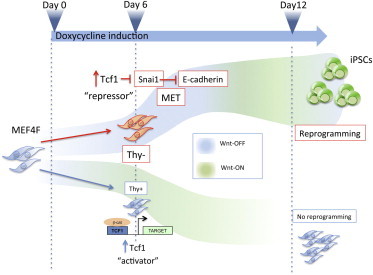
Summary
Cyclic activation of the Wnt/β-catenin signaling pathway controls cell fusion-mediated somatic cell reprogramming. TCFs belong to a family of transcription factors that, in complex with β-catenin, bind and transcriptionally regulate Wnt target genes. Here, we show that Wnt/β-catenin signaling needs to be off during the early reprogramming phases of mouse embryonic fibroblasts (MEFs) into iPSCs. In MEFs undergoing reprogramming, senescence genes are repressed and mesenchymal-to-epithelial transition is favored. This is correlated with a repressive activity of TCF1, which contributes to the silencing of Wnt/β-catenin signaling at the onset of reprogramming. In contrast, the Wnt pathway needs to be active in the late reprogramming phases to achieve successful reprogramming. In conclusion, continued activation or inhibition of the Wnt/β-catenin signaling pathway is detrimental to the reprogramming of MEFs; instead, temporal perturbation of the pathway is essential for efficient reprogramming, and the “Wnt-off” state can be considered an early reprogramming marker.
Graphical Abstract
Highlights
-
•
Time-dependent perturbation of Wnt pathway enhances reprogramming
-
•
TCF1 acts as repressor in cells undergoing reprogramming
-
•
TCF1 represses senescence genes and promotes MET
-
•
“Wnt-off” state is an early reprogramming marker
TCF1 is an effector of the Wnt/β-catenin pathway that can act as an activator or as a repressor. In this study, Lluis, Cosma, and colleagues show that the repressive activity of TCF1 or the inhibition of the Wnt/β-catenin at early stages of the reprogramming process is essential to generate induced pluripotent stem cells (iPSCs) from mouse embryonic fibroblasts (MEFs).
Introduction
The activation of canonical Wnt/β-catenin pathway controls embryo development and early differentiation events (MacDonald et al., 2009). However, it can also control the self-renewal and pluripotency of stem cells (Kühl and Kühl, 2013; Sato et al., 2004; Sokol, 2011). The activation of this pathway is due to the inhibition of the β-catenin destruction complex formed by APC, GSK3, and AXIN, resulting in β-catenin stabilization. Consequently, β-catenin can then translocate into the nucleus and activate target genes via its association with the TCF factors (Cadigan and Liu, 2006; Hoppler and Kavanagh, 2007; Moon et al., 2004). TCF proteins belong to a family of transcription factors, which include TCF1, LEF1, TCF3, and TCF4. TCF1 and LEF1 can bind β-catenin and activate target genes when the Wnt pathway is active (Hoppler and Kavanagh, 2007; Hurlstone and Clevers, 2002; Willert and Jones, 2006). In contrast, when the Wnt pathway is not active, all the TCF factors can recruit repressive complexes and function as repressors of target genes (Brantjes et al., 2001; Daniels and Weis, 2005).
Embryonic stem cells (ESCs) cultured in 2i medium, which contains GSK3 and MEK inhibitors, can be propagated in a pluripotent ground state (Silva et al., 2008). Pluripotent ground state is also established by Tcf3 deletion in ESCs (Cole et al., 2008; Tam et al., 2008; Wray et al., 2011; Yi et al., 2008, 2011). It is interesting to note that the GSK3 inhibitor in 2i medium stabilizes β-catenin. This suggests that the pluripotent ground state of ESCs can be maintained by derepression of TCF3 but also by activation of the Wnt pathway via stabilization of β-catenin. Moreover, activation of Wnt signaling prevents differentiation of ESCs into epiblast stem cells (epiSCs), through regulation of the transition between the ground and primed states (ten Berge et al., 2011).
The Wnt/β-catenin pathway can also activate somatic cell reprogramming to pluripotency. Mouse embryonic fibroblasts (MEFs) transduced with retroviruses carrying Oct4, Klf4, and Sox2 and cultured in medium containing Wnt3a can generate induced pluripotent stem cell (iPSC) colonies with enhanced efficiency in absence of c-Myc (Marson et al., 2008). Furthermore, activation of the Wnt pathway in ESCs enables them to reprogram neural precursor cells after fusion (Lluis et al., 2008). Finally, the deletion of Tcf3 greatly enhances cell-fusion-mediated reprogramming, as well as the production of induced pluripotent stem cells (iPSCs) (Lluis et al., 2011; Ombrato et al., 2012).
In addition to the Wnt-mediated control of ESC pluripotency and somatic cell reprogramming, Wnt signaling is also a driver of differentiation during early developmental phases (Tam and Loebel, 2009). Anterior-posterior axis specification in the mouse embryo occurs through the activity of Wnt signaling (Merrill et al., 2004; Sokol, 2011). In particular, Wnt signaling activity is essential for establishment of the primitive streak and anterior-posterior polarity, i.e., for epithelial-to-mesenchymal transition of epiblast cells in the primitive streak (Kalluri and Weinberg, 2009; Murry and Keller, 2008; Tanaka et al., 2011; ten Berge et al., 2008).
These apparently opposite roles of the Wnt signaling pathway are therefore a conundrum; on one hand, Wnt activity controls ESC pluripotency, and on the other hand, it regulates early developmental differentiation events. To reconcile these opposite functions, one reasonable hypothesis is based on the level of activation of the Wnt pathway in time. It is well known that Wnt signaling oscillates during development and that its target genes have an oscillatory behavior (Sokol, 2011; van Amerongen and Nusse, 2009). At the same time, cyclic activation of the Wnt/β-catenin pathway is essential for enhancing somatic cell reprogramming (Lluis and Cosma, 2009; Lluis et al., 2008). If β-catenin activity is either high or very low, reprogramming does not take place. We therefore wondered whether the activation of Wnt signaling activity mediated by TCF factors is essential in a specific phase of the reprogramming of MEFs into iPSCs.
TCF3 and TCF1 share a similar DNA binding domain, and they represent the most highly expressed TCF family factors in ESCs (Lluis et al., 2011; Pereira et al., 2006). TCF3 acts as a repressor of Wnt target genes, and in contrast TCF1 can activate or repress Wnt-targets via its association with β-catenin (Brantjes et al., 2001; Hikasa et al., 2010). However, little is known about TCF1 function in ESCs, and here we investigated the role of TCF1 in the reprogramming process of MEFs into iPSCs. Surprisingly, in this context, we found that the activity of the Wnt/β-catenin pathway needs to be switched off during the first days and that the cells undergoing reprogramming have low levels of stabilized β-catenin. Remarkably, sorted Wnt “off” MEFs generate a very high number of NANOG-positive iPSCs. Interestingly, during the early phases of four-factor-induced reprogramming, TCF1 functions as a repressor of Wnt signaling, and this activity correlates with downregulation of senescent genes, such as p21, p19, and p16, and activation of mesenchymal-to-epithelial transition (MET) genes. The activity of the Wnt/β-catenin pathway is instead necessary during the late phases of the reprogramming process.
Results
TCF1 Does Not Control Self-Renewal or Differentiation of ESCs
TCF3 is a repressor of pluripotency in ESCs, and its deletion drives the ESCs into a pluripotent ground state (Wray et al., 2011; Yi et al., 2011). Silencing of Tcf1 does not affect ESC self-renewal (Yi et al., 2011). Here, we investigated whether TCF1 has a role in controlling mouse ESC differentiation. With this aim, we silenced Tcf1 in ESCs by infecting the cells with two different lentiviral vectors that carry short hairpins for Tcf1 (ESCs-shTcf1A, ESCs-shTcf1B) (Figures S1A and S1B available online) and observed that Wnt signaling activity was impaired (Figure S1C). The Wnt signaling activity was measured after activating the pathway with the GSK3 inhibitor BIO in ESCs-shTcf1A, ESCs-shTcf1B, and ESCs-shScr, which were previously infected with a lentivirus carrying the 7xTcf-eGFP reporter (7TGP) for TCF/β-catenin activity (Figure S1C) (Fuerer and Nusse, 2010). The reporter allowed the measurement of GFP as a Wnt signaling activation readout (Fuerer and Nusse, 2010). The viruses carrying the short hairpins also carried a hygromicin selection cassette, to allow the selection of the infected cells. Then we analyzed the expression of stem cell genes and self-renewal in ESCs-shTcf1A and ESCs-shTcf1B (Figure 1A). Expression of Nanog, Stella, and Oct4 was not decreased after silencing of Tcf1, with respect to cells infected with a virus carrying a scrambled short hairpin (ESCs-shScr), which indicated that the expression of stem cell factors was not affected (Figure 1A). Finally, we performed clonogenic assays by culturing the same number of ESCs infected with shTcf1A, shTcf1B, and shScr and counting the number of alkaline-phosphatase-positive (AP+) colonies, as a marker of pluripotency. The same number of colonies was counted under all three conditions, and their morphologies were indistinguishable (Figure 1B).
Figure 1.
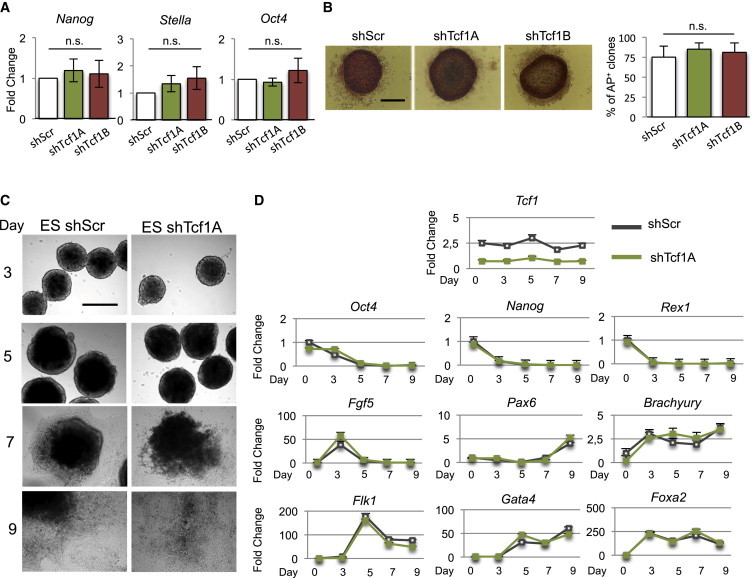
Silencing of Tcf1 in Mouse ESCs Does Not Affect Pluripotency
(A) Quantitative PCR expression analysis of pluripotent markers Nanog, Stella, and Oct4 in ESCs infected with lentiviruses carrying shScr or shTcf1A/B, as indicated. (n = 3, independent experiments).
(B) Representative colonies of ESCs-shScr and ESCs-shTcf1A/B showing alkaline phosphatase staining (left panels). Quantification on the right shows percentages of alkaline-phosphatase-positive (AP+) colonies with respect to all (AP+ and AP−) colonies (n = 3 independent experiments). Scale bar, 300 μm.
(C) ESCs-shScr and ESCs-shTcf1A were subjected to differentiation through aggregation into embryoid bodies. Representative phase-contrast images at 3, 5, 7, and 9 days during embryoid body differentiation. Scale bar, 400 μm.
(D) Representative quantitative real-time PCR experiment (out of three independent experiments) for the detection of Tcf1, stem cell (Oct-4, Nanog, and Rex1), ectoderm (Fgf5, Pax6), mesoderm (Brachyury, Flk1), and endoderm (Gata4 and Foxa2) marker genes in embryoid bodies at different times pi. The levels are normalized to Gapdh.
All pooled data are represented as means ± SD. See also Figure S1.
Next, we analyzed the possible defects in the differentiation potential of ESCs silenced for Tcf1. For this, we generated embryoid bodies using ESCs-shScr, ESCs-shTcf1A, and ESCs-shTcf1B. We analyzed the expression of pluripotent genes and of mesoderm, ectoderm, and endoderm markers at 3, 5, 7, and 9 days during embryoid body development. Embryoid bodies from both ESCs-shScr and ESCs-shTcf1A developed with the expected morphology at all time points analyzed (Figure 1C). Pluripotency genes, such as Oct4, Nanog, and Rex1, decreased with the same efficiency and kinetics in both the ESCs-shScr and ESCs-shTcf1A embryoid bodies. Furthermore, the mesoderm markers Brachyury and Flk1, the ectoderm markers Fgf5 and Pax6, and the endoderm markers Gata4 and Foxa2 were expressed with the expected timing at 3, 5, 7, and 9 days of both ESCs-shScr and ESCs-shTcf1A embryoid body development (Figure 1D). Comparable efficiency of the differentiation potential was also obtained after analyzing the expression of pluripotent genes and of the mesoderm, ectoderm, and endoderm markers during embryoid body development for ESCs-shScr and ESCs-shTcf1B (Figures S1D and S1E).
In all, these data clearly show that silencing of Tcf1 in mouse ESCs impairs neither self-renewal nor differentiation potential.
Continuous Tcf1 Silencing Impairs Reprogramming of MEFs to Pluripotency
Previous studies, including our own, have shown that the activation of Wnt signaling enhances cell-fusion-mediated reprogramming of somatic cells to pluripotency and the efficiency of generation of iPSCs in the absence of c-Myc (Lluis et al., 2008; Marson et al., 2008). Furthermore, derepression of Tcf3 greatly enhances the reprogramming efficiency of neural precursor cells to pluripotency (Lluis et al., 2011). We were next interested in studying the function of TCF1 in the control of somatic cell reprogramming. In addition, because TCF1 controls target genes through its association with β-catenin, in parallel, we investigated β-catenin activity in the regulation of reprogramming.
Thus, we infected doxycycline-inducible four-factor (Oct4, Klf4, Sox2, and c-Myc) MEFs (Carey et al., 2010) with shTcf1A, with shTcf1B, with shβ-catenin (shβ-cat), and with control shScr (Figures S2A and S2B). Infected MEFs were selected with hygromycin, replated at equal numbers, and subsequently induced with doxycycline for 12 days. Finally, doxycycline was removed in the last 4 days, and iPSC colonies were analyzed at day 16, by counting the NANOG-positive (NANOG+) colonies (Figure 2A). The number of NANOG+ colonies was strongly reduced after both Tcf1 and β-catenin silencing, with respect to the control (Figures 2B, S2C, and S2D). Furthermore, the few colonies that were formed after Tcf1 silencing were very small and with little NANOG expression (Figure 2C). In contrast, shScr-derived iPSCs formed with high efficiency, were positive to alkaline phosphatase (AP), expressed stem cell markers (such as SSEA-1, NANOG, OCT4, SOX2, Sall1, and Rex1), and differentiated into all three germ layers (Figures 2D S2E, and S2F). Finally, to exclude adaptation of the MEFs to the loss of Tcf1, we infected MEFs with shTcf1A and used doxycyline to induce the four factors at the same time. Also, in this case, we observed a decrease in the NANOG+ colonies, although this effect was weaker, as the MEFs were not hygromycin selected (Figure S2G). These data show that continuous silencing of β-catenin or Tcf1 for 16 days impairs reprogramming efficiency.
Figure 2.
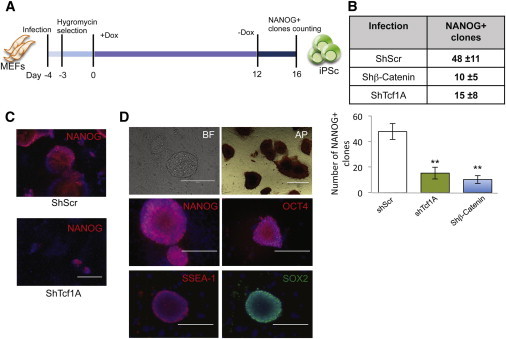
Silencing of Tcf1 in Four-Factor-Induced MEFs Impairs Reprogramming
(A) Experimental scheme for iPSC generation. Four-factor MEFs were infected with lentiviral vectors carrying shScr, shTcf1A, and shβ-catenin. Infected cells were hygromycin selected for 3 days. Doxycycline was applied from 0 to 12 days, to activate expression of Oct4, Klf4, Sox2, and c-Myc. Then doxycycline was removed, and cells were allowed to grow for 4 days. At day 16, immunostaining against NANOG was performed, to count the number of reprogrammed clones.
(B) Number of NANOG-positive (NANOG+) clones obtained according to each treatment, as indicated (n = 4 independent experiments).
(C) Representative image of a NANOG+ clone obtained from shScr- and shTcf1-infected MEFs analyzed at day 16.
(D) Images of immunostaining of iPSC clones obtained from four-factor MEFs infected with shScr. Scale bars, 200 μm (NANOG, OCT4, SSEA1, and SOX2) and 500 μm (AP). Nuclei were stained with DAPI.
All pooled data are represented as means ± SD. The asterisks indicate statistical significance by t test analysis (n.s., not significant; ∗p < 0.05; ∗∗p < 0.01). See also Figure S2.
Inhibition of the Wnt Pathway during the Early Phases Enhances MEF Reprogramming via TCF1 Activity
As TCF1 did not appear to have any role in the maintenance of ESC self-renewal, we hypothesized that TCF1 might control reprogramming onset, i.e., the early steps of the process. Thus, we investigated Tcf1 expression at different time points after doxycycline induction.
THY1 can be followed as an early marker of reprogramming (Brambrink et al., 2008; Stadtfeld et al., 2008). Cells negative for the expression of this marker 3 days after four-factor induction are enriched in the population undergoing reprogramming (Polo et al., 2012). So four-factor MEFs were induced with doxycycline and sorted by fluorescence-activated cell sorting (FACS) 3, 6, and 9 days after induction for the presence or absence of THY1 (Figures 3A and S3A). Enrichment of Thy expression in sorted cells was confirmed by real-time PCR (Figure S3B). Cells undergoing reprogramming embark into MET (Esteban et al., 2012; Li et al., 2010; Samavarchi-Tehrani et al., 2010). Thus, we analyzed expression of epidermal markers, Epcam and Cdh1, and mesenchymal markers, Snai1, Slug, and Vimentin (ten Berge et al., 2008). We observed, as previously shown (Polo et al., 2012), that THY1-negative cells undergo efficient MET, as they upregulated Epcam and Cdh1 and downregulated Snai1, Slug, and Vimentin. In contrast, THY1-positive cells maintained the mesenchymal phenotype (Figure 3B). In addition, the senescence genes p21, p16, and p19, which have been shown to be a barrier to the reprogramming of MEFs into iPSCs (Banito et al., 2009; Hong et al., 2009; Kawamura et al., 2009; Li et al., 2009; Marión et al., 2009; Utikal et al., 2009), were downregulated in the THY1-negative cells (Figure 3B). Finally, the THY1-negative cells gave rise to fully reprogrammed iPSCs colonies, whereas the THY1-positive cells did not (Figure 3C).
Figure 3.
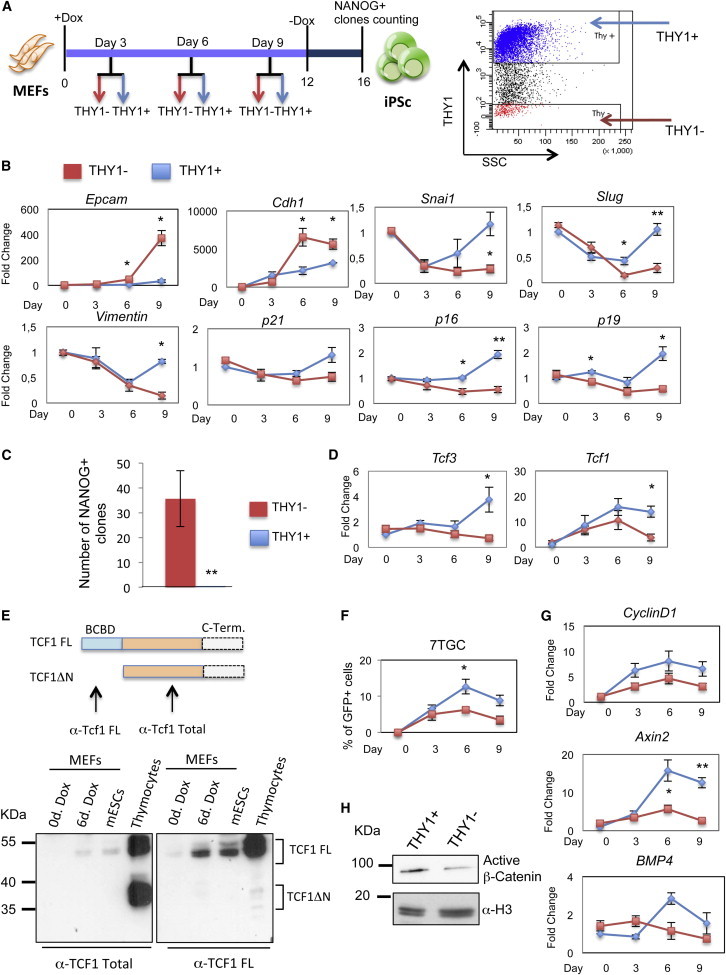
Time-Dependent Activation of Wnt/β-Catenin Pathway Controls Reprogramming of Four-Factor-Induced MEFs
(A) Experimental scheme of time course analysis of FACS-sorted cells. Four-factor MEFs were induced with doxycycline as in Figure 2A. At days 3, 6, and 9, the cells were FACS-sorted for THY1-positive (THY1+; blue) and THY1-negative (THY1−; red) expression (right panel) and analyzed.
(B) Time-dependent expression of MET and senescence markers. Relative expression of epithelial markers (Epcam, Cdh1), mesenchymal markers (Snai1, Slug, and Vimentin), and senescent markers (p21, p16, and p19) in THY1-positive (blue) and THY1-negative (red) sorted cells (n = 3 independent experiments).
(C) An equal number of THY1-positive (blue) and THY1-negative (red) cells were sorted and plated on feeders at day 6 after doxycycline induction. The number of reprogrammed clones was analyzed using NANOG immunostaining (n = 3 independent experiments).
(D) Expression of Tcf3 and Tcf1 was analyzed in sorted THY1-positive (blue) and THY1-negative (red) cells using quantitative real-time PCR at the indicated days after doxycycline induction (n = 3 independent experiments).
(E) Schematic representation of the two main TCF1 isoforms. TCF1FL contains a N-terminal β-catenin binding domain (BCBD), whereas TCF1ΔN lacks this domain. Representative western blotting of four-factor MEF extracts on day 0 and day 6 after doxycycline induction. Extracts from ESCs and thymocytes were used as controls. Antibodies against the total TCF1 isoforms and against the full-length TCF1 were used.
(F) MEFs were infected with 7TGP reporter construct and induced with doxycycline as in Figure 2A. At days 3, 6, and 9, the cells were analyzed by GFP expression by FACS (n = 3 independent experiments).
(G) Expression of Wnt/β-catenin target genes (Axin2, CyclinD1, and BMP4) were analyzed in sorted THY1-positive (blue) and THY1-negative (red) cells using quantitative real-time PCR at the indicated days after doxycycline induction (n = 3 independent experiments).
(H) Representative western blot of active β-catenin in nuclear extracts of THY1-positive and THY1-negative sorted cells 6 days after doxycycline induction.
All pooled data are represented as means ± SD. The asterisks indicate statistical significance by t test analysis (∗ p < 0.05; ∗∗ p < 0.01). See also Figure S3.
We found that, while in the THY1-negative cells, Tcf3 expression remained almost constant at all time points of the reprogramming process, it increased from 6 to 9 days in the THY1-positive cells. This indicates that as expected, cells undergoing reprogramming indeed maintain low levels of Tcf3, the repressor of pluripotency (Lluis et al., 2011). In contrast, Tcf1 expression levels increased progressively from 0 to 6 days in both THY1-positive and THY1-negative cells, before decreasing again in both populations 9 days postinduction (pi) (Figure 3D).
Tcf1 has several isoforms, including full-length and ΔN isoforms (FLTcf1 and ΔNTcf1, respectively), which have been extensively studied (Reya and Clevers, 2005; Van de Wetering et al., 1996). ΔNTCF1 was shown to be a repressor, as it lacks the β-catenin binding domain (Roose et al., 1999; Waterman, 2004). We therefore investigated which Tcf1 isoform increases during reprogramming of MEFs. FLTCF1 was greatly increased 6 days pi, as seen using an antibody against FLTCF1 (Figure 3E). FLTCF1 is also the isoform that is expressed in ESCs. In contrast, ΔNTCF1, which is highly expressed in thymocytes (Ioannidis et al., 2001; Yu et al., 2010), was not detected during the reprogramming process (Figure 3E).
Although the expression profile of Tcf1 was similar in the THY1-positive and THY1-negative cells 6 days pi, the activation of the Wnt signaling pathway was different. Activation of the Wnt pathway, through analysis of the 7TGC reporter activity (Fuerer and Nusse, 2010), was higher in the THY1-positive cells with respect to the THY1-negative cells (Figure 3F). Furthermore, expression of the Wnt/β-catenin targets CyclinD1, Axin2, and BMP4 was increased in the THY1-positive cells with respect to the THY1-negative cells, with a peak at 6 days pi (Figure 3G), indicating that Wnt activity is much higher in the THY1-positive cells, which do not undergo reprogramming.
To confirm these observations, we analyzed β-catenin levels in THY1-positive and THY1-negative cells at 6 days pi. Here, active and total β-catenin accumulated more in the THY1-positive cells, clearly indicating that β-catenin can efficiently activate the pathway along with TCF1 in the THY1-positive cells only (Figures 3H and S3C). In contrast, in the THY1-negative cells, Tcf1 appeared to act as a repressor, as at 6 days postinduction, the Wnt pathway was inactive and β-catenin accumulated much less than in the THY1-positive cells (Figure 3H). Indeed, when β-catenin does not accumulate, TCF1 acts as a repressor, by binding to corepressor factors, such as members of the Groucho-related family (Reya et al., 2003; Roose et al., 1998).
To further investigate the repressive activity of TCF1, we overexpressed Tcf1 in MEFs and analyzed target genes. In particular, for this purpose, we overexpressed FLTcf1, which increased during reprogramming, and as a control, ΔNTcf1. Axin2 was drastically downregulated after both ΔNTcf1 and FLTcf1 expression respect to the empty vector (E.V.), which indicated that both isoforms repress the Wnt signaling pathway (Figure 4A). The repressive activity of TCF1 was converted into an activation of the Wnt pathway when Chiron, a GSK3 inhibitor, or Wnt3a were added to the FLTcf1 infected MEFs. Axin2 was upregulated upon pathway activation (Figures S4A and S4B).
Figure 4.
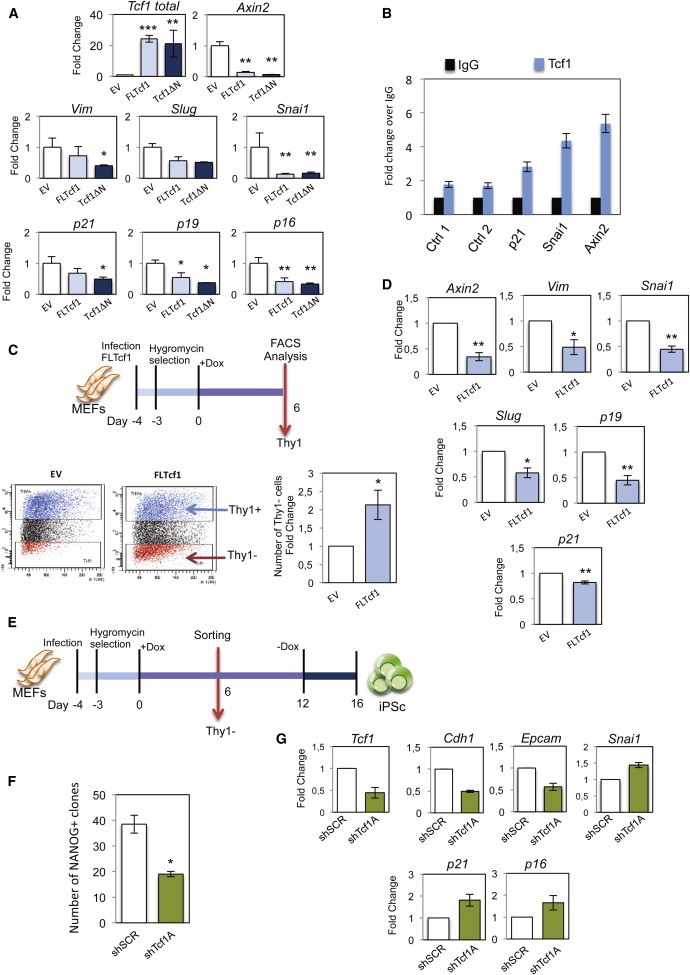
TCF1 Is a Repressor of Senescent and Mesenchymal during the First Days of Reprogramming
(A) Four-factor MEFs were infected with lentiviruses overexpressing either FLTcf1 or ΔN-Tcf1. Infected cells were selected with hygromycin. Four days after infection, the expression of Wnt/β-catenin pathway target (Axin2), mesenchymal markers (Vimentin, Slug, and Snai1), and senescence markers (p21, p19, and p16) were analyzed (n = 3 independent experiments). EV, empty vector control.
(B) Quantitative ChIP assay of the TCF1 targets Axin2, Snai1, and p21 in the four-factor MEFs. ChIP was performed using rabbit immunoglobulin G or a specific antibody against TCF1 (n = 2 independent experiments).
(C) Four-factor MEFs were infected with lentivirus overexpressing FLTcf1 or with an empty vector (EV) and selected with hygromycin. Six days after doxycycline induction, cells were analyzed by THY1 expression by FACS (n = 3 independent experiments).
(D) Quantitative real-time PCR expression analysis of empty vector (EV) or FLTcf1-overexpressing cells at 6 days after doxycycline induction (n = 3 independent experiments).
(E) Experimental scheme: four-factor MEFs were infected and treated as in Figure 2A. THY1-negative cells (THY1−) were sorted at day 6 after doxycycline induction, as indicated.
(F) The same number of THY1-negative cells was sorted at day 6 after doxycycline induction of shScr or shTcf1A-infected four-factor MEFs and plated on feeders. The number of NANOG+ clones was determined under each treatment (n = 3 independent experiments).
(G) Quantitative real-time PCR expression analysis of shScr and shTcf1A-infected, THY1−, four-factor MEFs at day 6 after doxycycline induction. The mesenchymal (Snai1),; epithelial (Cdh1, Epcam), and senescence (p21, p16) genes are analyzed (n = 2 independent experiments).
All pooled data are represented as means ± SD. The asterisks indicate statistical significance by t test analysis (∗ p < 0.05; ∗∗ p < 0.01). See also Figure S4.
We observed that the mesenchymal genes Vim, Slug, and Snai1 were downregulated by overexpression of both isoforms of Tcf1 (Figure 4A). The senescence genes are upregulated in ESCs that express very high levels of β-catenin, resulting in an impairment of cell-fusion-mediated reprogramming (Lluis et al., 2010). Therefore, we analyzed their expression after Tcf1 upregulation. p21, p19, and p16 were all downregulated upon FLTcf1 and ΔNTcf1 overexpression (Figure 4A). In contrast, senescence genes and Snai1 do not change in their expression when the Wnt pathway is activated in shTcf1A-MEFs (Figure S4C). Finally, to determine whether TCF1 directly regulates MET and senescence genes, we performed chromatin immunoprecipitation (ChIP) assays in MEFs, which demonstrated binding of TCF1 to Snai1, p21, and Axin2 promoters (Figure 4B).
Overall, these data indicated that TCF1 can act as a repressor of the Wnt/β-catenin pathway in MEFs, and this results in the repression of some of the mesenchymal and senescent genes.
Next, we investigated whether overexpression of Tcf1 increased the number of cells undergoing reprogramming. Indeed, we observed a higher number of THY1-negative cells 6 days after infection of four-factor MEFs, compared to EV. (Figure 4C). Also, these cells showed downregulation of Axin2 as well as the mesenchymal genes Vim, Snai1, and Slug and the senescence markers p19 and p21 (Figure 4D).
Next, because Tcf1 increased 6 days pi, we investigated further whether perturbation of Tcf1 at this time point was essential in the reprogramming process. For this purpose, we infected four-factor MEFs with shTcf1A, shTcf1B, and shScr, selected with hygromycin, induced with doxycycline, FACS sorted the THY1-negative cells 6 days after induction, and plated these cells in equal numbers (Figure 4E). We observed a reduction in the number of NANOG+ clones from the shTcf1-infected MEFs when compared to the control (Figures 4F and S4D). This shows that silencing the repressive activity of Tcf1 in THY1-negative cells inhibits the reprogramming process.
To determine whether this effect was dependent on the activation of the MET and/or senescence genes, we analyzed gene expression 6 days after induction in the THY1-negative cells. Interestingly, there was an upregulation of the mesenchymal gene Snai1 and a downregulation of the epithelial genes Cdh1 and Epcam (Figure 4G). Moreover, p21 and p16 were upregulated 6 days pi in the shTcf1-infected MEFs (Figure 4G). These data indicate that silencing the repressive activity of TCF1 in the THY1-negative cells in the early phases of reprogramming blocks MET and increases senescence barrier.
Wnt “Off” State Is an Early Reprogramming Marker
To further monitor whether Wnt “off” cells in early reprogramming phases embark upon efficient reprogramming, we infected four-factor MEFs with a lentivirus carrying the 7xTcf-eGFP reporter (7TGC) for TCF/β-catenin activity and mCherry under the constitutive SV40 promoter (Figure S5A) (Fuerer and Nusse, 2010). Among the cells that were mCherry positive, four populations of cells with different GFP fluorescence levels were FACS sorted 6 days after doxycycline induction (Figures 5A and S5B). Axin2 was analyzed to separate these populations of sorted cells accordingly to the Wnt activity (Figure S5B). GFP+ and GFP− cells (fractions P5 and P6), which had the Wnt pathway “on” and “off,” respectively, were analyzed for the expression of pluripotency and for MET and senescent genes. As expected, Axin2, CyclinD1, and BMP4 were more expressed in the GFP+ cells than in GFP− cells. The GFP− cells underwent efficient MET, as they upregulated Epcam and Cdh1 and expressed low levels of Snai1, Vimentin, and Slug with respect to GFP+ cells. Also, the senescent gene p16 and p19 were downregulated in GFP− cells. Furthermore, the pluripotency markers Rex1, Nanog, Oct4, and Sall1 were efficiently expressed in GFP− cells (Figure 5B). Finally, when replated and cultured in presence of doxycycline, only GFP− cells formed a very high number of NANOG+ or AP+ clones, whereas GFP+ cells did not (Figures 5C, S5C, and S5D). We also analyzed GFP+ cells using immunofluorescence and showed that these cells expressed Nestin (Figure 5D), which suggested they were more differentiated. These results clearly indicate that in the initial steps of the reprogramming process, i.e., up to 6 days after doxycycline induction, the Wnt “off” state is a robust marker of reprogramming.
Figure 5.
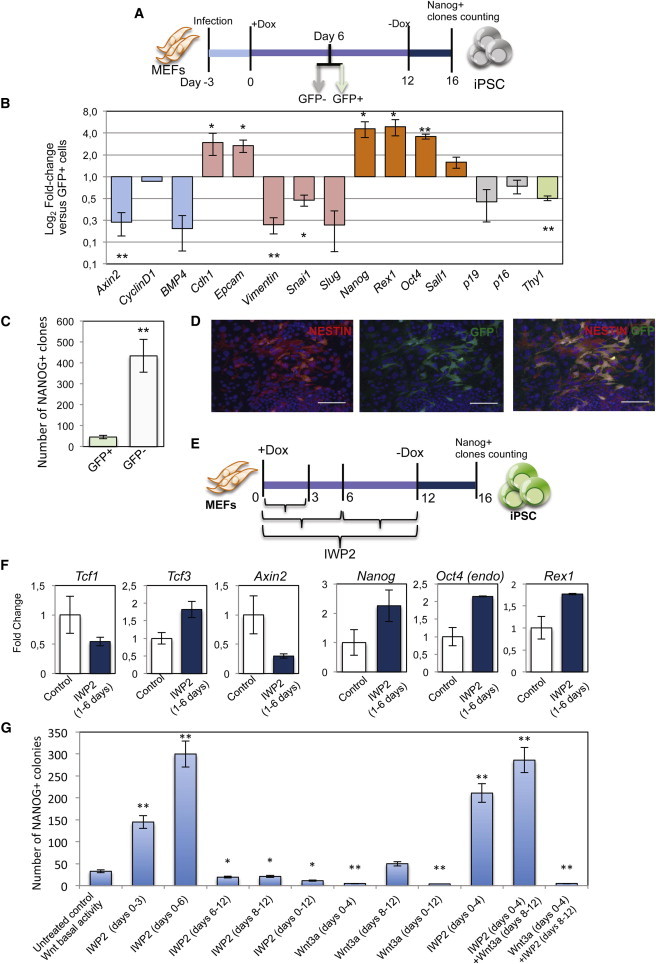
Inhibition of the Wnt/β-Catenin Pathway during the First Days of the Reprogramming Process Increases Reprogramming Efficiency
(A) Four-factor MEFs were infected with lentiviruses carrying 7TGP reporter. GFP+ and GFP− cells were sorted 6 days after doxycycline induction. Doxycycline was removed at day 12, and colonies were analyzed at day 16.
(B) GFP+ and GFP− cells were sorted 6 days pi and analyzed by quantitative real-time PCR for the indicated markers (n = 3 independent experiments).
(C) The number of NANOG-positive clones in GFP+- and GFP−-sorted cells at day 16 (n = 3 independent experiments).
(D) GFP+ cells were FACS-sorted at day 6 pi, and cells were analyzed by immunofluorescence for the expression of Nestin. Representative images of the immunofluorescence are shown. Scale bars, 100 μm.
(E) Experimental scheme: four-factor MEFs were treated with doxycycline for 12 days and with IWP2 to inhibit Wnt secretion for the indicated days. At day 16, after doxycycline removal, the colonies were stained for NANOG expression and counted.
(F) Four-factor MEFs were treated with IWP2 for 6 days. Quantitative real-time PCR of control and treated cells (n = 3 independent experiments).
(G) The number of NANOG-positive clones in the control and four-factor MEFs treated with IWP2 for the first 3, 4, 6, or 12 days or for the intervals from 6 to 12 or 8 to 12 days as indicated, Wnt3a for the first 4 or 12 days or for the intervals from 8 to 12 days as indicates, or with combination of IWP2 and Wnt3a at the indicated times (n = 3 independent experiments).
All pooled data are represented as means ± SD. The asterisks indicate statistical significance by t test analysis (n.s., not significant; ∗ p < 0.05; ∗∗ p < 0.01). See also Figure S5.
To further confirm these data, we treated four-factor MEFs for different times with IWP2, an inhibitor of Wnt secretion (Figure 5E) (ten Berge et al., 2011). IWP2 treatment for the first 6 days decreased Tcf1 expression and increased Tcf3 expression in the whole-cell population. Along with its effects on Wnt secretion, this resulted in inhibition of the activation of the Wnt/β-catenin pathway, as demonstrated by the repression of Axin2 (Figure 5F). Together with IWP2, doxycycline induction was applied for 12 days, and the iPSC colonies were analyzed at day 16 (4 days after doxycycline removal) (Figure 5E). MEFs treated with IWP2 for 6 days expressed high levels of Nanog, Rex1, and endogenous Oct4 (Figure 5F). Furthermore, the number of iPSC colonies was strongly increased when MEFs were treated for the first 3 or 6 days with IWP2 (Figure 5G). In addition, the silencing of p21 after IWP2 treatment for 6 days did not result in an increase of reprogramming, further confirming that p21 is an important effector of the Wnt pathway inhibition in the reprogramming process (Figures S5E and S5F). In contrast, treatment of IWP2 from day 6 to day 12 or continuous inhibition of the Wnt pathway with IWP2 strongly reduced the reprogramming efficiency (Figure 5G). This indicated that the pathway should be inhibited during the first phase of reprogramming and should have a basal activation level during the second phase of reprogramming. To confirm these data, we transfected MEFs with shTcf1 (Figure S5G). In this way, we obtained that shTcf1 was lost within 6 days after induction (Figure S5H), whereas in the infected cells, the silencing was constant for all the reprogramming process (Figures S5J and S5K). We found that whereas the constant inhibition of Tcf1 resulted in a decreased number of NANOG+ colonies (Figure 2B), Tcf1 transient inhibition resulted in increased efficiency of reprogramming (Figure S5I).
Activation of the pathway by adding Wnt3a early during the reprogramming process inhibits its efficiency (Figures 5G and S5L), whereas the addition of Wnt3a late during the reprogramming process only resulted in a slight increase in the reprogramming efficiency (Figure 5G). In addition, the combined inhibition and activation with IWP2 and Wnt3a during the early and late phases of reprogramming, respectively, moderately increased the efficiency of reprogramming as compared to early inhibition alone (Figure 5G). This indicates that inhibition of Wnt signaling in the early phase is essential and that a basal activation of Wnt pathway is sufficient during late phases of the reprogramming process (Figure 5G). Finally, we used two additional inhibitors of the Wnt pathway to treat the MEFs during early reprogramming phases: Dkk1, which acts as Wnt ligand antagonist (Katoh and Katoh, 2007; Niida et al., 2004), and ICRT3, which blocks the interaction between β-catenin and T cell factors (Faunes et al., 2013; Gonsalves et al., 2011). We further confirmed that early inhibition of the pathway results in increased reprogramming efficiency (Figure S5L). Overall, these findings showed that time-dependent perturbation of the Wnt pathway is fundamental to enhance reprogramming efficiency.
Discussion
Earlier we showed that cyclic activation of the Wnt signaling pathway is necessary to enhance cell-fusion-mediated reprogramming (Lluis et al., 2008). Also, we showed that finely tuned levels of the Wnt pathway are essential for enhancing somatic cell reprogramming. Too high or too low levels of β-catenin activity result in an impaired reprogramming efficiency (Lluis et al., 2008). Here, we demonstrate that, to achieve reprogramming, Wnt signaling needs to be repressed during the early phase of the process and to be active at the late steps; indeed, in a recent interesting study, a similar conclusion was reached (Ho et al., 2013). However, whereas Ho et al. studied prevalently the function of TCF3 in the regulation of reprogramming and focused on the analysis of the whole MEFs population, here we have extensively studied the function of TCF1, demonstrating its essential role during the reprogramming process. In addition, we have characterized the activity of TCF1 in the THY-negative cells, which are the cells that undergo reprogramming. We have shown that TCF1 acts as a repressor of the Wnt signaling pathway at the onset of reprogramming. Furthermore, it promotes repression of senescent genes and activation of MET (by inhibition of transcription of mesenchymal genes and by activation of epidermal genes) in the THY-negative cells, whereas TCF1 acts as an activator in the THY-positive cells that do not undergo reprogramming.
Of note, during the reprogramming process, THY-positive cells represent the majority of the total cell population; therefore, the repressive activity of TCF1 in the THY-negative cells cannot be identified without isolating them.
Finally, by sorting the “Wnt-off” cells, we obtained a striking increase in the number of reprogrammed NANOG+ clones, thus clearly identifying the “Wnt-off” state as an early reprogramming marker.
It is interesting to note how different the roles of TCF3 and TCF1 are in the control of somatic cell reprogramming. We showed previously that deletion of Tcf3 induces increased AcH3 and a decreased number of H3K9me3 heterochromatin foci. These epigenome modifications ultimately enhance reprogramming efficiency (Lluis et al., 2011). Here, we showed that the role of TCF1 during the early reprogramming phase is correlated with the repression of senescence genes and with the activation of MET. Ho et al. (2013) provided evidence that TCF3 and TCF4 promote early reprogramming events by repressing Wnt pathway target genes, including TCF1 and LEF1. However, they also postulated that TCF3 and TCF4 could control other targets that are independent of TCF1 and LEF1 or of the Wnt pathway activation (Ho et al., 2013). It is tempting to speculate that the latter mentioned might be activated by the epigenetic modifications induced by Tcf3 derepression.
Very little is known about TCF1 activity in ESCs, although it has been demonstrated that in medium lacking LIF, TCF1 contributes to the effect of Wnt3a stimulation to ESC self-renewal (Yi et al., 2011). Here, we have revealed that in medium containing LIF and serum, wild-type ESCs and ESCs silenced for Tcf1 do not show differences in self-renewal and differentiation, indicating that TCF1 does not control either process. TCF1 is instead a key regulator of the reprogramming process. This led to the interesting observation that some key transcription factors do not always control both somatic cell reprogramming processes and ESC self-renewal, as the effects on these two processes can be distinct.
Both MET and the downregulation of senescence genes are essential processes to achieve somatic cell reprogramming (Esteban et al., 2012; Mahmoudi and Brunet, 2012). Our data show that TCF1 is an important regulator of these processes. Senescent cells are characterized by cell-cycle arrest due to p16INK4a induction. Cell proliferation is crucial in the reprogramming process, and thus cell-cycle arrest is a major barrier to the efficiency of this process. Indeed, mouse fibroblasts cannot be reprogrammed efficiently when two antiproliferative genes, p16INK4a and p19ARF, which are part of the lnk4/Arf locus, are highly expressed. Accordingly, silencing of the lnk4/Arf locus restores reprogramming efficiency in senescent cells. Furthermore, knockdown of p53 and p21 also accelerates reprogramming of human and mouse fibroblasts (Banito et al., 2009; Hong et al., 2009; Kawamura et al., 2009; Li et al., 2009; Marión et al., 2009; Utikal et al., 2009). Thus, senescence is a barrier to the reprogramming activity, and this can be overcome by TCF1 action, which maintains low expression of p21 and p19 during reprogramming.
During MET, SOX2, OCT4, and C-MYC have been shown to supress Snai1 and TGF-β signaling, whereas KLF4 has been shown to upregulate E-cadherin (Li et al., 2010; Samavarchi-Tehrani et al., 2010). Here, we found that TCF1 activity is correlated not only with repression of senescence genes, but also with MET activation. Embryoid bodies derived from ESCs undergo epithelial-to-mesenchymal transition in a Wnt-dependent process, with the Wnt activity inducing upregulation of Snai1 and repression of E-cadherin (ten Berge et al., 2008). It is therefore intriguing that during reprogramming, TCF1 enhances MET by transcriptionally repressing Vim, Slug, and Snai1 and upregulating E-cadherin. Furthermore, we observed that TCF1 directly represses Snai1 by binding to its promoter. SNAI1 has been shown to be a transcriptional repressor of E-cadherin (Batlle et al., 2000). Thus, it might well be that E-cadherin is derepressed as a consequence of TCF1-mediated repression of Snai1.
However, we cannot definitively conclude here what the prevalent role of TCF1 is during reprogramming, i.e., if the main function of TCF1 is to modulate senescence genes and MET or also to activate other factors downstream of the Wnt pathway. Finally, it will be particularly interesting to investigate if the Wnt “off” state mediated by the repressive activity of TCF1 is one of the most efficient early reprogramming markers.
Experimental Procedures
iPSC Induction
The reprogramming experiments were routinely conducted in gelatinized plates, in the ESC culture medium supplemented with 2 μg/ml doxycycline, as described in Carey et al. (2010). Briefly, after doxycycline withdrawal, the cultures were grown in the ESC medium supplemented with 15% FBS. Colonies with ESC morphology were picked 4–6 days after doxycycline withdrawal and grown as iPSC colonies on a feeder layer of MEFs that was inactivated with mitomycin C.
Virus Preparation and MEF Infection
Lentiviruses were packaged in HEK293T cells. Briefly, the HEK293T cells were cultured in Dulbecco’s modified Eagle’s medium supplied with 10% FBS (Life Technologies), 10 U/ml penicillin, 10 μg/ml streptomycin, 2 mM glutamine, 1 mM sodium pyruvate, and 1 × nonessential amino acids. For the transfection, 5 × 106 HEK293T cells were seeded onto 10 cm plates and transfected with 10 μg pLKO-shTcf1A, pLKO-shTcf1B, or pLKO-shScr, 6.5 μg pCMV-dR8.9 dvpr, and 3.5 μg pCMV-VSV-G packaging plasmids. After 48 and 72 hr, the supernatants were collected and centrifuged at 20,000 rpm for 1.5 hr at 20°C. The pellet with the viruses was resuspended in PBS (Life Technologies) and stored at −80°C.
For the reprogramming experiments, 2 × 105 MEFs were infected with either pLKO-shTcf1A, pLKO-shTcf1B, or pLKO-shScr in the MEF culture medium. After 48 hr, hygromycin selection (20 μg/ml) was applied for 3–4 days. For iPSC induction, the cells were then plated in equal numbers (5 × 104) onto gelatin-coated six-well plates in the ESC culture medium supplemented with 2 μg/ml of doxycycline.
All animal procedures were approved by the local ethics committee, and met the guidelines of the local and European regulations.
Author Contributions
F.A., I.T., L.O., F.L., and M.P.C. planned the experiments; F.A., I.T., L.O., and F.L. performed research and acquired the data; F.A., I.T., L.O., F.L., and M.P.C. analyzed data; F.L. and M.P.C. supervised the project; and M.P.C. wrote the paper.
Acknowledgments
We would like to thank Karthik Arumugam, Lucia Marucci, and Elisa Pedone for critically reading the manuscript; Bruno Di Stefano for suggestions to iPSC generation experiments; and Vanessa Chigancas and Neus Romo for technical support. We thank Luigi Naldini for providing lentiviral vectors, Hans Clevers for providing the Tcf1 constructs, and Rudolf Jaenisch for providing the inducible four-factor mouse model. We are grateful for support from an European Research Council grant (242630-RERE) (to M.P.C.), an HFSP grant (to M.P.C.), the Ministerio de Ciencia e Inovación (SAF2011-28580) (to M.P.C.), Fundacio’ La Marato’ de TV3 (to M.P.C.), the AXA Research Fund (to M.P.C.), the “Miguel Servet” Project of the Instituto de Salud Carlos III (CP10/00445) (to F.L.), the Ministerio de Ciencia e Innovación FPI (to F.A.), and the Ingenium ITN Marie Curie Program (to I.T.). We acknowledge support of the Spanish Ministry of Economy and Competitiveness, “Centro de Excelencia Severo Ochoa 2013-2017” (SEV-2012-0208).
Footnotes
This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/3.0/).
Contributor Information
Frederic Lluis, Email: frederic.lluis@crg.es.
Maria Pia Cosma, Email: pia.cosma@crg.es.
Supplemental Information
References
- Banito A., Rashid S.T., Acosta J.C., Li S., Pereira C.F., Geti I., Pinho S., Silva J.C., Azuara V., Walsh M. Senescence impairs successful reprogramming to pluripotent stem cells. Genes Dev. 2009;23:2134–2139. doi: 10.1101/gad.1811609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batlle E., Sancho E., Francí C., Domínguez D., Monfar M., Baulida J., García De Herreros A. The transcription factor snail is a repressor of E-cadherin gene expression in epithelial tumour cells. Nat. Cell Biol. 2000;2:84–89. doi: 10.1038/35000034. [DOI] [PubMed] [Google Scholar]
- Brambrink T., Foreman R., Welstead G.G., Lengner C.J., Wernig M., Suh H., Jaenisch R. Sequential expression of pluripotency markers during direct reprogramming of mouse somatic cells. Cell Stem Cell. 2008;2:151–159. doi: 10.1016/j.stem.2008.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brantjes H., Roose J., van De Wetering M., Clevers H. All Tcf HMG box transcription factors interact with Groucho-related co-repressors. Nucleic Acids Res. 2001;29:1410–1419. doi: 10.1093/nar/29.7.1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadigan K.M., Liu Y.I. Wnt signaling: complexity at the surface. J. Cell Sci. 2006;119:395–402. doi: 10.1242/jcs.02826. [DOI] [PubMed] [Google Scholar]
- Carey B.W., Markoulaki S., Beard C., Hanna J., Jaenisch R. Single-gene transgenic mouse strains for reprogramming adult somatic cells. Nat. Methods. 2010;7:56–59. doi: 10.1038/nmeth.1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole M.F., Johnstone S.E., Newman J.J., Kagey M.H., Young R.A. Tcf3 is an integral component of the core regulatory circuitry of embryonic stem cells. Genes Dev. 2008;22:746–755. doi: 10.1101/gad.1642408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels D.L., Weis W.I. Beta-catenin directly displaces Groucho/TLE repressors from Tcf/Lef in Wnt-mediated transcription activation. Nat. Struct. Mol. Biol. 2005;12:364–371. doi: 10.1038/nsmb912. [DOI] [PubMed] [Google Scholar]
- Esteban M.A., Bao X., Zhuang Q., Zhou T., Qin B., Pei D. The mesenchymal-to-epithelial transition in somatic cell reprogramming. Curr. Opin. Genet. Dev. 2012;22:423–428. doi: 10.1016/j.gde.2012.09.004. [DOI] [PubMed] [Google Scholar]
- Faunes F., Hayward P., Descalzo S.M., Chatterjee S.S., Balayo T., Trott J., Christoforou A., Ferrer-Vaquer A., Hadjantonakis A.K., Dasgupta R., Arias A.M. A membrane-associated β-catenin/Oct4 complex correlates with ground-state pluripotency in mouse embryonic stem cells. Development. 2013;140:1171–1183. doi: 10.1242/dev.085654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuerer C., Nusse R. Lentiviral vectors to probe and manipulate the Wnt signaling pathway. PLoS ONE. 2010;5:e9370. doi: 10.1371/journal.pone.0009370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonsalves F.C., Klein K., Carson B.B., Katz S., Ekas L.A., Evans S., Nagourney R., Cardozo T., Brown A.M., DasGupta R. An RNAi-based chemical genetic screen identifies three small-molecule inhibitors of the Wnt/wingless signaling pathway. Proc. Natl. Acad. Sci. USA. 2011;108:5954–5963. doi: 10.1073/pnas.1017496108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hikasa H., Ezan J., Itoh K., Li X., Klymkowsky M.W., Sokol S.Y. Regulation of TCF3 by Wnt-dependent phosphorylation during vertebrate axis specification. Dev. Cell. 2010;19:521–532. doi: 10.1016/j.devcel.2010.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho R., Papp B., Hoffman J.A., Merrill B.J., Plath K. Stage-specific regulation of reprogramming to induced pluripotent stem cells by Wnt signaling and T cell factor proteins. Cell Rep. 2013;3:2113–2126. doi: 10.1016/j.celrep.2013.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong H., Takahashi K., Ichisaka T., Aoi T., Kanagawa O., Nakagawa M., Okita K., Yamanaka S. Suppression of induced pluripotent stem cell generation by the p53-p21 pathway. Nature. 2009;460:1132–1135. doi: 10.1038/nature08235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoppler S., Kavanagh C.L. Wnt signalling: variety at the core. J. Cell Sci. 2007;120:385–393. doi: 10.1242/jcs.03363. [DOI] [PubMed] [Google Scholar]
- Hurlstone A., Clevers H. T-cell factors: turn-ons and turn-offs. EMBO J. 2002;21:2303–2311. doi: 10.1093/emboj/21.10.2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ioannidis V., Beermann F., Clevers H., Held W. The beta-catenin—TCF-1 pathway ensures CD4(+)CD8(+) thymocyte survival. Nat. Immunol. 2001;2:691–697. doi: 10.1038/90623. [DOI] [PubMed] [Google Scholar]
- Kalluri R., Weinberg R.A. The basics of epithelial-mesenchymal transition. J. Clin. Invest. 2009;119:1420–1428. doi: 10.1172/JCI39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh M., Katoh M. WNT signaling pathway and stem cell signaling network. Clin. Cancer Res. 2007;13:4042–4045. doi: 10.1158/1078-0432.CCR-06-2316. [DOI] [PubMed] [Google Scholar]
- Kawamura T., Suzuki J., Wang Y.V., Menendez S., Morera L.B., Raya A., Wahl G.M., Izpisúa Belmonte J.C. Linking the p53 tumour suppressor pathway to somatic cell reprogramming. Nature. 2009;460:1140–1144. doi: 10.1038/nature08311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kühl S.J., Kühl M. On the role of Wnt/β-catenin signaling in stem cells. Biochim. Biophys. Acta. 2013;1830:2297–2306. doi: 10.1016/j.bbagen.2012.08.010. [DOI] [PubMed] [Google Scholar]
- Li H., Collado M., Villasante A., Strati K., Ortega S., Cañamero M., Blasco M.A., Serrano M. The Ink4/Arf locus is a barrier for iPS cell reprogramming. Nature. 2009;460:1136–1139. doi: 10.1038/nature08290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R., Liang J., Ni S., Zhou T., Qing X., Li H., He W., Chen J., Li F., Zhuang Q. A mesenchymal-to-epithelial transition initiates and is required for the nuclear reprogramming of mouse fibroblasts. Cell Stem Cell. 2010;7:51–63. doi: 10.1016/j.stem.2010.04.014. [DOI] [PubMed] [Google Scholar]
- Lluis F., Cosma M.P. Somatic cell reprogramming control: signaling pathway modulation versus transcription factor activities. Cell Cycle. 2009;8:1138–1144. doi: 10.4161/cc.8.8.8206. [DOI] [PubMed] [Google Scholar]
- Lluis F., Pedone E., Pepe S., Cosma M.P. Periodic activation of Wnt/beta-catenin signaling enhances somatic cell reprogramming mediated by cell fusion. Cell Stem Cell. 2008;3:493–507. doi: 10.1016/j.stem.2008.08.017. [DOI] [PubMed] [Google Scholar]
- Lluis F., Pedone E., Pepe S., Cosma M.P. The Wnt/β-catenin signaling pathway tips the balance between apoptosis and reprograming of cell fusion hybrids. Stem Cells. 2010;28:1940–1949. doi: 10.1002/stem.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lluis F., Ombrato L., Pedone E., Pepe S., Merrill B.J., Cosma M.P. T-cell factor 3 (Tcf3) deletion increases somatic cell reprogramming by inducing epigenome modifications. Proc. Natl. Acad. Sci. USA. 2011;108:11912–11917. doi: 10.1073/pnas.1017402108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald B.T., Tamai K., He X. Wnt/beta-catenin signaling: components, mechanisms, and diseases. Dev. Cell. 2009;17:9–26. doi: 10.1016/j.devcel.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahmoudi S., Brunet A. Aging and reprogramming: a two-way street. Curr. Opin. Cell Biol. 2012;24:744–756. doi: 10.1016/j.ceb.2012.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marión R.M., Strati K., Li H., Murga M., Blanco R., Ortega S., Fernandez-Capetillo O., Serrano M., Blasco M.A. A p53-mediated DNA damage response limits reprogramming to ensure iPS cell genomic integrity. Nature. 2009;460:1149–1153. doi: 10.1038/nature08287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marson A., Foreman R., Chevalier B., Bilodeau S., Kahn M., Young R.A., Jaenisch R. Wnt signaling promotes reprogramming of somatic cells to pluripotency. Cell Stem Cell. 2008;3:132–135. doi: 10.1016/j.stem.2008.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrill B.J., Pasolli H.A., Polak L., Rendl M., García-García M.J., Anderson K.V., Fuchs E. Tcf3: a transcriptional regulator of axis induction in the early embryo. Development. 2004;131:263–274. doi: 10.1242/dev.00935. [DOI] [PubMed] [Google Scholar]
- Moon R.T., Kohn A.D., De Ferrari G.V., Kaykas A. WNT and beta-catenin signalling: diseases and therapies. Nat. Rev. Genet. 2004;5:691–701. doi: 10.1038/nrg1427. [DOI] [PubMed] [Google Scholar]
- Murry C.E., Keller G. Differentiation of embryonic stem cells to clinically relevant populations: lessons from embryonic development. Cell. 2008;132:661–680. doi: 10.1016/j.cell.2008.02.008. [DOI] [PubMed] [Google Scholar]
- Niida A., Hiroko T., Kasai M., Furukawa Y., Nakamura Y., Suzuki Y., Sugano S., Akiyama T. DKK1, a negative regulator of Wnt signaling, is a target of the beta-catenin/TCF pathway. Oncogene. 2004;23:8520–8526. doi: 10.1038/sj.onc.1207892. [DOI] [PubMed] [Google Scholar]
- Ombrato L., Lluis F., Cosma M.P. Regulation of self-renewal and reprogramming by TCF factors. Cell Cycle. 2012;11:39–47. doi: 10.4161/cc.11.1.18759. [DOI] [PubMed] [Google Scholar]
- Pereira L., Yi F., Merrill B.J. Repression of Nanog gene transcription by Tcf3 limits embryonic stem cell self-renewal. Mol. Cell. Biol. 2006;26:7479–7491. doi: 10.1128/MCB.00368-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polo J.M., Anderssen E., Walsh R.M., Schwarz B.A., Nefzger C.M., Lim S.M., Borkent M., Apostolou E., Alaei S., Cloutier J. A molecular roadmap of reprogramming somatic cells into iPS cells. Cell. 2012;151:1617–1632. doi: 10.1016/j.cell.2012.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reya T., Clevers H. Wnt signalling in stem cells and cancer. Nature. 2005;434:843–850. doi: 10.1038/nature03319. [DOI] [PubMed] [Google Scholar]
- Reya T., Duncan A.W., Ailles L., Domen J., Scherer D.C., Willert K., Hintz L., Nusse R., Weissman I.L. A role for Wnt signalling in self-renewal of haematopoietic stem cells. Nature. 2003;423:409–414. doi: 10.1038/nature01593. [DOI] [PubMed] [Google Scholar]
- Roose J., Molenaar M., Peterson J., Hurenkamp J., Brantjes H., Moerer P., van de Wetering M., Destrée O., Clevers H. The Xenopus Wnt effector XTcf-3 interacts with Groucho-related transcriptional repressors. Nature. 1998;395:608–612. doi: 10.1038/26989. [DOI] [PubMed] [Google Scholar]
- Roose J., Huls G., van Beest M., Moerer P., van der Horn K., Goldschmeding R., Logtenberg T., Clevers H. Synergy between tumor suppressor APC and the beta-catenin-Tcf4 target Tcf1. Science. 1999;285:1923–1926. doi: 10.1126/science.285.5435.1923. [DOI] [PubMed] [Google Scholar]
- Samavarchi-Tehrani P., Golipour A., David L., Sung H.K., Beyer T.A., Datti A., Woltjen K., Nagy A., Wrana J.L. Functional genomics reveals a BMP-driven mesenchymal-to-epithelial transition in the initiation of somatic cell reprogramming. Cell Stem Cell. 2010;7:64–77. doi: 10.1016/j.stem.2010.04.015. [DOI] [PubMed] [Google Scholar]
- Sato N., Meijer L., Skaltsounis L., Greengard P., Brivanlou A.H. Maintenance of pluripotency in human and mouse embryonic stem cells through activation of Wnt signaling by a pharmacological GSK-3-specific inhibitor. Nat. Med. 2004;10:55–63. doi: 10.1038/nm979. [DOI] [PubMed] [Google Scholar]
- Silva J., Barrandon O., Nichols J., Kawaguchi J., Theunissen T.W., Smith A. Promotion of reprogramming to ground state pluripotency by signal inhibition. PLoS Biol. 2008;6:e253. doi: 10.1371/journal.pbio.0060253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokol S.Y. Maintaining embryonic stem cell pluripotency with Wnt signaling. Development. 2011;138:4341–4350. doi: 10.1242/dev.066209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadtfeld M., Maherali N., Breault D.T., Hochedlinger K. Defining molecular cornerstones during fibroblast to iPS cell reprogramming in mouse. Cell Stem Cell. 2008;2:230–240. doi: 10.1016/j.stem.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam P.P., Loebel D.A. Specifying mouse embryonic germ cells. Cell. 2009;137:398–400. doi: 10.1016/j.cell.2009.04.016. [DOI] [PubMed] [Google Scholar]
- Tam W.L., Lim C.Y., Han J., Zhang J., Ang Y.S., Ng H.H., Yang H., Lim B. T-cell factor 3 regulates embryonic stem cell pluripotency and self-renewal by the transcriptional control of multiple lineage pathways. Stem Cells. 2008;26:2019–2031. doi: 10.1634/stemcells.2007-1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka S.S., Kojima Y., Yamaguchi Y.L., Nishinakamura R., Tam P.P. Impact of WNT signaling on tissue lineage differentiation in the early mouse embryo. Dev. Growth Differ. 2011;53:843–856. doi: 10.1111/j.1440-169X.2011.01292.x. [DOI] [PubMed] [Google Scholar]
- ten Berge D., Koole W., Fuerer C., Fish M., Eroglu E., Nusse R. Wnt signaling mediates self-organization and axis formation in embryoid bodies. Cell Stem Cell. 2008;3:508–518. doi: 10.1016/j.stem.2008.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ten Berge D., Kurek D., Blauwkamp T., Koole W., Maas A., Eroglu E., Siu R.K., Nusse R. Embryonic stem cells require Wnt proteins to prevent differentiation to epiblast stem cells. Nat. Cell Biol. 2011;13:1070–1075. doi: 10.1038/ncb2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utikal J., Polo J.M., Stadtfeld M., Maherali N., Kulalert W., Walsh R.M., Khalil A., Rheinwald J.G., Hochedlinger K. Immortalization eliminates a roadblock during cellular reprogramming into iPS cells. Nature. 2009;460:1145–1148. doi: 10.1038/nature08285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Amerongen R., Nusse R. Towards an integrated view of Wnt signaling in development. Development. 2009;136:3205–3214. doi: 10.1242/dev.033910. [DOI] [PubMed] [Google Scholar]
- Van de Wetering M., Castrop J., Korinek V., Clevers H. Extensive alternative splicing and dual promoter usage generate Tcf-1 protein isoforms with differential transcription control properties. Mol. Cell. Biol. 1996;16:745–752. doi: 10.1128/mcb.16.3.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterman M.L. Lymphoid enhancer factor/T cell factor expression in colorectal cancer. Cancer Metastasis Rev. 2004;23:41–52. doi: 10.1023/a:1025858928620. [DOI] [PubMed] [Google Scholar]
- Willert K., Jones K.A. Wnt signaling: is the party in the nucleus? Genes Dev. 2006;20:1394–1404. doi: 10.1101/gad.1424006. [DOI] [PubMed] [Google Scholar]
- Wray J., Kalkan T., Gomez-Lopez S., Eckardt D., Cook A., Kemler R., Smith A. Inhibition of glycogen synthase kinase-3 alleviates Tcf3 repression of the pluripotency network and increases embryonic stem cell resistance to differentiation. Nat. Cell Biol. 2011;13:838–845. doi: 10.1038/ncb2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi F., Pereira L., Merrill B.J. Tcf3 functions as a steady-state limiter of transcriptional programs of mouse embryonic stem cell self-renewal. Stem Cells. 2008;26:1951–1960. doi: 10.1634/stemcells.2008-0229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi F., Pereira L., Hoffman J.A., Shy B.R., Yuen C.M., Liu D.R., Merrill B.J. Opposing effects of Tcf3 and Tcf1 control Wnt stimulation of embryonic stem cell self-renewal. Nat. Cell Biol. 2011;13:762–770. doi: 10.1038/ncb2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Q., Sharma A., Sen J.M. TCF1 and beta-catenin regulate T cell development and function. Immunol. Res. 2010;47:45–55. doi: 10.1007/s12026-009-8137-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


