Abstract
Cigarette smoke produces a molecular “field of injury” in epithelial cells lining the respiratory tract. However, the specific signaling pathways that are altered in the airway of smokers and the signaling processes responsible for the transition from smoking-induced airway damage to lung cancer remain unknown. In this study, we use a genomic approach to study the signaling processes associated with tobacco smoke exposure and lung cancer. First, we developed and validated pathway-specific gene expression signatures in bronchial airway epithelium that reflect activation of signaling pathways relevant to tobacco-exposure including ATM, BCL2, GPX1, NOS2, IKBKB, and SIRT1. Using these profiles and four independent gene expression datasets, we found that SIRT1 activity is significantly up-regulated in cytologically normal bronchial airway epithelial cells from active smokers compared to non-smokers. In contrast, this activity is strikingly down-regulated in non-small cell lung cancer. This pattern of signaling modulation was unique to SIRT1, and down-regulation of SIRT1 activity is confined to tumors from smokers. Decreased activity of SIRT1 was validated using genomic analyses of mouse models of lung cancer and biochemical testing of SIRT1 activity in patient lung tumors. Together, our findings indicate a role of SIRT1 in response to smoke and a potential role in repressing lung cancer. Further, our findings suggest that the airway gene-expression signatures derived in this study can provide novel insights into signaling pathways altered in the “field of inury” induced by tobacco smoke and thus may impact strategies for prevention of tobacco-related lung cancer.
Introduction
Cigarette smoking is a causative factor for chronic obstructive pulmonary disease (COPD) and lung cancer, with 10–20% of smokers developing these diseases (1). It is critical to understand the physiologic response to cigarette smoke, and the signaling events that protect smokers from lung cancer development so that we can develop effective lung cancer prevention strategies. Cigarette smoke creates a field of injury in the epithelial cells that line the respiratory tract (2-8). Our group and others have characterized smoking-related gene and microRNA expression alterations in the cytologically normal large and small bronchial airway epithelium (2, 9-15) and have categorized these by their degree of reversibility upon smoking cessation (12, 14). We have shown that lung cancer also significantly alters the airway transcriptome and have developed a gene expression-based biomarker for the detection of lung cancer using cells collected from the mainstem bronchus during bronchoscopy that are cytologically normal and distant from the primary tumor(11, 16).
Microarray studies are often employed to identify gene expresion alterations between groups of interest and use these gene sets to infer imporant disease-associated pathways or biomarkers. In contrast, a reverse genomic strategy using gene expression signatures of specific pathway activation has been used to identify patterns of activation of oncogenic pathways in tumors that are associated with disease outcome (17). As an example, using these oncogenic pathway activation signatures we previously found that phosphatidylinositiol 3-kinase (PI3K) pathway activation is increased in the cytologically normal airway epithelium of smokers with lung cancer and high-risk smokers with premalignant airway lesions (mild to moderate dysplasia). Further, we demonstrated that treatment of these high-risk smokers with myo-inositol, a PI3K inhibitor and a potential chemopreventive agent, resulted in reduced PI3K pathway activity in the airway which was associated with response to therapy as measured by regression of the dysplasic airway lesion (18).
In this study, we have extended this pathway approach to explore additional pathways that may be activated within the airway field of injury associated with tobacco smoke exposure and the development of lung cancer. Using primary human bronchial epithelial cells collected at bronchoscopy, we have perturbed signaling pathways important in regulation of response to tobacco smoke exposure and cancer development: ATM, BCL2, GPX1, NOS2, IKBKB, and SIRT1 (19-28). Differential pathway activation in intrathoracic epithelial cells from smoker and non-smokers and lung tumor and adjacent normal tissue identified the SIRT1 (sirtuin 1, an NAD+-dependent deacetylase) pathway as being activated in response to tobacco smoke exposure. Further, SIRT1 activity was consistently down-regulated following lung cancer development, an observation that was validated in primary tumors, using levels of H4K16, a biomarker of SIRT1 activity. Overall, these data suggest a role of SIRT1 signaling in response to stressors such as cigarette smoke, and in preventing lung cancer development. More broadly, our findings suggest that we have successfully developed, for the first time, pathway-specific gene expression signatures in bronchial airway epithelium that reflect activation of signaling pathways relevant to tobacco-related lung disease.
Materials and Methods
Patient Population
We recruited three never smoker volunteers at Boston University Medical Center to undergo flexible bronchoscopy, and endoscopic brushings were obtained from the right mainstem bronchus. The airway epithelial cells obtained from the brushings were cultured and used to generate the pathway signatures. Patients undergoing lung nodule resection for suspicion of lung cancer were recruited, and lung tissue from the nodule and an adjacent normal area of the lung (from the resection margin) was collected from 8 patients and used to measure H4K16 levels (see Supplemental Table 2 for patient information). Both studies were approved by the Institutional Review Board and the participants provided written informed consent.
Culture of Primary Airway Epithelial Cells
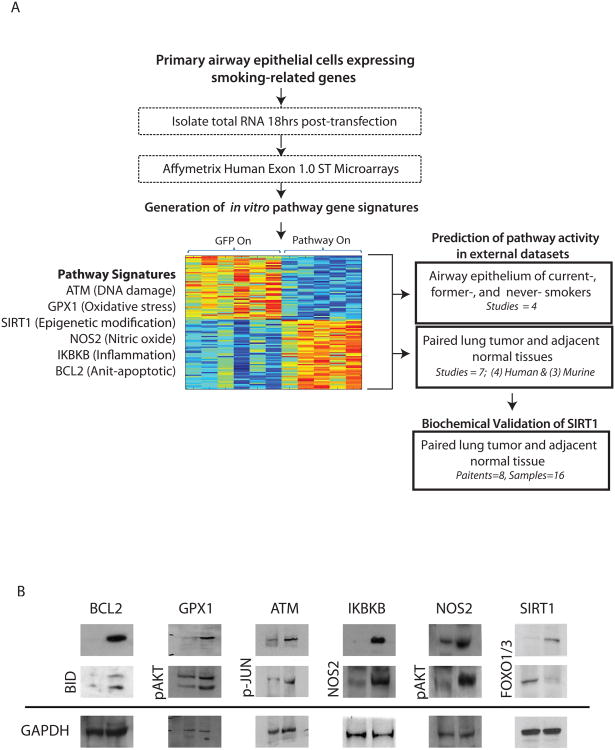
Cultures from early passage airway epithelial cells were grown using procedures previously described (17). Briefly, normal airway epithelial cells were obtained from mainstream bronchial brushings in 3 never smoke smoker volunteers and pooled in order to obtain enough viable cells for culture and subsequent development of the pathway signatures. Cells were dislodged into bronchial epithelial growth medium (BEGM), and frozen down at -80° C in media (BEGM, serum, and 10% glycerol) after centrifugation. Cells were subsequently grown on collagen-coated tissue-culture dish in BEGM. Adenoviruses were used to infect cells with constructs that express key genes from six different smoking related pathways – GPX1, NOS2, ATM, IKBKB, and SIRT1. Cells infected with an adenovirus containing GFP was used as the control, and RNA was isolated 18 hours after pathway activation, which is the time at which primary pathway targets were activated. Biochemical validation of pathway activation and relevant down-stream gene targets of each pathway was assessed using standard Western blotting (Figure 1B).
Figure 1.
A. Study design. Normal airway epithelial cells were obtained from mainstream bronchial brushings in 3 never smoke smoker volunteers. The cells were cultured and adenoviruses were used to infect cells with key genes from six different smoking related pathways – GPX1 (glutathione peroxidase 1, oxidative stress pathway), NOS2(nitric oxide synthase 2, inducible, nitric oxide pathway), ATM (ataxia telangiectasia mutated, DNA damage pathway), IKBKB (inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase beta, inflammatory pathway), SIRT1 (sirtuin 1, epigenetic modification), and BCL2 (B-cell CLL/lymphoma 2, anti-apoptotic pathway). RNA from each pathway perturbation and from a GFP control was isolated, processed, and hybridized to microarrays. Gene expression signatures specific to each pathway were generated and used to predict the pathway activation in human and murine smoking and lung cancer datasets. Dysregulation of the SIRT1 pathway was consistent and statistically significant in these datasets. Biochemical validation of the SIRT1 dysregulation in human lung tumor tissue and adjacent normal tissue was performed to confirm in silico findings. B. Biochemical validation of pathway activation. Expression of genes and their downstream effectors were assessed by Western Blot.
Hybridization to Microarrays
Total RNA isolated from multiple independent infections was processed and hybridized in 4 batches to Affymetrix Human Exon 1.0 ST microarrays (n=52) as described previously(29) (n=7 for ATM, n=8 for GPX1, n=7 for SIRT1, n=7 for NOS2, n=7 for IKBKB, n=8 for BCL2, and n=8 for GFP).
Generation of Gene Expression Signatures for each Pathway
The CEL files were processed using RMA and the ENTREZ Gene CDF file (version 11.0.1)(30). using R statistical software(31) and the aroma affymetrix package from Bioconductor. Processing the data with the ENTREZ Gene CDF file yields a single expression value for each ENTREZ Gene ID interrogated on the microarray. RMA-derived expression values were transformed to log base 2 scale for further analysis. Thirteen samples were removed either because of a low Pearson correlation between samples (n=1 SIRT1 sample) or a strong batch effect (n=2 ATM, n=2 BCL2, n=2 GFP, n=2 GPX1, n=1 IKBKB, n=2 NOS2, n=1 SIRT1 samples) as determined by PCA (Supplemental Figure 1). Gene expression signatures for each pathway were developed based on the remaining 39 samples.
Initially, for a pathway p, genes with low variance or low expression values across samples from pathway p and the GFP control were removed. Specially, genes are ranked both by their expression range and maximum expression value, and genes (n=23,536) that were among the lower 25% based on either parameter across the pathway p and GFP control samples were removed. Next, for each gene passing the variability filter, a Pearson-like correlation measure was computed to correlate gene expression across pathway p and the GFP control with a class vector (1 for pathway p samples and 0 for GFP control samples). The correlation metric ranks genes according to their differential expression between pathway p samples and the GFP control. The correlation metric, r, was calculated as follows were grp is the class vector and x is the gene expression: . Finally, an additional filter was applied to select for genes specifically differentially expressed in pathway p (i.e. not in common to other pathways). For each gene passing the variability filter in pathway p, a distance filter was created that retained pathway p genes that had the highest absolute r value compared to the other pathways and where the absolute r value was at least a distance of 0.025 greater than the absolute r value of the other pathways (see io.genetics.utah.edu/Jbeane for code). For each pathway, the top 300 genes that met the above criteria were used for statistical modeling.
To examine pathway activation across independent datasets, the gene signatures for each pathway were generated will all genes on the Affymetrix Exon 1.0 ST microarray and the intersection set of genes between each independent platforms were utilized.
Pathway Activation Probability Calculation
The expression of the gene signatures for each pathway was summarized as a single metagene by singular value decomposition across the training set of samples (samples from pathway p and GFP controls) and a binary probit regression model was estimated using Bayesian methods (see io.genetics.utah.edu/Jbeane for input files, code and output files)(32). The algorithm selects 100 genes (out of the 300genes differentially expressed for each specific pathway described above) to predict pathway activity in external samples (see Supplemental Table 1 for gene signatures derived for each microarray platform). The gene weights associated with each model were examined for outliers, and outliers were removed from the gene list of 300 to exclude them from the final signature (only NOS2 and GPX1 pathways had outliers). In order to predict the pathway activation of samples in other datasets, distance weighted discrimination (33) (DWD) was applied to correct batch effects between the pathway signature microarray samples and the samples in the new dataset. Only control samples (GFP samples) were used to correct for batch effects. For each test dataset, only genes in common to the test and training datasets were used to derive the gene signature and correct for batch effects. The regression models developed based on each gene signature were applied to samples in the new datasets. Each sample is assigned a probability representing how strongly the pathway was activated based on its gene expression pattern. Wilcoxon rank sum tests were used to assess pathway activation differences between two groups within a dataset, and a p-value < 0.05 was considered significant.
Processing of Test Microarray Datasets
The following test datasets were used to examine the behavior of each pathway in the intrathoracic and the extrathoracic epithelium and lung tissue tumor samples with paired adjacent normal tissue from non-smokers and smokers: GSE16008 (only airway samples, n=26), GSE7895, GSE10135 (subset of samples run on Affymetrix HGU133 Plus 2.0 microarray), GSE11952, GSE7670, GSE12236, GSE10072, GSE19804, GSE19188, GSE28571, GSE3141, and the Cancer Cell Line Encyclopedia (CCLE) which includes gene expression (measured by Affymetrix HGU133 Plus 2.0 microarrays) data across 180 lung cancer cell lines (991 cancer cell lines total). The following datasets were used as controls: ATM--GSE20549; GPX1—GSE23895; IKBKB—GSE18047; SIRT1--GSE9008, GSE13459, and GSE13309. The CEL files from the test datasets were processed using RMA and the ENTREZ Gene CDF file corresponding to the Affymetrix microarray used (version 11.0.1(34)) using R statistical software and the aroma affymetrix package from Bioconductor RMA-derived expression estimates were transformed to log base 2 scale for further analysis. For the CCLE data, RMA expression estimates as well as pathway predictions were computed separately for each of the 14 batches of CCLE CEL files. Correlations were computed between the CCLE pathway predictions for SIRT1 and MYC (derived as described above using data from Bild et al.(17)) and the copy number values(35). Somatic mutations in SIRT1 were assessed using the CCLE hybrid capture sequencing data(36).
Three additional murine datasets (GSE14277, GSE13963, and GSE2514) were used to examine the behavior of the SIRT1 pathway in mouse models of lung adenocarcinoma. These datasets were processed using RMA and the platform specific ENTREZ Gene CDF files version 11.0.1. The mouse ENTREZ Gene IDs were converted to human ENTREZ Gene IDs using the NCBI HomoloGene mappings. As described above, gene signatures for each pathway were generated using the intersection set of genes between microarray platforms. The pathway activation probability calculation was performed as described above except instead of DWD standardization, the standardization of the datasets is incorporated into the binary regression model as described previously (17). In this manner, the normalization of the dataset is performed based on the first principal component across datasets, and loss of information due to cross-platform analysis is minimized. In GSE14277, the phenotypes of samples GSM351070 and GSM357142 were switched when the samples were deposited in GEO (correspondence with Astrid Rohrbeck). This was corrected in the analysis of differences in pathway activation between samples.
One of the control datasets (GSE23895) used Affymetrix Rat Genome 230 2.0 microarrays. The same preprocessing and methodology used for the murine datasets was applied to the analysis of GSE23895.
All input files, code, and output files are posted at: io.genetics.utah.edu/Jbeane. The CEL files for each pathway are deposited in GEO under accession GSE37058.
Histone isolation from patient samples
Lung biopsies of patients with benign lesions or adenocarcinoma and their corresponding adjacent normal samples (n=8 patients, n=16 samples) were collected and dissociated using ultrasonication (Ultrasonic Processor Model GEX, Cole-Parmer, Vernon Hills, IL). Samples were sonicated 4 times for 5 seconds on ice at 35% amplitude and left in histone extraction buffer (Histone Purification Mini Kit, Active Motif, Carlsbad, CA) for 2 hours at 4°C. Core histones were purified and histone proteins were precipitated following the manufacturer's protocol. Total core histone proteins were quantified by measuring the absorbance at 230 nm using a spectrophotometer (NanoDrop ND-1000, Thermo Scientific, Wilmington, DE). H4K16 levels in benign lesions or tumors and their corresponding adjacent normal samples were assessed using Western blot analysis where equivalent amounts of histone proteins were resolved on 12% SDS-PAGE gels and transferred to PVDF membranes. Levels of H4K16 were detected by monoclonal primary antibody anti-acetyl histone H4 (lys16) (Millipore, Temecula, CA) and horseradish peroxidase secondary antibody anti-mouse IG (GE Healthcare, UK). Anti-Histone H3 (tri methyl K4) antibody (Abcam, Cambridge, MA) and horseradish peroxidase anti-rabbit IgG (GE healthcare, UK) were used as a loading control. Western blots were developed with SuperSignal West Dura Extended Duration Substrate (Thermo Scientific, Rockford, IL).
Results
Generation and Validation of Pathway Signatures
Past studies have uncovered signal transduction pathways activated upon exposure to cigarette smoke. These stress responses serve to protect cells from injury; however, long-term activation potentially contributes to the pathogenesis of COPD and lung cancer. Our goal is to map the critical signaling events that occur in response to smoking in a physiologically relevant setting (i.e. in vivo) and examine if these signaling events are then dysregulated upon lung cancer development. We have focused our studies on pathways related to smoke exposure and have generated gene expression profiles reflective of each pathway's activity. The key pathways are 1) GPX1, glutathione peroxidase 1, a key antioxidant gene involved in the response to oxidative stress; 2) NOS2, nitric oxide synthase 2, capable of generating nitric oxide that is involved in many immunomodulatory and anti-tumoral mechanisms; 3) ATM, ataxia telangiectasia mutated, an important cell cycle checkpoint kinase involved in DNA repair; 4) IKBKB, inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase beta, capable of inhibiting the NFKB complex activated by cytokines, oxidant-free radicals, and other stimuli; 5) BCL2, B-cell CLL/lymphoma 2, a regulator of apoptosis; and 6) SIRT1, sirtuin 1, a class III NAD+-dependent histone/protein deacetylase that regulates epigenetic gene silencing. We have leveraged publically available gene expression data of airway and lung tissue from smokers and lung cancer patients to investigate signaling events in relation to smoking and lung cancer development. Therefore, this approach enables us to simultaneously study the role of multiple signaling pathways in response to airway injury and during tumorigenesis in the host.
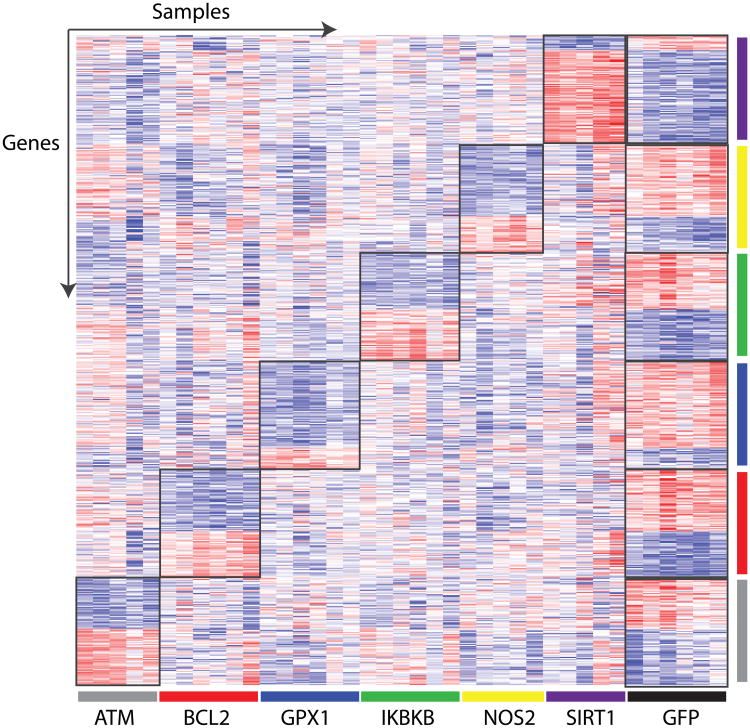
In order to generate gene expression profiles representative of a pathway's activation, the key genes listed above were perturbed in vitro in cultured primary human airway epithelial cells from never smokers. Western blot analysis of the expressed gene and of downstream targets for each of the pathways confirms pathway activation (Figure 1B). RNA from biological replicates of each pathway and control (GFP) was isolated and hybridized to microarrays. In order to isolate gene expression differences highly specific for each pathway perturbation, signatures were derived for each pathway by selecting genes differentially expressed between the pathway of interest and GFP that were not strongly differentially expressed when the other pathways were perturbed. Figure 2 shows a supervised heatmap of the expression of the genes in each pathway signature (100 genes per signature) across all of the samples demonstrating that the gene signatures are pathway specific. Pathway specific gene expression signatures were derived for each type of Affymetrix microarray platform as described in the Methods (Supplemental Table 1).
Figure 2.
Pathway specific gene expression signatures. Supervised heatmap of gene expression signatures specific to each pathway perturbation (n=100 genes/pathway). Samples are organized across the x-axis: ATM (gray), BCL2 (red), GPX1 (blue), IKBKB (green), NOS2 (yellow), SIRT1 (purple), and GFP (black). Genes are organized across the y-axis, and the sample colors also correspond to the pathway specific gene signature. Red represents high gene expression and blue represents low gene expression.
Previously published and independent datasets were used to confirm in silico that the gene expression signatures could accurately predict pathway activity (Supplemental Figure 2A-F) using our previously published approach (17, 18) This algorithm gives a probability of pathway activity based on the relationship between the pathway-specific gene expression signature and the pattern of gene expression observed in each test sample. The ATM pathway signature was tested using GSE20549 and ATM pathway activation increased in H1299 cells and transiently decreased and then restored in H460 cells (Supplemental Figure 2A). The pattern of ATM pathway activation is mirrored by the expression levels of MRE11A, ATM, ATR, p53BP1, CHK2, TOPBP1, CDC7, Claspin and CDC25C measured by Kim et al(37) demonstrating our genomic ATM profile accurately predicts ATM activity in this study. The GPX1 pathway signature was tested using GSE23895 and GPX1 pathway activation, as measured by the genomic profile for GPX1, decreased in the liver of rats fed selenium deficient diets and was highly correlated with GPX1 activity measured in the liver (Supplemental Figure 2 B) (38)). GSE18047 was used to test our IKBKB signature and IKBKB pathway activation was increased in cells infected with IKBKB as described by Demchenko et al (Supplemental Figure 2C) (39). In addition, SIRT1 pathway activation was confirmed in 3 datasets. In GSE9008, A549 cells treated with 25 microM resveratrol (n=3), a SIRT1 activator(40), had higher predicted SIRT1 pathway activity than cells treated with an ethanol control (n=1) (Supplemental Figure 2D). In GSE13459, MCF-7 cells treated with a shRNA to knockdown SIRT1 (n=3) had lower predicted SIRT pathway activity compared to a shRNA directed against Luciferase (n=3) (Supplemental Figure 2E). Finally, in GSE13309 A549 cells or Calu-6 cells were cultured with and without tobacco smoke condensate (TSC) and predicted SIRT1 pathway activation was increased in cells treated with TSC (Supplemental Figure 2F). (28). Prediction of SIRT1 activity in all three datasets using our gene expression signatures show expected patterns of pathway activation. Lastly, we also show that our approach is not sensitive to the number of genes in the signature across these control datasets (Supplemental Figure 3). In summary, the validation analyses for ATM, GPX1, IKBKB, and SIRT1 demonstrate that we were able to generate gene expression signatures that reflect the activity of pathways potentially involved in response to smoking and tumorigenesis.
Activation of smoking-related pathways in airway epithelial cells from non-smokers and smokers
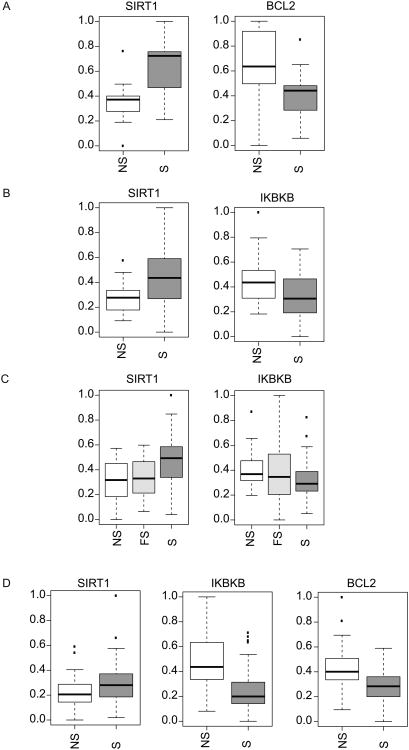
With the validated pathway signatures, we first investigated whether active exposure to tobacco smoke in vivo consistently alters the activity of these pathways. To explore patterns of pathway activation in cytologically normal airway epithelial cells from non-smokers and smokers we used 4 previously published datasets where airway samples were collected by brushing the airway during bronchoscopy. Out of the six pathways tested, SIRT1, BCL2, and IKBKB were found to have significant differential pathway activation (p<0.05) between non-smokers and smokers (Figure 3). SIRT1 activity was the most consistently and significantly up-regulated in smokers compared to non-smokers in all 4 datasets. While SIRT1 was activity correlated to smoking status, SIRT1 pathway activation was not significantly correlated with pack-years among smokers (p > 0.05; Spearman). Therefore, independent of cumulative exposure, SIRT1 activity is consistently up-regulated in smokers. This increase in SIRT1 activity may serve as a protective effect against oxidative stress and DNA damage induced by smoking.
Figure 3.
Significant differential pathway activation in airway epithelial cells between smokers and non-smokers. A. Large airway epithelial cells from non-smokers (NS, n=13) and smokers (S, n=13) (GSE16008) hybridized to microarrays. SIRT1 was the only pathway that show a significant difference in pathway activation between the NS and S samples (SIRT1 pathway activation increased in S samples, p<0.001). B. Large airway epithelial cells from non-smokers (NS, n=22) and smokers (S, n=37) from dataset GSE10135. SIRT1 showed significant pathway activation (p=0.004) and IKBKB showed significant pathway de-activation in S samples (p=0.017). C. Large airway epithelial cells from non-smokers (NS, n=21), former smokers (FS, n=31) and smokers (S, n=52) from dataset GSE7895. SIRT1 showed significant pathway activation (p=0.001) and IKBKB showed significant pathway de-activation in S samples (p=0.021). C. Small airway epithelial cells from non-smokers (NS, n=38) and smokers (S, n=45) from dataset GSE11952. SIRT1 showed significant pathway activation (p=0.044), IKBKB showed significant pathway de-activation (p<0.001), and BCL2 showed significant pathway de-activation in S samples (p<0.001).
SIRT1 pathway activation in non-small cell lung cancer
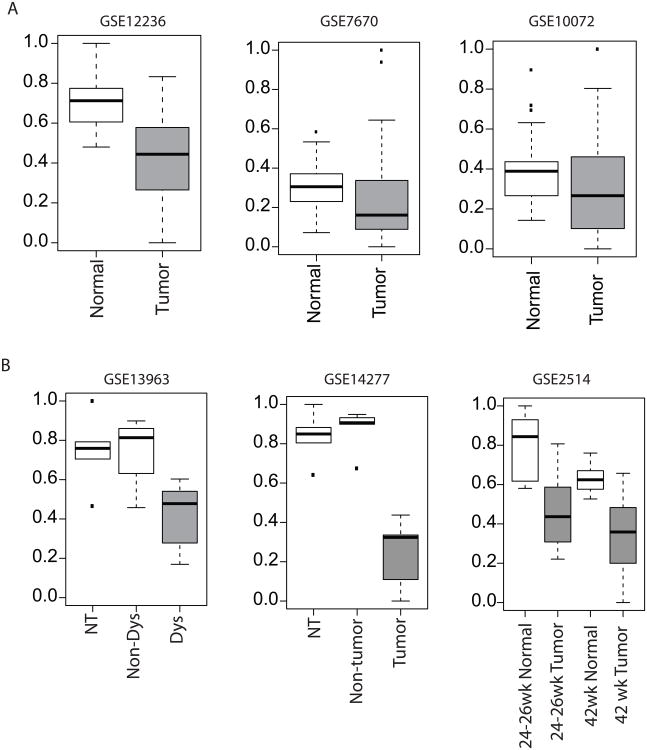
Given the elevated activity of the SIRT1 pathway in the airway epithelium of smokers, we wanted to explore SIRT1 pathway activation in smoking-related lung cancer. For this analysis, we used three previously published datasets containing lung adenocarcinoma and paired adjacent normal lung tissue. Strikingly, SIRT1 pathway activation was significantly decreased in lung tumor tissue compared to normal lung tissue in 2 of the datasets (p<0.05) and trended down in the third (p=0.062) (Figure 4A). ATM was the only other pathway that was significantly activated (p<0.05) in tumor tissue in all three lung cancer datasets. BCL was significantly activated in the tumor tissue in 2 out of 3 datasets (p<0.05) and trended down in the third (p=0.054).
Figure 4.
SIRT1 pathway activation in lung tumor tissue. Panels A and B show human datasets and panel C shows murine datasets. A. Left panel. Lung adenocarcinoma tumor tissue (n=20) and paired adjacent normal lung tissue (n=20) from dataset GSE12236. SIRT1 pathway activation was significantly decreased in lung tumor tissue (p<0.001). Middle panel. Lung adenocarcinoma tumor tissue (n=28) and paired adjacent normal lung tissue (n=28) from dataset GSE7670. SIRT1 pathway activation was significantly decreased in lung tumor tissue (p=0.033). Right panel. Lung adenocarcinoma tumor tissue (n=33) and paired adjacent normal lung tissue (n=33) from dataset GSE10072, samples without paired normal lung tissue were excluded. SIRT1 pathway activation was decreased in lung tumor tissue (p=0.062). B. Left panel. Laser microdissection pressure catapulting (LMCP) dysplastic lung epithelial cells from SP-C/c-raf-1 transgenic mice (Dys, n=5), LMCP unaltered lung epithelial cells from SP-C/c-raf-1 transgenic mice (Non-Dys, n=4), and LMCP lung epithelial cells from non-transgenic mice (NT, n=5) from dataset GSE13963. SIRT1 pathway activation was decreased in dysplastic cells versus non-dysplastic cells from transgenic mice (p=0.085). Middle panel. Laser microdissection pressure catapulting (LMCP) lung tumor epithelial cells from SP-C/c-raf transgenic mice (Tumor, n=5), LMCP unaltered lung epithelial cells from SP-C/c-raf transgenic mice (Non-Tumor, n=5), and LMCP lung epithelial cells from non-transgenic mice (NT, n=5) from dataset GSE14277. SIRT1 pathway activation was decreased in tumor cells versus non-tumor cells from transgenic mice (p=0.008). Right panel. Lung tissue from A/J mouse-urethane model of lung adenocarcinoma harvested 24-26 weeks after treatment (adjacent normal lung tissue (n=8) and tumor lung tissue (n=12)) and after 42 weeks of treatment (adjacent normal lung tissue (n=7) and tumor lung tissue at (n=17)) from dataset GSE2514. SIRT1 pathway activation was decreased in tumor tissue at both time points (p=0.002 and p=0.001, respectively).
Significant differences in SIRT1 pathway activation were not observed in a dataset of never smokers with lung cancer (primarily adenocarcinoma) and matched adjacent normal tissue (GSE19804) (Supplemental Figure 4A), and among never smokers in GSE10072 SIRT1 pathway activation trended down in lung tumor tissue, but was not significant. Additional datasets are needed to assess differences in SIRT1 signaling in never smokers. Finally, the extent of SIRT1 pathway dysregulation varies between adjacent normal and the different subtypes of non-small cell lung cancer (GSE19188, Supplemental Figure 4B) and in datasets with only tumor tissue of different subtypes (GSE10245, GSE28571, GSE3141, and CCLE dataset, Supplemental Figure 4B). Overall SIRT1 pathway activation is lower in adenocarcinomas compared to squamous cell carcinoma compared to adenocarcinoma in multiple datasets; however, the behavior of SIRT1 dysregulation in tumor subtypes compared to paired adjacent normal tissue needs to be confirmed when additional datasets become available.
Decreased SIRT1 pathway activation in lung adenocarcinoma was confirmed using mouse models of lung adenocarcinoma. Lung samples in GSE13963 and GSE14277 were obtained using a transgenic mouse model where targeted expression of c-Raf in respiratory epithelium results in dysplasia and subsequent adenocarcinoma (41, 42). Dataset GSE13963 examines differences in dysplastic epithelium compared to normal epithelium and GSE14277 compares cancer cells to normal epithelium. SIRT1 pathway activation decreased in dysplastic cells and decreased significantly in tumor cells compared to normal (p=0.085 and p=0.008, respectively, Figure 4C). Therefore, SIRT1 activity is decreased in mouse models of lung adenocarcinoma and is consistent with our finding in human disease. Additionally, in an exposure-induced mouse model (A/J mouse-urethane model) of lung adenocarcinoma (GSE2514) (43), SIRT1 pathway activation was decreased in tumor tissue compared to normal tissue at both time points sampled (p=0.002 and p=0.001 for 24-26 weeks and 42 weeks, respectively, Figure 4C). Together, these results are consistent with a loss of SIRT1 activity in the lung tumors of smokers, highlighting SIRT1's potential tumor suppressive role in lung cancer.
SIRT1 signaling is down-regulated in lung tumors
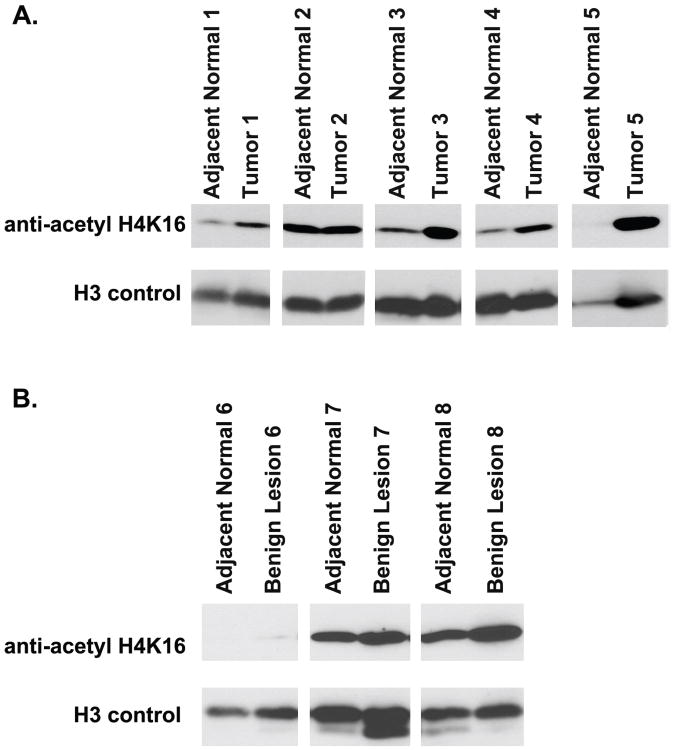
SIRT1 is recruited to chromatin by interaction with transcription factors and co-regulators, where it deacetylates specific residues on histones, including H4K16. SIRT1's recruitment to chromatin and subsequent deacetylation effects can be used as one biomarker of its activity (56-59). As shown in Figure 5, four out of five paired normal-tumor samples (all lung adenocarcinomas, see Supplemental Table 2 for patient information) show increased H4K16 acetylation, indicative of decreased histone deacetylase activity of SIRT1 in tumors. In contrast to tumor samples, we see no difference between matched normal-benign lesion samples, indicating that decreased SIRT1 activity is specific to tumor cells. Overall, these results confirm our genomic analyses, which show down-regulation of SIRT1 in lung adenocarcinomas.
Figure 5.
Western blot analysis of H4K16 acetylation in lung tissue. A. Five paired tumor and adjacent normal lung tissues. Four (1, 3, 4, and 5) samples show increased H4K16 acetylation B. Three paired benign nodules and adjacent normal lung tissue. The paired samples have similar levels of H4K16 acetylation.
Discussion
We have generated a set of gene expression signatures for pathways implicated in the response to tobacco smoke exposure. A number of external datasets were used to confirm, in silico, that these gene expression signatures could accurately predict pathway activity. Together, this signature compendium enables us to examine activity of a number of pathways potentially important for mediating the cellular response to cigarette smoke and whose dysregulation might contribute to lung cancer development. As it is difficult to comprehensively study pathway activity in human airway due to limited tissue availability, a genomic approach enabling highthroughput pathway analysis from a single sample is an important addition to the biologist's toolkit.
Four different datasets of epithelial cells obtained via brushing of the large or small airways from non-smokers and smokers undergoing bronchoscopy were used to assess differences in pathway activity in response to tobacco smoke exposure. SIRT1 was the most consistently and significantly increased in smokers compared to non-smokers across all datasets. This increase in SIRT1 activity could serve as a protective effect against oxidative stress and DNA damage induced by smoking, and supports the role of SIRT1 as a tumor suppressor. Consistent with this hypothesis, when we examined adjacent normal and lung adenocarcinoma tissue for SIRT1 pathway activation we found that SIRT1 pathway activation was decreased in the lung adenocarcinomas in all three datasets examined. SIRT1-pathway activity was also decreased in multiple mouse models of lung cancer. In humans, the decrease in SIRT1 pathway activation appears to be prevalent in smoking-associated lung adenocarcinoma, and further investigation of SIRT1 pathway activation is needed in lung tumors of never smokers as differences may be related to race, gender, or the molecularly defined subsets of cancer (presence of EGFR or KRAS mutations, etc.). Our conclusions and biochemical validation focus on adenocarcinoma, although we have evidence that SIRT1 pathway activation may vary between lung tumor cell types (Supplemental Figure 4B). Further studies are needed to explore the mechanisms leading to activation of the SIRT1 pathway in the setting of active exposure to tobacco smoke and its downstream impact on the ability of airway epithelium to mount a protective response oxidative stress and DNA damage, and to investigate SIRT1 dysregulation in lung cancer subtypes.
The function of SIRT1 is complex and its effects on various processes including stress response, metabolism, cancer, and aging is currently an active area of investigation. There are conflicting studies showing overexpression of homologues of SIRT1 in yeast, worms, and flies increase lifespan(44). Likewise, the role the SIRT1 expression in cancer remains to be elucidated with various studies indicating oncogenic and as well as tumor suppressor activity that may depend upon the status of p53(40). Several studies, especially in mouse models, support the role of SIRT1 as a tumor suppressor demonstrating that SIRT1 deficiency causes chromosomal instability and impaired DNA damage repair. Wang et al. demonstrated that Sirt1+/-; p53+/- mice develop spontaneous tumors in multiple tissues, and found that SIRT1 levels were lower in many human cancers (including the lung) compared to normal controls (45). Conversely, SIRT1-overexpressing mice had lower levels of DNA damage, decreased expression of p16Ink4a, and fewer spontaneous cancers. In addition, SIRT1-overexpressing mice in a liver cancer model were protected from metabolic syndrome associated liver carcinogenesis through increased DNA damage protection and reduced diet-induced inflammation(46). SIRT1 has also been shown to suppress intestinal tumor formation by deacetylation of beta-catenin (47).
The findings in this study are consistent with the behavior of SIRT1 as a tumor suppressor. Likewise, the decreased pathway activation of SIRT1 in lung tumor tissue compared with adjacent normal lung tissue indicates that decreased SIRT1 activity commonly accompanies and might contribute to lung carcinogenesis, at least for smoking-associated adenocarcinomas. The mechanism of SIRT1 dysregulation remains unclear as results from the Cancer Cell Line Encyclopedia (CCLE) that includes both gene expression and DNA copy number data suggest that decreases in SIRT1 pathway activation are not due to copy loss (Supplemental Figure 6). In addition, sequencing of SIRT1 in these lung cancer cell lines found coding mutations in SIRT1 in only 3 of the lung cancer cell lines with unclear functional significance. The low prevalence of SIRT1 somatic mutations in the lung cancer cell lines suggests that it is unlikely to be sole mechanism of the more widespread SIRT1 dysregulation observed in smoking and lung cancer. The decrease in SIRT1 pathway activation in lung tumor tissue was not reflected in histologically normal airway epithelial cells from current and former smokers undergoing bronchoscopy for suspicion of lung cancer (GSE4115, data not shown). Specifically, SIRT1 pathway activation did not decrease significantly in smokers with lung cancer compared to those without lung cancer. Dysregulation of SIRT1, therefore, does not appear to be an indicator in the airway epithelium of the presence of lung cancer. This result is in contrast to the pattern of PI3K pathway activation we have observed in normal airway epithelial cells from patients with lung cancer (Supplemental Figure 5) (18). Based on finding higher levels of PI3K-pathway activity in individuals with airway dysplasia, we hypothesize that increased PI3K levels may predispose individuals to lung cancer development or is deregulated early in the process of lung carcinogenesis, whereas SIRT1 activation following smoke exposure may provide a protective effect that is lost late in the process of lung carcinogenesis, only upon frank cancer development. Further study of both pathways is needed to better delineate the relative timing of their dysregulation during lung carcinogenesis.
In summary, we have successfully developed and validated pathway-specific gene expression signatures in bronchial airway epithelium that reflect activation of signaling pathways relevant to tobacco-exposure and lung disease. Application of these pathway signatures to airway and lung tissue gene-expression datasets has demonstrated dysregulation of the SIRT1 pathway in response to tobacco smoke exposure and in lung cancer tissue. These studies have provided novel insights into the molecular “field of injury” induced by tobacco smoke and thus hold the potential to impact strategies for prevention of tobacco-related lung cancer.
Supplementary Material
Acknowledgments
This study was supported by NIH/NIGMS R01 GM085601 (AHB), NIH/NCI R01CA 124640 (AS) and NIH/NIEHS U01ES016035 as part of the Genes and Environment Health Initiative (AS and AHB). We thank Frank Schembri for assistance in collecting bronchial airway cells and Jessica Vick for assistance with Figure 1A.
Footnotes
The authors have no conflicts of interest.
Author contributions: JB, AHB, and AS conceived and designed the study. JB designed and performed all microarray data analyses. MEL supported the data analysis. AB, LC, and RS generated the pathway signatures and performed the biochemical validation studies. XZ and GL performed the microarray processing. CA contributed patient samples to the study. JB, AS, and AHB wrote the manuscript.
References
- 1.Shields PG. Molecular epidemiology of lung cancer. Ann Oncol. 1999;10(5):S7–11. doi: 10.1093/annonc/10.suppl_5.s7. [DOI] [PubMed] [Google Scholar]
- 2.Spira A, Beane J, Shah V, Liu G, Schembri F, Yang X, et al. Effects of cigarette smoke on the human airway epithelial cell transcriptome. Proc Natl Acad Sci U S A. 2004;101:10143–8. doi: 10.1073/pnas.0401422101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Miyazu YM, Miyazawa T, Hiyama K, Kurimoto N, Iwamoto Y, Matsuura H, et al. Telomerase expression in noncancerous bronchial epithelia is a possible marker of early development of lung cancer. Cancer Res. 2005;65:9623–7. doi: 10.1158/0008-5472.CAN-05-0976. [DOI] [PubMed] [Google Scholar]
- 4.Guo M, House MG, Hooker C, Han Y, Heath E, Gabrielson E, et al. Promoter hypermethylation of resected bronchial margins: a field defect of changes? Clin Cancer Res. 2004;10:5131–6. doi: 10.1158/1078-0432.CCR-03-0763. [DOI] [PubMed] [Google Scholar]
- 5.Franklin WA, Gazdar AF, Haney J, Wistuba II, La Rosa FG, Kennedy T, et al. Widely dispersed p53 mutation in respiratory epithelium. A novel mechanism for field carcinogenesis. J Clin Invest. 1997;100:2133–7. doi: 10.1172/JCI119748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wistuba II, Lam S, Behrens C, Virmani AK, Fong KM, LeRiche J, et al. Molecular damage in the bronchial epithelium of current and former smokers. J Natl Cancer Inst. 1997;89:1366–73. doi: 10.1093/jnci/89.18.1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Powell CA, Klares S, O'Connor G, Brody JS. Loss of heterozygosity in epithelial cells obtained by bronchial brushing: clinical utility in lung cancer. Clin Cancer Res. 1999;5:2025–34. [PubMed] [Google Scholar]
- 8.Auerbach O, Hammond EC, Kirman D, Garfinkel L. Effects of cigarette smoking on dogs. II. Pulmonary neoplasms. Arch Environ Health. 1970;21:754–68. doi: 10.1080/00039896.1970.10667329. [DOI] [PubMed] [Google Scholar]
- 9.Hackett NR, Heguy A, Harvey BG, O'Connor TP, Luettich K, Flieder DB, et al. Variability of antioxidant-related gene expression in the airway epithelium of cigarette smokers. Am J Respir Cell Mol Biol. 2003;29:331–43. doi: 10.1165/rcmb.2002-0321OC. [DOI] [PubMed] [Google Scholar]
- 10.Harvey BG, Heguy A, Leopold PL, Carolan BJ, Ferris B, Crystal RG. Modification of gene expression of the small airway epithelium in response to cigarette smoking. J Mol Med. 2007;85:39–53. doi: 10.1007/s00109-006-0103-z. [DOI] [PubMed] [Google Scholar]
- 11.Spira A, Beane JE, Shah V, Steiling K, Liu G, Schembri F, et al. Airway epithelial gene expression in the diagnostic evaluation of smokers with suspect lung cancer. Nat Med. 2007;13:361–6. doi: 10.1038/nm1556. [DOI] [PubMed] [Google Scholar]
- 12.Beane J, Sebastiani P, Liu G, Brody JS, Lenburg ME, Spira A. Reversible and permanent effects of tobacco smoke exposure on airway epithelial gene expression. Genome Biol. 2007;8:R201. doi: 10.1186/gb-2007-8-9-r201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schembri F, Sridhar S, Perdomo C, Gustafson AM, Zhang X, Ergun A, et al. MicroRNAs as modulators of smoking-induced gene expression changes in human airway epithelium. Proc Natl Acad Sci U S A. 2009;106:2319–24. doi: 10.1073/pnas.0806383106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chari R, Lonergan KM, Ng RT, Macaulay C, Lam WL, Lam S. Effect of active smoking on the human bronchial epithelium transcriptome. BMC Genomics. 2007;8:297. doi: 10.1186/1471-2164-8-297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Beane J, Vick J, Schembri F, Anderlind C, Gower A, Campbell J, et al. Characterizing the impact of smoking and lung cancer on the airway transcriptome using RNA-Seq. Cancer Prev Res (Phila) 2011;4:803–17. doi: 10.1158/1940-6207.CAPR-11-0212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Beane J, Sebastiani P, Whitfield TH, Steiling K, Dumas YM, Lenburg ME, et al. A prediction model for lung cancer diagnosis that integrates genomic and clinical features. Cancer Prev Res (Phila) 2008;1:56–64. doi: 10.1158/1940-6207.CAPR-08-0011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bild AH, Yao G, Chang JT, Wang Q, Potti A, Chasse D, et al. Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature. 2006;439:353–7. doi: 10.1038/nature04296. [DOI] [PubMed] [Google Scholar]
- 18.Gustafson AM, Soldi R, Anderlind C, Scholand MB, Qian J, Zhang X, et al. Airway PI3K pathway activation is an early and reversible event in lung cancer development. Sci Transl Med. 2010;2:26ra25. doi: 10.1126/scitranslmed.3000251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tanaka T, Huang X, Jorgensen E, Gietl D, Traganos F, Darzynkiewicz Z, et al. ATM activation accompanies histone H2AX phosphorylation in A549 cells upon exposure to tobacco smoke. BMC Cell Biol. 2007;8:26. doi: 10.1186/1471-2121-8-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang P, Bamlet WR, Ebbert JO, Taylor WR, de AM. Glutathione pathway genes and lung cancer risk in young and old populations. Carcinogenesis. 2004;25:1935–44. doi: 10.1093/carcin/bgh203. [DOI] [PubMed] [Google Scholar]
- 21.Jorgensen ED, Zhao H, Traganos F, Albino AP, Darzynkiewicz Z. DNA damage response induced by exposure of human lung adenocarcinoma cells to smoke from tobacco- and nicotine-free cigarettes. Cell Cycle. 2010;9:2170–6. doi: 10.4161/cc.9.11.11842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jin Z, Gao F, Flagg T, Deng X. Tobacco-specific nitrosamine 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone promotes functional cooperation of Bcl2 and c-Myc through phosphorylation in regulating cell survival and proliferation. J Biol Chem. 2004;279:40209–19. doi: 10.1074/jbc.M404056200. [DOI] [PubMed] [Google Scholar]
- 23.Gresner P, Gromadzinska J, Jablonska E, Kaczmarski J, Wasowicz W. Expression of selenoprotein-coding genes SEPP1, SEP15 and hGPX1 in non-small cell lung cancer. Lung Cancer. 2009;65:34–40. doi: 10.1016/j.lungcan.2008.10.023. [DOI] [PubMed] [Google Scholar]
- 24.Kanwar JR, Kanwar RK, Burrow H, Baratchi S. Recent advances on the roles of NO in cancer and chronic inflammatory disorders. Curr Med Chem. 2009;16:2373–94. doi: 10.2174/092986709788682155. [DOI] [PubMed] [Google Scholar]
- 25.Dwyer-Nield LD, Srebernak MC, Barrett BS, Ahn J, Cosper P, Meyer AM, et al. Cytokines differentially regulate the synthesis of prostanoid and nitric oxide mediators in tumorigenic versus non-tumorigenic mouse lung epithelial cell lines. Carcinogenesis. 2005;26:1196–206. doi: 10.1093/carcin/bgi061. [DOI] [PubMed] [Google Scholar]
- 26.Comhair SA, Thomassen MJ, Erzurum SC. Differential induction of extracellular glutathione peroxidase and nitric oxide synthase 2 in airways of healthy individuals exposed to 100% O(2) or cigarette smoke. Am J Respir Cell Mol Biol. 2000;23:350–4. doi: 10.1165/ajrcmb.23.3.4076. [DOI] [PubMed] [Google Scholar]
- 27.Akca H, Demiray A, Tokgun O, Yokota J. Invasiveness and anchorage independent growth ability augmented by PTEN inactivation through the PI3K/AKT/NFkB pathway in lung cancer cells. Lung Cancer. 2011;73:302–9. doi: 10.1016/j.lungcan.2011.01.012. [DOI] [PubMed] [Google Scholar]
- 28.Hussain M, Rao M, Humphries AE, Hong JA, Liu F, Yang M, et al. Tobacco smoke induces polycomb-mediated repression of Dickkopf-1 in lung cancer cells. Cancer Res. 2009;69:3570–8. doi: 10.1158/0008-5472.CAN-08-2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang X, Liu G, Lenburg ME, Spira A. Comparison of smoking-induced gene expression on Affymetrix Exon and 3′-based expression arrays. Genome Inform. 2007;18:247–57. [PubMed] [Google Scholar]
- 30.ENTREZ CDF File. 2012 http://brainarray.mbni.med.umich.edu/Brainarray/Database/CustomCDF/11.0.1/entrezg.download/HuEx10stv2_Hs_ENTREZG_11.0.1.zip. Ref Type: Online Source.
- 31.R statistical software. 2012 http://www.r-project.org/ Ref Type: Online Source.
- 32.Cohen AL, Soldi R, Zhang H, Gustafson AM, Wilcox R, Welm BE, et al. A pharmacogenomic method for individualized prediction of drug sensitivity. Mol Syst Biol. 2011;7:513. doi: 10.1038/msb.2011.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Benito M, Parker J, Du Q, Wu J, Xiang D, Perou CM, et al. Adjustment of systematic microarray data biases. Bioinformatics. 2004;20:105–14. doi: 10.1093/bioinformatics/btg385. [DOI] [PubMed] [Google Scholar]
- 34.Custom CDF Files. 2012 http://brainarray.mbni.med.umich.edu/Brainarray/Database/CustomCDF/genomic_curated_CDF.asp. Ref Type: Online Source.
- 35.CCLE Copy Number Data. 2012 http://www.broadinstitute.org/ccle/downloadFile/DefaultSystemRoot/exp_10/ds_20/CCLE_copynumber_byGene_2012-04-06.txt.gz?downloadff=true&fileId=3041. Ref Type: Online Source.
- 36.CCLE hybrid capture Data. 2012 http://www.broadinstitute.org/ccle/downloadFile/DefaultSystemRoot/exp_10/ds_26/CCLE_hybrid_capture1650_hg19_NoCommonSNPs_NoNeutralVariants_CDS_2012.05.07.maf?downloadff=true&fileId=4225. Ref Type: Online Source.
- 37.Kim KH, Yoo HY, Joo KM, Jung Y, Jin J, Kim Y, et al. Time-course analysis of DNA damage response-related genes after in vitro radiation in H460 and H1229 lung cancer cell lines. Exp Mol Med. 2011;43:419–26. doi: 10.3858/emm.2011.43.7.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Raines AM, Sunde RA. Selenium toxicity but not deficient or super-nutritional selenium status vastly alters the transcriptome in rodents. BMC Genomics. 2011;12:26. doi: 10.1186/1471-2164-12-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Demchenko YN, Glebov OK, Zingone A, Keats JJ, Bergsagel PL, Kuehl WM. Classical and/or alternative NF-kappaB pathway activation in multiple myeloma. Blood. 2010;115:3541–52. doi: 10.1182/blood-2009-09-243535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brooks CL, Gu W. How does SIRT1 affect metabolism, senescence and cancer? Nat Rev Cancer. 2009;9:123–8. doi: 10.1038/nrc2562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rohrbeck A, Muller VS, Borlak J. Molecular characterization of lung dysplasia induced by c-Raf-1. PLoS One. 2009;4:e5637. doi: 10.1371/journal.pone.0005637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rohrbeck A, Borlak J. Cancer genomics identifies regulatory gene networks associated with the transition from dysplasia to advanced lung adenocarcinomas induced by c-Raf-1. PLoS One. 2009;4:e7315. doi: 10.1371/journal.pone.0007315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stearman RS, Dwyer-Nield L, Zerbe L, Blaine SA, Chan Z, Bunn PA, Jr, et al. Analysis of orthologous gene expression between human pulmonary adenocarcinoma and a carcinogen-induced murine model. Am J Pathol. 2005;167:1763–75. doi: 10.1016/S0002-9440(10)61257-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Herranz D, Serrano M. SIRT1: recent lessons from mouse models. Nat Rev Cancer. 2010;10:819–23. doi: 10.1038/nrc2962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang RH, Sengupta K, Li C, Kim HS, Cao L, Xiao C, et al. Impaired DNA damage response, genome instability, and tumorigenesis in SIRT1 mutant mice. Cancer Cell. 2008;14:312–23. doi: 10.1016/j.ccr.2008.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Herranz D, Munoz-Martin M, Canamero M, Mulero F, Martinez-Pastor B, Fernandez-Capetillo O, et al. Sirt1 improves healthy ageing and protects from metabolic syndrome-associated cancer. Nat Commun. 2010;1:3. doi: 10.1038/ncomms1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Firestein R, Blander G, Michan S, Oberdoerffer P, Ogino S, Campbell J, et al. The SIRT1 deacetylase suppresses intestinal tumorigenesis and colon cancer growth. PLoS One. 2008;3:e2020. doi: 10.1371/journal.pone.0002020. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.