Abstract
Soil nitrification plays an important role in the reduction of soil fertility and in nitrate enrichment of groundwater. Various ammonia-oxidizing archaea (AOA) are considered to be members of the pool of ammonia-oxidizing microorganisms in soil. This study reports the discovery of a chemolithoautotrophic ammonia oxidizer that belongs to a distinct clade of nonmarine thaumarchaeal group I.1a, which is widespread in terrestrial environments. The archaeal strain MY2 was cultivated from a deep oligotrophic soil horizon. The similarity of the 16S rRNA gene sequence of strain MY2 to those of other cultivated group I.1a thaumarchaeota members, i.e., Nitrosopumilus maritimus and “Candidatus Nitrosoarchaeum koreensis,” is 92.9% for both species. Extensive growth assays showed that strain MY2 is chemolithoautotrophic, mesophilic (optimum temperature, 30°C), and neutrophilic (optimum pH, 7 to 7.5). The accumulation of nitrite above 1 mM inhibited ammonia oxidation, while ammonia oxidation itself was not inhibited in the presence of up to 5 mM ammonia. The genome size of strain MY2 was 1.76 Mb, similar to those of N. maritimus and “Ca. Nitrosoarchaeum koreensis,” and the repertoire of genes required for ammonia oxidation and carbon fixation in thaumarchaeal group I.1a was conserved. A high level of representation of conserved orthologous genes for signal transduction and motility in the noncore genome might be implicated in niche adaptation by strain MY2. On the basis of phenotypic, phylogenetic, and genomic characteristics, we propose the name “Candidatus Nitrosotenuis chungbukensis” for the ammonia-oxidizing archaeal strain MY2.
INTRODUCTION
The first step of nitrification, i.e., the oxidation of ammonia, was long considered to be performed exclusively by ammonia-oxidizing bacteria (AOB). However, the theory of soil and marine nitrification changed significantly after the discovery of ammonia-oxidizing archaea (AOA) (1). AOA outnumber AOB in many soil environments (2, 3), and the ammonia-oxidizing activity of Archaea in various soil habitats has been demonstrated indirectly based on the detection of in situ-expressed archaeal amoA genes (3, 4). The predominance of thaumarchaeal group I.1b over I.1a has been demonstrated for most soils (5, 6). However, phylogenetic clades of AOA that are taxonomically related to thaumarchaeal group I.1a frequently become more abundant with increasing nitrification activity in soil microcosms (7–9). Key issues related to soil AOA that need to be addressed include the contribution to ammonia oxidation of AOA relative to AOB in various soil environments, the biochemical mechanism of ammonia oxidation, and the evolutionary history and ecological niche partitioning of AOA. Cultivation of various soil AOA and analysis of their genomic sequences may provide further insight into these issues, but limited information is currently available.
Several enrichment cultures (10–14) and a pure culture (15) of soil AOA have been used to elucidate the phylogenetic and physiological properties of soil AOA. Furthermore, genomes of soil and marine AOA (16–21) and several fosmid clones (4, 22, 23) have revealed that AOA utilize energy and carbon metabolism processes that are different from those of AOB. Gene homologs of hydroxylamine oxidoreductase and cytochrome genes (c552 and c554), which play key roles in ammonia oxidation and electron transport in AOB, are absent from all known AOA genomes, whereas putative genes for the specific 3-hydroxypropionate/4-hydroxybutyrate carbon fixation pathway were detected (16–19, 21).
Most cultivation and microcosm studies of soil AOA have been conducted with soils from organic-rich solum layers (O to B horizons). Microcosm studies have identified AOA activity in the B horizon (50 to 60 cm) of soil (3, 24, 25), but AOA have not been identified in deeper, oligotrophic soil horizons, such as the parent rock (C horizon) and bedrock (R horizon). These deeper soil horizons are affected little by soil weathering processes and are composed of partially weathered bedrock, which underlies the solum layers at the base of the soil profile (26). Here we report the physiological and genomic characteristics of a chemolithoautotrophic, ammonia-oxidizing thaumarchaeon, which was cultivated from the oligotrophic C horizon of an agricultural soil. This study helps to explain the role of AOA in deeper soil horizons, which may provide insights into the niche distribution of AOA in terrestrial ecosystems.
MATERIALS AND METHODS
Sampling site description.
Samples of a deep soil horizon were collected at depths of 1.5 to 1.6 m (C horizon) from plots planted with Caragana sinica at the experimental agricultural station of Chungbuk National University, Republic of Korea (127°27′18.5″E, 36°37′29.8″N). Bulk soils were collected and transported to the laboratory, where they were stored at 4°C before use for inoculation. The properties of the soil were as follows: loam texture (sand, 51%; silt, 33%; and clay, 16%); water content, 4.2%; pH, 4.8; total organic carbon, <0.1 g kg−1; total nitrogen, <0.01%; total ammonia, 6.0 mg kg−1; total phosphate, 174.8 mg kg−1; and cation exchange capacity, 12.1 cmol kg−1.
Cultivation of AOA.
The ammonia-oxidizing archaeon strain MY2 was grown aerobically without shaking in artificial freshwater medium (AFM) as described by Jung et al. (10, 27). Initial enrichment cultures were set up using allylthiourea (ATU) (20 μM) and chlorate (50 μM) to selectively inhibit the growth of AOB and nitrite-oxidizing bacteria (NOB) (10), respectively. The enriched AOA cultures contained heterotrophic bacteria, and attempts were made to further enrich AOA by successive cultivation in AFM containing a mixture of ampicillin and penicillin G (each at 50 μg ml−1). After ca. 3 years of triweekly transfer, the cultures were serially diluted (10-fold) to extinction, and the highest dilution with nitrifying activity was selected to isolate a single archaeal strain. All of the cultures were incubated in the dark at 25°C with unmodified ambient air in the headspace. The cultures were routinely supplemented with 1 mM ammonium chloride as the sole energy source. The pH of the medium was adjusted to the optimum growth pH values by using 1 N NaOH or HCl. After ammonia oxidation was completed (within ca. 3 weeks), 5% of the total culture was routinely transferred to fresh medium. The ammonia concentration was determined using the method described by Jung et al. (10), and the nitrite concentration was determined colorimetrically (28). For inhibition of ammonia oxidation, chlorite (50 μM) (10, 14, 29) or dicyandiamide (500 μM) (10, 24, 30) was supplied to the medium. For inhibition of AOA and AOB, ATU in the range of 10 to 500 μM was used.
Quantification of gene copy numbers by using real-time PCR.
To identify AOA in the cultures, the copy numbers of the amoA and 16S rRNA genes were determined using specific primers (see Table S1 in the supplemental material). Genomic DNA was extracted using the protocol described by Jung et al. (10), and the DNA concentration was determined using a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE). Archaeal 16S rRNA and amoA gene copies were quantified using a MiniOpticon real-time PCR detection system with built-in Opticon Monitor software, version 3.1 (Bio-Rad Laboratories, Hercules, CA). The real-time PCR efficiencies of the archaeal 16S rRNA and amoA gene assays were 90 to 95% and 87 to 93%, respectively, with r2 values of ≥0.99 for all assays. Thermal cycling parameters of 15 min at 95°C and 40 cycles of 95°C for 20 s, 55°C for 20 s, and 72°C for 20 s were used to amplify the 16S rRNA and amoA genes, with readings taken between each cycle. Standard curves were prepared using reference genes (for archaeal 16S rRNA gene, accession no. HQ331116; and for amoA, accession no. HQ331117) and their cycle threshold (CT) values, as described previously (10). The specificity of real-time PCR was assessed by analyzing the melting curves and checking the sizes of the PCR products by using gel electrophoresis. PCRs using amplification primers for various other bacterial and archaeal genes (see Table S1) were also applied.
Phylogenetic analysis of amoA genes and cloning.
The archaeal 16S rRNA and amoA genes were amplified by PCR using the primers shown in Table S1 in the supplemental material. The PCR conditions used for the 16S rRNA and amoA genes were as follows: 94°C for 5 min; 30 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 45 s; and 72°C for 7 min. Clone libraries were constructed using the PCR products, which were purified with a PCR purification kit (Solgent, Republic of Korea), ligated using a T&A cloning kit (Real Biotech Corporation, Taiwan), and transformed into Escherichia coli DH5α cells according to the manufacturers' instructions. Both strands of the gene were sequenced for each clone, using the corresponding PCR primers (see Table S1). The gene sequences of related taxa were obtained from the GenBank database for phylogenetic analysis. Multiple-sequence alignments of 16S rRNA gene sequences were performed with the online alignment tool SINA, using the database SILVA (http://www.arb-silva.de/aligner) (31), based on a consideration of the secondary structures of rRNA genes. The shared regions of amoA (560 bp) gene sequences were aligned using CLUSTALX (32). Phylogenetic analyses were conducted using MEGA, version 5.0 (33), and a neighbor-joining tree was constructed using Kimura's two-parameter model (34) with 1,000 replicates to generate the bootstrap values.
[13C]bicarbonate incorporation experiment.
To analyze the incorporation of bicarbonate by the AOA enrichment culture during ammonia oxidation, a small amount of 13C-labeled bicarbonate (99%; Cambridge Isotope Laboratories, Andover, MA) was added to the cultivation medium, resulting in an 8% 13C content of bicarbonate. To determine the degree of 13C incorporation into thaumarchaeal membrane lipids, total cells were harvested by centrifugation, freeze-dried, and analyzed according to procedures described by Pitcher et al. (35). Archaeal glycerol dialkyl glycerol tetraethers (GDGTs) were extracted from the freeze-dried cells by use of a modified Bligh-Dyer extraction method, and the attached polar head groups were removed by acid hydrolysis. The archaeal core GDGTs were analyzed with an Agilent (Palo Alto, CA) 1100 series LC/MSD SL chromatograph using selective ion monitoring, as described previously (36).
FISH and electron microscopy analyses.
For fluorescent in situ hybridization (FISH) analysis, cultures were mixed by vortexing and the cells were fixed in 4% paraformaldehyde before being filtered using 0.2-μm polycarbonate GTTP membranes (Millipore) (37). The paraformaldehyde-fixed samples were hybridized with a Cy3-labeled Archaea-specific probe (Arc915) (38) and a 6-carboxyfluorescein (FAM)-labeled Bacteria-specific probe (EUB338) (39). 4′,6-Diamidino-2-phenylindole (DAPI) was used to visualize the total cells. The samples were observed with an AxioScope A1 microscope (Carl Zeiss, Germany) using an oil-immersion objective.
For scanning electron microscopy (SEM) analysis, the cells were harvested, immersed in 4% (vol/vol) glutaraldehyde in 0.1 M sodium phosphate buffer (pH 7.2) for 24 h at 4°C, and dehydrated using a graded ethanol series (70 to 100%). The samples were examined with a Zeiss DSM 940 electron microscope (Carl Zeiss). For transmission electron microscopy (TEM) (Tecnai G2 Sprite; FEI, the Netherlands; installed at the Korean Basic Science Institute) analyses of AOA cells from the enrichment cultures, the cells were collected from 10 ml of culture and negatively stained with 1% (wt/vol) phosphotungstic acid. Ultrathin sections were produced with an Ultracute (Leica, Austria) ultramicrotome, stained with uranyl acetate and lead citrate, and examined with a CM 20 electron microscope (Philips, the Netherlands) (40).
Genome sequencing and analysis.
High-molecular-weight genomic DNA was extracted from a 1-liter culture of strain MY2, which was harvested using 0.22-μm-pore-size filters (Millipore, Billerica, MA) and a vacuum pump. The total genomic DNA was extracted from the frozen pellets and filters by using a previously described protocol (41), except for the phenol-chloroform-isoamyl alcohol extraction, which was performed before the chloroform-isoamyl alcohol purification step to avoid any carryover of residual phenol. Subsequently, the genomic sequences were obtained using GS FLX Titanium (Roche, Basel, Switzerland) and Illumina (Illumina, San Diego, CA) sequencing systems. A 1/8 picotiter plate Titanium run with an 8-kb paired-end library yielded 185,572 sequence reads, which were assembled using gsAssembler 2.6 (Roche). The 6,798,106 sequence reads (100 bp) obtained from the Illumina sequencing run were used to correct any Titanium sequencing errors. The sequences generated by Illumina sequencing were assembled with CLC Genomics Workbench 5.5 (CLCbio). Open reading frames (ORFs) were predicted using Glimmer 3.02. tRNAs and rRNAs were predicted with tRNAscan-SE (42) and HMMER, respectively. The predicted proteins were searched for in various databases, including the enzyme profile database CatFam (43), which predicts catalytic functions described by Enzyme Commission (EC) numbers; the Clusters of Orthologous Groups (COG) protein database (http://www.ncbi.nlm.nih.gov/COG) (44); the National Center for Biotechnology Information (NCBI) reference sequence database (RefSeq) (45); and the SEED database (Subsystem Technology) (46). Conserved regions within proteins, such as motifs and domains, were searched using InterProScan in databases including the InterPro, TIGRFam, and Pfam databases. The average nucleotide identity (ANI) was calculated according to the method of Konstantinidis and Tiedje (47). ORFs from the query genome were considered to be conserved if they had a BLAST match of ≥60% of the overall sequence identity and 70% of the length of the query ORF.
Nucleotide sequence accession numbers.
The sequences from the whole-genome shotgun project were deposited in DDBJ/EMBL/GenBank under accession number AVSQ00000000. The version described in this study is version AVSQ01000000.
RESULTS AND DISCUSSION
Enrichment of an ammonium-oxidizing archaeon from soil.
An archaeal strain was enriched from the C horizon of a soil sample collected from an experimental agricultural plot. The initial soil inoculum (1 g in 100 ml AFM) catalyzed the conversion of ammonia to nitrite. This nitrifying enrichment culture was repeatedly transferred to fresh AFM. Analysis of the archaeal composition showed that the initially complex archaeal community became refined and eventually uniarchaeal, according to the single band detected by denaturing gradient gel electrophoresis analysis (data not shown). The resulting AOA enrichment culture was shown to be free of bacterial ammonia oxidizers by nested PCR using specific primers for AOB (see Table S1 in the supplemental material). However, it still contained heterotrophic bacteria, and attempts were made to purify the AOA strain by successive culturing in AFM containing antibiotics (ampicillin and penicillin G) (see Fig. S1) and by inoculating 5% (vol/vol) of the cultures after filtration (0.45-μm filter). Finally, the culture was serially diluted to extinction in 10-fold steps, and the highest dilution with nitrifying activity was selected as a clonal AOA culture, which was designated “strain MY2.”
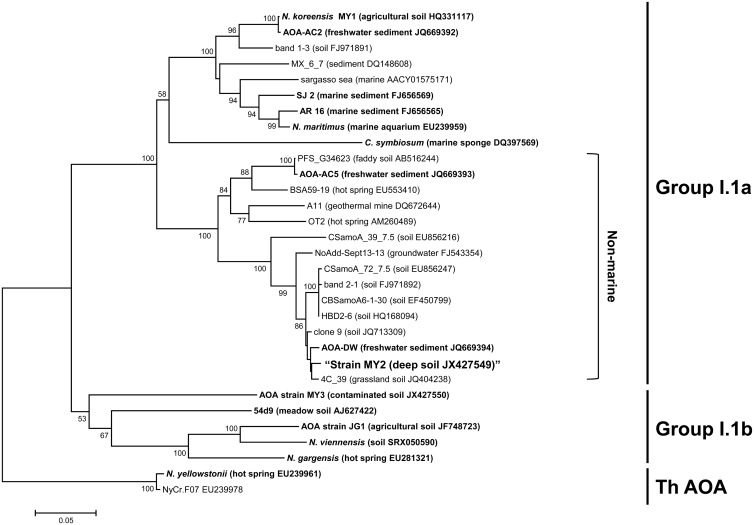
FISH analysis demonstrated archaeal dominance in the final enrichment; it contained 91% (3.2 × 108 ± 1.0 × 108 ml−1) archaeal cells, based on the ratio of the cell counts obtained using archaeal and bacterial probes. This was confirmed by real-time PCR using domain-specific 16S rRNA gene primers (3.5 × 108 ± 0.4 × 108 ml−1 for Archaea and 4.7 × 107 ± 0.2 × 107 ml−1 for Bacteria). The archaeal clone libraries obtained from the final enrichment culture revealed that there was only one unique sequence each (>99.7% identity, which was within the error rate expected for PCR amplification [1, 48]) for both the archaeal 16S rRNA and amoA genes (27). Based on the 16S rRNA and amoA gene sequences, strain MY2 belongs to the nonmarine thaumarchaeal group I.1a (Fig. 1; see Fig. S2 in the supplemental material), which also contains Nitrosopumilus maritimus, “Candidatus Nitrosoarchaeum koreensis,” and “Ca. Nitrosotenuis uzonensis” (72), but is only distantly related to other AOA (e.g., “Ca. Nitrososphaera gargensis” and Nitrososphaera viennensis [see Table S2]).
FIG 1.
Phylogenetic analysis of the archaeal amoA gene sequence (ca. 560 bp) obtained from strain MY2. The archaeal amoA genes were amplified using primers AamoAF and AamoAR. Branching patterns supported by >50% bootstrap values (1,000 iterations) according to the neighbor-joining method are denoted by their respective bootstrap values. The cluster groups are shown on the right, based on the origins of the reference sequences. ThAOA, thermophilic AOA lineage. The scale bar represents 5% estimated sequence divergence. Enriched or isolated AOA among the reference sequences are indicated in bold.
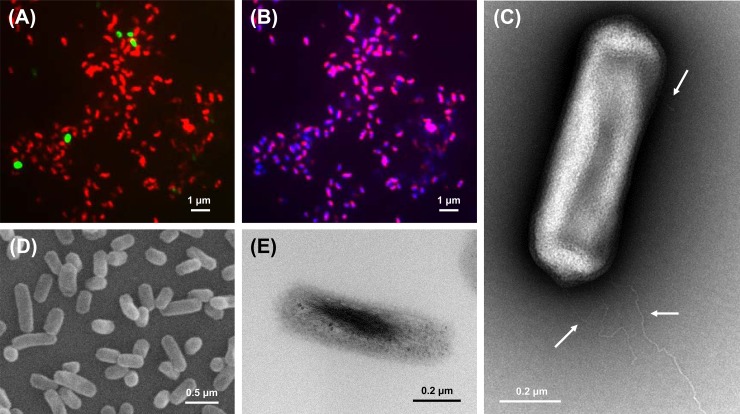
Strain MY2 was identified as straight, rod-shaped cells by SEM and TEM analyses (Fig. 2C to E). The cells appeared to have an average diameter of 0.2 μm and a length of 0.7 μm and were Gram negative. The FISH image in Fig. 2B shows that the chromosomes of strain MY2 were localized mostly to the sides of the cells and the ribosomes were not concentrated at the poles. Strain MY2 had peritrichous flagella which were not as smooth in shape as those of “Ca. Nitrososphaera gargensis” but similar in shape to those of “Ca. Nitrosoarchaeum limnia” (Fig. 2C).
FIG 2.
Morphology of strain MY2, based on FISH, TEM, and SEM analyses. (A) Merged image with a Cy3-labeled Archaea-specific probe (Arc915; red) and a FAM-labeled Bacteria-specific probe (EUB338; green). (B) Merged image with a Cy3-labeled Archaea-specific probe (Arc915) and DAPI. Magenta indicates the archaeal cells. (C) Transmission electron micrograph of strain MY2. The arrows indicate the archaeal flagella. (D) Scanning electron micrograph of strain MY2 cells. (E) Transmission electron micrograph of ultrathin section of strain MY2 cells.
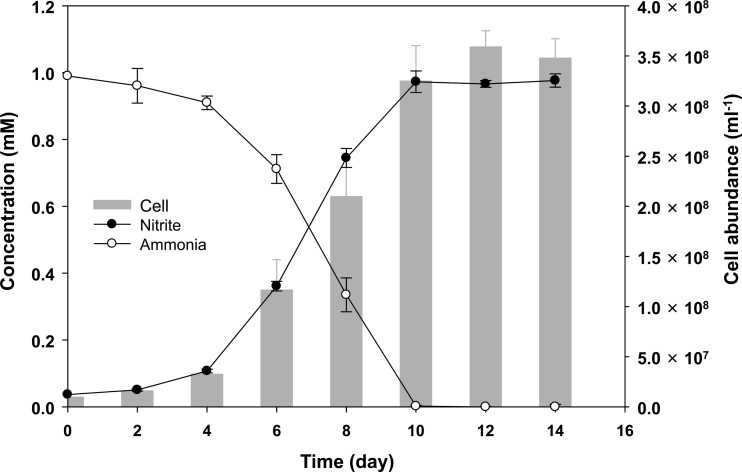
Ammonia oxidation by strain MY2 was evident from the decrease of the ammonia concentration coupled to exponential increases in the nitrite concentration and archaeal cell abundance as measured by archaeon-specific FISH counts (Fig. 3). No archaeal growth was observed in ammonia-free AFM. The cell yield from ammonia oxidation was estimated to be 3.4 × 105 cells ml−1 μM ammonia−1, which is higher than the yield reported for other group I.1a AOA (for N. maritimus, 5.0 × 104 cells ml−1 μM ammonia−1; and for “Ca. Nitrosoarchaeum koreensis,” 1.1 × 105 cells ml−1 μM ammonia−1) (1, 10) but slightly lower than that of an acidophilic AOA, “Ca. Nitrosotalea devanaterra” (4.5 × 105 cells ml−1 μM ammonia−1) (11).
FIG 3.
Relationships between ammonia oxidation, nitrite production, and archaeal growth in the enrichment culture of strain MY2. The ammonia and nitrite concentrations were determined colorimetrically. Archaeal cells were counted by FISH. The cell density in the initial inoculum was ca. 8.7 × 106 cells ml−1. The error bars represent the standard deviations based on triplicate experiments.
Analysis of the 16S rRNA gene sequences in the bacterial clone library of the MY2 enrichment culture showed that most of the bacterial sequences belonged to Acinetobacter (88.4%) (GenBank accession number EU834256) and Pseudomonas (9.3%) (GenBank accession number JX966440) species (27). The MY2 enrichment culture was stable but obligatorily dependent on these co-occurring bacteria. Bacterial cell-free archaeal suspensions were obtained by filtration using a 0.2-μm filter and serially diluted to extinction, but they failed to oxidize ammonia and grow. The interaction factor for the soil AOA N. viennensis, i.e., pyruvate (15), did not stimulate the growth of the bacterial cell-free archaeal suspensions. In addition, other simple (lactate, acetate, formate, glyoxylate, oxaloacetate, citrate, fumarate, and urea, each at 0.2 μM) or complex (yeast extract, tryptone, peptone, and tryptic soy broth, each at 20 mg liter−1) organic compounds also did not stimulate the oxidation of ammonia by bacterial cell-free suspensions of strain MY2. This kind of interaction has also been observed between AOB and cocultured bacteria (49, 50). Furthermore, tight interactions between autotrophs, such as cyanobacteria (51, 52), and heterotrophic bacteria are widely observed.
Autotrophic growth and further physiological characterization of strain MY2.
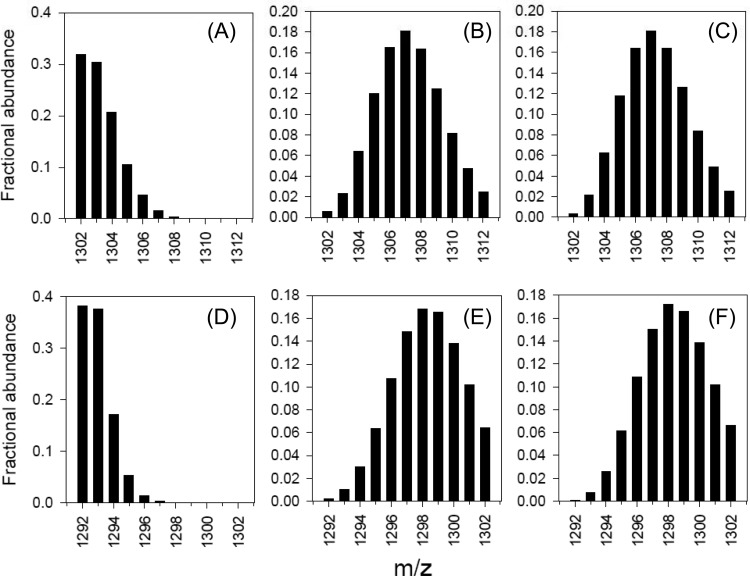
The carbon fixation capacity of strain MY2 was investigated using a 13C tracer experiment. Ammonia was oxidized by strain MY2 in the presence of 13C-labeled bicarbonate (8% 13C enriched), and label incorporation into membrane-specific lipids was determined. Strain MY2 produces two major GDGTs: GDGT-0 (GDGT with no internal rings; 24.8% of total GDGTs) and crenarchaeol (28.4%). Both GDGTs are produced by AOA, including those enriched from soils (53, 54). During the [13C]bicarbonate-amended growth experiments, these GDGTs became significantly enriched in 13C (Fig. 4B and E), and modeling of the isotope distribution of the molecular weights of these GDGTs indicated a 13C content of 6 to 8% (Fig. 4C and F). This is close to the labeling percentage of bicarbonate in the medium (8%), indicating that bicarbonate is the single carbon source of strain MY2 during growth, establishing its chemoautotrophic physiology.
FIG 4.
Mass spectra of GDGTs showing distributions of MH+ ions in the ranges of m/z 1302 to 1312 and 1292 to 1302 for GDGT-0 (A to C) and crenarchaeol (D to F), respectively, measured by liquid chromatography-mass spectrometry (LC-MS) analysis of these restricted m/z ranges. Panels B and E show the spectra obtained for strain MY2 cultivated with exposure to 13C-labeled bicarbonate. For reference, panels A and D show the spectra obtained for “Ca. Nitrosoarchaeum koreensis” cultivated with nonlabeled bicarbonate (10). Panels C and F show the modeled distributions of MH+ ions obtained using 13C contents of 6.3% (GDGT-0) and 7.8% (crenarchaeol), respectively. The 13C enrichment in the bicarbonate was 8.0%.
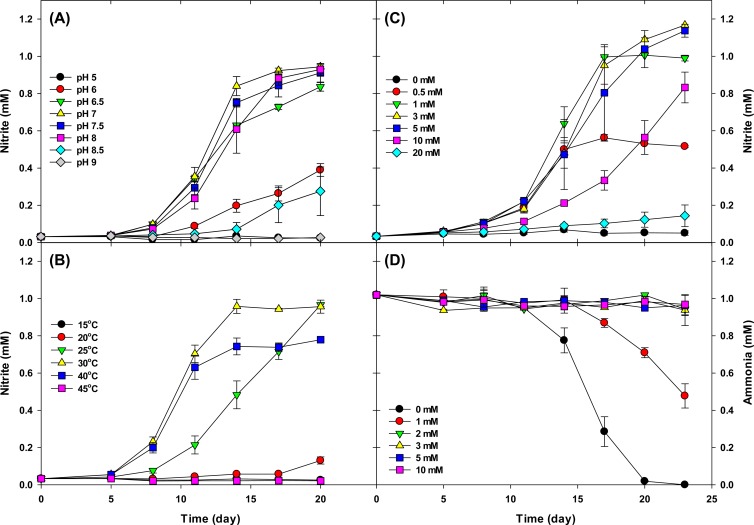
The effect of pH was investigated using batch cultures of strain MY2 at an initial pH of 5 to 9. Growth was restricted to the pH range of 6.5 to 8 (Fig. 5A) when cultures were supplied with 1 mM ammonium. Optimum growth occurred at pH 7, and the lag time increased at pH 6 and 8.5 (Fig. 5A). Strain MY2 grew in the temperature range of 25 to 40°C, with an optimum temperature of 30°C. Growth of strain MY2 was repressed at 40°C, and oxidation of 1 mM ammonia was not completed after prolonged incubation (Fig. 5B). Therefore, the organism can be considered to be neutrophilic and mesophilic. However, we need to verify these properties with an isolated culture, since they could have been affected by the activity of interacting cocultured bacteria.
FIG 5.
Effects of pH (A), temperature (B), ammonia concentration (C), and nitrite concentration (D) on the ammonia oxidation activity of strain MY2. For the experiments shown in panels A, B, and C, the ammonia oxidation activity was determined by nitrite accumulation. For the experiments shown in panel D, ammonia consumption was determined to study nitrite inhibition. The cell density in the initial inoculum was ca. 3.2 × 106 cells ml−1. The error bars represent the standard deviations based on triplicate experiments.
Ammonia oxidation was not inhibited significantly in the presence of up to 5 mM ammonia and was still observed at 10 mM, although an increase in the lag phase was observed at this concentration (Fig. 5C). In the presence of 20 mM ammonia, only slight ammonia oxidation was observed after 23 days of inoculation. Complete conversion of ammonia to nitrite was observed only at initial ammonium concentrations of 0.5 mM and 1 mM, and maximum nitrite production occurred with approximately 1.2 mM. This suggests that the accumulation of nitrite above this concentration may inhibit ammonia oxidation. To test this, strain MY2 was grown in medium with 1 mM ammonium and nitrite at various initial concentrations (Fig. 5D). Ammonium consumption did not occur at initial nitrite concentrations of >1 mM, and an increase in the lag phase was observed at an initial nitrite concentration of 1 mM. This result indicates that strain MY2 is more sensitive to accumulated nitrite than “Ca. Nitrosoarchaeum koreensis” (10) and “Ca. Nitrososphaera” strain JG1 (14). This experiment confirms the proposed inhibitory effect of accumulating nitrite during our experiments with different ammonium concentrations.
Sensitivity to nitrification inhibitors is a key physiological property of AOA. Differential sensitivity of ammonia-oxidizing microorganisms to inhibitors is frequently harnessed in environmental studies. Nitrite production and the increase in MY2 cell numbers were strongly inhibited in the presence of 50 μM chlorite or 500 μM dicyandiamide, both of which are commonly used inhibiters of autotrophic ammonia oxidation by AOA and AOB. Inhibition of strain MY2 was also observed at ATU concentrations of 50 μM and higher (see Fig. S3 in the supplemental material). These concentrations are higher than those for AOB (<10 μM) (10) and the acidophilic AOA “Ca. Nitrosotalea devanatera” (ca. 10 μM) (55) but lower than those for other enriched or isolated AOA (>500 μM for “Ca. Nitrosoarchaeum koreensis” and >100 μM for N. viennensis) (10, 56). ATU is known to be a Cu chelator, and AOA are believed to depend greatly on copper. Unlike AOB, AOA lack cytochromes and may have an alternative electron transfer mechanism, because copper-containing proteins are abundant in the strain MY2 genome as well as all published AOA genomes (see below) (17, 18, 20, 21). The affinity of AOA proteins for copper may be higher than that of AOB proteins. The present study suggests that AOB can be inhibited selectively by using a controlled concentration of ATU during microcosm or enrichment studies of AOA.
Genomic properties.
An almost complete genome of “Candidatus Nitrosotenuis chungbukensis” strain MY2 was reconstructed by high-throughput sequencing. The assembled sequence comprised 1.76 Mb in 24 contigs, which were organized using one scaffold (see Fig. S4 in the supplemental material). The G+C content of strain MY2 was 42%, which differs from those of other soil and marine strains in thaumarchaeal group I.1a (33 to 34%) (Table 1) and a hot spring strain in thaumarchaeal group I.1b (“Ca. Nitrososphaera gargensis”) (48%) (21).
TABLE 1.
Genomic features of “Candidatus Nitrosotenuis chungbukensis” MY2 and other thaumarchaeal group I.1a organisms
| Feature or parameter | Description or value |
||
|---|---|---|---|
| MY2 | Nitrosopumilus maritimus | “Ca. Nitrosoarchaeum koreensis” | |
| Isolation site | Oligotrophic deep agricultural soil | Marine aquarium | Nutritious agricultural topsoil |
| Temp range | Mesophilic | Mesophilic | Mesophilic |
| Origin of genome sequences | Enrichment culture | Isolate | Enrichment culture |
| Genome size (Mb) | 1.76 | 1.65 | 1.61 |
| G+C content (%) | 41.8 | 34.2 | 32.7 |
| No. of genomic objects (ORFs, partial ORFs, rRNAs, tRNAs) | 2,166 | 2,043 | 1,989 |
| No. of ORFs | 2,126 | 1,997 | 1,945 |
| No. of 16S-23S rRNA genes | 1 | 1 | 1 |
| No. of tRNA genes | 38 | 44 | 42 |
| % coding ORFs | 88.9 | 91.9 | 89.4 |
| ORF density (no. of ORFs/kb) | 1.21 | 1.19 | 1.21 |
| Avg ORF length (bp) | 738 | 757 | 739 |
Approximately 62% of the coding DNA sequence of the genome of strain MY2 was assigned, revealing 66 clusters of orthologous groups (COGs) (see Fig. S4 and Table S3 in the supplemental material), which is slightly higher than the numbers for the genomes of “Ca. Nitrosoarchaeum koreensis” and “Ca. Nitrososphaera gargensis” but similar to that for N. maritimus (see Table S3). Most of the genes in the strain MY2 genome were syntenic with those of soil and marine thaumarchaeal group I.1a genomes but not with those of the “Ca. Nitrososphaera gargensis” genome (see Fig. S5). The genome of strain MY2 has an average nucleotide identity (ANI) of the shared genes of 68.5% with both N. maritimus and “Ca. Nitrosoarchaeum koreensis.” ANI values of ∼94% correspond to the traditional 70% DNA-DNA reassociation standard currently used for species definition (47). This indicates that strain MY2 is evolutionarily distinct from N. maritimus and “Ca. Nitrosoarchaeum koreensis.” The genes that encode proteins for selected metabolic pathways and traits are summarized in Table S4.
Comparative genome analysis of strain MY2 with two other group I.1a strains (N. maritimus and “Ca. Nitrosoarchaeum koreensis”) indicated 1,241 unique ORFs in the total of 2,126 ORFs (see Fig. S6 in the supplemental material). The COG classification of the unique ORFs of strain MY2 showed a higher representation of genes involved in cell cycle control and mitosis (COG initial D), cell wall/membrane/envelope biogenesis (COG initial M), motility (COG initial N), and signal transduction (COF initial T) than that in conserved ORFs (see Fig. S6). In particular, signal transduction- and cell motility-associated ORFs were more prominent among the unique ORFs of strain MY2 than among those of other strains (see Table S3 and Fig. S6). Major signal transduction-associated ORFs of strain MY2 corresponded to three proteins: histidine kinase-like proteins (n = 21), response regulator-containing CheY-like receiver protein (n = 22), and universal stress protein (UspA) and related nucleotide-binding protein (n = 15). Most of these three signal transduction-associated proteins were encoded by unique ORFs (19/21, 19/22, and 12/15 ORFs, respectively). These three signal transduction-associated proteins were less represented in “Ca. Nitrosoarchaeum koreensis” and N. maritimus (n = 30 and n = 31, respectively). The histidine kinase- and response regulator-encoding genes are involved in two-component signal transduction pathways. The two-component regulatory system serves as a basic stimulus-response coupling mechanism to allow organisms to sense and respond to changes in environmental conditions (57). Archaeal flagellum- and chemotaxis-associated genes contributed to the high representation of motility-associated ORFs (COG initial N) among the unique ORFs of strain MY2. The relationship between the high representation of signal transduction and chemotaxis genes in the genome of strain MY2 and its niche differentiation is intriguing and warrants further research. In addition, the defense mechanism-associated ORFs (conferring resistance to antimicrobial chemicals) (n = 4) of strain MY2 were less abundant than those of other AOA (for N. maritimus, n = 13; for “Ca. Nitrosoarchaeum koreensis,” n = 14; and for “Ca. Nitrososphaera gargensis,” n = 15) (see Table S3).
Ammonia oxidation and carbon fixation.
Strain MY2 was shown to grow chemolithoautotrophically by using ammonia oxidation (Fig. 3) and inorganic carbon fixation (Fig. 4). The genomic analysis of strain MY2 supports this chemolithoautotrophic lifestyle. The homologs of the amoA, -B, and -C genes of strain MY2 were arranged in the same order as that in the genomes of other archaea in the thaumarchaeal group I.1a cluster (4, 22, 58), and an additional ORF, referred to as amoX, occurred between amoA and amoC. This arrangement of the amo gene cluster differed from that of thaumarchaeal group I.1b (see Fig. S7 in the supplemental material). Strain MY2 lacked the genes required to utilize potential energy sources other than ammonia, i.e., urease and/or cyanate hydratase, which have been found in the genomes of a marine AOA strain, “Ca. Nitrosopumilus sediminis” (59), as well as N. viennensis (15), Cenarchaeum symbiosum (16), and “Ca. Nitrososphaera gargensis” (21).
The exact mechanism of ammonia oxidation in AOA is still unclear, and hydroxylamine oxidoreductase (HAO) homologs of AOB have not been identified, despite the conservation of amo genes in all known AOA (including strain MY2) genomes (16–19, 21). Vajrala et al. (60) recently reported hydroxylamine-induced oxygen consumption and ATP production by the marine AOA strain N. maritimus, suggesting the presence of noncanonical HAO genes in AOA. Alternatively, multicopper proteins of AOA were suggested to be involved in ammonia oxidation (18, 22). Based on these observations, an alternative novel ammonia oxidation pathway involving the reactive intermediate nitroxyl (HNO) has been proposed (18). In agreement with this, ammonia oxidation and cell growth of strain MY2 were not observed in the presence of 100 μM 2-phenyl-4,4,5,5-tetramethylimidazoline-1-oxyl 3-oxide (PTIO) (27), a scavenger of nitroxyl and the free radical nitric oxide (NO) (61–63). Strain MY2 encodes two conserved multicopper proteins: an NO-forming nitrite reductase (NirK) homolog and a multicopper oxidase type 3 (see Table S4 in the supplemental material). Expression of the archaeal nirK gene was observed during ammonia oxidation and N2O production in a soil AOA culture (10). Strain MY2 was found to produce the greenhouse gas nitrous oxide (N2O) via nitrification and denitrification (27). NirK might play a key role in archaeal nitrification and/or N2O production, and further research into the role of the nirK gene is warranted.
The genome of strain MY2 carried putative carbon fixation genes of the 3-hydroxypropionate/4-hydroxybutyrate pathway (64), in good agreement with other published thaumarchaeal genomes (16–19, 21). Genes coding for proteins catalyzing two carboxylation reactions that transform acetyl-coenzyme A (acetyl-CoA) into succinyl-CoA and a multistep sequence that converts succinyl-CoA into two molecules of acetyl-CoA were detected (see Table S4 in the supplemental material). In addition to the genes encoding the 3-hydroxypropionate/4-hydroxybutyrate pathway, the genome of strain MY2 contained all the genes of the tricarboxylic acid cycle. Strain MY2 contained all of the genes necessary for gluconeogenesis. During autotrophic growth, gluconeogenesis might begin with succinyl-CoA or acetyl-CoA, generated by carbon fixation. Succinyl-CoA could be oxidized to malate and/or oxaloacetate, and oxaloacetate could be converted into phosphoenolpyruvate (PEP) by the ATP-dependent PEP carboxykinase. It remains unclear whether acetyl-CoA can be used for gluconeogenesis via the classical glyoxylate shunt, because the genome of strain MY2 lacked the gene required for the first step (isocitrate lyase) of the glyoxylate shunt, which has been found in all other AOA genomes (17, 18, 65).
Electron transfer, transport system, and other genomic traits.
Strain MY2 contained most of the essential genes proposed for electron transfer in N. maritimus (18). Strain MY2 encoded a complete respiratory chain of complex I-V (see Table S4 in the supplemental material), which is used for energy generation and reverse electron transport. Strain MY2 and all published thaumarchaeal genomes possess 11 genes that encode subunits of type I NADH dehydrogenase. NADH production requires the reverse operation of complex I (NuoABCDHIJKMLN) as a quinol oxidase, which is driven by a proton motive force (PMF). However, complex I lacked close homologs of the peripheral subunits E, F, and G, which are generally required as the canonical electron input module. It is proposed that the electrons extracted by either a CuHAO or NXOR protein (and transferred into the quinone pool) generate a PMF via complexes III and IV, thereby driving the generation of ATP through an FoF1-type ATP synthase (complex V) (18).
Various genes coding for proteins acting as organic transporters of different amino acids, oligo- and dipeptides, glycerol, sulfonate, and aminopeptidase were present in the genome of strain MY2. These genes have frequently been reported for the genomes of other AOA (18, 20, 21). Recently, it was found that pyruvate is required for ammonia oxidation in N. viennensis (clade I.1b) (15). Although genomes of all thaumarchaeal group I.1a members, including strain MY2, carry genes for organic transporters, potential mixotrophism was not supported by isotopic and physiological evidence from cultivated group I.1a AOA. A high-affinity, high-activity phosphate uptake operon (pstSCAB) was observed in the genome of strain MY2 (see Table S4 in the supplemental material) and has been identified previously only in the genome of a marine archaeon, N. maritimus (18).
The N. maritimus genome contains archaeal-type genes for ectoine biosynthesis, which might be involved in the regulation of cellular osmotic pressure. However, the genomes of all nonmarine thaumarchaeal group I.1a AOA (17, 19, 20), including strain MY2, lack genes related to ectoine biosynthesis. Instead, the genomes of nonmarine thaumarchaeal group I.1a AOA, including strain MY2, possess genes for a proline/betaine transporter and both large- and small-conductance mechanosensitive channel proteins (see Table S4). In a wide variety of Bacteria, proline/betaine is accumulated as an effective response to osmotic stress (66–68). The C. symbiosum and N. maritimus genomes appear to lack genes for large-conductance mechanosensitive channels, although they each encode at least one small-conductance mechanosensitive channel protein (16, 18). This indicates that strain MY2 and other nonmarine thaumarchaeal group I.1a AOA may have a different strategy to combat osmotic stress compared with that of marine thaumarchaeal group I.1a AOA, which might reflect the niche differentiation of soil AOA.
Similar to “Ca. Nitrosoarchaeum limnia” and “Ca. Nitrososphaera gargensis,” strain MY2 contained numerous motility-associated genes (see Table S4). Strain MY2 possessed all of the genes for chemotaxis, which are carried in a single gene cluster next to the flagellar genes, as found in “Ca. Nitrosoarchaeum limnia” and “Ca. Nitrososphaera gargensis.” The organization of the motility-associated genes in the genome of strain MY2 was more similar to that in “Ca. Nitrosoarchaeum limnia” than to that in “Ca. Nitrososphaera gargensis” (see Fig. S8). This result agrees with the morphological similarity of the flagella of strain MY2 and “Ca. Nitrosoarchaeum limnia.” Motility-associated genes are absent from the genomes of marine AOA, including “Ca. Nitrosopumilus koreensis” (69), “Ca. Nitrosopumilus sediminis,” N. maritimus, and C. symbiosum. Motility might be an important property for nonmarine AOA to be able to respond to various substrate and/or oxygen concentrations in soil aggregates (20), which could be verified when more genomic information from soil and marine AOA strains becomes available.
Environmental significance.
Strain MY2 was enriched from the same agricultural plot as “Ca. Nitrosoarchaeum koreensis” (10), but the depth and properties of the soil used for the inoculum were clearly distinct, i.e., the C horizon for strain MY2 and the A horizon for “Ca. Nitrosoarchaeum koreensis.” The A-horizon soil was rich in organic matter and ammonia (organic carbon, 1.6%; total nitrogen, 0.05%; and ammonium concentration, 134 mg/kg) (10), in contrast with the C-horizon soil (see Materials and Methods). This suggests that strain MY2 is more adapted to oligotrophic environments, although we did not test it at ammonia levels of <0.5 mM.
Environmental amoA and 16S rRNA gene sequences suggest that the isolated strain MY2 is environmentally significant. AOA sequences closely related to that of strain MY2 are commonly detected in soil (7–9, 70). The amoA gene sequence of strain MY2 is also closely related to a sequence (AOA-DW; 98.7%) from near-shore sediment samples from Lakes Acton and Delaware (13) and a sequence (4C_39; 98.9%) from Icelandic grassland soils affected by long-term N fertilization and geothermal heating (71). In the latter study, it was demonstrated that strain MY2-like archaea dominate (20 to 90% of total AOA amoA operational taxonomic units [OTUs]) the archaeal communities in Icelandic grassland soils. Soil microcosm studies revealed that growth of strain MY2-like AOA was especially prominent at lower ammonia concentrations than those seen with AOB (7). The results provide direct evidence for a role for strain MY2-like AOA in soil ammonia oxidation.
Conclusions.
This study reports the physiological and genomic characterization of a new archaeal chemolithoautotrophic ammonia oxidizer, strain MY2, which was enriched from a deep oligotrophic soil horizon. Phylogenetic analyses of the 16S rRNA and amoA genes placed strain MY2 in thaumarchaeal group I.1a, and the following candidate status is proposed: “Candidatus Nitrosotenuis chungbukensis.”
Taxonomy. (i) Etymology.
The taxonomy for “Candidatus Nitrosotenuis chungbukensis” sp. nov. is as follows: chungbukensis (N.L. masc. adj. chungbukensis, from Chungbuk, Republic of Korea).
(ii) Locality.
Temperate agricultural soil, Chungbuk, Republic of Korea.
(iii) Diagnosis.
A chemolithoautotrophic ammonia oxidizer in the kingdom Thaumarchaeota, which is straight and rod-shaped, with a diameter of approximately 0.2 μm and a length of 0.7 μm, an optimum pH of 7, and an optimum temperature of 30°C; it is Gram negative. Carbon dioxide is used as the sole carbon source. It belongs to nonmarine thaumarchaeal group I.1a. AOA with almost identical 16S rRNA and amoA gene sequences have been detected in various terrestrial environments, including soil and groundwater.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Basic Science Research Program (grant 2012R1A1A2A10039384) and the Mid-Career Researcher Program (grant NRF-2013R1A2A2A05006754) through the National Research Foundation of Korea, funded by the Ministry of Education, Science, and Technology, and by the Korea Polar Research Institute (grant PP13020). This work was partially supported by the Energy Efficiency & Resources Core Technology Program of the Korea Institute of Energy Technology Evaluation and Planning (KETEP), granted financial resources from the Ministry of Trade, Industry and Energy, Republic of Korea (grant 20132020000170).
Footnotes
Published ahead of print 4 April 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03730-13.
REFERENCES
- 1.Könneke M, Bernhard AE, de la Torre JR, Walker CB, Waterbury JB, Stahl DA. 2005. Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature 437:543–546. 10.1038/nature03911 [DOI] [PubMed] [Google Scholar]
- 2.Chen XP, Zhu YG, Xia Y, Shen JP, He JZ. 2008. Ammonia-oxidizing archaea: important players in paddy rhizosphere soil? Environ. Microbiol. 10:1978–1987. 10.1111/j.1462-2920.2008.01613.x [DOI] [PubMed] [Google Scholar]
- 3.Leininger S, Urich T, Schloter M, Schwark L, Qi J, Nicol GW, Prosser JI, Schuster SC, Schleper C. 2006. Archaea predominate among ammonia-oxidizing prokaryotes in soils. Nature 442:806–809. 10.1038/nature04983 [DOI] [PubMed] [Google Scholar]
- 4.Treusch AH, Leininger S, Kletzin A, Schuster SC, Klenk HP, Schleper C. 2005. Novel genes for nitrite reductase and Amo-related proteins indicate a role of uncultivated mesophilic crenarchaeota in nitrogen cycling. Environ. Microbiol. 7:1985–1995. 10.1111/j.1462-2920.2005.00906.x [DOI] [PubMed] [Google Scholar]
- 5.Auguet JC, Barberan A, Casamayor EO. 2010. Global ecological patterns in uncultured archaea. ISME J. 4:182–190. 10.1038/ismej.2009.109 [DOI] [PubMed] [Google Scholar]
- 6.Ochsenreiter T, Selezi D, Quaiser A, Bonch-Osmolovskaya L, Schleper C. 2003. Diversity and abundance of Crenarchaeota in terrestrial habitats studied by 16S RNA surveys and real time PCR. Environ. Microbiol. 5:787–797. 10.1046/j.1462-2920.2003.00476.x [DOI] [PubMed] [Google Scholar]
- 7.Verhamme DT, Prosser JI, Nicol GW. 2011. Ammonia concentration determines differential growth of ammonia-oxidising archaea and bacteria in soil microcosms. ISME J. 5:1067–1071. 10.1038/ismej.2010.191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang LM, Offre PR, He JZ, Verhamme DT, Nicol GW, Prosser JI. 2010. Autotrophic ammonia oxidation by soil thaumarchaea. Proc. Natl. Acad. Sci. U. S. A. 107:17240–17245. 10.1073/pnas.1004947107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Offre P, Prosser JI, Nicol GW. 2009. Growth of ammonia-oxidizing archaea in soil microcosms is inhibited by acetylene. FEMS Microbiol. Ecol. 70:99–108. 10.1111/j.1574-6941.2009.00725.x [DOI] [PubMed] [Google Scholar]
- 10.Jung MY, Park SJ, Min D, Kim JS, Rijpstra WI, Sinninghe Damste JS, Kim GJ, Madsen EL, Rhee SK. 2011. Enrichment and characterization of an autotrophic ammonia-oxidizing archaeon of mesophilic crenarchaeal group I.1a from an agricultural soil. Appl. Environ. Microbiol. 77:8635–8647. 10.1128/AEM.05787-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lehtovirta-Morley LE, Stoecker K, Vilcinskas A, Prosser JI, Nicol GW. 2011. Cultivation of an obligate acidophilic ammonia oxidizer from a nitrifying acid soil. Proc. Natl. Acad. Sci. U. S. A. 108:15892–15897. 10.1073/pnas.1107196108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hatzenpichler R, Lebedeva EV, Spieck E, Stoecker K, Richter A, Daims H, Wagner M. 2008. A moderately thermophilic ammonia-oxidizing crenarchaeote from a hot spring. Proc. Natl. Acad. Sci. U. S. A. 105:2134–2139. 10.1073/pnas.0708857105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.French E, Kozlowski JA, Mukherjee M, Bullerjahn G, Bollmann A. 2012. Ecophysiological characterization of ammonia-oxidizing archaea and bacteria from freshwater. Appl. Environ. Microbiol. 78:5773–5780. 10.1128/AEM.00432-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim JG, Jung MY, Park SJ, Rijpstra WI, Sinninghe Damste JS, Madsen EL, Min D, Kim JS, Kim GJ, Rhee SK. 2012. Cultivation of a highly enriched ammonia-oxidizing archaeon of thaumarchaeotal group I.1b from an agricultural soil. Environ. Microbiol. 14:1528–1543. 10.1111/j.1462-2920.2012.02740.x [DOI] [PubMed] [Google Scholar]
- 15.Tourna M, Stieglmeier M, Spang A, Konneke M, Schintlmeister A, Urich T, Engel M, Schloter M, Wagner M, Richter A, Schleper C. 2011. Nitrososphaera viennensis, an ammonia oxidizing archaeon from soil. Proc. Natl. Acad. Sci. U. S. A. 108:8420–8425. 10.1073/pnas.1013488108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hallam SJ, Konstantinidis KT, Putnam N, Schleper C, Watanabe Y-I, Sugahara J, Preston C, de la Torre J, Richardson PM, DeLong EF. 2006. Genomic analysis of the uncultivated marine crenarchaeote Cenarchaeum symbiosum. Proc. Natl. Acad. Sci. U. S. A. 103:18296–18301. 10.1073/pnas.0608549103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim BK, Jung MY, Yu DS, Park SJ, Oh TK, Rhee SK, Kim JF. 2011. Genome sequence of an ammonia-oxidizing soil archaeon, “Candidatus Nitrosoarchaeum koreensis” MY1. J. Bacteriol. 193:5539–5540. 10.1128/JB.05717-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Walker CB, de la Torre JR, Klotz MG, Urakawa H, Pinel N, Arp DJ, Brochier-Armanet C, Chain PS, Chan PP, Gollabgir A, Hemp J, Hugler M, Karr EA, Konneke M, Shin M, Lawton TJ, Lowe T, Martens-Habbena W, Sayavedra-Soto LA, Lang D, Sievert SM, Rosenzweig AC, Manning G, Stahl DA. 2010. Nitrosopumilus maritimus genome reveals unique mechanisms for nitrification and autotrophy in globally distributed marine crenarchaea. Proc. Natl. Acad. Sci. U. S. A. 107:8818–8823. 10.1073/pnas.0913533107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mosier AC, Allen EE, Kim M, Ferriera S, Francis CA. 2012. Genome sequence of “Candidatus Nitrosoarchaeum limnia” BG20, a low-salinity ammonia-oxidizing archaeon from the San Francisco Bay estuary. J. Bacteriol. 194:2119–2120. 10.1128/JB.00007-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Blainey PC, Mosier AC, Potanina A, Francis CA, Quake SR. 2011. Genome of a low-salinity ammonia-oxidizing archaeon determined by single-cell and metagenomic analysis. PLoS One 6:e16626. 10.1371/journal.pone.0016626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Spang A, Poehlein A, Offre P, Zumbragel S, Haider S, Rychlik N, Nowka B, Schmeisser C, Lebedeva EV, Rattei T, Bohm C, Schmid M, Galushko A, Hatzenpichler R, Weinmaier T, Daniel R, Schleper C, Spieck E, Streit W, Wagner M. 2012. The genome of the ammonia-oxidizing Candidatus Nitrososphaera gargensis: insights into metabolic versatility and environmental adaptations. Environ. Microbiol. 14:3122–3145. 10.1111/j.1462-2920.2012.02893.x [DOI] [PubMed] [Google Scholar]
- 22.Bartossek R, Nicol GW, Lanzen A, Klenk HP, Schleper C. 2010. Homologues of nitrite reductases in ammonia-oxidizing archaea: diversity and genomic context. Environ. Microbiol. 12:1075–1088. 10.1111/j.1462-2920.2010.02153.x [DOI] [PubMed] [Google Scholar]
- 23.Bartossek R, Spang A, Weidler G, Lanzen A, Schleper C. 2012. Metagenomic analysis of ammonia-oxidizing archaea affiliated with the soil group. Front. Microbiol. 3:208. 10.3389/fmicb.2012.00208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Di HJ, Cameron KC, Shen JP, Winefield CS, O'Callaghan M, Bowatte S, He JZ. 2010. Ammonia-oxidizing bacteria and archaea grow under contrasting soil nitrogen conditions. FEMS Microbiol. Ecol. 72:386–394. 10.1111/j.1574-6941.2010.00861.x [DOI] [PubMed] [Google Scholar]
- 25.Jia Z, Conrad R. 2009. Bacteria rather than Archaea dominate microbial ammonia oxidation in an agricultural soil. Environ. Microbiol. 11:1658–1671. 10.1111/j.1462-2920.2009.01891.x [DOI] [PubMed] [Google Scholar]
- 26.Brady NC, Weil RR. 2007. The nature and properties of soils, 14th ed. Pearson Prentice Hall, Upper Saddle River, NJ [Google Scholar]
- 27.Jung MY, Well R, Min D, Giesemann A, Park SJ, Kim JG, Kim SJ, Rhee SK. 14 November 2013. Isotopic signatures of N2O produced by ammonia-oxidizing archaea from soils. ISME J. 10.1038/ismej.2013.205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shinn MB. 1941. Colorimetric method for determination of nitrate. Ind. Chem. Anal. Engl. Ed. 13:33–35. 10.1021/i560089a010 [DOI] [Google Scholar]
- 29.Hynes RK, Knowles R. 1984. Production of nitrous oxide by Nitrosomonas europaea: effects of acetylene, pH, and oxygen. Can. J. Microbiol. 30:1397–1404. 10.1139/m84-222 [DOI] [Google Scholar]
- 30.Di HJ, Cameron KC, Shen JP, Winefield CS, O'Callaghan M, Bowatte S, He JZ. 2009. Nitrification driven by Bacteria and not Archaea in nitrogen-rich grassland soils. Nat. Geosci. 2:621–624. 10.1038/ngeo613 [DOI] [Google Scholar]
- 31.Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J, Glockner FO. 2007. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 35:7188–7196. 10.1093/nar/gkm864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876–4882. 10.1093/nar/25.24.4876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28:2731–2739. 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kimura M. 1980. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16:111–120. 10.1007/BF01731581 [DOI] [PubMed] [Google Scholar]
- 35.Pitcher A, Rychlik N, Hopmans EC, Spieck E, Rijpstra WI, Ossebaar J, Schouten S, Wagner M, Damste JS. 2010. Crenarchaeol dominates the membrane lipids of Candidatus Nitrososphaera gargensis, a thermophilic group I.1b archaeon. ISME J. 4:542–552. 10.1038/ismej.2009.138 [DOI] [PubMed] [Google Scholar]
- 36.Schouten S, Huguet C, Hopmans EC, Kienhuis MV, Sinninghe Damsté JS. 2007. Analytical methodology for TEX86 paleothermometry by high-performance liquid chromatography/atmospheric pressure chemical ionization-mass spectrometry. Anal. Chem. 79:2940–2944. 10.1021/ac062339v [DOI] [PubMed] [Google Scholar]
- 37.DeLong EF, Taylor LT, Marsh TL, Preston CM. 1999. Visualization and enumeration of marine planktonic archaea and bacteria by using polyribonucleotide probes and fluorescent in situ hybridization. Appl. Environ. Microbiol. 65:5554–5563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Alm EW, Oerther DB, Larsen N, Stahl DA, Raskin L. 1996. The oligonucleotide probe database. Appl. Environ. Microbiol. 62:3557–3559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Amann RI, Binder BJ, Olson RJ, Chisholm SW, Devereux R, Stahl DA. 1990. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl. Environ. Microbiol. 56:1919–1925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reynolds ES. 1963. The use of lead citrate at high pH as an electron-opaque stain in electron microscopy. J. Cell Biol. 17:208–212. 10.1083/jcb.17.1.208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhou J, Bruns MA, Tiedje JM. 1996. DNA recovery from soils of diverse composition. Appl. Environ. Microbiol. 62:316–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lowe TM, Eddy SR. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25:955–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yu C, Zavaljevski N, Desai V, Reifman J. 2009. Genome-wide enzyme annotation with precision control: catalytic families (CatFam) databases. Proteins 74:449–460. 10.1002/prot.22167 [DOI] [PubMed] [Google Scholar]
- 44.Tatusov RL, Galperin MY, Natale DA, Koonin EV. 2000. The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 28:33–36. 10.1093/nar/28.1.33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pruitt KD, Tatusova T, Maglott DR. 2007. NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 35:D61–D65. 10.1093/nar/gkl842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Overbeek R, Begley T, Butler RM, Choudhuri JV, Chuang HY, Cohoon M, de Crecy-Lagard V, Diaz N, Disz T, Edwards R, Fonstein M, Frank ED, Gerdes S, Glass EM, Goesmann A, Hanson A, Iwata-Reuyl D, Jensen R, Jamshidi N, Krause L, Kubal M, Larsen N, Linke B, McHardy AC, Meyer F, Neuweger H, Olsen G, Olson R, Osterman A, Portnoy V, Pusch GD, Rodionov DA, Ruckert C, Steiner J, Stevens R, Thiele I, Vassieva O, Ye Y, Zagnitko O, Vonstein V. 2005. The subsystems approach to genome annotation and its use in the project to annotate 1000 genomes. Nucleic Acids Res. 33:5691–5702. 10.1093/nar/gki866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Konstantinidis KT, Tiedje JM. 2005. Genomic insights that advance the species definition for prokaryotes. Proc. Natl. Acad. Sci. U. S. A. 102:2567–2572. 10.1073/pnas.0409727102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.von Wintzingerode F, Gobel UB, Stackebrandt E. 1997. Determination of microbial diversity in environmental samples: pitfalls of PCR-based rRNA analysis. FEMS Microbiol. Rev. 21:213–229. 10.1111/j.1574-6976.1997.tb00351.x [DOI] [PubMed] [Google Scholar]
- 49.Gundersen K. 1955. Effects of B-vitamins and ammo-acids on nitrification. Physiol. Plant. 8:136–141. 10.1111/j.1399-3054.1955.tb08966.x [DOI] [Google Scholar]
- 50.Jones RD, Hood M. 1980. Effects of temperature, pH, salinity, and inorganic nitrogen on the rate of ammonium oxidation by nitrifiers isolated from wetland environments. Microb. Ecol. 6:339–347. 10.1007/BF02010496 [DOI] [PubMed] [Google Scholar]
- 51.Cole JJ. 1982. Interactions between Bacteria and Algae in aquatic ecosystems. Annu. Rev. Ecol. Syst. 13:291–314. 10.1146/annurev.es.13.110182.001451 [DOI] [Google Scholar]
- 52.Hube AE, Heyduck-Söller B, Fischer U. 2009. Phylogenetic classification of heterotrophic bacteria associated with filamentous marine cyanobacteria in culture. Syst. Appl. Microbiol. 32:256–265. 10.1016/j.syapm.2009.03.001 [DOI] [PubMed] [Google Scholar]
- 53.Pitcher A, Hopmans EC, Mosier AC, Park SJ, Rhee SK, Francis CA, Schouten S, Damste JS. 2011. Core and intact polar glycerol dibiphytanyl glycerol tetraether lipids of ammonia-oxidizing archaea enriched from marine and estuarine sediments. Appl. Environ. Microbiol. 77:3468–3477. 10.1128/AEM.02758-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sinninghe Damsté JS, Rijpstra WIC, Hopmans EC, Jung M-Y, Kim J-G, Rhee S-K, Stieglmeier M, Schleper C. 2012. Intact polar and core glycerol dibiphytanyl glycerol tetraether lipids of group I.1a and I.1b thaumarchaeota in soil. Appl. Environ. Microbiol. 78:6866–6874. 10.1128/AEM.01681-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lehtovirta-Morley LE, Verhamme DT, Nicol GW, Prosser JI. 2013. Effect of nitrification inhibitors on the growth and activity of Nitrosotalea devanaterra in culture and soil. Soil Biol. Biochem. 62:129–133. 10.1016/j.soilbio.2013.01.020 [DOI] [Google Scholar]
- 56.Shen T, Stieglmeier M, Dai J, Urich T, Schleper C. 2013. Responses of the terrestrial ammonia-oxidizing archaeon Ca. Nitrososphaera viennensis and the ammonia-oxidizing bacterium Nitrosospira multiformis to nitrification inhibitors. FEMS Microbiol. Lett. 344:121–129. 10.1111/1574-6968.12164 [DOI] [PubMed] [Google Scholar]
- 57.Stock AM, Robinson VL, Goudreau PN. 2000. Two-component signal transduction. Annu. Rev. Biochem. 69:183–215. 10.1146/annurev.biochem.69.1.183 [DOI] [PubMed] [Google Scholar]
- 58.Schleper C, Jurgens G, Jonuscheit M. 2005. Genomic studies of uncultivated archaea. Nat. Rev. Microbiol. 3:479–488. 10.1038/nrmicro1159 [DOI] [PubMed] [Google Scholar]
- 59.Park SJ, Kim JG, Jung MY, Kim SJ, Cha IT, Ghai R, Martin-Cuadrado AB, Rodriguez-Valera F, Rhee SK. 2012. Draft genome sequence of an ammonia-oxidizing archaeon, “Candidatus Nitrosopumilus sediminis” AR2, from Svalbard in the Arctic Circle. J. Bacteriol. 194:6948–6949. 10.1128/JB.01869-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vajrala N, Martens-Habbena W, Sayavedra-Soto LA, Schauer A, Bottomley PJ, Stahl DA, Arp DJ. 2013. Hydroxylamine as an intermediate in ammonia oxidation by globally abundant marine archaea. Proc. Natl. Acad. Sci. U. S. A. 110:1006–1011. 10.1073/pnas.1214272110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Amano F, Noda T. 1995. Improved detection of nitric oxide radical (NO·) production in an activated macrophage culture with a radical scavenger, carboxy PTIO and Griess reagent. FEBS Lett. 368:425–428. 10.1016/0014-5793(95)00700-J [DOI] [PubMed] [Google Scholar]
- 62.Ellis A, Lu H, Li CG, Rand MJ. 2001. Effects of agents that inactivate free radical NO (NO·) on nitroxyl anion-mediated relaxations, and on the detection of NO· released from the nitroxyl anion donor Angeli's salt. Br. J. Clin. Pharmacol. 134:521–528. 10.1038/sj.bjp.0704287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Samuni U, Samuni Y, Goldstein S. 2010. On the distinction between nitroxyl and nitric oxide using nitronyl nitroxides. J. Am. Chem. Soc. 132:8428–8432. 10.1021/ja101945j [DOI] [PubMed] [Google Scholar]
- 64.Berg IA, Kockelkorn D, Buckel W, Fuchs G. 2007. A 3-hydroxypropionate/4-hydroxybutyrate autotrophic carbon dioxide assimilation pathway in Archaea. Science 318:1782–1786. 10.1126/science.1149976 [DOI] [PubMed] [Google Scholar]
- 65.Mosier AC, Lund MB, Francis CA. 2012. Ecophysiology of an ammonia-oxidizing archaeon adapted to low-salinity habitats. Microb. Ecol. 64:955–963. 10.1007/s00248-012-0075-1 [DOI] [PubMed] [Google Scholar]
- 66.Amin US, Lash TD, Wilkinson BJ. 1995. Proline betaine is a highly effective osmoprotectant for Staphylococcus aureus. Arch. Microbiol. 163:138–142. 10.1007/BF00381788 [DOI] [PubMed] [Google Scholar]
- 67.Roth WG, Leckie MP, Dietzler DN. 1985. Osmotic stress drastically inhibits active transport of carbohydrates by Escherichia coli. Biochem. Biophys. Res. Commun. 126:434–441. 10.1016/0006-291X(85)90624-2 [DOI] [PubMed] [Google Scholar]
- 68.Martin DD, Ciulla RA, Roberts MF. 1999. Osmoadaptation in Archaea. Appl. Environ. Microbiol. 65:1815–1825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Park SJ, Kim JG, Jung MY, Kim SJ, Cha IT, Kwon K, Lee JH, Rhee SK. 2012. Draft genome sequence of an ammonia-oxidizing archaeon, “Candidatus Nitrosopumilus koreensis” AR1, from marine sediment. J. Bacteriol. 194:6940–6941. 10.1128/JB.01857-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tourna M, Freitag TE, Nicol GW, Prosser JI. 2008. Growth, activity and temperature responses of ammonia-oxidizing archaea and bacteria in soil microcosms. Environ. Microbiol. 10:1357–1364. 10.1111/j.1462-2920.2007.01563.x [DOI] [PubMed] [Google Scholar]
- 71.Daebeler A, Abell GC, Bodelier PL, Bodrossy L, Frampton DM, Hefting MM, Laanbroek HJ. 2012. Archaeal dominated ammonia-oxidizing communities in Icelandic grassland soils are moderately affected by long-term N fertilization and geothermal heating. Front. Microbiol. 3:352. 10.3389/fmicb.2012.00352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lebedeva EV, Hatzenpichler R, Pelletier E, Schuster N, Hauzmayer S, Bulaev A, Grigor'eva NV, Galushko A, Schmid M, Palatinszky M, Le Paslier D, Daims H, Wagner M. 2013. Enrichment and genome sequence of the group I.1a ammonia-oxidizing archaeon “Ca. Nitrosotenuis uzonensis” representing a clade globally distributed in thermal habitats. PLoS One 8:e80835. 10.1371/journal.pone.0080835 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.