Abstract
Amyotrophic lateral sclerosis (ALS) is a devastating, rapidly progressing disease of the central nervous system that is characterized by motor neuron degeneration in the brain stem and the spinal cord. We employed time of flight secondary ion mass spectrometry (ToF-SIMS) to profile spatial lipid- and metabolite- regulations in post mortem human spinal cord tissue from ALS patients to investigate chemical markers of ALS pathogenesis. ToF-SIMS scans and multivariate analysis of image and spectral data were performed on thoracic human spinal cord sections. Multivariate statistics of the image data allowed delineation of anatomical regions of interest based on their chemical identity. Spectral data extracted from these regions were compared using two different approaches for multivariate statistics, for investigating ALS related lipid and metabolite changes. The results show a significant decrease for cholesterol, triglycerides, and vitamin E in the ventral horn of ALS samples, which is presumably a consequence of motor neuron degeneration. Conversely, the biogenic mediator lipid lysophosphatidylcholine and its fragments were increased in ALS ventral spinal cord, pointing towards neuroinflammatory mechanisms associated with neuronal cell death. ToF-SIMS imaging is a promising approach for chemical histology and pathology for investigating the subcellular mechanisms underlying motor neuron degeneration in amyotrophic lateral sclerosis.
Amyotrophic Lateral Sclerosis (ALS) is a very progressive, fatal disease that is characterized by irreversible degeneration of motor neurons in the spinal cord, brain stem, and cortex resulting in increasing muscle weakness and muscle atrophy1. ALS has a prevalence of 2–4 cases per 100 000 a year with a common onset age between age 40 and 60 and an average life expectancy three to five years after diagnosis2. Most ALS cases are idiopathic in nature; however, 10% have been found to be caused by genetic factors2. The main mechanisms underlying ALS are still unknown, although a couple of findings concerning molecular ALS pathology have been reported mainly focusing on genetic causes3,4,5. The limited knowledge of the disease results in further drawbacks such as the inaccuracy and time delay for diagnosis, which highlights the need for further investigation of the disease on a molecular level.
The limited understanding of molecular ALS pathobiology relates directly to the lack of appropriate analytical technologies that feature the necessary sensitivity and specificity. Studying complex biological and pathological processes requires acquisition of molecular images, since these processes involve the translocation and interaction of a wide range of chemical species.
Mass spectrometry based molecular imaging (IMS) is an emerging technique for probing the molecular architecture of biological tissue in an anatomical context6. In contrast to common molecular biology techniques, IMS does not require any a priori knowledge of the potential target species and allows direct and comprehensive identification and spatial profiling of biomolecular species in situ, while maintaining high molecular specificity7,8,9,10. A major advantage of ToF-SIMS based imaging MS is the possibility to robustly acquire high-resolution images at the cellular length scales (1–5 μm), without compromising sample treatment procedures11,12,13. These features make SIMS a powerful technology for spatially profiling lipids and metabolites at the single cell level14. This is of particular relevance for studying complex and heterogeneous samples, such as neuronal tissue and cells, which arguably constitute the most complex and least understood systems in the human body7,15. ToF-SIMS based molecular imaging represents therefore a promising approach for investigating spinal cord biochemistry.
Several studies on spinal cord using various imaging mass spectrometry techniques have been reported that have provided important data on spinal cord pathology16,17,18,19,20,21,22. In a recent study, we probed the native distribution of lipids and metabolites in human spinal cord using ToF-SIMS for high spatial resolution imaging23. In addition, different strategies for multivariate analysis of whole image datasets for tissue segmentation were evaluated that in turn facilitate identification of anatomical regions of interest based on their chemical identity.
Here we report the use of ToF-IMS based analysis of post mortem human spinal cord tissue samples in conjunction with multivariate data analysis (MVA) to identify ALS related changes in spinal cord lipid chemistry and metabolism in situ. This work should gain further insight in molecular mechanisms underlying ALS, particularly with respect to lower motor neuron degeneration.
Results
Multivariate image analysis for identification of anatomical regions of interest in human spinal cord
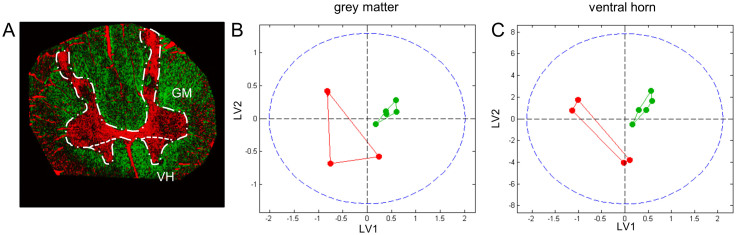
For each individual (n = 5; 2 control/3 ALS), subsets of two consecutive tissue sections were analyzed with ToF-SIMS in both positive and negative ion mode. The datasets were calibrated internally and reconstructed with a common mass interval list (binning) for data acquired in both positive- and negative ion mode. This resulted in a total of 20 imaging datasets (10 pos.- and 10 neg. mode), each at least 3 GB in size. Imaging data from the 20 datasets were subjected to unsupervised multivariate statistical image analysis by means of maximum autocorrelation factor analysis (MAF)24,25. This resulted in generation of eigenvalue (score) images that depict the chemical variance captured in the respective factors of the MAF analysis (Figure 1). This image analysis approach revealed characteristic outlines of the grey and white matter (Figure 1). The molecular species that outline the histological regions and cellular environments can then be deduced from the loadings (eigenvectors) and provide further information on local lipid biochemistry and metabolism.
Figure 1. Image Data Analysis for Tissue Segmentation.
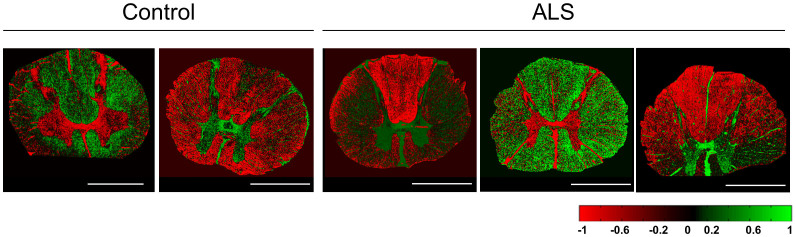
Multivariate statistical analysis of imaging MS data by means of maximum autocorrelation factor analysis (MAF) reveals anatomical regions of human spinal cord including grey and white matter in both control and ALS samples. Composition images of corresponding factors (i.e. components, MA-factors) allow identification of anatomical regions of interest. Each pixel intensity represents the respective score (eigenvalue, see scale). Scale bar = 4 mm.
Multivariate analysis of region of interest data allows discrimination of ALS and control groups
Two different multivariate approaches, partial least square discriminant analysis (PLS-DA) and “Statistical Analysis of Microarray Data” (SAM), were employed for statistical analysis of spectral data extracted from anatomical regions of interest (ROI). These histological regions including the whole grey matter (GM) as well as the ventral horn of the grey matter (VH) were assigned in accordance with the image analysis data (Figure 2A). The ROI data of the GM and the VH of each patient were normalized to the total ion count (TIC), log2 converted, and different ionization modes were pooled for each patient. This gave two sample/variable matrices for both the GM ROI and the VH ROI that were then evaluated by means of multivariate analysis using two different strategies. Here, as a first approach for MVA, multivariate analysis by means of PLS-DA was performed. PLS-DA is a linear regression extension of principal component analysis (PCA) and well suited for comparing two different sample groups. PLS-DA was performed after mean centering and Poisson scaling and resulted in distinct discrimination of the two groups (Control and ALS) both for the grey matter region and even more pronounced for the ventral horn data (Figure 2B, C). From the corresponding loading plots the individual variables (m/z values) were deduced that have the greatest impact on the score and thereby separation (SI Figure 1A, B).
Figure 2. Multivariate statistics of ROI spectral data using PLS DA.
MS spectral data from previously outlined anatomical regions (A), including the grey matter (GM) (B) and ventral horn (VH) (C), were subjected to multivariate analysis using PLS DA. (B,C) PLS DA score plots demonstrate grouping of ALS samples (green) and controls (red).
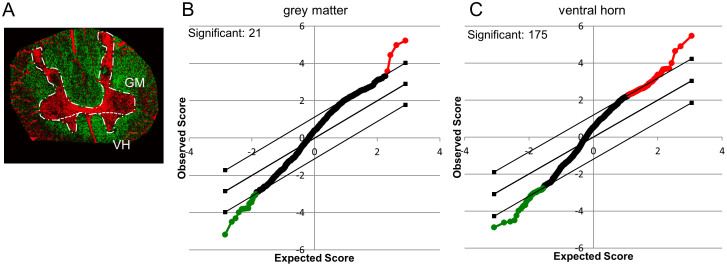
As a second approach, the “Statistical Analysis of Microarray data” tool26 was used for grouped unpaired t-statistics of the ALS and control group data for each ROI (GM and VH) (Figure 3A). For the whole grey matter region the results show significant intensity changes (p < 0.05) for in total 21 m/z values. Here, four mass peaks were significantly increased in ALS (red), while a number of 17 species were higher in the control group (green, Figure 3B). For the ventral horn region; however, the data show significant changes (p < 0.05) for 175 m/z values that were found either increased (130) or decreased (45) in the ALS samples compared to the controls (Figure 3C).
Figure 3. Statistical Analysis of Spectral Data using SAM.
Spectral data of previously outlined regions as revealed by image analysis were subjected to statistical analysis of microarray data (SAM). Spectra acquired in positive and negative mode for the GM- and VH-areas (A) were analyzed. (B, C) SAM plots illustrate relative changes of ALS versus control in the respective areas as revealed by unpaired t-statistics. The x-axis illustrates the degree of significance, while the y-axis refers to the observed degree of change. Red illustrates a relative increase in ALS compared to control, while green illustrates a relative decrease in ALS to control. The borders indicate the significance threshold (p < 0.05).
For verification of these findings, SAM results for the respective ROI were compared to the results obtained from PLS-DA. Here, the PLS-DA derived variables (loadings) that exhibited the greatest contribution to the corresponding score of the individual samples were investigated. Only, m/z compounds that were found significant in SAM and displayed a large PLS-DA loading value were considered. For the grey matter region all 21 compounds that were found significantly changed in between ALS and controls were also found to have a large loading value in PLS-DA. In contrast, of the 175 significant observed in SAM analysis for the VH region, only 40 were found to have a large loading value. These were then further verified using non-parametric unpaired statistics (Kruskal-Wallis). This resulted in a number of 38 compounds that were found significantly changes in the ventral horn in between ALS and control samples. A number of 15 peaks were increased and 23 peaks were decreased in ALS ventral horn as compared to controls. Here, for the decreased compounds all peaks were detected in negative mode, while for increased compounds 7 were detected in positive and 8 in negative mode.
Regional changes of spinal cord lipids associated with ALS
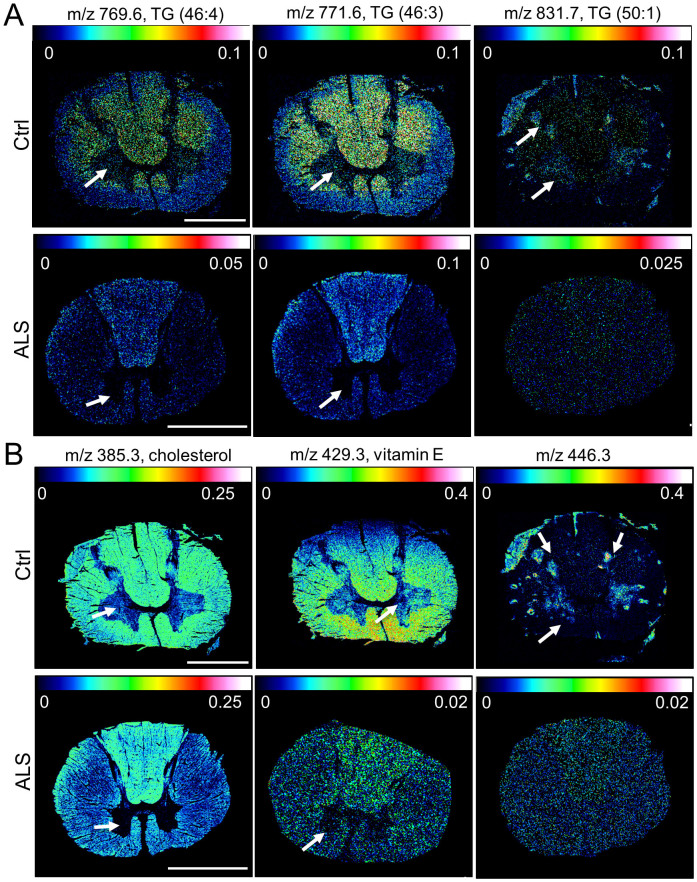
The most prominent signals were tentatively assigned based on accurate mass matching and comparison to values reported in the literature27,28,29 as well as by matching the experimental data to predicted isotopic patterns. Multivariate analysis of the ROI spectral data resulted in 6 molecular species that were found decreased in the grey matter as well as in the ventral horn of ALS spinal cord. Five of these peaks could be assigned to triglyceride species (TG, Figure 4A)28,29, while one compound (m/z 446.3) could not be identified. Further mass peak results obtained for the whole GM ROI could not be verified by manual inspection of the single ion images. By confining comparative analysis to the VH region more species were found significantly changed in between ALS and controls. Among the most prominent compounds that displayed an ALS associated decrease, cholesterol (C27H45O: [M − H]− 385.3) and vitamin E (C29H49O2: [M − H]− 429.3) could be identified27. For both species a significant decrease was observed in the ventral horn region of controls as compared to ALS samples (Figure 4B).
Figure 4. Single Ion Images of Chemical Species decreased in ALS.
Multivariate statistics of ROI spectral data using SAM and PLS DA resulted in detection of multiple molecular species displaying ALS associated decrease in the grey matter. Only compounds observed in negative mode displayed a decrease in ALS spinal cord. (A) Here, a decrease throughout the whole grey matter (arrows) was observed for various triglycerides (TG (46:4), [M − H]− 769.6; TG (46:3), [M − H]− 771.6; TG (50:1), [M − H]− 831.7). (B) Similarly, an unidentified peak at m/z 446.3 was found decreased in both ventral horn and whole grey matter (arrows). In contrast, cholesterol (C27H45O, [M − H]− 385.3) and vitamin E (C29H49O2, [M − H]− 429.3) were found consistently decreased in the ventral horn of ALS spinal cord but not in the whole grey matter. Scale bar = 4 mm.
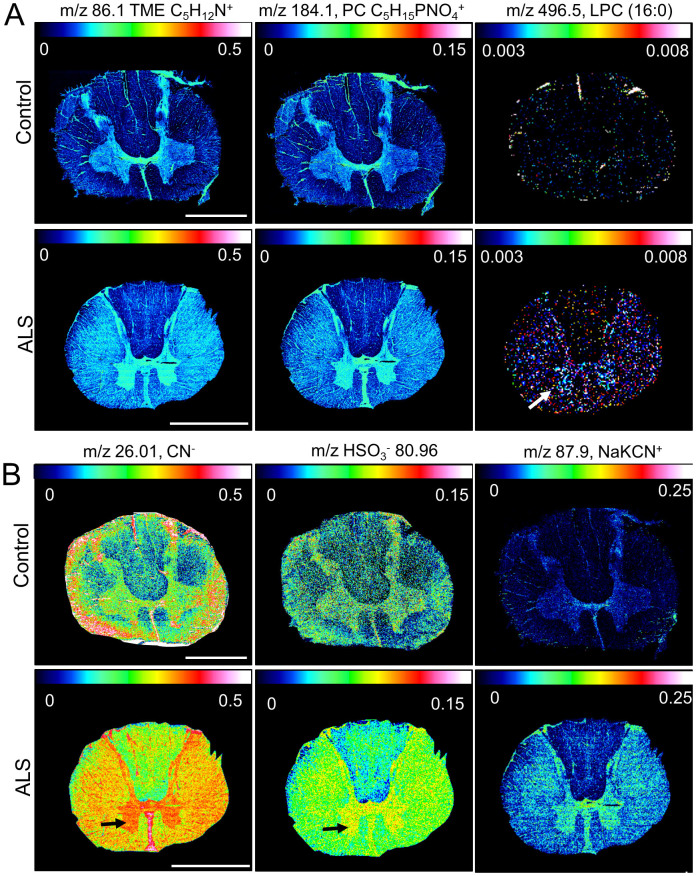
Conversely, a number of compounds were found increased in the ventral horn of ALS tissue as compared to controls. This included phosphatidylcholine (PC) -lipid fragments (TME, trimethylethylimine: m/z 86.1, [C5H12N+]; PC-headgroup: m/z 184.1 [C5H15PO4N+]) as well as intact lysophosphatidylcholine (LPC (16:0), m/z 496.5 [M + H]+) (Figure 5A). Furthermore, protein associated signals (m/z 26.01, [CN−]; m/z 80.96 [NaKCN+]) and hydrogensulfite [HSO3−] were found elevated in the ventral horn of ALS spinal cord samples (Figure 5B).
Figure 5. Single Ion Images of Chemical Species elevated in ALS spinal cord.
Multivariate statistics of ROI spectral data using SAM and PLS DA resulted in detection of several molecular species displaying ALS associated increase in the grey matter. (A) These included lysophosphatidylcholine (LPC (16:0), [M + H]+ 496.5, arrow) as well as its fragments, PC-headgroup (PC) and trimethylethylimine (TME), observed in pos. ion mode. (B) In addition, protein fragments (m/z 26.01, [CN−] and m/z 87.9, [NaKCN+]) as well as hydrogensulfite (m/z 80.96, [HSO3−]) were found elevated in the ventral horn of ALS spinal cord (arrow). Scale bar = 4 mm.
Discussion
The spinal cord constitutes the interface of the peripheral and the central nervous system comprising essential circuits in motor control as well as somatosensory function. Anatomically, the tissue is generally separated into the white matter and grey matter. Nervous tissue is characterized by its high complexity and diversity. This will eventually be reflected in the spatial intensity distribution pattern of biochemical species and in turn affect signal intensity values based on changes in the biochemical background. Therefore comparison between heterogeneous tissue specimens requires identification and annotation of distinct anatomical regions so that histological- and biochemical similar areas can be compared in between different individuals. For comparative analysis, we employed a ToF-SIMS imaging data guided approach for tissue segmentation of spinal cord samples. The aim was to outline anatomical regions of interest based on their chemical identity. This approach was based on multivariate analysis of whole scan image data using maximum autocorrelation factor analysis. The suitability of this approach was evaluated in a recent study by our group, where we compared the performance of PCA and MAF for image analysis with respect to contrast and their ability to identify anatomically relevant regions of interest23. Here, a MAF based image analysis guided strategy for ROI assignment was found to be a robust and straightforward approach for tissue segmentation23. MAF employs an implemented correction to the noise covariance matrix as achieved by an iterative comparison to adjacent pixel spectra. In contrast to PCA, this approach is therefore not dependent on data preprocessing (e.g. scaling, mean centering) and thus provides better contrast and sensitivity that allow catching slight intra tissue heterogeneities, although the technique is computational demanding. This highlights an important consideration for bulk-image data analysis with respect to sample size and data handling. Given the large data file size obtained from the SIMS and other IMS experiments (up to 10 GB), tradeoffs with respect to pixel resolution and number of peak assignments (bins) have to be made. Given the fact that most commercial solutions for image data analysis, as provided by the manufacturers, are still limited to 32-bit systems, the complexity of downstream data analysis approaches have to be considered in advance. In the present study, MAF facilitated exact annotation of these regions and was employed for subsequent extraction of ROI spectral data for comparative analysis of the localized chemistry in ALS tissue compared to controls. Using PLS-DA as well as SAM based statistics, the most significant changes were observed by confined comparative analysis of the ventral horn region (Figure 2 and 3). These observations further demonstrate that targeted ROI data analysis is mandatory to reveal local, disease related, biochemical changes. Indeed, ALS is characterized by degeneration of spinal cord motor neurons in the ventral horn. Spatially confined changes in lipid and metabolite expression are easily convoluted, if the ROI is assigned too widely, which further illustrates the relevance for targeted imaging data analysis when analyzing heterogeneous biological tissue.
A decrease of triglyceride species was observed throughout the whole grey matter (Figure 4A). This is consistent with previously reported results showing an ALS associated increase in clearance of peripheral lipids, including triglycerides30. In that work, an altered energy metabolism was observed in experimental ALS (mSOD1 mice) that resulted in increased TG metabolism. This shift in energy metabolism relates to an increased demand of nutrients in muscle tissue as a consequence of dysfunctional skeletal muscle metabolism30.
While 21 m/z peaks were found statistically changed between ALS and control spinal cords for the whole GM region, they could not be verified by manual inspection of the single ion images. This in turn highlights the importance of strict validation of MVA results by subsequent inspection of the imaging raw data. As mentioned previously, further refinement of the GM-ROI to the ventral horn yielded a significantly higher number of ALS associated molecular species (Figure 3C). Here, the data show that cholesterol (C27H45O: [M − H]− 385.3) and vitamin E (C29H49O2: [M − H]− 429.3) were increased in the ventral horn region of controls as compared to ALS samples (Figure 4B). Cholesterol and vitamin E are major constituents of the myelin sheath surrounding ascending and descending axons that constitute the white matter but also innervate the grey matter regions. These results are consistent with the fact that ALS is characterized by neuronal degeneration in the ventral horn. Decreased levels of the myelin sheath constituents correspond to decreased innervation as a result of neuronal cell death. Moreover, the degree of decreased white matter compounds may serve as a neuropathological hallmark of neurodegenerative mechanisms underlying ALS.
An ALS associated increase of lipids and metabolites was also observed and verified in the ventral horn (Figure 5). Here, SAM and PLS-DA revealed an increase in PC-lipid fragments (PC-headgroup and TME, Figure 5A) in the ventral horn of the grey matter in ALS patients (Figure 5A). Similarly, LPC (16:0) was significantly increased in the same region in ALS spinal cord tissue, while no other phospholipids displayed any significant change (Figure 5A). PC-headgroup and TME are most likely fragments of LPC resulting from in-source fragmentation. The signal can however also originate from sphingomyelin31. A mass peak at m/z 224.1 corresponding to the PC-lipid specific fragment C8H19PNO4+ (PC-headgroup + C3H4) displayed similar distribution pattern and regulation in intensity as the other PC fragments (SI Figure 2)31. This verifies that the PC attributed mass peaks (m/z 184.1; m/z 86.1) do indeed originate from PC-lipids, rather than sphingomyelin.
LPC is a precursor for lysophosphatitic acid (LPA), a biogenic lipid. This lipid mediator is synthesized from LPC by autotaxin (ATX) that was demonstrated to possess lysophospholipase D activity32. Interestingly, in cerebrospinal fluid (CSF) the levels of LPC and LPA are very low despite significant amounts of ATX, which indicates that LPC production would be a rate-limiting step for LPA production33. LPA production and activity in the spinal cord has been described before, where LPA was found to exert its activity through LPA1-receptor signaling, which initiates neuropathic pain and underlying mechanisms including demyelination34. Elevated LPA levels are a consequence of serum leakage through tissue damage but also result from abnormally elevated LPA biosynthesis35. LPA/LPA1-receptor signaling in the spinal cord was found to initiate e.g. neuropathic pain through different demyelination pathways35. These include phosphoinositol-bi/tri-phosphate (PIP2/PIP3) second messenger mediated genomic regulation of myelin protein expression36 as well as inositoltriphosphate (IP3) mediated myelin degradation by calpaein37. This is consistent with the observations presented here, where decreased cholesterol and vitamin E values indicate demyelination. In addition to these observations, a significant increase of protein specific fragments (m/z 26.01, [CN−]) was observed in the ventral of the ALS spinal cord (Figure 5B). Similar to other neurodegenerative diseases, ALS is characterized by progressive accumulation and aggregation of misfolded proteins into insoluble protein aggregates. Increased protein signals therefore indicate impaired protein turnover. Again, this is consistent with previously reported observations, where a diminished proteasome activity has been observed in the ventral horn of ALS patients38,39. We also observed increased levels of hydrogen sulfite (m/z 80.9, [HSO3−]) in the ventral horn of ALS spinal cord. Sulfite is a metabolite of cysteine, which accumulates when one or more of the enzymes needed for glutathione (GSH) synthesis are defective40. Sulfite exerts its neurotoxic potential through direct inhibition of glutamate dehydrogenase thereby impacting ATP biosynthesis40. The present observations on spinal cord sulfite accumulation indicate cysteine and sulfite mediated neurotoxicity as a consequence of disrupted GSH metabolism, as has been discussed earlier41.
Conclusions
The potential of ToF-SIMS imaging for interrogating the biochemical architecture of the human spinal cord in ALS is demonstrated. Multivariate statistics used for image data guided tissue segmentation facilitate accurate outlining of histological regions of interest based on their chemical identity. Spectral data analysis of these ROI's revealed spatially confined changes in lipid and metabolite regulation associated with ALS pathogenesis. ALS implicated decrease of cholesterol and vitamin E in the ventral horn was observed presumably associated with demyelination and motor neuron degeneration. In addition, ventral horn triglyceride clearance indicates ALS related changes in energy metabolism, while an accumulation of hydrogen sulfite and LPC in the ventral horn point to metabolic neurotoxicity and biogenic lipid induced neuroinflammation, respectively. The data presented reveal novel targets for investigating ALS pathology in spinal cord that have not been presented before. ToF-SIMS can be used to reveal disturbances in the neurochemical interplay affecting neuronal functionality and integrity and suggests a novel approach for future investigations concerning motor neuron degeneration.
Methods
Chemicals and Reagents
All chemicals were of pro-analysis grade and purchased from Sigma Aldrich (St. Louis, USA). TissueTek optimial cutting tool (OCT) was purchased from Sakura Finetek (AJ Alphen aan den Rijn,The Netherlands). Water was purified with a Milli-Q (Millipore, Bedford, MA, USA) purification system.
Clinical Samples
Post-mortem spinal cord tissue samples of thoracic level from two controls and three ALS patients were used in the study. The cause of death in the two control subjects was cardiac failure in one case and myocardial infarction in the other. At autopsy, the spinal cords were dissected, removed from the dura mater, and cut into 5-mm sections. The tissue was then immediately frozen on a metal plate maintained in liquid nitrogen and stored at −72°C. Autopsy was performed with written approval from the family in accordance with Swedish rules and regulations.
Sample Preparation
Thoracic spinal cord sections (12 μm) were collected using a cryostate microtome (Thermo Scientific, Waltham, MA) and thaw-mounted on conductive glass slides (indium tin oxide, ITO, Bruker Daltonics, Bremen, Germany). The thaw mounted sections were dried under vacuum for 10 min and stored at −80°C until further use. For SIMS experiments, 2 consecutive sections from each subject were analyzed.
ToF-SIMS Analysis
All ToF SIMS experiments were performed on an IONTOF V instrument (IONTOF GmbH, Münster, Germany) equipped with a Bi3+ cluster ion gun as primary ion source. For whole section analysis, data were acquired in high current bunched mode (HBC)42 with a primary ion current of 0.25 pA at 25 keV and a maximum ion dose was 1.5 109/cm2. Scans were acquired in positive and negative ion mode, using the “stage scan macro raster function” with 10 shots per pixel on 0.4 × 0.4 mm areas with 200 measurements per mm resulting in a pixel resolution of 5 μm. This acquisition mode comprises stepwise acquisition of 0.4 × 0.4 mm patches. Each patch was acquired in electrostatic raster mode where the beam was moved over the field of view. The whole tissue area was analyzed by stepwise acquisition of these patches as achieved by stepwise movement of the sample stage. The mass resolution in high current bunch mode was about M/ΔM = 5000.
Data Processing
All data were acquired and processed with the Surface Lab software (v. 6.3 IONTOF). All spectra were calibrated internally to signals of [C]+, [CH]+, [CH2]+, [CH3]+, [C5H15PNO4]+ and [C27H45]+ that were used as calibration points in positive mode and [C]−, [CH]−, [O]−, [C16H31O2]−, [C18H35O2]− and [C27H45O]− in negative ion mode. Two mass interval lists were created (one for each ion polarity), which contained the list of m/z values to be included in multivariate analysis. Mass interval lists were created by peak search of all individual samples according to the following search parameters: S/N > 3, width 0.8 Da. The assigned mass peaks were collected in the same mass interval list and redundant peak assignments removed. Image data were exported into the *.bif6 format and spectral data were exported directly from the peak statistics tab into Excel (v. 2010).
Data Analysis
For statistical analysis two approaches were employed comprising intact image data analysis as well as statistical analysis of spectral data of distinct regions of interests. For image analysis, *.bif6 files were loaded into PLS-Toolbox (v. 7.02, Eigenvector Research Inc., Wenatchee, WA) running in MatLab (v.12a, The MathWorks Inc., Natick, MA). Multivariate image analysis was performed by means of maximum autocorrelation factor analysis (MAF). For comparative statistics on spectral data, relevant anatomical regions of interest including the grey matter as well as the ventral horn of the grey matter were outlined followed by reconstruction of the respective ROI using the corresponding feature in the Surface Lab software (IONTOF). Two different approaches were employed for comparative, statistical analysis of ROI spectral data. These included 1) partial least square discriminant analysis (PLS-DA) as well as 2) “Statistical Analysis of Microarray data” (SAM)26. Peak area values were log2 normalized followed by SAM based grouped unpaired statistical analysis (t-statistics) using the corresponding macro in Excel26. PLS-DA was performed on the spectral data using PLS-Toolbox (Eigenvector Research). Here, data were mean centered and Poisson scaled. Further analysis of single mass peaks and comparisons between groups was performed with non-parametric ANOVA (Kuskal- Wallis) with 95% confidence interval. Identification of mass peaks was done using tentative assignment by comparing the accurate mass to previously reported values in the literature27 as well as matching of isotopic distribution.
Author Contributions
The study was conceived by J.H. and A.G.E. J.H. designed and performed all the experiments and analyzed the data. J.H. and A.G.E. discussed the data and wrote the manuscript.
Supplementary Material
Supplementary Information
Acknowledgments
This work was supported by the European Research Council (AGE ERC advanced grant), the Wallenberg Foundation (AGE, Wallenberg Fellow), the Swedish Research Council VR (AGE, JH) and the USA National Institutes of Health (AGE). The authors thank Dr. Titti Ekegren and Prof Håkan Askmark at the Dept. of Neurology at Uppsala University for providing the samples.
References
- Kiernan M. C. et al. Amyotrophic lateral sclerosis. Lancet 377, 942–955 (2011). [DOI] [PubMed] [Google Scholar]
- Hirtz D. et al. How common are the “common” neurologic disorders? Neurology 68, 326–337 (2007). [DOI] [PubMed] [Google Scholar]
- Clement A. M. et al. Wild-type nonneuronal cells extend survival of SOD1 mutant motor neurons in ALS mice. Science 302, 113–117 (2003). [DOI] [PubMed] [Google Scholar]
- Cristini J. Misdiagnosis and missed diagnoses in patients with ALS. JAAPA 19, 29–35 (2006). [PubMed] [Google Scholar]
- Chio A. Risk factors in the early diagnosis of ALS: European epidemiological studies. Amyotroph Lateral Scler Other Motor Neuron Disord 1, S13–S18 (2000). [DOI] [PubMed] [Google Scholar]
- Cornett D. S., Reyzer M. L., Chaurand P. & Caprioli R. M. MALDI imaging mass spectrometry: molecular snapshots of biochemical systems. Nat Methods 4, 828–833 (2007). [DOI] [PubMed] [Google Scholar]
- Hanrieder J., Phan N. T. N., Kurczy M. E. & Ewing A. G. Imaging Mass Spectrometry in Neuroscience. ACS Chem Neurosci 4, 666–679 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ljungdahl A., Hanrieder J., Bergquist J. & Andersson M. Analysis of neuropeptides by MALDI imaging mass spectrometry. Methods Mol Biol 1023, 121–136 (2013). [DOI] [PubMed] [Google Scholar]
- Ljungdahl A., Hanrieder J., Falth M., Bergquist J. & Andersson M. Imaging mass spectrometry reveals elevated nigral levels of dynorphin neuropeptides in L-DOPA-induced dyskinesia in rat model of Parkinson's disease. PLoS ONE 6, e25653, 1–12 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanrieder J. et al. L-DOPA-induced dyskinesia is associated with regional increase of striatal dynorphin peptides as elucidated by imaging mass spectrometry. Mol Cell Proteomics 10, M111:009308, 1–14 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher J. S. & Vickerman J. C. Secondary ion mass spectrometry: characterizing complex samples in two and three dimensions. Anal Chem 85, 610–639 (2013). [DOI] [PubMed] [Google Scholar]
- Fletcher J. S., Lockyer N. P. & Vickerman J. C. Molecular SIMS imaging; spatial resolution and molecular sensitivity: Have we reached the end of the road? Is there light at the end of the tunnel? Surf Interface Anal 43, 253–256 (2011). [Google Scholar]
- Fletcher J. S., Vickerman J. C. & Winograd N. Label free biochemical 2D and 3D imaging using secondary ion mass spectrometry. Curr Opin Chem Biol 15, 733–740 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher J. S. Cellular imaging with secondary ion mass spectrometry. Analyst 134, 2204–2215 (2009). [DOI] [PubMed] [Google Scholar]
- Lazar A. N. et al. Time-of-flight secondary ion mass spectrometry (TOF-SIMS) imaging reveals cholesterol overload in the cerebral cortex of Alzheimer disease patients. Acta Neuropathol 125, 133–144 (2013). [DOI] [PubMed] [Google Scholar]
- Monroe E. B. et al. SIMS and MALDI MS imaging of the spinal cord. Proteomics 8, 3746–3754 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van de Plas R. et al. Prospective exploration of biochemical tissue composition via imaging mass spectrometry guided by principal component analysis. Pac Symp Biocomput 12, 458–469 (2007). [PubMed] [Google Scholar]
- Becker J. S. et al. Mass spectrometry imaging (MSI) of metals in mouse spinal cord by laser ablation ICP-MS. Metallomics 4, 284–288 (2012). [DOI] [PubMed] [Google Scholar]
- Hanrieder J., Ekegren T., Andersson M. & Bergquist J. MALDI imaging of post-mortem human spinal cord in amyotrophic lateral sclerosis. J Neurochem 124, 695–707 (2013). [DOI] [PubMed] [Google Scholar]
- Girod M., Shi Y., Cheng J.-X. & Cooks R. G. Mapping Lipid Alterations in Traumatically Injured Rat Spinal Cord by Desorption Electrospray Ionization Imaging Mass Spectrometry. Anal Chem 83, 207–215 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girod M., Shi Y., Cheng J. X. & Cooks R. G. Desorption electrospray ionization imaging mass spectrometry of lipids in rat spinal cord. J Am Soc Mass Spectrom 21, 1177–1189 (2010). [DOI] [PubMed] [Google Scholar]
- Tucker K. R., Serebryannyy L. A., Zimmerman T. A., Rubakhin S. S. & Sweedler J. V. The modified-bead stretched sample method: development and application to MALDI-MS imaging of protein localization in the spinal cord. Chem Sci 2, 785–795 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanrieder J., Malmberg P., Lindberg O. R., Fletcher J. S. & Ewing A. G. Time-of-flight secondary ion mass spectrometry based molecular histology of human spinal cord tissue and motor neurons. Anal Chem 85, 8741–8748 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson A., Fletcher J. S. & Vickerman J. C. A comparison of PCA and MAF for ToF-SIMS image interpretation. Surf Interface Anal 41, 666–674 (2009). [Google Scholar]
- Tyler B. J., Rayal G. & Castner D. G. Multivariate analysis strategies for processing ToF-SIMS images of biomaterials. Biomaterials 28, 2412–2423 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tusher V. G., Tibshirani R. & Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA 98, 5116–5121 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passarelli M. K. & Winograd N. Lipid imaging with time-of-flight secondary ion mass spectrometry (ToF-SIMS). Biochim Biophys Acta 1811, 976–990 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunelle A., Touboul D. & Laprevote O. Biological tissue imaging with time-of-flight secondary ion mass spectrometry and cluster ion sources. J Mass Spectrom 40, 985–999 (2005). [DOI] [PubMed] [Google Scholar]
- Touboul D., Kollmer F., Niehuis E., Brunelle A. & Laprevote O. Improvement of biological time-of-flight-secondary ion mass spectrometry imaging with a bismuth cluster ion source. J Am Soc Mass Spectrom 16, 1608–1618 (2005). [DOI] [PubMed] [Google Scholar]
- Fergani A. et al. Increased peripheral lipid clearance in an animal model of amyotrophic lateral sclerosis. J Lipid Res 48, 1571–1580 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng L., McQuaw C. M., Ewing A. G. & Winograd N. Sphingomyelin/phosphatidylcholine and cholesterol interactions studied by imaging mass spectrometry. J Am Chem Soc 129, 15730–15731 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki J., Inoue A. & Okudaira S. Two pathways for lysophosphatidic acid production. Biochim Biophys Acta 1781, 513–518 (2008). [DOI] [PubMed] [Google Scholar]
- Nakamura K. et al. Autotaxin enzyme immunoassay in human cerebrospinal fluid samples. Clin Chim Acta 405, 160–162 (2009). [DOI] [PubMed] [Google Scholar]
- Ma L. et al. Lysophosphatidic acid-3 receptor-mediated feed-forward production of lysophosphatidic acid: an initiator of nerve injury-induced neuropathic pain. Mol Pain 5, 61–69 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda H. Peripheral mechanisms of neuropathic pain - involvement of lysophosphatidic acid receptor-mediated demyelination. Mol Pain 4, 1–13 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner J. A., Fukushima N., Contos J. J., Scherer S. S. & Chun J. Regulation of Schwann cell morphology and adhesion by receptor-mediated lysophosphatidic acid signaling. J Neurosci 21, 7069–7078 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie W. et al. Calpain-mediated down-regulation of myelin-associated glycoprotein in lysophosphatidic acid-induced neuropathic pain. J Neurochem 113, 1002–1011 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi H. et al. Involvement of cathepsin B in the motor neuron degeneration of amyotrophic lateral sclerosis. Acta Neuropathol 105, 462–468 (2003). [DOI] [PubMed] [Google Scholar]
- Lowe J. New Pathological Findings in Amyotrophic Lateral Sclerosis. J Neurol Sci 124, 38–51 (1994). [DOI] [PubMed] [Google Scholar]
- Zhang X., Vincent A. S., Halliwell B. & Wong K. P. A mechanism of sulfite neurotoxicity - Direct inhibition of glutamate dehydrogenase. J Biol Chem 279, 43035–43045 (2004). [DOI] [PubMed] [Google Scholar]
- Woolsey P. B. E. Cysteine, Sulfite, and Glutamate Toxicity: A Cause of ALS? J Altern Complement Med 14, 1159–1164 (2008). [DOI] [PubMed] [Google Scholar]
- Sodhi R. N. S. Time-of-flight secondary ion mass spectrometry (ToF-SIMS): versatility in chemical and imaging surface analysis. Analyst 129, 483–487 (2004). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information