Abstract
The nuclear envelope links the cytoskeleton to structural components of the nucleus. It functions to coordinate nuclear migration and anchorage, organize chromatin, and aid meiotic chromosome pairing. Forces generated by the cytoskeleton are transferred across the nuclear envelope to the nuclear lamina through a nuclear-envelope bridge consisting of SUN (Sad1 and UNC-84) and KASH (Klarsicht, ANC-1 and Syne/Nesprin homology) proteins. Some KASH-SUN combinations connect microtubules, centrosomes, actin filaments, or intermediate filaments to the surface of the nucleus. Other combinations are used in cell cycle control, nuclear import, or apoptosis. Interactions between the cytoskeleton and the nucleus also affect global cytoskeleton organization. SUN and KASH proteins were identified through genetic screens for mispositioned nuclei in model organisms. Knockouts of SUN or KASH proteins disrupt neurological and muscular development in mice. Defects in SUN and KASH proteins have been linked to human diseases including muscular dystrophy, ataxia, progeria, lissencephaly, and cancer.
Keywords: nuclear positioning, outer nuclear membrane, laminopathies, microtubule motor targeting, Syne, Nesprin
INTRODUCTION
The nuclear envelope is a specialized extension of the endoplasmic reticulum (ER) that compartmentalizes the genome. It also provides physical rigidity to the nucleus, organizes chromatin, functions in meiotic chromosome pairing, and positions the nucleus within the cell. Interactions between the cytoskeleton and the nuclear envelope are central to all of these functions. The same molecular mechanisms are used in these processes to transfer forces generated in the cytoskeleton to the structural components inside the nucleus. Here we review how the nucleus interacts with the cytoskeleton, how forces are transferred across the nuclear envelope, and how these processes relate to human disease.
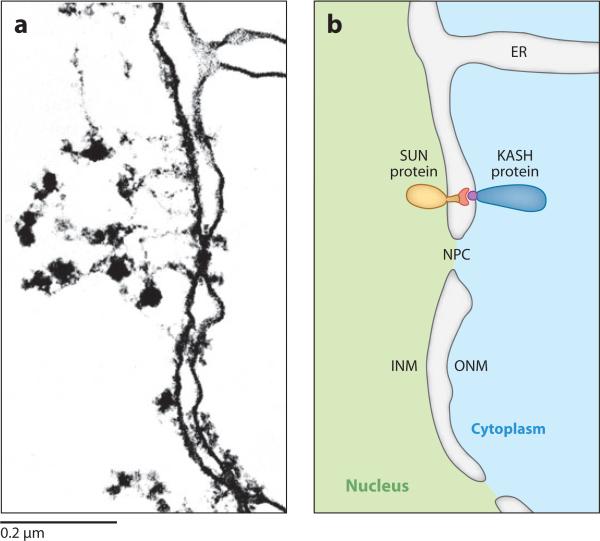
The nuclear envelope consists of two parallel membranes, the inner nuclear membrane (INM) and outer nuclear membrane (ONM), that are contiguous at the nuclear pore (Figure 1a). The ONM is also contiguous with the ER (reviewed in Franke et al. 1981). The membranes of the nuclear envelope are supported by the nuclear lamina that immediately underlies the INM. In metazoans, the lamina consists of membrane proteins, the intermediate filament lamin, and proteins associated with chromatin (reviewed in Gruenbaum et al. 2005). To form a nuclear-envelope bridge, membrane proteins are trafficked from the ER membrane either to the INM to interact with the lamina, or to the ONM to interact with the cytoskeleton. To complete the bridge, INM and ONM proteins interact with each other in the perinuclear space. The current KASHSUN nuclear-envelope bridging model, also referred to as the linker of nucleoskeleton and cytoskeleton (LINC) complex model, posits that SUN (Sad1 and UNC-84) proteins in the INM interact with KASH (Klarsicht, ANC-1, and Syne/Nesprin homology) proteins of the ONM to span the nuclear envelope (Figure 1b).
Figure 1.
Klarsicht, ANC-1, and Syne/Nesprin homology (KASH) and Sad1 and UNC-84 (SUN) proteins bridge the two membranes of the nuclear envelope. (a) A transmission electron microscopy (TEM) image and (b) a schematic representation of the nuclear envelope in an amphibian oocyte showing how the inner nuclear membrane (INM), outer nuclear membrane (ONM), and endoplasmic reticulum (ER) are contiguous. NPC, nuclear pore complex. A generic SUN and KASH protein bridge is drawn with conserved SUN (red ) and KASH (purple) domains in the perinuclear space of the nuclear envelope. TEM image reproduced with permission from J. Cell Biol. (Franke et al. 1981).
DISCOVERY OF KASH AND SUN PROTEINS AND THE FORMATION OF THE NUCLEAR-ENVELOPE BRIDGE
Discovery of SUN Proteins
Our current understanding of the molecular mechanisms of the SUN-KASH nuclear-envelope bridge is based on characterizations of genetic mutants in Caenorhabditis elegans and Drosophila with mispositioned nuclei (reviewed in Starr & Fischer 2005, Starr & Han 2005, Wilhelmsen et al. 2006). SUN proteins were discovered by molecular analysis of C. elegans unc-84. Alleles of unc-84 were originally isolated because of their defects in P cell nuclear migration, which results in missing neurons and vulval cells, leading to uncoordinated and egg-laying defective phenotypes (Figure 2a, Horvitz & Sulston 1980, Sulston & Horvitz 1981). unc-84 alleles also cause nuclear migration defects in epidermal precursors (Figure 2b, Horvitz & Sulston 1980, Sulston & Horvitz 1981). The molecular cloning of UNC-84 (Malone et al. 1999) in effect founded the SUN-KASH field.
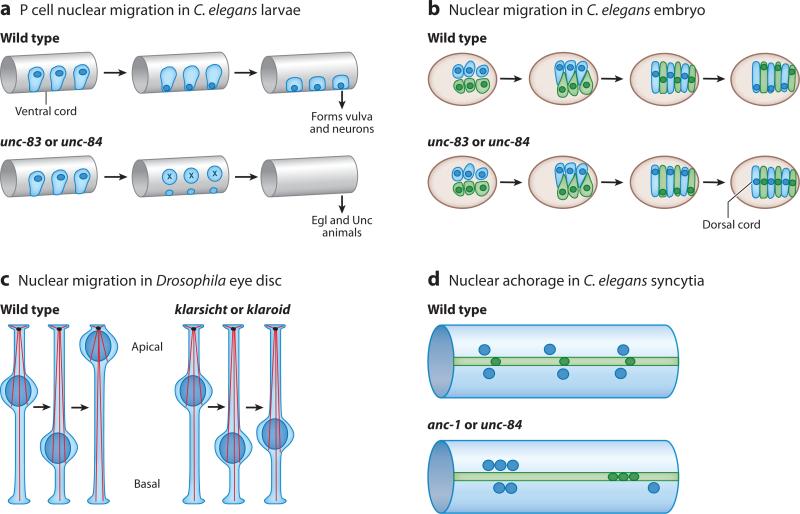
Figure 2.
Genetic models for studying nuclear positioning. (a) Nuclear migration in hypodermal P cells during the first larval stage of Caenorhabditis elegans. Nuclei (dark blue) migrate from a lateral to a ventral position through the cytoplasm (light blue). P cells normally go on to form the vulva and neurons in the ventral cord. Mutations in unc-83 or unc-84 disrupt nuclear migration and the P cells die, resulting in egg-laying defective (Egl) and uncoordinated (Unc) animals. (b) Nuclear migration in C. elegans embryonic hypodermal cells. Right (light blue) and left (light green) hyp7 precursors align along the dorsal cord of a pre-elongation embryo (brown circle, dorsal view, anterior to the left) and intercalate to form a row of column-shaped cells spanning the dorsal midline. Nuclei (dark blue and dark green) then migrate the length of the cell from right to left (blue) or left to right (green). Mutations in unc-83 and unc-84 disrupt nuclear migration, and all nuclei end up in the dorsal cord instead of their normal, lateral positions. (c) Nuclear migration in the Drosophila eye disc. After the morphogenetic furrow passes, nuclei (dark blue) migrate from a basal position to an apical one as they develop into photoreceptors. In klarsicht or klaroid mutants, nuclei abnormally remain basal, disrupting the development of the eye, whereas centrosomes (black ovals) organize microtubules (red strands) from an apical position. (d ) Nuclear anchorage in the adult C. elegans syncytial hypodermis. Three large hypodermal syncytia, the hyp7 (light blue) syncytium and the lateral seam-cell syncytia (light green), are used to study nuclear anchorage. Normally nuclei (dark blue and dark green) are anchored and spaced evenly apart. In anc-1 or unc-84 mutant animals, nuclei are unanchored and often associate in clusters.
The C terminus of UNC-84 was found to be conserved with the C termini of Schizosaccharomyces pombe Sad1 and two human proteins; each contains a ~175 amino acid domain termed the SUN domain (Malone et al. 1999). Sad1 is an essential component of the spindle pole body required for normal spindle architecture (Hagan & Yanmagida 1995). Overex-pressed Sad1::green fluorescent protein (GFP) accumulates at the nuclear envelope, suggesting an additional role for Sad1 in nuclear positioning during interphase (Goshima et al. 1999, Tran et al. 2001). UNC-84 also localizes to the nuclear envelope (Lee et al. 2002, Malone et al. 1999). Proteins with SUN domains are referred to as SUN proteins and localize to the INM.
SUN Proteins Constitute the Inner Nuclear Membrane Half of the Nuclear-Envelope Bridge
SUN proteins are conserved across eukaryotes including fungi, plants, animals, and basal eukaryotes such as Giardia (Graumann et al. 2009, Jaspersen et al. 2006, Malone et al. 1999, Mans et al. 2004, Starr 2009). Many higher eukaryotes have multiple SUN proteins that are often expressed at different times in development (Crisp et al. 2006, Ding et al. 2007, Graumann et al. 2009, Hodzic et al. 2004, Kracklauer et al. 2007, Lu et al. 2008, Malone et al. 2003, Padmakumar et al. 2005, Shao et al. 1999). SUN proteins have many common features. They have at least one trans-membrane domain that enables most to localize in the nuclear membrane (reviewed in Starr 2009, Tzur et al. 2006b, Worman & Gundersen 2006). Specifically, data suggest that SUN proteins are components of the INM with the SUN domain in the perinuclear space (Chikashige et al. 2006, Crisp et al. 2006, Haque et al. 2006, Hodzic et al. 2004, Jaspersen et al. 2002, McGee et al. 2006, Padmakumar et al. 2005). Most SUN proteins contain short coiled-coil regions in their perinuclear domains that aid in dimerization or multimerization (Crisp et al. 2006, Haque et al. 2006, Lu et al. 2008, Malone et al. 1999).
The nucleoplasmic domains of SUN proteins are not conserved. Although the N-terminal nucleoplasmic domains contain the signals needed for nuclear envelope localization, how they are targeted to the nucleoplasm is not understood (Haque et al. 2010, Hasan et al. 2006, Hodzic et al. 2004). Some SUN proteins contain classical nuclear localization signals, but mutating the mammalian Sun1 classical nuclear localization signal has no effect on nuclear envelope localization (Hodzic et al. 2004). Other SUN proteins have predicted INM-sorting motifs (Braunagel et al. 2004), but their function in targeting to the INM remains to be determined. Many SUN proteins interact with lamins, but only some SUN proteins require a functional lamin for localization (Fridkin et al. 2004; Haque et al. 2006, 2010; Hasan et al. 2006; Lee et al. 2002). Once in the nucleoplasm, the N termini of SUN proteins likely interact with the lamina or chromatin and have been hypothesized to regulate gene expression (Chi et al. 2007, King et al. 2008, Oza et al. 2009, Wang et al. 2009). In S. pombe, which has no lamins, the SUN protein Sad1 interacts with centromeres and heterochromatin in a network that is stabilized by the INM protein Ima1 (NET5, for nuclear envelope targeted, in mammals) (King et al. 2008).
The evidence supports a model in which SUN proteins form the INM half of a nuclear-envelope bridge (Figures 1 and 3). In this role, SUN proteins anchor the bridge to the nuclear lamina. Central to this model is that SUN proteins also recruit the second half of the bridge, KASH proteins, to the ONM.
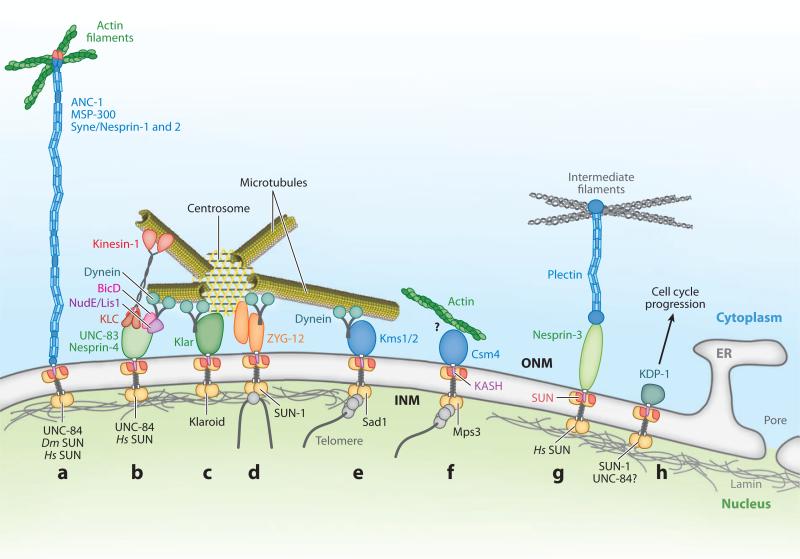
Figure 3.
Functions of KASH-SUN nuclear-envelope bridges. KASH proteins are in the outer nuclear membrane (ONM). SUN protein dimers (gold circles) are located in the inner nuclear membrane (INM) with their SUN domains (red ) in the perinuclear space, where they interact with KASH domains (purple) to bridge the nuclear envelope. (a) Giant KASH proteins (blue) tether nuclei to actin filaments (green). (b) UNC-83 and Nesprin-4 (green) function as nucleus-specific cargo adaptors for microtubule motors kinesin-1 (red ) through the kinesin light chain (KLC, dark red ) and, at least for UNC-83, dynein (teal) through BicD and NudE/Lis1 complexes (pink and purple). (c) Klarsicht (Klar, green) is thought to interact with dynein for nuclear migration. (d-f ) Nucleoplasmic domains of SUN proteins interact with meiotic chromosomes (gray lines) through adaptors (gray circles) to aid in proper homolog pairing. (d ) Worm ZYG-12 (orange) interacts with a KASH-less isoform of itself and dynein to tether the centrosome to the nucleus, to position the nucleus, and to move chromosomes in meiosis. (e) Fission yeast Kms1 and 2 (blue) recruit dynein to move nuclei and telomeres in meiosis. (f) Budding yeast Csm4 (blue) links actin filaments to the nucleus to move telomeres through unknown intermediates (?). (g) Nesprin-3 interacts with intermediate filaments (gray) through plectin (blue). (h) Worm KDP-1 promotes cell-cycle progression. Dm, Drosophila melanogaster; Hs, Homo sapiens.
Discovery of KASH Proteins
Drosophila Klarsicht, C. elegans ANC-1, and the mammalian paralogs Syne/Nesprin-1 and -2 are the founding KASH (Klarsicht, ANC-1, and Syne homology) proteins. The molecular cloning of ANC-1 led to the recognition that all of these proteins contain a conserved C-terminal KASH domain (Starr & Han 2002).
Mutations in Drosophila klarsicht (originally called marbles) were isolated in screens for eye morphogenesis defects (Fischer-Vize & Mosley 1994). Normally nuclei migrate basally in the pseudostratified neuroepithelium of the developing eye disc before migrating apically to differentiate (Tomlinson 1985). In klarsicht mutant animals, nuclei remain basal although centrosomes are in their normal apical position (Figure 2c, Fischer-Vize & Mosley 1994, Mosley-Bishop et al. 1999, Patterson et al. 2004). Klarsicht localization to the nuclear envelope requires lamin and the SUN protein Klaroid (Kracklauer et al. 2007, Patterson et al. 2004). Klarsicht has been proposed to coordinate the activity of microtubule motors during nuclear migration (Figure 3c; Patterson et al. 2004, Welte 2004, Welte et al. 1998).
Once a nucleus migrates to the proper position in the cell, mechanisms must exist to anchor it in place. Most of an adult C. elegans is covered with four large hypodermal syncytia containing more than 100 nuclei in total. Normally these nuclei are evenly spaced and anchored in place. However, in anc-1 or unc-84 mutant animals, nuclei float freely in the cytoplasm, often associating in large clusters (Figures 2d and 4a, Hedgecock & Thomson 1982, Malone et al. 1999). ANC-1 is a giant protein of more than 8500 residues with homology to Drosophila MSP-300, a protein required for muscle development, and mammalian Syne/Nesprin-1 and -2 (Starr & Han 2002). Like Klarsicht, ANC-1 localizes to the nuclear envelope via a SUN protein, UNC-84 (Starr & Han 2002).
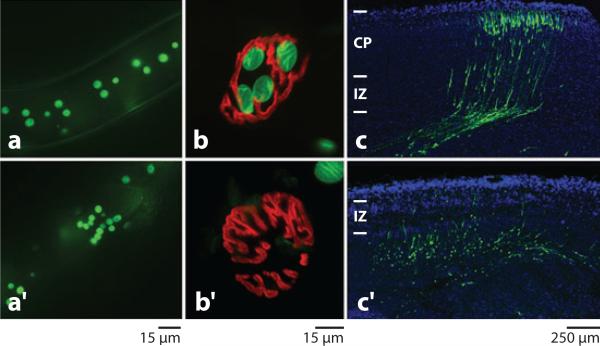
Figure 4.
ANC-1 and Syne/Nesprin-1 and -2 tether nuclei to the actin cytoskeleton. (a) In wild-type adult C. elegans, large syncytial nuclei [green fluorescent protein (GFP) positive] are evenly spaced. (a′ ) In an anc-1 mutant animal, nuclei are unanchored and are pushed around by underlying tissues and frequently cluster together. (b) A mouse neuromuscular junction (NMJ) (red ) from a heterozygous control with four muscle nuclei (green) clustered underneath. (b′ ) In a Syne/Nesprin-1 KASH knockout mouse, nuclei fail to anchor underneath the NMJ. Reproduced with permission from Development (Zhang et al. 2007b). (c) A coronal section of an E18.5 heterozygous control mouse showing neuronal migration. Green cells were transfected with a GFP construct at E14.5. In that time, neurons migrated through the intermediate zone (IZ) to the cortical plate (CP). (c′ ) In an analogously prepared slice of a Syne/Nesprin-1 and -2 homozygous double KASH knockout brain, most nuclear migrations failed. Adapted with permission from Neuron (Zhang et al. 2009).
Mammalian Syne/Nesprin-1 was first cloned as a component of the postsynaptic apparatus in the neuromuscular junction as a two-hybrid interacting partner of the muscle-specific receptor tyrosine kinase (MuSK) and named Syne-1 (synaptic nuclear envelope-1) (Apel et al. 2000). Subsequently, Syne/Nesprin-1 and/or -2 were isolated as markers of vascular smooth muscle cells and termed Nesprin (nuclear envelope spectrin repeat) (Zhang et al. 2001), as novel spectrin repeat-containing proteins called Myne (myocyte nuclear envelope) (Mislow et al. 2002b), by homology to C. elegans ANC-1 (Starr & Han 2002), as a Golgi complex–specific spectrin protein (Gough et al. 2003), and as a plasticity-related protein called CPG2 (Cottrell et al. 2004). The giant, full-length Syne/Nesprin-1 and -2 transcripts (Enaptin and NUANCE) were identified by their N-terminal actin-binding domains having similarity to α-actinin (Padmakumar et al. 2004, Zhen et al. 2002). Multiple antibodies against Syne/Nesprin-1 and -2 localize to the nuclear envelope (Apel et al. 2000, Mislow et al. 2002b, Zhang et al. 2001, Zhang et al. 2007b, Zhen et al. 2002), and this localization is dependent on SUN proteins (Crisp et al. 2006, Lei et al. 2009, Padmakumar et al. 2005).
KASH Proteins Interact with SUN Proteins to Complete the Nuclear-Envelope Bridge
KASH proteins have multiple common features (reviewed in Starr & Fischer 2005). First, they contain a C-terminal conserved KASH domain consisting of a membrane-spanning region followed by fewer than 35 residues before the C terminus. The KASH domains of the founding members of the family are highly similar, but other, more distantly related KASH domains have been identified (McGee et al. 2009, Starr 2009). KASH proteins are likely tail-anchored proteins inserted into the ER membrane post-translationally by Asna1-GET3 (Mateja et al. 2009, Rabu et al. 2009, Schuldiner et al. 2008, Stefanovic & Hegde 2007). Second, KASH proteins localize to the ONM with their conserved KASH domain inserted into the perinuclear lumen and their large, divergent N-terminal domains in the cytoplasm. A KASH domain is necessary and sufficient to target a protein to the ONM (Fischer et al. 2004, Grady et al. 2005, Guo et al. 2005, McGee et al. 2006, Meyerzon et al. 2009a, Starr & Han 2002, Zhang et al. 2001, Zhang et al. 2007b). All known integral membrane proteins that localize specifically to the ONM are KASH proteins (Figure 3 and Supplemental Table 1; follow the Supplemental Material link from the Annual Reviews home page at http://www.annualreviews.org).
A valuable tool for studying KASH proteins is a dominant-negative construct overex-pressing a KASH domain. When the ANC-1 KASH domain is overexpressed in C. elegans, endogenous ANC-1 is displaced from the nuclear envelope, causing a strong nuclear positioning phenotype (Starr & Han 2002). Analogous approaches have been used to study vertebrate Syne/Nesprin-1 and -2 (Grady et al. 2005, Tsujikawa et al. 2007). However, the approach is nonspecific, and all KASH proteins are likely displaced from the ONM, which makes it difficult to assign the resulting phenotypes to a single KASH protein.
Central to the nuclear-envelope bridge model is that KASH proteins require SUN proteins for localization to the ONM (Crisp et al. 2006, Lei et al. 2009, Padmakumar et al. 2005, Starr & Han 2002). In support of this genetic requirement, KASH and SUN domains directly interact (Crisp et al. 2006, Haque et al. 2010, McGee et al. 2006, Minn et al. 2009, Ostlund et al. 2009, Padmakumar et al. 2005, Stewart-Hutchinson et al. 2008). Once the nuclear-envelope bridge is formed, the divergent cytoplasmic domains of KASH proteins are localized to the outer surface of the nuclear envelope, where they perform a variety of functions.
KASH-SUN interactions are dynamic. For example, the SUN protein UNC-84 functions with the KASH protein ANC-1 during nuclear anchorage, but then functions through UNC-83 during nuclear migration (Starr & Han 2002, Starr et al. 2001). The best candidate for a regulator of KASH-SUN interactions is the chap-eronin TorsinA (Naismith et al. 2004). TorsinA localizes to the ER and perinuclear lumens and physically interacts with KASH domains (Goodchild & Dauer 2004, Nery et al. 2008). Furthermore, changing the amount of TorsinA at the nuclear envelope displaces KASH-SUN complexes and alters the structure of the nuclear envelope (Naismith et al. 2004, Nery et al. 2008, Vander Heyden et al. 2009).
FUNCTIONS OF KASH PROTEINS AT THE OUTER NUCLEAR MEMBRANE IN POSITIONING NUCLEI
Anchoring Nuclei to the Actin Cytoskeleton
The giant proteins C. elegans ANC-1 (8546 residues for the largest predicted isoform), Drosophila MSP-300 (8204), and mammalian Syne/Nesprin-1 and -2 (8739 and 6885) are orthologs (Starr & Han 2002, Zhang et al. 2002). Dictyostelium Interaptin, although smaller and less similar, might also be an ortholog (Rivero et al. 1998). All of them contain a highly similar C-terminal KASH domain that is sufficient for targeting to the ONM (Starr & Han 2002, Yu et al. 2006, Zhang et al. 2001). Additionally, their N termini are homologous with calponin, bind actin in vitro, and colocalize with actin filaments (Starr & Han 2002, Volk 1992, Zhen et al. 2002). Finally, they have extended middle domains that at least in the cases of MSP-300 and Syne/Nesprin-1 and -2 are related to spectrin, making these proteins related to dystrophin (Starr & Han 2003).
Phenotypic analyses demonstrate that C. elegans ANC-1 and mouse Syne/Nesprin-1 and -2 anchor nuclei within the cell (Figure 4a,b; Starr & Han 2002, Zhang et al. 2007b). The role of MSP-300 in nuclear anchorage is not clear (Technau & Roth 2008, Xie & Fischer 2008, Yu et al. 2006). These large KASH proteins are thought to function as ropes, extending away from the nuclear envelope to tether actin filaments to the ONM (Figure 3a). An alternative hypothesis is that Syne/Nesprin-1 and -2 form a spectrin-like filamentous basket surrounding and supporting the ONM (Schneider et al. 2008).
Centrosome Attachment to the Nuclear Envelope during Pronuclear Migration
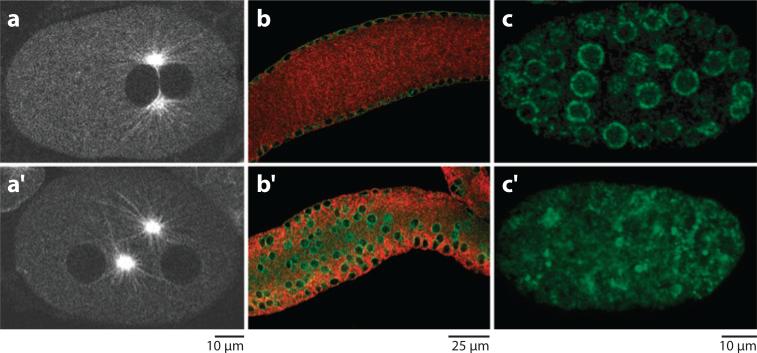
In most cells, at least during part of the cell cycle, centrosomes are closely associated with the nuclear envelope (Bornens 1977, Starr 2009). However, little is known about how this interaction is maintained. The KASH protein ZYG-12 and the SUN protein SUN-1/matefin are essential for pronuclear migration in C. elegans. Normally the female pronucleus migrates toward the male pronucleus along microtubules emanating from centrosomes that are closely associated with the male pronucleus (Reinsch & Gonczy 1998). Mutations in zyg-12, originally isolated as a zygotic lethal mutation (Wood et al. 1980), or sun-1/matefin, as found in a genome-wide RNAi screen, disrupt centrosome attachment to the nuclear envelope, blocking pronuclear migration (Figure 5a, Malone et al. 2003).
Figure 5.
Functions of ZYG-1 and SUN-1/matefin. (a) A wild-type one-cell C. elegans embryo expressing GFP::tubulin. The male and female pronuclei (dark holes in background) have completed their migration, and both centrosomes remain closely attached to the male pronucleus. (a′ ) In a zyg-12 mutant embryo, the centrosomes fail to attach to the male pronucleus, and pronuclear migration fails. Reproduced with permission from Cell (Malone et al. 2003). (b) The midsection of a wild-type adult C. elegans syncytial gonad is shown. ZYG-12 (green) marks the nuclear envelope, and tubulin is shown in red. Nuclei are anchored at the periphery of the syncytial gonad. (b′ ) In a zyg-12(ct350) mutant, nuclei fall into the center of the syncytial gonad. Reproduced with permission from The Journal of Cell Biology (Zhou et al. 2009). (c) CED-4 (green) is recruited to the nuclear envelope (rings) in a C. elegans embryo. (c′ ) In a similarly staged sun-1/mtf-1 mutant embryo, CED-4 fails to be recruited to the nuclear envelope. Reproduced with permission from the Proceedings of the National Academy of Sciences, U.S.A. (Tzur et al. 2006).
ZYG-12 is a KASH protein with an N-terminal cytoplasmic domain similar to that of Hook proteins (Malone et al. 2003), which are hypothesized to link membrane compartments to microtubules (Walenta et al. 2001). SUN-1/matefin recruits ZYG-12 to the ONM (Malone et al. 2003, Minn et al. 2009), where it interacts with the dynein light-intermediate chain DLI-1 to recruit dynein heavy chain to the cytoplasmic surface of the nuclear envelope (Malone et al. 2003). This allows the growing male pronucleus to quickly recapture microtubule asters that might have drifted away from it (Malone et al. 2003, Meyerzon et al. 2009b). However, disruption of dynein heavy chain by RNAi causes only approximately 15% of centrosomes to be detached from the male pronucleus (Gonczy et al. 1999). To completely attach the centrosome to the nuclear envelope, ZYG-12 in the ONM interacts with a KASH-less isoform of ZYG-12 independently recruited to the centrosome (Figure 3d, Malone et al. 2003).
In other tissues and systems, the role of KASH-SUN bridges in centrosome attachment to the nucleus is less clear. For example, KASH and SUN proteins are not required for centrosome attachment during nuclear migration events later in C. elegans development (Lee et al. 2002, Starr et al. 2001). However, in Dictyostelium, a SUN protein is required for centrosome attachment (Xiong et al. 2008), and mouse Syne/Nesprin-1 and -2 double knockout cells have a centrosome detachment pheno-type (Zhang et al. 2009). Other nuclear envelope components including emerin and lamin attach centrosomes to nuclei in tissue culture fibroblasts (Lee et al. 2007, Salpingidou et al. 2007).
Nuclear Anchorage to Microtubules through Dynein
Nuclei are in an orderly arrangement at the periphery of the syncytial C. elegans gonad (Hubbard & Greenstein 2005). ZYG-12 and SUN-1/matefin are required to maintain the even spacing of nuclei and to prevent them from falling into the nucleus-free center of the syncytial gonad via, surprisingly, a centrosome-independent process (Zhou et al. 2009). Both zyg-12 alleles, zyg-12(ct350) and zyg-12(or577), disrupt centrosome attachment during pronuclear migration, but only the zyg-12(ct350) allele disrupts nuclear positioning in the gonad (Figure 5b, Zhou et al. 2009). The zyg-12(ct350) allele disrupts the ZYG-12-DLI-1 interaction and ZYG-12 dimerization, whereas the zyg-12(or577) allele disrupts only dimerization (Malone et al. 2003, Zhou et al. 2009). Therefore, ZYG-12 is hypothesized to function through dynein, but not the centrosome, to anchor nuclei to micro-tubules in the gonad (Zhou et al. 2009). The extent to which this mechanism is conserved in other tissues and organisms remains to be determined.
Targeting and Coordination of Kinesin-1 and Dynein at the Nuclear Surface
The KASH protein UNC-83 and the SUN protein UNC-84 function together during nuclear migration in a variety of C. elegans tissues (Figure 2, Malone et al. 1999, Starr et al. 2001). The mechanisms through which UNC-83 generates forces to move nuclei were recently elucidated. The cytoplasmic domain of UNC-83 binds the KLC-2 light chain of kinesin-1 and three regulators of dynein (Fridolfsson et al. 2010, Meyerzon et al. 2009a). Kinesin-1 mutant animals have severe nuclear migration defects (Meyerzon et al. 2009a). Similarly, mammalian Nesprin-4 interacts with kinesin light chain (Roux et al. 2009). Nesprin-4 expression is limited to secretory endrocrine cells, making it difficult to study its role in nuclear migration (Roux et al. 2009). However, when expressed in a heterologous HeLa system, Nesprin-4 recruits kinesin-1 to the nuclear envelope and induces the nucleus to move away from the centrosome toward the plus ends of micro-tubules, suggesting that it plays a role in nuclear positioning (Roux et al. 2009). Syne/Nesprin-2 interacts with dynein and kinesin in the neuroepithelium, suggesting that different KASH proteins play similar functions in different tissues (Zhang et al. 2009). Drosophila Klarsicht has also been proposed to function through kinesin-1 (Shubeita et al. 2008, Welte et al. 1998). Although they do not contain any stretches of obvious similarity outside of their KASH domains, UNC-83, Nesprin-4, and Klarsicht may be functional homologs. All of them appear to function as cargo adaptors at the nuclear envelope to recruit kinesin-1 to the cytoplasmic face of the nucleus (Figure 3b, Meyerzon et al. 2009a, Roux et al. 2009). In support of this model, a hybrid KLC-2::KASH construct effectively targets to the ONM in transgenic C. elegans and rescues the unc-83 nuclear migration defect (Meyerzon et al. 2009a).
UNC-83 also interacts with two dynein-regulating complexes (Fridolfsson et al. 2010). Mutations in any of these components or in dynein heavy chain cause nuclear migration defects. The defects are much less severe than unc-83 or kinesin-1 mutants, suggesting that dynein plays a regulatory role in hypodermal nuclear migration (Fridolfsson et al. 2010). We propose that UNC-83 is a nuclear-specific cargo adaptor for both motors and functions to coordinate bidirectional movements leading to a net migration toward the plus ends of micro-tubules (Figure 3b). Whereas kinesin-1 provides the major force, dynein ensures that the migration proceeds normally (Fridolfsson et al. 2010).
Attachment of Nuclei to Intermediate Filaments
Intermediate filaments have long been proposed to play a role in nuclear positioning. Vimentin is often associated with nuclei, and defects in vimentin disrupt nuclear morphology (Toivola et al. 2005). Additionally, mice with knockouts of desmin have severe nuclear anchorage defects in skeletal muscles (Ralston et al. 2006). Thus, it is not surprising that a KASH protein associates with intermediate filaments (Figure 3g). Plectin is a plakin family member that cross-links actin filaments to intermediate filaments (Wiche 1998). A yeast two-hybrid screen with the actin-binding domain of plectin identified the N terminus of the KASH protein Nesprin-3 (Wilhelmsen et al. 2005). The KASH domain of Nesprin-3 and Sun1 and/or Sun2 are necessary for localization of Nesprin-3 to the ONM (Ketema et al. 2007). Overexpression of Nesprin-3 recruits intermediate filaments to the nuclear envelope (Wilhelmsen et al. 2005). However, a mouse knockout of Nesprin-3 has no obvious morphological phenotypes (K. Lei and R. Xu, personal communication). Thus, the role of Nesprin-3 in nuclear positioning remains to be characterized.
KASH AND SUN PROTEINS REGULATE THE GLOBAL CYTOSKELETON
ANC-1 and Syne/Nesprin-1 and -2 have additional functions in the regulation of the global cytoskeleton and subsequent localization of other organelles. For example, anc-1 mutant animals have a mitochondrial positioning defect (Hedgecock & Thomson 1982, Starr & Han 2002). Syne/Nesprin-1 plays roles in the structure of the Golgi complex, cytokinesis, formation of a perinuclear actin cap, and vesicle transport (Fan & Beck 2004, Gough & Beck 2004, Gough et al. 2003, Khatau et al. 2009). In dominant-negative KASH overexpression 3T3 cells, the mechanical stiffness of the cytoskeleton far away from the nucleus is disrupted (Stewart-Hutchinson et al. 2008). Additionally, mechanically pulling on beads attached to the surfaces of cells through integrins causes immediate reorganization of the nucleoplasm, suggesting that the extra-cellular matrix is physically connected to the nucleoplasm (Maniotis et al. 1997). These data lead to a hypothesis that KASH and SUN proteins function in mechanotransduction of physical signals from the extracellular matrix directly to chromatin ( Jaalouk & Lammerding 2009, Wang et al. 2009).
FUNCTIONS OF KASH-SUN BRIDGES IN MEIOTIC CHROMOSOME MOVEMENTS
The initial characterization of KASH-SUN bridges came from studying their roles in the positioning of whole nuclei. Another interesting problem is how forces generated in the cytoskeleton are used to move objects within the nucleus. A major discovery in the field was the demonstration that KASH-SUN bridges are also used to move telomeres within the nucleus during meiosis in S. pombe (Figures 3e and 6c; Chikashige et al. 2006).
Figure 6.
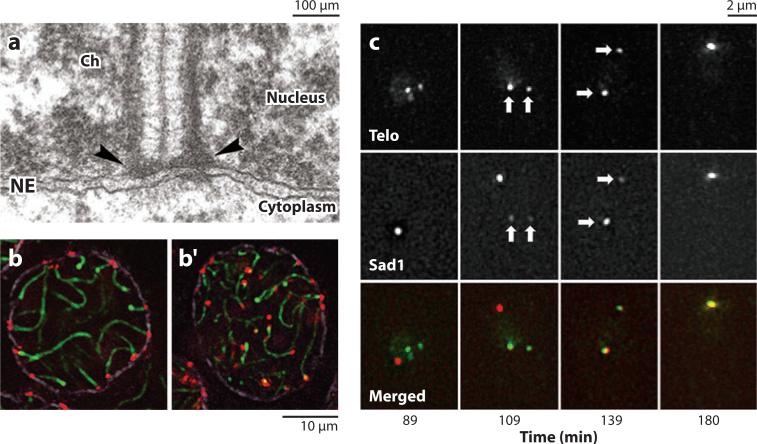
Roles of KASH-SUN bridges in moving meiotic chromosomes. (a) Transmission electron microscopy image from a mouse spermatocyte showing a telomere (arrowheads) of a synapsed meiotic bivalent (extending up from telomere) attached to the inner nuclear membrane (INM) of the nuclear envelope (NE). Ch, chromatin. Reproduced with permission from The Proceedings of the National Academy of Sciences, U.S.A. (Schmitt et al. 2007). (b) A heterozygous control mouse spermatocyte showing telomeres (red ) of bivalents (green) attached to the nuclear envelope (purple). (b′ ) A spermatoycyte from a sterile Sun1 homozygous knockout mouse showing that telomeres fail to attach to the nuclear envelope. Reproduced with permission from Developmental Cell (Ding et al. 2007). (c) Sad1 (red in merge) localizes predominantly to the spindle pole body (89 min). During meiosis, some Sad1 leaves the spindle pole body, associates with telomeres (Telo, green in merge) on the INM (109 min), and moves the telomeres to the spindle pole body to form the bouquet formation (180 min). Reproduced with permission from Cell (Chikashige et al. 2006).
In many systems, meiotic chromosomes dramatically rearrange such that telomeres associate with the INM and cluster together to form a bouquet, which aids in homolog pairing (reviewed in Harper et al. 2004). The forces to move meiotic chromosomes are generated by dynein on microtubules in S. pombe and C. elegans (Figure 3d,e; Chikashige et al. 2006, Sato et al. 2009) but by actin dynamics in Saccharomyces cerevisiae (Figure 3f, Conrad et al. 2008, Koszul et al. 2008). KASH-SUN bridges then transfer these forces across the nuclear envelope to the meiotic chromosomes (reviewed in Hiraoka & Dernburg 2009, Starr 2009). SUN proteins are required for the association of telomeres (or pairing centers in C. elegans) to the INM (Figure 6, Bupp et al. 2007, Conrad et al. 2007, Ding et al. 2007, Penkner et al. 2007). KASH proteins Kms1 and Kms2 in S. pombe and ZYG-12 in C. elegans interact with dynein to move along microtubules (Chikashige et al. 2006, Miki et al. 2004, Penkner et al. 2009, Sato et al. 2009). The S. cerevisiae KASH protein Csm4 interacts with the actin cytoskeleton (Conrad et al. 2008, Koszul et al. 2008, Starr 2009). Defects in the KASH or SUN proteins involved in meiotic chromosome movements lead to aberrant synapsis and a reduction in recombination, but do not block recombination (reviewed in Hiraoka & Dernburg 2009, Koszul & Kleckner 2009).
The finding that KASH-SUN bridges are used to move meiotic chromosomes opens up the question as to what their role might be in chromosome movements in interphase cells. Moving loci within the nucleoplasm affects transcription (reviewed in Akhtar & Gasser 2007). Therefore, and in agreement with the mechanotransduction hypothesis, KASH and SUN proteins are likely to play active roles in transcription control. In one example, the S. pombe KASH proteins Kms1 and Kms2 and the SUN protein Sad1 are involved in anchoring centromeres and heterochromatin at the INM (King et al. 2008).
OTHER FUNCTIONS OF KASH AND SUN PROTEINS
To this point, we have focused on the structural roles KASH-SUN bridges play in transferring forces generated in the cytoplasm to the structural components of the nucleus. Other KASH proteins recruited to the ONM are used to regulate diverse cellular processes.
C. elegans KDP-1 Regulates the Progression of the Cell Cycle
KDP-1 (for KASH-domain protein), which was identified in a membrane-bound yeast two-hybrid screen with the SUN domain of UNC-84 as bait, localizes to the nuclear envelope in a SUN-1/matefin dependent manner (Figure 3h, McGee et al. 2009). Disruption of kdp-1 by RNAi causes multiple phenotypes in the early embryo, larvae, and germ line that are characteristic of a delay in the cell cycle exiting S-phase or entering M-phase (McGee et al. 2009). We hypothesize that as a KASH protein, KDP-1 is perfectly positioned on the outer surface of the nuclear envelope to receive a signal from the nucleoplasm that S-phase is complete and then to relay the message to cdk/cyclin complexes. It will be interesting to see whether this role in cell cycle regulation is conserved.
WIPs Recruit RanGAP to the Outer Nuclear Membrane
The establishment of a Ran GTP gradient, with Ran GDP in the cytoplasm, controls the trafficking of soluble proteins between the nucleoplasm and cytoplasm (Cook et al. 2007, Meier et al. 2008). The mammalian nuclear pore component Nup358/RanBP2 targets RanGAP to the cytoplasmic surface of the pore to ensure that Ran GTP is quickly hydrolyzed to GDP upon exiting the nucleus (Mahajan et al. 1997). Alternatively, in Arabidopsis RanGAP is targeted to the ONM by a plant-specific family of integral ONM proteins called WIPs (for WP-domain interacting proteins) (Xu et al. 2007). WIP1, 2, and 3 are C-tail-anchored integral membrane proteins; the C terminus and the transmembrane domain are necessary and sufficient for ONM localization (Xu et al. 2007). Although the role of plant SUN proteins has not been tested in this process, the WIPs are likely KASH proteins.
Recruitment of CED-4 to the Nuclear Envelope to Mediate Apoptosis
During the initiation of programmed cell death in C. elegans, the caspase activator and proapoptotic factor CED-4 (the Apaf-1 homolog) translocates from the mitochondria to the nuclear envelope (Chen et al. 2000); SUN-1/matefin is required for this process (Figure 5c, Tzur et al. 2006a). Furthermore, sun-1/matefin(RNAi) embryos have significantly fewer apoptotic events, suggesting that the translocation of CED-4 to the nuclear envelope is an important step in apoptosis (Tzur et al. 2006a). A KASH protein associated with SUN-1/matefin would be ideally situated in the ONM as a CED-4 receptor. However, no KASH protein has been implicated in the translocation of CED-4 to the nuclear envelope. SUN-1/matefin binds CED-4 in vitro, raising the possibility that activated CED-4 enters the nucleoplasm and interacts with SUN-1/matefin at the INM (Tzur et al. 2006a).
FUNCTIONS OF KASH-SUN PROTEINS IN MAMMALS: MOUSE MODELS AND HUMAN DISEASES
The studies described to this point demonstrate that links between the nucleus and the cytoskeleton are essential to a wide variety of cellular processes. Because these studies were carried out in model organisms or tissue culture systems, there are limitations to interpreting how these results relate to human development and disease. Thus, it is gratifying that recent findings connect SUN and KASH proteins to human diseases and developmental processes in mouse models. Mutations in KASH or SUN proteins contribute to muscular, neurological, skin, and premature-aging disorders.
Nuclear Positioning in Muscles and Muscular Dystrophies
Mouse Syne/Nesprin-1 and -2 and their partners SUN1 and SUN2 anchor nuclei. In mammalian skeletal muscles, most nuclei are evenly spaced throughout the syncytial myotube, whereas 3–6 functionally specialized nuclei are anchored beneath the neuromuscular junction (NMJ). Overexpression of either the Syne/Nesprin-1 or -2 KASH domain in muscles displaces most of the endogenous Syne/Nesprin-1 and -2 and partially disrupts synaptic nuclear clustering (Figure 4b, Grady et al. 2005, Zhang et al. 2007b). Mouse knockouts of KASH domain–containing isoforms of both genes, individually or simultaneously, provide a more complete understanding of the functions of Syne/Nesprin-1 and -2 (Zhang et al. 2007b). In Syne/Nesprin-1 but not Syne/Nesprin-2 single KASH knockouts, synaptic and nonsynaptic nuclei are completely unanchored (Zhang et al. 2007b). Syne/Nesprin-1 and -2 have partially overlapping functions because mice lacking both proteins die shortly after birth (Zhang et al. 2007b). SUN1 single knockout mice have defects in nuclear anchorage at the NMJ, but SUN1/2 double knockout mice have more severe synaptic nuclear positioning defects as well as nonsynaptic nuclear positioning defects and die shortly after birth (Lei et al. 2009). Thus, mouse KASH and SUN proteins function in muscle development, but the question remains as to how they relate to human muscular diseases.
Emery-Dreifuss muscular dystrophy (EDMD) is a neuromuscular condition characterized by progressive skeletal muscle degeneration with associated cardiomyopathy that originally was found to be caused by mutations in the nuclear-envelope proteins emerin or lamin (Bione et al. 1994, Bonne et al. 1999). EDMD is one of a broad spectrum of more than 30 diseases known as laminopathies that are caused by mutations in components of the nuclear envelope (reviewed in Dauer & Worman 2009, Ellis 2006, Worman & Bonne 2007). Although the proteins underlying laminopathies are ubiquitously expressed, patients develop disease symptoms in a tissue-specific manner. The diseases may be caused by defective nuclei acquiring an increased sensitivity to mechanical stress or a misregulation of genes required for development (reviewed in Dauer & Worman 2009, Ellis 2006, Worman & Bonne 2007). The majority of EDMD patients do not have mutations in either lamin or emerin (Bonne et al. 2003). Syne/Nesprin-1 and -2 interact in vitro with both emerin and lamin, and this interaction is altered by mutations that cause EDMD (Mislow et al. 2002a, Wheeler et al. 2007). Therefore, Syne/ Nesprin-1 and -2 may contribute to maintaining the integrity of myonuclei during mechanical stress by forming complexes with emerin and lamin. In support of this hypothesis, Syne/Nesprin-1 and -2 sequence variants have been found in EDMD patients that do not have either emerin or lamin mutations (Zhang et al. 2007a). Syne/Nesprin-1 mutations have also been linked to myogenic autosomal recessive arthrogryposis, further implicating Syne mutations in human muscle pathogenesis (Attali et al. 2009).
Unlike the Syne/Nesprin-1 KASH knockout mouse described in Zhang et al. (2007b), a second Syne/Nesprin-1 KASH knockout mouse is a potential EDMD model. Mice with this Syne/Nesprin-1 mutation are peri-natal lethal, and only approximately half survive birth (Puckelwartz et al. 2009). Surviving mice display EDMD-like phenotypes including an abnormal curvature of the spine, muscle pathology, and cardiac conduction defects that develop with age. They also have myonu-clear anchorage defects, but Syne/Nesprin-1, SUN2, emerin, and lamin localize normally (Puckelwartz et al. 2009). These phenotypes are likely due to a dominant-negative effect of incorporating mutant Syne/Nesprin-1 into complexes at the nuclear envelope. Consistent with this, the KASH-SUN interaction is disrupted, indicating that an uncoupling of the nucleoskeleton and the cytoskeleton may be responsible for the muscle defects (Puckelwartz et al. 2009). In the mice in Zhang et al. (2007b), Syne/Nesprin-1 does not localize to the nuclear envelope, and there is no lethality or EDMD-like phenotypes. The differences in these two Syne/Nesprin-1 KASH-deleted mutant lines could be explained by the different mouse backgrounds used to generate the knockout lines or the different C termini generated by the slightly different KASH domain deletions (Puckelwartz et al. 2009, Zhang et al. 2007b).
Lamin A/C or lamin B knockout mice phenocopy EDMD, have defects in the development of the central nervous system and the phrenic nerve, and disrupt nuclear positioning at the NMJ (De Sandre-Giovannoli et al. 2002, Mejat et al. 2009, Nikolova et al. 2004, Sullivan et al. 1999, Vergnes et al. 2004). These pheno-types are strikingly similar to Syne/Nesprin and SUN knockout mice. Because lamins recruit SUN proteins to the nuclear envelope, many of the defects in lamin knockout mice likely are due to defects in recruiting SUN and KASH proteins to the nuclear envelope.
Syne/Nesprin and SUN proteins also have significant functions in other tissues. The phrenic nerve of the diaphragm in Syne/Nesprin double knockout mice displays longer branches than in wild-type mice, suggesting that Syne/Nesprin-1 and -2 mutations disrupt more than just myonuclear anchorage (Zhang et al. 2007b). Furthermore, the lethality of SUN double knockout mice is rescued by expressing SUN1 specifically in neurons (Lei et al. 2009). In these rescued mice, Syne/Nesprin localization in muscles remains disrupted, indicating that the lack of muscle functions of Syne/Nesprin and SUN proteins is not responsible for the lethality of the knockout mice (Lei et al. 2009).
Neuronal Functions of SUN-KASH Proteins
Syne/Nesprin-1 and -2 play important roles in neuronal development. In the developing neuroepithelium, nuclear migration events are required for both neurogenesis and neuronal migration (reviewed in Baye & Link 2008). Neurons proliferate in the neuroepithelium while undergoing interkinetic nuclear migration, during which centrosomes remain at the apical surface, while the nucleus migrates between the apical and basal surfaces in conjunction with cell cycle progression (Frade 2002). It has been proposed that dynein moves the nucleus to the apical surface, whereas kinesin moves the nucleus toward the basal surface (reviewed in Baye & Link 2008). Syne/Nesprin proteins genetically interact with dynein to move the nucleus during proliferative interkinetic nuclear migration and nuclear positioning events in photoreceptor cells in the developing retina of zebrafish (Del Bene et al. 2008, Tsujikawa et al. 2007). Lissencephaly, a severe mental retardation disease, is caused by a nuclear migration defect in neurons radially migrating out from the neuroepithelium into the outer layers of the cortex during development (reviewed in Lambert de Rouvroit & Goffinet 2001, Morris et al. 1998). In these neurons, the centrosome moves at a constant rate toward the leading edge of the cell, whereas the nucleus moves in a saltatory manner behind it (Bellion et al. 2005, Schaar & McConnell 2005, Tsai et al. 2007). Syne/Nesprin-dependent targeting of dynein to the nuclear envelope apparently pulls on centrosomal microtubules to move the nucleus (Tsai et al. 2007, Zhang et al. 2009), providing a possible link between KASH and SUN proteins and lissencephaly.
Double knockout mouse lines show that SUN and KASH proteins play essential functions in the nervous system. Loss of either SUN1 and SUN2 or Syne/Nesprin-1 and -2 leads to lethality, reduced brain size, malformed cortices, enlarged lateral ventricles, a smaller corpus callosum, and multiple brain regions with severe laminary defects (Zhang et al. 2009). Syne/Nesprin-2 alone is required for proper laminary formation in the hippocampus and cerebral cortex, whereas Syne/Nesprin-1 and -2 have redundant roles in other brain regions. Although Syne/Nesprin-2 KASH knockout mice are viable, they have working memory defects (Zhang et al. 2009). The laminary defects seen in the cerebral cortex of SUN double knockout and Syne/Nesprin-2 knockout mice are the result of failed radial migrations caused by failure of the nucleus to migrate (Figure 4c). This is likely due to dissociation of the centrosome from the nucleus, which supports the model that Syne/Nesprin proteins couple microtubules to the nucleus through dynein (Zhang et al. 2009). Consistent with this, Syne/Nesprin-1 and -2 colocalize at the nuclear envelope with dynein (Zhang et al. 2009). The small brain size in the double knockout mice is likely caused by a reduction in the number of progenitor cells caused by disrupted interkinetic nuclear migration, as Syne/Nesprin-2 colocalizes and interacts with kinesin-1 at the nuclear envelope in the neuroepithelium (Zhang et al. 2009).
Consistent with a role for Syne/Nesprin proteins in human neural development, Syne/Nesprin-1 mutations cause auto-somal recessive cerebellar ataxia type 1 (ARCA1) (Gros-Louis et al. 2007). Ataxias are characterized by a lack of coordination of gait and limbs along with neurological symptoms; ARCA1 is late onset (Gros-Louis et al. 2007). ARCA1 patients have misplaced nuclei at the NMJ, phenocopying Syne/Nesprin-1 knockout mice (Gros-Louis et al. 2007). The relationship between this nuclear positioning phenotype and the progression of ARCA1 remains unclear. Dystonia DYT1, a neurological disease of involuntary movements caused by deletion of a single residue in TorsinA (Ozelius et al. 1999, Tanabe et al. 2009) may also be the result of disrupted KASH-SUN complexes. Because TorsinA has been proposed to function as a regulator of KASH-SUN interactions, the role of TorsinA and KASH proteins in dystonia merits further investigation.
SUN-KASH Proteins in Aging
One of the more exciting issues in the field is the role of the nuclear envelope in normal aging and diseases of premature aging. Mutations in lamin A/C cause multiple laminopathies (reviewed in Dauer & Worman 2009, Prokocimer et al. 2009, Worman & Bonne 2007). One, Hutchinson-Gilford progeria syndrome (HGPS), is characterized by loss of subcutaneous fat, severe hair loss, restrictive joint mobility, bone abnormalities, cardiovascular disease, and progressive arteriosclerosis—all characteristics of premature aging (Hennekam 2006). Fibroblasts from HGPS patients and a lamin A/C HGPS-mouse model have altered mechanical properties that lead to abnormal nuclear architecture and morphology (Broers et al. 2004, Dahl et al. 2006). Furthermore, fibroblasts from HGPS mice have defects in the localization of KASH and SUN proteins to the nuclear envelope (Crisp et al. 2006, Libotte et al. 2005). The amount of Syne/Nesprin-2 at the nuclear envelope is inversely proportional to the HGPS nuclear morphology and chromatin organization defects (Kandert et al. 2007). This suggests that Syne/Nesprin-2 function at the nuclear membrane offsets the harmful effects of the HGPS lamin A/C mutation and may function in the prevention of normal aging.
SUN-KASH Proteins in Skin Development, Ciliogenesis, and Cancer
SUN and KASH proteins are ubiquitously expressed in human tissues (Zhang et al. 2001; Crisp et al. 2006), and whereas their functions in muscle and neurons are becoming evident, what role Syne/Nesprin and SUN proteins have in other tissues remains less clear. Nuclear positioning in the epidermis is essential for epidermal stratification during development (Lechler & Fuchs 2005), and mice lacking the largest isoform of Syne/Nesprin-2 in the skin show a thickening of the epidermis (Luke et al. 2008). These mice, which were created by deleting the actin-binding domain of Syne/Nesprin-2, also exhibit epidermal nuclear morphology defects (Luke et al. 2008), which suggests important roles for SUN and KASH proteins in skin development.
Meckel-Gruber Syndrome (MKS) is characterized by bilateral renal cystic dysplasia and central nervous system developmental defects caused by mutations in genes that contribute to building cilia (Kyttala et al. 2006). Syne/Nesprin-2 is required for ciliogenesis in cell culture, interacts with components of the primary cilium, and appears to be required for centrosome migration to the apical cell surface during the early stages of ciliogenesis (Dawe et al. 2009). In addition, MKS patient cells show a redistribution of Syne/Nesprin-2 and a reduced centrosome-nucleus distance (Dawe et al. 2009). Thus, SUN and KASH proteins apparently function in ciliogenesis.
Recent studies have linked Syne/Nesprin-1 and -2 to cancer. Mutations in Syne/Nesprin-1 and -2 frequently accumulate in colorectal and breast cancer tumors, respectively (Sjoblom et al. 2006). Furthermore, expression of Syne/Nesprin-1 is downregulated 20–180-fold in a variety of early tumors (Marme et al. 2008). Finally, a large epidemiological study found a potential association between a polymorphism in Syne/Nesprin-1 and an increased risk of invasive ovarian cancer (Doherty et al. 2010). Together these studies raise the exciting possibility that KASH and SUN proteins play roles in cancer progression and therefore warrant further studies.
SUMMARY POINTS.
Essential functions of KASH and SUN proteins were initially discovered and characterized by nonbiased forward genetic screens in model yeast and invertebrates.
SUN proteins are components of the INM; their N termini interact with the nuclear lamina, and their conserved C-terminal SUN domains extend into the perinuclear space.
SUN proteins recruit KASH proteins specifically to the ONM to complete a bridge across the nuclear envelope.
Divergent cytoplasmic domains of KASH proteins mediate interactions with micro-tubules, centrosomes, and actin filaments to position nuclei within the cell.
KASH-SUN nuclear-envelope bridges also function in meiosis to attach chromosomes to the INM and move them into the bouquet formation, aiding homolog pairing.
Mouse knockout studies show that functions of KASH and SUN proteins are conserved from single-cell eukaryotes all the way to mammalian systems, where they play essential roles in neuromuscular development.
KASH and SUN proteins have been linked to cancer and implicated in the progression of human diseases including a variety of laminopathies and neurological disorders.
FUTURE ISSUES.
It remains to be determined how the interactions between KASH and SUN domains are regulated and remodeled during developmental switches, such as between nuclear anchorage and migration.
Researchers need to identify the complete array of nucleoplasmic proteins that interact with SUN proteins, cytoplasmic proteins that interact with KASH proteins, and their corresponding functions.
The model that mechanical stimuli are transferred from the outside of the cell directly to chromatin through KASH-SUN bridges needs to be tested.
Researchers need to fully elucidate the mechanisms of how KASH and SUN proteins function in the progression of cancer, laminopathies, and other diseases.
ER: endoplasmic reticulum
INM: inner nuclear membrane
ONM: outer nuclear membrane
Nuclear-envelope bridge: a complex of SUN proteins at the INM and KASH proteins at the ONM that transfer forces generated in the cytoplasm to the nuclear lamina
LINC: linker of nucleoskeleton and cytoskeleton
Sad1 and UNC-84 (SUN) domain: a conserved domain of an INM protein that recruits KASH proteins to the ONM
Klarsicht, ANC-1, and Syne homology (KASH) domain: a transmembrane span followed by 6–30 residues at the C terminus of a protein that targets proteins to the ONM
Nuclear positioning:
the act of a nucleus migrating to a specific location within a cell and the process used to anchor it there
Microtubule motors:
ATPases that move cargo along microtubule tracks; dynein moves toward the minus ends and kinesin-1 toward the plus ends
Neuromuscular junction (NMJ): a synapse between the myotube and the neuron where 3–6 muscle nuclei associate and become transcriptionally specialized
EDMD: Emery-Dreifuss muscular dystrophy
Laminopathies: a broad range of human diseases caused by mutations in proteins associated with the nuclear envelope
Progeria: a human disease of premature, rapid aging
HGPS: Hutchinson-Gilford progeria syndrome
First report that SUN-KASH bridges function in meiosis. Dynein forces are transferred across the bridge to move chromosomes within the nucleoplasm.
Demonstrated that SUN and KASH proteins transmit forces from cytoskeletal actin to move meiotic chromosomes in budding yeast.
Tests many aspects of the KASH-SUN bridge model and shows that it is required to maintain spacing of the nuclear envelope.
Using forward human genetics, this is the first demonstration of a KASH protein causing a neuromuscular disease.
Field-defining paper that discovers SUN domains and shows that they function in nuclear positioning.
Demonstrates a role for the ZYG-12 KASH protein in the attachment of centrosomes to the nuclear envelope.
Demonstrates that UNC-83 mediates nuclear migration by acting as a nuclear-specific cargo-adaptor of kinesin-1.
Mouse knockout of the Syne/Nesprin-1 KASH domain has nuclear positioning defects and phenotypes similar to EDMD.
Defines KASH, demonstrates dependency of SUN proteins to recruit KASH proteins, and describes function of ANC-1 in nuclear anchorage.
Describes the essential roles of KASH and SUN proteins in nuclear migration during mouse neurological development.
ACKNOWLEDGMENTS
We apologize to those whose studies were not discussed in detail because of space limitations. We thank E. Tapley and Y.-T. Chang (University of California, Davis), W. Hanna-Rose (Penn State), and D. Hodzic (Washington University) for helpful comments on the manuscript. We thank R. Xu (Fudan University) for sharing unpublished data and R. Xu, W. Hanna-Rose, Y. Hiraoka (Osaka University), M. Alsheimer (Wuezburg University), and Y. Gruenbaum (Hebrew University) for pictures used in the figures. Our research is supported by grant 5R01GM073874 from the NIH. H.N.F. was supported by NIH training grant 5T32GM007377.
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
The Annual Review of Cell and Developmental Biology is online at cellbio.annualreviews.org
LITERATURE CITED
- Akhtar A, Gasser SM. The nuclear envelope and transcriptional control. Nat. Rev. Genet. 2007;8:507–17. doi: 10.1038/nrg2122. [DOI] [PubMed] [Google Scholar]
- Apel ED, Lewis RM, Grady RM, Sanes JR. Syne-1, a dystrophin- and Klarsicht-related protein associated with synaptic nuclei at the neuromuscular junction. J. Biol. Chem. 2000;275:31986–95. doi: 10.1074/jbc.M004775200. [DOI] [PubMed] [Google Scholar]
- Attali R, Warwar N, Israel A, Gurt I, McNally E, et al. Mutation of SYNE-1, encoding an essential component of the nuclear lamina, is responsible for autosomal recessive arthrogryposis. Hum. Mol. Genet. 2009;18:3462–69. doi: 10.1093/hmg/ddp290. [DOI] [PubMed] [Google Scholar]
- Baye LM, Link BA. Nuclear migration during retinal development. Brain Res. 2008;1192:29–36. doi: 10.1016/j.brainres.2007.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellion A, Baudoin JP, Alvarez C, Bornens M, Metin C. Nucleokinesis in tangentially migrating neurons comprises two alternating phases: forward migration of the Golgi/centrosome associated with centrosome splitting and myosin contraction at the rear. J. Neurosci. 2005;25:5691–99. doi: 10.1523/JNEUROSCI.1030-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bione S, Maestrini E, Rivella S, Mancini M, Regis S, et al. Identification of a novel X-linked gene responsible for Emery-Dreifuss muscular dystrophy. Nat. Genet. 1994;8:323–27. doi: 10.1038/ng1294-323. [DOI] [PubMed] [Google Scholar]
- Bonne G, Di Barletta MR, Varnous S, Becane HM, Hammouda EH, et al. Mutations in the gene encoding lamin A/C cause autosomal dominant Emery-Dreifuss muscular dystrophy. Nat. Genet. 1999;21:285–88. doi: 10.1038/6799. [DOI] [PubMed] [Google Scholar]
- Bonne G, Yaou RB, Beroud C, Boriani G, Brown S, et al. 108th ENMC International Workshop, 3rd Workshop of the MYO-CLUSTER project: EUROMEN, 7th International Emery-Dreifuss Muscular Dystrophy (EDMD) Workshop, 13–15 September 2002, Naarden, The Netherlands. Neuromuscul. Disord. 2003;13:508–15. doi: 10.1016/s0960-8966(03)00063-4. [DOI] [PubMed] [Google Scholar]
- Bornens M. Is the centriole bound to the nuclear membrane? Nature. 1977;270:80–82. doi: 10.1038/270080a0. [DOI] [PubMed] [Google Scholar]
- Braunagel SC, Williamson ST, Saksena S, Zhong Z, Russell WK, et al. Trafficking of ODV-E66 is mediated via a sorting motif and other viral proteins: facilitated trafficking to the inner nuclear membrane. Proc. Natl. Acad. Sci. USA. 2004;101:8372–77. doi: 10.1073/pnas.0402727101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broers JL, Peeters EA, Kuijpers HJ, Endert J, Bouten CV, et al. Decreased mechanical stiffness in LMNA−/− cells is caused by defective nucleo-cytoskeletal integrity: implications for the development of laminopathies. Hum. Mol. Genet. 2004;13:2567–80. doi: 10.1093/hmg/ddh295. [DOI] [PubMed] [Google Scholar]
- Bupp JM, Martin AE, Stensrud ES, Jaspersen SL. Telomere anchoring at the nuclear periphery requires the budding yeast Sad1-UNC-84 domain protein Mps3. J. Cell Biol. 2007;179:845–54. doi: 10.1083/jcb.200706040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, Hersh BM, Conradt B, Zhou Z, Riemer D, et al. Translocation of C. elegans CED-4 to nuclear membranes during programmed cell death. Science. 2000;287:1485–89. doi: 10.1126/science.287.5457.1485. [DOI] [PubMed] [Google Scholar]
- Chi YH, Haller K, Peloponese JM, Jr, Jeang KT. Histone acetyltransferase hALP and nuclear membrane protein hsSUN1 function in decondensation of mitotic chromosomes. J. Biol. Chem. 2007;282:27447–58. doi: 10.1074/jbc.M703098200. [DOI] [PubMed] [Google Scholar]
- Chikashige Y, Tsutsumi C, Yamane M, Okamasa K, Haraguchi T, Hiraoka Y. Meiotic proteins bqt1 and bqt2 tether telomeres to form the bouquet arrangement of chromosomes. Cell. 2006;125:59–69. doi: 10.1016/j.cell.2006.01.048. [DOI] [PubMed] [Google Scholar]
- Conrad MN, Lee CY, Chao G, Shinohara M, Kosaka H, et al. Rapid telomere movement in meiotic prophase is promoted by NDJ1, MPS3, and CSM4 and is modulated by recombination. Cell. 2008;133:1175–87. doi: 10.1016/j.cell.2008.04.047. [DOI] [PubMed] [Google Scholar]
- Conrad MN, Lee CY, Wilkerson JL, Dresser ME. MPS3 mediates meiotic bouquet formation in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA. 2007;104:8863–68. doi: 10.1073/pnas.0606165104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook A, Bono F, Jinek M, Conti E. Structural biology of nucleocytoplasmic transport. Annu. Rev. Biochem. 2007;76:647–71. doi: 10.1146/annurev.biochem.76.052705.161529. [DOI] [PubMed] [Google Scholar]
- Cottrell JR, Borok E, Horvath TL, Nedivi E. CPG2: a brain- and synapse-specific protein that regulates the endocytosis of glutamate receptors. Neuron. 2004;44:677–90. doi: 10.1016/j.neuron.2004.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crisp M, Liu Q, Roux K, Rattner JB, Shanahan C, et al. Coupling of the nucleus and cytoplasm: role of the LINC complex. J. Cell Biol. 2006;172:41–53. doi: 10.1083/jcb.200509124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahl KN, Scaffidi P, Islam MF, Yodh AG, Wilson KL, Misteli T. Distinct structural and mechanical properties of the nuclear lamina in Hutchinson-Gilford progeria syndrome. Proc. Natl. Acad. Sci. USA. 2006;103:10271–76. doi: 10.1073/pnas.0601058103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dauer WT, Worman HJ. The nuclear envelope as a signaling node in development and disease. Dev. Cell. 2009;17:626–38. doi: 10.1016/j.devcel.2009.10.016. [DOI] [PubMed] [Google Scholar]
- Dawe HR, Adams M, Wheway G, Szymanska K, Logan CV, et al. Nesprin-2 interacts with meckelin and mediates ciliogenesis via remodelling of the actin cytoskeleton. J. Cell Sci. 2009;122:2716–26. doi: 10.1242/jcs.043794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Sandre-Giovannoli A, Chaouch M, Kozlov S, Vallat JM, Tazir M, et al. Homozygous defects in LMNA, encoding lamin A/C nuclear-envelope proteins, cause autosomal recessive axonal neuropathy in human (Charcot-Marie-Tooth disorder type 2) and mouse. Am. J. Hum. Genet. 2002;70:726–36. doi: 10.1086/339274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Bene F, Wehman AM, Link BA, Baier H. Regulation of neurogenesis by interkinetic nuclear migration through an apical-basal notch gradient. Cell. 2008;134:1055–65. doi: 10.1016/j.cell.2008.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding X, Xu R, Yu J, Xu T, Zhuang Y, Han M. SUN1 is required for telomere attachment to nuclear envelope and gametogenesis in mice. Dev. Cell. 2007;12:863–72. doi: 10.1016/j.devcel.2007.03.018. [DOI] [PubMed] [Google Scholar]
- Doherty JA, Rossing MA, Cushing-Haugen KL, Chen C, Van Den Berg DJ, et al. ESR1/SYNE1 polymorphism and invasive epithelial ovarian cancer risk: an Ovarian Cancer Association Consortium study. Cancer Epidemiol. Biomarkers Prev. 2010;19:245–50. doi: 10.1158/1055-9965.EPI-09-0729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis JA. Emery-Dreifuss muscular dystrophy at the nuclear envelope: 10 years on. Cell Mol. Life Sci. 2006;63:2702–9. doi: 10.1007/s00018-006-6247-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J, Beck KA. A role for the spectrin superfamily member Syne-1 and kinesin II in cytokinesis. J. Cell Sci. 2004;117:619–29. doi: 10.1242/jcs.00892. [DOI] [PubMed] [Google Scholar]
- Fischer JA, Acosta S, Kenny A, Cater C, Robinson C, Hook J. Drosophila klarsicht has distinct subcellular localization domains for nuclear envelope and microtubule localization in the eye. Genetics. 2004;168:1385–93. doi: 10.1534/genetics.104.028662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer-Vize JA, Mosley KL. Marbles mutants: uncoupling cell determination and nuclear migration in the developing Drosophila eye. Development. 1994;120:2609–18. doi: 10.1242/dev.120.9.2609. [DOI] [PubMed] [Google Scholar]
- Frade JM. Interkinetic nuclear movement in the vertebrate neuroepithelium: encounters with an old acquaintance. Prog. Brain Res. 2002;136:67–71. doi: 10.1016/s0079-6123(02)36007-2. [DOI] [PubMed] [Google Scholar]
- Franke WW, Scheer U, Krohne G, Jarasch ED. The nuclear envelope and the architecture of the nuclear periphery. J. Cell Biol. 1981;91:39s–50s. doi: 10.1083/jcb.91.3.39s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridkin A, Mills E, Margalit A, Neufeld E, Lee KK, et al. Matefin, a Caenorhabditis elegans germ line-specific SUN-domain nuclear membrane protein, is essential for early embryonic and germ cell development. Proc. Natl. Acad. Sci. USA. 2004;101:6987–92. doi: 10.1073/pnas.0307880101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridolfsson HN, Ly N, Meyerzon M, Starr DA. UNC-83 coordinates kinesin-1 and dynein activities at the nuclear envelope during nuclear migration. Dev. Biol. 2010;338:237–50. doi: 10.1016/j.ydbio.2009.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonczy P, Pichler S, Kirkham M, Hyman AA. Cytoplasmic dynein is required for distinct aspects of MTOC positioning, including centrosome separation, in the one cell stage Caenorhabditis elegans embryo. J. Cell Biol. 1999;147:135–50. doi: 10.1083/jcb.147.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodchild RE, Dauer WT. Mislocalization to the nuclear envelope: an effect of the dystonia-causing torsinA mutation. Proc. Natl. Acad. Sci. USA. 2004;101:847–52. doi: 10.1073/pnas.0304375101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima G, Saitoh S, Yanagida M. Proper metaphase spindle length is determined by centromere proteins Mis12 and Mis6 required for faithful chromosome segregation. Genes Dev. 1999;13:1664–77. doi: 10.1101/gad.13.13.1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gough LL, Beck KA. The spectrin family member Syne-1 functions in retrograde transport from Golgi to ER. Biochim. Biophys. Acta. 2004;1693:29–36. doi: 10.1016/j.bbamcr.2004.04.004. [DOI] [PubMed] [Google Scholar]
- Gough LL, Fan J, Chu S, Winnick S, Beck KA. Golgi localization of Syne-1. Mol. Biol. Cell. 2003;14:2410–24. doi: 10.1091/mbc.E02-07-0446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grady RM, Starr DA, Ackerman GL, Sanes JR, Han M. Syne proteins anchor muscle nuclei at the neuromuscular junction. Proc. Natl. Acad. Sci. USA. 2005;102:4359–64. doi: 10.1073/pnas.0500711102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graumann K, Runions J, Evans DE. Characterization of SUN-domain proteins at the higher plant nuclear envelope. Plant J. 2009;61:134–44. doi: 10.1111/j.1365-313X.2009.04038.x. [DOI] [PubMed] [Google Scholar]
- Gros-Louis F, Dupre N, Dion P, Fox MA, Laurent S, et al. Mutations in SYNE1 lead to a newly discovered form of autosomal recessive cerebellar ataxia. Nat. Genet. 2007;39:80–85. doi: 10.1038/ng1927. [DOI] [PubMed] [Google Scholar]
- Gruenbaum Y, Margalit A, Goldman RD, Shumaker DK, Wilson KL. The nuclear lamina comes of age. Nat. Rev. Mol. Cell Biol. 2005;6:21–31. doi: 10.1038/nrm1550. [DOI] [PubMed] [Google Scholar]
- Guo Y, Jangi S, Welte MA. Organelle-specific control of intracellular transport: distinctly targeted isoforms of the regulator Klar. Mol. Biol. Cell. 2005;16:1406–16. doi: 10.1091/mbc.E04-10-0920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagan I, Yanagida M. The product of the spindle formation gene sad1+ associates with the fission yeast spindle pole body and is essential for viability. J. Cell Biol. 1995;129:1033–47. doi: 10.1083/jcb.129.4.1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haque F, Lloyd DJ, Smallwood DT, Dent CL, Shanahan CM, et al. SUN1 interacts with nuclear lamin A and cytoplasmic nesprins to provide a physical connection between the nuclear lamina and the cytoskeleton. Mol. Cell Biol. 2006;26:3738–51. doi: 10.1128/MCB.26.10.3738-3751.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haque F, Mazzeo D, Patel JT, Smallwood DT, Ellis JA, et al. Mammalian SUN protein networks at the inner nuclear membrane and their role in laminopathy disease processes. J. Biol. Chem. 2010;285:3487–98. doi: 10.1074/jbc.M109.071910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper L, Golubovskaya I, Cande WZ. A bouquet of chromosomes. J. Cell Sci. 2004;117:4025–32. doi: 10.1242/jcs.01363. [DOI] [PubMed] [Google Scholar]
- Hasan S, Guttinger S, Muhlhausser P, Anderegg F, Burgler S, Kutay U. Nuclear envelope localization of human UNC84A does not require nuclear lamins. FEBS Lett. 2006;580:1263–68. doi: 10.1016/j.febslet.2006.01.039. [DOI] [PubMed] [Google Scholar]
- Hedgecock EM, Thomson JN. A gene required for nuclear and mitochondrial attachment in the nematode C. elegans. Cell. 1982;30:321–30. doi: 10.1016/0092-8674(82)90038-1. [DOI] [PubMed] [Google Scholar]
- Hennekam RC. Hutchinson-Gilford progeria syndrome: review of the phenotype. Am. J. Med. Genet. A. 2006;140:2603–24. doi: 10.1002/ajmg.a.31346. [DOI] [PubMed] [Google Scholar]
- Hiraoka Y, Dernburg AF. The SUN rises on meiotic chromosome dynamics. Dev. Cell. 2009;17:598–605. doi: 10.1016/j.devcel.2009.10.014. [DOI] [PubMed] [Google Scholar]
- Hodzic DM, Yeater DB, Bengtsson L, Otto H, Stahl PD. Sun2 is a novel mammalian inner nuclear membrane protein. J. Biol. Chem. 2004;279:25805–12. doi: 10.1074/jbc.M313157200. [DOI] [PubMed] [Google Scholar]
- Horvitz HR, Sulston JE. Isolation and genetic characterization of cell-lineage mutants of the nematode Caenorhabditis elegans. Genetics. 1980;96:435–54. doi: 10.1093/genetics/96.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard EJA, Greenstein D. Introduction to the germ line. WormBook. 2005;2005:1–4. doi: 10.1895/wormbook.1.18.1. http://dev.wormbook.org/chapters/www_introgermline/introgermline.html. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaalouk DE, Lammerding J. Mechanotransduction gone awry. Nat. Rev. Mol. Cell Biol. 2009;10:63–73. doi: 10.1038/nrm2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaspersen SL, Giddings TH, Jr, Winey M. Mps3p is a novel component of the yeast spindle pole body that interacts with the yeast centrin homologue Cdc31p. J. Cell Biol. 2002;159:945–56. doi: 10.1083/jcb.200208169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaspersen SL, Martin AE, Glazko G, Giddings TH, Jr, Morgan G, et al. The Sad1-UNC-84 homology domain in Mps3 interacts with Mps2 to connect the spindle pole body with the nuclear envelope. J. Cell Biol. 2006;174:665–75. doi: 10.1083/jcb.200601062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandert S, Luke Y, Kleinhenz T, Neumann S, Lu W, et al. Nesprin-2 giant safeguards nuclear envelope architecture in LMNA S143F progeria cells. Hum. Mol. Genet. 2007;16:2944–59. doi: 10.1093/hmg/ddm255. [DOI] [PubMed] [Google Scholar]
- Ketema M, Wilhelmsen K, Kuikman I, Janssen H, Hodzic D, Sonnenberg A. Requirements for the localization of nesprin-3 at the nuclear envelope and its interaction with plectin. J. Cell Sci. 2007;120:3384–94. doi: 10.1242/jcs.014191. [DOI] [PubMed] [Google Scholar]
- Khatau SB, Hale CM, Stewart-Hutchinson PJ, Patel MS, Stewart CL, et al. A perinuclear actin cap regulates nuclear shape. Proc. Natl. Acad. Sci. USA. 2009;106:19017–22. doi: 10.1073/pnas.0908686106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King MC, Drivas TG, Blobel G. A network of nuclear envelope membrane proteins linking centromeres to microtubules. Cell. 2008;134:427–38. doi: 10.1016/j.cell.2008.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koszul R, Kim KP, Prentiss M, Kleckner N, Kameoka S. Meiotic chromosomes move by linkage to dynamic actin cables with transduction of force through the nuclear envelope. Cell. 2008;133:1188–201. doi: 10.1016/j.cell.2008.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koszul R, Kleckner N. Dynamic chromosome movements during meiosis: a way to eliminate unwanted connections? Trends Cell Biol. 2009;19:716–24. doi: 10.1016/j.tcb.2009.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kracklauer MP, Banks SML, Xie X, Wu Y, Fischer JA. Drosophila klaroid encodes a SUN domain protein required for Klarsicht localization to the nuclear envelope and nuclear migration in the eye. Fly. 2007;1:75–85. doi: 10.4161/fly.4254. [DOI] [PubMed] [Google Scholar]
- Kyttala M, Tallila J, Salonen R, Kopra O, Kohlschmidt N, et al. MKS1, encoding a component of the flagellar apparatus basal body proteome, is mutated in Meckel syndrome. Nat. Genet. 2006;38:155–57. doi: 10.1038/ng1714. [DOI] [PubMed] [Google Scholar]
- Lambert de Rouvroit C, Goffinet AM. Neuronal migration. Mech. Dev. 2001;105:47–56. doi: 10.1016/s0925-4773(01)00396-3. [DOI] [PubMed] [Google Scholar]
- Lechler T, Fuchs E. Asymmetric cell divisions promote stratification and differentiation of mammalian skin. Nature. 2005;437:275–80. doi: 10.1038/nature03922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JS, Hale CM, Panorchan P, Khatau SB, George JP, et al. Nuclear lamin A/C deficiency induces defects in cell mechanics, polarization, and migration. Biophys. J. 2007;93:2542–52. doi: 10.1529/biophysj.106.102426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KK, Starr DA, Cohen M, Liu J, Han M, et al. Lamin-dependent localization of UNC-84, a protein required for nuclear migration in C. elegans. Mol. Biol. Cell. 2002;13:892–901. doi: 10.1091/mbc.01-06-0294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei K, Zhang X, Ding X, Guo X, Chen M, et al. SUN1 and SUN2 play critical but partially redundant roles in anchoring nuclei in skeletal muscle cells in mice. Proc. Natl. Acad. Sci. USA. 2009;106:10207–12. doi: 10.1073/pnas.0812037106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libotte T, Zaim H, Abraham S, Padmakumar VC, Schneider M, et al. Lamin A/C–dependent localization of Nesprin-2, a giant scaffolder at the nuclear envelope. Mol. Biol. Cell. 2005;16:3411–24. doi: 10.1091/mbc.E04-11-1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu W, Gotzmann J, Sironi L, Jaeger VM, Schneider M, et al. Sun1 forms immobile macromolecular assemblies at the nuclear envelope. Biochim. Biophys. Acta. 2008;1783:2415–26. doi: 10.1016/j.bbamcr.2008.09.001. [DOI] [PubMed] [Google Scholar]
- Luke Y, Zaim H, Karakesisoglou I, Jaeger VM, Sellin L, et al. Nesprin-2 Giant (NUANCE) maintains nuclear envelope architecture and composition in skin. J. Cell Sci. 2008;121:1887–98. doi: 10.1242/jcs.019075. [DOI] [PubMed] [Google Scholar]
- Mahajan R, Delphin C, Guan T, Gerace L, Melchior F. A small ubiquitin-related polypeptide involved in targeting RanGAP1 to nuclear pore complex protein RanBP2. Cell. 1997;88:97–107. doi: 10.1016/s0092-8674(00)81862-0. [DOI] [PubMed] [Google Scholar]
- Malone CJ, Fixsen WD, Horvitz HR, Han M. UNC-84 localizes to the nuclear envelope and is required for nuclear migration and anchoring during C. elegans development. Development. 1999;126:3171–81. doi: 10.1242/dev.126.14.3171. [DOI] [PubMed] [Google Scholar]
- Malone CJ, Misner L, Le Bot N, Tsai MC, Campbell JM, et al. The C. elegans hook protein, ZYG-12, mediates the essential attachment between the centrosome and nucleus. Cell. 2003;115:825–36. doi: 10.1016/s0092-8674(03)00985-1. [DOI] [PubMed] [Google Scholar]
- Maniotis AJ, Chen CS, Ingber DE. Demonstration of mechanical connections between integrins, cytoskeletal filaments, and nucleoplasm that stabilize nuclear structure. Proc. Natl. Acad. Sci. USA. 1997;94:849–54. doi: 10.1073/pnas.94.3.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mans BJ, Anantharaman V, Aravind L, Koonin EV. Comparative genomics, evolution and origins of the nuclear envelope and nuclear pore complex. Cell Cycle. 2004;3:1612–37. doi: 10.4161/cc.3.12.1316. [DOI] [PubMed] [Google Scholar]
- Marme A, Zimmermann HP, Moldenhauer G, Schorpp-Kistner M, Muller C, et al. Loss of Drop1 expression already at early tumor stages in a wide range of human carcinomas. Int. J. Cancer. 2008;123:2048–56. doi: 10.1002/ijc.23763. [DOI] [PubMed] [Google Scholar]
- Mateja A, Szlachcic A, Downing ME, Dobosz M, Mariappan M, et al. The structural basis of tail-anchored membrane protein recognition by Get3. Nature. 2009;461:361–66. doi: 10.1038/nature08319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGee MD, Rillo R, Anderson AS, Starr DA. UNC-83 is a KASH protein required for nuclear migration and is recruited to the outer nuclear membrane by a physical interaction with the SUN protein UNC-84. Mol. Biol. Cell. 2006;17:1790–801. doi: 10.1091/mbc.E05-09-0894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGee MD, Stagljar I, Starr DA. KDP-1 is a nuclear envelope KASH protein required for cell-cycle progression. J. Cell Sci. 2009;122:2895–905. doi: 10.1242/jcs.051607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier I, Xu XM, Brkljacic J, Zhao Q, Wang HJ. Going green: plants’ alternative way to position the Ran gradient. J. Microsc. 2008;231:225–33. doi: 10.1111/j.1365-2818.2008.02038.x. [DOI] [PubMed] [Google Scholar]
- Mejat A, Decostre V, Li J, Renou L, Kesari A, et al. Lamin A/C-mediated neuromuscular junction defects in Emery-Dreifuss muscular dystrophy. J. Cell Biol. 2009;184:31–44. doi: 10.1083/jcb.200811035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyerzon M, Fridolfsson HN, Ly N, McNally FJ, Starr DA. UNC-83 is a nuclear-specific cargo adaptor for kinesin-1-mediated nuclear migration. Development. 2009a;136:2725–33. doi: 10.1242/dev.038596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyerzon M, Gao Z, Liu J, Wu JC, Malone CJ, Starr DA. Centrosome attachment to the C. elegans male pronucleus is dependent on the surface area of the nuclear envelope. Dev. Biol. 2009b;327:433–46. doi: 10.1016/j.ydbio.2008.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki F, Kurabayashi A, Tange Y, Okazaki K, Shimanuki M, Niwa O. Two-hybrid search for proteins that interact with Sad1 and Kms1, two membrane-bound components of the spindle pole body in fission yeast. Mol. Genet. Genomics. 2004;270:449–61. doi: 10.1007/s00438-003-0938-8. [DOI] [PubMed] [Google Scholar]
- Minn IL, Rolls MM, Hanna-Rose W, Malone CJ. SUN-1 and ZYG-12, mediators of centrosome-nucleus attachment, are a functional SUN/KASH pair in Caenorhabditis elegans. Mol. Biol. Cell. 2009;20:4586–95. doi: 10.1091/mbc.E08-10-1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mislow JM, Holaska JM, Kim MS, Lee KK, Segura-Totten M, et al. Nesprin-1α self-associates and binds directly to emerin and lamin A in vitro. FEBS Lett. 2002a;525:135–40. doi: 10.1016/s0014-5793(02)03105-8. [DOI] [PubMed] [Google Scholar]
- Mislow JM, Kim MS, Davis DB, McNally EM. Myne-1, a spectrin repeat transmembrane protein of the myocyte inner nuclear membrane, interacts with lamin A/C. J. Cell Sci. 2002b;115:61–70. doi: 10.1242/jcs.115.1.61. [DOI] [PubMed] [Google Scholar]
- Morris SM, Albrecht U, Reiner O, Eichele G, Yu-Lee L. The lissencephaly gene product Lis1, a protein involved in neuronal migration, interacts with a nuclear movement protein, NudC. Curr. Biol. 1998;8:603–6. doi: 10.1016/s0960-9822(98)70232-5. [DOI] [PubMed] [Google Scholar]
- Mosley-Bishop KL, Li Q, Patterson L, Fischer JA. Molecular analysis of the klarsicht gene and its role in nuclear migration within differentiating cells of the Drosophila eye. Curr. Biol. 1999;9:1211–20. doi: 10.1016/s0960-9822(99)80501-6. [DOI] [PubMed] [Google Scholar]
- Naismith TV, Heuser JE, Breakefield XO, Hanson PI. TorsinA in the nuclear envelope. Proc. Natl. Acad. Sci. USA. 2004;101:7612–17. doi: 10.1073/pnas.0308760101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nery FC, Zeng J, Niland BP, Hewett J, Farley J, et al. TorsinA binds the KASH domain of nesprins and participates in linkage between nuclear envelope and cytoskeleton. J. Cell Sci. 2008;121:3476–86. doi: 10.1242/jcs.029454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolova V, Leimena C, McMahon AC, Tan JC, Chandar S, et al. Defects in nuclear structure and function promote dilated cardiomyopathy in lamin A/C-deficient mice. J. Clin. Investig. 2004;113:357–69. doi: 10.1172/JCI19448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostlund C, Folker ES, Choi JC, Gomes ER, Gundersen GG, Worman HJ. Dynamics and molecular interactions of linker of nucleoskeleton and cytoskeleton (LINC) complex proteins. J. Cell Sci. 2009;122:4099–108. doi: 10.1242/jcs.057075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oza P, Jaspersen SL, Miele A, Dekker J, Peterson CL. Mechanisms that regulate localization of a DNA double-strand break to the nuclear periphery. Genes Dev. 2009;23:912–27. doi: 10.1101/gad.1782209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozelius LJ, Page CE, Klein C, Hewett JW, Mineta M, et al. The TOR1A (DYT1) gene family and its role in early onset torsion dystonia. Genomics. 1999;62:377–84. doi: 10.1006/geno.1999.6039. [DOI] [PubMed] [Google Scholar]
- Padmakumar VC, Abraham S, Braune S, Noegel AA, Tunggal B, et al. Enaptin, a giant actin-binding protein, is an element of the nuclear membrane and the actin cytoskeleton. Exp. Cell Res. 2004;295:330–39. doi: 10.1016/j.yexcr.2004.01.014. [DOI] [PubMed] [Google Scholar]
- Padmakumar VC, Libotte T, Lu W, Zaim H, Abraham S, et al. The inner nuclear membrane protein Sun1 mediates the anchorage of Nesprin-2 to the nuclear envelope. J. Cell Sci. 2005;118:3419–30. doi: 10.1242/jcs.02471. [DOI] [PubMed] [Google Scholar]
- Patterson K, Molofsky AB, Robinson C, Acosta S, Cater C, Fischer JA. The functions of klarsicht and nuclear lamin in developmentally regulated nuclear migrations of photoreceptor cells in the Drosophila eye. Mol. Biol. Cell. 2004;15:600–10. doi: 10.1091/mbc.E03-06-0374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penkner A, Tang L, Novatchkova M, Ladurner M, Fridkin A, et al. The nuclear envelope protein Matefin/SUN-1 is required for homologous pairing in C. elegans meiosis. Dev. Cell. 2007;12:873–85. doi: 10.1016/j.devcel.2007.05.004. [DOI] [PubMed] [Google Scholar]
- Penkner AM, Fridkin A, Gloggnitzer J, Baudrimont A, Machacek T, et al. Meiotic chromosome homology search involves modifications of the nuclear envelope protein Matefin/SUN-1. Cell. 2009;139:920–33. doi: 10.1016/j.cell.2009.10.045. [DOI] [PubMed] [Google Scholar]
- Prokocimer M, Davidovich M, Nissim-Rafinia M, Wiesel-Motiuk N, Bar D, et al. Nuclear lamins: key regulators of nuclear structure and activities. J. Cell Mol. Med. 2009;13:1059–85. doi: 10.1111/j.1582-4934.2008.00676.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puckelwartz MJ, Kessler E, Zhang Y, Hodzic D, Randles KN, et al. Disruption of nesprin-1 produces an Emery Dreifuss muscular dystrophy-like phenotype in mice. Hum. Mol. Genet. 2009;18:607–20. doi: 10.1093/hmg/ddn386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabu C, Schmid V, Schwappach B, High S. Biogenesis of tail-anchored proteins: the beginning for the end? J. Cell Sci. 2009;122:3605–12. doi: 10.1242/jcs.041210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralston E, Lu Z, Biscocho N, Soumaka E, Mavroidis M, et al. Blood vessels and desmin control the positioning of nuclei in skeletal muscle fibers. J. Cell Physiol. 2006;209:874–82. doi: 10.1002/jcp.20780. [DOI] [PubMed] [Google Scholar]
- Reinsch S, Gonczy P. Mechanisms of nuclear positioning. J. Cell Sci. 1998;111:2283–95. doi: 10.1242/jcs.111.16.2283. [DOI] [PubMed] [Google Scholar]
- Rivero F, Kuspa A, Brokamp R, Matzner M, Noegel AA. Interaptin, an actin-binding protein of the α-actinin superfamily in Dictyostelium discoideum, is developmentally and cAMP-regulated and associates with intracellular membrane compartments. J. Cell Biol. 1998;142:735–50. doi: 10.1083/jcb.142.3.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux KJ, Crisp ML, Liu Q, Kim D, Kozlov S, et al. Nesprin 4 is an outer nuclear membrane protein that can induce kinesin-mediated cell polarization. Proc. Natl. Acad. Sci. USA. 2009;106:2194–99. doi: 10.1073/pnas.0808602106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salpingidou G, Smertenko A, Hausmanowa-Petrucewicz I, Hussey PJ, Hutchison CJ. A novel role for the nuclear membrane protein emerin in association of the centrosome to the outer nuclear membrane. J. Cell Biol. 2007;178:897–904. doi: 10.1083/jcb.200702026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato A, Isaac B, Phillips CM, Rillo R, Carlton PM, et al. Cytoskeletal forces span the nuclear envelope to coordinate meiotic chromosome pairing and synapsis. Cell. 2009;139:907–19. doi: 10.1016/j.cell.2009.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaar BT, McConnell SK. Cytoskeletal coordination during neuronal migration. Proc. Natl. Acad. Sci. USA. 2005;102:13652–57. doi: 10.1073/pnas.0506008102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt J, Benavente R, Hodzic D, Hoog C, Stewart CL, Alsheimer M. Transmembrane protein Sun2 is involved in tethering mammalian meiotic telomeres to the nuclear envelope. Proc. Natl. Acad. Sci. USA. 2007;104:7426–31. doi: 10.1073/pnas.0609198104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider M, Noegel AA, Karakesisoglou I. KASH-domain proteins and the cytoskeletal landscapes of the nuclear envelope. Biochem. Soc. Trans. 2008;36:1368–72. doi: 10.1042/BST0361368. [DOI] [PubMed] [Google Scholar]
- Schuldiner M, Metz J, Schmid V, Denic V, Rakwalska M, et al. The GET complex mediates insertion of tail-anchored proteins into the ER membrane. Cell. 2008;134:634–45. doi: 10.1016/j.cell.2008.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao X, Tarnasky HA, Lee JP, Oko R, Van Der Hoorn FA. Spag4, a novel sperm protein, binds outer dense-fiber protein Odf1 and localizes to microtubules of manchette and axoneme. Dev. Biol. 1999;211:109–23. doi: 10.1006/dbio.1999.9297. [DOI] [PubMed] [Google Scholar]
- Shubeita GT, Tran SL, Xu J, Vershinin M, Cermelli S, et al. Consequences of motor copy number on the intracellular transport of kinesin-1-driven lipid droplets. Cell. 2008;135:1098–107. doi: 10.1016/j.cell.2008.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjoblom T, Jones S, Wood LD, Parsons DW, Lin J, et al. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314:268–74. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- Starr DA. A nuclear-envelope bridge positions nuclei and moves chromosomes. J. Cell Sci. 2009;122:577–86. doi: 10.1242/jcs.037622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr DA, Fischer JA. KASH ’n Karry: the KASH domain family of cargo-specific cytoskeletal adaptor proteins. Bioessays. 2005;27:1136–46. doi: 10.1002/bies.20312. [DOI] [PubMed] [Google Scholar]
- Starr DA, Han M. Role of ANC-1 in tethering nuclei to the actin cytoskeleton. Science. 2002;298:406–9. doi: 10.1126/science.1075119. [DOI] [PubMed] [Google Scholar]
- Starr DA, Han M. ANChors away: an actin based mechanism of nuclear positioning. J. Cell Sci. 2003;116:211–16. doi: 10.1242/jcs.00248. [DOI] [PubMed] [Google Scholar]
- Starr DA, Han M. A genetic approach to study the role of nuclear envelope components in nuclear positioning. Novartis Found. Symp. 2005;264:208–19. discussion 19–30. [PubMed] [Google Scholar]
- Starr DA, Hermann GJ, Malone CJ, Fixsen W, Priess JR, et al. unc-83 encodes a novel component of the nuclear envelope and is essential for proper nuclear migration. Development. 2001;128:5039–50. doi: 10.1242/dev.128.24.5039. [DOI] [PubMed] [Google Scholar]
- Stefanovic S, Hegde RS. Identification of a targeting factor for posttranslational membrane protein insertion into the ER. Cell. 2007;128:1147–59. doi: 10.1016/j.cell.2007.01.036. [DOI] [PubMed] [Google Scholar]
- Stewart-Hutchinson PJ, Hale CM, Wirtz D, Hodzic D. Structural requirements for the assembly of LINC complexes and their function in cellular mechanical stiffness. Exp. Cell Res. 2008;314:1892–905. doi: 10.1016/j.yexcr.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan T, Escalante-Alcalde D, Bhatt H, Anver M, Bhat N, et al. Loss of A-type lamin expression compromises nuclear envelope integrity leading to muscular dystrophy. J. Cell Biol. 1999;147:913–20. doi: 10.1083/jcb.147.5.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulston JE, Horvitz HR. Abnormal cell lineages in mutants of the nematode Caenorhabditis elegans. Dev. Biol. 1981;82:41–55. doi: 10.1016/0012-1606(81)90427-9. [DOI] [PubMed] [Google Scholar]
- Tanabe LM, Kim CE, Alagem N, Dauer WT. Primary dystonia: molecules and mechanisms. Nat. Rev. Neurol. 2009;5:598–609. doi: 10.1038/nrneurol.2009.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Technau M, Roth S. The Drosophila KASH domain proteins Msp-300 and Klarsicht and the SUN domain protein Klaroid have no essential function during oogenesis. Fly. 2008;2:82–91. doi: 10.4161/fly.6288. [DOI] [PubMed] [Google Scholar]
- Toivola DM, Tao GZ, Habtezion A, Liao J, Omary MB. Cellular integrity plus: organelle-related and protein-targeting functions of intermediate filaments. Trends Cell Biol. 2005;15:608–17. doi: 10.1016/j.tcb.2005.09.004. [DOI] [PubMed] [Google Scholar]
- Tomlinson A. The cellular dynamics of pattern formation in the eye of Drosophila. J. Embryol. Exp. Morphol. 1985;89:313–31. [PubMed] [Google Scholar]
- Tran PT, Marsh L, Doye V, Inoue S, Chang F. A mechanism for nuclear positioning in fission yeast based on microtubule pushing. J. Cell Biol. 2001;153:397–411. doi: 10.1083/jcb.153.2.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai JW, Bremner KH, Vallee RB. Dual subcellular roles for LIS1 and dynein in radial neuronal migration in live brain tissue. Nat. Neurosci. 2007;10:970–79. doi: 10.1038/nn1934. [DOI] [PubMed] [Google Scholar]
- Tsujikawa M, Omori Y, Biyanwila J, Malicki J. Mechanism of positioning the cell nucleus in vertebrate photoreceptors. Proc. Natl. Acad. Sci. USA. 2007;104:14819–24. doi: 10.1073/pnas.0700178104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzur YB, Margalit A, Melamed-Book N, Gruenbaum Y. Matefin/SUN-1 is a nuclear envelope receptor for CED-4 during Caenorhabditis elegans apoptosis. Proc. Natl. Acad. Sci. USA. 2006a;103:13397–402. doi: 10.1073/pnas.0604224103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzur YB, Wilson KL, Gruenbaum Y. SUN-domain proteins: ‘Velcro’ that links the nucleoskeleton to the cytoskeleton. Nat. Rev. Mol. Cell Biol. 2006b;7:782–88. doi: 10.1038/nrm2003. [DOI] [PubMed] [Google Scholar]
- Vander Heyden AB, Naismith TV, Snapp EL, Hodzic D, Hanson PI. LULL1 retargets TorsinA to the nuclear envelope revealing an activity that is impaired by the DYT1 dystonia mutation. Mol. Biol. Cell. 2009;20:2661–72. doi: 10.1091/mbc.E09-01-0094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vergnes L, Peterfy M, Bergo MO, Young SG, Reue K. Lamin B1 is required for mouse development and nuclear integrity. Proc. Natl. Acad. Sci. USA. 2004;101:10428–33. doi: 10.1073/pnas.0401424101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volk T. A new member of the spectrin superfamily may participate in the formation of embryonic muscle attachments in Drosophila. Development. 1992;116:721–30. doi: 10.1242/dev.116.3.721. [DOI] [PubMed] [Google Scholar]
- Walenta JH, Didier AJ, Liu X, Kramer H. The Golgi-associated hook3 protein is a member of a novel family of microtubule-binding proteins. J. Cell Biol. 2001;152:923–34. doi: 10.1083/jcb.152.5.923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N, Tytell JD, Ingber DE. Mechanotransduction at a distance: mechanically coupling the extra-cellular matrix with the nucleus. Nat. Rev. Mol. Cell Biol. 2009;10:75–82. doi: 10.1038/nrm2594. [DOI] [PubMed] [Google Scholar]
- Welte MA. Bidirectional transport along microtubules. Curr. Biol. 2004;14:R525–37. doi: 10.1016/j.cub.2004.06.045. [DOI] [PubMed] [Google Scholar]
- Welte MA, Gross SP, Postner M, Block SM, Wieschaus EF. Developmental regulation of vesicle transport in Drosophila embryos: forces and kinetics. Cell. 1998;92:547–57. doi: 10.1016/s0092-8674(00)80947-2. [DOI] [PubMed] [Google Scholar]
- Wheeler MA, Davies JD, Zhang Q, Emerson LJ, Hunt J, et al. Distinct functional domains in nesprin-1α and nesprin-2β bind directly to emerin and both interactions are disrupted in X-linked Emery-Dreifuss muscular dystrophy. Exp. Cell Res. 2007;313:2845–57. doi: 10.1016/j.yexcr.2007.03.025. [DOI] [PubMed] [Google Scholar]
- Wiche G. Role of plectin in cytoskeleton organization and dynamics. J. Cell Sci. 1998;111(Pt. 17):2477–86. doi: 10.1242/jcs.111.17.2477. [DOI] [PubMed] [Google Scholar]
- Wilhelmsen K, Ketema M, Truong H, Sonnenberg A. KASH-domain proteins in nuclear migration, anchorage and other processes. J. Cell Sci. 2006;119:5021–29. doi: 10.1242/jcs.03295. [DOI] [PubMed] [Google Scholar]
- Wilhelmsen K, Litjens SH, Kuikman I, Tshimbalanga N, Janssen H, et al. Nesprin-3, a novel outer nuclear membrane protein, associates with the cytoskeletal linker protein plectin. J. Cell Biol. 2005;171:799–810. doi: 10.1083/jcb.200506083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood WB, Hecht R, Carr S, Vanderslice R, Wolf N, Hirsh D. Parental effects and phenotypic characterization of mutations that affect early development in Caenorhabditis elegans. Dev. Biol. 1980;74:446–69. doi: 10.1016/0012-1606(80)90445-5. [DOI] [PubMed] [Google Scholar]
- Worman HJ, Bonne G. “Laminopathies”: a wide spectrum of human diseases. Exp. Cell Res. 2007;313:2121–33. doi: 10.1016/j.yexcr.2007.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worman HJ, Gundersen GG. Here come the SUNs: a nucleocytoskeletal missing link. Trends Cell Biol. 2006;16:67–69. doi: 10.1016/j.tcb.2005.12.006. [DOI] [PubMed] [Google Scholar]
- Xie X, Fischer JA. On the roles of the Drosophila KASH domain proteins Msp-300 and Klarsicht. Fly. 2008;2:74–81. doi: 10.4161/fly.6108. [DOI] [PubMed] [Google Scholar]
- Xiong H, Rivero F, Euteneuer U, Mondal S, Mana-Capelli S, et al. Dictyostelium Sun-1 connects the centrosome to chromatin and ensures genome stability. Traffic. 2008;9:708–24. doi: 10.1111/j.1600-0854.2008.00721.x. [DOI] [PubMed] [Google Scholar]
- Xu XM, Meulia T, Meier I. Anchorage of plant RanGAP to the nuclear envelope involves novel nuclear-pore-associated proteins. Curr. Biol. 2007;17:1157–63. doi: 10.1016/j.cub.2007.05.076. [DOI] [PubMed] [Google Scholar]
- Yu J, Starr DA, Wu X, Parkhurst SM, Zhuang Y, et al. The KASH domain protein MSP-300 plays an essential role in nuclear anchoring during Drosophila oogenesis. Dev. Biol. 2006;289:336–45. doi: 10.1016/j.ydbio.2005.10.027. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Bethmann C, Worth NF, Davies JD, Wasner C, et al. Nesprin-1 and -2 are involved in the pathogenesis of Emery Dreifuss muscular dystrophy and are critical for nuclear envelope integrity. Hum. Mol. Genet. 2007a;16:2816–33. doi: 10.1093/hmg/ddm238. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Ragnauth C, Greener MJ, Shanahan CM, Roberts RG. The nesprins are giant actin-binding proteins, orthologous to Drosophila melanogaster muscle protein MSP-300. Genomics. 2002;80:473–81. [PubMed] [Google Scholar]
- Zhang Q, Skepper JN, Yang F, Davies JD, Hegyi L, et al. Nesprins: a novel family of spectrin-repeat-containing proteins that localize to the nuclear membrane in multiple tissues. J. Cell Sci. 2001;114:4485–98. doi: 10.1242/jcs.114.24.4485. [DOI] [PubMed] [Google Scholar]
- Zhang X, Lei K, Yuan X, Wu X, Zhuang Y, et al. SUN1/2 and Syne/Nesprin-1/2 complexes connect centrosome to the nucleus during neurogenesis and neuronal migration in mice. Neuron. 2009;64:173–87. doi: 10.1016/j.neuron.2009.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Xu R, Zhu B, Yang X, Ding X, et al. Syne-1 and Syne-2 play crucial roles in myonuclear anchorage and motor neuron innervation. Development. 2007b;134:901–8. doi: 10.1242/dev.02783. [DOI] [PubMed] [Google Scholar]
- Zhen YY, Libotte T, Munck M, Noegel AA, Korenbaum E. NUANCE, a giant protein connecting the nucleus and actin cytoskeleton. J. Cell Sci. 2002;115:3207–22. doi: 10.1242/jcs.115.15.3207. [DOI] [PubMed] [Google Scholar]
- Zhou K, Rolls MM, Hall DH, Malone CJ, Hanna-Rose W. A ZYG-12-dynein interaction at the nuclear envelope defines cytoskeletal architecture in the C. elegans gonad. J. Cell Biol. 2009;186:229–41. doi: 10.1083/jcb.200902101. [DOI] [PMC free article] [PubMed] [Google Scholar]