Abstract
Despite recent technological advances in heterologous expression, stabilization and crystallization of membrane proteins (MPs), their structural studies remain difficult and require new transformative approaches. During the past two years, crystallization in lipidic cubic phase (LCP) has started gaining a widespread acceptance, owing to the spectacular success in high-resolution structure determination of G protein-coupled receptors (GPCRs) and to the introduction of commercial instrumentation, tools and protocols. The recent appearance of X-ray free-electron lasers (XFELs) has enabled structure determination from substantially smaller crystals than previously possible with minimal effects of radiation damage, offering new exciting opportunities in structural biology. The unique properties of LCP material have been exploited to develop special protocols and devices that have established a new method of serial femtosecond crystallography of MPs in LCP (LCP-SFX). In this method, microcrystals are generated in LCP and streamed continuously inside the same media across the intersection with a pulsed XFEL beam at a flow rate that can be adjusted to minimize sample consumption. Pioneering studies that yielded the first room temperature GPCR structures, using a few hundred micrograms of purified protein, validate the LCP-SFX approach and make it attractive for structure determination of difficult-to-crystallize MPs and their complexes with interacting partners. Together with the potential of femtosecond data acquisition to interrogate unstable intermediate functional states of MPs, LCP-SFX holds promise to advance our understanding of this biomedically important class of proteins.
Keywords: lipidic cubic phase, X-ray free-electron laser, G protein-coupled receptor, membrane proteins, serial femtosecond crystallography
1. Introduction
Membrane proteins (MPs) comprise about 30% of the human proteome and are targeted by 60% of all approved drugs [1], thus underscoring the importance of their structural and functional studies. MPs represent essential components of biological membranes and play crucial cellular roles including (i) transporting ions, metabolites, and large molecules such as proteins and RNA across the membranes, (ii) sensing and propagating environmental signals, (iii) mediating cell–cell interactions and cellular attachments to the extracellular matrix and maintaining cellular shape, and (iv) catalysing chemical reactions. Unlike soluble proteins, MPs reside in a lipid bilayer environment that presents unique challenges to researchers attempting to isolate, purify and crystallize them for structural studies.
Since 1985, when the first structure of a MP was solved [2], significant efforts have been focused on developing protocols and technologies to address barriers in expression, stabilization, crystallization and structure determination of MPs, with the goal of improving our understanding of their functional mechanisms [3]. By the end of 2013, over 1300 coordinate entries, representing 436 unique MPs (http://blanco.biomol.uci.edu/mpstruc/), have been deposited in the Protein Data Bank (http://www.pdb.org). Despite recent successes in solving the structures of several classes of challenging human MPs, including G protein-coupled receptors (GPCRs) [4], the overall progress in structural studies of MPs continues to lag strongly behind that of soluble proteins, emphasizing the need for innovative approaches and development of new disruptive technologies.
In this article, we review recent advances in the use of lipidic cubic phase (LCP) for crystallizing MPs and discuss the new exciting opportunities that have opened up with development of X-ray free-electron laser (XFEL) sources; technologies that begin to show promise in accelerating the rate of high-resolution MP structure determination. Key issues in combining the use of these two approaches are described, followed by a summary of our studies which led to the first high-resolution room temperature MP structure solved using this new method. These initial results demonstrate that XFELs enable high-quality structure determination of MPs crystallized and delivered in LCP, using substantially smaller crystals than those required for traditional microcrystallography at synchrotron sources, with the advantage of being able to obtain the structure at room temperature with minimal radiation damage while using only a few hundred micrograms of purified protein.
2. Crystallization of membrane proteins in lipidic cubic phase
Although recent advancements in miniaturization and automation have reduced the need for performing the onerous tasks associated with setting-up and tracking experiments, crystallization of MPs continues to be a significant barrier to structure determination with success rates remaining below 1%; significantly lower than the 10% observed in structural genomics studies targeting soluble proteins [5]. The most likely reason for this low success rate is the need to handle MPs removed from their native membrane environment to carry out crystallization studies. Hence, maintaining protein stability while searching for conditions leading to formation of diffraction quality crystals is of paramount importance.
Two general approaches are used for MP crystallization: the first, referred to as in surfo, is carried out with the MP solubilized in detergents forming a protein–detergent complex (PDC) [6]; the second, referred to as in meso, is performed in a membrane mimetic environment of lipidic mesophases or bicelles [7–9]. PDCs can, in general, be handled similarly to soluble proteins, and most crystallization methods developed for soluble proteins, such as vapour diffusion, batch method and free-interface diffusion [10], can be applied. Detergents, however, are often destabilizing to MPs, and the micelle envelope often interferes with the formation of crystal contacts. The choice of the right detergent is, therefore, very important for the success of crystallization. Crystals of PDCs typically have type II packing, where crystal contacts occur only between soluble parts of MPs, and their hydrophobic parts are shielded from protein–protein interactions by detergent micelles. Such crystals in many cases have large solvent content and suffer from accumulations of defects leading to poor diffraction.
As first demonstrated by Landau & Rosenbusch in 1996 [11], the LCP matrix supports growth of MP crystals with type I crystal packing, in which MPs contact each other through both hydrophobic and hydrophilic surfaces, and thus such crystals in our experience typically have lower solvent content and higher order. LCP crystallization was initially applied to solve the first high-resolution structure of bacteriorhodopsin [12]. Subsequent attempts to expand this approach to other MPs, however, ran into technical difficulties associated with handling the gel-like and sticky lipidic mesophase and with detecting small colourless crystals. It was not until the introduction of new tools, simplified protocols and instrumentation that LCP started to show its potential of delivering high-resolution structures of MPs from different families, in particular, human GPCRs [7]. Some of the most important and influential developments during the past 15 years include (i) lipid syringe mixer for fast and efficient mixing of lipids with MP solution allowing for the reconstitution of MPs in LCP within minutes [13]; (ii) LCP crystallization robot that automates and miniaturizes LCP crystallization set-up [14]; (iii) glass sandwich plates that improve detection of small colourless crystals growing in LCP [14,15]; (iv) new LCP host lipids for tailoring lipid bilayer properties, such as thickness and curvature, towards specific MP properties, and to facilitate crystallization at specific conditions, such as low temperatures [16,17]; (v) high-throughput fluorescence recovery after photobleaching (LCP-FRAP) pre-crystallization assay to assess diffusion properties of MPs in LCP at different conditions and guide subsequent crystallization trials [18]; (vi) thermostability assay, LCP-Tm, to compare stability of MPs directly in LCP, enabling the selection of the most stabilizing host lipids, lipid additives, protein constructs and ligands, in order to increase the likelihood of successful crystallization [19]; and (vii) second-order nonlinear imaging of chiral crystals (SONICC), which is used to detect submicrometre-sized protein crystals [20]. Commercial availability of many of these tools and instruments, as well as published detailed protocols [21,22] and video demonstrations [23–25], have enabled relatively straightforward adaptation of these technologies in different structural biology laboratories, resulting in the increased number of MP structures determined by this method.
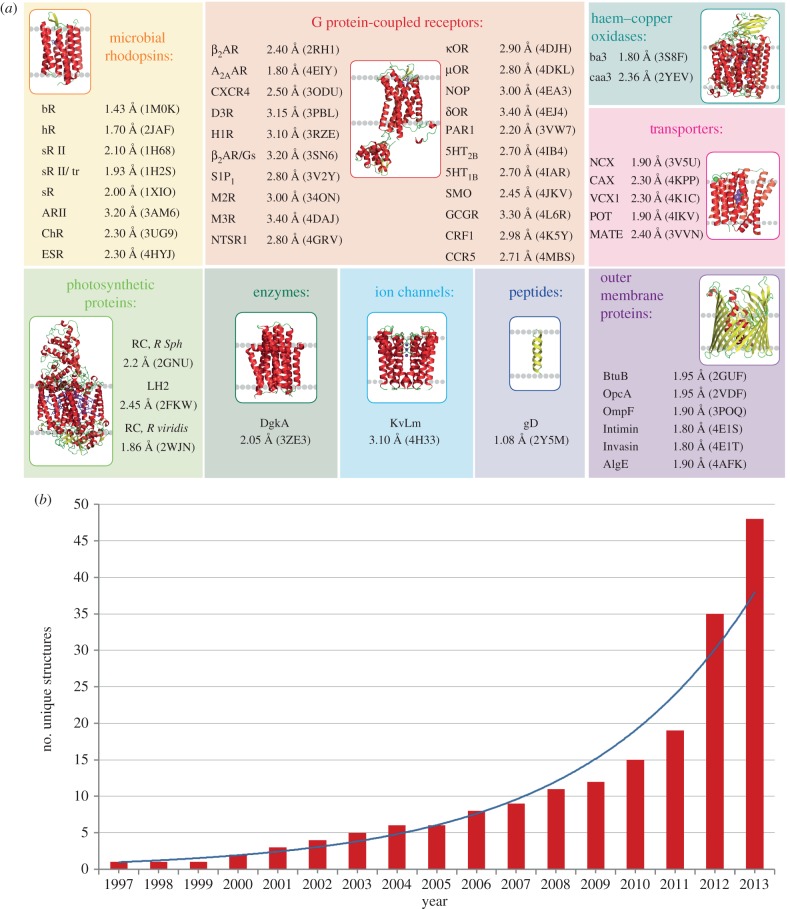
By the end of 2013, 48 unique MPs (about 11% of currently known MPs structures) from eight different classes, including those from both α-helical and β-barrel families, and covering bacterial as well as mammalian MPs, have been obtained using LCP technology (figure 1) [7,8]. During the past 2 years, contributions from this method have been rapidly increasing (in 2012–2013 25% of all new unique MP structures were determined by the LCP method), suggesting that it may likely soon become the preferred technique of MP crystallization for high-resolution structure determination.
Figure 1.
Progress in MP structure determination using LCP crystallization. (a) Gallery of MPs, structures of which were solved using LCP crystallization. For each unique MP, the highest resolution and corresponding PDB ID are shown. (b) Increase in the number of unique MPs structures obtained by LCP crystallization with time. Continuous curve represents an exponential fit.
The success of LCP crystallization can be attributed to several factors. First, the membrane-like environment of LCP confers higher stability on reconstituted MPs compared with the environment of detergent micelles [19]. Second, LCP crystallization supports type I packing that exhibits extensive interactions between transmembrane segments, leading to higher level ordering and high-resolution diffraction [7]. Finally, the LCP matrix acts as an effective size filter that removes large-size impurities and protein aggregates from contaminating growing crystals [26]. All these factors result in better diffraction quality crystals for the same proteins crystallized in meso when compared with in surfo [27]. However, owing to the higher nucleation rates and slower MP diffusion rates in LCP, crystals growing in LCP are on average smaller than their counterparts grown in surfo. The small crystal size imposes significant challenges at various steps used in standard crystallographic protocols, including those for detecting and harvesting crystals, as well as collecting high-resolution diffraction data and overcoming radiation damage. Thus, the success of LCP crystallization has greatly benefited from the development of cryocooling and micro-crystallographic techniques.
3. Microcrystallography at synchrotron sources
Before the appearance of third-generation synchrotron sources, most protein structures were obtained by collecting diffraction data at room temperature [28]. The detrimental effects of X-ray radiation damage, however, had been recognized relatively early during the development of macromolecular crystallography [29]. The advantage of cooling protein crystals during data collection was first examined in the late 1960s and 1970s [30–32]. It was demonstrated that cryocooling substantially reduced the diffusion rate of free radical products responsible for secondary damage, thereby increasing the tolerance of the crystal to radiation dose and usually resulting in higher resolution and better quality data. Widespread adoption of cryocrystallography in the late 1990s allowed for more efficient use of unattenuated X-ray beams at the third-generation synchrotron sources.
At the same time, the high brilliance of third-generation synchrotron sources triggered the development of microcrystallography [33]. Improvements in beam stability, goniometers, detectors and X-ray optics enabled structural studies of more challenging proteins and complexes that only form very small or highly inhomogeneous crystals. Dedicated microcrystallography beamlines with the beam sizes ranging between 1 and 20 μm are currently available at most modern synchrotron sources [33]. Matching the beamsize to the size of crystals reduces background from surrounding solvent and dramatically increases the signal-to-noise ratio, especially for high-resolution reflections [34]. Microfocus beams also help to address problems with inhomogeneous crystals by allowing selective collection of data from better ordered crystal regions [34].
Achievements in hardware design at the microfocus beamlines led to additional technological developments aimed at handling microcrystals and optimizing data collection, culminating in a series of biomedically important structures, such as virus polyhedra [35], amyloid fibrils [36] and GPCRs [37], among others. Additional challenges were encountered for microcrystals of GPCRs and other MPs, grown and harvested directly from LCP. Such crystals are often invisible through the beamline optics owing to the opacity of the cryocooled lipidic mesophase surrounding them. New methods and protocols have now been developed to address these issues, including automatic centring of optically invisible crystals by diffraction rastering [38,39] and SONICC [40], as well as new protocols for data collection from multiple microcrystals and assembling them together in a complete dataset [38].
Despite all these remarkable advancements in microfocus cryocrystallography, radiation damage remains a fundamental limitation for high-resolution data collection on well-ordered but small crystals. The recent emergence of new XFEL sources provides an opportunity to overcome this barrier, and thus should accelerate structural studies of difficult to crystallize MPs.
4. X-ray free-electron lasers and femtosecond crystallography
XFELs generate ultrabright and ultrashort pulses of coherent X-rays with tunable energy [41]. The peak brilliance of an XFEL beam delivered within a femtosecond pulse exceeds the brilliance of the most powerful synchrotron sources by a billion times. Such unique X-ray beam properties create new research opportunities across a variety of disciplines ranging from material science to medicine. In structural biology, XFELs promise data collection from much smaller crystals than previously possible, potentially down to the size of single molecules. Short XFEL pulses allow probing conformational changes in macromolecules with femtosecond resolution, providing essential information about the mechanisms of protein function. Although the energy within an XFEL pulse is so high that it can vapourize any material with which it interacts, the pulse duration is so short that it allows scattered photons to exit the sample before the appearance of damage initiated by photoionization [42]. Thus, diffraction can be recorded from the original, essentially undamaged molecules, and cryoconditions are not required, thereby enabling data collection at room temperature.
This ‘diffraction before destruction’ principle represents the basis for serial femtosecond crystallography (SFX), which relies on a continuous supply of microcrystals intersecting the XFEL beam at random orientations [43]. Following the rapid collection of several hundred thousand diffraction snapshots, Monte Carlo integration is applied to transform highly redundant partial reflections into structure factors [44]. The successful application of SFX was first demonstrated experimentally at low resolution, limited by the X-ray energy, on MPs crystallized in detergent solution [43] and in a liquid-like lipidic sponge phase [45]. When later applied to soluble proteins, it yielded high-resolution structures [46,47].
Efficient delivery of microcrystals within their native environment to the intersection with an XFEL beam with minimal sample waste and high hit rate is essential for successful SFX experiments. Such delivery can be accomplished either by using a fixed target approach, where crystals are deposited on an X-ray transparent support, or by streaming crystals using special injectors [48]. To date, two major types of injectors have been developed and used to deliver suspensions of microcrystals in liquid solutions for SFX data collection. The first SFX experiments used a gas dynamic virtual nozzle (GDVN) injector [49], which produces a continuous liquid jet stream focused by a gas sheath to a diameter of a few micrometres. Such a narrow and continuous stream guarantees a low background scattering and a high hit rate; however, its disadvantage is a high speed of about 10 m s−1 (a flow rate of about 7 μl min−1) and an associated high sample consumption. With the maximal pulse rate of 120 Hz at the Linac Coherent Light Source (LCLS), the sample stream travels several centimetres between X-ray pulses, resulting in less than one out of tens of thousands of crystals being probed by the beam and the rest being wasted between pulses. This translates into the requirement of several millilitres of dense suspension of microcrystals, equivalent to 10–100 mg of crystallized protein, for acquiring a complete dataset. Such quantities of purified protein are beyond reach for most eukaryotic MPs and other proteins with low expression yields. The GDVN injector was nevertheless successfully used to collect SFX data and solve several structures of soluble and MPs [43,45–47]. Further increases in the repetition pulse rate of XFELs and progress in the development of fast detectors should render the GDVN injector more efficient and more attractive in future.
To address the problem of the fast flowing jet, another injector that delivers microcrystals in a narrow liquid stream using an electrospinning principle was developed [50]. This injector can produce a jet stream a few micrometres in diameter inside a vacuum chamber with a flow rate of 0.17–3.1 μl min−1 by the application of an 3.4–5.4 kV cm−1 electric field. At these slow flow rates, the electrospun jet is prone to freezing owing to evaporative cooling and thus requires the addition of a cryoprotectant, such as 25–40% of glycerol. Despite the slower flow rate than that provided by the GDVN injector, recent experiments with the electrospun injector used similar amounts of protein sample per number of indexed images, likely owing to a lower crystal hit rate [51]. Clearly, further injector developments are needed in order to take advantage of the current XFEL capabilities while keeping sample consumption at a minimum.
5. Serial femtosecond crystallography in lipidic cubic phase
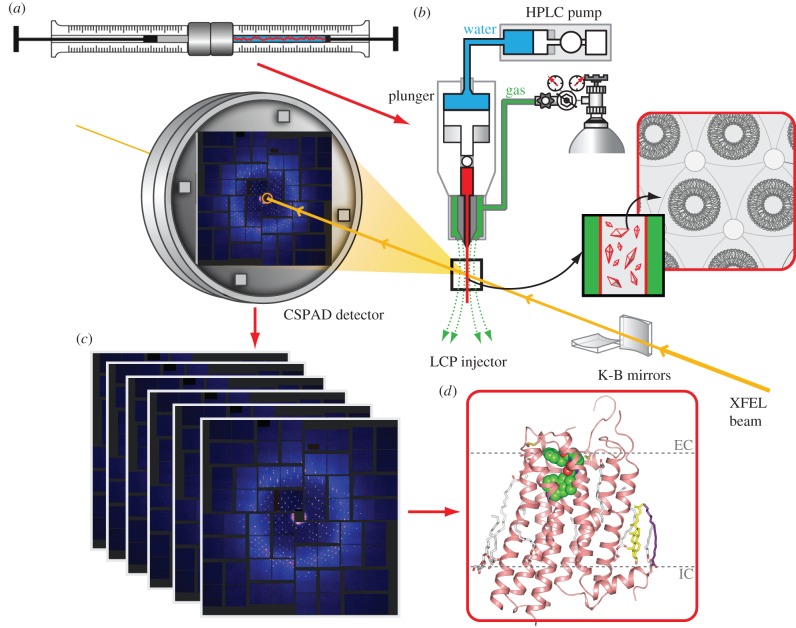
As discussed in §2, LCP crystallization often yields small but well-ordered crystals. Typical hits from initial crystallization screening appear as high density microcrystals, and it often takes substantial time and effort to optimize conditions for growing adequately sized crystals for data collection at a microfocus synchrotron beamline [38]. Thus, application of the SFX approach to LCP-grown microcrystals could have strong potential to accelerate MP structural studies through elimination of optimization and crystal harvesting steps. Additionally, it is conceivable that the gel-like texture and the high viscosity of LCP could allow for a better control of the flow rate and more efficient sample usage. These considerations prompted development of an LCP-SFX approach (figure 2).
Figure 2.
A schematic workflow associated with the LCP-SFX method. (a) MP microcrystals are grown in LCP (red) inside a syringe mixer. (b) LCP with microcrystals is streamed from an LCP injector and intersects with an XFEL beam focused to about 1.5 μm diameter. Diffraction snapshots are recorded at 120 Hz by the Cornell-SLAC pixel array detector. (c) Millions of snapshots are processed to extract patterns with crystal diffraction and index these patterns, followed by integration and merging of individual reflections to derive structure factors. (d) These structure factors are then used to solve and refine the MP structure.
Because none of the existing liquid injectors was compatible with streaming LCP, a special injector for LCP microextrusion was designed [52]. This LCP injector consists of a 20 μl sample reservoir, and a hydraulic plunger that amplifies pressure supplied via a system liquid by 34 times to extrude LCP through a 10–50 μm diameter capillary (figure 2b). LCP exits the injector nozzle as a continuous and straight column stabilized by a co-flowing gas (helium or nitrogen, supplied at 300–500 psi). LCP is extruded in a vacuum (approx. 10−3 torr), maintenance of which minimizes background scattering of an XFEL beam. The injector can operate either at a constant pressure or constant flow rate mode. In the constant pressure mode, the system liquid is pressurized by a gas, allowing the instrument to achieve flow rates as slow as a few hundred picolitres per minute. It is, however, difficult to maintain a consistent flow rate in this mode. In the constant flow rate mode, the injector is driven by an HPLC pump, allowing adjustment of the flow rate in the range 3–300 nl min−1. During data collection, the flow rate is therefore tuned to minimize sample waste, so that the stream advances only a sufficient distance between the XFEL pulses in order to remove the damaged material from the previous shot and expose fresh crystals for the next shot. This distance depends on the beam size, X-ray energy and flux density, and was estimated to be approximately 20 μm for 9.5 keV X-ray beam with pulse energy 50 μJ focused to a 1.5 μm diameter spot [52]. Therefore, in the case of the 120 Hz pulse repetition rate at LCLS, the most efficient sample consumption is achieved when the LCP jet is streamed at 2.4 mm s−1 velocity, which is equivalent to a flow rate of 10–300 nl min−1 for capillary nozzles ranging from 10 to 50 μm in diameter. The LCP injector is designed to withstand internal pressures up to 10 000 psi, which are required to extrude LCP through the 10 μm capillary. Extrusion through 10–30 μm capillaries, however, is highly prone to clogging, requiring implementation of special ‘clean room’ techniques during sample preparations. When using a 50 μm capillary nozzle, typical pressures inside the injector range between 2000 and 3000 psi.
Use of the LCP injector required the development and optimization of new microcrystal preparation protocols. High-throughput LCP crystallization trials are usually performed in 96-well glass sandwich plates using 20–50 nl volumes of protein-laden LCP boluses per well [14]. LCP-SFX data collection requires tens of microlitres of LCP that is uniformly filled with microcrystals at high density. Therefore, after a suitable crystallization condition is found in glass sandwich plates, LCP crystallization trials should be scaled up about 1000 times. Implementing this scale-up as a simple increase in the LCP bolus volume does not work, because the relatively slow diffusion of precipitants through the LCP matrix results in large concentration gradients across the drop and produces a highly inhomogeneous distribution in the size and density of crystals. To mimic the conditions encountered in glass sandwich plates, and for convenience in manipulations of LCP after crystals are grown, crystallization trials for LCP-SFX experiments are set up inside Hamilton gas-tight syringes. Several microlitres of protein-laden LCP are carefully injected as a continuous long column of 400 μm diameter into a syringe filled with a precipitant solution, allowing for fast equilibration and uniform growth of microcrystals within the whole LCP volume (figure 2a).
Initial tests of the LCP injector were performed in early 2012, with microcrystal LCP samples of the β2-adrenergic receptor [37] and A2A adenosine receptor (A2AAR) [53] generated using 90% monoolein (9.9 MAG)/10% cholesterol as the host LCP lipid. Both samples showed diffraction up to the detector edge at approximately 2.5 Å, even though the XFEL beam was attenuated by a factor of 20–30 to prevent oversaturation and damage of the detector owing to the appearance of strong sharp powder diffraction rings. These unexpected diffraction rings were attributed to lipidic lamellar crystalline, Lc, phase, which likely formed because of evaporative cooling upon injection of the 9.9 MAG LCP into a vacuum chamber. The equilibrium Lc to cubic-Pn3 m phase transition temperature for 9.9 MAG is 18°C [54], which is only a few degrees below the room temperature during data collection (20–21°C). Changing co-flowing gas from helium to nitrogen reduced Lc phase formation, but did not entirely prevent it. The problem was subsequently overcome by identifying two homologous lipids with shorter chains, 7.9 MAG [16] and 9.7 MAG (monopalmitolein), both of which form LCP that does not transform into Lc phase upon injection into vacuum. 7.9 MAG was specifically designed for low temperature LCP crystallization, as its equilibrium Lc-to-cubic Pn3m phase transition temperature of 6°C is among the lowest for the MAG series. It should be noted, however, that lipidic meshophases are prone to undercooling; therefore the equilibrium transition temperatures do not provide reliable references in such highly non-equilibrium conditions as those inside an LCP jet, and that it would be important, although technically difficult, to measure the actual temperature of microcrystals at the XFEL interrogation point. Finally, it was established that Lc phase formation upon injection could also be prevented by adding 7.9 MAG to LCP made of 9.9 MAG (the most successful LCP crystallization host lipid to date) post-crystal growth [52], relieving the requirement for a specific LCP host lipid, and thus making this method more generally applicable.
6. First applications of the LCP-SFX method
Because traditional crystallography and LCP-SFX are fundamentally different in how the data are both collected and processed, a comprehensive evaluation of the new method and comparison between these two approaches were called for. The first MP that was chosen to test and validate the new LCP-SFX method was the human serotonin (5-hydroxytryptamine, 5-HT) 5-HT2B receptor in complex with the agonist ergotamine. Just before the experiments at LCLS, a 2.7 Å resolution structure of 5-HT2B/ergotamine was solved by traditional crystallography [55] (PDB ID: 4IB4), and thus it served as a yardstick for comparison with the XFEL structure.
A high-quality 5-HT2B/ergotamine dataset was collected at the coherent X-ray imaging (CXI) beamline at LCLS using microcrystals with an average size of 5 × 5 × 5 μm3 (about 100 times smaller in volume than their counterparts used for synchrotron data collection) delivered with an LCP injector at room temperature [56]. Over four million diffraction snapshots were recorded within about 10 h using only 0.3 mg of purified protein. Over 30 000 of these snapshots were indexed yielding a complete dataset at 2.8 Å resolution with multiplicity of over 1000 (table 1). Optimization of the density of crystals and improvements in the data processing software should further decrease the time and the amount of protein required for collection of a similar quality dataset.
Table 1.
Data collection and refinement statistics for 5-HT2B/ergotamine structures obtained at XFEL and synchrotron sources. The most important differences are highlighted in italics. Data for the high-resolution shell are shown in parentheses.
| data collection | 5-HT2B-XFEL | 5-HT2B-SYN |
|---|---|---|
| temperature (K) | 294 | 100 |
| wavelength (Å) | 1.3 | 1.032 |
| beam size (µm) | 1.5 | 10 |
| average crystal size (µm) | 5 × 5 × 5 | 80 × 20 × 10 |
| number of crystals | 32 819 | 17 |
| max. dose per crystal (MGy) | 25 | 20 |
| space group | C2221 | C2221 |
| unit cell (Å) | 61.5, 122.2, 168.5 | 60.57, 119.75, 170.61 |
| oscillation (°)/exposure (s) | 0/5 × 10−14 | 1.0/1.0–3.0 |
| no. collected images | 4 217 508 | 91 |
| no. hits/indexed images | 152 651/32 819 | 91/91 |
| no. total/unique reflections | 18 515 376/16 052 | 51 559/16 041 |
| resolution (Å) | 35–2.8 (2.9–2.8) | 50–2.7 (2.8–2.7) |
| completeness (%) | 100 (100) | 90.5 (92.2) |
| multiplicity | 1150 (1035.6) | 3.2 (3.1) |
| CC* | 0.998 (0.74) | 0.992 (0.77) |
| Rsplit (XFEL) or Rmerge (SYN) (%) | 9.5 (161.9) | 15.0 (91.4) |
| refinement | ||
| no. reflections/test set | 16 025/814 | 15 818/823 |
| Rwork/Rfree (%) | 22.7/27.0 | 22.7/26.6 |
| no. atoms | ||
| protein/ligand/other | 2856/43/224 | 2854/43/170 |
| B-factors (Å2) | ||
| Wilson B/overall B | 115.7/98.7 | 72.1/80.0 |
| receptor/ligand | 88.4/68.1 | 67.2/57.7 |
| R.m.s bonds (Å)/angles (°) | 0.002/0.60 | 0.009/0.98 |
| Ramachandran plot stats (%) | ||
| favoured/allowed/disallowed | 96.4/3.6/0.0 | 98.1/1.9/0.0 |
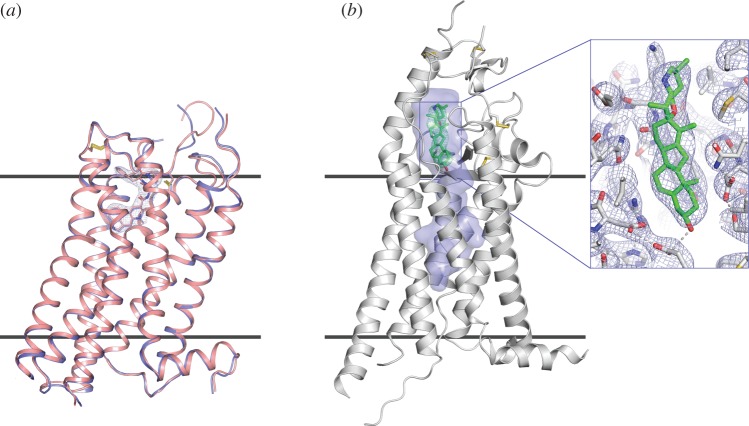
The 5-HT2B-XFEL structure was solved by molecular replacement and revealed excellent electron density for most receptor residues as well as for the ligand ergotamine, cholesterol and several lipids, which were omitted from the molecular replacement model [56]. The final 5-HT2B-XFEL structure is remarkably similar to the structure previously obtained by traditional crystallography at a synchrotron source, 5-HT2B-SYN (receptor Cα root mean square deviation (RMSD) = 0.45 Å), attesting that the LCP–SFX method produces reliable data (figure 3a). No obvious effects of radiation damage were apparent in either of the structures. Despite the overall agreement, several notable differences are observed between them: (i) the unit cell volume for 5-HT2B-XFEL is 2.1% larger, in agreement with a typical shrinkage of protein crystal lattice upon cryocooling [57]; (ii) the backbone of the N-terminus, several loops and extracellular part of helix II show substantial deviations, likely because of intrinsic flexibility of these regions; (iii) several sidechains display different rotamer conformations, consistent with partial remodelling of side chain conformational distribution upon cryocooling, previously observed in soluble proteins [57,58] and (iv) the average B-factor of the 5-HT2B-XFEL structure is about 20 Å2 higher compared with the 5-HT2B-SYN structure, consistent with increased thermal motions at higher temperature and possible effects of Bragg termination during the XFEL pulse [59]. The distribution of B-factors highlights a more rigid core of the seven transmembrane helices in comparison with loops, with more pronounced B-factor deviations observed in the room temperature 5-HT2B-XFEL structure.
Figure 3.
First GPCR structures obtained by LCP-SFX. (a) Comparison between XFEL (light red) and synchrotron (blue) structures of 5-HT2B/ergotamine. Ergotamine is shown as sticks along with 2mFo-DFc electron density map around it, contoured at 1σ. (b) Smoothened receptor in complex with cyclopamine. A long cavity inside the receptor is highlighted in blue. Zoomed insert shows the cyclopamine binding site with 2mFo-DFc electron density map contoured at 1σ. Horizontal lines indicate approximate membrane boundaries.
Overall, the XFEL data resulted in a very similar structure with comparable quality to that obtained at a synchrotron source despite the use of much smaller crystals. The observed structural differences are most likely related to different temperatures at which structures were solved, although potential effects of radiation damage as well as differences in data acquisition and processing cannot be fully ruled out and should be further investigated. Given that dynamics are an integral part of GPCR function and that LCP-SFX data are collected at room temperature, the 5-HT2B-XFEL structure likely provides a more accurate representation of the conformational ensemble of this receptor under native conditions.
After successful validation of the LCP-SFX approach with the 5-HT2B structure, the next target studied was the human Smoothened (SMO) receptor in complex with the naturally occurring teratogen cyclopamine. The SMO receptor belongs to the class Frizzled GPCRs and is implicated in embryonic development and carcinogenesis [60,61]. SMO is an orphan receptor as no native ligands that activate it have been established, although a number of small molecules have been identified that bind and modulate SMO receptor activity [62–64]. A structure of SMO bound to an anti-tumour drug candidate was recently published by Wang et al. [65]; however, our extensive efforts to obtain the SMO/cyclopamine structure by traditional microcrystallography failed owing to poor quality crystals suffering from high mosaicity and low isomorphism. We hypothesized that the poor crystal quality was associated with accumulation of defects during crystal growth and that smaller, sub-10 μm crystals could have better quality and allow for acquisition of a complete dataset using the LCP-SFX approach. Indeed, the SMO/cyclopamine diffraction patterns collected at LCLS were successfully indexed, integrated and merged by the CrystFEL software [66], resulting in a good-quality dataset, albeit at a moderate and anisotropic resolution of 3.2–4.0 Å. The molecular replacement solution revealed a clear density for cyclopamine bound to the receptor inside a long and narrow cavity starting in the extracellular loops and going through the transmembrane helices region (figure 3b) [52]. This structure, along with structures of other SMO/ligand complexes will provide new biological insights into modulations of this receptor by small molecules and on the effects of chemoresistance mutations in cancer patients [67].
7. Summary and future outlook
The remarkable progress achieved during the past few years in the development of X-ray lasers, instrumentation and data processing software has led to the establishment of a number of SFX approaches that minimize radiation damage and enable structure determination from micrometre- and submicrometre-sized crystals of soluble and MPs. The LCP-SFX method builds upon a synergy among the unique properties of LCP that facilitates MP crystallization and enables efficient delivery of microcrystals, and the advantages of XFEL. It allows for the collection of high-quality room temperature structural data with negligible radiation damage, reduces time and resources for optimizing crystals to sizes required for synchrotron microcrystallography, and substantially decreases the amounts of crystallized protein needed for data collection compared with liquid injectors. Further improvements in XFEL sources, detectors and data processing should expand the limits of achievable resolution, improve the quality of structural models, and allow working with even smaller crystals.
While the LCP-SFX method has been shown to work, further developments are required to make it more efficient, robust, routine and user-friendly. We summarize here some of the highlights and general issues that have to be addressed in the next few years to achieve the vision and related goals.
The first room temperature GPCR structures determined by LCP-SFX represent a significant and important milestone. It is highly desirable, however, to achieve a sub-2 Å resolution, which can allow for resolving multiple side-chain conformations and detecting waters, ions and lipid molecules bound to the receptor. Recently, structures of two GPCRs, A2AAR and δ-opioid receptors, were solved at 1.8 Å resolution by traditional microcrystallography, revealing a network of internal water molecules and a sodium ion that are essential for ligand binding and signal transduction [53,68]. Similar high-resolution structures determined at room temperature will shed light on the strength and dynamics of these interactions at close to native conditions.
The LCP-SFX approach may also serve as a foundation for an efficient structure-based drug design (SBDD) platform targeting MPs. SBDD has proved highly successful for soluble proteins, such as kinases and proteases [69,70]; however, its application to GPCRs and other MPs has been hampered by the difficulty of preparing large amounts of homogeneous and stable samples, and growing sufficiently large crystals for a large number of different protein–ligand complexes for high-resolution structure determination at synchrotron sources. LCP-SFX could help to streamline the process by reducing the time and effort spent on crystal optimization, harvesting and diffraction screening. Additionally, micrometre-size crystals could allow for faster and more efficient ligand soaking and exchange. Room temperature structures could also provide better templates for ligand docking studies.
Most published SFX structures so far have been solved using molecular replacement. Recently, SFX de novo phasing has been demonstrated on a test soluble protein, lysozyme [71]. Clearly, there is a need to perform experimental phasing of SFX data for more challenging MPs, possibly through the use of multiwave anomalous diffraction (MAD) or single wave anomalous diffraction (SAD) [72]. Such approaches are routine in traditional crystallography; however, they require sufficiently large crystals to accurately measure Friedel pairs for each reflection and avoid radiation damage decay as much as possible. The first successful experimental phasing of LCP-grown MP crystals was reported in 2012 [27,73,74]. LCP-grown GPCR microcrystals were recently used for phasing by SAD after an overnight soaking with a tantalum bromide cluster [65,75]. Similar approaches can be tested using LCP-SFX, where the key to successful extraction of phase information will be the accuracy of the structure factors that is related to the multiplicity with which each reflection is measured.
SFX experiments with soluble proteins and MPs crystallized in detergent solutions are currently hampered by high protein consumption because of a fast flow rate and/or low efficiency of the available liquid injectors. Owing to the low crystal consumption achieved with the LCP injector, LCP can be considered as a medium for delivery of microcrystals of soluble proteins. For this purpose, soluble proteins could be crystallized directly in LCP [76,77]. In cases when the protein only crystallizes in solution but not in LCP, a slurry of protein crystals obtained in solution can be mixed with an LCP-host lipid. This method may also work for MP crystals obtained in detergent solutions, but because detergents can partition in the lipid bilayer of LCP, compatibility of such crystals with LCP needs to be tested.
Along with many advantages of the LCP injector, there is a limitation on the minimal diameter of the capillary nozzle (10–15 μm) through which LCP can be extruded. An XFEL beam passing through such a relatively thick LCP stream scatters X-rays, producing a relatively strong background which can overwhelm high-resolution diffraction from submicrometre-sized crystals. It is therefore desirable to reduce the thickness of LCP down to a few micrometres, which could be potentially achieved using a fixed target approach. LCP with grown-in microcrystals could be squeezed down to a few micrometre thickness layer between two transparent to X-rays windows, or alternatively, microcrystals could be grown inside special chips and then rastered in situ by an XFEL beam. Both these approaches will require developments of wet sample cells to preserve crystal hydration under vacuum and fast rastering methods to collect data at 120 Hz with a high hit rate.
Finally, one of the most exciting opportunities for XFELs in structural biology is the prospect of recording molecular movies [78]. Femtosecond XFEL pulses can be used to probe short-lived intermediate conformational states of macromolecules as they perform their function inside small crystals. Photoactivated proteins and microcrystal injectors are ideally suited for this application, as the activation by a laser flash can be easily timed with the structural interrogation by the XFEL beam with variable time-delay ranging from femtoseconds to milliseconds. The first experiments in this direction using liquid injectors have been started [79,80]. Translation of these technologies for use in conjunction with LCP-SFX will require reducing fluctuations in the LCP flow rate that are relatively high in the current version of the LCP injector.
Although clearly in its infancy, the current rapid development of LCP-SFX protocols holds the promise to increase the success rate of structural studies on challenging MPs and their complexes, endowing the biomedical community with a powerful tool for developing a detailed understanding of the mechanisms of their actions, as well as for designing new and more selective therapeutics.
Acknowledgements
This work was supported in part by the National Institutes of Health grants no. P50 GM073197 and U54 GM094618, Jiangsu Provincial Natural Science Foundation of China (grant no. BK20130405), the Natural Science Foundation of the Jiangsu Higher Education Institutions of China (grant no. 13KJB180002) and the Priority Academic Programme Development of Jiangsu Higher Education Institutions of China. We thank K. Kadyshevskaya for assistance with figure preparation and A. Walker for assistance with manuscript preparation.
References
- 1.Yildirim MA, Goh KI, Cusick ME, Barabasi AL, Vidal M. 2007. Drug–target network. Nat. Biotechnol. 25, 1119–1126. ( 10.1038/nbt1338) [DOI] [PubMed] [Google Scholar]
- 2.Deisenhofer J, Epp O, Miki K, Huber R, Michel H. 1985. Structure of the protein subunits in the photosynthetic reaction centre of Rhodopseudomonas viridis at 3 Å resolution. Nature 318, 618–624. ( 10.1038/318618a0) [DOI] [PubMed] [Google Scholar]
- 3.Kang HJ, Lee C, Drew D. 2013. Breaking the barriers in membrane protein crystallography. Int. J. Biochem. Cell. Biol. 45, 636–644. ( 10.1016/j.biocel.2012.12.018) [DOI] [PubMed] [Google Scholar]
- 4.Katritch V, Cherezov V, Stevens RC. 2013. Structure-function of the G protein-coupled receptor superfamily. Annu. Rev. Pharmacol. Toxicol. 53, 531–556. ( 10.1146/annurev-pharmtox-032112-135923) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kloppmann E, Punta M, Rost B. 2012. Structural genomics plucks high-hanging membrane proteins. Curr. Opin. Struct. Biol. 22, 326–332. ( 10.1016/j.sbi.2012.05.002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Prive GG. 2007. Detergents for the stabilization and crystallization of membrane proteins. Methods 41, 388–397. ( 10.1016/j.ymeth.2007.01.007) [DOI] [PubMed] [Google Scholar]
- 7.Cherezov V. 2011. Lipidic cubic phase technologies for membrane protein structural studies. Curr. Opin. Struct. Biol. 21, 559–566. ( 10.1016/j.sbi.2011.06.007) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Caffrey M. 2009. Crystallizing membrane proteins for structure determination: use of lipidic mesophases. Annu. Rev. Biophys. 38, 29–51. ( 10.1146/annurev.biophys.050708.133655) [DOI] [PubMed] [Google Scholar]
- 9.Ujwal R, Bowie JU. 2011. Crystallizing membrane proteins using lipidic bicelles. Methods 55, 337–341. ( 10.1016/j.ymeth.2011.09.020) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wiener MC. 2004. A pedestrian guide to membrane protein crystallization. Methods 34, 364–372. ( 10.1016/j.ymeth.2004.03.025) [DOI] [PubMed] [Google Scholar]
- 11.Landau EM, Rosenbusch JP. 1996. Lipidic cubic phases: a novel concept for the crystallization of membrane proteins. Proc. Natl Acad. Sci. USA 93, 14 532–14 535. ( 10.1073/pnas.93.25.14532) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pebay-Peyroula E, Rummel G, Rosenbusch JP, Landau EM. 1997. X-ray structure of bacteriorhodopsin at 2.5 angstroms from microcrystals grown in lipidic cubic phases. Science 277, 1676–1681. ( 10.1126/science.277.5332.1676) [DOI] [PubMed] [Google Scholar]
- 13.Cheng A, Hummel B, Qiu H, Caffrey M. 1998. A simple mechanical mixer for small viscous lipid-containing samples. Chem. Phys. Lipids 95, 11–21. ( 10.1016/S0009-3084(98)00060-7) [DOI] [PubMed] [Google Scholar]
- 14.Cherezov V, Peddi A, Muthusubramaniam L, Zheng YF, Caffrey M. 2004. A robotic system for crystallizing membrane and soluble proteins in lipidic mesophases. Acta Crystallogr. D Biol. Crystallogr. 60, 1795–1807. ( 10.1107/S0907444904019109) [DOI] [PubMed] [Google Scholar]
- 15.Cherezov V, Caffrey M. 2003. Nano-volume plates with excellent optical properties for fast, inexpensive crystallization screening of membrane proteins. J. Appl. Crystallogr. 36, 1372–1377. ( 10.1107/S002188980301906x) [DOI] [Google Scholar]
- 16.Misquitta Y, Cherezov V, Havas F, Patterson S, Mohan JM, Wells AJ, Hart DJ, Caffrey M. 2004. Rational design of lipid for membrane protein crystallization. J. Struct. Biol. 148, 169–175. ( 10.1016/j.jsb.2004.06.008) [DOI] [PubMed] [Google Scholar]
- 17.Li D, Shah ST, Caffrey M. 2013. Host lipid and temperature as important screening variables for crystallizing integral membrane proteins in lipidic mesophases. Trials with diacylglycerol kinase. Cryst. Growth Des. 13, 2846–2857. ( 10.1021/cg400254v) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu F, Liu W, Hanson MA, Stevens RC, Cherezov V. 2011. Development of an automated high throughput LCP-FRAP assay to guide membrane protein crystallization in lipid mesophases. Cryst. Growth Des. 11, 1193–1201. ( 10.1021/cg101385e) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu W, Hanson MA, Stevens RC, Cherezov V. 2010. LCP-Tm: an assay to measure and understand stability of membrane proteins in a membrane environment. Biophys. J. 98, 1539–1548. ( 10.1016/j.bpj.2009.12.4296) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kissick DJ, Gualtieri EJ, Simpson GJ, Cherezov V. 2010. Nonlinear optical imaging of integral membrane protein crystals in lipidic mesophases. Anal. Chem. 82, 491–497. ( 10.1021/ac902139w) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Caffrey M, Cherezov V. 2009. Crystallizing membrane proteins using lipidic mesophases. Nat. Protoc. 4, 706–731. ( 10.1038/nprot.2009.31) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cherezov V, Abola E, Stevens RC. 2010. Recent progress in the structure determination of GPCRs, a membrane protein family with high potential as pharmaceutical targets. Methods Mol. Biol. 654, 141–168. ( 10.1007/978-1-60761-762-4_8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu W, Cherezov V. 2011. Crystallization of membrane proteins in lipidic mesophases. J. Vis. Exp. 49, e2501 ( 10.3791/2501) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Caffrey M, Porter C. 2010. Crystallizing membrane proteins for structure determination using lipidic mesophases. J. Vis. Exp. 45, e1712 ( 10.3791/1712) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li D, Boland C, Aragao D, Walsh K, Caffrey M. 2012. Harvesting and cryo-cooling crystals of membrane proteins grown in lipidic mesophases for structure determination by macromolecular crystallography. J. Vis. Exp. 67, e4001 ( 10.3791/4001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kors CA, Wallace E, Davies DR, Li L, Laible PD, Nollert P. 2009. Effects of impurities on membrane-protein crystallization in different systems. Acta Crystallogr. D Biol. Crystallogr. 65, 1062–1073. ( 10.1107/S0907444909029163) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liao J, Li H, Zeng W, Sauer DB, Belmares R, Jiang Y. 2012. Structural insight into the ion-exchange mechanism of the sodium/calcium exchanger. Science 335, 686–690. ( 10.1126/science.1215759) [DOI] [PubMed] [Google Scholar]
- 28.Garman E, Schneider T. 1997. Macromolecular cryocrystallography. J. Appl. Crystallogr. 30, 211–237. ( 10.1107/S0021889897002677) [DOI] [Google Scholar]
- 29.Low BW, Chen CC, Berger JE, Singman L, Pletcher JF. 1966. Studies of insulin crystals at low temperatures: effects on mosaic character and radiation sensitivity. Proc. Natl Acad. Sci. USA 56, 1746–1750. ( 10.1073/pnas.56.6.1746) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Haas DJ. 1968. Preliminary studies on the denaturation of cross-linked lysozyme crystals. Biophys. J. 8, 549–555. ( 10.1016/S0006-3495(68)86507-5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Haas DJ, Rossmann MG. 1970. Crystallographic studies on lactate dehydrogenase at −75 degrees C. Acta Crystallogr. B 26, 998–1004. ( 10.1107/S0567740870003485) [DOI] [PubMed] [Google Scholar]
- 32.Petsko GA. 1975. Protein crystallography at sub-zero temperatures: cryo-protective mother liquors for protein crystals. J. Mol. Biol. 96, 381–392. ( 10.1016/0022-2836(75)90167-9) [DOI] [PubMed] [Google Scholar]
- 33.Smith JL, Fischetti RF, Yamamoto M. 2012. Micro-crystallography comes of age. Curr. Opin. Struct. Biol. 22, 602–612. ( 10.1016/j.sbi.2012.09.001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sanishvili R, Nagarajan V, Yoder D, Becker M, Xu S, Corcoran S, Akey DL, Smith JL, Fischetti RF. 2008. A 7mum mini-beam improves diffraction data from small or imperfect crystals of macromolecules. Acta Crystallogr. D Biol. Crystallogr. 64, 425–435. ( 10.1107/S0907444908001741) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Coulibaly F, Chiu E, Ikeda K, Gutmann S, Haebel PW, Schulze-Briese C, Mori H, Metcalf P. 2007. The molecular organization of cypovirus polyhedra. Nature 446, 97–101. ( 10.1038/nature05628) [DOI] [PubMed] [Google Scholar]
- 36.Nelson R, Sawaya MR, Balbirnie M, Madsen AO, Riekel C, Grothe R, Eisenberg D. 2005. Structure of the cross-beta spine of amyloid-like fibrils. Nature 435, 773–778. ( 10.1038/nature03680) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cherezov V, et al. 2007. High-resolution crystal structure of an engineered human beta2-adrenergic G protein-coupled receptor. Science 318, 1258–1265. ( 10.1126/science.1150577) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cherezov V, et al. 2009. Rastering strategy for screening and centring of microcrystal samples of human membrane proteins with a sub-10 microm size X-ray synchrotron beam. J. R. Soc. Interface 6(Suppl. 5), S587–S597. ( 10.1098/rsif.2009.0142.focus) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hilgart MC, Sanishvili R, Ogata CM, Becker M, Venugopalan N, Stepanov S, Makarov O, Smith JL, Fischetti RF. 2011. Automated sample-scanning methods for radiation damage mitigation and diffraction-based centering of macromolecular crystals. J. Synchrotron Radiat. 18, 717–722. ( 10.1107/S0909049511029918) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Madden JT, et al. 2013. Integrated nonlinear optical imaging microscope for on-axis crystal detection and centering at a synchrotron beamline. J. Synchrotron Radiat. 20, 531–540. ( 10.1107/S0909049513007942) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Spence JC, Weierstall U, Chapman HN. 2012. X-ray lasers for structural and dynamic biology. Rep. Prog. Phys. 75, 102601 ( 10.1088/0034-4885/75/10/102601) [DOI] [PubMed] [Google Scholar]
- 42.Neutze R, Wouts R, van der Spoel D, Weckert E, Hajdu J. 2000. Potential for biomolecular imaging with femtosecond X-ray pulses. Nature 406, 752–757. ( 10.1038/35021099) [DOI] [PubMed] [Google Scholar]
- 43.Chapman HN, et al. 2011. Femtosecond X-ray protein nanocrystallography. Nature 470, 73–77. ( 10.1038/nature09750) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kirian RA, et al. 2011. Structure-factor analysis of femtosecond microdiffraction patterns from protein nanocrystals. Acta Crystallogr. A 67, 131–140. ( 10.1107/S0108767310050981) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Johansson LC, et al. 2012. Lipidic phase membrane protein serial femtosecond crystallography. Nat. Methods 9, 263–265. ( 10.1038/nmeth.1867) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Boutet S, et al. 2012. High-resolution protein structure determination by serial femtosecond crystallography. Science 337, 362–364. ( 10.1126/science.1217737) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Redecke L, et al. 2013. Natively inhibited Trypanosoma brucei cathepsin B structure determined by using an X-ray laser. Science 339, 227–230. ( 10.1126/science.1229663) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Weierstall U, Spence JC, Doak RB. 2012. Injector for scattering measurements on fully solvated biospecies. Rev. Sci. Instrum. 83, 035108 ( 10.1063/1.3693040) [DOI] [PubMed] [Google Scholar]
- 49.DePonte DP, Weierstall U, Starodub D, Schmidt K, Spence JC, Doak RB. 2008. Gas dynamic virtual nozzle for generation of microscopic droplet streams. J. Phys. D 41, 195505 ( 10.1088/0022-3727/41/19/195505) [DOI] [Google Scholar]
- 50.Sierra RG, et al. 2012. Nanoflow electrospinning serial femtosecond crystallography. Acta Crystallogr. D Biol. Crystallogr. 68, 1584–1587. ( 10.1107/S0907444912038152) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kern J, et al. 2012. Room temperature femtosecond X-ray diffraction of photosystem II microcrystals. Proc. Natl Acad. Sci. USA 109, 9721–9726. ( 10.1073/pnas.1204598109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Weierstall U, et al. 2014. Lipidic cubic phase injector facilitates membrane protein serial femtosecond crystallography. Nat. Commun. 5, 3309 ( 10.1038/ncomms4309) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu W, et al. 2012. Structural basis for allosteric regulation of GPCRs by sodium ions. Science 337, 232–236. ( 10.1126/science.1219218) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Qiu H, Caffrey M. 2000. The phase diagram of the monoolein/water system: metastability and equilibrium aspects. Biomaterials 21, 223–234. ( 10.1016/S0142-9612(99)00126-X) [DOI] [PubMed] [Google Scholar]
- 55.Wacker D, et al. 2013. Structural features for functional selectivity at serotonin receptors. Science 340, 615–619. ( 10.1126/science.1232808) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu W, et al. 2013. Serial femtosecond crystallography of G protein-coupled receptors. Science 342, 1521–1524. ( 10.1126/science.1244142) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fraser JS, van den Bedem H, Samelson AJ, Lang PT, Holton JM, Echols N, Alber T. 2011. Accessing protein conformational ensembles using room-temperature X-ray crystallography. Proc. Natl Acad. Sci. USA 108, 16 247–16 252. ( 10.1073/pnas.1111325108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dunlop KV, Irvin RT, Hazes B. 2005. Pros and cons of cryocrystallography: should we also collect a room-temperature data set? Acta Crystallogr. D Biol. Crystallogr. 61, 80–87. ( 10.1107/S0907444904027179) [DOI] [PubMed] [Google Scholar]
- 59.Barty A, et al. 2012. Self-terminating diffraction gates femtosecond X-ray nanocrystallography measurements. Nat. Photonics 6, 35–40. ( 10.1038/nphoton.2011.297) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ingham PW, McMahon AP. 2001. Hedgehog signaling in animal development: paradigms and principles. Genes Dev. 15, 3059–3087. ( 10.1101/gad.938601) [DOI] [PubMed] [Google Scholar]
- 61.Pasca di Magliano M, Hebrok M. 2003. Hedgehog signalling in cancer formation and maintenance. Nat. Rev. Cancer 3, 903–911. ( 10.1038/nrc1229) [DOI] [PubMed] [Google Scholar]
- 62.Frank-Kamenetsky M, et al. 2002. Small-molecule modulators of Hedgehog signaling: identification and characterization of Smoothened agonists and antagonists. J. Biol. 1, 10 ( 10.1186/1475-4924-1-10) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chen JK, Taipale J, Young KE, Maiti T, Beachy PA. 2002. Small molecule modulation of Smoothened activity. Proc. Natl Acad. Sci. USA 99, 14 071–14 076. ( 10.1073/pnas.182542899) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mahindroo N, Punchihewa C, Fujii N. 2009. Hedgehog-Gli signaling pathway inhibitors as anticancer agents. J. Med. Chem. 52, 3829–3845. ( 10.1021/jm801420y) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang C, et al. 2013. Structure of the human smoothened receptor bound to an antitumour agent. Nature 497, 338–343. ( 10.1038/nature12167) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.White A, Kirian RA, Martin AV, Aquila A, Nass K, Barty A, Chapman HN. 2012. CrystFEL: a software suite for snapshot serial crystallography. J. Appl. Crystallogr. 45, 335–341. ( 10.1107/S0021889812002312) [DOI] [Google Scholar]
- 67.Yauch RL, et al. 2009. Smoothened mutation confers resistance to a hedgehog pathway inhibitor in medulloblastoma. Science 326, 572–574. ( 10.1126/science.1179386) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fenalti G, Giguere PM, Katritch V, Huang XP, Thompson AA, Cherezov V, Roth BL, Stevens RC. 2014. Molecular control of delta-opioid receptor signalling. Nature 506, 191–196. ( 10.1038/nature12944) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Scapin G. 2002. Structural biology in drug design: selective protein kinase inhibitors. Drug Discov. Today 7, 601–611. ( 10.1016/S1359-6446(02)02290-0) [DOI] [PubMed] [Google Scholar]
- 70.Wlodawer A, Erickson JW. 1993. Structure-based inhibitors of HIV-1 protease. Annu. Rev. Biochem. 62, 543–585. ( 10.1146/annurev.bi.62.070193.002551) [DOI] [PubMed] [Google Scholar]
- 71.Barends TR, et al. 2014. De novo protein crystal structure determination from X-ray free-electron laser data. Nature 505, 244–247. ( 10.1038/nature12773) [DOI] [PubMed] [Google Scholar]
- 72.Son SK, Chapman HN, Santra R. 2011. Multiwavelength anomalous diffraction at high X-ray intensity. Phys. Rev. Lett. 107, 218 102–218 106. ( 10.1103/PhysRevLett.107.218102) [DOI] [PubMed] [Google Scholar]
- 73.Kato HE, et al. 2012. Crystal structure of the channelrhodopsin light-gated cation channel. Nature 482, 369–374. ( 10.1038/nature10870) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Fairman JW, et al. 2012. Crystal structures of the outer membrane domain of intimin and invasin from enterohemorrhagic E. coli and enteropathogenic Y. pseudotuberculosis. Structure 20, 1233–1243. ( 10.1016/j.str.2012.04.011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Siu FY, et al. 2013. Structure of the human glucagon class B G-protein-coupled receptor. Nature 499, 444–449. ( 10.1038/nature12393) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Landau EM, Rummel G, Cowan-Jacob SW, Rosenbusch JP. 1997. Crystallization of a polar protein and small molecules from the aqueous compartment of lipidic cubic phases. J. Phys. Chem. B 101, 1935–1937. ( 10.1021/jp963347q) [DOI] [Google Scholar]
- 77.Aherne M, Lyons JA, Caffrey M. 2012. A fast, simple and robust protocol for growing crystals in the lipidic cubic phase. J. Appl. Crystallogr. 45, 1330–1333. ( 10.1107/S0021889812037880) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Barty A, Küpper J, Chapman C. 2013. Molecular imaging using X-ray free-electron lasers. Annu. Rev. Phys. Chem. 64, 415–435. ( 10.1146/annurev-physchem-032511-143708) [DOI] [PubMed] [Google Scholar]
- 79.Aquila A, et al. 2012. Time-resolved protein nanocrystallography using an X-ray free-electron laser. Opt. Express. 20, 2706–2716. ( 10.1364/OE.20.002706) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kern J, et al. 2013. Simultaneous femtosecond X-ray spectroscopy and diffraction of photosystem II at room temperature. Science 340, 491–495. ( 10.1126/science.1234273) [DOI] [PMC free article] [PubMed] [Google Scholar]