Abstract
During the biogenesis of bacterial cell-wall polysaccharides, such as peptidoglycan, cytoplasmic synthesized precursors should be trafficked across the plasma membrane. This essential process requires a dedicated lipid, undecaprenyl-phosphate that is used as a glycan lipid carrier. The sugar is linked to the lipid carrier at the inner face of the membrane and is translocated toward the periplasm, where the glycan moiety is transferred to the growing polymer. Undecaprenyl-phosphate originates from the dephosphorylation of its precursor undecaprenyl-diphosphate, with itself generated by de novo synthesis or by recycling after the final glycan transfer. Undecaprenyl-diphosphate is de novo synthesized by the cytosolic cis-prenyltransferase undecaprenyl-diphosphate synthase, which has been structurally and mechanistically characterized in great detail highlighting the condensation process. In contrast, the next step toward the formation of the lipid carrier, the dephosphorylation step, which has been overlooked for many years, has only started revealing surprising features. In contrast to the previous step, two unrelated families of integral membrane proteins exhibit undecaprenyl-diphosphate phosphatase activity: BacA and members of the phosphatidic acid phosphatase type 2 super-family, raising the question of the significance of this multiplicity. Moreover, these enzymes establish an unexpected link between the synthesis of bacterial cell-wall polymers and other biological processes. In the present review, the current knowledge in the field of the bacterial lipid carrier, its mechanism of action, biogenesis, recycling, regulation, and future perspective works are presented.
The Universal Glycan Lipid Carrier
The use of long and linear-chain polyprenyl-phosphate lipids that facilitate the translocation of a sugar or glycan strand across a biological membrane is an extraordinary conserved process encountered in all kingdoms of life.39,101 They are involved in protein glycosylation in eukaryotic,10 archeal, and bacterial cells52 as well as for the biogenesis of bacterial cell-wall polysaccharides such as peptidoglycan,7 lipopolysaccharide (LPS) O-antigen,93,110 wall teichoic acids,8 capsular polysaccharides,120 common enterobacterial antigen,88 membrane-derived oligosaccharides,118 and exopolysaccharides. In mammals, defects in protein glycosylation referred to as congenital disorders of glycosylation119 engender severe malformations and developmental retards with outcomes ranging from death in infancy to late-onset diseases. In bacteria, the inhibition of the biosynthesis or recycling of the undecaprenyl-phosphate carrier lipid provokes the arrest of peptidoglycan synthesis and, consequently, cell lysis.7 This essential chemical carrier takes part in critical cellular processes, and, therefore, its metabolism represents a potential target for therapeutics such as new antibacterial agents.
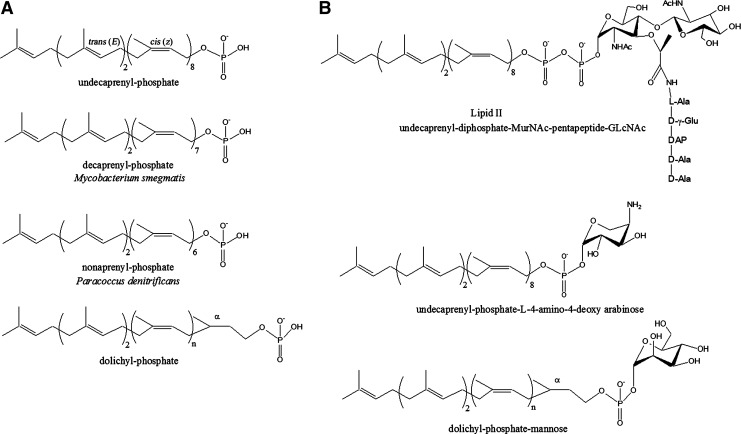
These long carbon chain lipids are generated by sequential condensations of several isoprene units (the 5-carbon building block, isopentenyl-diphosphate).39,101 Among the living organisms, the structure of the lipid carrier varies, to some extent, in both the carbon chain length (number of polymerized isoprene units) and stereochemistry. In bacteria, the 55-carbon chain length generated by condensation of 11 isoprene units yielding undecaprenyl-phosphate (C55-P) (Fig. 1) is the common rule; however, lower-sized decaprenyl-phosphate (C50-P)56 and nonaprenyl-phosphate (C45-P)48 homologues were found to assist glycan translocation in some mycobacterial species and Paracoccus denitrificans, respectively. The cis or trans configuration of the double bonds in the polyisoprenoid chain is another source of diversity. Typically, the isoprene units are added in a cis configuration on a short all-trans oligoprene precursor comprising one to three subunits. Finally, but not the least, the status of hydrogenation of the terminal isoprene unit proximal to the phosphate group (the α-position) appears either unsaturated, as found in bacteria, or saturated, as found in eukaryotes and archea, with the latter form thus requiring an additional reduction reaction, leading to the so-called dolichyl-phosphate (Fig. 1). In all organisms, these lipids fulfill a similar function, which is to facilitate translocation of a polar head group (mono- or oligosaccharides) across a highly hydrophobic environment; therefore, the driving force that presides over this observed structural diversity still remains unexplained.
FIG. 1.
Structures of different polyprenyl-phosphate carrier lipids (A) and lipid intermediates from various glycoconjugate biosynthetic pathways (B).
The way in which these lipids carry out their function is apparently very well conserved.39 The sugar/glycan is first transferred from a nucleotide-diphosphate activated donor to the lipid carrier, leading to the formation of a membrane-bound intermediate with a polyprenyl-diphosphate-glycan structure (Figs. 1 and 2). In some cases, the membrane intermediate displays a polyprenyl-monophosphate-glycan structure, as, for instance, in undecaprenyl-phosphate-L-4-amino-4-deoxy-arabinose that is involved in lipid A modification in Gram-negative bacteria,107 or in dolichyl-phosphate-mannose which is responsible for protein mannosylation96 (Fig. 1). The reason that there is such a divergence in the linkage nature is not understood. The initial step of glycan transfer occurs on the cytoplasmic side of the membrane, the endoplasmic reticulum membrane in eukaryotes, or the cytoplasmic membrane in prokaryotes. The resulting lipid intermediate is then translocated across the membrane so that the glycan moiety becomes accessible to enzymes catalyzing subsequent steps of glycan transfer or polymerization on the outer side of the membrane (Fig. 2). The fact that the sugar is linked to the lipid carrier is not sufficient per se to overcome the thermodynamic barrier which represents the translocation of a polar group across a lipid bilayer, and this event requires a dedicated flippase.94 Nevertheless, given that polyprenyl-phosphate lipids are the preferred membrane-bound glycan carriers found in nature, the polyprenol-based structure, its length, geometry, and membrane orientation, may play an active role in the flippase-catalyzed translocation process. Several biophysical studies have revealed that the presence of polyprenyl-phosphate or its derivatives (polyprenol, polyprenyl-diphosphate, or polyprenyl-sugar) in model membrane vesicles significantly increases their fluidity and ion permeability.109,112,117 It is reasonable to figure out that local destabilization of the membrane may, thus, be a determinant to overcome the translocation energetic barrier.
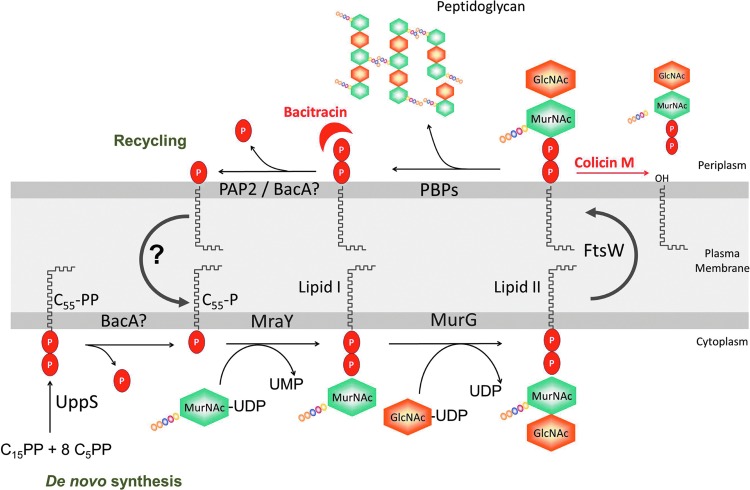
FIG. 2.
C55-P metabolism and membrane steps of peptidoglycan biosynthesis. C55-PP is de novo synthesized by undecaprenyl-diphosphate synthase (UppS) enzyme in the cytoplasm, from which it partitions in the inner side of the plasma membrane. Subsequently, C55-PP should be dephosphorylated to function as a lipid carrier in various polysaccharide biosynthetic pathways, such as the peptidoglycan synthesis. The enzymes MraY and MurG catalyze the successive transfers of the phospho-MurNAc-pentapeptide and GlcNAc motifs from the nucleotide precursors to the lipid carrier C55-P, generating the lipid I and lipid II intermediates, respectively. Lipid II is then translocated by the flippase FtsW to the periplasmic side of the membrane, where polymerization reactions of peptidoglycan that are catalyzed by the penicillin-binding proteins (PBPs) occur. The lipid carrier is released in its pyrophosphate form (C55-PP), which should be dephosphorylated and shuttled back to the inner face of the membrane to be reused. C55-P metabolism is the target of bacitracin that sequestrates C55-PP, thereby inhibiting its dephosphorylation, and of colicin M which cleaves lipid II in dead-end products, C55-OH and 1-pyrophospho-MurNAc(-peptide)-GlcNAc.
Several flippases have been identified, emphasizing the existence of a sharp specificity between the flippase and the nature of the glycan to be translocated.20,41,42,51,89,123 This underlies the fact that the flippase should recognize the sugar moiety at the membrane interface in the first place. In bacteria, the identification of flippases was facilitated, as their genes were generally clustered with genes involved in a particular polysaccharide biosynthesis pathway (FtsW for peptidoglycan,75 Wzx for O-antigen,50 etc.). Then, most flippases have been identified by genetic approaches and through the observation that their inactivation typically provoked the accumulation of a specific membrane-bound intermediate and the concomitant arrest of the biosynthesis of the corresponding polysaccharide. However, the function of these membrane proteins still requires biochemical validation via the reconstitution of the purified proteins in model membrane vesicles and the development of specific flippase assays. The flippase activity of FtsW toward the peptidoglycan lipid II intermediate [undecaprenyl-diphosphate-MurNAc-(pentapeptide)-GlcNAc] was recently addressed by using such an appropriate biochemical approach.75 The lipid II translocation mechanism is considered ATP independent, with the subsequent polymerization step being considered the driving force for unidirectional translocation; however, the mechanism per se is absolutely not established and highlights will now arise from structural studies of these polyprenol-based flippases. Whether the flippase binds the polyprenol chain is not clear. It is possible that the C55 aliphatic chain remains largely embedded in the phospholipid bilayer, providing a part of the driving force by local destabilization in order to elicit the transmembrane movement. Biophysical studies led to the hypothesis that alcohol derivatives (polyprenol) may be oriented parallel to the membrane plan near the center of the bilayer, whereas polyprenyl-phosphate and other derivatives should be oriented perpendicular to the membrane plan with the charged head groups protruding at the hydrophilic surface.125,126 It is then possible that on binding of the diphosphate-sugar moiety by the flippase, reorientation of the aliphatic chain parallel to the membrane plan is induced, thereby favoring translocation, while the head group would diffuse alongside a hydrophilic flippase groove. Once translocated, the membrane-bound intermediate will act as an activated sugar donor, playing the same role as the typical nucleotide-diphosphate donor in the cytoplasmic space. The sugar is finally transferred from this membrane-bound donor to a specific acceptor molecule such as the peptidoglycan polymer growing chains, the lipid A-core for O-antigen LPS synthesis, or other nascent polymers.
As a by-product of the final extracytoplasmic transfer reaction, the lipid carrier is released in a diphosphate form (Fig. 2), except when the membrane-bound intermediate possesses a monophosphate linkage, in which case the lipid carrier is directly released in its active form. The released lipid will then be recycled, providing what is considered an important supply of the lipid carrier.92 It should be dephosphorylated and flipped back to the cytoplasmic side of the membrane to initiate another cycle of glycan translocation, a sequence of events that will be considered further next. The mechanism by which the flipping of the free-lipid carrier occurs is unknown; nevertheless, it was shown that the rate of polyprenyl-phosphate translocation across a protein-free phospholipid bilayer was too slow to satisfy the high rate of polysaccharide synthesis.71,72 About 5,000 lipid II molecules should be flipped per second to match the high rate at which the peptidoglycan is polymerized in Escherichia coli growing cells, thus implying a high rate of lipid recycling.73,113 It was then assumed that polyprenyl-phosphate flipping occurs via a protein-assisted mechanism, even though no specific flippase has yet been identified. It was recently hypothesized that the glycan-derivative flippase itself may carry out the flip of the polyprenyl-phosphate back to the inner side of the membrane once the sugar moiety has been transferred to the final extracytoplasmic acceptor molecule.95 This hypothesis is based on the identification of a dolichyl-phosphate-mannosyl flippase activity in endoplasmic reticulum membranes that was found to be inhibited by dolichyl-phosphate. These data provided evidence that the flippase binds to the isoprenoid chain itself, at least in part, raising the possibility that recycling of the free lipid may also be facilitated by the same flippase in a bidirectional process. In the proposed model, the flippase would display two binding sites located on opposite sides of the membrane and the glycan-charged polyprenol and free isoprenoid would be flipped in opposite directions. We can even hypothesize that the polyprenyl-phosphate would not be released after its translocation, thereby enabling a new sugar unit to be directly charged on the flippase-bound lipid carrier to initiate another cycle of transport. This model implies the existence of multienzyme complexes involving glycosyltransferases on both sides of the membrane, with the polyprenyl-diphosphate phosphatase and the flippase acting in a concerted manner. Biosynthetic complexes have been underlined for the biogenesis of specific polymers, as, for instance, in the peptidoglycan biosynthesis pathway,22 thus supporting such a model. This model of lipid carrier regeneration is highly speculative and other possible scenarios exist, which will be considered further next, but it offers a framework for future studies.
Synthesis of Bacterial Undecaprenyl-Diphosphate
The de novo synthesis of bacterial undecaprenyl-phosphate (C55-P) proceeds in two sequential steps: (1) the biosynthesis of its precursor undecaprenyl-diphosphate (C55-PP) via eight consecutive condensation reactions of the building block, isopentenyl-diphosphate (C5-PP), onto farnesyl-diphosphate (C15-PP) and (2) the dephosphorylation of the latter precursor, yielding the monophosphate “active” form. All isoprenoids (linear isoprenoids, carotenoids, retinoids, ubiquinones, etc.) are synthesized by sequential head-to-tail 1′-4 condensations of the five-carbon building block, the homoallylic substrate, to an allylic diphosphate substrate of varying carbon chain length.16 These reactions are catalyzed by prenyltransferases, which belong to two families according to the stereochemical outcome, cis (Z-prenyltransferases) or trans (E-prenyltransferases), of the double bond in the newly added isoprene.80,116 Each prenyltransferase is specific to the allylic substrate it elongates as well as to the length of the final product (i.e., the number of consecutive condensation reactions). All-trans-farnesyl-diphosphate (C15-PP) results from the condensation of one isoprene unit on dimethylallyl-diphosphate, yielding geranyl-diphosphate (C10-PP), followed by a second condensation reaction catalyzed by the farnesyl-diphosphate synthase, which is the prototype of the trans-prenyltransferase family.64,79 The synthesis of C15-PP is not specific to the metabolism of C55-P, as this compound is also used as a precursor for the formation of a great variety of isoprenoids.
The cis-prenyltransferase undecaprenyl-diphosphate synthase (UppS) catalyzes the elongation of C15-PP with eight isoprene units, yielding di-trans, octa-cis-undecaprenyl-diphosphate.104 The UppS-encoding gene was identified in 1998 in Micrococcus luteus, thereby identifying the first cis-prenyltransferase enzyme.98 This was achieved by screening E. coli clones transformed by a genomic DNA library of M. luteus for increased expression of prenyltransferase activity. UppS enzymes from other bacteria were then identified by sequence homology, and this gene was shown to be essential in E. coli and Streptococcus pneumoniae.2,55 UppS is a soluble cytoplasmic protein that synthesizes a membrane-embedded lipid. The dissociation rate of C55-PP from UppS was shown in vitro to be much faster in the presence of detergent Triton X-100,68 suggesting that in vivo the enzyme should be in close proximity to the cytosolic face of the membrane to enable the rapid release of the product and its concomitant partition in the phospholipid bilayer. UppS activity was also shown to be dependent on the presence of the divalent cation Mg2+ for C5-PP binding and for catalysis, but not for initial C15-PP binding.13,83 Several UppS three-dimensional (3D) structures, with13,14,37 or without29,59 bound substrates, were solved, which along with intensive mutagenesis30,31,58 and kinetic studies68,103 provided great knowledge on the functioning of this biosynthetic step.
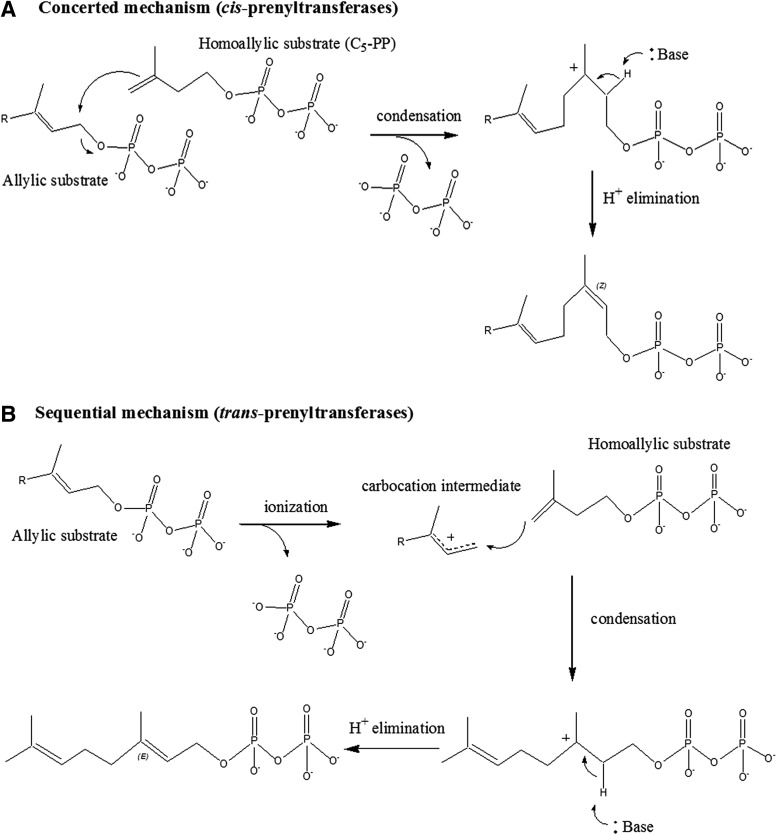
The condensation reaction catalyzed by prenyltransferases occurs via a nucleophilic attack of the C1-atom from the allylic substrate bearing the leaving phosphate group by the C4-atom of C5-PP (Fig. 3).79 The attack is followed by the stereospecific removal of a proton from the C5-PP C2-atom to generate the new double bond. This can take place through a concerted or a sequential mechanism, depending on whether the release of the pyrophosphate group and the nucleophilic attack happen simultaneously or not, with a carbocation intermediate being formed only in the sequential mechanism. Data from Liang and collaborators strongly suggested that UppS may display a concerted mechanism; these authors, indeed, showed that it was impossible to trap a carbocation intermediate during UppS catalysis by using an electron-withdrawing approach to slow down the reaction.67,68 In contrast, trans-prenyltransferases such as the farnesyl-diphosphate synthase obviously catalyze the condensation reaction through a sequential mechanism and formation of an isolable allylic carbocation intermediate species. In the latter process, the allylic substrate first undergoes an ionization, leading to the release of the pyrophosphate group, which is followed by the condensation event and finally, by proton elimination (Fig. 3).
FIG. 3.
Mechanisms of C5-PP condensation on an allylic substrate by cis-prenyltransferases (A) and trans-prenyltransferases (B).
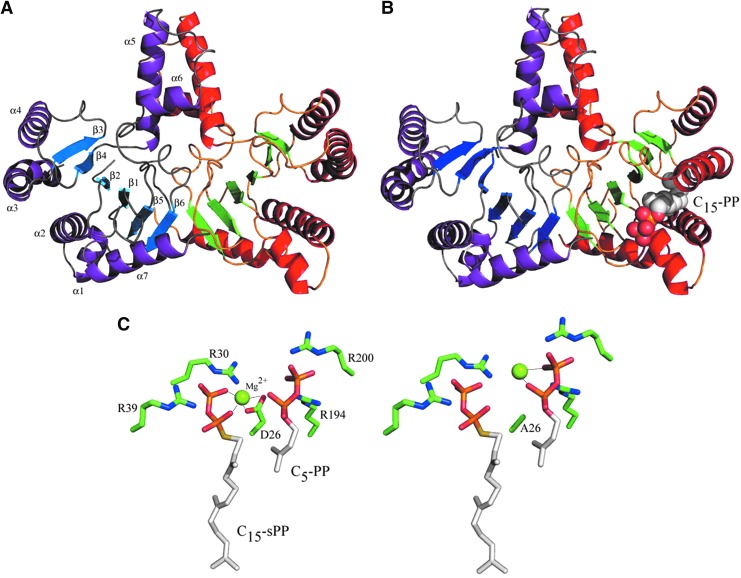
UppS forms a homodimer in which the two 29-kDa subunits are tightly associated via a coiled-coil structure between two long α-helices (α5) arising from each monomer and the edges of a central β-strand (β6) and small α-helices (α6) (Fig. 4A).29 The overall structure shows a central β-sheet core surrounded by five out of seven α-helices. Most of the conserved residues among UppS enzymes are located within two long α-helices (α2 and α3) and four β-strands (β1–β4), all together delineating a large 30Å-deep hydrophobic cleft that was shown to accommodate the elongating isoprenoid carbon tail (Fig. 4B).13 The entrance of the active-site tunnel, in which the binding of negatively charged pyrophosphate groups from the substrates and the catalysis occur, displays several positively charged residues along with an acidic aspartate residue (Asp26) that was shown to be critical for catalysis, and an important flexible loop connecting α3 to β2, the so-called entrance loop.
FIG. 4.
Structures of UppS enzyme from Escherichia coli. Overall structure of UppS (1UEH atomic coordinate): one dimer shown with purple α-helices and blue β-strands and the other monomer shown with red α-helices and green β-strands (A); structure of UppS in complex with C15-PP homoallylic substrate (represented with spheres) (1V7U atomic coordinate), highlighting conformational changes occurring in the α3-helix on substrate binding (B). Details of the ternary complex of UppS with Mg2+, C15-sPP, and C5-PP of wild-type enzyme (left) and a D26A mutant (right) (1X06, 1X07, 1X08, and 1X09 atomic coordinates) emphasizing the condensation mechanism and the role played by the aspartate 26 residue in this process (C).
UppS structures revealed conformational changes occurring on C15-PP binding,13 providing better insights into the molecular mechanism of the enzyme. These changes occurred on the tunnel α3 helix that was straight in the apo-enzyme (the open form), and appeared kinked in the C15-PP-bound structure (the closed form) with tighter space in the tunnel for better binding of the allylic substrate. The closed conformation likely ensures the correct positioning of the allylic substrate, enabling its nucleophilic attack by C5-PP later on. In addition, in contrast to the apo form in which the structure of the entrance loop could not be determined, likely due to its high degree of flexibility, the electron density of the loop was clearly visualized in the C15-PP-bound structure. The structure of a ternary complex composed of UppS, C5-PP, and a C15-PP analog (C15-sPP, in which the oxygen atom connecting the farnesyl and pyrophosphate groups was replaced by a sulfur atom to compromise catalysis) has revealed the central role played by the Asp26 residue and the metal ion in the catalytic process (Fig. 4C).37 In the latter complex, the Mg2+ was not bound to C5-PP to which it is primarily bound (in fact, UppS binds C5-PP in complex with Mg2+), but was, instead, coordinated by the C15-sPP pyrophosphate group and Asp26 side chain. In contrast, in an inactive UppS mutant in which the aspartate residue was replaced by an alanine, the Mg2+ ion remained associated to C5-PP and no Mg2+ was associated to C15-sPP. It was then hypothesized that the Asp26 residue plays a key role in catalysis by controlling the migration of the metal ion from C5-PP to C15-PP, thereby facilitating the allylic pyrophosphate group dissociation which was concomitant to the nucleophilic attack. This structure-based hypothesis also strongly supported the concerted mechanism that had already been suggested by chemical and biochemical approaches.
It was hypothesized that once it has reached its final length after the successive condensation reactions, the lipid product pushes away the entrance loop, a movement which then drives the relaxation of the binding site tunnel to adopt its open conformation, thus releasing C55-PP and enabling a new cycle of synthesis.59 The length of the final isoprenoid tail was shown to be controlled via a molecular ruler mechanism. The bottom of the tunnel is covered by bulky residues that should block a further elongation reaction via steric hindrance. The leucine residue Leu137 in E. coli UppS appeared particularly critical in such a process, as its replacement by an alanine residue resulted in the synthesis of a longer-chain product.59
As an essential bacterial enzyme, UppS represents a potential target for the search of new antibacterial agents. Several reports have already described the identification of UppS inhibitors by using either a rational design or high-throughput screening, validating the use of this enzyme as a drug target. For instance, various bisphosphonate compounds mimicking diphosphate substrates have been found to inhibit UppS via a competition with C15-PP for enzyme binding.36,127 Three dimensional structures of UppS-inhibitor complexes showed that several of these compounds occupied the enzyme active-site tunnel. High-throughput screening also provided several leads that were found to inhibit the growth of S. pneumoniae with satisfactory minimum inhibitory concentrations as a result of UppS activity inhibition.21,61,85
Dephosphorylation of Undecaprenyl-Diphosphate
The dephosphorylation of C55-PP constitutes the ultimate step for the formation of the lipid carrier. This essential reaction is required not only for de novo synthesis but also for the recycling of the lipid carrier when it is released in a diphosphate form after each round of polymerization of a cell wall component (Fig. 2). For several decades, the C55-PP dephosphorylation step was known to be the target of bacitracin, an antibiotic composed of a mixture of small cyclic peptides produced by Bacillus licheniformis.99 The most potent of these compounds, bacitracin A, tightly binds to C55-PP. Though the sequestration of the lipid carrier precursor, bacitracin blocks the formation of C55-P, thereby interrupting the synthesis of peptidoglycan and provoking cell death. In 1993, Cain et al. reported the identification of a gene from E. coli whose overexpression conferred increased bacitracin resistance; for this reason, the gene was named bacA.9 Due to its high degree of hydrophobicity, the 30-kDa BacA protein was predicted to be an integral membrane protein, which was further confirmed when BacA was purified from membrane extracts.25 Using appropriate enzymatic assays, it was then unambiguously established that BacA catalyzed the dephosphorylation of C55-PP to C55-P. This was the first time that such an activity was identified to date. The resistance toward bacitracin observed on bacA overexpression then likely arose from a competition between the enzyme and the antibiotic for C55-PP. Homologues of the bacA gene were then found in all sequenced bacterial genomes, except in the small-sized genome of Helicobacter pylori, and they were rarely found in eukaryotes. Surprisingly, although C55-PP dephosphorylation constitutes an essential step for the biogenesis of the lipid carrier, the inactivation of the chromosomal bacA gene could be readily obtained in several bacteria such as Staphylococcus aureus, S. pneumoniae,12 Mycobacterium smegmatis,90 and E. coli,25 without loss of cell viability. However, inactivation of this gene increased the susceptibility to bacitracin of all these species, and it was also found to attenuate the virulence of S. aureus and S. pneumoniae in the mouse model of infection and to reduce the ability of M. smegmatis to form biofilms. The way in which bacA is involved in these processes was not established. A common pool of C55-P likely participates in the synthesis of various cell wall carbohydrates other than the essential peptidoglycan, such as the teichoic acids and the LPS O-antigen in Gram-positive and Gram-negative bacteria, respectively. Although these structures are not essential for growth, they are known to modulate the virulence of pathogenic bacteria by increasing their host colonization capacities or resistance toward host defenses.8,19 Given that BacA contributes to the formation of the lipid carrier, the availability of C55-P pool might be altered in a bacA null mutant, thus likely compromising the synthesis of nonessential cell wall components; however, this assumption needs further investigations to be validated. To explain the nonessentiality of bacA, the C55-PP phosphatase activity was measured in membrane extracts from an E. coli K12 bacA null mutant, revealing a residual activity of about 25% compared with that in the wild-type strain extracts.25 This finding indicated that BacA accounted for 75% of the total wild-type activity and that another enzyme(s) was(were) responsible for the remaining activity. It should be noted that this residual activity was sufficient to sustain normal growth of E. coli in laboratory conditions.
The mechanisms by which certain bacteria are naturally resistant to bacitracin provided valuable insights into the identity of the other C55-PP phosphatases. In bacitracin-resistant bacteria, such as the antibiotic producer itself (B. licheniformis)86 or certain strains of Bacillus subtilis82 and Enterococcus faecalis,69 the resistance is conferred by the expression of three genes, namely bcrA, bcrB, and bcrC, which could be organized in operon (B. licheniformis and E. faecalis) or not (B. subtilis). In B. subtilis and E. faecalis, the expression of these genes is induced by a transcriptional regulator that senses the presence of bacitracin in the milieu.69,81 Together, bcrA and bcrB encode an ABC transporter that pumps out bacitracin and bcrC was initially thought to encode a subunit of the bacitracin efflux pump itself. However, bcrC was found to confer bacitracin resistance in a way that was not dependent on the pump activity,5 raising the question of the BcrC mode of action. In E. faecalis, bcrC codes for a homologue of BacA, revealing that the expression of a C55-PP phosphatase is specifically used as a means to resist bacitracin. In contrast, BcrC from B. licheniformis and B. subtilis was not related to the BacA family, but to another large superfamily of phosphatases named PAP2 (phosphatidic acid phosphatases of type 2).78,100 In 1999, Harel and collaborators revealed that E. coli cells' resistance to bacitracin was modulated by the expression of the YbjG protein,38 which is a homologue of the BcrC protein from Bacillus species. The disruption of the chromosomal ybjG gene resulted in bacitracin susceptibility, whereas its overexpression conferred antibiotic resistance. Altogether, these findings suggested that these PAP2 proteins might display C55-PP phosphatase activity, similar to BacA. The BcrC membrane protein from B. subtilis was purified and its C55-PP phosphatase activity was clearly demonstrated.4 PAP2 enzymes from Bacillus species are likely devoted to bacitracin resistance via the depletion of the C55-PP pool under the pressure of the antibiotic. Whether other PAP2 proteins encoded by housekeeping genes were also involved in the basic formation of C55-P required further investigations.
A search for PAP2 encoding genes in E. coli underlined the existence of two proteins in addition to YbjG: the phosphatidylglycerol-phosphate phosphatase PgpB and a protein of unknown function, YeiU.26 All three proteins were predicted to be integral membrane proteins with four to six transmembrane segments. The three corresponding chromosomal genes could be disrupted individually without an effect on cell viability and growth.26 In contrast, the co-inactivation of ybjG and pgpB along with bacA was lethal, as demonstrated by the construction of a conditional triple mutant (ΔybjG, ΔpgpB, and ΔbacA) expressing a copy of bacA gene on a temperature-sensitive plasmid. After a shift at the restrictive temperature, the latter mutant was shown to accumulate soluble peptidoglycan nucleotide precursors (UDP-GlcNAc and UDP-MurNAc-pentapeptide) and then to lyse as a result of the arrest of peptidoglycan synthesis. This lethal phenotype was endorsed to a depletion of the C55-P pool due to the absence of the three phosphatases. The data indicated that BacA and the PAP2 enzymes, except YeiU, were able to complement each other in supplying C55-P for normal cell growth, raising the question of the relevance of such a redundancy. Noteworthy, Waechter, and collaborators identified a gene from Saccharomyces cerevisiae, called CWH8, which encodes an endoplasmic reticulum transmembrane protein exhibiting a dolichyl-diphosphate phosphatase activity that yields the active form of the eukaryotic lipid carrier, dolichyl-phosphate.27 Mutations in CWH8 resulted in the accumulation of dolichyl-diphosphate and defects in protein glycosylation. These authors further identified a homologue of the yeast CWH8 gene in mammalian cells that was able to complement the CWH8 mutation.91 Therefore, the two eukaryotic proteins CWH8 and DOLPP1 belonging to the PAP2 superfamily likely carried out a similar function as the prokaryotic BacA, YbjG, and PgpB proteins in carrier lipid (re)generation.
The PAP2 phosphatases are characterized by a signature sequence displaying three distinct motifs designed C1, C2, and C3: K(X6)RP-(X12–54)-PSGH-(X31–54)-SR(X5)H(X3)D, and members of this family are widespread among the three domains of life.100 The homology between PAP2 members is relatively low and generally restricted to the signature sequence. The PAP2 superfamily comprises different types of enzymes: mammalian glucose-6-phosphatases, nonspecific acid phosphatases, various lipid phosphatases, vanadium peroxidases, and several uncharacterized enzymes. Interestingly, this family contains cytoplasmic soluble proteins as well as integral membrane proteins78 such as the identified bacterial C55-PP phosphatases. The 3D structure of a soluble PAP2 enzyme, the nonspecific acid phosphatase from Shimwellia blattae (formerly known as Escherichia blattae), was reported in complex with molybdate, giving insights into the architecture of the active site and the catalytic mechanism of this class of enzyme.49 The active site of these histidine phosphatases is shaped by the three signature motifs. The catalysis occurs via a nucleophilic attack of the phosphate ester bound by a catalytic histidine residue, leading to the covalent binding of a phosphate group to a nitrogen atom of the imidazole ring.33,34,49 From the phosphoenzyme intermediate structure, it was deduced that this catalytic histidine residue belonged to the C3 motif. The formation of this phosphoenzyme intermediate is believed to be established via a charge-relay system involving the aspartate residue from motif C3 and the protonation of the substrate leaving group (in our case C55-P) by the histidine residue from motif C2. In a second step, the phosphoenzyme intermediate should be hydrolyzed to release the enzyme and the phosphate group. By acting this time as a base, the C2-motif histidine may facilitate a nucleophilic attack of the phosphate group by water (releasing inorganic phosphate) or another acceptor molecule (in a phosphotransfer reaction). These catalytic triad residues HisC2-AspC3-HisC3 are conserved in E. coli PAP2 proteins, whereas the other signature residues are mostly variant, suggesting that the basic catalytic mechanism is conserved in all PAP2 members, in particular in membrane PAP2 hydrolases.
The fact that the dephosphorylation of C55-PP is an essential step for the (re)generation of the lipid carrier makes this reaction an attractive target for new antibacterial agents. However, now that we established that this activity can be ascribed to three distinct proteins, any one of which being sufficient to sustain normal cell growth, this implies that a potent inhibitor would have to inactivate all three proteins to display an antibacterial activity. The fact that BacA and PAP2 enzymes belong to two unrelated protein families suggests they have different structures and catalytic mechanisms, which may render the discovery of an effective inhibitor of the C55-PP dephosphorylation step even trickier.
Recycling Versus De Novo Synthesis
As already mentioned, the dephosphorylation of C55-PP is performed in the course of de novo synthesis after the UppS reaction, as well as for the recycling of the lipid carrier after the final glycosyltransferase reaction (Fig. 2). While de novo synthesized C55-PP likely partitions in the inner leaflet of the plasma membrane, the periplasmic oriented glycosyltransferases release C55-PP in the outer leaflet. Given that multiple C55-PP phosphatases have been identified, this raised the question of their respective sites of reaction and involvement in either de novo synthesis or recycling (or both). The membrane topology of E. coli PAP2 enzymes was addressed by the construction of hybrid proteins in which they were fused to reporter proteins being active at only one side of the membrane (β-lactamase, alkaline phosphatase, or Green Fluorescent Protein). PgpB was shown to contain six transmembrane segments and to have its active site residues facing the periplasm.105 Similarly, YbjG and YeiU signature residues were found to be oriented toward the periplasmic space.102 It should be noted that the eukaryotic PAP2 homologue CWH8 was also found to be active on the extracytoplasmic side of the membrane, that is, the endoplasmic reticulum lumen, similar to the prokaryotic PAP2 enzymes.27 In contrast, the membrane topology of the BacA protein, which lacks any feature of a typical phosphatase, remains to be explored. It has been speculated that, according to their topology, the PAP2 enzymes may be exclusively involved in recycling, whereas BacA would participate in de novo synthesis by displaying a cytoplasmic-oriented active site. Nevertheless, this simple model presents a paradox in the fact that any one of the BacA, YbjG, or PgpB proteins can sustain normal cell growth, which suggests that each of them can perform C55-PP dephosphorylation in the course of de novo synthesis as well as recycling, as both processes are supposed to be essential. One way to explain this paradox is that C55-PP and C55-P could be translocated indifferently from one side of the membrane to the other in order to be dephosphorylated and glycosylated at the respective appropriate sites. Another possibility is that the three phosphatases BacA, PgpB, and YbjG are oriented likewise toward the periplasmic space, which then easily explains why they complement each other. In this case, another nonidentified cytoplasmic-oriented C55-PP phosphatase should exist; if not, dephosphorylation of the de novo synthesized C55-PP should also occur at the periplasmic side, which implies that C55-PP has first to be flipped toward the periplasm to be dephosphorylated and then flipped back to the cytoplasm to be glycosylated. The fact that synthesis and dephosphorylation of C55-PP would occur on opposite sides of the membrane would have the benefit to confer a driving force for unidirectional transbilayer movement of the lipid; that is, an accumulation of C55-PP in the inner leaflet would favor its translocation toward the outer leaflet, with the opposite way for C55-P. In any case, the identity of a C55-PP and/or C55-P flippase remains a central question. Not only it was hypothesized that the polyprenol-sugar flippase could play this role but it is also possible that the integral membrane C55-PP phosphatases participate in this process with or without the aid of a flippase.
Links Between C55-PP Metabolism and Other Cellular Functions
The E. coli PgpB protein was first identified in 1983 by Raetz and collaborators as an enzyme participating in the biosynthesis of phosphatidylglycerol, an essential lipid representing about 25% of total E. coli cell membrane phospholipids.47 It is synthesized by dephosphorylation of phosphatidylglycerol-phosphate, a precursor present at extremely low levels in E. coli cell membranes (∼0.1%).15 Three enzymes called PgpA, PgpB, and PgpC that do not exhibit significant sequence similarity were shown to catalyze this dephosphorylation step.32,66 While single and double pgp mutants grew quite normally and contained wild-type levels of phosphatidylglycerol in their membranes, the co-inactivation of all three genes was found to be lethal. Given the apparent role of PgpB in two distinct metabolisms, the substrate specificity of the purified PgpB enzyme was assessed in vitro. The activity of this enzyme toward C55-PP was low (∼100-fold lower) compared with that measured for other substrates such as the diacylglycerol-diphosphate phospholipid or the shorter C15-PP isoprenoid, raising the question of the physiological function of this protein.105 Nevertheless, we showed that the dephosphorylation rate of C55-PP in detergent micelles was increased by the addition of phospholipids, which was not the case for the other substrates. This suggested that the long-chain isoprenoid lipid may adopt in detergent micelles a conformation that prevents maximal binding and/or hydrolysis by PgpB. Therefore, the apparent kinetic constants might be largely underestimated relative to physiological constants. Moreover, the fact that chromosomally expressed PgpB was able to sustain normal cell growth when the other enzymes involved in either C55-PP dephosphorylation26 or phosphatidylglycerol66 synthesis were absent clearly indicated that PgpB displays a dual physiological function and that this promiscuous protein establishes a link between two distinct essential pathways.
Among E. coli PAP2 enzymes, YeiU was initially considered as being unable to catalyze the formation of C55-P in vivo, as the expression of its gene from either the chromosome or a high-copy number plasmid did not restore the viability of the conditional triple mutant.26 In fact, not only E. coli YeiU but also its homologues from Salmonella typhimurium and Salmonella enterica were found to catalyze the specific transfer of the distal phosphate group from C55-PP to the lipid A moiety of LPS, yielding C55-P and lipid A 1-diphosphate (Fig. 5).54,106,111 The identification of C55-PP as the physiological phosphate donor was clearly established by demonstrating that the antibiotic bacitracin, which specifically binds C55-PP, inhibited the formation of lipid A 1-diphosphate in vivo.106 Thus, YeiU also displayed a C55-PP phosphatase activity that was coupled to a phosphotransfer reaction, creating an unexpected link between C55-P metabolism and the lipid A remodeling pathway. YeiU was renamed LpxT according to the nomenclature of proteins involved in lipid A synthesis/modification processes. Interestingly, the C55-PP phosphatase activity of LpxT was not dependent on the presence of lipid A when assayed in vitro. Thus, the reason that LpxT was unable to complement the conditional triple mutant remains to be elucidated.
FIG. 5.
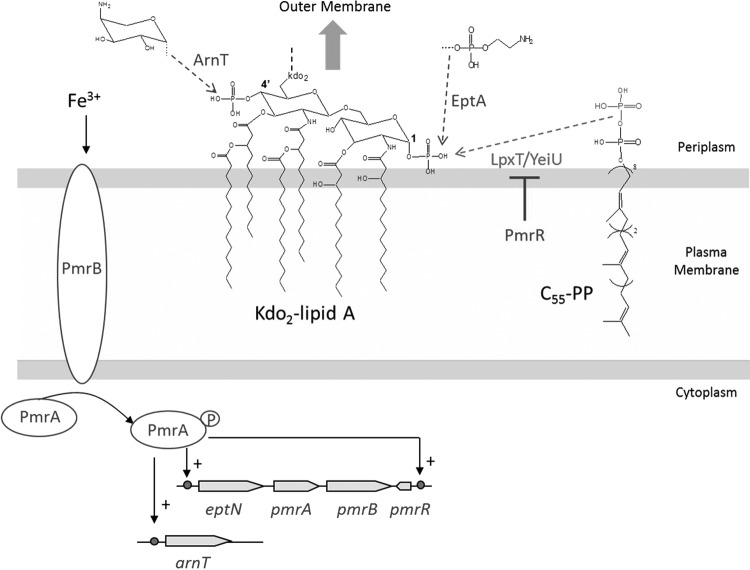
Phosphorylation of lipid A by LpxT in E. coli, Salmonella enterica, and Salmonella typhimurium. LpxT transfers the distal phosphate group from C55-PP to the 1-phosphate group of nascent lipid A at the periplasmic side of the plasma membrane, releasing lipid A-1-diphosphate and C55-P. The 6′ position of lipid A is linked to two Kdo (3-deoxy-D-manno-octulosonic acid) residues from the core region. Subsequently, the core-lipid A (followed or not by the outermost O-antigen depending on the bacterial species) is exported toward the outer membrane and forms the outer leaflet. In rich media, the modification, whose role is yet to be discovered, occurs in about one third of total lipid A molecules. The activation of the PmrA/PmrB two-component system, by high extracellular concentrations of Fe3+, induces the expression of a small-membrane peptide, PmrR, which inhibits LpxT; along with ArnT and EptA, which catalyze the modification of lipid A with 4-amino-4-deoxy-L-arabinose (to the 4′-phosphate group) and phosphoethanolamine (to the 1-phosphate group), respectively. The latter lipid A decorations confer increased resistance to Fe3+ and cationic antibacterial peptides (CAMPs) and attenuate the recognition of lipid A by the host immune TLR4 system.
Lipid A is the essential hydrophobic moiety of the LPS, constituting the outer leaflet of the outer membrane of all Gram-negative bacteria.87 Its predominant structure consists of a hexa-acylated disaccharide of glucosamine that is phosphorylated at both the 1 and 4′ positions. This basic structure undergoes multiple modifications that are regulated by environmental conditions. Although not essential for growth, these modifications are critical for resistance to certain bactericidal agents and for evasion of host immune defenses.77 In rich medium, about one third of E. coli lipid A is modified by the addition of another phosphate group at the 1-position, yielding a lipid A 1-diphosphate species, and this modification is totally abolished in a lpxT null mutant.106 Given that E. coli cells contain about 106 lipid A molecules, LpxT should, therefore, contribute notably to the pool of C55-P. The physiological relevance of this lipid A modification, which increases the negative charge of the bacterial surface, is still unknown, as lpxT mutants from E. coli, S. typhimurium, and S. enterica did not display any apparent phenotype as compared with parental strains. Under certain circumstances, such as the presence of a high concentration of Fe3+ in the extracellular medium, the two-component system PmrA/PmrB induces the expression of ArnT and EptA, which in contrast to LpxT neutralize lipid A phosphate negative charges through the addition of positively charged amino groups.17,77 Indeed, ArnT107 and EptA60 specifically catalyze the addition of 4-amino-4-deoxy-L-arabinose (L-Ara4N) and phosphoethanolamine (pEtN) to the monophosphate groups at the 4′ and 1 positions of lipid A, respectively. These modifications confer resistance to Fe3+ and cationic antibacterial peptides (CAMPs) such as polymyxin B by limiting their binding to the cell surface. They were also shown to decrease the recognition of lipid A by the host immune TLR4 signaling system, thereby enabling bacteria to escape host defenses.84 Thus, EptA and LpxT accomplish two different modifications, that is, the addition of a pEtN or a phosphate group, at the same (1-phosphate) position of the lipid A. While EptA transcription is increased under PmrA/PmrB-induction conditions, LpxT activity was recently demonstrated to be post-translationally repressed via the binding of a small membrane peptide called PmrR,54 whose expression is induced by phosphorylated PmrA, thus favoring the addition of the pEtN group instead of a phosphate group in these conditions. It was hypothesized that this mode of regulation of LpxT by PmrR was more efficient than a transcriptional repression to enable a rapid fine-tuning of cell surface properties (charges content). Interestingly, a search for PmrA/PmrB-regulated genes revealed the ybjG gene, whose transcription appeared induced by phosphorylated PmrA.70 This led to the hypothesis that YbjG expressed in these conditions may compensate for the lack of LpxT in the (re)generation of the C55-P carrier lipid.17
In Cupriavidus metallidurans CH34, a PAP2 phosphatase-encoding gene, named pbrB, was detected within a gene cluster known to participate in a lead resistance mechanism.46 This cluster, pbrTRABCD, was carried on a megaplasmid and consisted of two divergently transcribed operons: pbrTR and pbrABCD.6 The expression of the latter operon was regulated by the zinc/cadmium/lead-responsive element PbrR.76 PbrB and pbrC formed a single gene encoding a putative membrane PAP2 enzyme that was fused to a putative signal peptidase. The precise function of the pbrABCD encoded proteins in the heavy metal resistance mechanism of this species was investigated.46 It was demonstrated that lead resistance was achieved through the cooperation of a zinc/cadmium/lead-efflux pump P-type ATPase, PbrA, and a C55-PP phosphatase, PbrB, the activity of which was not dependent on the presence of the PbrC moiety. The PbrA protein exports the Pb2+ ions to the periplasm and, subsequently, inorganic phosphate groups released by PbrB-dependent dephosphorylation of C55-PP precipitate lead, thus preventing its re-entry into the cell.46 The precipitation of lead phosphate as a means of resistance to this heavy metal had been previously observed in different bacterial species62,74; however, the molecular determinants had not been determined. It should be noted that the pbrB gene was able to complement the conditional E. coli mutant strain in which all identified C55-PP phosphatase encoding genes (ybjG, pgpB, bacA, and lpxT) had been inactivated, and the C55-PP phosphatase activity of the PbrB protein was clearly established in vitro.46 Here, the metabolism of C55-P is linked to a mechanism of lead resistance based on the overexpression of a specific C55-PP phosphatase, which is reminiscent of the process by which certain bacteria are resistant to bacitracin. These different mechanisms rely on the existence of an available pool of C55-PP that can be mobilized in specific situations. As is the case for the phosphorylation of lipid A, C55-PP could be used for purposes that are beyond the basic formation of the lipid carrier. We highlighted the unexpected fact that C55-PP constitutes a donor of the phosphate group in the periplasm, a compartment which does not contain nucleotide-phosphate compounds that usually play this role in the cytoplasm. We can, therefore, envisage the existence of a periplasmic network of phosphorylations involving C55-PP as a phosphate donor and diverse acceptor molecules. The different C55-PP phosphatases could then be implied in specific phosphotransfer reactions, explaining their multiplicity. Periplasmic C55-PP could also represent a supply of energy. We can hypothesize that the C55-PP phosphatases could use the energy released through C55-PP hydrolysis to catalyze, by themselves or with a dedicated flippase, the translocation of the carrier lipid back to the cytoplasmic side of the membrane. This hypothesis is supported by the fact that all the C55-PP phosphatases are integral membrane proteins, which is an essential pattern for a flippase.
Formation and Role of C55-P Derivatives
In eukaryotes and archea, the de novo biosynthesis pathway leading to dolichyl-phosphate includes the formation of dolichol, the alcohol derivative of the lipid carrier.11,23 Such an alcohol intermediate is not found in the C55-PP pathway in prokaryotes. However, a significant pool of undecaprenol (C55-OH) has been observed in many Gram-positive bacteria,3,35,44,108 whose origin and function are not clearly established. In eukaryotes, dolichol is generated by dephosphorylation of the de novo synthesized polyprenyl-diphosphate followed by a reduction of its α-isoprene unit. While the enzymes responsible for the removal of the pyrophosphate group of polyprenyl-diphosphate have not yet been identified, the second step was recently shown to be catalyzed by the steroid 5α-reductase type 3 (SDR5A3) in humans.11 Interestingly, the dolichol derivative in mammalian cells can account for as much as 90% of the total polyprenol pool.57 The role of this derivative is obviously to generate the lipid carrier, but it possibly plays additional cellular functions. In order to generate the active form of the lipid carrier, a dolichol kinase subsequently catalyzes its phosphorylation. The SEC59 gene from yeast was the first gene shown to encode a CTP-dependent dolichol kinase,43 and orthologues with a similar function were identified in mammalians.28
The scenario differs greatly in bacteria where no C55-OH is generated during de novo synthesis of the lipid carrier, although this derivative could account for approximately 90% of the total polyprenol pool in certain bacteria45,108 (S. aureus, E. faecalis, Listeria plantarum, and Listeria monocytogenes). C55-OH was not detectable in other bacteria such as in E. coli,3 leading to the hypothesis that its presence was a characteristic of Gram-positive bacteria; however, further studies are required before drawing definitive conclusions. It was speculated that C55-OH could not only constitute a reserve pool of lipid carrier which can be mobilized in emergency or specific conditions but it may also play unidentified functions, as in eukaryotes. C55-OH can arise from the dephosphorylation of C55-PP and/or C55-P, or can be a by-product of a biosynthetic pathway that is yet to be discovered. None of the bacterial C55-PP phosphatases identified to date catalyzed the formation of C55-OH,26,105 that is, exhibited C55-P phosphatase activity. The potential use of this derivative as a source of lipid carrier relies on the detection of a C55-OH kinase activity in bacteria that produced C55-OH. In B. subtilis and Streptococcus mutant, this kinase was shown to be the product of the dgkA gene.53,65 A DgkA enzyme was first described as an ATP-dependent diacylglycerol kinase in E. coli.115 Thus, dgkA-encoded enzymes likely evolved toward two divergent functions in bacteria, as the E. coli DgkA enzyme is specific for the diacylglycerol substrate, while the DgkA enzyme from B. subtilis and S. pneumoniae are specific for C55-OH. The fact that E. coli DgkA does not phosphorylate C55-OH is consistent with the absence of this derivative in this bacterium. The DgkA enzymes constitute a family of small-membrane proteins (∼15-kDa) whose catalytic mechanism has not been yet determined. They do not share any common features with water-soluble kinases or eukaryotic dolichol kinases. Most studies relevant to this family of enzymes were performed on diacylglycerol kinases and yet little is known about the specific properties of C55-OH kinases.115 The 3D structure of the E. coli DgkA protein was determined by solution NMR spectroscopy114 and more recently, by crystallography.63 To what extent the structure and the catalytic mechanism of Gram-positive DgkA enzymes differ from their E. coli homologue remains to be established. Interestingly, Gram-negative DgkA enzymes recycle diacylglycerol into phosphatidic acid that will re-enter the glycerophospholipid biosynthesis pathway.115 The diacylglycerol appears at the outer leaflet of the plasma membrane after the transfer of a phosphoglycerol group from phosphatidylglycerol to decorate nascent LPS molecules or membrane-derived oligosaccharides (periplasmic MDO are produced in low osmolarity conditions). Then, diacylglycerol should be flipped toward the inner leaflet of the plasma membrane to be subsequently phosphorylated by DgkA. E. coli DgkA displays a homotrimeric structure with each subunit forming a bundle of three transmembrane helices preceded by an N-terminal amphiphilic surface helix protruding at the inner face of the plasma membrane. DgkA presents three active site pockets that are likely shaped by the surface helix emerging from one subunit along with the edges of the three transmembrane segments from another subunit, as suggested by 3D structures.
As already mentioned, the precise role of C55-OH in Gram-positive bacteria is unknown. Although the dgkA gene is not essential for growth, the conversion of C55-OH into C55-P was shown to be determinant under certain circumstances. For instance, a dgkA-null mutant of the oral pathogen S. mutans was more susceptible toward bacitracin than the parental strain,65 suggesting that C55-OH phosphorylation constitutes a bypass of the biosynthetic or recycling pathway when the latter is inhibited. For less evident reasons, the latter mutant was also affected for growth at low pH,122 the biosynthesis of certain lantibiotics,18 the formation of biofilms,124 as well as for virulence.97 Likewise, DgkA from B. subtilis was shown to be required for sporulation by a mechanism that is yet to be clearly established.1 During sporulation, the bacterium synthesizes a peptidoglycan layer called the spore cortex which contributes greatly to the stability of the spore toward a wide range of harsh conditions. In the dgkA mutant, the cortex was found to degenerate after a short period of time after the entry in sporulation. Therefore, the C55-OH pool may play a significant role in the formation of this specific peptidoglycan layer and transition from an actively growing cell to a dormant and stress-resistant spore. Interestingly, the chromosomal dgkA gene from the Gram-positive genus Clostridium was found to be fused to another gene encoding a phosphatase belonging to the PAP2 family. This intriguing finding suggests that clostridia DgkA are bifunctional enzymes which could catalyze the formation of C55-P by either C55-OH phosphorylation or C55-PP dephosphorylation.
While E. coli cells do not usually present any detectable amount of C55-OH within their membranes, the accumulation of this compound was found to occur when these cells were subjected to a bacteriocin called colicin M (ColM)24 (Fig. 2). ColM was known for several decades to target the biosynthesis of peptidoglycan and the recycling of the lipid carrier, thus causing cell lysis. The mechanism of action of ColM was deciphered in 2006, highlighting a hydrolytic activity raised against the lipid II peptidoglycan membrane intermediate and causing the release of C55-OH and 1-pyrophospho-MurNAc(-peptide)-GlcNAc as dead-end products. Interestingly, the overexpression of the cbrA gene (formely yidS), whose function is unknown, was recently demonstrated to confer increased resistance of E. coli toward ColM.40 Although the mechanism of action of CbrA in this process still remains to be established, it was interesting to find out that CbrA is an FAD-bound protein belonging to the geranylgeranyl reductase family. Members of this family are widespread in bacteria and plants, where they are involved not only in the biosynthesis of pigments, but also in archea for the biosynthesis of archeal membrane lipids through the FAD-dependent reduction of isoprenoid-chain double bonds.121 Thus, the E. coli CbrA protein may likely catalyze a similar reaction. Given that CbrA may prevent the hydrolytic activity of ColM against lipid II, it was hypothesized that CbrA should catalyze the hydrogenation of the lipid II, most likely that of the double bond which is proximal to the pyrophosphate linkage. It is also conceivable that the lipid carrier itself or its precursors may be substrates. In any case, this reaction would generate a saturated form of lipid carrier but attempts to detect such derivatives in membranes of CbrA-overproducing cells have failed, raising the question of the specific role of CbrA. Whether the action of CbrA toward lipid II or the lipid carrier is physiological or fortuitous and only due to its overproduction remains unclear. If this function was to be confirmed in future studies, this would imply the existence of saturated derivatives of the lipid carrier in bacteria, which has never been emphasized. The fact that the expression of cbrA is induced under stress conditions would suggest a potential implication of these derivatives in adaptation to harsh conditions.
Conclusion
The ubiquitous polyprenyl-phosphate lipids play a critical function in the formation of extracytoplasmic glycoconjugates by enabling the portage of glycosyl units across the plasma membrane. The translocation process also requires a flippase that specifically recognizes a sugar-linked lipid carrier and terminates the traffic. While the identities of these molecular determinants are now known, at least for certain pathways, the mechanism of translocation is not established. This point will now constitute a central research issue, and the role played by these very peculiar and conserved lipids will have to be decrypted. Once the sugar moiety has been transferred to its final acceptor, the released lipid carrier is shuttled back to the inner side of the membrane to be recycled. This latter translocation process is also not known and, thus, represents another important issue.
In most bacteria, C55-P accomplishes this essential task for the biosynthesis of the different cell wall elements. The first step in C55-P metabolism, that is, the formation of C55-PP, has been particularly well deciphered in the last two decades with the characterization of the essential UppS enzyme in great detail. More recently, the second step, that is, the dephosphorylation of C55-PP, has revealed interesting features, such as the existence of several enzymes, from two different families, that catalyze this single reaction in one bacterium, questioning us about the significance of this redundancy. Perhaps, a clue to this question was given by the discovery that one of these enzymes transfers a phosphate group from C55-PP to a specific extracytoplasmic acceptor molecule, that is, the lipid A. This finding now opens the way to the investigation of other potential phosphorylation reactions that could be important in fine-tuning extracytoplasmic activities and explain the enzyme multiplicity. No mechanistic or structural informations are yet known about the two kinds of C55-PP phosphatases. Therefore, these integral membrane proteins should now be the object of intensive investigations.
Being central to the synthesis of the bacterial cell wall and since a part of it occurs in the extracytoplasmic space, the metabolism of C55-P, its biosynthesis and recycling, represents an interesting target for the development of new antibacterials. A full knowledge of this pathway will likely open the way to new specific molecular targets.
Acknowledgments
This work was supported by grants from the Agence Nationale de la Recherche (BACTOPRENYL project, ANR-11-BSV3-002-01), the Partenariat Hubert Curien (PHC-Tournesol project N° 25434VB), and in part by the Belgian State—Federal Science Policy (Interuniversity Attraction Poles program, project P7/44), the Centre National de la Recherche Scientifique (CNRS), and the University Paris Sud.
Disclosure Statement
All authors report no conflicts of interest relevant to this article.
References
- 1.Amiteye S., Kobayashi K., Imamura D., Hosoya S., Ogasawara N., and Sato T.2003. Bacillus subtilis diacylglycerol kinase (DgkA) enhances efficient Sporulation. J. Bacteriol. 185:5306–5309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Apfel C.M., Takacs B., Fountoulakis M., Stieger M., and Keck W. 1999. Use of genomics to identify bacterial undecaprenyl pyrophosphate synthetase: cloning, expression, and characterization of the essential uppS Gene. J. Bacteriol. 181:483–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barreteau H., Magnet S., El Ghachi M., Touzé T., Arthur M., Mengin-Lecreulx D., and Blanot D.2009. Quantitative high-performance liquid chromatography analysis of the pool levels of undecaprenyl phosphate and its derivatives in bacterial membranes. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 877:213–220 [DOI] [PubMed] [Google Scholar]
- 4.Bernard R., El Ghachi M., Mengin-Lecreulx D., Chippaux M., and Denizot F.2005. BcrC from Bacillus subtilis acts as an undecaprenyl pyrophosphate phosphatase in bacitracin resistance. J. Biol. Chem. 280:28852–28857 [DOI] [PubMed] [Google Scholar]
- 5.Bernard R., Joseph P., Guiseppi A., Chippaux M., and Denizot F.2003. YtscD and YwoA, two independent systems that confer bacitracin resistance to Bacillus subtilis. FEMS Microbiol. Lett. 228:93–97 [DOI] [PubMed] [Google Scholar]
- 6.Borremans B., Hobman J.L., Provoost A., Brown N.L., and van Der Lelie D. 2001. Cloning and functional analysis of the Pbr lead resistance determinant of Ralstonia metallidurans CH34. J. Bacteriol. 183:5651–5658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bouhss A., Trunkfield A.E., Bugg T.D., and Mengin-Lecreulx D. 2008. The biosynthesis of peptidoglycan lipid-linked intermediates. FEMS Microbiol. Rev. 32:208–233 [DOI] [PubMed] [Google Scholar]
- 8.Brown S., Santa Maria J.P., Jr., and Walker S.2013. Wall teichoic acids of Gram-positive bacteria. Annu. Rev. Microbiol. 67:313–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cain B.D., Norton P.J., Eubanks W., Nick H.S., and Allen C.M. 1993. Amplification of the bacA gene confers bacitracin resistance to Escherichia coli. J. Bacteriol. 175:3784–3789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cantagrel V. and Lefeber D.J.2011. From glycosylation disorders to dolichol biosynthesis defects: a new class of metabolic diseases. J. Inherit. Metab. Dis. 34:859–867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cantagrel V., Lefeber D.J., Ng B.G., Guan Z., Silhavy J.L., Bielas S.L., et al. 2010. SRD5A3 is required for converting polyprenol to dolichol and is mutated in a congenital glycosylation disorder. Cell. 142:203–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chalker A.F., Ingraham K.A., Lunsford R.D., Bryant A.P., Bryant J., Wallis N.G., Broskey J.P., Pearson S.C., and Holmes D.J. 2000. The bacA gene, which determines bacitracin susceptibility in Streptococcus pneumoniae and Staphylococcus aureus, is also required for virulence. Microbiology. 146:1547–1553 [DOI] [PubMed] [Google Scholar]
- 13.Chang S.Y., Ko T.P., Chen A.P., Wang A.H., and Liang P.H. 2004. Substrate binding mode and reaction mechanism of undecaprenyl pyrophosphate synthase deduced from crystallographic studies. Protein. Sci. 13:971–978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chang S.Y., Ko T.P., Liang P.H., and Wang A.H. 2003. Catalytic mechanism revealed by the crystal structure of undecaprenyl pyrophosphate synthase in complex with sulfate, magnesium, and triton. J. Biol. Chem. 278:29298–29307 [DOI] [PubMed] [Google Scholar]
- 15.Chang Y.Y., and Kennedy E.P. 1967. Phosphatidyl glycerophosphate phosphatase. J. Lipid Res. 8:456–462 [PubMed] [Google Scholar]
- 16.Chaykin S., Law J., Phillips A.H., Tchen T.T., and Bloch K. Proc. Natl. Acad. Sci. U S A. 44:998–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen H.D., and Groisman E.A. 2013. The biology of the PmrA/PmrB two-component system: the major regulator of lipopolysaccharide modifications. Annu. Rev. Microbiol. 67:83–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen P., Novak J., Qi F., and Caufield P.W.1998. Diacylglycerol kinase is involved in regulation of expression of the lantibiotic mutacin II of Streptococcus mutans. J. Bacteriol. 180:167–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Comstock L.E. and Kasper D.L. 2006. Bacterial glycans: key mediators of diverse host immune responses. Cell. 126:847–850 [DOI] [PubMed] [Google Scholar]
- 20.Dong C., Beis K., Nesper J., Brunkan-Lamontagne A.L., Clarke B.R., Whitfield C., and Naismith J.H. 2006. Wza the translocon for Escherichia coli capsular polysaccharides defines a new class of membrane protein. Nature. 444:226–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Durrant J.D., Cao R., Gorfe A.A., Zhu W., Li J., Sankovsky A., Oldfield E., and McCammon J.A. 2011. Non-bisphosphonate inhibitors of isoprenoid biosynthesis identified via computer-aided drug design. Chem. Biol. Drug Des. 78:323–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Egan A.J. and Vollmer W. 2013. The physiology of bacterial cell division. Ann. N. Y. Acad. Sci. 1277:8–28 [DOI] [PubMed] [Google Scholar]
- 23.Ekstrom T.J., Chojnacki T., and Dallner G. 1984. Metabolic labeling of dolichol and dolichyl phosphate in isolate hepatocytes. J. Biol. Chem. 259:10460–10468 [PubMed] [Google Scholar]
- 24.El Ghachi M., Bouhss A., Barreteau H., Touzé T., Auger G., Blanot D., and Mengin-Lecreulx D.2006. Colicin M exerts its bacteriolytic effect via enzymatic degradation of undecaprenyl phosphate-linked peptidoglycan precursors. J. Biol. Chem. 281:22761–22772 [DOI] [PubMed] [Google Scholar]
- 25.El Ghachi M., Bouhss A., Blanot D., and Mengin-Lecreulx D.2004. The bacA Gene of Escherichia coli encodes an undecaprenyl pyrophosphate phosphatase activity. J. Biol. Chem. 279:30106–30113 [DOI] [PubMed] [Google Scholar]
- 26.El Ghachi M., Derbise A., Bouhss A., and Mengin-Lecreulx D.2005. Identification of multiple genes encoding membrane proteins with undecaprenyl pyrophosphate phosphatase (UppP) activity in Escherichia coli. J. Biol. Chem. 280:18689–18695 [DOI] [PubMed] [Google Scholar]
- 27.Fernandez F., Rush J.S., Toke D.A., Han G.S., Quinn J.E., Carman G.M., Choi J.Y., Voelker D.R., Aebi M., and Waechter C.J. 2001. The CWH8 gene encodes a dolichyl pyrophosphate phosphatase with a luminally oriented active site in the endoplasmic reticulum of Saccharomyces cerevisiae. J. Biol. Chem. 276:41455–41464 [DOI] [PubMed] [Google Scholar]
- 28.Fernandez F., Shridas P., Jiang S., Aebi M., and Waechter C.J.2002. Expression and characterization of a human cDNA that complements the temperature-sensitive defect in dolichol kinase activity in the yeast Sec59-1 mutant: the enzymatic phosphorylation of dolichol and diacylglycerol are catalyzed by separate CTP-mediated kinase activities in Saccharomyces cerevisiae. Glycobiology. 12:555–562 [DOI] [PubMed] [Google Scholar]
- 29.Fujihashi M., Zhang Y.W., Higuchi Y., Li X.Y., Koyama T., and Miki K. 2001. Crystal structure of cis-prenyl chain elongating enzyme, undecaprenyl diphosphate synthase. Proc. Natl. Acad. Sci. U S A. 98:4337–4342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fujikura K., Zhang Y.W., Fujihashi M., Miki K., and Koyama T.2003. Mutational analysis of allylic substrate binding site of Micrococcus luteus B-P 26 undecaprenyl diphosphate synthase. Biochemistry. 42:4035–4041 [DOI] [PubMed] [Google Scholar]
- 31.Fujikura K., Zhang Y.W., Yoshizaki H., Nishino T., and Koyama T.2000. Significance of Asn-77 and Trp-78 in the catalytic function of undecaprenyl diphosphate synthase of Micrococcus luteus B-P 26. J. Biochem. 128:917–922 [DOI] [PubMed] [Google Scholar]
- 32.Funk C.R., Zimniak L., and Dowhan W. 1992. The pgpA and pgpB genes of Escherichia coli are not essential: evidence for a third phosphatidylglycerophosphate phosphatase. J. Bacteriol. 174:205–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ghosh A., Shieh J.J., Pan C.J., and Chou J.Y. 2004. Histidine 167 is the phosphate acceptor in glucose-6-phosphatase-β forming a phosphohistidine enzyme intermediate during catalysis. J. Biol. Chem. 279:12479–12483 [DOI] [PubMed] [Google Scholar]
- 34.Ghosh A., Shieh J.J., Pan C.J., Sun M.S., and Chou J.Y. 2002. The catalytic center of glucose-6-phosphatase. His176 is the nucleophile forming the phosphohistidine-enzyme intermediate during catalysis. J. Biol. Chem. 277:32837–32842 [DOI] [PubMed] [Google Scholar]
- 35.Gough D.P., Kirby A.L., Richards J.B., and Hemming F.W. 1970. The characterization of undecaprenol of Lactobacillus plantarum. Biochem. J. 118:167–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guo R.T., Cao R., Liang P.H., Ko T.P., Chang T.H., Hudock M.P., Jeng W.Y., Chen C.K., Zhang Y., Song Y., Kuo C.J., Yin F., Oldfield E., and Wang A.H. 2007. Bisphosphonates target multiple sites in both cis- and trans-prenyltransferases. Proc. Natl. Acad. Sci. U S A. 104:10022–10027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guo R.T., Ko T.P., Chen A.P., Kuo C.J., Wang A.H., and Liang P.H. 2005. Crystal structures of undecaprenyl pyrophosphate synthase in complex with magnesium, isopentenyl pyrophosphate, and farnesyl thiopyrophosphate: roles of the metal ion and conserved residues in catalysis. J. Biol. Chem. 280:20762–20774 [DOI] [PubMed] [Google Scholar]
- 38.Harel Y.M., Bailone A., and Bibi E. 1999. Resistance to bacitracin as modulated by an Escherichia coli homologue of the bacitracin ABC transporter BcrC subunit from Bacillus licheniformis. J. Bacteriol. 181:6176–6178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hartley M.D. and Imperiali B. 2012. At the membrane frontier: a prospectus on the remarkable evolutionary conservation of polyprenols and polyprenyl-phosphates. Arch. Biochem. Biophys. 517:83–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Helbig S., Hantke K., Ammelburg M., and Braun V.2012. CbrA is a flavin adenine dinucleotide protein that modifies the Escherichia coli outer membrane and confers specific resistance to colicin M. J. Bacteriol. 194:4894–4903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Helenius J., and Aebi M.2002. Transmembrane movement of dolichol linked carbohydrates during N-glycoprotein biosynthesis in the endoplasmic reticulum. Semin. Cell. Dev. Biol. 13:171–178 [DOI] [PubMed] [Google Scholar]
- 42.Helenius J., Ng D.T., Marolda C.L., Walter P., Valvano M.A., and Aebi M. 2002. Translocation of lipid-linked oligosaccharides across the ER membrane requires Rft1 Protein. Nature. 415:447–450 [DOI] [PubMed] [Google Scholar]
- 43.Heller L., Orlean P., and Adair W.L., Jr.1992. Saccharomyces cerevisiae sec59 cells are deficient in dolichol kinase activity. Proc. Natl. Acad. Sci. U S A. 89:7013–7016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Higashi Y., Siewert G., and Strominger J.L.1970. Biosynthesis of the peptidoglycan of bacterial cell walls. XIX. isoprenoid alcohol phosphokinase. J. Biol. Chem. 245:3683–3690 [PubMed] [Google Scholar]
- 45.Higashi Y., Strominger J.L., and Sweeley C.C.1970. Biosynthesis of the peptidoglycan of bacterial cell walls. XXI. Isolation of free C55-isoprenoid alcohol and of lipid intermediates in peptidoglycan synthesis from Staphylococcus aureus. J. Biol. Chem. 245:3697–3702 [PubMed] [Google Scholar]
- 46.Hynninen A., Touzé T., Pitkanen L., Mengin-Lecreulx D., and Virta M.2009. An efflux transporter PbrA and a phosphatase PbrB cooperate in a lead-resistance mechanism in bacteria. Mol. Microbiol. 74:384–394 [DOI] [PubMed] [Google Scholar]
- 47.Icho T., and Raetz C.R.1983. Multiple genes for membrane-bound phosphatases in Escherichia coli and their action on phospholipid precursors. J. Bacteriol. 153:722–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ishii K., Sagami H., and Ogura K.1986. A novel prenyltransferase from Paracoccus denitrificans. Biochem. J. 233:773–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ishikawa K., Mihara Y., Gondoh K., Suzuki E., and Asano Y.2000. X-ray structures of a novel acid phosphatase from Escherichia blattae and its complex with the transition-state analog molybdate. EMBO J. 19:2412–2423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Islam S.T., Eckford P.D., Jones M.L., Nugent T., Bear C.E., Vogel C., and Lam J.S. 2013. Proton-dependent gating and proton uptake by Wzx support O-antigen-subunit antiport across the bacterial inner membrane. MBio. 4:e00678–e00713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Islam S.T., and Lam J.S. 2013. Wzx flippase-mediated membrane translocation of sugar polymer precursors in bacteria. Environ. Microbiol. 15:1001–1015 [DOI] [PubMed] [Google Scholar]
- 52.Iwashkiw J.A., Vozza N.F., Kinsella R.L., and Feldman M.F. 2013. Pour some sugar on it: the expanding world of bacterial protein O-linked glycosylation. Mol. Microbiol. 89:14–28 [DOI] [PubMed] [Google Scholar]
- 53.Jerga A., Lu Y.J., Schujman G.E., de Mendoza D., and Rock C.O. 2007. Identification of a soluble diacylglycerol kinase required for lipoteichoic acid production in Bacillus subtilis. J. Biol. Chem. 282:21738–21745 [DOI] [PubMed] [Google Scholar]
- 54.Kato A., Chen H.D., Latifi T., and Groisman E.A.2012. Reciprocal control between a bacterium's regulatory system and the modification status of its lipopolysaccharide. Mol. Cell. 47:897–908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kato J., Fujisaki S., Nakajima K., Nishimura Y., Sato M., and Nakano A.1999. The Escherichia coli homologue of yeast RER2, a key enzyme of dolichol synthesis, is essential for carrier lipid formation in bacterial cell wall synthesis. J. Bacteriol. 181:2733–2738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kaur D., Brennan P.J., and Crick D.C.2004. Decaprenyl diphosphate synthesis in Mycobacterium tuberculosis. J. Bacteriol. 186:7564–7570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Keller R.K., and Nellis S.W. 1986. Quantitation of dolichyl phosphate and dolichol in major organs of the rat as a function of age. Lipids. 21:353–355 [DOI] [PubMed] [Google Scholar]
- 58.Kharel Y., Zhang Y.W., Fujihashi M., Miki K., and Koyama T.2001. Identification of significant residues for homoallylic substrate binding of Micrococcus luteus B-P 26 undecaprenyl diphosphate synthase. J. Biol. Chem. 276:28459–28464 [DOI] [PubMed] [Google Scholar]
- 59.Ko T.P., Chen Y.K., Robinson H., Tsai P.C., Gao Y.G., Chen A.P., Wang A.H., and Liang P.H. 2001. Mechanism of product chain length determination and the role of a flexible loop in Escherichia coli undecaprenyl-pyrophosphate synthase catalysis. J. Biol. Chem. 276:47474–47482 [DOI] [PubMed] [Google Scholar]
- 60.Lee H., Hsu F.F., Turk J., and Groisman E.A.2004. The PmrA-Regulated pmrC gene mediates phosphoethanolamine modification of lipid A and polymyxin resistance in Salmonella enterica. J. Bacteriol. 186:4124–4133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lee L.V., Granda B., Dean K., Tao J., Liu E., Zhang R., Peukert S., Wattanasin S., Xie X., Ryder N.S., Tommasi R., and Deng G. 2010. Biophysical investigation of the mode of inhibition of tetramicacids, the allosteric inhibitors of undecaprenyl pyrophosphate synthase. Biochemistry. 49:5366–5376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Levinson H.S., and Mahler I. 1998. Phosphatase activity and lead resistance in Citrobacter freundii and Staphylococcus aureus. FEMS Microbiol. Lett. 161:135–138 [DOI] [PubMed] [Google Scholar]
- 63.Li D., Lyons J.A., Pye V.E., Vogeley L., Aragao D., Kenyon C.P., Shah S.T., Doherty C., Aherne M., and Caffrey M. Nature. 497:521–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liang P.H., Ko T.P., and Wang A.H. 2002. Structure, mechanism and function of prenyltransferases. Eur. J. Biochem. 269:3339–3354 [DOI] [PubMed] [Google Scholar]
- 65.Lis M., and Kuramitsu H.K.2003. The stress-responsive dgk gene from Streptococcus mutans encodes a putative undecaprenol kinase activity. Infect. Immun. 71:1938–1943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lu Y.H., Guan Z., Zhao J., and Raetz C.R. 2011. Three phosphatidylglycerol-phosphate phosphatases in the inner membrane of Escherichia coli. J. Biol. Chem. 286:5506–5518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lu Y.P., Liu H.G., and Liang P.H. 2009. Different reaction mechanisms for cis- and trans-prenyltransferases. Biochem. Biophys. Res. Commun. 379:351–355 [DOI] [PubMed] [Google Scholar]
- 68.Lu Y.P., Liu H.G., Teng K.H., and Liang P.H. 2010. Mechanism of cis-prenyltransferase reaction probed by substrate analogues. Biochem. Biophys. Res. Commun. 400:758–762 [DOI] [PubMed] [Google Scholar]
- 69.Manson J.M., Keis S., Smith J.M., and Cook G.M. 2004. Acquired bacitracin resistance in Enterococcus faecalis is mediated by an ABC transporter and a novel regulatory protein, BcrR. Antimicrob. Agents Chemother. 48:3743–3748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Marchal K., De Keersmaecker S., Monsieurs P., van Boxel N., Lemmens K., Thijs G., Vanderleyden J., and De Moor B.2004. In silico identification and experimental validation of PmrAB targets in Salmonella typhimurium by regulatory motif detection. Genome Biol. 5:R9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.McCloskey M.A., and Troy F.A. 1980. Paramagnetic isoprenoid carrier lipids. 1. chemical synthesis and incorporation into model membranes. Biochemistry. 19:2056–2060 [DOI] [PubMed] [Google Scholar]
- 72.McCloskey M.A., and Troy F.A. 1980. Paramagnetic isoprenoid carrier lipids. 2. dispersion and dynamics in lipid membranes. Biochemistry. 19:2061–2066 [DOI] [PubMed] [Google Scholar]
- 73.Mengin-Lecreulx D., and van Heijenoort J.1985. Effect of growth conditions on peptidoglycan content and cytoplasmic steps of its biosynthesis in Escherichia coli. J. Bacteriol. 163:208–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mire C.E., Tourjee J.A., O'Brien W.F., Ramanujachary K.V., and Hecht G.B. 2004. Lead precipitation by Vibrio harveyi: evidence for novel quorum-sensing interactions. Appl. Environ. Microbiol. 70:855–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mohammadi T., van Dam V., Sijbrandi R., Vernet T., Zapun A., Bouhss A., Diepeveen-de Bruin M., Nguyen-Disteche M., de Kruijff B., and Breukink E.2011. Identification of FtsW as a transporter of lipid-linked cell wall precursors across the membrane. EMBO J. 30:1425–1432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Monchy S., Benotmane M.A., Wattiez R., van Aelst S., Auquier V., Borremans B., Mergeay M., Taghavi S., van der Lelie D., and Vallaeys T.2006. Transcriptomic and proteomic analyses of the pMOL30-encoded copper resistance in Cupriavidus metallidurans strain CH34. Microbiology. 152:1765–1776 [DOI] [PubMed] [Google Scholar]
- 77.Needham B.D., and Trent M.S. 2013. Fortifying the barrier: the impact of lipid A remodelling on bacterial pathogenesis. Nat. Rev. Microbiol. 11:467–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Neuwald A.F.1997. An unexpected structural relationship between integral membrane phosphatases and soluble haloperoxidases. Protein Sci. 6:1764–1767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ogura K., and Koyama T.1998. Enzymatic aspects of isoprenoid chain elongation. Chem. Rev. 98:1263–1276 [DOI] [PubMed] [Google Scholar]
- 80.Ogura K., Koyama T., and Sagami H.1997. Polyprenyl diphosphate synthases. Subcell. Biochem. 28:57–87 [DOI] [PubMed] [Google Scholar]
- 81.Ohki R., Giyanto , Tateno K., Masuyama W., Moriya S., Kobayashi K., and Ogasawara N.2003. The BceRS two-component regulatory system induces expression of the bacitracin transporter, BceAB, in Bacillus subtilis. Mol. Microbiol. 49:1135–1144 [DOI] [PubMed] [Google Scholar]
- 82.Ohki R., Tateno K., Okada Y., Okajima H., Asai K., Sadaie Y., Murata M., and Aiso T.2003. A bacitracin-resistant Bacillus subtilis gene encodes a homologue of the membrane-spanning subunit of the Bacillus licheniformis ABC transporter. J. Bacteriol. 185:51–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Pan J.J., Yang L.W., and Liang P.H. 2000. Effect of site-directed mutagenesis of the conserved aspartate and glutamate on Escherichia coli undecaprenyl pyrophosphate synthase catalysis. Biochemistry. 39:13856–13861 [DOI] [PubMed] [Google Scholar]
- 84.Park B.S., Song D.H., Kim H.M., Choi B.S., Lee H., and Lee J.O. 2009. The structural basis of lipopolysaccharide recognition by the TLR4-MD-2 complex. Nature. 458:1191–1195 [DOI] [PubMed] [Google Scholar]
- 85.Peukert S., Sun Y., Zhang R., Hurley B., Sabio M., Shen X., Gray C., Dzink-Fox J., Tao J., Cebula R., and Wattanasin S.2008. Design and structure-activity relationships of potent and selective inhibitors of undecaprenyl pyrophosphate synthase (UppS): tetramic, tetronic acids and dihydropyridin-2-ones. Bioorg. Med. Chem. Lett. 18:1840–1844 [DOI] [PubMed] [Google Scholar]
- 86.Podlesek Z., Comino A., Herzog-Velikonja B., Zgur-Bertok D., Komel R., and Grabnar M.1995. Bacillus licheniformis bacitracin-resistance ABC transporter: relationship to mammalian multidrug resistance. Mol. Microbiol. 16:969–976 [DOI] [PubMed] [Google Scholar]
- 87.Raetz C.R., Reynolds C.M., Trent M.S., and Bishop R.E. 2007. Lipid A modification systems in Gram-negative bacteria. Annu. Rev. Biochem. 76:295–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Rahman A., Barr K., and Rick P.D.2001. Identification of the structural gene for the TDP-Fuc4NAc:lipid II Fuc4NAc transferase involved in synthesis of enterobacterial common antigen in Escherichia coli K-12. J. Bacteriol. 183:6509–6516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Rick P.D., Barr K., Sankaran K., Kajimura J., Rush J.S., and Waechter C.J. 2003. Evidence that the wzxE gene of Escherichia coli K-12 encodes a protein involved in the transbilayer movement of a trisaccharide-lipid intermediate in the assembly of enterobacterial common antigen. J. Biol. Chem. 278:16534–16542 [DOI] [PubMed] [Google Scholar]
- 90.Rose L., Kaufmann S.H., and Daugelat S.2004. Involvement of Mycobacterium smegmatis undecaprenyl phosphokinase in biofilm and smegma formation. Microbes Infect. 6:965–971 [DOI] [PubMed] [Google Scholar]
- 91.Rush J.S., Cho S.K., Jiang S., Hofmann S.L., and Waechter C.J. 2002. Identification and characterization of a cDNA encoding a dolichyl pyrophosphate phosphatase located in the endoplasmic reticulum of mammalian cells. J. Biol. Chem. 277:45226–45234 [DOI] [PubMed] [Google Scholar]
- 92.Rush J.S., Gao N., Lehrman M.A., and Waechter C.J. 2008. Recycling of dolichyl monophosphate to the cytoplasmic leaflet of the endoplasmic reticulum after the cleavage of dolichyl pyrophosphate on the lumenal monolayer. J. Biol. Chem. 283:4087–4093 [DOI] [PubMed] [Google Scholar]
- 93.Samuel G., and Reeves P.2003. Biosynthesis of O-antigens: genes and pathways involved in nucleotide sugar precursor synthesis and O-antigen assembly. Carbohydr. Res. 338:2503–2519 [DOI] [PubMed] [Google Scholar]
- 94.Sanyal S., and Menon A.K.2009. Flipping lipids: why an’ what's the reason for?. ACS Chem. Biol. 4:895–909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Sanyal S., and Menon A.K.2010. Stereoselective transbilayer translocation of mannosyl phosphoryl dolichol by an endoplasmic reticulum flippase. Proc. Natl. Acad. Sci. U S A. 107:11289–11294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Schenk B., Fernandez F., and Waechter C.J.2001. The ins(ide) and outs(ide) of dolichyl phosphate biosynthesis and recycling in the endoplasmic reticulum. Glycobiology. 11:61R–70R [DOI] [PubMed] [Google Scholar]
- 97.Shibata Y., van der Ploeg J.R., Kozuki T., Shirai Y., Saito N., Kawada-Matsuo M., Takeshita T., and Yamashita Y.2009. Kinase activity of the dgk gene product is involved in the virulence of Streptococcus mutans. Microbiology. 155:557–565 [DOI] [PubMed] [Google Scholar]
- 98.Shimizu N., Koyama T., and Ogura K.1998. Molecular cloning, expression, and purification of undecaprenyl diphosphate synthase. No sequence similarity between E- and Z-prenyl diphosphate synthases. J. Biol. Chem. 273:19476–19481 [DOI] [PubMed] [Google Scholar]
- 99.Siewert G., and Strominger J.L.1967. Bacitracin: an inhibitor of the dephosphorylation of lipid pyrophosphate, an intermediate in the biosynthesis of the peptidoglycan of bacterial cell wall. Proc. Natl. Acad. Sci. U S A. 57:767–773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Stukey J., and Carman G.M.1997. Identification of a novel phosphatase sequence motif. Protein Sci. 6:469–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Surmacz L., and Swiezewska E.2011. Polyisoprenoids—secondary metabolites or physiologically important superlipids?. Biochem. Biophys. Res. Commun. 407:627–632 [DOI] [PubMed] [Google Scholar]
- 102.Tatar L.D., Marolda C.L., Polischuk A.N., van Leeuwen D., and Valvano M.A. 2007. An Escherichia coli undecaprenyl-pyrophosphate phosphatase implicated in undecaprenyl phosphate recycling. Microbiology. 153:2518–2529 [DOI] [PubMed] [Google Scholar]
- 103.Teng K.H., and Liang P.H. 2012. Structures, mechanisms and inhibitors of undecaprenyl diphosphate synthase: a cis-prenyltransferase for bacterial peptidoglycan biosynthesis. Bioorg. Chem. 43:51–57 [DOI] [PubMed] [Google Scholar]
- 104.Teng K.H., and Liang P.H. 2012. Undecaprenyl diphosphate synthase, a cis-prenyltransferase synthesizing lipid carrier for bacterial cell wall biosynthesis. Mol. Membr. Biol. 29:267–273 [DOI] [PubMed] [Google Scholar]
- 105.Touzé T., Blanot D., and Mengin-Lecreulx D.2008. Substrate specificity and membrane topology of Escherichia coli PgpB, an undecaprenyl pyrophosphate phosphatase. J. Biol. Chem. 283:16573–16583 [DOI] [PubMed] [Google Scholar]
- 106.Touzé T., Tran A.X., Hankins J.V., Mengin-Lecreulx D., and Trent M.S. 2008. Periplasmic phosphorylation of lipid A is linked to the synthesis of undecaprenyl phosphate. Mol. Microbiol. 67:264–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Trent M.S., Ribeiro A.A., Lin S., Cotter R.J., and Raetz C.R. 2001. An inner membrane enzyme in Salmonella and Escherichia coli that transfers 4-Amino-4-deoxy-L-arabinose to lipid A: induction on polymyxin-resistant mutants and role of a novel lipid-linked donor. J. Biol. Chem. 276:43122–43131 [DOI] [PubMed] [Google Scholar]
- 108.Umbreit J.N., Stone K.J., and Strominger J.L. 1972. Isolation of polyisoprenyl alcohols from Streptococcus faecalis. J. Bacteriol. 112:1302–1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Valtersson C., van Duyn G., Verkleij A.J., Chojnacki T., de Kruijff B., and Dallner G. 1985. The influence of dolichol, dolichol esters, and dolichyl phosphate on phospholipid polymorphism and fluidity in model membranes. J. Biol. Chem. 260:2742–2751 [PubMed] [Google Scholar]
- 110.Valvano M.A.2003. Export of O-specific lipopolysaccharide. Front. Biosci. 8:s452–s471 [DOI] [PubMed] [Google Scholar]
- 111.Valvano M.A.2008. Undecaprenyl phosphate recycling comes out of age. Mol. Microbiol. 67:232–235 [DOI] [PubMed] [Google Scholar]
- 112.van Duijn G., Valtersson C., Chojnacki T., Verkleij A.J., Dallner G., and de Kruijff B. 1986. Dolichyl phosphate induces non-bilayer structures, vesicle fusion and transbilayer movement of lipids: a model membrane study. Biochim. Biophys. Acta. 861:211–223 [DOI] [PubMed] [Google Scholar]
- 113.van Heijenoort Y., Gomez M., Derrien M., Ayala J., and van Heijenoort J.1992. Membrane intermediates in the peptidoglycan metabolism of Escherichia coli: possible roles of PBP 1b and PBP 3. J. Bacteriol. 174:3549–3557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Van Horn W.D., Kim H.J., Ellis C.D., Hadziselimovic A., Sulistijo E.S., Karra M.D., Tian C., Sonnichsen F.D., and Sanders C.R. 2009. Solution nuclear magnetic resonance structure of membrane-integral diacylglycerol kinase. Science. 324:1726–1729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Van Horn W.D., and Sanders C.R. 2012. Prokaryotic diacylglycerol kinase and undecaprenol kinase. Annu. Rev. Biophys. 41:81–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Wang K.C., and Ohnuma S. 2000. Isoprenyl diphosphate synthases. Biochim. Biophys. Acta. 1529:33–48 [DOI] [PubMed] [Google Scholar]
- 117.Wang X., Mansourian A.R., and Quinn P.J.2008. The effect of dolichol on the structure and phase behaviour of phospholipid model membranes. Mol. Membr. Biol. 25:547–556 [DOI] [PubMed] [Google Scholar]
- 118.Weissborn A.C., Rumley M.K., and Kennedy E.P. 1991. Biosynthesis of membrane-derived oligosaccharides. Membrane-bound glucosyltransferase system from Escherichia coli requires polyprenyl phosphate. J. Biol. Chem. 266:8062–8067 [PubMed] [Google Scholar]
- 119.Welti M. 2013. Regulation of dolichol-linked glycosylation. Glycoconj. J. 30:51–56 [DOI] [PubMed] [Google Scholar]
- 120.Whitfield C. 2006. Biosynthesis and assembly of capsular polysaccharides in Escherichia coli. Annu. Rev. Biochem. 75:39–68 [DOI] [PubMed] [Google Scholar]
- 121.Xu Q., Eguchi T., Mathews I.I., Rife C.L., Chiu H.J., Farr C.L., Feuerhelm J., Jaroszewski L., Klock H.E., Knuth M.W., Miller M.D., Weekes D., Elsliger M.A., Deacon A.M., Godzik A., Lesley S.A., and Wilson I.A. 2010. Insights into substrate specificity of geranylgeranyl reductases revealed by the structure of digeranylgeranylglycerophospholipid reductase, an essential enzyme in the biosynthesis of archaeal membrane lipids. J. Mol. Biol. 404:403–417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Yamashita Y., Takehara T., and Kuramitsu H.K.1993. Molecular characterization of a Streptococcus mutans mutant altered in environmental stress responses. J. Bacteriol. 175:6220–6228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Yan A., Guan Z., and Raetz C.R.2007. An undecaprenyl phosphate-aminoarabinose flippase required for polymyxin resistance in Escherichia coli. J. Biol. Chem. 282:36077–36089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Yoshida A., and Kuramitsu H.K.2002. Multiple Streptococcus mutans genes are involved in biofilm formation. Appl. Environ. Microbiol. 68:6283–6291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Zhou G.P., and Troy F.A., 2nd 2005. NMR studies on how the binding complex of polyisoprenol recognition sequence peptides and polyisoprenols can modulate membrane structure. Curr. Protein Pept. Sci. 6:399–411 [DOI] [PubMed] [Google Scholar]
- 126.Zhou G.P., and Troy F.A., 2nd 2005. NMR study of the preferred membrane orientation of polyisoprenols (dolichol) and the impact of their complex with polyisoprenyl recognition sequence peptides on membrane structure. Glycobiology. 15:347–359 [DOI] [PubMed] [Google Scholar]
- 127.Zhu W., Zhang Y., Sinko W., Hensler M.E., Olson J., Molohon K.J., Lindert S., Cao R., Li K., Wang K., Wang Y., Liu Y.L., Sankovsky A., de Oliveira C.A., Mitchell D.A., Nizet V., McCammon J.A., and Oldfield E. 2013. Antibacterial drug leads targeting isoprenoid biosynthesis. Proc. Natl. Acad. Sci. U S A. 110:123–128 [DOI] [PMC free article] [PubMed] [Google Scholar]