SUMMARY
Thermogenic UCP1-positive cells, which include brown and beige adipocytes, transform chemical energy into heat and increase whole body energy expenditure. Using a selective translational profiling approach, we present a comprehensive molecular description of brown and beige gene expression in vivo. This UCP1-TRAP dataset demonstrates striking similarities and important differences between these cell types, including a smooth muscle-like signature expressed by beige, but not classical brown adipocytes. In vivo fate mapping demonstrates that at least a subset of beige cells arise from a smooth muscle-like origin. Finally, ectopic expression of PRDM16 converts bona fide vascular smooth muscle cells into Ucp1-positive adipocytes in vitro. These results establish a portrait of brown and beige adipocyte gene expression in vivo and identify a previously unrecognized origin for beige cells.
INTRODUCTION
Obesity and its related metabolic disorders, including diabetes, cardiovascular disease and hypertension, are growing in prevalence worldwide. A promising and emerging avenue for obesity treatment is to increase energy expenditure by augmenting the number or the activity of thermogenic adipocytes (Nedergaard et al., 2011; Wu et al., 2013). These specialized cells are characterized by having multiple lipid droplets, high mitochondrial density, and the expression of uncoupling protein 1 (UCP1), an inner mitochondrial membrane protein that dissipates the proton gradient to uncouple fuel oxidation from ATP synthesis (Cannon and Nedergaard, 2004; Nicholls et al., 1978). In rodents, there are at least two distinct types of UCP1-positive cells (Wu et al., 2012). The classical brown fat is found both in the interscapular region as well as in the perirenal area. Beige adipocytes are found interspersed in various white fat depots and their formation can be stimulated by cold exposure or by β-adrenergic receptor agonists (Cousin et al., 1992; Young et al., 1984). That brown or beige fat activity confers metabolic benefit is now well established in mice (Cederberg et al., 2001; Cohen et al., 2014; Feldmann et al., 2009; Harms and Seale, 2013; Seale et al., 2011). Imaging and histological analyses have confirmed the existence of UCP1-positive fat in both newborn and adult humans and indicate the presence of both classical brown fat and beige fat (Cypess et al., 2009; Cypess et al., 2013; Lidell et al., 2013; Sharp et al., 2012; van Marken Lichtenbelt et al., 2009; Virtanen et al., 2009).
Classical brown and beige adipocytes both express UCP1 and share many morphological and biochemical characteristics, including a well-characterized β-adrenergic receptor/cAMP-dependent pathway that regulates thermogenic gene expression. However, multiple lines of evidence have demonstrated that brown and beige adipocytes are in fact distinct cell types. First, classical brown, but not beige adipocytes, arise from a Myf5/Pax7 skeletal muscle lineage, indicating that there are separate developmental origins for these two cell types (Lepper and Fan, 2010; Sanchez-Gurmaches et al., 2012; Seale et al., 2008). Second, classical brown and beige fat are differentially responsive to various hormonal stimuli or genetic manipulations (Bostrom et al., 2012; Harms and Seale, 2013). Third, classical brown, beige, and white adipocytes possess distinct gene expression signatures in cell culture (Wu et al., 2012). Despite this progress, a complete molecular description of UCP1-positive adipocytes in vivo is still lacking. This is in large part because UCP1-positive adipocytes generally, and beige adipocytes in particular, occur within a complex, heterogeneous mixture of other cell types in adipose tissues.
To address the cellular heterogeneity issue, we have adapted translating ribosome affinity purification (TRAP) technology (Heiman et al., 2008; Sanz et al., 2009). This method, originally developed for neuroscience applications, allows for genetic tagging and isolation of polysomes in a highly cell type-specific manner. Using this approach, we have selectively isolated polysomes of UCP1-positive cells directly from whole adipose tissues in mice. Our studies demonstrate striking similarities and surprising differences between brown and beige cells in vivo, including a smooth muscle-like gene expression signature in beige, but not classical brown cells. In vivo fate mapping and primary smooth muscle culture experiments provide additional evidence for a smooth muscle-like origin of some beige cells. These results reveal a hitherto unappreciated link between beige fat and smooth muscle-like cells, and provide a powerful foundation for elucidating additional aspects of thermogenic adipocyte function in vivo.
RESULTS
Generation and characterization of the UCP1-TRAP mouse
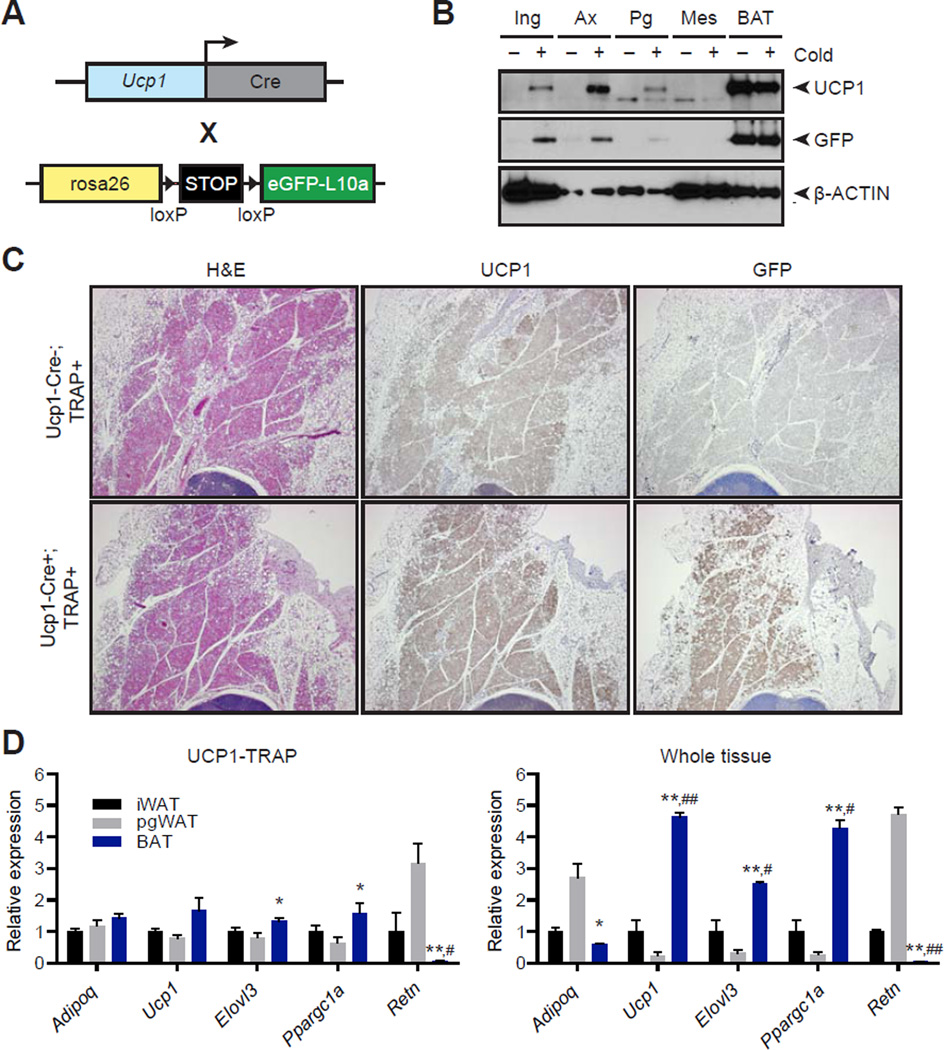
To characterize UCP1-positive adipocytes in vivo, we first generated a TRAP mouse. This animal expresses a fusion protein of eGFP and the 60S ribosomal subunit L10a downstream of a lox-stop-lox cassette knocked into the Rosa26 locus. The TRAP mouse was then crossed to a BAC transgenic strain expressing Ucp1-driven Cre to produce UCP1-TRAP mice (Fig. 1A). In this way, the TRAP protein (eGFP-L10a) is only produced in those cells that also possess sufficiently high levels of Ucp1 promoter activity to induce Cre expression. Because the recombination event is irreversible and heritable, recombination was minimized in the immediate postnatal period by breeding UCP1-TRAP mice at thermoneutrality and then weaning litters at room temperature. In four-week old mice at room temperature, both UCP1 and eGFP-L10a proteins were principally detected in the classical brown adipose tissue (BAT) but not in subcutaneous or visceral white adipose tissues (Fig. 1B). Next, mice were housed at 4°C for an additional two weeks to “brown” the white fat depots. Under these conditions, induction of both UCP1 and GFP-L10a proteins occurred in the inguinal and axillary depots, and to a lesser extent, in the perigonadal visceral depots (Fig. 1B). Neither UCP1 nor eGFP-L10a protein was observed in the mesenteric visceral depot at any temperature, consistent with the known reduced prevalence of beige cells in visceral adipose depots.
Fig. 1. Characterization of the UCP1-TRAP mouse.
(A) Schematic of the cross to generate UCP1-TRAP mice. A BAC transgenic expressing Ucp1-Cre was crossed with a TRAP mouse expressing a fusion protein of eGFP and the ribosomal subunit L10a (eGFP-L10a, referred to as the TRAP allele) under the control of a lox-stop-lox cassette in the rosa26 locus. (B) Western blot analysis of UCP1, GFP, and β-actin loading control from various fat depots in 6-week old UCP1-TRAP mice at room temperature (−) or after two weeks cold exposure (+) at 4°C. Ing, inguinal WAT; Ax, axillary WAT; Pg, perigonadal WAT; Mes, mesenteric WAT. (C) Representative H&E, UCP1, and GFP immunohistochemistry from 6-week old male UCP1-TRAP (Cre-positive, TRAP-positive) or control mice (Cre-negative, TRAP-positive) after two weeks cold exposure at 4°C. (D) Relative mRNA expression of the indicated genes from various fat depots in UCP1-TRAP samples (left) or whole tissues (right). Tissues were harvested from 6-week old female mice after two weeks cold exposure at 4°C. Data are presented as means ± standard error; n = 3–5/group. *, ** p < 0.05 or < 0.01, respectively, in BAT versus pgWAT samples; #, ## p < 0.05 or < 0.01, respectively, in BAT versus iWAT samples.
Immunohistochemical analysis of the inguinal depots from UCP1-TRAP mice is consistent with recombination occurring in a substantial portion of UCP1-positive cells in multiple fat depots (Fig. 1C and Fig. S1A). Lastly, we also observed colocalization of GFP and UCP1 protein at the cellular level (Fig. S1B). Taken together, these data indicate that TRAP expression in the UCP1-TRAP mice recapitulates the endogenous expression of UCP1 in a substantial subset of cells.
Following this two-week cold exposure protocol, the expression of genes related to thermogenesis was compared from whole-tissues and also from immunoaffinity purified mRNA samples (UCP1-TRAP samples). In principle, the UCP1-TRAP samples should provide a readout of gene expression only in the UCP1-positive cells from different fat pads. Consistent with previous reports (Seale et al., 2011), the thermogenesis-related genes Ucp1, Elovl3, and Ppargc1a (PGC-1α) were highly expressed in the BAT depot, at an intermediate level in the subcutaneous iWAT depot, and at the lowest levels in the pgWAT depot (Fig. 1D). Importantly, the expression of these genes from UCP1-TRAP samples was much more similar in magnitude across the depots (Fig. 1D). These data strongly suggest that the basic thermogenic gene program of classical brown and beige cells is quite similar. Moreover, the differences observed in expression of these genes on the tissue level are likely because UCP1-positive cells are diluted by the presence of other cell types, especially white adipocytes. Interestingly, not all adipocyte genes were “normalized” in the UCP1-TRAP samples. Resistin (Retn), a marker of white adipocytes, was highly expressed in white depots at a pad level as well as in the UCP1-TRAP samples (Fig. 1D). These data indicate that UCP1-positive beige cells in vivo retain some white-like characteristics that are completely absent in UCP1-positive cells of the classical BAT. Finally, we surveyed the expression of a panel of gene expression events that were previously annotated to be selective for different fat depots or fat cell types (Gesta et al., 2006; Walden et al., 2012; Wu et al., 2012). Many of these genes, including Tbx1, Hoxc10, and Zic1, were seen in our UCP1-TRAP samples (Fig. S1B and S1C), further demonstrating that this translational profiling approach can validate previous findings relating to gene expression differences between fat depots.
Identification of the common gene expression program from brown and beige adipocytes in vivo
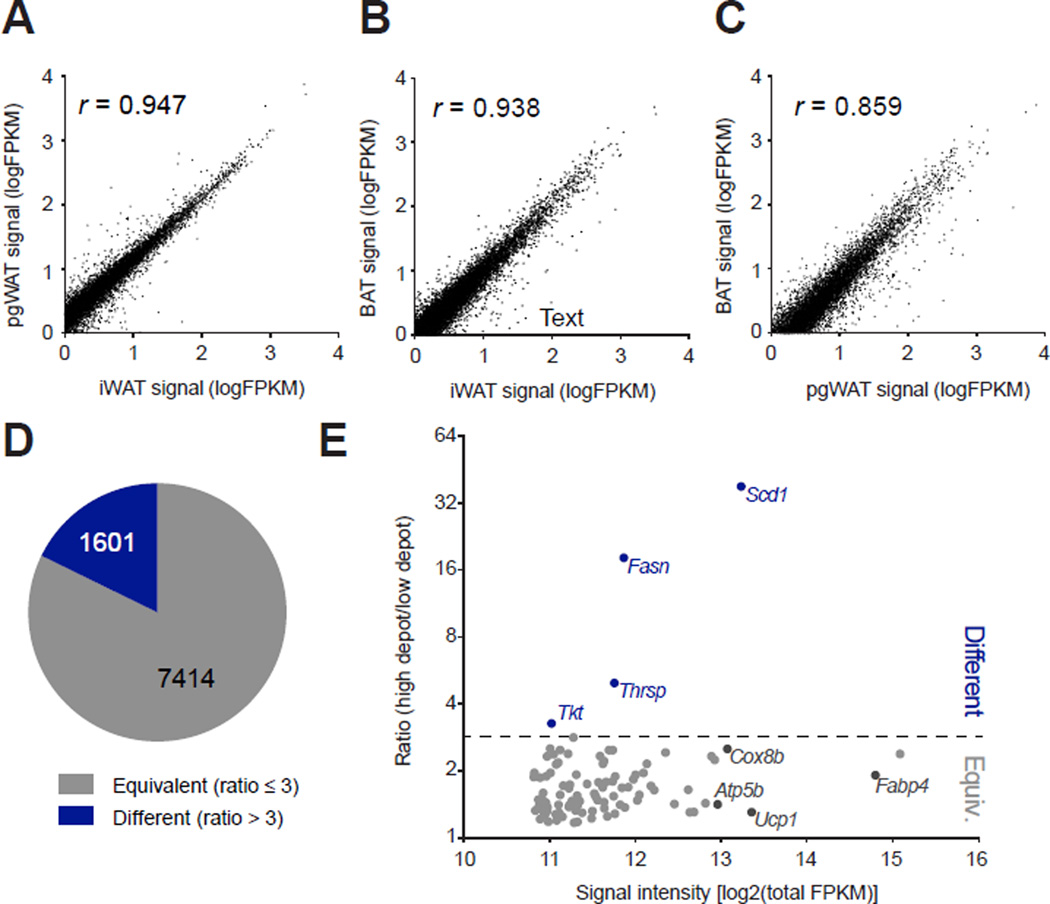
These initial experiments suggest that the TRAP methodology provided a unique gene expression signature of UCP1-positive cells that would be otherwise obscured by other large differences in whole fat pad gene expression. To obtain an unbiased portrait of the gene expression pattern of brown and beige cells in vivo, UCP1-TRAP samples from the iWAT, pgWAT, and BAT were analyzed by RNA-Seq (Table S1). At a global level, the expression signature of UCP1-positive cells from these distinct fat pads was remarkably similar (Fig. 2). This similarity was quantified by two different methods. First, pair-wise comparisons between UCP1-TRAP samples from distinct depots are shown in Fig. 2A–C. Genes that map along the diagonal are equally expressed in UCP1-positive cells between the two depots, whereas genes that fall off the diagonal are enriched in UCP1-positive cells from one depot versus the other. By this analysis, a majority of genes are coordinately expressed between depots (Pearson’s r > 0.85). Second, we devised a “ratio” metric so that UCP1-TRAP gene expression could be evaluated from all three depots simultaneously (see Experimental Procedures and Table S2). A ratio greater than 3 indicates at least a three-fold difference in the expression of a particular gene between the highest and lowest expressing depot in UCP1-TRAP samples. Using this metric, only 1601 of the 9015 detected genes (18%) are differentially expressed in UCP1-TRAP samples; the remaining 7414 genes (82%) are common and equivalently expressed by UCP1-positive cells, regardless of depot of origin (Fig. 2D). As a specific example, the one hundred most abundant genes in the UCP1-TRAP-Seq dataset are shown in Fig. 2E. Many of the highly abundant genes in the “equivalent” category include well known genes of thermogenesis and mitochondria (i.e., Ucp1, Cox8b, Atp5b), as well as genes critical for the function of all adipocytes (i.e., Fabp4) (Fig. 2E and Fig. S2A). In contrast, Scd1 and Fasn were in a small group of genes that were differentially expressed owing to their enrichment in beige versus classical BAT cells (Fig. S2B). Taken together, these two analyses demonstrate that >80% of the gene expression in classical brown and beige cells converge onto a common and shared thermogenesis program in vivo following cold exposure (Fig. S2C, and for a more detailed analysis see Table S3 and Experimental Procedures).
Fig. 2. Identification of the common brown and beige gene expression signature in vivo.
(A–C) The average signal intensities (FPKM) for genes in the UCP1-TRAP-Seq dataset are pair-wise compared between depots. Pearson’s correlation coefficient (r) for each comparison is shown as an insert. n = 2 for pgWAT and n = 3 for iWAT and BAT. UCP1-TRAP-Seq samples were obtained from 6-week old female mice following two weeks cold exposure at 4°C. (D) Pie chart showing the fraction of total detected genes from UCP1-TRAP-Seq classified as either “equivalent” (ratio ≤ 3) or “different” (ratio > 3) (also see Experimental Procedures). (E) Scatter plot showing the ratio metric versus average signal intensity for the top one hundred most abundant UCP1-TRAP-Seq genes. Abundance was determined by total FPKM detected across all UCP1-TRAP-Seq samples.
UCP1-positive cells have a signature of the anatomical location in which they reside
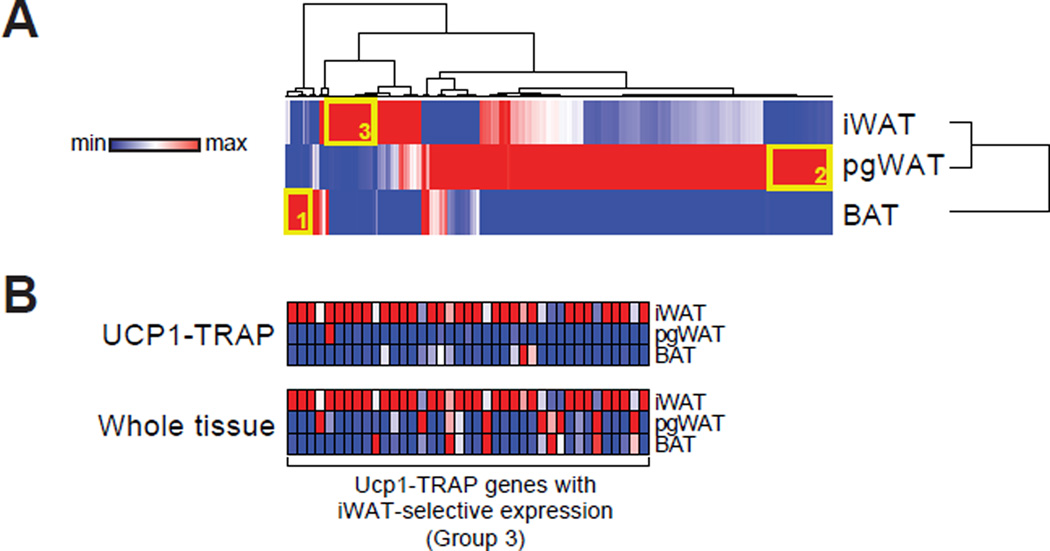
We next analyzed the differentially expressed genes from our UCP1-TRAP-Seq dataset. Hierarchical clustering of the differentially expressed genes (ratio > 3) from the UCP1-TRAP-Seq gene set is shown in Fig. 3A. Interestingly, the majority of differentially expressed genes are depot-specific, as shown by low degree of overlapping red rows in the heat map. To further study these differences, the most abundant and selectively expressed genes from each depot of the UCP1-TRAP-Seq data were identified (Groups 1, 2, and 3 in Fig. 3A; see also Experimental Procedures and Table S4) and their depot-selective expression was validated by qPCR in an independent cohort of UCP1-TRAP mice (Fig. S3).
Fig. 3. Analysis of differentially expressed genes.
(A) Hierarchical clustering of the differentially expressed genes (ratio > 3) from the UCP1-TRAP-Seq dataset. Rows and columns were each clustered using the one minus Pearson correlation distance metric. Clustering was performed with the average signal intensity for each depot. n = 2 for pgWAT and n = 3 for iWAT and BAT. (B) Relative expression by qPCR of most abundant iWAT-selective UCP1-TRAP genes (Group 3) from both UCP1-TRAP samples and from whole tissues across depots. n = 3/group; the average signal intensity is shown. For (A) and (B), red and blue indicate relative high and low expression, respectively, for the column.
Initially, we hypothesized that these differentially expressed genes could directly serve as novel in vivo markers for distinct populations of UCP1-positive cells. However, a straightforward application of this simplistic logic proved difficult because many of these genes also convey information about the anatomical location. As a specific example, the most abundantly expressed and iWAT-selective genes from UCP1-TRAP samples (Group 3 from Fig. 3A) are also highly expressed at the whole tissue level in iWAT pads versus other fat pads (Fig. 3B). Genes in this iWAT-enriched set include transcriptional components (e.g., Irx1) as well as cell surface proteins (e.g., Ltb). This striking similarity implies that UCP1-positive cells share the gene expression of adipose pads in which they reside. Consequently, many differentially expressed UCP1-TRAP genes do not provide direct information regarding enrichment within the UCP1-positive versus UCP1-negative cells of a particular depot. Rather, the dissociation of anatomical gene expression information from true markers of classical brown or beige cells required further refinements of the UCP1-TRAP gene signatures.
Identification of “anatomy-independent” molecular markers for brown and beige adipocytes in vivo
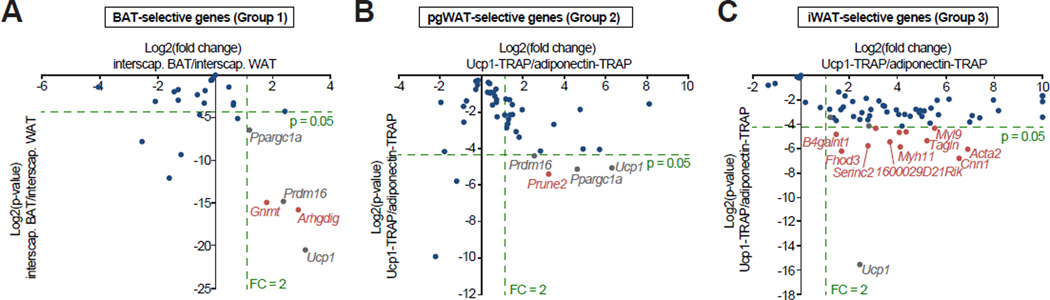
We devised two different methods for analyzing UCP1-positive and UCP1-negative cells within a single anatomical depot. As demonstrated below, these methods can identify anatomy-independent markers for brown or beige cells in vivo. First, we posited that true BAT-enriched genes should also be more highly expressed in the interscapular BAT compared to the neighboring interscapular white fat. UCP1-TRAP-Seq BAT-selective genes (Group 1 in Fig. 3A) not enriched in this manner may simply be more highly expressed in the interscapular region compared to other parts of the body. Such a comparison for BAT-selective UCP1-TRAP genes is shown in Fig. 4A. As expected, the known thermogenic genes Ucp1, Ppargc1a, and Prdm16 were 2- to 8-fold enriched in the interscapular BAT versus interscapular WAT (Fig. 4A). The UCP1-TRAP-Seq BAT-selective gene set (Group 1) was roughly equally distributed between enrichment in interscapular BAT or enrichment in interscapular WAT [median log2(fold change) = −0.1]. This distribution confirms that anatomical information is a significant contributor to the expression signature of UCP1-positive cells within the classical BAT. Of the BAT-selective UCP1-TRAP (Group 1) genes analyzed by qPCR, only Gnmt and Arhgdig were enriched in the expected manner for a true BAT marker (Fig. 4A). Thus, these two genes may serve as novel molecular markers that distinguish classical brown from beige cells in mice in an anatomy-independent manner. Gnmt and Arhgdig encode for an N-methyltransferase and a GDP dissociation inhibitor, respectively. Whether these proteins contribute to classical BAT-selective functions remains to be determined.
Fig. 4. Identification of anatomy-independent markers for UCP1-positive cells from each depot.
(A) Scatter plot of the most BAT-selective genes (Group 1 in Fig. 3A) between interscapular WAT or interscapular BAT tissues. (B and C) Scatter plot of the most pgWAT-selective (Group 2) or iWAT-selective (Group 3) genes between UCP1-TRAP and Adipoq-TRAP samples from the pgWAT (B) or iWAT depot (C). The bottom right quadrant, as marked by the dashed green lines, denote genes with p < 0.05 and FC > 2 for the indicated comparison; genes from groups 1–3 in this category are highlighted in red. As positive controls, the expression of Ucp1 Prdm16, and Ppargc1a are shown in grey. Blue dots indicate those genes that are not statistically significantly enriched or those with FC < 2 for the indicated comparison. n = 4–6/group.
For the iWAT and pgWAT depots, dissection of neighboring UCP1-positive and UCP1-negative cells is not technically feasible as all beige cells are embedded within white fat tissues. As an alternative strategy, we compared UCP1-positive cells of the iWAT or pgWAT with all adipocytes in the corresponding pad. Anatomical markers should be equally expressed between these two groups, whereas true iWAT- or pgWAT-specific markers of UCP1-positive cells should be appropriately enriched. To obtain the proper comparison group, the TRAP mice were crossed with Adiponectin-Cre to generate Adipoq-TRAP mice (Eguchi et al., 2011); in these animals TRAP protein is expressed in mature adipocytes, irrespective of Ucp1 expression (Fig. S4). Immunopurified RNA from cold-acclimated Adipoq-TRAP mice was then used in a direct comparison with UCP1-TRAP samples (Fig. 4B and C). The expression of Ucp1 was ~6-fold or ~80-fold enriched in UCP1-TRAP versus Adipoq-TRAP in the iWAT and pgWAT depots, respectively (Fig. 4B and C). Of the UCP1-TRAP pgWAT-selective genes (Group 2), only Prune2 was also significantly enriched in UCP1-positive cells versus all adipocytes of the pgWAT (Fig. 4B). Prune2 is also highly expressed in the central nervous system (Li et al., 2011) but its function remains poorly characterized, and a role for Prune2 in adipocytes has not been previously described. In contrast to both the BAT and pgWAT comparisons, we found many more UCP1-TRAP-Seq iWAT-selective genes (Group 3) enriched in UCP1-TRAP versus Adipoq-TRAP (Fig. 4C, also see Table S5 for a full list). Similar to the BAT and pgWAT, many of these iWAT-enriched genes (e.g., Serinc2) are poorly characterized and have not been previously linked to adipocyte function.
Beige but not brown adipocytes express a “smooth muscle-like” signature
In addition to the large-scale analyses presented above, the UCP1-TRAP dataset also provides a powerful platform for investigating novel aspects of brown or beige adipocyte biology in vivo. Towards this end, we focused on the unusual constellation of enriched UCP1-TRAP genes (Fig. 4C; Acta2, Tagln, Myh11, Myl9, and Cnn1) that have classically been associated with smooth muscle cells (SMCs) (Miano et al., 1994; Owens et al., 2004) or smooth muscle-like cells (Lazard et al., 1993; Li et al., 2006) (Fig. 4C). This smooth muscle-like signature is enriched in UCP1-TRAP samples from both the inguinal and axillary subcutaneous depots versus BAT (Fig. S5A), indicating that this signature is not specific to the inguinal fat pad alone. Lastly, endothelial cells and pericyte markers such as Cspg4, Pdgfrb, and Cdh5 were equally expressed in UCP1-TRAP across all the depots, demonstrating a specificity for the smooth muscle-like expression pattern versus other vasculature cells (Fig. S5B).
Beige cells can arise from a smooth muscle-like origin
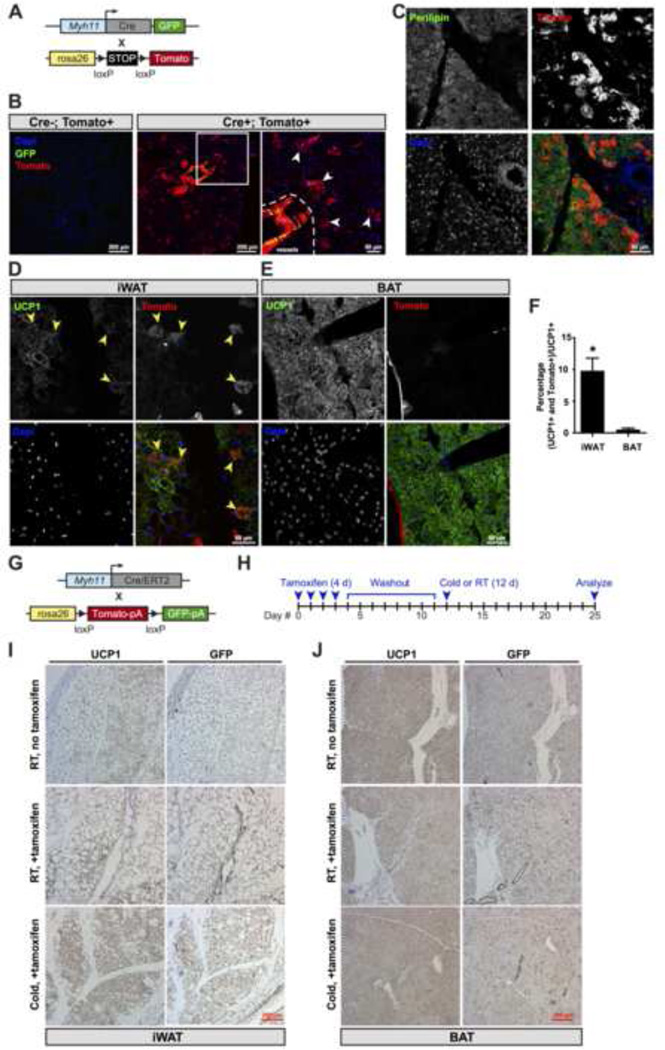
To date, the cellular origins of beige cells in vivo remain enigmatic, though skeletal muscle precursors (Seale et al., 2008) and existing adipocytes (Wang et al., 2013) have been excluded as sources. Based on the UCP1-TRAP expression signature, we investigated the possibility that the precursors for some beige cells might be smooth muscle-like cells. To do this, two genetic fate mapping approaches were employed. The tracing of SMCs in vivo is generally difficult owing to the heterogeneity of this population and the lack of single genetic markers that can precisely and concurrently differentiate SMCs (e.g., vascular SMCs), related smooth muscle-like cells (e.g., myoepithelial cells), and non-smooth muscle cells (e.g., pericytes). In the first approach, we selected a constitutive Myh11 promoter-driven Cre because Myh11 is generally considered to be the most selective marker for bona fide mature smooth muscle and other smooth muscle-like cells (Miano et al., 1994) and the specificity of this transgenic line had been previously validated by lacZ tracing (Xin et al., 2002). Transgenic mice expressing a bicistronic Cre recombinase and eGFP cassette under the control of the Myh11 promoter were crossed with reporter mice expressing a lox-stop-lox tdTomato cassette (Fig. 5A). The Myh11-GFP/tdTomato mouse model provides independent fluorescence signals for both both recombination (tdTomato) and Myh11 expression (GFP). Thus, cells currently expressing high levels of Myh11 (GFP+; tdTomato+) can be distinguished from cells that have arisen from SMCs that had once expressed Myh11 (GFP-; tdTomato+).
Fig. 5. Smooth muscle-like origin of beige cells.
(A) Schematic of the cross to generate Myh11-GFP/tdTomato reporter mice. Transgenic mice expressing a bicistronic transgene consisting of Cre and eGFP under the control of 16 kb of the Myh11 promoter were crossed with tdTomato reporter mice. (B) GFP immunofluorescence and endogenous tomato fluorescence in the iWAT pad. Representative reporter mice (Cre-positive, tomato-positive) and control mice (Cre-negative, tomato-positive) are shown. (C–E) Perilipin (C) or UCP1 (D and E) immunofluorescence and endogenous Tomato fluorescence in iWAT (C and D) or BAT (E). (F) Quantification of double tdTomato+; UCP1+ cells as a percentage of total UCP1-positive cells from iWAT and BAT of Myh11-GFP/tdTomato reporter mice. Data are shown as means ± standard error; n = 3 mice per group. * p < 0.05 for iWAT versus BAT. (G) Schematic of the cross to generate Myh11-CreERT2/GFP reporter mice. BAC transgenic mice expressing a CreERT2 allele under the control of the Myh11 promoter were crossed to GFP (ROSAmT/mG) reporter mice. (H) Cartoon depicting the time course for tamoxifen injections, the washout period, and cold exposure. Tamoxifen was injected at 2 mg/mouse/day for four consecutive days prior to the washout period. (I, J) Immunohistochemical staining of tissues from day 25 for UCP1 (left panels) or GFP (right panels) in the iWAT (I) or BAT (J) of Myh11-CreERT2/GFP reporter mice. For both reporter experiments, mice were heterozygous for Cre and heterozygous for the reporter gene. For B–F, representative images were taken in 6-week old female Myh11-GFP/tdTomato reporter mice following two weeks cold exposure at 4°C. For I–J, representative images were taken in 8-week old male Myh11-CreERT2/GFP reporter mice following the treatment scheme indicated in (H).
Following a two-week cold exposure protocol, robust tdTomato immunofluorescence signal was observed in the iWAT of Cre-positive, but not Cre-negative mice (Fig. 5B). In addition to tdTomato labeling of SMCs in the vessels, strong tdTomato signal was also observed in many cells that displayed morphological characteristics similar to multilocular adipocytes (Fig. 5C). While the Myh11-positive cells surrounding lymph vessels expressed high levels of GFP, the tdTomato-positive extra-lymphatic cells were GFP-negative, indicating that they arise from a once Myh11-expressing precursor. These data suggest that the smooth muscle-like expression signature in beige cells, while detectable, is still significantly lower compared to vascular SMCs or smooth muscle-like cells. Co-staining with the markers perilipin and UCP1 unambiguously established the identity of these extra-lymphatic cells as mature, thermogenic adipocytes (Fig. 5C and D). Lastly, we quantified the number of the double positive population (tdTomato+; UCP1+) cells relative to all UCP1-positive cells in the iWAT and BAT depots. Though we can detect a small number of double positive adipocytes in the BAT, these cells constitute a much higher fraction of total UCP1-positive in the iWAT (Fig. 5E and F).
In the second approach, we sought to pulse-chase existing Myh11-positive cells in adult mice and map their fate upon cold exposure. Towards this goal, we crossed transgenic mice expressing an inducible Myh11-driven Cre (Myh11-CreERT2) with GFP (ROSAmT/mG) reporter mice (Fig. 5G). Myh11-CreERT2/GFP mice were treated with tamoxifen daily for four days to permanently mark Myh11-expressing smooth muscle and smooth muscle-like cells. After a one week tamoxifen washout period, mice were either left at room temperature or housed at 4°C (Fig. 5H). By stopping tamoxifen prior to the cold exposure, only smooth muscle and smooth muscle-like cells at the time of the injection, as well as their progency, are labeled with GFP throughout the experiment. Immunohistochemical staining for GFP in the iWAT depot at room temperature indicated that recombination and labeling occurred only in the smooth muscle cells but not in the existing beige adipocytes (Fig. 5I). Cold exposure profoundly increased the number of beige adipocytes in the iWAT, and a majority of these cells were GFP-positive (Fig. 5I). Importantly, in the classical BAT depot we observed GFP-positivity only in the vasculature surrounding the brown adipocytes, but not in the brown adipocytes themselves (Fig. 5J). Taken together, these fate mapping experiments, together with the UCP1-TRAP data, strongly suggest that mature Myh11-expressing cells suppress, but do not extinguish, their smooth muscle gene signature during differentiation into mature beige adipocytes.
Lastly, it is now well established that the many adipose precursors in vivo reside near the vasculature (Gupta et al., 2012; Tang et al., 2008). These populations have been characterized by both genetic (Berry and Rodeheffer, 2013; Gupta et al., 2012; Lee et al., 2013; Lee et al., 2012; Sanchez-Gurmaches et al., 2012) or prospective (Rodeheffer et al., 2008; Schulz et al., 2011) methods. To better understand the relationship between these previously identified adipocyte precursors and the smooth muscle-like population identified here, we analyzed Tomato-positive and Tomato-negative cells from the stromal vascular fraction of Myh11-GFP/tdTomato mice. Tomato-positive cells were de-enriched in markers for pericytes, endothelial cells, and hematopoietic cells, and enriched in some (e.g., Sca1, Pdgfra, Zfp423) but not all (e.g., Pparg) of the previously identified preadipocyte markers (Fig. S6). Thus the smooth muscle lineage may constitute a portion of previously identified preadipocyte populations.
In vitro conversion of smooth muscle cells into adipocytes with a thermogenic gene signature
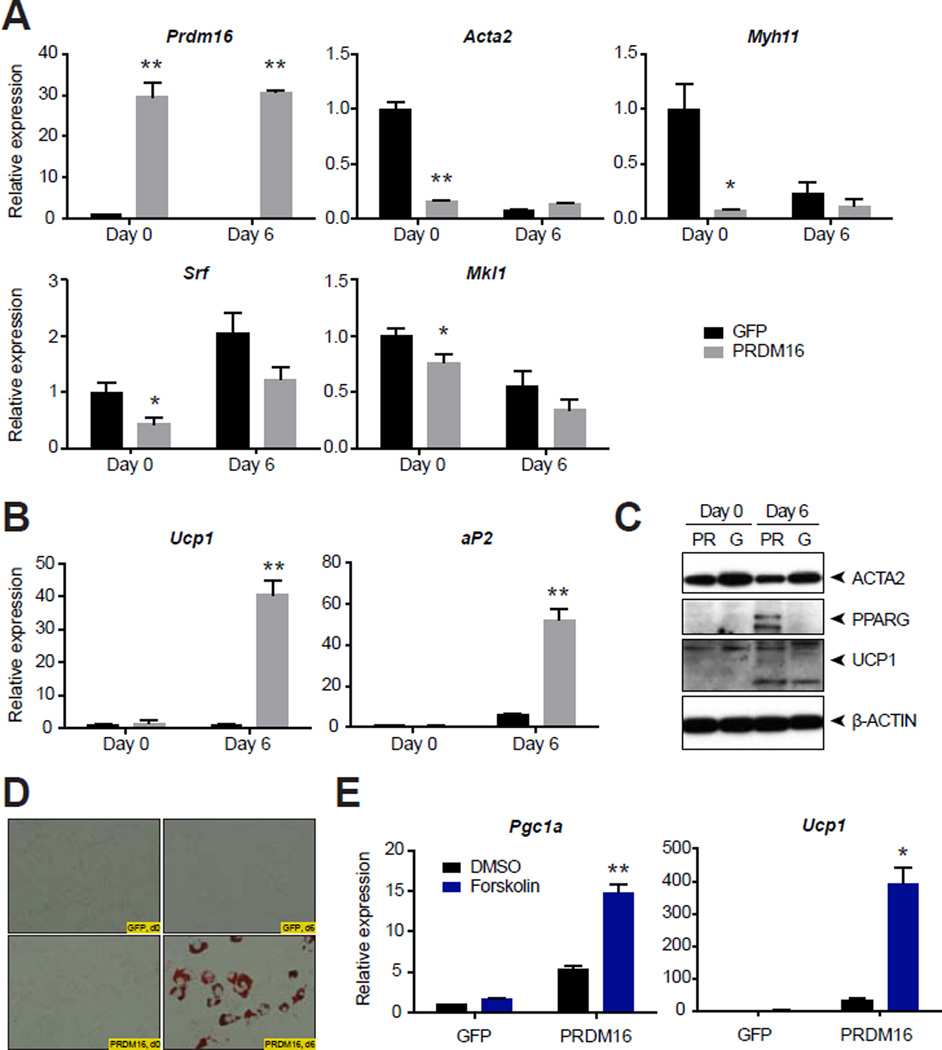
Some evidence indicates that a stem cell population in adipose tissues can differentiate into either SMCs or adipocytes (Rodriguez et al., 2006; Yin et al., 2012), but there are no investigations of a direct phenotypic switch between SMCs or SMC-like cells into adipocytes. Since the development of a beige phenotype requires the coregulatory protein PRDM16 (Cohen et al., 2014), we asked whether this factor might promote adipogenesis with a thermogenic signature, even in mature SMCs. Ectopic expression of PRDM16 markedly suppressed smooth muscle gene expression in primary murine aortic SMCs compared to GFP control, including the expression of the master smooth muscle transcription factor Srf and a principal non-cardiac co-activator, Mkl1 (MRTF-A) (Fig. 6A). Subjecting these cells to the adipogenic cocktail for two days, followed by a longer-term treatment with insulin, rosiglitazone, and triiodothyronine, produced lipid droplet-containing adipocytes in PRDM16-overexpressing SMCs that were UCP1-positive (Fig. 6B–D). Notably, in these SMCs derived adipocytes, the thermogenic genes Ucp1 and Ppargc1a were responsive to the cAMP-signaling agent forskolin, indicating that these cells are bona fide UCP1-positive adipocytes (Fig. 6E). Taken together, these data indicate that mature SMC cells can give rise to adipocytes with a thermogenic gene expression signature, at least under these experimental conditions.
Fig. 6. Conversion of primary aortic SMCs into thermogenic adipocytes.
(A and B) Relative expression of the indicated genes in primary murine aortic SMCs. SMCs were transduced with retrovirus overexpressing PRDM16 or GFP control and then selected with puromycin. Day 0 indicates the gene expression before differentiation. Day 6 indicates gene expression after two days of an adipogenic cocktail followed by five days of insulin, rosiglitazone, and triiodothyronine. Data are presented as means ± standard error; n = 3–5/group. *, ** p < 0.05 or < 0.01, respectively, in GFP- versus PRDM16-overexpressing samples at the same time point. (C) Western blot analysis of ACTA2, PPARG, UCP1, and β-actin loading control from primary aortic murine SMCs at the indicated times. G, GFP; PR, PRDM16. (D) Oil red O staining of primary murine aortic SMCs with the indicated overexpressing construct and at the indicated times. (E) Relative expression of the indicated genes in primary murine aortic SMCs at day 6, after 4 h treatment with DMSO or forskolin (10 µM). Data are presented as means ± standard error; n = 3–5/group. *, ** p < 0.05 or < 0.01, respectively, in forskolin-treated versus DMSO-treated within the same overexpressing group.
DISCUSSION
Here we provide a comprehensive and detailed molecular description of brown and beige adipocytes in vivo. Several insights of potential importance emerge: first, a large fraction of the gene expression shared by brown and beige fat cells represents a common and core thermogenesis gene signature, independent of anatomical location and cell type. The equivalent thermogenic potential of beige and classical brown adipocytes has been suggested in cell culture (Wu et al., 2012) and in humans (Lidell et al., 2013), and here we provide direct evidence for this hypothesis in rodents in vivo. By virtue of their co-expression with well-established thermogenic factors, the poorly annotated genes within this “core” signature are likely to have some type of functional role in the adaptive thermogenesis. These factors include transcriptional components (e.g., Zfp768), potential secreted proteins (e.g., BC028528), and cell-surface receptors (e.g., Il17rc).
Second, many differentially expressed genes between UCP1-positive adipocytes reflect the anatomical location in which they reside. We can also identify anatomy-independent markers that distinguish brown and beige fat in mice. These data suggest that the beige adipocyte population is perhaps more heterogeneous than previously recognized and could provide a rational for why some genetic models show preferential browning in either the subcutaneous (Seale et al., 2011) or visceral (Kiefer et al., 2012) depots. Regarding the gene expression events previously identified as being brown or beige specific, the analyses presented here point to limitations of our approach and also potential re-interpretation of the existing literature. For some genes [e.g., Zic1; (Seale et al., 2007; Timmons et al., 2007)], anatomical information simply cannot be dissociated from differential expression in classical brown versus beige fat cells. Our inability to detect others [e.g., Tmem26; (Wu et al., 2012)] suggests that actively translating mRNAs do not completely reflect total mRNAs in brown or beige cells. Regardless, in light of these observations, it is of paramount importance for future human studies that comparisons are done from neighboring white and brown/beige regions in approximately the same anatomical location.
Third and perhaps most importantly, we provide evidence for a close relationship between smooth muscle-like cells and beige fat cells but not classical brown adipocytes. Three independent lines of evidence support this hypothesis: 1) UCP1-TRAP mice identified a smooth muscle-like gene expression signature in beige cells from multiple fat depots; 2) two independent fate mapping approaches showed that beige adipocytes arise from Myh11-expressing, smooth muscle-like cells in vivo; and 3) ectopic expression of PRDM16 converted primary murine aortic SMCs into UCP1-positive adipocytes in vitro. Interestingly, differentiated SMCs, unlike skeletal or cardiac muscle cells, are known to possess a remarkable degree of phenotypic plasticity even in their mature state. For instance, during vascular calcification, SMCs of the aorta transdifferentiate into osteochondrogenic precursors and chondrocyte-like cells (Speer et al., 2009). In cell culture, cholesterol-loaded SMCs lose their SMC features and adopt a gene expression and phenotype that is characteristic of macrophages (Rong et al., 2003). The data presented here suggest that a SMC-to-adipocyte conversion is also possible in vivo, and ectopic expression of PRDM16 can promote this conversion in vitro. Combined with the previous recognition of at least eight distinct cellular lineages that give rise to various SMC or smooth muscle-like populations (Majesky, 2007), these data imply a potential mosaic of developmental origins for beige adipocytes in vivo.
In summary, the UCP1-TRAP mouse and corresponding UCP1-TRAP dataset provides a powerful platform for elucidating the function of brown and beige fat in the in vivo context. Projecting forward, functional interrogation of the common or differentially expressed gene sets, as well as the metabolic characterization of mice with perturbed or enhanced smooth muscle function, should afford further insights into the “browning” process in vivo. These studies may also illuminate novel pharmacologically tractable pathways for manipulating thermogenic adipocytes in humans.
EXPERIMENTAL PROCEDURES
General Animal Information
Animal experiments were performed according to procedures approved by the Dana-Farber Cancer Institute IACUC. Myh11-Cre (stock #007742), Myh11-Cre/ERT2 (stock #019079), tdTomato reporter mice (stock #007914), and ROSAmT/mG reporter mice (stock #007676) were obtained from Jackson Laboratories. The generation of TRAP and Ucp1-Cre transgenic mice is described in detail in the Supplemental Experimental Procedures. UCP1-TRAP breeding pairs were housed at 30°C and litters were weaned to room temperature at 3 weeks. Unless otherwise stated, for cold exposure experiments 4 week-old female UCP1-TRAP mice were individually housed at 4°C for two weeks.
Molecular Studies
qPCR and western blotting were done according to standard methods. For qPCR from whole tissues or TRAP samples, all values were normalized by the ΔΔCt method to Tbp or to Actb, respectively. RNA-Seq was performed by the Dana-Farber Cancer Institute Center for Cancer Computational Biology Sequencing Facility (see Supplemental Experimental Procedures for details). The following antibodies were used: UCP1 (catalog #ab10983, Abcam), GFP (catalog #ab290, Abcam, for immunoaffinity purifications and Western blotting), GFP (catalog #NB-100–1678, Novus, for immunohistochemistry), perilipin (catalog #ab61682, Abcam) β-actin (catalog #ab20272, Abcam), and PPARG (catalog #2435, Cell Signaling).
TRAP RNA isolation
Immunoaffinity purification of TRAP RNA from adipose tissues was carried out similar to the protocol previously described (Sanz et al., 2009) but with some minor modifications as described in the Supplemental Experimental Procedures.
Bioinformatic analyses
The “ratio” metric (Fig. 2 and Table S2) was calculated as follows. In the UCP1-TRAP-Seq dataset, a detected gene was defined by FPKM > 1 in at least two out of the eight samples; this filtering yields 9015 detected genes. For each gene from this list, the average FPKM for each depot was calculated. The “ratio” for a given gene is then the quotient of the depot with the highest average expression to the depot with the lowest average expression. In this manner, the ratio metric is a measure of the normalized range for a particular gene and is always greater than 1. As a cutoff, genes with ratios > 3 or ≤ 3 were identified as differentially or equivalently expressed, respectively. Hierarchical clustering (Fig. 3A) was performed using GENE-E software (Broad Institute). One minus Pearson correlation was used as the distance metric for clustering of rows and columns.
For the identification of the differentially expressed genes from each UCP1-TRAP depot (Groups 1, 2, and 3 in Fig. 3A), slightly more generous initial criteria were used to minimize false negatives. For the identification of iWAT-selective genes, the filtering criteria were: 10-fold enrichment in the average iWAT signal versus the combined average of the pgWAT and BAT signals; iWAT p-value < 0.2 versus the average signal from the pgWAT and BAT; and iWAT average FPKM > 0.5. For the identification of pgWAT-selective genes, the filtering criteria were: 5-fold enrichment in the average pgWAT signal versus the combined average of the iWAT and BAT signals; pgWAT p-value < 0.2 versus the average signal from the pgWAT and BAT; and pgWAT average FPKM > 0.5. For the identification of BAT-selective genes, the filtering criteria were 5-fold enrichment in the average BAT signal versus the combined average iWAT and pgWAT signals; BAT p-value < 0.2 versus the average signal from the pgWAT and BAT; and BAT average FPKM > 0.2.
Details regarding the analysis of the “core” thermogenesis set can be found in the Supplemental Experimental Procedures.
Immunofluorescence and immunohistochemistry
Immunofluorescence and immunohistochemical stainings were done according to standard protocols. Details can be found in the Supplemental Experimental Procedures. ImageJ software was used for quantification of tdTomato- or UCP1-positivity. Briefly, the percentage co-localized (tdTomato-positive; UCP1-positive) was calculated by calculating the area of co-localization divided by the total UCP1-positive area.
Cell Culture
Retrovirus production and infection from pMSCV-PRDM16 or pMSCV-GFP was performed as described previously (Seale et al., 2008). To generate stable cells, primary murine aortic SMCs were infected with the indicated retroviruses and selected with 3 µg/ml puromycin. Adipogenic differentiation was induced by addition of a hormone cocktail in media for two days [isobutylmethylxanthine (Sigma), 0.5 mM; dexamethasone (Sigma), 5 µM; rosiglitazone (Cayman), 1 µM; insulin (Sigma), 5 µM; triiodothyronine (Sigma), 1 nM; indomethacin (Sigma), 125 µM] followed by maintenance media until time of harvest [rosiglitazone (Cayman), 1 µM; insulin (Sigma), 5 µM; triiodothyronine (Sigma)]. Where indicated, cells were treated with forskolin (10 µM, Sigma) for 4 h. A detailed protocol for the isolation of primary aortic SMCs can be found in the Supplemental Experimental Procedures.
Statistics
The Student’s t-test was used for pair-wise comparisons. Unless otherwise specified, stastistical significance was set at p < 0.05
Supplementary Material
Highlights.
Ribosomal profiling provides a molecular portrait of thermogenic adipocytes in vivo
Beige, but not classical brown cells, express smooth muscle-like genes
Smooth muscle-like cells are precursors for beige adipocytes in vivo
ACKNOWLEDGEMENTS
We thank members of the Spiegelman and Rosen laboratories, S. Nallamshetty, J. Plutzky, L. Chang, and Y. E. Chen for helpful discussions. We are grateful for technical assistance from the Rodent Histopathology Core at the Dana-Farber/Harvard Cancer Center and the Histology Core at the Beth Israel Deaconess Medical Center. Imaging was performed at the Nikon Imaging Center of Harvard Medical School. JZL is supported by a joint postdoctoral fellowship from the American and Canadian Diabetes Associations (PF-3-12-3927-JL). KJS is supported by the Swedish Research Council international post-doctoral fellowship. This work was supported by NIH grants DK085171 and DK017690 (to EDR) and by NIH grant DK031405 (BMS).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Berry R, Rodeheffer MS. Characterization of the adipocyte cellular lineage in vivo. Nature cell biology. 2013;15:302–308. doi: 10.1038/ncb2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bostrom P, Wu J, Jedrychowski MP, Korde A, Ye L, Lo JC, Rasbach KA, Bostrom EA, Choi JH, Long JZ, et al. A PGC1-alpha-dependent myokine that drives brown-fat-like development of white fat and thermogenesis. Nature. 2012;481:463–468. doi: 10.1038/nature10777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon B, Nedergaard J. Brown adipose tissue: function and physiological significance. Physiological reviews. 2004;84:277–359. doi: 10.1152/physrev.00015.2003. [DOI] [PubMed] [Google Scholar]
- Cederberg A, Gronning LM, Ahren B, Tasken K, Carlsson P, Enerback S. FOXC2 is a winged helix gene that counteracts obesity, hypertriglyceridemia, and diet-induced insulin resistance. Cell. 2001;106:563–573. doi: 10.1016/s0092-8674(01)00474-3. [DOI] [PubMed] [Google Scholar]
- Cohen P, Levy JD, Zhang Y, Frontini A, Kolodin DP, Svensson KJ, Lo JC, Zeng X, Ye L, Khandekar MJ, et al. Ablation of PRDM16 and Beige Adipose Causes Metabolic Dysfunction and a Subcutaneous to Visceral Fat Switch. Cell. 2014;156:304–316. doi: 10.1016/j.cell.2013.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cousin B, Cinti S, Morroni M, Raimbault S, Ricquier D, Penicaud L, Casteilla L. Occurrence of brown adipocytes in rat white adipose tissue: molecular and morphological characterization. Journal of cell science. 1992;103(Pt 4):931–942. doi: 10.1242/jcs.103.4.931. [DOI] [PubMed] [Google Scholar]
- Cypess AM, Lehman S, Williams G, Tal I, Rodman D, Goldfine AB, Kuo FC, Palmer EL, Tseng YH, Doria A, et al. Identification and importance of brown adipose tissue in adult humans. The New England journal of medicine. 2009;360:1509–1517. doi: 10.1056/NEJMoa0810780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cypess AM, White AP, Vernochet C, Schulz TJ, Xue R, Sass CA, Huang TL, Roberts-Toler C, Weiner LS, Sze C, et al. Anatomical localization, gene expression profiling and functional characterization of adult human neck brown fat. Nature medicine. 2013;19:635–639. doi: 10.1038/nm.3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eguchi J, Wang X, Yu S, Kershaw EE, Chiu PC, Dushay J, Estall JL, Klein U, Maratos-Flier E, Rosen ED. Transcriptional control of adipose lipid handling by IRF4. Cell metabolism. 2011;13:249–259. doi: 10.1016/j.cmet.2011.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldmann HM, Golozoubova V, Cannon B, Nedergaard J. UCP1 ablation induces obesity and abolishes diet-induced thermogenesis in mice exempt from thermal stress by living at thermoneutrality. Cell metabolism. 2009;9:203–209. doi: 10.1016/j.cmet.2008.12.014. [DOI] [PubMed] [Google Scholar]
- Gesta S, Bluher M, Yamamoto Y, Norris AW, Berndt J, Kralisch S, Boucher J, Lewis C, Kahn CR. Evidence for a role of developmental genes in the origin of obesity and body fat distribution. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:6676–6681. doi: 10.1073/pnas.0601752103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta RK, Mepani RJ, Kleiner S, Lo JC, Khandekar MJ, Cohen P, Frontini A, Bhowmick DC, Ye L, Cinti S, et al. Zfp423 expression identifies committed preadipocytes and localizes to adipose endothelial and perivascular cells. Cell metabolism. 2012;15:230–239. doi: 10.1016/j.cmet.2012.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harms M, Seale P. Brown and beige fat: development, function and therapeutic potential. Nature medicine. 2013;19:1252–1263. doi: 10.1038/nm.3361. [DOI] [PubMed] [Google Scholar]
- Heiman M, Schaefer A, Gong S, Peterson JD, Day M, Ramsey KE, Suarez-Farinas M, Schwarz C, Stephan DA, Surmeier DJ, et al. A translational profiling approach for the molecular characterization of CNS cell types. Cell. 2008;135:738–748. doi: 10.1016/j.cell.2008.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiefer FW, Vernochet C, O'Brien P, Spoerl S, Brown JD, Nallamshetty S, Zeyda M, Stulnig TM, Cohen DE, Kahn CR, et al. Retinaldehyde dehydrogenase 1 regulates a thermogenic program in white adipose tissue. Nature medicine. 2012;18:918–925. doi: 10.1038/nm.2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazard D, Sastre X, Frid MG, Glukhova MA, Thiery JP, Koteliansky VE. Expression of smooth muscle-specific proteins in myoepithelium and stromal myofibroblasts of normal and malignant human breast tissue. Proceedings of the National Academy of Sciences of the United States of America. 1993;90:999–1003. doi: 10.1073/pnas.90.3.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YH, Petkova AP, Granneman JG. Identification of an adipogenic niche for adipose tissue remodeling and restoration. Cell metabolism. 2013;18:355–367. doi: 10.1016/j.cmet.2013.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YH, Petkova AP, Mottillo EP, Granneman JG. In vivo identification of bipotential adipocyte progenitors recruited by beta3-adrenoceptor activation and high-fat feeding. Cell metabolism. 2012;15:480–491. doi: 10.1016/j.cmet.2012.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepper C, Fan CM. Inducible lineage tracing of Pax7-descendant cells reveals embryonic origin of adult satellite cells. Genesis (New York, NY) 2010;48:424–436. doi: 10.1002/dvg.20630. 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Chang S, Qi X, Richardson JA, Olson EN. Requirement of a myocardin-related transcription factor for development of mammary myoepithelial cells. Molecular and cellular biology. 2006;26:5797–5808. doi: 10.1128/MCB.00211-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Itoh M, Ohta K, Ueda M, Mizuno A, Ohta E, Hida Y, Wang MX, Takeuchi K, Nakagawa T. The expression and localization of Prune2 mRNA in the central nervous system. Neuroscience letters. 2011;503:208–214. doi: 10.1016/j.neulet.2011.08.037. [DOI] [PubMed] [Google Scholar]
- Lidell ME, Betz MJ, Dahlqvist Leinhard O, Heglind M, Elander L, Slawik M, Mussack T, Nilsson D, Romu T, Nuutila P, et al. Evidence for two types of brown adipose tissue in humans. Nature medicine. 2013;19:631–634. doi: 10.1038/nm.3017. [DOI] [PubMed] [Google Scholar]
- Majesky MW. Developmental basis of vascular smooth muscle diversity. Arteriosclerosis, thrombosis, and vascular biology. 2007;27:1248–1258. doi: 10.1161/ATVBAHA.107.141069. [DOI] [PubMed] [Google Scholar]
- Miano JM, Cserjesi P, Ligon KL, Periasamy M, Olson EN. Smooth muscle myosin heavy chain exclusively marks the smooth muscle lineage during mouse embryogenesis. Circulation research. 1994;75:803–812. doi: 10.1161/01.res.75.5.803. [DOI] [PubMed] [Google Scholar]
- Nedergaard J, Bengtsson T, Cannon B. New powers of brown fat: fighting the metabolic syndrome. Cell metabolism. 2011;13:238–240. doi: 10.1016/j.cmet.2011.02.009. [DOI] [PubMed] [Google Scholar]
- Nicholls DG, Bernson VS, Heaton GM. The identification of the component in the inner membrane of brown adipose tissue mitochondria responsible for regulating energy dissipation. Experientia Supplementum. 1978;32:89–93. doi: 10.1007/978-3-0348-5559-4_9. [DOI] [PubMed] [Google Scholar]
- Owens GK, Kumar MS, Wamhoff BR. Molecular regulation of vascular smooth muscle cell differentiation in development and disease. Physiological reviews. 2004;84:767–801. doi: 10.1152/physrev.00041.2003. [DOI] [PubMed] [Google Scholar]
- Rodeheffer MS, Birsoy K, Friedman JM. Identification of white adipocyte progenitor cells in vivo. Cell. 2008;135:240–249. doi: 10.1016/j.cell.2008.09.036. [DOI] [PubMed] [Google Scholar]
- Rodriguez LV, Alfonso Z, Zhang R, Leung J, Wu B, Ignarro LJ. Clonogenic multipotent stem cells in human adipose tissue differentiate into functional smooth muscle cells. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:12167–12172. doi: 10.1073/pnas.0604850103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rong JX, Shapiro M, Trogan E, Fisher EA. Transdifferentiation of mouse aortic smooth muscle cells to a macrophage-like state after cholesterol loading. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:13531–13536. doi: 10.1073/pnas.1735526100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Gurmaches J, Hung CM, Sparks CA, Tang Y, Li H, Guertin DA. PTEN loss in the Myf5 lineage redistributes body fat and reveals subsets of white adipocytes that arise from Myf5 precursors. Cell metabolism. 2012;16:348–362. doi: 10.1016/j.cmet.2012.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz E, Yang L, Su T, Morris DR, McKnight GS, Amieux PS. Cell-type-specific isolation of ribosome-associated mRNA from complex tissues. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:13939–13944. doi: 10.1073/pnas.0907143106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz TJ, Huang TL, Tran TT, Zhang H, Townsend KL, Shadrach JL, Cerletti M, McDougall LE, Giorgadze N, Tchkonia T, et al. Identification of inducible brown adipocyte progenitors residing in skeletal muscle and white fat. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:143–148. doi: 10.1073/pnas.1010929108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seale P, Bjork B, Yang W, Kajimura S, Chin S, Kuang S, Scime A, Devarakonda S, Conroe HM, Erdjument-Bromage H, et al. PRDM16 controls a brown fat/skeletal muscle switch. Nature. 2008;454:961–967. doi: 10.1038/nature07182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seale P, Conroe HM, Estall J, Kajimura S, Frontini A, Ishibashi J, Cohen P, Cinti S, Spiegelman BM. Prdm16 determines the thermogenic program ofsubcutaneous white adipose tissue in mice. The Journal of clinical investigation. 2011;121:96–105. doi: 10.1172/JCI44271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seale P, Kajimura S, Yang W, Chin S, Rohas LM, Uldry M, Tavernier G, Langin D, Spiegelman BM. Transcriptional control of brown fat determination by PRDM16. Cell metabolism. 2007;6:38–54. doi: 10.1016/j.cmet.2007.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp LZ, Shinoda K, Ohno H, Scheel DW, Tomoda E, Ruiz L, Hu H, Wang L, Pavlova Z, Gilsanz V, et al. Human BAT possesses molecular signatures that resemble beige/brite cells. PloS one. 2012;7:e49452. doi: 10.1371/journal.pone.0049452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speer MY, Yang HY, Brabb T, Leaf E, Look A, Lin WL, Frutkin A, Dichek D, Giachelli CM. Smooth muscle cells give rise to osteochondrogenic precursors and chondrocytes in calcifying arteries. Circulation research. 2009;104:733–741. doi: 10.1161/CIRCRESAHA.108.183053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W, Zeve D, Suh JM, Bosnakovski D, Kyba M, Hammer RE, Tallquist MD, Graff JM. White fat progenitor cells reside in the adipose vasculature. Science (New York, NY) 2008;322:583–586. doi: 10.1126/science.1156232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmons JA, Wennmalm K, Larsson O, Walden TB, Lassmann T, Petrovic N, Hamilton DL, Gimeno RE, Wahlestedt C, Baar K, et al. Myogenic gene expression signature establishes that brown and white adipocytes originate from distinct cell lineages. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:4401–4406. doi: 10.1073/pnas.0610615104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Marken Lichtenbelt WD, Vanhommerig JW, Smulders NM, Drossaerts JM, Kemerink GJ, Bouvy ND, Schrauwen P, Teule GJ. Cold-activated brown adipose tissue in healthy men. The New England journal of medicine. 2009;360:1500–1508. doi: 10.1056/NEJMoa0808718. [DOI] [PubMed] [Google Scholar]
- Virtanen KA, Lidell ME, Orava J, Heglind M, Westergren R, Niemi T, Taittonen M, Laine J, Savisto NJ, Enerback S, et al. Functional brown adipose tissue in healthy adults. The New England journal of medicine. 2009;360:1518–1525. doi: 10.1056/NEJMoa0808949. [DOI] [PubMed] [Google Scholar]
- Walden TB, Hansen IR, Timmons JA, Cannon B, Nedergaard J. Recruited vs. nonrecruited molecular signatures of brown, "brite," and white adipose tissues. American journal of physiology Endocrinology and metabolism. 2012;302:E19–E31. doi: 10.1152/ajpendo.00249.2011. [DOI] [PubMed] [Google Scholar]
- Wang QA, Tao C, Gupta RK, Scherer PE. Tracking adipogenesis during white adipose tissue development, expansion and regeneration. Nature medicine. 2013;19:1338–1344. doi: 10.1038/nm.3324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Bostrom P, Sparks LM, Ye L, Choi JH, Giang AH, Khandekar M, Virtanen KA, Nuutila P, Schaart G, et al. Beige adipocytes are a distinct type of thermogenic fat cell in mouse and human. Cell. 2012;150:366–376. doi: 10.1016/j.cell.2012.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Cohen P, Spiegelman BM. Adaptive thermogenesis in adipocytes: is beige the new brown. Genes & development. 2013;27:234–250. doi: 10.1101/gad.211649.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin HB, Deng KY, Rishniw M, Ji G, Kotlikoff MI. Smooth muscle expression of Cre recombinase and eGFP in transgenic mice. Physiological genomics. 2002;10:211–215. doi: 10.1152/physiolgenomics.00054.2002. [DOI] [PubMed] [Google Scholar]
- Yin JW, Liang Y, Park JY, Chen D, Yao X, Xiao Q, Liu Z, Jiang B, Fu Y, Bao M, et al. Mediator MED23 plays opposing roles in directing smooth muscle cell and adipocyte differentiation. Genes & development. 2012;26:2192–2205. doi: 10.1101/gad.192666.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young P, Arch JR, Ashwell M. Brown adipose tissue in the parametrial fat pad of the mouse. FEBS letters. 1984;167:10–14. doi: 10.1016/0014-5793(84)80822-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.