Abstract
The tea geometrid (Ectropis obliqua Prout, Lepidoptera: Geometridae) is a dominant chewing insect endemic in most tea-growing areas in China. Recently some E. obliqua populations have been found to be resistant to the nucleopolyhedrovirus (EoNPV), a host-specific virus that has so far been found only in E. obliqua. Although the resistant populations are morphologically indistinguishable from susceptible populations, we conducted a nationwide collection and examined the genetic divergence in the COI region of the mtDNA in E. obliqua. Phylogenetic analyses of mtDNA in 17 populations revealed two divergent clades with genetic distance greater than 3.7% between clades and less than 0.7% within clades. Therefore, we suggest that E. obliqua falls into two distinct groups. Further inheritance analyses using reciprocal single-pair mating showed an abnormal F1 generation with an unbalanced sex ratio and the inability to produce fertile eggs (or any eggs) through F1 self-crossing. These data revealed a potential cryptic species complex with deep divergence and reproductive isolation within E. obliqua. Uneven distribution of the groups suggests a possible geographic effect on the divergence. Future investigations will be conducted to examine whether EoNPV selection or other factors prompted the evolution of resistance.
Introduction
Tea, Camellia sinensis (L.) O. Kuntze, originated from China and is now grown in almost sixty countries. The diversified regional climate, planting history, tea varieties, cultivation management, pest management model, tea production characteristics and environmental factors affect the distribution of insects in tea plantations worldwide. China, the largest tea growing country and with a long history of tea planting, processing and consumption, has more than 2,600 cultivars and six major categories of tea across three climatic zones and four tea ecological areas. Consequently, the pest fauna in tea fields is significantly diversified.
Loopers or inchworm moths (Lepidoptera: Geometridae) are one of the largest insect families, composed of nearly 23,000 species worldwide [1]–[3]. The tea geometrid (Ectropis obliqua Prout, Lepidoptera: Geometridae) was named by Prout in the year of 1915 [4]–[6]. E. obliqua has 4–5 generations a year in most areas, and it is now the most important chewing pest in tea plantations in China. This pest infests thousands of hectares a year, especially in the lower reaches of the Yangtze River which is a key conventional green tea producing area, including Zhejiang, Jiangsu and Anhui provinces [2]. E. obliqua, a voracious insect that feeds on tea leaves and tender buds, severely reduces tea production in the summer and autumn, causing growth recession and further impacting the tea production of the following year. It has been known for a long time that chemical treatment is the most effective method to control E. obliqua. However, because tea is being accepted by more and more consumers as a healthy beverage, its safety and quality are also garnering more attention. Therefore, the massive use of chemicals is seriously affecting not only the safety of the tea but also the ecosystems and environment in which tea is grown [2], [7]. Nowadays biological control measures are widely accepted in China to produce safer (contaminant-free) or organic tea [7].
Biological control can be an environmentally sound and effective means of reducing or mitigating pests and pest effects through the use of natural enemies, and is a widely accepted method in agriculture systems worldwide. E. obliqua nucleopolyhedrovirus (EoNPV) is an effective natural biological agent that has been successfully developed as a bio-insecticide in China for the control of the tea geometrid [7], [8]. Recently, we found that some tea geometrid populations were insensitive to the virus. To explain this unusual phenomenon, we collected tea geometrid populations in eight main tea production provinces from 2008 to 2011 and performed a feeding test with EoNPV after morphological validation of the species. We found that some E. obliqua populations were susceptible to this host-specific virus, but others were insensitive. The virus was more than 700 times more virulent to the susceptible population from Yixing (YX) Jiangsu province compared to the insensitive population of Quzhou (QZ) Zhejiang province [8]. Initially, the development of resistance to EoNPV, possibly caused by selection pressure in some populations, was considered as an explanation [9]. However, there were no heavy applications of EoNPV in any of our seventeen collection sites. Because NPV is a highly host-specific virus [2], [8], the significant variations in susceptibility to EoNPV in non-EoNPV-selected geometrid populations indicates that the insect might have already developed genetic divergence before EoNPV was used. In addition, we hypothesize that this divergence was induced by geographic, ecological or other factors, rather than EoNPV selection pressure.
Examining divergence levels by means of molecular markers and inheritance analysis is the first step toward exploring a possible mechanism. Mitochondrial DNA (mtDNA) has been widely used in insect taxonomy and molecular systematics to elucidate the phylogenetic relationships of insects [10]–[12], especially among some sibling species and cryptic species that are difficult to distinguish solely by morphology [11], [13]. For example, partial cytochrome oxidase I (COI) sequences reveal that Helicoverpa. armigera and H. zea are genetically separate, albeit by a short distance [14], [15]. Combined COI and COII were used to distinguish three subspecies of the hemlock looper (Lambdina fiscellaria, Gn.) [16]. The widely distributed neotropical skipper butterfly (Astraptes fulgerator) was formerly recognized as at least 10 species in northwestern Costa Rica, but morphological analysis and a DNA barcoding study of COI showed that whilst A. fulgerator is a species complex the actual number of separate taxa is smaller than it has usually been thought to be in this region [17].
In this study, we collected seventeen populations from eight main tea-producing provinces of China. To reveal genetic divergence among the populations, phylogenetic analysis of the mtDNA COI sequences was conducted. Single-pair cross analysis was also conducted to verify any species-level diversification and to understand the biological effects of divergence among populations.
Materials and Methods
Ethics Statement
No specific permits were required for this study. E. obliqua is an agricultural pest and is not an endangered or protected species. All samples were collected in open tea plantations and not from any national parks or protected areas.
Sample collection and preparation
E. obliqua specimens were collected by sweep netting from eight major tea producing provinces: Zhejiang, Anhui, Jiangsu, Jiangxi, Hubei, Fujian, Henan and Hunan, located in southern China (Figure 1). Two outgroup species of Buzura suppressaria Guenee and Scopula subpunctaria Herrich-Schaeffer were collected from a tea plantation in Hangzhou, Zhejiang province. All insect larvae were collected in summer in 2011 and 2012, rinsed in 70% ethanol three times, and stored at −80°C in 100% ethanol until DNA was extracted. All insect specimens were first identified under a stereo microscope following the current keys based on characters such as antennae, forewing and venation before further analysis [2], [5], [6]. The insects and DNA samples were deposited in the Tea Research Institute, Chinese Academy of Agricultural Sciences, Hangzhou, China.
Figure 1. The location of sampled E. obliqua populations.
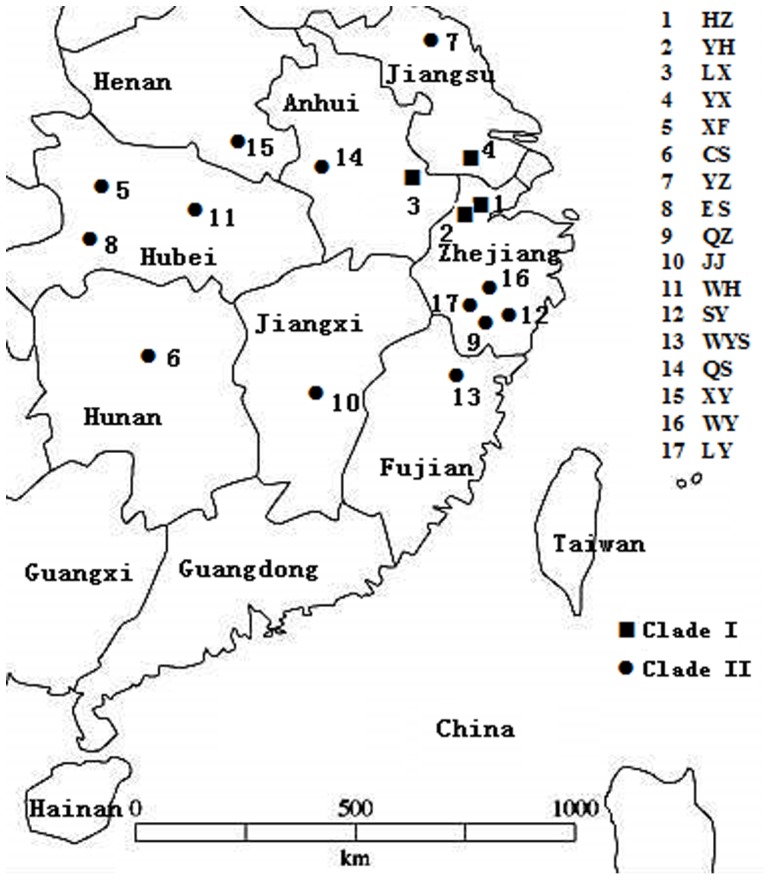
Four populations of Clade I (square) were sampled from Hangzhou (HZ), Yuhang (YH), Langxi (LX) and Yixing (YX); thirteen populations of Clade II (closed circle) were sampled from Xiangfan (XF), Changsha (CS), Yangzhou (YZ), Enshi (ES), Quzhou (QZ), Jiujiang (JJ), Wuhan (WH), Songyang (SY), Wuyishan (WYS), Qianshan (QS), Xinyang (XY), Wuyi (WY) and Longyou (LY) in Chinese tea production areas.
DNA extraction, amplification and sequencing
A total of 171 specimens were sequenced for this study, representing 17 different populations (Table 1). Total DNA was extracted from individual larvae after air-drying for 20 min. The specimen tissue was first ground with a flame-sealed pipette tip in a 1.5 ml microcentrifuge tube, and then, DNA from each individual was extracted using the DNeasy Tissue kit (Qiagen). All genomic DNA was dissolved in 100 µl deionized and sterilized water and stored at −80°C until use. Polymerase chain reactions were carried out on an ABI Veriti thermocycler in a 50 µl total volume containing 5 ng DNA, 1.5 mM MgCl2, 0.25 mM dNTPs, 0.2 µM of each primer and 0.5 units taq NEB DNA polymerase. We amplified the COI genes with primers 5′-GGTCAACAAATCATAAAGATATTG-3′ and 5′-TAAACTTCAGGGTGACCAAAAAAT-3′ [18]. The typical amplification profile for COI was melting at 94°C for 4 min, followed by 35 cycles of melting at 94°C for 30 sec, annealing at 50°C for 1 min, extension at 72°C for 1 min, followed by a final extension at 72°C for 10 min. The PCR products were detected by electrophoresis on a 1.2% agarose gel stained with ethidium bromide, purified with the Gel DNA extraction kit (Axygen), and introduced into pGEM T-easy vector (Promega). Three clones for each product were sequenced bidirectionally by an ABI 3730 automated sequencer (ABI) with the universal primers M13F: 5′-CGCCAGGGTTTTCCCAGTCACGAC-3′ and M13R: 5′-AGCGGATAACAATTTCACACAGGA-3′.
Table 1. Samples and collection information used in this study.
| Populations | Code | Collecting locality | Longitude°(E)/Latitude°(N) | Collecting date (M/Y) | Clade (I/II) |
| Hangzhou | HZ | Hangzhou/Zhejiang | 120.10/30.16 | 6/2011 | I |
| Yuhang | YH | Yuhang/Zhejiang | 119.90/30.39 | 5/2012 | I |
| Langxi | LX | Langxi/Anhui | 119.13/30.98 | 6/2011 | I |
| Yixing | YX | Yixing/Jiangsu | 119.67/31.34 | 6/2011 | I |
| Quzhou | QZ | Quzhou/Zhejiang | 118.88/28.97 | 6/2011 | II |
| Songyang | SY | Songyang/Zhejiang | 119.42/28.49 | 5/2012 | II |
| Longyou | LY | Longyou/Zhejiang | 119.17/29.03 | 5/2011 | II |
| Wuyi | WY | Wuyi/Zhejiang | 119.85/28.96 | 5/2012 | II |
| Qianshan | QS | Qianshan/Anhui | 116.57/30.64 | 5/2011 | II |
| Yangzhou | YZ | Yangzhou/Jiangsu | 119.26/32.23 | 5/2011 | II |
| Wuhan | WH | Wuhan/Hubei | 114.31/30.55 | 5/2012 | II |
| Xiangfan | XF | Xiangfan/Hubei | 112.13/32.02 | 5/2012 | II |
| Enshi | ES | Enshi/Hubei | 109.42/29.89 | 5/2012 | II |
| Xinyang | XY | Xinyang/Henan | 114.08/32.12 | 6/2012 | II |
| Wuyishan | WYS | Wuyishan/Fujian | 117.09/27.07 | 6/2012 | II |
| Jiujiang | JJ | Jiujiang/Jiangxi | 115.99/29.55 | 6/2012 | II |
| Changsha | CS | Changsha/Hunan | 113.26/28.27 | 5/2011 | II |
Genetic diversity and phylogenetic analysis
Genetic diversity was assessed using DnaSP v5.10 to estimate haplotype (Hd) diversity [19], and the outgroup species (B. suppressaria and S. subpunctaria) were excluded from these analyses. Phylogenetic tree searches were conducted using neighbor-joining (NJ) and maximum-parsimony (MP) methods by MEGA 5 program with 1000 bootstrap replications. The software MEGA 5 was also used to estimate mean sequence divergence among populations based on the Kimura 2-Parameter (K2-P) model [20]. The software Arlequin 3.11 was used to analyze genetic differentiation [21], gene flow and molecular variance between populations. The evolutionary network was constructed via a median joining method as implemented in the Network 4.6.0.0 software.
Propagation of Populations
All the tea geometrid populations used were first inspected using a microscope for morphological analysis. Taxonomic keys including antennae, forewing, wing, and genitalia characters were used for diagnosing specimens as E. obliqua [5], [6]. All collected populations were reared under the same conditions, feeding on fresh tea leaves, in a temperature-controlled room at 24±1°C, 75% relative humidity (RH), and a 12-hr light/12-hr dark photoperiod. Three populations susceptible or insensitive to virus were randomly selected for cross-breeding and maintained at least two generations prior to the start of cross-breeding and propagated in the laboratory as Xi et al. (2011) described [8]. The male and female pupae of each population were isolated and reared for mating.
Cross-breeding of Populations
Based on the genetic divergence and molecular phylogenetic analysis of the mitochondrial COI gene, the populations were divided into two distinct groups. We randomly selected several populations from each group for further crossing experiments. The populations of HZ, YH, YX and LX were classified into one group, and the other thirteen populations were classified into another group. The YH, YX, LX populations from the first group and the SY, WH, QS populations from the second group were used in cross-breeding experiments. Two pairs from the YX and LX, and QS and WH populations were crossed for inter-population and intra-group studies, and YH and SY was the cross pair for inter-group population studies.
All reciprocal crosses were conducted simultaneously and under the same laboratory conditions as the parental colonies were maintained [22], [23]. Each cross was set up between one virgin female and one male in a transparent colorless glass cage, and intra-population mating was used as the control treatment. Two pairs were used for one repeat and twelve experimental repeats were for all cross-breeding or mating tests. Each pair was observed daily to record egg number, hatching rate, survival to adult, the proportion of normal adults, and the sex-ratio. The F1 offspring were self-crossed using the conditions mentioned above. The self-crosses of the intra-population of the F1 generation were the same as for the population in the control treatment, and the eggs, hatching rate and larvae development of F2 were simultaneously recorded. The sex-ratios across replicates were calculated by dividing the total number of females by the overall number of males in each treatment. All the other parameters were compared amongst treatments by one-way ANOVA analysis using SPSS statistical software 21.0 and the equality of means in the same column was tested by Duncan's multiple range test.
Results
PCR amplifications and sequences analysis
We cloned and sequenced the COI gene for all 171 specimens from seventeen tea geometrid populations and 20 samples from two outgroup populations. The COI fragments are 658 bp in length with an A+T content of 70.6%. The sequence alignment contains 157 polymorphic sites and 115 parsimony informative sites. A total of 50 COI haplotypes were observed. Sequences of 50 haplotypes for E. obliqua and 6 haplotypes for the two outgroups of B. suppressaria and S. subpunctaria were deposited in GenBank (Accession Nos. KF748178-KF748227, KF748228-KF748230, and KF748231-KF748233). The overall haplotype diversity index (Hd) was 0.73. The mean nucleotide diversity index (Pi) and mean gene flow (Nm) among the 17 populations were 0.018 and 0.050 respectively, and the Fst index of population differentiation was 0.877. Only two haplotypes QZ1 and HZ2 were shared between populations: HZ2 was shared by 21 samples from four populations of HZ, YH, LX and YX, and QZ1 was shared by 86 samples from thirteen populations of QZ, JJ, XY, QS, CS, WH, XF, LY, YZ, SY, WY, ES and WYS. The other 48 haplotypes were each found in only one population. Among the 17 populations, the LY population had just one haplotype, and other sixteen populations had more than one haplotype. The populations with the highest haplotype diversity were WH, which had seven haplotypes, and LX, which had eight haplotypes. The mean genetic divergences (D) and nucleotide diversity (Pi) differed significantly between the two groups (Table 2). In addition, the neutrality-test values (Tajima's D, Fu and Li's D and Fu and Li's F) were all significantly less than zero.
Table 2. Comparison of the COI gene evolution of two E. obliqua clades.
| n | h | S | Pi | D | Tajima's D | Fu and Li's D | Fu and Li's F | |
| Clade I | 53 | 17 | 21 | 0.00453 | −2.068547 | −2.16308** | −3.22719** | −3.38123** |
| Clade II | 118 | 33 | 36 | 0.00332 | −2.714675 | −2.71467*** | −5.25702** | −5.21491** |
The letters n, h, S, Pi and D are the no. of E. obliqua samples, no. of mitochondrial DNA haplotype, no. of segregating sites, nucleotide diversity and genetic divergence respectively.
*: P<0.05;
**: P<0.02;
***: P<0.01.
Genetic distance
The genetic distances among seventeen populations were calculated using the Kimura 2-parameter model in MEGA 5 (Table 3). The genetic divergences for the COI genes among populations of E. obliqua were 0.0%–4.3%, which was a large interval. They were obviously less than those between E. obliqua and either outgroup, which were 10.7%–12.9%. We found that the genetic distances between each pair of populations from the group containing HZ, YH, LX and YX were 0.0%–0.3%, and those between each pair of populations from the group containing QZ, JJ, XY, QS, CS, WH, XF, LY, YZ, SY, WY, ES and WYS were 0.0%–0.7%. The pairwise distances between populations from two groups were 3.7 to 4.3%. These results showed that the seventeen E. obliqua populations contained two deeply divergent groups, one including the populations of HZ, YH, LX and YX, and the other including the populations of QZ, JJ, XY, QS, CS, WH, XF, LY, YZ, SY, WY, ES and WYS.
Table 3. The average pairwise genetic distance of the populations of E. obliqua based on the Kimura 2-parameter model.
| Population code | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | |
| 1 | HZ | |||||||||||||||||||
| 2 | YH | 0.001 | ||||||||||||||||||
| 3 | LX | 0.003 | 0.002 | |||||||||||||||||
| 4 | YX | 0.002 | 0.002 | 0.003 | ||||||||||||||||
| 5 | XF | 0.037 | 0.037 | 0.037 | 0.037 | |||||||||||||||
| 6 | CS | 0.042 | 0.042 | 0.042 | 0.043 | 0.007 | ||||||||||||||
| 7 | YZ | 0.042 | 0.042 | 0.042 | 0.043 | 0.007 | 0.001 | |||||||||||||
| 8 | ES | 0.042 | 0.042 | 0.042 | 0.043 | 0.007 | 0.001 | 0.001 | ||||||||||||
| 9 | QZ | 0.042 | 0.042 | 0.042 | 0.043 | 0.007 | 0.001 | 0.001 | 0.001 | |||||||||||
| 10 | JJ | 0.042 | 0.042 | 0.042 | 0.043 | 0.007 | 0.001 | 0.001 | 0.001 | 0.001 | ||||||||||
| 11 | WH | 0.043 | 0.043 | 0.043 | 0.043 | 0.007 | 0.001 | 0.002 | 0.002 | 0.002 | 0.002 | |||||||||
| 12 | SY | 0.042 | 0.042 | 0.042 | 0.043 | 0.008 | 0.001 | 0.001 | 0.001 | 0.001 | 0.001 | 0.001 | ||||||||
| 13 | WYS | 0.042 | 0.042 | 0.042 | 0.043 | 0.007 | 0.001 | 0.001 | 0.001 | 0.001 | 0.001 | 0.002 | 0.001 | |||||||
| 14 | QS | 0.042 | 0.042 | 0.042 | 0.043 | 0.007 | 0.001 | 0.001 | 0.001 | 0.001 | 0.001 | 0.001 | 0.001 | 0.001 | ||||||
| 15 | XY | 0.042 | 0.042 | 0.042 | 0.042 | 0.007 | 0.000 | 0.001 | 0.001 | 0.001 | 0.001 | 0.001 | 0.000 | 0.001 | 0.001 | |||||
| 16 | WY | 0.042 | 0.042 | 0.042 | 0.043 | 0.007 | 0.001 | 0.001 | 0.001 | 0.001 | 0.001 | 0.001 | 0.001 | 0.001 | 0.001 | 0.000 | ||||
| 17 | LY | 0.042 | 0.042 | 0.042 | 0.042 | 0.007 | 0.000 | 0.001 | 0.001 | 0.001 | 0.001 | 0.001 | 0.001 | 0.001 | 0.000 | 0.000 | 0.000 | |||
| 18 | Ss | 0.107 | 0.107 | 0.109 | 0.108 | 0.107 | 0.107 | 0.107 | 0.107 | 0.107 | 0.107 | 0.108 | 0.107 | 0.107 | 0.107 | 0.107 | 0.107 | 0.107 | ||
| 19 | Bs | 0.116 | 0.116 | 0.115 | 0.117 | 0.122 | 0.123 | 0.123 | 0.123 | 0.124 | 0.123 | 0.123 | 0.123 | 0.123 | 0.123 | 0.123 | 0.123 | 0.123 | 0.129 | |
Bs (S. subpunctaria) and Ss (B. suppressaria) are the outgroups.
Phylogenetic analysis
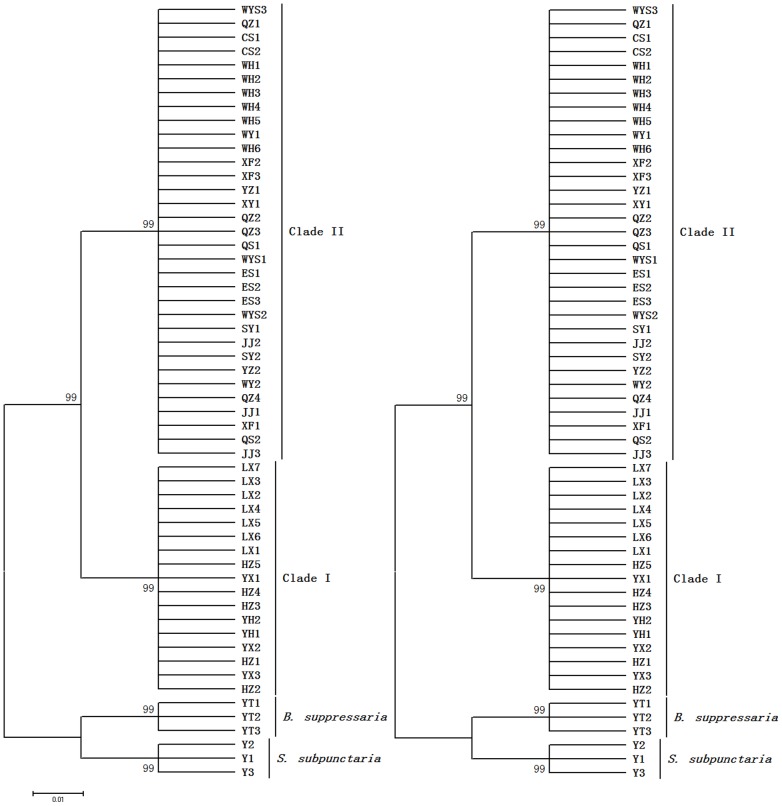
Based on the 50 haplotypes and two outgroups of B. suppressaria and S. subpunctaria, we analyzed the phylogenetic relationships among the seventeen populations using the NJ and MP methods in MEGA 5 (Figure 2). The phylogenetic trees were highly consistent, and both showed that all haplotypes within any population fell into a single group. The E. obliqua populations of HZ, YH, LX and YX clustered into one group we named Clade I, while the populations of QZ, JJ, XY, QS, CS, WH, XF, LY, YZ, SY, WY, ES and WYS clustered into another group we named Clade II. The haplotypes found in multiple populations (QZ1 and HZ2) were restricted to Clade II or Clade I populations, respectively. Because two haplotypes of QZ1 and HZ2 distributed universally within their respective clade, and the analysis of median joining network also supported they were predominant ones, they may be the original haplotypes. The phylogenetic results were consistent with the genetic distance analysis that concluded that E. obliqua has two deeply divergent clades in China.
Figure 2. The NJ tree (L) and MP tree (R) of E. obliqua and two outgroup species based on COI sequence haplotypes.
Numbers above the nodes indicate bootstrap support. The outgroups used were B. suppressaria and S. subpunctaria. The QZ1 haplotype was shared by all thirteen populations insensitive to the EoNPV virus within Clade II; the HZ2 haplotype was shared by all four populations susceptible to the virus within Clade I; and other haplotypes were unique.
Inheritance analysis
Three populations were selected from each clade, the YH, YX and LX populations from Clade I, and the SY, WH and QS populations from Clade II. The cross of YH and SY was inter-clade, and the two pairs of YX and LX and WH and QS were intra-clade. All the crossed pairs either inter-clade or intra-clade, produced eggs. Significantly fewer eggs were observed in the cross-breeding of YH♀×SY♂ compared with the control self-cross of the SY population: the number of eggs laid per female was 357±17.31, which was less than 442±20.30 (P<0.01) (Table 4). All eggs produced in the intra-population and inter-population intra-clade crosses were fertile. The hatchability, survival to adult and percentage normal adults of the two cross-breeding combinations were all much lower than for intra-population self-crosses (P<0.01). Moreover, the F1 generation sex ratio (♀:♂) after cross-breeding between the YH and SY populations was highly unbalanced, at 1∶4 for the cross of YH♀×SY♂ and 1∶27 for the cross of SY♀×YH♂.
Table 4. The cross-breeding for the YH and SY populations of E. obliqua.
| Cross | Replications | No. eggs laid/female | Hatchability | Survival to adult | Percentage of normal adults | Sex ratio (♀:♂) |
| YH♀×SY♂ | 12 | 357±15.58 B | 0.01±0.00 C | 0.17±0.02 B | 18.23±12.25 B | 1∶4 |
| SY♀×YH♂ | 12 | 394±19.35 AB | 0.54±0.05 B | 0.19±0.01 B | 24.70±10.06 B | 1∶27 |
| YH♀×YH♂ | 12 | 396±8.90 AB | 0.74±0.09 A | 0.80±0.03 A | 93.49±2.05 A | 1∶1 |
| SY♀×SY♂ | 12 | 442±13.80 A | 0.66±0.02 A | 0.77±0.02 A | 94.16±0.62 A | 1∶1 |
Data in the table are mean ± SE. The letters following the SEs group observations in the same column that are not significantly different (P>0.01, Duncan's multiple range test. Means with no common letters were significantly different. Note that the values of the sex ratio (♀:♂) were not analysed in this manner.
To detect reproductive isolation level between the two E. obliqua clades, we conducted intra-population self-crosses of the F1 generation. Highly unbalanced sex-ratios and few emerged normal adults reduced the opportunities for selfcrosses and backcrosses of the F1 hybrids. We found that the YS offspring of the F1 generation of YH♀×SY♂ couldn't emerge within three days and therefore produced no eggs with no hatching, and the SY' offspring of the F1 generation of SY♀×YH♂ produced extremely few eggs with no hatching (P<0.01) (Table 5). Therefore, the inter-clade cross-breeding for E. obliqua could not produce an F2 generation. In addition to using the self-cross intra-population as controls, we also conducted one intra-clade cross. Compared with the self-cross controls, the F1 generations from cross-breeding the YX×LX populations in Clade I and the WH×QS populations in Clade II exhibited no difference in egg production, hatchability, emergence or sex ratio (Table 6, 7). Furthermore, their F2 and F3 generations also developed and produced normal offspring after self-crosses.
Table 5. The self-crosses of F1 generation from cross-breeding of the YH and SY populations.
| Treatment | Replications | No. eggs laid/female | Hatchability |
| F1(YS)♀×F1(YS)♂ | 0 | — | — |
| F1(SY′)♀×F1(SY′)♂ | 1 | 315 | — |
| F1(YH)♀×F1(YH)♂ | 8 | 465±16.62 | 0.82±0.03 |
| F1(SY)♀×F1(SY)♂ | 8 | 453±37.99 | 0.72±0.10 |
Data shown in the table are mean ± SE. There were no significant differences at the 0.01 level between mean values within columns. The YS is the F1 generation from cross-breeding of YH♀×SY♂, and the SY′ is the F1 generation from cross-breeding of SY♀×YH♂. “—” means none, that is the YS couldn't emerge within three days and therefore produced no eggs with no hatching, and the SY′ produced extremely few eggs with no hatching.
Table 6. The cross-breeding of the YX and LX populations within Clade I.
| Treatment | Replications | No. eggs laid/female | Hatchability | Adult emergence rate | Sex ratio (♀:♂) |
| YX♀×LX♂ | 8 | 146±18.25 | 0.72±0.08 | 0.62±0.05 | 1: 1.33 |
| LX♀×YX♂ | 8 | 172±22.65 | 0.68±0.14 | 0.56±0.01 | 1∶1.38 |
| YX♀×YX♂ | 8 | 235±34.47 | 0.82±0.01 | 0.51±0.02 | 1∶1.57 |
| LX♀×LX♂ | 8 | 235±54.20 | 0.74±0.08 | 0.52±0.05 | 1∶1.09 |
Data shown in the table are mean ± SE. There were no significant differences at the 0.01 level between mean values within columns. The values of sex ratio (♀:♂) were not included in this analysis.
Table 7. The cross-breeding of the WH and QS populations within Clade II.
| Treatment | Replications | No. eggs laid/female | Hatchability | Adult emergence rate | Sex ratio (♀:♂) |
| WH♀×QS♂ | 8 | 115±25.47 | 0.54±0.04 | 0.95±0.04 | 1∶1.47 |
| QS♀×WH♂ | 8 | 109±26.47 | 0.60±0.06 | 0.89±0.07 | 1∶1.54 |
| WH♀×WH♂ | 8 | 121±17.98 | 0.65±0.01 | 0.89±0.02 | 1∶1.05 |
| QS♀×QS♂ | 8 | 139±17.31 | 0.63±0.09 | 0.85±0.15 | 1∶1.42 |
Data shown in the table were mean ± SE. There were no significant differences at the 0.01 level between mean values within columns. The values of sex ratio (♀:♂) were not included in this analysis.
Discussion
Tea is an evergreen bush with a long growing season from March to September. Growers have to spray several times to minimize herbivore damage. The development of resistance to chemical insecticides is very common in many tea insects [3], [24], possibly driven by selection pressure [9]. Although EoNPV is not a chemical insecticide, resistance may still develop in target pest populations. Our previous study showed significant variations (up to 700-fold) in susceptibility to EoNPV in field populations of E. obliqua [5], causing us to question whether resistance has evolved in this insect in a similar manner to the well-documented development of Bt resistance due to extensive use of Bt or transgenic Bt crops or to newly arising baculovirus resistance in moths [9], [25]. Therefore, we surveyed the level of EoNPV used in those locations. Because the only EoNPV pesticide product used in China was developed by our institute and an agricultural chemical company, we have a clear application record of this virus in tea plantation. Surprisingly, there was no record of EoNPV for E. obliqua control in the locations of the studied populations. Hence, EoNPV may not have had a direct influence on the susceptibility changes in E. obliqua populations. Considering the high specificity of the virus and the vast geographic variation in tea ecosystems, our second hypothesis for this study was that E. obliqua may have evolved deep genetic divergence to adapt to unique and localized tea ecosystems.
In this study, we first conducted phylogenetic analysis of the COI region of E. obliqua mtDNA, which is a useful tool for identifying sibling species that lack morphological characters [17], [26]–[28]. All 50 haplotypes (assembled from 171 sequences) from seventeen E. obliqua populations fell into two clear clades. Within a clade, populations showed close relationships, with genetic distances less than 0.7%. In contrast, the genetic divergences between clades were 3.7%–4.3%, well beyond the criterion of 2% for insect species differentiation in general or the 3% specifically suggested for lepidopteran species differentiation [29], [30]. These molecular data strongly indicated that E. obliqua is a cryptic complex including two deeply divergent clades. Moreover, the lack of shared haplotypes also indicated a certain degree of reproductive isolation between the clades [17].
To test species divergence and reproductive isolation, we selected three populations from each clade and conducted inter-clade crossing and F1 self-crossing tests. Although the number of eggs produced by inter-clade crossing was not significantly different from the in-clade self-crossed control, the egg hatchability, survival rates to adult and percentage of normal adults were significantly lower (along with an abnormal sex ratio) than those of the self-crossing controls. The selfcross of the F1 generation from inter-clade hybrids produced sterile eggs or no eggs, while the control group F1, F2 and F3 generations obtained from crossings between populations within a clade could grow, develop and produce normal progenies. The cross-breeding results further support the suggestion of reproductive isolation between two clades of E. obliqua.
Cryptic species complexes, virtually identical in appearance but nonetheless having reproductive isolation, are very common in nature [17], [31]–[33]. The biological characteristics of the tea plant and decreasing artificial disturbance after initial planting turn tea plantations into a shaded and stable ecosystem that is consequently a permanent habitat for herbivores. Thus, not all insects in tea plantations migrate long distances, and thus, these insects can easily colonize and develop diversified evolution in adaptive surroundings [34], [35]. Furthermore, few tea plant varieties require replacement during production due to their long life, which also decreases the genetic exchanges among E. obliqua populations. Many tea plantations are also located on mountains with highly diversified geography and climate, which might drive the divergence. The locations of the four populations within Clade I (HZ, YH, YX and LX) are close to each other and possess the same geographic characteristics and climate, while the Clade II locations are in the mountains with diversified geography and changeable climate within each of them. As a result, these possible factors may drive the divergent evolution in relatively isolated E. obliqua populations.
In summary, the detection of strong genetic divergence and reproductive isolation in E. obliqua provides valuable information for guiding effective control of this pest. However, more research is needed to understand the distribution of these cryptic species in tea-growing areas. Future studies also need to include clarification of the mechanism of reduced or lost sensitivity to EoNPV and should explore the potential application of the reproductive incompatibility of the cryptic species for strategic pest management.
Acknowledgments
The authors wish to thank all the reviewers for their comments that help improve the manuscript and all the people for collecting specimens. Thanks Dr. Nan Jiang at the Institute of Zoology, Chinese Academy of Sciences (IOZ, CAS) for kind help of morphological diagnosing. Zhi-Jun Yuan and Gui-Hua Zhang are post-graduate students at the Tea Research Institute, Chinese Academy of Agricultural Sciences, (TRI, CAAS). This work is part of their academic dissertations.
Funding Statement
This work was supported by the National Science and Technology Support Program Projects (2011BAD01B02), the Fundamental Research Funds of National Science and Technology (2013FY113200), the Key Innovation Team of Zhejiang Province (Grant No. 2011R09027-13), the National Natural Science Foundation of China (Grant No. 31100503) and the public technology application research of Zhejiang Province (2013C32086). The funders had no role in study design, samples collection and data analysis, decision to publish, or manuscript preparation.
References
- 1.Scoble M, Hausmann A ([updated 2007]) Online list of valid and available names of the Geometridae of the World Available: http://wwwlepbarcodingorg/geometridae/species_checklistsphp. Accessed 2013 Feb 2.
- 2.Zhang HH, Tan JC (2004) Tea pests and non-pollution management in China. Anhui Science and Technology Publishing House: 3–9.
- 3. Hazarika LK, Bhuyan M, Hazarika BN (2009) Insect pests of tea and their management. Ann Rev Entomol 54: 267–284. [DOI] [PubMed] [Google Scholar]
- 4. Warren W (1894) New genera and species of Geometridae. Novit zool 1: 366–466. [Google Scholar]
- 5. Prout LB (1912–1916) The Palaearctic Geometrae. In: Seitz A (Ed) The Macrolepidoptera of the World 4: 1–479. [Google Scholar]
- 6. Prout LB (1930) On the Japanese Geometridae of the Aigner collection. Novit zool 35: 289–377. [Google Scholar]
- 7. Ye GY, Xiao Q, Chen M, Chen XX, Yuan ZJ, et al. (2014) Tea: Biological control of insect and mite pests in China. Biol Control 68: 73–91. [Google Scholar]
- 8. Xi Y, Xiao Q, Yin KS (2011) The Susceptibility Difference against EoNPV in Different Geographic Populations of Tea Geometrid (Ectropis obliqua Prout). J Tea Sci 31: 100–104. [Google Scholar]
- 9. Asser-Kaiser S, Fritsch E, Undorf-Spahn K, Kienzle J, Eberle KE, et al. (2007) Rapid emergence of baculovirus resistance in codling moth due to dominant, sex-linked inheritance. Science 317: 1916–1918. [DOI] [PubMed] [Google Scholar]
- 10. Simon C, Frati F, Beckenbach A, Crespi B, Liu H, et al. (1994) Evolution, weighting, and phylogenetic utility of mitochondrial gene sequences and a compilation of conserved polymerase chain reaction primers. Ann Entomol Soc Am 87: 651–701. [Google Scholar]
- 11. Monti MM, Nappo AG, Giorgini M (2005) Molecular characterization of closely related species in the parasitic genus Encarsia (Hymenoptera: Aphelinidae) based on the mitochondrial cytochrome oxidase subunit I gene. Bull Entomol Res 95: 401–408. [DOI] [PubMed] [Google Scholar]
- 12. Boykin LM, Shatters RG, Hall DG, Burns RE, Franqui RA (2006) Analysis of host preference and geographical distribution of Anastrepha suspensa (Diptera: Tephritidae) using phylogenetic analyses of mitochondrial cytochrome oxidase I DNA sequence data. Bull Entomol Res 96: 457–469. [PubMed] [Google Scholar]
- 13. Polaszek A, Manzari S, Quicke DLJ (2004) Morphological and molecular taxonomic analysis of the Encarsia meritoria species-complex (Hymenoptera, Aphelinidae), parasitoids of whiteflies (Hemiptera, Aleyrodidae) of economic importance. Zool Scr 33: 403–421. [Google Scholar]
- 14. Behere GT, Tay WT, Russell DA, Heckel DG, Appleton BR, et al. (2007) Mitochondrial DNA analysis of field populations of Helicoverpa armigera (Lepidoptera: Noctuidae) and of its relationship to H. zea . BMC Evol Biol 14: 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Beltran M, Jiggins CD, Bull V, Linares M, Mallet J, et al. (2002) Phylogenetic discordance at the species boundary: comparative gene genealogies among rapidly radiating Heliconius butterflies. Mol Biol Evol 19: 2176–2190. [DOI] [PubMed] [Google Scholar]
- 16. Sperling FA, Raske AG, Otvos IS (1999) Mitochondrial DNA sequence variation among populations and host races of Lambdina fiscellaria (Gn.) (Lepidoptera: Geometridae). Insect Mol Biol 8: 97–106. [DOI] [PubMed] [Google Scholar]
- 17. Hebert PDN, Penton EH, Burns JM, Janzen DH, Hallwachs W (2004) Ten species in one: DNA barcoding reveals cryptic species in the neotropical skipper butterfly Astraptes fulgerator . PNAS 101: 14812–14817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yin XB, Hao JS, Xu L, Zhu GP, Huang DY, et al. (2007) Phylogeny of butterflies of the subfamily Elymninae and Satyrinae (Lepidoptera: Satyridae) based on mitochondrial COI and Cytb gene sequences. Acta Entomol Sinica 50: 1263–1271. [Google Scholar]
- 19. Librado P, Rozas J (2009) DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25: 1451–1452. [DOI] [PubMed] [Google Scholar]
- 20. Kumar S, Nei M, Dudley J, Tamura K (2008) MEGA: a biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief Bioinform 9: 299–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Excoffier L, Estoup A, Cornuet JM (2005) Bayesian analysis of an admixture model with mutations and arbitrarily linked markers. Genetics 169: 1727–1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Famah Sourassou N, Hanna R, Zannou I, Breeuwer JA, de Moraes G, et al. (2012) Morphological, molecular and cross-breeding analysis of geographic populations of coconut-mite associated predatory mites identified as Neoseiulus baraki: evidence for cryptic species? Exp Appl Acarol 57: 15–36. [DOI] [PubMed] [Google Scholar]
- 24. Saha D, Mukhopadhyay A (2013) Insecticide resistance mechanisms in three sucking insect pests of tea with reference to North-East India: an appraisal. Int J Trop Insect Sci 33: 46–70. [Google Scholar]
- 25. Liu FY, Xu ZP, Zhu YC, Huang FN, Wang YH, et al. (2010) Evidence of field-evolved resistance to Cry1Ac-expressing Bt cotton in Helicoverpa armigera (Lepidoptera: Noctuidae) in northern China. Pest Manag Sci 66: 155–161. [DOI] [PubMed] [Google Scholar]
- 26. Pages N, Munoz-Munoz F, Talavera S, Sarto V, Lorca C, et al. (2009) Identification of cryptic species of Culicoides (Diptera: Ceratopogonidae) in the subgenus Culicoides and development of species-specific PCR assays based on barcode regions. Vet Parasitol 165: 298–310. [DOI] [PubMed] [Google Scholar]
- 27. Blair CP, Abrahamson WG, Jackman JA, Tyrrell L (2005) Cryptic speciation and host-race formation in a purportedly generalist tumbling flower beetle. Evolution 59: 304–316. [PubMed] [Google Scholar]
- 28. Qin L, Wang J, Bing XL, Liu SS (2013) Identification of nine cryptic species of Bemisia tabaci (Hemiptera: Aleyrodidae) from China by using the mtCOI PCR-RFLP technique. Acta Entomol Sinica 56: 186–194. [Google Scholar]
- 29. Hebert PD, Ratnasingham S, deWaard JR (2003) Barcoding animal life: cytochrome c oxidase subunit 1 divergences among closely related species. Proc Biol Sci 270 Suppl 1S96–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hebert PDN, Cywinska A, Ball SL, DeWaard JR (2003) Biological identifications through DNA barcodes. P Roy Soc B-Biol Sci 270: 313–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Dinca V, Lukhtanov VA, Talavera G, Vila R (2011) Unexpected layers of cryptic diversity in wood white Leptidea butterflies. Nat Commun 2: 324. [DOI] [PubMed] [Google Scholar]
- 32. Ndo C, Simard F, Kengne P, Awono-Ambene P, Morlais I, et al. (2013) Cryptic genetic diversity within the Anopheles nili group of malaria vectors in the equatorial forest area of Cameroon (Central Africa). PLoS One 8: e58862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Smith MA, Woodley NE, Janzen DH, Hallwachs W, Hebert PD (2006) DNA barcodes reveal cryptic host-specificity within the presumed polyphagous members of a genus of parasitoid flies (Diptera: Tachinidae). PNAS 103: 3657–3662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. deWaard J, Hebert P D, Humble L (2011) A Comprehensive DNA Barcode Library for the Looper Moths (Lepidoptera: Geometridae) of British Columbia, Canada. PLoS One 6: e18290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Doak P (2000) Population consequences of restricted dispersal for an insect herbivore in a subdivided habitat. Ecology 81: 1828–1841. [Google Scholar]