Highlights
-
•
miRNA expression arrays and RNA Seq were employed for unbiased spatial analyses of miRNAs.
-
•
Small RNomics of subcellular compartments revealed the presence of miRNAs in the nucleolus.
-
•
Several miRNAs were detected at low abundance in cancer cell nucleoli.
-
•
The nucleolar abundance of miR-31 was dependent on CRM1 export factor.
Abbreviations: ActD, Actinomycin D; CRM1, chromosomal maintenance 1; LNA, locked nucleic acid; pre, precursor; qPCR, quantitative real-time PCR; Pol I, RNA polymerase I; rRNA, ribosomal RNA; siRNA, short interfering RNA; snoRNA, small nucleolar RNA
Keywords: Small RNA, Nucleolus, Deep sequencing, miRNA expression profiling
Abstract
Micro RNAs (miRNA) are non-coding RNAs expressed in the cytoplasm as their mature, 21–22-nucleotide short forms. More recently, mature miRNAs have also been detected in the nucleus, raising the possibility that their spatial distribution may be more complex than anticipated. Here we undertook comprehensive systematic analyses of miRNA distribution in several subcellular compartments of human cancer cells. In particular, we focused on the potential presence of miRNAs in the nucleolus, which contains an abundance of small non-coding RNAs. We employed two miRNA expression array platforms and small RNA deep sequencing of small RNAs isolated from cells, nuclei, cytoplasm and the nucleoli. We developed an assay to compare RNAs of isolated nucleoli before and after denaturation and used Northern hybridization to verify the presence of miRNAs in the subcellular compartments. Consistently, we found more than 10 miRNAs associated with the nucleolar preparations. Several miRNAs had greater relative abundance in the nucleolus compared to the other compartments. The nucleolar presence of miRNAs was independent of Dicer and the main activity of the nucleolus, RNA polymerase I transcription, but was dependent on CRM1 previously associated with nucleolar trafficking of small nucleolar RNAs. These results highlight the complexity of miRNA spatial arrangement and regulation.
1. Introduction
MicroRNAs (miRNAs) are evolutionary conserved single-stranded ∼22 nt RNA species implicated in a broad range of physiological and developmental processes [1,2]. MiRNAs are transcribed mainly by RNA polymerase II as long primary transcripts, processed by Drosha and DGCR8 microprocessor complex, and transported to the cytoplasm by the nuclear export factor exportin-5 [3]. The cytoplasmic precursor (pre) miRNA is further cleaved by Dicer to ∼22 nt RNA duplex and loaded by Argonaute proteins to generate the RISC silencing complex [3]. MiRNAs recognize their target mRNAs in their 3′UTRs and thus act in the cytoplasm to repress mRNA translation and to accelerate mRNA decay [1]. The canonical miRNA processing pathway is extensively posttranscriptionally modified by proteins that interfere or augment the microprocessor or Dicer activity and miRNA stability [4–6]. Many of these processes intersect with regulation and synthesis of other short RNAs, most notably short interfering RNAs (siRNAs) and mirtrons [3]. MiRNAs are expressed in tissue-specific manner and depend on developmental stage, but their regulation is frequently abnormal in human disease [7–11].
Based on their known functions, mature miRNAs have been mainly considered cytoplasmic. However, miRNA nuclear localization signals have been identified [12], and RNA deep sequencing and expression analyses have indicated that mature forms co-exist both in the cytoplasm and the nucleus [13]. Further, a few miRNAs have been identified with presumably nuclear functions [14,15]. MiRNAs have been described in the nucleoli of rat myoblasts, mouse Sertoli cells and human cancer cell lines [16–19], and presence of precursor miRNA forms have been proposed, but not confirmed, in the nucleoli [17,20,21]. MiRNA-binding and processing proteins are detected in multiple cellular compartments raising the possibility that miRNAs may display more complex cellular locations, and possibly, associated functions, than anticipated [22–24].
The nucleolus has a uniquely rich RNA content composed mainly of ribosomal (r) RNAs and small nucleolar (sno) RNAs. RNA polymerase I (Pol I) transcription is the main metabolic activity of the nucleolus that entails transcription of a long rRNA precursor, and its processing, maturation and modification of the 5.8S, 18S and 28S rRNAs assembled in the small and large 40S and 60S ribosomes, respectively [25–27]. The nucleolus has an exquisite concentration of double-stranded RNA-binding proteins, several of which exhibit nucleocytoplasmic shuttling and are involved in nuclear export of their main cargo, ribosomes. In addition, nucleolar proteomics, imaging and spatial analyses have demonstrated highly dynamic features of the nucleolus [28,29]. These are evident firstly by the high output of the nucleolus in terms of ribosome production in response to normal extracellular cues and growth stimuli [27], and secondly, rapid changes in proteome and RNA content in response to transcription and viral stresses [28,29].
The processing and editing of rRNA in the nucleolus involves the activity of small non-coding RNAs. SnoRNAs are small abundant RNAs that function in rRNA processing [30,31]. Box C/D and box H/ACA snoRNAs have short (3–7 nt) consensus motifs that direct their nucleolar localization and protein interaction [30,32]. They are typically transcribed in the nucleoplasm, transported to the nucleoli either directly or through subnucleolar Cajal bodies in a manner that surprisingly requires the nuclear export protein exportin-1 (CRM1) [33]. Interestingly, CRM1, besides its nucleoplasmic location, localizes also to nucleoli where it recognizes its cargo [34–39]. SnoRNAs fold to specific secondary structures and assemble with a defined set of proteins to fulfill their functions as guide RNAs to assist rRNA maturation [31]. SnoRNAs and miRNAs have interesting parallels. Recently, miRNA precursors have been found to resemble snoRNAs, display H/ACA like structures and bind to dyskerin, present in the H/ACA ribonucleoprotein complex [20,21,40–43]. Processing of these snoRNA-derived miRNAs require Dicer, but are independent of Drosha [21,44]. Whether the miRNA-like snoRNAs are transported to cytoplasm for the Dicer-mediated processing is not know.
Given the previous findings on miRNA-like properties of snoRNAs and findings of isolated miRNAs in the nucleolus, we considered relevant to address the nucleolar small RNAome contents. However, no large-scale small RNA deep sequencing studies have been conducted to date to provide a comprehensive outlook. For this purpose, we analyzed subcellular localization of miRNAs, and specifically, their presence in the nucleolus. We analyzed small RNA content in total, cytoplasmic, nuclear and nucleolar RNA isolates using expression array platforms, deep sequencing and Northern analysis in two human cancer cell lines. These analyses revealed that several miRNAs were frequently, and consistently, detected in small RNA nucleolar fractions. These findings suggest that the nucleolus represents a small RNA depot that includes miRNAs.
2. Materials and methods
2.1. Cell line and reagents
HeLa cervical adenocarcinoma cells (CCL-2, ATCC) and HCT116 cells (wt and DICER−/−) were maintained in DMEM and 10% FCS. The HCT116 DICER−/− cells were a kind gift of Dr. Velculescu (Johns Hopkins University, Baltimore, MD). MCF7 breast adenocarcinoma cells (HTB22, ATCC) were maintained in MEM supplemented with 10% FCS and non-essential amino acids. All cell culture reagents were obtained from Invitrogen. The following drugs were used; Actinomycin D (Sigma–Aldrich), Leptomycin B (Sigma–Aldrich).
2.2. Fractionation of cells and isolation of RNA
Nuclear, nucleolar and cytoplasmic fractions were prepared essentially as in our previous study [45]. RNA was extracted from subcellular compartments using TRIzol Reagent (Invitrogen). From a starting population of about 1.8 × 108 cells, 6% of the nuclei were used for extraction of nuclear RNA (typically yielding 120 μg RNA), and the rest (94%) was used for extraction of nucleoli and nucleolar RNA (typically yielding 40–160 μg RNA). Of the cytoplasmic fraction corresponding to 1.8 × 108 cells, 1.6% was used for extraction of cytoplasmic RNA (typically yielding 240 μg RNA). This leads to a RNA output ratio of 1:10:80 per nucleolus/nucleus/cytoplasm.
2.3. NCode™ microRNA profiling
Total RNA was extracted from whole cells and nucleoli using the TRIzol Reagent (Invitrogen) according to the manufacturer’s instructions. Small RNAs (<200 nt) were further enriched using mirVana kit (Ambion). The small RNAs were poly-A tailed and tagged with the sequence tag for either Alexa Fluor® 5 fluorophor or Alexa Fluor® 3 fluorophor using the NCode™ miRNA Labeling System (Invitrogen). The samples were hybridized in triplicate to NCode™ Multi-Species miRNA Microarray (Invitrogen) overnight according to the instructions by the manufacturer. The microarrays were then hybridized with Alexa Fluor® 3 and Alexa Fluor® 5 capture reagents and washed. Hybridization was performed on a MAUI hybridization station (BioMicro Systems Inc). Each array was subsequently scanned using an Agilent DNA microarray scanner, and images were processed using GenePix Pro software (Molecular Devices). Data analyses were conducted using R statistical program. Nucleolar and cellular miRNA intensities were normalized according to intensities of 6–7 snoRNAs included in the arrays. HeLa and MCF7 cellular intensities were normalized by fold difference of their averaged cellular intensity.
2.4. Heat-treatment of nucleoli and quantitative RT-PCR
Equal amount of freshly purified HeLa nucleolus suspension was either heated to 95 °C (“heat-denatured nucleolus”) for 5 min or incubated on ice for 5 min (intact nucleolus). The nucleoli were then subjected directly to reverse transcription using SuperScript® III Reverse Transcriptase (Invitrogen) and random hexamers according to the manufacturer’s instructions. QPCR was performed using SYBR Green (Atila Biosystem) and specific primer pairs. Amplification was conducted for 38 cycles at 94 °C for 10 s and at 60 °C for 1 min each using an ABI7900 thermocycler (Applied Biosystems). ΔCt represents Ct heat-denaturated nucleoli – Ct non-denatured nucleoli. The primer sequences used were as follows: RNU44: 5′-CCTGGATGATGATAGCAAATGC-3′ and 5′-GAGCTAATTAAGACCTTCATGTT-3′; RNU48: 5′-AGTGATGATGACCCCAGGTAA-3′ and 5′-GTGATGGCATCAGCGACACA-3′; HBII-239: 5′-GAAGCAGTGGGAGTGGAGAA-3′ and 5′-TCAGCAGTTTGAGTGTCAGCA-3′; hY1: 5′-GGCTGGTCCGAAGGTAGTGA-3′ and 5′-GCAGTAGTGAGAAGGGGGGA-3′; hY3: 5′-GGCTGGTCCGAGTGCAGTG-3′ and 5′-GAAGCAGTGGGAGTGGAGAA-3′; U1: 5′-ATACTTACCTGGCAGGGGAGA-3′ and 5′-CAGGGGGAAAGCGCGAACGCA-3′; U2: 5′-ATCGCTTCTCGGCCTTTTTG-3′ and 5′-TCCTATTCCATCTCCCTGCTC-3′. Primers for miR-206, miR-23a and miR-100 were from Applied Biosystems (TaqMan miRNA Assays ID 000510, ID 000399 and ID 000437).
2.5. TaqMan microRNA array
Comparisons of global miRNA expression profiles in intact and heat-treated nucleoli were performed using the TaqMan Array Human MicroRNA Panel v2.0 (Applied Biosystems), which includes Cards A and B in a 384-well format. These contain assays for quantification of 664 human miRNAs and 7 control RNAs. The experimental procedures were according to the manufacturer’s instructions. In brief, suspension of intact and heat-treated nucleoli were directly subjected to reverse-transcription using the Multiplex RT pool set (Applied Biosystems) and the High-Capacity cDNA Archive Kit (Applied Biosystems), whereby a stem-loop RT primer specifically binds to its corresponding miRNA and initiates its reverse-transcription. The reaction products were subsequently amplified with sequence-specific primers using the Applied Biosystems 7900 HT Real-Time PCR system. The reactions were incubated in a 384-well plate at 95 °C for 10 min followed by 40 cycles at 95 °C for 15 s and at 60 °C for 1 min. The data were collected and processed using the Plate Utility and Automation Controller software (Applied Biosystems).
2.6. RNA isolation and generation of cDNA libraries
Total RNA was extracted from whole cells and the isolated cellular compartments using TRIzol Reagent according to manufacturer’s instructions as outlined in our previous study [45] and manuscript submitted. Approximately 400 μg total RNA from each compartment was used to prepare cDNA libraries. Briefly, small RNAs (less than 200 nt) were isolated from the total RNA samples using mirVana Isolation Kit (Ambion) and separated on denaturing 15% polyacrylamide gel (15% PAA, 19:1 acrylamide/bis, 7 M urea) to purify RNAs in the approximate range of 10–40 nt. The small RNA size range was confirmed and the RNA was quantified using Bioanalyzer and the Agilent Small RNA kit (Agilent). cDNA libraries were constructed using the Ion Torrent RNA-seq kit v1 for small RNA libraries according to manufacturer’s instructions, and their purity and concentration was confirmed using Bioanalyzer with the Agilent DNA kit. Small RNA library sequencing was performed using a 314 chip on an Ion Torrent sequencing platform (Life Technologies, Invitrogen) at the Sidney Kimmel Comprehensive Cancer Center Next Generation Sequencing Core. Raw data is deposited to GEO: GSE50057.
2.7. Northern hybridization
Northern blotting was carried out according to Ref. [12] with some modifications. Briefly, 20 μg RNA were resolved on denaturing polyacrylamide gels in 1X TBE, transferred onto a positively charged nylon membrane (Roche Diagnostics) using semi-dry electroblotting (Bio-Rad), and immobilized by UV irradiation (Stratagene Crosslinker 120 mJ/cm2). The membrane was pre-hybridized with hybridization buffer at 37 °C for 1 h, and then hybridized overnight at 37 °C with specific oligodeoxynucleotides. The probes were labeled with Digoxigenin using the DIG oligonucleotide 3′END labeling kit (Roche) and detected using the DIG Nucleic Acid Detection Kit (Roche). The following probes were used: RNU44: 5′-AGTTAGAGCTAATTAAGACCT-3′; tRNA-Ile, 5′-UGGUGGCCCGUACGGGGAUCGA-3′; U11: 5′-TCTTGATGTCGATTCCGCACGCAGAGCAATCGAGTTGCCC-3′; hY1: 5′-AAGGGGGGAAAGAGTAGAACA-3′. The probes were synthesized at the Johns Hopkins Genetic Resources Core Facility. The miRCURY locked nucleic acid (LNA) Digoxigenin-labeled probes for detection of miR-19b, miR-21, miR-23a, miR-24, miR-30c, miR-31, miR-99a and miR-191 were purchased from Exiqon.
3. Results
3.1. MiRNA expression arrays of nucleolar RNA
We conducted miRNA expression profiling of HeLa cervical and MCF7 breast adenocarcinoma cells. For this purpose, we isolated nucleoli and purified small RNAs from the nucleolar preparation and from total cellular RNA. The nucleoli were visually verified for 99% purity under phase contrast and by staining the nucleoli for nucleolar marker protein nucleophosmin (not shown). Analysis of small RNAs by 16% denaturing PAGE showed that the RNAs were distinct (Fig. 1A). The RNAs were dual-color labeled and hybridized to NCode Multispecies miRNA array v.2 using three technical repeats. Signals of over 60% miRNAs present in the array were detected in both cell lines. The nucleolar signal intensities were normalized according to several snoRNAs present in the array (e.g. U46, U47, U49, U50, HBII-13, HBII-239 and HBII-436), and data for high-ranking miRNAs are shown in Supplementary Table S1. As shown in Fig. 1B and C, high-intensity signals were detected in both cell lines in the total cellular and nucleolar RNA preparations. Scatter plot graphs showed that cellular miRNA content correlated with nucleolar signal intensity (Pearson correlation coefficiency, r = 0.843) in HeLa cells, whereas modest correlation was observed in MCF7 cells (r = 0.567). In HeLa and MCF7 nucleolar RNA isolates, 78% of top-ranking 46 miRNAs were identical, and their intensities were highly associated (Fig. 1D and E). However, the intensities of cellular miRNAs in the cell lines were not (Fig. 1F). Data derived from HeLa cells was suggestive that highly expressed miRNAs were detected also in the nucleolar RNA preparation.
Fig. 1.
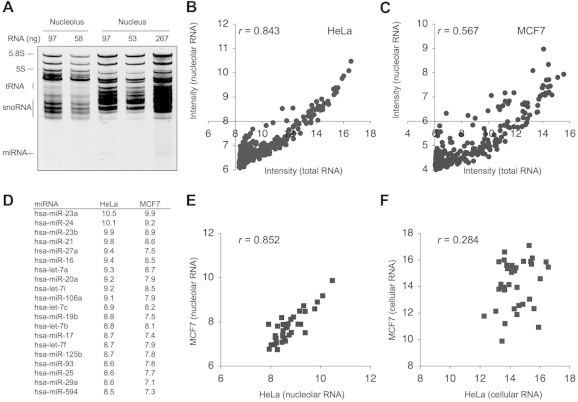
MiRNA expression profiling of nucleolar RNA. (A) Small RNA isolation and analysis by 16% denaturing PAGE. Amount of RNA loaded is indicated at the top. (B, C) MiRNA expression analysis using NCode Multi-Species array. (B) HeLa (N = 41 miRNA); (C) MCF7 (N = 48 miRNA). Scatter plot of nucleolar and total RNA intensities are shown. The nucleolar intensities were adjusted according to snoRNA expression in the respective total cellular and nucleolar fraction. Scatter plots are thresholded at 8 (HeLa) and 6 (MCF7) to exlude low intensity miRNAs. (D) Top-ranking nucleolar miRNAs in HeLa and MCF7 cells. (E, F) Scatter plots of HeLa and MCF7 nucleolar (E) and cellular (F) miRNA intensities (N = 36 miRNA). Pearson correlation coefficiencies (r) are indicated in each plot.
However, cellular RNAs present in the nucleolar preparation may confound this interpretation. We hence decided to test an approach that benefits from the highly compact structure of the nucleolus. We reasoned that if non-nucleolar RNAs are present in the nucleolar preparation there would be a substantial difference in the detection rates if isolated, intact nucleoli as compared to nucleoli subjected to denaturing conditions are used as source for miRNA profiling. For this and subsequent analyses we decided to use HeLa cells due to the abundance of miRNAs present, and our previous comparative analyses on cellular and nucleolar proteins and RNA in this cell line [39,45]. Accordingly, we subjected isolated HeLa nucleoli to heat-denaturation (95 °C, 5 min) or not, followed by qPCR using TaqMan assay designed for detection of mature miRNAs (Fig. 2A). Analysis of abundant, largely cytoplasmic RNAs (hY1, hY3) or nuclear RNAs (U1, U2) showed that there was no difference in the amplification cycles (Ct) between the denatured vs. non-denatured nucleoli (ΔCt), whereas the ΔCt of snoRNAs RNU44 and RNU48 were close to 6 (Fig. 2B). In a further repeat of this assay, RNU44 and RNU48 had ΔCt-values of ⩾7, representing over 128-fold higher detection rate in the denatured nucleolar sample (Fig. 2C). Of three miRNAs tested in the same assay, ΔCt of miR-100 was similar to the snoRNAs (ΔCt 7.8), and miR-23a was somewhat lower (ΔCt 5.4) (Fig. 2C). There was no change in miR-206, included into the analysis based on a previous publication [16], but which was present in HeLa at very low level (Fig. 2C). We concluded that this approach provided a feasible way to enrich and detect nucleolus-associated small RNAs.
Fig. 2.
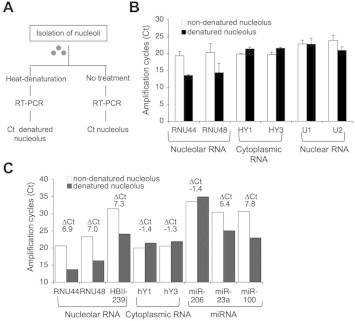
Differential nucleolar RNA qPCR analysis protocol. (A) Scheme of nucleolar denaturation and RT-PCR protocol. (B) Small RNA qPCR controls of denatured and non-denatured nucleoli. Bars represent mean (N = 3 biological experiments) and error bars SD. (C) qPCR for heat-denatured and non-denatured nucleoli for the indicated small RNAs and miRNAs. ΔCt represents Ct denaturated nucleoli – Ct non-denatured nucleoli.
We then conducted TaqMan miRNA array profiling of isolated nucleoli using the comparative heat-denaturation protocol. To further increase robustness, we limited the amplification cycles to <38 to exclude rare and potentially contaminating signals. Of a total of 664 miRNAs represented by the array, 92 individual miRNAs were detected (Fig. 3A). Of these, 32 miRNAs had ΔCt ⩾ 7, that is, were 128-fold more abundant in the heat-denatured nucleolus (Fig. 3B). All snoRNA controls included in the array had ΔCt of over 10, whereas small nuclear RNAs had <4 (Fig. 3B). Based on these results we believe that the assay represents with good predictability the presence of miRNAs in the nucleolar RNA preparation. The identified nucleolar miRNAs included members of four miRNA families, namely miR-17-92, miR-106a-363, miR-23a-24 and miR-30 (Fig. 3C). This may be an indicator that these miRNAs share regulatory pathways or functional activities relevant for their nucleolar presence.
Fig. 3.
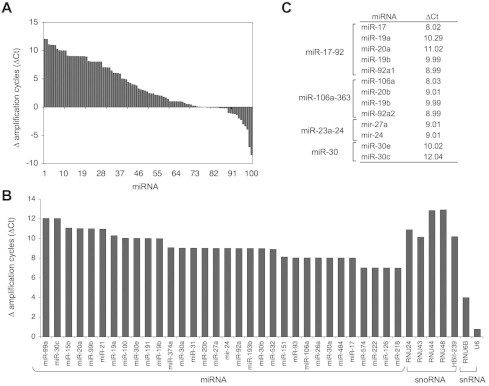
MiRNA profiling using TaqMan miRNA expression array. (A) Waterfall plot of detectable nucleolar miRNAs. ΔCt represents Ct denaturated nucleoli – Ct non-denatured nucleoli. (B) MiRNA nucleolar abundance as shown by ranking order of the ΔCt. Small nuclear (snRNA) and snoRNAs present in the TaqMan array are shown as controls. (C) MiRNA families represented in the dataset.
3.2. Northern hybridization of miRNA in subcellular compartments
To ascertain the presence of miRNAs in the nucleolar RNA preparations we conducted Northern hybridization of seven representative miRNAs. We assessed the presence of the miRNAs in the nucleolar, nuclear, cytoplasmic and total cellular fractions. Cellular fractionation and RNA isolation was performed as in our previous studies [39,45] and manuscript submitted (Bai et al., submitted). Equal amounts of RNA (25 μg) was loaded from each subcellular fraction. The purity of the RNA preparations was confirmed using probes against several small RNAs. These included RNU44, a snoRNA abundantly expressed in the nucleolus, U11, a small nuclear RNA with mostly nucleoplasmic localization, cyto-nucleoplasmic tRNA, and hY1, a mostly cytoplasmic RNA. According to the Northern analysis, and analyses shown in Refs. [39,45], the nucleolar RNA preparations were judged to be free of nucleoplasmic and cytoplasmic RNA (Fig. 4A). To further test the purity of RNA isolates we conducted qPCR using primers for cytoplasmic hY1 and nucleolar RNU44. The qPCR for hY1 and RNU44 confirmed their enrichment in the compartment-specific RNA preparations (Fig. 4C). As shown in Fig. 4B, several miRNAs (miR-19b, miR-21, miR-24, miR-30c, miR-31, miR-99a, miR-191) were detected in the nucleolar fraction in their mature 20–23 nt forms, but were proportionally much lower as compared to the total cellular RNA fraction. Mature miRNAs were also detected in the nuclear RNA fraction. This is consistent with recent deep sequencing of small RNAs, which showed nuclear presence of over 60 mature miRNAs including all miRNAs analyzed here [13].
Fig. 4.
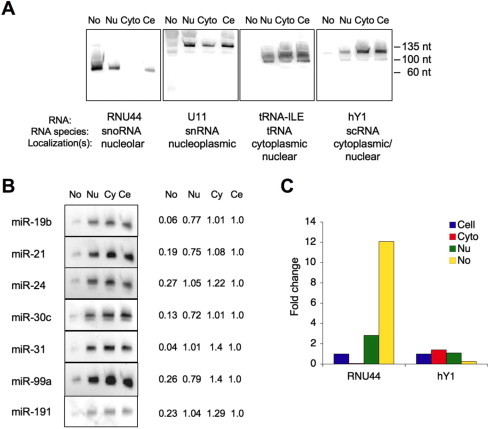
Northern analysis of miRNA presence in cellular compartments. RNA was isolated from nucleolar (No), nuclear (Nu) or cytoplasmic (Cyto) fractions or from total cellular RNA (Ce). RNA (20 μg) was separated on 15% gel. The blots were probed for (A) known small RNAs or (B) miRNAs, as indicated. Size markers are indicated to the right. Relative proportion of miRNAs is expressed as compared to total cellular RNA set as 1. (C) RNAs isolated from respective subcellular compartments were amplified using qPCR with primers for RNU44 and hY1.
3.3. Small RNA deep sequencing of the subcellular compartments
To provide a comprehensive and quantitative analysis of miRNA abundance in the HeLa subcellular fractions, we used small RNA deep sequencing. Following subcellular fractionation, RNA isolation and separation, small RNAs (10–40 nt) were gel purified and used to generate cDNA libraries followed by deep sequencing using Ion Torrent platform. Full data set is deposited as #GSE50057, and detailed description of the small RNA annotation is provided in a manuscript submitted (Bai et al., submitted). The analysis showed that the nucleolar small RNome was unique and predominantly composed of snoRNA-derived small RNAs. Approximately 200,000 miRNA reads were detected in the total cellular, cytoplasmic and nuclear fraction each, and 2100 reads in the nucleolar RNA library (Table 1 and Supplementary Table S2). Eleven miRNAs were present in the nucleolar small RNA fraction at read numbers ⩾10. Of these, eight were present in the TaqMan array (miR-19b, miR-21, miR-24, miR-31, miR-93, miR-151, miR-532, miR-574) and scored positive in the nucleolus-denaturation protocol (Fig. 3B). Overall, the miRNA reads between cytoplasmic and total RNA and nuclear and total RNA showed linear distribution as displayed in scatter plots, and very high correlation (Pearson correlation coefficiency 0.998 and 0.997, respectively) (Supplementary Fig. S1A and B). Scatter plot analyses of the distribution between nucleolar and cytoplasmic or nuclear miRNAs were more diverse, although a general trend of correlation between the read abundances was maintained (Supplementary Fig. S1C and D). The Pearson correlations between nucleolar and cytoplasmic or nuclear reads were 0.634 and 0.735, respectively.
Table 1.
Ion Torrent miRNA reads and frequencies in cellular compartments. miRNAs present in the nucleolus at reads ⩾ 0 are shown.
| Cellular | %Cellular | Cytoplasm | %Cytoplasm | Nuclear | %Nuclear | Nucleolus | %Nucleolar | |
|---|---|---|---|---|---|---|---|---|
| miR-24 | 20,998 | 11.12 | 18,051 | 9.02 | 34,499 | 17.45 | 972 | 46.26 |
| miR-21 | 101,664 | 53.83 | 120,398 | 60.19 | 118,190 | 59.77 | 691 | 32.89 |
| miR-93 | 1535 | 0.81 | 1072 | 0.54 | 1241 | 0.63 | 113 | 5.38 |
| miR-31 | 456 | 0.24 | 243 | 0.12 | 377 | 0.19 | 49 | 2.33 |
| miR-19B | 994 | 0.53 | 727 | 0.36 | 1075 | 0.54 | 34 | 1.62 |
| miR-1307 | 151 | 0.08 | 92 | 0.05 | 97 | 0.05 | 38 | 1.81 |
| miR-574 | 42 | 0.02 | 38 | 0.02 | 87 | 0.04 | 24 | 1.14 |
| miR-151 | 618 | 0.33 | 398 | 0.20 | 657 | 0.33 | 19 | 0.90 |
| miR-532 | 174 | 0.09 | 100 | 0.05 | 179 | 0.09 | 17 | 0.81 |
| miR-148 | 132 | 0.07 | 75 | 0.04 | 122 | 0.06 | 15 | 0.71 |
| miR-18A | 2283 | 1.21 | 1436 | 0.72 | 2463 | 1.25 | 12 | 0.57 |
| Sum | 129,047 | 68.3 | 142,630 | 71.3 | 158,987 | 80.4 | 1984 | 94.4 |
| All miRNAs | 188,852 | 100 | 200,033 | 100 | 197,731 | 100 | 2101 | 100 |
Given that small RNA deep sequencing of cytoplasmic and nuclear fractions have indicated that mature miRNAs are frequently present in the nucleus consistent with our results here, we queried for this property by comparing reads of all miRNAs present in both compartments at reads ⩾10. For this purpose, we normalized the numbers of nuclear and cytoplasmic reads against the cellular reads, and then compared the relative ratios of nuclear and cytoplasmic reads. 17 miRNAs were 2-fold more abundant in the nucleus than in the cytoplasm (Fig. 5A), and included e.g. miR-29b previously shown to be stabilized in HeLa cell nuclei [12]. Conversely, those miRNAs with higher relative cytoplasmic abundances included seven members of miR-let-7 family and miR-34a (Fig. 5A).
Fig. 5.
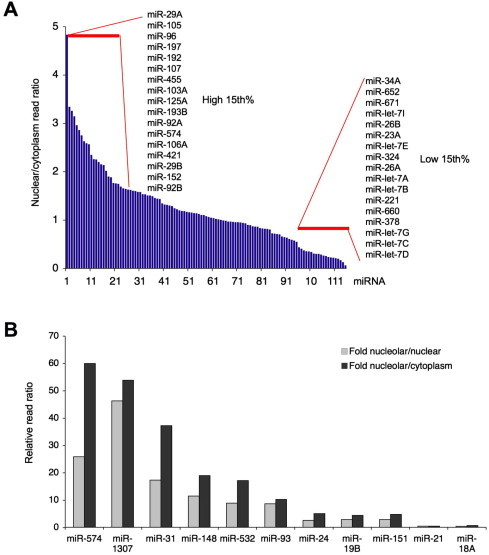
Relative abundance of miRNAs in the subcellular compartments as analyzed by deep sequencing. (A) Comparison of miRNA reads between the nucleus and cytoplasm. MiRNAs with reads ⩾10 in the nucleus, cytoplasm and total cellular fractions were identified (N = 127) and nuclear and cytoplasmic reads were normalized against the total number of cellular reads. The nuclear/cytoplasmic read ratio is shown. Insets display miRNAs in the highest and lowest 15% ratio. (B) Relative frequencies of nucleolar reads as compared to nuclear (gray bar) and cytoplasmic (black bars) reads are shown.
We then compared the relative frequencies of the eleven nucleolar miRNA reads to their frequencies in the other cellular fractions. This analysis showed that the frequencies of several miRNAs were substantially higher in the nucleolus than in the nuclear and cytoplasmic compartments (Fig. 5B). MiR-574, miR-1307, miR-31, miR-148, miR-532 and miR-93 were proportionally ⩾8-fold more abundant in the nucleolus than in the other respective compartments (Fig. 5B). These findings suggested that several miRNAs showed compartment-specific enrichment.
3.4. Dicer inactivation does not affect miRNA nucleolar accumulation
Dicer has been suggested to affect processing of certain snoRNAs to miRNA-sized fragments [40,41]. To address whether Dicer affects miRNA nucleolar presence, we analyzed small RNAs from Dicer−/− HCT116 cells and the parent Dicer-proficient HCT116 cells. It has been previously shown that miR-21 processing is compromised in the Dicer−/− HCT116 cells, whereas miR-31 processing to mature form is less affected [46]. We hence used subcellular RNA fractions of the HCT116 parent and Dicer-null derivative cells to analyze for miR-21 and miR-31 by Northern blotting. As shown in Supplementary Fig. S2, miR-21 was proportionally decreased in the Dicer-null cells in all cellular compartments. Similarly, the decrease in abundance of miR-31 was proportional. Hence, ablation of Dicer did not alter the nucleolar presence of miR-21.
3.5. MiR-21 and miR-31 nucleolar association increases following CRM1 inhibition
As shown by numerous studies, Pol I transcription blocks cause nucleolar disintegration and reorganization of the nucleolar proteome and RNome [28,39,47,48]. We therefore used Actinomycin D (ActD) to inhibit Pol I to ask whether nucleolar miRNA abundance would be affected. Interestingly, although the presence of hY1 was increased in the nucleolar fraction of ActD-treated cells, that of miR-31 was unaffected (Fig. 6A). This suggests that ongoing Pol I transcription may not be required for the nucleolar presence of miRNAs.
Fig. 6.
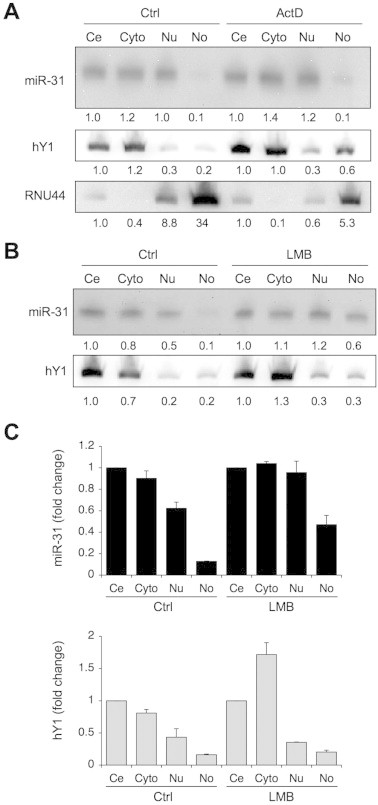
Effect of CRM1 and Pol I transcription inhibition on nucleolar miRNA abundance. Cells were treated with ActD (50 ng/ml) (A) or leptomycin B (LMB) (10 μM) (B) for 3 h, fractionated and RNA was isolated. RNA (25 μg) was separated on 15% gel and hybridized to respective LNA probes. Relative proportion of miRNAs is expressed as compared to total cellular RNA set as 1. (C) Quantitative analysis of (B) for miR-31 and hY1. Results represent the mean and SEM of two biological experiments.
CRM1/NMD3 export/adaptor complex extensively accumulates in the nucleolus when Pol I is inhibited [35,39]. This is especially interesting as CRM1 is involved in the nucleolar trafficking of snoRNA [33], and has been implicated in the trafficking of miRNAs [49]. We treated the cells with a CRM1-specific inhibitor, leptomycin B. Subcellular fractionation and analysis of miR-31 by Northern hybridization indicated that the nucleolar abundance of miR-31 was increased by 3-fold in leptomycin B-treated cells (Fig. 6B and C).
4. Discussion
This is the first small RNA deep sequencing study that addresses small RNome of the nucleolus with specific reference to presence of miRNAs in this subcellular compartment. We consistently detect several miRNAs in the nucleolar RNA preparations using different methodological approaches including two separate miRNA expression arrays, Northern analysis and small RNA deep sequencing. We also established a new method to detect nucleolus-associated small RNAs in the isolated nucleoli. Collectively, these results showed that mature forms of over ten miRNAs were detected in the nucleolus. MiRNA nucleolar localization was independent of Dicer activity, but was dependent on CRM1. These data indicate that small RNAs, including miRNA are discernible in the nucleolus and expand the aspects of miRNA regulation and potential functions.
There are no previous systematic unbiased approaches that address the extent of human miRNAs in the nucleoli. Here we find the presence of several miRNAs in the nucleoli using different experimental approaches. The subcellular preparations used here have been previously extensively analyzed for compartment-specific expression of both protein and RNA which validate their purity in addition to assays shown here [39,45, Bai et al., submitted]. Yet, it is possible that the miRNAs are retained in the perinucleolar area rich in heterochromatin, or that the RNA preparations contain other co-purifying RNA-rich complexes like those present in Cajal bodies, or in cytoplasmic and nuclear RNAs. To address these concerns, we designed and used an approach that employed heat-denaturation of the isolated intact nucleoli and used this as a source for qPCR. This approach indicated a significant difference in the ability to amplify nucleolar snoRNAs but not small nuclear RNAs that were used as controls. Using this approach with a highly restrictive cut-off at over 128-fold difference, 32 miRNAs were identified as nucleolus-associated. Of these, 84% were present in the NCode miRNA array at high intensity. Further, the presence of miR-19b, miR-21, miR-24, miR-30c, miR-31, miR-99a and miR-191 was confirmed by Northern hybridization in the nucleolar RNA fractions. RNA Seq of nucleolar small RNome identified eleven miRNAs in the nucleolus (miR-24, miR-21, miR-93, miR-31, mir-19B, miR-1307, miR-574, miR-151, miR-532, miR-148, miR-18A) out of which eight were also highly differentially expressed in the nucleolar denaturation assay (except for miR-1307, miR-148 and miR-18a either absent in the assay or amplifiable only in the denatured nucleoli). These results strongly support the interpretation that miRNAs can be detected in the nucleoli although their abundance is low compared to their presence in the cytoplasm. Overall, this study identified several overlapping nucleolar miRNAs to that of Li et al. [18] including miR-191, miR-484, miR-193b, miR-93 and miR-574. Collectively, the data is consistent with the interpretation that miRNAs are detectable in the nucleolar fractions.
We tested whether the regulation of nucleolar activity affects miRNA nucleolar localization. Inhibition of Pol I transcription which causes segregation of nucleolar structures had no effect on miR-31 spatial distribution suggesting that it may be retained in the nucleolar remnants. While XPO5 exports miRNAs to the cytoplasm, CRM1, involved both in ribosome and mRNA export [50], is also a nucleolar resident [39], and mediates transfer of snoRNAs to the nucleolus [33]. For these reasons we tested whether CRM1 could affect miRNA nucleolar abundance. We found here that inhibition of CRM1 increased the presence of miR-31 in the nucleolus. This is consistent with the finding of increased miRNA nuclear accrual by Castanotto et al. [49]. As CRM1 inhibition has been shown to affect nucleolar transit of U3 RNAs, it is possible that CRM1 is involved in trafficking of several small RNAs.
The relevance of miRNA nucleolar localization is currently unknown. The typical miRNA targets are absent from the nucleolus. The study by Li et al. [18] suggested that miRNA nucleolar localization could be affected by exposure to DNA and ds RNA. Given that RNA editing, especially A-to-I takes place in the nucleolus [32,51], and that A-to-I editing has been observed in a high-throughput sequencing study [52], we specifically searched for this in the nucleolar deep sequencing data. However, no indication of A-to-I editing among the nucleolar miRNA reads was detected (not shown). Small RNAs such as siRNAs direct methylation of histones and DNA thereby altering chromatin states and regulate the activity of transposons and genomic areas rich in heterochromatin and repetitive elements such as centromeres [53]. The perinucleolar area is highly enriched in heterochromatin and rDNA genes have a unique arrangement of active and epigenetically silenced genes. The possibility that nucleolar miRNAs could be involved in controlling nucleolar chromatin states is intriguing. In plants, siRNAs regulate rRNA gene promoter activity and govern nucleolar dominance [54]. Although non-coding RNAs are critical regulators of rRNA transcription in mammals [55], miRNA-sized RNAs have not been ascribed with similar activities.
The nucleolus is highly dynamic and its activity reflects the cellular needs for protein synthesis. Many nucleolar proteins have high mobility and constantly shuttle between the different cellular compartments, and this dynamic mobility relates to shuttling of rRNA components as well as small RNAs. Argonaute complexes contain a large number of RNA metabolism-associated proteins including gemins, helicases and hnRNP-proteins and co-sediment with ribosomes [56,57]. Interestingly, many of the Argonaute-complex associated proteins are also nucleolar. MiRNAs have been detected in association with ribosomes, and nuclear import of ribosomal proteins is coordinated [58]. Therefore it is possible that miRNAs shuttle with binding proteins or ribosomal particles to the nucleolus. Hence, the contents of nucleolar proteome or RNome cannot be strictly defined, but rather reflect the present state of cellular activity. Based on these dynamic properties we believe it is likely that there is also great variability in the miRNA content of the nucleolus that depends on the physiological state and cell type in question.
Disclosures
The authors have no conflicts of interest to disclose.
Acknowledgements
We thank our colleagues for discussions and helpful suggestions. We thank Dr. Sarah Wheelan and Dr. Vasan Yegnasubramanian and the Johns Hopkins Deep Sequencing and Microarray Core Facility and the Sidney Kimmel Comprehensive Cancer Center Next Generation Sequencing Core for the high-throughput analyses, discussions and support. This work was supported by the National Institutes of Health [P30 CA006973] and Johns Hopkins University start-up funds (ML).
Contributor Information
Baoyan Bai, Email: bby54@hotmail.com.
Hester Liu, Email: hester@jhmi.edu.
Marikki Laiho, Email: mlaiho1@jhmi.edu.
Appendix A. Supplementary data
References
- 1.Bartel D.P. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 3.Kim V.N., Han J., Siomi M.C. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol. 2009;10:126–139. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- 4.Siomi H., Siomi M.C. Posttranscriptional regulation of microRNA biogenesis in animals. Mol Cell. 2010;38:323–332. doi: 10.1016/j.molcel.2010.03.013. [DOI] [PubMed] [Google Scholar]
- 5.Newman M.A., Hammond S.M. Emerging paradigms of regulated microRNA processing. Genes Dev. 2010;24:1086–1092. doi: 10.1101/gad.1919710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Krol J., Loedige I., Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet. 2010;11:597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- 7.Croce C.M. Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet. 2009;10:704–714. doi: 10.1038/nrg2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dimmeler S., Nicotera P. MicroRNAs in age-related diseases. EMBO Mol Med. 2013;5:180–190. doi: 10.1002/emmm.201201986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu N., Olson E.N. MicroRNA regulatory networks in cardiovascular development. Dev Cell. 2010;18:510–525. doi: 10.1016/j.devcel.2010.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.O’Connell R.M., Rao D.S., Chaudhuri A.A., Baltimore D. Physiological and pathological roles for microRNAs in the immune system. Nat Rev Immunol. 2010;10:111–122. doi: 10.1038/nri2708. [DOI] [PubMed] [Google Scholar]
- 11.Eacker S.M., Dawson T.M., Dawson V.L. Understanding microRNAs in neurodegeneration. Nat Rev Neurosci. 2009;10:837–841. doi: 10.1038/nrn2726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hwang H.W., Wentzel E.A., Mendell J.T. A hexanucleotide element directs microRNA nuclear import. Science. 2007;315:97–100. doi: 10.1126/science.1136235. [DOI] [PubMed] [Google Scholar]
- 13.Liao J.Y., Ma L.M., Guo Y.H., Zhang Y.C., Zhou H., Shao P., Chen Y.Q., Qu L.H. Deep sequencing of human nuclear and cytoplasmic small RNAs reveals an unexpectedly complex subcellular distribution of miRNAs and tRNA 3′ trailers. PLoS ONE. 2010;5:e10563. doi: 10.1371/journal.pone.0010563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim D.H., Saetrom P., Snøve O., Jr., Rossi J.J. MicroRNA-directed transcriptional gene silencing in mammalian cells. Proc Natl Acad Sci U S A. 2008;105:16230–16235. doi: 10.1073/pnas.0808830105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Place R.F., Li L.C., Pookot D., Noonan E.J., Dahiya R. MicroRNA-373 induces expression of genes with complementary promoter sequences. Proc Natl Acad Sci U S A. 2008;105:1608–1613. doi: 10.1073/pnas.0707594105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Politz J.C., Zhang F., Pederson T. MicroRNA-206 colocalizes with ribosome-rich regions in both the nucleolus and cytoplasm of rat myogenic cells. Proc Natl Acad Sci U S A. 2006;103:18957–18962. doi: 10.1073/pnas.0609466103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Politz J.C., Hogan E.M., Pederson T. MicroRNAs with a nucleolar location. RNA. 2009;15:1705–1715. doi: 10.1261/rna.1470409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li Z.F., Liang Y.M., Lau P.N., Shen W., Wang D.K., Cheung W.T., Xue C.J., Poon L.M., Lam Y.W. Dynamic localisation of mature microRNAs in Human nucleoli is influenced by exogenous genetic materials. PLoS ONE. 2013;8:e70869. doi: 10.1371/journal.pone.0070869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marcon E., Babak T., Chua G., Hughes T., Moens P.B. MiRNA and piRNA localization in the male mammalian meiotic nucleus. Chromosome Res. 2008;16:243–260. doi: 10.1007/s10577-007-1190-6. [DOI] [PubMed] [Google Scholar]
- 20.Scott M.S., Avolio F., Ono M., Lamond A.I., Barton G.J. Human miRNA precursors with box H/ACA snoRNA features. PLoS Comput Biol. 2009;5:e1000507. doi: 10.1371/journal.pcbi.1000507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ono M., Scott M.S., Yamada K., Avolio F., Barton G.J., Lamond A.I. Identification of human miRNA precursors that resemble box C/D snoRNAs. Nucleic Acids Res. 2011;39:3879–3891. doi: 10.1093/nar/gkq1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ohrt T., Muetze J., Svoboda P., Schwille P. Intracellular localization and routing of miRNA and RNAi pathway components. Curr Top Med Chem. 2012;12:79–88. doi: 10.2174/156802612798919132. [DOI] [PubMed] [Google Scholar]
- 23.Pickering B.F., Yu D., Van Dyke M.W. Nucleolin protein interacts with microprocessor complex to affect biogenesis of microRNAs 15a and 16. J Biol Chem. 2011;286:44095–44103. doi: 10.1074/jbc.M111.265439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jeffries C.D., Fried H.M., Perkins D.O. Nuclear and cytoplasmic localization of neural stem cell microRNAs. RNA. 2011;17:675–686. doi: 10.1261/rna.2006511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haag J.R., Pikaard C.S. RNA polymerase I: a multifunctional molecular machine. Cell. 2007;131:1224–1225. doi: 10.1016/j.cell.2007.12.005. [DOI] [PubMed] [Google Scholar]
- 26.Russell J., Zomerdijk J.C. The RNA polymerase I transcription machinery. Biochem Soc Symp. 2006:203–216. doi: 10.1042/bss0730203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fatica A., Tollervey D. Making ribosomes. Curr Opin Cell Biol. 2002;14:313–318. doi: 10.1016/s0955-0674(02)00336-8. [DOI] [PubMed] [Google Scholar]
- 28.Boulon S., Westman B.J., Hutten S., Boisvert F.M., Lamond A.I. The nucleolus under stress. Mol Cell. 2010;40:216–227. doi: 10.1016/j.molcel.2010.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hernandez-Verdun D. Nucleolus: from structure to dynamics. Histochem Cell Biol. 2006;125:127–137. doi: 10.1007/s00418-005-0046-4. [DOI] [PubMed] [Google Scholar]
- 30.Kiss T., Fayet-Lebaron E., Jady B.E. Box H/ACA small ribonucleoproteins. Mol Cell. 2010;37:597–606. doi: 10.1016/j.molcel.2010.01.032. [DOI] [PubMed] [Google Scholar]
- 31.Watkins N.J., Bohnsack M.T. The box C/D and H/ACA snoRNPs: key players in the modification, processing and the dynamic folding of ribosomal RNA. Wiley Interdiscip Rev RNA. 2012;3:397–414. doi: 10.1002/wrna.117. [DOI] [PubMed] [Google Scholar]
- 32.Kiss T. SnoRNP biogenesis meets pre-mRNA splicing. Mol Cell. 2006;23:775–776. doi: 10.1016/j.molcel.2006.08.023. [DOI] [PubMed] [Google Scholar]
- 33.Boulon S., Verheggen C., Jady B.E., Girard C., Pescia C., Paul C., Ospina J.K., Kiss T., Matera A.G., Bordonne R. PHAX and CRM1 are required sequentially to transport U3 snoRNA to nucleoli. Mol Cell. 2004;16:777–787. doi: 10.1016/j.molcel.2004.11.013. [DOI] [PubMed] [Google Scholar]
- 34.Fornerod M., van Deursen J., van Baal S., Reynolds A., Davis D., Murti K.G., Fransen J., Grosveld G. The human homologue of yeast CRM1 is in a dynamic subcomplex with CAN/Nup214 and a novel nuclear pore component Nup88. EMBO J. 1997;16:807–816. doi: 10.1093/emboj/16.4.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thomas F., Kutay U. Biogenesis and nuclear export of ribosomal subunits in higher eukaryotes depend on the CRM1 export pathway. J Cell Sci. 2003;116:2409–2419. doi: 10.1242/jcs.00464. [DOI] [PubMed] [Google Scholar]
- 36.Trotta C.R., Lund E., Kahan L., Johnson A.W., Dahlberg J.E. Coordinated nuclear export of 60S ribosomal subunits and NMD3 in vertebrates. EMBO J. 2003;22:2841–2851. doi: 10.1093/emboj/cdg249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ernoult-Lange M., Wilczynska A., Harper M., Aigueperse C., Dautry F., Kress M., Weil D. Nucleocytoplasmic traffic of CPEB1 and accumulation in Crm1 nucleolar bodies. Mol Biol Cell. 2009;20:176–187. doi: 10.1091/mbc.E08-09-0904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Latonen L., Moore H.M., Bai B., Jaamaa S., Laiho M. Proteasome inhibitors induce nucleolar aggregation of proteasome target proteins and polyadenylated RNA by altering ubiquitin availability. Oncogene. 2011;30:790–805. doi: 10.1038/onc.2010.469. [DOI] [PubMed] [Google Scholar]
- 39.Bai B., Moore H.M., Laiho M. CRM1 and its ribosome export adaptor NMD3 localize to the nucleolus and affect rRNA synthesis. Nucleus. 2013;4:315–325. doi: 10.4161/nucl.25342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ender C., Krek A., Friedlander M.R., Beitzinger M., Weinmann L., Chen W., Pfeffer S., Rajewsky N., Meister G. A human snoRNA with microRNA-like functions. Mol Cell. 2008;32:519–528. doi: 10.1016/j.molcel.2008.10.017. [DOI] [PubMed] [Google Scholar]
- 41.Saraiya A.A., Wang C.C. SnoRNA, a novel precursor of microRNA in Giardia lamblia. PLoS Pathog. 2008;4:e1000224. doi: 10.1371/journal.ppat.1000224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Taft R.J., Glazov E.A., Lassmann T., Hayashizaki Y., Carninci P., Mattick J.S. Small RNAs derived from snoRNAs. RNA. 2009;15:1233–1240. doi: 10.1261/rna.1528909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brameier M., Herwig A., Reinhardt R., Walter L., Gruber J. Human box C/D snoRNAs with miRNA like functions: expanding the range of regulatory RNAs. Nucleic Acids Res. 2011;39:675–686. doi: 10.1093/nar/gkq776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Scott M.S., Ono M. From snoRNA to miRNA: dual function regulatory non-coding RNAs. Biochimie. 2011;93:1987–1992. doi: 10.1016/j.biochi.2011.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bai B., Laiho M. Efficient sequential recovery of nucleolar macromolecular components. Proteomics. 2012;12:3044–3048. doi: 10.1002/pmic.201200071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cummins J.M., He Y., Leary R.J., Pagliarini R., Diaz L.A., Jr., Sjoblom T., Barad O., Bentwich Z., Szafranska A.E., Labourier E. The colorectal microRNAome. Proc Natl Acad Sci U S A. 2006;103:3687–3692. doi: 10.1073/pnas.0511155103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Moore H.M., Bai B., Boisvert F.M., Latonen L., Rantanen V., Simpson J.C., Pepperkok R., Lamond A.I., Laiho M. Quantitative proteomics and dynamic imaging of the nucleolus reveal distinct responses to UV and ionizing radiation. Mol Cell Proteomics. 2011;10(M111):009241. doi: 10.1074/mcp.M111.009241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Turner A.J., Knox A.A., Watkins N.J. Nucleolar disruption leads to the spatial separation of key 18S rRNA processing factors. RNA Biol. 2012;9:175–186. doi: 10.4161/rna.18811. [DOI] [PubMed] [Google Scholar]
- 49.Castanotto D., Lingeman R., Riggs A.D., Rossi J.J. CRM1 mediates nuclear-cytoplasmic shuttling of mature microRNAs. Proc Natl Acad Sci U S A. 2009;106:21655–21659. doi: 10.1073/pnas.0912384106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kohler A., Hurt E. Exporting RNA from the nucleus to the cytoplasm. Nat Rev Mol Cell Biol. 2007;8:761–773. doi: 10.1038/nrm2255. [DOI] [PubMed] [Google Scholar]
- 51.Vitali P., Basyuk E., Le Meur E., Bertrand E., Muscatelli F., Cavaillé J., Huttenhofer A. ADAR2-mediated editing of RNA substrates in the nucleolus is inhibited by C/D small nucleolar RNAs. J Cell Biol. 2005;169:745–753. doi: 10.1083/jcb.200411129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chiang H.R., Schoenfeld L.W., Ruby J.G., Auyeung V.C., Spies N., Baek D., Johnston W.K., Russ C., Luo S., Babiarz J.E. Mammalian microRNAs: experimental evaluation of novel and previously annotated genes. Genes Dev. 2010;24:992–1009. doi: 10.1101/gad.1884710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.van Wolfswinkel J.C., Ketting R.F. The role of small non-coding RNAs in genome stability and chromatin organization. J Cell Sci. 2010;123:1825–1839. doi: 10.1242/jcs.061713. [DOI] [PubMed] [Google Scholar]
- 54.Pontes O., Li C.F., Costa Nunes P., Haag J., Ream T., Vitins A., Jacobsen S.E., Pikaard C.S. The Arabidopsis chromatin-modifying nuclear siRNA pathway involves a nucleolar RNA processing center. Cell. 2006;126:79–92. doi: 10.1016/j.cell.2006.05.031. [DOI] [PubMed] [Google Scholar]
- 55.Grummt I., Längst G. Epigenetic control of RNA polymerase I transcription in mammalian cells. Biochim Biophys Acta. 2013;1829:393–404. doi: 10.1016/j.bbagrm.2012.10.004. [DOI] [PubMed] [Google Scholar]
- 56.Höck J., Weinmann L., Ender C., Rüdel S., Kremmer E., Raabe M., Urlaub H., Meister G. Proteomic and functional analysis of Argonaute-containing mRNA–protein complexes in human cells. EMBO Rep. 2007;8:1052–1060. doi: 10.1038/sj.embor.7401088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mourelatos Z., Dostie J., Paushkin S., Sharma A., Charroux B., Abel L., Rappsilber J., Mann M., Dreyfuss G. MiRNPs: a novel class of ribonucleoproteins containing numerous microRNAs. Genes Dev. 2002;16:720–788. doi: 10.1101/gad.974702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kressler D., Bange G., Ogawa Y., Stjepanovic G., Bradatsch B., Pratte D., Amlacher S., Strauß D., Yoneda Y., Katahira J. Synchronizing nuclear import of ribosomal proteins with ribosome assembly. Science. 2012;338:666–671. doi: 10.1126/science.1226960. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


