Significance
Ecological and evolutionary investigations require accurate and high-resolution biodiversity information. Conventional morphological approaches to identifying species in species-rich tropical ecosystems are often unavailable or incapable of timely, cost-effective identification. We show that next-generation sequencing (NGS) of cytochrome c oxidase subunit I (COI) DNA barcodes can accurately detect 83.5% of individually sequenced species (corresponding to 91% of individuals) in a bulk sample of terrestrial arthropods from a Costa Rican species-rich site. Additionally, the 16S and 18S ribosomal DNA gene regions obtained also provide an assessment of the bacteria and protozoa in the bulk sample. This metasystematic approach provides the initial infrastructure for a next generation of biodiversity assessment and environmental monitoring. It can lead to more effective understanding, appreciation, and management of complex ecosystems.
Keywords: cytochrome c oxidase subunit I, Costa Rica, insect, Malaise trap, NGS
Abstract
Conventional assessments of ecosystem sample composition are based on morphology-based or DNA barcode identification of individuals. Both approaches are costly and time-consuming, especially when applied to the large number of specimens and taxa commonly included in ecological investigations. Next-generation sequencing approaches can overcome the bottleneck of individual specimen isolation and identification by simultaneously sequencing specimens of all taxa in a bulk mixture. Here we apply multiple parallel amplification primers, multiple DNA barcode markers, 454-pyrosequencing, and Illumina MiSeq sequencing to the same sample to maximize recovery of the arthropod macrobiome and the bacterial and other microbial microbiome of a bulk arthropod sample. We validate this method with a complex sample containing 1,066 morphologically distinguishable arthropods from a tropical terrestrial ecosystem with high taxonomic diversity. Multiamplicon next-generation DNA barcoding was able to recover sequences corresponding to 91% of the distinguishable individuals in a bulk environmental sample, as well as many species present as undistinguishable tissue. 454-pyrosequencing was able to recover 10 more families of arthropods and 30 more species than did conventional Sanger sequencing of each individual specimen. The use of other loci (16S and 18S ribosomal DNA gene regions) also added the detection of species of microbes associated with these terrestrial arthropods. This method greatly decreases the time and money necessary to perform DNA-based comparisons of biodiversity among ecosystem samples. This methodology opens the door to much cheaper and increased capacity for ecological and evolutionary studies applicable to a wide range of socio-economic issues, as well as a basic understanding of how the world works.
Ecological and evolutionary investigations, as well as many socio-economic applications from human health to agriculture and environmental assessments, require access to accurate, high-resolution biodiversity information. Species-level diversity within a sample can be a measure of the ecological status of the biodiversity universe sampled. Conventional biodiversity measures rely on morphology-based or DNA-based identification of individual specimens. The labor-intensive nature of morphological sorting and identification, with either method, can be a major hindrance to large-scale, high-throughput biodiversity studies. In one recent study of a single tropical sampling site, more than 6,000 species of arthropods were identified morphologically, but required an accumulated 24,000 d of trapping and laboratory identification to document (1). Even this concerted effort is not likely to have recorded the cryptic species that would have been exposed only by molecular means. To overcome the limitations imposed by solely morphology-based identification, DNA barcodes (2) are now used in many biodiversity and taxonomic studies (3–8). Sorting and tissue sampling of individual specimens from bulk samples before single-specimen sequencing, however, is slow and labor-intensive. If conclusions are to be drawn rapidly about the present and changing status of ecosystems, multitaxon biodiversity assessments need to move beyond the problems and high costs associated with single specimen identification.
A “DNA metasystematic” framework advocates sequencing standardized DNA markers from a wide taxonomic range of organisms present in mixed environmental samples (9). For example, the mitochondrial protein-coding gene cytochrome c oxidase subunit I (COI) is the standard DNA barcode for the identification of animal specimens (2). The 16S ribosomal DNA gene region (16S) is the comparable DNA marker for bacteria and archaea (10), whereas the 18S ribosomal DNA gene region (18S) is commonly used as a DNA marker for microbial eukaryotes (11).
DNA sequence data gathered from bulk environmental samples have been analyzed for a variety of environments. Most studies have focused on discovering microbial biodiversity using PCR amplification followed by next-generation sequencing (NGS) of a segment of 16S rDNA. A variety of habitats and environmental conditions have been targeted with this “microbiome” approach, including soil (12), human gut contents (13), and oil sands tailings pond sediments (14). Similar approaches have been attempted with “macrobiome” samples, including both benthic (6, 15) and terrestrial (16, 17) mixtures of arthropods. To date, studies targeting arthropods have sequenced assemblages containing a relatively small number of individuals and species. The initial attempt to recover DNA from mixed arthropod tissue recovered 74% of the 23 species present in a benthic sample (15). An NGS analysis of a constructed mixture of terrestrial arthropod tissue recovered 76% of the species present (16). These previous approaches used only one amplification primer set to amplify the COI gene region. A subsequent analysis of a small sample (37 species, 73 individuals) of terrestrial arthropods used ultradeep sequencing with no amplification and was able to recover 89% of the species present (17). By using a combination of three PCR-amplification primers sets, Hajibabaei et al. (6) were able to recover 87% of the individuals present in a mixed benthic sample using only the preservative fluid and not the sample tissue.
Arthropod species can carry, both internally and phoretically, a large number of microbes that are pathogenic, beneficial, and commensal to humans, other animals, and plants (18). Despite this clear affiliation, biodiversity data from both arthropods and arthropod-associated microbes are not included in simultaneous and conventional analyses of terrestrial biodiversity. Microbes present in bulk arthropod samples appear to have not been previously sampled, identified, or included in biodiversity research.
The use of universal PCR amplification primers when sequencing mixed environmental samples can lead to greater recovery of the DNA markers of some species and the exclusion or nonperception of others (19–21). In particular, DNA sequences with lower GC content are likely to be overrepresented in amplified environmental mixtures, resulting in an overestimation of abundance for some species (19). The binding energies of the amplification primers themselves can generate this bias (20). Using a single amplification primer set can result in as much as 50% of the target DNA sequences being missed in a mixed environmental sample (21). One of the key recommendations of previous studies using NGS with mixed DNA templates has been the development and use of multiple amplification primers before sequencing (6, 16).
We developed multiple primer sets for the COI DNA barcode region to use in NGS of mixed tissue samples. We hypothesize that, through parallel PCR amplifications, these multiple primer sets will amplify target specimens regardless of their relative abundance in the mixture. In this way, we will avoid the potential primer bias of a single universal primer set, and the amplification of all kinds of DNA barcodes in the mixture will be made more feasible. We chose three different gene regions (COI, 16S, and 18S) and two NGS platforms (Roche 454 and Illumina MiSeq). Rather than focusing on a narrow taxonomic group, we used a mixed environmental sample of arthropods captured in a Malaise trap (22) that is representative of a wide variety of ecological samples.
Results
Morphological and Sanger-Based Identification.
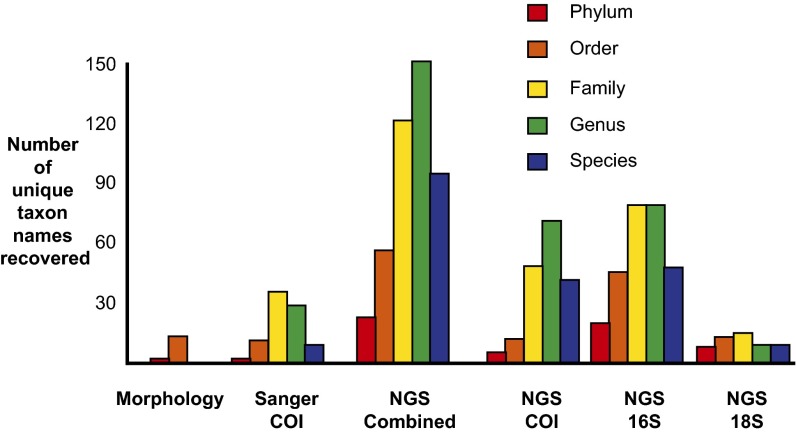
A total of 1,066 individuals from the same Malaise trap sample were isolated before tissue homogenization and all were morphologically identified to order. A total of 14 orders of Arthropoda were represented in the mixture, with between 1 and 235 individuals represented per order (Fig. 1). A total of 699 COI sequences (65.6%) were successfully Sanger sequenced (>300 bp) from the isolated individuals. When clustered at 98% similarity, 357 operational taxonomic units (OTUs) were generated. All sequences were assigned to the order level, and all order assignments matched the morphological identification. The overall rate of assignment to a single family, genus, and species for each sequence was progressively lower (51.2%, 19.3%, and 5.9%, respectively). A total of 12 orders, 37 families, 30 genera, and 11 species of Arthropoda were recovered (Fig. 1 and SI Appendix, Tables S1–S3).
Fig. 1.
Number of unique taxon names recovered by three different methods: morphological identification, Sanger sequencing of individuals, and NGS of COI, 16S, and 18S gene regions. See SI Appendix, Table S1 for complete taxon names.
Analysis of the COI 454-Pyrosequencing Data.
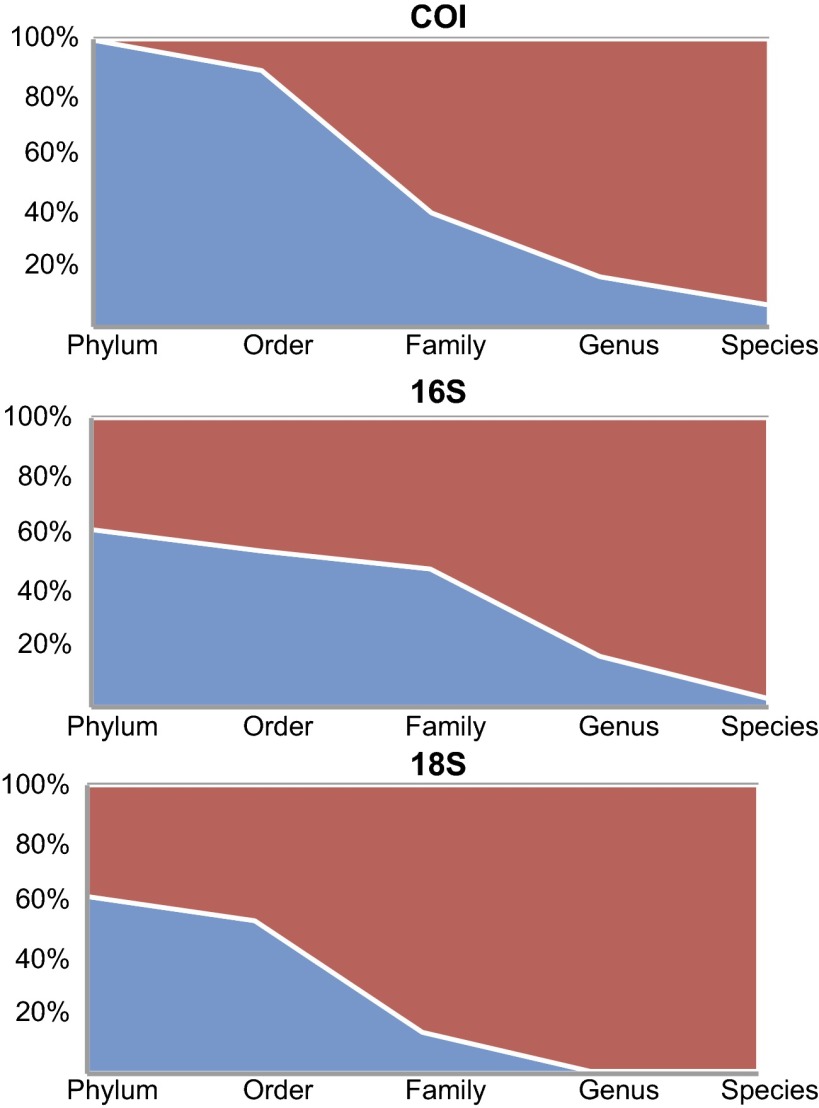
A total of 274,238 454-pyrosequencing reads were generated. Following trimming, dereplication, denoising, and removal of chimeras, short sequences, singletons, and doubletons, 110,584 (40.3%) sequences were used for our analyses. Of these sequences, 110,205 (99.7%) were assigned to a single phylum through comparison with GenBank sequences. Generally, for all analyzed markers, the overall rate of assignment to a single order, family, genus, and species for each sequence was progressively lower (Fig. 2). A total of 3 phyla, 12 orders, 47 families, 70 genera, and 41 species were recovered (Fig. 1 and SI Appendix, Tables S1–S3).
Fig. 2.
Percent of sequences generated from three sources (COI pyrosequences, three regions of 16S combined, and 18S) assigned at the phylum, order, family, genus, and species level. Sequence assignments were generated through a BLASTn similarity search against the GenBank public database followed by lowest common ancestor parsing of results. Blue represents sequences successfully assigned at a given level and red represents sequences not assigned.
Analysis of the Illumina MiSeq Data.
For 16Sv3, 325,486 sequences were generated. Following trimming, dereplication, denoising, and removal of chimeras, short sequences, singletons, and doubletons, 273,887 (84.1%) sequences were used for our analyses. Of these sequences, 181,667 sequences (66.3%) were assigned to the phylum level. A total of 13 phyla, 29 orders, 44 families, 35 genera, and 9 species were recovered (Fig. 1 and SI Appendix, Tables S1–S3).
For 16Sv4, 1,282,084 sequences were generated. Following trimming, dereplication, denoising, and removal of chimeras, short sequences, singletons, and doubletons, 422,244 (32.9%) sequences were used for analysis. Of these sequences, 257,768 sequences (61%) were assigned to a single phylum through comparison with GenBank sequences. A total of 14 phyla, 34 orders, 54 families, 41 genera, and 7 species were recovered (Fig. 1 and SI Appendix, Tables S1–S3).
For 16Sv6, 1,282,084 sequences were generated. Following trimming, dereplication, denoising, and removal of chimeras, short sequences, singletons, and doubletons, 268,237 (20.9%) sequences were used for our analyses. Of these sequences, 154,216 sequences (57.5%) were assigned to a single phylum through comparison with GenBank sequences. A total of 18 phyla, 43 orders, 67 families, 61 genera, and 36 species were recovered (Fig. 1 and SI Appendix, Tables S1–S3).
Sequences generated from the two primer sets for 18S were pooled together for analysis; 2,014,046 sequences were generated. Following trimming, dereplication, denoising, and removal of chimeras, short sequences, singletons, and doubletons, 1,127,798 (56%) sequences were used for our analyses. Of these sequences, 693,103 sequences (61.5%) were assigned to a single phylum through comparison with GenBank sequences (Fig. 2). A total of 8 phyla, 15 orders, 18 families, 8 genera, and 8 species were recovered (Fig. 1 and SI Appendix, Tables S1–S3).
Validation of Individual Specimen Recovery.
Multiple parallel PCR amplification and 454-pyrosequencing recovered 298 of the 357 (83.5%) sequence clusters detected by Sanger sequencing. The clusters recovered represented 634 of the 699 (90.7%) individuals present in the Sanger library. Furthermore, 103 of the 108 (95.4%) barcode clusters containing more than one individual in the mixture were successfully recovered. Of the 90 taxonomic identifications at all levels (order, family, genus, or species) within the Sanger library, 86 (95.6%) were recovered by NGS. The names not recovered appear to represent a family and genus of mites (Trombidiformes: Lebertiidae: Lebertia) and a family (Hymenoptera: Mymaridae) and genus (Hymenoptera: Scelionidae: Baeus) of parasitoid wasps.
Validation of Improved Recovery by Multiple Amplification Primers.
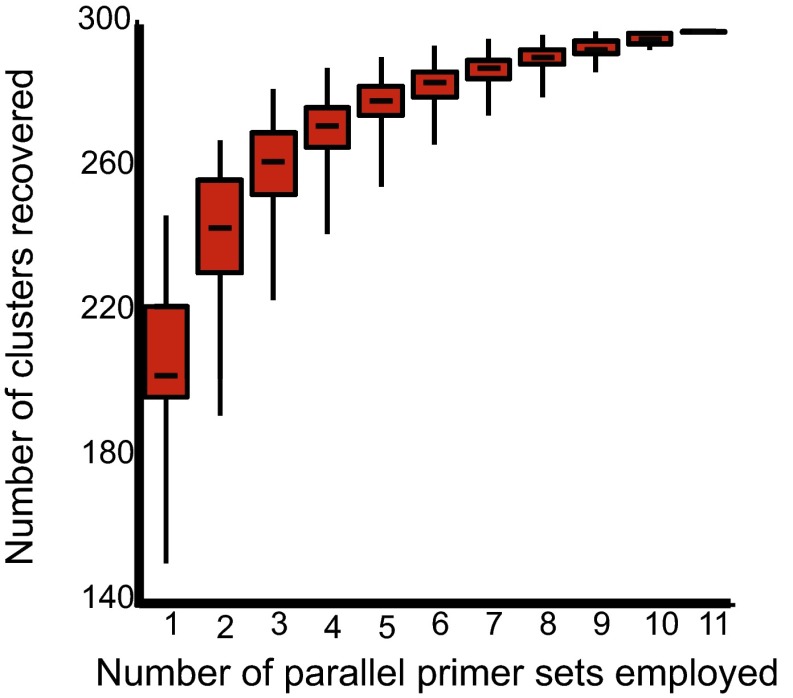
The overall recovery rate of COI barcode sequence clusters by any individual primer set ranged from 42.2% to 69.2%—all much lower than the 83.5% recovery rate of all 11 primer sets combined. A rarefaction curve of sequence cluster recovery vs. number of primer sets used (Fig. 3) indicates that the median two most successful primer sets recovered nearly the same number of sequence clusters as any single primer set alone. Furthermore, the least successful five primer sets combined recovered more sequence clusters than the best single primer set. The most successful single primer sets and combination of two primers sets differ for the total taxon set and for targeted subsets of arthropods (Table 1).
Fig. 3.
Number of Sanger sequence-generated sequence clusters successfully recovered by each possible combination of 11 primer sets in a multiple-PCR 454-pyrosequencing protocol using mixed tissue.
Table 1.
Single primer sets and combinations of two primer sets with the highest number of recovered sequence clusters for all included taxa and the four most abundant arthropod orders
| Group | Best primer set | Best duo | All 11 primers |
| All taxa combined | ArF1xArR3 (247) | ArF1xArR2 | 298 |
| ArF10xArR3 (268) | |||
| Coleoptera | ArF1xArR3 (52) | ArF1xArR3 | 55 |
| ArF1xArR6 (55) | |||
| Diptera | ArF4xArR5 (65) | ArF1xArR2 | 77 |
| ArF4xArR5 (74) | |||
| Hymenoptera | ArF10xArR3 (55) | ArF10xArR3 | 69 |
| ArF10xArR5 (60) | |||
| Lepidoptera | ArF1xArR3 (49) | ArF1xArR3 | 51 |
| ArF1xArR6 | |||
| or | |||
| ArF1xArR6 | |||
| ArF10xArR3 (50) |
Number of recovered sequence clusters are in brackets. See SI Appendix, Table S3 for primer sequences.
Discussion
Success of NGS Processing of Environmental Samples.
With the conventional Sanger sequencing approach, despite using a commonly used universal PCR amplification primer set, 367 of the 1,066 individuals present in the mixture either did not successfully amplify or else produced DNA sequences that were short or unalignable. This 34% failure rate is not unusual for taxonomically large-scale DNA barcoding projects (3, 7). Aside from presumably unsuccessful primer binding, failure to successfully recover DNA barcodes using universal primers and Sanger sequencing may be due to failed sequencing reactions due to cross-contamination from other individuals in the mixture or the presence of competing COI sequence information (e.g., heteroplasmy and endosymbiotic bacteria) within individuals. Shokralla et al. (23) examined this subject and found that as many as 41% of individuals in a mixture may produce multiple, although very similar, COI sequences, thus preventing successful Sanger sequencing. Another explanation may lie in the inadequacy of single universal primers. More than 50% of the individuals that were not successfully Sanger sequenced were representatives of just six arthropod orders (Neuroptera, Orthoptera, Psocoptera, Thysanoptera, Trichoptera, and Trombidiformes). Within each of these orders, the success rate for Sanger sequencing was between 0% and 36%. This result supports the conclusion of past research that universal primers are not as suitable for some arthropod orders as others (6, 15, 24, 25).
NGS of COI was able to recover 91% of the sequence data and 96% of the taxonomic data generated by Sanger sequencing of individuals. This frequency of success was demonstrated across a wide taxonomic range of arthropods. The use of multiple PCR primers contributed greatly to the overall success of this approach. The use of BLAST searching combined with lowest common ancestor (LCA) summaries of results allows for some level of taxonomic assignment to be given to between 57.5% and 99.7% of the sequences generated. Although all markers displayed a declining frequency of assignment at more exclusive taxonomic levels (Fig. 2), each marker had a unique pattern of assignment. The relatively higher frequency of assignment for COI is likely due to the large number of identified COI barcode sequences publically available for arthropods. The low (7.6%) frequency of assignment at the species level is likely due to the lack of DNA barcodes for Costa Rican species of all arthropod orders. The different regions of 16S showed a similar pattern of assignment; however, each did recover unique lists of families, genera, and species. The overall lack of taxonomic assignment for the 18S sequences reflects a shortcoming of the use of 18S for metasystematic analyses. It has been noted that 18S sequences are not suited to determining species-level richness of environmental samples (26). Our recovery of species or genus assignments for only 0.8% of the 18S sequences generated recapitulates this conclusion. The greatly increased number of publically available identified COI and 16S sequences is also likely a factor in the increased assignment rate compared with 18S. The continuation of barcode library-building enterprises will only increase the effectiveness of the current method.
Increased Biodiversity Recovery.
In addition to the specimens identified through individual Sanger sequencing, a further 4 orders, 21 families, 40 genera, and 19 species were detected through NGS of COI from mixed tissue (Fig. 1). Although some of these taxa (e.g., Orthoptera: Gryllidae, Diptera: Stratiomyidae, and Coleoptera: Buprestidae) represent specimens that were isolated, but not successfully Sanger sequenced, some taxa represent specimens that could not have been isolated morphologically. Fragments of arthropods, gut contents, and phoretic individuals cannot easily be isolated and identified, but are readily detected with the present method. DNA sequences obtained from gut contents can be used to identify the diets of predators (27) and also the initial hosts of parasitoid wasps (28). Large, easily isolated insects, such as Megaloptera and many species of Lepidoptera, are likely present in COI NGS sequences, but not present as intact specimens in the Malaise trap, due to the inclusion of the gut contents of predators and parasitoids. The intracellular endosymbiotic bacterium, Wolbachia (Proteobacteria: Rickettsiales: Anaplasmataceae), has been the target of much research both on its obscurative impact on barcoding efforts (29) and its potential impact on insect vectors of pathogens (30). Using our multiple-primer approach, we were able to detect Wolbachia in the mixed tissue sample by using COI sequences as well as all three 16S regions.
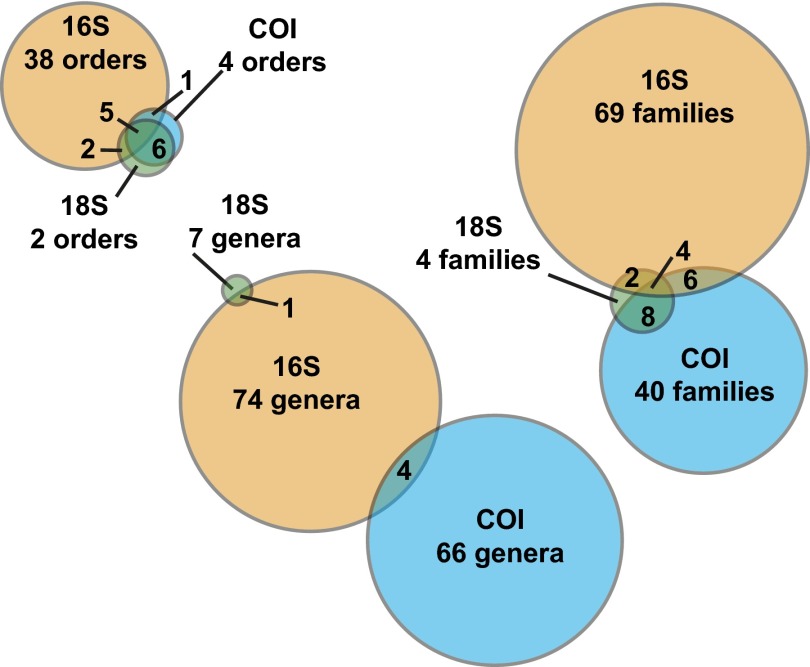
The inclusion of COI, 16S, and 18S sequence data gathered from arthropod-focused sampling allows the simultaneous detection of both insect and microbial taxa (Fig. 4). Stewart’s wilt is a major disease of agricultural crops in North America. It is caused by the bacterium Pantoea stewartii (Proteobacteria: Enterobacteriales: Enterobacteriaceae) and is vectored by a chrysomelid beetle (Chaetocnema pulicaria) in the United States and Canada (31), although, of course, it may be vectored by other species in other regions. Although 16S sequences matching public records of the pathogen were detected in our sample, the known vector species of insect was not. These results suggest not only the presence of a potentially important crop disease organism in Costa Rica, but also the expected presence of an alternate vector. NGS of individual ants from across the globe has revealed a number of associated bacterial endosymbionts, including the genera Wolbachia, Spiroplasma (Tenericutes: Entomoplasmatales: Spiroplasmataceae), and Asaia (Proteobacteria: Rhodospiralles: Acetobacteraceae) (32). Our sample included COI sequences that appear to be from two of these ant genera (Camponotus and Pseudomyrmex) and 16S sequences matching all three bacterial genera previously studied. Although Wolbachia and Spiroplasma are relatively well studied, the recovery of Asaia is important as it is suspected to be an endosymbiotic, nitrogen-fixing bacterium associated with only three genera of ants worldwide. Two of the bacterial genera detected in our samples, Pseudonocardia (Actinobacteria: Actinomycetales: Pseudonocardiaceae) and Blattabacterium (Proteobacteria: Flavobactertiales: Blattabacteriaceae), are exclusively associated with specific insect groups, leaf-cutter ants and cockroaches, respectively (33, 34). An important contribution from the 18S data was the detection of the phylum Apicomplexa. This phylum of protozoans includes important arthropod-vectored pathogens such as the causative agents of Malaria and Babesiosis.
Fig. 4.
Number of names recovered at three taxonomic levels (order, family, genus) recovered by each gene region with NGS of mixed tissue. Sequence assignments were generated through a BLASTn similarity search against the GenBank public database followed by lowest common ancestor parsing of results.
In addition to the DNA markers included here, the present approach could be used with the inclusion of other DNA markers. The nuclear internal transcribed spacer (ITS) ribosomal DNA gene region is the preferred DNA barcode for fungi (35), whereas the rbcL and matK gene regions are used as DNA barcodes for plants (36). Any or all of these markers could be included in the processing of mixed tissues to investigate the plants or fungi that are phoretic internally or externally on the sampled arthropods. The use of multiple primer sets for multiple recoveries of taxa from a single sample also allows for the potential of other future applications. The inclusion of species-specific markers targeting endangered, invasive, or pest species could easily be implemented within the current protocol.
Decreased Cost of Biodiversity Assessment.
The so-called taxonomic impediment is, in large part, a description of the cost of the labor, time, and complexity of performing detailed biodiversity assessments, as well as assigning species boundaries and names (37). The specialized training and microscope time required to morphologically identify all of the specimens in a highly diverse environmental sample are prohibitively large and show no indication of decreasing. Conversely, NGS costs per sample are dramatically decreasing, whereas throughput rates are increasing (38, 39). Using an approximately equal investment of laboratory time for each, we performed three separate analyses of the biodiversity content of a single environmental sample. The amount of biological detail of certain kinds extracted from the sample is far greater for the NGS analysis than for either the morphological or Sanger-based analysis (Fig. 1). Family-, genus-, and even species-level resolution could have been approximated for the morphological analysis, but only with an exponential increase in time investment. Further to this, no amount of microscope time could have detected organisms present only as fragments, microbes, or gut contents. Although the cost of an NGS sequencing run may be prohibitive for some projects today, DNA sequences from multiple environmental samples can be combined before sequencing, effectively dividing the per-sample cost.
Conclusion.
The investment of the equivalent of 66 scientist-years (1) into an arthropod biodiversity tally at one tropical site is not feasible for most scientific goals. For many purposes, it is necessary to dramatically increase the pace of biodiversity discovery and assessment, both per individual and per multitaxon sample, while simultaneously reducing the cost of such efforts (40, 41). This necessity is especially important when biodiversity information is required for socio-economic applications such as environmental assessment (42). Our method helps to meet this challenge on both fronts. As public sequence libraries become better populated with sequences from well-characterized species, identifications made from DNA analysis of bulk samples will provide richer taxonomic information. By using our method of DNA metasystematic analysis (9), a taxonomically comprehensive picture of biodiversity and its changes in response to natural and anthropogenic events is possible and financially feasible. Our analysis shows the significance of using multiple amplification primers in alleviating single amplification biases in sequence recovery. We also demonstrate the increased taxonomic detection made possible by using different DNA markers on a single mixed environmental sample. This method could easily be combined with previously established methods using sample preservative (6). This combination would allow for the retention of permanent morphological vouchers. By using multiple DNA barcode markers, useful for identifying different dimensions of biodiversity, interactions between the macrobiome and microbiome can be investigated with NGS.
Materials and Methods
COI Primer Design.
A total of 128,269 publically available [Barcode of Life Data System (BOLD); ref. 43], individual, identified arthropod COI sequences were separated into 23 orders and aligned. Representatives of all four orders of the Class Collembola (44) were combined into the order Collembola for analysis. Individual sequences included 559 total families of arthropods, representing between 5% and 100% (mean, 47%) of the family-level taxonomic diversity available to us for each order. From these matrices, primer sets were designed to be a consensus of all representatives of each matrix. All primer sets were designed to amplify the same fragment of the COI DNA barcode region. A total of 11 unique PCR amplification primer sets were generated. The amplified region represents a 310-bp fragment within the standard COI barcode region. This sequence allows full-length sequencing of the 310-bp target amplicon on multiple NGS platforms. For primer sequences, please see SI Appendix, Table S3.
Morphological and Sanger-Based DNA Identification.
All 1,066 individuals from a single Malaise trap sample in Area de Conservación Guanacaste, northwestern Costa Rica (Bosque Humedo; latitude, 10.85145; longitude, −85.60801; 290 m; January 24–31, 2011) were isolated, identified to order morphologically, photographed, and tissue subsampled for individual DNA extraction. Tissue subsamples (i.e., a leg from each arthropod) were DNA extracted using a Nucleospin Tissue kit (Macherey-Nagel). The standard 5′ end of the COI region was amplified using the primers LCO1490 and HCO2198 (45). Amplicon sequences were obtained using an ABI 3730XL sequencer (Applied Biosystems). Sequences were edited and assembled using CodonCode Aligner v 3.7.1.1 (CodonCode).
DNA Extraction and COI Amplification for NGS.
All remaining tissue from each individual was pooled and homogenized using an MP FastPrep-24 Instrument (MP Biomedicals; speed 6; 40 s). Total DNA was extracted with a Nucleospin tissue kit (Macherey-Nagel) and eluted in 70 μL of molecular biology-grade water. A fragment of the COI DNA barcode region was amplified using the 11 newly designed primer sets in parallel PCR reactions (SI Appendix, Table S3). A second round of PCR used the same 11 primer sets with hybrid 454 fusion-tailed primers and specifically designed multiplex identifier (MID) tags. Each PCR contained 2 μL DNA template, 17.5 μL molecular biology-grade water, 2.5 μL 10× reaction buffer, 1 μL 50× MgCl2 (50 mM), 0.5 μL dNTPs mix (10 mM), 0.5 μL forward primer (10 mM), 0.5 μL reverse primer (10 mM), and 0.5 μL Invitrogen Platinum Taq polymerase (5 U/μL) in a total volume of 25 μL. PCR conditions were 95 °C for 5 min; 15 cycles of 94 °C for 40 s, 46 °C for 1 min, and 72 °C for 30 s; and 72 °C for 5 min. Amplicons were purified with Qiagen’s MiniElute PCR purification columns and eluted in 50 μL molecular biology-grade water. The purified amplicons from the first PCR round were used as templates in the second PCR round using 454 fusion-tailed and MID-tagged primers in a 30-cycle amplification regime. An Eppendorf Mastercycler ep gradient S thermal cycler was used for all PCR reactions. Negative controls were included in all experiments.
454-Pyrosequencing.
All amplicons from the pooled and homogenized tissue were purified and fluorometrically quantified. Equimolar amounts of the MID-generated amplicons were combined and sequenced on a 454 Genome Sequencer FLX System (Roche Diagnostics) following the amplicon sequencing protocol with GS Titanium chemistry. Amplicons of the bulk tissue sample were bidirectionally sequenced in one-half of a full sequencing run (70 × 75 picotiter plate). Details of the 454-pyrosequencing run are available by request from the corresponding author.
16S and 18S Amplification and Illumina MiSeq Sequencing.
Three fragments of the 16S gene region (v3, v4, and v6) and one fragment of the 18S gene region were amplified for the pooled and homogenized tissue using four primer sets in parallel PCR reactions for the bulk sample [16Sv4F, TGCCAGCAGCCGCGGTAA; 16Sv6R, ACGAGCTGACGACARCCATG; 16S v3F, ACTCCTACGGGAGGCAGCAG; 16Sv3R, GGACTACARGGTATCTAAT (46); 18SEukF, ATGTCAGAGGTTCGAAGGCG; 18SEukR, TGATCCTTCCGCAGGTTCACC (47); 18SNemF, TGCTYIICYCAAAGATTAAGCC; 18SNemR, ATGCCTGCTGCYITCCTTRGA (11)]. Each PCR contained 2 μL DNA template, 17.5 μL molecular biology-grade water, 2.5 μL 10× reaction buffer, 1 μL 50× MgCl2 (50 mM), 0.5 μL dNTPs mix (10 mM), 0.5 μL forward primer (10 mM), 0.5 μL reverse primer (10 mM), and 0.5 μL Invitrogen Platinum Taq polymerase (5 U/μL) in a total volume of 25 μL. PCR conditions were 95 °C for 5 min; 25 cycles of 94 °C for 40 s, 46 °C for 1 min, and 72 °C for 30 s; and 72 °C for 5 min. Amplicons were purified with Qiagen’s MiniElute PCR purification columns and eluted in 50 μL molecular biology-grade water. The purified amplicons from the first PCR round were used as templates in the second PCR round using Illumina adaptors-tailed primers in a 10-cycle amplification regime. An Eppendorf Mastercycler ep gradient S thermal cycler was used for all PCR reactions. Negative controls were included in all experiments. Equimolar amounts of the generated libraries were dual-indexed, combined, and sequenced on an Illumina MiSeq platform using MiSeq Reagent kits (300 cycles) following the 2 × 300-bp paired-end sequencing protocol. Details of the sequencing run are available by request from the corresponding author.
Bioinformatic Processing and Sequence Identification.
All sequencing reads, including Sanger-based, 454-pyrosequences, and Illumina MiSeq sequences, were analyzed using the in-house pipelines of the corresponding author, as follows. For the Illumina-generated 16Sv3 and 18S reads, separate paired-end sequences were reassembled using SEQPREP software (available from https://github.com/jstjohn/SeqPrep) with default settings, a minimum sequence quality of Phred 20, and a minimum overlap of 25 bp. Primer sequences were trimmed from paired Illumina sequences using CUTADAPT v1.1 with default settings (48). CUTADAPT was run three times: first to remove forward primers, second to remove reverse primers, and third to filter out sequence regions with a minimum Phred score of 20 and reads less than 100 bp in length. Trimmed Illumina sequences were then clustered at 97% sequence identity using the cd-hit-est algorithm available from the CD-HIT v4.6 package (49). Using USEARCH (50), clustered reads were sorted by decreasing read abundance, and putative chimeric sequences were removed using the de novo algorithm. Nonchimeric reads were then filtered to exclude singletons. All 454-pyrosequences were trimmed from the 3′ end for the minimum Phred score of 20 (window size = 10, sliding step = 5) using PRINSEQ (51). MID tags were used to separate sequences into primer set-specific sets. After removing primers and tags, sequence reads were dereplicated using PRINSEQ. 454-pyrosequences were also denoised in 99% similarity clusters using USEARCH (50). All resulting sequence clusters were screened for chimeras using a de novo search in UCHIME (52). Short sequences (<300 bp for COI and <100 bp for 16Sv3, 16Sv4, 16Sv6, and 18S) and clusters represented by fewer than three sequences were excluded.
Processed sequences were subjected to a megablast (BLAST+ v2.2.26) (53) search against a local installation of the GenBank nucleotide database (accessed October–November 2013) with an expected e-value of 1e−20 for Illumina reads and 1e−10 for 454-pyrosequences, retaining the top 100 matches. Taxonomic assignments were generated by parsing the top BLAST matches with MEGAN 4.70.4 (54). LCA parameters were adjusted for each amplicon (COI: minimum bitscore 175, top percent 8, minimum support 2; all 16S and 18S: minimum bitscore 250, top percent 8, minimum support 2). Taxonomic assignments at the phylum, order, family, genus, and species level were then tabulated for each sequence.
The sequences generated have been deposited in GenBank and the Sequence Read Archive (SRA).
Supplementary Material
Acknowledgments
We thank Claudia Bertrand for help in sample collection; Stéphanie Boilard for assistance with processing Sanger sequencing; M. Alex Smith for the use of photographic equipment; Hamid Nikbakht for assistance with bioinformatic analysis; and Angela Holliss and Jeff Gross for Sanger-based DNA sequencing. We are grateful to Area de Conservación Guanacaste for protecting the forest habitat that we sampled. This project was funded by the Government of Canada through Genome Canada and the Ontario Genomics Institute through the Biomonitoring 2.0 project (OGI-050) (to M.H.), National Science Foundation Grant Division of Environmental Biology 0515699 (to D.H.J.), and the JRS Biodiversity Foundation and the Wege Foundation of Grand Rapids, MI, to the Guanacaste Dry Forest Conservation Fund. J.G. is also funded by a Natural Sciences and Engineering Research Council of Canada postdoctoral fellowship.
Footnotes
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. KJ435325–KJ436376) and the Sequence Read Archive (SRA).
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1406468111/-/DCSupplemental.
References
- 1.Basset Y, et al. Arthropod diversity in a tropical forest. Science. 2012;338(6113):1481–1484. doi: 10.1126/science.1226727. [DOI] [PubMed] [Google Scholar]
- 2.Hebert PDN, Cywinska A, Ball SL, deWaard JR. Biological identifications through DNA barcodes. Proc R Soc B Biol Sci. 2003;270(1512):313–321. doi: 10.1098/rspb.2002.2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hajibabaei M, Janzen DH, Burns JM, Hallwachs W, Hebert PDN. DNA barcodes distinguish species of tropical Lepidoptera. Proc Natl Acad Sci USA. 2006;103(4):968–971. doi: 10.1073/pnas.0510466103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Janzen DH, et al. Integration of DNA barcoding into an ongoing inventory of complex tropical biodiversity. Mol Ecol Resour. 2009;9(Suppl s1):1–26. doi: 10.1111/j.1755-0998.2009.02628.x. [DOI] [PubMed] [Google Scholar]
- 5.Janzen DH, et al. Reading the complex skipper butterfly fauna of one tropical place. PLoS ONE. 2011;6(8):e19874. doi: 10.1371/journal.pone.0019874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hajibabaei M, Spall JL, Shokralla S, van Konynenburg S. Assessing biodiversity of a freshwater benthic macroinvertebrate community through non-destructive environmental barcoding of DNA from preservative ethanol. BMC Ecol. 2012;12:28. doi: 10.1186/1472-6785-12-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Laforest BJ, et al. Insights into biodiversity sampling strategies for freshwater microinvertebrate faunas through bioblitz campaigns and DNA barcoding. BMC Ecol. 2013;13:13. doi: 10.1186/1472-6785-13-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chacón IA, Janzen DH, Hallwachs W, Hajibabaei M, Hajibabaei M. J Bolling Sullivan Cryptic species within cryptic moths: New species of Dunama Schaus (Notodontidae, Nystaleinae) in Costa Rica. Zookeys. 2013;264(264):11–45. doi: 10.3897/zookeys.264.4440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hajibabaei M. The golden age of DNA metasystematics. Trends Genet. 2012;28(11):535–537. doi: 10.1016/j.tig.2012.08.001. [DOI] [PubMed] [Google Scholar]
- 10.Tringe SG, Hugenholtz P. A renaissance for the pioneering 16S rRNA gene. Curr Opin Microbiol. 2008;11(5):442–446. doi: 10.1016/j.mib.2008.09.011. [DOI] [PubMed] [Google Scholar]
- 11.Creer S, et al. Ultrasequencing of the meiofaunal biosphere: Practice, pitfalls and promises. Mol Ecol. 2010;19(Suppl 1):4–20. doi: 10.1111/j.1365-294X.2009.04473.x. [DOI] [PubMed] [Google Scholar]
- 12.Roesch LFW, et al. Pyrosequencing enumerates and contrasts soil microbial diversity. ISME J. 2007;1(4):283–290. doi: 10.1038/ismej.2007.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang H, et al. Human gut microbiota in obesity and after gastric bypass. Proc Natl Acad Sci USA. 2009;106(7):2365–2370. doi: 10.1073/pnas.0812600106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yergeau E, et al. Next-generation sequencing of microbial communities in the Athabasca River and its tributaries in relation to oil sands mining activities. Appl Environ Microbiol. 2012;78(21):7626–7637. doi: 10.1128/AEM.02036-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hajibabaei M, Shokralla S, Zhou X, Singer GAC, Baird DJ. Environmental barcoding: A next-generation sequencing approach for biomonitoring applications using river benthos. PLoS ONE. 2011;6(4):e17497. doi: 10.1371/journal.pone.0017497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu DW, et al. Biodiversity soup: Metabarcoding of arthropods for rapid biodiversity assessment and biomonitoring. Methods Ecol Evol. 2012;3(4):613–623. [Google Scholar]
- 17.Zhou X, et al. Ultra-deep sequencing enables high-fidelity recovery of biodiversity for bulk arthropod samples without PCR amplification. Gigascience. 2013;2(1):4. doi: 10.1186/2047-217X-2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Clark EL, Karley AJ, Hubbard SF. Insect endosymbionts: Manipulators of insect herbivore trophic interactions? Protoplasma. 2010;244(1-4):25–51. doi: 10.1007/s00709-010-0156-2. [DOI] [PubMed] [Google Scholar]
- 19.Suzuki MT, Giovannoni SJ. Bias caused by template annealing in the amplification of mixtures of 16S rRNA genes by PCR. Appl Environ Microbiol. 1996;62(2):625–630. doi: 10.1128/aem.62.2.625-630.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Polz MF, Cavanaugh CM. Bias in template-to-product ratios in multitemplate PCR. Appl Environ Microbiol. 1998;64(10):3724–3730. doi: 10.1128/aem.64.10.3724-3730.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hong S, Bunge J, Leslin C, Jeon S, Epstein SS. Polymerase chain reaction primers miss half of rRNA microbial diversity. ISME J. 2009;3(12):1365–1373. doi: 10.1038/ismej.2009.89. [DOI] [PubMed] [Google Scholar]
- 22.Malaise R. A new insect trap. Entomol Tidskr. 1937;58:148–160. [Google Scholar]
- 23.Shokralla S, et al. Mol Ecol Res. 2014. Next-generation barcoding: Using next-generation sequencing to enhance and accelerate DNA barcode capture from single specimens. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Park DS, Suh SJ, Oh HW, Hebert PD. Recovery of the mitochondrial COI barcode region in diverse Hexapoda through tRNA-based primers. BMC Genomics. 2010;11:423. doi: 10.1186/1471-2164-11-423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carew ME, Pettigrove VJ, Metzeling L, Hoffmann AA. Environmental monitoring using next generation sequencing: Rapid identification of macroinvertebrate bioindicator species. Front Zool. 2013;10(1):45. doi: 10.1186/1742-9994-10-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tang CQ, et al. The widely used small subunit 18S rDNA molecule greatly underestimates true diversity in biodiversity surveys of the meiofauna. Proc Natl Acad Sci USA. 2012;109(40):16208–16212. doi: 10.1073/pnas.1209160109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leray M, et al. A new versatile primer set targeting a short fragment of the mitochondrial COI region for metabarcoding metazoan diversity: Application for characterizing coral reef fish gut contents. Front Zool. 2013;10(1):34. doi: 10.1186/1742-9994-10-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rougerie R, et al. Molecular analysis of parasitoid linkages (MAPL): Gut contents of adult parasitoid wasps reveal larval host. Mol Ecol. 2011;20(1):179–186. doi: 10.1111/j.1365-294X.2010.04918.x. [DOI] [PubMed] [Google Scholar]
- 29.Smith MA, et al. Wolbachia and DNA barcoding insects: Patterns, potential, and problems. PLoS ONE. 2012;7(5):e36514. doi: 10.1371/journal.pone.0036514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hoffmann AA, Turelli M. Facilitating Wolbachia introductions into mosquito populations through insecticide-resistance selection. P Roy Soc B – Biol Sci. 2013;280:20130371. doi: 10.1098/rspb.2013.0371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nadarasah G, Stavrinides J. Insects as alternative hosts for phytopathogenic bacteria. FEMS Microbiol Rev. 2011;35(3):555–575. doi: 10.1111/j.1574-6976.2011.00264.x. [DOI] [PubMed] [Google Scholar]
- 32.Kautz S, Rubin BER, Moreau CS. Bacterial infections across the ants: Frequency and prevalence of Wolbachia, Spiroplasma, and Asaia. Psyche. 2013;2013(936341):1–11. [Google Scholar]
- 33.Gier HT. The morphology and behavior of the intracellular bacteroids of roaches. Biol Bull. 1936;71(3):433–452. [Google Scholar]
- 34.Cafaro MJ, Currie CR. Phylogenetic analysis of mutualistic filamentous bacteria associated with fungus-growing ants. Can J Microbiol. 2005;51(6):441–446. doi: 10.1139/w05-023. [DOI] [PubMed] [Google Scholar]
- 35.Schoch CL, et al. Fungal Barcoding Consortium Fungal Barcoding Consortium Author List Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proc Natl Acad Sci USA. 2012;109(16):6241–6246. doi: 10.1073/pnas.1117018109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hollingsworth PM, et al. CBOL Plant Working Group A DNA barcode for land plants. Proc Natl Acad Sci USA. 2009;106(31):12794–12797. doi: 10.1073/pnas.0905845106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Weeks PJD, Gaston KJ. Image analysis, neural networks, and the taxonomic impediment to biodiversity studies. Biodivers Conserv. 1997;6(2):263–274. [Google Scholar]
- 38.Glenn TC. Field guide to next-generation DNA sequencers. Mol Ecol Resour. 2011;11(5):759–769. doi: 10.1111/j.1755-0998.2011.03024.x. [DOI] [PubMed] [Google Scholar]
- 39.Shokralla S, Spall JL, Gibson JF, Hajibabaei M. Next-generation sequencing technologies for environmental DNA research. Mol Ecol. 2012;21(8):1794–1805. doi: 10.1111/j.1365-294X.2012.05538.x. [DOI] [PubMed] [Google Scholar]
- 40.Stuart SN, Wilson EO, McNeely JA, Mittermeier RA, Rodríguez JP. Ecology. The barometer of life. Science. 2010;328(5975):177. doi: 10.1126/science.1188606. [DOI] [PubMed] [Google Scholar]
- 41.Laurance WF, et al. Averting biodiversity collapse in tropical forest protected areas. Nature. 2012;489(7415):290–294. doi: 10.1038/nature11318. [DOI] [PubMed] [Google Scholar]
- 42.Baird DJ, Hajibabaei M. Biomonitoring 2.0: A new paradigm in ecosystem assessment made possible by next-generation DNA sequencing. Mol Ecol. 2012;21(8):2039–2044. doi: 10.1111/j.1365-294x.2012.05519.x. [DOI] [PubMed] [Google Scholar]
- 43.Ratnasingham S, Hebert PDN. bold: The Barcode of Life Data System ( http://www.barcodinglife.org) Mol Ecol Notes. 2007;7(3):355–364. doi: 10.1111/j.1471-8286.2007.01678.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Janssens F, Christiansen KA. 2011 Class Collembola Lubbock, 1870. In Zhang Z-Q (Ed.) Animal biodiversity: An outline of higher-level classification and survey of taxonomic richness. Zootaxa 3148:192–194. [Google Scholar]
- 45.Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol Mar Biol Biotechnol. 1994;3(5):294–299. [PubMed] [Google Scholar]
- 46.Sundquist A, et al. Bacterial flora-typing with targeted, chip-based Pyrosequencing. BMC Microbiol. 2007;7:108. doi: 10.1186/1471-2180-7-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wu T, et al. Molecular profiling of soil animal diversity in natural ecosystems: Incongruence of molecular and morphological results. Soil Biol Biochem. 2009;41(4):849–857. [Google Scholar]
- 48.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011;17(1):10–12. [Google Scholar]
- 49.Li W, Godzik A. Cd-hit: A fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006;22(13):1658–1659. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- 50.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26(19):2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 51.Schmieder R, Edwards R. Quality control and preprocessing of metagenomic datasets. Bioinformatics. 2011;27(6):863–864. doi: 10.1093/bioinformatics/btr026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. 2011;27(16):2194–2200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 54.Huson DH, Mitra S, Ruscheweyh H-J, Weber N, Schuster SC. Integrative analysis of environmental sequences using MEGAN4. Genome Res. 2011;21(9):1552–1560. doi: 10.1101/gr.120618.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.