Abstract
Protein misfolding and aggregation are inevitable but detrimental cellular processes. Cells therefore possess protein quality control mechanisms based on chaperones and proteases that (re)fold or hydrolyze unfolded, misfolded, and aggregated proteins. Besides these conserved quality control mechanisms, the spatial organization of protein aggregates (PAs) inside the cell has been proposed as an important additional strategy to deal with their cytotoxicity. In the bacterium Escherichia coli, however, it remained unclear how this spatial organization is established and how this process of assembling PAs in the cell poles affects cellular physiology. In this report, high hydrostatic pressure was used to transiently reverse protein aggregation in living E. coli cells, allowing the subsequent (re)assembly of PAs to be studied in detail. This approach revealed PA assembly to be dependent on intracellular energy and metabolic activity, with the resulting PA structure being confined to the cell pole by nucleoid occlusion. Moreover, a correlation could be observed between the time needed for PA reassembly and the individual lag time of the cells, which might prevent symmetric segregation of cytotoxic PAs among siblings to occur and ensure rapid spatial clearance of molecular damage throughout the emerging population.
INTRODUCTION
Protein misfolding is an inevitable process in cellular life, often aggravated by genetic defects and/or a number of stresses encountered in the environment (1–6). Since misfolded proteins typically expose hydrophobic residues that are normally buried within their native structure, they tend to aggregate with each other into larger insoluble structures termed protein aggregates (PAs) (5). Misfolded and aggregated proteins not only reduce the concentration of functional proteins and squander the cellular time and energy invested in their translation (7) but also have been shown to be cytotoxic themselves by mediating aberrant interactions with other proteins (8), sequestering aids to cellular folding away from other (essential) proteins (9), and possibly even affecting the integrity of lipid membranes (10). As such, this interference can (progressively) compromise cellular fitness (3, 11) and even extrapolate into debilitating neurodegenerative diseases in humans (12–14).
Due to the link with general cellular degeneracy, protein misfolding and aggregation are intensively studied processes in both prokaryotes and eukaryotes (15). Using a combination of biochemical, biophysical, structural, and genetic approaches, this has led to an extensive knowledge of protein quality control mechanisms and aggregation in vitro, as well as the identification of a number of genes involved in these processes (16–19). These quality control mechanisms are highly conserved and essentially consist of chaperones and proteases that, respectively (re)fold or hydrolyze unfolded, misfolded, and aggregated proteins (17–22). In spite of this elaborate protein homeostasis network, however, the accumulation and aggregation of misfolded proteins to some extent remain inevitable (5, 23), and recent studies have forwarded spatial organization as another important strategy of dealing with PAs inside the cell (24).
Recently, it became possible to monitor the emergence and fate of PAs inside living cells, using reporter systems based on fluorescently labeled heat shock proteins (e.g., IbpA of Escherichia coli) or thermolabile proteins, which themselves interact or associate with PAs (2, 11, 25). These reporter systems have in turn revealed the specific sequestration and asymmetric inheritance of cellular PAs during growth and division in bacteria (2, 11, 25), yeast (4), and higher eukaryotes (26), altogether suggesting a general rejuvenation strategy through which cytotoxic PAs preferentially remain linked to the older compartment upon cell division (24). In the Escherichia coli model system, for example, the polar localization and subsequent asymmetric segregation of cellular PAs were shown to be at least partially responsible for the previously observed pattern of aging in clonal lineages of this bacterium. In fact, cells inheriting the older and PA-bearing pole were shown to display a diminished growth rate, reduced production of offspring, and a higher death rate (2, 11, 27). How E. coli manages to sequester PAs to one of the cell poles, however, remains controversial, and active as well as passive mechanisms have been proposed. More specifically, one study claimed this process to be dependent on the actions of the proton motive force, cytoskeleton (MreB), and chaperones (DnaK and DnaJ) (25), while others provide evidence for nucleoid occlusion as a passive mechanism that is responsible and sufficient for the polar sequestration of PAs (2, 28).
In order to further elaborate on the intracellular organization and dynamics of PAs, we devised a new model system in which high hydrostatic pressure (HP) was used as a perturbant to reverse PA assembly in living E. coli cells, based on the ability of HP to thermodynamically interfere with the ionic and hydrophobic interactions that hold PAs together (29–32). Aside from convincingly demonstrating that PA reassembly is an energy-dependent (i.e., active) process and that PAs are spatially constrained by nucleoid occlusion, this model system could also reveal the existence of a clear link between PA assembly and growth resumption, suggesting a novel cellular strategy that prevents growth as long as dispersed PAs have not been reassembled and positioned sufficiently.
MATERIALS AND METHODS
Strain construction and growth conditions.
The ibpA-yfp locus of E. coli MGAY (11) was P1 transduced into E. coli MG1655 and its HP-resistant derivative, LMM1010 (33), resulting in MG1655 ibpA-yfp and LMM1010 ibpA-yfp, respectively. Different deletion strains of LMM1010 ibpA-yfp (ΔdnaK, ΔdnaJ, ΔclpB, Δlon, ΔhslU, ΔhslV, ΔclpX, and ΔrecA) were constructed based on the protocol of Datsenko and Wanner (34) and using an amplicon prepared on pKD13 using the primers listed in the study by Baba et al. (35). This procedure replaced the genes of interest with an frt-flanked kanamycin resistance cassette, which could subsequently be excised by transiently equipping this strain with plasmid pCP20 expressing the Flp site-specific recombinase (36), resulting in the desired deletion strain.
For culturing of bacteria, lysogeny broth (LB) medium (37, 38) was used as either a broth or solid medium after the addition of 2% agarose (for agar pads intended for microscopy). Stationary-phase cultures were obtained by growing E. coli overnight for approximately 15 h in LB broth at 37°C under well-aerated conditions (200 rpm on an orbital shaker). When appropriate, the following chemicals (Applichem, Darmstadt, Germany and Sigma-Aldrich) were added to the medium at the indicated final concentrations: chloramphenicol (30 μg/ml), DAPI (4′,6-diamidino-2-phenylindole) (1 μg/ml), and CCCP (carbonyl cyanide m-chlorophenyl hydrazine) (1 to 20 μM).
HP treatment.
HP treatment was performed essentially as described previously (39). Briefly, cells from a stationary-phase culture were harvested by centrifugation (4,000 × g, 5 min) and resuspended in an equal volume of 0.85% KCl. Subsequently, a 200-μl portion of resuspended cells was heat sealed in a sterile polyethylene bag after exclusion of the air bubbles and subjected to 300 MPa for 15 min in an 8-ml pressure vessel (HPIU-10000, 95/1994; Resato, Roden, The Netherlands), held at 20°C with an external water jacket connected to a cryostat. Finally, at the end of the holding time, decompression was almost instantaneous. After HP treatment, populations were aseptically retrieved from the polyethylene bags, transferred to LB agarose pads placed on a microscopy slide, and subjected to microscopy as described below.
In cases where the same cells were microscopically examined before and after HP shock, cells were first mounted on a microscopy slide as described below and analyzed microscopically while their spatial coordinates on the slide were noted. Subsequently, the slide as a whole was subjected to HP shock (300 MPa, 15 min, 20°C) using a 590-ml vessel (warm isostatic press, SO.5-7422-0EPSI; EPSI, Temse, Belgium), after which the spatial coordinates were used to trace back and microscopically follow-up the same cells on the pressurized slide.
Fluorescence microscopy and image analysis.
For imaging, cell suspensions were transferred to agarose pads placed on a microscopy slide and mounted with a cover glass as described before (40). A Gene Frame (Thermo Scientific) was used to physically fix the cover glass to the microscopy slide. Subsequently, fluorescence microscopy and time-lapse fluorescence microscopy were performed with a temperature-controlled (Okolab, Ottaviano, Italy) Ti-Eclipse inverted microscope (Nikon, Champigny-sur-Marne, France) equipped with a 60× objective, a TI-CT-E motorized condenser, a yellow fluorescent protein (YFP) filter (excitation, [Ex] 500/24; dichroic mirror [DM], 520; emission, [Em], 542/27), a DAPI filter (Ex, 377/50; DM, 409; Em, 447/60), and a CoolSnap HQ2 FireWire charge-coupled device (CCD) camera. Images were acquired using NIS-Elements software (Nikon), and the resulting pictures were further handled with the open source software ImageJ. For further analysis, cell meshes were obtained from the original images using the open source, MATLAB-based software MicrobeTracker (41), and fluorescent spots were manually detected using the SpotFinder tool from MicrobeTracker.
Determination of viability.
Cellular viability was determined either through spot-plating experiments or time-lapse microscopy. In spot-plating experiments, the appropriate dilutions of a sample were prepared in 0.85% KCl and subsequently spot-plated (5 μl) on LB agar. After 24 h of incubation at 37°C, the plates were counted, and the number of survivors in CFU per ml was determined. (Please note that the detection limit for these spot-plated samples was 200 CFU/ml.) When cellular viability was determined via time-lapse microscopy, the number of cells (out of 100 to 300 randomly chosen cells per independent experiment) that were able to grow and divide within a 3-h time frame were scored. (Please note that the detection limit for viability in the time-lapse microscopy samples varies between experiments and depends on the number of randomly chosen cells, from 1/100 cells or 1% to 1/300 cells or 0.33%.)
Energy depletion experiments.
To investigate the energy requirements of PA reassembly, different concentrations of CCCP (1 to 20 μM) were added to nutrient-free agarose pads used for microscopy. In cases where PA reassembly was monitored after CCCP treatment, cells were first treated with CCCP (for 3 h) in a nutrient-free environment, washed, and subsequently placed on LB agarose pads to allow PA reassembly to be monitored under the microscope.
Data analysis.
The distribution of fluorescent PA foci was obtained by determining their relative localization along the cell axis using the MicrobeTracker software (41). PA reassembly was determined via time-lapse microscopy, by visually scoring the number of cells that were able to assemble their PAs after HP exposure. Reassembly was considered completed when a cell was able to reassemble its PAs into one or two larger PAs (located in the cell poles) within a time frame of 3 h.
Lag-phase measurements were based on the measured area of the meshes generated by the MicrobeTracker program (41). First, an initial area was calculated as the mean of the first three measurements (corresponding to the first 9 min after time-lapse recording had begun) for each individual cell. The area of a cell in the subsequent frames was then compared to this initial area, and the length of the lag phase was defined as the time corresponding to the frame where a cell's area had increased over 10% compared to its initial area, plus the time between the end of HP exposure and the beginning of recording (typically around 15 min). This 10% increase in initial area was taken as a threshold to prevent random measurement fluctuations from influencing the results and to ensure that only lag times of cells that had fully committed to growth were measured. During lag time measurements, the assembly of fluorescent PA foci was also tracked using the SpotFinder tool (41). This allowed the simultaneous comparison of PA reassembly and growth in individual cells.
RESULTS AND DISCUSSION
HP-mediated reversal of protein aggregation in E. coli.
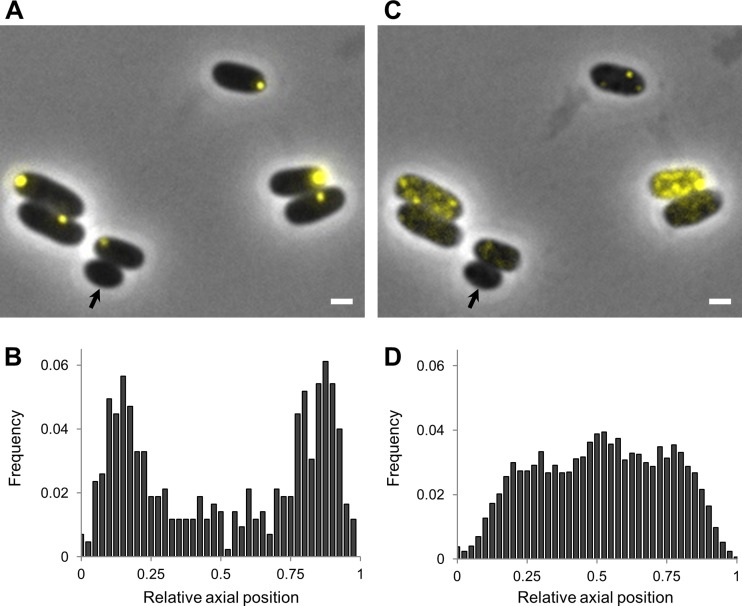
In order to examine the impact of HP exposure on intracellular PAs, we exposed E. coli MG1655 ibpA-yfp (in which the IbpA-Yfp fusion protein labels the existing PAs as fluorescent foci [11]) to 300 MPa for 15 min, an HP intensity shown in vitro to be able to dissociate oligomeric proteins into subunits (29). Interestingly, while PAs were typically singular and positioned in one of the cell poles before HP shock (Fig. 1A and B) (as described previously [11]), a number of smaller PA foci that were randomly distributed across the cytoplasm appeared after HP shock (Fig. 1C and D). Further analysis confirmed that these smaller PAs arose from the disassembly of the larger and polarly located preexisting PA rather than from the de novo emergence of new PAs, since (i) cells devoid of PAs before HP exposure also remained devoid of PAs after HP (Fig. 1A and B), and (ii) the average cellular fluorescence (and thus IbpA-Yfp concentration) within each cell was not increased after HP shock (see Fig. S1A in the supplemental material). Interestingly, the extent to which the polar PA became dispersed was very heterogeneous across the population, with the final number of smaller PAs generated in pressurized cells ranging from 1 to 12 (with an average of 5.45 per cell) (Fig. 1 and see Fig. S1B). Since the absence of HP gradients during pressurization can be reasonably assumed, this observation might be indicative for the heterogeneity in biophysical properties among polar PAs.
FIG 1.
In vivo HP exposure leads to PA disassembly in E. coli MG1655 ibpA-yfp. (A and C) Microscopic images show the same MG1655 ibpA-yfp cells before (A) and after (C) HP exposure (300 MPa, 20°C, 15 min). Phase-contrast images are superimposed with YFP epifluorescence images (reporting PAs), and the scale bar corresponds to 1 μm. The black arrow indicates a cell devoid of PAs. (B and D) Binned histograms show PA distribution along the relative axial position of the cells as detected in untreated (B) and HP-exposed (300 MPa, 20°C, 15 min) (D) MG1655 ibpA-yfp cells. The average numbers of PA foci per cell were 1.00 for control cells (n = 427) and 5.45 for HP-exposed cells (n = 640).
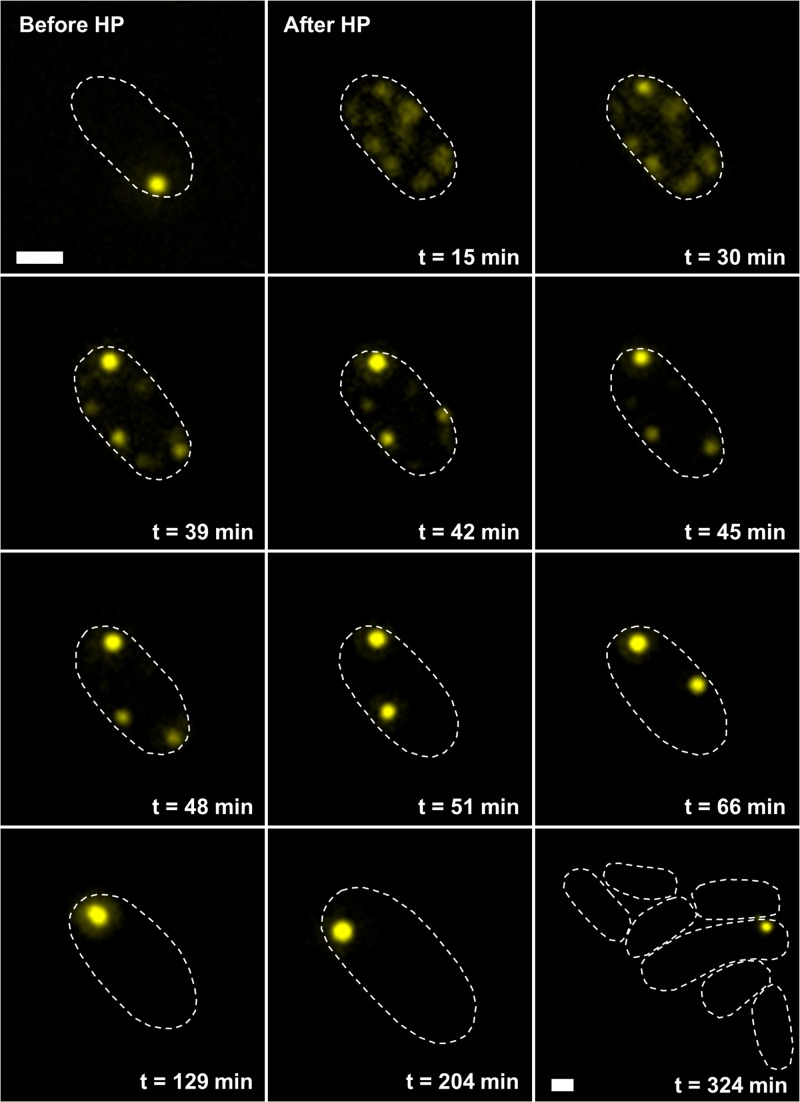
Surprisingly, although their smaller size should in principle have increased their diffusibility, the mobility of HP-dispersed cellular PAs seemed to be abolished (see Fig. S1C in the supplemental material). Since HP exposure killed the vast majority of MG1655 cells (see Fig. S1D), this observation suggested that viability is required to sustain intracellular PA mobility. The ibpA-yfp PA reporter locus was therefore transduced into LMM1010 (an HP-resistant derivative of MG1655 [33]), after which the resulting strain was similarly exposed to HP. While PAs in pressurized LMM1010 ibpA-yfp cells were also dispersed (see Fig. S2A to C in the supplemental material), most of them (ca. 75%) (see Fig. S1D) survived HP exposure and were able to (re)assemble their dispersed PAs into their cell pole (Fig. 2), thus restoring the situation before HP shock.
FIG 2.
HP-induced disassembly and reassembly of PAs in E. coli LMM1010 ibpA-yfp. Shown are representative images of a time-lapse fluorescence microscopy image sequence of the same cell before and after HP exposure (300 MPa, 20°C, 15 min), showing the reassembly of dispersed PAs into one larger polar PA and subsequent outgrowth of the cell. YFP epifluorescence images (reporting PAs) in combination with cell outlines are shown at the indicated times after HP exposure. The scale bars correspond to 1 μm.
Cellular requirements for PA assembly and localization.
Subsequently, this model system allowed a closer examination of the structural and physiological requirements underlying intracellular PA dynamics during (re)assembly. To examine the possible role of a number of chaperones and proteases, the effect of their deletion on PA organization before and PA reassembly after HP exposure was scored (Table 1). In unexposed cells, none of these deletions appeared to affect PA organization, leaving cellular PAs typically singular and positioned in one of the cell poles. In HP-exposed cells, the dnaK or dnaJ deletions rendered the cells highly sensitive toward the HP treatment and decreased cellular viability dramatically (Table 1) with concomitant abolition of PA mobility. In contrast to the dnaK mutant, where this reduced viability made it impossible to study the role of this chaperone in PA reassembly, the few surviving cells of the dnaJ mutant were also able to reassemble their PAs. Taken together, these observations indicate that, in contrast to what was previously reported (25), the chaperone DnaJ is not absolutely required for PA reassembly. The same holds for all the other tested chaperones and proteases: while the survival of their respective deletion strains was not significantly altered, PA reassembly occurred normally in these strains, indicating that these proteins exhibited no significant role in proper PA reassembly (Table 1).
TABLE 1.
Overview of the effects of different chaperone or protease deletions on PA assembly and survival in HP-treated populations of LMM1010 ibpA-yfp cellsa
| Type of strain | Effect of HP treatment onb: |
|
|---|---|---|
| PA assembly | Survival | |
| Wild type | 0.77 (0.12) | 0.65 (0.12) |
| Mutant | ||
| ΔdnaK | <0.01* | <0.01* |
| ΔdnaJ | 0.01 (0.01) | 0.01 (0.01) |
| ΔclpB | 0.66 (0.12) | 0.55 (0.09) |
| Δlon | 0.57 (0.16) | 0.48 (0.14) |
| ΔhslU | 0.76 (0.02) | 0.65 (0.04) |
| ΔhslV | 0.76 (0.14) | 0.59 (0.11) |
| ΔclpX | 0.83 (0.04) | 0.61 (0.03) |
Note that all the strains typically displayed a single polar PA before HP exposure.
The results represent relative fractions of cells able to engage in PA assembly or to survive and are presented as averages and standard deviations (in parentheses) from three independent experiments. *, value was below the detection limit of 0.01.
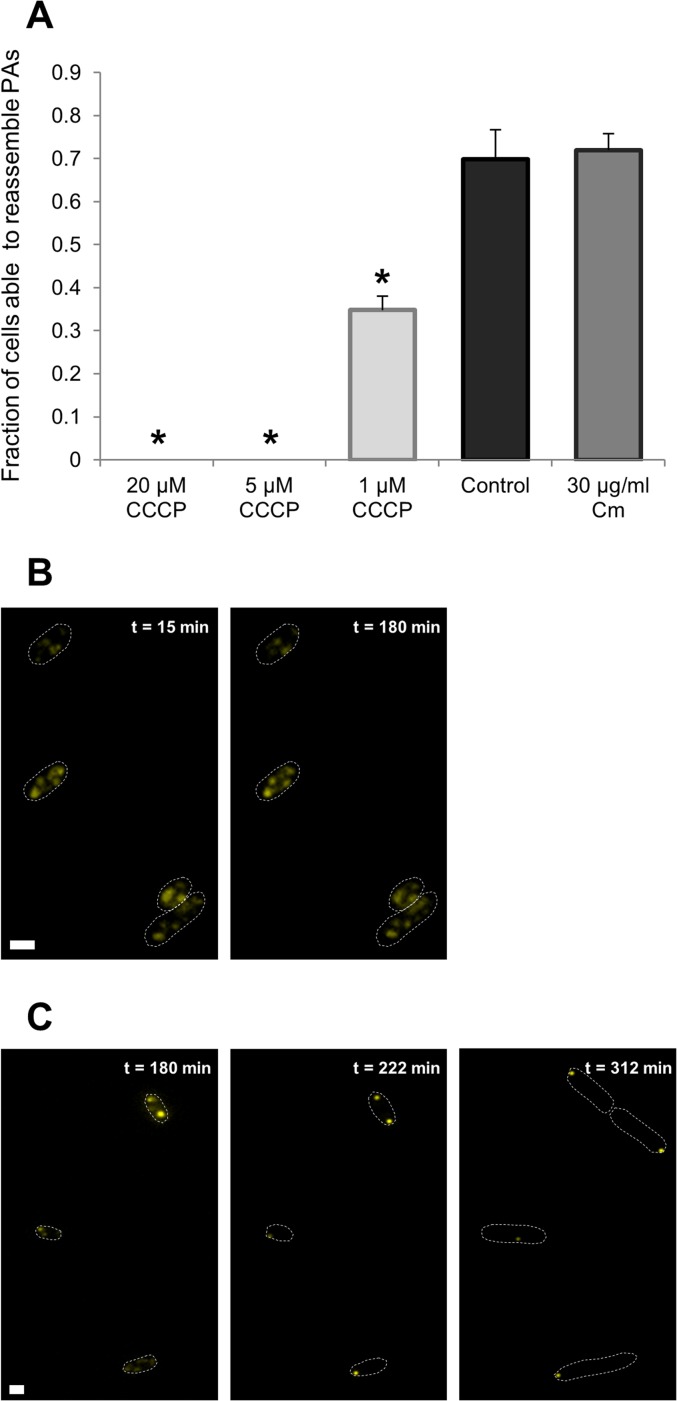
Because none of the tested chaperones or proteases were clearly found to be required for PA assembly, we set out to identify other required cellular factors. One of such factors, although currently still controversial, is the possible dependence of PA assembly on intracellular energy. To investigate this, we exposed LMM1010 ibpA-yfp cells after HP treatment to CCCP (carbonyl cyanide m-chlorophenyl hydrazine), an oxidative phosphorylation-uncoupling agent that was previously shown to reduce cellular energy (2). Moreover, by exposing these cells to CCCP in an environment without nutrients, we also attenuate other cellular sources of energy, such as substrate-level phosphorylation in glycolysis. Compared to pressurized cells without CCCP, the resulting cells displayed a strongly reduced mobility of their dispersed PAs and a concomitantly reduced capacity to reassemble them, with this effect being proportional to the CCCP concentration used (Fig. 3A). Importantly, none of the treatments with CCCP affected cellular viability (see Fig. S3 in the supplemental material), indicating that the observed decrease in PA mobility and assembly was not due to a decrease in cellular survival but to the presence and action of the compound itself. Moreover, when HP-exposed cells were treated with CCCP (for 3 h), washed, and subsequently placed on LB agarose pads, PA reassembly, which was halted until then due to the action of CCCP, again proceeded normally (Fig. 3B and C). These findings indicate that the energy depletion effect of the CCCP uncoupler is reversible, and energy repletion occurs when fresh nutrients are supplied. It is therefore important to continuously expose the cells to CCCP or other ATP-depleting compounds in a nutrient-free environment (an approach that contrasts those previously used in studies where the energy dependence of PA assembly in E. coli was investigated [2, 25]), in order to be able to truly investigate the energy dependency of this process. Although these results demonstrate PA reassembly to be an energy-dependent process, the synthesis of novel proteins did not account for this dependence, since a translation inhibitor, such as chloramphenicol, did not prevent PA reassembly in HP-exposed cells (Fig. 3A).
FIG 3.
PA reassembly after HP exposure is an energy-dependent process not dependent on the synthesis of novel proteins. (A) Treatment of an LMM1010 ibpA-yfp population with indicated concentrations of CCCP after HP exposure (300 MPa, 20°C, 15 min) decreases the fraction of cells (n ≥ 115 per independent experiment) that are able to assemble their PAs. Asterisks indicate statistically significant differences from the control (i.e., no addition of CCCP) (Student's t test, α = 0.01, P = 0.003 for 20 μM and 5 μM CCCP, and P = 0.005 for 1 μM CCCP). Addition of chloramphenicol (30 μg/ml) to the growth medium of an HP-exposed (300 MPa, 20°C, 15 min) LMM1010 ibpA-yfp population does not influence the ability of cells (n ≥ 120 per independent experiment) to reassemble their PAs (Student's t test, P = 0.28). Reassembly was considered completed when a cell was able to reassemble its PAs into one or two larger PAs (located in the cell poles) within 3 h after HP exposure. The means of three independent experiments are shown, with error bars representing the standard deviation. (B and C) Representative images of two time-lapse fluorescence image sequences indicating the reversibility of CCCP-induced energy depletion. (B) PA assembly does not occur after HP exposure (300 MPa, 20°C, 15 min) in the presence of CCCP (20 μM, for 180 min). (C) PA assembly resumes normally after CCCP is washed away (after 180 min) and cells are supplied with fresh nutrients. YFP epifluorescence images (reporting PAs) in combination with cell outlines are shown at the indicated times after HP exposure. The scale bar corresponds to 1 μm.
Interestingly, while an active PA assembly process might at first sight contradict previous studies that have shown and modeled PA movement and assembly to be strictly diffusion based (2, 28), a unifying explanation might stem from the most recently observed glass-like properties of the bacterial cytoplasm (42). More specifically, by tracking the intracellular movement of protein filaments, plasmids, storage granules, and foreign particles of different sizes, Parry et al. (42) found that the bacterial cytoplasm displays glass-like properties that restrict the mobility of cytoplasmic components in a size-dependent fashion. Strikingly, they also found that cellular metabolism fluidizes the cytoplasm, allowing larger components, normally increasingly constrained with increasing size, to escape their local environment and explore larger regions of the cytoplasm. As such, PAs might be seen as macromolecules of which the diffusive movement not only requires cellular energy (in order for metabolic activity to be able to fluidize the cytoplasm) but where the amount of energy needed is also dependent on macromolecular crowding and PA size. This in turn offers an explanation for studies in which protein aggregation was reported to be energy independent and their movement strictly diffusive, since these studies mainly focused on smaller PAs (2, 28).
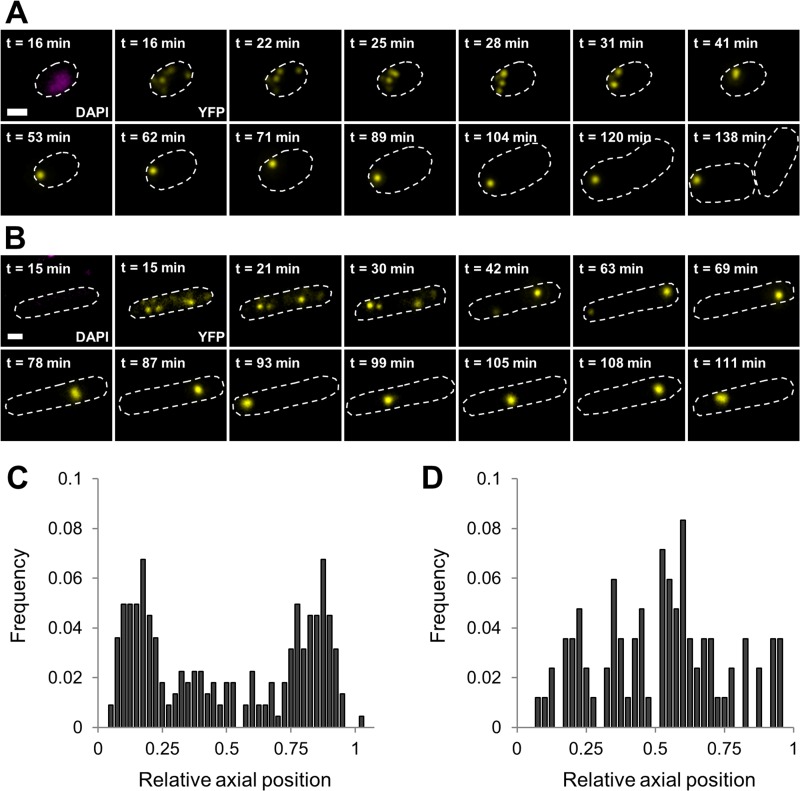
The impact of the nucleoid on assembling and localizing cellular PAs to one of the poles has been debated as well (2, 25, 28), and to examine this in our model system, a ΔrecA derivative of LMM1010 ibpA-yfp was constructed because of its natural propensity to frequently produce anucleate cells (43). When the latter cells of such an HP-exposed population were examined, their behavior was similar to that of wild-type nucleated cells in terms of reassembling dispersed PAs into a singular PA (Fig. 4A). In strong contrast to the situation in nucleated cells, however, this PA was not confined to the cell pole anymore but instead roamed freely throughout the cytoplasm (Fig. 4B). In addition, when looking at the relative localization of PAs in nucleate and anucleate cells after PA reassembly, we found that while PAs in nucleated cells were again preferentially located in the cell pole, PAs in anucleate cells displayed a more random localization (Fig. 4C and D). In support of these findings, PAs in anucleate control cells, in both LMM1010 and MG1655 ibpA-yfp ΔrecA cells, also appeared to roam freely throughout the cell without being restricted to a specific (polar) location (see Fig. S4A and B in the supplemental material). These observations clearly prove that the presence of the nucleoid enforces a polar localization on assembled PAs, although it does not per se interfere with the assembly process itself.
FIG 4.
Polar positioning of PAs is imposed by the nucleoid. (A and B) Representative images of a time-lapse fluorescence microscopy image sequence of a nucleoid-bearing LMM1010 ibpA-yfp cell (A) and an anucleate LMM1010 ibpA-yfp ΔrecA cell (B) after HP exposure (300 MPa, 20°C, 15 min). DAPI (reporting the nucleoid) or YFP (reporting PAs) epifluorescence images in combination with cell outlines are shown at the indicated times after HP exposure. The scale bar corresponds to 1 μm. (C and D) Binned histograms show the PA distribution along the relative axial position of the cells as detected in nucleoid-bearing LMM1010 ibpA-yfp (C) and anucleate LMM1010 ibpA-yfp ΔrecA (D) cells after PA reassembly after HP exposure (300 MPa, 20°C, 15 min) was completed. The average numbers of PA foci per cell were 1.18 for nucleoid-bearing cells (n = 224) and 1.00 for anucleate cells (n = 84).
Dispersed PAs suspend growth resumption.
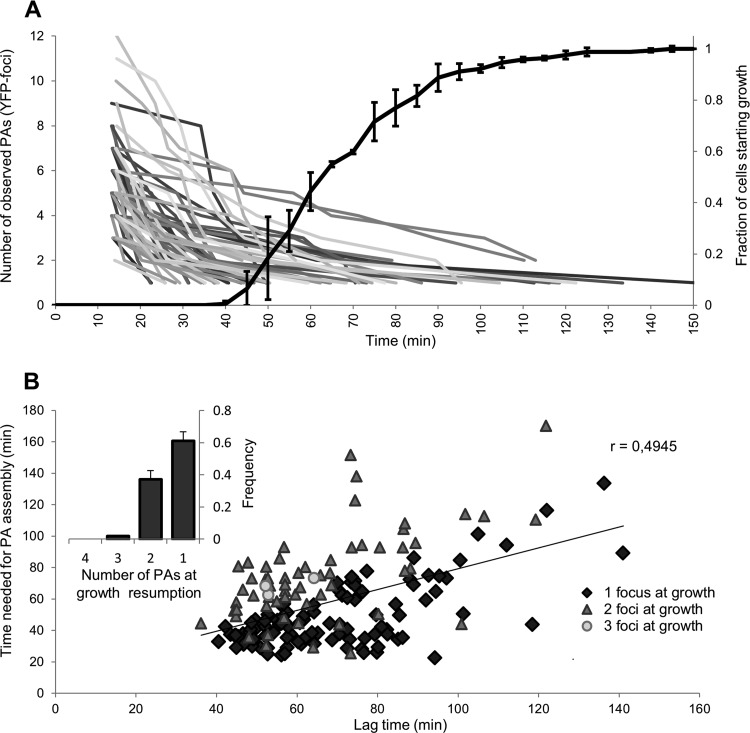
Upon further analyzing cellular PA reassembly in an HP-exposed LMM1010 ibpA-yfp population more quantitatively (Fig. 5), we noticed that cells never seemed to commit to growth (defined here as a microscopically detectable increase in cell surface) as long as intracellular PAs were still dispersed. In fact, regardless of the extent of HP-induced PA dispersion (which ranged from 1 to 12 PAs per cell) (Fig. 5A; see Fig. S2D in the supplemental material), cells did not resume growth until the reassembly process yielded one (in 61.5% of cases [104/169]), two (in 36.7% of cases [62/169]), or—exceptionally—three (in 1.8% of cases [3/169]) PAs (Fig. 5B), of which the location seemed to match the three stable PA repositories in the cell (i.e., both poles and midcell) reported earlier (2, 11). In addition, a good correlation was also found between individual cellular lag times (defined as the time needed for a cell to resume growth after HP exposure) and the corresponding times needed to reassemble their dispersed PAs (Fig. 5B). Although it remains to be established whether there is a causal relationship between these two phenomena, it could be hypothesized that dispersed PAs functionally interfere with essential cellular processes responsible for growth (e.g., because the cytotoxic PA surface area exposed in the cytoplasmic milieu increases significantly upon dispersion) or even that such processes might be deliberately halted until the proper assembly of PAs allows their asymmetric segregation (and subsequent riddance) among siblings. Either way, such behavior would prevent cytotoxic PAs from being randomly partitioned among siblings and would ensure a rapid spatial clearance of molecular damage throughout the emerging population, which has previously been proposed to be most effective at maintaining high growth rates (44–46). Nevertheless, repair or other resuscitation processes different from PA assembly are likely to contribute to cellular lag times after HP stress as well.
FIG 5.
Growth resumption is correlated with PA reassembly. (A) Cumulative lag time distribution of LMM1010 ibpA-yfp cells after HP exposure (300 MPa, 20°C, 15 min) was combined with the PA reassembly process in the same cells (n = 169; derived from three independent experiments). The time individual cells needed to resume growth after HP exposure was determined and binned to create the cumulative lag time distribution. The mean cumulative lag time distribution of three independent experiments is shown with error bars representing the standard deviation per bin. At the same time, the number of individual PAs (visible as IbpA-YFP foci) inside these cells was monitored. Every line represents the PA assembly process of a single cell. Because of visibility, not all of the monitored cells are included. (B) Correlation between the time a cell needs to reassemble its PAs and its corresponding lag time, based on the data set from panel A. A subdivision was made depending on the number of larger PAs a cell assembles its dispersed PAs into prior to resumption of growth. The linear regression line (trend line) and Pearson's correlation coefficient (r = 0.4945, P = 2.35 × 10−11) are based on all cells together. (Inset) Fraction of cells resuming growth after HP treatment when PAs are reassembled into one, two, or three larger PAs. The means of three independent experiments are shown with error bars representing the standard deviation.
In a related context, it has most recently been shown in Caulobacter crescentus that proteotoxic stress stimulates the production and activity of the Lon protease, in turn leading to the degradation of the DnaA replication initiator and a subsequent arrest in chromosome replication (47). Although it remains to be established whether the PA-based growth arrest suggested in this study and the Lon/DnaA effect revealed by Jonas et al. (47) are mechanistically similar, both could support a more universal cellular strategy in which cellular proliferation is halted until the proper spatial alleviation or structural repair of protein damage.
Conclusion.
In summary, we have elaborated a unique model system to study the assembly of PAs and its consequences on cellular physiology, based on the thermodynamically enforced in vivo dispersion of preexisting PAs inside the cell by high hydrostatic pressure. Using this model system, we were able to clearly establish that the assembly of PAs is dependent on intracellular energy and metabolic activity, with the resulting (larger) PA being confined to one of the cell poles by nucleoid occlusion. Moreover, by closely monitoring the PA assembly process and its impact on the timing of resuscitation, we provide possible evidence for a cellular strategy that postpones growth and proliferation until proper PA assembly is achieved.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by a doctoral fellowship of the Flemish Agency for Innovation by Science and Technology (IWT-Vlaanderen to S.K.G.) and by grants from the Research Foundation of Flanders (FWO-Vlaanderen; grant G.0580.11) and the KU Leuven Research Fund (grants STRT1/10/036 and IDO/10/012).
We thank Ariel Lindner for providing the MGAY strain and Dietrich Vanlint and Nele Rutten for technical assistance.
Footnotes
Published ahead of print 14 March 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.01549-14.
REFERENCES
- 1.Pakula AA, Sauer RT. 1989. Genetic analysis of protein stability and function. Annu. Rev. Genet. 23:289–310. 10.1146/annurev.ge.23.120189.001445 [DOI] [PubMed] [Google Scholar]
- 2.Winkler J, Seybert A, Konig L, Pruggnaller S, Haselmann U, Sourjik V, Weiss M, Frangakis AS, Mogk A, Bukau B. 2010. Quantitative and spatio-temporal features of protein aggregation in Escherichia coli and consequences on protein quality control and cellular ageing. EMBO J. 29:910–923. 10.1038/emboj.2009.412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Geiler-Samerotte KA, Dion MF, Budnik BA, Wang SM, Hartl DL, Drummond DA. 2011. Misfolded proteins impose a dosage-dependent fitness cost and trigger a cytosolic unfolded protein response in yeast. Proc. Natl. Acad. Sci. U. S. A. 108:680–685. 10.1073/pnas.1017570108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aguilaniu H, Gustafsson L, Rigoulet M, Nystrom T. 2003. Asymmetric inheritance of oxidatively damaged proteins during cytokinesis. Science 299:1751–1753. 10.1126/science.1080418 [DOI] [PubMed] [Google Scholar]
- 5.Dobson CM. 2003. Protein folding and misfolding. Nature 426:884–890. 10.1038/nature02261 [DOI] [PubMed] [Google Scholar]
- 6.Lindner AB, Demarez A. 2009. Protein aggregation as a paradigm of aging. Biochim. Biophys. Acta 1790:980–996. 10.1016/j.bbagen.2009.06.005 [DOI] [PubMed] [Google Scholar]
- 7.Stoebel DM, Dean AM, Dykhuizen DE. 2008. The cost of expression of Escherichia coli lac operon proteins is in the process, not in the products. Genetics 178:1653–1660. 10.1534/genetics.107.085399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Olzscha H, Schermann SM, Woerner AC, Pinkert S, Hecht MH, Tartaglia GG, Vendruscolo M, Hayer-Hartl M, Hartl FU, Vabulas RM. 2011. Amyloid-like aggregates sequester numerous metastable proteins with essential cellular functions. Cell 144:67–78. 10.1016/j.cell.2010.11.050 [DOI] [PubMed] [Google Scholar]
- 9.Gidalevitz T, Ben-Zvi A, Ho KH, Brignull HR, Morimoto RI. 2006. Progressive disruption of cellular protein folding in models of polyglutamine diseases. Science 311:1471–1474. 10.1126/science.1124514 [DOI] [PubMed] [Google Scholar]
- 10.Lashuel HA, Lansbury PT., Jr 2006. Are amyloid diseases caused by protein aggregates that mimic bacterial pore-forming toxins? Q. Rev. Biophys. 39:167–201. 10.1017/S0033583506004422 [DOI] [PubMed] [Google Scholar]
- 11.Lindner AB, Madden R, Demarez A, Stewart EJ, Taddei F. 2008. Asymmetric segregation of protein aggregates is associated with cellular aging and rejuvenation. Proc. Natl. Acad. Sci. U. S. A. 105:3076–3081. 10.1073/pnas.0708931105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bucciantini M, Giannoni E, Chiti F, Baroni F, Formigli L, Zurdo J, Taddei N, Ramponi G, Dobson CM, Stefani M. 2002. Inherent toxicity of aggregates implies a common mechanism for protein misfolding diseases. Nature 416:507–511. 10.1038/416507a [DOI] [PubMed] [Google Scholar]
- 13.Ross CA, Poirier MA. 2004. Protein aggregation and neurodegenerative disease. Nat. Med. 10(Suppl):S10–S17. 10.1038/nm1066 [DOI] [PubMed] [Google Scholar]
- 14.Stefani M, Dobson CM. 2003. Protein aggregation and aggregate toxicity: new insights into protein folding, misfolding diseases and biological evolution. J. Mol. Med. 81:678–699. 10.1007/s00109-003-0464-5 [DOI] [PubMed] [Google Scholar]
- 15.Tyedmers J, Mogk A, Bukau B. 2010. Cellular strategies for controlling protein aggregation. Nat. Rev. Mol. Cell Biol. 11:777–788. 10.1038/nrm2993 [DOI] [PubMed] [Google Scholar]
- 16.Martin JB. 1999. Molecular basis of the neurodegenerative disorders. N. Engl. J. Med. 340:1970–1980. 10.1056/NEJM199906243402507 [DOI] [PubMed] [Google Scholar]
- 17.Gottesman S, Wickner S, Maurizi MR. 1997. Protein quality control: triage by chaperones and proteases. Genes Dev. 11:815–823. 10.1101/gad.11.7.815 [DOI] [PubMed] [Google Scholar]
- 18.Wickner S, Maurizi MR, Gottesman S. 1999. Posttranslational quality control: folding, refolding, and degrading proteins. Science 286:1888–1893. 10.1126/science.286.5446.1888 [DOI] [PubMed] [Google Scholar]
- 19.Hartl FU, Hayer-Hartl M. 2002. Molecular chaperones in the cytosol: from nascent chain to folded protein. Science 295:1852–1858. 10.1126/science.1068408 [DOI] [PubMed] [Google Scholar]
- 20.Bukau B, Weissman J, Horwich A. 2006. Molecular chaperones and protein quality control. Cell 125:443–451. 10.1016/j.cell.2006.04.014 [DOI] [PubMed] [Google Scholar]
- 21.Mogk A, Deuerling E, Vorderwulbecke S, Vierling E, Bukau B. 2003. Small heat shock proteins, ClpB and the DnaK system form a functional triade in reversing protein aggregation. Mol. Microbiol. 50:585–595. 10.1046/j.1365-2958.2003.03710.x [DOI] [PubMed] [Google Scholar]
- 22.Doyle SM, Wickner S. 2009. Hsp104 and ClpB: protein disaggregating machines. Trends Biochem. Sci. 34:40–48. 10.1016/j.tibs.2008.09.010 [DOI] [PubMed] [Google Scholar]
- 23.Maisonneuve E, Fraysse L, Moinier D, Dukan S. 2008. Existence of abnormal protein aggregates in healthy Escherichia coli cells. J. Bacteriol. 190:887–893. 10.1128/JB.01603-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nystrom T. 2011. Spatial protein quality control and the evolution of lineage-specific ageing. Philos. Trans. R. Soc. Lond. B Biol. Sci. 366:71–75. 10.1098/rstb.2010.0282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rokney A, Shagan M, Kessel M, Smith Y, Rosenshine I, Oppenheim AB. 2009. E. coli transports aggregated proteins to the poles by a specific and energy-dependent process. J. Mol. Biol. 392:589–601. 10.1016/j.jmb.2009.07.009 [DOI] [PubMed] [Google Scholar]
- 26.Rujano MA, Bosveld F, Salomons FA, Dijk F, van Waarde MA, van der Want JJ, de Vos RA, Brunt ER, Sibon OC, Kampinga HH. 2006. Polarised asymmetric inheritance of accumulated protein damage in higher eukaryotes. PLoS Biol. 4:e417. 10.1371/journal.pbio.0040417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stewart EJ, Madden R, Paul G, Taddei F. 2005. Aging and death in an organism that reproduces by morphologically symmetric division. PLoS Biol. 3:e45. 10.1371/journal.pbio.0030045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Coquel AS, Jacob JP, Primet M, Demarez A, Dimiccoli M, Julou T, Moisan L, Lindner AB, Berry H. 2013. Localization of protein aggregation in Escherichia coli is governed by diffusion and nucleoid macromolecular crowding effect. PLoS Comput. Biol. 9:e1003038. 10.1371/journal.pcbi.1003038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.St. John RJ, Carpenter JF, Randolph TW. 1999. High pressure fosters protein refolding from aggregates at high concentrations. Proc. Natl. Acad. Sci. U. S. A. 96:13029–13033. 10.1073/pnas.96.23.13029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aertsen A, Meersman F, Hendrickx ME, Vogel RF, Michiels CW. 2009. Biotechnology under high pressure: applications and implications. Trends Biotechnol. 27:434–441. 10.1016/j.tibtech.2009.04.001 [DOI] [PubMed] [Google Scholar]
- 31.Seefeldt MB, Rosendahl MS, Cleland JL, Hesterberg LK. 2009. Application of high hydrostatic pressure to dissociate aggregates and refold proteins. Curr. Pharm. Biotechnol. 10:447–455. 10.2174/138920109788488914 [DOI] [PubMed] [Google Scholar]
- 32.Silva JL, Foguel D, Royer CA. 2001. Pressure provides new insights into protein folding, dynamics and structure. Trends Biochem. Sci. 26:612–618. 10.1016/S0968-0004(01)01949-1 [DOI] [PubMed] [Google Scholar]
- 33.Hauben KJ, Bartlett DH, Soontjens CC, Cornelis K, Wuytack EY, Michiels CW. 1997. Escherichia coli mutants resistant to inactivation by high hydrostatic pressure. Appl. Environ. Microbiol. 63:945–950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640–6645. 10.1073/pnas.120163297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2:2006.0008. 10.1038/msb4100050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cherepanov PP, Wackernagel W. 1995. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene 158:9–14. 10.1016/0378-1119(95)00193-A [DOI] [PubMed] [Google Scholar]
- 37.Miller JH. 1992. A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor Laboratory Press, Plainview, NY [Google Scholar]
- 38.Bertani G. 2004. Lysogeny at mid-twentieth century: P1, P2, and other experimental systems. J. Bacteriol. 186:595–600. 10.1128/JB.186.3.595-600.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vanlint D, Rutten N, Govers SK, Michiels CW, Aertsen A. 2013. Exposure to high hydrostatic pressure rapidly selects for increased RpoS activity and general stress-resistance in Escherichia coli O157:H7. Int. J. Food Microbiol. 163:28–33. 10.1016/j.ijfoodmicro.2013.02.001 [DOI] [PubMed] [Google Scholar]
- 40.Cenens W, Mebrhatu MT, Makumi A, Ceyssens PJ, Lavigne R, Van Houdt R, Taddei F, Aertsen A. 2013. Expression of a novel P22 ORFan gene reveals the phage carrier state in Salmonella typhimurium. PLoS Genet. 9:e1003269. 10.1371/journal.pgen.1003269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sliusarenko O, Heinritz J, Emonet T, Jacobs-Wagner C. 2011. High-throughput, subpixel precision analysis of bacterial morphogenesis and intracellular spatio-temporal dynamics. Mol. Microbiol. 80:612–627. 10.1111/j.1365-2958.2011.07579.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Parry BR, Surovtsev IV, Cabeen MT, O'Hern CS, Dufresne ER, Jacobs-Wagner C. 2014. The bacterial cytoplasm has glass-like properties and is fluidized by metabolic activity. Cell 156:183–194. 10.1016/j.cell.2013.11.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Capaldo FN, Barbour SD. 1975. DNA content, synthesis and integrity in dividing and non-dividing cells of rec− strains of Escherichia coli K12. J. Mol. Biol. 91:53–66. 10.1016/0022-2836(75)90371-X [DOI] [PubMed] [Google Scholar]
- 44.Watve M, Parab S, Jogdand P, Keni S. 2006. Aging may be a conditional strategic choice and not an inevitable outcome for bacteria. Proc. Natl. Acad. Sci. U. S. A. 103:14831–14835. 10.1073/pnas.0606499103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Erjavec N, Cvijovic M, Klipp E, Nystrom T. 2008. Selective benefits of damage partitioning in unicellular systems and its effects on aging. Proc. Natl. Acad. Sci. U. S. A. 105:18764–18769. 10.1073/pnas.0804550105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Escusa-Toret S, Vonk WI, Frydman J. 2013. Spatial sequestration of misfolded proteins by a dynamic chaperone pathway enhances cellular fitness during stress. Nat. Cell Biol. 15:1231–1243. 10.1038/ncb2838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jonas K, Liu J, Chien P, Laub MT. 2013. Proteotoxic stress induces a cell-cycle arrest by stimulating Lon to degrade the replication initiator DnaA. Cell 154:623–636. 10.1016/j.cell.2013.06.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.