Abstract
The phyllosphere is colonized by complex microbial communities, which are adapted to the harsh habitat. Although the role and ecology of nonpathogenic microorganisms of the phyllosphere are only partially understood, leaf microbiota could have a beneficial role in plant growth and health. Pesticides and biocontrol agents are frequently applied to grapevines, but the impact on nontarget microorganisms of the phyllosphere has been marginally considered. In this study, we investigated the effect of a chemical fungicide (penconazole) and a biological control agent (Lysobacter capsici AZ78) on the leaf microbiota of the grapevine at three locations. Amplicons of the 16S rRNA gene and of the internal transcribed spacer were sequenced for bacterial and fungal identification, respectively. Pyrosequencing analysis revealed that the richness and diversity of bacterial and fungal populations were only minimally affected by the chemical and biological treatments tested, and they mainly differed according to grapevine locations. Indigenous microbial communities of the phyllosphere are adapted to environmental and biotic factors in the areas where the grapevines are grown, and they are resilient to the treatments tested. The biocontrol properties of phyllosphere communities against downy mildew differed among grapevine locations and were not affected by treatments, suggesting that biocontrol communities could be improved with agronomic practices to enrich beneficial populations in vineyards.
INTRODUCTION
Plants support a complex micro-ecosystem, and they host distinct bacterial communities on and inside various plant organs (1). The aerial part of plants (phyllosphere) is normally colonized by a variety of bacteria, filamentous fungi and yeasts (2). Microbial phyllosphere communities are complex and composed by many uncultured microorganisms (2, 3), which are adapted to the harsh environmental conditions (4, 5). In particular, microbial epiphytes of the phyllosphere are exposed to the atmosphere and must deal with direct UV radiation, wide fluctuations in temperature, low water availability, and limited access to nutrients (2, 6). Therefore, the composition of phyllosphere communities could be affected by environmental factors, such as UV radiation, air pollution, and nitrogen fertilization, as well as by biotic factors, such as plant species and invading microorganisms (6–8). Moreover, the phyllosphere is an open system and microbes can invade plant leaves by migration from the atmosphere, soil, other plants, insects, and animals (9).
The phyllosphere has been less intensively studied than the rhizosphere and has received considerable attention in recent years (1). The interest in phyllosphere microbiology was initially driven by investigations into plant pathogens, but most phyllosphere-colonizing microorganisms live as commensals and/or mutualistic symbionts on their host plants (2, 4, 6). Phyllosphere communities are involved in functional processes as large in scale as the carbon cycle, nitrogen fixation, and degradation of organic pollutants, pesticide residues included (2, 4). Phyllosphere communities are also thought to be relevant for plant development and health as biofertilizers, phytostimulators, and biopesticides to protect against invading pathogens (1, 2, 6). As a consequence, it is not surprising that axenic plants are more susceptible to infection (10), suggesting that a barrier effect is conferred by the plant microbiota against pathogens (6). However, the role and ecology of nonpathogenic microorganisms on phyllosphere is only partially understood (2, 11), and many practical applications of leaf microbiota may result from studies on the interaction of microbes with plants and among themselves (4). In particular, better understanding of community structure and multitrophic interactions in the phyllosphere will be the key to develop new strategies for plant protection (6). A prominent area of applied phyllosphere microbiology is the improvement of plant health to increase plant biomass production and reduce pathogen infections by the use of beneficial microbial communities (6).
Chemicals such as fertilizers and pesticides are frequently used in agriculture. Many active molecules have a wide spectrum of activity and might affect nontarget microorganisms within the ecosystem, including those on the plant (12). The impact of chemical treatments has been evaluated on the microbial structure of soil (13, 14) and aquatic ecosystems (15), while only a few studies have addressed the impact of pesticides on nontarget microorganisms of the phyllosphere (9, 11, 12). The use of biological control agents offers a promising alternative or supplement to chemical fungicides for the control of crop diseases. In terms of safety, biocontrol agents should not have any effects on nontarget organisms (16). Successful colonization of the phyllosphere by immigrating microorganisms may be highly dependent on the competitiveness of biocontrol agents, and competitive biocontrol agents might eventually harm not only pathogenic microorganisms but also nontarget organisms (17, 18). However, the nontarget effects of biocontrol agents have mainly been investigated on soil microbial communities (17, 19), and the impact on indigenous leaf microbiota has only recently been analyzed (18, 20, 21). Therefore, chemical and biological treatments could potentially affect taxonomic structures and functional properties of the phyllosphere microbiota, and deeper knowledge of different crops and environmental conditions is required to better understand the impact on indigenous leaf communities. The grapevine (Vitis vinifera L.) is one of the major fruit crops worldwide, and it is susceptible to a large spectrum of pathogens, which are mainly controlled by the frequent use of chemical fungicides. Concerns about the environmental impact of pesticide overuse have sparked increasing interest in alternatives to chemical treatments (22). However, the impact of commercial fungicides and/or promising biocontrol agents on the phyllosphere microbiota has not yet been investigated in grapevine. Phyllosphere communities of the grapevine have been analyzed mainly in terms of bacterial composition in relation to populations of the rhizosphere, bark, and berries (23, 24). The objective of the present study was to investigate the effect of a chemical fungicide based on sterol biosynthesis inhibition (penconazole) and a biocontrol agent, Lysobacter capsici AZ78 (AZ78), on the taxonomic structure and functional properties of indigenous microbial communities on grapevine leaves at three different locations.
MATERIALS AND METHODS
Grapevine treatments and locations.
Grapevine plants were treated with penconazole (Support 10 EC, 10.2% penconazole; Cheminova Agro Italia, Bergamo, Italy) at a concentration of 0.3 ml/liter or with the biocontrol agent AZ78 at a concentration of 106 CFU/ml, while other plants were left untreated. Penconazole is a widely used chemical fungicide, principally adopted against powdery mildews (25, 26), and it has a broad range of activity against ascomycetes and basidiomycetes (27). Penconazole was chosen in our experiments because it is not active against the oomycetes (27); thus, it is not expected to affect the efficacy tests against Plasmopara viticola on leaf disks. AZ78 is a promising biocontrol agent against grapevine downy mildew (28), and it is phylogenetically related to L. capsici YC5194 and L. capsici PG4 (28), which showed broad range of activity against fungi and bacteria (29, 30).
Treatments were applied in a randomized block design with three replicates of four plants for each treatment. Treatments were applied weekly on 29 May and on 5 and 12 June 2012 using a compressed-air hand sprayer (with a volume of 500 liters/ha). In order to control possible downy mildew infection before starting the experiment, phosphites (Fito-fos; Sunchemical, Crespellano, Italy) were applied to all plants at a concentration of 1.5 ml/liter on 8, 15, and 22 May 2012.
Grapevine plants of the Vitis vinifera cultivar Pinot gris were analyzed at three locations in northern Italy. A vineyard located in Udine (UD; latitude, N46.081460; longitude, E13.228974; altitude, 113 m; 23-year-old plants) and a vineyard located in San Michele all'Adige (SM1; latitude: N46.184391, longitude: E11.124499, altitude: 228 m, 11-year-old plants) were used. At a second location in S. Michele all'Adige (SM2; latitude, N46.190723; longitude, E11.135518; altitude, 228 m), we used 2-year-old plants grown in 2.5-liter pots containing a peat and pumice mixture (3:1), protected from rain by a glass roof during the experiment.
Sample collection and isolation of phyllosphere microorganisms.
For each treatment and location, asymptomatic leaves were randomly collected 1 day after the last treatment and three replicates (named from A to C) of 50 leaves were obtained. Leaves were placed in sterile plastic boxes and washed with 1-liter isotonic solution containing 0.01% Tween 80 by manual shaking for 15 min. The leaf-washing suspension was filtered with sterile cheesecloth, and 100 ml of suspension per each replicate were immediately used for the efficacy tests against P. viticola on grapevine leaf discs. The remaining part (900 ml) was centrifuged at 4,000 × g for 20 min at 4°C, the supernatant was discarded, and a pellet of each replicate was stored at −20°C before DNA extraction.
The viability of the phyllosphere microorganisms was determined using the classical plating method. Basically, 1 ml of each leaf-washing suspension was 10-fold serially diluted, and 0.2 ml of each dilution was plated on both nutrient agar (NA), in order to quantify total cultivable bacteria, and potato dextrose agar (PDA) with 0.1 g of chloramphenicol/liter to determine total cultivable fungi and yeasts. Plates were incubated at 25°C, and after 24 and 48 h the CFU/ml of leaf-washing suspension was determined for bacteria and fungi, respectively.
DNA extraction and amplification.
DNA was extracted from the pellets of leaf-washing suspensions using the Meta-G-Nome isolation kit (Epicentre; Illumina) according to the manufacturer's instructions for the isolation of metagenomic DNA from soil.
For identification of bacteria, amplicons of the 16S rRNA gene were obtained by PCR using the fusion primer pair specific for the V5-to-V9 region: 799-forward (31) and 1520-reverse (32). The forward fusion primer contained the Lib-L Primer A sequence specific for the Lib-L chemistry of unidirectional 454 sequencing technology (Roche, Branford, CT), the key sequence TCAG, the barcode multiplex identifier (MID) sequence specific for each DNA sample, and the 799-forward sequence (see Table S1 in the supplemental material). The reverse primer contained the Lib-L Primer B sequence (Roche), the key sequence TCAG, and the 1520-reverse sequence.
For the identification of fungi, amplicons of the internal transcribed spacer (ITS) were obtained by PCR using the fusion primer pairs ITS5-forward and ITS4-reverse (33–35). The forward fusion primer contained the Lib-L Primer A sequence for the Lib-L chemistry of unidirectional 454 sequencing technology (Roche), the key sequence TCAG, the MID sequence specific for each DNA sample, and the ITS5-forward sequence (see Table S1 in the supplemental material). The reverse primer contained the Lib-L Primer B sequence (Roche), the key sequence TCAG, and the ITS4-reverse sequence.
PCR products were generated by amplifying 5 μl of extracted DNA using the FastStart High-Fidelity PCR system (Roche) with 0.25 mM each deoxynucleoside triphosphate (dNTP), 0.5 mg of bovine serum albumin (BSA)/ml, 4% (vol/vol) dimethyl sulfoxide, a 0.3 μM concentration of each primer, and 2.5 U of FastStart High-Fidelity DNA polymerase (Roche). The PCR protocol consisted of denaturation at 95°C for 5 min, followed by 30 cycles at 95°C for 30 s, annealing 1 min at 53 and 51°C for bacteria and fungi, respectively, and extension at 72°C for 2 min, followed by a final extension at 72°C for 10 min.
Library construction and pyrosequencing.
The PCR products were analyzed by gel electrophoresis and cleaned using an AMPure XP bead kit (Beckman Coulter, Brea, CA) according to the manufacturer's instructions. The PCR products of the different 27 DNA samples (three replicates of three treatments at three locations) were quantified with quantitative PCR using a Roche 454 Titanium library quantification kit (KAPA Biosystems, Boston, MA) and pooled in equimolar amounts in a final amplicon library for each specific amplicon (16S and ITS library). 454 pyrosequencing was carried out on a GS FLX+ system (Roche) using XL+ chemistry (Roche) according to the manufacturer's instructions.
Bioinformatics analysis and data processing.
Raw sequences were preprocessed using Mothur (36), and sequence quality was checked using PRINSEQ (37). Sequence analysis was carried out using the quantitative insights into microbial ecology (QIIME) pipeline (38). For sequence filtering, reads shorter than 200 bases or longer than 1,000 bases were discarded, sequences with homopolymer runs longer than six bases or more than six ambiguous bases were also discarded, whereas one barcode correction and two primer mismatches were accepted. Reads with an average of the Phred quality score lower than 25 in a sliding window of 50 bases were discarded. Barcodes and tags were removed and sequences were denoised using the built-in Denoiser algorithm (39). Chimeras were removed using the UCHIME program (40), according to the USEARCH pipeline (41). Specifically, the strategy for the chimera search was both a de novo and reference-based approach for 16S rRNA sequences and a de novo approach for ITS sequences. For the reference-based approach, the ChimeraSlayer database was used as a gold standard for 16S rRNA sequences (42). Operational taxonomic units (OTU) were determined using the UCLUST algorithm (41) at 97% sequence similarity. The taxonomy assignment of the bacterial OTU was carried out using the Naive Bayesian RDP classifier with a minimum confidence of 0.8 (43) against the Greengenes database downloaded from October 2012 (44). Fungal OTU were assigned using BLAST search against the UNITE database downloaded from November 2012 (45), with an E value lower than 0.001. Singleton OTU were removed for statistical analysis. To correct for sampling effort, randomly selected subsets of bacterial and fungal data were obtained using the QIIME pipeline (38), on the basis of the lowest number of 16S and ITS sequences in any sample, and these data were further analyzed.
Assessment of the biocontrol activity of leaf microbial communities against Plasmopara viticola.
For efficacy tests against P. viticola, leaf discs (18 mm in diameter) were obtained from the third and fourth leaves from the top of grapevine shoots (Pinot gris cultivar) grown under controlled greenhouse conditions at 25 ± 1°C with a photoperiod of 16 h of light and a relative humidity of 60% ± 10% for 2 months. To remove leaf microbial communities, the leaf discs were surface sterilized by incubation in 1% hypochlorite for 10 min and then washed four times with sterile water (46). Leaf discs were dried using sterile absorbent paper and transferred (lower surface uppermost) onto sterilized moist filter paper (three foils) in 90-mm petri dishes (five discs for each dish). P. viticola inoculum was grown and propagated on grapevine plants as previously described (47), and sporangia were collected by washing the abaxial surfaces carrying freshly sporulating lesions. The inoculum suspension was adjusted to a concentration of 104 sporangia/ml by counting with a hemocytometer.
To evaluate the efficacy of leaf microbial communities from the different locations and treatments in controlling grapevine downy mildew, each leaf-washing suspension (100 ml) was centrifuged at 4,000 × g for 20 min at 4°C. The pellets were resuspended in 10 ml of 0.01% Tween 80, and 1 ml was then centrifuged at 4,000 × g for 20 min at 4°C. Pellets were resuspended in 1 ml of the P. viticola suspension (104 sporangia/ml), and the pure P. viticola suspension was used as a control.
Leaf discs were inoculated with 40 μl of each suspension, and two petri dishes were analyzed for each leaf-washing suspension. Dishes were incubated in the dark at 25 ± 1°C overnight and then maintained under controlled greenhouse conditions. At 7 days after inoculation, the severity of downy mildew disease was assessed visually as the proportion (percentage from 0 to 100) of abaxial leaf area covered by white sporulation of P. viticola, according to the standard guidelines of the European and Mediterranean Plant Protection Organization (48).
Statistical analysis.
Alpha-diversity metrics were calculated after rarefaction by random subsampling of the OTU table in the QIIME pipeline (38), and an OTU-based analysis was performed to calculate the richness, diversity, evenness, and coverage at 97% sequence similarity. Rarefaction curves were obtained for each sample by random resampling without replacement in Mothur (36) based on the lowest number of 16S and ITS sequences. Sample richness was estimated using the Chao1 estimator (49) and the abundance-based coverage estimator (ACE) (50). The diversity within each sample was estimated using the Shannon diversity index (51), and the Good's coverage (52) was calculated to estimate the percentage of total taxa sequenced in each sample.
The microbial community structure and diversity among samples were calculated using the QIIME pipeline (38) with a Bray-Curtis dissimilarity matrix (53) on jackknifed read abundance data at the deepest level possible. The results were displayed using principal coordinate analysis (PCoA) (54) and plotted with the KiNG software (55). Differences in community structure related to the specific categories were displayed by means of constrained ordination technique using the Vegan R package (56). To detect significant differences among treatments and locations, a PERMANOVA analysis (57) was carried out with the Vegan R package (56) using the ADONIS function on the Bray-Curtis distance matrix for multidimensional scaling ordination.
The data of efficacy tests on leaf discs, OTU, Shannon index, and microbial relative abundance were analyzed using Statistica 9 software (StatSoft, Tulsa, OK). After validation of normal distribution (K-S test, P > 0.05) and variance homogeneity (Levene's test, P > 0.05) of the data, analysis of variance (ANOVA) was carried out using Tukey's test (α = 0.05) to detect significant differences among treatments and locations. CFU data were normalized by log10 transformation, and relative abundance data were normalized by root square transformation. To detect significant differences in microbial abundances among treatments and growth conditions, ANOVA was applied using Tukey's test (α = 0.05) (58, 59). Spearman's rank correlations (P < 0.01) (60, 61) were used to compare relative microbial abundances with the efficacy against downy mildew on leaf discs and with the environmental parameters of rainfall (mm), maximum, minimum, and mean temperatures (°C) at each location.
BioProject accession number.
Pyrosequencing data obtained in this study were submitted to NCBI under BioProject number PRJNA237062.
RESULTS
Identification of leaf microbial communities.
The composition of microbial communities was analyzed on leaves treated with penconazole and AZ78, and on untreated leaves, located in Udine (UD), San Michele all'Adige (SM1), and San Michele all'Adige protected from rain (SM2). The viabilities of cultivable phyllosphere microorganisms were comparable in the cases of untreated, AZ78-treated, and penconazole-treated leaves for each grapevine location (see Fig. S1 in the supplemental material). Bacterial and fungal populations were 2 orders of magnitude greater in UD than in the two SM locations.
After filtering out low-quality reads and short sequences, 30,515 and 134,172 reads of bacteria and fungi were obtained, respectively. The total number of filtered reads for each replicate ranged from 140 to 3,294 for bacteria and from 1,353 to 9,303 for fungi (see Table S2 in the supplemental material). The average sequence lengths of the pyrosequencing data were 750 and 650 bases for bacterial 16S and fungal ITS amplicons, respectively (BioProject accession number PRJNA237062). Good's coverage was used to estimate the completeness of sampling with a probability calculation based on randomly selected sequences, and it ranged from 69.3 to 97.9% for bacterial data and from 98.3 to 99.7% for fungal data (see Table S2 in the supplemental material). Significant (P < 0.01) positive correlations between replicates were obtained (see Table S3 in the supplemental material), and rarefaction curves reached saturation, merging the results of triplicates (see Fig. S2 in the supplemental material). Good's coverage of merged replicates ranged from 93.0 to 98.3% for bacteria and was >99.6% for fungal sampling (Table 1), which means that more than 250 extra reads [1/(1 − 0.996)] would need to be sequenced before detecting a new fungal OTU.
TABLE 1.
Richness and diversity of phyllosphere microbiota based on pyrosequencing of bacterial (16S rRNA gene) and fungal (ITS fragment) communities collected from grapevine leaves
| Leaf community and samplea | Identified OTUb | Good's coverage | Estimated OTU richness |
H′ | |
|---|---|---|---|---|---|
| Chao1 | ACE | ||||
| Bacteria | |||||
| UD-UNT | 106 | 96.9 | 91 | 95 | 4.30 |
| UD-PEN | 78 | 97.0 | 87 | 92 | 4.23 |
| UD-AZ78 | 51 | 98.3 | 70 | 56 | 4.06 |
| SM1-UNT | 201 | 94.1 | 173 | 186 | 6.01 |
| SM1-PEN | 215 | 93.0 | 202 | 202 | 6.17 |
| SM1-AZ78 | 201 | 95.1 | 158 | 162 | 5.96 |
| SM2-UNT | 114 | 94.8 | 159 | 151 | 3.19 |
| SM2-PEN | 87 | 96.9 | 132 | 139 | 2.16 |
| SM2-AZ78 | 49 | 96.5 | 99 | 117 | 3.05 |
| Fungi | |||||
| UD-UNT | 110 | 99.7 | 102 | 99 | 3.42 |
| UD-PEN | 123 | 99.7 | 127 | 122 | 4.51 |
| UD-AZ78 | 121 | 99.7 | 121 | 116 | 3.54 |
| SM1-UNT | 124 | 99.7 | 140 | 121 | 2.46 |
| SM1-PEN | 142 | 99.6 | 139 | 140 | 2.67 |
| SM1-AZ78 | 125 | 99.8 | 117 | 116 | 3.02 |
| SM2-UNT | 135 | 99.7 | 145 | 143 | 5.09 |
| SM2-PEN | 110 | 99.7 | 123 | 127 | 4.62 |
| SM2-AZ78 | 117 | 99.8 | 112 | 113 | 4.75 |
Leaf samples were collected from untreated plants (UNT) or plants treated with penconazole (PEN) or with Lysobacter capsici AZ78 (AZ78) grown in Udine (UD), San Michele all'Adige (SM1), and San Michele all'Adige protected from rain (SM2). Data of three replicates for each sample (reported in Table S2 in the supplemental material) were merged.
Number of operational taxonomic units (OTU) identified with the UCLUST algorithm (41) at 97% sequence similarity.
Richness and diversity of leaf microbial communities.
Chao1 and ACE indexes revealed that more than 50 and 42% of the estimated bacterial richness was covered by the sequencing effort, respectively (Table 1). Specifically, adequate saturation of taxonomic richness was obtained for the leaf communities of untreated plants of UD and for all treatments in SM1. The richness estimators also showed adequate saturation (>87%) of the fungal richness present on grapevine leaves (Table 1).
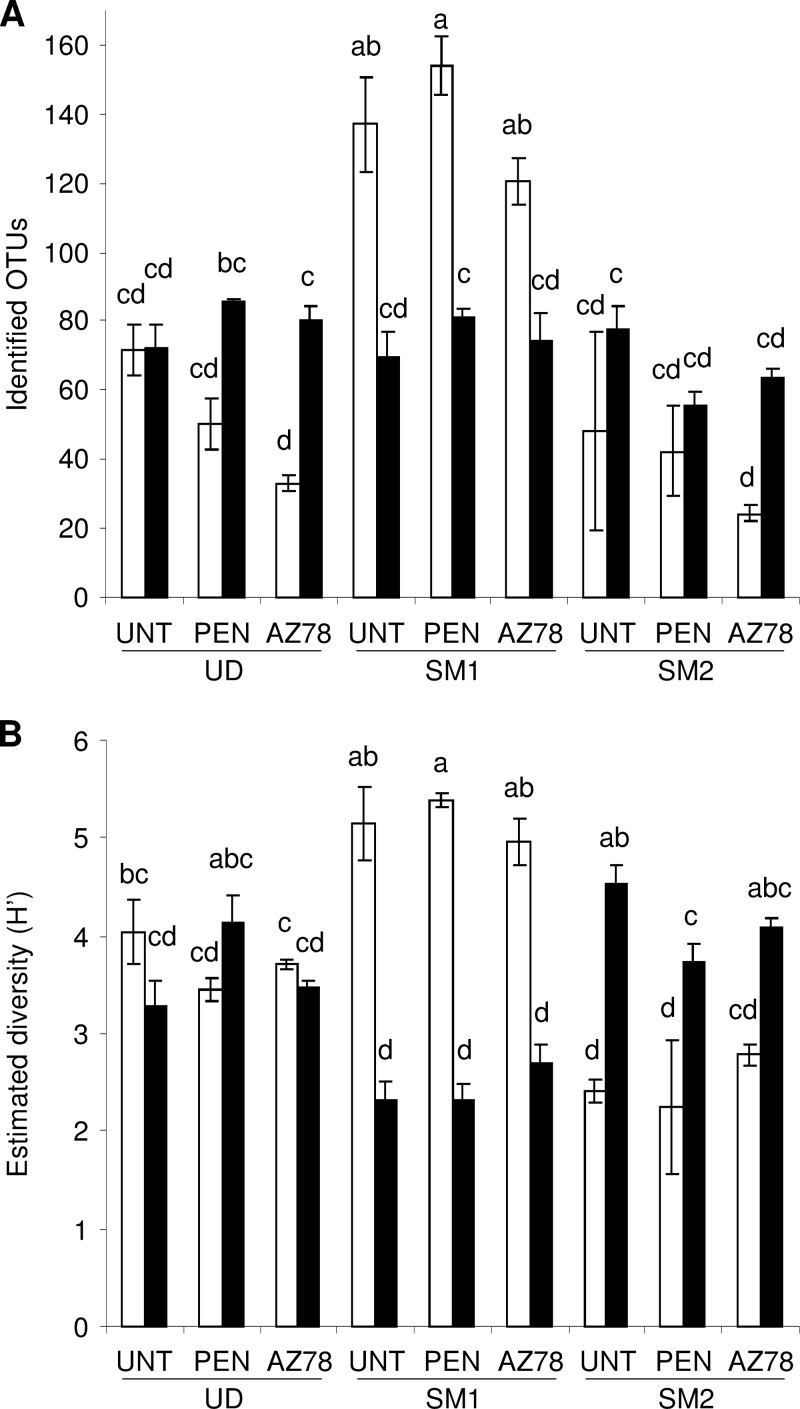
The ecological diversity of microbial communities was estimated using the Shannon index (H′), which ranged from 2.16 to 6.17 for bacteria and from 2.46 to 5.09 for fungi (Table 1). The richness and diversity of bacterial communities was greater in SM1 than in UD and SM2 (Fig. 1). Fungal diversity was lower in SM1 than in SM2. The microbial diversity of penconazole- and AZ78-treated plants was comparable to that of untreated plants at each grapevine location.
FIG 1.
Richness (A) and diversity (B) of microbial populations on grapevine leaves. Operational taxonomic units (OTUs) and Shannon index (H′) were determined for bacteria (white) and fungi (black) identified on the leaves of untreated plants (UNT) or of plants treated with penconazole (PEN) or Lysobacter capsici AZ78 (AZ78) in Udine (UD), San Michele all'Adige (SM1), and San Michele all'Adige protected from rain (SM2). The means and standard errors of three replicates are presented for each sample. Different letters indicate significant differences according to Tukey's test (α = 0.05).
Distribution of bacterial OTU among grapevine treatments and locations.
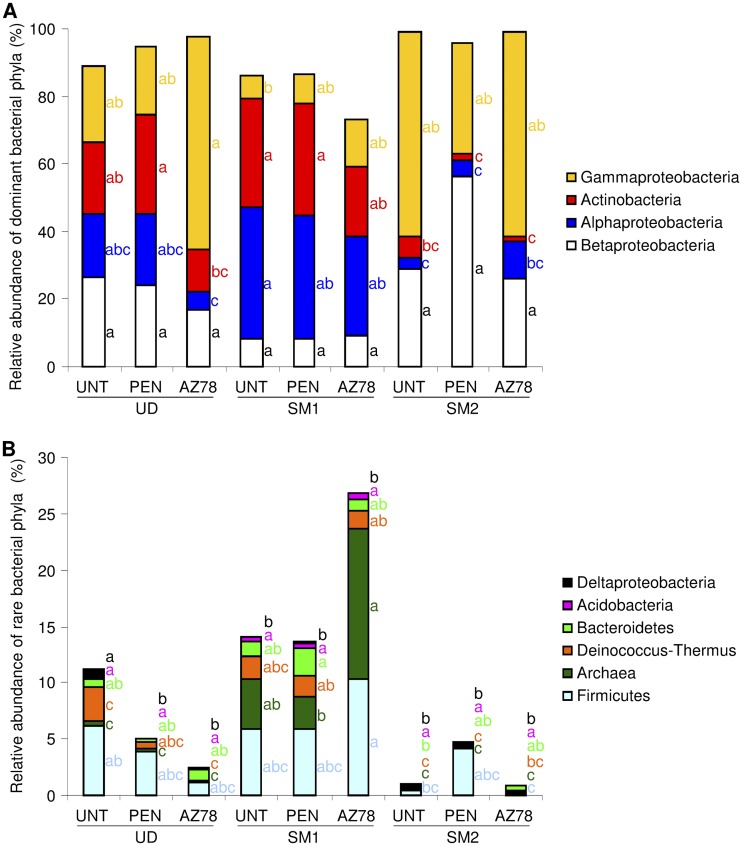
Of all of the bacterial reads, 61.8% were attributed to a bacterial genus (1,312 OTU), while 38.1 and 0.1% were assigned to taxa at family (716 OTU) and order level (15 OTU), respectively (see Table S4 in the supplemental material). Four dominant and six rare phyla were identified (Fig. 2). The proportions of bacterial phyla were not affected by treatments, but they differed according to grapevine locations. Comparing untreated leaves, the relative abundance of Actinobacteria, Alphaproteobacteria, and Archaea was greater in SM1 than in SM2. Moreover, the abundance of Archaea was greater in SM1 than in UD, whereas the abundance of Deltaproteobacteria was greater in UD than in SM1.
FIG 2.
Relative abundance of bacterial phyla and proteobacterial classes on grapevine leaves. Percentages of relative abundance of dominant (A) and rare (B) bacterial phyla and proteobacterial classes were determined for bacteria of leaves of untreated plants (UNT) or of plants treated with penconazole (PEN) or Lysobacter capsici AZ78 (AZ78) in Udine (UD), San Michele all'Adige (SM1) and San Michele all'Adige protected from rain (SM2). The means of three replicates are presented for each sample. For each phylum indicated with the color given in the legend, different letters indicate significant differences according to Tukey's test (α = 0.05).
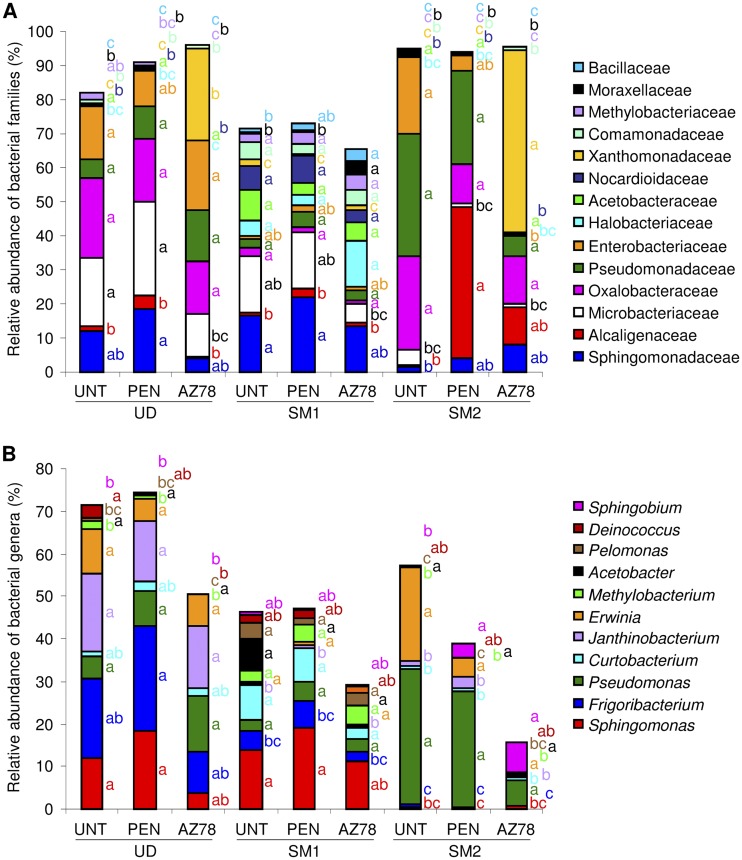
The relative abundance of bacterial families revealed greater diversity in SM1 than in UD and SM2 (Fig. 3A). The proportions of bacterial families were minimally affected by penconazole treatment, which increased only the abundance of Alcaligenaceae on plants of SM2. AZ78 treatment reduced the abundance of Microbacteriaceae and Methylobacteriaceae in UD, increased Moraxellaceae and Bacillaceae in SM1 and reduced Enterobacteriaceae in SM2.
FIG 3.
Relative abundance of dominant bacterial families (A) and genera (B) on grapevine leaves. Percentages of relative abundance of dominant (relative abundance >1%) families and genera were determined for bacteria of leaves of untreated plants (UNT) or of plants treated with penconazole (PEN) or with Lysobacter capsici AZ78 (AZ78) in Udine (UD), San Michele all'Adige (SM1), and San Michele all'Adige protected from rain (SM2). The means of three replicates are presented for each sample. For each taxa indicated with the color given in the legend, different letters indicate significant differences according to Tukey's test (α = 0.05).
The genera Pseudomonas, Erwinia, and Acetobacter were found in all samples, and they showed no significant changes in abundance (Fig. 3B), representing the core bacterial community. Conversely, the distribution of the dominant genera Sphingomonas, Janthinobacterium, Methylobacterium, and Pelomonas varied among the grapevine locations. Penconazole did not affect the abundance of the dominant genera, and AZ78 reduced the relative abundance of Deinococcus in UD and increased Sphingobium in SM2.
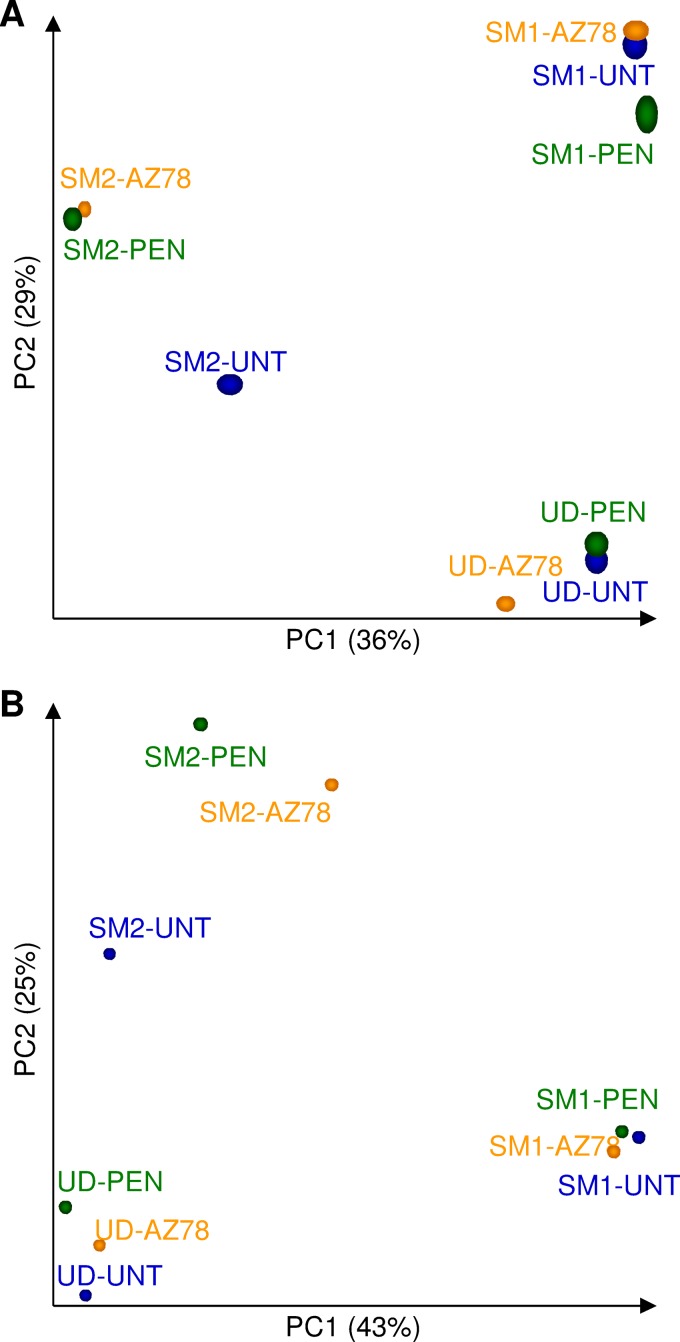
OTUs belonging to the order of Xanthomonadales, which comprises the Lysobacter genus, were removed to avoid bias due to AZ78 sequences (see Table S2 in the supplemental material), and the effect of treatments and locations was examined using principal coordinate analysis (PCoA) (Fig. 4A). Leaf bacterial communities were clustered according to grapevine locations. The first principal component discriminated samples of UD and SM1 vineyards from samples of SM2, whereas the second axis highlighted differences between SM1 and UD. PERMANOVA analysis revealed significant differences in bacterial populations among grapevine locations (P = 0.001) but not among treatments (P = 0.112). However, sample distribution in the PCoA suggested a partial effect of penconazole and AZ78 treatment on SM2 bacterial communities.
FIG 4.
Principal coordinate analysis (PCoA) of bacterial (A) and fungal (B) communities on grapevine leaves. PCoA was obtained with the Vegan R package using a Bray-Curtis dissimilarity matrix on data of untreated plants (UNT; blue) or of plants treated with penconazole (PEN; green) or Lysobacter capsici AZ78 (AZ78; orange) in Udine (UD), San Michele all'Adige (SM1), and San Michele all'Adige protected from rain (SM2). PERMANOVA was performed using ADONIS function and a Bray-Curtis dissimilarity matrix. Significant differences were detected among grapevine locations for both bacterial (P = 0.001) and fungal (P = 0.007) data but not among treatments (P = 0.112 and P = 0.542 for bacteria and fungi, respectively).
Distribution of fungal OTU among grapevine treatments and locations.
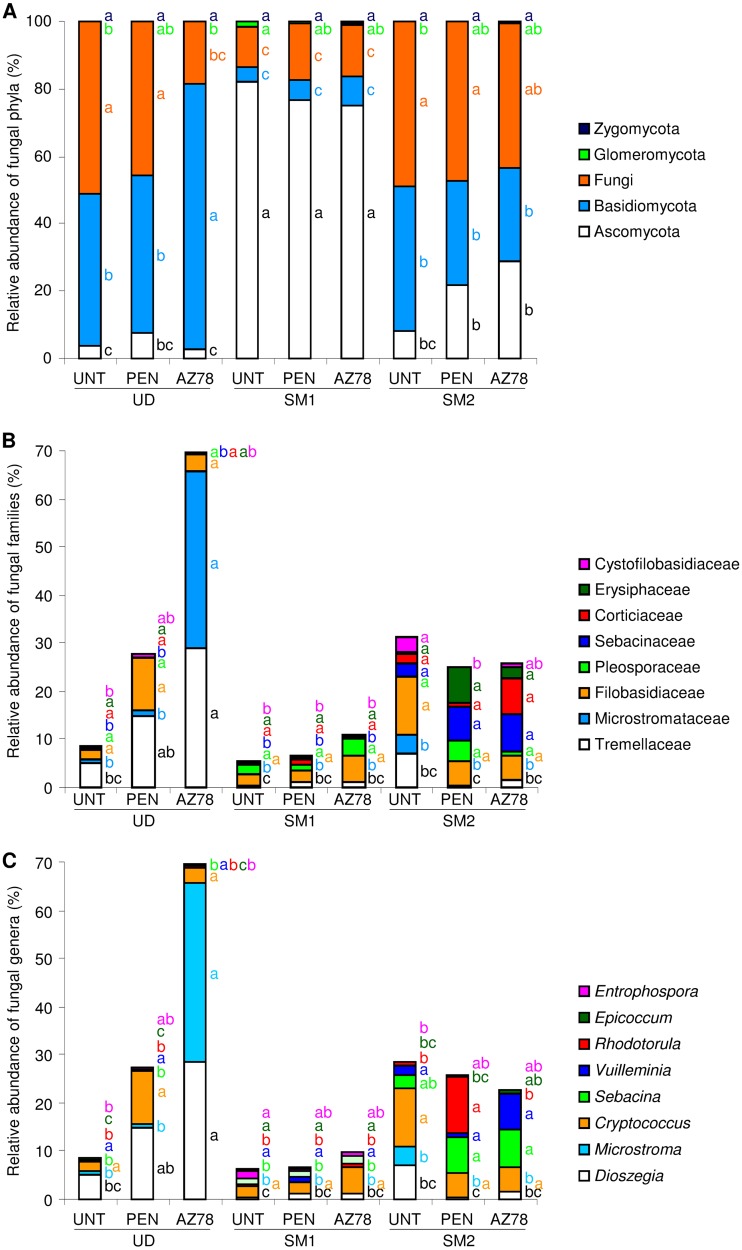
Of fungal reads, 25.1, 23.9, and 26.5% were assigned to taxa at the genus (130 OTU), family (117 OTU), and order (150 OTU) levels, respectively (see Table S5 in the supplemental material). The OTU identified were affiliated with four phyla, and reads attributed to unannotated OTU were assigned to an undefined group, named Fungi (Fig. 5A). The proportions of fungal phyla were not affected by treatments and they differed according to grapevine locations. The relative abundance of ascomycetes was greater in SM1 than in UD and SM2. Conversely, the relative abundances of Basidiomycota and Fungi were greater in UD and SM2 than in SM1. Likewise, the relative abundance of dominant families differed according to grapevine locations (Fig. 5B). In particular, the relative abundances of Sebacinaceae and Cystofilobasidiaceae were greater in SM2 than in UD and SM1. On leaves of UD, AZ78 treatment increased only the abundance of Tremellaceae and Microstromataceae, while penconazole increased the abundance of Tremellaceae. At the level of fungal genera, the abundance of Epicoccum and Entrophospora was greater in SM1 than in UD and SM2 (Fig. 5C). AZ78 treatment increased the abundance of Dioszegia and Microstroma in UD, and penconazole increased the abundance of Rodotolura in SM2.
FIG 5.
Relative abundance of fungal phyla (A), dominant family (B), and dominant genera (C) on grapevine leaves. Fungal operational taxonomic units (OTU) were identified at 97% sequence similarity and grouped according to the taxonomy. Percentages of relative abundance were calculated for leaves of untreated plants (UNT) or of plants treated with penconazole (PEN) or Lysobacter capsici AZ78 (AZ78) in Udine (UD), San Michele all'Adige (SM1), and San Michele all'Adige protected from rain (SM2). Unannotated fungal OTU are grouped in the general category of Fungi (A), and OTU affiliated to undefined or minor families (relative abundance, <0.3% [B]) or to rare genera (relative abundance, <0.3% [C]) are not shown. The means of three replicates are presented for each sample. For each taxa indicated with the color given in the legend, different letters indicate significant differences according to Tukey's test (α = 0.05).
PCoA revealed that fungal communities clustered according to grapevine locations (Fig. 4B). The first component highlighted differences between fungal communities in UD and SM1, while the second axis discriminated samples of SM2 from UD and SM1. Distribution of SM2 samples suggested a partial effect of penconazole and AZ78 treatment on the community structure. However, PERMANOVA analysis revealed significant differences in fungal populations among grapevine locations (P = 0.007) but not among treatments (P = 0.542).
Functional characterization of leaf microbial communities.
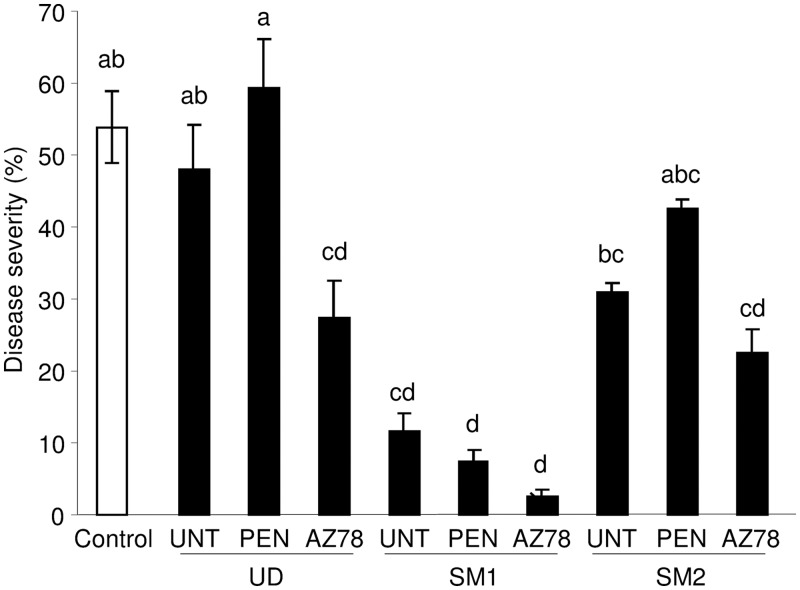
Leaf microbial communities collected from untreated leaves of SM1 significantly reduced downy mildew symptoms on surface-sterilized leaf discs (Fig. 6). The disease severity was not affected by the microbial communities of untreated leaves of UD and SM2. For each location, communities of AZ78-treated leaves partially reduced downy mildew severity compared to communities of untreated leaves, and this effect was significant for UD populations. However, the disease severity of discs treated with microbial communities of penconazole-treated plants was comparable to untreated plants at the same location, indicating that the fungicide did not alter the properties of the indigenous community against the pathogen tested.
FIG 6.
Downy mildew severity of leaf discs treated with microbial communities of grapevine leaves. Surface-sterilized leaf discs were treated with each leaf-washing suspension (10× concentrated) mixed with a P. viticola inoculum suspension (104 sporangia/ml), and the pure P. viticola suspension was used as a control (Control). After incubation under controlled greenhouse conditions, disease severity was assessed as the percentage of leaf area covered by sporulation. Leaf-washing suspensions were obtained from grapevine leaves of untreated plants (UNT) or of plants treated with penconazole (PEN) or Lysobacter capsici AZ78 (AZ78) in Udine (UD), San Michele all'Adige (SM1), and San Michele all'Adige protected from rain (SM2). The mean scores and standard errors of six replicates are presented for each sample. Different letters indicate significant differences according to Tukey's test (α = 0.05).
Correlation analysis of leaf microbiota, grapevine locations, and disease control.
The taxonomic structure of microbial communities were mainly related to the grapevine location, and the relative abundance of bacterial and fungal taxa correlated with the rainfall and temperature (Table 2). Rainfall was positively correlated with the abundance of Alphaproteobacteria, Actinobacteria, and Bacteroidetes, whereas it was negatively correlated with Betaproteobacteria. Maximum and mean temperatures were positively correlated with Ascomycota and Zygomycota and negatively correlated with Basidiomycota. In particular, the bacterial and fungal genera strongly correlated with rainfall were Agrobacterium (r = 0.949), Curtobacterium (r = 0.948), Stenotrophomonas (r = 0.958), Rothia (r = 0.908), Sebacina (r = −0.899), Cystofilobasidium (r = −0.843), and Drechslera (r = 0.845). The maximum and mean temperatures of the grapevine locations correlated with Frigoribacterium (r = −0.822), Janthinobacterium (r = −0.819), Microbacterium (r = 0.825), Kondoa (r = −0.976), Rhodotorula (r = −0.985), and Hannaella (r = −0.905).
TABLE 2.
Correlations among bacterial and fungal OTU identified on grapevine leaves with environmental conditions and functional properties against downy mildewa
| Leaf community and phylum | Correlation of relative abundance with indicated environmental parameter |
Correlation of relative abundance with downy mildew severity | |||
|---|---|---|---|---|---|
| Tmax | Tmin | Tavg | Rainfall | ||
| Bacteria | |||||
| Betaproteobacteria | –0.09 | 0.09 | –0.09 | –0.90 | 0.70 |
| Alphaproteobacteria | 0.09 | –0.09 | 0.09 | 0.90 | –0.50 |
| Actinobacteria | –0.18 | 0.18 | –0.18 | 0.84 | –0.18 |
| Gammaproteobacteria | –0.27 | 0.27 | –0.27 | –0.79 | 0.37 |
| Firmicutes | 0.00 | 0.00 | 0.00 | 0.74 | –0.30 |
| Archaea | 0.37 | –0.37 | 0.37 | 0.74 | –0.55 |
| Deinococcus-Thermus | 0.00 | 0.00 | 0.00 | 0.63 | –0.18 |
| Bacteroidetes | 0.00 | 0.00 | 0.00 | 0.84 | –0.67 |
| Acidobacteria | 0.41 | –0.41 | 0.41 | 0.72 | –0.79 |
| Deltaproteobacteria | –0.25 | 0.25 | –0.25 | 0.29 | 0.07 |
| Fungi | |||||
| Ascomycota | 0.82 | –0.82 | 0.82 | 0.47 | –0.73 |
| Basidiomycota | –0.82 | 0.82 | –0.82 | –0.47 | 0.75 |
| Fungi | –0.37 | 0.37 | –0.37 | –0.74 | 0.82 |
| Glomeromycota | 0.65 | –0.65 | 0.65 | 0.59 | –0.73 |
| Zygomycota | 0.82 | –0.82 | 0.82 | 0.05 | –0.75 |
Spearman's rank correlation coefficients are based on the relative abundances of bacterial and fungal OTU identified on grapevine leaves and the effect of microbial communities against downy mildew (Fig. 6) and the environmental parameters of maximum temperature (Tmax), minimum temperature (Tmin), mean temperature (Tavg), and rainfall (mm). Significant (P < 0.01) correlations are indicated in boldface. Maximum temperatures (Tmax): 28.5°C at Udine (UD), 31.5°C at San Michele (SM1), and 31.5°C at San Michele protected from rain (SM2). Minimum temperatures (Tmin): 12.1°C at UD, 9.7°C at SM1, and 9.7°C at SM2. Mean temperatures (Tavg): 20.0°C at UD, 21.1°C at SM1, and 21.1°C at SM2. Rainfall: 49.2 mm at UD, 56.6 mm at SM1, and 0 mm at SM2.
The relative abundance of bacterial and fungal genera related to the biocontrol of downy mildew differed among grapevine locations (Table 3). Greater abundance of Bacillus was found in SM1 than in SM2 and UD. Conversely, the highest proportion of Alternaria was found on untreated leaves of UD. Microbial communities included genera negatively correlated with the disease severity on leaf discs, indicating possible biocontrol properties of some strains against downy mildew. In particular, negative correlations were found for bacteria of Haemophilus (r = −0.842), Swaminathania (r = −0.814), Paracoccus (r = −0.817), Roseomonas (r = −0.842), Kineosporia (r = −0.822) and Porphyromonas (r = −0.851), and fungi of Epicoccum (r = −0.929), Teratosphaeria (r = −0.888), Exophiala (r = −0.858), Claviceps (r = −0.913), and Chalastospora (r = −0.858).
TABLE 3.
Relative abundance of bacterial and fungal genera which comprise known biocontrol agents against grapevine downy mildew
| Genusa | Mean relative abundance (%) of genera ± SEb |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| UD-UNT | UD-PEN | UD-AZ78 | SM1-UNT | SM1-PEN | SM1-AZ78 | SM2-UNT | SM2-PEN | SM2-AZ78 | |
| Erwinia | 10.4 ± 0.2ab | 5.5 ± 0.9ab | 7.8 ± 2.4ab | 0.5 ± 0.1ab | 1.0 ± 0.5ab | 0.3 ± 0.2b | 22.3 ± 6.2a | 4.6 ± 3.7a | 0.2 ± 0.2b |
| Bacillus | 0 ± 0c | 0 ± 0c | 0 ± 0c | 0.8 ± 0.4ab | 1.1 ± 0.5a | 2.5 ± 0.6a | 0 ± 0c | 0.1 ± 0c | 0 ± 0c |
| Pseudomonas | 5.6 ± 2a | 8.2 ± 1.0a | 13.0 ± 2.6a | 2.7 ± 1.2a | 4.5 ± 3.4a | 3.2 ± 1.0a | 31.9 ± 2.8a | 27.2 ± 2.5a | 5.9 ± 0.5a |
| Stenotrophomonas | 0.3 ± 0.3a | 0.2 ± 0.1a | 0.6 ± 0.3a | 1.2 ± 0.4a | 0.6 ± 0.1a | 1.0 ± 0.5a | 0 ± 0a | 0 ± 0a | 0 ± 0a |
| Aureobasidium | 0.1 ± 0a | 0.1 ± 0.1a | 0.2 ± 0.1a | 0.1 ± 0a | 0.1 ± 0.1a | 0.3 ± 0.2a | 0.2 ± 0.1a | 0 ± 0a | 0 ± 0a |
| Fusarium | 0 ± 0a | 0 ± 0a | 0 ± 0a | 0 ± 0a | 0 ± 0a | 0 ± 0a | 0 ± 0a | 0 ± 0a | 0 ± 0a |
| Alternaria | 0.3 ± 0.1a | 0 ± 0b | 0 ± 0b | 0 ± 0b | 0 ± 0b | 0 ± 0b | 0 ± 0b | 0 ± 0b | 0 ± 0b |
Genera comprising known biocontrol agents against grapevine downy mildew: i.e., Erwinia herbicola, Bacillus spp., Pseudomonas spp., Stenotrophomonas maltophilia (22), Aureobasidium pullulans (75), Fusarium proliferatum (76), and Alternaria alternata (77).
The relative abundance (%) of genera with possible biocontrol properties was determined at 97% sequence similarity for bacteria on the leaves of untreated plants (UNT) or of plants treated with penconazole (PEN) or with Lysobacter capsici AZ78 (AZ78) in Udine (UD), San Michele all'Adige (SM1), and San Michele all'Adige protected from rain (SM2). The mean values and standard errors (SEs) of three replicates are presented for each sample. For each genus, different superscript letters indicate significant differences according to Tukey's test (α = 0.05).
DISCUSSION
The structure and dynamics of indigenous bacterial and fungal populations of the phyllosphere are important aspects, due to significant effects on the promotion of plant growth and protection against pathogens (5, 6). As regards the phyllosphere, it is well known that environmental factors, including radiation, pollution, and nitrogen fertilization, as well as biotic factors such as plant species and the presence of other microorganisms, affect the microbial community structure (6, 8, 62). However, little is known about the effect of plant protection strategies on the equilibrium of indigenous microbial communities on the leaves of crops. We investigated here the impact of treatments with a biocontrol agent and a commercial fungicide on the phyllosphere microbiota of the grapevine at three different locations, and we analyzed bacterial and fungal populations in terms of taxonomic structure and functional properties against downy mildew.
Bacteria belonging to the Betaproteobacteria, Alphaproteobacteria, Actinobacteria, and Gammaproteobacteria phyla predominated on untreated grapevine leaves, accordingly to previous findings (24, 63). Ascomycota and Basidiomycota were the most abundant fungal phyla, and a substantial part of sequenced reads was assigned to unannotated fungal OTU, which possibly represent environmental sequences of uncultured fungi. The dominant bacterial genera were Sphingomonas, Frigoribacterium, Pseudomonas, and Curtobacterium, as previously reported for grapevine leaves (23, 24, 63). Sphingomonas and Pseudomonas are widespread on plant leaves (64, 65), while Curtobacterium are commonly found in soil, and they possibly spread by aerial migration (24).
According to the PERMANOVA analysis, treatments with the biocontrol agent and the chemical fungicide did not affect the structure of indigenous bacterial and fungal communities on grapevine leaves, demonstrating the resilience of indigenous communities to the treatments tested. However, the distribution of bacterial and fungal data in the PCoA suggested a partial effect of the biological and chemical treatments on communities of plants protected from rain (SM2). In particular, AZ78 treatment decreased the abundance of the Enterobacteriaceae family and increased the abundance of the Sphingobium genus on plants of SM2. Negligible changes in microbial composition have been also observed after the introduction of fungal and bacterial biocontrol agents on strawberry leaves in field conditions (18, 21). Likewise, treatments with Bacillus thuringiensis and a parathyroid insecticide only minimally altered the leaf microflora of Brassica oleracea in field conditions (66). However, significant changes in leaf communities have been shown for greenhouse-grown pepper after B. thuringiensis treatment (20). Thus, indigenous communities of the phyllosphere in fields seem to be more stable than those on leaves under controlled conditions, suggesting that exogenous microbes should compete strongly with indigenous communities to occupy leaf niches. The biocontrol agents introduced could compete with indigenous microorganisms for nutrients and space (21, 67), and successful colonization could be affected by the resilience of the leaf community. AZ78 treatment increased the abundance of Xanthomonadaceae (which comprises AZ78 reads) on the leaves of plants of SM2, but not in the vineyard in a very closed location (SM1), suggesting a different level of AZ78 immigration based on pyrosequencing analysis. Leaf communities of the SM1 vineyard showed high bacterial diversity and high biocontrol properties to downy mildew, suggesting greater stability and antagonistic properties to exogenous microbes. However, further investigations on the interaction between exogenous and indigenous microorganisms in the phyllosphere are required to better evaluate the proper establishment of the biocontrol agent AZ78 on above-ground plant parts.
The chemical fungicide partially affected fungal (Rhodotorula and Cystofilobasidium) and bacterial (Alcaligenaceae family) communities on plants of SM2. Penconazole treatment reduced the abundance of Deltaproteobacteria of UD populations, but did not significantly modify the structure of SM1 populations. Chemical treatments used in the grapevine protection program affected the microbial populations on and inside plant leaves during the growing season, and modified the balance between pathogenic and beneficial microorganisms (63). Likewise, the fungicides enostroburin and metalaxyl caused changes in the bacterial communities of the wheat and pepper phyllosphere (9, 11), as well as the insecticides cypermethrin, abamectin, and imidacloprid in the communities of the pepper, cucumber, and broccoli phyllosphere (11, 12). However, the impacts of chemical and biological treatments on the phyllosphere microbiota are dependent on the dosage and frequency of applications and the mechanisms of action of the products tested, as well as the types of indigenous microorganisms and the weather conditions.
Under the conditions tested, the phyllosphere microbiota of the grapevine was mainly influenced by the field site rather than by plant treatments, as shown for indigenous populations of the lettuce rhizosphere and endophyllosphere (68). Vineyards of UD and SM1 differed significantly in terms of the abundance of Archaea, Deltaproteobacteria, Ascomycota, and Basidiomycota. Moreover, the proportions of Actinobacteria and Alphaproteobacteria were lower on plants of SM2 than in vineyards, whereas the proportion of Cystofilobasidiaceae and Sebacinaceae showed the opposite pattern. The abundance of these bacterial and fungal taxa correlated significantly with the environmental parameters of grapevine locations, such as temperature and the amount of rainfall. However, plant age and soil were different in the locations tested, suggesting that leaf communities could be influenced by other biotic and abiotic factors of the areas where plants are grown.
The benefits provided by the plant include a supply of nutrients, but the advantages provided by phyllosphere inhabitants to their host plants are not necessarily as apparent (6). Epiphytes are involved in processes affecting plant growth and health (4). Because of the importance of phyllosphere microbial inhabitants for the plant, there are likely to be many practical applications from a better understanding of the interaction of microbes with plants and among themselves (4). In the human gut, many community members that were previously considered commensals are now regarded as beneficial symbionts because of their contribution to the host metabolism and immunity (69). The barrier effect against pathogens conferred by human gut microbiota (70) might similarly affect the outcome of plant-pathogen interactions in the phyllosphere (6). Microbial populations of untreated plants of SM1 reduced downy mildew severity on sterilized leaf discs, whereas populations of UD and SM2 did not. In particular, the relative abundances of indigenous bacteria (Haemophilus, Swaminathania, Paracoccus, Roseomonas, Kineosporia, and Porphyromonas) and fungi (Epicoccum, Teratosphaeria, Exophiala, Claviceps, and Chalastospora) was negatively correlated with downy mildew severity, suggesting that some strains of these genera could have a role as biocontrol agents. Indigenous microbes also play a key role in fruit quality, yield, and the flavors produced during alcoholic fermentation (71–73). Thus, beneficial communities could be seen as potential plant probiotic agents (74), which could defend the host, promote its growth, and improve fruit quality. In agreement with pyrosequencing results, functional analysis of leaf communities against downy mildew revealed the resilience of the indigenous communities and only limited perturbation after treatment with the fungicide or the biocontrol agent. Further system-level analysis of the complex interaction that governs outcomes among community members in the context of the plant host is required, in order to identify beneficial interaction and selection processes for beneficial communities in specific environmental conditions and pathogen pressures. In the present study, we attempted to link functional traits (biocontrol against P. viticola) of phyllosphere communities with pyrosequencing analysis of these populations, and we identified beneficial microbial communities, which could represent a new tool for crop protection. In particular, natural microbial communities with beneficial properties could be manipulated with agronomic practices to restore the beneficial microbiota for plant defense.
Supplementary Material
ACKNOWLEDGMENTS
This research was supported by the Envirochange project funded by the autonomous province of Trentino and the EU project CO-FREE (theme KBBE.2011.1.2-06, grant agreement 289497). M.P. was supported by the Mecagrafic project funded by the autonomous province of Trentino.
We thank Carmela Sicher and Denise Ress for technical support, Andrea Campisano for advice on the fusion primer of pyrosequencing, Andrea Cattani for informatics support, and the technical staff of the FEM Sequencing Platform.
Footnotes
Published ahead of print 28 March 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.00415-14.
REFERENCES
- 1.Bulgarelli D, Schlaeppi K, Spaepen S, van Themaat EVL, Schulze-Lefert P. 2013. Structure and functions of the bacterial microbiota of plants. Annu. Rev. Plant Biol. 64:807–838. 10.1146/annurev-arplant-050312-120106 [DOI] [PubMed] [Google Scholar]
- 2.Müller T, Ruppel S. 2014. Progress in cultivation-independent phyllosphere microbiology. FEMS Microbiol. Ecol. 87:2–17. 10.1111/1574-6941.12198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang C-H, Crowley DE, Borneman J, Keen NT. 2001. Microbial phyllosphere populations are more complex than previously realized. Proc. Natl. Acad. Sci. U. S. A. 98:3889–3894. 10.1073/pnas.051633898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lindow SE, Brandl MT. 2003. Microbiology of the phyllosphere. Appl. Environ. Microbiol. 69:1875–1883. 10.1128/AEM.69.4.1875-1883.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rastogi G, Coaker GL, Leveau JHJ. 2013. New insights into the structure and function of phyllosphere microbiota through high-throughput molecular approaches. FEMS Microbiol. Lett. 348:1–10. 10.1111/1574-6968.12225 [DOI] [PubMed] [Google Scholar]
- 6.Vorholt JA. 2012. Microbial life in the phyllosphere. Nat. Rev. Microbiol. 10:828–840. 10.1038/nrmicro2910 [DOI] [PubMed] [Google Scholar]
- 7.Whipps JM, Hand P, Pink DAC, Bending GD. 2008. Human pathogens and the phyllosphere. Adv. Appl. Microbiol. 64:183–221. 10.1016/S0065-2164(08)00407-3 [DOI] [PubMed] [Google Scholar]
- 8.Williams TR, Moyne AL, Harris LJ, Marco ML. 2013. Season, irrigation, leaf age, and Escherichia coli inoculation influence the bacterial diversity in the lettuce phyllosphere. PLoS One 8:e68642. 10.1371/journal.pone.0068642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gu L, Bai Z, Jin B, Hu Q, Wang H, Zhuang G, Zhang H. 2010. Assessing the impact of fungicide enostroburin application on bacterial community in wheat phyllosphere. J. Environ. Sci. 22:134–141. 10.1016/S1001-0742(09)60084-X [DOI] [PubMed] [Google Scholar]
- 10.Innerebner G, Knief C, Vorholt JA. 2011. Protection of Arabidopsis thaliana against leaf-pathogenic Pseudomonas syringae by Sphingomonas strains in a controlled model system. Appl. Environ. Microbiol. 77:3202–3210. 10.1128/AEM.00133-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moulas C, Petsoulas C, Rousidou K, Perruchon C, Karas P, Karpouzas DG. 2013. Effects of systemic pesticides imidacloprid and metalaxyl on the phyllosphere of pepper plants. Biomed. Res. Int. 10.1155/2013/969750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang B, Bai Z, Hoefel D, Tang L, Wang X, Li B, Li Z, Zhuang G. 2009. The impacts of cypermethrin pesticide application on the non-target microbial community of the pepper plant phyllosphere. Sci. Total Environ. 407:1915–1922. 10.1016/j.scitotenv.2008.11.049 [DOI] [PubMed] [Google Scholar]
- 13.Johnsen K, Jacobsen C, Torsvik V, Sørensen J. 2001. Pesticide effects on bacterial diversity in agricultural soils: a review. Biol. Fertil. Soils 33:443–453. 10.1007/s003740100351 [DOI] [Google Scholar]
- 14.Imfeld G, Vuilleumier S. 2012. Measuring the effects of pesticides on bacterial communities in soil: a critical review. Eur. J. Soil Biol. 49:22–30. 10.1016/j.ejsobi.2011.11.010 [DOI] [Google Scholar]
- 15.DeLorenzo ME, Scott GI, Ross PE. 2001. Toxicity of pesticides to aquatic microorganisms: a review. Environ. Toxicol. Chem. 20:84–98. 10.1002/etc.5620200108 [DOI] [PubMed] [Google Scholar]
- 16.Cook RJ, Bruckart WL, Coulson JR, Goettel MS, Humber RA, Lumsden RD, Maddox JV, McManus ML, Moore L, Meyer SF, Quimby JPC, Stack JP, Vaughn JL. 1996. Safety of microorganisms intended for pest and plant disease control: a framework for scientific evaluation. Biol. Control 7:333–351. 10.1006/bcon.1996.0102 [DOI] [Google Scholar]
- 17.Brimner TA, Boland GJ. 2003. A review of the non-target effects of fungi used to biologically control plant diseases. Agric. Ecosyst. Environ. 100:3–16. 10.1016/S0167-8809(03)00200-7 [DOI] [Google Scholar]
- 18.Sylla J, Alsanius BW, Kruger E, Reineke A, Strohmeier S, Wohanka W. 2013. Leaf microbiota of strawberries as affected by biological control agents. Phytopathology 103:1001–1011. 10.1094/PHYTO-01-13-0014-R [DOI] [PubMed] [Google Scholar]
- 19.Scheepmaker JWA, van de Kassteele J. 2011. Effects of chemical control agents and microbial biocontrol agents on numbers of non-target microbial soil organisms: a meta-analysis. Biocontrol Sci. Technol. 21:1225–1242. 10.1080/09583157.2011.594952 [DOI] [Google Scholar]
- 20.Zhang B, Bai Z, Hoefel D, Tang L, Yang Z, Zhuang G, Yang J, Zhang H. 2008. Assessing the impact of the biological control agent Bacillus thuringiensis on the indigenous microbial community within the pepper plant phyllosphere. FEMS Microbiol. Lett. 284:102–108. 10.1111/j.1574-6968.2008.01178.x [DOI] [PubMed] [Google Scholar]
- 21.Sylla J, Alsanius B, Krüger E, Reineke A, Bischoff-Schaefer M, Wohanka W. 2013. Introduction of Aureobasidium pullulans to the phyllosphere of organically grown strawberries with focus on its establishment and interactions with the resident microbiome. Agronomy 3:704–731. 10.3390/agronomy3040704 [DOI] [Google Scholar]
- 22.Gessler C, Pertot I, Perazzolli M. 2011. Plasmopara viticola: a review of knowledge on downy mildew of grapevine and effective disease management. Phytopathol. Mediterr. 50:3–44 http://www.fupress.net/index.php/pm/article/view/9360 [Google Scholar]
- 23.Leveau JHJ, Tech JJ. 2011. Grapevine microbiomics: bacterial diversity on grape leaves and berries revealed by high-throughput sequence analysis of 16S rRNA amplicons. Acta Hortic. 905:31–42 [Google Scholar]
- 24.Martins G, Lauga B, Miot-Sertier C, Mercier A, Lonvaud A, Soulas ML, Soulas G, Masneuf-Pomarede I. 2013. Characterization of epiphytic bacterial communities from grapes, leaves, bark and soil of grapevine plants grown, and their relations. PLoS One 8:e73013. 10.1371/journal.pone.0073013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oliva J, Navarro S, Navarro G, Cámara MA, Barba A. 1999. Integrated control of grape berry moth (Lobesia botrana), powdery mildew (Uncinula necator), downy mildew (Plasmopara viticola) and grapevine sour rot (Acetobacter spp.). Crop Prot. 18:581–587. 10.1016/S0261-2194(99)00064-2 [DOI] [Google Scholar]
- 26.Čuš F, Česnik HB, Bolta ŠV, Gregorčič A. 2010. Pesticide residues in grapes and during vinification process. Food Control 21:1512–1518. 10.1016/j.foodcont.2010.04.024 [DOI] [Google Scholar]
- 27.Worthington P. 2012. Sterol biosynthesis inhibiting triazole fungicides, p 129–145 In Lamberth C, Dinges J. (ed), Bioactive heterocyclic compound classes. Wiley-VCH Verlag GmbH, Berlin, Germany [Google Scholar]
- 28.Puopolo G, Giovannini O, Pertot I. 2013. Lysobacter capsici AZ78 can be combined with copper to effectively control Plasmopara viticola on grapevine. Microbiol. Res. 10.1016/j.micres.2013.09.013 [DOI] [PubMed] [Google Scholar]
- 29.Park JH, Kim R, Aslam Z, Jeon CO, Chung YR. 2008. Lysobacter capsici sp. nov., with antimicrobial activity, isolated from the rhizosphere of pepper, and emended description of the genus Lysobacter. Int. J. Syst. Evol. Microbiol. 58:387–392. 10.1099/ijs.0.65290-0 [DOI] [PubMed] [Google Scholar]
- 30.Puopolo G, Raio A, Zoina A. 2010. Identification and characterization of Lysobacter capsici strain PG4: a new plant health-promoting rhizobacterium. J. Plant Pathol. 92:157–164 http://sipav.org/main/jpp/index.php/jpp/article/view/25 [Google Scholar]
- 31.Chelius MK, Triplett EW. 2001. The diversity of archaea and bacteria in association with the roots of Zea mays L. Microb. Ecol. 41:252–263 [DOI] [PubMed] [Google Scholar]
- 32.Massol-Deya AA, Odelson DA, Hickey RF, Tiedje JM. 1995. Bacterial community fingerprinting of amplified 16S and 16-23S ribosomal DNA gene sequences and restriction endonuclease analysis (ARDRA), p 289–296 In Akkermans AL, Elsas J, Bruijn F. (ed), Molecular microbial ecology manual. Springer, Dordrecht, Netherlands [Google Scholar]
- 33.White T, Bruns T, Lee S, Taylor J. 1990. Amplification and direct sequencing of fungal rRNA genes for phylogenetics, p 315–322 In Innis M, Gelfand D, Shinsky J, White T. (ed), PCR protocols: a guide to methods and applications. Academic Press, Inc, New York, NY [Google Scholar]
- 34.Tedersoo L, Nilsson RH, Abarenkov K, Jairus T, Sadam A, Saar I, Bahram M, Bechem E, Chuyong G, Kõljalg U. 2010. 454 pyrosequencing and Sanger sequencing of tropical mycorrhizal fungi provide similar results but reveal substantial methodological biases. New Phytol. 188:291–301. 10.1111/j.1469-8137.2010.03373.x [DOI] [PubMed] [Google Scholar]
- 35.Toju H, Tanabe AS, Yamamoto S, Sato H. 2012. High-coverage ITS primers for the DNA-based identification of ascomycetes and basidiomycetes in environmental samples. PLoS One 7:e40863. 10.1371/journal.pone.0040863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF. 2009. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. 75:7537–7541. 10.1128/AEM.01541-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schmieder R, Edwards R. 2011. Quality control and preprocessing of metagenomic datasets. Bioinformatics 27:863–864. 10.1093/bioinformatics/btr026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7:335–336. 10.1038/nmeth.f.303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reeder J, Knight R. 2010. Rapid denoising of pyrosequencing amplicon data: exploiting the rank-abundance distribution. Nat. Methods 7:668–669. 10.1038/nmeth0910-668a [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. 2011. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27:2194–2200. 10.1093/bioinformatics/btr381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Edgar RC. 2010. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461. 10.1093/bioinformatics/btq461 [DOI] [PubMed] [Google Scholar]
- 42.Haas BJ, Gevers D, Earl AM, Feldgarden M, Ward DV, Giannoukos G, Ciulla D, Tabbaa D, Highlander SK, Sodergren E, Methe B, DeSantis TZ, Petrosino JF, Knight R, Birren BW. 2011. Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res. 21:494–504. 10.1101/gr.112730.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang Q, Garrity GM, Tiedje JM, Cole JR. 2007. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73:5261–5267. 10.1128/AEM.00062-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McDonald D, Price MN, Goodrich J, Nawrocki EP, DeSantis TZ, Probst A, Andersen GL, Knight R, Hugenholtz P. 2012. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 6:610–618. 10.1038/ismej.2011.139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Abarenkov K, Nilsson RH, Larsson K-H, Alexander IJ, Eberhardt U, Erland S, Høiland K, Kjøller R, Larsson E, Pennanen T, Sen R, Taylor AFS, Tedersoo L, Ursing BM, Vrålstad T, Liimatainen K, Peintner U, Kõljalg U. 2010. The UNITE database for molecular identification of fungi: recent updates and future perspectives. New Phytol. 186:281–285. 10.1111/j.1469-8137.2009.03160.x [DOI] [PubMed] [Google Scholar]
- 46.Palmieri MC, Perazzolli M, Matafora V, Moretto M, Bachi A, Pertot I. 2012. Proteomic analysis of grapevine resistance induced by Trichoderma harzianum T39 reveals specific defence pathways activated against downy mildew. J. Exp. Bot. 63:6237–6251. 10.1093/jxb/ers279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Perazzolli M, Moretto M, Fontana P, Ferrarini A, Velasco R, Moser C, Delledonne M, Pertot I. 2012. Downy mildew resistance induced by Trichoderma harzianum T39 in susceptible grapevines partially mimics transcriptional changes of resistant genotypes. BMC Genomics 13:660. 10.1186/1471-2164-13-660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.European and Mediterranean Plant Protection Organization. 2001. Guidelines for the efficacy evaluation of fungicides: Plasmopara viticola. EPPO Bull. 31:313–317. 10.1111/j.1365-2338.2001.tb01000.x [DOI] [Google Scholar]
- 49.Chao A. 1984. Nonparametric estimation of the number of classes in a population. Scand. J. Stat. 11:265–270 [Google Scholar]
- 50.Chazdon RL, Colwell RK, Denslow JS, Guariguata MR. 1998. Statistical methods for estimating species richness of woody regeneration in primary and secondary rain forests of northeastern Costa Rica, p 285–309 In Dallmeier F, Comiskey JA. (ed), Forest biodiversity research, monitoring and modeling: conceptual background and Old World case studies. Parthenon Publishing, Paris, France [Google Scholar]
- 51.Shannon CE. 1948. A mathematical theory of communication. Bell Syst. Tech. J. 27:379–423. 10.1002/j.1538-7305.1948.tb01338.x [DOI] [Google Scholar]
- 52.Good IJ. 1953. The population frequencies of species and the estimation of population parameters. Biometrika 40:237–264. 10.1093/biomet/40.3-4.237 [DOI] [Google Scholar]
- 53.Bray JR, Curtis JT. 1957. An ordination of the upland forest communities of southern Wisconsin. Ecol. Monogr. 27:325–349. 10.2307/1942268 [DOI] [Google Scholar]
- 54.Gower JC, Blasius J. 2005. Multivariate prediction with nonlinear principal components analysis: theory. Qual. Quant. 39:359–372. 10.1007/s11135-005-3005-1 [DOI] [Google Scholar]
- 55.Chen VB, Davis IW, Richardson DC. 2009. KING (Kinemage, Next Generation): a versatile interactive molecular and scientific visualization program. Protein Sci. 18:2403–2409. 10.1002/pro.250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dixon P. 2003. VEGAN, a package of R functions for community ecology. J. Veg. Sci. 14:927–930. 10.1111/j.1654-1103.2003.tb02228.x [DOI] [Google Scholar]
- 57.Anderson MJ. 2001. A new method for non-parametric multivariate analysis of variance. Austral Ecol. 26:32–46. 10.1111/j.1442-9993.2001.01070.pp.x [DOI] [Google Scholar]
- 58.Nacke H, Thurmer A, Wollherr A, Will C, Hodac L, Herold N, Schoning I, Schrumpf M, Daniel R. 2011. Pyrosequencing-based assessment of bacterial community structure along different management types in German forest and grassland soils. PLoS One 6:e17000. 10.1371/journal.pone.0017000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bodenhausen N, Horton MW, Bergelson J. 2013. Bacterial communities associated with the leaves and the roots of Arabidopsis thaliana. PLoS One 8:e56329. 10.1371/journal.pone.0056329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jones RT, Robeson MS, Lauber CL, Hamady M, Knight R, Fierer N. 2009. A comprehensive survey of soil acidobacterial diversity using pyrosequencing and clone library analyses. ISME J. 3:442–453. 10.1038/ismej.2008.127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wu N, Yang X, Zhang R, Li J, Xiao X, Hu Y, Chen Y, Yang F, Lu N, Wang Z, Luan C, Liu Y, Wang B, Xiang C, Wang Y, Zhao F, Gao GF, Wang S, Li L, Zhang H, Zhu B. 2013. Dysbiosis signature of fecal microbiota in colorectal cancer patients. Microb. Ecol. 66:462–470. 10.1007/s00248-013-0245-9 [DOI] [PubMed] [Google Scholar]
- 62.Whipps JM, Hand P, Pink D, Bending GD. 2008. Phyllosphere microbiology with special reference to diversity and plant genotype. J. Appl. Microbiol. 105:1744–1755. 10.1111/j.1365-2672.2008.03906.x [DOI] [PubMed] [Google Scholar]
- 63.Pinto C, Pinho D, Sousa S, Pinheiro M, Egas C, Gomes CA. 2014. Unravelling the diversity of grapevine microbiome. PLoS One 9:e85622. 10.1371/journal.pone.0085622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Delmotte N, Knief C, Chaffron S, Innerebner G, Roschitzki B, Schlapbach R, von Mering C, Vorholt JA. 2009. Community proteogenomics reveals insights into the physiology of phyllosphere bacteria. Proc. Natl. Acad. Sci. U. S. A. 106:16428–16433. 10.1073/pnas.0905240106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rastogi G, Sbodio A, Tech JJ, Suslow TV, Coaker GL, Leveau JHJ. 2012. Leaf microbiota in an agroecosystem: spatiotemporal variation in bacterial community composition on field-grown lettuce. ISME J. 6:1812–1822. 10.1038/ismej.2012.32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Russell D, Chard J, McKinlay R. 1999. Effect of Bacillus thuringiensis and a pyrethroid insecticide on the leaf microflora of Brassica oleracea. Lett. Appl. Microbiol. 28:359–362. 10.1046/j.1365-2672.1999.00548.x [DOI] [Google Scholar]
- 67.Elad Y, Kirshner B. 1993. Survival in the phylloplane of an introduced biocontrol agent (Trichoderma harzianum) and populations of the plant pathogen Botrytis cinerea as modified by abiotic conditions. Phytoparasitica 21:303–313. 10.1007/BF02981048 [DOI] [Google Scholar]
- 68.Scherwinski K, Grosch R, Berg G. 2008. Effect of bacterial antagonists on lettuce: active biocontrol of Rhizoctonia solani and negligible, short-term effects on nontarget microorganisms. FEMS Microbiol. Ecol. 64:106–116. 10.1111/j.1574-6941.2007.00421.x [DOI] [PubMed] [Google Scholar]
- 69.Hooper LV. 2009. Do symbiotic bacteria subvert host immunity? Nat. Rev. Microbiol. 7:367–374. 10.1038/nrmicro2114 [DOI] [PubMed] [Google Scholar]
- 70.Guarner F, Malagelada J-R. 2003. Gut flora in health and disease. Lancet 361:512–519. 10.1016/S0140-6736(03)12489-0 [DOI] [PubMed] [Google Scholar]
- 71.Martins G, Miot-Sertier C, Lauga B, Claisse O, Lonvaud-Funel A, Soulas G, Masneuf-Pomarede I. 2012. Grape berry bacterial microbiota: impact of the ripening process and the farming system. Int. J. Food Microbiol. 158:93–100. 10.1016/j.ijfoodmicro.2012.06.013 [DOI] [PubMed] [Google Scholar]
- 72.Setati ME, Jacobson D, Andong UC, Bauer FF. 2012. The vineyard yeast microbiome, a mixed model microbial map. PLoS One 7:e52609. 10.1371/journal.pone.0052609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bokulich NA, Thorngate JH, Richardson PM, Mills DA. 2014. Microbial biogeography of wine grapes is conditioned by cultivar, vintage, and climate. Proc. Natl. Acad. Sci. U. S. A. 111:E139–E148. 10.1073/pnas.1317377110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Berlec A. 2012. Novel techniques and findings in the study of plant microbiota: search for plant probiotics. Plant Sci. 193–194:96–102. 10.1016/j.plantsci.2012.05.010 [DOI] [PubMed] [Google Scholar]
- 75.Harm A, Kassemeyer H-H, Seibicke T, Regner F. 2011. Evaluation of chemical and natural resistance inducers against downy mildew (Plasmopara viticola) in grapevine. Am. J. Enol. Vitic. 62:184–192. 10.5344/ajev.2011.09054 [DOI] [Google Scholar]
- 76.Falk SP, Pearson RC, Gadoury DM, Seem RC, Sztejnberg A. 1996. Fusarium proliferatum as a biocontrol agent against grape downy mildew. Phytopathology 86:1010–1017. 10.1094/Phyto-86-1010 [DOI] [Google Scholar]
- 77.Musetti R, Vecchione A, Stringher L, Borselli S, Zulini L, Marzani C, D'Ambrosio M, di Toppi LS, Pertot I. 2006. Inhibition of sporulation and ultrastructural alterations of grapevine downy mildew by the endophytic fungus Alternaria alternata. Phytopathology 96:689–698. 10.1094/PHYTO-96-0689 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.