Abstract
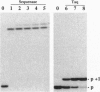
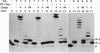
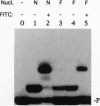
Enzymatic incorporation of 2',3'-dideoxynucleotides into DNA results in chain termination. We report that 3'-esterified 2'-deoxynucleoside 5'-triphosphates (dNTPs) are false chain-terminator substrates since DNA polymerases, including human immunodeficiency virus reverse transcriptase, can incorporate them into DNA and, subsequently, use this new 3' end to insert the next correctly paired dNTP. Likewise, a DNA substrate with a primer chemically esterified at the 3' position can be extended efficiently upon incubation with dNTPs and T7 DNA polymerase lacking 3'-to-5' exonuclease activity. This enzyme is also able to use dTTP-bearing reporter groups in the 3' position conjugated through amide or thiourea bonds and cleave them to restore a DNA chain terminated by an amino group at the 3' end. Hence, a number of DNA polymerases exhibit wide catalytic versatility at the 3' end of the nascent DNA strand. As part of the polymerization mechanism, these capabilities extend the number of enzymatic activities associated with these enzymes and also the study of interactions between DNA polymerases and nucleotide analogues.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abboud M. M., Sim W. J., Loeb L. A., Mildvan A. S. Apparent suicidal inactivation of DNA polymerase by adenosine 2',3'-riboepoxide 5'-triphosphate. J Biol Chem. 1978 May 25;253(10):3415–3421. [PubMed] [Google Scholar]
- Beese L. S., Steitz T. A. Structural basis for the 3'-5' exonuclease activity of Escherichia coli DNA polymerase I: a two metal ion mechanism. EMBO J. 1991 Jan;10(1):25–33. doi: 10.1002/j.1460-2075.1991.tb07917.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boosalis M. S., Petruska J., Goodman M. F. DNA polymerase insertion fidelity. Gel assay for site-specific kinetics. J Biol Chem. 1987 Oct 25;262(30):14689–14696. [PubMed] [Google Scholar]
- Canard B., Sarfati R. S. DNA polymerase fluorescent substrates with reversible 3'-tags. Gene. 1994 Oct 11;148(1):1–6. doi: 10.1016/0378-1119(94)90226-7. [DOI] [PubMed] [Google Scholar]
- Catalano C. E., Benkovic S. J. Inactivation of DNA polymerase I (Klenow fragment) by adenosine 2',3'-epoxide 5'-triphosphate: evidence for the formation of a tight-binding inhibitor. Biochemistry. 1989 May 16;28(10):4374–4382. doi: 10.1021/bi00436a038. [DOI] [PubMed] [Google Scholar]
- Chidgeavadze Z. G., Beabealashvilli R. S., Atrazhev A. M., Kukhanova M. K., Azhayev A. V., Krayevsky A. A. 2',3'-Dideoxy-3' aminonucleoside 5'-triphosphates are the terminators of DNA synthesis catalyzed by DNA polymerases. Nucleic Acids Res. 1984 Feb 10;12(3):1671–1686. doi: 10.1093/nar/12.3.1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chidgeavadze Z. G., Beabealashvilli R. Sh, Krayevsky A. A., Kukhanova M. K. Nucleoside 5'-triphosphates with modified sugars as substrates for DNA polymerases. Biochim Biophys Acta. 1986 Nov 13;868(2-3):145–152. doi: 10.1016/0167-4781(86)90017-5. [DOI] [PubMed] [Google Scholar]
- Cretton E. M., Xie M. Y., Bevan R. J., Goudgaon N. M., Schinazi R. F., Sommadossi J. P. Catabolism of 3'-azido-3'-deoxythymidine in hepatocytes and liver microsomes, with evidence of formation of 3'-amino-3'-deoxythymidine, a highly toxic catabolite for human bone marrow cells. Mol Pharmacol. 1991 Feb;39(2):258–266. [PubMed] [Google Scholar]
- Davies J. F., 2nd, Almassy R. J., Hostomska Z., Ferre R. A., Hostomsky Z. 2.3 A crystal structure of the catalytic domain of DNA polymerase beta. Cell. 1994 Mar 25;76(6):1123–1133. doi: 10.1016/0092-8674(94)90388-3. [DOI] [PubMed] [Google Scholar]
- Delarue M., Poch O., Tordo N., Moras D., Argos P. An attempt to unify the structure of polymerases. Protein Eng. 1990 May;3(6):461–467. doi: 10.1093/protein/3.6.461. [DOI] [PubMed] [Google Scholar]
- Gibson K. J., Benkovic S. J. Synthesis and application of derivatizable oligonucleotides. Nucleic Acids Res. 1987 Aug 25;15(16):6455–6467. doi: 10.1093/nar/15.16.6455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klenow H., Henningsen I. Selective elimination of the exonuclease activity of the deoxyribonucleic acid polymerase from Escherichia coli B by limited proteolysis. Proc Natl Acad Sci U S A. 1970 Jan;65(1):168–175. doi: 10.1073/pnas.65.1.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohlstaedt L. A., Wang J., Friedman J. M., Rice P. A., Steitz T. A. Crystal structure at 3.5 A resolution of HIV-1 reverse transcriptase complexed with an inhibitor. Science. 1992 Jun 26;256(5065):1783–1790. doi: 10.1126/science.1377403. [DOI] [PubMed] [Google Scholar]
- Kutateladze T. V., Kritzyn A. M., Florentjev V. L., Kavsan V. M., Chidgeavadze Z. G., Beabealashvilli R. S. 3'-Hydroxymethyl 2'-deoxynucleoside 5'-triphosphates are inhibitors highly specific for reverse transcriptase. FEBS Lett. 1986 Oct 27;207(2):205–212. doi: 10.1016/0014-5793(86)81489-2. [DOI] [PubMed] [Google Scholar]
- Metzker M. L., Raghavachari R., Richards S., Jacutin S. E., Civitello A., Burgess K., Gibbs R. A. Termination of DNA synthesis by novel 3'-modified-deoxyribonucleoside 5'-triphosphates. Nucleic Acids Res. 1994 Oct 11;22(20):4259–4267. doi: 10.1093/nar/22.20.4259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsuya H., Weinhold K. J., Furman P. A., St Clair M. H., Lehrman S. N., Gallo R. C., Bolognesi D., Barry D. W., Broder S. 3'-Azido-3'-deoxythymidine (BW A509U): an antiviral agent that inhibits the infectivity and cytopathic effect of human T-lymphotropic virus type III/lymphadenopathy-associated virus in vitro. Proc Natl Acad Sci U S A. 1985 Oct;82(20):7096–7100. doi: 10.1073/pnas.82.20.7096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ollis D. L., Brick P., Hamlin R., Xuong N. G., Steitz T. A. Structure of large fragment of Escherichia coli DNA polymerase I complexed with dTMP. 1985 Feb 28-Mar 6Nature. 313(6005):762–766. doi: 10.1038/313762a0. [DOI] [PubMed] [Google Scholar]
- Pandey V. N., Modak M. J. Affinity labeling of Escherichia coli DNA polymerase I by 5'-fluorosulfonylbenzoyladenosine. Identification of the domain essential for polymerization and Arg-682 as the site of reactivity. J Biol Chem. 1988 May 5;263(13):6068–6073. [PubMed] [Google Scholar]
- Pandey V. N., Williams K. R., Stone K. L., Modak M. J. Photoaffinity labeling of the thymidine triphosphate binding domain in Escherichia coli DNA polymerase I: identification of histidine-881 as the site of cross-linking. Biochemistry. 1987 Dec 1;26(24):7744–7748. doi: 10.1021/bi00398a031. [DOI] [PubMed] [Google Scholar]
- Pelletier H., Sawaya M. R., Kumar A., Wilson S. H., Kraut J. Structures of ternary complexes of rat DNA polymerase beta, a DNA template-primer, and ddCTP. Science. 1994 Jun 24;264(5167):1891–1903. [PubMed] [Google Scholar]
- Smith L. M., Fung S., Hunkapiller M. W., Hunkapiller T. J., Hood L. E. The synthesis of oligonucleotides containing an aliphatic amino group at the 5' terminus: synthesis of fluorescent DNA primers for use in DNA sequence analysis. Nucleic Acids Res. 1985 Apr 11;13(7):2399–2412. doi: 10.1093/nar/13.7.2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa R., Chung Y. J., Rose J. P., Wang B. C. Crystal structure of bacteriophage T7 RNA polymerase at 3.3 A resolution. Nature. 1993 Aug 12;364(6438):593–599. doi: 10.1038/364593a0. [DOI] [PubMed] [Google Scholar]
- Tabor S., Richardson C. C. Effect of manganese ions on the incorporation of dideoxynucleotides by bacteriophage T7 DNA polymerase and Escherichia coli DNA polymerase I. Proc Natl Acad Sci U S A. 1989 Jun;86(11):4076–4080. doi: 10.1073/pnas.86.11.4076. [DOI] [PMC free article] [PubMed] [Google Scholar]