Summary
Therapeutic approaches for “sick sinus syndrome” rely on electrical pacemakers, which lack hormone responsiveness and bear hazards such as infection and battery failure. These issues may be overcome via “biological pacemakers” derived from pluripotent stem cells (PSCs). Here, we show that forward programming of PSCs with the nodal cell inducer TBX3 plus an additional Myh6-promoter-based antibiotic selection leads to cardiomyocyte aggregates consisting of >80% physiologically and pharmacologically functional pacemaker cells. These induced sinoatrial bodies (iSABs) exhibited highly increased beating rates (300–400 bpm), coming close to those found in mouse hearts, and were able to robustly pace myocardium ex vivo. Our study introduces iSABs as highly pure, functional nodal tissue that is derived from PSCs and may be important for future cell therapies and drug testing in vitro.
Highlights
-
•
TBX3 plus Myh6-promoter antibiotic selection yields pacemaker cells from PSCs
-
•
Induced sinoatrial bodies (iSABs) consist of >80% functional pacemaker cells
-
•
iSABs showed highly increased beating rates and were able to pace myocardium ex vivo
-
•
iSABs represent highly pure functional nodal tissue derived from PSCs
David and colleagues describe how forward programming of PSCs with TBX3 plus additional Myh6-promoter-based antibiotic selection yielded cardiomyocyte aggregates consisting of >80% physiologically and pharmacologically functional pacemaker cells. These induced sinoatrial bodies (iSABs) exhibited highly increased beating rates (300–400 bpm) and were able to pace myocardium ex vivo. Their study introduces iSABs as highly pure functional nodal tissue derived from PSCs.
Introduction
“Sick sinus syndrome” is used as a collective term to describe a number of diseases caused by perturbed function of the sinus node, which represents the cardiac pacemaker. This structure is composed of specialized cardiomyocytes innervated by autonomic nerves. The diseases include pathological and/or symptomatic sinus bradycardia, sinoatrial (SA) block, sinus arrest, and tachycardia-bradycardia syndrome. Sick sinus syndrome is often accompanied by general cardiac diseases such as ischemic heart disease, cardiomyopathy, and myocarditis. These result in either disturbed pacemaking within the sinus node or disturbed conduction of the electric impulse from the sinus node to the atrium (also known as SA conduction disturbance). At present, therapeutic approaches for sick sinus syndrome rely on the implantation of artificial pacemakers, which are insensitive to hormone stimulation. Furthermore, the hazards of infection and battery failure are potential limitations. Therefore, patients with an implanted pacemaker generally bear a risk of developing severe complications during their remaining lifespan.
To avoid these hazards, the availability of functional SA node (SAN) cells for transplantation or the de novo generation of such cells in vivo is required. Correspondingly, to obtain biological pacemaker cells for future therapy, two possible approaches are being addressed. One approach is to convert beating myocardium into pacemaker cells in situ via genetic manipulations (i.e., direct reprogramming). In this regard, the early key transcription factor TBX3 led to cells with incomplete pacemaker characteristics (Bakker et al., 2012). More recently, viral overexpression of TBX18, a member of the same transcription factor family, was reported to enable reprogramming of chamber myocardium toward nodal cells (Kapoor et al., 2013). Thus far, however, this approach requires integration of viruses, prohibiting a spatially and timely controllable expression of TBX18, which would mimic the in vivo situation in the developing embryo. TBX18 is only transiently expressed in the head part of the developing sinus node, whereas TBX3 is permanently expressed in the entire node in vivo (Wiese et al., 2009). In addition, the efficiency of Tbx18-based direct reprogramming is low (Kapoor et al., 2013).
Another approach relies on the transplantation of in-vitro-generated “biological pacemakers” derived from pluripotent stem cells (PSCs) such as embryonic stem cells (ESCs) (Wobus et al., 1991; Kehat et al., 2002). In this regard, Kleger et al. (2010) postulated that the small-molecule compound EBIO enhances the formation of nodal cells from murine ESCs to some extent. However, they did not address the actual ability of the cells to pace ventricular myocardium, and again the cells’ beating rates were low. Moreover, on the electrophysiological level, they did not discriminate between relatively mature pacemaker cells and similar spontaneously contracting early/intermediate cell types (Kleger et al., 2010; Maltsev et al., 1994; David and Franz, 2012).
More recently, a cell population purified based on Alcam (CD166) expression was described to give rise to nodal cells (Scavone et al., 2013). It will be interesting to see whether this approach can be transferred to the human system, as surface marker expression specificity often differs among species.
As an alternative, in this work we further developed our previous method for cardiomyocyte-subtype forward programming of ESCs via single transcription factors (David et al., 2008, 2009; David and Franz, 2012). We applied this approach to TBX3 and found that it led to a doubling of functional pacemaker cells, but alone was still insufficient to obtain pure populations of these cells. This may be related to the insufficiency of TBX3 as a single factor for direct reprogramming (Bakker et al., 2012).
However, when we combined our approach with Myh6 promoter-based antibiotic selection (Klug et al., 1996), we achieved cell aggregates that consisted exclusively of spontaneously beating cardiomyocytes, with beats per minute (bpm) rates close to those of a mouse heart. Of these cells, >80% corresponded to the desired pacemaker phenotypes bearing full functionality at the levels of protein expression, electrophysiological parameters, and Ca2+ signaling characteristics, as well as potency to robustly pace murine myocardium ex vivo. Therefore, we provide and example of highly enriched, stem cell-derived pacemaker cell populations that possess all of the properties characteristic of that cell type, which may be an important prerequisite for future cell therapies and in vitro drug evaluation.
Results
Relying on the high conservation of vertebrate TBX3 proteins, we used human TBX3 in murine ESCs due to its specific traceability. We inserted human TBX3 cDNA into the pEF-DEST51 vector (Invitrogen) for EF1α-promoter-based stable and constitutive overexpression in ESCs. Out of 20 independent clones, we selected four clones that represented the entire spectrum, ranging from low to high levels of TBX3 mRNA overexpression based on quantitative RT-PCR (qRT-PCR; Figure 1A). These overexpression levels were confirmed on the protein level (Figure 1B). Fluorescence-activated cell sorting (FACS) analyses showed no influence of TBX3 overexpression on the numbers of pluripotency marker Oct-4/Pou5f1 and Sox2-positive cells (Figure 1C). This corresponds to normal undifferentiated colony growth in leukemia inhibitory factor (LIF)-containing medium (Figure 1B) and is in agreement with our previous findings for MESP1 and NKX2-5 overexpression (David et al., 2008, 2009; David and Franz, 2012). We concluded that similar to what was found for those factors, TBX3 alone was not sufficient to induce differentiation of ESCs.
Figure 1.
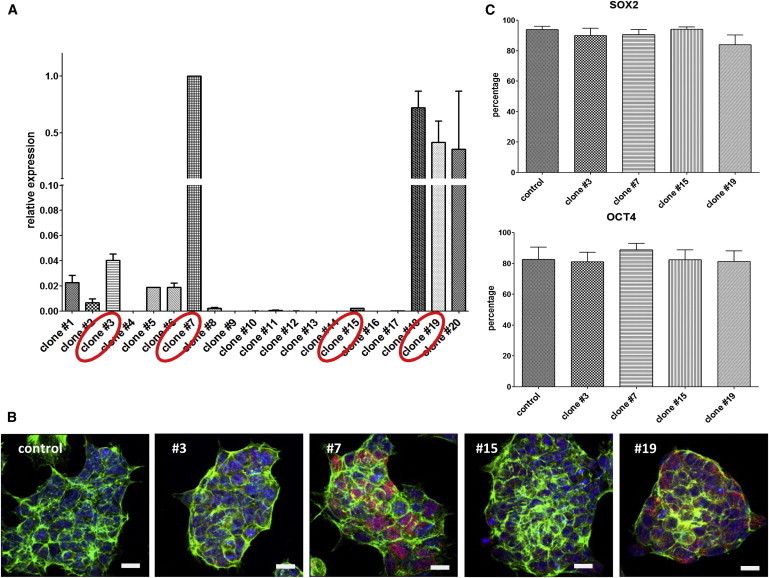
Functionality of the TBX3 Overexpression Construct in ESCs
(A) Twenty independent cell clones were stably transfected with an overexpression construct bearing the human TBX3-cDNA. Overexpression levels were analyzed via qRT-PCR. Four representative clones were selected for further analysis. The data represent two biological replicates and are presented as means ± SD.
(B) Immunostaining of overexpressed TBX3 (red), Actin (green), and nuclei (blue) in the four selected clones confirm overexpression levels of TBX3. Scale bar, 10 μm.
(C) FACS analyses of OCT-4/POU5f1 and SOX2 reveal no influence of TBX3 overexpression on pluripotency under LIF administration. The data represent five independent experiments and are presented as means ± SEM.
During differentiation, highly and moderately TBX3-overexpressing ESC clones started to contract earlier and showed approximately 5- to 10-fold more contracting areas than controls (Figure 2A). This enhanced cardiac differentiation resembles the effect of MESP1 and NKX2-5 overexpression described in our previous studies (David et al., 2008, 2009; David and Franz, 2012). Correspondingly to their beating activity, Tbx3-overexpressing cardiomyocytes showed a normal pattern of the sarcomeric marker MYH6 (Figure 2B).
Figure 2.
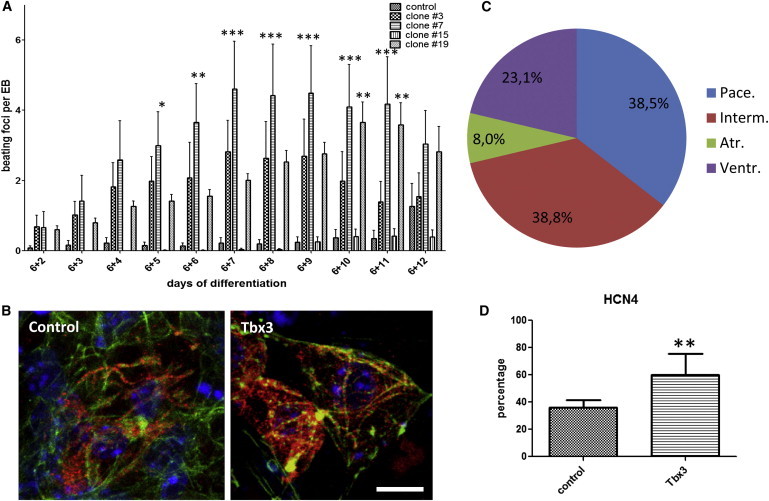
Dominant Appearance of Pacemaker-like Cardiomyocytes in TBX3-Programmed ESCs
(A) Spontaneous beating activity is increased in independent TBX3 clones and control ESCs (GSES). The data represent five independent experiments and are presented as means ± SEM; ∗p < 0.05, ∗∗p < 0.01 ∗∗∗p < 0.001; number of EBs > 100.
(B) Confocal analysis of MYH6 expression in control and TBX3-overexpressing cardiomyocytes (red). Counterstaining against Actin (green) and nuclei (blue). Scale bar, 10 μm.
(C) Distribution of cardiomyocyte subtypes and examples of the four APs as shown by single-cell patch-clamping analyses. In a total of 39 cells, 38.5% of pacemaker-like cells were obtained. Ventr., ventricular-like (23.1%); Atr., atrial-like (8%); Pace., SAN-like (38.5%); Interm., intermediate/early type (38.8%).
(D) HCN4-expressing cells are increased significantly in Tbx3 clones at day 18. The data represent five independent experiments and are presented as means ± SD; ∗∗p < 0.01.
To further verify the functionality and subtype content of the cardiomyocytes, we analyzed their electrophysiological properties via single-cell patch clamping and “funny channel” density measurements at day 6+12 as previously described (David et al., 2008, 2009, 2013).
In general, three cell types that have been described for isolated beating cardiomyocytes obtained from embryoid body (EB) development, namely, ventricle-like, atrial-like, and SA/AV (pacemaker-like) cells, plus intermediate cells were present in preparations from TBX3 cell clones (Figure 2C). The action potentials (APs) generated by the various cell types were typical with respect to their distinct parameters, including the maximum diastolic potential (MDP), diastolic depolarization rate (DDR), overshoot or AP/plateau duration, and cycle length (Figure 2C; Figure S1 and Table S1 available online). However, we found an unusually high proportion of the pacemaker-like subtype in TBX3 programmed cells, representing 38.5% of all cardiomyocytes. These cells lacked a plateau phase, had a positive MDP of >−50 mV, and high DDR values typically exceeding 100 mV/s. In addition, they had the smallest positive overshoot, with a maximum of ∼+20 mV. In contrast, within control cells, only 20% of these pacemaker-like cells were found (Table S2). In accordance with the high proportion of pacemaker cells, cell numbers expressing HCN4 (representing the funny channel) were enhanced among the Tbx3 programmed cardiomyocytes (Figure 2D).
To further enhance the yield of the SA/AV subtype achieved via TBX3 programming, we chose to combine this approach with MYH6-promoter-based antibiotic selection (Klug et al., 1996). This was recently described as directed pacemaker cell enrichment (Kensah et al., 2013; Otsuji et al., 2010). We therefore additionally introduced the plasmid containing the Myh6-Neomycin cassette (Klug et al., 1996) into our TBX3 clones. As expected, administration of the antibiotic during differentiation led to an enrichment of beating tissue in Myh6-promoter antibiotic-selected controls. Likewise, Myh6-TBX3 double-transgenic clones were enriched for beating foci (Movies S1 and S2). However, in the latter, spontaneous beating rates were approximately doubled (Figure 3A). After introduction of an additional dissociation step, beating rates increased in Myh6-promoter antibiotic-selected controls, leading to antibiotic-selected cardiac bodies (aCaBs), as well as in Myh6-Tbx3 double-transgenic clones. In the latter, we often found cellular aggregates contracting at 300–400 bpm, which is ∼6-fold faster than wild-type cells. These beating rates come close to murine heart rates (∼500 bpm), indicating the formation of induced SA bodies (iSABs; Figure 3A; Movie S3). The cells within iSABs were positive for HCN4 as well as for Connexins CX45 and CX30.2 (Figure 3B) which is characteristic for pacemaker cells (Kreuzberg et al., 2005; Verheijck et al., 2001).
Figure 3.
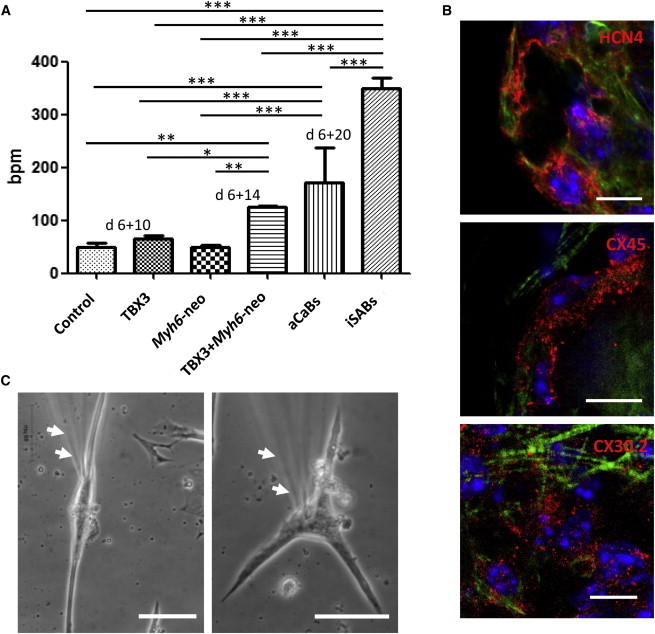
Phenotypic Appearance and Marker Expression of Pacemaker-like Cardiomyocytes Obtained from iSABs
(A) Beating frequencies during the generation of aCaBs and iSABs; 300–400 bpm were achieved via a combination of TBX3 programming, antibiotic selection, and an additional dissociation step (iSABs). The data represent five independent experiments and are presented as means ± SD; ∗p < 0.05, ∗∗p < 0.01 ∗∗∗p < 0.001. See also Movies S1, S2, S3, S4, and S5.
(B) Expression of HCN4 (left panel), Cx45 (middle panel), and Cx30.2 (right panel) in iSABs (red stainings). Counterstaining for Actin (green) and nuclei (blue). Scale bar, 10 μm.
(C) Typical spindle and spider cellular shape of nodal cells derived from iSABs and subjected to electrophysiological analysis. Arrows indicate patch-clamp pipette (out of focus). Scale bar, 50 μm. See also Movie S5.
Further cultivation of iSABs for 3 weeks on gelatin-coated dishes led to attached, highly synchronized cell layers beating at >350 bpm (Movie S4). The cells exhibited the typical shape of nodal cells (i.e., spindle and spider-shaped cells; Figure 3C; Movie S5).
To address the electrophysiological parameters of iSAB-derived cardiomyocytes, we performed single-cell patch clamping again. Out of 65 cells analyzed, 53 cells (81.5%) now corresponded to the regular and fast-beating pacemaker subtype, and only 12 cells (18.5%) represented the irregular and slowly beating myocardial subtypes. Moreover, 43 of the 53 cells classified as the pacemaker-system subtype showed AP parameters of mature pacemaker cells, whereas ten of these spontaneously and regularly beating cells were still immature (Figures 4A and S2; Table S3). The latter exhibited a short plateau phase and a more negative MDP, which distinguishes them from fully mature pacemaker cells. Yet, they also showed typical pacemaker features, such as regular spontaneous AP generation with a fast DDR, and expressed the typical pacemaker current If comparably to the cells classified as mature pacemaker cells (Figures 4B–4E). Likewise, the iSAB-derived cells responded to isoproterenol and carbachol with an acceleration and deceleration, respectively, of AP rates (Figure 4F). Thus, the beating rates were able to reach >550 bpm.
Figure 4.
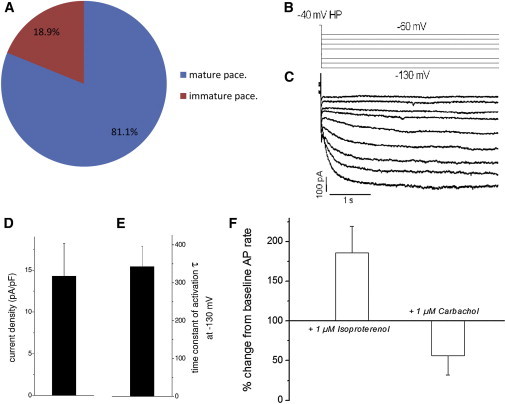
Electrophysiological Parameters of Pacemaker-like Cardiomyocytes Obtained from iSABs
(A) Distribution of pacemaker cells as shown by single-cell patch-clamp and funny-channel measurements: >81% of pacemaker-like cells were achieved; 19% of these represented immature pacemaker cells and the rest were mature pacemaker cells (n = 65). The data represent seven independent experiments and are presented as means ± SD. See also Tables S1–S3.
(B) Patch-clamp protocol. Voltage was applied to elicit the hyperpolarization-activated current for If recordings from isolated iSAB cells.
(C) Example of If current, recorded from an isolated iSAB-derived cell.
(D and E) Current density (D) and time constant of activation (E) at –130 mV, demonstrating a robust If expression with slow activation kinetics typical of the HCN4 channel subtype and mature SA nodal If (n = 17). The data represent three independent experiments and are presented as means ± SD.
(F) Reaction to β-adrenergic (isoprotenerol) and muscarinic (carbachol) stimulation leads to typical accelerated versus decelerated AP rates. iSAB-derived cells show a pronounced response to isoprotenerol, with beating rates reaching up to 560 bpm. iSAB/Iso: n = 9; iSAB/Carb: n = 5. The data represent seven independent experiments and are presented as means ± SD.
See also Figures S1 and S2.
To further verify the pacemaker cell identity, we characterized the Ca2+ flux from the extracellular space and intracellular Ca2+ stores, which can be mediated by sarcolemmal Ca2+ channels or by Ca2+ release from the stores. Electrogenic features of the sarcolemmal channels also determine the frequency of the Ca2+ transients (Movie S6). As a characteristic of pacemaker cells, we confirmed that hyperpolarization-activated, cyclic nucleotide-gated (HCN) channels modulated the rate of the Ca2+ transients. The application of the HCN channel blocker ZD 7288 time dependently reduced the frequency of Ca2+ transients in iSABs (Figure 5A). In addition, the frequencies of Ca2+ transients are also based on the activity of T-type and L-type voltage-sensitive Ca2+ channels. Correspondingly, the spontaneous frequencies of the Ca2+ transients were dramatically reduced after inhibition of L-type Ca2+ channels with nifedipine, and moderately by inhibition of T-type Ca2+ channels with mibefradil in iSAB-derived cells (Figure 5B).
Figure 5.
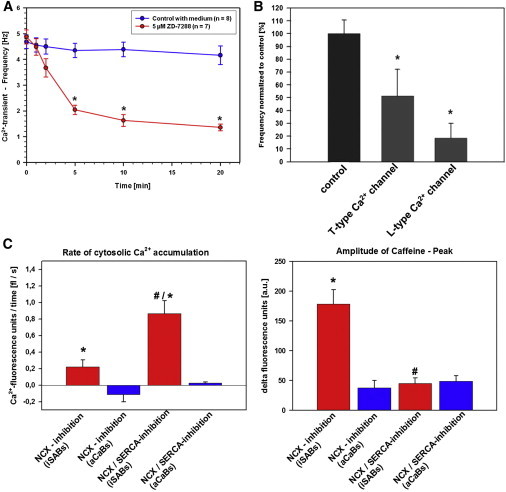
Calcium Characteristics of Pacemaker-like Cardiomyocytes Obtained from iSABs
(A) Frequencies of spontaneous Ca2+ transients in iSAB cells significantly decrease after inhibition of HCN channels via ZD7288. The data represent four independent experiments and are presented as means ± SEM; n ≥ 7.
(B) Frequencies of spontaneous Ca2+ transients in iSAB cells significantly decrease after inhibition of T-type and L-type Ca2+ channels via mibefradil and nifedipine. The data represent four independent experiments and are presented as means ± SEM; n > 12; ∗p < 0.05. See also Movie S6.
(C) Analysis of SR Ca2+ efflux as a characteristic of pacemaker-active cells reveals an enhanced rate of intracellular Ca2+ accumulation in iSAB-derived cells (red) in comparison with aCaBs (blue). Na+/Ca2+ exchanger blockade plus inhibition of SERCA caused an intracellular Ca2+ accumulation comparable to the reduction of the caffeine peak in iSAB-derived cells. The rate of cytosolic Ca2+ accumulation and amplitude of the caffeine peak in aCaBs do not show a difference. The data represent five independent experiments and are presented as means ± SEM; #p < 0.05 versus no SERCA inhibition; ∗p < 0.05 versus controls.
See also Figure S3.
A functional sarcoplasmic reticulum (SR) characterizes the maturation state of cardiomyocytes. Therefore, in pacemaker cells, Ca2+ from SR plays an important role in spontaneous activity. The impact of Ca2+ from the SR on spontaneous Ca2+ transients becomes evident if a complete release of Ca2+ from SR with caffeine or an inhibition of SERCA with thapsigargin is induced. In iSAB-derived cells, caffeine-induced SR-Ca2+ release increased the diastolic Ca2+ level comparable to a Ca2+ peak, but with recognizable spontaneous Ca2+ transients in the time course of the Ca2+ peak and unchanged systolic Ca2+ values (Figure S3A). A similar caffeine-induced effect on the SR-Ca2+ release could not be detected in aCaB-derived cells. Blocking Ca2+ reuptake into SR by thapsigargin resulted in an increase of the diastolic Ca2+ level only in iSAB-derived cells (Figure S3B).
The main proportion of Ca2+ underlying the Ca2+ transients is derived from the extracellular space. Therefore, replacement of extracellular Ca2+ leads to an abolishment of Ca2+ transients. When sarcolemmal Ca2+ flux was eliminated by blocking the Na+/Ca2+ exchanger and the Ca2+ channels, only intracellular Ca2+ cycling could be detected. In this case, the application of caffeine induced a Ca2+ peak in iSAB-derived cells that was 4-fold greater than in aCaB-derived cells (Figures 5C and S3C). Additional inhibition of SERCA under these conditions revealed a leaky SR in iSAB-derived cells. The rate of intracellular Ca2+ accumulation was increased 4-fold compared with no SERCA inhibition. After addition of caffeine 5 min after thapsigargin incubation, the Ca2+ peak was reduced by a factor of 4. In contrast, the caffeine-induced Ca2+ peak was identical with and without SERCA inhibition in control cells (Figures 5C and S3C), where an intracellular Ca2+ accumulation also could not be detected.
To ultimately test the functional pacemaker activity of the iSABs, we took advantage of the ex vivo model based on cultivated ventricular slices of murine hearts (Halbach et al., 2006; Figure 6A). Although such slices typically have spontaneous beating activity immediately after preparation, they can be induced to robustly contract at up to ∼60 bpm via electrodes and are therefore an ideal system for testing the functionality of our iSABs. Correspondingly, when seeded on the slices, the iSABs were able to attach and persist on the slices as shown by labeling with DiI (d.n.s.). To address the effect of iSAB seeding, we first carefully quantified spontaneous slice activity in naive slices.
Figure 6.
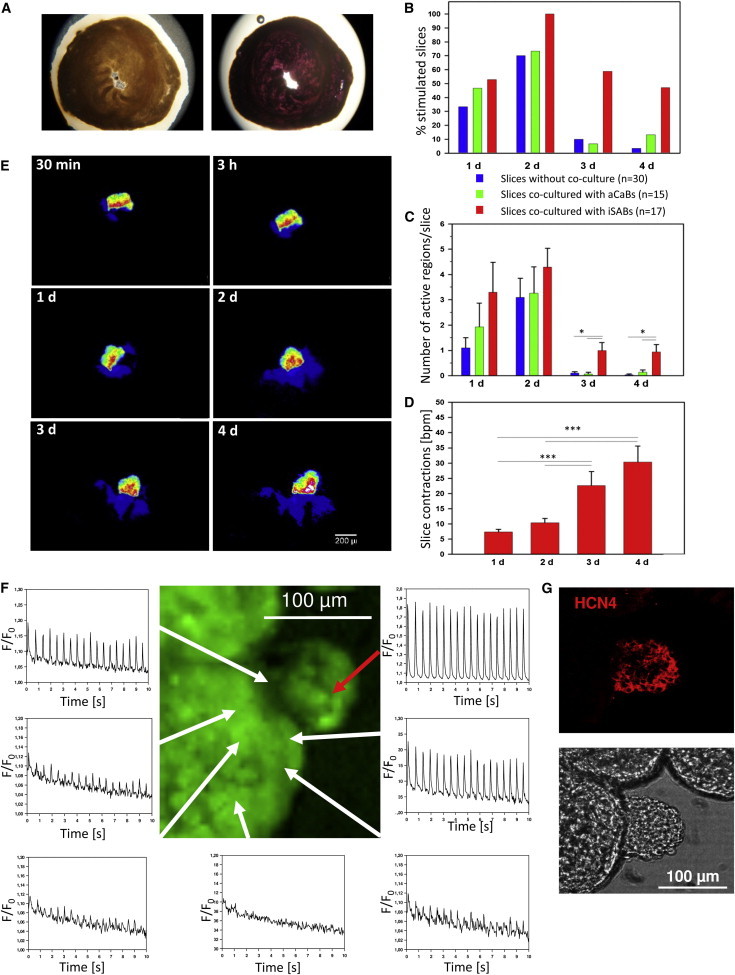
Functionality of iSABs in an Ex Vivo Model of Cultivated Murine Ventricular Slices
(A) Ventricular slices were cultivated for 5 days (left) and MTT staining was used to assess viability (right).
(B) Percentage distribution of iSAB-seeded, aCaB-seeded, and nonseeded slices containing at least one contracting region over time. Spontaneous activity ceased after day 2, whereas activity in iSAB-seeded slices was greatly retained (n ≥ 15).
(C) Average number of active regions per iSAB-seeded, aCaB-seeded, and unseeded slices over time. The data represent three independent experiments and are presented as means ± SEM; n ≥ 15; ∗p < 0.05.
(D) Increase of beating frequencies of iSAB-seeded slices from day 1 until day 4. The data represent three independent experiments and are presented as means ± SEM; n ≥ 16; ∗∗∗p < 0.05. See also Movie S7.
(E) Transmission of calcein dye from an iSAB to the recipient slice over time. Scale bar, 200 μm.
(F) Stimulation of slice regions (white arrows) in the proximity of an iSAB (red arrow) is accompanied by highly synchronized Ca2+ transients.
(G) The identical iSAB/slice coculture as shown in (F) was stained for HCN4 expressing the nodal cell marker in the iSAB. See also Movie S8.
On day 2 after slice preparation, a maximum of 70% of slices containing at least one area of spontaneous beating activity was observed. This value decreased dramatically on days 3 and 4 (Figure 6B). Yet, whereas coculturing slices with aCaBs did not significantly alter the number of beating areas or beating parameters compared with naive slices, seeding slices with iSABs resulted in a ∼1.5-fold increased beating activity as early as days 1 and 2. Moreover, only slices cocultured with iSABs preserved their spontaneous beating activity and multiple beating sites also on days 3 and 4 (Figures 6B and 6C). Furthermore, the beating rates of iSAB-seeded slices continuously and significantly increased over 4-fold from day 1 until day 4 (Figure 6D), which we did not observe for unseeded and aCaB seeded slices.
Loading the iSABs with calcein before transferring them to the slice confirmed the formation of syncytia between iSABs and slice cells, as shown by transmission of the dye over time (Figure 6E). The same iSAB is shown in Figure 6E and Movie S7, demonstrating pacing of the host slice within the respective area. Furthermore, functional coupling is evident from synchronized Ca2+ transients between iSAB and slice myocardium. Therefore, peaks of Ca2+ transients within the slice are smaller but highly synchronous to spontaneous iSAB activity and can be detected within a radius of ∼200 μm (Figure 6F; Movie S8). Thus, the signals were derived from the slice myocardium, especially since no significant outgrowths of iSABs into the slice were observed in staining for the pacemaker cell marker HCN4 (Figure 6G).
Discussion
The ability to produce de novo, highly enriched, stem cell-derived populations of cardiac pacemaker cells that bear all the functional parameters of mature nodal cells is of great interest for future cell-based therapies. Such cells might be able to reconstitute a proper cardiac rhythm in the sense of a biological pacemaker. In addition, drug testing in vitro would benefit from the availability of such purified nodal cells. PSCs are the focus of these goals because they have been shown to give rise to any cell type of the mammalian organism, including spontaneously beating cardiomyocytes with molecular and functional properties characteristic of SAN/pacemaker cells (Maltsev et al., 1994; He et al., 2003; Yanagi et al., 2007; Barbuti et al., 2009; Ma et al., 2011; David and Franz, 2012). Typically, however, the cell populations within EBs are highly heterogeneous, which inevitably leads to the requirement of reliable selection and isolation strategies. This applies in particular to the very rare cell type of cardiac nodal cells.
Cardiac nodal cells are characterized by a low membrane potential, diastolic depolarization, and low upstroke velocities. A number of currents contribute to diastolic depolarization and to the AP in the SAN, including the pacemaker current If. This current is carried by the HCN channel. The cyclic-AMP binding site on the HCN channel permits catecholamines to modulate activation, and this property might regulate the autonomic responsiveness of the pacemaker mechanism (Rosen, 2005). There are four genes representing the four known isoforms, termed HCN1–HCN4. In the SAN, the predominant isoform by far is HCN4 (Ishii et al., 1999; Stieber et al., 2004).
In this regard, murine ESC derivatives bearing EGFP transcriptionally controlled via the promoter of the HCN4 gene were reported to coexpress EGFP, HCN4, and other cardiac markers in spontaneously beating foci (Morikawa et al., 2010). Yet, after the EGFP-positive fraction was subjected to cell sorting, spontaneous APs were rarely observed and, interestingly, most of the cells were positive for nestin, a marker for neurons (Morikawa et al., 2010). This corresponds to the observation that HCN4 is a marker not only for spontaneously active cardiomyocytes but also for nerve cells (Garcia-Frigola et al., 2003).
Therefore, alternative promising approaches based on endogenous surface markers (Scavone et al., 2013) and pharmacological administration of small molecules (Wiese et al., 2011; Kleger et al., 2010) have been pursued in addition to direct reprogramming strategies (Bakker et al., 2012; Kapoor et al., 2013).
Here, we describe a protocol that combines ESC forward programming using TBX3 as a key transcription factor with Myh6-based antibiotic selection (Klug et al., 1996). This approach consistently led to highly effective generation of nodal-like pacemaker cell aggregates characterized by spontaneously contracting cardiomyocytes with highly synchronized beating rates of 300–400 bpm, approximating those of a mouse heart. Most (>80%) of the cardiomyocytes within these clusters clearly represented the nodal cell type, and, to our knowledge, such high purities have not been achieved before (Kleger et al., 2010; Wiese et al., 2011; Scavone et al., 2013).
Although it is technically almost impossible to discriminate between murine SAN and AV node cells, especially on the electrophysiological level, Marger et al. (2011) showed that the two cell types differ in a more easily accessible feature, namely, their spontaneous beating rates after isolation. They observed rates of ∼260 bpm in isolated SAN cells and ∼175 bpm in AV node cells in culture. Isoproterenol stimulation led to beating rates of ∼350 bpm in SAN cells and ∼225 bpm in AV node cells. Our iSAB-derived cells showed rates of ∼305 bpm and >550 bpm without and with isoproterenol stimulation, respectively, thus exceeding the values for SAN cells reported by Marger et al. (2011). Moreover, as our cells provide all other properties required for full functionality (i.e., protein expression patterns and electrophysiological and Ca2+ signaling parameters), we propose to call these engineered pacemaker aggregates induced sinoatrial bodies (iSABs). Furthermore, our iSABs are functional in the sense that they can robustly pace working myocardium ex vivo.
Our initial experiments with TBX3-based programming alone led to merely a doubling of functional pacemaker cell numbers in cell aggregates with significantly lower beating frequencies. This may correspond to the insufficiency of TBX3 for fully direct reprogramming of ventricular myocardium to pacemaker cells (Bakker et al., 2012).
Yet, the effect of TBX3 in ESCs was drastically further enhanced when we introduced the additional Myh6 promoter-based antibiotic selection plus an additional dissociation step (Figure 7). Our rationale for this was based on recent reports describing the enrichment of ventricular and nodal cardiomyocytes via Myh6-based selection (Klug et al., 1996; Kensah et al., 2013; Otsuji et al., 2010), although this method was originally thought to enrich cardiomyocyte subtypes in an undirected manner.
Figure 7.
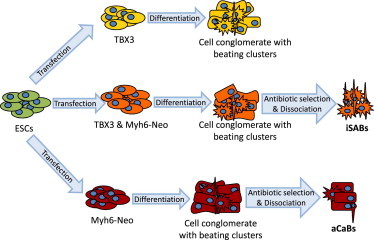
Flowchart for iSAB Generation
Cartoon displaying sequential steps for the generation of iSABs, combining TBX3 programming with Myh6-promoter selection and an additional cell-dissociation step.
Our approach differs from those described in previous studies, which simulated biological pacemakers by manipulating terminal effector molecules underlying the sarcolemmal electrophysiology instead of de novo generating fully functional nodal cells (Johns et al., 1995; Nuss et al., 1999). In contrast, our protocol leads to cells that exhibit not only electrical oscillations but also the subtle electrophysiological and calcium signaling characteristics and distinctive morphological features of endogenous pacemaker cells. Our technology thus may become important for future alternatives to electronic pacing devices.
However, although our pacemaker cells formed a synchronized and rapidly beating syncytium, and were capable of electrically connecting to and pacing ventricular slice cultures ex vivo, their potential to robustly engraft intramyocardially and to pace at physiological beating rates needs to be analyzed. Likewise, the transferability of our protocol to human PSCs (i.e., human ESCs and induced PSCs) (Gerecht-Nir et al., 2004; Takahashi and Yamanaka, 2006; Takahashi et al., 2007; Mauritz et al., 2008) remains to be addressed. Combining this protocol with DNA-free approaches such as RNA or protein transfections and suitable endogenous surface markers may then bring it closer to practical application.
Experimental Procedures
qRT-PCR
qRT-PCR was performed using RNA isolated via the RNeasy kit (QIAGEN). First-strand cDNA was synthesized at 37°C from 2 μg RNA with AMV reverse transcriptase (Amersham) and random hexamer primers. Real-time PCR was performed with an iCycler and the MyiQ detection system (Bio-Rad) using the IQ Syber Green Super Mix kit (Bio-Rad). Primers were designed using the DNA Star software and the specificity of each primer pair was confirmed by agarose gel electrophoresis. The annealing temperature was 57°C for all primer pairs and the amplified murine cDNA fragments corresponded to bp 812–934 of Gapdh (forward primer: 5′-TCTTGGGCTACACTGAGGAC-3′; reverse primer: 5′-ACCAGGAAATGAGCTTGACA-3′) and to bp 287–429 of TBX3 (forward primer: 5′-AAGAAGAGGTGGAGGACGAC-3′; reverse primer: 5′- CAGCCCAGAACATCTCACTT-3′). All samples were analyzed in duplicate, and total RNA pooled from undifferentiated and differentiated ESCs as well as murine hearts was used as control and to generate a standard curve for markers analyzed. As a negative control, total RNA from each of the samples was run without reverse transcriptase. No signal was detected after 40 PCR cycles in the absence of reverse transcriptase, indicating that all samples were free of DNA contamination. In addition, no signal was detected when reverse transcriptase was added but RNA template was not, indicating that there was no contamination from exogenous RNA or DNA. The standard curve for all genes displayed an increase of one threshold cycle for each bisection of template concentration. Evaluation of relative gene-expression levels was performed based on the ΔΔCT method. Factors of changes in relative mRNA expression levels were calculated using Gapdh as a reference gene, and the expression value in control cells was defined as one.
Electrophysiological Analyses
Spontaneously beating cardiac cells were isolated from EBs for electrophysiological analysis as previously described (David et al., 2009, 2013). Briefly, beating areas of the EBs were excised under a microscope and transferred to 37°C prewarmed isolation buffer containing 116 mM NaCl, 5 mM KCl, 0.8 mM MgSO4, 1 mM NaH2PO4, 20 mM HEPES, and 5.5 mM glucose. The pH was adjusted to 7.3 with NaOH. For enzymatic release of single cells, the tissue pieces were incubated for 15 min at 37°C and with slight shaking in isolation buffer with the addition of 0.5 mg/ml collagenase B (Roche) and 1 mg/ml pankreatin (GIBCO). Immediately after incubation, the tissue pieces were mechanically dispersed in isolation buffer/medium (Dulbecco’s modified Eagle’s medium [DMEM] + 10% fetal calf serum [FCS; PAA], 1:1 v/v) and plated on polylysated glass coverslips. iSAB-derived cells for patch clamping were obtained via dissociation with 100% Accutase (Affimetrix) for 15 min. In each case, the cells were allowed to settle for 5 min before medium was added. Cells were kept at 37°C in 5% CO2 for 12–24 hr before analyses.
Spontaneous APs and currents were recorded from contracting cardiomyocytes at 37°C in the perforated patch configuration using a MultiClamp 700B amplifier and pClamp10 software (Molecular Devices). Offline data analysis was done with Clampfit software (Molecular Devices) or Origin 6.0 software (Microcal). Patch pipettes were pulled from borosilicate glass and heat polished. They had a resistance of 2–5 MΩ when filled with intracellular solution containing 10 mM NaCl, 130 mM potassium aspartate, 0.04 mM CaCl2, 3 mM Mg-ATP, 10 mM HEPES, and 200 μg/ml amphotericin B, pH adjusted to 7.2 with KOH. The extracellular (bath) solution contained 140 mM NaCl, 5.4 mM KCl, 1 mM MgCl2, 1.8 mM CaCl2, 5 mM HEPES, 5.5 mM glucose, pH adjusted to 7.4 with NaOH. For better If recordings, 2 mM BaCl2 and 0.3 mM CdCl2 were sometimes added to the bath solution to block IKI and ICa. These measurements were done at room temperature (RT). If was measured by stepping from a holding potential of –40 mV to test potentials between –130 mV and +20 mV. The current amplitude after 3 s during the –130 mV pulse was divided by the cell capacitance to calculate the current (If) density. To determine If activation kinetics, the time constant of activation (τ) was obtained by fitting the current trace of the −130 mV step after the initial lag with the sum of two exponential functions: y = A1e(−x/τ1) + A2e(−x/τ2), where τ1 and τ2 are the fast and slow time constants of activation, respectively; τ1 is consequently referred to as τ because the slow component (A2) of HCN channel activation generally accounts for <10% of the current amplitude. Isoproterenol or carbachol (Sigma) were directly dissolved in bath solution on the day of the experiment and applied to the cells via a fast-exchange superfusion system. APs were recorded with a 10 kHz sampling rate. Analyses were done on the original traces. The slope of the linear fit of the distance from the MDP to the threshold potential is the DDR. AP duration is the time from threshold potential to MDP.
Ca2+ Imaging
All Ca2+ transients were measured by fluorescence imaging microscopy (Visitron Systems) and analyzed with VisiView imaging software. Ca2+ signals were recorded at an emission wavelength of 525/50 nm with an excitation wavelength of 470/40 nm (50 ms) using a cooled digital CCD camera in 4 × 4 binning mode. iSABs and aCaBs were loaded with 2.5 μM Fluo-4/acetoxymethyl ester (Fluo-4/AM) in differentiation medium at 37°C for 30 min. After a second medium change, measurements were carried out at a temperature of 28°C. Ca2+ transients in ESCs are expressed as acute value of fluorescence intensity (F) normalized to minimal fluorescence intensity (Fo) during analyzed time.
Spontaneous Ca2+ transients were influenced by inhibition of HCN channels with 5 μM ZD 7288 (Sigma-Aldrich), inhibition of T-type voltage-sensitive Ca2+ channels with 1 μM mibefradil (Sigma-Aldrich), and inhibition of L-type voltage-sensitive Ca2+ channels with 1 μM nifedipine (Sigma-Aldrich). For experiments involving blocking of sarcolemmal Ca2+ transport, the culture medium was replaced by a tyrode solution in a composition of (in mM) LiCl 140, KCl 6, MgCl 1, glucose 10, EGTA 1, HEPES 5, pH 7.4 (adjusted with KOH). The influence of the Ca2+ stores on the Ca2+ transients was investigated by applying caffeine (10 mM; Sigma-Aldrich) to open the ryanodine-receptor-coupled SR-Ca2+ channels, and thapsigargin (2 μM; Sigma-Aldrich) to inhibit the SERCA. ZD 7288, nifedipine, and Fluo-4/AM were dissolved in DMSO (final concentration < 0.1%) and thapsigargin in ethanol (final concentration 0.2%). Mibefradil and caffeine were dissolved in H2O.
Slice Preparation and Culture
The experimental protocols for animal experiments were approved by the Bavarian Institutional Animal Care and Use Committee and were conducted in accordance with the Guide for the Care and Use of Laboratory Animals (NIH publication no. 85-23, revised 1996). Hearts of adult mice from both sexes were rapidly removed and transferred to ice-cold modified Tyrode’s solution containing (in mM) NaCl 136, KCl 5.4, MgCl2 1, CaCl2 0.9, NaH2PO4 0.33, glucose 10, 2,3-butanedione-monoxime 30, HEPES 5, pH 7.4 (adjusted with NaOH). Ventricles were dissected from the atria, valves, and vessels, and embedded in 4% low-melt agarose gel dissolved in modified Tyrode’s solution without glucose. The agarose block containing the heart was glued to the sample holder of the vibratome (VT1200S; Leica) and quickly covered with ice-cold Tyrode’s solution. The hearts were sectioned parallel to the valve plane into 300-μm-thick tissue slices with steel blades (Wilkinson), resulting in ring-shaped slices of the ventricular myocardium.
After an incubation of 30 min in ice-cold Tyrode’s solution, the slices were transferred onto a Biopore membrane of tissue culture inserts (Millicell; Millipore) for culturing at an air-media interface. Tissue culture inserts were placed in Petri dishes containing culture medium M199 supplemented with 1% insulin-transferrin-selenium (GIBCO) and 1% penicillin/streptomycin (Sigma). Slices were cultured in the incubator (37°C, 5% CO2) until coculturing. For routine determination of slice viability, slices were incubated with thiazolyl blue tetrazolium bromide (MTT, 0.5 mg/ml; Sigma) for 40 min at 37°C and analyzed by light microscopy.
Coculture of iSABs with Cardiac Slices
iSABs were labeled with calcein red (AAT Bioquest) to monitor living cells. iSABs were incubated with 10 mg/ml calcein red/AM in differentiation medium for 30 min at 37°C in the incubator. After centrifugation (1,000 rpm, 5 min) and resuspension in differentiation medium, iSABs were placed on the cardiac murine slices with a pipette under the microscope. iSABs and cardiac slices were cocultured in the incubator (37°C, 5% CO2). The beating frequencies of iSABs and slice regions were recorded daily with a Sony NEX-5N camera. Coupling between iSABs and slices was detected by transmission of calcein red from iSABs into the cells of slices using fluorescence imaging (Ex/Em 646/659 nm; Visitron Systems). Contracting regions of iSABs with slices were recorded every day and the distribution of the calcein red fluorescence was analyzed visually.
Ca2+ Imaging in Contracting Regions of iSABs with Slices
Slices with iSABs were loaded with 5 μM Fluo-4/AM (Invitrogen) in M199 supplemented with 1% insulin-transferrin-selenium (GIBCO) and 1% penicillin/ streptomycin (Sigma) at 37°C for 30 min. Ca2+ signals were recorded by fluorescence imaging microscopy (Visitron Systems) at an emission wavelength of 525/50 nm with an excitation wavelength of 470/40 nm (exposition time 30 ms) using a cooled digital CCD camera in 4 × 4 binning mode. All Ca2+ transients were analyzed with VisiView imaging software and expressed as acute value of fluorescence intensity (F) normalized to minimal fluorescence intensity (Fo) during analyzed time (20 s) in the respective regions.
Cell Culture
ESC lines were derived from the murine line GSES (David et al., 2005) and grown in high-glucose DMEM with stable glutamine (GIBCO) containing 10% FBS Superior (Biochrom), 100 μM nonessential amino acids (GIBCO), 1% penicillin/streptomycin (GIBCO), and 100 μM β-mercaptoethanol (Sigma) in the presence of 1,000 U/ml of LIF (Milllipore). Differentiation was performed according to standard protocols in Iscove’s basal medium (Biochrom) containing 10% FBS (Biochrom), 100 μM nonessential amino acids (GIBCO), 1% penicillin/streptomycin (GIBCO), and 450 μM 1-thioglycerol (David et al., 2008). The cells were passaged by trypsin/EDTA (GIBCO) at 70% confluence, which was usually achieved after 2–3 days. Typically, three or four passages after cell-thawing differentiation were induced. Transfections with 15 μg plasmid DNA were performed on 10 cm tissue culture dishes using JetPEI (Peqlab), with subsequent selection via 10 μg/ml blasticidin (Invivogen) or 250 μg/ml hygromycin (Invitrogen). Stable clones were picked manually, further cultivated, and tested by qRT-PCR and differentiation assays (David et al., 2008). Positively evaluated clones were subjected to antibiotic treatment for cardiomyocyte enrichment as previously described (Klug et al., 1996).
Generation of aCaBs and iSABs
Formation of aCaBs and iSABs was achieved as follows: Differentiation was consistently performed in hanging-drop culture for 2 days using 1,000 cells as the starting material for one EB in Iscove’s basal medium (Biochrom) containing 10% FBS (Biochrom), 100 μM nonessential amino acids (GIBCO), 1% penicillin/streptomycin (GIBCO), and 450 μM 1-thioglycerol. For an additional 4 days, the cells were differentiated in suspension culture, and at day 6 of differentiation, consistently 15 EBs were seeded on one well of a 24-well plate. Antibiotic selection with 400 μg/ml G418 (Biochrom) was initiated at day 8 after seeding and 4 days later, iSABs and aCaBs were isolated via treatment with 6,000 U/ml collagenase IV (GIBCO) for 30 min.
To obtain single cells for subsequent experiments, the bodies were further dissociated with 100% Accutase (Affimetrix) for 15 min.
To ensure successful generation of aCaBs and iSABs, potential mycoplasma contamination was routinely controlled twice a week using the PCR-based MycoSPY kit system (Biontex).
For quantification of spontaneously beating foci per EB, the numbers of EBs successfully attached to the well were scored 1 day after plating. Based on this, beating foci per EB were counted daily. Single beating foci were defined either as being clearly spatially separated from each other or as having different beating frequencies.
FACS Analysis
Cells were detached with trypsin/EDTA (GIBCO), fixed in 0.25% formalin (Sigma), permeabilized in 0.1% Triton-X (Sigma), and blocked with 2% BSA (Roth). Primary antibodies (SOX2, 1:200 [Abcam]; OCT4, 1:200 [Abcam]; and HCN4; 1:50 [Alamone Labs]) and secondary antibodies (anti-mouse PE, 1:500 [Abnova]; anti-rabbit Alexa Fluor 488, 1:500 [Life Technologies]) were incubated in blocking solution containing 0.01% saponin for 1 hr at RT. FACS analyses were performed with an Epics XL Flow Cytometer (Beckman Coulter).
Immunostaining
Cells on coverslips were fixed in 4% paraformaldehyde (Sigma) for 1 hr at 4°C, permeabilized with 0.1% Triton-X (Sigma), and blocked with 0.2% cold-water fish skin gelatin (Sigma)/10% FCS (GIBCO). All antibodies were incubated in blocking solution plus 0.01% saponin (Sigma), primary antibodies (TBX3, 1:200 [Abnova]; α-MHC, 1:200; [Abcam]; HCN4, 1:100 [Alomone Labs]; Connexin 30.2, 1:200 [Abcam]; and Connexin 45, 1:200 [Abcam]) for 1 hr at RT, secondary antibodies (anti-sheep Alexa Fluor 633, 1:500 [Life Technologies]; anti-mouse Alexa Fluor 647, 1:500 [Abcam]; and anti-rabbit Alexa Fluor 647, 1:500 [Abcam]) for 45 min at RT with additional phalloidin (1:500; Enzo Life Science) and bisBenzimide (DAPI, 1:2,000; Sigma). The coverslips were mounted with Dako mounting medium (DAKO) to glass slides and analyzed by fluorescence microscopy using a Leica SP-5 confocal laser scanning microscope (Leica Microsystems) or an ELYRA PS.1 LSM 780 microscope (Carl Zeiss).
Data analysis
The data represent at least three biological replicates and are presented as means ± SD unless stated otherwise. Graphs and calculations were prepared using GraphPad Prism 5.0 software. Means were compared using the two-tailed Student’s t test. Statistical significance was defined as p < 0.05 (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001).
Acknowledgments
We thank Keyence, Germany, for assisting with the live microscopy. We are grateful to Christiane Gross, Judith Arcifa, and Susanne Gabriel for expert technical assistance and Andrea Agli for performing the patch-clamp recordings. A.D. and W.M.F. wish to thank the German Center for Cardiovascular Research (DZHK), partner site Munich Heart Alliance, Munich, Germany, for its support. R.D. and W.-M.F. were supported by the BMBF (01GN0960) and the DFG (DA 1296/2-1 and FR 705/14-2). Additional funding was provided by the Else Kröner Stiftung and the Fritz Bender Stiftung.
Footnotes
This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/3.0/).
Supplemental Information
Beating rate: 194 bpm.
Controls related to Movie S3. Beating rate: 86 bpm.
Beating rate: 325 bpm.
Synchronization was analyzed with the use of Keyence VW9000 Motion Analyser software.
The identical iSAB is shown in Figure 6E demonstrating transmission of calcein dye over time; ∼30 bpm are achieved in the host slice as compared with a maximum of ∼60 bpm via electrode stimulation.
Please refer to Figures 6F and 6G, which show the identical iSAB, for detailed analysis.
References
- Bakker M.L., Boink G.J., Boukens B.J., Verkerk A.O., van den Boogaard M., den Haan A.D., Hoogaars W.M., Buermans H.P., de Bakker J.M., Seppen J. T-box transcription factor TBX3 reprogrammes mature cardiac myocytes into pacemaker-like cells. Cardiovasc. Res. 2012;94:439–449. doi: 10.1093/cvr/cvs120. [DOI] [PubMed] [Google Scholar]
- Barbuti A., Crespi A., Capilupo D., Mazzocchi N., Baruscotti M., DiFrancesco D. Molecular composition and functional properties of f-channels in murine embryonic stem cell-derived pacemaker cells. J. Mol. Cell. Cardiol. 2009;46:343–351. doi: 10.1016/j.yjmcc.2008.12.001. [DOI] [PubMed] [Google Scholar]
- David R., Franz W.M. From pluripotency to distinct cardiomyocyte subtypes. Physiology (Bethesda) 2012;27:119–129. doi: 10.1152/physiol.00044.2011. [DOI] [PubMed] [Google Scholar]
- David R., Groebner M., Franz W.M. Magnetic cell sorting purification of differentiated embryonic stem cells stably expressing truncated human CD4 as surface marker. Stem Cells. 2005;23:477–482. doi: 10.1634/stemcells.2004-0177. [DOI] [PubMed] [Google Scholar]
- David R., Brenner C., Stieber J., Schwarz F., Brunner S., Vollmer M., Mentele E., Müller-Höcker J., Kitajima S., Lickert H. MesP1 drives vertebrate cardiovascular differentiation through Dkk-1-mediated blockade of Wnt-signalling. Nat. Cell Biol. 2008;10:338–345. doi: 10.1038/ncb1696. [DOI] [PubMed] [Google Scholar]
- David R., Stieber J., Fischer E., Brunner S., Brenner C., Pfeiler S., Schwarz F., Franz W.M. Forward programming of pluripotent stem cells towards distinct cardiovascular cell types. Cardiovasc. Res. 2009;84:263–272. doi: 10.1093/cvr/cvp211. [DOI] [PubMed] [Google Scholar]
- David R., Schwarz F., Rimmbach C., Nathan P., Jung J., Brenner C., Jarsch V., Stieber J., Franz W.M. Selection of a common multipotent cardiovascular stem cell using the 3.4-kb MesP1 promoter fragment. Basic Res. Cardiol. 2013;108:312. doi: 10.1007/s00395-012-0312-2. [DOI] [PubMed] [Google Scholar]
- Garcia-Frigola C., Shi Y., Evans S.M. Expression of the hyperpolarization-activated cyclic nucleotide-gated cation channel HCN4 during mouse heart development. Gene Expr. Patterns. 2003;3:777–783. doi: 10.1016/s1567-133x(03)00125-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerecht-Nir S., Fishman B., Itskovitz-Eldor J. Cardiovascular potential of embryonic stem cells. Anat. Rec. A Discov. Mol. Cell. Evol. Biol. 2004;276:58–65. doi: 10.1002/ar.a.10136. [DOI] [PubMed] [Google Scholar]
- Halbach M., Pillekamp F., Brockmeier K., Hescheler J., Müller-Ehmsen J., Reppel M. Ventricular slices of adult mouse hearts—a new multicellular in vitro model for electrophysiological studies. Cell. Physiol. Biochem. 2006;18:1–8. doi: 10.1159/000095132. [DOI] [PubMed] [Google Scholar]
- He J.Q., Ma Y., Lee Y., Thomson J.A., Kamp T.J. Human embryonic stem cells develop into multiple types of cardiac myocytes: action potential characterization. Circ. Res. 2003;93:32–39. doi: 10.1161/01.RES.0000080317.92718.99. [DOI] [PubMed] [Google Scholar]
- Ishii T.M., Takano M., Xie L.H., Noma A., Ohmori H. Molecular characterization of the hyperpolarization-activated cation channel in rabbit heart sinoatrial node. J. Biol. Chem. 1999;274:12835–12839. doi: 10.1074/jbc.274.18.12835. [DOI] [PubMed] [Google Scholar]
- Johns D.C., Nuss H.B., Chiamvimonvat N., Ramza B.M., Marban E., Lawrence J.H. Adenovirus-mediated expression of a voltage-gated potassium channel in vitro (rat cardiac myocytes) and in vivo (rat liver). A novel strategy for modifying excitability. J. Clin. Invest. 1995;96:1152–1158. doi: 10.1172/JCI118103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor N., Liang W., Marbán E., Cho H.C. Direct conversion of quiescent cardiomyocytes to pacemaker cells by expression of Tbx18. Nat. Biotechnol. 2013;31:54–62. doi: 10.1038/nbt.2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehat I., Gepstein A., Spira A., Itskovitz-Eldor J., Gepstein L. High-resolution electrophysiological assessment of human embryonic stem cell-derived cardiomyocytes: a novel in vitro model for the study of conduction. Circ. Res. 2002;91:659–661. doi: 10.1161/01.res.0000039084.30342.9b. [DOI] [PubMed] [Google Scholar]
- Kensah G., Roa Lara A., Dahlmann J., Zweigerdt R., Schwanke K., Hegermann J., Skvorc D., Gawol A., Azizian A., Wagner S. Murine and human pluripotent stem cell-derived cardiac bodies form contractile myocardial tissue in vitro. Eur. Heart J. 2013;34:1134–1146. doi: 10.1093/eurheartj/ehs349. [DOI] [PubMed] [Google Scholar]
- Kleger A., Seufferlein T., Malan D., Tischendorf M., Storch A., Wolheim A., Latz S., Protze S., Porzner M., Proepper C. Modulation of calcium-activated potassium channels induces cardiogenesis of pluripotent stem cells and enrichment of pacemaker-like cells. Circulation. 2010;122:1823–1836. doi: 10.1161/CIRCULATIONAHA.110.971721. [DOI] [PubMed] [Google Scholar]
- Klug M.G., Soonpaa M.H., Koh G.Y., Field L.J. Genetically selected cardiomyocytes from differentiating embronic stem cells form stable intracardiac grafts. J. Clin. Invest. 1996;98:216–224. doi: 10.1172/JCI118769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreuzberg M.M., Söhl G., Kim J.S., Verselis V.K., Willecke K., Bukauskas F.F. Functional properties of mouse connexin30.2 expressed in the conduction system of the heart. Circ. Res. 2005;96:1169–1177. doi: 10.1161/01.RES.0000169271.33675.05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J., Guo L., Fiene S.J., Anson B.D., Thomson J.A., Kamp T.J., Kolaja K.L., Swanson B.J., January C.T. High purity human-induced pluripotent stem cell-derived cardiomyocytes: electrophysiological properties of action potentials and ionic currents. Am. J. Physiol. Heart Circ. Physiol. 2011;301:H2006–H2017. doi: 10.1152/ajpheart.00694.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maltsev V.A., Wobus A.M., Rohwedel J., Bader M., Hescheler J. Cardiomyocytes differentiated in vitro from embryonic stem cells developmentally express cardiac-specific genes and ionic currents. Circ. Res. 1994;75:233–244. doi: 10.1161/01.res.75.2.233. [DOI] [PubMed] [Google Scholar]
- Marger L., Mesirca P., Alig J., Torrente A., Dubel S., Engeland B., Kanani S., Fontanaud P., Striessnig J., Shin H.S. Pacemaker activity and ionic currents in mouse atrioventricular node cells. Channels (Austin) 2011;5:241–250. doi: 10.4161/chan.5.3.15264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauritz C., Schwanke K., Reppel M., Neef S., Katsirntaki K., Maier L.S., Nguemo F., Menke S., Haustein M., Hescheler J. Generation of functional murine cardiac myocytes from induced pluripotent stem cells. Circulation. 2008;118:507–517. doi: 10.1161/CIRCULATIONAHA.108.778795. [DOI] [PubMed] [Google Scholar]
- Morikawa K., Bahrudin U., Miake J., Igawa O., Kurata Y., Nakayama Y., Shirayoshi Y., Hisatome I. Identification, isolation and characterization of HCN4-positive pacemaking cells derived from murine embryonic stem cells during cardiac differentiation. Pacing Clin. Electrophysiol. 2010;33:290–303. doi: 10.1111/j.1540-8159.2009.02614.x. [DOI] [PubMed] [Google Scholar]
- Nuss H.B., Marbán E., Johns D.C. Overexpression of a human potassium channel suppresses cardiac hyperexcitability in rabbit ventricular myocytes. J. Clin. Invest. 1999;103:889–896. doi: 10.1172/JCI5073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsuji T.G., Minami I., Kurose Y., Yamauchi K., Tada M., Nakatsuji N. Progressive maturation in contracting cardiomyocytes derived from human embryonic stem cells: Qualitative effects on electrophysiological responses to drugs. Stem Cell Res. (Amst.) 2010;4:201–213. doi: 10.1016/j.scr.2010.01.002. [DOI] [PubMed] [Google Scholar]
- Rosen M.R. 15th annual Gordon K. Moe Lecture. Biological pacemaking: in our lifetime? Heart Rhythm. 2005;2:418–428. doi: 10.1016/j.hrthm.2004.12.016. [DOI] [PubMed] [Google Scholar]
- Scavone A., Capilupo D., Mazzocchi N., Crespi A., Zoia S., Campostrini G., Bucchi A., Milanesi R., Baruscotti M., Benedetti S. Embryonic stem cell-derived CD166+ precursors develop into fully functional sinoatrial-like cells. Circ. Res. 2013;113:389–398. doi: 10.1161/CIRCRESAHA.113.301283. [DOI] [PubMed] [Google Scholar]
- Stieber J., Hofmann F., Ludwig A. Pacemaker channels and sinus node arrhythmia. Trends Cardiovasc. Med. 2004;14:23–28. doi: 10.1016/j.tcm.2003.09.006. [DOI] [PubMed] [Google Scholar]
- Takahashi K., Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Takahashi K., Tanabe K., Ohnuki M., Narita M., Ichisaka T., Tomoda K., Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Verheijck E.E., van Kempen M.J., Veereschild M., Lurvink J., Jongsma H.J., Bouman L.N. Electrophysiological features of the mouse sinoatrial node in relation to connexin distribution. Cardiovasc. Res. 2001;52:40–50. doi: 10.1016/s0008-6363(01)00364-9. [DOI] [PubMed] [Google Scholar]
- Wiese C., Grieskamp T., Airik R., Mommersteeg M.T., Gardiwal A., de Gier-de Vries C., Schuster-Gossler K., Moorman A.F., Kispert A., Christoffels V.M. Formation of the sinus node head and differentiation of sinus node myocardium are independently regulated by Tbx18 and Tbx3. Circ. Res. 2009;104:388–397. doi: 10.1161/CIRCRESAHA.108.187062. [DOI] [PubMed] [Google Scholar]
- Wiese C., Nikolova T., Zahanich I., Sulzbacher S., Fuchs J., Yamanaka S., Graf E., Ravens U., Boheler K.R., Wobus A.M. Differentiation induction of mouse embryonic stem cells into sinus node-like cells by suramin. Int. J. Cardiol. 2011;147:95–111. doi: 10.1016/j.ijcard.2009.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wobus A.M., Wallukat G., Hescheler J. Pluripotent mouse embryonic stem cells are able to differentiate into cardiomyocytes expressing chronotropic responses to adrenergic and cholinergic agents and Ca2+ channel blockers. Differentiation. 1991;48:173–182. doi: 10.1111/j.1432-0436.1991.tb00255.x. [DOI] [PubMed] [Google Scholar]
- Yanagi K., Takano M., Narazaki G., Uosaki H., Hoshino T., Ishii T., Misaki T., Yamashita J.K. Hyperpolarization-activated cyclic nucleotide-gated channels and T-type calcium channels confer automaticity of embryonic stem cell-derived cardiomyocytes. Stem Cells. 2007;25:2712–2719. doi: 10.1634/stemcells.2006-0388. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Beating rate: 194 bpm.
Controls related to Movie S3. Beating rate: 86 bpm.
Beating rate: 325 bpm.
Synchronization was analyzed with the use of Keyence VW9000 Motion Analyser software.
The identical iSAB is shown in Figure 6E demonstrating transmission of calcein dye over time; ∼30 bpm are achieved in the host slice as compared with a maximum of ∼60 bpm via electrode stimulation.
Please refer to Figures 6F and 6G, which show the identical iSAB, for detailed analysis.


