Abstract
Glioblastoma (GBM) is a highly lethal brain tumor presenting as one of two subtypes with distinct clinical histories and molecular profiles. The primary GBM subtype presents acutely as high-grade disease that typically harbors EGFR, Pten and Ink4a/Arf mutations, and the secondary GBM subtype evolves from the slow progression of low-grade disease that classically possesses PDGF and p53 events1–3. Here, we show that concomitant CNS-specific deletion of p53 and Pten in the mouse CNS generates a penetrant acute-onset high-grade malignant glioma phenotype with striking clinical, pathological and molecular resemblance to primary GBM in humans. This genetic observation prompted p53 and Pten mutational analysis in human primary GBM, demonstrating unexpectedly frequent inactivating mutations of p53 as well the expected Pten mutations. Integrated transcriptomic profiling, in silico promoter analysis and functional studies of murine neural stem cells (NSCs) established that dual, but not singular, inactivation of p53 and Pten promotes an undifferentiated state with high renewal potential and drives elevated c-Myc levels and its associated signature. Functional studies validated increased c-Myc activity as a potent contributor to the impaired differentiation and enhanced renewal of p53-Pten null NSCs as well as tumor neurospheres (TNSs) derived from this model. c-Myc also serves to maintain robust tumorigenic potential of p53-Pten null TNSs. These murine modeling studies, together with confirmatory transcriptomic/promoter studies in human primary GBM, validate a pathogenetic role of a common tumor suppressor mutation profile in human primary GBM and establish c-Myc as a key target for cooperative actions of p53 and Pten in the regulation of normal and malignant stem/progenitor cell differentiation, self-renewal and tumorigenic potential.
High-grade malignant glioma, the most common intrinsic brain tumor, is uniformly fatal despite maximum treatment3. A wealth of molecular genetic studies have established central roles of the RTK-PI3K-PTEN, ARF-MDM2-p53, and INK4a-RB pathways in gliomagenesis3, 4. To explore the role of p53 and Pten in glioma, we utilized the hGFAP-Cre transgene5, 6 to delete p53 alone or in combination with Pten in all CNS lineages using conditional p537 and Pten alleles (Supplementary Fig. 1, 2a-c). Since broad CNS deletion of Pten results in lethal hydrocephalus in early postnatal life (data not shown), modeling efforts henceforth emphasized the Ptenlox/+ genotype.
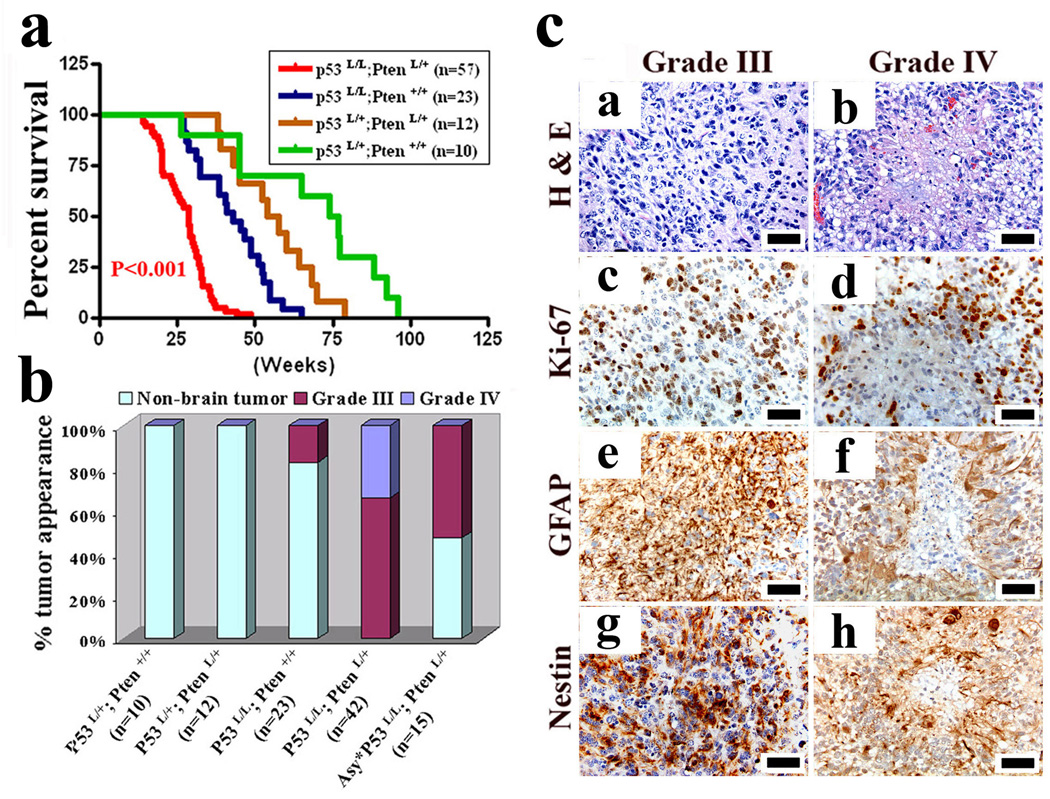
Clinically, between ages 15 to 40 weeks, 42/57 (73%) of hGFAP-Cre+;P53lox/lox;Ptenlox/+ mice presented with acute-onset neurological symptoms – seizure, ataxia, and/or paralysis (Fig. 1a). Histopathologically, all 42 neurologically symptomatic mice harbored malignant gliomas that, based on WHO criteria8, were classified as anaplastic astrocytomas (WHO III, n=28, 66%) or GBM (WHO IV, n=14, 34%) (Fig. 1b) with classical features of pseudopalisading necrosis, marked cellular pleomorphism, and highly infiltrative spread including perineuronal and perivascular satellitosis as well as subpial spread in the cerebral cortex (Supplementary Fig. 3a). Occasional tumors exhibited abnormal vessels suggestive of microvascular proliferation. All tumors exhibited increased mitoses (Ki67 staining) and expression of classical human glioma markers, GFAP and Nestin (Fig. 1c). Necropsy of 15 neurologically asymptomatic mice showed no cases of incipient low-grade glioma disease but rather 8 with high-grade pathology including very small lesions with anaplastic features of nuclear atypia, multinucleated tumor cells, and/or high cellularity (Supplementary Fig. 3b). For the remaining genotypes, 4/23 hGFAP-Cre;P53lox/lox mice developed anaplastic astrocytoma (WHO III); whereas 19/23 hGFAP-Cre;P53lox/lox, 12/12 hGFAP-Cre;P53lox/+;Ptenlox/+, and 10/10 hGFAP-Cre;P53lox/+) had no CNS pathology and developed only non-CNS tumors (data not shown).
Figure 1. p53 and Pten inactivation cooperate to induce high-grade malignant gliomas.
a, Kaplan-Meier tumor-free survival curves for mice of indicated genotypes as a function of weeks.
b, Graph shows frequency and grade of gliomas vs. non-CNS malignancies observed in end-stage of indicated mice from a. Asy* indicates neurological asymptomatic hGFAP-Cre;P53lox/lox;Ptenlox/+ mice (n=15) sacrificed for non-CNS malignancies.
c, H&E histology and immunohistochemical staining of sections of WHO grade III and grade IV malignant gliomas from hGFAP-Cre;P53lox/lox;Ptenlox/+ mice with antibodies against Ki67, GFAP, and Nestin. Scale bar, 50 µm.
Historically, p53 inactivation has been considered a classical lesion in low-grade astrocytomas and secondary GBM, but infrequent in primary GBM1, 9. The remarkable clinical and histological resemblance of this model to the primary GBM subtype in humans prompted p53 and Pten re-sequencing in human primary GBM. Of 35 clinically annotated human primary GBM samples, 10/35 (29%) tumors registered prototypical p53 mutations and 14/35 (40%) tumors possessed Pten missense mutations, insertions, deletions, or splicing mutations (Supplementary Table 1). Moreover, 6/10 tumors with p53 mutations harbored concomitant Pten mutations or homozygous deletion. Encouragingly, our mutational data agrees with The Cancer Genome Atlas data reporting p53 and Pten as the two most commonly mutated tumor suppressor genes (http://tcga-data.nci.nih.gov/tcga/findArchives.htm). These results, together with recent population-based studies10, 11, indicate that p53 is a key tumor suppressor for both GBM subtypes.
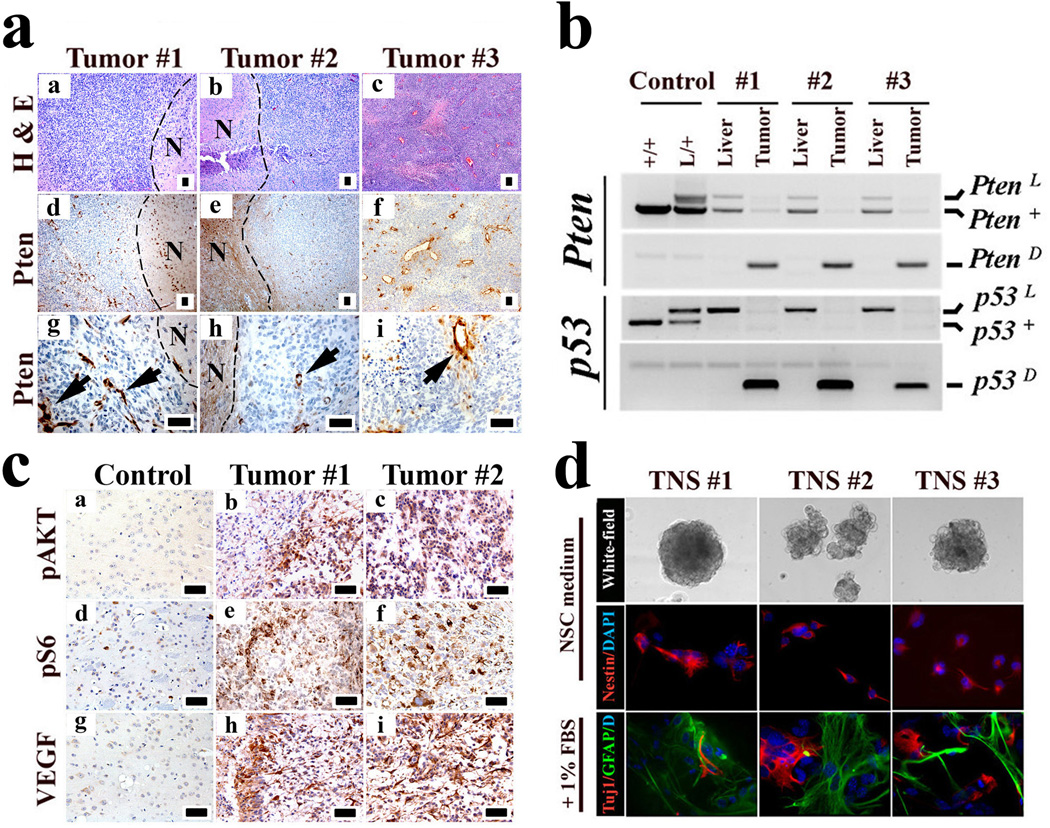
Consistent with frequent Pten LOH (60–70%) in human high-grade glioma3, 16/16 mouse high-grade glimoas showed no Pten expression in tumor cells but robust signal in surrounding non-malignant cells and intratumoral vessels (Fig. 2a). PCR geneotyping indicated that 6/7 tested tumors sustained loss of the wild-type Pten allele (Fig. 2b). The Pten reduction to homozygosity, and documented Cre-mediated deletion of both p53 floxed alleles indicate that inactivation of both genes is required for gliomagenesis. Loss of Pten expression correlated with activation of key PI3K signaling surrogates, AKT and S6-Kinase (Fig. 2c). In accordance with human high-grade disease, 8/8 malignant murine gliomas expressed high VEGF levels relative to normal brain tissue (Fig. 2c). Multi-RTK co-activation in human primary GBM12 was also evident in the murine tumors with robust PDGFRα expression overlapping with strong regional activation of EGFR (Supplementary Fig. 4a-d)
Figure 2. hGFAP-Cre;P53lox/lox;Ptenlox/+ gliomas mirror key features of human malignant gliomas.
a, Pten expression is completely extinguished in tumor cells. Sections of three independent malignant gliomas were stained with H&E, and an anti-Pten antibody. Note “N” indicates the adjacent normal regions of the tumor cells; the arrows point to Pten-positive vascular cells embedded in the tumor.
b, Wild-type Pten allele is lost in glioma cells. Genomic DNA isolated from liver tissues and brain tumor cells were subjected to PCR-based assays for genotyping Pten and p53 alleles. Note “+” designates the Pten wild-type allele; “L” the conditional allele; and “D” the inactivated form of the conditional allele after Cre-mediated recombination.
c, Immunohistochemical staining of mouse normal brain or glioma sections with antibodies against activated p-AKT, p-S6 kinase, and VEGF.
d, TNS lines isolated from independent malignant gliomas were cultured in NSC medium or differentiation medium and immuno-stained for Nestin, GFAP and Tuj1 as indicated. Scale bar, 50 µm.
A classical feature of human high-grade malignant glioma is a significant degree of inter- and intra-tumoral morphological and lineage heterogeneity, hence the moniker glioblastoma ‘mutliforme’. This characteristic plasticity was evident in the hGFAP-Cre;P53lox/lox;Ptenlox/+ gliomas wherein occasional tumors (5/50) presented with both astrocytic and oligodendroglial histopathological features (Supplementary Fig. 5). The basis for morphological variability is not known and may relate, among many possibilities, to acquisition of an immature developmental state with multipotency and/or differentiative plasticity. Consistent with this notion, all murine tumors express stem or lineage progenitor markers including Nestin, GFAP, and Olig2 similar to human glioma profiles13, but negative for mature neuronal and oligodendrocyte markers, NeuN and MBP (Supplementary Fig. 6a-b). This stem/progenitor marker profile is in accord with the ability of all murine tumors tested to readily generate tumor neurospheres (TNS) with (i) strong tumor-initiating potential with secondary tumors faithfully retaining the primary tumor’s histological features (Supplementary Fig. 7) (ii) robust NSC marker Nestin expression, and (iii) limited capacity to differentiate into astrocytic and neuronal lineages upon exposure to differentiation agents (Fig. 2d). As NSC/progenitor cells have been proposed to be the preferred cell-of-origin for GBM6, the immature marker profile and varied morphological presentation of our murine tumors prompted us to posit that Pten and p53 deficiencies might contribute to gliomagenesis by affecting NSC self-renewal and differentiation potential.
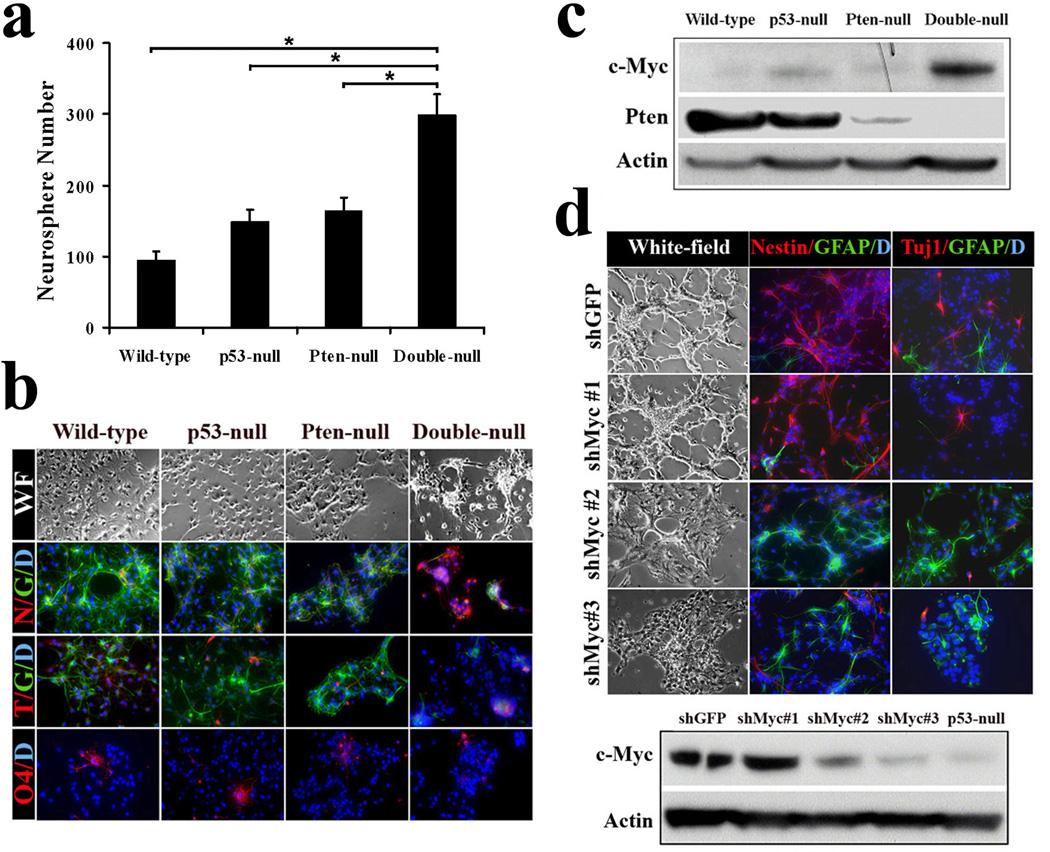
To explore this hypothesis, we characterized primary murine E13.5 NSC cultures singly or doubly null for p53 and Pten. Compared to Pten or p53 null NSCs which show only modestly increased proliferation and self-renewal reflected by neurosphere formation capacity14–16, NSCs null for p53 and Pten exhibited significant proliferation and self-renewal activity (Fig. 3a; Supplementary Fig. 8a). This impact on NSC renewal, coupled with the aforementioned varied tumor histology, suggest that combined p53 and Pten loss might cooperate in tumorigenesis by impairing NSC differentiation potential. When NSCs were continuously cultured in NSC medium, all gentoypes showed similar robust expression of NSC/progenitor markers (Nestin) and minimal expression of differentiated lineage markers (Supplementary Fig. 8b). Upon exposure to differentiation-inducing medium, wild-type and single-null NSC cultures differentiated into GFAP-positive astrocytes, Tuj1-positive neurons, or O4-positive oligodendrocytes. In contrast, NSCs null for both p53 and Pten failed to respond to these differentiation cues and retained stem cell-like morphology and lineage marker (Nestin) expression (Fig. 3b; Supplementary Fig. 9a). Similar differentiation defect and abnormal self-renewal potential were also observed in adult NSCs that were deleted for Pten and p53 postnatally (Supplementary Fig. 9b, c). The contribution of Pten deficiency in maintaining impaired differentiation was further verfied by the ability of the AKT inhibitor, Triciribine17, to enable differentiation of Pten/p53 null NSCs (Supplementary Fig. 10a, b).
Figure 3. p53 and Pten coordinately regulate c-Myc protein level as well as NSC self-renewal and differentiation.
a, The number of neurospheres formed by p53/Pten double-null NSCs in culture is significantly increased as compared to wild-type or singly null NSCs (*P < 0.001; n=3). Values represent mean ± s.d. from three experiments.
b, The multi-lineage differentiation potential was impaired in double-null NSCs. Note “WF” designates for white-field; N for Nestin (red); G for GFAP (green); T for Tuj-1 (red); O4 (red); D for DAPI (blue).
c, Combined inactivation of p53 and Pten in NSCs stimulates c-Myc protein expression.
d, Knockdown of c-Myc expression restores p53/Pten double-null NSCs differentiation capacity. Lower panel, western blot of double-null NSC c-Myc protein expression after infected with indicated lenti-shRNA. Note c-Myc expression in shMyc #2 and #3 infected double-null cells is comparable to that in p53-null cells, and shMyc #1 as a control shows no knockdown.
To understand the molecular basis of impaired differentiation capacity, we performed transcriptome comparisons of p53-null and p53/Pten double-null murine NSCs at 1 day post exposure to the differentiation inducer. Among the 410 genes exhibiting significant differential expression (Supplementary Table 2), promoter analysis identified E2F and MYC motifs as two of the most enriched promoter binding elements (BE) (1.7x and 1.4x, respectively; P < 10−4). Remarkably, promoter analysis using 69 pre-treatment human primary GBM cases in the TCGA database showed strong enrichment of Myc binding elements – 10 p53/Pten mutant tumors versus the 59 remaining tumors (1.40x, p=2.20x10−3) or versus the 12 p53 only mutant tumors (1.46x, p=1.54x10−3).
c-MYC is well-known for its roles in cell cycle progression and apoptosis18 as well as stem cell self-renewal and differentiation during development and oncogenic processes19–22. It is also notable that both p53 and Pten/PI3K pathways can directly regulate c-Myc with p53 repressing c-MYC transcription through direct binding to c-MYC promoter23, while downstream PI3K pathway arms can modulate c-MYC translation and protein degradation24, 25. In agreement, c-Myc protein levels were substantially increased in the murine double-null NSCs, but only marginally elevated in single p53-null or Pten-null NSCs when compared to wild-type controls (Fig. 3c), raising the possibility that p53 and Pten cooperate to regulate c-Myc levels which in turn could control NSC self-renewal and differentiation.
To test this hypothesis, we examined the impact of c-Myc knockdown on murine p53/Pten-null NSC differentiation potential and observed that c-Myc shRNAs (#2 and #3), which reduced c-Myc levels to those in p53-null NSCs, largely restored their differentiation capacity (Fig. 3d; Supplementary Fig. 11a). Conversely, enforced c-Myc expression in p53-null NSCs repressed their differentiation and enabled retention of stem/progenitor marker expression (Nestin and Sox2) (Supplementary Fig. 11b), indicating that concomitant loss of p53 and Pten elevates c-MYC activity to impede NSC differentiation capacity.
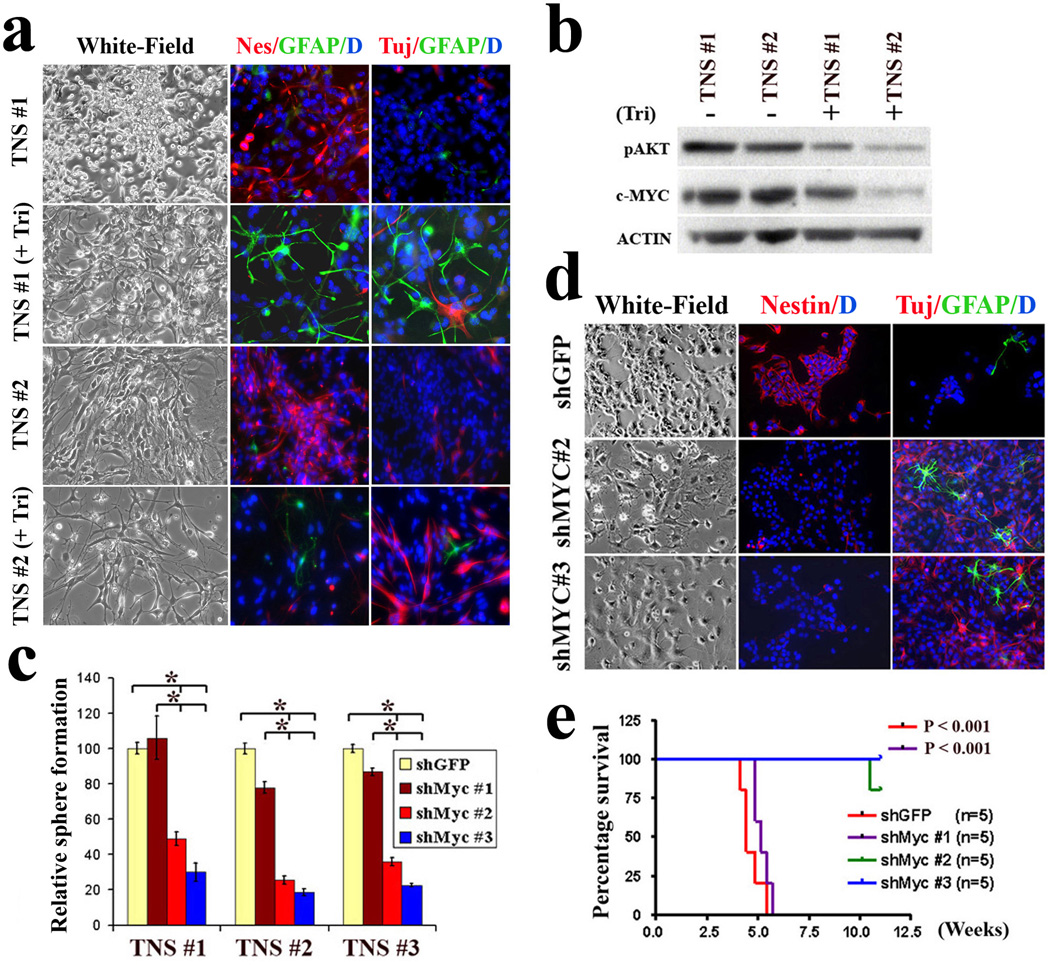
The strong pleiotropic activities attributed to c-MYC demands tight control of its expression to avoid development of diverse human malignancies, including gliomas21, 26. Our finding that concomitant loss of p53 and Pten compromised NSC differentiation capacity via elevated c-Myc levels prompted further assessment of its relevance in the so-called ‘brain cancer stem cells’ in our model. Using murine TNSs which are enriched for such tumor initiating cells (TICs), AKT inhibitor treatment strongly reduced c-MYC protein and promoted differentiation (Fig. 4a, b; Supplementary Fig. 12a). Correspondingly, c-Myc knockdown in TNS cells not only markedly reduced their proliferation and self-renewal capacity (Fig. 4c; Supplementary Fig. 12b, c), but also strongly sensitized them to differentiation induction (Fig. 4d; Supplementary Fig. 12d). Importantly, while 10/10 intracranial injections of vector-transduced murine TNSs resulted in lethal infiltrating gliomas within 1 month, 9/10 mice injected with c-Myc knockdown TNSs survived for more than 3 months (Fig. 4e). Thus, c-MYC plays a key role in the maintenance of impaired differentiation and potent tumorigenic potential of TNSs inactivated for p53 and Pten (Supplementary Fig. 13).
Figure 4. Attenuated c-Myc expression restores hGFAP-Cre;P53lox/lox;Ptenlox/+ TNS differentiation potential and reduces tumorigenic potential.
a, Inhibition of the AKT pathway by triciribine induces TNS cell differentiation. Two independent TNS lines were cultured in 1% FBS in the absence or presence of triciribine (5 µM) for 7 days before being subjected to immuno-staining with antibodies against Nestin (red), GFAP (Green) and Tuj1 (red).
b, Inhibition of the AKT pathway in TNS cells with triciribine attenuates their cellular c-Myc expression.
c, Knockdown of c-Myc expression in TNS cells reduces their self-renewal potential assessed by sphere formation (*P < 0.001, n=3). Values represent mean ± s.d. from three experiments.
d, ShRNA-mediated reduction of c-Myc expression in TNS cells sensitizes cells to differentiation stimuli. Cells infected with control and indicated shRNA were incubated with differentiation medium before subjected to indicated lineage marker analysis.
e, ShRNA-mediated reduction of c-Myc expression represses TNS tumorigenic potency in orthotopically transplanted SCID mice.
The identification of TICs with stem-like properties in diverse human cancers including GBM represents an important conceptual advance in cancer biology with therapeutic implications27, 28. These TICs appear to constitute a reservoir of self-sustaining cells with potent tumorigenic potential. However, unlike normal NSCs which readily differentiate along a developmental hierarchy into lineage-restricted differentiated progenies, TNSs derived from p53/Pten-null malignant gliomas show resistance towards differentiation cues. The diminished tumorigenicity of these TICs upon restoration of differentiation potential, along with recent reports supporting pro-differentiation as a potential strategy to inhibit GBM-derived TICs29, 30, encourages the identification and testing of agents targeting these differentiation pathways including c-Myc in the treatment of primary GBM in humans.
Full Methods
Mice
P53L and hGFAP-Cre mice have been described previously5, 7. PtenL mice were generated using a standard knock-in approach in which Pten exon 5 is flanked by loxP sites (details on targeting construct and procedures are available upon request). Mice were interbred and maintained on FvB/C57Bl6 hybrid background in pathogen-free conditions at Dana-Farber Cancer Institute, monitored for signs of ill-health every other day, and euthanized and necropsied when moribund. All manipulations were performed with IACUC approval.
Histology and Immunohistochemistry
At time of sacrifice, mice were perfused with 4% paraformaldehyde (PFA), and brains were dissected, followed by overnight post-fixation in 4% PFA at 4°C. Serial sections were prepared at 5 µm for paraffin sections or 10 µm for cryostat sections with every tenth slide stained by haematoxylin and eosin (DF/HCC Research Pathology Cores). Tumor grading was determined by K.L.L and H.Z. based upon the WHO grading system for malignant astrocytoma8. Immunohistochemical and immunofluorescence analyses were performed as described31. The PDGFRα/pEGFR double immunohistochemical staining was performed using DakoCytomation EnVision doublestain system (K1395, Dako) following manufacturer instructions. The primary antibodies used were: Ki67 (VP-RM04, Vector), GFAP (Z0334, DAKO), GFAP (556330, BD Pharmingen), Nestin (MAB353, Chemicon; specifically for mouse), Nestin (MAB5326, Chemicon; specifically for human), Pten (9559, Cell Signaling), Phospho-AktSer473 (3787, Cell Signaling), phospho-S6 ribosomal protein (2215, Cell Signaling), Cyclin D1 (18-0220, ZYMED), VEGF (sc-152, Santa Cruz), PDGFRα (3174, Cell Signaling), Phospho-PDGFRaY754 (sc-12911, Santa Cruz), EGFR (IHC-00005, Bethyl), Phospho-EGFRY1068 (ab40815, Abcam), Phospho-EGFRY1173 (sc-12351, Santa Cruz), Olig-2 (AB9610, Chemicon), TUJ-1 (MMS-435P, Covance), O4 (MAB1326, R&D), NeuN (MAB377, Millipore), MBP (ab7349, Abcam), c-Myc (ABcam, ab39688), and Cre (69050-3, Novagen). Images were captured using a Leica DM1400B microsystem and Leica FW4000 version 1.2.1.
Cell culture
Primary NSCs were isolated from the brain subventricular zone (SVZ) of E13.5 embryos or 1 month old mice with the indicated genotype as previously described31, 32. NSCs were maintained in NSC proliferation media (05702, StemCell) supplemented with 20 ng/mL EGF (E4127, Sigma) and 10ng/mL bFGF (F0291, Sigma). To generate primary TNS cells, tumor samples from freshly dissected mouse brains were subjected to mechanical and enzymatic dissociation. Single-cell suspensions were cultured in NSC proliferation media. Tumor spheres formed were then disaggregated and used for indicated assays. NSC differentiation assays were carried out by plating the indicated cells in culture wells on coverslips pre-coated with 15 µg/mL poly-L-ornithine (P3655, Sigma) and 1 µg/mL fibronectin (F1141, Sigma); the cells were incubated in neurobasal medium supplemented with 1% FBS for 7–10 days, and the differentiation capacities examined under either light or fluorescence microscope (Nikon). For TNS cell differentiation, cells were incubated in differentiation media with varying doses of triciribine (BioMol) or vehicle (DMSO, Sigma). Knockdown of mouse c-Myc was performed by infecting the indicated cells with lentivirus containing shMyc constructs (provided by Dr. William Hahn, Dana Farber Cancer Institute). The shRNA constructs shMyc #1, #2 and #3 correspond to clone ID#s TRCN000000 54856, 42517 and 42513, respectively (The DFCI-Broad RNAi Consortium, commercially available from Sigma-Aldrich).
Western, cell growth and self-renewal assays
Western blot assays were performed as previously described31 with antibodies against c-Myc (sc-42, Santa Cruz), Phospho-AKTSer473 (9271, Cell signaling), Pten (9569, Cell Signaling) and Actin (sc-1615, Santa Cruz). For in vitro cell growth assays, NSCs or TNS cells (10,000) were plated in triplicate in 96-well format and incubated in NSC proliferation media for 5 days, and growth was quantified using Luminescence ATP detection assay system (PerkinElmer). Self-renewal capacity was measured by plating 1,000 cells/well (6-well plate) in NSC proliferation media containing EGF/bFGF with 0.3% agarose (A9049, Sigma). The number of neurospheres or tumor neurospheres that formed subsequently per well was quantified after 10–14 days and relative sphere formation was plotted versus indicated control. Three replicates were performed for each. All experiments were conducted at cell passage <5.
Orthotopic transplants
Female SCID (Charles River) aged 6–8 weeks were anesthetized and placed into stereotactic apparatus equipped with a Z axis (Stoelting). A small hole was bored in the skull 0.5 mm anterior and 3.0 mm lateral to the Bregma using a dental drill. 2×104 cells in Hanks Buffered Salt Solution was injected into the right caudate nucleus 3 mm below the surface of the brain using a 10 µl Hamilton syringe with an unbeveled 30 gauge needle. The scalp was closed using a 9 mm Autoclip Applier. Animals were followed daily for development of neurological deficits.
Mutation screening
Frozen tumor specimens were obtained from the Memorial Sloan Kettering Cancer Center tumor bank. Genomic DNA was prepared from frozen primary GBM tumour samples using the Qiagen genomic purification kit. Coding exons were PCR amplified and sequenced using standard protocols at the Harvard Partners Center for Genetics and Genomics as previously described33. All known single-nucleotide polymorphisms and synonymous mutations were removed from the analysis in the current study. This study was approved by the Institutional Review Board of the hospital.
Microarray analysis
Early passage wild-type and indicated mutant NSCs were incubated with NSC proliferation media or differentiation media for 18 hours. RNA was isolated using Trizol (Invitrogen) and the RNeasy mini kit (Qiagen). Gene expression profiling was performed utilizing the Affymetrix 430 2.0 chips at DFCI Microarray core facility.
Promoter Analysis
Gene expressions were modeled using dChip software34. Sets of genes differentially expressed pre- and post- differentiation induction were generated using the SAM statistic35, with a cutoff of +/− 2.0. Promoter analysis on both these gene sets used the CisGenome software (http://biogibbs.stanford.edu/~jihk/CisGenome/index.htm) to scan the 8 kb upstream to 2kb downstream regions of these genes for the ~550 motifs in the TRANSFAC 12.1 database. Enrichment was measured against control regions at a comparable distance from the transcription start sites of random genes.
Statistical analysis
Tumor-free survivals were analyzed using Graphpad Prism4. Statistical analyses were performed using nonparametric Mann–Whitney test. Significance of enrichment in the promoter analysis was computed based on Poisson distribution with Bonferroni correction. Comparisons of cell growth, self-renewal and differentiation were performed using the unpaired Student's t-test. For all experiments with error bars, standard deviation was calculated to indicate the variation within each experiment and data, and values represent mean ± s.d.
Supplementary Material
Acknowledgements
We thank A. Berns for providing p53L mice; S. Zhou and S. Jiang for excellent mouse husbandry and care; R. T. Bronson for helpful discussion on pathology analysis; K. Montgomery for discussion on sequencing; and Y. -H. Xiao, B. Feng, and J. Zhang for bioinformatic help. H.Z. was supported by Helen Hay Whitney Foundation. H.Y. is a recipient of the Marsha Mae Moeslein Fellowship from the American Brain Tumor Association. A.C.K. is a recipient of the Leonard B. Holman Research Pathway Fellowship. Z.D is supported by the Damon Runyon Cancer Research Foundation. J.M.S is supported by a Ruth L. Kirschstein National Research Service Award Fellowship. R.W. is supported by a Mildred Scheel Fellowship (Deutsche Krebshilfe). Grant support comes from the Goldhirsh Foundation (R.A.D.), and NIH grants U01 CA84313 (R.A.D.), RO1CA99041 (L.C.) and 5P01CA95616 (R.A.D., L.C., W. H. W., C.B. and K.L.L.). R.A.D. is an American Cancer Society Research Professor supported by the Robert A. and Renee E. Belfer Foundation Institute for Innovative Cancer Science.
Footnotes
Author Contributions
H.Z. and H.Y. performed the experiments and contributed equally as first authors. R.A.D. supervised experiments and contributed as senior author. M.J.Y. generated the PtenL mouse allele. D.J.H., W.H.W. and G.T. conducted the microarray and promoter analyses. K.L.L, H.Z. and G.C.C. provided the pathology analyses. H.Y., A.C.K., A.J.C., S.R.P., Z.D., J.M.S, K.L.D and R.W. performed experiments. C.B. contributed patient samples and pathologic information. L.C. and Y.A.W. contributed to the writing of the manuscript.
The microarray data are available at the Gene Expression Array Omnibus website (http://www.ncbi.nlm.nih.gov/geo/) under accession number (GSE12694).
References
- 1.Kleihues P, Ohgaki H. Primary and secondary glioblastomas: from concept to clinical diagnosis. Neuro Oncol. 1999;1:44–51. doi: 10.1093/neuonc/1.1.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhu Y, Parada LF. The molecular and genetic basis of neurological tumours. Nat Rev Cancer. 2002;2:616–626. doi: 10.1038/nrc866. [DOI] [PubMed] [Google Scholar]
- 3.Furnari FB, et al. Malignant astrocytic glioma: genetics, biology, and paths to treatment. Genes Dev. 2007;21:2683–2710. doi: 10.1101/gad.1596707. [DOI] [PubMed] [Google Scholar]
- 4.Wiedemeyer R, et al. Feedback circuit among INK4 tumor suppressors constrains human glioblastoma development. Cancer Cell. 2008;13:355–364. doi: 10.1016/j.ccr.2008.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhuo L, et al. hGFAP-cre transgenic mice for manipulation of glial and neuronal function in vivo. Genesis. 2001;31:85–94. doi: 10.1002/gene.10008. [DOI] [PubMed] [Google Scholar]
- 6.Zhu Y, et al. Early inactivation of p53 tumor suppressor gene cooperating with NF1 loss induces malignant astrocytoma. Cancer Cell. 2005;8:119–130. doi: 10.1016/j.ccr.2005.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jonkers J, et al. Synergistic tumor suppressor activity of BRCA2 and p53 in a conditional mouse model for breast cancer. Nat Genet. 2001;29:418–425. doi: 10.1038/ng747. [DOI] [PubMed] [Google Scholar]
- 8.Louis DN, Ohgaki H, Wiestler OD, Cavenee WK. In: WHO Classification of Tumours of the Central Nervous System, Fourth Edition. Louis DN, Ohgaki H, Wiestler OD, Cavenee WK, editors. Lyon, France: World Health Organization; 2007. [Google Scholar]
- 9.Watanabe K, et al. Overexpression of the EGF receptor and p53 mutations are mutually exclusive in the evolution of primary and secondary glioblastomas. Brain Pathol. 1996;6:217–223. doi: 10.1111/j.1750-3639.1996.tb00848.x. discussion 23-4. [DOI] [PubMed] [Google Scholar]
- 10.Ohgaki H, et al. Genetic pathways to glioblastoma: a population-based study. Cancer Res. 2004;64:6892–6899. doi: 10.1158/0008-5472.CAN-04-1337. [DOI] [PubMed] [Google Scholar]
- 11.Fukushima T, et al. Genetic alterations in primary glioblastomas in Japan. J Neuropathol Exp Neurol. 2006;65:12–18. doi: 10.1097/01.jnen.0000196132.66464.96. [DOI] [PubMed] [Google Scholar]
- 12.Stommel JM, et al. Coactivation of receptor tyrosine kinases affects the response of tumor cells to targeted therapies. Science. 2007;318:287–290. doi: 10.1126/science.1142946. [DOI] [PubMed] [Google Scholar]
- 13.Ligon KL, et al. Olig2-regulated lineage-restricted pathway controls replication competence in neural stem cells and malignant glioma. Neuron. 2007;53:503–517. doi: 10.1016/j.neuron.2007.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Groszer M, et al. PTEN negatively regulates neural stem cell self-renewal by modulating G0-G1 cell cycle entry. Proc Natl Acad Sci U S A. 2006;103:111–116. doi: 10.1073/pnas.0509939103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.GilPerotin S, et al. Loss of p53 induces changes in the behavior of subventricular zone cells: implication for the genesis of glial tumors. J Neurosci. 2006;26:1107–1116. doi: 10.1523/JNEUROSCI.3970-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meletis K, et al. p53 suppresses the self-renewal of adult neural stem cells. Development. 2006;133:363–369. doi: 10.1242/dev.02208. [DOI] [PubMed] [Google Scholar]
- 17.Yang L, et al. Akt/protein kinase B signaling inhibitor-2, a selective small molecule inhibitor of Akt signaling with antitumor activity in cancer cells overexpressing Akt. Cancer Res. 2004;64:4394–4399. doi: 10.1158/0008-5472.CAN-04-0343. [DOI] [PubMed] [Google Scholar]
- 18.Patel JH, Loboda AP, Showe MK, Showe LC, McMahon SB. Analysis of genomic targets reveals complex functions of MYC. Nat Rev Cancer. 2004;4:562–568. doi: 10.1038/nrc1393. [DOI] [PubMed] [Google Scholar]
- 19.Cartwright P, et al. LIF/STAT3 controls ES cell self-renewal and pluripotency by a Myc-dependent mechanism. Development. 2005;132:885–896. doi: 10.1242/dev.01670. [DOI] [PubMed] [Google Scholar]
- 20.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 21.Ben-Porath I, et al. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat Genet. 2008;40:499–507. doi: 10.1038/ng.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wong DJ, et al. Module map of stem cell genes guides creation of epithelial cancer stem cells. Cell Stem Cell. 2008;2:333–344. doi: 10.1016/j.stem.2008.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ho JS, Ma W, Mao DY, Benchimol S. p53-Dependent transcriptional repression of c-myc is required for G1 cell cycle arrest. Mol Cell Biol. 2005;25:7423–7431. doi: 10.1128/MCB.25.17.7423-7431.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gera JF, et al. AKT activity determines sensitivity to mammalian target of rapamycin (mTOR) inhibitors by regulating cyclin D1 and c-myc expression. J Biol Chem. 2004;279:2737–2746. doi: 10.1074/jbc.M309999200. [DOI] [PubMed] [Google Scholar]
- 25.Sears R, et al. Multiple Ras-dependent phosphorylation pathways regulate Myc protein stability. Genes Dev. 2000;14:2501–2514. doi: 10.1101/gad.836800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bredel M, et al. Functional network analysis reveals extended gliomagenesis pathway maps and three novel MYC-interacting genes in human gliomas. Cancer Res. 2005;65:8679–8689. doi: 10.1158/0008-5472.CAN-05-1204. [DOI] [PubMed] [Google Scholar]
- 27.Clarke MF, et al. Cancer stem cells--perspectives on current status and future directions: AACR Workshop on cancer stem cells. Cancer Res. 2006;66:9339–9344. doi: 10.1158/0008-5472.CAN-06-3126. [DOI] [PubMed] [Google Scholar]
- 28.Lobo NA, Shimono Y, Qian D, Clarke MF. The biology of cancer stem cells. Annu Rev Cell Dev Biol. 2007;23:675–699. doi: 10.1146/annurev.cellbio.22.010305.104154. [DOI] [PubMed] [Google Scholar]
- 29.Piccirillo SG, et al. Bone morphogenetic proteins inhibit the tumorigenic potential of human brain tumour-initiating cells. Nature. 2006;444:761–765. doi: 10.1038/nature05349. [DOI] [PubMed] [Google Scholar]
- 30.Lee J, et al. Epigenetic-mediated dysfunction of the bone morphogenetic protein pathway inhibits differentiation of glioblastoma-initiating cells. Cancer Cell. 2008;13:69–80. doi: 10.1016/j.ccr.2007.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bachoo RM, et al. Epidermal growth factor receptor and Ink4a/Arf: convergent mechanisms governing terminal differentiation and transformation along the neural stem cell to astrocyte axis. Cancer Cell. 2002;1:269–277. doi: 10.1016/s1535-6108(02)00046-6. [DOI] [PubMed] [Google Scholar]
- 32.Rietze RL, Reynolds BA. Neural stem cell isolation and characterization. Methods Enzymol. 2006;419:3–23. doi: 10.1016/S0076-6879(06)19001-1. [DOI] [PubMed] [Google Scholar]
- 33.Maser RS, et al. Chromosomally unstable mouse tumours have genomic alterations similar to diverse human cancers. Nature. 2007;447:966–971. doi: 10.1038/nature05886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li C, Wong WH. Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc Natl Acad Sci U S A. 2001;98:31–36. doi: 10.1073/pnas.011404098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.