Abstract
In the study of bacterial community composition, 16S rRNA gene amplicon sequencing is today among the preferred methods of analysis. The cost of nucleotide sequence analysis, including requisite computational and bioinformatic steps, however, takes up a large part of many research budgets. High-resolution melt (HRM) analysis is the study of the melt behavior of specific PCR products. Here we describe a novel high-throughput approach in which we used HRM analysis targeting the 16S rRNA gene to rapidly screen multiple complex samples for differences in bacterial community composition. We hypothesized that HRM analysis of amplified 16S rRNA genes from a soil ecosystem could be used as a screening tool to identify changes in bacterial community structure. This hypothesis was tested using a soil microcosm setup exposed to a total of six treatments representing different combinations of pesticide and fertilization treatments. The HRM analysis identified a shift in the bacterial community composition in two of the treatments, both including the soil fumigant Basamid GR. These results were confirmed with both denaturing gradient gel electrophoresis (DGGE) analysis and 454-based 16S rRNA gene amplicon sequencing. HRM analysis was shown to be a fast, high-throughput technique that can serve as an effective alternative to gel-based screening methods to monitor microbial community composition.
INTRODUCTION
The increased availability of sequencing facilities and the relative reduction in sequencing costs have made nucleotide sequencing a common tool in research. Despite this, the costs of sequencing and subsequent bioinformatic analyses still represent a substantive expense in many project budgets. A cheap and efficient initial screening of samples before sequencing is therefore an attractive way to discriminate samples and thus potentially reduce costs by focusing the effort on the most interesting ones. Traditional fingerprinting methods such as denaturing gradient gel electrophoresis (DGGE) (1) or terminal restriction fragment length polymorphism (T-RFLP) have previously been used extensively to study diversity changes in complex environmental samples. However, these methods require considerable technical experience and are laborious and time-consuming (2). An alternative to these gel-based methods is desirable, and we suggest here that high-resolution melt (HRM) analysis (3) as a screening tool could provide a superior alternative. The thermal stability of a PCR product is determined by its GC content, sequence length, and primary structure (4). This stability is used in melt curve analysis, in which specific PCR products are denatured under tightly controlled conditions and their melting behavior observed. Melt curve analysis of a PCR product was initially developed in conjunction with the quantitative PCR (qPCR) approach for quantifying a particular amplicon product (4). With this method it is possible to identify nonspecific PCR products or primer dimers. To examine PCR products in further detail, HRM analysis was developed (5). The bases of HRM analysis include a high-quality (saturating) double-stranded DNA (dsDNA) dye and an HRM machine capable of precise temperature increments of 0.1°C or less (6). This technology has been used to study single nucleotide polymorphisms (SNPs) and to identify genotypes and the presence of heterozygotes in individuals (7). HRM studies targeting the 16S rRNA gene and using the melt curve as molecular fingerprint for species identification were first done by Cheng et al. (8). They were able to distinguish between 25 different pathogenic bacteria with 94% accuracy. Since then, others have used this technique to identify different species of Chlamydiaceae (9), Bartonella (10), and a wide array of pathogenic bacteria (11). Similar experiments have been done using denaturing high-performance liquid chromatography (DHPLC), which also succeeded in discriminating between a wide range of pure bacterial cultures (12).
Melting profiles of PCR amplicons can be chosen as a terminal integrated part of any qPCR protocol following amplification on many modern quantitative thermal cyclers. By combination of qPCR and software for HRM analysis, a fast and simple means to screen metagenomic DNA or metatranscriptomic RNA samples for compositional differences, prior to the commitment and expense of deep sequencing (e.g., of 16S rRNA gene amplicons), exists.
The aim of the current study was to examine the use of HRM analysis of 16S rRNA gene amplicons from metagenomic DNA and RNA as a screening method for changes in bacterial community composition using a mixed-culture approach. We added different pesticides and ammonium sulfate, alone and in combination, to a suite of soil microcosms and used HRM analysis to examine the resultant alterations in soil bacterial community structure during the time course of the experiment. To verify the findings from the HRM analysis, selected samples were also subjected to PCR-DGGE and to 16S rRNA gene amplicon pyrosequencing.
MATERIALS AND METHODS
Soil.
The soil was an agricultural clayey sand soil (37% coarse sand, 42% fine sand, 10% silt, 9% clay, and 2% organic C) from the experimental station in Askov, Denmark (latitude, 55°28′20N, and longitude, 9°6′36E). The soil contained 1% total C, 19.9 mg kg−1 of NO3−-N, and 1.53 mg kg−1 of NH4+-N and had a pH of 6.4 and a C/N ratio of 4.7. Soil analysis was performed by the OK Laboratorium for Jordbrug (Viborg, Denmark), according to Danish procedures (13). We sieved the soil (4-mm mesh) and stored it at 5°C in the dark for 8 months prior to the experiment.
Microcosm setup.
The soil was air dried at room temperature in the dark for 3 days and then homogenized by sieving (2-mm mesh). The microcosms were prepared by adding 10 g of sieved soil into 50-ml propylene tubes (for a total of 90 tubes). We then prepared a full factorial design with six treatments: Basamid GR, Tridex DG, and no pesticide, each with and without ammonium sulfate. The treatment with neither pesticide nor fertilizer addition served as a negative control. The two formulated pesticides Basamid GR and Tridex DG were added to the system in concentrations of 266 mg kg−1 and 13.3 mg kg−1, corresponding to one and five times the field dose, respectively. Ammonium sulfate (100 mg N kg−1 of soil) was added to stimulate microbial growth and simulated fertilizer addition in an agricultural setting. Sterile H2O was added to a final soil moisture content of 60% of water holding capacity (WHC). All microcosms were covered with polyvinyl chloride film and incubated in the dark at 20°C. Each treatment was prepared in triplicate and sampled destructively after 0, 3, 12, 20, and 28 days. Samples were frozen in liquid nitrogen and stored at −80°C until extraction of nucleic acids.
Extraction of nucleic acids.
DNA and RNA were coextracted from a 500-mg subsample using a phenol-chloroform protocol (14) with the following modifications: to inhibit DNA sorption to clay particles, 0.5 ml of G2 blocking agent (GEUS, Copenhagen, Denmark) was added to 1.4-mm ceramic bead tubes (Mo Bio Laboratories, Inc., Carlsbad, CA), and the tubes were freeze-dried prior to use. Hexadecyltrimethylammonium bromide (CTAB) and phenol-chloroform were added to the bead tubes together with the frozen soil samples. Then bead beating was performed at speed 5 for 20 s in a FastPrep FP120 (BIO 101, Farmingdale, NY). Two bead beating runs were performed with a 1-min cooling step on ice in between. The aqueous phase was separated by centrifugation for 10 min (16,000 × g) at 4°C. After removal of residual phenol with chloroform-isoamyl alcohol extraction using standard techniques, 1 μl of glycogen (20 mg/ml) (Roche, Basel, Switzerland) was added instead of polyethylene glycol, and the samples were placed on ice for 2 h to facilitate nucleic acid precipitation. Samples were kept on ice during all steps of the extraction. Precipitated DNA and RNA were purified using the RNA purification kit (NucleoSpin RNA Cleanup XS kit; Macherey-Nagel GmbH & Co. KG, Düren, Germany) and eluted in 20 μl of RNase-free H2O. The samples were then split into two parts. One part was diluted 10 times with RNase-free H2O and stored at −80°C until DNA analysis. The second part was immediately subjected to DNase treatment using the RTS DNase kit (Mo Bio) according to the manufacturer's protocol. A subsample of the DNase-treated sample was used as a template for cDNA production using random hexamer primers (Fermentas, Vilnius, Lithuania) and the RevertAid Premium RT kit (Fermentas) in a reverse transcriptase PCR (RT-PCR) procedure according to the manufacturer's protocol. Extraction, DNase treatment, and conversion to cDNA by reverse transcription were done in one working day. The quantity and quality of extracted RNA were checked on an Agilent 2100 bioanalyzer with the prokaryote total RNA picochip, which uses the relative signal intensity of 16S rRNA and 23S rRNA to calculate the RNA integrity number (RIN).
qPCR.
The numbers of 16S rRNA gene copies and 16S rRNA molecules were quantified by qPCR (CFX96 real-time system; Bio-Rad, USA) (Fig. 1A). Genomic DNA from Escherichia coli K-12 was used as a standard (15). All qPCR samples were run in technical duplicates. The master mix consisted of 2 μl of bovine serum albumin (BSA) (20 mg/ml; BIORON, Ludwigshafen, Germany), 10 μl of SsoFast EvaGreen Supermix (Bio-Rad), 0.8 μl of forward primer 341f (5′-CCTAYGGGRBGCASCAG-3′; 10 μM) (16), 0.8 μl of reverse primer 806r (5′-GGACTACNNGGGTATCTAAT-3′; 10 μM) (16), 1 μl of 10× diluted template, and distilled H2O (dH2O) to a total of 20 μl. PCR conditions were 98°C for 15 min, followed by 35 cycles of 98°C for 30 s, 56°C for 30 s, and 72°C for 30 s (with fluorescence measurements) and ending with 72°C for 7 min. The PCR efficiencies for the assay were 87.98% (standard error [SE] = 1.91), and R2 values were 0.984 (SE = 0.003).
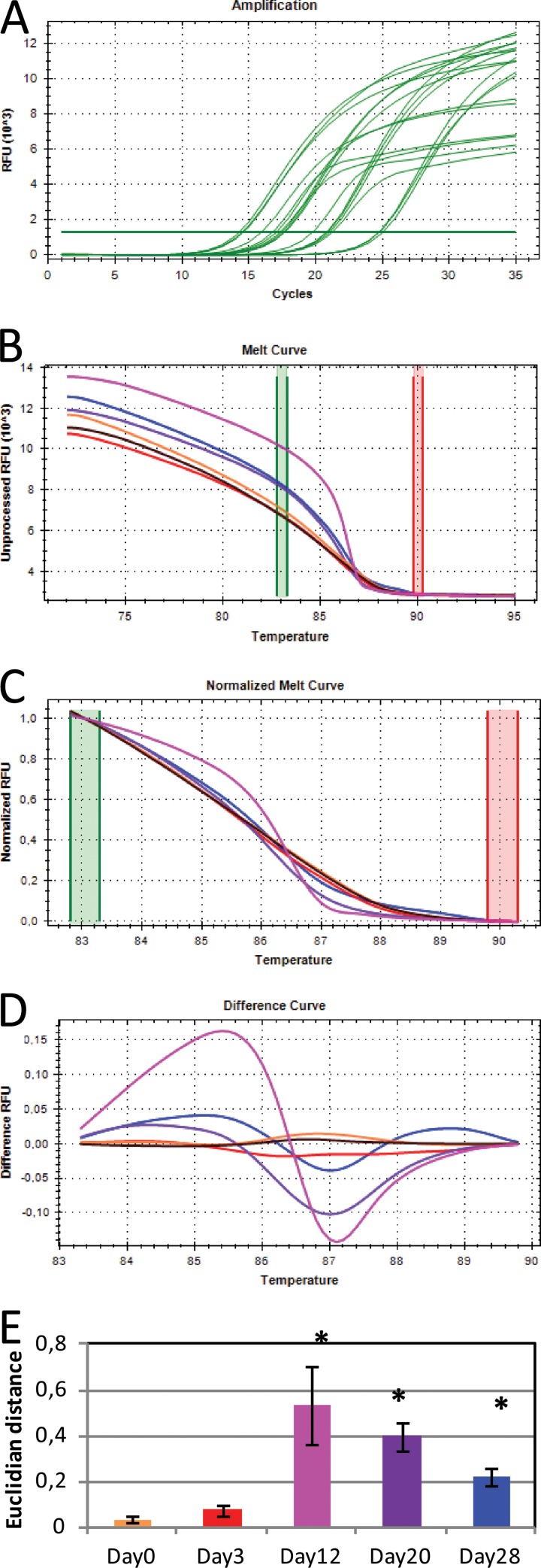
FIG 1.
Overview of the steps in 16S rRNA gene HRM analysis. Amplification and quantification of the 16S rRNA gene (A), melting of the PCR product in 0.1°C increments (B), normalization of the melt curves (C), conversion into difference curves in relation to control sample (black curve) (D), and average Euclidian distance between the different samples and the control (E) are shown. Note that the control sample was the H2O day 0 sample. Bars represent the means of triplicates from the Basamid GR samples. Error bars represent the standard deviations. In panels A to D, data for only one of the triplicate samples are shown for easier visualization. Asterisks represent sample means that were statistically different from the H2O day 0 samples (Tukey honestly significant difference [HSD]).
High-resolution melt analysis.
Following amplification and quantification of the 16S rRNA gene, a high-resolution melting curve analysis was performed at the end of the PCR protocol. This procedure melted the amplified 16S rRNA gene products starting at 72°C and ending at 95°C, with fluorescence measurements taken at every 0.1°C increment (Fig. 1B). Melting curves were normalized to relative fluorescence units (RFU) in a specified “melt region” (83.5°C to 89.5°C), thereby negating the effect of absolute RFU values (Fig. 1C). This melt region was autocalled by the melt analysis software (Precision Melt Analysis; Bio-Rad). Only RFU values of the melt region were used for downstream analysis. The control sample (H2O day 0) was included as a standard in all PCR runs, and all other melting curves are shown in relation to this, producing a so-called “difference curve” (Fig. 1D). This approach made graphical representation of the data easier to interpret and facilitated comparison of curves from different runs. In order to perform statistical analysis on the melt curves, the Euclidian distance between the standard and each sample was calculated using the statistical program R (17) (Fig. 1E). This was done by calculating the squared difference between the RFU values at each 0.1°C temperature point (83.5°C, 83.6°C, and so forth). These were then summarized and the square root was taken, thus resulting in larger Euclidian distances for more dissimilar melt curves. The Euclidian distance was subsequently used to test if the samples were significantly different from the control (standard).
HRM sensitivity assay.
Pure cultures of Bacillus thuringiensis and Corynebacterium glutamicum were grown in LB broth for 24 h at 37°C with rotary shaking at 120 rpm. DNA was then extracted using the UltraClean microbial DNA isolation kit (Mo Bio) according to the manufacturer's protocol. DNA concentrations were measured on a Qubit (Invitrogen, Carlsbad, CA). Based on the reported genome size and number of 16S rRNA genes from B. thuringiensis (18, 19) and C. glutamicum (20, 21), the DNA extracts were diluted to the same number of 16S rRNA gene copies per unit volume. To estimate the sensitivity of the HRM analysis, the 16S rRNA genes from the two bacteria were mixed in different ratios (see Fig. S1 in the supplemental material) and subjected to the same PCR and HRM protocol as described above. The sensitivity could then be determined as the mix with the lowest ratio having a Euclidian distance significantly different from that of the pure cultures (normalized to be 100%).
DGGE.
DNA samples from all sampling days from the control (only water), Tridex, and Basamid treatments of the microcosm experiment were amplified with a master mix containing 4 μl of 5× Phusion HF buffer, 0.4 μl of 10 mM deoxynucleoside triphosphate (dNTP) mixture, 0.2 μl of Phusion Hot Start DNA polymerase (2 units/μl; Finnzymes), 1 μl of GC-clamped forward primer 341f-clamp (5′-CGCCCGCCGCGCCCCGCGCCCGTCCCGCCGCCCCCGCCCGCCTAYGGGRBGCASCAG-3′; 10 μM), 1 μl of reverse primer 806r (10 μM), 1 μl of template (undiluted DNA), and dH2O to a total of 20 μl. PCR conditions were 98°C for 30 s, followed by 30 cycles of 98°C for 5 s, 56°C for 20 s, and 72°C for 20 s and ending with 72°C for 5 min. DGGE was performed using the Dcode system (Bio-Rad). The PCR products were loaded into the wells of an 8% acrylamide gel containing a gradient of 35 to 60% denaturant (urea and deionized formamide). The gels were run at 100 V for 17 h in 1× Tris-acetate-EDTA (TAE) buffer. Gels were stained with SYBR gold nucleic acid gel stain (Invitrogen) for 45 min before being photographed.
454 sequencing.
The results of the HRM screening showed the largest aberration in diversity composition in the samples from day 12. Thus, the samples selected for sequencing were control treatment from day 0 (only water) and all treatments from day 12, for a total of 39 samples. The samples were amplified with a master mix containing 4 μl of 5× Phusion HF buffer (Thermo Scientific, Waltham, MA), 0.4 μl of 10 mM dNTP mixture, 0.2 μl of Phusion Hot Start II DNA polymerase (2 U/μl; Thermo Scientific), 1 μl of forward primer 341f (10 μM), 1 μl of reverse primer 806r (10 μM), 1 μl of template (10× dilution), and dH2O to a total of 20 μl. PCR conditions were 98°C for 30 s, followed by 30 cycles of 98°C for 5 s, 56°C for 20 s, and 72°C for 20 s, with a final extension at 72°C for 5 min. The samples were then run on a 1.25% agarose gel, and specific bands were cut out and purified using the Montage DNA gel extraction kit (Millipore, Billerica, MA). Individual tags were added to the purified products, in a second PCR, using identical primers but with 10-bp-long individual multiplex identifiers (MIDs) (Roche) attached. Purified PCR products were used as the template, and only 15 PCR cycles were performed. Conditions and master mix were otherwise the same as in the first-round PCR. PCR products with MIDs were purified as before and DNA concentrations measured on a Qubit (Invitrogen). Samples were then mixed together (i.e., multiplexed) to create an equimolar mixture to a total of 1 μg DNA. Adapter ligation, emulsion PCR, and 454 sequencing were done by Beckman Coulter Genomics (Brea, CA) on a 454 GS FLX Titanium apparatus (Roche) (1/2 titanium plate) producing, on average, 5,229 raw sequences per sample.
454 sequence analysis.
The sequences were analyzed with the bioinformatics tool Qiime v.1.6.0 (22). First, low-quality reads, defined as having a quality score (QC) below 25 in a sliding window of 50 bp, were removed from the data set. The Roche 454 sequencing technology has been shown to introduce errors in the sequences obtained, which artificially inflates the sequence diversity (23). To reduce these errors, the Denoiser algorithm (24) was applied. Chimeric sequences were removed using the script ChimeraSlayer (25). The sequences were then mapped to a reference 16S rRNA gene database (Greengenes v.12.10). Singletons were deleted to remove artificial sequences not picked up by the Denoiser algorithm. The remaining sequences (∼2,500 sequences per sample) were used for construction of taxonomic tables and phylogenetic trees using the Qiime tutorial on 454 sequence data analysis.
RESULTS
The sensitivity of the HRM analysis applied in this study was assessed using a simple composite system consisting of only two bacterial strains. This assay was used to elucidate the degree of community change that was required in order to be detectable by our HRM assay. We found that an introduction of 10% of B. thuringiensis genomic DNA into 90% C. glutamicum genomic DNA was needed to be able to observe a significant difference in Euclidian distance from the pure culture of C. glutamicum when comparing the 16S rRNA genes by HRM analysis (data not shown).
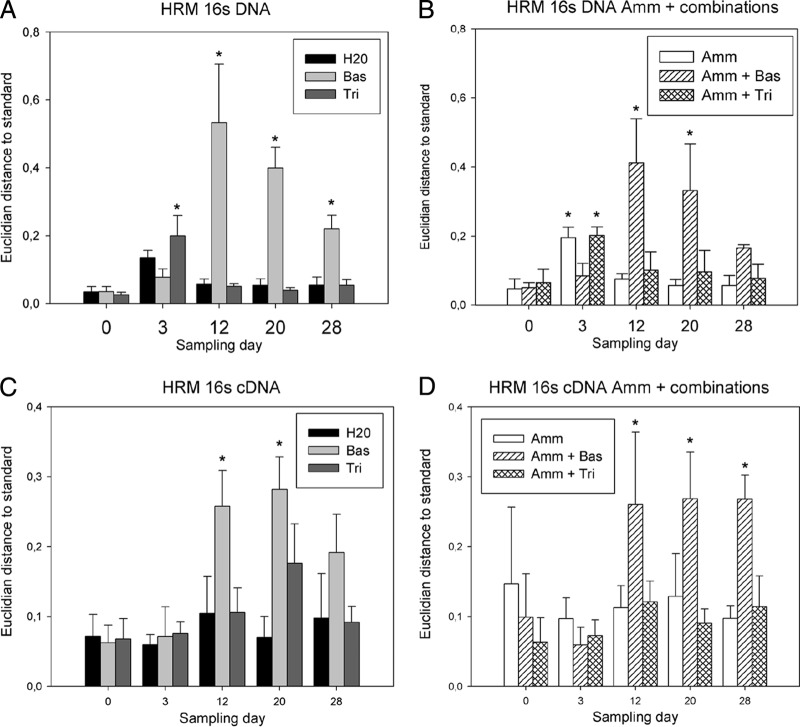
The bacterial community compositions of the six microcosm treatments were compared by HRM analysis and following conversion into Euclidian distances. This screening approach identified the Basamid HRM profiles as being statistically different from the treatment with only water (Fig. 2). This was observed in treatments with Basamid alone and in combination with ammonium sulfate. The most pronounced change in 16S rRNA gene composition caused by the Basamid treatments was observed at day 12 (Fig. 2A and B). A similar result was seen for the cDNA samples, but the effect was less distinct as shown by the lower Euclidian distances (Fig. 2C and D). Ammonium sulfate and Tridex either alone or in combination had no significant effect on the HRM profiles, except for a small deviation at day 3.
FIG 2.
HRM analysis of the bacterial community compositions of DNA samples (A and B) and cDNA samples (C and D). Treatments either did (B and D) or did not (A and C) include ammonium sulfate amendment. The data presented are the means and standard deviations of three replicates. The Euclidian distance, shown on the y axis, is the distance between the H2O day 0 samples and the respective sample. Asterisks represent sample means that were statistically different from the H2O day 0 samples (Tukey HSD). Bas, Basamid GR; Tri, Tridex DG; Amm, ammonium sulfate.
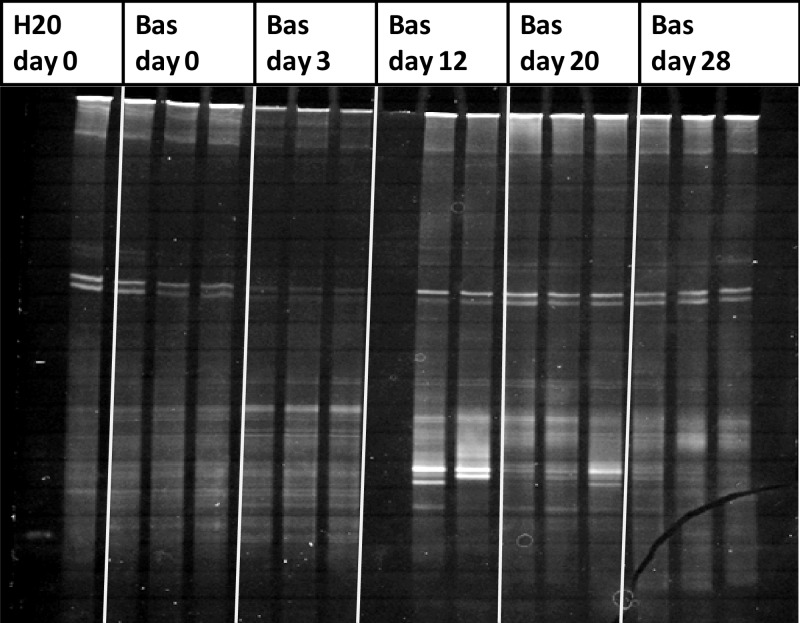
In agreement with the HRM analysis, the DGGE fingerprints of DNA samples exposed to Basamid revealed a minor change compared to the control at day 3 while showing a clear change in band pattern at day 12. This change was predominantly caused by the appearance of two new dominant bands, which were still visible at day 20 but less dominant (Fig. 3). Large variations appear among the biological replicates of the Basamid-treated soil, as are also reflected in the large error bars of the Euclidian distance calculation when analyzing the DNA using HRM (Fig. 2A and B). In correspondence with the HRM analysis, no changes in DGGE band patterns were seen for the control and Tridex treatments (see Fig. S2 and S3 in the supplemental material).
FIG 3.
DGGE analysis of PCR amplified 16S rRNA gene fragments originating from the Basamid GR-treated microcosm. The H2O day 0 control sample in the far left lane is used as a standard. The DNA of the Bas day 12 sample A was not extracted successfully and therefore produced no bands on the DGGE gel.
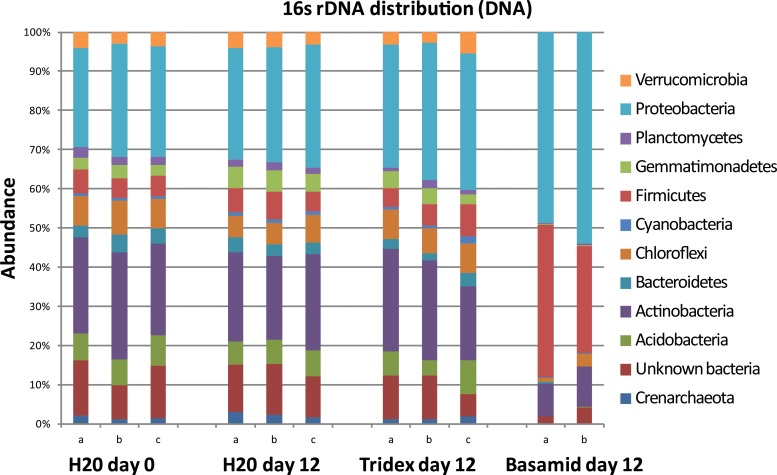
The 454 16S rRNA gene amplicon sequencing showed a distinct alteration of the bacterial community composition in the Basamid-treated samples from day 12 (Fig. 4). These samples showed a large relative increase in the phyla Proteobacteria and Firmicutes, more specifically in the orders Burkholderiales and Bacillales, respectively. A similar shift was observed in the treatment including both Basamid and ammonium sulfate (data not shown). There were no effects on the bacterial community composition of the treatments including Tridex (Fig. 4) or ammonium sulfate alone or in combination (data not shown).
FIG 4.
Relative abundance of the 12 most abundant phyla within the microcosm day 12 samples and H2O at day 0 control. One of the Basamid GR day 12 triplicates was not sequenced due to low quality of the extraction.
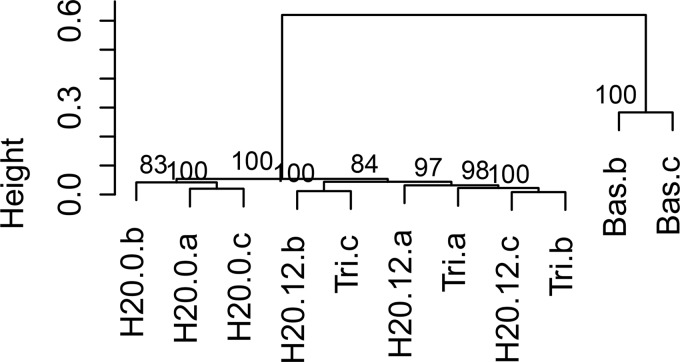
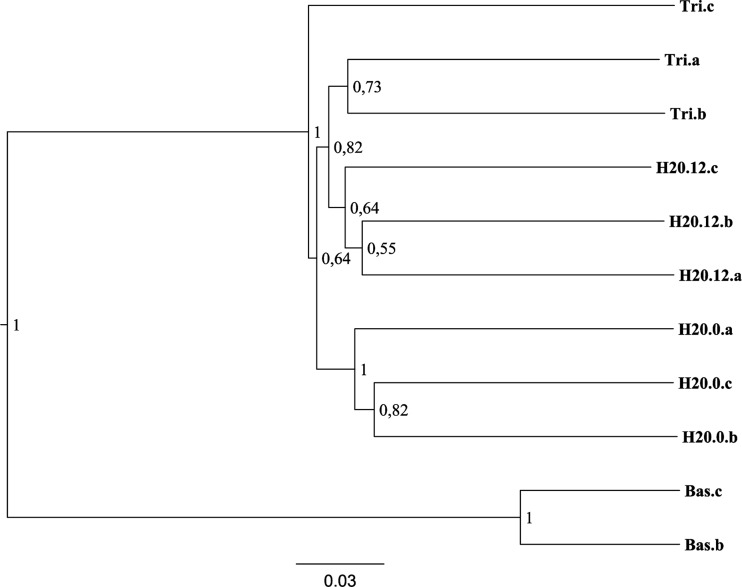
In our experimental setup, the use of Euclidian distance calculation was very useful to compare the effects of different treatments in relation to a control sample, where only water was added. In other studies where no such control or standard exists, a direct clustering of the normalized melt curves may be preferable. We tested the result of direct clustering on a subset of the HRM samples and found the existence of two clusters. The first cluster contained the Basamid samples and the second cluster included the remaining samples (Fig. 5). The same samples were also clustered on the basis of the 454 sequence results, which resulted in an identical cluster pattern (Fig. 6).
FIG 5.
Hierarchical clustering of HRM normalized curves (pv.clust package, R V. 3.02). Distance measure was set to “Euclidian,” and agglomerative method was set to “average.” Node numbers represent approximately unbiased (AU) multiscale bootstrap value with 1,000 bootstrap repetitions. The DNA of the Bas.a sample was not extracted successfully and is therefore not included.
FIG 6.
UPGMA clustering of 16S rRNA gene sequences from the microcosm 454 day 12 DNA samples without ammonium sulfate and H2O day 0 samples. Node labels represent confidence level of clustering based on 10 jackknife replicates. The DNA of the Bas.a sample was not extracted successfully and is therefore not included.
DISCUSSION
In this study, we successfully validated the HRM analysis of 16S rRNA gene amplicons as a robust method for directly comparing total bacterial community composition. The HRM analysis was done in the same workflow as the qPCR quantification of the 16S rRNA gene, thus requiring little extra work. The changes in bacterial community composition were confirmed with 454 amplicon sequencing and DGGE using the same primer set as in the HRM analysis, eliminating potential primer biases.
The HRM melt curves do not provide any phylogenetic information themselves. Yet, we propose a strategy whereby samples with differing bacterial community structures can be rapidly identified by HRM and then those samples of interest subjected to further analysis (e.g., via 16S rRNA gene amplicon sequencing) to elucidate bacterial community composition in detail. The use of a 96-well qPCR system and the capability of running three sets of reactions per day facilitate both quantification and HRM screening of >250 samples per day. In comparison, it is our experience that the DGGE method can be used to screen around 30 samples in a full 2-day workload, an observation also discussed by Green (2).
In our experiments, we used the strong soil fumigant Basamid GR, which targets biological thiols and amines and thus has a broad mechanism of action and a strong bactericidal effect (26). The HRM analysis clearly showed that soil samples that had been exposed to Basamid were significantly different from the control samples. This was observed in both the DNA and cDNA from these samples, representing bacterial abundance and activity, respectively (Fig. 2). We also observed large changes in specific taxa at the phylum level, as shown by the 454 sequencing results in Fig. 4. Using the HRM analysis in an initial screening step, we were able to target our sequencing strategy, which, in turn, increased our understanding of the bacterial community response to soil fumigants. Admittedly, fumigant treatment likely represents an extreme perturbation producing large changes in bacterial community composition, while many other studies might have smaller perturbation and resultant compositional differences between treatments. The sensitivity of the HRM assay will be of importance in such studies. It is also worth mentioning that melting curves of some closely related bacterial species have been shown to be indistinguishable by standard HRM analysis (8). Consequently, small phylogenetic shifts in the bacterial community might not be detectable by HRM analysis. In that respect, HRM analysis, as used in this study, might not be suitable for discriminating between highly similar bacterial communities. This is observed in the lack of discrimination between the Tridex and control samples from day 12 in the HRM clustering (Fig. 5), compared to the 454 amplicon clustering (Fig. 6). Similar sensitivity problems also exists for DGGE however, where bands of related species may overlap (27), and in some respect for next-generation sequencing, where the number of amplicons typically is in the range of 104 to 105 per sample, whereas the number of bacterial cells typically is several orders of magnitude higher (28).
This topic of sensitivity was addressed with our genomic DNA mixing experiment indicating that a 10% change in 16S rRNA genes was necessary before it was detectable using HRM. The sensitivity of the particular assay is, of course, dependent on the difference in melt behavior of the specific sequences used. The two bacterial strains used in this assay were chosen because of the large difference in the GC contents of their 16S rRNA genes, which resulted in large differences in melt behavior. If the chosen 16S rRNA genes had come from more closely related species, the sensitivity would probably be higher than 10%. Additionally, the numbers of 16S rRNA gene copies/genome differ widely among bacteria (29); thus, they might correspond to a higher or lower change in bacterial cell number. A similar sensitivity (12.5%) has previously been reported for determination of the ratio between two different PCR fragments using HRM analysis (30), although the two fragments had a much higher similarity than the 16S rRNA gene sequences used in this experiment.
Degenerate primers, as used in this study, can maximize the number of different PCR products obtained (31), leading to an increase in the melt curve variation and thus lower sensitivity. An optimization of the 16S rRNA gene HRM assay, including the use of nondegenerate primers or more specific primers (e.g., for specific groups or taxa or functional genes), probably would result in increased sensitivity. Furthermore, targeting shorter gene fragments (3) and using the high-sensitivity dye LCGreen Plus (32) for fluorescence labeling of the PCR products could be applied for improvement of the assay.
In conclusion, the HRM analysis is an efficient screening tool for discrimination of microbial community variations among samples. The analysis can, e.g., be used for identification of major breakpoints of xenobiotic compound effects in natural microbial environments. Furthermore, the analysis can be used to screen large sample sets for the more interesting samples prior to deep sequencing, thus saving time and money on sequencing cost and bioinformatics. HRM analysis is fast, requires limited technical training, and is within the economical reach of most laboratories in regard to HRM-compatible qPCR instrument access or purchase.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Oticon Foundation, the Center for Permafrost (CENPERM), the GENEPEASE project funded by the Danish EPA, and the ASHBACK project funded by the Danish Council for Strategic Research.
Morten Schostag Nielsen is thanked for performing the DNA and RNA extractions and for help in initiating the Qiime analysis.
Footnotes
Published ahead of print 7 March 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03923-13.
REFERENCES
- 1.Muyzer G, De Waal EC, Uitterlinden AG. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59:695–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Green JS. 2005. A guide to denaturing gradient gel electrophoresis. http://www.planta.cn/forum/files_planta/dggehelpv2_106.pdf [Google Scholar]
- 3.Tindall EA, Petersen DC, Woodbridge P, Schipany K, Hayes VM. 2009. Assessing high-resolution melt curve analysis for accurate detection of gene variants in complex DNA fragments. Hum. Mutat. 30:876–883. 10.1002/humu.20919 [DOI] [PubMed] [Google Scholar]
- 4.Ririe KM, Rasmussen RP, Wittwer CT. 1997. Product differentiation by analysis of DNA melting curves during the polymerase chain reaction. Anal. Biochem. 245:154–160. 10.1006/abio.1996.9916 [DOI] [PubMed] [Google Scholar]
- 5.Wittwer CT, Reed GH, Gundry CN, Vandersteen JG, Pryor RJ. 2003. High-resolution genotyping by amplicon melting analysis using LCGreen. Clin. Chem. 49:853–860. 10.1373/49.6.853 [DOI] [PubMed] [Google Scholar]
- 6.Herrmann MG, Durtschi JD, Wittwer CT, Voelkerding KV. 2007. Expanded instrument comparison of amplicon DNA melting analysis for mutation scanning and genotyping. Clin. Chem. 53:1544–1548. 10.1373/clinchem.2007.088120 [DOI] [PubMed] [Google Scholar]
- 7.Montgomery J, Wittwer CT, Palais R, Zhou L. 2007. Simultaneous mutation scanning and genotyping by high-resolution DNA melting analysis. Nat. Protoc. 2:59–66. 10.1038/nprot.2007.10 [DOI] [PubMed] [Google Scholar]
- 8.Cheng JC, Huang CL, Lin CC, Chen CC, Chang YC, Chang SS, Tseng CP. 2006. Rapid detection and identification of clinically important bacteria by high-resolution melting analysis after broad-range ribosomal RNA real-time PCR. Clin. Chem. 52:1997–2004. 10.1373/clinchem.2006.069286 [DOI] [PubMed] [Google Scholar]
- 9.Robertson T, Bibby S, O'Rourke D, Belfiore T, Lambie H, Noormohammadi AH. 2009. Characterization of Chlamydiaceae species using PCR and high resolution melt curve analysis of the 16S rRNA gene. J. Appl. Microbiol. 107:2017–2028. 10.1111/j.1365-2672.2009.04388.x [DOI] [PubMed] [Google Scholar]
- 10.Morick D, Baneth G, Avidor B, Kosoy MY, Mumcuoglu KY, Mintz D, Eyal O, Goethe R, Mietze A, Shpigel N, Harrus S. 2009. Detection of Bartonella spp. in wild rodents in Israel using HRM real-time PCR. Vet. Microbiol. 139:293–297. 10.1016/j.vetmic.2009.06.019 [DOI] [PubMed] [Google Scholar]
- 11.Šimenc J, Potočnik U. 2011. Rapid differentiation of bacterial species by high resolution melting curve analysis. Appl. Biochem. Microbiol. 47:256–263. 10.1134/S0003683811030136 [DOI] [PubMed] [Google Scholar]
- 12.Hurtle W, Shoemaker D, Henchal E, Norwood D. 2002. Denaturing HPLC for identifying bacteria. Biotechniques 33:386–391 [DOI] [PubMed] [Google Scholar]
- 13.Plantedirektoratet. 1994. Fælles arbejdsmetoder for jordbundsanalyser. Plantedirektoratet, Lyngby, Denmark [Google Scholar]
- 14.Griffiths RI, Whiteley AS, O'Donnell AG, Bailey MJ. 2000. Rapid method for coextraction of DNA and RNA from natural environments for analysis of ribosomal DNA- and rRNA-based microbial community composition. Appl. Environ. Microbiol. 66:5488–5491. 10.1128/AEM.66.12.5488-5491.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blattner FR. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453–1462. 10.1126/science.277.5331.1453 [DOI] [PubMed] [Google Scholar]
- 16.Hansen CHF, Krych L, Nielsen DS, Vogensen FK, Hansen LH, Sørensen SJ, Buschard K, Hansen A. 2012. Early life treatment with vancomycin propagates Akkermansia muciniphila and reduces diabetes incidence in the NOD mouse. Diabetologia 55:2285–2294. 10.1007/s00125-012-2564-7 [DOI] [PubMed] [Google Scholar]
- 17.R Development Core Team. 2008. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria [Google Scholar]
- 18.Challacombe JF, Altherr MR, Xie G, Bhotika SS, Brown N, Bruce D, Campbell CS, Campbell ML, Chen J, Chertkov O, Cleland C, Dimitrijevic M, Doggett NA, Fawcett JJ, Glavina T, Goodwin LA, Green LD, Han CS, Hill KK, Hitchcock P, Jackson PJ, Keim P, Kewalramani AR, Longmire J, Lucas S, Malfatti S, Martinez D, McMurry K, Meincke LJ, Misra M, Moseman BL, Mundt M, Munk AC, Okinaka RT, Parson-Quintana B, Reilly LP, Richardson P, Robinson DL, Saunders E, Tapia R, Tesmer JG, Thayer N, Thompson LS, Tice H, Ticknor LO, Wills PL, Gilna P, Brettin TS. 2007. The complete genome sequence of Bacillus thuringiensis Al Hakam. J. Bacteriol. 189:3680–3681. 10.1128/JB.00241-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.He J, Wang J, Yin W, Shao X, Zheng H, Li M, Zhao Y, Sun M, Wang S, Yu Z. 2011. Complete genome sequence of Bacillus thuringiensis subsp. chinensis strain CT-43. J. Bacteriol. 193:3407–3408. 10.1128/JB.05085-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kalinowski J, Bathe B, Bartels D, Bischoff N, Bott M, Burkovski A, Dusch N, Eggeling L, Eikmanns BJ, Gaigalat L, Goesmann A, Hartmann M, Huthmacher K, Krämer R, Linke B, McHardy AC, Meyer F, Möckel B, Pfefferle W, Pühler A, Rey DA, Rückert C, Rupp O, Sahm H, Wendisch VF, Wiegräbe I, Tauch A. 2003. The complete Corynebacterium glutamicum ATCC 13032 genome sequence and its impact on the production of l-aspartate-derived amino acids and vitamins. J. Biotechnol. 104:5–25. 10.1016/S0168-1656(03)00154-8 [DOI] [PubMed] [Google Scholar]
- 21.Lv Y, Liao J, Wu Z, Han S, Lin Y, Zheng S. 2012. Genome sequence of Corynebacterium glutamicum ATCC 14067, which provides insight into amino acid biosynthesis in coryneform bacteria. J. Bacteriol. 194:742–743. 10.1128/JB.06514-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7:335–336. 10.1038/nmeth.f.303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Quince C, Lanzén A, Curtis TP, Davenport RJ, Hall N, Head IM, Read LF, Sloan WT. 2009. Accurate determination of microbial diversity from 454 pyrosequencing data. Nat. Methods 6:639–641. 10.1038/nmeth.1361 [DOI] [PubMed] [Google Scholar]
- 24.Reeder J, Knight R. 2010. Rapidly denoising pyrosequencing amplicon reads by exploiting rank-abundance distributions. Nat. Methods 7:668–669. 10.1038/nmeth0910-668b [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haas BJ, Gevers D, Earl AM, Feldgarden M, Ward DV, Giannoukos G, Ciulla D, Tabbaa D, Highlander SK, Sodergren E, Methé B, DeSantis TZ, The Human Microbiome Consortium. Petrosino JF, Knight R, Birren BW. 2011. Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res. 21:494–504. 10.1101/gr.112730.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Macalady JL, Fuller ME, Scow KM. 1998. Effects of Metam sodium fumigation on soil microbial activity and community structure. J. Environ. Qual. 27:54–63. 10.2134/jeq1998.00472425002700010009x [DOI] [Google Scholar]
- 27.Vallaeys T, Topp E, Muyzer G, Macheret V, Laguerre G, Rigaud A, Soulas G. 1997. Evaluation of denaturing gradient gel electrophoresis in the detection of 16S rDNA sequence variation in rhizobia and methanotrophs. FEMS Microbiol. Ecol. 24:279–285. 10.1111/j.1574-6941.1997.tb00445.x [DOI] [Google Scholar]
- 28.Vogel TM, Simonet P, Jansson JK, Hirsch PR, Tiedje JM, Van Elsas JD, Bailey MJ, Nalin R, Philippot L. 2009. TerraGenome: a consortium for the sequencing of a soil metagenome. Nat. Rev. Microbiol. 7:252. 10.1038/nrmicro2119 [DOI] [Google Scholar]
- 29.Vĕtrovský T, Baldrian P. 2013. The variability of the 16S rRNA gene in bacterial genomes and its consequences for bacterial community analyses. PLoS One 8:e57923. 10.1371/journal.pone.0057923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aten E, White S, Kalf M, Vossen R, Thygesen H, Ruivenkamp C, Kriek M, Breuning M, Den Dunnen J. 2008. Methods to detect CNVs in the human genome. Cytogenet. Genome Res. 123:313–321. 10.1159/000184723 [DOI] [PubMed] [Google Scholar]
- 31.Kowalchuk GA, Stephen JR, De Boer W, Prosser JI, Embley TM, Woldendorp JW. 1997. Analysis of ammonia-oxidizing bacteria of the beta subdivision of the class Proteobacteria in coastal sand dunes by denaturing gradient gel electrophoresis and sequencing of PCR-amplified 16S ribosomal DNA fragments. Appl. Environ. Microbiol. 63:1489–1497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Herrmann MG, Durtschi JD, Bromley LK, Wittwer CT, Voelkerding KV. 2006. Amplicon DNA melting analysis for mutation scanning and genotyping: cross-platform comparison of instruments and dyes. Clin. Chem. 52:494–503. 10.1373/clinchem.2005.063438 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.