Highlights
-
•
A truncated kinase suppressor of Ras 2 (T-KSR2) mRNA was identified.
-
•
T-KSR2 is expressed exclusively in mouse testes and sperm.
-
•
Analysis of T-KSR2 may enhance understanding of spermatogenesis and fertility.
Abbreviations: KSR, kinase suppressor of Ras; ERK, extracellular signal-regulated protein kinase; T-KSR2, testes-kinase suppressor of Ras 2; MAPK, mitogen activated protein kinase; CA, conserved area; SH2, Src homology 2; RACE, rapid amplification of cDNA ends; NDK-1, nucleoside diphosphate kinase 1
Keywords: KSR2, Scaffold, Sperm
Abstract
The kinase suppressor of Ras 2 (KSR2) is a scaffold protein for the extracellular signal-regulated protein kinase (ERK) signaling pathway. KSR2 mediates germline mpk-1 (Caenorhabditis elegans ERK) phosphorylation in C. elegans and has been implicated the regulation of meiosis. KSR2−/− mice exhibit metabolic abnormalities and are reproductively impaired. The role of KSR2 in meiosis and fertility in mice has yet to be elucidated. Here, we describe a novel truncated KSR2 mRNA identified in mouse testes (T-KSR2). Further analysis demonstrates T-KSR2 is specific to mouse testes and mature sperm cells. The detection of T-KSR2 may enhance our understanding of mechanisms controlling spermatogenesis and fertility.
1. Introduction
Kinase suppressor of Ras 1 and 2 (KSR1 and KSR2) are scaffold proteins for the Raf/MEK/ERK Mitogen Activated Protein Kinase (MAPK) signaling pathway [1–3]. KSR1−/− mice are overtly normal but resistant to Ras-induced tumors [4,5]. KSR2−/− adult mice are profoundly obese and insulin resistant [6,7]. Similarly, humans with KSR2 mutations have early onset obesity and severe insulin resistance [8]. The extent of murine KSR1 and KSR2 homology is contained within five conserved areas (CA). The N-terminus contains the CA1 domain, a domain unique to the KSR family; the CA2 domain is proline-rich and contains a Src homology 2 (SH2) domain; the CA3 domain is cysteine-rich and responsible for mediating the translocation of KSR protein to the plasma membrane; the CA4 is a serine/threonine-rich domain that contains the ERK binding motif; and the CA5 domain, which is located in the C-terminus end of KSR proteins, is a kinase-like domain and mediates the interaction with MEK [1]. In Caenorhabditis elegans, ksr1 and ksr2 are required for Ras-mediated signaling [3]. Although the two members coordinately regulate Ras signaling, these genes have distinct effects on fertility in C. elegans. Disruption of ksr1 in C. elegans results in fertile offspring while ksr2 disruption causes sterility. ksr2 is specifically required for Ras-mediated signaling during germline meiotic progression in C. elegans. Without KSR2, oogenesis is arrested at the pachytene stage [3]. Proteins that interact with and phosphorylate KSR1 and KSR2 regulate Ras/MAPK activity to regulate C. elegans development. Nucleoside diphosphate kinase, NDK-1, regulates vulva development in C. elegans by direct physical interaction with KSR1 and KSR2 [9–11].
KSR1−/− mice are also fertile and develop normally, but do exhibit enlarged adipocytes, altered hair follicles, and modest defects in T cell activation [4,5,12]. However, KSR2 plays a larger role in reproduction, as male and female KSR2−/− mice exhibit impaired fertility [6]. KSR2−/− females begin estrous cycles later than WT females and have impaired mammary development, while KSR2−/− males have reduced sex drive and copulate infrequently (unpublished observations). These studies suggest KSR2 plays an important role in regulating fertility and metabolism in mammalian animals.
KSR1 protein is expressed in the brain, spleen, bladder, ovary, testis, and lung. However, a variant form of KSR1, B-KSR1, has been identified in brain tissue [13,14]. KSR1 functions in mediating Ras-induced cell proliferation, cell transformation, and survival. B-KSR1, which has a longer CA4 domain and a truncated C-terminus relative to KSR1, is critical in mediating Ras-dependent signaling to promote neurite growth and to maintain neuronal differentiation [14]. The paralog ksr2 gene was first discovered in C. elegans and was found to have two alternative spliced forms, with one variant having shorter CA1 and CA4 domains [3]. Human KSR2 was also found to have two alternative spliced forms that varied from the full-length 950 amino acids. One variant lacks the first 29 amino acids (hKSR2ΔN29) and the second identified variant (hKSR2ΔCA1) lacks the CA1 domain. The hKSR2ΔCA1 cDNA clones were obtained from kidney and testes cDNA libraries. Northern blot analysis revealed hKSR2ΔCA1 mRNA expression in human brain and kidney tissue [15]. hKSR2ΔCA1 has been described as a regulator of proto-oncogene Cot-induced MAPK signaling. In mice, KSR2 protein is detected in the brain and pancreas [6,16]. A murine homolog of hKSR2ΔCA1 cDNA was detected in a mouse kidney cDNA library [15]. These data suggest that mouse KSR2 can be alternatively spliced and variant KSR2 expression is tissue-dependent.
In this study, we describe and characterize a truncated KSR2 mRNA in mouse testis. This variant form of KSR2 lacks the CA1 and CA2 domains, encoding a predicted 598 amino acids. We determined that this truncated mRNA leads to stable protein expression in vitro.
2. Materials and methods
2.1. Animals and tissue collection
Mice were housed in pathogen-free conditions and experiments were carried out under a protocol approved by the University of Nebraska Medical Center Institutional Animal Care and Use Committee (University of Nebraska Medical Center, Omaha, NE). Mice were maintained on a 12 h light/dark schedule with free access to laboratory chow (ad libitum) and water. Mice were sacrificed by the administration of CO2 followed by cervical dislocation. Tissues from 8 to 12 week old C57BL/6J mice were dissected, frozen on dry ice, and stored at −80 °C until used.
2.2. Sperm purification
Sperm were collected as previously described [17]. Briefly, the caudal epididymis was minced and the sperm were allowed to swim out in phosphate buffered saline (PBS). The sperm-containing PBS was gently aspirated and collected by centrifugation at 800×g for 5 min at room temperature. Sperm were then lysed in TRI reagent (Molecular Research Center Ins, TR118). Sperm were 99% pure as assessed by light microscope. To remove any potential somatic cells, the sperm were centrifuged at 800×g for 5 min and the pellet was treated with a hypotonic buffer (0.1% SDS, 0.5% Triton X100 in deionised water) for half hour, as previously described [18]. The sample was centrifuged at 600×g for 15 min at 4 °C. The supernatant was removed and the sample washed twice with PBS, then centrifuged at 600×g for 5 min at 4 °C.
2.3. RNA isolation and cDNA synthesis
Total RNA from mouse tissues was isolated with an RNeasy mini kit (Qiagen) according to the manufacturer’s protocol with modification on the lysis step as previously described [19]. Blood samples were lysed in Tri reagent BD (MRC, TB126). Due to the low level of RNA in hypotonic buffer-treated sperm, yeast tRNA was added as carrier RNA during lysis. RNA was treated with DNase I (Ambion, AM1906) before cDNA synthesis. cDNA from total RNA was produced with M-MLV reverse transcriptase (Ambion, AM2043). PCR was done with Herculase II DNA polymerase (Agilent, 600675-5).
2.4. Rapid amplification of cDNA 5′ ends (5′ RACE)
Ten micrograms of total RNA was used as template for the FirstChoice RLM-RACE kit (Ambion, AM1700). Briefly, RNA was treated by Calf Intestine Alkaline Phosphatase (CIP), followed by Tobacco Acid Pyrophosphatase (TAP) and adaptor ligation. A no TAP (–TAP) control was used to ensure the 5′ RACE products are from full-length mRNA. A primary and a nested PCR were performed with 5́ RACE adapter primers provided by the kit (modified by changing the BamH1 restriction enzyme site into an EcoR1 site) and gene specific reverse primers. The PCR products were cloned into the vector pcDNA3.1(−) and several clones were isolated for sequencing.
Primer sequences (Kpn1 and EcoR1 sites in bold, PYO tag in Italic):
Gene-specific RACE inner primer R460: 5′-GAT TAT CCA CAG AGG AGA CCC GGT ACC GG-3′
Gene specific RACE outer primer R490: 5′-GTC AGA CTC TCC CCA AAA CC-3′
KSR2 F610: 5′-CC GAA TTC CAA CCT CCG AGA ACG AAG AG-3′
KSR2 Rstop: 5′-GC GGT ACC TCA CAG CTC TGC AGA CTT CCA GAA ATG TCC-3′
T-KSR2 5UTR: 5′-CG GAA TTC AAT GTA TCA GGC GCT TTG CCG AAC AC-3′
KSR2 F9: 5′-CGA AAA GCG AAG AGC AGC AAC-3′
KSR2 R207: 5′-CG GAA TTC GGC TGG TAG GAC AGA AGT GC-3′
GAPDH F: 5′-AGG CCG GTG CTG AGT ATG TC-3′
GAPDH R: 5′-TGC CTG CTT CAC CAC CTT CT-3′
PYO-KSR2 R: 5′-GC GGT ACC TCA CTC CAT TGG CAT GTA CTC CAT CTC CAT TGG CAT GTA CTC CAT CAG CTC TGC AGA CTT CCA GAA ATG TCC-3′
Protamine-1 F: 5′-AGC AAA GCA GGA GCA GAT G-3′
Protamine-1 R: 5′-GGC GAG ATG CTC TTG AAG TC-3′
E-cadherin F: 5′-CAG CTC CTT CCC TGA GTG TG-3′
E-cadherin R: 5′-TGC ACC CAC ACC AAG ATA CC-3′
c-kit F: 5′-AAC GAT GTG GGC AAG AGT TC-3′
c-kit R: 5′- CCT CGA CAA CCT TCC ATT GT-3′
2.5. Cell culture and transfection
HEK293T cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% fetal bovine serum. Cells were passaged every 2–3 days. Both T-KSR2 and full-length KSR2 were infused with polyoma virus-derived (PYO) epitope tag and were cloned into pcDNA3(−) vectors. Two copies of the PYO epitope tag (amino acids MEYMPME) were included in the 3′ primer immediately upstream of the stop codon. Transfections were performed utilizing Lipofectamine 2000 (Life Technologies) according to the manufacturer’s protocol.
2.6. Western blots
Western blots were done as described previously [20] with slight modifications. Briefly, cells were lysed in buffer containing 1% Igepal, 20 mM Tris (pH 8), 137 mM NaCl, 10% glycerol, 10 μg/ml aprotinin, 20 nM leupeptin, 0.5 mM sodium orthovanadate, 2 mM EDTA, 10 mM sodium fluoride, and 1 mM phenylmethylsulfonyl fluoride (PMSF). Equal amounts of protein were resolved using sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS–PAGE) and transferred to nitrocellulose membranes. Primary PYO antibody was obtained from cultured hybridoma cells as described previously [21]. Proteins were detected using Odyssey imaging system (LI-COR Biosciences).
3. Results
3.1. A truncated KSR2 transcript in testes
By analyzing a number of 5′ RACE products from testes, an alternate KSR2 mRNA (T-KSR2) (GenBank: KJ719253) was detected exclusively in testes (Fig. 1A). The predicted first exon of T-KSR2 (exon1t) resides within intron 5 of full-length KSR2, contains a predicted 62 base pair (bp) 5′ UTR region, and is followed by a start code and a predicted novel 55 bp coding sequence in the same reading frame as the full-length KSR2 mRNA (Fig. 1C and D). Using the exon1t-specific forward primer (T-KSR2 5UTR) and a reverse primer targeting the last exon of KSR2 (Rstop), the T-KSR2 was confirmed by PCR and sequencing to contain exons 6–20 of full-length KSR2 mRNA (Fig. 1B). In a search of NCBI databases, no human genomic sequences identical to the unique exon1t of T-KSR2 were found. However, we did detect an alternative human KSR2 RNA (KSR2b) expressed specifically in testes (Fig. 1C) (GenBank: AK098831.1). The first two exons of human KSR2b are located in intron 4, and the last exon located in intron 14. This RNA encodes sequences similar to T-KSR2, but with a shorter predicted CA5 kinase-like domain (Fig. 3B).
Fig. 1.
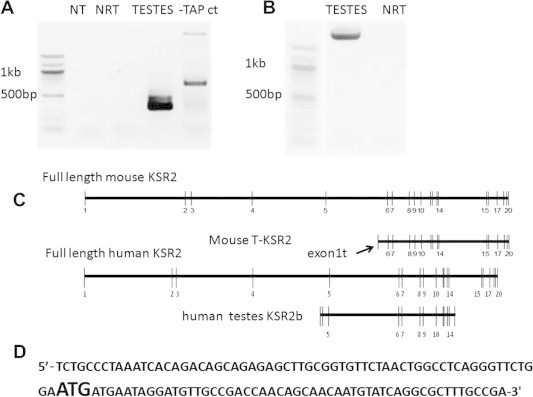
T-KSR2 is expressed in testes. (A) RT-PCR from the nested PCR with 5′RACE inner primer and R460. NT, no temperature control; NRT, No reverse transcriptase control; testes, testes RNA sample; -TAP ct, no Tobacco Acid Pyrophosphatase (TAP) control. (B) RT-PCR with T-KSR2 using F5UTR and Rstop from testes RNA. (C) The genomic structure of mouse KSR2, T-KSR2, human KSR2, human testes specific KSR2b. Truncated human testes KSR2b sequence is from NCBI gene bank AK098831.1. (D) The sequence of the 5′UTR and exon1t of T-KSR2. The initiation codon is in bold large font.
Fig. 3.
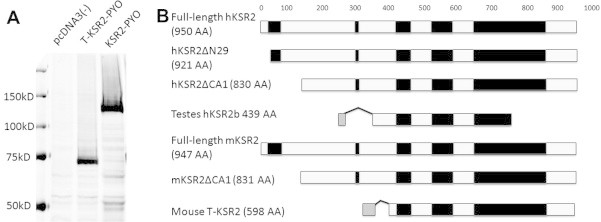
T-KSR2 can be expressed as protein in vitro. (A) Pyo-tagged full-length mouse KSR2, T-KSR2, and vector only were overexpressed in HEK293T cells and examined for protein expression by blotting with Pyo tag antibody. (B) Schematic illustration of the human and mouse KSR2 homologs. Five conserved domains (CA1–CA5) are represented in black. The novel domains of T-KSR2 and human testes KSR2 are in gray. hKSR2: human KSR2; mKSR2: mouse KSR2.
3.2. T-KSR2 transcript is expressed only in testes and sperm
Since KSR2 mRNA is also detected in other tissues such as kidney and ovary (Fig. 2B), we sought to identify other tissues in which T-KSR2 might be expressed. Primers targeting sequences encoding the N-terminus of KSR2 (F9 and R207), the C-terminus of KSR2 (F610 and R860), and T-KSR2 (F5UTR and 3R) were used to probe for full-length KSR2 only, both forms, and T-KSR2 RNA only respectively in 16 different tissues (Fig. 2A). T-KSR2 RNA expression was only detected in testes (Fig. 2B). These data suggest that T-KSR2 RNA expression is limited to testes.
Fig. 2.
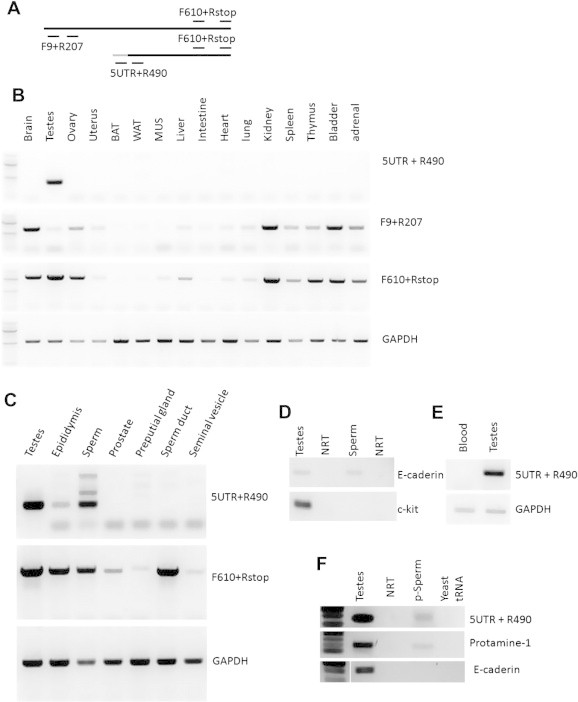
T-KSR2 is expressed exclusively in testes and mature sperm. (A) Primer pairs used for detecting KSR2 mRNA. F9 and 207R selectively detect full-length KSR2. F5UTR and R490 primer pairs selectively detect T- KSR2. F610 and Rstop primer pairs detect both mRNAs. T-KSR2 exon1t which can be recognized only by F5UTR primer is in gray. (B) RT-PCR was performed with primers described in panel A. RNA was isolated from the indicated tissues from 8 to 12 week old wild type C57BL/6 mice. BAT: brown adipose tissue, WAT: white adipose tissue, MUS: Quadriceps muscle. (C) Male reproductive tracts were dissected from 12 week old mice. RNA was purified and RT-PCR was done with the primer pairs illustrated in panel A. (D) RT-PCR was performed with primers detecting somatic cell marker E-cadherin and round germ cell marker c-kit. NRT: no reverse transcriptase control for the previous sample. (E) Blood samples were tested for T-KSR2 expression. (F) RT-PCR was performed with purified sperm (p-Sperm) treated with hypotonic buffer for detecting T-KSR2, protamine-1, and E-cadherin expression. Yeast tRNA was used as a negative control. NRT: no reverse transcriptase control for testes.
We next assessed whether T-KSR2 mRNA expression extended to other tissues of the male reproductive system including the epididymis, preputial gland, seminal vesicle, and sperm duct. Mature sperm were isolated from the epididymis by cutting the epididymis caudally and allowing the sperm to swim out. RNA was purified from the above tissues and the RT-PCR results show that in addition to the testes, mature sperm also express T-KSR2 mRNA (Fig. 2C). The epididymis shows a low level of T-KSR2 mRNA, which may result from residual sperm left contaminating the epididymal tubes. However other tissues within the male reproductive tract do not express T-KSR2 mRNA.
Since somatic cells have much higher RNA content than the sperm cells, trace amount of contamination may affect the results. Contamination of sperm preparations consists of fat cells, blood cells, and round germ cells [22]. To test for contamination of the sperm samples, E-cadherin (detected in all three cell types) and c-kit (detected in round germ cells) were used as markers of somatic cells and round germ cells. E-cadherin contamination was detected, but c-kit was not be detected (Fig. 2C). Our results suggest that the sperm sample may contain blood and fat cell, but do not have round germ cell contamination. T-KSR2 is not expressed in fat tissue (Fig. 2A). We further tested RNA purified from blood. As Fig. 2D shown, RNA purified from blood does not contain T-KSR2. These observations indicate that T-KSR2 is expressed selectively in mature sperm. To further confirm that T-KSR2 is expressed in mature sperm cells, we treated collected sperm with hypotonic buffer to remove any somatic cells. As shown in Fig. 2E, purified sperm (p-Sperm) does not have somatic cell contamination, as E-Cadherin is not detected. Protamine1, one of the abundant RNAs in sperm is readily detected with T-KSR2 in purified mature sperm cells.
3.3. T-KSR2 transcript can be expressed in vitro
Since T-KSR2 mRNA is expressed in testes and mature sperm, we next asked if the transcript could be translated into protein. To determine protein expression, HEK293T cells were transfected with a PYO-tagged in-frame fused T-KSR2. After cell lysis, western blot analysis was performed using an antibody that recognizes the PYO tag to detect a protein of the size predicted for T-KSR2 (Fig. 3A). We assessed the expression of KSR2 protein in mouse testes. Western blot analysis was performed in whole cell lysates with a KSR2 polyclonal antibody whose antigen encompasses a region in between the CA3 and CA5 domain of full-length KSR2 and T-KSR2. Using this antibody, we did not detect T-KSR2 protein expression in mouse testes (data not shown). These data suggested that T-KSR2 is either not expressed in testes or is expressed at levels below the sensitivity of available antibodies, which is the case for expression of KSR2 in tissues other than brain [6]. The predicted protein of the known human and mouse KSR2 alternative spliced mRNA is shown in Fig. 3B.
4. Discussion
We reveal a novel truncated RNA of KSR2, T-KSR2, which is specifically expressed in mouse testes and mature sperm. Humans also have a similar truncated form of KSR2 (KSR2b) expressed in testes. Instead of containing exon1t, KSR2b contains 2 extra exons from intron 4 of full-length human KSR2, and the last exon of KSR2b is from intron 14 of full-length human KSR2. Despite the differences between human KSR2b and mouse T-KSR2 RNA at the 3′- and 5′-terminal sequences, they contain similar domains as predicted proteins, with KSR2b having a shortened CA5 domain.
Based on the conserved structural similarity to KSR1 and a KSR2/MEK co-crystal structure [23], the KSR2 CA5 domain constitutively binds MEK and regulates MEK activity [23]. After growth factor stimulation, KSR1 and KSR2 are recruited to the plasma membrane, where KSR proteins interact with B-Raf mediated by the CA5 domain [24,25]. The B-Raf/KSR interaction will promote MEK and ERK phosphorylation, activating the kinase cascade. To ensure temporal regulation of ERK signaling, ERK phosphorylates KSR1 at T260, T274, S320, S443 localized in the region between CA1 and CA2 to CA4 domains to promote B-Raf/KSR dissociation, which leads to pathway inactivation [24]. The phosphorylated KSR proteins will promote the dissociation of KSR protein from the plasma membrane and translocation to the cytosol. The predicted T-KSR2 protein lacks 3 of the 4 ERK phosphorylation sites, which may affect the localization and change the duration and intensity of the ERK pathway. However, this hypothetical mechanism will only apply if T-KSR2 binds MEK and ERK, and translocates to the plasma membrane, T-KSR2 lacks the Coiled Coil-Sterile α Motif (CC-SAM), located between the CA1 and CA2 domains, and found to be necessary for localization to the plasma membrane [25]. It is also possible that T-KSR2 functions as an inhibitor to full-length KSR2 by occupying MEK and prevents the localization of full-length KSR2 to the plasma membrane.
Nucleoside diphosphate kinase 1 (NDK-1) was reported to involve vulva development by interacting with KSR2 in C. elegans [9]. T-KSR2 was detected in mouse testes and sperm. We do not know if C. elegans also expresses a truncated KSR2. T-KSR2 contains sites phosphorylated by NDK-1 homolog, NM23-H1, and NDK-1 was reported to be involved in vulva development by interacting with KSR2. However, our inability to detect T-KSR2 in ovary and uterus suggests T-KSR2 is not related to female infertility.
Messenger RNA in mature spermatozoa is low in humans and is thought to be a remnant of untranslated mRNA during spermatogenesis [26]. However, accumulated mRNA in mature human ejaculates constitute stable transcripts that are consistently expressed from individual men. Most spermatozoan RNAs encode proteins participating in signal transduction, oncogenesis, and cell proliferation, in which KSR2 and its related pathway also function [27]. Selective retention of particular RNA species has been proposed and may suggest the function of these mRNAs [28]. The observation that sperm RNA was detected in zygotes at 30 min and 3 h post-fertilization further support that sperm RNA may have important roles in early zygotic and embryonic development [22,29]. T-KSR2 mRNA can be translated into protein in HEK293T cells, where cells have active translation. Although we cannot detect the T-KSR2 protein in testes, the presence of T-KSR2 mRNA in mature sperm may imply T-KSR2 mRNA is translated during early zygotic development, when translation is active. KSR2 mediates Ras-to-ERK signaling due to calcium influx. Calcium influx promotes the dephosphorylation of Ser471 on KSR2 by calcineurin, which promotes the translocation of KSR2 to the plasma membrane [16]. The predicted murine T-KSR2 protein contains the calcineurin LXVP binding motif and it is predicted to interact with calcineurin. Therefore, it is conceivable that T-KSR2 affects Ca++-induced ERK signaling in the germline.
In summary, we identified a translatable truncated KSR2 mRNA in mouse testes and mature sperm. The data suggest that this variant of KSR2 may play a unique role in male reproduction. Identifying the specific RNA expressed in spermatozoa may facilitate investigation into the mechanisms that cause infertility by using high throughput methods such as microarray technology.
Acknowledgements
We thank Dr. Mario Fernandez for his help in preparing the manuscript and Dr. MaLinda Henry for help in sperm collection. This research was supported by the NIH grant R01 CA090400 and U.S. Army award W081XWH-10-1-0139.
References
- 1.Therrien M. KSR, a novel protein kinase required for RAS signal transduction. Cell. 1995;83(6):879–888. doi: 10.1016/0092-8674(95)90204-x. [DOI] [PubMed] [Google Scholar]
- 2.Michaud N.R. KSR stimulates Raf-1 activity in a kinase-independent manner. Proc. Natl. Acad. Sci. U.S.A. 1997;94(24):12792–12796. doi: 10.1073/pnas.94.24.12792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ohmachi M. C. elegans ksr-1 and ksr-2 have both unique and redundant functions and are required for MPK-1 ERK phosphorylation. Curr. Biol. 2002;12(5):427–433. doi: 10.1016/s0960-9822(02)00690-5. [DOI] [PubMed] [Google Scholar]
- 4.Lozano J. Deficiency of kinase suppressor of Ras1 prevents oncogenic ras signaling in mice. Cancer Res. 2003;63(14):4232–4238. [PubMed] [Google Scholar]
- 5.Nguyen A. Kinase suppressor of Ras (KSR) is a scaffold which facilitates mitogen-activated protein kinase activation in vivo. Mol. Cell Biol. 2002;22(9):3035–3045. doi: 10.1128/MCB.22.9.3035-3045.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Costanzo-Garvey D.L. KSR2 is an essential regulator of AMP kinase, energy expenditure, and insulin sensitivity. Cell Metab. 2009;10(5):366–378. doi: 10.1016/j.cmet.2009.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Revelli J.P. Profound obesity secondary to hyperphagia in mice lacking kinase suppressor of ras 2. Obesity (Silver Spring) 2011;19(5):1010–1018. doi: 10.1038/oby.2010.282. [DOI] [PubMed] [Google Scholar]
- 8.Pearce L.R. KSR2 mutations are associated with obesity, insulin resistance, and impaired cellular fuel oxidation. Cell. 2013;155(4):765–777. doi: 10.1016/j.cell.2013.09.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Masoudi N. The NM23-H1/H2 homolog NDK-1 is required for full activation of Ras signaling in C. elegans. Development. 2013;140(16):3486–3495. doi: 10.1242/dev.094011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hartsough M.T. Nm23-H1 metastasis suppressor phosphorylation of kinase suppressor of Ras via a histidine protein kinase pathway. J. Biol. Chem. 2002;277(35):32389–32399. doi: 10.1074/jbc.M203115200. [DOI] [PubMed] [Google Scholar]
- 11.Salerno M. Nm23-H1 metastasis suppressor expression level influences the binding properties, stability, and function of the kinase suppressor of Ras1 (KSR1) Erk scaffold in breast carcinoma cells. Mol. Cell Biol. 2005;25(4):1379–1388. doi: 10.1128/MCB.25.4.1379-1388.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kortum R.L. The molecular scaffold kinase suppressor of Ras 1 (KSR1) regulates adipogenesis. Mol. Cell Biol. 2005;25(17):7592–7604. doi: 10.1128/MCB.25.17.7592-7604.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Giblett S.M. Expression of kinase suppressor of Ras in the normal adult and embryonic mouse. Cell Growth Differ. 2002;13(7):307–313. [PubMed] [Google Scholar]
- 14.Muller J. Identification of B-KSR1, a novel brain-specific isoform of KSR1 that functions in neuronal signaling. Mol. Cell Biol. 2000;20(15):5529–5539. doi: 10.1128/mcb.20.15.5529-5539.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Channavajhala P.L. Identification of a novel human kinase supporter of Ras (hKSR-2) that functions as a negative regulator of Cot (Tpl2) signaling. J. Biol. Chem. 2003;278(47):47089–47097. doi: 10.1074/jbc.M306002200. [DOI] [PubMed] [Google Scholar]
- 16.Dougherty M.K. KSR2 is a calcineurin substrate that promotes ERK cascade activation in response to calcium signals. Mol. Cell. 2009;34(6):652–662. doi: 10.1016/j.molcel.2009.06.00. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cao W. Sorbitol can fuel mouse sperm motility and protein tyrosine phosphorylation via sorbitol dehydrogenase. Biol. Reprod. 2009;80(1):124–133. doi: 10.1095/biolreprod.108.068882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goodrich R., Johnson G., Krawetz S.A. The preparation of human spermatozoal RNA for clinical analysis. Arch. Androl. 2007;53(3):161–167. doi: 10.1080/01485010701216526. [DOI] [PubMed] [Google Scholar]
- 19.Guo L., Eviatar-Ribak T., Miskimins R. Sp1 phosphorylation is involved in myelin basic protein gene transcription. J. Neurosci. Res. 2010;88(15):3233–3242. doi: 10.1002/jnr.22486. [DOI] [PubMed] [Google Scholar]
- 20.Fernandez M.R., Henry M.D., Lewis R.E. Kinase suppressor of Ras 2 (KSR2) regulates tumor cell transformation via AMPK. Mol. Cell Biol. 2012;32(18):3718–3731. doi: 10.1128/MCB.06754-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McKay M.M., Morrison D.K. Proteomic analysis of scaffold proteins in the ERK cascade. Methods Mol. Biol. 2010;661:323–334. doi: 10.1007/978-1-60761-795-2_19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hamatani T. Human spermatozoal RNAs. Fertil. Steril. 2012;97(2):275–281. doi: 10.1016/j.fertnstert.2011.12.035. [DOI] [PubMed] [Google Scholar]
- 23.Brennan D.F. A Raf-induced allosteric transition of KSR stimulates phosphorylation of MEK. Nature. 2011;472(7343):366–369. doi: 10.1038/nature09860. [DOI] [PubMed] [Google Scholar]
- 24.McKay M.M., Ritt D.A., Morrison D.K. Signaling dynamics of the KSR1 scaffold complex. Proc. Natl. Acad. Sci. U.S.A. 2009;106(27):11022–11027. doi: 10.1073/pnas.0901590106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koveal D. A CC-SAM, for coiled coil-sterile alpha motif, domain targets the scaffold KSR-1 to specific sites in the plasma membrane. Sci. Signal. 2012;5(255):ra94. doi: 10.1126/scisignal.2003289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lalancette C. Identification of human sperm transcripts as candidate markers of male fertility. J. Mol. Med. (Berl) 2009;87(7):735–748. doi: 10.1007/s00109-009-0485-9. [DOI] [PubMed] [Google Scholar]
- 27.Dadoune J.P. Identification of transcripts by macroarrays, RT-PCR and in situ hybridization in human ejaculate spermatozoa. Mol. Hum. Reprod. 2005;11(2):133–140. doi: 10.1093/molehr/gah137. [DOI] [PubMed] [Google Scholar]
- 28.Moldenhauer J.S. Diagnosing male factor infertility using microarrays. J. Androl. 2003;24(6):783–789. doi: 10.1002/j.1939-4640.2003.tb03122.x. [DOI] [PubMed] [Google Scholar]
- 29.Gur Y., Breitbart H. Mammalian sperm translate nuclear-encoded proteins by mitochondrial-type ribosomes. Genes Dev. 2006;20(4):411–416. doi: 10.1101/gad.367606. [DOI] [PMC free article] [PubMed] [Google Scholar]


