Abstract
The mechanisms by which air pollution has multiple systemic effects in humans are not fully elucidated, but appear to include inflammation and thrombosis. This study examines whether concentrations of ozone and components of fine particle mass are associated with changes in methylation on tissue factor (F3), interferon gamma (IFN-γ), interleukin 6 (IL-6), toll-like receptor 2 (TLR-2), and intercellular adhesion molecule 1 (ICAM-1). We investigated associations between air pollution exposure and gene-specific methylation in 777 elderly men participating in the Normative Aging Study (1999–2009). We repeatedly measured methylation at multiple CpG sites within each gene’s promoter region and calculated the mean of the position-specific measurements. We examined intermediate-term associations between primary and secondary air pollutants and mean methylation and methylation at each position with distributed-lag models. Increase in air pollutants concentrations was significantly associated with F3, ICAM-1, and TLR-2 hypomethylation, and IFN-γ and IL-6 hypermethylation. An interquartile range increase in black carbon concentration averaged over the four weeks prior to assessment was associated with a 12% reduction in F3 methylation (95% CI: -17% to -6%). For some genes, the change in methylation was observed only at specific locations within the promoter region. DNA methylation may reflect biological impact of air pollution. We found some significant mediated effects of black carbon on fibrinogen through a decrease in F3 methylation, and of sulfate and ozone on ICAM-1 protein through a decrease in ICAM-1 methylation.
Keywords: air pollution, traffic, gene-specific DNA methylation, effect modification, mediation analysis, elderly
Introduction
Although levels of most air pollutants have decreased over the last decade, air pollution remains an important public health issue. Moreover, the US population is aging and the elderly constitutes a population susceptible to air pollution exposure. Particulate pollution is known to increase cardiovascular morbidity and mortality1 and the relative contribution of particle components is still unclear. Animal and human studies have linked high air pollution levels to thrombosis and systemic inflammation.2-5
Recent research has identified a new biological mechanism to explain adverse health effects from air pollution: epigenetics.6,7 Epigenetics refers to chromosome changes that do not modify the genetic code, but influence its expression. The most frequently examined epigenetic mechanism is called DNA methylation because it involves methylation of cytosine in CpG pairs. A recent study suggested that DNA methylation is a mechanism that cells use to control gene expression in a switch-like manner.8 DNA methylation has been associated with health outcomes9-12 that have, in turn, been related to particles and ozone exposure.1 This raises the question of whether DNA methylation plays a role in the air pollution adverse effects on cardiovascular diseases.
Changes in methylation have been associated with exposure to lead13,14 and air pollution.15-19 Short- and intermediate-term exposures to black carbon and particles over one week have been associated with inducible nitric oxide synthase hypomethylation.16,20-22 In addition, black carbon exposure over weeks or months has been related to Long Interspersed Nucleotide Element 1 (LINE-1) hypomethylation.17,23 However, the literature on the effects of air pollution on methylation remains limited.
We investigated whether air pollution exposures are related to methylation on genes related to two cardiovascular pathways: coagulation and inflammation in the elderly. We focused on five genes: tissue factor (F3), interferon gamma (IFN-γ), interleukin 6 (IL-6), toll-like receptor 2 (TLR-2), and intercellular adhesion molecule 1 (ICAM-1). We examined whether the association between air pollution and gene-specific methylation varied by participants’ characteristics. We also investigated whether the association between air pollution and cardiovascular-related biomarkers was mediated via a change in gene-specific methylation.
Results
Descriptive statistics
At baseline, the median age of the study population was 72 years (Table 1). Also 27% of the participants were obese, 14% were diabetics, and only 4% were current smokers. Participants’ characteristics varied according to their total number of visits: individuals with more visits seemed “healthier” than participants with fewer visits.
Table 1. Demographical characteristics of the NAS participants across visits.
| Age (years) [5th, 50th, 95th percentiles] |
% of neutrophils [5th, 50th, 95th percentiles] |
% of lymphocytes [5th, 50th, 95th percentiles] |
Obesity | Statin user | Diabetics | Smoking [Never, Former, Current] |
|
|---|---|---|---|---|---|---|---|
| Baseline (n = 777) | [62, 72, 84] | [48, 62, 74] | [15, 26, 38] | 27% | 36% | 14% | [29%, 67%, 4%] |
| Nmissing | 0 | 22 | 22 | 0 | 0 | 0 | 0 |
| Among participants having one visit (n1 = 221) | |||||||
| Visit 1 | [64, 76, 88] | [48, 63, 77] | [13, 25, 37] | 30% | 40% | 18% | [26%, 70%, 4%] |
| Among participants having two visits (n1 = 217) | |||||||
| Visit 1 | [60, 73, 83] | [47, 62, 74] | [15, 25, 40] | 28% | 35% | 16% | [26%, 69%, 5%] |
| Visit 2 | [66, 77, 86] | [48, 64, 75] | [14, 24, 37] | 27% | 54% | 19% | [26%, 70%, 4%] |
| Among participants having three visits (n3 = 216) | |||||||
| Visit 1 | [62, 71, 82] | [47, 62, 72] | [16, 26, 39] | 25% | 36% | 9% | [29%, 68%, 3%] |
| Visit 2 | [66, 74, 86] | [48, 62, 74] | [15, 26, 38] | 26% | 52% | 13% | [28%, 69%, 3%] |
| Visit 3 | [69, 78, 89] | [48, 62, 76] | [13, 25, 39] | 25% | 62% | 17% | [27%, 71%, 2%] |
| Among participants having four visits (n4 = 120) | |||||||
| Visit | [60, 69, 77] | [49, 61, 74] | [15, 26, 36] | 22% | 29% | 10% | [38%, 58%, 4%] |
| Visit 2 | [63, 72, 81] | [46, 62, 78] | [13, 25, 40] | 22% | 42% | 11% | [38%, 58%, 4%] |
| Visit 3 | [66, 75, 84] | [47, 61, 76] | [13, 26, 37] | 18% | 59% | 16% | [38%, 59%, 3%] |
| Visit 4 | [70, 78, 87] | [50, 63, 76] | [12, 25, 37] | 17% | 65% | 18% | [38%, 60%, 2%] |
| Among participants having five visits (n5 = 3) | |||||||
| Visit 1 | [62, 66, 66] | [49, 58, 67] | [18, 25, 33] | 33% | 33% | 0% | [33%, 67%, 0%] |
| Visit 2 | [65, 68, 70] | [59, 64, 70] | [18, 22, 26] | 33% | 33% | 0% | [33%, 67%, 0%] |
| Visit 3 | [68, 70, 72] | [16, 54, 68] | [18, 29, 78] | 33% | 0% | 0% | [33%, 67%, 0%] |
| Visit 4 | [71, 73, 74] | [14, 60, 67] | [17, 18, 83] | 33% | 0% | 0% | [33%, 67%, 0%] |
| Visit 5 | [73, 76, 77] | [15, 55, 75] | [13, 24, 82] | 33% | 33% | 0% | [33%, 67%, 0%] |
NAS, Normative Aging Study.
Boston has a continental climate with direct influences from the ocean. While it is mostly cold and dry in winter, it is usually warm and humid in summer. Ambient air pollutants levels in Boston are mostly below EPA standards. Over the 1999–2009 study period, the 24-h PM2.5 mass concentrations exceeded the daily standard of 35 µg/m3 only during 13 days in summer 2002. Summary statistics of the weather and air pollution distributions during the study period are presented in Table S1. Ozone was negatively correlated with particle number (Correlation = -0.39) and black carbon (Correlation = -0.14) (Table S2). We observed a decrease over time of the correlations between the weekly averaged air pollutant concentrations (Table S3).
The distributions of the gene-specific methylation varied according to genes: IFN-γ methylation distribution was wider compared with that of F3 methylation (Table S4). The gene-specific distributions were concentrated around low or high values, except for IL-6 methylation.
Main associations
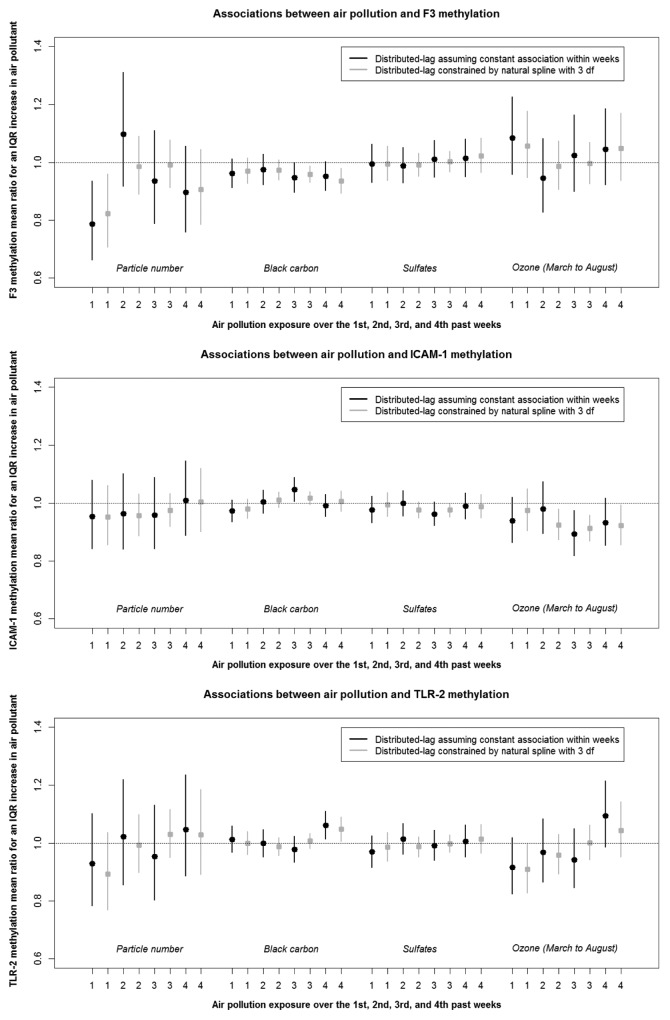
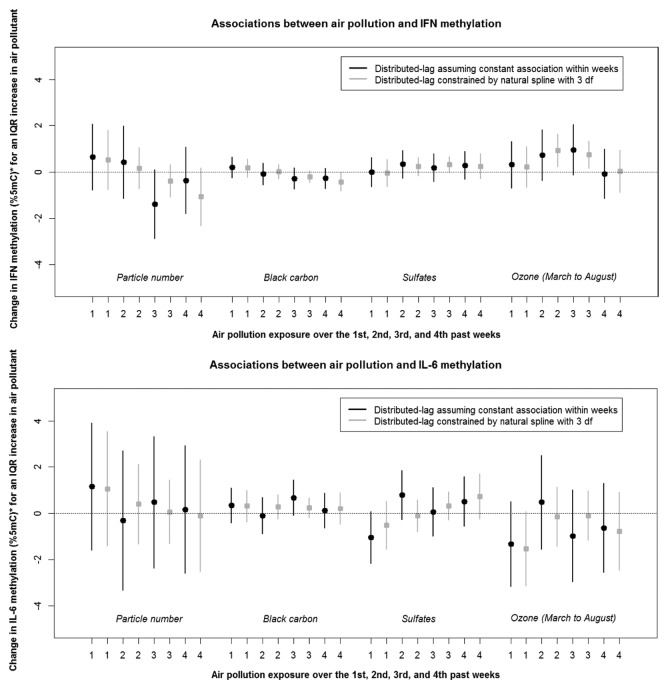
Air pollutants levels were associated with significant changes in gene-specific methylation (Fig. 1A and Fig. 1B). We observed significant negative associations between markers of traffic such as particle number (1st week) and black carbon (3rd and 4th weeks) and F3 methylation. For instance, an interquartile range (IQR) increase in particle number (which corresponds approximately to an increase of 15 000 particles per cm3) over the 1st week preceding each participant’s methylation measurement was associated with an 18% reduction in F3 methylation (95% confidence interval (CI): –29% to –4%). In contrast, the secondary pollutants sulfate and ozone were not associated with F3 methylation. Ozone exposures over the 2nd to 4th weeks were negatively related to ICAM-1 methylation. In addition, black carbon and ozone exhibited opposite associations with ICAM-1, IFN-γ, and IL-6 methylation.
Figure 1A. Associations between air pollution and F3, ICAM-1, and TLR-2 DNA methylation across the 1st to 4th weeks of exposure (Estimates and associated 95% CI). Variables included in the models: f1(air pollutant), f2(temperature), f3(relative humidity), age, body mass index, smoking status, diabetes status, statin use, percentage of neutrophils and lymphocytes in blood count, seasonal sine and cosine, season, and batch.
Figure 1B. Associations between air pollution and IFN-γ, and IL-6 DNA methylation across the 1st to 4th weeks of exposure (Estimates and associated 95% CI). *%5mC is the unit of DNA methylation and corresponds to the percentage of methylated cytosines over the sum of methylated and unmethylated cytosines at position 5. Variables included in the models: f1(air pollutant), f2(temperature), and f3(relative humidity) represent the distributed-lag functions with sets of coefficients constrained by a natural spline (with 3 degrees of freedom) that correspond to the air pollution, temperature, and relative humidity associations at lags 0 and 27 days.
Our main analysis indicated temporal variation across lags in the association between air pollution exposure and DNA methylation. For instance, we observed an earlier decrease in F3 methylation for particle number (1st week) compared with black carbon (3rd and 4th weeks), as well as a negative association between ozone and ICAM-1 methylation after the 2nd week of exposure.
Air pollutants associations over the 4-week period preceding each medical visit are presented in Table 2. For example, an interquartile range increase in black carbon concentration was associated with a 12% reduction in F3 methylation (95%CI: –17% to –6%).
Table 2. Association between air pollution exposure over the 4-week period preceding medical examination and gene-specific DNA methylation.
| Methylation mean ratio for an interquartile range increase in air pollution | ||||||||
|---|---|---|---|---|---|---|---|---|
| Particle number | Black carbon | Sulfate | Ozone (March to August) |
|||||
| Estimate | 95% CI | Estimate | 95% CI | Estimate | 95% CI | Estimate | 95% CI | |
| F3 | 0.725 | 0.655, 0.803 | 0.877 | 0.825, 0.932 | 1.008 | 0.930, 1.093 | 1.081 | 0.885, 1.322 |
| ICAM-1 | 0.889 | 0.824, 0.961 | 1.009 | 0.965, 1.055 | 0.948 | 0.896, 1.003 | 0.759 | 0.664, 0.868 |
| TLR-2 | 0.943 | 0.851, 1.044 | 1.032 | 0.979, 1.088 | 0.987 | 0.920, 1.058 | 0.917 | 0.775, 1.086 |
| Change in methylation (% 5mC) for an interquartile range increase in air pollution | ||||||||
| Particle number | Black carbon | Sulfate |
Ozone (March to August) |
|||||
| Estimate | 95% CI | Estimate | 95% CI | Estimate | 95% CI | Estimate | 95% CI | |
| IFN-γ | –0.845 | –1.704, 0.013 | –0.363 | –0.872, 0.145 | 0.589 | –0.198, 1.376 | 1.898 | 0.244, 3.552 |
| IL-6 | 1.412 | –0.245, 3.068 | 0.806 | –0.077, 1.688 | 0.317 | –1.055, 1.688 | –2.486 | –5.424, 0.452 |
Variables included in the models: f1(air pollutant)b, f2(temperature)b, f3(relative humidity)b, age, body mass index, smoking status, diabetes status, statin use, % neutrophils in blood count, % lymphocytes in blood count, seasonal sine and cosine, season, and methylation batch. b) f1(air pollutant), f2(temperature), and f3(relative humidity) represent the distributed-lag functions with sets of coefficients constrained by a natural spline (with 3 degrees of freedom) that correspond to the air pollution, temperature, and relative humidity associations at lags 0 and 27 days.
We also examined whether some participants’ characteristics were related to gene-specific methylation (Table S5). Compare with individuals who never smoked, former smokers had higher IL-6 methylation and lower TLR-2 and IFN-γ methylation. Age was also positively associated with TLR-2 methylation.
Analyses of effect modification and mediation
We did not observe any effect modification by smoking status, except for the association between particle number and IL-6 methylation which was stronger in participants who never smoked (methylation mean ratio for an IQR increase in particle number = 2.877 [95% CI: 1.221, 4.534]), compare with former and current smokers (methylation mean ratio = 0.886 [95% CI: -0.770, 2.543]) (Table 3). The association between air pollution and gene-specific methylation did not vary by obesity status (Table S6) or age category (Table S7). The air pollution association with gene-specific methylation was fairly similar according to baseline participants’ levels of LINE-1 and Alu methylation, except for ozone, for which results were not consistent (Table S8).
Table 3. Association between air pollution exposure over the 4-week period preceding medical examination on gene-specific DNA methylation according to the smoking status.
| Methylation mean ratio for an interquartile range increase in air pollution | ||||||||
|---|---|---|---|---|---|---|---|---|
| Particle number | Black carbon | Sulfate | Ozone (March to August) |
|||||
| Estimate | 95% CI | Estimate | 95% CI | Estimate | 95% CI | Estimate | 95% CI | |
| F3 | ||||||||
| Never smokers | 0.739 | 0.648, 0.0.844 | 0.863 | 0.793, 0.939 | 0.936 | 0.828, 1.058 | 1.032 | 0.773, 1.377 |
| Former and current smokers | 0.719 | 0.644, 0.801 | 0.882 | 0.825, 0.943 | 1.037 | 0.948, 1.135 | 1.106 | 0.882, 1.387 |
| ICAM-1 | ||||||||
| Never smokers | 0.910 | 0.826, 1.002 | 1.005 | 0.943, 1.071 | 0.976 | 0.899, 1.059 | 0.779 | 0.640, 0.948 |
| Former and current smokers | 0.880 | 0.810, 0.957 | 1.011 | 0.964, 1.060 | 0.933 | 0.877, 0.993 | 0.747 | 0.642, 0.868 |
| TLR-2 | ||||||||
| Never smokers | 0.911 | 0.800, 1.037 | 1.016 | 0.943, 1.094 | 0.939 | 0.848, 1.039 | 0.864 | 0.681, 1.098 |
| Former and current smokers | 0.958 | 0.859, 1.069 | 1.040 | 0.981, 1.102 | 1.007 | 0.932, 1.088 | 0.953 | 0.789, 1.152 |
| Change in methylation (% 5mC) for an interquartile range increase in air pollution | ||||||||
| Particle number | Black carbon | Sulfate |
Ozone (March to August) |
|||||
| Estimate 95% CI | Estimate 95% CI | Estimate 95% CI | Estimate 95% CI | |||||
| IFN-γ | ||||||||
| Never smokers | –0.805 –1.664, 0.053 | –0.651 –1.159, –0.143 | 1.197 0.411, 1.984 | 2.734 1.080, 4.388 | ||||
| Former and current smokers | –0.870 –1.729, –0.012 | –0.288 –0.796, 0.220 | 0.377 –0.410, 1.163 | 1.401 –0.253, 3.055 | ||||
| IL-6 | ||||||||
| Never smokers | 2.877 1.221, 4.534 | 0.553 –0.329, 1.436 | –0.367 –1.738, 1.005 | –3.536 –6.474, –0.598 | ||||
| Former and current smokers | 0.886 –0.770, 2.543 | 0.896 0.014, 1.779 | 0.662 –0.710, 2.034 | –2.028 –4.966, 0.910 | ||||
Variables included in the models: f1(air pollutant)b, f2(temperature)b, f3(relative humidity)b, smoking status*f1(air pollutant)b, age, body mass index, smoking status, diabetes status, statin use, % neutrophils in blood count, % lymphocytes in blood count, seasonal sine and cosine, season, and methylation batch. f1(air pollutant), f2(temperature), and f3(relative humidity) represent the distributed-lag functions with sets of coefficients constrained by a natural spline (with 3 degrees of freedom) that correspond to the air pollution, temperature, and relative humidity associations at lags 0 and 27 days.
We conducted mediation analyses and found some significant mediated effects of black carbon on fibrinogen through a decrease in F3 methylation (Estimate: 0.012, 95% CI: [0.000, 0.024]), and of sulfate and ozone on ICAM-1 protein through a decrease in ICAM-1 methylation (Estimates: 0.007, 95% CI: [0.000, 0.014] and 0.037, 95% CI: [0.003, 0.072], respectively) (Table 4).
Table 4. Mediation analysis: the mediated effect of air pollution on cardiovascular-related biomarkers through a change in gene-specific methylation.
| Estimates correspond to a standard deviation increase from the mean | ||||
|---|---|---|---|---|
| Particle number | Black carbon | Sulfate | Ozone (March to August) |
|
| γ1β2 estimate [95% CI] |
γ1β2 estimate [95% CI] |
γ1β2 estimate [95% CI] |
γ1β2 estimate [95% CI] |
|
| F3 methylation and fibrinogen protein |
(–0.267)(–0.032) = 0.009 [–0.013, 0.030] |
(–0.142)(–0.084) = 0.012 [0.000, 0.024] |
(–0.054)(–0.099) = 0.005 [–0.003, 0.014] |
(0.023)( –0.088) = –0.002 [–0.021, 0.017] |
| ICAM-1 methylation and ICAM-1 protein | (–0.053)( –0.077) = 0.004 [–0.004, 0.012] |
(0.022)(–0.069) = –0.001 [–0.007, 0.004] |
(-0.090)(–0.075) = 0.007 [0.000, 0.014] |
(–0.441)(–0.085) = 0.037 [0.003, 0.072] |
| TLR-2 methylation and CRP protein | (–0.002)(0.023) = 0.000 [–0.002, 0.002] |
(0.034)( –0.023) = –0.001 [–0.003, 0.002] |
(–0.015)(–0.010) = 0.000 [–0.001, 0.001] |
(–0.140)(–0.130) = 0.018 [–0.010, 0.047] |
| IFN-γ methylation and CRP protein |
(–0.101)(0.008) = –0.001 [–0.008, 0.006] |
(–0.050)(0.039) = –0.002 [–0.006, 0.002] |
(0.021)(0.017) = 0.000 [–0.001, 0.002] |
(0.163)(0.050) = 0.008 [–0.006, 0.023] |
| IL-6 methylation and IL-6 protein | (0.037)(–0.071) = –0.003 [–0.009, 0.003] |
(0.056)( –0.046) = –0.003 [–0.007, 0.001] |
(0.003)(-0.046) = 0.000 [–0.003, 0.002] |
(–0.128)(-0.024) = 0.003 [–0.008, 0.014] |
Variables included in the models: air pollutantb, temperatureb, relative humidityb, age, body mass index, smoking status, diabetes status, statin use, % neutrophils in blood count, % lymphocytes in blood count, seasonal sine and cosine, season, and methylation batch. Air pollutant, temperature, and relative humidity represent the moving average between lags 0 and 27 days.
Sensitivity analyses
We found similar results when we assumed a distributed-lag model with constant lag associations within weeks for air pollution, temperature, and relative humidity (Fig. 1A and B) vs. a distributed-lag model constrained by natural splines with 3 degrees of freedom.
The magnitude and significance of the air pollution associations with gene-specific methylation differed by CpG positions where the methylation was measured (Table S9). For example, the signals of particle number, black carbon, and sulfate were the strongest at position 4 within the promoter region of F3, whereas the signal was the greatest at position 2 for ozone.
In addition, we examined the robustness of our main findings considering additional statistical approaches (Table S10). After fitting co-pollutant models, negative associations between traffic-related pollutants and F3 methylation remained significant after adjusting for ozone. The ozone association estimate with F3 methylation became significant when we controlled for particle number. Similarly, the co-pollutant models suggested that changes in ICAM-1 methylation were related to particle number but not to ozone. Moreover, our results were not affected by residual confounding of smoking intensity captured by cigarette pack-years, daily alcohol consumption, and the number of years of education.
We obtained slightly greater estimates when we fitted the the generalized estimating equations (GEE) model for the association between sulfate and ICAM-1 methylation, and between particle number, black carbon, and ozone and IL-6 methylation. When we included censoring weights, our results were similar, except that we observed slightly stronger associations between ozone and methylation on TLR-2 and IL-6.
Finally, the analysis restricted to days with 24-h PM2.5 mass concentrations below 35 µg/m3 showed no difference in significance and magnitude of our results, except that we obtained a significant association (over a monthly period) between sulfate and ICAM-1 methylation. An interquartile range increase in sulfate concentration was associated with a 6% reduction in ICAM-1 methylation (95% CI: -11% to 0%).
Discussion
We have previously reported positive associations between air pollution and levels of plasma fibrinogen, C-reactive protein (CRP), ICAM-1, and vascular cell adhesion protein 1 (VCAM-1) in this cohort; and our current results linking air pollution exposure to methylation on genes related to coagulation and inflammation pathways are consistent with those associations. We did not identify susceptible subgroups in which the association between air pollution and gene-specific methylation was stronger, except that we observed a greater effect of particle number on IL-6 methylation in non-smokers compare with former and current smokers. The mediation analysis provided further insight into potential importance of the association between air pollution and gene-specific methylation because it suggests that the relationship between air pollution and protein levels are in part mediated by a change in gene-specific methylation.
Particle number and black carbon were negatively associated with F3 methylation. Since hypomethylation is usually related to gene expression,8 this result is consistent with higher fibrinogen levels observed after exposures to particle number and black carbon in the same cohort;24 which is what the mediation analysis confirmed. F3, also known as tissue factor, is a key player in the coagulation cascade that results in fibrinogen production. Hypercoagulable states have been associated with high levels of tissue factor and fibrinogen.25-27 F3 expression and fibrinogen have also been shown to increase in response to the intrusion of fine and ultrafine particles.28-30 Exposure to fresh local and transported traffic emissions may therefore induce F3 hypomethylation leading to increased tissue factor and fibrinogen production.
Similarly, we have previously shown that sulfate concentrations were associated with higher ICAM-1 protein levels in the same cohort.24 In the present study, higher sulfate and ozone concentrations were related to ICAM-1 hypomethylation. In our data, a 1%5mC decrease in ICAM-1 methylation was associated with a 0.7% increase in ICAM-1 protein (95% CI: 0.0% to 1.4%). Our analyses of association and mediation suggested that exposure to sulfate and ozone decreased ICAM-1 methylation, which could cause ICAM-1 gene de-silencing and ICAM-1 protein overexpression.
In our secondary analysis across CpG positions, we observed a positive association between traffic-related pollutants and IL-6 methylation and a negative association between ozone and TLR-2 methylation. In the same cohort, TLR-2 hypomethylation and IL-6 hypermethylation were also associated with lower lung function12 and ozone was positively related to CRP.24 TLR-2, IL-6, and CRP are related to each other: they participate in innate immune responses and consist of early defense mechanisms against infections. A recent study linked TLR-2 to IL-6 when they observed greater air pollution associations on IL-6 for wild type mice compared with knockout mice for TLR-2.31 High IL-6 levels, in turn, increase CRP.32 We conclude that black carbon and ozone exposures may cause changes in TLR-2 and IL-6 methylation, which would be causing higher TLR-2, IL-6, and CRP levels. Our sensitivity analysis also suggests that air pollution affects methylation at specific positions within a gene’s promoter region differently. Although taking the average methylation across specific position could give a more stable estimate of methylation on a certain gene’s promoter region, this could be misleading.
We observed a positive association between ozone and IFN-γ methylation. Ozone exposure has been shown to increase IFN-γ protein levels.33 IFN-γ is a cytokine that plays an important role in innate and adaptive immune responses against the intrusion of a foreign compound.34 The direction of this association is also consistent with a previous study relating IFN-γ hypermethylation to lower lung function.12 Ozone exposure could cause changes in IFN-γ methylation that would in turn regulate IFN-γ production.
Black carbon and particle number, which are associated with traffic emissions, were associated with F3 and IL-6 methylation, whereas sulfate and ozone, which are secondary pollutants that were transported to Boston, were related to changes in ICAM-1, TLR-2, and IFN-γ methylation. These findings suggest that air pollutants from distinct sources may affect intermediary cardiovascular-related blood markers differently. Even though our results do not prove that these methylation pathways are the primary reason for the associations between air pollution and plasma blood markers reported previously, they do suggest a plausible role in the exacerbation of cardiovascular morbidity and mortality due to air pollution.
Our analyses of effect modification did not reveal that participants’ characteristics, such as smoking and obesity status, age, and baseline methylation levels of LINE-1 and Alu elements identify subsets of individuals with stronger molecular responses to air pollution. We previously found in the same cohort that the association between air pollution and cardiovascular-related biomarkers was modified by methylation levels on LINE-1 and Alu elements at baseline.24 Cigarette smoking is known to modify DNA methylation levels.35 Even though air pollution was related to gene-specific methylation in the overall sample containing never, current, and former smokers, we also found significant associations among never smokers.
Limitations and strengths
Our approach in this study is subject to some limitations. We did not compare our main findings to estimates obtained from a model that would multiply impute the missing values of the exposures, the covariates, and the outcomes. Although our assumption of Berkson measurement error for air pollution exposures assessed at a central site is supported by a previous study,36 correction for measurement error may yield less biased estimates for spatially heterogeneous traffic-related air pollutants. In our analysis, we adjusted for potential confounding variables. We did not examine confounding due to other variables such as diet, although they are unlikely to be associated with intermediate-term air pollution. Our analysis is also limited to five genes. Methylation on other genes and other epigenetics mechanisms such as histone modifications and microRNA might be important variables to consider. We measured gene-specific methylation but not the levels of all associated proteins, which limit our power to detect effects in the mediation analysis. We measured levels of methylation and protein at the same visit. The potential for reverse causation is a limitation of our mediation findings. Potential risk factors or confounding variables included in our models, such as diabetes status and obesity, may also be intermediate variables. Our study population consists of white and elderly men and we cannot provide evidence for similar air pollution associations with gene-specific methylation in a different population.
Our approach has several strengths too. The prospective study design with repeated outcome measurements permitted us to perform a well-powered analysis. Our distributed-lag approach limited the number of tests and therefore the false positive rate, while assuring that complex lag structures are flexibly modeled. High precision pyrosequencing yields more accurate methylation measurements than are available from array data. DNA methylation constitutes a more stable biomarker than mRNA or protein expression, and therefore, represents a somewhat longer time window in biological responses. In addition, we checked for model misspecification by allowing the dose-response relationships between methylation and air pollution, temperature, and relative humidity to be non-linear. The similarity of the estimates we obtained using two different distributed-lag functions showed the robustness of our results. Our sensitivity analysis allowed us to identify positions within promoter regions of relevant genes that may play an important role in air pollution health effects. Our sensitivity analysis also presented stable results when we adjusted for additional potential confounding variables as well as when we considered GEE models with robust variance and censoring weights. The GEE estimator does not assume a normal distribution for the random intercepts and therefore provides more robust confidence intervals than the mixed-effects model, as well as population interpretation for non-Gaussian distributions. The directed acyclic diagram (DAG) we constructed also presented a setting with no time-varying confounding. Therefore, if our assumptions hold, the estimates we obtained by fitting association models should have a causal interpretation. The mediation analysis we performed allowed us to conclude that the air pollution effect on gene-specific methylation is likely to be followed by a change in cardiovascular-related proteins.
Materials and Methods
Study population
This prospective cohort study included participants from the Normative Aging Study, an investigation established in Boston in 1963 by the US. Veterans Administration.24 We measured DNA methylation on blood samples collected after an overnight fast and smoking abstinence during the period 1999–2009. A total of 777 participants had their methylation assessed one to five times with intervals of three to five years. We excluded observations with CRP levels over 10 mg/L so that the results are not confounded by infection state.37 This study was approved by the institutional review boards of all participating institutions.
DNA methylation
We collected participant’s blood at every visit and isolated DNA to assess gene-specific DNA methylation using highly quantitative methods based on bisulfite polymerase chain reaction pyrosequencing.38 The degree of methylation was expressed as the percentage of methylated cytosines over the sum of methylated and unmethylated cytosines at position 5 (%5mC).
We focused on genes that are expressed in blood leukocytes. We measured F3, ICAM-1, TRL-2, and IFN-γ methylation levels at two to five CpG positions within each gene’s promoter region and calculated the mean values of the position-specific measurements. IL-6 methylation was quantified outside the gene’s promoter region. Exact positions within promoter regions have been previously described.24
Air pollution
The relevant exposure window for the air pollution association with methylation is unknown. Previous studies suggested an association spread over several weeks.16,17,23 In a recent study, we observed weekly associations between air pollution and coagulation and inflammation markers in this cohort.24 Therefore, we chose to explore a similar intermediate-term exposure window and focused on air pollution concentrations measured during the monthly period preceding each participant’s methylation assessment.
Particulate concentrations were measured at the Harvard supersite located near downtown Boston and approximately 1 km from the examination center. Since the study participants lived in the Greater Boston area with a median distance of 20 km, we assumed that the ambient air pollutant concentrations could serve as surrogates of their exposures.
We measured hourly particle number per cm3 (0.007–3 µm) with a Condensation Particle Counter (TSI Inc., Model 3022A) and hourly PM2.5 black carbon concentrations using a Tapered Element Oscillation Microbalance (Model 1400A, Rupprecht and Pastashnick) and an Aethalometer (Magee Scientific Co., Model AE-16). We measured daily sulfate (SO42-) concentrations with a Sulfate Particulate Analyzer (Thermo Electron Co., Model 5020) from 2000 to 2003. After this time period, SO42- levels were estimated from elemental sulfur, measured by X-Ray Fluorescence analysis of Teflon filters. We did not measure ozone at the central monitoring site, but obtained hourly levels by averaging concentrations measured by two local state monitors operated by the Massachusetts Department of Environmental Protection.
Whereas particle number is a marker for fresh local traffic emissions, black carbon originates from both local and transported traffic emissions. Sulfates and ozone are secondary regional pollutants that were transported to Boston.
Statistical methods
Assumptions and directed acyclic graph (DAG)
Because we had repeated methylation measures for 71% of the participants, we fit generalized mixed-effects models with random intercepts to investigate whether air pollution levels weekly averaged over the 4-week period before the jth visit of the ith participant were associated with its methylation level at visit j (Yij). Because F3, ICAM-1, and TLR-2 methylation had a point mass at zero and their residuals’ distribution showed important deviation from a normal density, we assumed a Tweedie distribution (Fig. S1) for these outcomes with a log-link and reported associations on the multiplicative scale. For IFN-γ and IL-6 methylation, we assumed a Gaussian distribution for the regression residuals and presented our results on the additive scale.
We adjusted for potential confounders (C1) such as: temperature, relative humidity, seasonal sine and cosine, season (Winter/Spring-Fall/Summer), and batch of methylation measurement. We also controlled for factors likely to influence methylation (C2) such as: age, race, diabetes, body mass index, smoking status, statin use, as well as percentages of neutrophils and lymphocytes in differential blood count. We included C2 in the models for efficiency and blocking any potential back-door path through unmeasured variables that would be a common cause of air pollution and C2.39 We thus assumed no unmeasured confounding between air pollution and methylation, given the random intercept and the C1 and C2 vectors (Fig. 2). Moreover, we assumed the missing mechanisms of the exposures and outcomes to be at random conditional on the covariates, and the air pollution measurement error to be primarily Berkson.
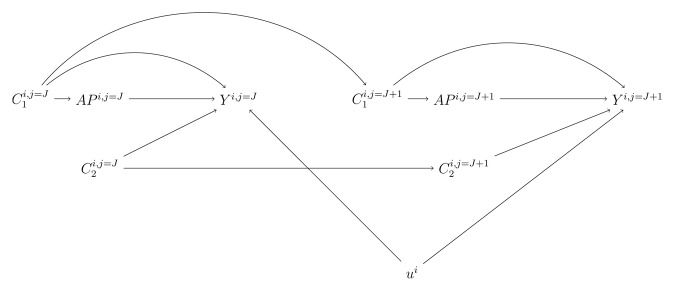
Figure 2. Directed acyclic graph (DAG) at visits j = J and j = J+1. We present the directed acyclic graph (DAG) at adjacent visits j = J and j = J+1 to illustrate the relationships we assumed between the variables included in the regression models. APi,j represents 1st, 2nd, 3rd, and 4th weeks averages of air pollutant concentration before the jth visit of the ith participant. Yi,j represents the ith participant gene-specific DNA methylation at visit j. C1i,j and C2i,j corresponds to the potential confounding variables and risk factors of DNA methylation for participant ith at visit j, respectively. ui represents the random intercept of participant i.
We checked for non-linear dose-response relationships between methylation and air pollution, temperature, and relative humidity using generalized additive models and cubic splines. We found no deviation from linear dose-response relationships with respect to methylation. We explored the nature of the air pollution association with methylation over time using distributed-lag linear models (lags 0 to 27 days) and examined how the association between the methylation outcomes and lagged exposure changes across lags. This methodology, previously developed for the analysis of time series data,40 is extended here in the context of individual longitudinal data. We chose natural splines with three degrees of freedom to model the non-linear shape of the distributed-lag. Because the association between methylation and air pollution exposure varied over the exposure lags we examined, we calculated from the distributed-lag model cumulative associations over four weekly periods (lags 0 to 6, 7 to 13, 14 to 20, and 21 to 27 days) preceding the jth visit of the ith participant, as well as cumulative associations over the entire monthly period.
Main regression models
Distributed-lag model for F3, ICAM-1, and TLR-2 (multiplicative scale)
log E[Yij] = (γ0+ui)+f1(γ1,APij)+f2(γ2,Temperatureij)+f3(γ3,Relative humidityij)+∑k γ4k C1kij+
∑k γ5k C2kij
with Yij~Tweedie and ui ~N(0,σu2)
Distributed-lag linear models for IFN-γ and IL-6 (additive scale)
Yij = (γ0+ui)+f1(γ1,APij)+f2(γ2,Temperatureij)+f3(γ3,Relative humidityij)+
∑k γ4k C1kij+∑k γ5k C2kij+εij
with εij~N(0, σ2) and ui ~N(0,σu2)
f1, f2, and f3 represent distributed-lag functions with sets of coefficients γ1, γ2, and γ3 constrained by a natural spline (with 3 degrees of freedom) that correspond to the air pollution, temperature, and relative humidity associations with methylation at lags 0 to 27 days. C1 and C2 represent sets of variables for which we adjusted.
Analyses of effect modification and mediation
In order to detect any differences in the relationship between air pollution and gene-specific methylation in susceptible subgroups, we used interaction terms to examine whether the association between air pollution and methylation differed according to participants’ smoking status, obesity status, age, as well as baseline levels of LINE-1 and Alu methylation.
We also conducted mediation analyses and calculated the mediated effect of air pollution (APij) on relevant inflammatory and coagulation markers (Yij) through a change in DNA methylation (Mij). We fit simultaneously two linear mixed-effects models:41
Mij = (γ0+ui)+ γ1APij +∑k γ2k C1kij+∑k γ3k C2kij+εij
with εij~N(0, σ2) and ui ~N(0,σu2)
Yij = (β0+g0i)+ β1 APij+ (β2+g2) Mij +∑k β3k C1kij+∑k β4k C2kij+ηij
with ηij~N(0, σ2), g0i ~N(0,σ0u2), and g2i ~N(0,σ2u2)
C1 and C2 represent sets of variables for which we adjusted. The mediated effect is given by the product formula γ1β2. The delta method allowed us to approximate the variance of the mediated effect by Var(β2) γ12 + 2 Cov(γ1, β2) γ1 β2 + Var(γ1)β22.
Sensitivity analyses
We considered another choice of distributed-lag functions that assume constant lag association within weeks. This assumption is equivalent to fitting a model that simultaneously includes four consecutive weekly moving averages of air pollution, temperature, and relative humidity. For simplicity and to limit the number of tests, our additional secondary analyses examined only cumulative associations over the entire monthly period.
Our main analysis considered the average methylation of CpG sites in each gene’s promoter region. However, methylation at specific positions within a gene’s promoter region may affect gene expression differently. Therefore, we examined whether the air pollution associations we observed in the main analysis were specific to certain positions within the gene’s promoter region. Exact positions within promoter region have been described in a previous paper.24
Because of the negative correlations between ozone and the traffic-related air pollutants (particle number and black carbon) as well as between sulfate and particle number in our data, we also fit co-pollutant models including either ozone and particle number, ozone and black carbon, or sulfate and particle number. To confirm that our results were not subject to residual confounding, we further adjusted for cigarette pack-years, more than two alcohol drinks a day, and years of education. We also checked the robustness of the confidence intervals obtained in the main analysis by fitting generalized estimating equations (GEE) allowing an exchangeable correlation structure and a sandwich variance. In addition, we further explored the effect of censoring due to incomplete follow-up by using inverse probability censoring weights (IPCW) in the same GEE framework. The construction of the censoring weights has already been described.12 This methodology assumes that participants with complete and incomplete follow-up are exchangeable, conditional on the covariates. The probability of each participant to providing complete data at all visits is first estimated. The analysis is then restricted to the complete data and each participant is assigned a weight inversely proportional to his estimated probability of complete follow-up. We finally excluded observations for which the 24 h PM2.5 mass concentrations exceeded the daily standard of 35 µg/m3 and examined the robustness of our results.
Supplementary Material
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
This study was supported by the US Environmental Protection Agency grants EPA RD-827353 and RD-832416; National Institute of Environmental Health Sciences (NIEHS) grants RO1-ES015172, 2RO1-ES015172, ES014663, ES00002, PO1-ES008925, Clean Air Act (CLARC) grant RD83479701, and Medical Research Council-UK G1002296. The VA Normative Aging Study is supported by the Cooperative Studies Program/Epidemiology Research and Information Center of the US. Department of Veterans Affairs and is a component of the Massachusetts Veterans Epidemiology Research and Information Center, Boston, Massachusetts.
Glossary
Abbreviations:
- CRP
C-reactive protein
- CpG
Cytosine-phosphodiester-guanine
- DAG
Directed acyclic graph
- GEE
Generalized estimating equations
- ICAM-1
Intercellular adhesion molecule 1
- IFN-γ
Interferon gamma
- IL-6
Interleukin 6
- IPCW
Inverse probability censoring weights
- LINE-1
Long Interspersed Nucleotide Element 1
- %5mC
Percent of 5-methylated cytosine
- F3
Tissue factor
- TLR-2
Toll-like receptor 2
- VCAM-1
Vascular cell adhesion protein 1
References
- 1.Brook RD, Rajagopalan S, Pope CA, 3rd, Brook JR, Bhatnagar A, Diez-Roux AV, Holguin F, Hong Y, Luepker RV, Mittleman MA, et al. American Heart Association Council on Epidemiology and Prevention, Council on the Kidney in Cardiovascular Disease, and Council on Nutrition, Physical Activity and Metabolism Particulate matter air pollution and cardiovascular disease: An update to the scientific statement from the American Heart Association. Circulation. 2010;121:2331–78. doi: 10.1161/CIR.0b013e3181dbece1. [DOI] [PubMed] [Google Scholar]
- 2.Baccarelli A, Martinelli I, Zanobetti A, Grillo P, Hou LF, Bertazzi PA, Mannucci PM, Schwartz J. Exposure to particulate air pollution and risk of deep vein thrombosis. Arch Intern Med. 2008;168:920–7. doi: 10.1001/archinte.168.9.920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nemmar A, Hoet PH, Dinsdale D, Vermylen J, Hoylaerts MF, Nemery B. Diesel exhaust particles in lung acutely enhance experimental peripheral thrombosis. Circulation. 2003;107:1202–8. doi: 10.1161/01.CIR.0000053568.13058.67. [DOI] [PubMed] [Google Scholar]
- 4.Alexeeff SE, Coull BA, Gryparis A, Suh H, Sparrow D, Vokonas PS, Schwartz J. Medium-term exposure to traffic-related air pollution and markers of inflammation and endothelial function. Environ Health Perspect. 2011;119:481–6. doi: 10.1289/ehp.1002560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yamawaki H, Iwai N. Mechanisms underlying nano-sized air-pollution-mediated progression of atherosclerosis: carbon black causes cytotoxic injury/inflammation and inhibits cell growth in vascular endothelial cells. Circ J. 2006;70:129–40. doi: 10.1253/circj.70.129. [DOI] [PubMed] [Google Scholar]
- 6.Bollati V, Baccarelli A. Environmental epigenetics. Heredity (Edinb) 2010;105:105–12. doi: 10.1038/hdy.2010.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hou L, Zhang X, Wang D, Baccarelli A. Environmental chemical exposures and human epigenetics. Int J Epidemiol. 2012;41:79–105. doi: 10.1093/ije/dyr154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang Y, Rohde C, Tierling S, Jurkowski TP, Bock C, Santacruz D, Ragozin S, Reinhardt R, Groth M, Walter J, et al. DNA methylation analysis of chromosome 21 gene promoters at single base pair and single allele resolution. PLoS Genet. 2009;5:e1000438. doi: 10.1371/journal.pgen.1000438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Castro R, Rivera I, Struys EA, Jansen EE, Ravasco P, Camilo ME, Blom HJ, Jakobs C, Tavares de Almeida I. Increased homocysteine and S-adenosylhomocysteine concentrations and DNA hypomethylation in vascular disease. Clin Chem. 2003;49:1292–6. doi: 10.1373/49.8.1292. [DOI] [PubMed] [Google Scholar]
- 10.Kim M, Long TI, Arakawa K, Wang R, Yu MC, Laird PW. DNA methylation as a biomarker for cardiovascular disease risk. PLoS One. 2010;5:e9692. doi: 10.1371/journal.pone.0009692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stenvinkel P, Karimi M, Johansson S, Axelsson J, Suliman M, Lindholm B, Heimbürger O, Barany P, Alvestrand A, Nordfors L, et al. Impact of inflammation on epigenetic DNA methylation - a novel risk factor for cardiovascular disease? J Intern Med. 2007;261:488–99. doi: 10.1111/j.1365-2796.2007.01777.x. [DOI] [PubMed] [Google Scholar]
- 12.Lepeule J, Baccarelli A, Motta V, Cantone L, Litonjua AA, Sparrow D, Vokonas PS, Schwartz J. Gene promoter methylation is associated with lung function in the elderly: the Normative Aging Study. Epigenetics. 2012;7:261–9. doi: 10.4161/epi.7.3.19216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wright RO, Schwartz J, Wright RJ, Bollati V, Tarantini L, Park SK, Hu H, Sparrow D, Vokonas P, Baccarelli A. Biomarkers of lead exposure and DNA methylation within retrotransposons. Environ Health Perspect. 2010;118:790–5. doi: 10.1289/ehp.0901429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Schneider JS, Kidd SK, Anderson DW. Influence of developmental lead exposure on expression of DNA methyltransferases and methyl cytosine-binding proteins in hippocampus. Toxicol Lett. 2013;217:75–81. doi: 10.1016/j.toxlet.2012.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Soberanes S, Gonzalez A, Urich D, Chiarella SE, Radigan KA, Osornio-Vargas A, Joseph J, Kalyanaraman B, Ridge KM, Chandel NS, et al. Particulate matter Air Pollution induces hypermethylation of the p16 promoter Via a mitochondrial ROS-JNK-DNMT1 pathway. Sci Rep. 2012;2:275. doi: 10.1038/srep00275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Salam MT, Byun HM, Lurmann F, Breton CV, Wang X, Eckel SP, Gilliland FD. Genetic and epigenetic variations in inducible nitric oxide synthase promoter, particulate pollution, and exhaled nitric oxide levels in children. J Allergy Clin Immunol. 2012;129:232–, e1-7. doi: 10.1016/j.jaci.2011.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Madrigano J, Baccarelli A, Mittleman MA, Wright RO, Sparrow D, Vokonas PS, Tarantini L, Schwartz J. Prolonged exposure to particulate pollution, genes associated with glutathione pathways, and DNA methylation in a cohort of older men. Environ Health Perspect. 2011;119:977–82. doi: 10.1289/ehp.1002773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baccarelli A, Wright RO, Bollati V, Tarantini L, Litonjua AA, Suh HH, Zanobetti A, Sparrow D, Vokonas PS, Schwartz J. Rapid DNA methylation changes after exposure to traffic particles. Am J Respir Crit Care Med. 2009;179:572–8. doi: 10.1164/rccm.200807-1097OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peluso M, Bollati V, Munnia A, Srivatanakul P, Jedpiyawongse A, Sangrajrang S, Piro S, Ceppi M, Bertazzi PA, Boffetta P, et al. DNA methylation differences in exposed workers and nearby residents of the Ma Ta Phut industrial estate, Rayong, Thailand. Int J Epidemiol. 2012;41:1753–60, discussion 1761-3. doi: 10.1093/ije/dys129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tarantini L, Bonzini M, Apostoli P, Pegoraro V, Bollati V, Marinelli B, Cantone L, Rizzo G, Hou L, Schwartz J, et al. Effects of particulate matter on genomic DNA methylation content and iNOS promoter methylation. Environ Health Perspect. 2009;117:217–22. doi: 10.1289/ehp.11898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Madrigano J, Baccarelli A, Mittleman MA, Sparrow D, Spiro A, 3rd, Vokonas PS, Cantone L, Kubzansky L, Schwartz J. Air pollution and DNA methylation: interaction by psychological factors in the VA Normative Aging Study. Am J Epidemiol. 2012;176:224–32. doi: 10.1093/aje/kwr523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Breton CV, Salam MT, Wang X, Byun HM, Siegmund KD, Gilliland FD. Particulate matter, DNA methylation in nitric oxide synthase, and childhood respiratory disease. Environ Health Perspect. 2012;120:1320–6. doi: 10.1289/ehp.1104439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baccarelli A, Wright RO, Bollati V, Tarantini L, Litonjua AA, Suh HH, Zanobetti A, Sparrow D, Vokonas PS, Schwartz J. Rapid DNA methylation changes after exposure to traffic particles. Am J Respir Crit Care Med. 2009;179:572–8. doi: 10.1164/rccm.200807-1097OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bind MA, Baccarelli A, Zanobetti A, Tarantini L, Suh H, Vokonas P, Schwartz J. Air pollution and markers of coagulation, inflammation, and endothelial function: associations and epigene-environment interactions in an elderly cohort. Epidemiology. 2012;23:332–40. doi: 10.1097/EDE.0b013e31824523f0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chu AJ. Tissue factor mediates inflammation. Arch Biochem Biophys. 2005;440:123–32. doi: 10.1016/j.abb.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 26.Chu AJ. Role of tissue factor in thrombosis. Coagulation-inflammation-thrombosis circuit. Front Biosci. 2006;11:256–71. doi: 10.2741/1796. [DOI] [PubMed] [Google Scholar]
- 27.Seaton A, MacNee W, Donaldson K, Godden D. Particulate air pollution and acute health effects. Lancet. 1995;345:176–8. doi: 10.1016/S0140-6736(95)90173-6. [DOI] [PubMed] [Google Scholar]
- 28.Emmerechts J, Jacobs L, Van Kerckhoven S, Loyen S, Mathieu C, Fierens F, Nemery B, Nawrot TS, Hoylaerts MF. Air pollution-associated procoagulant changes: the role of circulating microvesicles. J Thromb Haemost. 2012;10:96–106. doi: 10.1111/j.1538-7836.2011.04557.x. [DOI] [PubMed] [Google Scholar]
- 29.Kilinç E, Van Oerle R, Borissoff JI, Oschatz C, Gerlofs-Nijland ME, Janssen NA, Cassee FR, Sandström T, Renné T, Ten Cate H, et al. Factor XII activation is essential to sustain the procoagulant effects of particulate matter. J Thromb Haemost. 2011;9:1359–67. doi: 10.1111/j.1538-7836.2011.04280.x. [DOI] [PubMed] [Google Scholar]
- 30.Sun Q, Yue P, Kirk RI, Wang A, Moatti D, Jin X, Lu B, Schecter AD, Lippmann M, Gordon T, et al. Ambient air particulate matter exposure and tissue factor expression in atherosclerosis. Inhal Toxicol. 2008;20:127–37. doi: 10.1080/08958370701821482. [DOI] [PubMed] [Google Scholar]
- 31.Shoenfelt J, Mitkus RJ, Zeisler R, Spatz RO, Powell J, Fenton MJ, Squibb KA, Medvedev AE. Involvement of TLR2 and TLR4 in inflammatory immune responses induced by fine and coarse ambient air particulate matter. J Leukoc Biol. 2009;86:303–12. doi: 10.1189/jlb.1008587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Van Gaal LF, Mertens IL, De Block CE. Mechanisms linking obesity with cardiovascular disease. Nature. 2006;444:875–80. doi: 10.1038/nature05487. [DOI] [PubMed] [Google Scholar]
- 33.Bocci V, Paulesu L. Studies on the biological effects of ozone 1. Induction of interferon gamma on human leucocytes. Haematologica. 1990;75:510–5. [PubMed] [Google Scholar]
- 34.Lundborg M, Johansson A, Lâstbom L, Camner P. Ingested aggregates of ultrafine carbon particles and interferon-gamma impair rat alveolar macrophage function. Environ Res. 1999;81:309–15. doi: 10.1006/enrs.1999.3992. [DOI] [PubMed] [Google Scholar]
- 35.Lee KW, Pausova Z. Cigarette smoking and DNA methylation. Front Genet. 2013;4:132. doi: 10.3389/fgene.2013.00132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zeger SL, Thomas D, Dominici F, Samet JM, Schwartz J, Dockery D, Cohen A. Exposure measurement error in time-series studies of air pollution: concepts and consequences. Environ Health Perspect. 2000;108:419–26. doi: 10.1289/ehp.00108419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Simon L, Gauvin F, Amre DK, Saint-Louis P, Lacroix J. Serum procalcitonin and C-reactive protein levels as markers of bacterial infection: a systematic review and meta-analysis. Clin Infect Dis. 2004;39:206–17. doi: 10.1086/421997. [DOI] [PubMed] [Google Scholar]
- 38.Yang AS, Estécio MR, Doshi K, Kondo Y, Tajara EH, Issa JP. A simple method for estimating global DNA methylation using bisulfite PCR of repetitive DNA elements. Nucleic Acids Res. 2004;32:e38. doi: 10.1093/nar/gnh032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Greenland S, Pearl J, Robins JM. Causal diagrams for epidemiologic research. Epidemiology. 1999;10:37–48. doi: 10.1097/00001648-199901000-00008. [DOI] [PubMed] [Google Scholar]
- 40.Gasparrini A, Armstrong B, Kenward MG. Distributed lag non-linear models. Stat Med. 2010;29:2224–34. doi: 10.1002/sim.3940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bauer DJ, Preacher KJ, Gil KM. Conceptualizing and testing random indirect effects and moderated mediation in multilevel models: new procedures and recommendations. Psychol Methods. 2006;11:142–63. doi: 10.1037/1082-989X.11.2.142. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.