Abstract
MicroRNA (miRNA) are short sequences of RNA that function as post-transcriptional regulators by binding to target mRNA transcripts resulting in translational repression. A number of recent studies have identified miRNA as being involved in neurodegenerative disorders including Alzheimer’s disease (AD), Parkinson’s disease (PD) and Huntington’s disease (HD). However, the role of miRNA in Multiple System Atrophy (MSA), a progressive neurodegenerative disorder characterized by oligodendroglial accumulation of alpha-synuclein remains unexamined. In this context, this study examined miRNA profiles in MSA cases compared to controls and in transgenic (tg) models of MSA compared to non tg mice.
The results demonstrate a widespread dysregulation of miRNA in MSA cases, which is recapitulated in the murine models. The study employed a cross-disease, cross-species approach to identify miRNA that were either specifically dysregulated in MSA or were commonly dysregulated in neurodegenerative conditions such as AD, Dementia with Lewy bodies, Progressive Supranuclear Palsy and Corticobasal Degeneration or the tg mouse model equivalents of these disorders. Using this approach we identified a number of miRNA that were commonly dysregulated between disorders and those that were disease-specific.
Moreover, we identified miR-96 as being up regulated in MSA. Consistent with the up regulation of miR-96, mRNA and protein levels of members of the solute carrier protein family SLC1A1 and SLC6A6, miR-96 target genes, were down regulated in MSA cases and a tg model of MSA. These results suggest that miR-96 dysregulation may play a role in MSA and its target genes may be involved in the pathogenesis of MSA.
Keywords: Transgenic, neurodegeneration, mouse, human
INTRODUCTION
MicroRNA (miRNA) are short (∼ 22 nucleotide) RNA molecules that function as post-transcriptional regulators by binding to complementary sequences on target mRNA transcripts, typically resulting in translational repression or target degradation and gene silencing (Ambros, 2001; Moss, 2002; Berezikov, 2011). miRNA each have hundreds of target genes, therefore dysregulation in a single miRNA may have a widespread effect across a variety of cellular pathways and process. While miRNA have been studied in neurodegenerative disorders such as Alzheimer’s disease (AD) (Lukiw, 2007; Barbato et al., 2009; Sethi & Lukiw, 2009; Schonrock et al., 2010; Shioya et al., 2010; Wang et al., 2010; Delay & Hebert, 2011; Geekiyanage & Chan, 2011; Satoh, 2011), Parkinson’s disease (PD) (Barbato et al., 2009; Harraz et al., 2011; Margis & Rieder, 2011) and Huntington’s disease (HD) (Enciu et al., 2011; Lee et al., 2011), they remain unexamined in Multiple System Atrophy (MSA).
MSA is a progressive neurodegenerative disorder characterized clinically by symptoms such as autonomic dysfunction and motor abnormalities and neuropathologically by oligodendrocytic accumulation of alpha-synuclein (α-syn) (Lantos & Papp, 1994; Wakabayashi & Takahashi, 2006; Yoshida, 2007). In MSA, the motor, autonomic and non-motor deficits are not responsive to conventional antiparkinsonian treatments and there are no therapies available for MSA. MSA patients display considerable neuronal loss in the striatum, cerebellum, brainstem and cortex, accompanied by astrogliosis, microgliosis and myelin loss (Wakabayashi & Takahashi, 2006; Yoshida, 2007). Comparable neuropathological alterations have been observed in transgenic (tg) mice over expressing human α-syn (h-αsyn) under the control of an oligodendrocytic-specific promoter (MBP-myelin basic protein) (Shults et al., 2005b) and in other tg mice over expressing h-αsyn under the proteolipid protein (PLP) (Kahle et al., 2002) and the 2,' 3'-cyclic nucleotide 3'-phosphodiesterase (CNP) promoter (Yazawa et al., 2005).
Typically, previous studies of neurodegeneration have compared the miRNA profile of a single neurodegenerative disease state against control levels, however there remains a need for more detailed analysis of miRNA profiles across neurodegenerative disorders compared to each other and controls. This type of cross-disease analysis will enable identification of miRNA that are commonly dysregulated across a number of diseases and those that are disease-specific. Identification of such disease-specific miRNA profiles will help elucidate common disease-related pathways and those which may be central to the pathogenesis of a particular disorder and may also enable identification of gene and/or protein biomarkers for particular neurodegenerative disease states.
In this context, this study sought to examine the miRNA profile from humans diagnosed with MSA and to compare them to profiles from patients diagnosed with Dementia with Lewy Bodies (DLB), AD, Progressive Supranuclear Palsy (PSP) and corticobasal degeneration (CBD) to control samples to identify miRNA profiles that distinguished MSA from these other disorders. In addition, we conducted a parallel examination of miRNA profiles from two tg lines that model MSA (an intermediate and high expressor line), DLB/PD, AD and tauopathy respectively and compared these profiles to each other and to non tg mice. Furthermore, a human versus tg model comparison was conducted to identify disease-common and disease-specific miRNA that correlate between the tg mice models and human patients.
We demonstrate widespread miRNA dysregulation across neurodegenerative conditions in both humans and mice. Moreover, we show that our tg model of MSA recapitulates the altered miRNA profile observed in human MSA patients. Finally we identified an MSA-related miR-96 dysregulation in both tg mice and humans, and were able to demonstrate dysregulation of miR-96 target mRNA and protein in mice and humans.
MATERIAL AND METHODS
Human cases
All cases were from the Alzheimer Disease Research Center (ADRC) at the University of California, San Diego (UCSD). The materials were collected and utilized with the written consent of the subjects and the study conforms with The Code of Ethics of the World Medical Association (Declaration of Helsinki), printed in the British Medical Journal (18 July 1964). Cases selection for this study was based on neuropathological examination and determination of diagnosis of MSA, DLB, AD, PSP and CBD. Group demographics are presented in Supp. Table 1.
Animal samples
Mice from 5 different tg lines and age-matched non tg mice were in this study. A detailed summary of the models used in this study is provided in Supp. Table 2, briefly this study used tg mice expressing human (h) alpha-synuclein (asyn) under the control of the oligodendrocyte-specific Myelin Basic Protein (MBP) (Lines MBP1-hαsyn and MBP29-hαsyn (medium and high expressers respectively) (Shults et al., 2005a), tg mice expressing hαsyn under the control of the mThy1.2 promoter (mThy1-hαsyn tg) (Rockenstein et al., 2002), tg mice expressing human APP751 cDNA containing the London (V717I) and Swedish (K670M/N671L) mutations under the regulatory control of the murine (m)Thy-1 gene (mThy1-hAPP tg) (Rockenstein et al., 2001) and tg mice over-expressing TAU441 bearing the missense mutations V337M and R406W under the control of the brain specific murine Thy-1 promoter (mThy1-htau) (Kindly donated by JSW Life Sciences GmbH, 8020 Graz, Austria)(Flunkert et al., 2013). These tg models and non tg controls were used for the initial miRNA microarray profiling (n=4 for the non tg, MBP1-hαsyn tg, Mthy1-hαsyn tg, mThy1-hAPP tg and mTHy1-htau tg groups and n=3 for the MBP29-hαsyn tg group) and independent samples from a different cohort of mice were used for the subsequent qPCR validation (n=5 for each group). All mice used in this study were male. The MBP29 mice rapidly develop pathology and as such were used at an age of 3 months, all other tg mice used in this study were 6 months of age, an age representing full onset of pathology. Determination of mouse genotype was assessed via genomic DNA extracted from tail biopsies and analyzed by PCR amplification, as previously reported (Rockenstein et al., 2002). Following NIH guidelines for the humane treatment of animals, under anesthesia mice were sacrificed and brains removed. The right hemibrain was immersion-fixed in 4% paraformaldehyde in PBS pH 7.4 and the left hemibrain was kept at −80°C for and was subsequently used for miRNA isolation. UCSD is an Institutional Animal Care and Use Committee accredited institution and the UCSD Animal Subjects Committee approved the experimental protocol followed in all studies according to the Association for Assessment and Accreditation of Laboratory Animal Care International guidelines. Additionally, all experiments were carried out in accordance with the Guidelines laid down by the NIH regarding the care and use of animals for experimental procedures.
miRNA isolation and quality control
Total RNA containing miRNA was isolated from murine and human frontal cortex samples using the miRNAeasy kit (Qiagen) as per the manufacturer’s protocol. The concentration and quality of RNA were determined using a spectrophotometer and a Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA, USA). All samples used in this study had 260:280 ratios above 1.8 and RIN values of 8 or greater.
miRNA profiling in mouse and human
The miRNA profiles of the non tg and tg mice were analyzed using the miFinder RT² miRNA PCR Array system (SABiosciences), as per the manufacturer’s protocol. This array profiles the expression of the 88 most abundantly expressed and bestcharacterized miRNA sequences (based on miRBase version 14). Comparative miRNA profiling of human control (n=4) and MSA (n=3) samples was also conducted using this assay to provide a means of comparing profiles across the same platform. Analysis was performed using the web-based RT² Profiler™ PCR Array Data Analysis (http://pcrdataanalysis.sabiosciences.com/pcr/arrayanalysis.php). Detailed miRNA profiling across the human samples from controls, MSA, DLB, AD, CBD and PSP (all n=4) was on the OneArray® Human microRNA Microarray v3. This microarray contains 99.94% of Sanger miRBase version 17 human miRNA content and uses spotted probes to interrogate 1087 human miRNA. This analysis was out-sourced to Phalanx Biotech Group who performed the comparison. Phalanx Biotech Group also performed subsequent statistical analysis of the comparative miRNA expression levels identified using the OneArray system.
RT-qPCR validation of miR-96 levels
Independent validation of miR-96 levels across the murine and human samples was performed using miScript Primer Assays for murine and human miR-96 (Qiagen) as per the manufacture’s protocols. The PCR cycling parameters were: 95 °C for 10 min followed by 40 cycles of (95 °C for 15 sec; 60 °C for 35 sec and 72 °C for 30 sec). Finally, a dissociation protocol was also performed at the end of each run to verify the presence of a single product with the appropriate melting point temperature for each amplicon. The amount of studied cDNA in each sample was calculated by the 2^ddCT comparative threshold cycle method and expressed as fold change across groups compared to controls.
RT-qPCR analysis of miRNA machinery
Levels of mRNA for components of the miRNA processing machinery (Drosha, DiGeorge syndrome critical region (DGCR8), Exportin-5 (XPO5), Dicer1 and AGO2) in all the murine and human cases were analyzed by RT-qPCR. Primers were designed using OligoPerfect™ Designer online tool (Life Technologies). All reactions were run in triplicate on the BioRad iQ5 system and the mean expression values of each triplicate were used for data analysis. Gene expressions were normalized using Glyceraldehyde 3-phosphate dehydrogenase (GAPDH). Primer sets used for validation are provided in Supp Table 3. The PCR cycling parameters were: 50 °C for 2 min, 95 °C for 10 min, and 40 cycles of 94 °C for 15 s, 60 °C for 1 min. Finally, a dissociation protocol was also performed at the end of each run to verify the presence of a single product with the appropriate melting point temperature for each amplicon. The amount of studied cDNA in each sample was calculated by the 2^ddCT comparative threshold cycle method and expressed as fold change across groups compared to controls.
mRNA target identification and validation
Potential targets of mouse and human miR-96 were retrieved from three wellestablished target prediction programs: Targetscan (Lewis et al., 2005; Grimson et al., 2007), Pictar (Krek et al., 2005) and Microcosm(Rehmsmeier et al., 2004)). The top 50 targets from each database were analyzed and targets appearing across databases were chosen for subsequent RT-qPCR validation, primers sets for mRNA targets are provided in Supp Table 4. Primers were designed using OligoPerfect™ Designer online tool (Life Technologies). The PCR cycling parameters were: 50 °C for 2 min, 95 °C for 10 min, and 40 cycles of 94 °C for 15 s, 60 °C for 1 min. Finally, a dissociation protocol was also performed at the end of each run to verify the presence of a single product. The amount of studied cDNA in each sample was calculated by the 2^ddCT comparative threshold cycle method and expressed as fold change across groups compared to controls.
Luciferase reporter constructs and luciferase reporter assay validation of miRNA target proteins
The effect of miRNA on gene regulation was monitored by co-transfecting 300ng human and mouse mir-96 and the luciferase reporter gene into HEK293 cells. Reporter constructs were obtained from Genecopoeia (Maryland, USA). The 3′-UTR of SLC1A1 and SLC6A6 were cloned downstream of the luciferase open reading frame. In addition to human and mouse mir-96 that could bind to the corresponding UTR, negative control (scrambled human and mouse mir-96) was transfected with the corresponding luciferase reporter plasmid for each gene analyzed. The luciferase activity was measured 24 hrs after transfection and is expressed as percentage of control ± standard deviation.
Immunoblot analysis of target proteins
Briefly as previously described (Ubhi et al., 2010), twenty micrograms of protein per case, from the particulate fraction of the human (control vs MSA) or mouse (non transgenic vs MBP-α-syn transgenic) brain homogenates, was loaded onto 4–12% Bis-Tris (Invitrogen) SDS-PAGE gels, transferred onto Immobilon membranes, washed and blocked in BSA. After an overnight incubation with antibodies against SLC1A1 (Mouse monoclonal, 1:1000, Abcam, (ab78395)), SLC6A6 (rabbit polyclonal, 1:1000, Santa Cruz (sc-135257)) or α-syn (rabbit polyclonal, 1:1000, Millipore (AB5038)), membranes were incubated in appropriate secondary antibodies, reacted with ECL and developed on a VersaDoc gel-imaging machine (Bio-Rad, Hercules, CA). The samples were processed and analyzed blind coded and in duplicate to confirm the reproducibility of the results. For analysis of the bands in the western blots the Quantity One (Biorad) program was used. Briefly, the digital images were maximize at first followed by creating a box around the specific band with the volume tools and then image analysis while previously setting the volume report options that includes the integrated pixel intensity in the volume of interest. The value was corrected to the background gray value of the blot and then expressed as signal over beta-actin ratio (mouse monoclonal, 1:1000; Sigma) to correct for loading.
To confirm the specificity of the results, the same samples from the human and mouse brains were incubated with incubated with alternative antibodies against SLC1A1 (rabbit polyclonal, 1:1000, Alomone (AGC-023)) and SLC6A6 (rabbit polyclonal, 1:1000, Millipore (AB5414P)) from a different source.
Immunohistochemical analysis of target proteins
As previously described (Ubhi et al., 2010), vibratome sections (40µm) were immunolabeled overnight with antibodies against SLC1A1 (Mouse monoclonal, 1:100, Abcam, (ab78395)), SLC6A6 (rabbit polyclonal, 1:100, Santa Cruz, (sc-135257)), and total α-syn (rabbit polyclonal, 1:100, Millipore, AB5038) followed by incubation with species-appropriate secondary antibodies (1:2000, Vector Laboratories). Sections were reacted with 3,3’-Diaminobenzidine and transferred to SuperFrost slides (Fisher Scientific, Tustin, CA) and mounted under glass coverslips with anti-fading media (Vector Laboratories). The samples were processed and analyzed blind coded and in duplicate to confirm the reproducibility of the results.
A total of 10 digital images from the frontal cortex (1024 × 1024 pixels) per section were captured per case using the Olympus BX54 bright-field digital microscope at 400X magnification. Two serial tissue sections of the brains (20 digital images) were processed and analyzed per human and mouse case. Immunostained sections were analyzed with the NIH Program Image J. Briefly, the digital color images were converted to gray scale followed by applying a threshold over a region of interest and optical density determination to each of files. Black, gray and white box were used as reference point to estimate maximum and minimum values and calibrate the system. The individual values for the 10 images per section were averaged and expressed as corrected optical density per human or mouse case.
To confirm the specificity of the results, the same samples from the human and mouse brains were incubated with incubated with alternative antibodies against SLC1A1 (rabbit polyclonal, 1:1000, Alomone (AGC-023)) and SLC6A6 (rabbit polyclonal, 1:1000, Millipore (AB5414P)) from a different source.
Double immunolabeling and confocal microscopy studies
To determine the co-localization between SLC1A1 and SLC6A6 with α-syn, neuronal and glial markers double-labeling experiments were performed. For this purpose, vibratome sections were double immunolabeled with the SLCA1 or 6 antibodies and rabbit polyclonal antibodies against α-syn (Millipore, affinity purified polyclonal, 1:500 (AB5038P)) or monoclonal antibodies against NeuN (Millipore, neuronal marker (MAB377)) or GFAP (Millipore, astroglial markers (MAB3402)). The SLC1A1 and SLC6A6 immunoreactive cells were detected with the Tyramide Signal Amplification™-Direct (Red) system (1:100, NEN Life Sciences, Boston, MA) while asynuclein, GFAP and NeuN was detected with FITC tagged antibodies (Vector, 1:75). All sections were processed simultaneously under the same conditions and experiments were performed twice in order to assess the reproducibility of results. Sections were imaged with a Zeiss 63X (N.A. 1.4) objective on an Axiovert 35 microscope (Zeiss) with an attached MRC1024 LSCM system (BioRad)
Statistical analysis
Statistical analysis of the OneArray system was conduced by and provided by the Phalanx Biotech Group, who compared the expression levels of the specific probes to control probes and provided comparison tables comparing miRNA profiles across the human disease groups compared to controls (Supp. table 5). Complete details are found online athttp://www.phalanxbiotech.com/technology/microRNA_performance.php. As indicated above, statistical analysis of the murine groups and the initial human MSA samples was conducted using the web-based RT² Profiler™ PCR Array Data Analysis Tool (http://pcrdataanalysis.sabiosciences.com/pcr/arrayanalysis.php) (Supp. Table 6). For the purposes of miRNA profiling, a 2-fold up or down regulation with p<0.05 was deemed significant
All other data are presented as mean± SEM unless otherwise stated.. Data was analyzed and the graphs were generated using the PRISM software package (GraphPad).
RESULTS
Comparison of miRNA profiles between MSA and the MBP-hαsyn tg models
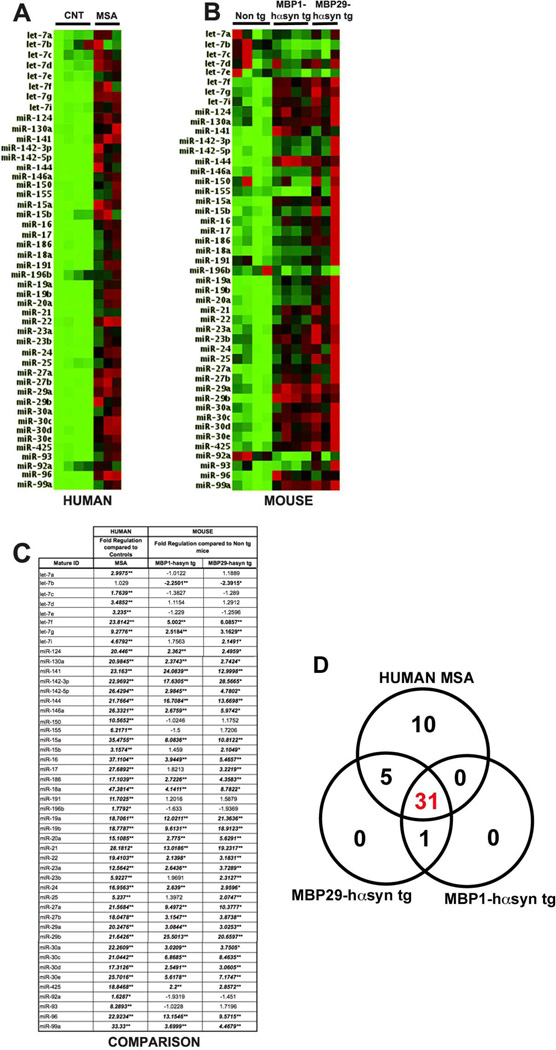
In order to examine miRNA profiles across species we isolated and compared miRNA profiles from the frontal cortex of human control and MSA samples and from transgenic mouse models of MSA. We compared the miRNA profiles from MSA cases (Figure 1A, C) with those in the MBP1hα-syn tg and MBP29-hαsyn tg mice (Figure 1B, C) using the miFinder RT² miRNA PCR Array system (SABiosciences). These assays are composed of the 88 most investigated miRNA and provide a good overview of miRNA dysregulation. Of the 88 miRNA present on the human plate, there was an overlap of 47 miRNA that were also present on the mouse plate, of these 47, 40 (85%) were similarly dysregulated in both humans and tg mice (Figure 1A–C). The greatest disparity was seen in members of the let-7 miRNA family, which appeared to be up regulated in the human cases compared to controls but down regulated or unchanged in the MSA tg mice compared to non tg mice (Figure 1C). Regressions analysis was conducted between the groups analyzed and demonstrated a slight but statistically significant correlation between the human MSA samples and the tg mouse models (human vs. MBP1-hasyn r2=0.2093, p=0.00122; human vs. MBP29-hαsyn r2=0.25197, p=0.00032) and strong correlation between the tg mouse models (r2=0.75081, p=3.5527×10(−15))
Figure 1. Comparison of miRNA profiles between MSA cases and transgenic models of MSA.
(A) miRNA profiles from control (n=4) and MSA (n=3) cases. (B) miRNA profiles from non tg (n=4), MBP1-hαsyn tg (n=4) and MBP29-hαsyn tg (n=3) mice. All samples were analyzed using the miFinder RT² miRNA PCR Array system (SABiosciences) in order to enable comparison across species. Of the 88 genes present on the human and mouse plates, 47 were identical, these 47 are depicted. The colors indicate the magnitude of fold change over the group average, green indicates down regulation while red indicates up regulation. (C) Comparative fold regulation of the 47 miRNA displayed. Changes that met our selection criteria of ± 2-fold regulation and p<0.05 are in bold and italicized * indicates p<0.05, ** indicates p<0.01 calculated based on a Student’s t-test of the replicate 2^ (- Delta Ct) values for each miRNA. (D) Venn Diagram showing overlapping miRNA dysregulation between the human MSA cases and the two tg mouse models. CNT = Control, MSA = Multiple System Atrophy
This comparison demonstrated that the tg mouse models of MSA recapitulate the altered miRNA signature observed in human cases, indicating that the MBP tg lines are a suitable model in which to investigate miRNA-related MSA mechanisms and pathways.
Comparative analysis of miRNA dysregulation in MSA and other neurodegenerative disorders in humans
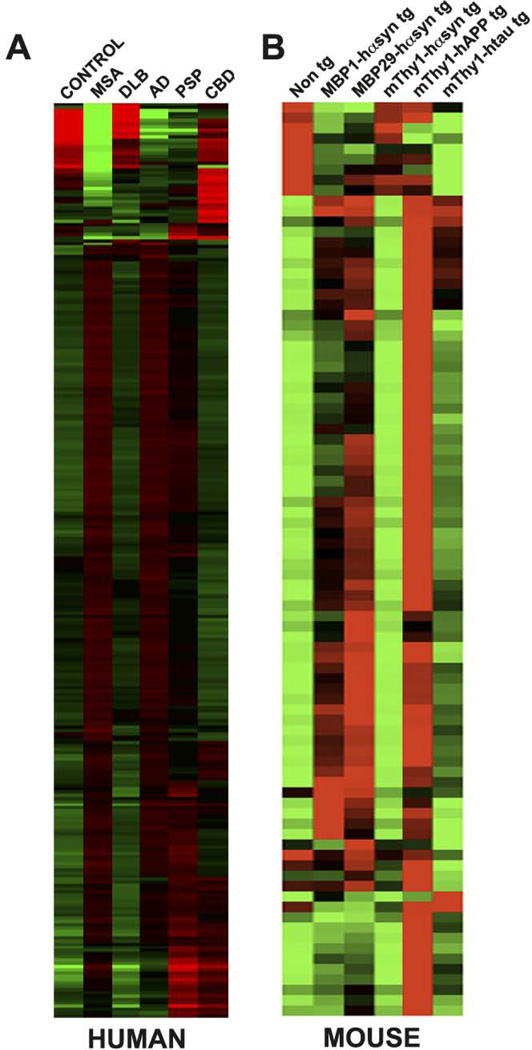
In order to examine the miRNA profiles across a variety of neurodegenerative disorders we isolated and examined miRNA from the frontal cortex of human samples of patients diagnosed with MSA, DLB, AD, PSP and CBD and from controls (Figure 2A). The rationale for comparing MSA with DLB is based on the shared characteristic α-syn accumulation observed in these disorders. AD, PSP and CBD were chosen as these disorders are associated with abnormal tau accumulation and alterations in tau aggregation have reported in MSA, albeit to a lesser extent than involved in frank tauopathies (Cairns et al., 1997; Nagaishi et al., 2011). Additionally, the clinical features of MSA, PSP and CBD overlap to some degree (Gilman et al., 1999; Kurata et al., 2011).
Figure 2. miRNA profiles in human cases and murine models of neurodegeneration.
(A) miRNA was isolated from the frontal cortex of MSA, DLB, AD, PSP, CBD cases and normal controls and analyzed using the OneArray® Human microRNA Microarray Phalanx Biotech Group. A total of 543 miRNA were analyzed and are presented as the mean miRNA levels in each group. (B) miRNA isolated from the frontal cortex of MBP1-hαsyn tg, MBP29-hαsyn tg, mThy1-hαsyn, mThy1-hAPP, mThy1-htau and non tg mice analyzed using the miFinder RT² miRNA PCR Array system (SABiosciences). A total of 88 miRNA were analyzed and are presented as the mean miRNA levels in each group. The colors indicate the magnitude of fold change over the group average, green indicates down regulation while red indicates up regulation. MSA = Multiple System Atrophy, DLB = Dementia with Lewy Bodies, AD = Alzheimer’s Disease, PSP = Progressive supranuclear palsy and CBD = Corticobasal degeneration.
Analyses of the miRNA profiles in MSA, DLB, AD, PSP and CBD identified a large number that were dysregulated in MSA, AD and PSP, while none of the miRNA examined in DLB or CBD fulfilled our criteria for statistical significance (2-fold up or down regulation with p<0.05). Notably all of the miRNA dysregulated in MSA, AD and PSP appeared to be up regulated in comparison to controls. Cross-group analysis also showed that many of these miRNA were altered in more that one disorder (Table 1, Supp. Table 5), suggesting that they may play a role in neurodegenerative pathways/processes that are common across disorders. A number of miRNA specific for each disorder were also identified. For MSA, of the 543 miRNA examined, 214 (39% of total) were up-regulated in comparison to control samples and of these 214 dysregulated miRNA, 83 (39% of total dysregulated) were specific to MSA (i.e. not altered in other disorders) − by definition this means 131 of the 214 miRNA dysregulated in MSA were also altered in other disorders. These results, and those for DLB, AD, PSP and CBD, compared to control samples are summarized in Table 1 and presented in full in Supp. Table 5.
Table 1.
Summary of miRNA dysregulation in human MSA, DLB, AD, PSP and CBD cases compared with controls
| MSA | DLB | AD | PSP | CBD | |
|---|---|---|---|---|---|
| Up-regulated | 214 | 0 | 125 | 49 | 0 |
| Down-regulated | 0 | 0 | 0 | 0 | 0 |
| Disease-specific | 83 | 0 | 2 | 9 | 0 |
miRNA that met selection criteria of ± 2-fold regulation and p<0.05 are included. MSA = Multiple System Atrophy, DLB = Dementia with Lewy Bodies, AD = Alzheimer’s Disease, PSP = Progressive supranuclear palsy and CBD = Corticobasal degeneration.
Hierarchical clustering of miRNA profiles across the human groups was consistent with the summary of miRNA dysregulation in human samples described in Table 1. MSA and AD cluster together as they display the greatest number of dysregulated miRNAs compared to controls, PSP samples exhibit a second level pairing with MSA and AD, while the apparent lack of statistically significant miRNA dysregulation in DLB and CBD causes these sets to group closer to control samples (Supp. Fig. 1)
Comparative analysis of miRNA dysregulation in the MBP-hαsyn tg mice and other tg models of neurodegeneration
In order to examine the miRNA profiles across a number of different tg mouse models of neurodegeneration we isolated and examined miRNA from the frontal cortex of tg models of MSA (MBP1- and MBP29-hαsyn tg, intermediate and high expression, respectively), DLB (mThy1-hαsyn), AD (mThy1-hAPP), and a model of tauopathy (PSP/CBD) (mThy1-htau) and from non tg mice (Figure 2B).
Analogous to the results in the human samples, we identified a large number of miRNA that were dysregulated in the MBP1- and MBP29-hαsyn tg mice, the mThy1-hAPPtg mice and the mThy1-htau, while no miRNA examined in the mThy1-hαsyn tg mice fulfilled our criteria for statistical significance. Similar to the results in the humans, cross-group analysis also showed that many miRNA were altered in more that one mouse model (Table 2, Supp. Table 6) again suggesting a role in disease-common neurodegenerative pathways/processes, however a number of miRNA specific for each mouse model were also identified.
Table 2.
Summary of miRNA dysregulation in MBP1-hasyn tg, mThy1-hαsyn, mThy1-hAPP and mThy1-htau compared with non-tg mice
| MBP1-hαsyn tg |
mThy1-αsyn tg |
mThy1-hAPP tg |
mThy1-htau tg |
|
|---|---|---|---|---|
| Up-regulated | 57 | 0 | 62 | 39 |
| Down-regulated | 1 | 0 | 0 | 5 |
| Disease-specific | 5 | 0 | 3 | 5 |
miRNA that met selection criteria of ± 2-fold regulation and p<0.05 are included.
For the MBP1hα-syn tg mice 57 of the 88 miRNA examined, (65% of total) were up-regulated in comparison to non tg samples, 1 (1.1% of total) was down-regulated and of these 58 dysregulated miRNA, 5 (8.6% of total dysregulated) (let-7b, miR-141, miR-182, miR-183 and miR-96) were specific to the MBP1hα-syn tg and MBP29hα-syn tg mice. Of the 58 miRNA dysregulated in the MBP1hα-syn tg, 55, (95%) were similarly altered in the MBP29-hαsyn tg mice (a higher expressor line), while an additional 5 (miR-15b, miR-17, miR-23b, miR-31 and miR-154) were only dysregulated in the MBP29hα-syn tg mice. These results, and those for the other tg mouse lines are summarized in Table 2 and presented in full in Supp. Table 6.
Preserved miRNA machinery in MSA cases and transgenic mice
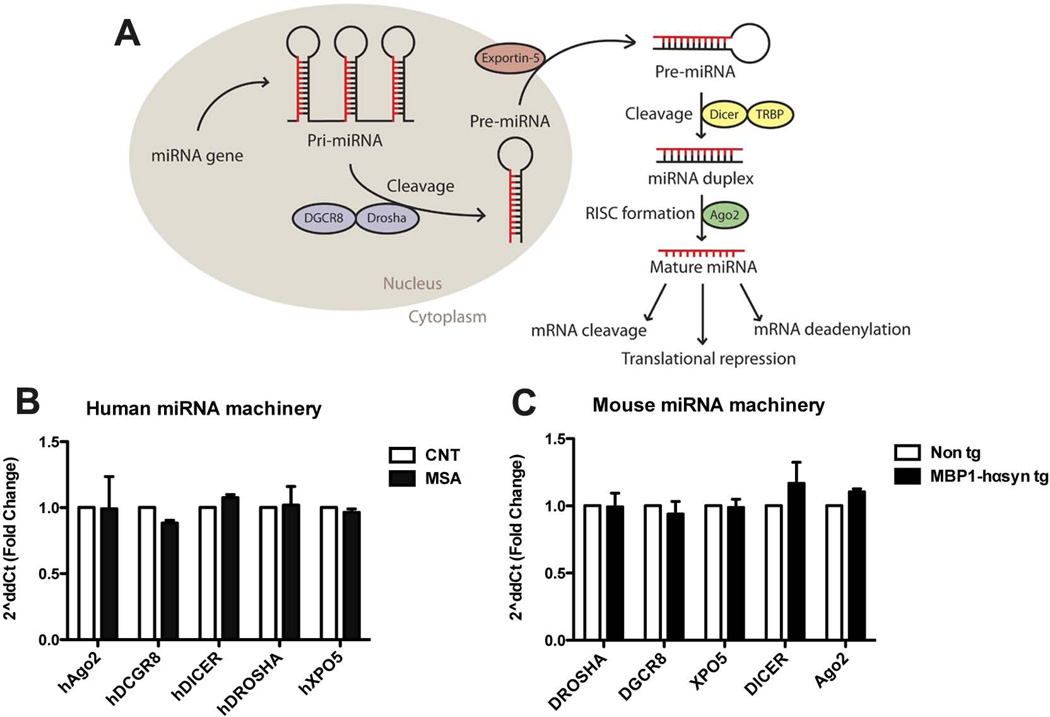
Having established that there is a widespread dysregulation in miRNA levels in the MSA cases and transgenic mice, we next examined components involved in miRNA processing in order to determine whether the miRNA dysregulation observed in these samples was due to a generalized alteration in miRNA processing.
miRNA processing is a multistage process that converts pre-miRNA into the functional mature forms and involves and number of different components (Figure 3A). Given the extensive miRNA dysregulation we observed in the MSA cases and MBP1hα-syn tg we sought to examine the levels of key components in the miRNA processing pathway as an alteration in miRNA processing machinery could lead to the observed alterations in miRNA expression profiles.
Figure 3. Preserved miRNA processing machinery in MSA cases and MBP1-hαsyn tg mice.
(A) Schematic of the miRNA processing pathway. In order to examine whether any components of the miRNA processing machinery were altered in the (B) MSA cases or (C) the MBP1-hαsyn tg mice, mRNA levels of Ago2, DGCR8, Dicer1, Drosha and Exportin5 (XPO5) were analyzed by qPCR in comparision to normal controls and non tg mice, respectively. MSA = Multiple System Atrophy. Error bars represent mean ± SEM.
In order to determine whether the altered miRNA expression profiles was due to a generalized alteration in components of the miRNA processing pathway, mRNA levels of Drosha, DGCR8, Exportin5, Dicer1 and Ago2 were analyzed by RT-qPCR in the human MSA samples compared to control samples (Figure 3B) and the MBP1-hαsyn tg mice in comparison to non tg mice (Figure 3C). There were no significant alterations at the transcriptional level of any of the miRNA processing components across any of the groups examined, indicating that the altered miRNA profiles observed in the human MSA samples and the MBP1-hαsyn tg mice are not due to a generalized deficit in the processing system but rather may reflect disease-related miRNA dysregulation.
Disease-specific miR-96 dysregulation in MSA
Although out initial comparison of miRNA profiles demonstrated that a number of miRNA were specifically dysregulated in the MSA cases (Table 1, Supp. Table 5) and the MBP-hαsyn tg mice (Table 2, Supp. Table 6), many of these were either specific for the MBP1-hαsyn tg mice versus other tg mice in comparison to controls or were specific for human MSA cases versus other disorders in comparison to controls. However, further detailed examination of miRNA that were disease-specific for MSA identified the miR-96 cluster (miR-96, mir-182 and mir-183) as being specifically dysregulated in both the human cases and the MBP1-hαsyn tg mice, additionally the miR-96 paralog miR-1271 (Jensen & Covault, 2011) was specifically dysregulated in the MSA cases in comparison to controls (Supp Tables 5, 6). Although let-7b and miR-141 were specifically dysregulated in the MBP1-hαsyn tg mice, they were not significantly altered in the MSA cases, however it was interesting to note that miR-141* (the miRNA precursor of miR-141) was significantly altered in the MSA cases (Supp Table 5, Table 6).
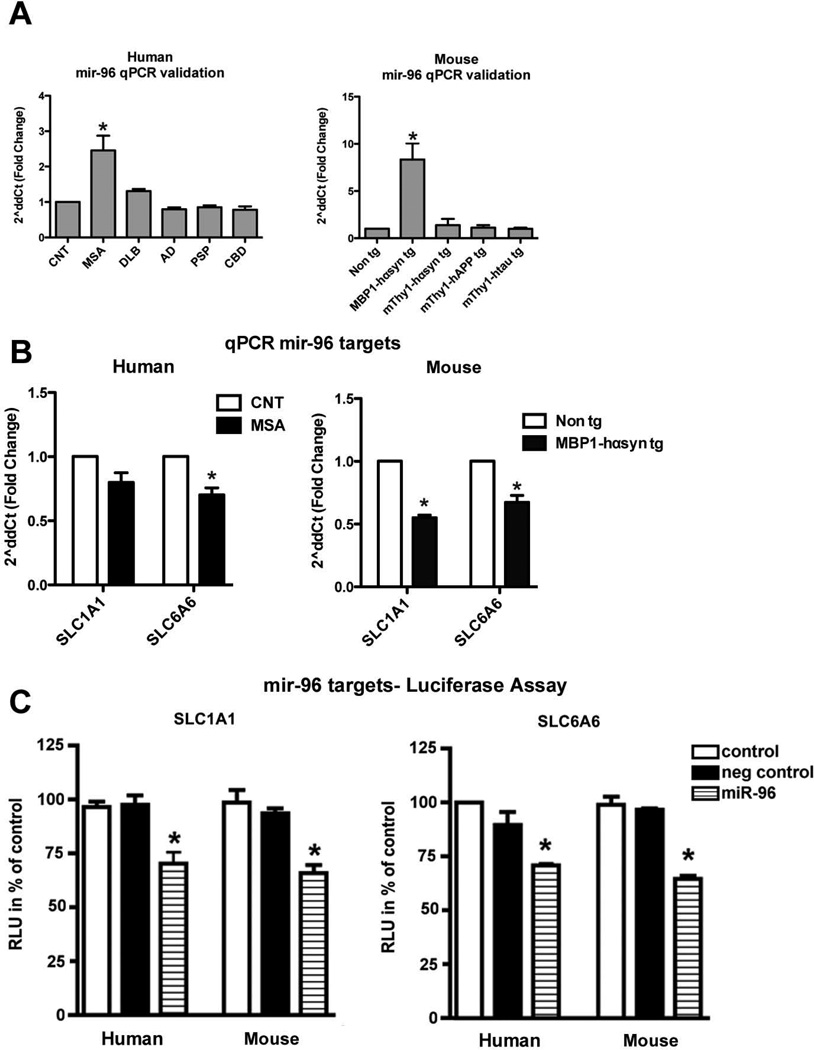
The dysregulation of miR-96 across the human (Figure 4A) and mouse (Figure 4B) samples was validated by RT-qPCR in independent murine samples and the human cases and demonstrated an 8-fold increase in the MBP1-hαsyn tg mice over non tg levels and other tg models of disease and a 2.5-fold increase in human MSA samples over levels in the control samples and other disorders.
Figure 4. Specific miR-96 dysregulation in MSA cases and MBP1-hαsyn tg mice.
(A) qPCR validation of miR-96 levels in human MSA, DLB, AD, PSP and CBD cases compared to normal controls and of miR-96 levels in MBP1-hαsyn tg, mThy1-hαsyn, mThy1-hAPP and mThy1-htau compared to non tg mice. MSA = Multiple System Atrophy, DLB = Dementia with Lewy Bodies, AD = Alzheimer’s Disease, PSP = Progressive supranuclear palsy and CBD = Corticobasal degeneration. Error bars represent mean ± SEM. * Indicates a significant difference (p< 0.05) between the MSA cases and other disease groups or a significant difference (p< 0.05) between the MBP1-hαsyn tg and other mouse groups analyzed by one-way ANOVA and Dunnett’s post hoc test. (B) qPCR validation of SLC1A1 and SLC6A6 in human MSA cases compared to controls and MBP1-hαsyn tg compared to non tg mice. (C) The effect of miRNA on gene regulation was monitored by co-transfecting 300ng human and mouse mir-96 and the luciferase reporter gene. The 3′-UTR of SLC1A1 and SLC6A6 were cloned downstream of the luciferase open reading frame. In addition to human and mouse mir-96 that could bind to the corresponding UTR, negative control (scrambled human and mouse mir-96) was transfected with the corresponding luciferase reporter plasmid for each gene analyzed. The luciferase activity was measured 24 hrs after transfection is expressed as percentage of control ± standard deviation shown. * Indicates statistically significant (p-value of <0.05) difference from control miRNA calculated using one-way ANOVA.
miR-96 mRNA target identification and characterization; members of the SLC-protein family are targets of miR-96 and are dysregulated in MSA
A single miRNA is predicted to potentially regulate over a thousand target genes; the identification of these target genes is based on degree of sequence complementarity between the seeding sequence of the miRNA and the mRNA sequence of the target gene (usually in the 3`UTR of mammalian genes). There a number of different online databases which predict mouse and human miRNA target genes using different algorithms, each identifying numerous hits, which are then ranked according to their likelihood as bona fide as, targets. We used three different online databases (Targetscan (Lewis et al., 2005; Grimson et al., 2007), Pictar (Krek et al., 2005) and Microcosm(Rehmsmeier et al., 2004)) to identify the top 50 predicted human and murine targets of miR-96. This analysis demonstrated that a number of target families occur frequently across the human and murine predicted target lists (Table 3).
Table 3.
High-frequency mRNA targets of miR-96
| Target | Human databases | Mouse databases |
|---|---|---|
| SLC family | 9 | 9 |
| FOXO family | 5 | 5 |
| SOX family | 3 | 3 |
| FYN | 1 | 2 |
| DCX | 0 | 1 |
| MSN | 1 | 1 |
| GRK6 | 2 | 0 |
| NLGN2 | 2 | 2 |
Predicted human and mouse mir-96 targets were members from particular gene families and a number appeared frequently across the databases and across species, these are summarized here.
Members of the solute carrier protein (SLC) family were the most frequently occurring family with members of the FOXO and SOX family also heavily represented. In addition to being particularly over-represented as targets of miR-96 (Table 5), the SLC proteins were highly ranked in Targetscan as targets of mmu-miR-182, mmu-miR-183 and hsa-miR-1271 (the human paralog of miR-96), (data not shown).
The SLC group of membrane transport proteins include over 300 members organized into 51 families typically located at the membrane of the cell or intracellular organelles which act as transporters allowing the transport of a variety of solutes through the membrane (Hediger et al., 2004; Ren et al., 2007; Featherstone, 2011). Of the SLC members predicted to be targets of miR-96, the neuronal/epithelial high affinity glutamate transporter, SLC1A1 (Kanai & Hediger, 2004a) and the taurine transporter, SLC6A6 (Anderson et al., 2009; Kristensen et al., 2011) appear on both the murine and human tables and may represent miR-96 target genes that are conserved across species.
Quantitative RT-PCR was performed in the MBP1-hαsyn tg mice and MSA cases (Figure 4B) in order to examine mRNA levels of SLC1A1 and SLC6A6 in these groups. The results demonstrate a modest but significant reduction in both mRNA transcripts in the MBP1-hαsyn tg mice compared to non tg controls (Figure 4B) and in SLC6A6 in the MSA cases compared to controls (Figure 4B), with a trend towards a decrease (p=0.08) in SLC1A1, suggesting that these are bona fide targets of miR-96.
The SLC1A1 and SLC6A6 targets were validated using a Luciferase reporter assay. HEK293 cells were co-transfected with human or mouse mir-96 and a luciferase reporter construct consisting of the 3′-UTR of human or mouse SLC1A1 and SLC6A6 cloned downstream of the luciferase open reading frame. There was a statistically significant decrease in luciferase activity when the 3′-UTR of human or mouse SLC1A1 and SLC6A6 was transfected with human or mouse mir-96 (Figure 4C), while transfection with the negative control (scrambled human or mouse mir-96) had no effect on luciferase activity.
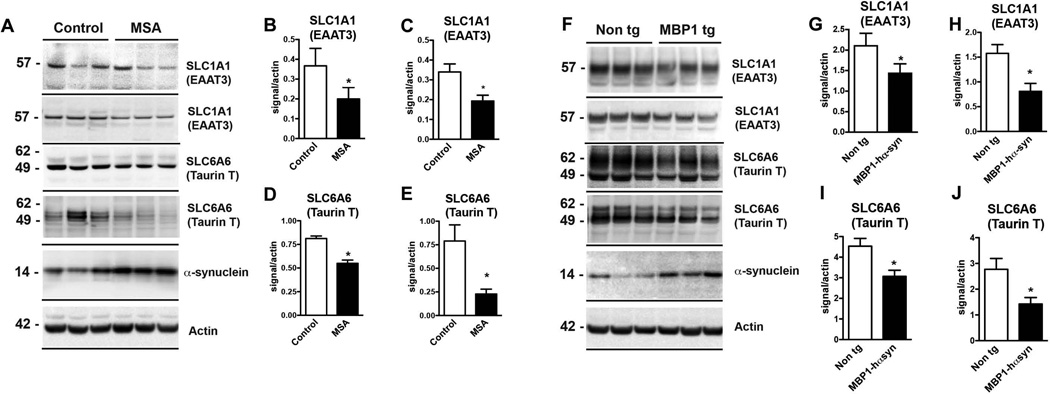
Immunoblot analysis of SLC1A1 and SLC6A6 protein levels demonstrated a significant decrease in SLC 1A1 and SLC6A6 in the homogenates from the frontal cortex of MSA cases compared to control (Figure 5A–E)) and dramatic decrease in the levels of both SLC proteins in the MBP1hα-syn tg mice compared to non tg controls (Figure 5F–J).
Figure 5. Reduced mRNA and protein levels of SLC-family proteins in MSA cases and MBP1-hαsyn tg mice.
(A, F) Representative immunoblots of SLC1A1 and SLC6A6 and α-syn protein levels in human MSA cases compared to controls and MBP1-hαsyn tg compared to non tg mice, respectively. (B) Analysis of SLC1A1 protein levels using the Abcam antibody (top bar in (A)). (C) Analysis of SLC1A1 protein levels using the Alomone antibody (second bar in (A)). (D) Analysis of SLC6A6 protein levels using the Santa Cruz antibody (third bar in (A)). (E) Analysis of SLC6A6 protein levels using the Millipore antibody (fourth bar in (A)). (G) Analysis of SLC1A1 protein levels using the Abcam antibody (top bar in (F)). (H) Analysis of SLC1A1 protein levels using the Alomone antibody (second bar in (F)). (I) Analysis of SLC6A6 protein levels using the Santa Cruz antibody (third bar in (F)). (J) Analysis of SLC6A6 protein levels using the Millipore antibody (fourth bar in (F)).MSA = Multiple System Atrophy. Error bars represent mean ± SEM. (C–D) * Indicates a significant difference (p< 0.05) between the MBP1-hαsyn tg and non tg mice. (F–H) * Indicates a significant difference (p< 0.05) between the MSA and control samples, analyzed by one-way ANOVA and Dunnett’s post hoc test.
As expected both MSA cases and the MBP1-hαsyn tg mice displayed a robust increase in levels of α-syn in comparison to controls and non tg mice, respectively (Figure 5A,F, respectively). In order to confirm the immunoblot results, two different commercial antibodies against SLCA1A and SLC6A6 were used for the analyses, both yielded comparable results. Immunoblot analysis of SLC6A6 was performed using the 49kDa band, bands were also observed at around 60 and 107kDa (Supp. Fig. 2 showing a full blot with the Santa Cruz antibody, comparable results were observed with the Millipore antibody).
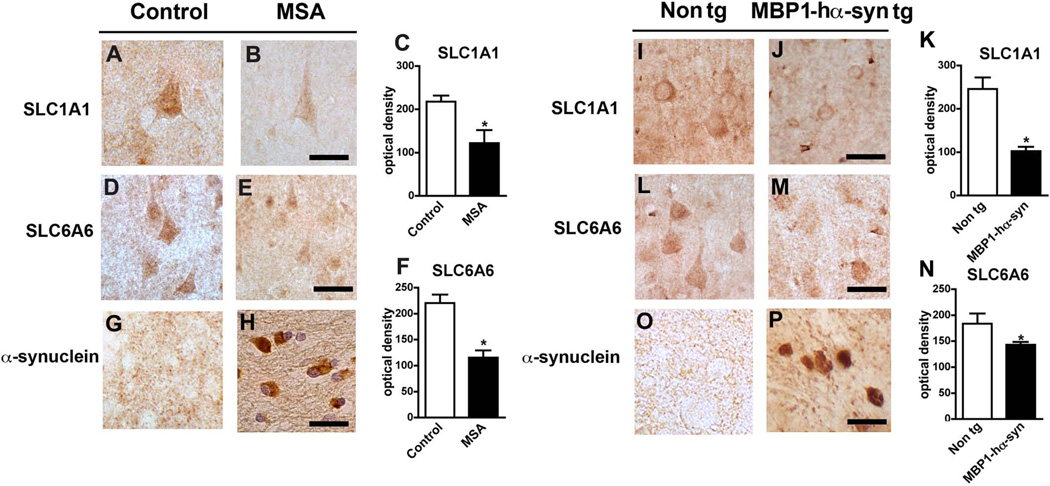
Consistent with the immunoblot results, immunohistochemical analysis demonstrated a robust decrease in SLC6A6 and SLC6A6 levels in the frontal cortex of human MSA cases in comparison to normal controls (Figure 6A–F). Levels of both SLC proteins were also significantly decreased in the frontal cortex of MBP1-hαsyn tg mice in comparison to non tg mice (Figure 6I–N). Again, as anticipated both MSA cases and the MBP1-hαsyn tg mice displayed a robust increase in levels of α-syn in the frontal cortex in comparison to controls and non tg mice, respectively (Figure 6G, H, O, P). In order to confirm the immunohistochemical results, two different commercial antibodies against SLCA1A and SLC6A6 were used for the analyses, both yielded comparable results (data from additional antibody not shown).
Figure 6. Immunohistochemical analysis of SLC-family proteins in MSA cases and MBP1-hαsyn tg mice.
(A, B) Representative image of SLC1A1 levels in control and MSA samples, respectively. (C) Analysis of SLC1A1 levels in control and MSA samples. (D, E) Representative image of SLC6A6 levels in control and MSA samples, respectively. (F) Analysis of SLC1A1 levels in control and MSA samples. (G, H) Representative image of α-syn levels in control and MSA samples, respectively. (I, J) Representative image of SLC1A1 levels in non tg and MBP1-hαsyn tg mice, respectively. (K) Analysis of SLC1A1 levels in non tg and MBP1-hαsyn tg mice. (L, M) Representative image of SLC6A6 levels in non tg and MBP1-hαsyn tg mice, respectively. (N) Analysis of SLC1A6 levels in non tg and MBP1-hαsyn tg mice. (O, P) Representative image of α-syn levels in non tg and MBP1-hαsyn tg mice, respectively. All images are from the frontal cortex. MSA = Multiple System Atrophy. Scale bar = 25µM. Error bars represent mean ± SEM. In (C, F) * indicates a significant difference (p< 0.05) between the MSA and control samples and in (K, N) * indicates a significant difference (p< 0.05) between the MBP1-hαsyn tg and non tg mice, analyzed by one-way ANOVA and Dunnett’s post hoc test.
Collectively these results demonstrate that the up regulation of miR-96 observed in the MSA cases and MBP1-hαsyn tg mice leads to the downstream transcriptional repression of target genes and the subsequent reduction of these targets at the protein level.
SLC proteins co-localize predominantly with neurons
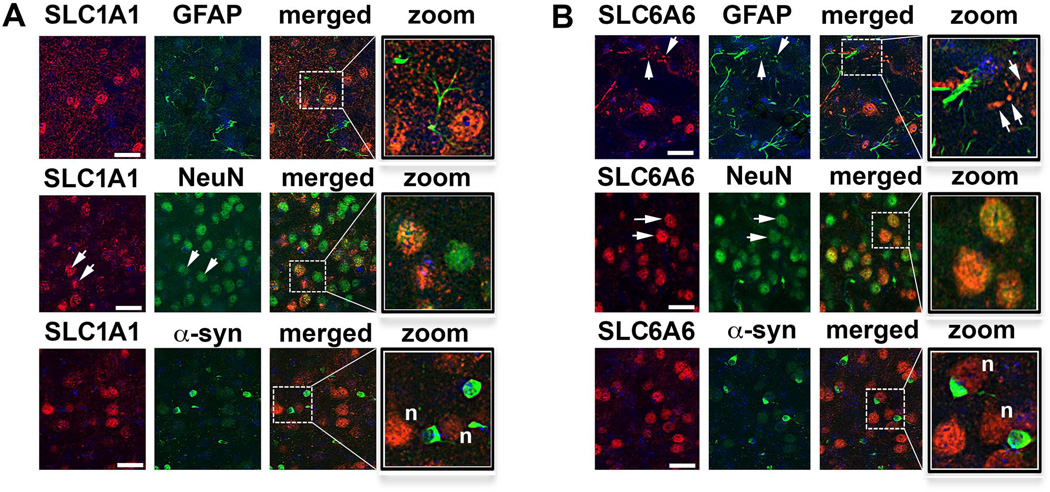
In order to further characterize the location of the SLC1A1 and SLC6A6 expression in the cortex of MBP1-hαsyn tg mice we conducted double-labeling studies with the astroglial marker GFAP and the neuronal marker NeuN. Consistent with previous studies, we demonstrate abundant SLC1A1 expression in the brain, including cortical neurons (Torp et al., 1997; Utsumi et al., 2001; Otis et al., 2004) (Fig. 7A, arrows) and neuronal SLC6A6 expression (Smith et al., 1992; Warskulat et al., 2007) (Fig. 7B, arrows). Furthermore we show α-syn positive oligodendrocytes in close proximity to the SLC1A1 annSLC6A6 positive neurons and glia (Fig. 7A, B, indicated by ‘n’).
Figure 7. Confocal co-localization analysis of SLC1A1 and SLC6A6 in the frontal cortex of MBP1-hαsyn tg mice.
(A) Colocalization of the SLC1A1 signal with astroglial (GFAP) and neuronal (NeuN) cell markers and with α-syn-positive oligodendrocytes. (B) Colocalization of the SLC6A6 signal with astroglial (GFAP) and neuronal (NeuN) cell markers and with α-syn-positive oligodendrocytes. Scale bar = 30µM. Arrows indicate cells in which the proteins of interest colocalized, ‘n’ indicates neighboring cells that are positive for α-syn and SLC1A1 or SLC6A6.
DISCUSSION
The present study sought to investigate miRNA dysregulation in both humans and tg models of neurodegeneration with the eventual aim of identifying disease-specific miRNA profiles in MSA. We showed that in MSA, AD and PSP cases and in the corresponding tg models there was widespread up regulation of miRNA, furthermore we identified an alteration in miR-96 in MSA.
Previous studies into miRNA dysregulation in neurodegeneration have typically compared a single disease state with control (Asikainen et al., 2010; Schonrock et al., 2010; Wang et al., 2010; Delay & Hebert, 2011; Harraz et al., 2011; Lee et al., 2011; Li et al., 2011; Margis & Rieder, 2011; Satoh, 2011), and while this is informative of the dysregulation of a particular miRNA in that disease it does not allow the evaluation of the role of that miRNA in neurodegeneration per se. We demonstrate a widespread dysregulation of miRNA across neurodegenerative conditions in both the humans and mice, and while a number of these were commonly dysregulated across a number of diseases and tg models, a particular subset were found to be specific for MSA and some of the other disorders examined.
The importance of a cross-disease approach to miRNA dysregulation is exemplified by the comparison of the results from this study with previous studies in AD that have reported up-regulated levels of miR-146a in tg mouse models of AD (Li et al., 2011; Wang et al., 2012a). Consistent with these results we also see a 4-fold up-regulation of miR-146a in our mThy1-hAPP tg mice, however we also observe murine miR-146a dysregulation in mouse models of MSA, DLB and tauopathy. Moreover, in the humans while miR-146a is up regulated in human AD samples (although it did not meet our stringent criteria for significance (±2-fold regulation and p<0.05)), a similar level of up-regulation was observed in MSA and CBD. Furthermore, levels of miR-146a* (the precursor of miR-146a) were significantly up regulated in MSA and PSP. Similarly, a recent study reported that levels of miR-34c are elevated in the hippocampus of AD patients and tg mouse AD models (Zovoilis et al., 2011), consistent with this study we also observe an up-regulation of miR-34c in our mThy1-hAPP tg model of AD, however we see a similar degree of up-regulation in the MBP and mThy1-htau tg models. miR-146a and miR-34c have been reported to target genes involved in immune and inflammatory signaling (Wang et al., 2012a; Wang et al., 2012b) and cell cycle progression, cellular senescence and apoptosis via SIRT1 and p53 (Yamakuchi & Lowenstein, 2009) respectively, these are all potential pathways that may have a relevance across a number of neurodegenerative conditions.
Of the 88 miRNA examined in the mice, 58 (66%) were found to occur in more than one disease model, of these disease-common miRNA, a significant overlap was seen in the mThy1-hAPP and mThy1-htau tg models (40 of the 88 miRNA examined (45%) were commonly dysregulated in these two models). Given the neuropathological overlap of these models (both display alterations in tau (Rockenstein et al., 2001) (Manfred Windisch, personal communication), is not unconceivable that should any miRNA play a role in tau-related neuropathology, it would be similarly altered in both models. It is noteworthy that in the humans 125 of the miRNA dysregulated in AD, 32 (25%) are also dysregulated in PSP.
miRNA have also been investigated in PD and a number of miRNA have been reported to be either involved in either α-syn repression (miR-7 and miR-153) (Junn et al., 2009; Doxakis, 2010), expressed in cells that are specifically lost in PD (miR-133b regulated maturation and function of midbrain dopaminergic neurons) (Kim et al., 2007) or associated with polymorphisms linked to PD risk (Wang et al., 2008; de Mena et al., 2010). While we observed no alterations in miR-7 or miR-153, miR-133b was increased in the MSA and AD cases. Interestingly in addition to miRNA that regulate α-syn, key PD related proteins have themselves been reported to alter miRNA. For example LRRK2, which when mutated causes PD type 8 (PARK8) (Seol, 2010), has been reported to interact with miR-184* and let-7 in drosophila (Gehrke et al., 2010). These miRNA are thought to regulate protein synthesis and levels of dp and e2f1 (targets of miR-184* and let-7, respectively) are reportedly critical for LRRK2 pathogenesis in the fly (Gehrke et al., 2010).
It was surprising that, despite some dysregulation in the mThy1-hαsyn tg mice compared to controls, none of the miRNA we examined reached significance according to our criteria. It likely that these results are a function of the assay used to analyze the mouse samples; the miFinder RT² miRNA PCR Array system (SABiosciences) had 88 of the ‘most well-documented’ miRNA, however most of these were cancer-related with a few that reported to be altered in AD, which may account for the relative abundance of miRNA observed to be dysregulated in the models of AD and tauopathy and the apparent dearth of miRNA dysregulation in the mThy1-hαsyn tg mice. However it is noteworthy that the analysis of human DLB cases also did not identify any miRNA that fulfilled our significance criteria, these results are consistent with a recent study of small RNAs from the cerebral neocortex of subjects with various forms of dementia (Hebert et al., 2013). However, a number of miRNA reportedly dysregulated in α-syn (A30P) tg mice (miR-10a, miR-10b and mir-32) (Gillardon et al., 2008) were also dysregulated in the MBP-hαyn tg mice (both MBP1-and MBP29-hαsyn), although they were also altered in the mThy1-hAPP tg mice – again highlighting the value of a cross-disease comparison.
In addition to identifying disease-common miRNA, this study identified miRNA that were specific for each disorder/model. Identification of such disease-specific miRNA is crucial in elucidating potential mechanisms that may be particular to a given disorder and which provide clues as to novel aspects of disease progression and pathology. Another key aspect of miRNA research is the extent to which miRNA profiles in tg animal models of a particular neurodegenerative disease replicate the miRNA profiles of human patients with that disorder. Given that miRNA-based therapies are a major area of academic and commercial focus (Roshan et al., 2009; Gambari et al., 2011; Junn & Mouradian, 2011; Kasinski & Slack, 2011), this knowledge will be invaluable when trying to distinguish disease-related miRNA and develop potential miRNA-based therapeutics.
We were particularly interested in MSA and the degree to which miRNA dysregulation in humans was recapitulated in theMBP1-hαsyn tg mice. We identified 5 miRNA that were specific for the MBP1- and MBP29-hαsyn tg mice (let-7b, miR-141, miR-182. miR-183 and miR-96), 3 of these 5 (miR-182, miR-183 and miR-96) were members of the miR-96 cluster (all within 10kb of each other, perhaps indicating a stretch of sequence specifically perturbed in MSA) and given that miR-96, and its paralog miR-1271 were also dysregulated in humans, we chose to focus on miR-96 for this study. Given a recent report that miR-1271 is more abundant in the human brain than miR-96 (Jensen & Covault, 2011), it is important that it is also overexpressed in MSA. A number of studies reported a role for miR-96 in cancers including those effecting the liver (Ladeiro et al., 2008; Chen et al., 2011), lungs (Ma et al., 2011; Nymark et al., 2011; Zhu et al., 2011), bladder (Han et al., 2011; Yamada et al., 2011; Wang et al., 2012b) and prostate (Navon et al., 2009; Schaefer et al., 2010a; Schaefer et al., 2010b) and it has also been implicated in diabetic retinopathy and retinal degeneration (Loscher et al., 2007; Xu et al., 2007; Loscher et al., 2008; Wu et al., 2011) and zinc homeostasis (Mihelich et al., 2011), however its role in neurodegeneration remain unexamined.
Having chosen to focus on miR-96, we sought to identify its mRNA targets. Traditionally this is done using databases that employ algorithms to predict targets based in the degree of complementarity between the seed sequence of the miRNA and the 3`UTR of putative targets and then rack the results from the strongest target, with the greatest degree of homology, to less likely targets. As different databases tend to use different predictive algorithms (Sethupathy et al., 2006; Xu, 2007), we chose to use three different databases, Targetscan (Lewis et al., 2005; Grimson et al., 2007), Pictar (Krek et al., 2005) and Microcosm(Rehmsmeier et al., 2004) to draw up a list of putative targets and to focus our analysis on the top 50 targets predicted by these database in both humans and mice. This cross-database, cross- species type of analysis provides strong leads to targets that are not only highly likely to be bona fide miR-96 targets but also identifies those that may be conserved from mouse to human. Using this approach we identified a number of interesting putative targets including FOXO, SOX, Fyn, and neuregulin, which have been previously implicated in neurodegeneration (Chin et al., 2005; Go et al., 2005; Kanao et al., 2010; Manolopoulos et al., 2010; Monsalve & Olmos, 2011; Roberson et al., 2011; Scales et al., 2011), furthermore we identified the SLC family of proteins as being highly over-represented as targets of miR-96, across the databases and in both humans and mice. Importantly SLC proteins are also highly ranked targets of miR-182, miR-183 and miR- 1271 underlining their potential importance as downstream targets of miR-96 dysregulation.
The SLC group of membrane transport proteins include over 300 members organized into 51 families based on a greater than 20–25% sequence homology (Hediger et al., 2004; Featherstone, 2011) to each other and are involved in the transport of a diverse group of solutes. Most members of the SLC group are located on the cell membrane but some members are located on intracellular organelles such as the mitochondria (Maynard et al., 2008; Iacobazzi et al., 2009; Morciano et al., 2009). Members of this family have been identified in other studies as being PD-relevant (Santosh et al., 2009). Of the SLCs predicted to be targets of miR-96, SLC1A1 and SLC6A6 were common in the human and mouse predictions.
Of these targets, we were particularly interested in SLC1A1 based on a number of pieces of evidence; firstly polymorphisms in family members of this protein have been associated with MSA (Soma et al., 2008) and have been linked to the hypothesis that oxidative stress is associated with the pathogenesis of MSA (Soma et al., 2008). We observed a decrease in SLC1A1 mRNA in both the humans and the mice, however the greater decrease in murine SLC1A1 mRNA compared to human is likely due to the greater fold increase in miR-96 observed in the MBP1-hαsyn tg mice compared to the humans cases (8-fold increase in the tg mice compared to the 2.5-fold increase in humans).
SLC1A1 codes for the neuronal/epithelial high affinity glutamate transporter (EAAT3/EAAC1) that is widely expressed in the hippocampus, basal ganglia and cerebellum (Beal et al., 2005) and selective loss of this transporter has been implicated in olivopontocerebellar atrophy (Dirson et al., 2002), the cerebellar form of MSA. EAAT3/EAAC1 uptake of cysteine provides substrate for neuronal glutathione synthesis (Aoyama et al., 2012) which plays a key role in both antioxidant defenses and intracellular zinc binding (Won et al., 2010) and a recent report has suggested that cysteine uptake by EAAC1 is important for zinc homeostasis and neuronal antioxidant function (Won et al., 2010). The link between EAAT3/EAAC1 and zinc levels is particularly relevant to α-synucleinopathies such as MSA, given that the 5' promoter region of α-syn contains two zinc-finger domains that regulate α-syn transcription (Clough et al., 2009), dysregulation of zinc may modulate α-syn transcription. Additionally, the neuronal double zinc finger protein (ZNF231) is enhanced in MSA (Hashida et al., 1998) and elevation of ZNF231 expression has been hypothesized to be involved in the pathogenesis of MSA (Hashida et al., 1998). Furthermore, EAAT3/EAAC1 have been implicated in excitotoxicity (Kanai & Hediger, 2004b) and may play a role in the pathogenesis of MSA (Perez-Navarro et al., 1999)
SLC6A6 is a taurine transporter(Chen et al., 2004; Tomi et al., 2008; Anderson et al., 2009; Kristensen et al., 2011) and taurine has been reported to have neuroprotective activity this targets, in addition to SLC1A1, is an interesting candidate for further, more detailed, studies aimed at elucidating potential mechanism that may linked alterations in these target with the specific pathogenesis of MSA. It is important to note the while we chose to focus on the SLC family; other interesting genes have been reported to be associated with this family, most notably Nuclear respiratory factor 1 (nrf1). The nrf1 gene encodes a transcription factor which activates the expression of genes involved in the regulation of cellular growth and nuclear genes required for respiration. Additionally nrf1 has been implicated in the transcription and replication of mitochondrial DNA and in neurite outgrowth (Goffart & Wiesner, 2003; Jaiswal, 2004; Pierce et al., 2008; Biswas & Chan, 2010) . In the present study, levels of SLC1AA and SLC6A6 were examined in models of neurodegeneration at an age that may represent late-stage pathology; therefore it is important to note that changes in these proteins may reflect mechanism related to a general degenerative process such as excitotoxicity in these mice. Further studies are necessary in order to elucidate the relationship between the expression of SLC1AA and SLC6A6 and cellular changes related to degeneration.
The colocalization studies were particularly revealing as they show SLC1A1 and SLC6A6 expression in neuronal and glial cells neighboring the α-syn-positive oligodendrocytes. While the full implications of these cellular expression profiles remains to be fully defined, it may suggest a possible feed back mechanism whereby miRNA dysregulation in one cell type may affect protein expression in neighboring cells. One hypothesis may be that this communication is mediated by miRNA circulating in exosomes (Hannafon & Ding, 2013; Turchinovich et al., 2013), a mechanism that has recently been reported to be involved in oligodendrocyte-neuronal communication (Fruhbeis et al., 2013).
While we chose to pursue a cross-disease and cross-species approach for our study, it is worth noting that a number of other parameters could have justifiably been chosen. For example we chose to focus on a single-region and single time point in our study and chose a time point at which all tg mice models were exhibiting the pathological signs of the disease, however examination of other time points (presymptomatic, early-stage or advanced pathological), or a comparison across time points, could also have been conducted and would have likely provided equally interesting comparisons. For example, previous studies have shown that neural miRNAs are involved at various stages of synaptic development, formation and maturation (Martino et al., 2009; Gao, 2010; Smith et al., 2010; Siegel et al., 2011). Obviously the time points of human tissue are not as easy to dictate however it can be supposed that these would also provide informative comparisons. We also chose to examine miRNA profiles from miRNA isolated from the frontal cortex of our mice, however recent studies have reported that miRNA profiles change across regions (Juhila et al., 2011; Pichardo-Casas et al., 2012). The authors recognize that the present study is underpowered as it utilized a small number of MSA samples. Further studies, with a larger sample size are necessary in order to clarify the role of miR-96 and its targets in MSA. Future studies would benefit from a non-biased assessment of miR-96 targets and the inclusion of microarray experiments to simultaneously analyze transcriptome-wide expression changes between the models and controls. Strategies to determine global microRNA targets using miR-96 knockdown and mimics followed by microarray analysis allowing comparison with in silico findings would also prove instructive.Although a cross-disease/species/time point and brain region approach would be the gold standard for miRNA research in neurodegeneration, it is clearly beyond the scope of a single study and such comparisons are likely to be the result of multi-center approaches to miRNA investigation.
CONCLUSIONS
The results from this study have important implications in understanding disease-related pathways and processes and the identification of target miRNA, modulation of which may have a therapeutic indications via the regulation of target gene expression profiles in neurodegenerative conditions. The findings of MSA-related changes in miR-96 suggest that miR-96 modulation may be a disease-specific biomarker and may have a potential therapeutic value in MSA in particular.
Supplementary Material
There is widespread miRNA dysregulation in mouse models of neurodegenerative disorders.
Analysis of miRNA profiles may help identify disease-specific and/or disease-common patterns of miRNA dysregulation
miRNA dysregulation in murine models of multiple system atrophy recapitulate the human profile.
Dysregulation of miR-96 and its target genes may play a role in multiple system atrophy.
ACKNOWLEDGEMENTS
The authors would also like to thank Dr Manfred Windisch for the generous donation of the mThy1-htau tg mice. This work was supported by the National Institutes of Health [grant numbers AG05131, AG18440, NS044233 and AG022074] and by the Donner Canadian Foundation. The authors have no conflicts of interest to report.
REFERENCES
- Ambros V. microRNAs: tiny regulators with great potential. Cell. 2001;107:823–826. doi: 10.1016/s0092-8674(01)00616-x. [DOI] [PubMed] [Google Scholar]
- Anderson CM, Howard A, Walters JR, Ganapathy V, Thwaites DT. Taurine uptake across the human intestinal brush-border membrane is via two transporters: H+coupled PAT1 (SLC36A1) and Na+ and Cl(-)-dependent TauT (SLC6A6) J Physiol. 2009;587:731–744. doi: 10.1113/jphysiol.2008.164228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoyama K, Watabe M, Nakaki T. Modulation of neuronal glutathione synthesis by EAAC1 and its interacting protein GTRAP3-18. Amino Acids. 2012;42:163–169. doi: 10.1007/s00726-011-0861-y. [DOI] [PubMed] [Google Scholar]
- Asikainen S, Rudgalvyte M, Heikkinen L, Louhiranta K, Lakso M, Wong G, Nass R. Global microRNA expression profiling of Caenorhabditis elegans Parkinson’s disease models. J Mol Neurosci. 2010;41:210–218. doi: 10.1007/s12031-009-9325-1. [DOI] [PubMed] [Google Scholar]
- Barbato C, Ruberti F, Cogoni C. Searching for MIND: microRNAs in neurodegenerative diseases. J Biomed Biotechnol. 2009:871313. doi: 10.1155/2009/871313. 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beal MF, Lang AE, Ludolph AC. Neurodegenerative diseases: neurobiology, pathogenesis, and therapeutics. Cambridge University Press; 2005. [Google Scholar]
- Berezikov E. Evolution of microRNA diversity and regulation in animals. Nat Rev Genet. 2011;12:846–860. doi: 10.1038/nrg3079. [DOI] [PubMed] [Google Scholar]
- Biswas M, Chan JY. Role of Nrf1 in antioxidant response element-mediated gene expression and beyond. Toxicol Appl Pharmacol. 2010;244:16–20. doi: 10.1016/j.taap.2009.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairns NJ, Atkinson PF, Hanger DP, Anderton BH, Daniel SE, Lantos PL. Tau protein in the glial cytoplasmic inclusions of multiple system atrophy can be distinguished from abnormal tau in Alzheimer's disease. Neuroscience letters. 1997;230:49–52. doi: 10.1016/s0304-3940(97)00474-6. [DOI] [PubMed] [Google Scholar]
- Chen NH, Reith ME, Quick MW. Synaptic uptake and beyond: the sodium- and chloride-dependent neurotransmitter transporter family SLC6. Pflugers Archiv : European journal of physiology. 2004;447:519–531. doi: 10.1007/s00424-003-1064-5. [DOI] [PubMed] [Google Scholar]
- Chen RX, Xia YH, Xue TC, Ye SL. Suppression of microRNA-96 expression inhibits the invasion of hepatocellular carcinoma cells. Mol Med Report. 2011 doi: 10.3892/mmr.2011.695. [DOI] [PubMed] [Google Scholar]
- Chin J, Palop JJ, Puolivali J, Massaro C, Bien-Ly N, Gerstein H, Scearce-Levie K, Masliah E, Mucke L. Fyn kinase induces synaptic and cognitive impairments in a transgenic mouse model of Alzheimer’s disease. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2005;25:9694–9703. doi: 10.1523/JNEUROSCI.2980-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough RL, Dermentzaki G, Stefanis L. Functional dissection of the alpha-synuclein promoter: transcriptional regulation by ZSCAN21 and ZNF219. Journal of neurochemistry. 2009;110:1479–1490. doi: 10.1111/j.1471-4159.2009.06250.x. [DOI] [PubMed] [Google Scholar]
- de Mena L, Cardo LF, Coto E, Miar A, Diaz M, Corao AI, Alonso B, Ribacoba R, Salvador C, Menendez M, Moris G, Alvarez V. FGF20 rs12720208 SNP and microRNA-433 variation: no association with Parkinson’s disease in Spanish patients. Neurosci Lett. 2010;479:22–25. doi: 10.1016/j.neulet.2010.05.019. [DOI] [PubMed] [Google Scholar]
- Delay C, Hebert SS. MicroRNAs and Alzheimer's Disease Mouse Models: Current Insights and Future Research Avenues. Int J Alzheimers Dis. 2011;2011:894938. doi: 10.4061/2011/894938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dirson G, Desjardins P, Tannenberg T, Dodd P, Butterworth RF. Selective loss of expression of glutamate GluR2/R3 receptor subunits in cerebellar tissue from a patient with olivopontocerebellar atrophy. Metab Brain Dis. 2002;17:77–82. doi: 10.1023/a:1015412027708. [DOI] [PubMed] [Google Scholar]
- Doxakis E. Post-transcriptional regulation of alpha-synuclein expression by mir-7 and mir-153. J Biol Chem. 2010;285:12726–12734. doi: 10.1074/jbc.M109.086827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enciu AM, Popescu BO, Gheorghisan-Galateanu A. MicroRNAs in brain development and degeneration. Mol Biol Rep. 2011 doi: 10.1007/s11033-011-0973-1. [DOI] [PubMed] [Google Scholar]
- Featherstone DE. Glial solute carrier transporters in Drosophila and mice. Glia. 2011;59:1351–1363. doi: 10.1002/glia.21085. [DOI] [PubMed] [Google Scholar]
- Flunkert S, Hierzer M, Loffler T, Rabl R, Neddens J, Duller S, Schofield EL, Ward MA, Posch M, Jungwirth H, Windisch M, Hutter-Paier B. Elevated levels of soluble total and hyperphosphorylated tau result in early behavioral deficits and distinct changes in brain pathology in a new tau transgenic mouse model. Neurodegener Dis. 2013;11:194–205. doi: 10.1159/000338152. [DOI] [PubMed] [Google Scholar]
- Fruhbeis C, Frohlich D, Kuo WP, Amphornrat J, Thilemann S, Saab AS, Kirchhoff F, Mobius W, Goebbels S, Nave KA, Schneider A, Simons M, Klugmann M, Trotter J, Kramer-Albers EM. Neurotransmitter-triggered transfer of exosomes mediates oligodendrocyte-neuron communication. PLoS Biol. 2013;11:e1001604. doi: 10.1371/journal.pbio.1001604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gambari R, Fabbri E, Borgatti M, Lampronti I, Finotti A, Brognara E, Bianchi N, Manicardi A, Marchelli R, Corradini R. Targeting microRNAs involved in human diseases: a novel approach for modification of gene expression and drug development. Biochem Pharmacol. 2011;82:1416–1429. doi: 10.1016/j.bcp.2011.08.007. [DOI] [PubMed] [Google Scholar]
- Gao FB. Context-dependent functions of specific microRNAs in neuronal development. Neural Dev. 2010;5:25. doi: 10.1186/1749-8104-5-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geekiyanage H, Chan C. MicroRNA-137/181c regulates serine palmitoyltransferase and in turn amyloid beta, novel targets in sporadic Alzheimer’s disease. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2011;31:14820–14830. doi: 10.1523/JNEUROSCI.3883-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehrke S, Imai Y, Sokol N, Lu B. Pathogenic LRRK2 negatively regulates microRNA-mediated translational repression. Nature. 2010;466:637–641. doi: 10.1038/nature09191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillardon F, Mack M, Rist W, Schnack C, Lenter M, Hildebrandt T, Hengerer B. MicroRNA and proteome expression profiling in early-symptomatic alpha-synuclein(A30P)-transgenic mice. Proteomics Clin Appl. 2008;2:697–705. doi: 10.1002/prca.200780025. [DOI] [PubMed] [Google Scholar]
- Gilman S, Low PA, Quinn N, Albanese A, Ben-Shlomo Y, Fowler CJ, Kaufmann H, Klockgether T, Lang AE, Lantos PL, Litvan I, Mathias CJ, Oliver E, Robertson D, Schatz I, Wenning GK. Consensus statement on the diagnosis of multiple system atrophy. J Neurol Sci. 1999;163:94–98. doi: 10.1016/s0022-510x(98)00304-9. [DOI] [PubMed] [Google Scholar]
- Go RC, Perry RT, Wiener H, Bassett SS, Blacker D, Devlin B, Sweet RA. Neuregulin-1 polymorphism in late onset Alzheimer's disease families with psychoses. American journal of medical genetics. Part B, Neuropsychiatric genetics : the official publication of the International Society of Psychiatric Genetics. 2005;139B:28–32. doi: 10.1002/ajmg.b.30219. [DOI] [PubMed] [Google Scholar]
- Goffart S, Wiesner RJ. Regulation and co-ordination of nuclear gene expression during mitochondrial biogenesis. Exp Physiol. 2003;88:33–40. doi: 10.1113/eph8802500. [DOI] [PubMed] [Google Scholar]
- Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y, Chen J, Zhao X, Liang C, Wang Y, Sun L, Jiang Z, Zhang Z, Yang R, Li Z, Tang A, Li X, Ye J, Guan Z, Gui Y, Cai Z. MicroRNA expression signatures of bladder cancer revealed by deep sequencing. PLoS One. 2011;6:e18286. doi: 10.1371/journal.pone.0018286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannafon BN, Ding WQ. Intercellular Communication by Exosome-Derived microRNAs in Cancer. Int J Mol Sci. 2013;14:14240–14269. doi: 10.3390/ijms140714240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harraz MM, Dawson TM, Dawson VL. MicroRNAs in Parkinson's disease. J Chem Neuroanat. 2011;42:127–130. doi: 10.1016/j.jchemneu.2011.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashida H, Goto J, Zhao N, Takahashi N, Hirai M, Kanazawa I, Sakaki Y. Cloning and mapping of ZNF231, a novel brain-specific gene encoding neuronal double zinc finger protein whose expression is enhanced in a neurodegenerative disorder, multiple system atrophy (MSA) Genomics. 1998;54:50–58. doi: 10.1006/geno.1998.5516. [DOI] [PubMed] [Google Scholar]
- Hebert SS, Wang WX, Zhu Q, Nelson PT. A study of small RNAs from cerebral neocortex of pathology-verified Alzheimer's disease, dementia with lewy bodies, hippocampal sclerosis, frontotemporal lobar dementia, and non-demented human controls. J Alzheimers Dis. 2013;35:335–348. doi: 10.3233/JAD-122350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hediger MA, Romero MF, Peng JB, Rolfs A, Takanaga H, Bruford EA. The ABCs of solute carriers: physiological, pathological and therapeutic implications of human membrane transport proteinsIntroduction. Pflugers Archiv : European journal of physiology. 2004;447:465–468. doi: 10.1007/s00424-003-1192-y. [DOI] [PubMed] [Google Scholar]
- Iacobazzi V, Infantino V, Convertini P, Vozza A, Agrimi G, Palmieri F. Transcription of the mitochondrial citrate carrier gene: identification of a silencer and its binding protein ZNF224. Biochem Biophys Res Commun. 2009;386:186–191. doi: 10.1016/j.bbrc.2009.06.003. [DOI] [PubMed] [Google Scholar]
- Jaiswal AK. Nrf2 signaling in coordinated activation of antioxidant gene expression. Free Radic Biol Med. 2004;36:1199–1207. doi: 10.1016/j.freeradbiomed.2004.02.074. [DOI] [PubMed] [Google Scholar]
- Jensen KP, Covault J. Human miR-1271 is a miR-96 paralog with distinct non-conserved brain expression pattern. Nucleic Acids Res. 2011;39:701–711. doi: 10.1093/nar/gkq798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juhila J, Sipila T, Icay K, Nicorici D, Ellonen P, Kallio A, Korpelainen E, Greco D, Hovatta I. MicroRNA expression profiling reveals miRNA families regulating specific biological pathways in mouse frontal cortex and hippocampus. PLoS One. 2011;6:e21495. doi: 10.1371/journal.pone.0021495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junn E, Lee KW, Jeong BS, Chan TW, Im JY, Mouradian MM. Repression of alpha-synuclein expression and toxicity by microRNA-7. Proc Natl Acad Sci U S A. 2009;106:13052–13057. doi: 10.1073/pnas.0906277106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junn E, Mouradian MM. MicroRNAs in neurodegenerative diseases and their therapeutic potential. Pharmacol Ther. 2011 doi: 10.1016/j.pharmthera.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahle PJ, Neumann M, Ozmen L, Muller V, Jacobsen H, Spooren W, Fuss B, Mallon B, Macklin WB, Fujiwara H, Hasegawa M, Iwatsubo T, Kretzschmar HA, Haass C. Hyperphosphorylation and insolubility of alpha-synuclein in transgenic mouse oligodendrocytes. EMBO Rep. 2002;3:583–588. doi: 10.1093/embo-reports/kvf109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanai Y, Hediger MA. The glutamate/neutral amino acid transporter family SLC1: molecular, physiological and pharmacological aspects. Pflugers Arch. 2004a;447:469–479. doi: 10.1007/s00424-003-1146-4. [DOI] [PubMed] [Google Scholar]
- Kanai Y, Hediger MA. The glutamate/neutral amino acid transporter family SLC1: molecular, physiological and pharmacological aspects. Pflugers Archiv : European journal of physiology. 2004b;447:469–479. doi: 10.1007/s00424-003-1146-4. [DOI] [PubMed] [Google Scholar]
- Kanao T, Venderova K, Park DS, Unterman T, Lu B, Imai Y. Activation of FoxO by LRRK2 induces expression of proapoptotic proteins and alters survival of postmitotic dopaminergic neuron in Drosophila. Human molecular genetics. 2010;19:3747–3758. doi: 10.1093/hmg/ddq289. [DOI] [PubMed] [Google Scholar]
- Kasinski AL, Slack FJ. Epigenetics and genetics. MicroRNAs en route to the clinic: progress in validating and targeting microRNAs for cancer therapy. Nat Rev Cancer. 2011;11:849–864. doi: 10.1038/nrc3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Inoue K, Ishii J, Vanti WB, Voronov SV, Murchison E, Hannon G, Abeliovich A. A MicroRNA feedback circuit in midbrain dopamine neurons. Science. 2007;317:1220–1224. doi: 10.1126/science.1140481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krek A, Grun D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M, Rajewsky N. Combinatorial microRNA target predictions. Nature genetics. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- Kristensen AS, Andersen J, Jorgensen TN, Sorensen L, Eriksen J, Loland CJ, Stromgaard K, Gether U. SLC6 neurotransmitter transporters: structure, function, and regulation. Pharmacol Rev. 2011;63:585–640. doi: 10.1124/pr.108.000869. [DOI] [PubMed] [Google Scholar]
- Kurata T, Kametaka S, Ohta Y, Morimoto N, Deguchi S, Deguchi K, Ikeda Y, Takao Y, Ohta T, Manabe Y, Sato S, Abe K. PSP as distinguished from CBD, MSA-P and PD by clinical and imaging differences at an early stage. Intern Med. 2011;50:2775–2781. doi: 10.2169/internalmedicine.50.5954. [DOI] [PubMed] [Google Scholar]
- Ladeiro Y, Couchy G, Balabaud C, Bioulac-Sage P, Pelletier L, Rebouissou S, Zucman-Rossi J. MicroRNA profiling in hepatocellular tumors is associated with clinical features and oncogene/tumor suppressor gene mutations. Hepatology. 2008;47:1955–1963. doi: 10.1002/hep.22256. [DOI] [PubMed] [Google Scholar]
- Lantos PL, Papp MI. Cellular pathology of multiple system atrophy: a review. J Neurol Neurosurg Psychiatry. 1994;57:129–133. doi: 10.1136/jnnp.57.2.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee ST, Chu K, Im WS, Yoon HJ, Im JY, Park JE, Park KH, Jung KH, Lee SK, Kim M, Roh JK. Altered microRNA regulation in Huntington’s disease models. Exp Neurol. 2011;227:172–179. doi: 10.1016/j.expneurol.2010.10.012. [DOI] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Li YY, Cui JG, Hill JM, Bhattacharjee S, Zhao Y, Lukiw WJ. Increased expression of miRNA-146a in Alzheimer's disease transgenic mouse models. Neuroscience letters. 2011;487:94–98. doi: 10.1016/j.neulet.2010.09.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loscher CJ, Hokamp K, Kenna PF, Ivens AC, Humphries P, Palfi A, Farrar GJ. Altered retinal microRNA expression profile in a mouse model of retinitis pigmentosa. Genome Biol. 2007;8:R248. doi: 10.1186/gb-2007-8-11-r248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loscher CJ, Hokamp K, Wilson JH, Li T, Humphries P, Farrar GJ, Palfi A. A common microRNA signature in mouse models of retinal degeneration. Exp Eye Res. 2008;87:529–534. doi: 10.1016/j.exer.2008.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukiw WJ. Micro-RNA speciation in fetal, adult and Alzheimer's disease hippocampus. Neuroreport. 2007;18:297–300. doi: 10.1097/WNR.0b013e3280148e8b. [DOI] [PubMed] [Google Scholar]
- Ma L, Huang Y, Zhu W, Zhou S, Zhou J, Zeng F, Liu X, Zhang Y, Yu J. An integrated analysis of miRNA and mRNA expressions in non-small cell lung cancers. PLoS One. 2011;6:e26502. doi: 10.1371/journal.pone.0026502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manolopoulos KN, Klotz LO, Korsten P, Bornstein SR, Barthel A. Linking Alzheimer's disease to insulin resistance: the FoxO response to oxidative stress. Molecular psychiatry. 2010;15:1046–1052. doi: 10.1038/mp.2010.17. [DOI] [PubMed] [Google Scholar]
- Margis R, Rieder CR. Identification of blood microRNAs associated to Parkinsonis disease. J Biotechnol. 2011;152:96–101. doi: 10.1016/j.jbiotec.2011.01.023. [DOI] [PubMed] [Google Scholar]
- Martino S, di Girolamo I, Orlacchio A, Datti A. MicroRNA implications across neurodevelopment and neuropathology. Journal of biomedicine & biotechnology. 2009;2009:654346. doi: 10.1155/2009/654346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maynard TM, Meechan DW, Dudevoir ML, Gopalakrishna D, Peters AZ, Heindel CC, Sugimoto TJ, Wu Y, Lieberman JA, Lamantia AS. Mitochondrial localization and function of a subset of 22q11 deletion syndrome candidate genes. Mol Cell Neurosci. 2008;39:439–451. doi: 10.1016/j.mcn.2008.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihelich BL, Khramtsova EA, Arva N, Vaishnav A, Johnson DN, Giangreco AA, Martens-Uzunova E, Bagasra O, Kajdacsy-Balla A, Nonn L. miR-183-96-182 Cluster Is Overexpressed in Prostate Tissue and Regulates Zinc Homeostasis in Prostate Cells. J Biol Chem. 2011;286:44503–44511. doi: 10.1074/jbc.M111.262915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monsalve M, Olmos Y. The complex biology of FOXO. Curr Drug Targets. 2011;12:1322–1350. doi: 10.2174/138945011796150307. [DOI] [PubMed] [Google Scholar]
- Morciano P, Carrisi C, Capobianco L, Mannini L, Burgio G, Cestra G, De Benedetto GE, Corona DF, Musio A, Cenci G. A conserved role for the mitochondrial citrate transporter Sea/SLC25A1 in the maintenance of chromosome integrity. Hum Mol Genet. 2009;18:4180–4188. doi: 10.1093/hmg/ddp370. [DOI] [PubMed] [Google Scholar]
- Moss EG. MicroRNAs: hidden in the genome. Curr Biol. 2002;12:R138–R140. doi: 10.1016/s0960-9822(02)00708-x. [DOI] [PubMed] [Google Scholar]
- Nagaishi M, Yokoo H, Nakazato Y. Tau-positive glial cytoplasmic granules in multiple system atrophy. Neuropathology : official journal of the Japanese Society of Neuropathology. 2011;31:299–305. doi: 10.1111/j.1440-1789.2010.01159.x. [DOI] [PubMed] [Google Scholar]
- Navon R, Wang H, Steinfeld I, Tsalenko A, Ben-Dor A, Yakhini Z. Novel rank-based statistical methods reveal microRNAs with differential expression in multiple cancer types. PLoS One. 2009;4:e8003. doi: 10.1371/journal.pone.0008003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nymark P, Guled M, Borze I, Faisal A, Lahti L, Salmenkivi K, Kettunen E, Anttila S, Knuutila S. Integrative analysis of microRNA, mRNA and aCGH data reveals asbestos- and histology-related changes in lung cancer. Genes Chromosomes Cancer. 2011;50:585–597. doi: 10.1002/gcc.20880. [DOI] [PubMed] [Google Scholar]
- Otis TS, Brasnjo G, Dzubay JA, Pratap M. Interactions between glutamate transporters and metabotropic glutamate receptors at excitatory synapses in the cerebellar cortex. Neurochem Int. 2004;45:537–544. doi: 10.1016/j.neuint.2003.11.007. [DOI] [PubMed] [Google Scholar]
- Perez-Navarro E, Arenas E, Marco S, Alberch J. Intrastriatal grafting of a GDNF-producing cell line protects striatonigral neurons from quinolinic acid excitotoxicity in vivo. Eur J Neurosci. 1999;11:241–249. doi: 10.1046/j.1460-9568.1999.00433.x. [DOI] [PubMed] [Google Scholar]
- Pichardo-Casas I, Goff LA, Swerdel MR, Athie A, Davila J, Ramos-Brossier M, Lapid-Volosin M, Friedman WJ, Hart RP, Vaca L. Expression profiling of synaptic microRNAs from the adult rat brain identifies regional differences and seizure-induced dynamic modulation. Brain research. 2012;1436:20–33. doi: 10.1016/j.brainres.2011.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce ML, Weston MD, Fritzsch B, Gabel HW, Ruvkun G, Soukup GA. MicroRNA-183 family conservation and ciliated neurosensory organ expression. Evol Dev. 2008;10:106–113. doi: 10.1111/j.1525-142X.2007.00217.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehmsmeier M, Steffen P, Hochsmann M, Giegerich R. Fast and effective prediction of microRNA/target duplexes. Rna. 2004;10:1507–1517. doi: 10.1261/rna.5248604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren Q, Chen K, Paulsen IT. TransportDB: a comprehensive database resource for cytoplasmic membrane transport systems and outer membrane channels. Nucleic acids research. 2007;35:D274–D279. doi: 10.1093/nar/gkl925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberson ED, Halabisky B, Yoo JW, Yao J, Chin J, Yan F, Wu T, Hamto P, Devidze N, Yu GQ, Palop JJ, Noebels JL, Mucke L. Amyloid-beta/Fyn-induced synaptic, network, and cognitive impairments depend on tau levels in multiple mouse models of Alzheimer's disease. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2011;31:700–711. doi: 10.1523/JNEUROSCI.4152-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockenstein E, Mallory M, Hashimoto M, Song D, Shults CW, Lang I, Masliah E. Differential neuropathological alterations in transgenic mice expressing alpha-synuclein from the platelet-derived growth factor and Thy-1 promoters. Journal of neuroscience research. 2002;68:568–578. doi: 10.1002/jnr.10231. [DOI] [PubMed] [Google Scholar]
- Rockenstein E, Mallory M, Mante M, Sisk A, Masliaha E. Early formation of mature amyloid-beta protein deposits in a mutant APP transgenic model depends on levels of Abeta(1–42) Journal of neuroscience research. 2001;66:573–582. doi: 10.1002/jnr.1247. [DOI] [PubMed] [Google Scholar]
- Roshan R, Ghosh T, Scaria V, Pillai B. MicroRNAs: novel therapeutic targets in neurodegenerative diseases. Drug Discov Today. 2009;14:1123–1129. doi: 10.1016/j.drudis.2009.09.009. [DOI] [PubMed] [Google Scholar]
- Santosh PS, Arora N, Sarma P, Pal-Bhadra M, Bhadra U. Interaction map and selection of microRNA targets in Parkinson's disease-related genes. J Biomed Biotechnol. 2009;2009:363145. doi: 10.1155/2009/363145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh JI. Molecular network of microRNA targets in Alzheimer’s disease brains. Exp Neurol. 2011 doi: 10.1016/j.expneurol.2011.09.003. [DOI] [PubMed] [Google Scholar]
- Scales TM, Derkinderen P, Leung KY, Byers HL, Ward MA, Price C, Bird IN, Perera T, Kellie S, Williamson R, Anderton BH, Reynolds CH. Tyrosine phosphorylation of tau by the SRC family kinases lck and fyn. Mol Neurodegener. 2011;6:12. doi: 10.1186/1750-1326-6-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer A, Jung M, Miller K, Lein M, Kristiansen G, Erbersdobler A, Jung K. Suitable reference genes for relative quantification of miRNA expression in prostate cancer. Exp Mol Med. 2010a;42:749–758. doi: 10.3858/emm.2010.42.11.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer A, Jung M, Mollenkopf HJ, Wagner I, Stephan C, Jentzmik F, Miller K, Lein M, Kristiansen G, Jung K. Diagnostic and prognostic implications of microRNA profiling in prostate carcinoma. Int J Cancer. 2010b;126:1166–1176. doi: 10.1002/ijc.24827. [DOI] [PubMed] [Google Scholar]
- Schonrock N, Ke YD, Humphreys D, Staufenbiel M, Ittner LM, Preiss T, Gotz J. Neuronal microRNA deregulation in response to Alzheimer's disease amyloid-beta. PLoS One. 2010;5:e11070. doi: 10.1371/journal.pone.0011070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seol W. Biochemical and molecular features of LRRK2 and its pathophysiological roles in Parkinson's disease. BMB Rep. 2010;43:233–244. doi: 10.5483/bmbrep.2010.43.4.233. [DOI] [PubMed] [Google Scholar]
- Sethi P, Lukiw WJ. Micro-RNA abundance and stability in human brain: specific alterations in Alzheimer's disease temporal lobe neocortex. Neurosci Lett. 2009;459:100–104. doi: 10.1016/j.neulet.2009.04.052. [DOI] [PubMed] [Google Scholar]
- Sethupathy P, Megraw M, Hatzigeorgiou AG. A guide through present computational approaches for the identification of mammalian microRNA targets. Nat Methods. 2006;3:881–886. doi: 10.1038/nmeth954. [DOI] [PubMed] [Google Scholar]
- Shioya M, Obayashi S, Tabunoki H, Arima K, Saito Y, Ishida T, Satoh J. Aberrant microRNA expression in the brains of neurodegenerative diseases: miR-29a decreased in Alzheimer disease brains targets neurone navigator 3. Neuropathol Appl Neurobiol. 2010;36:320–330. doi: 10.1111/j.1365-2990.2010.01076.x. [DOI] [PubMed] [Google Scholar]
- Shults CW, Rockenstein E, Crews L, Adame A, Mante M, Larrea G, Hashimoto M, Song D, Iwatsubo T, Tsuboi K, Masliah E. Neurological and neurodegenerative alterations in a transgenic mouse model expressing human alpha-synuclein under oligodendrocyte promoter: implications for multiple system atrophy. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2005a;25:10689–10699. doi: 10.1523/JNEUROSCI.3527-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shults CW, Rockenstein E, Crews L, Adame A, Mante M, Larrea G, Hashimoto M, Song D, Iwatsubo T, Tsuboi K, Masliah E. Neurological and neurodegenerative alterations in a transgenic mouse model expressing human alpha-synuclein under oligodendrocyte promoter: implications for multiple system atrophy. J Neurosci. 2005b;25:10689–10699. doi: 10.1523/JNEUROSCI.3527-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel G, Saba R, Schratt G. microRNAs in neurons: manifold regulatory roles at the synapse. Curr Opin Genet Dev. 2011;21:491–497. doi: 10.1016/j.gde.2011.04.008. [DOI] [PubMed] [Google Scholar]
- Smith B, Treadwell J, Zhang D, Ly D, McKinnell I, Walker PR, Sikorska M. Large-scale expression analysis reveals distinct microRNA profiles at different stages of human neurodevelopment. PLoS One. 2010;5:e11109. doi: 10.1371/journal.pone.0011109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith KE, Borden LA, Wang CH, Hartig PR, Branchek TA, Weinshank RL. Cloning and expression of a high affinity taurine transporter from rat brain. Mol Pharmacol. 1992;42:563–569. [PubMed] [Google Scholar]
- Soma H, Yabe I, Takei A, Fujiki N, Yanagihara T, Sasaki H. Associations between multiple system atrophy and polymorphisms of SLC1A4, SQSTM1, and EIF4EBP1 genes. Mov Disord. 2008;23:1161–1167. doi: 10.1002/mds.22046. [DOI] [PubMed] [Google Scholar]
- Tomi M, Tajima A, Tachikawa M, Hosoya K. Function of taurine transporter (Slc6a6/TauT) as a GABA transporting protein and its relevance to GABA transport in rat retinal capillary endothelial cells. Biochim Biophys Acta. 2008;1778:2138–2142. doi: 10.1016/j.bbamem.2008.04.012. [DOI] [PubMed] [Google Scholar]
- Torp R, Hoover F, Danbolt NC, Storm-Mathisen J, Ottersen OP. Differential distribution of the glutamate transporters GLT1 and rEAAC1 in rat cerebral cortex and thalamus: an in situ hybridization analysis. Anat Embryol (Berl) 1997;195:317–326. doi: 10.1007/s004290050051. [DOI] [PubMed] [Google Scholar]
- Turchinovich A, Samatov TR, Tonevitsky AG, Burwinkel B. Circulating miRNAs: cell-cell communication function? Front Genet. 2013;4:119. doi: 10.3389/fgene.2013.00119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ubhi K, Rockenstein E, Mante M, Inglis C, Adame A, Patrick C, Whitney K, Masliah E. Neurodegeneration in a transgenic mouse model of multiple system atrophy is associated with altered expression of oligodendroglial-derived neurotrophic factors. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2010;30:6236–6246. doi: 10.1523/JNEUROSCI.0567-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utsumi M, Ohno K, Onchi H, Sato K, Tohyama M. Differential expression patterns of three glutamate transporters (GLAST, GLT1 and EAAC1) in the rat main olfactory bulb. Brain Res Mol Brain Res. 2001;92:1–11. doi: 10.1016/s0169-328x(01)00098-5. [DOI] [PubMed] [Google Scholar]
- Wakabayashi K, Takahashi H. Cellular pathology in multiple system atrophy. Neuropathology. 2006;26:338–345. doi: 10.1111/j.1440-1789.2006.00713.x. [DOI] [PubMed] [Google Scholar]
- Wang G, van der Walt JM, Mayhew G, Li YJ, Zuchner S, Scott WK, Martin ER, Vance JM. Variation in the miRNA-433 binding site of FGF20 confers risk for Parkinson disease by overexpression of alpha-synuclein. Am J Hum Genet. 2008;82:283–289. doi: 10.1016/j.ajhg.2007.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Liu J, Zong Y, Xu Y, Deng W, Zhu H, Liu Y, Ma C, Huang L, Zhang L, Qin C. miR-106b aberrantly expressed in a double transgenic mouse model for Alzheimer's disease targets TGF-beta type II receptor. Brain Res. 2010;1357:166–174. doi: 10.1016/j.brainres.2010.08.023. [DOI] [PubMed] [Google Scholar]
- Wang LL, Huang Y, Wang G, Chen SD. The potential role of microRNA-146 in Alzheimer's disease: Biomarker or therapeutic target? Med Hypotheses. 2012a;78:398–401. doi: 10.1016/j.mehy.2011.11.019. [DOI] [PubMed] [Google Scholar]
- Wang Y, Luo H, Li Y, Chen T, Wu S, Yang L. hsa-miR-96 up-regulates MAP4K1 and IRS1 and may function as a promising diagnostic marker in human bladder urothelial carcinomas. Mol Med Report. 2012b;5:260–265. doi: 10.3892/mmr.2011.621. [DOI] [PubMed] [Google Scholar]
- Warskulat U, Heller-Stilb B, Oermann E, Zilles K, Haas H, Lang F, Haussinger D. Phenotype of the taurine transporter knockout mouse. Methods Enzymol. 2007;428:439–458. doi: 10.1016/S0076-6879(07)28025-5. [DOI] [PubMed] [Google Scholar]
- Won SJ, Yoo BH, Brennan AM, Shin BS, Kauppinen TM, Berman AE, Swanson RA, Suh SW. EAAC1 gene deletion alters zinc homeostasis and exacerbates neuronal injury after transient cerebral ischemia. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2010;30:15409–15418. doi: 10.1523/JNEUROSCI.2084-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu JH, Gao Y, Ren AJ, Zhao SH, Zhong M, Peng YJ, Shen W, Jing M, Liu L. Altered MicroRNA Expression Profiles in Retinas with Diabetic Retinopathy. Ophthalmic Res. 2011;47:195–201. doi: 10.1159/000331992. [DOI] [PubMed] [Google Scholar]
- Xu S, Witmer PD, Lumayag S, Kovacs B, Valle D. MicroRNA (miRNA) transcriptome of mouse retina and identification of a sensory organ-specific miRNA cluster. J Biol Chem. 2007;282:25053–25066. doi: 10.1074/jbc.M700501200. [DOI] [PubMed] [Google Scholar]
- Xu X. Same computational analysis, different miRNA target predictions. Nat Methods. 2007;4:p191. doi: 10.1038/nmeth0307-191a. author reply 191. [DOI] [PubMed] [Google Scholar]
- Yamada Y, Enokida H, Kojima S, Kawakami K, Chiyomaru T, Tatarano S, Yoshino H, Kawahara K, Nishiyama K, Seki N, Nakagawa M. MiR-96 and miR-183 detection in urine serve as potential tumor markers of urothelial carcinoma: correlation with stage and grade, and comparison with urinary cytology. Cancer Sci. 2011;102:522–529. doi: 10.1111/j.1349-7006.2010.01816.x. [DOI] [PubMed] [Google Scholar]
- Yamakuchi M, Lowenstein CJ. MiR-34, SIRT1 and p53: the feedback loop. Cell Cycle. 2009;8:712–715. doi: 10.4161/cc.8.5.7753. [DOI] [PubMed] [Google Scholar]
- Yazawa I, Giasson BI, Sasaki R, Zhang B, Joyce S, Uryu K, Trojanowski JQ, Lee VM. Mouse model of multiple system atrophy alpha-synuclein expression in oligodendrocytes causes glial and neuronal degeneration. Neuron. 2005;45:847–859. doi: 10.1016/j.neuron.2005.01.032. [DOI] [PubMed] [Google Scholar]
- Yoshida M. Multiple system atrophy: alpha-synuclein and neuronal degeneration. Neuropathology. 2007;27:484–493. doi: 10.1111/j.1440-1789.2007.00841.x. [DOI] [PubMed] [Google Scholar]
- Zhu W, Liu X, He J, Chen D, Hunag Y, Zhang YK. Overexpression of members of the microRNA-183 family is a risk factor for lung cancer: a case control study. BMC Cancer. 2011;11:393. doi: 10.1186/1471-2407-11-393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zovoilis A, Agbemenyah HY, Agis-Balboa RC, Stilling RM, Edbauer D, Rao P, Farinelli L, Delalle I, Schmitt A, Falkai P, Bahari-Javan S, Burkhardt S, Sananbenesi F, Fischer A. microRNA-34c is a novel target to treat dementias. The EMBO journal. 2011;30:4299–4308. doi: 10.1038/emboj.2011.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.