are shown.
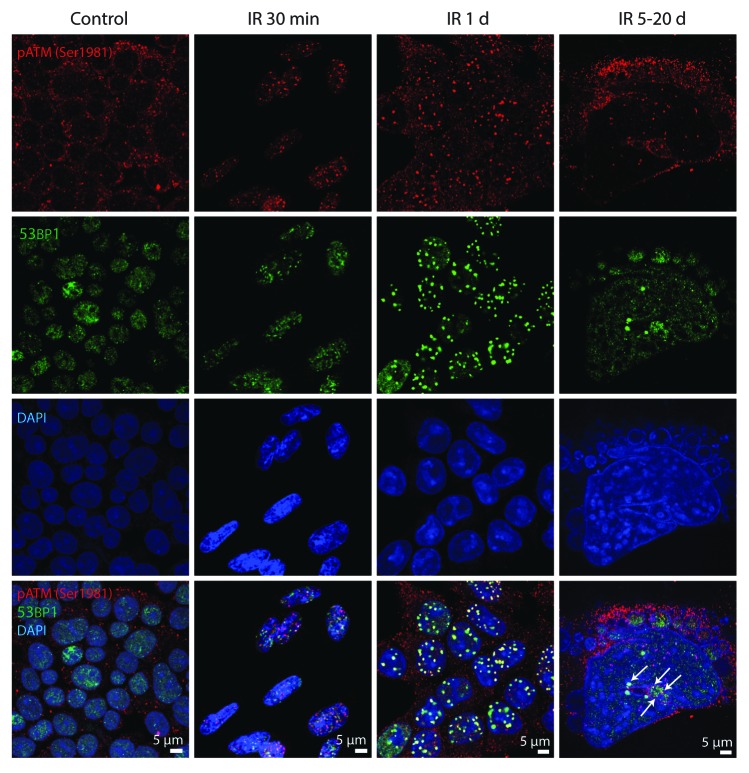
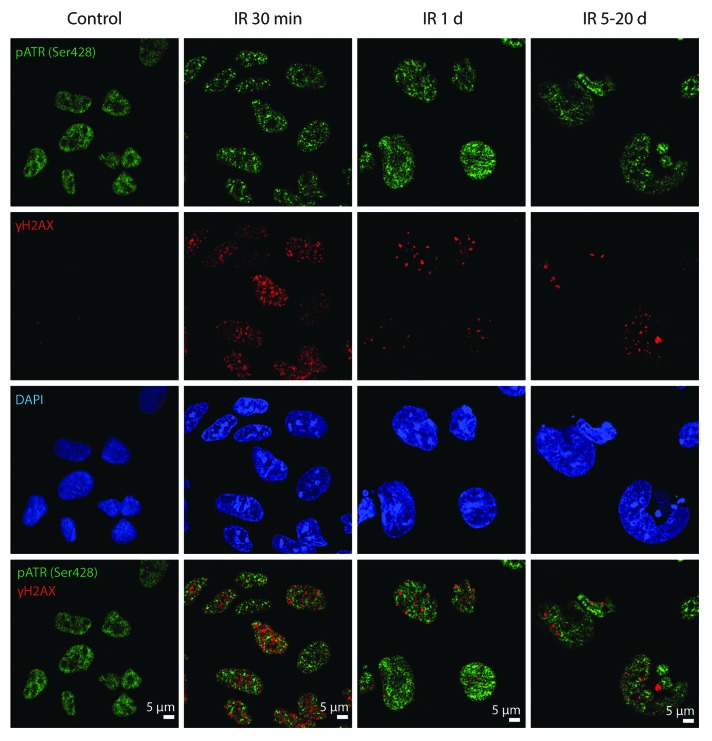
To reveal whether ATM and ATR kinases are the components of DDR foci in E1A + E1B cells, their colocalization with γH2AX and 53BP1 was analyzed. IR-activated pATMSer1981 accumulated in DDR foci within the minutes after exposure to IR and remained persistent showing distribution in the nuclei and micronuclei and colocalization with 53BP1 foci (Fig. 4). However, IR-induced pATRSer428 was detected neither in early nor in persistent DDR foci (Fig. 5). Our data suggest that sustained DDR signaling in E1A + E1B cells is mediated by ATM, but not ATR.
Figure 4. pATMSer1981 is a component of early and persistent DDR foci. E1A + E1B cells were irradiated or left untreated and stained with the antibodies against pATMSer1981 and 53BP1. Colocalization of pATMSer1981 and 53BP1 in giant cell is indicated with arrows. Confocal images are shown.
Figure 5. pATRSer428 does not colocolize with DDR foci in E1A + E1B cells. Irradiated and untreated cells were stained with the antibodies against pATRSer428 and γH2AX. Confocal images are shown.
The DDR foci persistent in E1A + E1B cells are the sites of DNA lesions
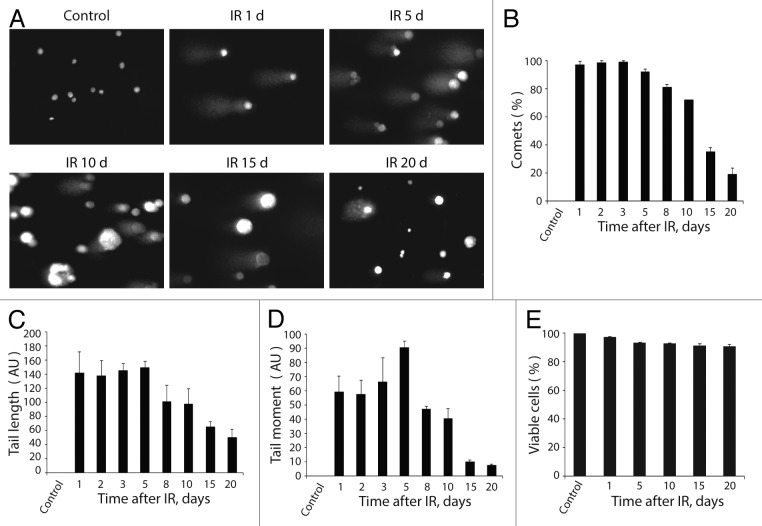
Rodier and colleagues have previously suggested that persistent DDR foci are distinct from the transient ones.15 Although they share common components, the persistent foci do not contain DNA repair factors and are not the sites of unscheduled DNA synthesis.15 To reveal whether the DDR foci that persisted in E1A + E1B cells are the sites of DNA breaks, we performed single-cell gel electrophoresis (comet assay).45,46 Formation of comet tails was found in virtually all irradiated cells until day 5 post-irradiation, when the percentage of cells forming the comets started to decrease (Fig. 6A and B). The number of cells with DNA breaks and the level of DNA damage as measured by comets’ tail length and tail moment remained high within 5 d after exposure to IR and then declined gradually (Fig. 6C and D). Taking into consideration that comets may arise due to the apoptotic cell death, we assayed DNA fragmentation and investigated cell viability. According to our data, not less than 94% of irradiated cells remained viable during all the period of experiment (Fig. 6E) and did not demonstrate any evidence of apoptotic cell death, including morphological features and nucleosomal DNA fragmentation (data not shown).
Figure 6. Analysis of DNA breaks persistence in E1A + E1B cells. (A) Untreated and irradiated cells were subjected to single-cell gel electrophoresis at the indicated time intervals after exposure to IR. Magnification 10 × 20. (B) Quantification of percentage of cells with DNA breaks in untreated and irradiated cells. Measurement of comet tail length (C), and comet tail moment (D), performed with CaspLab software. (E) Quantification of percentage of viable cells based on acridin orange and ethidium bromide staining. Mean data with standard deviation are shown for (B–E).
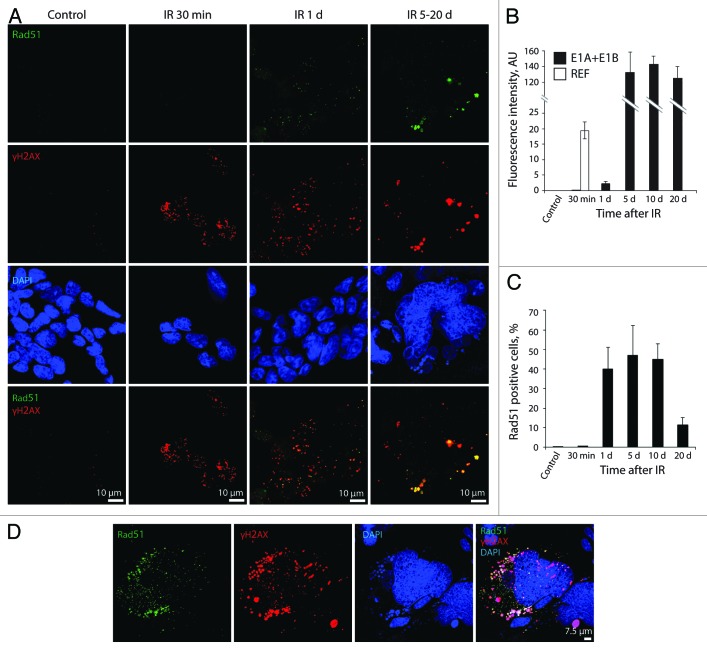
Further, we examined HR and NHEJ DNA repair by activation of Rad51 recombinase and DNA-dependent protein kinase catalytic subunit (DNA-PKcs) and their accumulation within the DDR foci. According to our results, E1A + E1B cells failed to activate HR repair immediately after exposure to IR (Fig. 7A–C), as revealed by the absence of Rad51 in the nuclei 30 min after irradiation. A weak activation of Rad51 was detected in 40% of cells only 1 d post-IR treatment (Fig. 7A–C), which correlated with accumulation of 53BP1 in the DDR foci (Fig. 3A, D and E). In contrast, REFs already demonstrated activation of HR repair 30 min after irradiation and completed DNA repair 1 d post-exposure to IR, as revealed by analysis of Rad51 accumulation within the DDR foci and measurement of its fluorescence intensity (Fig. 7B; Fig. S2A). Interestingly, the intensity of Rad51 fluorescence in E1A + E1B cells increased more than 50 times on day 5 after irradiation compared with day 1, and remained at this level until day 20 (Fig. 7B). Rad51 foci persisted in a significant number of E1A + E1B cells until day 20 post-irradiation (Fig. 7A and C). They were colocolized with γH2AX both in giant nuclei and micronuclei (Fig. 7A and D).
Figure 7. Irradiated E1A + E1B cells show delayed accumulation and persistence of Rad51 within the DDR foci. (A) Cells were left untreated or irradiated followed by staining with antibodies against Rad51 and γH2AX. Confocal images are shown. (B) Fluorescence intensity of Rad51 in untreated and irradiated cells was calculated as ratio of raw density to the cell surface measured with ImageJ software. Only cells expressing Rad51 were included in the analysis. (C) The percentage of cells containing Rad51 foci. (B and C) Mean data with standard deviation are shown. (D) Colocalization of Rad51 and γH2AX in the micronuclei indicate elimination of damaged DNA. Confocal images are shown.
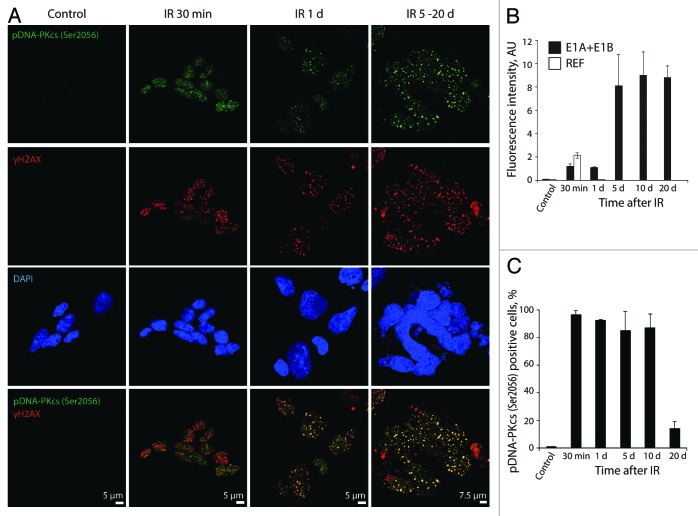
The DDR-dependent activation of DNA-PKcs by autophosphorylation on Ser2056 (pDNA-PKcsSer2056) and accumulation in the DDR foci were observed in all irradiated E1A + E1B cells already within the minutes after exposure to IR (Fig. 8A and C). They persisted and colocolized with γH2AX over the following 20 d (Fig. 8A). In addition, the number of pDNA-PKcsSer2056-positive cells did not decrease until day 10 post-exposure to IR (Fig. 8C). In contrast, in REFs, the pDNA-PKcsSer2056 foci appeared within minutes after treatment with IR and were not detected 1 d after irradiation, thus demonstrating that DNA repair is completed (Fig. S2B). The intensity of pDNA-PKcsSer2056 fluorescence 30 min post-irradiation was approximately twice lower in E1A + E1B cells than in REFs (Fig. 8B). It increased on day 5 after exposure of E1A + E1B cells to IR and remained at this level until day 20 (Fig. 8B). The number of cells positive for Rad51 and pDNA-PKcsSer2056 remained high until day 10 after irradiation then showed a dramatic decrease on day 20 post-treatment (Figs. 7C and 8C).
Figure 8. pDNA-PKcsSer2056 colocolizes with DDR foci within the minutes after irradiation and remains persistent. (A) Cells were irradiated or left untreated and stained with antibodies against pDNA-PKcsSer2056 and γH2AX. Confocal images are shown. (B) Fluorescence intensity of pDNA-PKcsSer2056 in untreated and irradiated cells was calculated as ratio of raw density to the cell surface measured with ImageJ software. Only cells that express pDNA-PKcsSer2056 were included in the analysis. (C) The percentage of cells containing pDNA-PKcsSer2056 foci. (B and C) Mean data with standard deviation are shown.
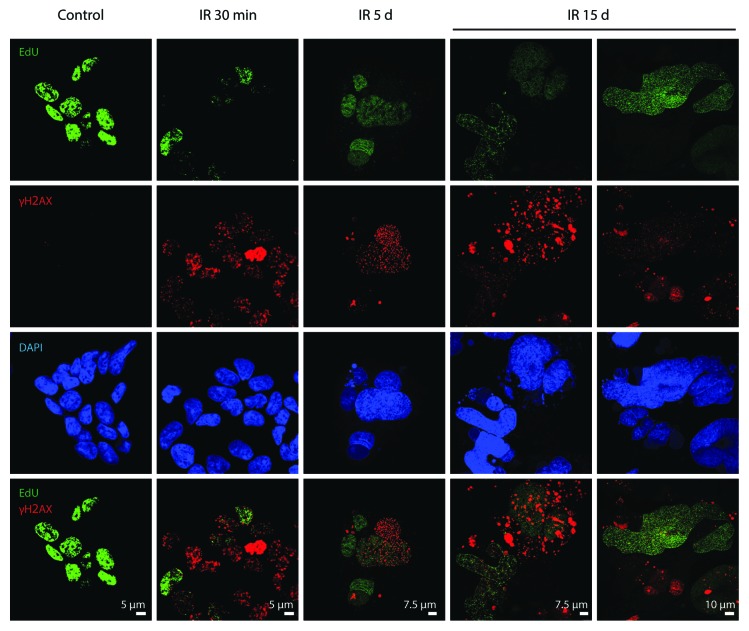
Despite the accumulation of pDNA-PKcsSer2056 in the DDR foci in E1A + E1B cells, already within minutes upon irradiation, only few γH2AX foci showed colocalization with EdU (Fig. 9). A time-course study revealed that both DNA replicating and non-replicating giant polyploid cells contained γH2AX foci (Fig. 9). In addition, we did not observe a difference in the intensity of γH2AX foci formation in EdU-incorporating and non-incorporating cells. A vast majority of γH2AX foci in giant polyploid cells did not colocolize with EdU, indicating the lack of DNA replication at the sites of lesions (Fig. 9).
Figure 9. Analysis of colocalization of DDR foci with the sites of DNA replication. Non-irradiated and IR-exposed cells were subjected to EdU incorporation assay by “click-it” method and stained with antibodies against γH2AX. Confocal images are shown.
Irradiated E1A + E1B cells undergo reversible senescence
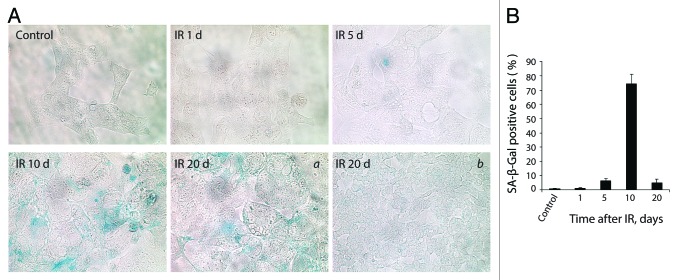
It was previously discussed that sustained DDR signaling tightly correlates with the establishment of senescence.47 Persistent DDR foci may arise from unrepaired lesions induced by genotoxic agents and stalled DNA replication, as well as may reflect a modified chromatin structure.28,44,48,49 The essential requirement for senescence is an irreversible arrest of cell cycle and proliferation. According to the growth curve assay, E1A + E1B cells did not proliferate until day 7 after treatment (Fig. 1C). However, they did not arrest DNA replication, which eventually resulted in the formation of polyploid cells (Figs. 1B and 2B). Polyploid cells showed the characteristic features of senescence, including enlarged flattened morphology (Fig. 2A), persistence of DDR foci (Figs. 3 and 4), and expression of SA-β-Gal (Fig. 10A and B). Therefore, we conclude that E1A + E1B cells activate senescence program.
Figure 10. E1A + E1B cells overpass senescence induced by IR. (A) SA-β-Gal staining of untreated and irradiated cells was performed. Images were acquired in transmitted light, magnification 10 × 40. Giant cells remain SA-β-Gal-positive (a), whereas cells of near-normal size are SA-β-Gal-negative (b). (B) Quantification of the percentage of senescent cells stained for SA-β-Gal detection. Mean values with standard deviation are shown.
However, the population of senescent cells showed an increase of the cell number starting from day 7 after irradiation (Fig. 1C). In turn, the percent of SA-β-Gal-positive cells dropped significantly in the period of 10–20 d after exposure to IR (Fig. 10B). Importantly, the expression of SA-β-Gal 20 d after IR-treatment was predominantly observed in giant cells, but not in the cells of near-normal size, which arose in the population (Fig. 10A). Notably, while the number of SA-β-Gal-positive cells decreased after day 10 post-exposure to IR, the population of irradiated cells demonstrated a fast proliferation starting at day 17 after irradiation (Fig. 1C). Moreover, the percent of cells with DDR foci (Fig. 3C and E) and DNA breaks, as well as the degree of DNA damage (Fig. 6B, C, and D) decreased significantly by day 20.
Cellular program switching is often accompanied by changes in chromatin organization. For example, enhanced heterochromatization, such as SAHF, is a characteristic of several types of senescence and reflects the silencing of proliferation genes.50 We revealed that irradiated E1A + E1B cells demonstrate alterations of chromatin organization such as formation of heterochromatin structures contrasted with the overall week DAPI staining (Fig. 2D), which, however, was distinct from the typical SAHF. Besides that, several nuclei of multinuclear cells showed the lack of DAPI staining, suggesting chromatin decompaction (Figs. 2D and 3A).
Reversion of senescence in E1A + E1B cells is associated with decrease of mTOR activity, induction of autophagy, and expression of stem cell markers Nanog and Oct3/4
mTOR is a master regulator of cellular senescence and autophagy. It is considered that elevated mTORC1 activity underlies the establishment of irreversible cellular senescence. Since irradiated E1A + E1B cells were shown to bypass the senescence, we examined the activity of mTOR by analyzing the phosphorylation of mTORC1 and mTORC2 downstream targets.
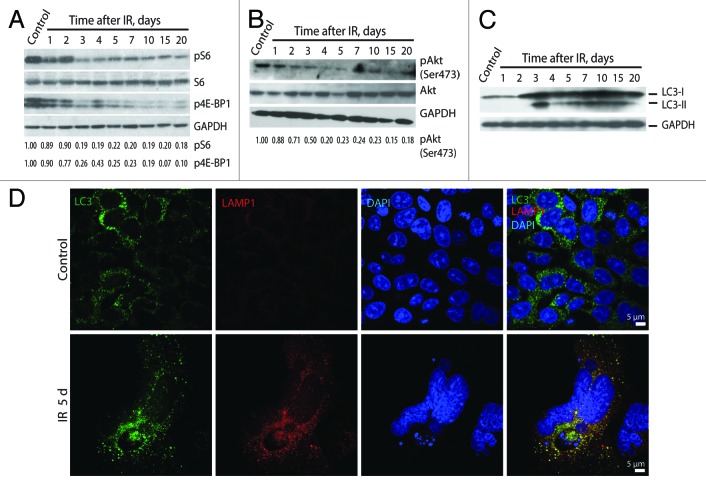
The suppression of mTORC1 activity was revealed in irradiated cells by analysis of phosphorylation of S6 ribosomal protein and repressor of translation initiation factor 4E-BP1. The phosphorylation of S6 ribosomal protein and 4E-BP1 remained high during 2 d post-irradiation and showed a 5-fold decrease on day 3 post-exposure to IR (Fig. 11A). Similarly, the activity of mTORC2 was also downregulated in cells exposed to IR as follows from a 5-fold decrease of the mTORC2-dependent phosphorylation of Akt on Ser473 (Fig. 11B).
Figure 11. Irradiated E1A + E1B cells show suppression of mTORC1 and mTORC2 and activation of autophagy. Western blot analysis of phosphorylation of S6 ribosomal protein and 4E-BP1 (A) and phosphorylation of Akt on Ser473 (B). The indicated numbers show the results of western blot densitometry. (C) Western blot analysis of LC3-I conversion to LC3-II. (D) Analysis of LC3 and LAMP1 colocalization in non-irradiated and IR-treated cells. Confocal images are shown.
Downregulation of mTOR leads to activation of autophagy.19 Indeed, autophagy was observed in irradiated E1A + E1B cells simultaneously with suppression of mTORC1 and mTORC2. Activation of autophagy was analyzed according to conversion of cytosolic MAP1-light chain protein LC3-I to LC3-II isoform, and colocalization of lysosomal-associated membrane protein LAMP1 with LC3. As a confirming evidence, both LC3-I to LC3-II conversion (Fig. 11C) and LAMP1/LC3 colocalization (Fig. 11D) were revealed in irradiated E1A + E1B cells simultaneously with a decrease of mTOR activity.
Though autophagy was reported to be an effector mechanism for senescence,18 recent data indicate that suppression of mTOR and activation of autophagy may facilitate reprogramming and favor the reversion of cellular senescence.51 The increasing body of evidence demonstrates that reversion of senescence in cancer cells and normal embryonic fibroblasts associates with expression of stem cell markers such as Oct3/4, Nanog, and Sox2.52,53 Therefore, we checked whether the establishment of reversible senescence in E1A + E1B cells correlates with the expression of stem cell markers. We revealed that both untreated and irradiated E1A + E1B cells expressed Nanog that localized in the nucleus and cytoplasm (Fig. 12). Unlike untreated cells, the vast majority of irradiated cells showed positive staining for Oct3/4 in the nuclei starting day 5 post-exposure to IR (Fig. 12).
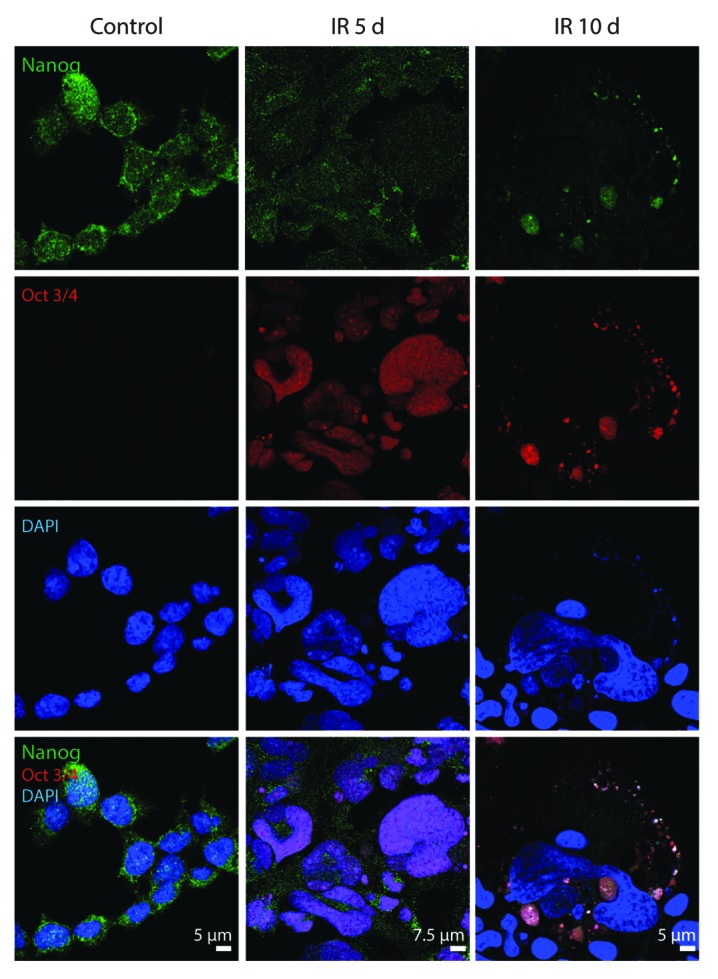
Figure 12. Expression of Nanog and Oct3/4 in E1A + E1B cells. Confocal images of immunofluorescent stained cells are shown.
Discussion
Here we studied the activation of senescence in apoptosis-resistant cells exposed to IR. We show that irradiation of E1A + E1B cells leads to the persistence of unrepaired DNA lesions and results in the induction of reversible senescence. A large number of works demonstrate that establishment and maintenance of various types of cellular senescence are associated with the activation of DDR signaling and persistence of DDR foci.1,11,15,28,54,55 The foci persistent in senescent cells may also reflect the chromatin rearrangement in the absence of DNA breaks48 or represent unrepaired DNA lesions.30,44 We revealed that in apoptosis-resistant E1A + E1B cells the sustained DDR signaling is provided by DNA breaks. The persistence of DNA lesions in E1A + E1B cells can be caused by delay in DNA repair, which, in turn, results from the impaired kinetics of DDR components activation. More precisely, the delayed accumulation of 53BP1 adaptor protein at the sites of DNA lesions may alter the recruitment of other DDR proteins and assembly of DNA repair molecular machinery. In addition, chromatin reorganization in irradiated E1A + E1B cells may impact the constitutively activated DDR signaling. As previously reported, chromatin relaxation in cells lacking histone H1 or treated with histone deacetylase inhibitors leads to enhanced H2AX phosphorylation in IR-exposed cells.56 From the other side, unrepaired lesions are probably not the only source of persistent DDR foci in E1A + E1B cells. As the DNA replication was not arrested in irradiated cells, and even the giant highly polyploid cells were able to replicate DNA, it may cause DNA replication stress. More specifically, the formation of multiple stalled replication forks could lead to DNA breaks.28
Irradiation of E1A + E1B cells induced the formation of giant highly polyploid cells due to ongoing DNA replication upon suppressed cell division. It was previously shown that increased DNA amount complicates the maintaining of genomic material, impairs DDR and DNA repair due to altered spatial chromatin organization,57 and thereby may contribute to the sustained DDR activation in E1A + E1B cells. Alternatively, polyploidy causes vast epigenetic changes57,58 and promotes overexpression of DNA repair genes upon replicative stress.59 Indeed, activation of DNA repair in E1A + E1B cells was observed only when the majority of cells in the population reached a highly polyploid state, thereby suggesting a new role for polyploidy in survival of apoptosis-resistant cells upon genotoxic stress.
The persistence of DNA lesions in irradiated E1A + E1B cells resulted in the activation of senescence program, which, however, was reversible. The sustained DDR activation is believed to be a driving force for the establishment and maintenance of senescence.47 Therefore, the question arises whether attenuation of DDR signaling reverses this process. Indeed, the escape of senescence in E1A + E1B cells was associated with a gradual decrease in the number of cells with DNA breaks and the degree of DNA damage as was shown by comet assay. The possible mechanisms for that may include the elimination of damaged DNA in the micronuclei and a delayed activation of DNA repair.
It should be noted that in E1A + E1B cells, initiation of the senescence program occurs upon high activity of mTOR, which then decreases. We do not know the mechanisms that regulate mTOR activity in E1A + E1B cells in response to irradiation; however, it was previously shown that IR treatment induces transient induction of mTOR via activation of ERK1/2 stress kinase.60 The subsequent downregulation of mTOR can be mediated by p53- or ATM-dependent activation of AMPK and mTOR inhibitor complex TSC1/2.61-64 The mTOR activity is involved in irreversible senescence, namely in conversion from quiescent to senescent state (geroconversion) associated with hypertrophic flattened phenotype.20 Inhibition of mTOR decelerates geroconversion, maintaining quiescence instead.35,36,65,66 Quiescent cells are able to resume proliferation later.36,65 Notably, proliferation restarts within a certain lag period upon removal of senescence-inducing factor. Similarly, the recovery of proliferation in IR-treated senescent E1A + E1B cells was also delayed. Besides, it was reported that suppression of mTOR and activation of autophagy potentiate somatic cells reprogramming.51,67 Therefore, we suggest that downregulation of mTOR in E1A + E1B cells exposed to IR predisposes the reversion of senescence and acquisition of stem cell-like characteristics.
Chromatin reorganization in E1A + E1B cells may facilitate cellular reprogramming. It was described that usage of chemical agents that cause chromatin modification enhances reprogramming.68 Besides that, recent findings demonstrate the important role of DNA repair factors in cellular reprogramming. For example, the components of HR repair, including BRCA1, BRCA2, and Rad51, are crucial for iPSCs generation,69 among which Rad51 is required not only for the induced pluripotent stem cells (iPSCs) conversion, but also for the maintenance of pluripotency in embryonic stem cells (ESCs).70 Moreover, cells deficient in NHEJ component DNA-PKcs show a decreased efficiency of iPSCs generation.71 Notably, untreated and irradiated E1A + E1B cells expressed the stem cell factor Nanog. However, the increase of pDNA-PKcsSer2056, and especially Rad51 protein level in polyploid E1A + E1B cells correlated with the expression of Oct3/4, thereby may imply a cross-talk between self-renewal and reversion of senescence.
The transcription factors Oct3/4 and Nanog are the key regulators of self-renewal and pluripotency of stem cells.72 Activation of stem cell factors in somatic cells promotes malignant transformation and acquirement of cancer stem cells properties.73-75 While the role of stem cell transcription factors in senescent cells remains unclear, their elevated expression is often observed in various types of tumors and associates with cancer progression, resistance to therapy, and poor prognosis.74,76-79
The survival of the irradiated population was provided by cells with the size and ploidy close to untreated E1A + E1B cells. We did not identify the source of those cells, but several hypothesis of their origin can be provided. For example, a small fraction of cells may be resistant to initial treatment with IR and provide regrowth of population. A number of observations also suggest that the novel cells may arise from the giant polyploid cells by multipolar division or depolyploidization caused by autophagic degradation of genetic material.80-82 Apparently, the resistance to apoptosis, provided by adenoviral E1B 19 kDa protein, a functional homolog of Bcl-2, allows E1A + E1B cells to remain viable and replicate DNA in the presence of unrepaired DNA, eventually acquiring a highly polyploid state. Resistance to apoptosis and high polyploid state increase the cellular plasticity, and enable various pro-survival strategies.
Together, our results indicate that exposure of E1A + E1B cells to IR induces cellular senescence, which is determined by the persistence of unrepaired DNA lesions and, therefore, sustained activation of DDR signaling. We have found that mechanisms of gerosuppression in apoptosis-resistant IR-treated cells associate with polyploidization, attenuation of DDR signaling, downregulation of mTOR, and expression of pluripotency markers Oct3/4 and Nanog. Reversion of IR-induced senescence in cells resistant to apoptosis results in the appearance of SA-β-Gal-negative cells of near normal size and ploidy, which exhibit high proliferative potential and restore the population.
Materials and Methods
Cell culture and treatment
Cells with stable expression of adenoviral E1A and E1B19 kDa proteins were selected from rat embryonic fibroblasts co-transfected with HindIII-G region of Ad5 viral DNA and pSV 2neo plasmid. Cells were cultured in DMEM supplemented with 10% fetal calf serum (FCS), penicillin, and streptomycin in 5% CO2 at 37 °C, irradiated in a dose of 6 Gy using X-ray machine Axiom Iconos R200 (Siemens) and analyzed up to 20 d after treatment.
Antibodies
Primary antibodies: BrDU (Millipore), E1A, 53BP1, pATMSer1981, pATRSer428, S6 ribosomal protein, pS6 ribosomal protein, p4E-BP1, Akt, pAktSer473, GAPDH, LAMP1, Nanog (all by Cell Signaling Technology); Rad51, Oct3/4 (all by Santa Cruz Biotechnology); γH2AX, pDNA-PKcsS2056 (all by Abcam); LC3 (MBL). Secondary antibodies: Alexa-fluor 488, Alexa-fluor 568 (all by Invitrogen); anti-mouse and anti-rabbit antibodies conjugated with horseradish peroxidase (Sigma).
Flow cytometry
To assay cell cycle distribution cells flow cytometry assay of propidium iodide-stained cells was performed as described before.83
Growth curves
Cells were seeded at the initial density of 3 × 104 cells per 30-mm dish in 3 repeats 24 h before the treatment. Cells were irradiated or left untreated and counted in cell counting chamber daily up to 20 d. The medium was replaced by the fresh one supplemented with 10% FCS every second day. The growth curve was made based on the data obtained in 3 independent experiments.
Morphological staining with hematoxylin and eosin
To analyze morphology of irradiated cells, E1A + E1B cells were grown on coverslips, fixed with −20 °C methanol for 5 min, and stained with hematoxylin and eosin as previously described.83
Feulgen DNA staining and integrated optical density measurement
For analysis of cell ploidy by DNA cytometry, cells were grown on coverslips, irradiated, or left untreated. Cells were fixed with methanol −20 °C for 5 min followed by hydrolysis with 5N HCl for 30 min at room temperature. Afterwards, the coverslips were immediately transferred into Schiff reagent and incubated for 1.5 h at room temperature in the dark. The samples were washed with fresh SO2 water 3 times, with ultrapure water 3 times, and then dehydrated with 96% ethanol. The coverslips were allowed to dry at room temperature and mounted on microscope slides prior to analysis. Images were acquired using Axioscope, DFC360 (Zeiss) microscope equipped with a digital camera. DNA content was measured as integrated optical density using software (VideoTesT); DNA content of non-irradiated cells in metaphase was taken as 4C. The ploidy of 100 cells per sample was analyzed.
Immunoblotting
Cells were lysed in a buffer containing 10 mM TRIS-HCl, pH 7.4, 150 mM NaCl, 1% Triton X-100, 0.5% Nonidet P-40, 20 mM β-glycerophosphate, 1 mM sodium orthovanadate, 5 mM EGTA, 10 mM sodium fluoride, 1 mM phenylmethylsulfonyl fluoride, and protease inhibitors cocktail (Roche). Extracts were subjected to SDS-polyacrilamyde gel electrophoresis (SDS-PAGE), transferred to PVDF membrane (Invitrogen), and immunoblotted with primary antibodies followed by incubation with horseradish peroxidase-conjugated secondary antibodies. Immunocomplexes were visualized by enhanced chemiluminescence (ECL, Thermo Fisher Scientific). Western blot densitometry was performed using ImageJ software (US National Institutes of Health).
Immunofluorescence and confocal microscopy
For immunofluorescence analysis, cells grown on coverslips were fixed with 3.7% paraformaldehyde in PBS for 15 min. Cells were washed with PBS containing 0.5% Tween 20 (PBST) and permerabilized with 0.1% Triton X-100 in PBS for 30 min followed by incubation in blocking solution (5% goat serum in PBST) for 1 h. Cells were incubated with primary antibodies diluted in blocking solution overnight at 4 °C, washed with PBST, and incubated with secondary antibodies Alexa-488 and Alexa-568 (Invitrogen) for 1 h at room temperature. Coverslips were mounted using ProLong Gold mounting medium containing 4,6-diamidino-2-phenylindole (DAPI) (Invitrogen). Cells were analyzed with Leica TCP SP5 scanning confocal microscope (Leica Microsystems). Confocal images were acquired using a Plan-Apochromat 40 × /1.4 oil immersion objective. Pinholes were set at 1 airy unit. The dynamics of γH2AX and 53BP1 foci accumulation, as well as percentage of IF-positive cells were calculated based on analysis of 200 cells in each sample in 3 independent experiments.
Fluorescence intensity measurment
The integrated density of Rad51 and pDNA-PKcsSer2056 fluorescence in the nuclei, mean fluorescence of background (outside the nuclei), and nuclei area were measured using ImageJ software (US National Institutes of Health). The fluorescence intensity was calculated as corrected total nuclei fluorescence intensity (CTNFI) in 100 cells in 2 independent experiments:
IntFluor = CTNFI = integrated density – (nucleus area × mean fluorescence of background).
BrdU incorporation assay
DNA replication was analyzed by BrdU incorporation. Cells were pulse-labeled with 10 µM of BrdU (BD Biosciences) for 1 h. The following procedures were performed as described previously.83 Images were acquired using Leica TCP SP5 scanning confocal microscope (Leica Microsystems).
Analysis of EdU and yH2AX colocalization
Untreated and irradiated cells were incubated with 10 µM of EdU (Click-iT EdU AlexaFluor 488 Imaging Kit, Invitrogen) for 1 h and proceeded to EdU detection and staining with the antibodies against γH2AX according to manufacturer’s instruction.
SA-β-Gal activity
To analyze senescence-associated SA-β-Gal expression, cells were grown on coverslips, fixed with 3.7% paraformaldehyde in PBS for 15 min, and SA-β-Gal staining was performed as previously described.83 The coverslips were washed with PBS and mounted on microscope slides using ProLong Gold mounting medium (Invitrogen). The images were acquired in transmitted light, magnification 10 × 40, using Zeiss Pascal microscope (Zeiss) equipped with digital camera and Adobe Photoshop software (Adobe Systems). To calculate the number of SA-β-Gal positive cells, 200 cells per sample were analyzed in 3 independent experiments.
Single-cell gel electrophoresis (comet assay)
Comet assay in alkaline conditions was performed as follows. The microscope slides were covered with 1% agarose and dried. The suspension containing 1.5 × 104 of living cells was prepared in 0.5% low melting agarose, 37 °C, placed on microscope slide, covered with cover glass and set at 4 °C for 10 min protected from light. Cells were covered with another layer of cell-free agarose and lysed overnight at 4 °C in a buffer containing 2.5 M NaCl, 0.1 M EDTA, 10 mM TRIS-HCl, 1% Triton X-100, pH 10.0. Slides were rinsed in electrophoresis buffer (0.3 M NaOH, 1 mM EDTA, pH 13.0) and subjected to electrophoresis at 4 °C in the dark. Following that, slides were rinsed with neutralizing solution (0.4 M TRIS-HCl, pH 7.5), stained with SYBR-green, and visualized using Zeiss Pascal fluorescent microscope (Zeiss) equipped with digital camera and Adobe Photoshop software (Adobe Systems). To calculate the number of comets, comet tail length and tail moment, 100 cells were analyzed in 3 independent experiments. Comet length and tail moment were measured using CaspLab software.
Cell viability assay
To determine cell viability, cells were stained with acridine orange/ethidium bromide mixture (1:1) in PBS. Cells growing on coverslips were washed with PBS 37 °C, the acridine orange and ethidium bromide solution was applied, and fluorescent microscopy was performed immediately using Leica TCP SP5 scanning confocal microscope (Leica Microsystems). The number of live cells was counted, and the percent of viable cells was calculated for 200 cells per each of 3 independent experiments.
Supplementary Material
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
This work was funded by Program of the Russian Academy of Sciences (MCB RAS), grant from Russian Foundation for Basic Research (13-04-00552) and by the Program of Saint Petersburg State University (1.38.247.2014).
References
- 1.Bartkova J, Rezaei N, Liontos M, Karakaidos P, Kletsas D, Issaeva N, Vassiliou LV, Kolettas E, Niforou K, Zoumpourlis VC, et al. Oncogene-induced senescence is part of the tumorigenesis barrier imposed by DNA damage checkpoints. Nature. 2006;444:633–7. doi: 10.1038/nature05268. [DOI] [PubMed] [Google Scholar]
- 2.Serrano M, Lin AW, McCurrach ME, Beach D, Lowe SW. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell. 1997;88:593–602. doi: 10.1016/S0092-8674(00)81902-9. [DOI] [PubMed] [Google Scholar]
- 3.Deng Q, Liao R, Wu BL, Sun P. High intensity ras signaling induces premature senescence by activating p38 pathway in primary human fibroblasts. J Biol Chem. 2004;279:1050–9. doi: 10.1074/jbc.M308644200. [DOI] [PubMed] [Google Scholar]
- 4.te Poele RH, Okorokov AL, Jardine L, Cummings J, Joel SP. DNA damage is able to induce senescence in tumor cells in vitro and in vivo. Cancer Res. 2002;62:1876–83. [PubMed] [Google Scholar]
- 5.Robles SJ, Adami GR. Agents that cause DNA double strand breaks lead to p16INK4a enrichment and the premature senescence of normal fibroblasts. Oncogene. 1998;16:1113–23. doi: 10.1038/sj.onc.1201862. [DOI] [PubMed] [Google Scholar]
- 6.Munro J, Barr NI, Ireland H, Morrison V, Parkinson EK. Histone deacetylase inhibitors induce a senescence-like state in human cells by a p16-dependent mechanism that is independent of a mitotic clock. Exp Cell Res. 2004;295:525–38. doi: 10.1016/j.yexcr.2004.01.017. [DOI] [PubMed] [Google Scholar]
- 7.Rebbaa A, Zheng X, Chu F, Mirkin BL. The role of histone acetylation versus DNA damage in drug-induced senescence and apoptosis. Cell Death Differ. 2006;13:1960–7. doi: 10.1038/sj.cdd.4401895. [DOI] [PubMed] [Google Scholar]
- 8.Hayflick L. The limited in vitro lifetime of human diploid cell strains. Exp Cell Res. 1965;37:614–36. doi: 10.1016/0014-4827(65)90211-9. [DOI] [PubMed] [Google Scholar]
- 9.Dimri GP, Lee X, Basile G, Acosta M, Scott G, Roskelley C, Medrano EE, Linskens M, Rubelj I, Pereira-Smith O, et al. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc Natl Acad Sci U S A. 1995;92:9363–7. doi: 10.1073/pnas.92.20.9363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hara E, Smith R, Parry D, Tahara H, Stone S, Peters G. Regulation of p16CDKN2 expression and its implications for cell immortalization and senescence. Mol Cell Biol. 1996;16:859–67. doi: 10.1128/mcb.16.3.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rodier F, Coppé JP, Patil CK, Hoeijmakers WA, Muñoz DP, Raza SR, Freund A, Campeau E, Davalos AR, Campisi J. Persistent DNA damage signalling triggers senescence-associated inflammatory cytokine secretion. Nat Cell Biol. 2009;11:973–9. doi: 10.1038/ncb1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kuilman T, Michaloglou C, Vredeveld LC, Douma S, van Doorn R, Desmet CJ, Aarden LA, Mooi WJ, Peeper DS. Oncogene-induced senescence relayed by an interleukin-dependent inflammatory network. Cell. 2008;133:1019–31. doi: 10.1016/j.cell.2008.03.039. [DOI] [PubMed] [Google Scholar]
- 13.Adams PD. Remodeling of chromatin structure in senescent cells and its potential impact on tumor suppression and aging. Gene. 2007;397:84–93. doi: 10.1016/j.gene.2007.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Narita M, Nũnez S, Heard E, Narita M, Lin AW, Hearn SA, Spector DL, Hannon GJ, Lowe SW. Rb-mediated heterochromatin formation and silencing of E2F target genes during cellular senescence. Cell. 2003;113:703–16. doi: 10.1016/S0092-8674(03)00401-X. [DOI] [PubMed] [Google Scholar]
- 15.Rodier F, Muñoz DP, Teachenor R, Chu V, Le O, Bhaumik D, Coppé JP, Campeau E, Beauséjour CM, Kim SH, et al. DNA-SCARS: distinct nuclear structures that sustain damage-induced senescence growth arrest and inflammatory cytokine secretion. J Cell Sci. 2011;124:68–81. doi: 10.1242/jcs.071340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim DH, Sarbassov DD, Ali SM, King JE, Latek RR, Erdjument-Bromage H, Tempst P, Sabatini DM. mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell. 2002;110:163–75. doi: 10.1016/S0092-8674(02)00808-5. [DOI] [PubMed] [Google Scholar]
- 17.Sarbassov DD, Ali SM, Kim DH, Guertin DA, Latek RR, Erdjument-Bromage H, Tempst P, Sabatini DM. Rictor, a novel binding partner of mTOR, defines a rapamycin-insensitive and raptor-independent pathway that regulates the cytoskeleton. Curr Biol. 2004;14:1296–302. doi: 10.1016/j.cub.2004.06.054. [DOI] [PubMed] [Google Scholar]
- 18.Young AR, Narita M, Ferreira M, Kirschner K, Sadaie M, Darot JF, Tavaré S, Arakawa S, Shimizu S, Watt FM, et al. Autophagy mediates the mitotic senescence transition. Genes Dev. 2009;23:798–803. doi: 10.1101/gad.519709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jung CH, Ro SH, Cao J, Otto NM, Kim DH. mTOR regulation of autophagy. FEBS Lett. 2010;584:1287–95. doi: 10.1016/j.febslet.2010.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Blagosklonny MV. Cell cycle arrest is not yet senescence, which is not just cell cycle arrest: terminology for TOR-driven aging. Aging (Albany NY) 2012;4:159–65. doi: 10.18632/aging.100443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem. 1998;273:5858–68. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- 22.Rappold I, Iwabuchi K, Date T, Chen J. Tumor suppressor p53 binding protein 1 (53BP1) is involved in DNA damage-signaling pathways. J Cell Biol. 2001;153:613–20. doi: 10.1083/jcb.153.3.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bakkenist CJ, Kastan MB. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature. 2003;421:499–506. doi: 10.1038/nature01368. [DOI] [PubMed] [Google Scholar]
- 24.Thompson LH. Recognition, signaling, and repair of DNA double-strand breaks produced by ionizing radiation in mammalian cells: the molecular choreography. Mutat Res. 2012;751:158–246. doi: 10.1016/j.mrrev.2012.06.002. [DOI] [PubMed] [Google Scholar]
- 25.Zou L. Single- and double-stranded DNA: building a trigger of ATR-mediated DNA damage response. Genes Dev. 2007;21:879–85. doi: 10.1101/gad.1550307. [DOI] [PubMed] [Google Scholar]
- 26.Fernandez-Capetillo O, Chen HT, Celeste A, Ward I, Romanienko PJ, Morales JC, Naka K, Xia Z, Camerini-Otero RD, Motoyama N, et al. DNA damage-induced G2-M checkpoint activation by histone H2AX and 53BP1. Nat Cell Biol. 2002;4:993–7. doi: 10.1038/ncb884. [DOI] [PubMed] [Google Scholar]
- 27.Jowsey P, Morrice NA, Hastie CJ, McLauchlan H, Toth R, Rouse J. Characterisation of the sites of DNA damage-induced 53BP1 phosphorylation catalysed by ATM and ATR. DNA Repair (Amst) 2007;6:1536–44. doi: 10.1016/j.dnarep.2007.04.011. [DOI] [PubMed] [Google Scholar]
- 28.Di Micco R, Fumagalli M, Cicalese A, Piccinin S, Gasparini P, Luise C, Schurra C, Garre’ M, Nuciforo PG, Bensimon A, et al. Oncogene-induced senescence is a DNA damage response triggered by DNA hyper-replication. Nature. 2006;444:638–42. doi: 10.1038/nature05327. [DOI] [PubMed] [Google Scholar]
- 29.Noda A, Hirai Y, Hamasaki K, Mitani H, Nakamura N, Kodama Y. Unrepairable DNA double-strand breaks that are generated by ionising radiation determine the fate of normal human cells. J Cell Sci. 2012;125:5280–7. doi: 10.1242/jcs.101006. [DOI] [PubMed] [Google Scholar]
- 30.d’Adda di Fagagna F, Reaper PM, Clay-Farrace L, Fiegler H, Carr P, Von Zglinicki T, Saretzki G, Carter NP, Jackson SP. A DNA damage checkpoint response in telomere-initiated senescence. Nature. 2003;426:194–8. doi: 10.1038/nature02118. [DOI] [PubMed] [Google Scholar]
- 31.Nakamura AJ, Chiang YJ, Hathcock KS, Horikawa I, Sedelnikova OA, Hodes RJ, Bonner WM. Both telomeric and non-telomeric DNA damage are determinants of mammalian cellular senescence. Epigenetics Chromatin. 2008;1:6. doi: 10.1186/1756-8935-1-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/S0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 33.Roninson IB, Broude EV, Chang BD. If not apoptosis, then what? Treatment-induced senescence and mitotic catastrophe in tumor cells. Drug Resist Updat. 2001;4:303–13. doi: 10.1054/drup.2001.0213. [DOI] [PubMed] [Google Scholar]
- 34.Wang Q, Wu PC, Dong DZ, Ivanova I, Chu E, Zeliadt S, Vesselle H, Wu DY. Polyploidy road to therapy-induced cellular senescence and escape. Int J Cancer. 2013;132:1505–15. doi: 10.1002/ijc.27810. [DOI] [PubMed] [Google Scholar]
- 35.Cao K, Graziotto JJ, Blair CD, Mazzulli JR, Erdos MR, Krainc D, Collins FS. Rapamycin reverses cellular phenotypes and enhances mutant protein clearance in Hutchinson-Gilford progeria syndrome cells. Sci Transl Med. 2011;3:89ra58. doi: 10.1126/scitranslmed.3002346. [DOI] [PubMed] [Google Scholar]
- 36.Pospelova TV, Leontieva OV, Bykova TV, Zubova SG, Pospelov VA, Blagosklonny MV. Suppression of replicative senescence by rapamycin in rodent embryonic cells. Cell Cycle. 2012;11:2402–7. doi: 10.4161/cc.20882. [DOI] [PubMed] [Google Scholar]
- 37.Beauséjour CM, Krtolica A, Galimi F, Narita M, Lowe SW, Yaswen P, Campisi J. Reversal of human cellular senescence: roles of the p53 and p16 pathways. EMBO J. 2003;22:4212–22. doi: 10.1093/emboj/cdg417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Whyte P, Buchkovich KJ, Horowitz JM, Friend SH, Raybuck M, Weinberg RA, Harlow E. Association between an oncogene and an anti-oncogene: the adenovirus E1A proteins bind to the retinoblastoma gene product. Nature. 1988;334:124–9. doi: 10.1038/334124a0. [DOI] [PubMed] [Google Scholar]
- 39.Chellappan SP, Hiebert S, Mudryj M, Horowitz JM, Nevins JR. The E2F transcription factor is a cellular target for the RB protein. Cell. 1991;65:1053–61. doi: 10.1016/0092-8674(91)90557-F. [DOI] [PubMed] [Google Scholar]
- 40.Bagchi S, Raychaudhuri P, Nevins JR. Adenovirus E1A proteins can dissociate heteromeric complexes involving the E2F transcription factor: a novel mechanism for E1A trans-activation. Cell. 1990;62:659–69. doi: 10.1016/0092-8674(90)90112-R. [DOI] [PubMed] [Google Scholar]
- 41.Hiebert SW, Chellappan SP, Horowitz JM, Nevins JR. The interaction of RB with E2F coincides with an inhibition of the transcriptional activity of E2F. Genes Dev. 1992;6:177–85. doi: 10.1101/gad.6.2.177. [DOI] [PubMed] [Google Scholar]
- 42.Johnson DG, Schwarz JK, Cress WD, Nevins JR. Expression of transcription factor E2F1 induces quiescent cells to enter S phase. Nature. 1993;365:349–52. doi: 10.1038/365349a0. [DOI] [PubMed] [Google Scholar]
- 43.Sedelnikova OA, Pilch DR, Redon C, Bonner WM. Histone H2AX in DNA damage and repair. Cancer Biol Ther. 2003;2:233–5. doi: 10.4161/cbt.2.3.373. [DOI] [PubMed] [Google Scholar]
- 44.Fumagalli M, Rossiello F, Clerici M, Barozzi S, Cittaro D, Kaplunov JM, Bucci G, Dobreva M, Matti V, Beausejour CM, et al. Telomeric DNA damage is irreparable and causes persistent DNA-damage-response activation. Nat Cell Biol. 2012;14:355–65. doi: 10.1038/ncb2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Olive PL, Banáth JP, Durand RE. Heterogeneity in radiation-induced DNA damage and repair in tumor and normal cells measured using the “comet” assay. 1990. Radiat Res. 2012;178:AV35–42. doi: 10.1667/RRAV04.1. [DOI] [PubMed] [Google Scholar]
- 46.Singh NP, McCoy MT, Tice RR, Schneider EL. A simple technique for quantitation of low levels of DNA damage in individual cells. Exp Cell Res. 1988;175:184–91. doi: 10.1016/0014-4827(88)90265-0. [DOI] [PubMed] [Google Scholar]
- 47.d’Adda di Fagagna F. Living on a break: cellular senescence as a DNA-damage response. Nat Rev Cancer. 2008;8:512–22. doi: 10.1038/nrc2440. [DOI] [PubMed] [Google Scholar]
- 48.Pospelova TV, Demidenko ZN, Bukreeva EI, Pospelov VA, Gudkov AV, Blagosklonny MV. Pseudo-DNA damage response in senescent cells. Cell Cycle. 2009;8:4112–8. doi: 10.4161/cc.8.24.10215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dimauro T, David G. Chromatin modifications: the driving force of senescence and aging? Aging (Albany NY) 2009;1:182–90. doi: 10.18632/aging.100023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kosar M, Bartkova J, Hubackova S, Hodny Z, Lukas J, Bartek J. Senescence-associated heterochromatin foci are dispensable for cellular senescence, occur in a cell type- and insult-dependent manner and follow expression of p16(ink4a) Cell Cycle. 2011;10:457–68. doi: 10.4161/cc.10.3.14707. [DOI] [PubMed] [Google Scholar]
- 51.Menendez JA, Vellon L, Oliveras-Ferraros C, Cufí S, Vazquez-Martin A. mTOR-regulated senescence and autophagy during reprogramming of somatic cells to pluripotency: a roadmap from energy metabolism to stem cell renewal and aging. Cell Cycle. 2011;10:3658–77. doi: 10.4161/cc.10.21.18128. [DOI] [PubMed] [Google Scholar]
- 52.Pospelova TV, Bykova TV, Zubova SG, Katolikova NV, Yartzeva NM, Pospelov VA. Rapamycin induces pluripotent genes associated with avoidance of replicative senescence. Cell Cycle. 2013;12:3841–51. doi: 10.4161/cc.27396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Salmina K, Jankevics E, Huna A, Perminov D, Radovica I, Klymenko T, Ivanov A, Jascenko E, Scherthan H, Cragg M, et al. Up-regulation of the embryonic self-renewal network through reversible polyploidy in irradiated p53-mutant tumour cells. Exp Cell Res. 2010;316:2099–112. doi: 10.1016/j.yexcr.2010.04.030. [DOI] [PubMed] [Google Scholar]
- 54.Sedelnikova OA, Horikawa I, Zimonjic DB, Popescu NC, Bonner WM, Barrett JC. Senescing human cells and ageing mice accumulate DNA lesions with unrepairable double-strand breaks. Nat Cell Biol. 2004;6:168–70. doi: 10.1038/ncb1095. [DOI] [PubMed] [Google Scholar]
- 55.Mallette FA, Gaumont-Leclerc MF, Ferbeyre G. The DNA damage signaling pathway is a critical mediator of oncogene-induced senescence. Genes Dev. 2007;21:43–8. doi: 10.1101/gad.1487307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Murga M, Jaco I, Fan Y, Soria R, Martinez-Pastor B, Cuadrado M, Yang SM, Blasco MA, Skoultchi AI, Fernandez-Capetillo O. Global chromatin compaction limits the strength of the DNA damage response. J Cell Biol. 2007;178:1101–8. doi: 10.1083/jcb.200704140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Comai L. The advantages and disadvantages of being polyploid. Nat Rev Genet. 2005;6:836–46. doi: 10.1038/nrg1711. [DOI] [PubMed] [Google Scholar]
- 58.Duelli D, Lazebnik Y. Cell fusion: a hidden enemy? Cancer Cell. 2003;3:445–8. doi: 10.1016/S1535-6108(03)00114-4. [DOI] [PubMed] [Google Scholar]
- 59.Zheng L, Dai H, Zhou M, Li X, Liu C, Guo Z, Wu X, Wu J, Wang C, Zhong J, et al. Polyploid cells rewire DNA damage response networks to overcome replication stress-induced barriers for tumour progression. Nat Commun. 2012;3:815. doi: 10.1038/ncomms1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Braunstein S, Badura ML, Xi Q, Formenti SC, Schneider RJ. Regulation of protein synthesis by ionizing radiation. Mol Cell Biol. 2009;29:5645–56. doi: 10.1128/MCB.00711-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Feng Z, Zhang H, Levine AJ, Jin S. The coordinate regulation of the p53 and mTOR pathways in cells. Proc Natl Acad Sci U S A. 2005;102:8204–9. doi: 10.1073/pnas.0502857102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Alexander A, Cai SL, Kim J, Nanez A, Sahin M, MacLean KH, Inoki K, Guan KL, Shen J, Person MD, et al. ATM signals to TSC2 in the cytoplasm to regulate mTORC1 in response to ROS. Proc Natl Acad Sci U S A. 2010;107:4153–8. doi: 10.1073/pnas.0913860107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Demidenko ZN, Korotchkina LG, Gudkov AV, Blagosklonny MV. Paradoxical suppression of cellular senescence by p53. Proc Natl Acad Sci U S A. 2010;107:9660–4. doi: 10.1073/pnas.1002298107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Korotchkina LG, Leontieva OV, Bukreeva EI, Demidenko ZN, Gudkov AV, Blagosklonny MV. The choice between p53-induced senescence and quiescence is determined in part by the mTOR pathway. Aging (Albany NY) 2010;2:344–52. doi: 10.18632/aging.100160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Demidenko ZN, Zubova SG, Bukreeva EI, Pospelov VA, Pospelova TV, Blagosklonny MV. Rapamycin decelerates cellular senescence. Cell Cycle. 2009;8:1888–95. doi: 10.4161/cc.8.12.8606. [DOI] [PubMed] [Google Scholar]
- 66.Kolesnichenko M, Hong L, Liao R, Vogt PK, Sun P. Attenuation of TORC1 signaling delays replicative and oncogenic RAS-induced senescence. Cell Cycle. 2012;11:2391–401. doi: 10.4161/cc.20683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chen T, Shen L, Yu J, Wan H, Guo A, Chen J, Long Y, Zhao J, Pei G. Rapamycin and other longevity-promoting compounds enhance the generation of mouse induced pluripotent stem cells. Aging Cell. 2011;10:908–11. doi: 10.1111/j.1474-9726.2011.00722.x. [DOI] [PubMed] [Google Scholar]
- 68.Feng B, Ng JH, Heng JC, Ng HH. Molecules that promote or enhance reprogramming of somatic cells to induced pluripotent stem cells. Cell Stem Cell. 2009;4:301–12. doi: 10.1016/j.stem.2009.03.005. [DOI] [PubMed] [Google Scholar]
- 69.González F, Georgieva D, Vanoli F, Shi ZD, Stadtfeld M, Ludwig T, Jasin M, Huangfu D. Homologous recombination DNA repair genes play a critical role in reprogramming to a pluripotent state. Cell Rep. 2013;3:651–60. doi: 10.1016/j.celrep.2013.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fong YW, Inouye C, Yamaguchi T, Cattoglio C, Grubisic I, Tjian R. A DNA repair complex functions as an Oct4/Sox2 coactivator in embryonic stem cells. Cell. 2011;147:120–31. doi: 10.1016/j.cell.2011.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Molina-Estevez FJ, Lozano ML, Navarro S, Torres Y, Grabundzija I, Ivics Z, Samper E, Bueren JA, Guenechea G. Brief report: impaired cell reprogramming in nonhomologous end joining deficient cells. Stem Cells. 2013;31:1726–30. doi: 10.1002/stem.1406. [DOI] [PubMed] [Google Scholar]
- 72.Loh YH, Wu Q, Chew JL, Vega VB, Zhang W, Chen X, Bourque G, George J, Leong B, Liu J, et al. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat Genet. 2006;38:431–40. doi: 10.1038/ng1760. [DOI] [PubMed] [Google Scholar]
- 73.Noh KH, Kim BW, Song KH, Cho H, Lee YH, Kim JH, Chung JY, Kim JH, Hewitt SM, Seong SY, et al. Nanog signaling in cancer promotes stem-like phenotype and immune evasion. J Clin Invest. 2012;122:4077–93. doi: 10.1172/JCI64057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jeter CR, Liu B, Liu X, Chen X, Liu C, Calhoun-Davis T, Repass J, Zaehres H, Shen JJ, Tang DG. NANOG promotes cancer stem cell characteristics and prostate cancer resistance to androgen deprivation. Oncogene. 2011;30:3833–45. doi: 10.1038/onc.2011.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chiou SH, Wang ML, Chou YT, Chen CJ, Hong CF, Hsieh WJ, Chang HT, Chen YS, Lin TW, Hsu HS, et al. Coexpression of Oct4 and Nanog enhances malignancy in lung adenocarcinoma by inducing cancer stem cell-like properties and epithelial-mesenchymal transdifferentiation. Cancer Res. 2010;70:10433–44. doi: 10.1158/0008-5472.CAN-10-2638. [DOI] [PubMed] [Google Scholar]
- 76.Chang CC, Shieh GS, Wu P, Lin CC, Shiau AL, Wu CL. Oct-3/4 expression reflects tumor progression and regulates motility of bladder cancer cells. Cancer Res. 2008;68:6281–91. doi: 10.1158/0008-5472.CAN-08-0094. [DOI] [PubMed] [Google Scholar]
- 77.Lu X, Mazur SJ, Lin T, Appella E, Xu Y. The pluripotency factor nanog promotes breast cancer tumorigenesis and metastasis. Oncogene. 2013 doi: 10.1038/onc.2013.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Watanabe M, Ohnishi Y, Inoue H, Wato M, Tanaka A, Kakudo K, Nozaki M. NANOG expression correlates with differentiation, metastasis and resistance to preoperative adjuvant therapy in oral squamous cell carcinoma. Oncol Lett. 2014;7:35–40. doi: 10.3892/ol.2013.1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ben-Porath I, Thomson MW, Carey VJ, Ge R, Bell GW, Regev A, Weinberg RA. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat Genet. 2008;40:499–507. doi: 10.1038/ng.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Erenpreisa J, Salmina K, Huna A, Kosmacek EA, Cragg MS, Ianzini F, Anisimov AP. Polyploid tumour cells elicit paradiploid progeny through depolyploidizing divisions and regulated autophagic degradation. Cell Biol Int. 2011;35:687–95. doi: 10.1042/CBI20100762. [DOI] [PubMed] [Google Scholar]
- 81.Puig PE, Guilly MN, Bouchot A, Droin N, Cathelin D, Bouyer F, Favier L, Ghiringhelli F, Kroemer G, Solary E, et al. Tumor cells can escape DNA-damaging cisplatin through DNA endoreduplication and reversible polyploidy. Cell Biol Int. 2008;32:1031–43. doi: 10.1016/j.cellbi.2008.04.021. [DOI] [PubMed] [Google Scholar]
- 82.Martin F, Puig PE, Ghiringhelli F, Chauffert B. DNA-damaged polyploid cancer cells can reverse to diploidy: an ordered, but little understood, process of genomic reduction [with reference to the previous comments of Forer (2008) and Wheatley (2008a and b)] Cell Biol Int. 2009;33:702–3. doi: 10.1016/j.cellbi.2009.02.016. [with reference to the previous comments of Forer (2008) and Wheatley (2008a and b)] [DOI] [PubMed] [Google Scholar]
- 83.Pospelova TV, Chitikova ZV, Pospelov VA. An integrated approach for monitoring cell senescence. Methods Mol Biol. 2013;965:383–408. doi: 10.1007/978-1-62703-239-1_26. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.