Abstract
Background
Within the genus Streptococcus, only Streptococcus thermophilus is used as a starter culture in food fermentations. Streptococcus macedonicus though, which belongs to the Streptococcus bovis/Streptococcus equinus complex (SBSEC), is also frequently isolated from fermented foods mainly of dairy origin. Members of the SBSEC have been implicated in human endocarditis and colon cancer. Here we compare the genome sequence of the dairy isolate S. macedonicus ACA-DC 198 to the other SBSEC genomes in order to assess in silico its potential adaptation to milk and its pathogenicity status.
Results
Despite the fact that the SBSEC species were found tightly related based on whole genome phylogeny of streptococci, two distinct patterns of evolution were identified among them. Streptococcus macedonicus, Streptococcus infantarius CJ18 and Streptococcus pasteurianus ATCC 43144 seem to have undergone reductive evolution resulting in significantly diminished genome sizes and increased percentages of potential pseudogenes when compared to Streptococcus gallolyticus subsp. gallolyticus. In addition, the three species seem to have lost genes for catabolizing complex plant carbohydrates and for detoxifying toxic substances previously linked to the ability of S. gallolyticus to survive in the rumen. Analysis of the S. macedonicus genome revealed features that could support adaptation to milk, including an extra gene cluster for lactose and galactose metabolism, a proteolytic system for casein hydrolysis, auxotrophy for several vitamins, an increased ability to resist bacteriophages and horizontal gene transfer events with the dairy Lactococcus lactis and S. thermophilus as potential donors. In addition, S. macedonicus lacks several pathogenicity-related genes found in S. gallolyticus. For example, S. macedonicus has retained only one (i.e. the pil3) of the three pilus gene clusters which may mediate the binding of S. gallolyticus to the extracellular matrix. Unexpectedly, similar findings were obtained not only for the dairy S. infantarius CJ18, but also for the blood isolate S. pasteurianus ATCC 43144.
Conclusions
Our whole genome analyses suggest traits of adaptation of S. macedonicus to the nutrient-rich dairy environment. During this process the bacterium gained genes presumably important for this new ecological niche. Finally, S. macedonicus carries a reduced number of putative SBSEC virulence factors, which suggests a diminished pathogenic potential.
Keywords: Streptococcus, Genome, Adaptation, Gene decay, Pseudogene, Horizontal gene transfer, Pathogenicity, Virulence factor, Milk, Niche
Background
Lactic acid bacteria (LAB) constitute a very important group of microorganisms for the food industry, as well as the health of humans and animals [1,2]. Several species in this group have a long history of safe use in fermented foods and thus belong to the very few bacteria that may qualify for the "generally regarded as safe" (GRAS) or the "qualified presumption of safety" (QPS) status according to FDA and EFSA, respectively [3]. Other LAB species are commensals of the skin, the oral cavity, the respiratory system, the gastrointestinal tract (GIT) and the genitals of mammals or other organisms. Furthermore, the presence of specific LAB strains, called "probiotics", in certain niches of the body is considered to promote the health of the host [2]. This benign nature of LAB, as well as their economic value, often obscure the existence of notorious LAB pathogens that are among the leading causes of human morbidity and mortality worldwide [4].
This oxymoron about the vast differences in the pathogenic potential within the LAB group is probably best exemplified by streptococci. The genus basically consists of commensals that include several severe pathogens, like group A streptococci (GAS), group B streptococci (GBS) and Streptococcus pneumoniae[5]. Streptococcal pathogens are implicated in a plethora of diseases, ranging from mild (e.g. pharyngitis) to invasive and life-threatening (e.g. necrotizing fasciitis) infections [6]. In contrast, Streptococcus thermophilus is one of the most frequent starter LAB consumed by humans in yogurt and cheese [7]. It is believed that this is the only streptococcal species that, during its adaptation to the nutrient-rich milk environment, underwent extensive genome decay, resulting in the loss of pathogenicity-related genes present in members of the genus [7,8].
Apart from S. thermophilus, other streptococci can grow in milk and milk products. Such streptococci mainly belong to the Streptococcus bovis/Streptococcus equinus complex (SBSEC) [9]. The exact route that would explain their presence in milk is yet unidentified. In theory, since some of them can naturally occur in the GIT or on the teat skin of lactating animals, they could be passively transmitted to raw milk. In addition, species of the SBSEC are known to be involved in human cases of endocarditis, meningitis, bacteremia and colon cancer [10-12]. However, Streptococcus macedonicus, which is a member of this specific complex, has been suggested to be adapted to milk and it has been hypothesized that it could be non pathogenic. These assumptions were based on the fact that the primary ecological niche of S. macedonicus appears to be naturally fermented foods, mostly of dairy origin similarly to S. thermophilus[13]. Initial in vitro and in vivo evaluation did not support virulence of S. macedonicus ACA-DC 198 [14]. PCR and Southern blotting analyses indicated the absence of several Streptococcus pyogenes pathogenicity genes. In addition, oral administration of the organism at high dosages (8.9 log cfu daily) for an extended period of time (12 weeks) to mice did not result in any observable adverse effects including inflammation in the stomach or translocation from the GIT to the organs of the animals [14]. Moreover, strains of S. macedonicus have been shown to present important technological properties of industrial cultures like the production of texturizing exopolysaccharides and anti-clostridial bacteriocins [13].
Streptococcus macedonicus was originally isolated from traditional Greek Kasseri cheese [15] and it is phylogenetically related to Streptococcus gallolyticus subsp. gallolyticus and Streptococcus pasteurianus (formerly known as S. bovis biotypes I and II.2, respectively), as well as to Streptococcus infantarius (formerly known as S. bovis biotype II.1). The inclusion of S. macedonicus and S. pasteurianus as subspecies of S. gallolyticus subsp. gallolyticus (from this point on S. gallolyticus) has been previously suggested [16], but this taxonomic reappraisal has not been formally accepted so far [17]. Streptococcus gallolyticus and S. pasteurianus are considered pathogenic. Preliminary investigations concerning the mechanisms by which S. gallolyticus causes endocarditis indicated that S. macedonicus may lack at least some of the pathogenic determinants implicated in this disease [18,19]. Furthermore, the recent study of the genome of S. infantarius subsp. infantarius CJ18 (from this point on S. infantarius) isolated from spontaneously fermented camel milk in Africa has indicated strain-dependent traits of adaptation to the dairy environment despite the fact that the species is considered as a putative pathogen [20]. Overall, the presence in fermented foods of SBSEC species with a currently unresolved pathogenicity status, such as S. macedonicus and S. infantarius, may represent an underestimated cause of concern in terms of food safety and public health, which needs to be addressed.
Here we present the first complete genome sequence of S. macedonicus in order to shed light on the biology of the species. We are particularly interested in assessing niche adaptation and in investigating the pathogenic potential of the strain analyzed based on comparative genomics against other complete genomes within the SBSEC. This is an important step to rationally deduce whether the bacterium is safe to be used as a starter or if extra technological measures are needed to avoid its presence in food fermentations.
Results and discussion
General features of Streptococcus macedonicus ACA-DC 198 genome
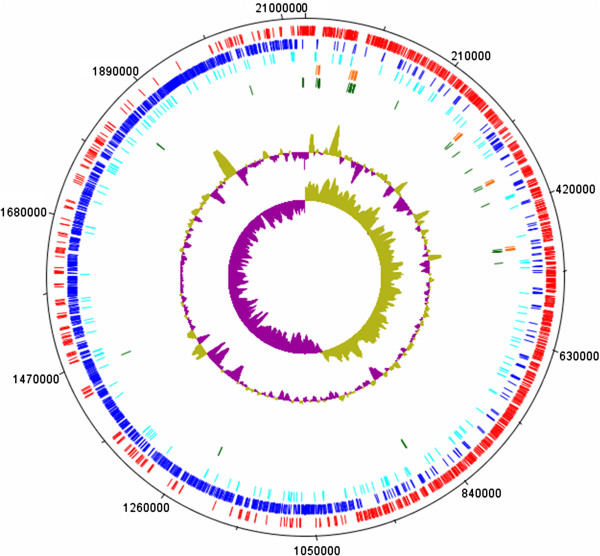
The circular chromosome of S. macedonicus ACA-DC 198 consists of 2,130,034 bp (Figure 1) with a G + C content of 37.6%, which is among the lowest values within the available complete streptococcal genomes (39.3% ± 1.7%, n = 95 by May 2013). A total of 2,192 protein coding DNA sequences (CDSs) were annotated, covering 87.3% of the S. macedonicus chromosome. Of these, 192 were identified as putative pseudogenes according to GenePRIMP [21] analysis followed by manual curation. The bacterium also carries 18 rRNA genes organized in 5 clusters co-localized with most of the 70 tRNA genes. The S. macedonicus genome was found to be 220–232 kb smaller and only 30 kb larger than the genomes of S. gallolyticus and S. pasteurianus, respectively. Streptococcus infantarius has one of the smallest genome sizes within the SBSEC reported up to now (i.e. 141 kb smaller than that of S. macedonicus). The percentage of potential pseudogenes in S. macedonicus was 8.7%, in S. pasteurianus 7.7% and in S. infantarius 4.9%. In contrast, the percentage of pseudogenes in at least two S. gallolyticus strains (i.e. strains UCN34 and ATCC 43143) has been found to be 2.1% or less. This analysis is in accordance with previous findings [9,22]. Based on the close phylogenetic relationship among the four species, these observations suggest that the genome of S. macedonicus, as well as those of S. pasteurianus and S. infantarius may be evolving under selective pressures that allow gene loss events and genome decay processes when compared to the S. gallolyticus genomes.
Figure 1.
The circular map of the genome of Streptococcus macedonicus ACA-DC 198. Genomic features appearing from the periphery to the centre of the map: 1. Forward CDSs (red); 2. Reverse CDSs (blue); 3. Putative pseudogenes (cyan); 4. rRNA genes (orange); 5. tRNA genes (green); 6.% GC plot; 7. GC skew.
Whole genome phylogeny, comparative genomics, and core genome analysis
A phylogenetic tree based on the currently available complete streptococcal genome sequences was constructed using the EDGAR software [23]. On this tree, S. gallolyticus, S. macedonicus, S. pasteurianus, as well as S. infantarius formed a single, monophyletic branch, providing strong evidence for the taxonomic integrity of the SBSEC (Additional file 1: Figure S1).
Subsequently, full chromosome alignments were performed using progressiveMAUVE [24]. The analysis revealed a mosaic pattern of homology organized in local collinear blocks (LCBs) among S. gallolyticus, S. macedonicus and S. pasteurianus (Figure 2A). Evidently, a significant portion of the genetic information has been overall conserved, as the majority of the LCBs are shared by all species. In addition, chromosomal rearrangements seem to have been rather minimal, as the number of LCBs showing a change in relative genomic position among the strains was low and their length short. Nevertheless, numerous differences were also detected. Some LCBs were common only among some of the strains, while some regions were identified as strain-specific (and hence not included within an LCB). The presence of such strain-specific regions suggests that, in addition to gene loss mentioned earlier, gene acquisition events mediated by horizontal gene transfer (HGT) may have played a role during the evolution of the three species (see below). Interestingly, the inclusion of the S. infantarius genome in the MAUVE analysis resulted in an increased number of LCBs with a decreased average length. As the level of sequence conservation of individual LCBs among the four species remains relatively high, this observation suggests that specific genome structure reorganization events occurred specifically in S. infantarius (Figure 2B). Analysis with the EDGAR software revealed a core genome of only 1,372 orthologous genes based on the sequence and the current annotation of S. gallolyticus, S. pasteurianus and S. macedonicus (Figure 3A, Additional file 2: Table S1) [23]. Once more, inclusion of S. infantarius increased the diversity, resulting in reduction of the core genome by more than 100 genes among the four species (Figure 3B, Additional file 3: Table S2). The significant percentage of variable genes within the four SBSEC species may underpin their adaptation to specific environments.
Figure 2.
Chromosome alignments of the Streptococcus bovis/Streptococcus equinus complex members as calculated by progressiveMauve. Chromosome alignments among Streptococcus gallolyticus, Streptococcus macedonicus and Streptococcus pasteurianus(A) and all the aforementioned streptococci and Streptococcus infantarius(B). Local collinear blocks (LCBs) of conserved sequences among the strains are represented by rectangles of the same colour. Connecting lines can be used to visualize synteny or rearrangement. LCBs positioned above or under the chromosome (black line) correspond to the forward and reverse orientation, respectively. The level of conservation is equivalent to the level of vertical colour filling within the LCBs (e.g. white regions are strain-specific). Sequences not placed within an LCB are unique for the particular strain.
Figure 3.
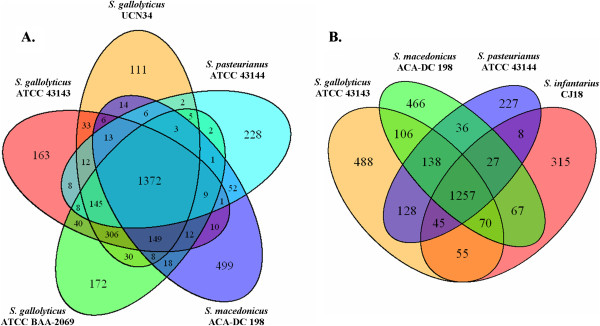
Core genome analysis of members of the Streptococcus bovis/Streptococcus equinus complex. Whole CDS Venn diagrams of Streptococcus gallolyticus, Streptococcus macedonicus and Streptococcus pasteurianus(A) or Streptococcus gallolyticus, Streptococcus infantarius, Streptococcus macedonicus and Streptococcus pasteurianus(B). In (B)Streptococcus gallolyticus ATCC 43143 was selected as a representative of the S. gallolyticus species, since it has the longest genome size among the three sequenced strains.
Genes involved in the survival in the GIT
It has been established that S. gallolyticus displays the notable ability to accumulate and metabolize a broad range of complex carbohydrates from plants when compared to other streptococci [25]. The necessity for this repertoire of carbohydrate-degrading activities has been considered to reflect the adaptation of S. gallolyticus to the rumen of herbivores [22,25]. Preliminary analysis indicated that at least some of the relevant genes are either entirely absent or they have been converted into pseudogenes in the genomes of S. macedonicus, S. pasteurianus and S. infantarius (Table 1). The presence of pseudogenes related to carbohydrate metabolism reinforces the notion that S. macedonicus, S. pasteurianus and S. infantarius have undergone genome decay processes during adaptation to their ecological niches. The entire glycobiome of the SBSEC members was further analyzed based on the data available in the CAZy database (Additional file 4: Table S3) [26]. Important differences in the distribution of enzymes among the SBSEC members were observed for all CAZy categories including glycoside hydrolases (GHs), glycosyl tranferases, polysaccharide lyases, carbohydrate esterases and carbohydrate-binding modules (CBMs). Streptococcus macedonicus and Streptococcus infantarius had the smallest glycobiome within the SBSEC. The two strains had only 24 and 23 GHs, while the rest SBSEC members had more than 40. Given that most of these GHs are potentially involved in plant and dietary carbohydrate catabolism (e.g. GH1, GH3, GH13, GH36 etc.) [27], it could be hypothesized that S. macedonicus and S. infantarius have a diminished necessity for such enzymes in their ecological niche. Streptococcus pasteurianus had the highest number of GHs, some of which were unique among SBSEC (i.e. GH35, GH78, GH79, GH85, GH92, GH125). This observation indicates differences in the range of carbohydrates the strain is able to catabolize in comparison to the other members of the complex. Interestingly, none of the SBSEC members were found to carry GHs that are implicated in the degradation of host derived oligosaccharides (e.g. GH33 and GH98) [27]. In contrast, Streptococcus gallolyticus strains, S. macedonicus and S. infantarius had hits in the CBM41 family, while S. pasteurianus in the CBM32 family, both of which have been associated with the recognition of host glycans [27,28].
Table 1.
Genes in the Streptococcus bovis / Streptococcus equinus complex potentially involved in adaptation to the rumen
| Function | Gene | S. gallolyticus UCN34 | S. gallolyticus ATCC 43143 | S. gallolyticus ATCC BAA-2069 | S. pasteurianus ATCC 43144 | S. macedonicus ACA-DC 198 | S. infantarius CJ18 |
|---|---|---|---|---|---|---|---|
| Pullulanase |
- (a) |
GALLO_1462 |
SGGB_1458 |
SGGBAA2069_c14850 |
SGPB_1362 (t) |
SMA_1464 (s) |
Sinf_1270 |
| |
|
|
|
|
|
SMA_1465 (s) |
|
| Pullulanase |
- |
GALLO_0781 |
SGGB_0764 |
SGGBAA2069_c07530 |
- |
SMA_0719 (p) |
- |
| |
|
|
|
|
|
SMA_0720 (r) |
|
| |
|
|
|
|
|
SMA_0721 (p) |
|
| α-amylase, neopullulanase |
- |
GALLO_0753 |
SGGB_0736 |
SGGBAA2069_c07260 |
- |
- |
- |
| Fructan hydrolase |
fruA |
GALLO_0112 |
SGGB_0110 |
SGGBAA2069_c01280 |
- |
- |
- |
| Beta-1,4-endoglucanase V (cellulase) |
- |
GALLO_0330 |
SGGB_0358 |
SGGBAA2069_c03180 |
- |
- |
- |
| Cinnamoyl ester hydrolase |
cinA |
GALLO_0140 |
SGGB_0137 |
SGGBAA2069_c01580 |
- |
- |
- |
| Mannanase |
- |
GALLO_0162 |
SGGB_0206 |
SGGBAA2069_c01800 |
- |
- |
Sinf_0174 (p) |
| Endo-beta-1,4-galactanase |
- |
GALLO_0189 |
SGGB_0233 |
SGGBAA2069_c02070 |
SGPB_0176 |
SMA_0214 (p) |
Sinf_0197 (p) |
| Pectate lyase |
- |
GALLO_1577 |
SGGB_1576 |
SGGBAA2069_c16050 |
- |
- |
Sinf_1418 |
| Pectate lyase |
- |
GALLO_1578 |
SGGB_1577 |
SGGBAA2069_c16060 |
SGPB_1461 (p) |
SMA_1582 (p) |
- |
| |
|
|
|
|
|
SMA_1583 (s) |
|
| |
|
|
|
|
|
SMA_1584 (s) |
|
| Malate transporter |
mleP |
GALLO_2048 |
SGGB_2031 |
SGGBAA2069_c20060 |
SGPB_1855 |
SMA_1945 |
Sinf_1750 |
| Malate dehydrogenase |
mleS |
GALLO_2049 |
SGGB_2032 |
SGGBAA2069_c20070 |
SGPB_1856 |
SMA_1946 |
Sinf_1751 |
| PTS system, mannitol-specific IIBC component |
mtlA |
GALLO_0993 |
SGGB_0982 |
SGGBAA2069_c09680 |
- |
SMA_0905 (p) |
- |
| Mannitol operon transcriptional antiterminator |
mtlR |
GALLO_0994 |
SGGB_0983 |
SGGBAA2069_c09690 |
- |
SMA_0906 (p) |
- |
| |
|
|
|
|
|
SMA_0907 |
|
| |
|
|
|
|
|
SMA_0908 |
|
| |
|
|
|
|
|
SMA_0909 |
|
| |
|
|
|
|
|
SMA_0910 |
|
| |
|
|
|
|
|
SMA_0911 |
|
| |
|
|
|
|
|
SMA_0912 |
|
| |
|
|
|
|
|
SMA_0913 |
|
| |
|
|
|
|
|
SMA_0914 |
|
| |
|
|
|
|
|
SMA_0915 |
|
| |
|
|
|
|
|
SMA_0916 |
|
| |
|
|
|
|
|
SMA_0917 |
|
| PTS system, mannitol-specific IIA component |
mtlF |
GALLO_0995 |
SGGB_0984 |
SGGBAA2069_c09700 |
- |
- |
- |
| Mannitol-1-phosphate 5-dehydrogenase |
mtlD |
GALLO_0996 |
SGGB_0985 |
SGGBAA2069_c09710 |
- |
- |
- |
| α-amylase |
- |
GALLO_0757 |
SGGB_0740 |
SGGBAA2069_c07300 |
- |
- |
- |
| α-amylase |
amyE |
GALLO_1632 |
SGGB_1646 |
SGGBAA2069_c16600 |
SGPB_1505 (p) |
SMA_1612 (t) |
Sinf_1443 |
| α-amylase |
- |
GALLO_1043 |
SGGB_1033 |
SGGBAA2069_c10200 |
SGPB_0905 |
SMA_0972 |
Sinf_0846 |
| tannase |
tanA |
GALLO_0933 |
SGGB_0917 |
SGGBAA2069_c09070 (s) |
- |
- |
- |
| |
|
|
|
SGGBAA2069_c09080 (s) |
|
|
|
| Tannase (similar to tanA) |
- |
GALLO_1609 |
SGGB_1624 |
SGGBAA2069_c16370 |
- |
- |
- |
| Phenolic acid decarboxylase |
padC |
GALLO_2106 |
SGGB_2089 |
SGGBAA2069_c21040 |
SGPB_1899 |
SMA_2074 |
- |
| Carboxymuconolactone decarboxylase |
- |
GALLO_0906 |
SGGB_0891 |
SGGBAA2069_c08850 |
SGPB_0775 |
- |
- |
| Bile salt hydrolase | bsh | GALLO_0818 | SGGB_0803 | SGGBAA2069_c07920 | SGPB_0678 | SMA_0753 (p) | Sinf_0639 |
(a) Not found; (t) Truncated; (s) Split CDSs corresponding to fragments of the original gene not yet characterized as pseudogenes; (p) Pseudogenes; (r) Transposase genes in italics.
Furthermore, S. gallolyticus can detoxify toxic compounds met in the rumen and other environments. Again, S. macedonicus, S. pasteurianus and S. infantarius miss some of the genes involved in detoxification (Table 1). None of them carry genes for tannin hydrolysis similar to GALLO_0933 or GALLO_1609. The potential to degrade additional phenolic compounds like gallic acid seems to be comparable between S. gallolyticus and S. pasteurianus. In contrast, S. infantarius has no orthologs of either PadC (GALLO_2106) or GALLO_0906, i.e. the two gallic acid decarboxylases found in S. gallolyticus UCN34, while S. macedonicus has retained only PadC. Furthermore, the bsh gene (GALLO_0818), coding for a bile salt hydrolase, is present in all four species with the exception of S. macedonicus, in which it appears as a pseudogene. Thus, our findings clearly suggest that not only S. macedonicus, but also S. pasteurianus and S. infantarius have deviated from S. gallolyticus in their potential to cope with the harsh environment of the GIT of herbivores.
Genes involved in the growth in milk or dairy products
Dairy LAB are considered fastidious microorganisms due to their adaptation to growth in milk that is particularly nutritious by nature. Lactose and milk proteins (both caseins and whey proteins) are characteristic of the dairy environment. LAB are able to ferment lactose to lactic acid and they have evolved a proteolytic system for the degradation of milk proteins down to amino acids [1,29].
All SBSEC species are able to utilize lactose and to catabolize galactose. Sequence similarity searches revealed a gene cluster (SMA_0197 – SMA_0211) dedicated to lactose metabolism with a unique organization in SBSEC when compared to those previously reported for other LAB (Table 2). The typical sequence of lac genes is interrupted in the majority of SBSEC strains by genes coding for the IIA, IIB and IIC components of a PEP-PTS (SMA_0202 – SMA _0204). Annotation of this PEP-PTS varies among the SBSEC species/strains and for this reason functional analysis is required to properly determine its exact function. In contrast to other SBSEC species, these three PTS genes are absent from S. infantarius. The lactose-specific PTS found at the end of the lac gene cluster (SMA_0206 – SMA _0210) is also inactivated in S. infantarius through disruption of the lacT antiterminator gene by transposases [20]. Interestingly, the lac gene cluster in S. macedonicus contains two 6-phospho-beta-galactosidase (lacG) genes that may be indicative of adaptation of this particular species to milk. Galactose can also be catabolized through the Leloir pathway and a galRKTE operon coding for the relevant enzymes was previously determined in S. infantarius[30]. The gal operon is conserved in all SBSEC species analyzed here (Table 2).
Table 2.
Genes in the Streptococcus bovis / Streptococcus equinus complex potentially involved in lactose and galactose metabolism
| Function | Gene | S. gallolyticus UCN34 | S. gallolyticus ATCC 43143 | S. gallolyticus ATCC BAA-2069 | S. pasteurianus ATCC 43144 | S. macedonicus ACA-DC 198 | S. infantarius CJ18 |
|---|---|---|---|---|---|---|---|
| Lactose-specific PTS system repressor |
lacR |
GALLO_0176 |
SGGB_0220 |
SGGBAA2069_c01940 |
SGPB_0163 |
SMA_0197 |
Sinf_0181 |
| Galactose-6-phosphate isomerase, LacA subunit |
lacA |
GALLO_0177 |
SGGB_0221 |
SGGBAA2069_c01950 |
SGPB_0164 |
SMA_0198 |
Sinf_0182 |
| Galactose-6-phosphate isomerase, LacB subunit |
lacB1 |
GALLO_0178 |
SGGB_0222 |
SGGBAA2069_c01960 |
SGPB_0165 |
SMA_0199 |
Sinf_0183 |
| Tagatose-6-phosphate kinase |
lacC |
GALLO_0179 |
SGGB_0223 |
SGGBAA2069_c01970 |
SGPB_0166 |
SMA_0200 |
Sinf_0184 |
| Tagatose 1,6-diphosphate aldolase |
lacD2 |
GALLO_0180 |
SGGB_0224 |
SGGBAA2069_c01980 |
SGPB_0167 |
SMA_0201 |
Sinf_0185 |
| Putative PTS system, IIA component |
- (a) |
GALLO_0181 |
SGGB_0225 |
SGGBAA2069_c01990 |
SGPB_0168 |
SMA_0202 |
- |
| Putative PTS system, IIB component |
- |
GALLO_0182 |
SGGB_0226 |
SGGBAA2069_c02000 |
SGPB_0169 |
SMA_0203 |
- |
| Putative PTS system, IIC component |
- |
GALLO_0183 |
SGGB_0227 |
SGGBAA2069_c02010 |
SGPB_0170 |
SMA_0204 |
- |
| Aldose 1-epimerase |
lacX |
GALLO_0184 |
SGGB_0228 |
SGGBAA2069_c02020 |
SGPB_0171 |
SMA_0205 |
Sinf_0186 |
| Transcriptional antiterminator |
lacT |
GALLO_0185 |
SGGB_0229 |
SGGBAA2069_c02030 |
SGPB_0172 |
SMA_0206 |
Sinf_0187 (p) |
| |
|
|
|
|
|
|
Sinf_0188 (r) |
| |
|
|
|
|
|
|
Sinf_0189 |
| |
|
|
|
|
|
|
Sinf_0190 (p) |
| 6-phospho-beta-galactosidase |
lacG |
GALLO_0186 |
SGGB_0230 |
SGGBAA2069_c02040 |
SGPB_0173 |
SMA_0207 |
- |
| Transcriptional antiterminator |
lacT |
- |
- |
- |
- |
SMA_0208 (p) |
- |
| Lactose-specific PTS system, IIA component |
lacF |
GALLO_0187 |
SGGB_0231 |
SGGBAA2069_c02050 |
SGPB_0174 |
SMA_0209 |
Sinf_0191 |
| Lactose-specific PTS system, IIBC component |
lacE |
GALLO_0188 |
SGGB_0232 |
SGGBAA2069_c02060 |
SGPB_0175 |
SMA_0210 |
Sinf_0192 |
| 6-phospho-beta-galactosidase |
lacG2 |
- |
- |
- |
- |
SMA_0211 |
Sinf_0193 (p) |
| |
|
|
|
|
|
|
Sinf_0194 |
| |
|
|
|
|
|
|
Sinf_0195 (p) |
| Galactose repressor |
galR |
GALLO_0197 |
SGGB_0241 |
SGGBAA2069_c02150 |
SGPB_0184 |
SMA_0222 |
Sinf_0205 |
| Galactokinase |
galK |
GALLO_0198 |
SGGB_0242 |
SGGBAA2069_c02160 |
SGPB_0185 |
SMA_0223 |
Sinf_0206 |
| Galactose-1-P-uridyl transferase |
galT |
GALLO_0199 |
SGGB_0243 |
SGGBAA2069_c02170 |
SGPB_0186 |
SMA_0224 |
Sinf_0207 |
| UDP-glucose 4-epimerase |
galE |
GALLO_0200 |
SGGB_0244 |
SGGBAA2069_c02180 |
SGPB_0187 |
SMA_0225 |
Sinf_0208 |
| Beta-galactosidase |
lacZ |
- |
- |
- |
SGPB_0344 |
- |
- |
| Glucokinase |
glcK |
GALLO_0594 |
SGGB_0562 |
SGGBAA2069_c05300 |
SGPB_0467 |
SMA_0546 |
Sinf_0470 |
| Beta-galactosidase |
lacZ |
- |
- |
- |
- |
- |
Sinf_0935 |
| Lactose and galactose permease |
lacS |
- |
- |
- |
- |
- |
Sinf_0936 |
| Aldose 1-epimerase |
galM |
- |
- |
- |
- |
- |
Sinf_0937 |
| UDP-glucose 4-epimerase |
galE1 |
- |
- |
- |
- |
- |
Sinf_0938 |
| Galactose-1-P-uridyl transferase |
galT |
- |
- |
- |
- |
- |
Sinf_0939 (p) |
| UDP-glucose 4-epimerase |
lacS |
- |
- |
- |
- |
- |
Sinf_1514 |
| Aldose 1-epimerase |
lacX |
- |
- |
- |
- |
SMA_1156 |
- |
| 6-phospho-beta-galactosidase |
lacG2 |
- |
- |
- |
- |
SMA_1157 |
- |
| Lactose-specific PTS system, IIBC component |
lacE |
- |
- |
- |
- |
SMA_1158 |
- |
| Lactose-specific PTS system, IIA component |
lacF |
- |
- |
- |
- |
SMA_1159 |
- |
| Tagatose 1,6-diphosphate aldolase |
lacD |
- |
- |
- |
- |
SMA_1160 |
- |
| Tagatose-6-phosphate kinase |
lacC |
- |
- |
- |
- |
SMA_1161 |
- |
| Galactose-6-phosphate isomerase, LacB subunit |
lacB |
- |
- |
- |
- |
SMA_1162 |
- |
| Galactose-6-phosphate isomerase, LacA subunit |
lacA1 |
- |
- |
- |
- |
SMA_1163 |
- |
| Glucokinase |
glcK |
- |
- |
- |
- |
SMA_1164 |
- |
| Lactose phosphotransferase system repressor |
lacR |
- |
- |
- |
- |
SMA_1165 |
- |
| Transcription antiterminator |
lacT |
GALLO_1046 |
SGGB_1036 |
SGGBAA2069_c10230 |
SGPB_0907 |
- |
- |
| Lactose-specific PTS system, IIA component |
lacF |
GALLO_1047 |
SGGB_1037 |
SGGBAA2069_c10240 |
SGPB_0908 |
- |
- |
| Lactose-specific PTS system, IIBC component |
lacE |
GALLO_1048 |
SGGB_1038 |
SGGBAA2069_c10250 |
SGPB_0909 |
- |
- |
| Phospho-beta-galactosidase |
lacG |
GALLO_1049 |
SGGB_1039 |
SGGBAA2069_c10260 |
SGPB_0910 |
- |
- |
| Aldose 1-epimerase |
galM |
GALLO_0137 |
SGGB_0134 |
SGGBAA2069_c01550 |
SGPB_0130 |
- |
- |
| UDP-glucose 4-epimerase | galE1 | GALLO_0728 | SGGB_0709 | SGGBAA2069_c06910 | SGPB_0601 | - | - |
(a) Not found; (p) Pseudogenes; (r) Transposase genes in italics.
A partial gal-lac operon galT(truncated)/galE1M/lacSZ with high sequence identity to S. thermophilus is also present in the genome of S. infantarius[30]. It has been demonstrated that the lactose and galactose permease (lacS) and the β-galactosidase (lacZ) are responsible for the uptake and initial hydrolysis of lactose in S. infantarius in a manner similar to that employed by S. thermophilus[20]. This gal-lac operon of S. infantarius is missing from the other SBSEC strains as a whole. A LacZ ortholog (SGPB_0344) is only present in S. pasteurianus and dispersed galE and galM genes can be found in the S. gallolyticus and S. pasteurianus genomes. Similarly to the presence of the extra gal-lac operon in S. infantarius, we detected a second lac gene cluster in S. macedonicus (SMA_1156 – SMA_1165), also suggesting adaptation to the milk environment. This second gene cluster is solely present in S. macedonicus and not in any other SBSEC member. Surprisingly, an additional lacTFEG region coding for a complete lactose PEP-PTS and a 6-phospho-beta-galactosidase is present in the genomes of S. gallolyticus and S. pasteurianus. This is an unexpected finding since S. gallolyticus and S. pasteurianus have hardly ever been related to milk up to now [9].
We then investigated the proteolytic system of S. macedonicus and the rest of the SBSEC members adapting the scheme previously described by Liu and co-workers (i.e. excluding housekeeping proteases or proteases involved in specific cellular processes other than the acquisition of amino acids) [29]. In milk, casein utilization by LAB is initiated after hydrolysis by a cell-envelope associated proteinase (CEP) releasing oligopeptides. The oligopeptides are then transferred intracellularly via specialized peptide transport systems where they are systematically degraded into amino acids by an array of intracellular peptidases. The four species have essentially the same proteolytic system, albeit showing some differences (Table 3). None of them has a typical PrtP CEP, but S. gallolyticus and S. infantarius carry a lactocepin coding gene. The lactocepin of the SBSEC shows ≥ 63% sequence similarity to the PrtS CEP involved in the degradation of milk proteins in S. thermophilus[31,32]. The exact role of lactocepin in SBSEC species needs to be experimentally examined. SBSEC strains like S. macedonicus may require CEP activity to be provided by other bacteria when growing in milk. This is a common strategy of nonstarter LAB that rely on starter CEP-producing strains for casein hydrolysis [33]. Streptococcus infantarius carries two oligopeptide transport systems (Opp) [20], but all the other SBSEC species have only one such system. All SBSEC strains own a proton motive force (PMF)-driven DtpT transporter for the transport of di- and tri-peptides and they all possess an entire repertoire of proteolytic enzymes including endopeptidases, general aminopeptidases and specialized peptidases (Table 3). They only lack enzymes of the PepE/PepG (endopeptidases) and the PepI/PepR/PepL (proline peptidases) superfamilies in accordance to previous observations for streptococci and lactococci [29]. The conservation of this proteolytic system among streptococci in the SBSEC despite their presumed adaptation to different ecological niches [20,22,25] indicates that it may somehow be essential. Furthermore, S. macedonicus and the other SBSEC members are autotrophs for several amino acids (data not shown) and only S. pasteurianus has been reported to be unable to synthesize tryptophan [22]. Thus, the preservation of an entire proteolytic system by SBSEC members while retaining the ability to synthesize most, if not all, amino acids is puzzling, especially when considering that some of them have obviously undergone extensive genome decay processes. It could be hypothesized that this property of SBSEC species may provide a competitive advantage in poor environments, but this needs to be further investigated.
Table 3.
Genes in the Streptococcus bovis / Streptococcus equinus complex potentially involved in proteolysis of milk proteins
| Function | Gene | S. gallolyticus UCN34 | S. gallolyticus ATCC 43143 | S. gallolyticus ATCC BAA-2069 | S. pasteurianus ATCC 43144 | S. macedonicus ACA-DC 198 | S . infantarius CJ18 |
|---|---|---|---|---|---|---|---|
| Lactocepin |
prtS |
GALLO_0748 |
SGGB_0730 |
SGGBAA2069_c07210 |
- |
- |
Sinf_0588 |
| Oligopeptide ABC transporter, substrate-binding protein |
oppA |
GALLO_0324 |
SGGB_0352 |
SGGBAA2069_c03120 |
SGPB_0276 |
SMA_0353 |
Sinf_0305 |
| |
|
GALLO_1412 |
SGGB_1406 |
SGGBAA2069_c14340 |
SGPB_1328 |
SMA_1347 |
Sinf_1225 |
| |
|
GALLO_1413 |
SGGB_1407 |
SGGBAA2069_c14350 |
|
|
Sinf_1226 |
| |
|
|
|
|
|
|
Sinf_1825 |
| Oligopeptide ABC transporter, permease protein |
oppB |
GALLO_0325 |
SGGB_0353 |
SGGBAA2069_c03130 |
SGPB_0277 |
SMA_0354 |
Sinf_0306 |
| |
|
|
|
|
|
|
Sinf_1824 |
| Oligopeptide ABC transporter, permease protein |
oppC |
GALLO_0326 |
SGGB_0354 |
SGGBAA2069_c03140 |
SGPB_0278 |
SMA_0355 |
Sinf_0307 |
| |
|
|
|
|
|
|
Sinf_1823 |
| Oligopeptide ABC transporter, ATP-binding protein |
oppD |
GALLO_0327 |
SGGB_0355 |
SGGBAA2069_c03150 |
SGPB_0279 |
SMA_0356 |
Sinf_0308 |
| |
|
|
|
|
|
|
Sinf_1822 |
| Oligopeptide ABC transporter, ATP-binding protein |
oppF |
GALLO_0328 |
SGGB_0356 |
SGGBAA2069_c03160 |
SGPB_0280 |
SMA_0357 |
Sinf_0309 |
| |
|
|
|
|
|
|
Sinf_1821 |
| Dipeptide/tripeptide permease |
dtpT |
GALLO_0638 |
SGGB_0613 |
SGGBAA2069_c05810 |
SGPB_0507 |
SMA_0600 |
Sinf_0519 |
| Cysteine aminopeptidase C |
pepC |
GALLO_0478 |
SGGB_0452 |
SGGBAA2069_c04140 |
SGPB_0379 |
SMA_0442 |
Sinf_0388 |
| Aminopeptidase N |
pepN |
GALLO_1143 |
SGGB_1134 |
SGGBAA2069_c11310 |
SGPB_1002 |
SMA_1066 |
Sinf_0984 |
| Methionine aminopeptidase |
pepM |
GALLO_0775 |
SGGB_0758 |
SGGBAA2069_c07470 |
SGPB_0642 |
SMA_0713 |
Sinf_0604 |
| Glutamyl aminopeptidase |
pepA |
GALLO_0101 |
SGGB_0101 |
SGGBAA2069_c01190 |
SGPB_0100 |
SMA_0113 |
Sinf_0111 |
| |
|
GALLO_0151 |
SGGB_0195 |
SGGBAA2069_c01680 |
SGPB_0141 |
|
|
| Endopeptidase |
pepO |
GALLO_2172 |
SGGB_2204 |
SGGBAA2069_c21680 |
SGPB_1933 |
SMA_2096 |
Sinf_1874 |
| Oligoendopeptidase |
pepF |
GALLO_0669 |
SGGB_0651 |
SGGBAA2069_c06210 |
SGPB_0551 |
SMA_0630 |
Sinf_0554 |
| |
|
GALLO_1516 |
SGGB_1511 |
SGGBAA2069_c15390 |
SGPB_1410 |
SMA_1526 |
Sinf_1335 |
| Dipeptidase |
pepD |
GALLO_0732 |
SGGB_0713 |
SGGBAA2069_c06950 |
SGPB_0605 |
SMA_0668 |
Sinf_1301 |
| Xaa-His dipeptidase |
pepV |
GALLO_0931 |
SGGB_0915 |
SGGBAA2069_c09050 |
SGPB_0797 |
SMA_0836 |
Sinf_0699 |
| Peptidase T |
pepT |
GALLO_1366 |
SGGB_1360 |
SGGBAA2069_c13560 |
SGPB_1287 |
SMA_1297 |
Sinf_1183 |
| X-prolyl-dipeptidyl aminopeptidase |
pepX |
GALLO_1959 |
SGGB_1942 |
SGGBAA2069_c19090 |
SGPB_1791 |
SMA_1862 |
Sinf_1676 |
| Aminopeptidase P |
pepP |
GALLO_1901 |
SGGB_1885 |
SGGBAA2069_c18550 |
SGPB_1732 |
SMA_1811 |
Sinf_1626 |
| Xaa-proline dipeptidase | pepQ | GALLO_1583 | SGGB_1582 | SGGBAA2069_c16110 | SGPB_1466 | SMA_1589 | Sinf_1424 |
Apart from amino acids, S. gallolyticus UCN34 also carries complete pathways for the synthesis of a number of vitamins including riboflavin, nicotine amide, pantothenate, pyridoxine, and folic acid, while the biosynthetic pathways for biotin and thiamine are partial [25]. The genes potentially involved in the de novo biosynthesis of pyridoxine in the SBSEC strains were determined based on the respective pathway of S. pneumoniae D39 [34]. The corresponding loci are conserved among S. gallolyticus strains but once more S. macedonicus, S. pasteurianus and S. infantarius appear to have undergone a heterogeneous gene loss process, indicating the necessity for exogenous supply of some of these vitamins (Table 4). For example, S. macedonicus misses the bioBDY, panBCD and ribDEAH loci involved in the biosynthesis of biotin, pantothenate and riboflavin, respectively. In addition, the presence of pseudogenes or truncated/split genes may have disrupted the biosynthesis of pyridoxine, nicotine amide and thiamine through the routes analyzed here. It is not uncommon for LAB to be auxotrophic for several vitamins [35], though milk and other dairy products may contain all essential vitamins to sustain the growth of these microorgansims.
Table 4.
Genes in the Streptococcus bovis / Streptococcus equinus complex potentially involved in the biosynthesis of vitamins
| Vitamin | Gene | S . gallolyticus UCN34 | S . gallolyticus ATCC 43143 | S. gallolyticus ATCC BAA-2069 | S. pasteurianus ATCC 43144 | S. macedonicus ACA-DC 198 | S . infantarius CJ18 |
|---|---|---|---|---|---|---|---|
| Biotin (B8, partial) |
bioB |
GALLO_1916 |
SGGB_1900 |
SGGBAA2069_c18670 |
SGPB_1745 |
- (a) |
- |
| |
bioD |
GALLO_1915 |
SGGB_1899 |
SGGBAA2069_c18660 |
SGPB_1744 |
- |
- |
| |
bioY |
GALLO_1914 |
SGGB_1898 |
SGGBAA2069_c18650 |
SGPB_1743 |
- |
- |
| |
pdxS |
GALLO_1189 |
SGGB_1183 |
SGGBAA2069_c11790 |
- |
SMA_1105 (s) |
Sinf_1022 |
| |
|
|
|
|
|
SMA_1106 (p) |
|
| |
pdxT |
GALLO_1188 |
SGGB_1182 |
SGGBAA2069_c11780 |
- |
SMA_1104 |
Sinf_1021 |
| |
pdxR |
GALLO_1111 |
SGGB_1101 |
SGGBAA2069_c10980 |
SGPB_0968 |
SMA_1031 |
Sinf_0955 |
| Folic acid (B9) |
folC |
GALLO_1233 |
SGGB_1227 |
SGGBAA2069_c12240 |
SGPB_1087 |
SMA_1137 |
Sinf_1067 |
| |
folE |
GALLO_1232 |
SGGB_1226 |
SGGBAA2069_c12230 |
SGPB_1086 |
SMA_1136 |
Sinf_1066 |
| |
folP |
GALLO_1231 |
SGGB_1225 |
SGGBAA2069_c12220 |
SGPB_1085 |
SMA_1135 |
Sinf_1065 |
| |
folB |
GALLO_1230 |
SGGB_1224 |
SGGBAA2069_c12210 |
SGPB_1084 |
SMA_1134 |
Sinf_1064 |
| |
folK |
GALLO_1229 |
SGGB_1223 |
SGGBAA2069_c12200 |
SGPB_1083 |
SMA_1133 |
Sinf_1063 |
| |
folD |
GALLO_0622 |
SGGB_0594 |
SGGBAA2069_c05620 |
SGPB_0494 |
SMA_0581 |
Sinf_0503 |
| Nicotine amide (NAD, B3) |
nadA |
GALLO_1937 |
SGGB_1920 |
SGGBAA2069_c18890 |
SGPB_1769 |
SMA_1844 (p) |
Sinf_1655 |
| |
nadB |
GALLO_1936 |
SGGB_1919 |
SGGBAA2069_c18880 |
SGPB_1768 |
SMA_1840 (s) |
Sinf_1654 |
| |
|
|
|
|
|
SMA_1841 (s) |
|
| |
|
|
|
|
|
SMA_1842 (s) |
|
| |
|
|
|
|
|
SMA_1843 (p) |
|
| |
nadC |
GALLO_1935 |
SGGB_1918 |
SGGBAA2069_c18870 |
SGPB_1767 |
SMA_1839 |
Sinf_1653 |
| |
nadE |
GALLO_0477 |
SGGB_0451 |
SGGBAA2069_c04130 |
SGPB_0377 (p) |
SMA_0441 |
Sinf_0387 |
| |
|
|
|
|
SGPB_0378 (p) |
|
|
| Pantothenate (B5) |
panB |
GALLO_0161 |
SGGB_0205 |
SGGBAA2069_c01790 |
- |
- |
- |
| |
panC |
GALLO_0160 |
SGGB_0204 |
SGGBAA2069_c01780 |
- |
- |
Sinf_0173 (t) |
| |
panD |
GALLO_0159 |
SGGB_0203 |
SGGBAA2069_c01770 |
- |
- |
Sinf_0172 |
| |
panE |
GALLO_0232 |
SGGB_0274 |
SGGBAA2069_c02470 |
SGPB_0217 |
SMA_0254 |
Sinf_0233 (p) |
| Riboflavin (B2) |
ribD |
GALLO_0692 |
SGGB_0673 |
SGGBAA2069_c06490 |
SGPB_0567 |
- |
Sinf_0572 |
| |
ribE |
GALLO_0693 |
SGGB_0674 |
SGGBAA2069_c06500 |
SGPB_0568 |
- |
Sinf_0573 |
| |
ribA |
GALLO_0694 |
SGGB_0675 |
SGGBAA2069_c06510 |
SGPB_0569 |
- |
Sinf_0574 |
| |
ribH |
GALLO_0695 |
SGGB_0676 |
SGGBAA2069_c06520 |
SGPB_0570 |
- |
Sinf_0575 |
| |
ribF |
GALLO_1160 |
SGGB_1152 |
SGGBAA2069_c11480 |
SGPB_1019 |
SMA_1086 |
Sinf_0999 |
| Thiamine (B1, partial) |
tenA |
GALLO_1181 |
SGGB_1175 |
SGGBAA2069_c11710 |
SGPB_1039 |
- |
Sinf_1014 |
| |
thiE |
GALLO_1178 |
SGGB_1172 |
SGGBAA2069_c11680 |
SGPB_1036 |
SMA_1100 (t) |
Sinf_1011 |
| |
thiM |
GALLO_1179 |
SGGB_1173 |
SGGBAA2069_c11690 |
SGPB_1037 |
- |
Sinf_1012 |
| |
thiD |
GALLO_1180 |
SGGB_1174 |
SGGBAA2069_c11700 |
SGPB_1038 |
- |
Sinf_1013 |
| |
thiI |
GALLO_1346 |
SGGB_1341 |
SGGBAA2069_c13350 |
SGPB_1268 |
SMA_1273 |
Sinf_1163 |
| thiN | GALLO_2003 | SGGB_1987 | SGGBAA2069_c19580 | SGPB_1830 | SMA_1899 | Sinf_1712 |
(a) Not found; (s) Split CDSs corresponding to fragments of the original gene not yet characterized as pseudogenes; (p) Pseudogenes; (t) Truncated.
Genomic islands (GIs) and unique genes of Streptococcus macedonicus
GIs are sites of HGT that can uncover important features of the plasticity of a bacterial genome and they are primarily linked to gene gain processes. We used the IslandViewer application [36] to identify GIs of the SBSEC members in parallel. Streptococcus macedonicus had 14 predicted GIs with an average length of 18,109 bp corresponding to a total sequence of 253,523 bp or 11.9% the size of the bacterium’s genome (Additional file 5: Figure S2). This percentage of externally acquired DNA is higher compared to the other SBSEC members, in which it ranged from 8.8% in S. gallolyticus ATCC BAA-2069 down to 5.9% in S. gallolyticus UCN34.
As could be expected, the highest degree of sequence conservation among GIs was observed in the S. gallolyticus strains (e.g. S. gallolyticus UCN34 GIs 2, 6, 7, 8 and 9). When different SBSEC species were compared, a number of GIs were only partially conserved (e.g. S. gallolyticus UCN34 GIs 1, 3, 6, 7, 8 and 9). Unique GIs were also present in most genomes analyzed (e.g. S. pasteurianus GIs 2, 4, 6 and 8). Partially conserved GIs may be remnants of GIs acquired before speciation events in the SBSEC and their subsequent gene decay may be the result of adaptation to diverged ecological niches. The existence of unique GIs among the SBSEC species, whose acquisition must have been more recent (i.e. most probably after speciation), also points to the same direction. Furthermore, our analysis suggests that S. macedonicus shares stretches of GI sequences exclusively with S. infantarius among the SBSEC members (e.g. in S. macedonicus GIs 1, 4, 5, 6, 7, 8 and 14) in accordance with previous findings [20]. Potential donors of GI sequences were identified from best BLASTN hits showing sequence identity > 90%. In several instances sequence segments within S. macedonicus GIs may have derived from more than one donor (Additional file 6: Figure S3). Potential donors of the S. macedonicus GIs were Streptococcus agalactiae, Streptococcus intermedius, Streptococcus suis, Streptococcus uberis, Enterococcus faecium, Lactococcus garvieae and Pediococcus pentosaceus. Most importantly, Lactococcus lactis or S. thermophilus were found among these donors in 9 out of 14 S. macedonicus GIs and the same applies for S. infantarius in 6 out of 12 GIs. None of the GI sequences of the other SBSEC members could be linked to L. lactis or S. thermophilus apart from the S. gallolyticus ATCC BAA-2069 GI 6 that exhibited a 96% identity over an approximately 3 kb genomic region of S. thermophilus JIM 8232 (data not shown). These observations constitute additional evidence that S. macedonicus and S. infantarius are the only members of the complex that have extensively interacted with the dairy L. lactis and S. thermophilus.
We then calculated the unique genes (also referred here as singleton genes) of S. macedonicus against the other SBSEC species twice, taking or not into account the genome of S. infantarius. Results from singleton gene analysis using EDGAR [23] were manually curated to relieve the set from the high numbers of transposable elements. There was an important overlap between the list of genes found in GIs of S. macedonicus and the singleton genes (Additional file 7: Table S4 and Additional file 8: Table S5). Again, S. macedonicus and S. infantarius were found to share a number of genes that are absent from the other SBSEC genomes (Additional file 8: Table S5).
According to the aforementioned analysis S. macedonicus carries the complete biosynthetic pathways for two lantibiotic bacteriocins, i.e. the macedocin and the macedovicin peptides [37,38]. The presence of both antimicrobials can provide an additional link between S. macedonicus and the milk environment. Production of macedocin has been observed only in milk up to now and proteolytic fragments of casein may trigger biosynthesis of this peptide [39]. In addition, the entire macedovicin gene cluster is practically identical (99% sequence identity over the entire length of the ~9.8 kb cluster) to the respective clusters of thermophilin 1277 and bovicin HJ50 found in the dairy isolates S. thermophilus SBT1277 and S. bovis HJ50, respectively [37]. The locus seems to have spread among the three strains by HGT and their common dairy origin increases the possibility that this exchange of genetic material has taken place in milk [37].
Another evident characteristic of the S. macedonicus genome was the presence of multiple restriction modification (RM) systems among the singleton genes (Additional file 9: Figure S4). Streptococcus macedonicus possesses the highest number of RM systems within the SBSEC and it is the only member of the group with all three types of RM systems. A yet unresolved difference in the number and the type of RM systems between commensal and dairy LAB has been previously observed [40,41]. As mentioned earlier, phages are present in milk and dairy products often in high numbers [42] and traditional practices (e.g. backslopping) may promote the selection of phage resistant strains [40,41]. In S. thermophilus RM systems are considered as important technological traits [8] and it has been previously suggested that genes of the type III RM system may provide a signature for milk adaptation [40]. Streptococcus macedonicus has two type III RM systems, one of which is inactive since it consists of pseudogenes. The increased number of RM systems of S. macedonicus compared to the other SBSEC members suggests that it should be particularly competent in resisting invading DNA. These findings coincide with the fact that S. macedonicus carries the highest number of spacers in its CRISPR (clustered regularly interspaced short palindromic repeats) locus within the SBSEC (Additional file 10: Table S6). Furthermore, BLASTN analysis of the spacers in the S. macedonicus CRISPR revealed that four of them, namely spacers 3, 5, 17 and 18, had hits in S. thermophilus phages (e.g. phages O1205, 7201, Abc2, etc.), S. thermophilus plasmids (e.g. pER36) or S. thermophilus CRISPR spacer sequences (data not shown). In contrast, among the 140 spacers of the different CRISPR found in the other SBSEC species, only one had a hit in L. lactis phage 1706 (spacer 35 in the CRISPR of S. pasteurianus). According to these findings the occurrence of S. macedonicus in the same habitat as that of S. thermophilus can be supported.
In addition, S. macedonicus contains singleton genes – several copies in some instances – coding for proteins involved in the transport and homeostasis of metal ions (Additional file 7: Table S4 and Additional file 8: Table S5). Some of these genes are also shared by S. infantarius, but not all. These genes may play a role in the transport of copper (e.g. copA and copB), cadmium (e.g. cadA and cadC), manganese (e.g. mntH) and magnesium (e.g. SMA_2044). Copper and cadmium are of no evident biological role for Lactobacillales[43] and thus transport systems for such metals in S. macedonicus should be perceived as a protective mechanism towards their deleterious effects (e.g. through oxidative stress). The presence of metal transport genes has been previously reported in several LAB including L. lactis and S. thermophilus strains [43-48]. In our opinion the high number of metal transport associated genes in S. macedonicus was an unexpected observation and further investigation is required regarding their physiological relevance.
Distribution of virulence factors (VFs) within species of the SBSEC
One of the main goals behind the genome sequencing of S. macedonicus was to clarify its pathogenic potential. Unfortunately, despite the well-known association of S. bovis with human disease, especially endocarditis and colon cancer, there is very little knowledge about the pathogenicity mechanisms employed by members of the SBSEC. In Table 5 we have gathered genes previously assigned as potential VFs in SBSEC. The available studies have shed some light on the ability of S. gallolyticus to colonize host tissues, a step that is considered as a prerequisite for the initiation of the infection by this bacterium. Streptococcus gallolyticus UCN34 contains three pilus gene clusters which may mediate binding to the extracellular matrix (ECM), similarly to the clinical isolate TX20005 whose genome is partially characterized [25,49]. The pil1 and pil3 of strain UCN34 have been found identical to the acb-sbs7-srtC1 and sbs15-sbs14-srtC3 loci of strain TX20005, respectively, but their additional predicted pilus gene cluster (i.e. pil2 vs. sbs12-sbs11-srtC2) was only distantly related [25]. While all three strains of S. gallolyticus carry the three pilus loci (as found in strain UCN34), S. macedonicus, S. pasteurianus and S. infantarius carry only the pil3 locus. Functional analysis indicated that pil1 is a crucial factor of S. gallolyticus UCN34 for binding to ECM, especially to collagen [18]. The preference of S. gallolyticus to bind to collagen is of particular importance, since it may allow the adherence of the bacterium to the collagen-rich surfaces of damaged heart valves and (pre)cancerous sites [50]. Besides the pilus loci, additional MSCRAMM (microbial surface recognizing adhesive matrix molecules) proteins have been predicted in S. gallolyticus, most of which are either absent or preudogenes in S. macedonicus, S. pasteurianus and S. infantarius (Table 5) [49]. The cell surface protein antigen c (PAc) also appears exclusively in the S. gallolyticus genomes, sometimes in more than one copy. Only the surface-exposed histone-like protein A (HlpA) and the autolysin (AtlA) are universally conserved in the SBSEC. HlpA has been shown to be a major heparin-binding protein regulating the ability of S. gallolyticus adherence to the heparan sulfate proteoglycans at the colon tumor cell surface [51]. AtlA is a fibronectin-binding protein which is a VF of S. mutans associated with infective endocarditis [52]. Furthermore, S. gallolyticus UCN34 carries loci for the biosynthesis of insoluble glucan polymers from sucrose and the synthesis of hemicellulose [25]. Insoluble glucan polymers may contribute to feedlot bloat in cattle [25], while hemicellulose could play a role in biofilm formation [53]. It is possible that the production of these polymers may vary among strains of S. gallolyticus (Table 5). Streptococcus macedonicus is devoid of the biosynthetic gene cluster of glucan, while the hemicellulose synthesis operon seems to be comprised of pseudogenes. Similarly, S. pasteurianus and S. infantarius seem to be also unable to synthesize both sugar polymers, either due to full or partial absence of the genetic loci.
Table 5.
Genes in the Streptococcus bovis / Streptococcus equinus complex identified as putative virulence factors
| Virulence factor | Gene | S. gallolyticus UCN34 | S . gallolyticus ATCC 43143 | S . gallolyticus ATCC BAA-2069 | S. pasteurianus ATCC 43144 | S. macedonicus ACA-DC 198 | S . infantarius CJ18 |
|---|---|---|---|---|---|---|---|
| Pilus 1 (pil1) |
acb |
GALLO_2179 |
SGGB_2211 |
SGGBAA2069_c21760 |
- (a) |
- |
- |
| |
sbs7 |
GALLO_2178 |
SGGB_2210 |
SGGBAA2069_c21750 |
SGPB_1938 (p) |
- |
- |
| |
srtC1 |
GALLO_2177 |
SGGB_2209 |
SGGBAA2069_c21740 |
- |
- |
Sinf_1876 |
| Pilus 2 (pil2) |
- |
GALLO_1570 |
SGGB_1568 |
SGGBAA2069_c15960 |
- |
- |
- |
| |
- |
GALLO_1569 |
SGGB_1567 |
SGGBAA2069_c15950 |
- |
- |
- |
| |
- |
GALLO_1568 |
SGGB_1566 |
SGGBAA2069_c15940 |
- |
- |
- |
| Pilus 3 (pil3) |
sbs15 |
GALLO_2040 |
SGGB_2022 |
SGGBAA2069_c19980 |
SGPB_1847 |
SMA_1939 |
Sinf_1744 |
| |
sbs14 |
GALLO_2039 |
SGGB_2021 |
SGGBAA2069_c19970 |
SGPB_1846 |
SMA_1938 |
Sinf_1743 |
| |
srtC3 |
GALLO_2038 |
SGGB_2020 |
SGGBAA2069_c19960 |
SGPB_1845 |
SMA_1937 |
Sinf_1742 |
| Cell envelope proteinase (lactocepin) |
sbs6 |
GALLO_0748 |
SGGB_0730 |
SGGBAA2069_c07210 |
- |
- |
Sinf_0588 |
| Fructan hydrolase |
sbs10 |
GALLO_0112 |
SGGB_0110 |
SGGBAA2069_c01280 |
- |
- |
- |
| Collagen adhesin |
sbs13 |
GALLO_2032 |
SGGB_2016 |
SGGBAA2069_c19910 |
SGPB_1839 (p) |
SMA_1932 (s) |
Sinf_1737 (p) |
| |
|
|
|
|
SGPB_1840 (p) |
SMA_1933 (p) |
|
| |
|
|
|
|
|
SMA_1934 (s) |
|
| Collagen adhesin |
sbs16 |
GALLO_0577 |
SGGB_0544 |
SGGBAA2069_c05110 |
- |
- |
- |
| Cell surface protein antigen C (PAc) |
- |
GALLO_1675 |
SGGB_0154 |
SGGBAA2069_c13880 |
- |
- |
- |
| |
|
|
SGGB_1687 |
SGGBAA2069_c20560 |
|
|
|
| Surrface-exposed histone-like protein A |
hlpA |
GALLO_0636 |
SGGB_0611 |
SGGBAA2069_c05790 |
SGPB_0505 |
SMA_0597 |
Sinf_0517 |
| Autolysin |
atlA |
GALLO_1368 |
SGGB_1362 |
SGGBAA2069_c13580 |
SGPB_1289 |
SMA_1299 |
Sinf_1186 (t) |
| Glucan biosynthesis gene cluster |
- |
GALLO_1052 |
- |
SGGBAA2069_c10370 |
- |
- |
- |
| |
- |
GALLO_1053 |
SGGB_1042 |
SGGBAA2069_c10380 |
- |
- |
- |
| |
rggA |
GALLO_1054 |
SGGB_1043 |
SGGBAA2069_c10390 |
- |
- |
Sinf_0876 |
| |
gtfA |
GALLO_1055 |
SGGB_1044 |
SGGBAA2069_c10400 |
- |
- |
Sinf_0877 |
| |
rggB |
GALLO_1056 |
SGGB_1045 |
SGGBAA2069_c10410 |
- |
- |
- |
| |
gtfB |
GALLO_1057 |
SGGB_1046 |
SGGBAA2069_c10420 |
- |
- |
- |
| |
sbs2/gbpC |
GALLO_1058 |
SGGB_1047 |
SGGBAA2069_c10430 |
- |
SMA_0989 (p) |
- |
| |
|
|
|
|
|
SMA_0990 (s) |
|
| |
|
|
|
|
|
SMA_0991 (s) |
|
| Hemicellulose biosynthesis gene cluster |
- |
GALLO_0364 |
SGGB_0392 |
SGGBAA2069_c03530 |
- |
SMA_0392 (p) |
Sinf_0344 |
| |
- |
GALLO_0365 |
SGGB_0393 (p) |
SGGBAA2069_c03540 (s) |
- |
SMA_0393 (p) |
- |
| |
|
|
SGGB_0394 (p) |
SGGBAA2069_c03550 (s) |
|
|
|
| |
- |
GALLO_0366 |
SGGB_0395 |
SGGBAA2069_c03560 |
- |
SMA_0394 (p) |
Sinf_0345 |
| |
|
|
|
|
|
|
Sinf_0346 (s) |
| |
- |
GALLO_0367 |
SGGB_0396 |
SGGBAA2069_c03570 |
- |
- |
- |
| Hemolysin TLY |
- |
GALLO_0630 |
SGGB_0605 |
SGGBAA2069_c05730 |
SGPB_0499 |
SMA_0591 |
Sinf_0511 |
| Hemolysin III |
- |
GALLO_1262 |
SGGB_1256 |
SGGBAA2069_c12530 |
SGPB_1172 |
SMA_1191 |
Sinf_1093 |
| Hemolysin A family protein |
- |
GALLO_1799 |
SGGB_1786 |
SGGBAA2069_c17570 |
SGPB_1603 |
SMA_1706 |
Sinf_1530 |
| Exfoliative toxin B |
- |
- |
- |
- |
- |
- |
Sinf_0933 |
| Macrophage infectivity potentiator protein | - | - | - | - | - | - | Sinf_0931 |
(a) Not found; (p) Pseudogenes; (s) Split CDSs corresponding to fragments of the original gene not yet characterized as pseudogenes; (t) Truncated.
More genes whose products may be implicated in other interactions with the host cells beyond adherence could be identified. Despite the fact that the SBSEC members are considered non-hemolytic (as members of the group D streptococci), S. gallolyticus ATCC BAA-2069 has been reported to cause alpha-hemolysis on Schaedler Agar with 5% sheep blood [54]. Three hemolysins are conserved among the SBSEC members (Table 5). Sequence analysis of Sinf_1513 and Sinf_1683, also annotated as hemolysin genes, was not supportive of a hemolysin protein product (data not shown). Apart from hemolysins, a putative exfoliative toxin B (Sinf_0933) and a macrophage infectivity potentiator protein (Sinf_0931) are present in the S. infantarius genome [20]. Similar genes can be found in S. thermophilus strains but not in the other SBSEC species and in our opinion functional analysis is required to verify these annotations.
In order to expand our investigation for putative pathogenicity traits, we screened the genomes of S. macedonicus and its related SBSEC species using the VFDB (virulence factors database) [55] and the genes determined to encode putative VFs during this analysis are presented in Additional file 11: Table S7. Current results of comparative pathogenomics have allowed the classification of available streptococcal VFs in nine categories, i.e. adhesion factors, DNases, exoenzymes, immune evasion factors, immunoreactive antigens, factors involved in metal transport, proteases, superantigens and toxins [56]. The general profile of VFs for the six streptococci under investigation was rather similar and we determined a number of previously unidentified potential VFs dispersed among all or some of the SBSEC members. Several of these genes coding for putative VFs like the agglutinin receptor, the fibronectin/fibrinogen-binding protein (fbp54/pavA), the lipoprotein rotamase A (slrA), the plasmin receptor/GAPDH multifunctional protein, the streptococcal enolase exoenzyme, the pneumococcal surface antigen A and specific proteases (i.e. cppA, htrA/degP and tig/ropA) have been experimentally correlated with the virulence of pathogenic streptococci beyond SBSEC members [57-67]. Some genes were also involved in the production of a capsule that enables bacterial cells to evade phagocytosis (Additional file 11: Table S7) [68]. According to our analysis, all SBSEC streptococci carry a main gene cluster spanning practically the same position in the chromosome that could be involved in the biosynthesis of a capsule (Additional file 12: Figure S5). Even though the cps clusters are identical between S. gallolyticus UCN34 and ATCC BAA-2069 [54], multiple sequence alignment indicates significant structural diversity in the rest of the strains. The existence of dispersed pseudogenes in the gene clusters of S. infantarius and S. macedonicus (e.g. SMA_0865 and SMA_0866) may prohibit the production of capsule substances. It should be emphasized that the strains of the SBSEC missed hits in several major categories of streptococcal VFs (e.g. DNases, immunoreactive antigens, superantigens and toxins) supporting a reduced pathogenic potential for the SBSEC in general.
Conclusions
In this study we presented the analysis of the first complete genome sequence of a dairy isolate of S. macedonicus. While comparative analysis among specific subgroups of the SBSEC species has been previously presented [20,22,25,54], comparative genomics of the six complete genome sequences was missing. Most importantly, the inclusion of S. macedonicus into this analysis provided a better opportunity to assess niche adaptation of the SBSEC species that was so far limited by the presence of only one dairy isolate (i.e. S. infantarius CJ18) among four clinical strains.
Our findings clearly support two distinct evolutionary patterns within the SBSEC. On the one hand, S. gallolyticus is a species without apparent genome decay and the available genomes suggest that it is a robust bacterium able to thrive in the rumen of herbivores. On the other hand, the remaining SBSEC species, i.e. S. macedonicus, S. pasteurianus and S. infantarius exhibit decreased genome sizes accompanied by increased percentages of potential pseudogenes due to extensive genome decay, suggesting adaptation to nutrient-rich environments. This does not necessarily mean that the environment to which the three species have been adapted is the same. The three species appear with a reduced ability to catabolize complex plant carbohydrates and to detoxify substances met in the rumen, which indicates that they must have deviated from this niche. It has been proposed that S. pasteurianus may now reside in the human gut [22], while S. infantarius presents adaptations to milk [20]. Streptococcus macedonicus also possesses traits that may contribute to growth in the dairy environment, like the extra lactose gene cluster and its proteolytic system. However, all SBSEC strains, including clinical isolates, seem to be competent in the metabolism of lactose and galactose or the degradation of milk proteins. Taking into account these shared characteristics of all SBSEC species, it is tempting to speculate that their common ancestor may have been able to grow in milk.
In our opinion, several genome traits per se suggest adaptation of S. macedonicus to milk. This hypothesis is also supported by the predicted interspecies interactions of S. macedonicus with other bacteria. As it has been recently reported for S. infantarius[20], the S. macedonicus genome may have acquired genes originating from L. lactis and S. thermophilus through HGT. The predicted exposure of S. macedonicus to S. thermophilus phages, based on our CRISPR sequence analysis, is also in favor of this theory. No such evidence was found for the rest of the SBSEC members apart from S. infantarius. These findings are in accordance with the frequent isolation of S. macedonicus from dairy products [13] and the prevalence of S. infantarius in certain African fermented milks [20]. One additional question that arises is whether S. macedonicus and S. infantarius are specialized dairy microbes like S. thermophilus. We believe that the available data does not support this idea. Traits of milk adaptation have been shown to be strain-specific in S. infantarius[20]. In addition, the genome size of S. macedonicus is significantly larger, containing a higher number of functional genes in comparison to S. thermophilus. Streptococcus macedonicus and S. infantarius may thus represent intermediate evolutionary stages analogous to those followed by the ancestors of S. thermophilus before it became today’s starter culture.
Thus, the safety concerns raised from the presence of SBSEC members in foods remain, even if reports implicating S. macedonicus with disease are rather scarce [69,70]. Our comparative genomic analysis showed that both S. macedonicus and S. infantarius miss several VFs that are highly conserved in S. gallolyticus. However, the interpretation of these findings becomes complicated as the available genome of the human blood isolate S. pasteurianus ATCC 43144 also exhibited diminished traits of pathogenicity similarly to the two dairy SBSEC members. Overall, our analysis provides evidence in agreement with the clinical perception that the members of the SBSEC are lower grade streptococcal pathogens [10]. In terms of food safety, the dairy SBSEC could thus constitute a risk factor similar to the presence of enterococci that are widely found in fermented products, but cause no major problem for the average healthy and adult consumer. Nevertheless, it is the correlation of the SBSEC microorganisms with human endocarditis and colon cancer in particular that may require special considerations. For example, it has been proposed that members of the SBSEC like S. gallolyticus may be part of the etiology of colon cancer by causing chronic inflammation [10]. In order to assess the pathogenicity of this group of streptococci, more research is needed on the specific mechanisms employed by SBSEC members to cause disease. More comparative and functional genomics studies comprising SBSEC genomes are necessary that will cover additional species of the complex, like the recently sequenced Streptococcus lutetiensis[71]. New clinico-epidemiological studies should also be undertaken in view of the most recent changes in the taxonomy of the SBSEC complex [72]. In the meantime, assuming the worse case scenario, we propose that the presence of SBSEC members including S. macedonicus and S. infantarius in foods should be avoided until their pathogenicity status is resolved.
Methods
Sequencing and annotation of the genome of Streptococcus macedonicus ACA-DC 198
The genome of S. macedonicus ACA-DC 198 was sequenced and annotated as described previously [19]. In brief, we employed a sequencing strategy involving shotgun/paired-end pyrosequencing and shotgun Illumina sequencing with the 454 GS-FLX (Roche Diagnostics, Basel, Switzerland) and the Hiseq 2000 (Illumina, San Diego, CA), respectively. Sequences were assembled in two contigs corresponding to the complete genome sequence and the pSMA198 plasmid of S. macedonicus. The hybrid assembly was validated against an NheI optical map of the S. macedonicus genome generated at OpGen Technologies, Inc. (Madison, WI). The genome was annotated using the RAST [73] and the Basys [74] pipelines. Predictions of the two pipelines were compiled into a single annotation file after manual curation in the Kodon software environment (Applied Maths N.V., Sint-Martens-Latem, Belgium). Final corrections and quality assessment of the annotation were performed with the GenePRIMP pipeline [21]. GenePRIMP was also used for the identification of putative pseudogenes. The circular map of the S. macedonicus genome was generated by the DNAPlotter software [75].
Comparative genomics of Streptococcus macedonicus ACA-DC 198 against related members of the SBSEC
The complete genome sequence of S. macedonicus was compared to those of S. gallolyticus strains UCN34, ATCC 43143 and ATCC BAA-2069, S. pasteurianus ATCC 43144 and S. infantarius CJ18 using a variety of tools. In order to visualize conserved genomic regions or chromosomal rearrangements, whole genome sequence alignments were performed by progressiveMAUVE [24]. Estimation of the differential gene content of the genomes, as well as whole genome phylogeny of streptococci was carried out within the EDGAR software framework [23]. Venn diagrams were designed with the VennDiagram package in R [76]. The glycobiome of the SBSEC members was determined based on the pre-computed data available in the CAZy database [26].
Additional analysis
Sequence similarity searches were performed with the BLAST suite [77]. Whenever necessary, protein sequences were analyzed in the CDD [78]. Figures showing similarity of gene clusters were constructed with the Easyfig comparison visualizer [79]. Potential VFs included in the VFDB [55] were identified in the SBSEC genomes with mpiBLAST, as implemented in the mGenomeSubtractor website [80]. In brief, the entire VFDB was uploaded as the reference sequence in the mGenomeSubtractor website and each genome was used as the query sequence. Only hits with H-value homology score > 0.6 were considered significant. CRISPRs were analyzed by the tools available in the CRISPRcompar web-service [81]. A general bit score cutoff value of 42.0 was applied during BLASTN of CRISPR spacers. GIs were identified and visualized by the IslandViewer application that utilizes three different prediction tools (i.e. IslandPick, SIGI-HMM and IslandPath-DIMOB) relying on either sequence composition or comparative genomics [36]. Genomic regions of RM systems were determined in the REBASE genomes database [82].
Availability of supporting data
The data set supporting the phylogenetic tree presented in Additional file 1: Figure S1 of this article is available in the [Dryad] repository, [unique persistent identifier doi:10.5061/dryad.7d039 and hyperlink to datasets in http://datadryad.org/]. Additional data sets supporting the results of this article are included within the article and its additional files.
Abbreviations
SBSEC: Streptococcus bovis/Streptococcus equinus complex; LAB: Lactic acid bacteria; GRAS: Generally regarded as safe; QPS: Qualified presumption of safety; GIT: Gastrointestinal tract; GAS: Group A streptococci; GBS: Group B streptococci; CDS: Coding DNA sequence; LCB: Local collinear block; HGT: Horizontal gene transfer; PEP-PTS: Phosphoenolpyruvate-dependent phosphotransferase system; CEP: Cell-envelope associated proteinase; PMF: Proton motive force; CRISPR: Clustered regularly interspaced short palindromic repeats; GI: Genomic island; RM: Restriction modification; VF: Virulence factor; ECM: Extracellular matrix; MSCRAMM: Microbial surface recognizing adhesive matrix molecule; CDD: Conserved domain database; VFDB: Virulence factors database.
Competing interests
SF is an employee of Genoscreen and PS is a consultant of the same company. BP is also an employee of Applied Maths NV.
Authors’ contributions
KP initiated the project, performed the genome analysis and comparative genomics and wrote the manuscript; RA isolated the genomic DNA and critically revised the manuscript; EM assisted in the genome analysis and helped to draft the manuscript; JB performed analysis with the EDGAR software; NCP helped with data interpretation and critically revised the manuscript; SJH helped with data interpretation and critically revised the manuscript; SF performed the sequencing of the genome and participated in the genome assembly; PR helped with data interpretation and critically revised the manuscript; PS helped with data interpretation and critically revised the manuscript; BP initiated the project, helped with data interpretation and critically revised the manuscript; ET initiated the project, helped with data interpretation and critically revised the manuscript. All authors have read and approved the final manuscript.
Supplementary Material
Whole genome phylogeny of the Streptococcus genus. The phylogenetic tree was constructed using the EDGAR tool based on complete genome sequences of streptococci. The branch of the members of the Streptococcus bovis/Streptococcus equinus complex (SBSEC) is delimited by a bracket.
Core genome analysis among Streptococcus gallolyticus UCN34, Streptococcus gallolyticus ATCC 43143, Streptococcus gallolyticus ATCC BAA-2069, Streptococcus macedonicus ACA-DC 198 and Streptococcus pasteurianus ATCC 43144 calculated using the EDGAR software.
Core genome analysis among Streptococcus gallolyticus ATCC 43143, Streptococcus infantarius CJ18, Streptococcus macedonicus ACA-DC 198 and Streptococcus pasteurianus ATCC 43144 calculated using the EDGAR software. In this analysis S. gallolyticus ATCC 43143 was selected as a representative of the S. gallolyticus species, since it has the longest genome size among the three sequenced strains.
Glycobiome analysis of Streptococcus gallolyticus UCN34, Streptococcus gallolyticus ATCC 43143, Streptococcus gallolyticus ATCC BAA-2069, Streptococcus pasteurianus ATCC 43144, Streptococcus macedonicus ACA-DC 198 and Streptococcus infantarius CJ18 using the CAZy database.
Circular maps of the Streptococcus bovis/Streptococcus equinus complex genomes highlighting the regions corresponding to genomic islands (GIs). GIs are coloured within the circular maps according to the tool that predicted each one of them: green, orange and blue were predicted with IslandPick, SIGI-HMM and IslandPath-DIMOB, respectively. The integrated GIs are presented at the periphery of the map in red colour. The black line plot represents the GC content (%) of the genomic sequences. Numbering of the GIs for each genome starts from the first GI found after position 0 of the genome in a clockwise direction.
Analysis of the genomic island (GI) 4 of Streptococcus macedonicus ACA-DC 198 presented as an example of a GI potentially originating from multiple donors. In the graphical summary of the BLASTN results arrows indicate the best BLASTN hits with > 90% sequence identity corresponding to: a. Streptococcus thermophilus MN-ZLW-002 genomic sequence (96% sequence identity); b. Lactococcus garvieae 21881 plasmid pGL3 sequence (98% sequence identity); c. Streptococcus intermedius B196 genomic sequence (96% sequence identity); d. Streptococcus thermophilus MN-ZLW-002 genomic sequence (99% sequence identity) and e. Streptococcus thermophilus MN-ZLW-002 genomic sequence (99% sequence identity).
Genes within each integrated GI of Streptococcus macedonicus ACA-DC 198 as determined by IslandViewer.
The singleton genes of Streptococcus macedonicus ACA-DC 198 calculated against the other members of the Streptococcus bovis/Streptococcus equinus complex (SBSEC) using the EDGAR software. The singleton genes of S. macedonicus were calculated twice, taking or not into account the genome of S. infantarius. Thus, genes shared only by S. macedonicus and S. infantarius among the SBSEC members also appear in the table.
Circular maps of the Streptococcus bovis/Streptococcus equinus complex genomes highlighting the regions corresponding to restriction modification systems (RMs). RMs are presented as predicted in the REBASE database. Colours and symbols are exemplified at the bottom of the figure.
Comparison of the CRISPR/Cas systems among members of the Streptococcus bovis/Streptococcus equinus complex using CRISPRcompar.
Genes in the Streptococcus bovis/Streptococcus equinus complex identified as virulence factors within the VFDB.
Multiple sequence alignment of the capsule biosynthetic gene cluster found in the genomes of the Streptococcus bovis/Streptococcus equinus complex after BLASTN analysis. Grey shading represents the % identity among the nucleotide sequences according to the colour gradient presented at the lower right corner of the figure. Potential pseudogenes are marked with a "p".
Contributor Information
Konstantinos Papadimitriou, Email: kpapadimitriou@aua.gr.
Rania Anastasiou, Email: ranastasiou@aua.gr.
Eleni Mavrogonatou, Email: elmavro@bio.demokritos.gr.
Jochen Blom, Email: jblom@cebitec.uni-bielefeld.de.
Nikos C Papandreou, Email: npapand@biol.uoa.gr.
Stavros J Hamodrakas, Email: shamodr@biol.uoa.gr.
Stéphanie Ferreira, Email: stephanie.ferreira@genoscreen.fr.
Pierre Renault, Email: pierre.renault@jouy.inra.fr.
Philip Supply, Email: philip.supply@ibl.fr.
Bruno Pot, Email: bruno.pot@ibl.fr.
Effie Tsakalidou, Email: et@aua.gr.
Acknowledgements
The present work was cofinanced by the European Social Fund and the National resources EPEAEK and YPEPTH through the Thales project.
References
- Konings WN, Kok J, Kuipers OP, Poolman B. Lactic acid bacteria: the bugs of the new millennium. Curr Opin Microbiol. 2000;15(3):276–282. doi: 10.1016/S1369-5274(00)00089-8. [DOI] [PubMed] [Google Scholar]
- Masood MI, Qadir MI, Shirazi JH, Khan IU. Beneficial effects of lactic acid bacteria on human beings. Crit Rev Microbiol. 2011;15(1):91–98. doi: 10.3109/1040841X.2010.536522. [DOI] [PubMed] [Google Scholar]
- Donohue DC, Gueimonde M. In: Lactic Acid Bacteria: Microbiological and Functional Aspects. 4. Lahtinen S, Salminen S, von Wright A, Ouwehand AC, editor. Boca Raton: CRC Press Taylor & Francis Group; 2012. Some Considerations for the Safety of Novel Probiotic Bacteria. [Google Scholar]
- Woodford N, Livermore DM. Infections caused by Gram-positive bacteria: a review of the global challenge. J Infect. 2009;15(Suppl 1):S4–S16. doi: 10.1016/S0163-4453(09)60003-7. [DOI] [PubMed] [Google Scholar]
- Facklam R. What happened to the streptococci: overview of taxonomic and nomenclature changes. Clin Microbiol Rev. 2002;15(4):613–630. doi: 10.1128/CMR.15.4.613-630.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobbs AH, Lamont RJ, Jenkinson HF. Streptococcus adherence and colonization. Microbiol Mol Biol Rev. 2009;15(3):407–450. doi: 10.1128/MMBR.00014-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolotin A, Quinquis B, Renault P, Sorokin A, Ehrlich SD, Kulakauskas S, Lapidus A, Goltsman E, Mazur M, Pusch GD, Fonstein M, Overbeek R, Kyprides N, Purnelle B, Prozzi D, Ngui K, Masuy D, Hancy F, Burteau S, Boutry M, Delcour J, Goffeau A, Hols P. Complete sequence and comparative genome analysis of the dairy bacterium Streptococcus thermophilus. Nat Biotechnol. 2004;15(12):1554–1558. doi: 10.1038/nbt1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hols P, Hancy F, Fontaine L, Grossiord B, Prozzi D, Leblond-Bourget N, Decaris B, Bolotin A, Delorme C, Dusko Ehrlich S, Guedon E, Monnet V, Renault P, Kleerebezem M. New insights in the molecular biology and physiology of Streptococcus thermophilus revealed by comparative genomics. FEMS Microbiol Rev. 2005;15(3):435–463. doi: 10.1016/j.femsre.2005.04.008. [DOI] [PubMed] [Google Scholar]
- Jans C, Lacroix C, Meile L. A novel multiplex PCR/RFLP assay for the identification of Streptococcus bovis/Streptococcus equinus complex members from dairy microbial communities based on the 16S rRNA gene. FEMS Microbiol Lett. 2012;15(2):144–150. doi: 10.1111/j.1574-6968.2011.02443.x. [DOI] [PubMed] [Google Scholar]
- Abdulamir AS, Hafidh RR, Abu Bakar F. The association of Streptococcus bovis/gallolyticus with colorectal tumors: the nature and the underlying mechanisms of its etiological role. J Exp Clin Cancer Res. 2011;15:11. doi: 10.1186/1756-9966-30-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galdy S, Nastasi G. Streptococcus bovis endocarditis and colon cancer: myth or reality? A case report and literature review. BMJ Case Rep. 2012;15:1–2. doi: 10.1136/bcr-2012-006961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrera P, Kwon YM, Ricke SC. Ecology and pathogenicity of gastrointestinal Streptococcus bovis. Anaerobe. 2009;15(1–2):44–54. doi: 10.1016/j.anaerobe.2008.11.003. [DOI] [PubMed] [Google Scholar]
- De Vuyst L, Tsakalidou E. Streptococcus macedonicus, a multi-functional and promising species for dairy fermentations. Int Dairy J. 2008;15(5):476–485. doi: 10.1016/j.idairyj.2007.10.006. [DOI] [Google Scholar]
- Maragkoudakis PA, Papadelli M, Georgalaki M, Panayotopoulou EG, Martinez-Gonzalez B, Mentis AF, Petraki K, Sgouras DN, Tsakalidou E. In vitro and in vivo safety evaluation of the bacteriocin producer Streptococcus macedonicus ACA-DC 198. Int J Food Microbiol. 2009;15(1–2):141–147. doi: 10.1016/j.ijfoodmicro.2009.05.012. [DOI] [PubMed] [Google Scholar]
- Tsakalidou E, Zoidou E, Pot B, Wassill L, Ludwig W, Devriese LA, Kalantzopoulos G, Schleifer KH, Kersters K. Identification of streptococci from Greek Kasseri cheese and description of Streptococcus macedonicus sp. nov. Int J Syst Bacteriol. 1998;15(Pt 2):519–527. doi: 10.1099/00207713-48-2-519. [DOI] [PubMed] [Google Scholar]
- Schlegel L, Grimont F, Ageron E, Grimont PA, Bouvet A. Reappraisal of the taxonomy of the Streptococcus bovis/Streptococcus equinus complex and related species: description of Streptococcus gallolyticus subsp. gallolyticus subsp. nov., S. gallolyticus subsp. macedonicus subsp. nov. and S. gallolyticus subsp. pasteurianus subsp. nov. Int J Syst Evol Microbiol. 2003;15(Pt 3):631–645. doi: 10.1099/ijs.0.02361-0. [DOI] [PubMed] [Google Scholar]
- Whiley RA, Kilian M. International committee on systematics of prokaryotes subcommittee on the taxonomy of staphylococci and streptococci: minutes of the closed meeting, 31 July 2002, paris France. Int J Syst Evol Microbiol. 2003;15(3):915–917. doi: 10.1099/ijs.0.02589-0. [DOI] [Google Scholar]
- Danne C, Entenza JM, Mallet A, Briandet R, Debarbouille M, Nato F, Glaser P, Jouvion G, Moreillon P, Trieu-Cuot P, Dramsi S. Molecular characterization of a Streptococcus gallolyticus genomic island encoding a pilus involved in endocarditis. J Infect Dis. 2011;15(12):1960–1970. doi: 10.1093/infdis/jir666. [DOI] [PubMed] [Google Scholar]
- Papadimitriou K, Ferreira S, Papandreou NC, Mavrogonatou E, Supply P, Pot B, Tsakalidou E. Complete genome sequence of the dairy isolate Streptococcus macedonicus ACA-DC 198. J Bacteriol. 2012;15(7):1838–1839. doi: 10.1128/JB.06804-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jans C, Follador R, Hochstrasser M, Lacroix C, Meile L, Stevens MJ. Comparative genome analysis of Streptococcus infantarius subsp. infantarius CJ18, an African fermented camel milk isolate with adaptations to dairy environment. BMC Genomics. 2013;15:200. doi: 10.1186/1471-2164-14-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pati A, Ivanova NN, Mikhailova N, Ovchinnikova G, Hooper SD, Lykidis A, Kyrpides NC. GenePRIMP: a gene prediction improvement pipeline for prokaryotic genomes. Nat Methods. 2010;15(6):455–457. doi: 10.1038/nmeth.1457. [DOI] [PubMed] [Google Scholar]
- Lin IH, Liu TT, Teng YT, Wu HL, Liu YM, Wu KM, Chang CH, Hsu MT. Sequencing and comparative genome analysis of two pathogenic Streptococcus gallolyticus subspecies: genome plasticity, adaptation and virulence. PLoS One. 2011;15(5):e20519. doi: 10.1371/journal.pone.0020519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blom J, Albaum SP, Doppmeier D, Puhler A, Vorholter FJ, Zakrzewski M, Goesmann A. EDGAR: a software framework for the comparative analysis of prokaryotic genomes. BMC Bioinforma. 2009;15:154. doi: 10.1186/1471-2105-10-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darling AE, Mau B, Perna NT. ProgressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One. 2010;15(6):e11147. doi: 10.1371/journal.pone.0011147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusniok C, Couve E, Da Cunha V, El Gana R, Zidane N, Bouchier C, Poyart C, Leclercq R, Trieu-Cuot P, Glaser P. Genome sequence of Streptococcus gallolyticus: insights into its adaptation to the bovine rumen and its ability to cause endocarditis. J Bacteriol. 2010;15(8):2266–2276. doi: 10.1128/JB.01659-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombard V, Golaconda Ramulu H, Drula E, Coutinho PM, Henrissat B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014;15(1):D490–D495. doi: 10.1093/nar/gkt1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crost EH, Tailford LE, Le Gall G, Fons M, Henrissat B, Juge N. Utilisation of mucin glycans by the human gut symbiont Ruminococcus gnavus is strain-dependent. PLoS One. 2013;15(10):e76341. doi: 10.1371/journal.pone.0076341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ficko-Blean E, Boraston AB. Insights into the recognition of the human glycome by microbial carbohydrate-binding modules. Curr Opin Struct Biol. 2012;15(5):570–577. doi: 10.1016/j.sbi.2012.07.009. [DOI] [PubMed] [Google Scholar]
- Liu M, Bayjanov JR, Renckens B, Nauta A, Siezen RJ. The proteolytic system of lactic acid bacteria revisited: a genomic comparison. BMC Genomics. 2010;15:36. doi: 10.1186/1471-2164-11-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jans C, Gerber A, Bugnard J, Njage PM, Lacroix C, Meile L. Novel Streptococcus infantarius subsp. infantarius variants harboring lactose metabolism genes homologous to Streptococcus thermophilus . Food Microbiol. 2012;15(1):33–42. doi: 10.1016/j.fm.2012.02.001. [DOI] [PubMed] [Google Scholar]
- Fernandez-Espla MD, Garault P, Monnet V, Rul F. Streptococcus thermophilus cell wall-anchored proteinase: release, purification, and biochemical and genetic characterization. Appl Environ Microbiol. 2000;15(11):4772–4778. doi: 10.1128/AEM.66.11.4772-4778.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dandoy D, Fremaux C, de Frahan MH, Horvath P, Boyaval P, Hols P, Fontaine L. The fast milk acidifying phenotype of Streptococcus thermophilus can be acquired by natural transformation of the genomic island encoding the cell-envelope proteinase PrtS. Microb Cell Fact. 2011;15(Suppl 1):S21. doi: 10.1186/1475-2859-10-S1-S21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calasso M, Gobbetti M. In: Encyclopedia of Dairy Sciences. 2. Fuquay JW, editor. San Diego: cademic Press; 2011. Lactic Acid Bacteria | Lactobacillus spp.: Other Species; pp. 125–131. [Google Scholar]
- El Qaidi S, Yang J, Zhang JR, Metzger DW, Bai G. The vitamin B(6) biosynthesis pathway in Streptococcus pneumoniae is controlled by pyridoxal 5′-phosphate and the transcription factor PdxR and has an impact on ear infection. J Bacteriol. 2013;15(10):2187–2196. doi: 10.1128/JB.00041-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeBlanc JG, Laino JE, del Valle MJ, Vannini V, van Sinderen D, Taranto MP, de Valdez GF, de Giori GS, Sesma F. B-group vitamin production by lactic acid bacteria–current knowledge and potential applications. J Appl Microbiol. 2011;15(6):1297–1309. doi: 10.1111/j.1365-2672.2011.05157.x. [DOI] [PubMed] [Google Scholar]
- Dhillon BK, Chiu TA, Laird MR, Langille MG, Brinkman FS. IslandViewer update: improved genomic island discovery and visualization. Nucleic Acids Res. 2013;15(Web Server issue):W129–W132. doi: 10.1093/nar/gkt394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgalaki M, Papadimitriou K, Anastasiou R, Pot B, Van Driessche G, Devreese B, Tsakalidou E. Macedovicin, the second food-grade lantibiotic produced by Streptococcus macedonicus ACA-DC 198. Food Microbiol. 2013;15(1):124–130. doi: 10.1016/j.fm.2012.09.008. [DOI] [PubMed] [Google Scholar]
- Georgalaki MD, Van Den Berghe E, Kritikos D, Devreese B, Van Beeumen J, Kalantzopoulos G, De Vuyst L, Tsakalidou E. Macedocin, a food-grade lantibiotic produced by Streptococcus macedonicus ACA-DC 198. Appl Environ Microbiol. 2002;15(12):5891–5903. doi: 10.1128/AEM.68.12.5891-5903.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgalaki M, Papadelli M, Chassioti E, Anastasiou R, Aktypis A, De Vuyst L, Van Driessche G, Devreese B, Tsakalidou E. Milk protein fragments induce the biosynthesis of macedocin, the lantibiotic produced by Streptococcus macedonicus ACA-DC 198. Appl Environ Microbiol. 2010;15(4):1143–1151. doi: 10.1128/AEM.00151-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Sullivan O, O’Callaghan J, Sangrador-Vegas A, McAuliffe O, Slattery L, Kaleta P, Callanan M, Fitzgerald GF, Ross RP, Beresford T. Comparative genomics of lactic acid bacteria reveals a niche-specific gene set. BMC Microbiol. 2009;15:50. doi: 10.1186/1471-2180-9-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szczepankowska AK, Górecki RK, Kołakowski P, Bardowski JK. In: Dairy Fermentation in R & D for Food, Health and Livestock Purposes. Kongo JM, editor. Croatia: InTech; 2013. Lactic Acid Bacteria Resistance to Bacteriophage and Prevention Techniques to Lower Phage Contamination. [Google Scholar]
- Marco MB, Moineau S, Quiberoni A. Bacteriophages and dairy fermentations. Bacteriophage. 2012;15(3):149–158. doi: 10.4161/bact.21868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solioz M, Mermod M, Abicht HKM, Mancini S. In: Stress Responses of Lactic Acid Bacteria. 1. Tsakalidou E, Papadimitriou K, editor. New York: Springer; 2011. Responses of Lactic Acid Bacteria to Heavy Metal Stress. [Google Scholar]
- Liu CQ, Khunajakr N, Chia LG, Deng YM, Charoenchai P, Dunn NW. Genetic analysis of regions involved in replication and cadmium resistance of the plasmid pND302 from Lactococcus lactis. Plasmid. 1997;15(2):79–90. doi: 10.1006/plas.1997.1301. [DOI] [PubMed] [Google Scholar]
- Magnani D, Barre O, Gerber SD, Solioz M. Characterization of the CopR regulon of Lactococcus lactis IL1403. J Bacteriol. 2008;15(2):536–545. doi: 10.1128/JB.01481-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirawski J, Hagens W, Fitzgerald GF, Van Sinderen D. Molecular characterization of cadmium resistance in Streptococcus thermophilus strain 4134: an example of lateral gene transfer. Appl Environ Microbiol. 2002;15(11):5508–5516. doi: 10.1128/AEM.68.11.5508-5516.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siezen RJ, Renckens B, van Swam I, Peters S, van Kranenburg R, Kleerebezem M, de Vos WM. Complete sequences of four plasmids of Lactococcus lactis subsp. cremoris SK11 reveal extensive adaptation to the dairy environment. Appl Environ Microbiol. 2005;15(12):8371–8382. doi: 10.1128/AEM.71.12.8371-8382.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fallico V, McAuliffe O, Fitzgerald GF, Ross RP. Plasmids of raw milk cheese isolate Lactococcus lactis subsp. lactis biovar diacetylactis DPC3901 suggest a plant-based origin for the strain. Appl Environ Microbiol. 2011;15(18):6451–6462. doi: 10.1128/AEM.00661-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sillanpaa J, Nallapareddy SR, Qin X, Singh KV, Muzny DM, Kovar CL, Nazareth LV, Gibbs RA, Ferraro MJ, Steckelberg JM, Weinstock GM, Murray BE. A collagen-binding adhesin, Acb, and ten other putative MSCRAMM and pilus family proteins of Streptococcus gallolyticus subsp. gallolyticus (Streptococcus bovis Group, biotype I) J Bacteriol. 2009;15(21):6643–6653. doi: 10.1128/JB.00909-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boleij A, Muytjens CM, Bukhari SI, Cayet N, Glaser P, Hermans PW, Swinkels DW, Bolhuis A, Tjalsma H. Novel clues on the specific association of Streptococcus gallolyticus subsp gallolyticus with colorectal cancer. J Infect Dis. 2011;15(8):1101–1109. doi: 10.1093/infdis/jiq169. [DOI] [PubMed] [Google Scholar]
- Boleij A, Schaeps RM, de Kleijn S, Hermans PW, Glaser P, Pancholi V, Swinkels DW, Tjalsma H. Surface-exposed histone-like protein a modulates adherence of Streptococcus gallolyticus to colon adenocarcinoma cells. Infect Immun. 2009;15(12):5519–5527. doi: 10.1128/IAI.00384-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung CJ, Zheng QH, Shieh YH, Lin CS, Chia JS. Streptococcus mutans autolysin AtlA is a fibronectin-binding protein and contributes to bacterial survival in the bloodstream and virulence for infective endocarditis. Mol Microbiol. 2009;15(4):888–902. doi: 10.1111/j.1365-2958.2009.06903.x. [DOI] [PubMed] [Google Scholar]
- Garcia B, Latasa C, Solano C, Garcia-del Portillo F, Gamazo C, Lasa I. Role of the GGDEF protein family in Salmonella cellulose biosynthesis and biofilm formation. Mol Microbiol. 2004;15(1):264–277. doi: 10.1111/j.1365-2958.2004.04269.x. [DOI] [PubMed] [Google Scholar]
- Hinse D, Vollmer T, Ruckert C, Blom J, Kalinowski J, Knabbe C, Dreier J. Complete genome and comparative analysis of Streptococcus gallolyticus subsp. gallolyticus, an emerging pathogen of infective endocarditis. BMC Genomics. 2011;15:400. doi: 10.1186/1471-2164-12-400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Xiong Z, Sun L, Yang J, Jin Q. VFDB 2012 update: toward the genetic diversity and molecular evolution of bacterial virulence factors. Nucleic Acids Res. 2012;15(Database issue):D641–D645. doi: 10.1093/nar/gkr989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VFDB. Comparison of pathogenomic composition of Streptococcus. [ http://www.mgc.ac.cn/cgi-bin/VFs/compvfs.cgi?Genus=Streptococcus]
- Demuth DR, Lammey MS, Huck M, Lally ET, Malamud D. Comparison of Streptococcus mutans and Streptococcus sanguis receptors for human salivary agglutinin. Microb Pathog. 1990;15(3):199–211. doi: 10.1016/0882-4010(90)90022-I. [DOI] [PubMed] [Google Scholar]
- Courtney HS, Li Y, Dale JB, Hasty DL. Cloning, sequencing, and expression of a fibronectin/fibrinogen-binding protein from group A streptococci. Infect Immun. 1994;15(9):3937–3946. doi: 10.1128/iai.62.9.3937-3946.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes AR, McNab R, Millsap KW, Rohde M, Hammerschmidt S, Mawdsley JL, Jenkinson HF. The pavA gene of Streptococcus pneumoniae encodes a fibronectin-binding protein that is essential for virulence. Mol Microbiol. 2001;15(6):1395–1408. doi: 10.1046/j.1365-2958.2001.02610.x. [DOI] [PubMed] [Google Scholar]
- Hermans PW, Adrian PV, Albert C, Estevao S, Hoogenboezem T, Luijendijk IH, Kamphausen T, Hammerschmidt S. The streptococcal lipoprotein rotamase A (SlrA) is a functional peptidyl-prolyl isomerase involved in pneumococcal colonization. J Biol Chem. 2006;15(2):968–976. doi: 10.1074/jbc.M510014200. [DOI] [PubMed] [Google Scholar]
- Terao Y, Yamaguchi M, Hamada S, Kawabata S. Multifunctional glyceraldehyde-3-phosphate dehydrogenase of Streptococcus pyogenes is essential for evasion from neutrophils. J Biol Chem. 2006;15(20):14215–14223. doi: 10.1074/jbc.M513408200. [DOI] [PubMed] [Google Scholar]
- Winram SB, Lottenberg R. The plasmin-binding protein Plr of group a streptococci is identified as glyceraldehyde-3-phosphate dehydrogenase. Microbiology. 1996;15(Pt 8):2311–2320. doi: 10.1099/13500872-142-8-2311. [DOI] [PubMed] [Google Scholar]
- Pancholi V, Fischetti VA. alpha-enolase, a novel strong plasmin(ogen) binding protein on the surface of pathogenic streptococci. J Biol Chem. 1998;15(23):14503–14515. doi: 10.1074/jbc.273.23.14503. [DOI] [PubMed] [Google Scholar]
- Tseng HJ, McEwan AG, Paton JC, Jennings MP. Virulence of Streptococcus pneumoniae: PsaA mutants are hypersensitive to oxidative stress. Infect Immun. 2002;15(3):1635–1639. doi: 10.1128/IAI.70.3.1635-1639.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angel CS, Ruzek M, Hostetter MK. Degradation of C3 by Streptococcus pneumoniae. J Infect Dis. 1994;15(3):600–608. doi: 10.1093/infdis/170.3.600. [DOI] [PubMed] [Google Scholar]
- Ibrahim YM, Kerr AR, McCluskey J, Mitchell TJ. Role of HtrA in the virulence and competence of Streptococcus pneumoniae. Infect Immun. 2004;15(6):3584–3591. doi: 10.1128/IAI.72.6.3584-3591.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyon WR, Caparon MG. Trigger factor-mediated prolyl isomerization influences maturation of the Streptococcus pyogenes cysteine protease. J Bacteriol. 2003;15(12):3661–3667. doi: 10.1128/JB.185.12.3661-3667.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadioglu A, Taylor S, Iannelli F, Pozzi G, Mitchell TJ, Andrew PW. Upper and lower respiratory tract infection by Streptococcus pneumoniae is affected by pneumolysin deficiency and differences in capsule type. Infect Immun. 2002;15(6):2886–2890. doi: 10.1128/IAI.70.6.2886-2890.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrero IA, Rouse MS, Piper KE, Alyaseen SA, Steckelberg JM, Patel R. Reevaluation of Streptococcus bovis endocarditis cases from 1975 to 1985 by 16S ribosomal DNA sequence analysis. J Clin Microbiol. 2002;15(10):3848–3850. doi: 10.1128/JCM.40.10.3848-3850.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malkin J, Kimmitt PT, Ou HY, Bhasker PS, Khare M, Deng Z, Stephenson I, Sosnowski AW, Perera N, Rajakumar K. Identification of Streptococcus gallolyticus subsp. macedonicus as the etiological agent in a case of culture-negative multivalve infective endocarditis by 16S rDNA PCR analysis of resected valvular tissue. J Heart Valve Dis. 2008;15(5):589–592. [PubMed] [Google Scholar]
- Jin D, Chen C, Li L, Lu S, Li Z, Zhou Z, Jing H, Xu Y, Du P, Wang H, Xiong Y, Zheng H, Bai X, Sun H, Wang L, Ye C, Gottschalk M, Xu J. Dynamics of fecal microbial communities in children with diarrhea of unknown etiology and genomic analysis of associated Streptococcus lutetiensis. BMC Microbiol. 2013;15:141. doi: 10.1186/1471-2180-13-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero B, Morosini MI, Loza E, Rodriguez-Banos M, Navas E, Canton R, Campo RD. Reidentification of Streptococcus bovis isolates causing bacteremia according to the new taxonomy criteria: still an issue? J Clin Microbiol. 2011;15(9):3228–3233. doi: 10.1128/JCM.00524-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, Meyer F, Olsen GJ, Olson R, Osterman AL, Overbeek RA, McNeil LK, Paarmann D, Paczian T, Parrello B, Pusch GD, Reich C, Stevens R, Vassieva O, Vonstein V, Wilke A, Zagnitko O. The RAST Server: rapid annotations using subsystems technology. BMC Genomics. 2008;15:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Domselaar GH, Stothard P, Shrivastava S, Cruz JA, Guo A, Dong X, Lu P, Szafron D, Greiner R, Wishart DS. BASys: a web server for automated bacterial genome annotation. Nucleic Acids Res. 2005;15(Web Server issue):W455–W459. doi: 10.1093/nar/gki593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carver T, Thomson N, Bleasby A, Berriman M, Parkhill J. DNAPlotter: circular and linear interactive genome visualization. Bioinformatics. 2009;15(1):119–120. doi: 10.1093/bioinformatics/btn578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Boutros PC. VennDiagram: a package for the generation of highly-customizable Venn and Euler diagrams in R. BMC Bioinforma. 2011;15:35. doi: 10.1186/1471-2105-12-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;15(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Marchler-Bauer A, Zheng C, Chitsaz F, Derbyshire MK, Geer LY, Geer RC, Gonzales NR, Gwadz M, Hurwitz DI, Lanczycki CJ, Lu F, Lu S, Marchler GH, Song JS, Thanki N, Yamashita RA, Zhang D, Bryant SH. CDD: conserved domains and protein three-dimensional structure. Nucleic Acids Res. 2013;15(Database issue):D348–D352. doi: 10.1093/nar/gks1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan MJ, Petty NK, Beatson SA. Easyfig: a genome comparison visualizer. Bioinformatics. 2011;15(7):1009–1010. doi: 10.1093/bioinformatics/btr039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao Y, He X, Harrison EM, Tai C, Ou HY, Rajakumar K, Deng Z. mGenomeSubtractor: a web-based tool for parallel in silico subtractive hybridization analysis of multiple bacterial genomes. Nucleic Acids Res. 2010;15(Web Server issue):W194–W200. doi: 10.1093/nar/gkq326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grissa I, Vergnaud G, Pourcel C. CRISPRcompar: a website to compare clustered regularly interspaced short palindromic repeats. Nucleic Acids Res. 2008;15(Web Server issue):W145–W148. doi: 10.1093/nar/gkn228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts RJ, Vincze T, Posfai J, Macelis D. REBASE–a database for DNA restriction and modification: enzymes, genes and genomes. Nucleic Acids Res. 2010;15(Database issue):D234–D236. doi: 10.1093/nar/gkp874. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Whole genome phylogeny of the Streptococcus genus. The phylogenetic tree was constructed using the EDGAR tool based on complete genome sequences of streptococci. The branch of the members of the Streptococcus bovis/Streptococcus equinus complex (SBSEC) is delimited by a bracket.
Core genome analysis among Streptococcus gallolyticus UCN34, Streptococcus gallolyticus ATCC 43143, Streptococcus gallolyticus ATCC BAA-2069, Streptococcus macedonicus ACA-DC 198 and Streptococcus pasteurianus ATCC 43144 calculated using the EDGAR software.
Core genome analysis among Streptococcus gallolyticus ATCC 43143, Streptococcus infantarius CJ18, Streptococcus macedonicus ACA-DC 198 and Streptococcus pasteurianus ATCC 43144 calculated using the EDGAR software. In this analysis S. gallolyticus ATCC 43143 was selected as a representative of the S. gallolyticus species, since it has the longest genome size among the three sequenced strains.
Glycobiome analysis of Streptococcus gallolyticus UCN34, Streptococcus gallolyticus ATCC 43143, Streptococcus gallolyticus ATCC BAA-2069, Streptococcus pasteurianus ATCC 43144, Streptococcus macedonicus ACA-DC 198 and Streptococcus infantarius CJ18 using the CAZy database.
Circular maps of the Streptococcus bovis/Streptococcus equinus complex genomes highlighting the regions corresponding to genomic islands (GIs). GIs are coloured within the circular maps according to the tool that predicted each one of them: green, orange and blue were predicted with IslandPick, SIGI-HMM and IslandPath-DIMOB, respectively. The integrated GIs are presented at the periphery of the map in red colour. The black line plot represents the GC content (%) of the genomic sequences. Numbering of the GIs for each genome starts from the first GI found after position 0 of the genome in a clockwise direction.
Analysis of the genomic island (GI) 4 of Streptococcus macedonicus ACA-DC 198 presented as an example of a GI potentially originating from multiple donors. In the graphical summary of the BLASTN results arrows indicate the best BLASTN hits with > 90% sequence identity corresponding to: a. Streptococcus thermophilus MN-ZLW-002 genomic sequence (96% sequence identity); b. Lactococcus garvieae 21881 plasmid pGL3 sequence (98% sequence identity); c. Streptococcus intermedius B196 genomic sequence (96% sequence identity); d. Streptococcus thermophilus MN-ZLW-002 genomic sequence (99% sequence identity) and e. Streptococcus thermophilus MN-ZLW-002 genomic sequence (99% sequence identity).
Genes within each integrated GI of Streptococcus macedonicus ACA-DC 198 as determined by IslandViewer.
The singleton genes of Streptococcus macedonicus ACA-DC 198 calculated against the other members of the Streptococcus bovis/Streptococcus equinus complex (SBSEC) using the EDGAR software. The singleton genes of S. macedonicus were calculated twice, taking or not into account the genome of S. infantarius. Thus, genes shared only by S. macedonicus and S. infantarius among the SBSEC members also appear in the table.
Circular maps of the Streptococcus bovis/Streptococcus equinus complex genomes highlighting the regions corresponding to restriction modification systems (RMs). RMs are presented as predicted in the REBASE database. Colours and symbols are exemplified at the bottom of the figure.
Comparison of the CRISPR/Cas systems among members of the Streptococcus bovis/Streptococcus equinus complex using CRISPRcompar.
Genes in the Streptococcus bovis/Streptococcus equinus complex identified as virulence factors within the VFDB.
Multiple sequence alignment of the capsule biosynthetic gene cluster found in the genomes of the Streptococcus bovis/Streptococcus equinus complex after BLASTN analysis. Grey shading represents the % identity among the nucleotide sequences according to the colour gradient presented at the lower right corner of the figure. Potential pseudogenes are marked with a "p".