Abstract
The outer membrane of Gram-negative bacteria is a unique asymmetric lipid bilayer composed of phospholipids (PLs) in the inner leaflet and lipopolysaccharides (LPSs) in the outer leaflet. Its function as a selective barrier is crucial for the survival of bacteria in many distinct environments, and it also renders Gram-negative bacteria more resistant to antibiotics than their Gram-positive counterparts. Here, we report the structural properties of a model of the Escherichia coli outer membrane and its interaction with outer membrane phospholipase A (OmpLA) utilizing molecular dynamics simulations. Our results reveal that given the lipid composition used here, the hydrophobic thickness of the outer membrane is ∼3 Å thinner than the corresponding PL bilayer, mainly because of the thinner LPS leaflet. Further thinning in the vicinity of OmpLA is observed due to hydrophobic matching. The particular shape of the OmpLA barrel induces various interactions between LPS and PL leaflets, resulting in asymmetric thinning around the protein. The interaction between OmpLA extracellular loops and LPS (headgroups and core oligosaccharides) stabilizes the loop conformation with reduced dynamics, which leads to secondary structure variation and loop displacement compared to that in a DLPC bilayer. In addition, we demonstrate that the LPS/PL ratios in asymmetric bilayers can be reliably estimated by the per-lipid surface area of each lipid type, and there is no statistical difference in the overall membrane structure for the outer membranes with one more or less LPS in the outer leaflet, although individual lipid properties vary slightly.
Introduction
The structure, function, and dynamics of proteins are intimately linked to the properties of their solvent. Knowing how membrane proteins interact with their cognate bilayers is critical for understanding how the bilayer may influence membrane protein behavior. Folding studies are one experimental approach that is useful in dissecting how membranes influence the formation of membrane protein structure. As a class, transmembrane β-barrel outer membrane proteins (OMPs) have been shown to be particularly amenable to such investigations (1,2). The water-to-bilayer folding stabilities are known for several transmembrane β-barrels (3,4), and there are folding kinetics of β-barrels from Escherichia coli, Fusobacterium nucleatum, and mitochondria (1,5). One of the most striking findings from this body of work is how profoundly the membrane affects the apparent folding kinetics. OMPs fold faster into bilayers that are generally considered to have relatively thin hydrophobic regions. For example, under otherwise identical experimental conditions, the folding of nine E. coli OMPs is fastest into vesicles with diC10 acyl chains, slower in diC11 acyl chains, and even slower in diC12 acyl chains (1). Furthermore, the apparent rates of OMP folding are strongly modulated by the bilayer geometry. Small unilamellar vesicles with high curvature support faster folding compared to equivalent large unilamellar vesicles with lower curvature (6). Although function is preserved in proteins folded into these synthetic vesicles (4), the extent to which these synthetic bilayer compositions and geometries in vitro recapitulate the influences of the biological outer membrane of Gram-negative bacteria is currently unknown.
The E. coli outer membrane is structurally quite distinct from the synthetic bilayers used in folding studies. Although the inner leaflet of this membrane is composed of phospholipids, the outer leaflet lacks phospholipids and is instead made up of a specialized, highly glycosylated amphipathic molecule known as lipopolysaccharide (LPS) that consists of lipid A, a core oligosaccharide, and an O-antigen polysaccharide. The presence of phospholipids on the outer leaflet only occurs in situations of bacterial stress, such as phage infection, and one function of some OMPs is to degrade phospholipids in the outer leaflet when such flipping occurs (7). Because there are not yet any suitable protocols for assembling the asymmetric structure of the outer membrane in solution, folding studies of OMPs using bacterial outer membranes in vitro are not currently possible.
One of the questions we sought to address about the many membrane properties was the hydrophobic thickness of the bacterial outer membrane. It is generally accepted that the hydrophobic thickness of a membrane protein should match the thickness of its native lipid bilayer. Any mismatch between these hydrophobic regions should be energetically costly, because it could result in exposure of nonpolar groups to the aqueous solution or lead to bilayer deformations. Because the E. coli fatty acid composition under normal growth conditions is enriched in lipids with C14 and C16 acyl chains, it might be concluded that the hydrophobic thickness of the outer membrane should be similar to that of the bacterial cytoplasmic (or inner) membrane. However, Lomize et al. have calculated the average hydrophobic thickness of all known OMP structures to be ∼24 Å, whereas the average thickness of plasma membrane protein structures is ∼29 Å (8). The OMPs themselves thus suggest that the hydrophobic thickness of the bacterial outer membrane should be less than that of the inner membrane. A thinner biological membrane could be one reason why OMP folding studies in vitro show faster apparent folding kinetics into synthetic lipid vesicles composed of relatively short chain lipids.
To understand how the properties of this asymmetric outer membrane may affect OMPs, or vice versa, we used all-atom molecular dynamics (MD) to simulate the E. coli outer membrane both alone and with an embedded OMP, outer membrane phospholipase A (OmpLA). In this study, we discuss 1), the microscopic structural properties of the E. coli outer membrane; 2), the protein-lipid interaction in the native outer membrane environment; 3), an explanation for the appropriateness of diC12 acyl chains for OMP folding experiments (hydrophobic matching); 4), the importance of using an asymmetric bilayer with LPS for OMP loop conformation refinement; and 5), a reliable method to build a complex outer membrane bilayer both alone and with OMPs.
Methods
Following the replacement protocol used to build a protein/membrane complex structure in Membrane Builder (9,10) in CHARMM-GUI (11), the OmpLA monomer structure from the Protein Data Bank (PDB ID 1QD5) (12) was embedded in an asymmetric bilayer to mimic the E. coli outer membrane (Fig. 1). The inner leaflet is a mixture of 1-palmitoyl(16:0)-2-palmitoleoyl(16:1 cis-9)-phosphatidylethanolamine (PPPE), 1-palmitoyl(16:0)-2-vacenoyl(18:1 cis-11)-phosphatidylglycerol (PVPG), and 1,1′-palmitoyl-2,2′-vacenoyl cardiolipin with a net charge of −2e (PVCL2) at a ratio of 15:4:1 (13). The corresponding lipid topology and atom charges were transferred by analogy based on the C36 lipid force field (14). The outer leaflet is composed of the rough LPS with lipid A and R1 core (i.e., without O-antigen polysaccharide), because its length is sufficient to immerse the protein loops (15). The lipid A of E. coli LPS consists of six amide/ester-linked fatty acids with a length of either 14 or 12 carbons (16). The initial LPS structure was constructed using CHARMM (17) and followed by sequential Langevin dynamics to reduce the molecular radius from a starting value of 16 Å to 5 Å to avoid bad contacts during membrane assembly (see Wu et al. (15) for details).
Figure 1.
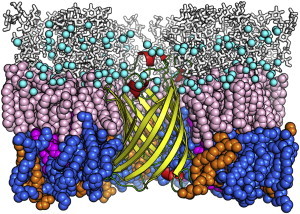
A snapshot of OmpLA (yellow, barrel; red, helix; green, loop and turn) embedded in an asymmetric E. coli outer membrane bilayer at 450 ns. Lipid A, R1 core, PPPE, PVPG, and PVCL2 are pink, white (sticks), blue, orange, and magenta spheres, respectively. Calcium ions are cyan spheres. Water molecules and KCl ions are omitted for clarity. To see this figure in color, go online.
The following step-by-step assembly protocol was used to build the OmpLA/outer membrane simulation system, which is similar to that used in CHARMM-GUI Membrane Builder. In Steps 1 and 2, an OmpLA structure preorientated with respect to the membrane normal (z axis) together with 28 crystal water molecules was read from the OPM database (18) and translated to the xy center. Then, the protein cross-sectional area profile was calculated along the z axis.
In Step 3, the system dimension in xy was estimated by fixing the total lipid number in the inner leaflet to 100, i.e., 75 PPPE, 20 PVPG, and 5 PVCL2. The number of LPS molecules in the outer leaflet was then determined based on the protein cross-sectional areas at the inner and outer leaflets and individual lipid areas (∼63 Å2 for PPPE, ∼62 Å2 for PVPG, ∼130 Å2 for PVCL2, and ∼180 Å2 for rough LPS (15)). Lipidlike pseudoatoms were distributed and packed around OmpLA using Langevin dynamics with planar harmonic restraints; to accurately represent their larger molecular size, we increased the atomic radii of the pseudoatoms representing LPS (9.4 Å) and cardiolipin (7.6 Å) relative to the radius of 5.4 Å for common phospholipids (PLs).
In Steps 4 and 5, the individual components, including bilayer, ions, and bulk water, were built and assembled. An asymmetric bilayer was generated with the replacement method (19,20) by substituting for each pseudoatom a lipid molecule that was randomly selected from a lipid conformer library, where each LPS molecule also has its corresponding Ca2+ ions for neutralization (the total charge of each LPS is −10e in this study). To remove bad contacts, a systematic rigid-body translation (in xy) and rotation (around the z axis) search was performed for each lipid molecule until the optimal orientation was found. After the lipid replacement, protein surface penetration by lipid tails was checked to prevent unstable simulations. The system size along the z axis was determined by adding 20 Å (for bulk water) on the top and bottom of the z maximum and minimum values of the assembled protein/LPS/PL structure. KCl (150 mM) was used for bulk ionic solution, and the initial K+ and Cl− positions were assigned by random placement without bad contact with protein and lipids.
In addition to the system with OmpLA in the E. coli LPS-PL outer membrane (OmpLA-LPS-PL; see Table S1), we constructed several other bilayer systems to compare their structural features with those of the OmpLA-LPS-PL system. These included 1), a neat asymmetric outer membrane without OmpLA but with the same lipid composition (LPS-PL[36,100]); 2), a neat, symmetric PL bilayer with only PPPE, PVPG, and PVCL2 (PL-only); and 3), dilauroylphophatidylcholine (DLPC) bilayers with and without OmpLA (OmpLA/DLPC and DLPC-only), because DLPC has been used previously in both experimental (4) and computational (21) studies of OmpLA. All the systems containing E. coli-type lipids were assembled with the five-step protocol described above. In contrast, the OmpLA/DLPC and DLPC-only systems were constructed through CHARMM-GUI Membrane Builder.
In addition, one important consideration when building the asymmetric bilayer is the possibility for a potential area mismatch between the inner and outer leaflets. This could occur as a consequence of the thermal fluctuations and the lack of a priori knowledge of detailed packing between lipids and between lipids and protein. For these reasons, a second asymmetric bilayer was built with one additional LPS molecule in the outer leaflet (LPS/PL[37,100]) to investigate the statistical difference between outer membranes with two different LPS/PL ratios. To improve sampling and to check simulation convergence, we constructed two independent versions of every system with different initial lipid displacement and packing, except in the case of the DLPC bilayer, for which only one system was constructed. Thus, a total of 11 systems were constructed and simulated (Table S1).
Equilibration simulations were performed for all membrane systems using CHARMM (17) with the C36 lipid (14) and carbohydrate (22–24) force fields, and the TIP3P water model (25). These simulations were 450 ps long for all systems except DLPC systems, for which they were 375 ps. To ensure gradual equilibration of the assembled system based on the equilibration steps used in CHARMM-GUI Membrane Builder, various planar and dihedral restraints (9,10,15) were applied to the LPS molecules, PLs, and water molecules, and the restraint forces were gradually reduced during the equilibration. Additional dihedral angle restraints were applied to restrain all the sugar rings to the pertinent chair conformation for the sugar residues in the LPS. These latter chair-conformation restraints were maintained during the production simulations, because NMR-based studies in conjunction with MD simulations show that common pyranose sugar residues are mainly present in a chair conformation on the submicrosecond timescale (26). The NVT (constant particle number, volume, and temperature) dynamics was used first and followed by the NPT (constant particle number, pressure, and temperature) dynamics during the equilibration. After equilibration, a 450 ns NPT production run was performed with NAMD (27) for one replica of all systems except OmpLA/DLPC and DLPC-only (200 ns production). The other replicas were simulated until equilibrium was apparent for at least 100 ns, as indicated by the membrane surface area. Total simulation times for these replicas were between 250 and 450 ns. The last 100 ns of each trajectory was analyzed to obtain average structural properties.
All simulations were performed according to the following protocol. We used 2-fs time steps with the SHAKE algorithm (28). The van der Waals interactions were smoothly switched off at 10–12 Å by a force-switching function (29), and the long-range electrostatic interactions were calculated using the particle-mesh Ewald method (30). The temperature and pressure were held at 310.15 K and 1 bar, respectively. In CHARMM simulations, Langevin temperature control was used for NVT dynamics. Temperature and pressure controls were achieved with a Hoover thermostat (31) and Langevin piston for NPT dynamics (32,33). For NAMD NPT simulations, Langevin dynamics was used to maintain constant temperature with a Langevin coupling coefficient set to 1 ps−1, and a Nosé-Hoover Langevin piston (34,35) was used to maintain constant pressure with a piston period of 50 fs and a piston decay time of 25 fs.
Results and Discussion
Structural properties of E. coli outer membranes
To investigate the structural properties of the E. coli outer membrane, we calculated the per-lipid surface areas, hydrophobic thickness, density profile of heavy atoms, and acyl chain order parameters and compared these values between LPS-PL[36,100] and LPS-PL[37,100], and also between asymmetric LPS-PL bilayers and PL-only bilayers.
Fig. S1 shows the time-series plots of the overall membrane surface areas of all the systems, which demonstrates that the time evolution of the simulations reached equilibrium states. Moreover, no significant variations between independent runs were observed, indicating that these simulations are well converged on the current timescale. In addition, no statistical difference of the overall surface area was found between LPS-PL[36,100] and LPS-PL[37,100]. Both surface areas match that of PL-only quite well, suggesting that the LPS/PL ratio used in the inner and outer leaflets is reasonable for the asymmetric outer membrane simulations. We noticed that LPS-PL membranes show less variability in the overall membrane area compared to PL-only (Fig. S1 B), which could be induced by the slow movement of the LPS molecules (see next section).
To obtain a general idea of lipid packing in the membrane plane, the per-lipid area of each lipid type was calculated and averaged over the last 100 ns for LPS-PL[36,100], LPS-PL[37,100], and PL-only bilayers with the approach of Pandit et al. (36), utilizing Voronoi tessellation (Fig. 2). In this study, only carbonyl carbon atoms were used to define an acyl chain of a lipid, i.e., two atoms each (C21 and C31) for PPPE and PVPG, four atoms (CA1, CB1, CC1, and CD1) for PVCL2, and six atoms (C11, C21, C31, C41, C51, and C61) for LPS lipid A. Although the surface areas of lipid A in both LPS-PL[36,100] and LPS-PL[37,100] (182 ± 1 Å2 and 177 ± 0.3 Å2, respectively) are close to that reported previously for LPS bilayers (180 ± 1 Å2) (15), there is an obvious reduction of surface area in the more crowded leaflet of LPS-PL[37,100], indicating possible lipid self-adjusting ability in different environments. For PLs, the statistical difference of per-lipid area between different bilayers is small. However, the observation that PPPE, PVPG, and PVCL2 exhibit slightly higher surface areas in asymmetric LPS-PL bilayers than in PL-only bilayers suggests a disturbance of the PL (inner) leaflet by the LPS (outer) leaflet, probably due to subtle area mismatch in the asymmetric bilayers.
Figure 2.
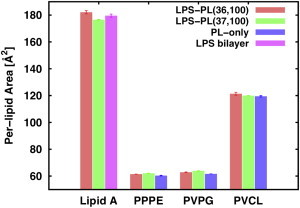
Average per-lipid area of each lipid type in LPS/PL[36,100], LPS/PL[37,100], PL-only, and LPS bilayers with standard errors over two replicas (four replicas for LPS bilayers (15)). To see this figure in color, go online.
Mismatch between the hydrophobic regions of a membrane and its cognate membrane proteins should be energetically costly and lead to structural adjustments to minimize the exposure of nonpolar groups to the aqueous phase (37). In addition, membrane thickness has been shown to profoundly affect the folding kinetics of transmembrane β-barrels in vitro (1). Because the average hydrophobic thickness of transmembrane β-barrels is smaller than that of plasma membrane proteins (8), we analyzed the various PL and LPS systems to determine how the LPS leaflet influences the membrane thickness. The hydrophobic thickness of the membrane systems was calculated by measuring the average distance between acyl chain C2 atoms (the carbon-bonded-to-carbonyl group) for the PL leaflet, and between C2 and C4 atoms for the LPS leaflet (15). As shown in Fig. 3, the calculated hydrophobic thickness of PL-only (28.3 Å) is significantly larger than those of the OmpLA-LPS-PL (24.4 Å), LPS-PL[36,100] (24.7 Å), and LPS-PL[37,100] (24.8 Å) asymmetric bilayers, reflecting that the bacterial outer membrane is much thinner than the PL bilayer composed of the inner-leaflet lipids. Notably, the hydrophobic thickness of this asymmetric bilayer is also much more compatible with that of OmpLA (∼24 Å) and with the average hydrophobic thickness of OMPs in general (8,38). The inclusion of an additional LPS molecule in the neat asymmetric systems had no effect on thickness, as the average hydrophobic thicknesses of LPS-PL[36,100] and LPS-PL[37,100] are statistically identical, with a difference of <0.15 Å. We observed a slightly reduced thickness of OmpLA-LPS-PL compared to the neat asymmetric bilayers, which indicates a local thinning effect occurring around the protein (discussed in detail in the next section). As expected, DLPC bilayers with or without OmpLA have the smallest hydrophobic thickness (20.4 and 20.2 Å) due to the shorter acyl chain. A consistent overall trend (Fig. S2) was observed when the hydrophobic thickness was measured based on the distance between the half-maximal values in the number density profile of the lipid carbon tail atoms (39).
Figure 3.
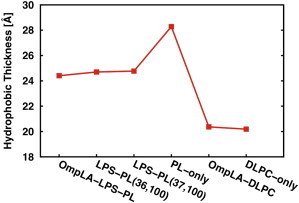
Average hydrophobic thickness of each lipid bilayer system. The standard errors over two replicas are <0.1. To see this figure in color, go online.
We further examined the lipid and protein distributions along the z axis. Heavy-atom number density profiles along the bilayer normal (z axis) for protein backbone atoms, lipid components (carbon tail, headgroups, LPS inner core, LPS outer core), and water are shown in Fig. 4 for OmpLA-LPS-PL and OmpLA-DLPC, and for LPS-PL[36,100] and LPS-PL[37,100]. Consistent with the aforementioned overall membrane surface and hydrophobic thickness comparisons, LPS-PL[36,100] and LPS-PL[37,100] have very similar membrane structure and spatial distribution of lipid components along the z axis. A subtle discrepancy on the height of inner core and outer core peaks arises from the increased number density with one more LPS in LPS-PL[37,100]. In addition, water molecules penetrate through the inner core region and deep into the hydrophobic carbon tail region in all the asymmetric bilayers, which agrees well with previous studies (15,40,41).
Figure 4.
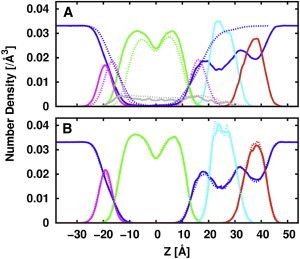
Density profiles of water (blue), PL headgroup (magenta), lipid carbon tail (green), LPS inner core (cyan), LPS outer core (red), and protein backbone atoms (gray) along the membrane normal (z axis) for OmpLA-LPS-PL (solid lines) and OmpLA-DLPC (dotted lines) (A) and LPS/PL[36,100] (solid lines) and LPS/PL[37,100] (dotted lines) (B). To see this figure in color, go online.
One of the most important bilayer properties, lipid deuterium order parameters (SCD), is a common metric for distinguishing a liquid-disordered bilayer phase from a liquid-ordered phase: , where is the time-dependent angle between the C-H bond vector and the bilayer normal (the z axis), and the angular bracket denotes a time and ensemble average (42). Fig. S3 shows the SCD of lipid A chain 1 (15) and sn-1 chains of PPPE, PVPG, PVCL2, and DLPC for all the membrane systems together with the previous symmetric LPS bilayer systems of Wu et al. (15). The general behaviors for these saturated acyl chains are in good accordance with other studies (14,43) that showed increasing disorder along the fatty acid chains toward the methyl groups. Furthermore, a good correlation is observed between the trend of calculated order parameter and per-lipid surface area (Fig. 2). For example, lipid A in LPS-PL[37,100], with smaller per-lipid surface area, tends to be more ordered than that in LPS-PL[36,100], suggesting a more compact packing. On the other hand, the acyl chains of PPPE, PVPG, and PVCL2 show increased disorder in asymmetric bilayers compared to PL-only bilayers, indicating less dense packing in asymmetric bilayers.
Structure and dynamics of OmpLA in E. coli outer membrane
The structural stability of OmpLA was evaluated by monitoring the root mean-square deviation (RMSD) of β-barrel backbone atoms from the crystal structure (PDB 1QD5) (12). Overall, the asymmetric bilayer imposed only minor structural changes on OmpLA. As shown in Fig. S4, RMSD rises up initially and reaches a plateau of <1.5 Å with little drift, which is indicative of the conformational similarity of our MD simulations to the crystal structure. As expected, the OmpLA barrel is very stable in both asymmetric bilayers and DLPC bilayers on the current simulation timescale. The relatively small RMSD and stable barrel structure have also been reported in other β-barrel membrane protein simulations (21,44–48).
The time-averaged root mean-square fluctuation (RMSF) provides a means to assess the relative flexibility of different regions of the protein (46). The calculated RMSF (Fig. 5) for the heavy atoms over the last 100 ns of trajectories shows that the residues in the loop region exhibit a more pronounced flexibility in the DLPC bilayer as compared to the asymmetric bilayer, in particular for loop L1 (residues 46–64) and loop L2 (residues 99–108) (12). This could be related to the fact that the interactions of the OmpLA loop residues with the LPS core oligosaccharide constrain the loop motions and thus lower their dynamics, whereas the loop residues are mainly exposed to the solvent environment in DLPC bilayers. This effect was also observed in other simulation studies (45,46) and could be functionally important for bacteria. Several loops contain additional α-helices, which increase the local rigidity, and thus, no apparent correlation was observed between the length of a loop and the magnitude of its RMSF. Consistent with the aforementioned RMSD results, the β-barrel residues are very rigid, with less fluctuation, and the integrity of the protein secondary structures was maintained well during the simulations.
Figure 5.
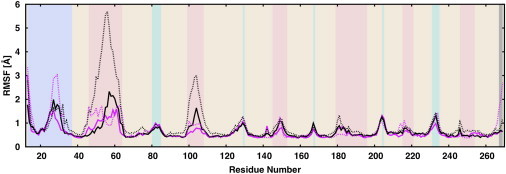
Root mean-square fluctuations (RMSF) of the OmpLA backbone atoms in replicas 1 (solid line) and 2 (dotted line) for the OmpLA-LPS-PL (magenta) and OmpLA-DLPC systems (black). Protein secondary structure is indicated by the background color: β-barrel (beige), loop (coral), turn (turquoise), N terminus (light blue), and C terminus (gray). To see this figure in color, go online.
Figs. 6 and S5 show the average structure of OmpLA in LPS-PL and DLPC bilayers during the last 100 ns of simulation. The membrane and periplasm views of the protein do not exhibit substantially diverging conformations. However, we noticed secondary structure changes (loops to α-helices) of the extracellular loop L2 (orange dashed boxes) in both replicas and displacement of L1 (green dashed box) in the extracellular view in one replica (Fig. 6), whereas in the other replica (Fig. S5), the additional helical structures are absent in both OmpLA-LPS-PL and OmpLA-DLPC. Loop L6 also exhibits a secondary structure change in one of the replicas, despite the fact that it interacts mostly with solvent (Fig. 7). As the two termini and the L1, L4, and L6 loops of OmpLA cover the interior of the barrel and prevent the pore function (12), the LPS oligosaccharide may play a crucial role in determining the location of the loops, which could be important for bacterial function and survival. Interestingly, the extra secondary-structure element observed in loop L2 exists in the dimer structure (PDB 1QD6) (12) as well, suggesting that the LPS-containing asymmetric bilayers favor the conformation found in the dimer for this loop. This could be biologically meaningful, as dimerization is required for the activation of this enzyme (12).
Figure 6.
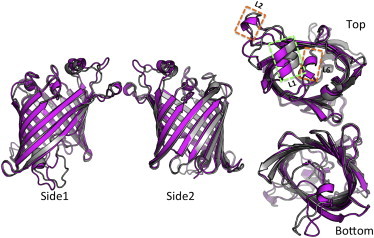
Membrane (Side 1 and Side 2), extracellular (top), and periplasmic (bottom) views of average structures from the last 100 ns of simulation of OmpLA-LPS-PL (magenta) and OmpLA-DLPC (gray) replica 1,. Fig. S5 shows the corresponding plots for replica 2. To see this figure in color, go online.
Figure 7.
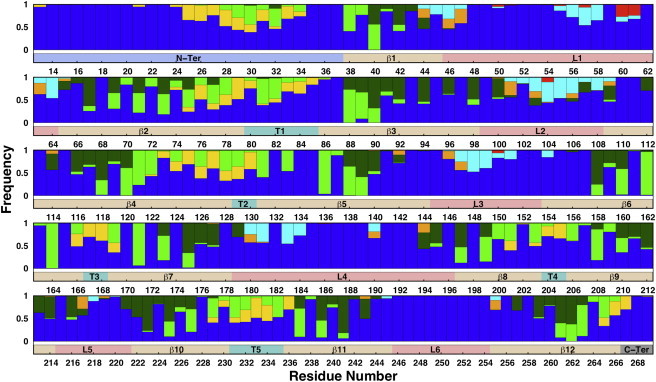
Patterns of interaction of protein residues with their environment in OmpLA-LPS-PL. The graph shows, for each residue, the frequency of occurrence within 4 Å of a water molecule (blue), a PL headgroup (yellow), a PL carbon tail (green), a lipid A tail (dark green), a lipid A headgroup (orange), the LPS inner core (cyan), or the LPS outer core (red). A contact is first counted when the distance of between the heavy atoms of a residue and those of its interacting partner is <4 Å, and normalized for each interacting partner. The bar below each set of patterns indicates protein secondary structure: β-barrel (beige), loop (coral), turn (turquoise), N terminus (light blue), and C terminus (gray). To see this figure in color, go online.
It is of interest to examine the interaction pattern of each protein residue with lipid molecules or water in the LPS-PL outer membrane. A cutoff distance of 4 Å was used to define a contact in this analysis, and the average result of two replicas is presented in Fig. 7. It is evident that an alternative pattern occurs with the 12 transmembrane β-strands that mainly interact with LPS or the PL tail, six extracellular loops that mostly interact with LPS core oligosaccharides and water, and five turns at the periplasmic side that mainly interact with PLs and water, which provides a clear picture of how the amphipathic β-strands traverse the membrane. It is interesting to note that some residues in the transmembrane β-strands (e.g., Leu88, Trp98, and Leu100 in β3) interact with the acyl chains of both leaflets, illustrating the flexibility of side-chain orientations as well as lipid A and PL tails. The aromatic belt residues (e.g., Trp78, Tyr114, and Phe128) are located at the membrane-water interface, interacting with water, lipid headgroups, and lipid tails. As would be expected from the structure of OmpLA, most of the inward-facing β-barrel residues also interact with water molecules, because the interior of the barrel is polar and partially water-filled with an intricate hydrogen-bonding network (12,44). Another interesting feature is that residues in the loop regions mainly interact with LPS headgroups and inner core sugars, whereas OmpLA residues rarely interact with the outer core sugars during the simulations. Only two residues (Glu60 and Asn61) in loop L1 show stable contact with outer core sugars. The interactions between protein loops and LPS oligosaccharide chains lead to effectively reduced loop dynamics in asymmetric LPS-PL bilayers compared to DLPC bilayers (Fig. 5). Furthermore, the restrained loop dynamics and flexibility could be essential for stabilizing the loop conformation and additional secondary-structure formation, leading to average structure differences between the loop regions of OmpLA in LPS-PL and those in DLPC bilayers (Figs. 6 and S5).
To better understand the interactions between the protein and lipids and to probe the impact of the protein on the membrane structure, two-dimensional (2D) hydrophobic thickness profiles were calculated utilizing the same atom selection as in the hydrophobic thickness calculations (Fig. 3) after the protein was rotated to the same orientation in different bilayers. The 2D plots were constructed with a grid spacing of 2.4 Å, which was determined by the closest C2-C2 distance (3.4 Å) based on a radial distribution function to ensure that each grid box covers one C2 atom. Figs. 8 and S6 show the 2D z-position distributions of the C2 and C4 atoms for lipid A and the C2 atoms for PLs, as well as the 2D thickness distribution of the full bilayer, for both OmpLA-LPS-PL and OmpLA-DLPC. It is particularly evident that the bilayer structure is disturbed, and local thinning is observed in the vicinity of protein in the asymmetric bilayers (Figs. 8 C and S6 C), which is likely a reflection of the system seeking a hydrophobic match (43,49,50). The hydrophobic thickness of the lipids proximal to the protein is ∼20 Å in the asymmetric bilayer, very close to the thickness of the DLPC bilayer (Figs. 3 and 8 F), which validates the rationality of using DLPC as an effective alternative to the outer membrane in OmpLA folding experiments in vitro (4).
Figure 8.
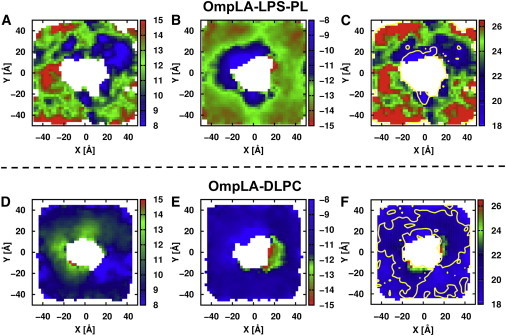
Two-dimensional z-position distributions of the C2 and C4 atoms of lipid A (A) and the acyl chain C2 atoms of PLs (B, D, and E), as well as the 2D thickness distribution of the full bilayer (C and F), for OmpLA-LPS-PL (upper row) and OmpLA-DLPC (lower row). Color keys indicate the distance from the bilayer center to C2 atoms along the z axis, where blue is closer to the bilayer center and red is farther away (A, B, D, and E), and total membrane thickness in the case of the full bilayer (C and F). The contours are drawn for 20 Å in yellow. Fig. S6 shows the corresponding plots for replica 2. To see this figure in color, go online.
A comparison between the hydrophobic interface locations for each leaflet in OmpLA-LPS-PL (Figs. 8, A and B, and S6, A and B) suggests that both leaflets contribute to the thinning around the protein; some areas are mainly from the PL leaflet and others are from the LPS leaflet. One important observation is that on the righthand side of the protein in the PL leaflet (Figs. 8 B and S6 B), as well as in the bottom leaflet of OmpLA-DLPC (Figs. 8 E and S6 E), there are areas of local thickening in both replicas. No lipid-type preference was observed in this region (Fig. S7), and PLs could exchange into and out of this region during the simulations, meaning that the thickening might be caused by some essential structural properties of OmpLA. In general, energetically unfavorable hydrophobic mismatch between a bilayer hydrophobic core and the protein transmembrane domain is relieved either by adaptation of the bilayer (i.e., local thinning or thickening) or by changes in protein orientation or conformation (43,49–52). To determine whether there is an intrinsic hydrophobic mismatch in OmpLA-LPS-PL, the thickness of the protein barrel was estimated by calculating the 2D average z distributions of the side-chain atoms of hydrophobic residues on the rim of each β-strand. The protein orientation is the same as in the above analysis (Fig. 6, Top, and Fig. 8). As expected, the protein barrel buried in the PL leaflet is clearly thicker on the righthand side than in other regions (Fig. S8 B). The thinning area at the top right corner of the LPS leaflet (Figs. 8 A and S6 A) is also reflected in the protein hydrophobic length (Fig. S8 A) in the corresponding region. On the other hand, the DLPC lipids adjacent to the protein barrel are lifted, with increased thickness in both leaflets in OmpLA-DLPC (Figs. 8, D and E, and S6, D and E), which is consistent with the increased phosphate-phosphate distance around FepA in a DLPC bilayer observed in the coarse-grained MD simulation by Scott et al. (53).
Although hydrophobic match provides a reasonable explanation for the 2D thickness profile around the protein, inconsistent patterns between the two replicas were observed in the LPS leaflet (Figs. 8 A and S6 A). Fig. S9 shows the 2D density profiles of the center of mass of each lipid type in OmpLA-LPS-PL with a grid spacing of 0.5 Å. Compared to the ubiquitous density of PLs in the inner leaflet (Fig. S9, B and D), LPS molecules (Fig. S9, A and C) are highly immobile and show little translocation on the simulation timescale used here. In addition, the distribution of the relative orientation between LPS and OmpLA was calculated for each LPS molecule (Fig. S10). The narrow distributions suggest that LPS molecules show minimal lateral diffusion and also limited rotation. Although such immobility is characteristic for LPS in the outer membranes, as many experiments suggest (54,55), the LPS orientation and location could depend strongly on its initial placement within the limited simulation time, and the variation of the thickness profiles of the LPS leaflets between the two replicas might be a result of the significantly lower translational or rotational diffusion rate of the LPS molecules. Although it may indicate a potential sampling issue in the simulation timescale used here, the rigidity and low mobility of the LPS leaflet is a reflection of its true nature and is mostly due to the divalent ion-mediated, cross-linking electrostatic interaction network in the core region (15,55).
Conclusions
The outer membrane of Gram-negative bacteria separates its periplasm from the external environment and serves as a protective barrier that controls the entry of many toxic molecules, such as antibiotics and bile salts, into the bacteria, which is crucial for bacterial survival in diverse/hostile environments (55,56). This unique membrane is highly asymmetric and composed of a mixture of PLs in the inner leaflet and LPS in the outer leaflet (57–62). OmpLA is constitutively expressed in this asymmetric bilayer in vivo and functions as an outer membrane phospholipase that degrades a wide variety of PLs as well as diglycerides under situations of bacterial stress (63). In this work, we present an MD simulation study of asymmetric bilayers containing LPS, PPPE, PVPG, and PVCL2 with and without OmpLA to explore the structural properties of the bacterial outer membrane and the structure and dynamics of OmpLA in its native environment. The results of this study are compared with those from DLPC bilayers, which have proven to be a useful PL environment for in vitro folding experiments of OmpLA. Furthermore, asymmetric bilayers with different LPS/PL ratios are compared to elucidate the impact of the area mismatch between the inner and outer leaflets on bilayer properties, which could be a technical issue in building asymmetric bilayers.
Our study shows that the overall bilayer structural properties, including the membrane area, density profile, and hydrophobic thickness, do not show a statistically significant difference between LPS/PL[36:100] and LPS/PL[37:100]. In fact, the overall areas of both asymmetric bilayers match that of the bilayer composed of PLs in both leaflets (PL-only), meaning that our approach of constructing asymmetric bilayers based simply on the estimation of per-lipid area of each lipid type is reliable. However, LPS exhibits a slightly reduced area per lipid and an increased acyl chain order in the more crowded environment (LPS/PL[37:100]). In addition, the PLs are more disordered in the inner leaflet of the asymmetric bilayers, with increased area per lipid compared to the corresponding PL-only bilayer. This is interesting, as it suggests that lipids are capable of self-adjusting their individual properties to a certain extent to improve the overall fit between two leaflets in an asymmetric bilayer. Thus, bacteria could take advantage of this lipid self-adjusting mechanism in the outer membrane by altering the lipid ratio to control the compact level of lipid packing or induce bilayer defects that facilitate protein folding, which might be functionally important.
A comparison of the hydrophobic thickness reveals that the asymmetric bacterial outer membrane is much thinner than its corresponding symmetric PL bilayer given the lipid acyl chain composition used in our study. This is expected, because, considering that the hydrophobic thickness of an LPS bilayer is ∼22 Å (15) and that of a PL bilayer ∼28 Å, the combination of one leaflet of each type of bilayer would result in an asymmetric bilayer with an intermediate thickness of ∼25 Å. It is important to point out that the LPS leaflet is the main determinant of the reduced thickness of the outer membrane; therefore, including LPS in the OMP simulation study is pivotal to mimicking the native environment. Moreover, independent analyses of the known structures of OMPs demonstrate that their average hydrophobic thickness is ∼24 Å (8,64), in good agreement with our value for the asymmetric bilayer. A possible reason for the decreased thickness of the LPS bilayer compared to 1,2-dimyristoyl-sn-glycero-3-phosphocholine (DMPC) bilayers (∼25 Å) (43), which have an acyl chain length similar to that of lipid A, might be the electrostatic repulsive interaction between LPS molecules due to the highly negatively charged headgroup. In addition, it might be also related to the conical shape of the LPS molecule due to its smaller headgroup relative to its bulky tail region (15).
Compared to the PL bilayer in this study, with acyl chains that mimic E. coli composition, the hydrophobic thickness of OmpLA is much better matched to that of the LPS asymmetric bilayer. Nevertheless, the presence of OmpLA perturbs the local membrane structure and induces a thinning effect in the asymmetric bilayer driven by a small hydrophobic mismatch. However, inconsistent thinning patterns in the LPS leaflets were found between two independent simulations due to immobilization of the LPS membrane by the calcium-mediated, cross-linking electrostatic interaction network. This low lateral diffusion significantly hinders the complete adaptability of the LPS leaflet to perturbation of the protein.
Our study further illustrates how OmpLA loop movement is constrained in asymmetric bilayers by interactions predominantly with LPS headgroups and oligosaccharides in the inner core region, which are clearly lacking in the DLPC bilayer. The reduced loop dynamics facilitates more rigid secondary-structure formation, as well as loop displacement, which could be vital to prevent pore formation across the barrel and avoid entry of toxic molecules. Since LPS clearly affects the structure and dynamics of the extracellular loop of OmpLA, structural refinement of OMPs—especially their loop conformations—needs to be performed in the native lipid environment. Thus, the information gained from this study should be beneficial in future studies for determining the OMP structure, exploring the interaction mechanism between the bacterial outer membrane and OMPs, and facilitating the discovery of new antibiotic drugs.
Acknowledgments
This work was supported in part by grants from the National Science Foundation (MCB-1157677, ABI-1145987, and XSEDE MCB070009 to W.I., and MCB-0919868 to K.G.F.), Human Frontier Science Program (RGP0064/2011 to W.I.), the National Institutes of Health (GM0790440 and XSEDE MCB120050 to K.G.F.), the National Institute of Supercomputing and Networking/Korea Institute of Science and Technology Information, including technical support (KSC-2013-C3-030 to M.S.Y.), and the Swedish Research Council and the Stockholm Center for Biomembrane Research/Swedish Foundation for Strategic Research (to G.W.).
Contributor Information
Karen G. Fleming, Email: Karen.Fleming@jhu.edu.
Wonpil Im, Email: wonpil@ku.edu.
Supporting Material
References
- 1.Burgess N.K., Dao T.P., Fleming K.G. Beta-barrel proteins that reside in the Escherichia coli outer membrane in vivo demonstrate varied folding behavior in vitro. J. Biol. Chem. 2008;283:26748–26758. doi: 10.1074/jbc.M802754200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stanley A.M., Fleming K.G. The process of folding proteins into membranes: challenges and progress. Arch. Biochem. Biophys. 2008;469:46–66. doi: 10.1016/j.abb.2007.09.024. [DOI] [PubMed] [Google Scholar]
- 3.Moon C.P., Zaccai N.R., Fleming K.G. Membrane protein thermodynamic stability may be the energy sink for sorting in the periplasm. Proc. Natl. Acad. Sci. USA. 2013;110:5285–5290. doi: 10.1073/pnas.1212527110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moon C.P., Fleming K.G. Side-chain hydrophobicity scale derived from transmembrane protein folding into lipid bilayers. Proc. Natl. Acad. Sci. USA. 2011;108:10174–10177. doi: 10.1073/pnas.1103979108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pocanschi C.L., Apell H.J., Kleinschmidt J.H. The major outer membrane protein of Fusobacterium nucleatum (FomA) folds and inserts into lipid bilayers via parallel folding pathways. J. Mol. Biol. 2006;355:548–561. doi: 10.1016/j.jmb.2005.10.060. [DOI] [PubMed] [Google Scholar]
- 6.Kleinschmidt J.H., Tamm L.K. Secondary and tertiary structure formation of the β-barrel membrane protein OmpA is synchronized and depends on membrane thickness. J. Mol. Biol. 2002;324:319–330. doi: 10.1016/s0022-2836(02)01071-9. [DOI] [PubMed] [Google Scholar]
- 7.Dekker N., Tommassen J., Verheij H.M. Bacteriocin release protein triggers dimerization of outer membrane phospholipase A in vivo. J. Bacteriol. 1999;181:3281–3283. doi: 10.1128/jb.181.10.3281-3283.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lomize A.L., Pogozheva I.D., Mosberg H.I. Anisotropic solvent model of the lipid bilayer. 2. Energetics of insertion of small molecules, peptides, and proteins in membranes. J. Chem. Inf. Model. 2011;51:930–946. doi: 10.1021/ci200020k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jo S., Kim T., Im W. Automated builder and database of protein/membrane complexes for molecular dynamics simulations. PLoS ONE. 2007;2:e880. doi: 10.1371/journal.pone.0000880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jo S., Lim J.B., Im W. CHARMM-GUI Membrane Builder for mixed bilayers and its application to yeast membranes. Biophys. J. 2009;97:50–58. doi: 10.1016/j.bpj.2009.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jo S., Kim T., Im W. CHARMM-GUI: a web-based graphical user interface for CHARMM. J. Comput. Chem. 2008;29:1859–1865. doi: 10.1002/jcc.20945. [DOI] [PubMed] [Google Scholar]
- 12.Snijder H.J., Ubarretxena-Belandia I., Dijkstra B.W. Structural evidence for dimerization-regulated activation of an integral membrane phospholipase. Nature. 1999;401:717–721. doi: 10.1038/44890. [DOI] [PubMed] [Google Scholar]
- 13.Vance D.E., Vance J.E. Elsevier; Amsterdam: 2002. Biochemistry of Lipids, Lipoproteins, and Membranes. [Google Scholar]
- 14.Klauda J.B., Venable R.M., Pastor R.W. Update of the CHARMM all-atom additive force field for lipids: validation on six lipid types. J. Phys. Chem. B. 2010;114:7830–7843. doi: 10.1021/jp101759q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu E.L., Engström O., Im W. Molecular dynamics and NMR spectroscopy studies of E. coli lipopolysaccharide structure and dynamics. Biophys. J. 2013;105:1444–1455. doi: 10.1016/j.bpj.2013.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grozdanov L., Zähringer U., Dobrindt U. A single nucleotide exchange in the wzy gene is responsible for the semirough O6 lipopolysaccharide phenotype and serum sensitivity of Escherichia coli strain Nissle 1917. J. Bacteriol. 2002;184:5912–5925. doi: 10.1128/JB.184.21.5912-5925.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brooks B.R., Brooks C.L., 3rd, Karplus M. CHARMM: the biomolecular simulation program. J. Comput. Chem. 2009;30:1545–1614. doi: 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lomize M.A., Lomize A.L., Mosberg H.I. OPM: orientations of proteins in membranes database. Bioinformatics. 2006;22:623–625. doi: 10.1093/bioinformatics/btk023. [DOI] [PubMed] [Google Scholar]
- 19.Woolf T.B., Roux B. Molecular dynamics simulation of the gramicidin channel in a phospholipid bilayer. Proc. Natl. Acad. Sci. USA. 1994;91:11631–11635. doi: 10.1073/pnas.91.24.11631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Woolf T.B., Roux B. Structure, energetics, and dynamics of lipid-protein interactions: a molecular dynamics study of the gramicidin A channel in a DMPC bilayer. Proteins. 1996;24:92–114. doi: 10.1002/(SICI)1097-0134(199601)24:1<92::AID-PROT7>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 21.Fleming P.J., Freites J.A., Fleming K.G. Outer membrane phospholipase A in phospholipid bilayers: a model system for concerted computational and experimental investigations of amino acid side chain partitioning into lipid bilayers. Biochim. Biophys. Acta. 2012;1818:126–134. doi: 10.1016/j.bbamem.2011.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guvench O., Greene S.N., Mackerell A.D., Jr. Additive empirical force field for hexopyranose monosaccharides. J. Comput. Chem. 2008;29:2543–2564. doi: 10.1002/jcc.21004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hatcher E., Guvench O., Mackerell A.D., Jr. CHARMM additive all-atom force field for aldopentofuranoses, methyl-aldopentofuranosides, and fructofuranose. J. Phys. Chem. B. 2009;113:12466–12476. doi: 10.1021/jp905496e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guvench O., Hatcher E.R., Mackerell A.D., Jr. CHARMM additive all-atom force field for glycosidic linkages between hexopyranoses. J. Chem. Theory Comput. 2009;5:2353–2370. doi: 10.1021/ct900242e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jorgensen W.L., Chandrasekhar J., Klein M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983;79:926–935. [Google Scholar]
- 26.Sattelle B.M., Almond A. Assigning kinetic 3D-signatures to glycocodes. Phys. Chem. Chem. Phys. 2012;14:5843–5848. doi: 10.1039/c2cp40071e. [DOI] [PubMed] [Google Scholar]
- 27.Phillips J.C., Braun R., Schulten K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ryckaert J.P., Ciccotti G., Berendsen H.J.C. Numerical integration of Cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J. Comput. Phys. 1977;23:327–341. [Google Scholar]
- 29.Steinbach P.J., Brooks B.R. New spherical cutoff methods for long-range forces in macromolecular simulation. J. Comput. Chem. 1994;15:667–683. [Google Scholar]
- 30.Essmann U., Perera L., Pedersen L.G. A smooth particle mesh Ewald method. J. Chem. Phys. 1995;103:8577–8593. [Google Scholar]
- 31.Hoover W.G. Canonical dynamics: equilibrium phase-space distributions. Phys. Rev. A. 1985;31:1695–1697. doi: 10.1103/physreva.31.1695. [DOI] [PubMed] [Google Scholar]
- 32.Nose S., Klein M.L. A study of solid and liquid carbon tetrafluoride using the constant pressure molecular-dynamics technique. J. Chem. Phys. 1983;78:6928–6939. [Google Scholar]
- 33.Andersen H.C. Molecular dynamics simulations at constant pressure and/or temperature. J. Chem. Phys. 1980;72:2384–2393. [Google Scholar]
- 34.Feller S.E., Zhang Y.H., Brooks B.R. Constant pressure molecular dynamics simulation: the Langevin piston method. J. Chem. Phys. 1995;103:4613–4621. [Google Scholar]
- 35.Martyna G.J., Tobias D.J., Klein M.L. Constant pressure molecular dynamics algorithms. J. Chem. Phys. 1994;101:4177–4189. [Google Scholar]
- 36.Pandit S.A., Vasudevan S., Scott H.L. Sphingomyelin-cholesterol domains in phospholipid membranes: atomistic simulation. Biophys. J. 2004;87:1092–1100. doi: 10.1529/biophysj.104.041939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rui H., Lee J., Im W. Comparative molecular dynamics simulation studies of protegrin-1 monomer and dimer in two different lipid bilayers. Biophys. J. 2009;97:787–795. doi: 10.1016/j.bpj.2009.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pogozheva I.D., Tristram-Nagle S., Lomize A.L. Structural adaptations of proteins to different biological membranes. Biochim. Biophys. Acta. 2013;1828:2592–2608. doi: 10.1016/j.bbamem.2013.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Klauda J.B., Kucerka N., Nagle J.F. Simulation-based methods for interpreting x-ray data from lipid bilayers. Biophys. J. 2006;90:2796–2807. doi: 10.1529/biophysj.105.075697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Abraham T., Schooling S.R., Katsaras J. Neutron diffraction study of Pseudomonas aeruginosa lipopolysaccharide bilayers. J. Phys. Chem. B. 2007;111:2477–2483. doi: 10.1021/jp066012+. [DOI] [PubMed] [Google Scholar]
- 41.Kucerka N., Papp-Szabo E., Katsaras J. Effect of cations on the structure of bilayers formed by lipopolysaccharides isolated from Pseudomonas aeruginosa PAO1. J. Phys. Chem. B. 2008;112:8057–8062. doi: 10.1021/jp8027963. [DOI] [PubMed] [Google Scholar]
- 42.Vermeer L.S., de Groot B.L., Czaplicki J. Acyl chain order parameter profiles in phospholipid bilayers: computation from molecular dynamics simulations and comparison with H-2 NMR experiments. Eur. Biophys. J. 2007;36:919–931. doi: 10.1007/s00249-007-0192-9. [DOI] [PubMed] [Google Scholar]
- 43.Kim T., Lee K.I., Im W. Influence of hydrophobic mismatch on structures and dynamics of gramicidin a and lipid bilayers. Biophys. J. 2012;102:1551–1560. doi: 10.1016/j.bpj.2012.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Baaden M., Meier C., Sansom M.S. A molecular dynamics investigation of mono and dimeric states of the outer membrane enzyme OMPLA. J. Mol. Biol. 2003;331:177–189. doi: 10.1016/s0022-2836(03)00718-6. [DOI] [PubMed] [Google Scholar]
- 45.Piggot T.J., Holdbrook D.A., Khalid S. Conformational dynamics and membrane interactions of the E. coli outer membrane protein FecA: a molecular dynamics simulation study. Biochim. Biophys. Acta. 2013;1828:284–293. doi: 10.1016/j.bbamem.2012.08.021. [DOI] [PubMed] [Google Scholar]
- 46.Straatsma T.P., Soares T.A. Characterization of the outer membrane protein OprF of Pseudomonas aeruginosa in a lipopolysaccharide membrane by computer simulation. Proteins. 2009;74:475–488. doi: 10.1002/prot.22165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Im W., Roux B. Ions and counterions in a biological channel: a molecular dynamics simulation of OmpF porin from Escherichia coli in an explicit membrane with 1 M KCl aqueous salt solution. J. Mol. Biol. 2002;319:1177–1197. doi: 10.1016/S0022-2836(02)00380-7. [DOI] [PubMed] [Google Scholar]
- 48.Rui H.A., Lee K.I., Im W. Molecular dynamics studies of ion permeation in VDAC. Biophys. J. 2011;100:602–610. doi: 10.1016/j.bpj.2010.12.3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Killian J.A. Hydrophobic mismatch between proteins and lipids in membranes. Biochim. Biophys. Acta. 1998;1376:401–415. doi: 10.1016/s0304-4157(98)00017-3. [DOI] [PubMed] [Google Scholar]
- 50.Niu S.L., Mitchell D.C., Litman B.J. Reduced G protein-coupled signaling efficiency in retinal rod outer segments in response to n-3 fatty acid deficiency. J. Biol. Chem. 2004;279:31098–31104. doi: 10.1074/jbc.M404376200. [DOI] [PubMed] [Google Scholar]
- 51.Kim T., Im W. Revisiting hydrophobic mismatch with free energy simulation studies of transmembrane helix tilt and rotation. Biophys. J. 2010;99:175–183. doi: 10.1016/j.bpj.2010.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rui H., Kumar R., Im W. Membrane tension, lipid adaptation, conformational changes, and energetics in MscL gating. Biophys. J. 2011;101:671–679. doi: 10.1016/j.bpj.2011.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Scott K.A., Bond P.J., Sansom M.S.P. Coarse-grained MD simulations of membrane protein-bilayer self-assembly. Structure. 2008;16:621–630. doi: 10.1016/j.str.2008.01.014. [DOI] [PubMed] [Google Scholar]
- 54.Nikaido H. Molecular basis of bacterial outer membrane permeability revisited. Microbiol. Mol. Biol. Rev. 2003;67:593–656. doi: 10.1128/MMBR.67.4.593-656.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ruiz N., Kahne D., Silhavy T.J. Transport of lipopolysaccharide across the cell envelope: the long road of discovery. Nat. Rev. Microbiol. 2009;7:677–683. doi: 10.1038/nrmicro2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Van Wielink J.E., Duine J.A. How big is the periplasmic space? Trends Biochem. Sci. 1990;15:136–137. doi: 10.1016/0968-0004(90)90208-s. [DOI] [PubMed] [Google Scholar]
- 57.Smit J., Kamio Y., Nikaido H. Outer membrane of Salmonella typhimurium: chemical analysis and freeze-fracture studies with lipopolysaccharide mutants. J. Bacteriol. 1975;124:942–958. doi: 10.1128/jb.124.2.942-958.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Brade H., Steven O.M., David M.C. Marcel Dekker; New York: 1999. Endotoxin in health and disease. [Google Scholar]
- 59.Silipo A., Castro D.C., Molinaro A. Lipopolysaccharides. In: König H., Claus H., Varma A., editors. Prokaryotic Cell Wall Compounds. Springer-Verlag; Berlin: 2010. pp. 133–153. [Google Scholar]
- 60.Wang X., Quinn P.J. Endotoxins: lipopolysaccharides of Gram-negative bacteria. In: Wang X., Quinn P.J., editors. Endotoxins: Structure, Function and Recognition, Subcellular Biochemistry. Springer; Dordrecht: 2010. pp. 3–25. [DOI] [PubMed] [Google Scholar]
- 61.Yuriy K.A., Miguel V.A. Springer-Verlag; Wien: 2011. Bacterial Lipopolysaccharides: Structure, Chemical Synthesis, Biogenesis and Interaction with Host Cells. [Google Scholar]
- 62.Kamio Y., Nikaido H. Outer membrane of Salmonella typhimurium: accessibility of phospholipid head groups to phospholipase c and cyanogen bromide activated dextran in the external medium. Biochemistry. 1976;15:2561–2570. doi: 10.1021/bi00657a012. [DOI] [PubMed] [Google Scholar]
- 63.Horrevoets A.J., Hackeng T.M., de Haas G.H. Kinetic characterization of Escherichia coli outer membrane phospholipase A using mixed detergent-lipid micelles. Biochemistry. 1989;28:1139–1147. doi: 10.1021/bi00429a031. [DOI] [PubMed] [Google Scholar]
- 64.Slusky J.S.G., Dunbrack R.L., Jr. Charge asymmetry in the proteins of the outer membrane. Bioinformatics. 2013;29:2122–2128. doi: 10.1093/bioinformatics/btt355. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


