Significance
It is well established that individuals are more similar to their spouses than other individuals on important traits, such as education level. The genetic similarity, or lack thereof, between spouses is less well understood. We estimate the genome-wide genetic similarity of spouses and compare the magnitude of this value to a comparable measure of educational similarity. We find that spouses are more genetically similar than two individuals chosen at random but this similarity is at most one-third the magnitude of educational similarity. Furthermore, social sorting processes in the marriage market are largely independent of genetic dynamics of sexual selection.
Keywords: homophily, random mating, genetic homogamy
Abstract
Understanding the social and biological mechanisms that lead to homogamy (similar individuals marrying one another) has been a long-standing issue across many fields of scientific inquiry. Using a nationally representative sample of non-Hispanic white US adults from the Health and Retirement Study and information from 1.7 million single-nucleotide polymorphisms, we compare genetic similarity among married couples to noncoupled pairs in the population. We provide evidence for genetic assortative mating in this population but the strength of this association is substantially smaller than the strength of educational assortative mating in the same sample. Furthermore, genetic similarity explains at most 10% of the assortative mating by education levels. Results are replicated using comparable data from the Framingham Heart Study.
Assortative mating occurs when individuals exhibit a preference for those who are either similar, (homogamy) or dissimilar (heterogamy) to themselves. Two expressions—“birds of a feather flock together” and “opposites attract”—are used to explain friendship and spousal pairings but denote opposite assumptions regarding the direction of selection. Critically, no existing research has quantified the degree to which individuals who select into a marriage are genetically similar to one another across the entire genome.
Quantifying genome-wide genetic assortative mating (GAM) in the population is important for methodological and substantive reasons. First, statistical models in genetic epidemiology, such as Hardy–Weinberg equilibrium, often assume random mating to forecast population allele frequencies, homozygosity rates, and other parameters of interest across generations (1) and behavior genetics models assume random mating to calculate heritability estimates (2). Second, social scientists have long studied the causes and consequences of assortative mating on a number of phenotypic measures such as height, education, religiosity, and political partisanship (3–5). Although there is research with a focus on the implications of genetic homogamy for phenotypic assortative mating (6), most studies of assortative mating have not considered the possibility that GAM may underlie phenotypic sorting. Social factors clearly limit opportunities to interact with people of different backgrounds (7, 8) but there is no study that simultaneously estimates educational assortative mating (EAM) and GAM in the population. Although much is known about changes in the nature of assortative mating over the past 50 y (5, 8, 9), little is known about the relationship between GAM and EAM.
We focus on EAM because it has received the largest amount of attention in the assortative mating literature (4) and, equally important, research has shown that educational attainment reflects genetic influences (10, 11). No existing study has used genome-wide data among spousal pairs to quantify GAM in the population. This observation coupled with the potential bias caused by GAM in traditional heritability estimates (12) makes this line of inquiry both substantively and methodologically important to a large group of biological and social scientists. In this paper we ask three related questions. First, is there any evidence of GAM in the population? Are genetically similar persons more likely to marry than genetically dissimilar persons both inclusive of and net of ethnic intramarriage? Or are spousal genotypes uncorrelated, as is sometimes assumed? Second, how does the magnitude of GAM compare with other phenotypically-based measures of assortative mating in the population—such as education? Third, to what extent is phenotypic assortative mating linked to GAM in the population?
Results
Estimates of EAM and GAM.
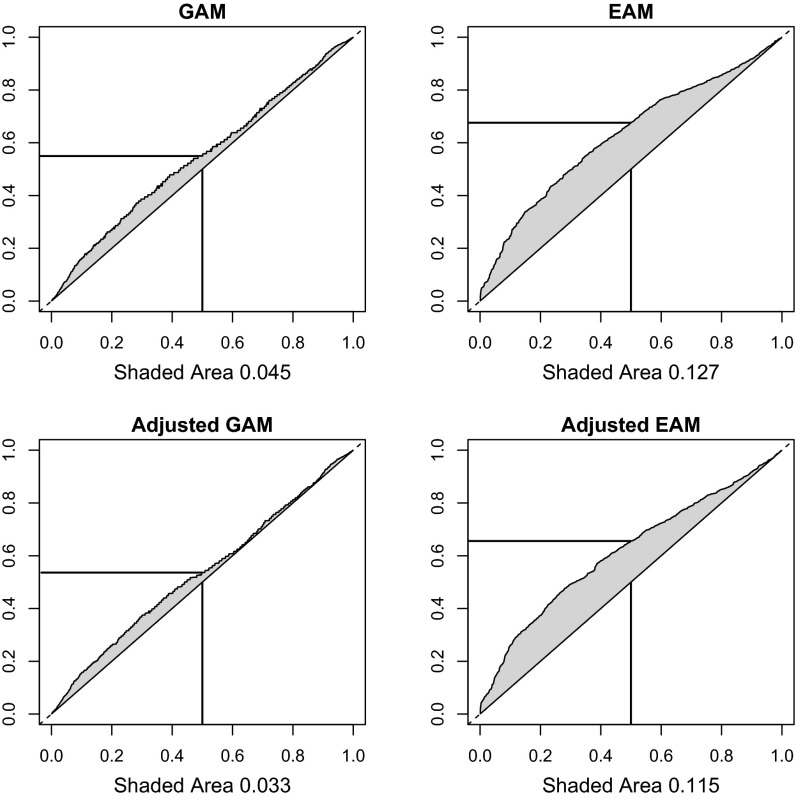
EAM and GAM estimates from the Health and Retirement Study (HRS) (13) are shown graphically in Fig. 1. Fig. 1, Upper addresses the first two research questions in our study; Upper Left presents a graphical representation of GAM. To illustrate the meaning of this curve, consider the point where the two lines intersect. This point indicates that the median value of genetic similarity among spouses corresponds to the 55th percentile (the horizontal line) in the general population of all possible pairs; spouses are more genetically similar than randomly generated pairs in the population. To assess the magnitude of the increased spousal genetic similarity, we focus on the area of the shaded region above the 45° line. This produces an estimate of GAM of 0.045 [95% confidence interval (CI): 0.026, 0.061]. This estimate of GAM includes GAM due to intraethnic marriage among non-Hispanic whites, which we attempt to remove in subsequent analyses.
Fig. 1.
Graphical representation of GAM and EAM. The y axis charts quantiles of the distribution of kinship or squared educational differences between all pairs. The x axis charts quantiles of the same distribution but restricted to just cross-sex white spousal pairs. The shaded area in each gives an estimate of assortative mating. The horizontal and vertical lines aid in interpretation. In Upper Left, one can observe that the genetic relatedness estimate at the 0.5 quantile of spousal pairs corresponds to the 0.55 quantile of all pairs. Adjusted GAM (Lower Left) includes control for same birth region (census division). Adjusted EAM (Lower Right) includes a control for kinship between the pairs.
To gauge the magnitude of this GAM coefficient, we performed the same analysis using years of completed education plus a small amount of noise (the rationale for the inclusion of the noise is included in SI Text, section S1). This graph is shown in Fig. 1, Upper Right. Our estimate of EAM is 0.127 (95% CI: 0.109, 0.144), an estimate that is 2.9 times as large as our estimate of GAM. Together, these results answer our first two questions. Namely, GAM exists in this sample but it is substantially smaller in magnitude than EAM.
Next, we investigated whether GAM and EAM have a specific common explanation through a small set of SNPs related to educational attainment. We tested this hypothesis by examining proxies for the SNPs that reached genome-wide significance in a recent genome-wide association study (GWAS) on educational attainment (11). In particular, we conducted a χ2 test using the sum of the risk alleles for the target SNPs for husbands and wives. The original SNPs were rs9320913, rs11584700, and rs4851266. We identified proxies using SNP Annotation and Proxy Search (14) that were correlated with the original SNP at no less than 0.8. The P values for the three tests were all above 0.35. Hence, we found little evidence that there was assortative mating based on these SNPs.
We conducted a replication analysis using data from the second generation of the Framingham Heart Study (15). It is important to note that the participants of this study are a group of predominantly white respondents from a geographically constrained area. In this secondary data set, we estimated GAM to be 0.025 (95% CI: 0.005, 0.046) based on 685 spousal pairs. Although we replicate the rejection of the null hypothesis of zero GAM in a second sample, we also note the decline in the magnitude of GAM compared with the estimate from HRS. Our estimated EAM in the Framingham sample was similar to the result from the HRS sample, 0.121 (95% CI: 0.102, 0.141).
Impact of Population Stratification on GAM.
The existence of population stratification, small differences in allele frequencies that may exist across socially defined racial and ethnic groups, complicates many genetic analyses. In this section, we consider the extent to which population stratification may be present in our sample and how it may influence our measure of GAM. To characterize genetic divisions among the sample of non-Hispanic whites, we computed principal components (PCs) (SI Text, section S2) based on the complete set of SNPs. These methods consider the correlation between all of the SNPs within a population and identify factors that account for the greatest amount of common genetic variance. These factors align strongly with self-reported race and ethnicity and provide continuous measures of ancestry that are important controls for population stratification. There is substantial variability in the first PC only. Although we do not have information on ethnicity aside from Hispanicity, the PCs are largely unassociated with birth region (as a proxy for ethnic mixture). Differences in PCs may be capturing the genetic similarity (unrelated to population stratification) that we hope to investigate in our GAM analysis. As it is unclear if these PCs are confounding our estimate of GAM or are themselves an interesting component of GAM, we do not focus on estimates that control for these differences. We instead consider three alternative methods of adjusting for these differences in population stratification (estimates based on direct controls for PCs are shown in SI Text, section S2).
First, we use a subsample of our respondents with less variability in the first, and subsequent, PC(s) that presumably have less ethnic variability than the full sample. This should in turn reduce the impact that ethnic intramarriage among whites would have on our estimates. We estimated GAM among only those respondents with PC 1 > −0.003 to be 0.021 (95% CI: 0.002, 0.041). Note that this is very similar to the value obtained from our estimate of GAM in the Framingham Heart Study (15), a geographically homogenous sample.
A second approach for controlling the impact of population stratification is to control for birth region in our estimate of GAM because individuals from the same birth region are more likely to come from the same ethnic group than two individuals sampled from the entire nation (SI Text, section S3 and ref. 16). In Fig. 1, Lower Left, we present an adjusted GAM estimate produced by residualizing kinship using a linear model with a dummy variable indicating whether a pair was born in the same census division. Based on this approach, we estimated an adjusted GAM of 0.033 (95% CI: 0.013, 0.049). This change suggests that some of our the initial GAM estimate is due to the fact that people from the same geographic area are more likely to marry one another than people from different areas (65% of the spousal pairs are from the same birth region compared with just 13% of nonspousal pairs) and that these geographic areas may capture subtle allele frequency differences across the population. That said, there is evidence for residual GAM, even with geographic controls. We note that this source of GAM is often not adjusted for in many estimates of heritability or demographic models of spousal assortative mating that use national (or international) samples, even if the samples are of non-Hispanic whites.
Finally, we also attempted to adjust for the influence of population stratification via direct manipulation of the genetic data. After computation of PCs, we identified SNPs that were most associated with the first five PCs (and thus potential ethnic markers) via GWAS. We then removed these SNPs from the genetic data and recalculated kinship (additional details on this process are in SI Text, section S4). Even after imposing extremely conservative restrictions that removed 70% of the SNPs (remaining SNPs were unrelated to any of the first five PCs), we estimated a GAM of 0.026 (95% CI: 0.005, 0.045). We discuss the relationship between the various estimates of GAM that control for population stratification in Discussion, but pause to note that several different approaches have converged on an estimate of residual GAM between 0.02 and 0.03.
Relationship Between GAM and EAM.
To answer the third research question, we estimated EAM after first regressing out genetic similarity (based on the kinship estimates). Fig. 1, Lower Right describes the results of this analysis. As shown in this figure, adjusting for GAM reduced EAM to 0.115 (95% CI: 0.102, 0.133). Given that the kinship values used for this analysis may be affected by population stratification, we view this as an upper bound. Hence, at most 10% of the variance in EAM is due to GAM. We also examine this relationship in reverse by computing a GAM coefficient based on the residualized kinship coefficients (kinship was regressed on the squared educational differences of a pair). This coefficient declined from 0.045 to 0.026, a reduction of 42%. We discuss our interpretation of this result in the following section.
Discussion
Spouses are more genetically similar than two individuals chosen at random. As described in SI Text, section S5, our unadjusted GAM result of 0.045 suggests that a 1-SD increase in genetic similarity increases the probability of marriage by roughly 15%. This association is confounded, in part, by intraethnic marriage among whites but we continue to observe GAM even after a series of models designed to eliminate this source of assortative mating. That is, after replication with an independent dataset that is geographically homogeneous, restriction of our analyses to a genetically homogeneous subsample of respondents, adjustment of kinships for common birth region, and elimination from genetic data of SNPs that capture population structure, we obtain estimates of GAM between 0.02 and 0.03. The lack of additional ethnicity information in HRS makes it difficult to understand the quantity of GAM that is due to ethnic homogamy alone but the additional analyses suggest that preference for intraethnic marriage accounts for roughly one-half of observed GAM among non-Hispanic whites. It is worth noting that other phenomena could be related to both marriage preference and genetic architecture. Religion, for example, could be a source of GAM in this respect. Future research could consider the proportion of GAM that is due to such factors.
Although GAM exists, an important finding in our analyses is that the magnitude of GAM is significantly smaller than the magnitude of EAM. Furthermore, similar genotype explains only a small fraction of EAM (less than 10%). Our attempt to understand the amount of EAM that could be explained by GAM is based on the hypothesis that a fraction of phenotypic similarity is due to genetic similarity. In short, that GAM causes EAM. However, it is important for us to acknowledge that there are alternative explanations. Education could structure GAM through gene–environment correlations (17). For example, previous research (18) suggests that genetic similarity among friends is higher in schools with higher levels of economic inequality, which emphasizes the need to consider structural differences in educational institutions as a precursor to genetic selection into friendships. Our results (in particular, the 42% decline in GAM after controlling for EAM) indicate that social institutions may segregate people on genotype (presumably unwittingly), which could be behind some of the GAM that we observe. We do not assess this hypothesis empirically but we encourage others to consider this possibility in future research.
It is also important to note that both understandings (EAM causes GAM or GAM causes EAM) do not consider that this relationship is contingent upon the mean level of education among the pairs. For example, Eckland (19) hypothesizes that spousal correlations for intelligence are higher when the intelligence of either spouse is either exceptionally high or exceptionally low. This nonlinear relationship in conjunction with the strong correlation between intelligence and years of completed education suggests that the direction and magnitude of the GAM–EAM relationship may vary across the educational spectrum. Eckland (19) and others (20) have argued that assortative mating and the genetic influences on status-related outcomes may change over time. Higher levels of social inequality reduce the likelihood that otherwise small genetic factors will significantly shape an individual’s socioeconomic attainment but historical changes in equality over time may provide or limit opportunities for these otherwise latent traits to manifest. Although it is unclear if the cohort range in the HRS is large enough to evaluate this hypothesis, we encourage future researchers to examine this possibility as well as the interactive hypothesis described above.
Our findings have important implications for a range of disciplines. Social scientists might gain additional understanding of assortative mating (or similar processes, such as friendship selection) by considering the role of genes. This is particularly important when one considers the significance of social factors that limit or enable two individuals to select into a relationship and how these factors differ across contexts and over time (18). Although it is beyond the scope of this paper, it is also important to consider the possibility that the intergenerational transmission of education may depend on the relative influence of EAM and GAM, which may change over time and context. That is, the influence of EAM on the intergenerational transmission of education may depend on the extent to which EAM is due to GAM. For example, if the proportion of EAM that is due to GAM is increasing over time, then it has important implications for our understanding of the intergenerational transmission of education. This perspective is not possible when one only examines EAM and offspring education.
Researchers presenting heritability estimates should consider including estimates of general assortative mating or trait specific genetic homogamy. Scientists have begun to interrogate the underlying assumptions of kinship based models that attempt to decompose the variation in a trait such as education into its additive genetic, common environmental, and unique environmental components. Recent work has used molecular approaches to test one major assumption: the equal environments assumption (21). The second key assumption, random mating with respect to the genetic architecture of the trait among the parental generation, has seen less investigation. Typically researchers use parental correlations in the phenotype as a rough estimate of nonrandom mating. However, of even greater value would be understanding the quantity of nonrandom mating that there is genetically with respect to the trait and how these associations have changed over time.
The results presented here only represent a first step in understanding the ways in which humans may assortatively mate with respect to their genome. For instance, an extensive literature (22) has emerged suggesting that heterosexual individuals find the odors of opposite sex persons more attractive if the test odor comes from someone who is genetically discordant on markers in the major histocompatiability complex area of chromosome six, which is thought to be under pressures of balancing selection. Such a region-specific, negative-assortative-mating dynamic may serve to depress overall (positive) GAM estimates. Thus, it may behoove future researchers to break apart the genome into parts that are relevant to specific pathways or processes that may be under different selective pressures to see if genome-wide GAM estimates mask a mixture of strong positive and negative dynamics with respect to different dimensions.
Our paper contributes to the literature on both GAM and EAM but has several limitations that we encourage others to consider. First, our results apply to opposite-sex non-Hispanic white pairs within the United States. For nonwhite pairs within the United States, different results might be obtained due to limited genetic variance among non-Hispanic whites compared with other groups (23) or because of different social contexts for non-Hispanic whites compared with others (e.g., the racial inequities that exist in the United States). That is, if individuals are selecting into a relationship because of genetic similarity, then we might expect GAM to be higher among non-Hispanic whites who are less likely than others to face limitations in terms of residential, educational, or occupational choices. Second, patterns of GAM and EAM might differ in same-sex couples. Third, differences may be changing over time. For example, recent research (24) suggests that there has been a rise in assortative mating which has contributed to a rise in income inequality. Fourth, we estimated genetic similarity using SNPs from across the genome. Future research could focus on SNPs known to be important for education (11) or those identified in other GWAS to examine homogamy at a finer level than our whole-genome approach. Given our results from the SNPs implicated in the education GWAS, it might be that analyses at levels finer than the entire genome but much larger than a single SNP, such as chromosomes, would be appropriate.
Materials and Methods
Data.
This paper uses data from the Health and Retirement Study (HRS) RAND fat files (13). Access to the genome-wide data was approved by National Center for Biotechnology Information Genotypes and Phenotypes Database (access no. 19335-3). Of the 9,429 individual with genetic data (described below), 4,584 were from the HRS cohort (five other cohorts are also included in the full data). Of the 4,584, there were 3,504 non-Hispanic whites. Of these, 1,763 individuals were in 862 spousal pairs (some individuals had more than one spouse). We focus on only those individuals (with complete data) in spousal pairs, 1,716 individuals in 825 spousal pairs, as there are differences between individuals in spousal pairs and those not in spousal pairs (e.g., spouses have roughly a quarter year of education more on average). These individuals were born during a large span of time (between 1920 and 1970) but the majority (59%) were born in the 1930s. To assess EAM, we used total years of education. In our sample, 14% had less than a high school education, 38% had a high school education, and the remainder had more than a high school education. We also used information on the respondent’s birthplace (coded as one of nine census divisions plus two categories for US birth with no additional information and foreign birth, 0.1% and 5.1% of the sample, respectively).
Genetic data for the HRS is based on DNA samples collected in two phases. The first phase was collected via buccal swabs in 2006 using the Qiagen Autopure method. The second phase used saliva samples collected in 2008 and extracted with Oragene. Genotype calls were then made based on a clustering of both data sets using the Illumina HumanOmni2.5-4v1 array (details on the quality control process can be found via ref. 25). After standard quality control procedures (e.g., removing SNPs that were missing in more than 5% of samples; minor allele frequencies below 1%; failure to meet Hardy–Weinberg equilibrium, violations of which suggest errors in the genotyping process), we retained 1,707,214 SNPs. We also performed replication analysis on data from the Framingham Heart Study (15) (a description of these data can be found in SI Text, section S6).
Measuring Genetic Similarity.
Quantifying GAM in the population relies on a valid and reliable measure of genetic relatedness between all individuals in the study. Genetic relatedness is a basic biological concept that undergirds quantitative genetic analyses (1). The bulk of this research relied on unmeasured genetic similarity among different types of relatives (e.g., siblings, twins, cousins, etc.) and recently this same conceptual approach used genome-wide data from related (26) and unrelated (27) individuals. These methods are similar in that they take advantage of naturally occurring variability in the degree to which two individuals’ genomes are more or less similar compared with others in the population. It is precisely this variability between unrelated individuals that we use here. There are a number of methods for estimating genetic similarity based on measured genotype but the properties of these various estimates differ. We experimented with a measure that is based on the assumption of a common allele frequency across a sample (28) but this measure was found to be highly sensitive to population stratification (details are shown in SI Text, section S7). Therefore, we use a measure of kinship that has been shown to be more robust to population stratification than previous estimates of genetic similarity across the genome (29). This procedure produces a matrix that describes the genetic similarity for all pairs of individuals in our sample.
Measuring GAM.
The traditional approach to measuring EAM is to analyze the correlation of spousal educational attainment. It is important to note that this approach is possible because each spouse has a level of education. In contrast, measures of genetic relatedness exist at the pair level because relatedness measures a quantity with respect to a specific alter, rather than an absolute level (e.g., years of completed schooling). Hence, a spousal pair would have only a single measure of genetic relatedness versus two measures of education, one for each spouse. The correlation approach is thus not a viable option for measuring GAM. We have instead chosen to concentrate on differences in the distributions of genetic relatedness between married and unmarried pairs of respondents. Although this approach is unique, we studied its behavior via a simulation study (SI Text, section S5), which demonstrated that the method is able to distinguish assortative mating from random mating in samples of this size.
Characterizing the presence and magnitude of genetic homogamy via a comparison of distributions is challenging because it requires a relevant comparison group. One approach would be to consider, for a focal individual, only those individuals with whom the individual is likely to marry given certain characteristics (e.g., age). Results based on such an approach would perhaps be unpersuasive given their potential sensitivity to the formation of the group of potential spouses for a person. To avoid this dilemma, we test GAM against the null hypothesis of random mating. As such, we make only minimal assumptions about the possible range of mates by restricting our comparisons of interest only to cross-sex, same-race individuals. We impose these sex and race restrictions due to limitations in existing data and methods. With respect to sex, we do not have data on same-sex couples. The restriction to same-race couples is done because the relatedness measures can be sensitive to population stratification that may exist across racial groups (additionally, there are relatively few cross-race couples in the data: only 6% of the spousal pairs from the 1,093 spousal pairs in the HRS cohort data discussed in SI Text, section S7).
For both EAM and GAM, our motivating counterfactual is that mates select at random into unions. As such, the distribution of educational or genetic differences among spousal pairs would be the same for all possible cross-sex and same-race pairs in the population. To test this assumption, we compute quantiles (0.001–0.999 in increments of 0.001) for the distribution of the differences among the spousal pairs. We then map these values among spousal pairs to the corresponding quantiles among nonspousal pairs (all cross-sex, same-race pairs). When such results are depicted graphically (Fig. 1), the 45° line indicates the null hypothesis that the similarity among spouses matches the similarity among nonspouses. If the similarity among spouses differs from the similarity of nonspouses, then this is captured by departure from the 45° line. EAM and GAM are estimated as the area between this curve and the 45° line. For key estimates, 95% CIs for the estimates were then created via 1,000 bootstrap replications.
When measuring EAM, we first standardize education within each sex. Our motivation for standardizing education with respect to sex is that more highly educated females will tend to marry more highly educated males. Because of the demographic composition of this cohort, “more education” might mean different things for males and females (e.g., “some college” for females versus a college degree for males). Without standardization, a monotonic relationship between the probability of marriage and educational differences cannot be assumed because there would be ambiguity about the region between 0 and the mean educational difference. That is, if the average difference in completed schooling between males and females is 2 y, a couple with the same level of schooling are not at the same point of their sex specific distribution of years of schooling, and are thus “different.” For education, our results are comparable with and without standardization because the distributions across the genders are similar (SI Text, section S1). However, standardization is a potentially important component of the methodology and would be an important consideration if analyzing phenotypes, such as height, whose distributions vary more across sex. We also multiply all educational differences by −1 so that, as with kinships, larger numbers mean more similar respondents.
Population Stratification.
Because racial/ethnic homogamy is already well known in the literature (30), we focus on residual GAM—GAM that remains within genetically stratified samples that may challenge the assumptions of random mating and intergenerational models in the social sciences. Thus, we only use a sample of non-Hispanic whites in the HRS. Intraethnic assortative mating among Americans of European descent is well documented (3) and small differences in allele frequencies across European ethnic groups are easily identified with genome-wide data (31). As such, the identification of GAM may simply show that Europeans with a similar ethnic background are more likely to marry one another than individuals from different ethnic backgrounds. For example, using data from the Framingham Heart Study, researchers decomposed total genetic variation into PCs that characterize these otherwise small genetic differences across European subpopulations and they calculate a spousal correlation of 0.58 for the first PC in this sample (32). Using similar methods, we estimated a comparable value (r = 0.54) for the first PC among non-Hispanic and white spouses in the HRS. To identify residual GAM, we describe the results from a series of analyses that introduce restrictions in an attempt to understand the extent to which GAM may simply arise from ethnic homogamy within non-Hispanic white couples. These models include the following adjustments: (i) restriction of the sample based on the first PC, (ii) including statistical controls for census division of birth as a proxy for ethnic background, and (iii) estimating GAM with a reduced set of SNPs that do not show any evidence of stratification in our sample.
Supplementary Material
Acknowledgments
This research uses data from the HRS, which is sponsored by the National Institute on Aging (Grants NIA U01AG009740, RC2AG036495, and RC4AG039029) and conducted by the University of Michigan. Research was supported by the Eunice Kennedy Shriver National Institute Of Child Health and Human Development (NICHD) of the National Institutes of Health (NIH) under Award R21HD078031. The authors also acknowledge cofunding from the NICHD and the Office of Behavioral and Social Sciences Research (1R21HD071884). Further support was provided by the NIH/NICHD-funded CU Population Center (R24HD066613). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1321426111/-/DCSupplemental.
References
- 1.Falconer DS, Mackay TFC. Introduction to Quantitative Genetics. 4th Ed. Essex, England: Pearson Education Limited; 1996. [Google Scholar]
- 2.Neale MC, Cardon LR. Methodology for Genetic Studies of Twins and Families. Dordrecht, The Netherlands: Kluwer Academic; 1992. [Google Scholar]
- 3.Schwartz CR. Trends and variation in assortative mating: Causes and consequences. Annu Rev Sociol. 2013;39:451–470. [Google Scholar]
- 4.Blossfeld HP. Educational assortative marriage in comparative perspective. Annu Rev Sociol. 2009;35:513–530. [Google Scholar]
- 5.Breen R, Salazar L. Educational assortative mating and earnings inequality in the United States. Am J Sociol. 2011;117(3):808–843. [Google Scholar]
- 6.Thiessen D, Gregg B. Human assortative mating and genetic equilibrium: An evolutionary perspective. Ethol Sociobiol. 1980;1:111–140. [Google Scholar]
- 7.Blau P. Structural Contexts of Opportunities. Chicago: Univ of Chicago Press; 1994. [Google Scholar]
- 8.Mare RD. Five decades of educational assortative mating. Am Sociol Rev. 1991;56(1):15–32. [Google Scholar]
- 9.Schwartz CR, Mare RD. Trends in educational assortative marriage from 1940 to 2003. Demography. 2005;42(4):621–646. doi: 10.1353/dem.2005.0036. [DOI] [PubMed] [Google Scholar]
- 10.Branigan AR, McCallum KJ, Freese J. Variation in the heritability of educational attainment: An international meta-analysis. Soc Forces. 2013;92(1):109–140. [Google Scholar]
- 11.Rietveld CA, et al. LifeLines Cohort Study GWAS of 126,559 individuals identifies genetic variants associated with educational attainment. Science. 2013;340(6139):1467–1471. doi: 10.1126/science.1235488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Plomin R, DeFries JC, Roberts MK. Assortative mating by unwed biological parents of adopted children. Science. 1977;196(4288):449–450. doi: 10.1126/science.850790. [DOI] [PubMed] [Google Scholar]
- 13. Center for the Study of Aging (2014) RAND Enhanced Fat Files (RAND Corporation, Santa Monica, CA). Available at www.rand.org/labor/aging/dataprod/enhanced-fat.html. Accessed April 29, 2014.
- 14.Johnson AD, et al. SNAP: A web-based tool for identification and annotation of proxy SNPs using HapMap. Bioinformatics. 2008;24(24):2938–2939. doi: 10.1093/bioinformatics/btn564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. National Center for Biotechnology Information dbGaP (2007) Framingham SNP Health Association Resource (National Center for Biotechnology Information, Bethesda, MD). Available at www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000007.v1.p1. Accessed April 29, 2014.
- 16.Lieberson S, Waters MC. From Many Strands: Ethnic and Racial Groups in Contemporary America. New York: Russell Sage Foundation; 1988. [Google Scholar]
- 17.Shanahan MJ, Hofer SM. Social context in gene-environment interactions: Retrospect and prospect. J Gerontol B Psychol Sci Soc Sci. 2005;60(Spec No 1) Special Issue:65–76. doi: 10.1093/geronb/60.special_issue_1.65. [DOI] [PubMed] [Google Scholar]
- 18.Boardman JD, Domingue BW, Fletcher JM. How social and genetic factors predict friendship networks. Proc Natl Acad Sci USA. 2012;109(43):17377–17381. doi: 10.1073/pnas.1208975109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eckland BK. Evolutionary consequences of differential fertility and assortative mating in man. Evol Biol. 1972;5:293–305. [Google Scholar]
- 20.Adkins DE, Guo G. Societal development and the shifting influence of the genome on status attainment. Res Soc Stratif Mobil. 2008;26:235–255. [Google Scholar]
- 21.Conley D, Rauscher E, Dawes C, Magnusson PKE, Siegal ML. Heritability and the equal environments assumption: Evidence from multiple samples of misclassified twins. Behav Genet. 2013;43(5):415–426. doi: 10.1007/s10519-013-9602-1. [DOI] [PubMed] [Google Scholar]
- 22.Milinski M. The major histocompatibility complex, sexual selection, and mate choice. Annu Rev Ecol Evol Syst. 2006;37:159–186. [Google Scholar]
- 23.Tishkoff SA, Gonder MK. Human Origins Within and Out of Africa. Cambridge, UK: Cambridge Univ Press; 2007. [Google Scholar]
- 24.Greenwood J, Guner N, Kocharkov G, Santos C. 2014. Marry Your Like: Assortative Mating and Income Inequality (National Bureau of Economic Research, Cambridge, MA), NBER Working Paper No. 19829.
- 25. National Center for Biotechnology Information dbGaP (2012) Health and Retirement Study (National Center for Biotechnology Information, Bethesda, MD). Available at www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000428.v1.p1. Accessed April 29, 2014.
- 26.Visscher PM, et al. Assumption-free estimation of heritability from genome-wide identity-by-descent sharing between full siblings. PLoS Genet. 2006;2(3):e41. doi: 10.1371/journal.pgen.0020041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang J, et al. Common SNPs explain a large proportion of the heritability for human height. Nat Genet. 2010;42(7):565–569. doi: 10.1038/ng.608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: A tool for genome-wide complex trait analysis. Am J Hum Genet. 2011;88(1):76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Manichaikul A, et al. Robust relationship inference in genome-wide association studies. Bioinformatics. 2010;26(22):2867–2873. doi: 10.1093/bioinformatics/btq559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lin KH, Lundquist J. Mate selection in cyberspace: The intersection of race, gender, and education. Am J Sociol. 2013;119:183–215. [Google Scholar]
- 31.Novembre J, et al. Genes mirror geography within Europe. Nature. 2008;456(7218):98–101. doi: 10.1038/nature07331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sebro R, Hoffman TJ, Lange C, Rogus JJ, Risch NJ. Testing for non-random mating: Evidence for ancestry-related assortative mating in the Framingham heart study. Genet Epidemiol. 2010;34(7):674–679. doi: 10.1002/gepi.20528. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.