Abstract
Methyl CpG binding protein 2 (MeCP2) is an X-linked, multifunctional epigenetic regulator that is best known for its role in the neurological disorder Rett Syndrome; however, it is also linked to multiple autoimmune disorders. We examined a potential role for MeCP2 in regulating the responses of CD4+ T cells to stimulation with antigen. We showed that MeCP2 was indispensable for the differentiation of naïve CD4+ T cells into T helper type 1 (TH1) and TH17 cells and for TH1- or TH17-mediated pathologies in vitro and in vivo. Loss of MeCP2 in CD4+ T cells impaired the expression of the microRNA (miR) miR-124, and consequently relieved miR-124-mediated repression of the translation of suppressor of cytokine signaling 5 (Socs5) mRNA. The resulting accumulation of SOCS5 protein led to inhibition of the cytokine-dependent activation of signal transducer and activator of transcription 1 (STAT1) and STAT3, which are necessary for the differentiation of TH1 and TH17 cells, respectively. Upon silencing of MeCP2, primary neurons and astrocytes also failed to respond properly to STAT3-dependent signaling stimulated by neurotrophic factors. Together, these findings suggest that the regulation of STAT3 signaling may represent a common etiology underpinning the roles of MeCP2 in both the nervous and immune systems.
Introduction
Methyl CpG binding protein 2 (MeCP2) was initially identified as a nuclear protein that binds to cytosine-methylated DNA within dinucleotide CpG elements (1, 2). In early studies, MeCP2 was described as a transcriptional silencer, because it appeared to maintain the dinucleotide methylation state of target genes and to recruit the corepressor Sin3A and histone deacetylase 1 (HDAC1) and HDAC2 to these methylated CpG sites (3, 4). However, biochemical and genomics data suggest that, rather than acting strictly a silencer, MeCP2 acts as a multifunctional regulator of gene transcription (5, 6), and that it is actively involved in RNA splicing (7), chromatin remodeling (8, 9), and transcriptional activation (10, 11). MeCP2 is especially relevant in biomedicine because of its role in Rett syndrome (RTT) (12, 13), a progressive neurodevelopmental disorder that manifests in young girls at a ratio of 1:10,000 (14, 15). Heterozygous loss-of-function mutations in MECP2 underlie the etiology for more than 95% of typical RTT patients (13, 16); however, the resultant molecular pathology remains largely elusive (5). The neurodegenerative phenotype of RTT is the result of the loss of MeCP2 specifically in neuronal cells (17, 18), and it is unlikely to rely on immune cell dysfunction (19, 20).
MeCP2 is not limited to the brain, and studies have implicated it in the regulation of immunological disorders. Specifically, polymorphisms in MECP2 in humans have been linked to increased susceptibility to autoimmune diseases, such as systemic lupus erythematosus (SLE) (21, 22) and primary Sjogren’s syndrome (pSS) (23). Moreover, MeCP2 associates with CpG elements within the regulatory regions of Foxp3 (24), which encodes a transcription factor required for the generation of regulatory T (Treg) cells, although the functional consequence of this association is yet to be examined. Thus, although RTT does not appear to be phenotypically linked to immune cell dysregulation, we postulate that the roles of MeCP2 in neuronal cells and in T cells might nonetheless be mechanistically linked by some common molecular pathways. We therefore generated mice that had a T cell–specific loss of Mecp2 to investigate the potential role of MeCP2 in T cell function and immune regulation. Mechanistically, our investigation identified the microRNA (miR) miR-124, which represses the translation of mRNA for suppressor of cytokine signaling 5 (Socs5), as a direct downstream effector of MeCP2-mediated epigenetic regulation. Functionally, the MeCP2–miR-124–SOCS5 axis was indispensable for the cytokine-dependent activation of signal transducer and activator of transcription 3 (STAT3) in naïve CD4+ T cells, and consequently for the generation of T helper type 17 (TH17) cells.
Results
MeCP2 is indispensable for the differentiation of naïve CD4+ T cells into TH17 cells
Because the association between MECP2 polymorphisms and autoimmune diseases was demonstrated by recent human genetic studies, we used the CD4-Cre transgene to induce specific deletion of Mecp2 in both natural Treg (nTreg) cells and conventional T (Tcon) cells in mice. Since Mecp2 resides on the X chromosome, male transgenic mice carry a single floxed allele. Examination of sorted T cells, B cells, as well as of the brain and lung tissues of these CD4-Cre+Mecp2f/y mice confirmed that loss of MeCP2 protein occurred specifically in T cells, but not other cell types (fig. S1A). Consistent with earlier reports (25, 26), we observed that compared to their wild-type CD4-Cre+Mecp2x/y littermate controls, CD4-Cre−Mecp2f/y mice carrying floxed Mecp2 alleles demonstrated hypomorphic MeCP2 abundance (reduced expression) in the brain and lung tissues (Fig. S1A). Nevertheless, such hypomorphism did not occur in the lymphoid compartments of T cells and B cells (fig. S1A). Therefore, both CD4-Cre−Mecp2 f/y mice and CD4-Cre+Mecp2 x/y mice served as controls in our experiments.
In young adult mice and in mice up to one year of age, the deletion of MeCP2 in T cells (Fig. 1A and fig. S1A) did not result in any overt T cell developmental irregularities (fig. S1, B and C) or in spontaneous T cell activation (fig. S2). To examine the intrinsic autoinflammatory potential of CD4+ Tcon cells, sorted CD4+CD25−CD45Rbhigh naïve Tcon cells from MeCP2-deficient or wild-type mice, in combination with wild-type nTreg cells, were transferred into lymphopenic Rag2−/− recipient mice at a ratio of 25:1 to induce autoimmune colitis, which is an effector T cell-mediated inflammation of the large intestine (27). Mice that received wild-type Tcon cells developed severe experimental colitis, which was evidenced by a continuous loss in body weight; however, the transfer of MeCP2-deficient T cells conferred the recipient mice with disease resistance rather than promoting inflammation (Fig. 1B). Further histopathological analysis of the colons of these mice illustrated a marked difference in the extent of infiltration by inflammatory leukocytes and in the integrity of the mucosal tissue architecture (Fig. 1C). In this transfer model, the aberrant generation of Th17 cells against host microbiota is the major cause of intestinal inflammation (28). Accordingly, in the mesenteric lymph nodes of mice bearing MeCP2-deficient Tcon cells, the proportion of CD4+ T cells that produced interleukin-17 (IL-17) was significantly reduced compared to that in mice that received wild-type Tcon cells (Fig. 1D).
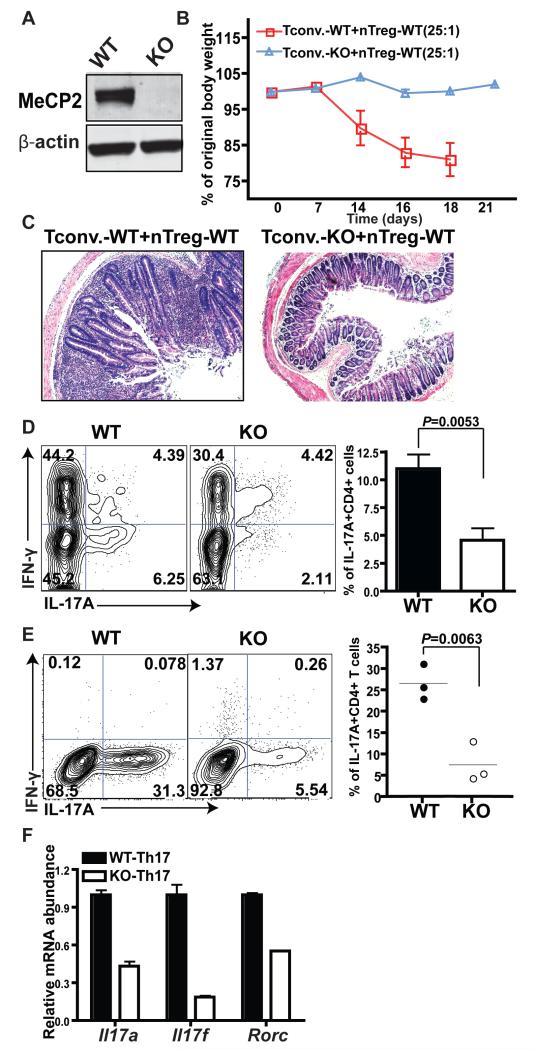
Fig. 1. MeCP2 is indispensable for the differentiation of naive CD4+ T cells into TH17 cells.
(A) Naïve CD4+CD25− T cells from CD4-Cre+Mecp2x/x [wild-type (WT)] and CD4-Cre+Mecp2f/f [(MeCP2 knockout (KO)] littermates were sorted by flow cytometry. Cell lysates were analyzed by western blotting to detect MeCP2 protein. Data shown represents three independent experiments. (B to D) CD4+CD25−CD45Rbhigh naïve T cells from WT or MeCP2 KO mice were mixed with WT nTreg cells at a ratio of 25:1 and were injected intraperitoneally (i.p.) into Rag2−/− recipient mice to induce IBD. (B) Changes in the body weights of the Rag2−/− recipient mice are presented as percentages of their original weights. Data are means ± SEM from five mice of each group from a single experiment and are representative of four independent experiments. (C) Histological sections of colon tissues obtained from the indicated mice were subjected to H&E staining. Images are representative of samples from four WT mice and eight KO mice. (D) Left: Flow cytometric analysis of the percentages of IL-17A-producing CD4+TCRβ+ cells in mesenteric lymph nodes. Right: Data are means ± SEM from four WT mice and eight KO mice from a single experiment and are representative of four independent experiments. (E and F) Lymphocytes from LLO118 TCR transgenic WT and KO littermates were cultured in vitro under TH17-polarizing conditions for 4 days. (E) Left: Flow cytometric analysis of the percentages of IL-17A-producing CD4+ T cells. Right: Quantification of flow cytometry data from three independent experiments. Data are means ± SEM. (F) CD4+ T cells were sorted by flow cytometry, and the relative amounts of Il17a, Il17f, and Rorc mRNAs were measured by quantitative PCR analysis. The abundances of the mRNAs of interest were normalized to that of succinate dehydrogenase complex, subunit A, flavoprotein (Fp) (Sdha) and are shown relative to those of the WT. Data are means ± SEM from three biological replicates and represent two independent experiments.
In response to specific immunization conditions, Tcon cells proliferate to increase cell numbers, undergo contraction to reduce cell numbers, and then differentiate into various TH cell lineages to orchestrate appropriate immune responses related to host defence and tolerance (29). Defects in any of these steps could account for the reduced size of the autoinflammatory Th17 cell population observed in mice that received MeCP2-deficient Tcon cells. To further dissect the intrinsic defect of MeCP2-deficient CD4+ T cells during inflammatory responses, we backcrossed CD4-Cre+Mecp2f/f or CD4-Cre+Mecp2f/y mice with strains carrying the LLO118 T cell receptor (TCR) transgene (30), which recognizes the dominant Listeria monocytogenes antigen (corresponding to amino acid residues 190 to 205 of the Listeriolysin O protein) in the context of the I-Ab major histocompatibility complex (MHC) class II molecule. Upon in vitro stimulation with antigen-presenting cells (APCs) that were loaded with LLO190-205 peptide, CD4+ Tcon cells proliferated and then contracted comparably in the presence or absence of MeCP2 protein (fig. S3). However, when we cultured these cells under Th17-polarizing conditions in vitro, MeCP2-deficient Tcon cells exhibited severe defects in IL-17A production (Fig. 1E). Consistent with this, the abundances of messenger RNAs (mRNAs) for Il-17a, Il-17f, and rorc, which encodes the master transcription factor for Th17 cells, RORγT, were markedly reduced in MeCP2-deficient Th17 cells compared to those in wild-type cells (Fig. 1F).
MeCP2 is indispensable for the commitment of naïve CD4+ T cells to the TH1 lineage
Interferon-γ (IFN-γ)–producing Th1 cells represent another critical TH cell subset that mediates the onset and progression of autoimmune diseases (31, 32). With our TCR transgenic mouse models, we examined whether MeCP2 played any role in the differentiation of naïve CD4+ T cells into TH1 cells. When naïve CD4+ T cells were cultured under Th1-polarizing conditions in vitro, the amount of IFN-γ produced by the MeCP2-deficicent CD4+ T cells was substantially reduced compared to that produced by wild-type CD4+ T cells (Fig. 2A). To validate this phenotype in vivo, we sorted and transferred wild-type or MeCP2-deficient LLO118 T cells into TCRα−/− recipient mice that lack endogenous αβ T cells, and then immunized the recipient mice subcutaneously with LLO190-205 peptide emulsified in complete Freund’s adjuavant (CFA). The effector T cells that were generated in vivo were subsequently rechallenged in vitro with the same antigen to assess their functional differentiation. As observed in vitro (Fig. 2A), MeCP2-deficient CD4+ T cells were markedly impaired in their ability to differentiate into IFN-γ-producing Th1 cells (Fig. 2B). To exclude the possibility that this phenotype was limited to the LLO118 TCR, we directly challenged wild-type or MeCP2-deficient mice with keyhole limpet hemocyanin (KLH) protein through the footpad to induce delayed-type hypersensitivity (DTH), a classical Th1-dominated immune response. Defects in Th1 responses were apparent in mice with a T cell–specific deletion of MeCP2, as characterized by limited footpad swelling (Fig. 2C), reduced infiltration by inflammatory cells (Fig. 2D), and reduced numbers of Th1 cells in the draining lymph node (Fig. 2E). Collectively, our data indicate that MeCP2 is an indispensable factor that drives the differentiation of naïve CD4+ T cells into Th1 and Th17 cells in vitro and in vivo.
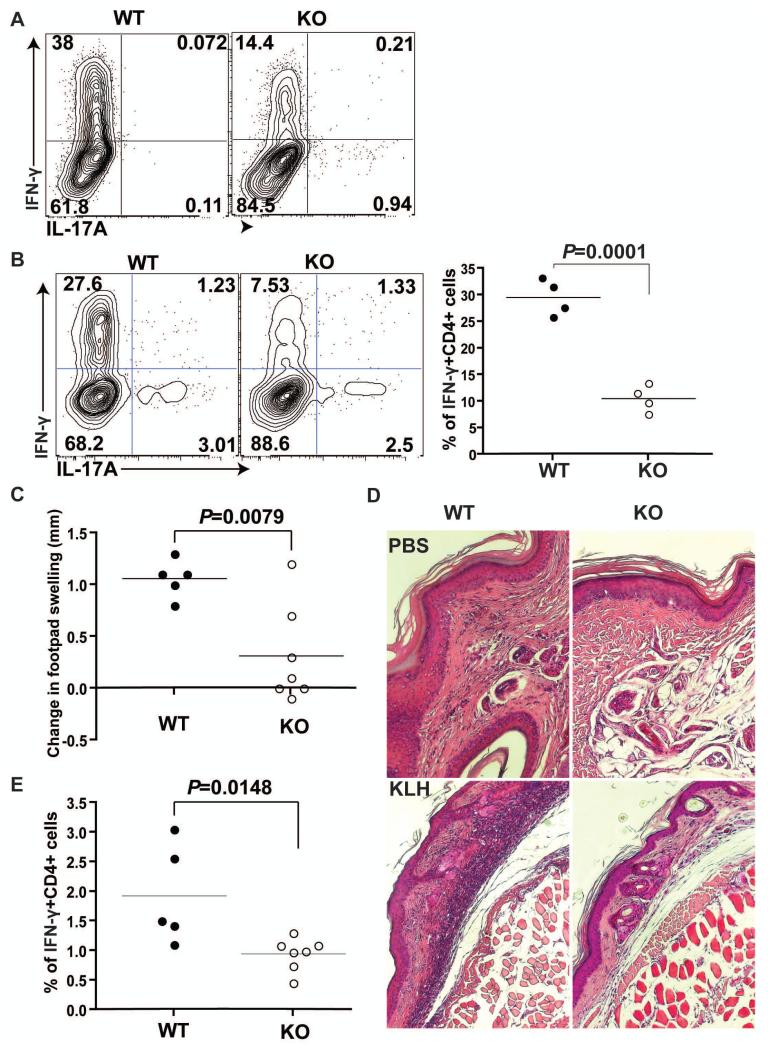
Fig. 2. MeCP2 is indispensable for IFN-γ production by TH1 cells.
(A) CD4+ T cells from LLO118 TCR transgenic WT and MeCP2 KO littermates were activated with LLO190-205 peptide under TH1-skewing conditions in vitro for 4 days. The percentages of IFN-γ–producing CD4+ T cells were determined by intracellular staining and flow cytometric analysis. Data are representative of three independent experiments. (B) Naïve CD4+CD25− T cells from LLO118 TCR transgenic WT or MeCP2 KO mice were transferred into TCRα−/− recipient mice, which were subsequently primed in vivo by subcutaneous immunization with LLO190-205 peptide emulsified in CFA. Five days after immunization, splenocytes of recipient mice were isolated and then challenged in vitro with 5 μM LLO190-205 peptide. Left: Forty-eight hours later, IFN-γ production in CD4+ T cells was detected by intracellular staining and flow cytometric analysis. Right: Data are means ± SEM from four mice of each genotype and are representative of two independent experiments. (C to E) Analysis of the DTH responses of WT and MeCP2 KO littermate mice that were immunized with KLH. (C) The extent of footpad swelling was measured. Data are means ± SEM from five WT mice and seven KO mice and represent two independent experiments. (D) The histology of footpad tissues was determined by H&E staining, Note the thickness difference of the dermis layer and the infiltration of inflammatory mononuclear cells into the epithelium and dermis upon KLH immunization and re-challenge. Images represent samples from five WT mice and seven KO mice .(E) The percentages of IFN-γ–producing CD4+ T cells in the popliteal lymph nodes were determined by flow cytometric analysis. Data are means ± SEM from five WT mice and seven KO mice and are representative of two independent experiments.
MeCP2 is indispensable for the activation of STAT3 and STAT1 in CD4+ T cells
To differentiate effector T cells into various subsets from naïve precursors, CD4+ T cells respond to signals from APCs and the cytokine environment by activating specific transcription factors, which then lead to chromatin remodeling surrounding the master cytokine genes (29, 33). Because MeCP2 primarily functions at the epigenetic level, we first examined whether the loss of MeCP2 directly impaired the accessibility of the Il17 and Ifng loci. With chromatin immunoprecipitation (ChIP) assays, we examined the acetylation status of histone H3 (HeAcy), the dimethylation status of Lys4 (K4) of histone H3 (H3K4me2), and the trimethylation status of He3K4 and H3K27 (H3K4me3 and H3K27me3) across critical regulatory regions of Il17 and Ifng in MeCP2-deleted T cells. Despite observing some minor differences within some regions, we could not identify unidirectional changes in the accessibility of these cytokine genes (fig. S4). Because the differentiation path of naïve CD4+ T cells is primarily determined by their response to different environmental cytokines, we next considered whether loss of MeCP2 affected cytokine signaling. Cytokines activate various transcription factors within the family of signal transducer and activator (STAT) proteins (29); in particular, the differentiation of naïve CD4+ T cells into Th17 cells requires STAT3 activation (34-36). In both naïve and antigen-stimulated (“primed”) CD4+ T cells, loss of MeCP2 did not alter the abundance or activity of STAT3 protein; however, it did dampen the IL-6–dependent phosphorylation of Tyr705 of STAT3, the hallmark of STAT3 activation (Fig. 3A). Similarly, in the context of stimulation of cells with IFN-γ, loss of MeCP2 substantially inhibited the activation of STAT1 (Fig. 3B), a signaling intermediate that is critical for the generation of Th1 cells. Together, these data suggest that the loss of MeCP2 results in the inhibition of multiple STAT signaling pathways.
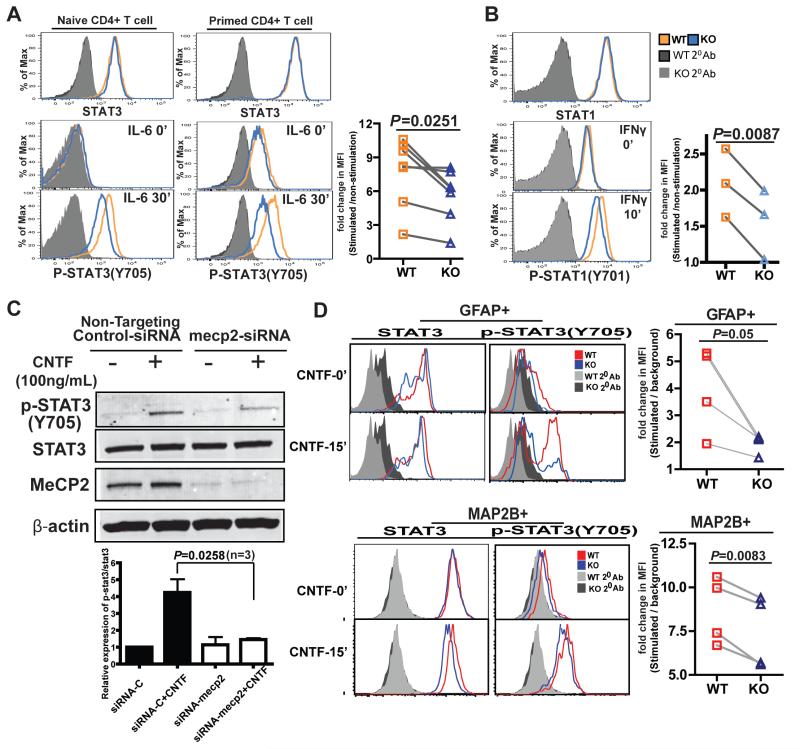
Fig. 3. MeCP2 is necessary to activate the STAT3 and STAT1 signaling pathways in CD4+ T cells.
(A) CD4+CD25− T cells sorted from WT and MeCP2 KO littermate mice were stimulated with IL-6 (50 ng/ml) for 30 min, and the amounts of total STAT3 and STAT3 phosphorylated at Tyr705 (pSTAT3) were determined by intracellular staining and flow cytometric analysis. Left: Naïve CD4+ T cells. Middle: CD4+ T cells that had been primed (activated) with anti-CD3 and anti-CD28 antibodies for 48 hours. Data are representative of three independent experiments for naïve T cells and four independent experiments for primed T cells. One pair of WT and KO mice were used in each independent experiment. Right: The plot on the right shows pooled results from seven independent experiments (Naïve T cells, n=3; primed T cells, n=4). For each pair of matched samples, the mean fluorescence intensity (MFI) of pSTAT3 staining at 30 min after stimulation was normalized to that of its unstimulated control (at time zero). (B) CD4+CD25− T cells sorted from WT and MeCP2 KO littermate mice were stimulated with IFN-γ (10 ng/ml) for 10 min. Total STAT1 and pSTAT1 (Tyr701) were then detected by intracellular staining and flow cytometric analysis. The plot on the right shows pooled results from three independent experiments. The MFI of pSTAT1 staining at 10 min after stimulation was normalized to that of unstimulated cells at time zero. (C) Human primary astrocytes were transfected with non-targeting control siRNA or with MeCP2-specific siRNA. Seventy-two hours later, the cells were stimulated with CNTF (100 ng/ml) for 15 min. The amounts of total STAT3 and pSTAT3 (Tyr705) were detected by Western blotting analysis. Blots are representative of three independent experiments. (D) Astrocytes (GFAP+) and neuron-like cells (MAP2B+) were induced from neural stem cells and progenitor cells in neurosphere cultures. Cells treated with CNTF (100 ng/ml) for 15 min were assessed for total STAT3 and pSTAT3 (Tyr705) by intracellular staining and flow cytometric analysis. Histograms are representative of four independent experiments. The plots on the right shows pooled results from four independent experiments. The MFI of pSTAT3 in cells at 15 min after stimulation was normalized to the background signal from the secondary antibody staining.
In addition to its proinflammatory role during immune responses, STAT3 is also highly abundant in the central and peripheral nervous systems, and the activation of STAT3 is essential for the survival, differentiation, and regeneration of neurons and glia cells (37). Based on our findings in T cells, we investigated whether the STAT3 signaling defect that resulted from a deficiency in MeCP2 was also evident in the nervous system. We knocked down MeCP2 in primary human astrocytes with specific small inhibitory RNA (siRNA), and then stimulated these cells with ciliary neurotrophic factor (CNTF), an important neuroprotective and neuropoietic cytokine that functions through the STAT3 pathway (38). The loss of MeCP2 attenuated STAT3 activation in human astrocytes (Fig. 3C). In addition, we crossed mice carrying conditional Mecp2 alleles with mice carrying an estrogen receptor (ER)-Cre transgene, isolated neural stem cells and progenitor cells in neurosphere cultures, and induced the Cre-mediated deletion of Mecp2 by treating the cells with tamoxifen in vitro. Stimulation of the primary neurosphere cultures with CNTF revealed similar deficiencies in the activation of STAT3 in cells expressing astrocyte-specific (GFAP+) or neuron-specific (MAP2B+) lineage markers (Fig. 3D). Together, these data suggest that MeCP2 is required for optimal STAT3 activation in both immune and neuronal cell types.
SOCS5 protein accumulates in MeCP2-deficient CD4+ T cells
Upon stimulation of cells with the appropriate cytokines, STAT3 signaling is tightly controlled by the activation of Janus-activated kinases (JAKs) and various negative feedback mechanisms, including those mediated by the SOCS family of proteins (39). Through quantitative polymerase chain reaction (PCR) analysis, we profiled the expression of 70 genes related to STAT signaling in naïve CD4+ T cells that were left untreated or were cultured under Th1-or Th17-polarizing conditions. These tests failed to identify any substantial differences between wild-type and MeCP2-deleted T cells at the mRNA level (fig. S5); however, at the protein level, we consistently observed a marked increase in the abundance of SOCS5 protein in naïve MeCP2-deficient CD4+ T cells (Fig. 4, A and B). Upon stimulation with antigen and IL-6, MeCP2-deficient T cells displayed a delay in the production of SOCS5 protein, which reiterated the defect in initial STAT3 signaling (Fig. 4C and fig. S6). Furthermore, in response to prolonged stimulation of cells, the loss of MeCP2 also resulted in an increase in the accumulation of SOCS5 protein (Fig. 4D and fig. S6), which spanned the duration of time required for Th17 cell generation.
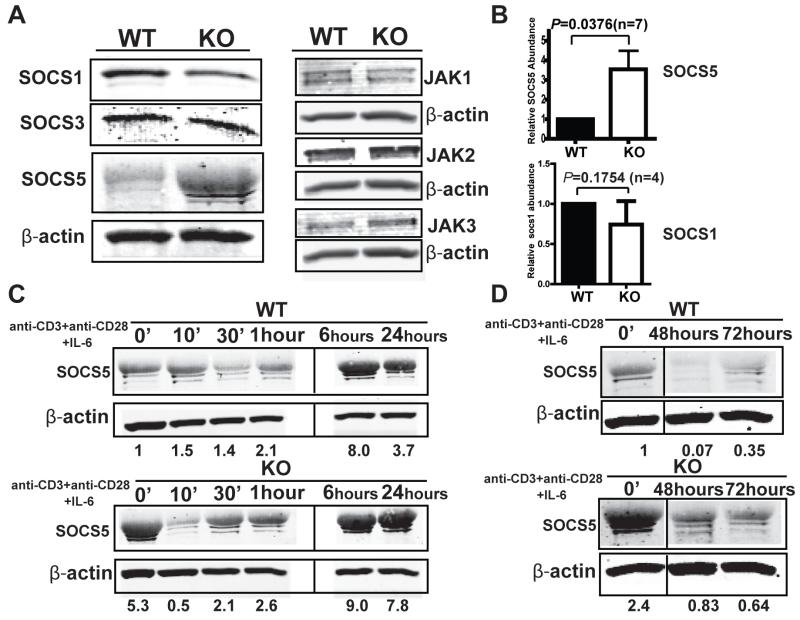
Fig. 4. SOCS5 accumulates in MeCP2-deficient CD4+ T cells.
(A and B) Analysis of the amounts of the indicated SOCS and JAK proteins in naïve CD4+CD25− T cells. Naïve CD4+CD25− T cells from WT and MeCP2 KO mice were analyzed by Western blotting with antibodies specific for the indicated proteins. (B) Quantification of SOCS5 and SOCS1 protein abundance. Western blots were subjected to densitometric analysis, and the intensities of the bands corresponding to SOCS5 and SOCS1 were normalized to those corresponding to β-actin and were calculated relative to those of WT cells. Data are means ± SEM from seven (for SOCS5) and four (for SOCS1) independent experiments. (C and D) CD4+CD25− T cells from WT and MeCP2 KO mice were left untreated or were activated in vitro with anti-CD3 and anti-CD28 antibodies together with IL-6 (50 ng/ml) for the indicated times. Cells were analyzed by Western blotting with antibodies specific for the indicated proteins. Western blots were subjected to densitometric analysis, and the intensities of the bands corresponding to SOCS5 were normalized to those corresponding to β-actin and were calculated relative to those of WT cells at time zero. The data are representative for three independent experiments. The samples from WT and MeCP2 KO CD4+ T cells that were activated up to 1 hour were analyzed on one gel, whereas those from samples stimulated for longer times were analyzed on a different gel.
SOCS5 inhibits STAT3 activation and the differentiation of naïve CD4+ T cells into Th17 cells
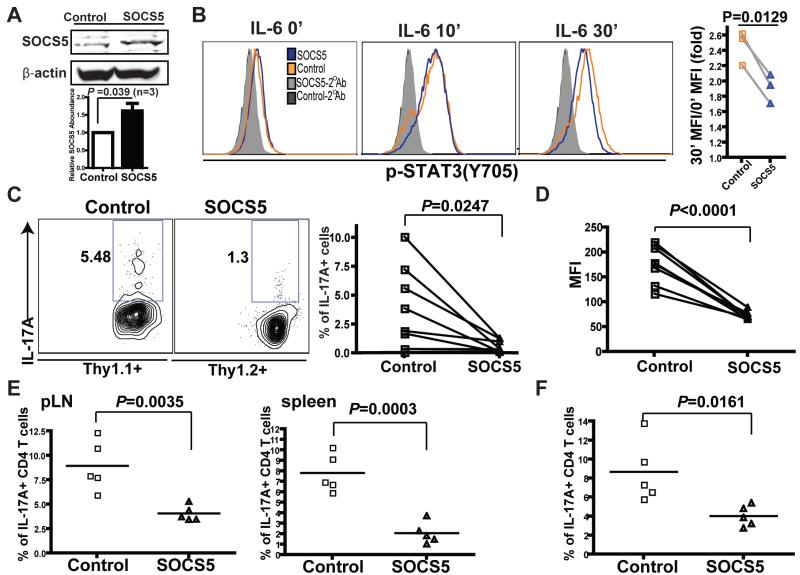
Unlike for SOCS1 and SOCS3, studies on the function of SOCS5 in lymphocytes remain limited (40, 41), and its effect on the generation of Th17 cells is unknown. However, SOCS5 is a potent inhibitor of STAT3 in M1 macrophages and human embryonic kidney (HEK) 293T cells (42). Through retroviral transduction, we ectopically expressed SOCS5 in primary LLO118 TCR transgenic T cells to try to mimic the effect of loss of MeCP2. With a relatively modest increase in SOCS5 abundance compared to that in cells transduced with control retrovirus (Fig. 5A), the responses of SOCS5-expressing T cells to IL-6 were impaired (Fig. 5B). To assess the role of SOCS5 in vivo, we competitively transferred equal numbers of green fluorescent protein (GFP)-expressing Thy1.1+ LLO118 T cells and SOCS5-overexpressing Thy1.2+ LLO118 T cells into TCRα−/− recipient mice, and activated these T cells by subcutaneous injection with the LLO190-205 peptide antigen. After 5 days of in vivo conditioning, we assessed the numbers of Th17 cells in these mice. Like their MeCP2-deficient counterparts, SOCS5-expressing T cells had a marked defect in the generation of Th17 cells (Fig. 5, C and D).
Fig. 5. SOCS5 inhibits STAT3 activation and the differentiation of naïve CD4+ T cells into TH17 cells.
(A and B) CD4+ T cells from LLO118 TCR transgenic mice were transduced with retroviruses that expressed either GFP (Control) or mouse SOCS5 and GFP (SOCS5). Seventy-two hours later, CD4+GFP+ cells were sorted by flow cytometry. (A) The relative abundance of SOCS5 protein in these cells was determined by Western blotting analysis. Data shows means ± SEM from three independent experiments. (B) The sorted cells were left untreated or were treated with IL-6 (50 ng/ml) for 10 or 30 min, as indicated, and pSTAT3 (Tyr705) was detected by intracellular staining and flow cytometry. The plot on the far right summarizes results from three independent experiments. For each group of samples, the MFI of pSTAT3 at the 30-min time point was normalized to that of unstimulated cells (time zero). (C and D) CD4+ T cells from LLO118 TCR transgenic mice with different Thy markers were transduced with retroviruses expressing either GFP (Thy1.1+) or SOCS5 (Thy1.2+). Six hours after competitive transfers of control and SOCS5 (CD4+GFP+) cells in a ratio of 1:1 to TCRα−/− recipient mice (n = eight mice), the recipient mice were immunized subcutaneously with LLO190-205 peptide emulsified in CFA. Five days after immunization, splenocytes from the recipient mice were rechallenged with LLO190-205 peptide and were cultured under TH17-skewing conditions in vitro. Forty-eight hours later, IL-17A–producing CD4+ T cells were enumerated by intracellular staining and flow cytometric analysis. (C) Percentages of IL-17A–producing CD4+ T cells. (D) MFIs of IL-17A. The data are representative for two independent experiments. (E and F) Control and SOCS5-expressing cells from LLO118 TCR transgenic mice were separately transferred into two groups of TCRα−/− recipient mice intravenously (n = 5 per group) and were immunized as described for (C) and (D). Seven days after immunization, the recipient mice were rechallenged by footpad injections of LLO190-205 peptide. (E) Forty-eight hours after rechallenge, the percentages of IL-17A–producing CD4+ cells in the peripheral lymph nodes and spleens were measured by intracellular staining and flow cytometric analysis. (F) Splenocytes from recipient mice were challenged with LLO190-205 peptide in vitro. Forty-eight hours later, the numbers of IL-17A–producing CD4+ T cells were measured by intracellular staining and flow cytometric analysis. The data represent two independent experiments.
To further determine whether this defect was - intrinsic or the result of competitive disadvantage, we transferred each of these two T cell populations into separate mice and challenged them with the same immunization protocol. In the setting of ex vivo examination (Fig. 5E) and in vitro rechallenge with antigen (Fig. 5F) we identified even more pronounced defects in the Th17 differentiation potential of SOCS5-expressing LLO T cells. Similarly, enforced expression of SOCS5 resulted in defective activation of STAT1 (fig. S7A) and generation of TH1 cells (fig. S7, B to D). To validate that the accumulation of SOCS5 was a causal factor for the defective differentiation of MeCP2 deficient T cells into TH17 cells, we transduced LLO T cells with a retrovirus expressing a short hairpin RNA (shRNA) against Socs5 mRNA (fig. S7E). This shRNA partially suppressed SOCS5 mRNA in MeCP2-deficient T cells, and partially rescued their differentiation into Th17 cells (fig. S7F). Thus, we propose that the impaired STAT signaling and constraints on T cell differentiation in MeCP2-deficient T cells are primarily a result of the aberrant accumulation of SOCS5.
MeCP2 enhances the expression of pri-mmu-miR-124-1 in CD4+ T cells
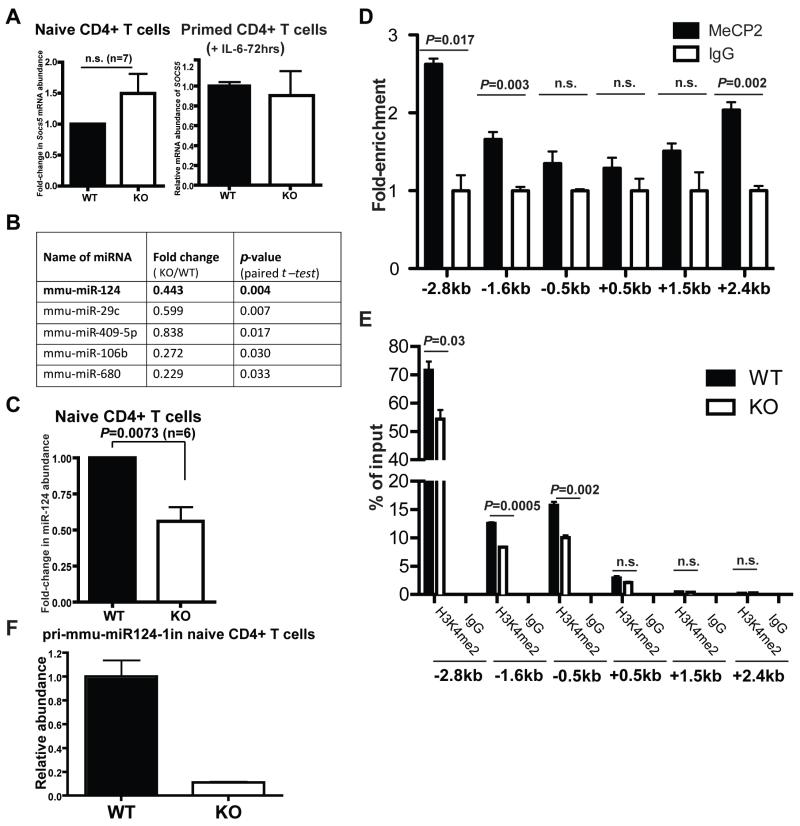
The next question was what was responsible for the accumulation of SOCS5 in MeCP2-deficient T cells. One possibility was that MeCP2 functioned as a transcriptional repressor of Socs5 expression. However, the effect of loss of MeCP2 on Socs5 expression in either naïve or cytokine-stimulated T cells was not statistically significant at the mRNA level (Fig. 6A). In agreement with this finding, epigenetic analysis across the Socs5 locus showed that MeCP2-deficient and wild-type CD4+ T cells did not substantially differ in either histone modification (H3 acetylation and H3K4 di- and tri-methylation) (fig. S8A) or DNA methylation (fig. S8B). Therefore, we speculated that MeCP2 regulated SOCS5 abundance in T cells indirectly at the posttranscriptional level.
Fig. 6. MeCP2 stimulates the transcription of pri-mmu-miR-124-1 in CD4+ T cells.
(A) Total RNA was extracted from naïve CD4+CD25− cells from WT and MeCP2 KO mice (left), and the amounts of Socs5 mRNA were determined by quantitative PCR analysis. The Data are means ± SEM from seven independent experiments. CD4+CD25− T cells from WT and MeCP2 KO mice were primed with anti-CD3 and anti-CD28 antibodies together with IL-6 (50 ng/ml) for 72 hours (right). Total RNA was extracted and the amounts of Socs5 mRNA were determined by quantitative PCR analysis. Data are means ± SEM from three replicates and represent two independent experiments. (B) List of miRNAs whose abundances were statistically significantly reduced in CD4+ T cells and primary human astrocytes upon deletion of MeCP2. Data are representative of three independent experiments. (C) Total RNA was extracted from naïve CD4+CD25− cells from WT and MeCP2 KO mice, and the amounts of miR-124 were determined by quantitative PCR analysis. Data are means ± SEM from six independent experiments. (D) ChIP analysis for the enrichment of MeCP2 at the pri-mmu-miR124-1 gene locus in naïve CD4+CD25− T cells. Data are means ± SEM from three replicated samples and represent two independent experiments. (E) Naïve CD4+ T cells from the indicated mice were subjected to ChIP analysis to determine the extent of dimethylation of Lys4 of histone H3 (H3K4me2) within the regulatory region of the pri-mmu-miR124-1 locus. Data are means ± SEM from three replicated samples and represent two independent experiments. (F) Naïve CD4+ T cells from WT and MeCP2 KO mice were subjected to quantitative PCR analysis to determine the relative amounts of pri-mmu-miR-124-1 transcripts. Data are means ± SEM from three from three replicated samples and represent three independent experiments.
MicroRNAs (miRNAs) are one of the principal molecular machineries responsible for posttranscriptional regulation (43). Although most miRNA-mediated repression involves at least moderate destabilization of target mRNAs, 11 to 16% of the regulation is attributed solely to translational inhibition (44). Based on the divergent abundances of SOCS5 mRNA and protein in MeCP2-dificient CD4+ T cells, we speculated that miRNAs might be a key player in the regulation of SOCS5. Through quantitative PCR analysis, we analyzed the expression profiles of ~400 miRNAs from wild-type and MeCP2-deficient cells. Because of the role of MeCP2 in neural cells, we narrowed our analysis by focusing on miRNAs that decreased in MeCP2-deficient mouse CD4+ T cells as well as in MeCP2-deficient primary human astrocytes (Fig. 6B). We found five miRNAs that were decreased in abundance in both cell types. Among these candidates, miR-124 was computationally predicted to target Socs5 mRNA at a highly conserved site fig. S9)
miR-124 is one of the most abundant miRNAs in the vertebrate central nervous system (45). Loss of miR-124 causes severe defects in neuronal maturation and survival (46). In addition, data from a clinical study suggest that the expression of miR-124 is dynamically modulated in human T cells from patients with sepsis, and an increase in miR-124 abundance limits the anti-inflammatory effects of steroids (47). To quantify the abundance of miR-124 in CD4+ T cells, we generated a standard curve from a synthesized RNA template (fig. S10A, left panel), and quantitated the copy number of miR-124. With this assay, we estimated miR-124 to be in the range of 4 × 107 copies/μg total RNA in naïve CD4+ T cells (fig. S10A, right panel). This is similar to the amount of miR-19b, a miRNA that regulates T cell effector responses (48). In the absence of MeCP2, the abundance of miR-124 was consistently reduced in naïve CD4+ T cells (Fig. 6C) and in T cells that were stimulated by antigen and cytokines (fig. S10B).
Mature miR-124 is generated from three distinct precursor transcripts that are coded within three highly conserved genomic loci (pri-miR-124-1 on chromosome 14, pri-miR-124-2 on chromosome 3, and pri-miR-124-3 on chromosome 2.). As was previously demonstrated in the nervous system (46), the pri-miR-124-1 locus is also the dominant precursor of mature miR-124 in T cells: the abundances of pri-miR-124-2 and pri-miR-124-3 were below the detection limit in our quantitative PCR assays. Based on information collected from the ENCODE project, the proximate regulatory regions for pri-miR-124-1 transcription are located within the −4.7 to +3.5 kb region that surrounds its transcription initiation site. Our ChIP assays showed that in naïve CD4+ T cells, MeCP2 readily associated with multiple CpG islands within the −2.8 to +2.4 kb region (Fig. 6D). Using H3K4 dimethylation as an epigenetic marker, we also examined the genomic accessibility and transcriptional capacity of these regions. In wild-type CD4+ T cells, the −2.8 kb and −1.6 kb regions, which were associated with enriched MeCP2 binding, constituted regions with higher accessibility. Indeed, loss of MeCP2 protein reduced the accessibility across this locus (Fig. 6E). Furthermore, when MeCP2 was lost, the reduced chromatin accessibility resulted in dampened transcription of pri-miR-124-1 in naïve CD4+ T cells (Fig. 6F), as well as in T cells subjected to inflammatory stimuli (fig. S10C). These data suggest a direct role for MeCP2 in promoting the transcription of pri-miR-124-1.
miR-124 inhibits the translation of Socs5 mRNA in CD4+ T cells
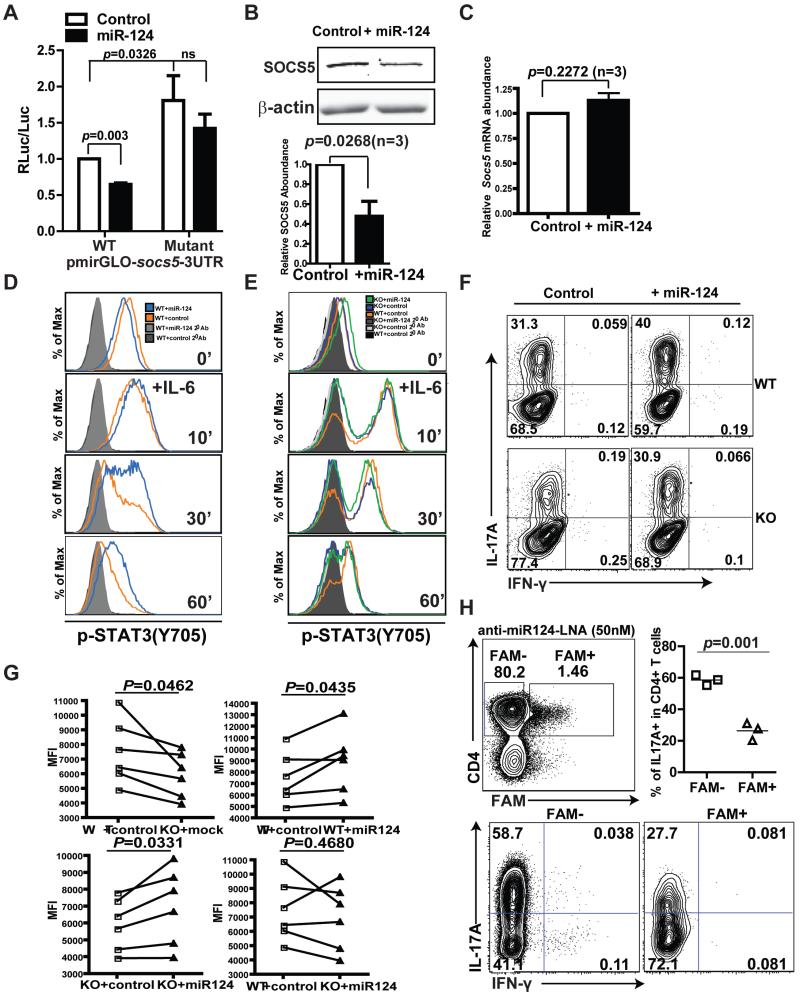
To determine whether miR-124 connected a deficiency in MeCP2 with the accumulation of SOCS5, we first used a luciferase-based reporter assay to verify that the predicted targeting site in the 3′ untranslated region (UTR) of Socs5 (fig. S9) was specifically targeted for suppression by miR-124 (Fig. 7A and fig. S11A). Furthermore, when miR-124 was ectopically introduced into antigen-primed LLO118 T cells (fig. S11B), the abundance of SOCS5 protein was reduced compared to that in the control cells (Fig. 7B), without there being any substantial effect on Socs5 mRNA abundance (Fig. 7C), which mirrored the pattern of expression seen in MeCP2-deficient T cells. In primed wild-type T cells stimulated with IL-6, the forced expression of miR-124 markedly delayed the kinetics of dephosphorylation of STAT3 (Fig. 7D and fig. S12A), reinforcing the idea that the primary target(s) of miR-124 are inhibitors of STAT3 signaling. Finally, we used a retroviral vector to restore miR-124 abundance in MeCP2-deficient T cells similarly to that in wild-type T cells (fig. S11C). The restoration of miR-124 partially rescued the activation of STAT3 in MeCP2-deficient T cells (Fig. 7E and fig. S12B). Phenotypically, the exogenous miR-124 substantially augmented the numbers of both wild-type and MeCP2-deficient LLO118 T cells that differentiated into TH17 cells. The restoration of miR-124 in MeCP2-deficient T cells fully rescued IL-17 production at both the population level and on a per-cell basis (Fig. 7F and G). In reciprocal loss-of-function studies, we sought to corroborate the idea that miR-124 promoted the differentiation of naïve CD4+ T cells into Th17 cells. To this end, we treated Th17-polarized LLO118 T cells with a locked nucleic acid (LNA) oligonucleotide capable of functionally inhibiting miR-124 (anti-miR-124 LNA) (49). Compared to untransfected cells in the same culture, Th17-polarized CD4+ T cells transfected with the anti-miR-124 LNA demonstrated reduced IL-17A production (Fig. 7H), effectively phenocopying the Th17 cell differentiation defect caused by the loss of MeCP2. Together, these data suggest that miR-124-mediated targeting of SOCS5 might account for the impaired differentiation of MeCP2-deficient T cells into TH17 cells.
Fig. 7. miR-124 inhibits the translation of Socs5 mRNA in CD4+ T cells.
(A) The entire 3′UTR of murine Socs5 (WT) or a 3′UTR containing mutations in the miR-124 seed region–binding sites were cloned into pmirGLO downstream of the firefly luciferase reporter gene. NIH 3T3 cells stably expressing miR-124 were transiently transfected with pmirGLO-mSOCS5-3UTR, and luciferase activity was measured 72 hours later. Data are means ± SEM of three independent experiments. (B and C) CD4+ T cells from LLO118 TCR transgenic mice were transduced with control retrovirus or with retrovirus expressing miR-124. Seventy-two hours later, CD4+GFP+ cells were sorted by flow cytometry and subjected to Western blotting and quantitative PCR analysis to determine the relative abundances of (B) SOCS5 protein and (C) Socs5 mRNA, respectively. Data are means ± SEM from three independent experiments. (D and E) CD4+ T cells from LLO118 TCR transgenic WT and MeCP2 KO littermate mice were transduced with control retrovirus or with retrovirus expressing miR-124. Previously primed T cells were stimulated with IL-6 (50 ng/ml), and the dynamics of STAT3 activation were monitored by flow cytometric analysis of Y705 phosphorylation. Analysis was performed on gated CD4+GFP+ cells. (D) The dynamics of STAT3 activation in control WT CD4+ T cells and in WT CD4+ T cells expressing miR-124. (E) The dynamics of STAT3 activation in WT CD4+ T cells and in MeCP2 KO CD4+ T cells that either did or did not express miR-124. (F and G) CD4+ T cells from LLO118 TCR transgenic WT and MeCP2 KO littermates transduced with either control retrovirus or with retrovirus expressing miR-124 were cultured under TH17-skewing conditions. Four to six days later, IL-17A production in GFP+CD4+ T cells was detected by intracellular staining and flow cytometry. (F) Percentages of IL-17A+ cells among GFP+CD4+ cells. Data are representative of six independent experiments. (G) MFI of IL-17A staining. (H) Lymphocytes from LLO118 TCR transgenic mice were cultured in vitro under TH17-skewing conditions in the presence of 50 nM 5′-6-fluorescein (6-FAM)–conjugated anti-miR-124 LNA for 4 days. Upper left: Gating strategy to identify CD4+ T cells that contain the anti-miR-124 LNA (FAM+). Bottom: Percentages of IL-17A–producing cells among FAM+ and FAM− CD4+ T cells. Upper right: Data are pooled results from three independent experiments. The horizontal line represents means.
Discussion
Since its identification as the causal factor of RTT, MeCP2 has been extensively studied in the nervous system; however the molecular mechanism by which MCP2 drives pathology remains largely elusive. Here, we dissected its function in T cells and determined that MeCP2 is indispensable for the differentiation of Tcon cells (fig. S13). We showed that MeCP2 plays a critical role in promoting multiple cytokine signaling pathways by supporting the expression of miR-124 and restraining negative feedback that targets STAT3. Blocking miR-124, or, forcing the expression of its target, SOCS5, in wild-type CD4+ T cells recapitulated, partially, the signaling and differentiation defects associated with MeCP2 deficiency. In addition, we found that a similar mechanism operated during CNTF signaling in primary neuronal and glial cells. STAT3 is a necessary inflammatory signal that stimulates the differentiation of naïve CD4+ T cells into Th17 cells, as well as being a vital neurotrophic transcription factor for neuronal survival and regeneration. Mutations in the gene encoding STAT3 cause a severe immunodeficiency disease, the Job/Buckley syndrome, one of the symptoms of which is scoliosis (50). Of note, RTT patients are also prone to the development of scoliosis as a comorbidity (51); coincidentally, mice with osteoblast- and osteocyte-specific loss of Stat3 develop a severe spinal deformity at 3 to 4 weeks of age (52). If this STAT3 signaling defect could be validated in the nervous systems of RTT patients, then it could provide a potential molecular target for the development of therapies, or guidance for current growth factor-based clinical trials.
Loss-of-function mutations in MECP2 lead to RTT in humans, whereas overexpression of MECP2 leads to MECP2 duplication syndrome (MDS), a neurological disorder with similar symptoms to those RTT (53). The neuropathological similarities between RTT and MDS could be overstated since their shared symptoms are common for many other neurodevelopmental disorders. Nevertheless, it is safe to conclude that the abundance of functional MECP2 is critical to the integrity of the central nervous system. Considering the CD4+ T cell, patients with MDS experience recurrent respiratory tract infections (55), and MeCP2-overexpressing mice are defective in mounting efficient TH1 cell responses against Leishmania major (56). Surprisingly, we found that similar to the effect carried out by MeCP2-overexpression, MeCP2-deficiency in mouse T cells also leads to impaired TH1 differentiation and TH1-mediated responses. With our current knowledge in T cell biology, it is difficult to interpret how the loss and gain of Mecp2 could result in the similar phenotype of inhibiting TH1 differentiation. However, the intrigue lies in the mechanistic aspect. We speculate that a critical set of immunoregulatory genes are modulated by MeCP2 in a delicate balance. This balance may depend on the divergent biochemical feature of this protein: for a specific regulatory region, MeCP2 can reinforce DNA methylation (repressive remodeling) (54) and elicit histone acetylation (activating remodeling) (10). Therefore, deletion of MeCP2 could result in both enhanced DNA demethylation and reduced histone acetylation, although the overall effect of loss of MeCP2 in a TH1-critical region would be predominantly inhibition of transcription. Overexpression of MeCP2, on the other hand, could potentially counteract local demethylation and result in a mechanistically distinct, but phenotypically similar, suppression of TH1-related gene transcription. Therefore, we speculate that, for certain genes, the overarching role of MeCP2 in transcriptional regulation could be switched from being supportive to being suppressive, depending on its concentration. However, immunodeficiency has not been found among RTT patients. We speculate that this apparent discrepancy results from differential degrees of loss-of-function of MeCP2. Immune function is intact in most RTT patients that bear heterozygous mutations in MECP2, and we also failed to detect substantial defects in the generation of Th1 and TH17 cells in mice with a heterozygous deletion of Mecp2 (fig. S14), whereas we observed that profound immunodeficiency ensued upon homozygous loss of Mecp2.
Because early studies identified MeCP2 as a transcriptional silencer, more recent work has largely focused on the role of MeCP2 in regulating gene expression at the mRNA level (10, 57, 58). However, this view is somewhat at odds with expression profiling studies on samples from RTT patients, which have shown that only a limited subset of genes are dysregulated at the mRNA level (59, 60). In experiments with Mecp2-deleted mouse models, expression differences were identified for additional genes, but most changes were very subtle (10, 61). In T cells, we found that the major defect associated with Mecp2 deletion was in STAT signaling, which was caused by the accumulation of SOCS5 protein; however, none of these changes was detectable at the mRNA level. Especially in wild-type naïve CD4+ T cells, the divergence between the amounts of SOCS5 mRNA and protein is striking: abundant mRNA did not result in even a modest increase in the amount of protein. This indicates that active posttranscriptional suppression of SOCS5 is required for optimal and sustained T-cell responses to cytokines. We identified miR-124 as the responsible suppressive factor, whose expression is tightly controlled by MeCP2. If a similar miRNA-mediated mechanism operates in the signaling networks of neuronal and glial cells, it would likely do so without marked changes in mRNA expression, and might therefore be more difficult to detect with traditional transcriptome analyses.
Materials and Methods
Mice
Mice homozygous for the floxed Mecp2 allele were purchased from The Jackson Laboratory (B6.129P2-Mecp2tm1Bird/J). Lck-Cre [B6.Cg-Tg (lck-cre)1Cwi N9] and Cd4-Cre mice [C57BL/6Tac-Tg(cd4-cre)N9] were purchased from Taconic. T cell–specific MeCP2- deficient C57BL/6 mice (CD4-Cre+Mecp2f/f or CD4-Cre+Mecp2f/y) were generated by crossing floxed Mecp2 mice with the cd4-cre or lck-cre mice for more than 10 generations.
Mouse models of IBD and the DTH reaction
For the IBD model, 5 × 105 naïve T cells (CD4+CD25−CD45Rbhigh) from wild-type or MeCP2 knockout (KO) littermates were mixed with 2 × 104 nTregs (CD4+CD25+) from wild-type mice, and injected intraperitoneally into Rag2−/− recipients (300 μl per mouse per injection). Recipient mice were weighed throughout the course of the experiments and were euthanized at the experimental endpoints for immunological and histological analysis. For the DTH reaction, wild-type and MeCP2 KO littermates were subcutaneously immunized with KLH protein (100 μg/mouse) in CFA. Seven days after immunization, mice were rechallenged with KLH (50 μg/mouse) or PBS in each lateral footpad. Forty-eight hours later, footpad swelling was measured before the mice were euthanized for immunological and histological analyses.
Induction of CD4+ T cell differentiation and analysis of cytokine production
Lymphocytes from the lymph nodes and spleens of LLO118 TCR transgenic mice were primed with 5 μM LLO190-205 peptide under various TH-skewing conditions for 4 days. Cytokine production by CD4+ T cells was determined by intracellular staining after 4 hours of stimulation with 0.9 nM PMA and ionomycin (0.5 μg/ml, Sigma) in the presence of brefeldin A (5 μg/ml, Sigma) and 2 μM monensin (eBioscience). CD4+ T cells were sorted on a flow cytometer based on surface CD4 expression and relative mRNA abundances were measured by quantitative PCR analysis. TH cell skewing conditions were as follows: Th1-skewing condition: recombinant mouse IL-12 (50 ng/ml, Peprotech), purified anti-IL-4 antibody (10 μg/ml, 11B11), and recombinant mouse IL-2 (50 U/ml, Peprotech); Th2-skewing condition: recombinant mouse IL-2 (50 U/ml), recombinant mouse IL-4 (50 ng/ml, Peprotech), purified anti-IFN-γ antibody (10 μg/ml, XNG1.2) and anti-IL-12 antibody (5 μg/ml, BD Biosciences); Th17-skewing condition: recombinant mouse IL-6 (20 ng/ml, Peprotech), recombinant TGF-β (4 ng/ml, Peprotech), purified anti-IFN-γ antibody (10 μg/ml, XNG1.2) and purified anti-IL-4 antibody (10 μg/ml, 11B11). Staining antibodies were purchased from Biolegend.
Loss-of-function study with anti-miR-124 LNA
T cells from LLO118 TCR transgenic mice were cultured in vitro under Th17- or Th1-skewing conditions in the presence of 50 nM 5′-6-fluorescein (6-FAM)–conjugated anti-miR-124 LNA (EXIQON) for 4 days. Cytokine production by CD4+ T cells was determined by flow cytometric analysis. The sequence for the anti-miR-124 LNA is: 5′-/56-FAM/LNAGLNAGLNACLNAALNATLNATLNACLNAALNACLNACLNAGLNACLNAGLNATLNAGLNACLNACLNATLNAT A-3′.
Quantitative PCR analysis
Total RNA was isolated with the miRVana extraction kit (Ambion) according to the manufacturer’s instructions. Reverse transcription was performed with qScript Flex cDNA Kit (Quanta Biosciences). Gene expression was quantified by SYBR Green–based quantitative PCR analysis. The quantitative PCR analysis of mature miRNA was performed as previously described (48).
Antibodies for Western blotting and intracellular staining
The following primary antibodies were used for detecting proteins by Western blotting analysis or by intracellular staining and flow cytometry: anti-STAT1, anti-STAT3, anti-pSTAT1 (Y701), anti-pSTAT3 (Y705), anti-SOCS1, anti-SOCS3, anti-SOCS5, anti-JAK1, anti-JAK2, anti-JAK3, anti-MeCP2 (all from Cell Signaling Technology), and anti-β-actin (Sigma). The primary anti-SOCS5 antibody used to analyze LLO118 TCR transgenic T cells was purchased from Santa Cruz Biotechnology. Alexa Fluor 680–conjugated anti-rabbit antibody and Alexa Fluor 800–conjugated anti-goat antibody (Invitrogen) were used as secondary antibodies, and fluorescence intensity was measured on an Odyssey system (Licor).
ChIP assay and DNA methylation analysis
Genomic DNA was purified with GenElute Mammalian Genomic DNA Miniprep Kit (Sigma, Cat. G1N79). Methylation analysis was quantified by sequencing of genomic DNA after bisulfite conversion with the MethylDetector kit (Active Motif), PCR amplification, and cloning. ChIP assays were performed with a standard protocol with anti-H3Ac, anti-H3K4me2, anti-H3K4me3, and anti-H3K27me3 rabbit polyclonal antibodies (Millipore), anti-MeCP2 (D4F3) XP rabbit monoclonal antibody (Cell Signaling), or a nonspecific rabbit anti-mouse immunoglobulin G (IgG, Jackson ImmunoResearch Laboratories). The amount of DNA immunoprecipitated by antibodies was quantified by quantitative PCR with primers specific for the indicated gene regulatory regions and normalized to the amount of input DNA before immunoprecipitation. For MeCP2 ChIP assays, the ratio of enrichment was determined by normalizing the amount of DNA immunoprecipitated with rabbit anti-MeCP2 antibody to that immunoprecipitated with nonspecific rabbit IgG.
miRNA target predictions, luciferase assays, and T cell transductions
miRNA target candidates were predicted by multiple methods assembled on the miRecords website (http://mirecords.biolead.org/) (62). The full-length 3′UTR of mouse Socs5 was amplified from a 3′ RACE-ready cDNA library generated from mouse total T cell RNA. Luciferase activity was determined 72 hours after transfection with a dual luciferase assay kit (Promega). For retroviral transductions, naïve CD4+ T cells from LLO118 TCR transgenic mice were activated with APCs loaded with LLO190-205 peptide (10μM) for 18 hours and then were subjected to spin-infection with retrovirus in 24-well plates at 1258g, 37°C.
Neurosphere cultures and primary human astrocyte cultures
Neurosphere cultures were prepared as described at http://www.nature.com/protocolexchange/protocols/77. Mice carrying conditional Mecp2 alleles were crossed with mice carrying ER-Cre transgenes to generate Mecp2f/f ER-Cre+ mice. The ventricle tissues of three-week-old Mecp2f/f ER-Cre+ mice were dissected and dissociated with the papain dissociation system (Worthington Biochemical Corporation) before being plated into 24-well plates for in vitro culture. The cells were first treated with 200 nM 4-hydroxytamoxifen (Sigma) for 3 days to induce Mecp2 deletion in neural stem cells and progenitor cells, and then were cultured in normal medium for an additional 3 weeks, during which time they were passaged every three days. Normal human astrocytes (NHA, CC-2565, Clonetics) were cultured in 6-well plates with astrocyte growth medium (CC-3186, Clonetics). NHA cells were transfected with siRNA targeting human MECP2 (L-013094-00-0005, Thermo Scientific Dharmacon) or with scrambled, non-targeting siRNA (D-001810-10-05, Thermo Scientific Dharmacon) with Lipofectamine RNAiMAX (13778150, Invitrogen). Seventy-two hours after transfection, cells were cultured in serum-free medium for 2 hours before they were stimulated with cytokines. Whole-cell lysates were then harvested for Western blotting analysis.
Histology
Footpads and colons were excised, inflated with 4% paraformaldehyde in PBS, fixed overnight at room temperature, placed in 70% ethanol, and embedded in paraffin before undergoing H&E staining.
Statistical analysis
Unpaired and paired two-tailed t tests were used to determine whether the difference between a given set of means was statistically significant. P < 0.05 was considered statistically significant.
Supplementary Material
Acknowledgments
We are grateful to F. Wang for technical support with the neurosphere cultures. We thank B. Hao and M. Krangel for technical support with the ChIP experiments. We thank P. J. R. Ebert and R. Lin for critical suggestions and reading of the manuscript.
Funding: Q.-J.L. is a Whitehead Family Foundation Scholar and is supported by grants from the American Cancer Society (RSG-10-157-01-LIB), the American Diabetes Association (1-10-JF-28), and the NIAID (R01AI091878). W.Y. is supported by the National Program on Key Basic Research Project in China (973 Program, 2010CB911800).
Footnotes
Author contributions: S.J. and C.L. designed and performed most of the experiments, analyzed data, and interpreted the results; G.M. performed miRNA expression profiling experiments; J.S. and S.L. provided technical support for animal studies; E.L., S.L., and Y.W. contributed critical reagents and technical support; Y.W. and Q.-J.L. conceptualized the project, designed experiments, supervised the work, and interpreted results; and S.J., C.L., and Q.-J.L. prepared the manuscript.
Competing interests: The authors declare that they have no competing interests.
Fig. S1. CD4+ T cell development is normal in mice with T cell–specific deletion of Mecp2.
Fig. S2. T cells in Mecp2-deficient mice do not undergo spontaneous activation.
Fig. S3. Loss of MeCP2 does not alter antigen-dependent proliferation or activation-induced cell death.
Fig. S4. Chromatin accessibility of the Il17 and Ifng loci in naïve CD4+CD25− T cells.
Fig. S5. Abundances of mRNAs for components of STATs signaling pathways.
Fig. S6. The dynamics of SOCS5 expression during CD4+ T cells activation in the presence of IL-6.
Fig. S7. SOCS5 inhibits the activation of STAT1 and the differentiation of naïve CD4+ T cells.
Fig. S8. The chromatin accessibility of the Socs5 locus in naïve CD4+CD25− T cells.
Fig. S9. miR-124 was computationally predicted to target Socs5 mRNA at a highly conserved site.
Fig. S10. Activated MeCP2-deficient CD4+ T cells have decreased amounts of pri-miR-124 and mature miR-124 compared to those of wild type CD4+ cells.
Fig. S11. Retroviral-mediated overexpression of miR-124 in 3T3 cells and CD4+ T cells.
Fig. S12. miR-124 promotes IL-6-induced STAT3 activation in CD4+ T cells.
Fig. S13.The MeCP2–miR-124–SOCS5 axis enables CD4+ T cell differentiation.
Fig. S14. Loss of a single copy of Mecp2 does not adversely affect the generation of TH1 and TH17 mouse T cells.
References and Notes
- 1.Lewis JD, Meehan RR, Henzel WJ, Maurer-Fogy I, Jeppesen P, Klein F, Bird A. Purification, sequence, and cellular localization of a novel chromosomal protein that binds to methylated DNA. Cell. 1992;69:905–914. doi: 10.1016/0092-8674(92)90610-o. [DOI] [PubMed] [Google Scholar]
- 2.Meehan RR, Lewis JD, Bird AP. Characterization of MeCP2, a vertebrate DNA binding protein with affinity for methylated DNA. Nucleic Acids Res. 1992;20:5085–5092. doi: 10.1093/nar/20.19.5085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nan X, Campoy FJ, Bird A. MeCP2 is a transcriptional repressor with abundant binding sites in genomic chromatin. Cell. 1997;88:471–481. doi: 10.1016/s0092-8674(00)81887-5. [DOI] [PubMed] [Google Scholar]
- 4.Nan X, Ng HH, Johnson CA, Laherty CD, Turner BM, Eisenman RN, Bird A. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature. 1998;393:386–389. doi: 10.1038/30764. [DOI] [PubMed] [Google Scholar]
- 5.Guy J, Cheval H, Selfridge J, Bird A. The role of MeCP2 in the brain. Annu Rev Cell Dev Biol. 2011;27:631–652. doi: 10.1146/annurev-cellbio-092910-154121. [DOI] [PubMed] [Google Scholar]
- 6.Hite KC, Adams VH, Hansen JC. Recent advances in MeCP2 structure and function. Biochem Cell Biol. 2009;87:219–227. doi: 10.1139/o08-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Young JI, Hong EP, Castle JC, Crespo-Barreto J, Bowman AB, Rose MF, Kang D, Richman R, Johnson JM, Berget S, Zoghbi HY. Regulation of RNA splicing by the methylation-dependent transcriptional repressor methyl-CpG binding protein 2. Proc Natl Acad Sci U S A. 2005;102:17551–17558. doi: 10.1073/pnas.0507856102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Georgel PT, Horowitz-Scherer RA, Adkins N, Woodcock CL, Wade PA, Hansen JC. Chromatin compaction by human MeCP2. Assembly of novel secondary chromatin structures in the absence of DNA methylation. J Biol Chem. 2003;278:32181–32188. doi: 10.1074/jbc.M305308200. [DOI] [PubMed] [Google Scholar]
- 9.Horike S, Cai S, Miyano M, Cheng JF, Kohwi-Shigematsu T. Loss of silent-chromatin looping and impaired imprinting of DLX5 in Rett syndrome. Nat Genet. 2005;37:31–40. doi: 10.1038/ng1491. [DOI] [PubMed] [Google Scholar]
- 10.Chahrour M, Jung SY, Shaw C, Zhou X, Wong ST, Qin J, Zoghbi HY. MeCP2, a key contributor to neurological disease, activates and represses transcription. Science. 2008;320:1224–1229. doi: 10.1126/science.1153252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yasui DH, Peddada S, Bieda MC, Vallero RO, Hogart A, Nagarajan RP, Thatcher KN, Farnham PJ, Lasalle JM. Integrated epigenomic analyses of neuronal MeCP2 reveal a role for long-range interaction with active genes. Proc Natl Acad Sci U S A. 2007;104:19416–19421. doi: 10.1073/pnas.0707442104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rett A. On a unusual brain atrophy syndrome in hyperammonemia in childhood. Wien Med Wochenschr. 1966;116:723–726. [PubMed] [Google Scholar]
- 13.Amir RE, Van den Veyver IB, Wan M, Tran CQ, Francke U, Zoghbi HY. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat Genet. 1999;23:185–188. doi: 10.1038/13810. [DOI] [PubMed] [Google Scholar]
- 14.Laurvick CL, de Klerk N, Bower C, Christodoulou J, Ravine D, Ellaway C, Williamson S, Leonard H. Rett syndrome in Australia: a review of the epidemiology. J Pediatr. 2006;148:347–352. doi: 10.1016/j.jpeds.2005.10.037. [DOI] [PubMed] [Google Scholar]
- 15.Hagberg B. Rett’s syndrome: prevalence and impact on progressive severe mental retardation in girls. Acta Paediatr Scand. 1985;74:405–408. doi: 10.1111/j.1651-2227.1985.tb10993.x. [DOI] [PubMed] [Google Scholar]
- 16.Neul JL, Fang P, Barrish J, Lane J, Caeg EB, Smith EO, Zoghbi H, Percy A, Glaze DG. Specific mutations in methyl-CpG-binding protein 2 confer different severity in Rett syndrome. Neurology. 2008;70:1313–1321. doi: 10.1212/01.wnl.0000291011.54508.aa. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen RZ, Akbarian S, Tudor M, Jaenisch R. Deficiency of methyl-CpG binding protein-2 in CNS neurons results in a Rett-like phenotype in mice. Nat Genet. 2001;27:327–331. doi: 10.1038/85906. [DOI] [PubMed] [Google Scholar]
- 18.Lioy DT, Garg SK, Monaghan CE, Raber J, Foust KD, Kaspar BK, Hirrlinger PG, Kirchhoff F, Bissonnette JM, Ballas N, Mandel G. A role for glia in the progression of Rett’s syndrome. Nature. 2011;475:497–500. doi: 10.1038/nature10214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Armstrong DD. Neuropathology of Rett syndrome. J Child Neurol. 2005;20:747–753. doi: 10.1177/08830738050200090901. [DOI] [PubMed] [Google Scholar]
- 20.Jellinger K, Seitelberger F. Neuropathology of Rett syndrome. Am J Med Genet Suppl. 1986;1:259–288. doi: 10.1002/ajmg.1320250528. [DOI] [PubMed] [Google Scholar]
- 21.Sawalha AH, Webb R, Han S, Kelly JA, Kaufman KM, Kimberly RP, Alarcon-Riquelme ME, James JA, Vyse TJ, Gilkeson GS, Choi CB, Scofield RH, Bae SC, Nath SK, Harley JB. Common variants within MECP2 confer risk of systemic lupus erythematosus. PLoS One. 2008;3:e1727. doi: 10.1371/journal.pone.0001727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Webb R, Wren JD, Jeffries M, Kelly JA, Kaufman KM, Tang Y, Frank MB, Merrill J, Kimberly RP, Edberg JC, Ramsey-Goldman R, Petri M, Reveille JD, Alarcon GS, Vila LM, Alarcon-Riquelme ME, James JA, Vyse TJ, Moser KL, Gaffney PM, Gilkeson GS, Harley JB, Sawalha AH. Variants within MECP2, a key transcription regulator, are associated with increased susceptibility to lupus and differential gene expression in patients with systemic lupus erythematosus. Arthritis Rheum. 2009;60:1076–1084. doi: 10.1002/art.24360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cobb BL, Fei Y, Jonsson R, Bolstad AI, Brun JG, Rischmueller M, Lester SE, Witte T, Illei G, Brennan M, Bowman S, Moser KL, Harley JB, Sawalha AH. Genetic association between methyl-CpG binding protein 2 (MECP2) and primary Sjogren’s syndrome. Ann Rheum Dis. 2010;69:1731–1732. doi: 10.1136/ard.2009.122903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lal G, Zhang N, van der Touw W, Ding Y, Ju W, Bottinger EP, Reid SP, Levy DE, Bromberg JS. Epigenetic regulation of Foxp3 expression in regulatory T cells by DNA methylation. J Immunol. 2009;182:259–273. doi: 10.4049/jimmunol.182.1.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Samaco RC, Fryer JD, Ren J, Fyffe S, Chao HT, Sun Y, Greer JJ, Zoghbi HY, Neul JL. A partial loss of function allele of methyl-CpG-binding protein 2 predicts a human neurodevelopmental syndrome. Hum Mol Genet. 2008;17:1718–1727. doi: 10.1093/hmg/ddn062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kerr B, Alvarez-Saavedra M, Saez MA, Saona A, Young JI. Defective body-weight regulation, motor control and abnormal social interactions in Mecp2 hypomorphic mice. Hum Mol Genet. 2008;17:1707–1717. doi: 10.1093/hmg/ddn061. [DOI] [PubMed] [Google Scholar]
- 27.Ostanin DV, Bao J, Koboziev I, Gray L, Robinson-Jackson SA, Kosloski-Davidson M, Price VH, Grisham MB. T cell transfer model of chronic colitis: concepts, considerations, and tricks of the trade. American journal of physiology. Gastrointestinal and liver physiology. 2009;296:G135–146. doi: 10.1152/ajpgi.90462.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saleh M, Elson CO. Experimental inflammatory bowel disease: insights into the host-microbiota dialog. Immunity. 2011;34:293–302. doi: 10.1016/j.immuni.2011.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhu J, Yamane H, Paul WE. Differentiation of effector CD4 T cell populations (*) Annu Rev Immunol. 2010;28:445–489. doi: 10.1146/annurev-immunol-030409-101212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weber KS, Li QJ, Persaud SP, Campbell JD, Davis MM, Allen PM. Distinct CD4+ helper T cells involved in primary and secondary responses to infection. Proc Natl Acad Sci U S A. 2012;109:9511–9516. doi: 10.1073/pnas.1202408109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sospedra M, Martin R. Immunology of multiple sclerosis. Annu Rev Immunol. 2005;23:683–747. doi: 10.1146/annurev.immunol.23.021704.115707. [DOI] [PubMed] [Google Scholar]
- 32.Luger D, Silver PB, Tang J, Cua D, Chen Z, Iwakura Y, Bowman EP, Sgambellone NM, Chan CC, Caspi RR. Either a Th17 or a Th1 effector response can drive autoimmunity: conditions of disease induction affect dominant effector category. J Exp Med. 2008;205:799–810. doi: 10.1084/jem.20071258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wilson CB, Rowell E, Sekimata M. Epigenetic control of T-helper-cell differentiation. Nat Rev Immunol. 2009;9:91–105. doi: 10.1038/nri2487. [DOI] [PubMed] [Google Scholar]
- 34.Yang XO, Panopoulos AD, Nurieva R, Chang SH, Wang D, Watowich SS, Dong C. STAT3 regulates cytokine-mediated generation of inflammatory helper T cells. J Biol Chem. 2007;282:9358–9363. doi: 10.1074/jbc.C600321200. [DOI] [PubMed] [Google Scholar]
- 35.Harris TJ, Grosso JF, Yen HR, Xin H, Kortylewski M, Albesiano E, Hipkiss EL, Getnet D, Goldberg MV, Maris CH, Housseau F, Yu H, Pardoll DM, Drake CG. Cutting edge: An in vivo requirement for STAT3 signaling in TH17 development and TH17-dependent autoimmunity. J Immunol. 2007;179:4313–4317. doi: 10.4049/jimmunol.179.7.4313. [DOI] [PubMed] [Google Scholar]
- 36.Korn T, Bettelli E, Oukka M, Kuchroo VK. IL-17 and Th17 Cells. Annu Rev Immunol. 2009;27:485–517. doi: 10.1146/annurev.immunol.021908.132710. [DOI] [PubMed] [Google Scholar]
- 37.Dziennis S, Alkayed NJ. Role of signal transducer and activator of transcription 3 in neuronal survival and regeneration. Rev Neurosci. 2008;19:341–361. doi: 10.1515/revneuro.2008.19.4-5.341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Taga T. Gp130, a shared signal transducing receptor component for hematopoietic and neuropoietic cytokines. J Neurochem. 1996;67:1–10. doi: 10.1046/j.1471-4159.1996.67010001.x. [DOI] [PubMed] [Google Scholar]
- 39.O’Shea JJ, Murray PJ. Cytokine signaling modules in inflammatory responses. Immunity. 2008;28:477–487. doi: 10.1016/j.immuni.2008.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Seki Y, Hayashi K, Matsumoto A, Seki N, Tsukada J, Ransom J, Naka T, Kishimoto T, Yoshimura A, Kubo M. Expression of the suppressor of cytokine signaling-5 (SOCS5) negatively regulates IL-4-dependent STAT6 activation and Th2 differentiation. Proc Natl Acad Sci U S A. 2002;99:13003–13008. doi: 10.1073/pnas.202477099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brender C, Columbus R, Metcalf D, Handman E, Starr R, Huntington N, Tarlinton D, Odum N, Nicholson SE, Nicola NA, Hilton DJ, Alexander WS. SOCS5 is expressed in primary B and T lymphoid cells but is dispensable for lymphocyte production and function. Mol Cell Biol. 2004;24:6094–6103. doi: 10.1128/MCB.24.13.6094-6103.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nicholson SE, Willson TA, Farley A, Starr R, Zhang JG, Baca M, Alexander WS, Metcalf D, Hilton DJ, Nicola NA. Mutational analyses of the SOCS proteins suggest a dual domain requirement but distinct mechanisms for inhibition of LIF and IL-6 signal transduction. EMBO J. 1999;18:375–385. doi: 10.1093/emboj/18.2.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annu Rev Biochem. 2010;79:351–379. doi: 10.1146/annurev-biochem-060308-103103. [DOI] [PubMed] [Google Scholar]
- 44.Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–840. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, Tuschl T. Identification of tissue-specific microRNAs from mouse. Curr Biol. 2002;12:735–739. doi: 10.1016/s0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]
- 46.Sanuki R, Onishi A, Koike C, Muramatsu R, Watanabe S, Muranishi Y, Irie S, Uneo S, Koyasu T, Matsui R, Cherasse Y, Urade Y, Watanabe D, Kondo M, Yamashita T, Furukawa T. miR-124a is required for hippocampal axogenesis and retinal cone survival through Lhx2 suppression. Nat Neurosci. 2011;14:1125–1134. doi: 10.1038/nn.2897. [DOI] [PubMed] [Google Scholar]
- 47.Ledderose C, Mohnle P, Limbeck E, Schutz S, Weis F, Rink J, Briegel J, Kreth S. Corticosteroid resistance in sepsis is influenced by microRNA-124--induced downregulation of glucocorticoid receptor-alpha. Critical care medicine. 2012;40:2745–2753. doi: 10.1097/CCM.0b013e31825b8ebc. [DOI] [PubMed] [Google Scholar]
- 48.Jiang S, Li C, Olive V, Lykken E, Feng F, Sevilla J, Wan Y, He L, Li QJ. Molecular dissection of the miR-17-92 cluster’s critical dual roles in promoting Th1 responses and preventing inducible Treg differentiation. Blood. 2011;118:5487–5497. doi: 10.1182/blood-2011-05-355644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Obad S, dos Santos CO, Petri A, Heidenblad M, Broom O, Ruse C, Fu C, Lindow M, Stenvang J, Straarup EM, Hansen HF, Koch T, Pappin D, Hannon GJ, Kauppinen S. Silencing of microRNA families by seed-targeting tiny LNAs. Nat Genet. 2011;43:371–378. doi: 10.1038/ng.786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Freeman AF, Holland SM. Clinical manifestations, etiology, and pathogenesis of the hyper-IgE syndromes. Pediatr Res. 2009;65:32R–37R. doi: 10.1203/PDR.0b013e31819dc8c5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Percy AK, Lee HS, Neul JL, Lane JB, Skinner SA, Geerts SP, Annese F, Graham J, McNair L, Motil KJ, Barrish JO, Glaze DG. Profiling scoliosis in Rett syndrome. Pediatr Res. 2010;67:435–439. doi: 10.1203/PDR.0b013e3181d0187f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhou H, Newnum AB, Martin JR, Li P, Nelson MT, Moh A, Fu XY, Yokota H, Li J. Osteoblast/osteocyte-specific inactivation of Stat3 decreases load-driven bone formation and accumulates reactive oxygen species. Bone. 2011;49:404–411. doi: 10.1016/j.bone.2011.04.020. [DOI] [PubMed] [Google Scholar]
- 53.Chahrour M, Zoghbi HY. The story of Rett syndrome: from clinic to neurobiology. Neuron. 2007;56:422–437. doi: 10.1016/j.neuron.2007.10.001. [DOI] [PubMed] [Google Scholar]
- 54.Kimura H, Shiota K. Methyl-CpG-binding protein, MeCP2, is a target molecule for maintenance DNA methyltransferase, Dnmt1. J Biol Chem. 2003;278:4806–4812. doi: 10.1074/jbc.M209923200. [DOI] [PubMed] [Google Scholar]
- 55.Friez MJ, Jones JR, Clarkson K, Lubs H, Abuelo D, Bier JA, Pai S, Simensen R, Williams C, Giampietro PF, Schwartz CE, Stevenson RE. Recurrent infections, hypotonia, and mental retardation caused by duplication of MECP2 and adjacent region in Xq28. Pediatrics. 2006;118:e1687–1695. doi: 10.1542/peds.2006-0395. [DOI] [PubMed] [Google Scholar]
- 56.Yang T, Ramocki MB, Neul JL, Lu W, Roberts L, Knight J, Ward CS, Zoghbi HY, Kheradmand F, Corry DB. Overexpression of Methyl-CpG Binding Protein 2 Impairs TH1 Responses. Sci Transl Med. 2012;4:163ra158. doi: 10.1126/scitranslmed.3004430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tudor M, Akbarian S, Chen RZ, Jaenisch R. Transcriptional profiling of a mouse model for Rett syndrome reveals subtle transcriptional changes in the brain. Proc Natl Acad Sci U S A. 2002;99:15536–15541. doi: 10.1073/pnas.242566899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ballestar E, Ropero S, Alaminos M, Armstrong J, Setien F, Agrelo R, Fraga MF, Herranz M, Avila S, Pineda M, Monros E, Esteller M. The impact of MECP2 mutations in the expression patterns of Rett syndrome patients. Hum Genet. 2005;116:91–104. doi: 10.1007/s00439-004-1200-0. [DOI] [PubMed] [Google Scholar]
- 59.Traynor J, Agarwal P, Lazzeroni L, Francke U. Gene expression patterns vary in clonal cell cultures from Rett syndrome females with eight different MECP2 mutations. BMC Med Genet. 2002;3:12. doi: 10.1186/1471-2350-3-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gibson JH, Slobedman B, K NH, Williamson SL, Minchenko D, El-Osta A, Stern JL, Christodoulou J. Downstream targets of methyl CpG binding protein 2 and their abnormal expression in the frontal cortex of the human Rett syndrome brain. BMC Neurosci. 2010;11:53. doi: 10.1186/1471-2202-11-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ben-Shachar S, Chahrour M, Thaller C, Shaw CA, Zoghbi HY. Mouse models of MeCP2 disorders share gene expression changes in the cerebellum and hypothalamus. Hum Mol Genet. 2009;18:2431–2442. doi: 10.1093/hmg/ddp181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xiao F, Zuo Z, Cai G, Kang S, Gao X, Li T. miRecords: an integrated resource for microRNA-target interactions. Nucleic Acids Res. 2009;37:D105–110. doi: 10.1093/nar/gkn851. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.