Abstract
The FlgM protein is secreted in response to flagellar hook-basal body secretion and can be used as a secretion signal to direct selected protein secretion via the flagellar type III secretion (T3S) system [H. M. Singer, M. Erhardt, A. M. Steiner, M. M. Zhang, D. Yoshikami, G. Bulaj, B. M. Olivera, and K. T. Hughes, mBio 3(3):e00115-12, 2012, http://dx.doi.org/10.1128/mBio.00115-12]. Conditions known to affect flagellar gene expression, FlgM stability, and flagellar T3S were tested either alone or in combination to determine their effects on levels of secreted FlgM. These conditions included mutations that affect activity of the flagellar FlhD4C2 master regulatory protein complex or the FlgM T3S chaperone σ28, the removal of Salmonella pathogenicity island 1 (Spi1), the removal of flagellar late secretion substrates that could compete with FlgM for secretion, and changes in the ionic strength of the growth medium. Conditions that enhanced FlgM secretion were combined in order to maximize levels of secreted FlgM. An optimized FlgM secretion strain was used to secrete and isolate otherwise difficult-to-produce proteins and peptides fused to the C terminus of FlgM. These include cysteine-rich, hydrophobic peptides (conotoxins δ-SVIE and MrVIA), nodule-specific, cysteine-rich antimicrobial peptides (NCR), and a malaria surface antigen domain of apical membrane antigen AMA-1.
INTRODUCTION
Many bacteria utilize flagella to move in a directed manner, either away from stressful environments or toward nutrients, O2, light, and other positive stimuli (1). The bacterial flagellum is a complex cellular machine that requires more than 30 gene products for its construction. For Salmonella enterica, there are currently more than 60 genes involved in the biogenesis and function of its flagella (2). These genes are organized into a transcriptional hierarchy of 3 promoter classes. At the top of the flagellar transcriptional hierarchy is the flhDC operon, encoding the master regulator proteins FlhD and FlhC, which form a heteromultimeric, transcriptional activation complex (3). The FlhD4C2 complex directs σ70 RNA polymerase to transcribe from class 2 flagellar promoters. Class 2 flagellar genes encode proteins required for the structure and assembly of a rotary motor called the hook-basal body (HBB), a key structural intermediate in flagellum assembly (4). The HBB includes the flagellar type III secretion (T3S) system, which exports flagellar proteins from the cytoplasm through the growing structure during assembly (5). In addition to HBB gene expression, flagellar class 2 transcription produces σ28 (FliA) and FlgM. These are regulatory proteins that couple transcription of the flagellar class 3 promoters to completion of the HBB. The σ28 protein is a flagellum-specific transcription factor that directs RNA polymerase to transcribe from the flagellar class 3 promoters (6). Class 3 genes include the structural genes of the flagellar filament and genes of the chemosensory signal transduction system, which controls the direction of flagellar rotation according to changing concentrations of extracellular ligands (7). Prior to HBB completion, FlgM binds σ28 and prevents flagellar class 3 promoter transcription (8). Upon HBB completion, a change in flagellar T3S substrate specificity results in FlgM secretion and initiation of σ28-dependent transcription from flagellar class 3 promoters (9, 10). The secretion signal requirements for T3S substrates remains poorly defined, but all substrates utilize an N-terminal peptide secretion signal that is disordered in structure and, unlike the case with type II secretion, is not cleaved during the secretion process (11). Substrate secretion is often facilitated by T3S chaperone-assisted delivery to the secretion apparatus (12). The FlgM protein is 97 amino acids in length, and its secretion is dependent on an N-terminal secretion signal. FlgM secretion is greatly enhanced by its secretion chaperone, σ28, which binds to the C-terminal half of FlgM (13–15).
The FlgM type III secretion system can be used to express and purify recombinant proteins (16). Here we have identified factors that improve yields of secreted FlgM. These factors include those controlling flagellar gene expression, ionic conditions, cell growth phase, and removal of cellular proteases or secretion competitors. This characterization allowed us to construct strains that produce high yields of secreted protein for the purposes of protein purification via flagellar T3S.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
All strains used in this study are derivatives of LT2, a wild-type strain of Salmonella enterica serovar Typhimurium, and are listed in Table 1. Strain construction utilized either P22-mediated generalized transduction or λ Red-mediated targeted mutagenesis (17–19). For strains with the flhDC operon expressed from its native promoter, flagellar gene expression was induced by 100-fold dilution from overnight stationary cultures into fresh LB medium (10 g Bacto tryptone, 5 g Bacto yeast extract, and 5 g NaCl [per liter]). For strains with the flhDC operon expressed from the tetA promoter of transposon Tn10 (ΔPflhDC8089::tetR-PtetA), flagellar gene expression was induced by addition of the tetracycline analog, anhydrotetracycline (1 μg/ml). For all strains used in this study, the arabinose utilization operon, araBAD, was replaced with the flgM+ gene or flgM gene fusion flgM-6His-TEV-δ-SVIE or flgM-6His-ETK-δ-SVIE (ΔaraBAD::flgM+, ΔaraBAD::flgM-6His-TEV-δ-SVIE, or ΔaraBAD::flgM-6His-ETK-δ-SVIEI, respectively). This allowed for the induction of FlgM, FlgM-6His-TEV-δ-SVIE, and FlgM-6His-ETK-δ-SVIE production from the ParaBAD promoter by the addition of l-arabinose to the growth medium. Arabinose was added to a 0.2% final concentration 2 h after the induction of the flhDC operon. After another 5 h, the cultures were centrifuged at 10,000 × g for 30 min to pellet the cells. The supernatant was filtered with a 0.2-μm low-protein-binding filter (Acrodisk syringe filter; Pall Life Sciences) to remove remaining cells. Secreted proteins in the filtered supernatant were precipitated by addition of TCA (trichloroacetic acid) to a 10% final concentration. The cell pellet was resuspended in cold PBS (phosphate-buffered saline) containing 5 mM PMSF (phenylmethylsulfonyl fluoride), followed by sonication to lyse the cell suspension. The cell lysate was either analyzed directly for whole-cell protein or separated into soluble and insoluble fractions for analysis by 30 min of centrifugation (15,000 × g) at 4°C. To test the effect of different concentrations of NaCl and KCl on FlgM secretion, LB medium was prepared without NaCl and either NaCl or KCl was added to the desired concentrations.
TABLE 1.
Strains used in this work
| Straina | Genotypeb |
|---|---|
| TH17831 | ΔaraBAD1124::flgM-6His-TEV-δ-SVIE |
| TH18353 | ΔaraBAD1124::flgM-6H-TEV-δ-SVIE ΔclpX::tetRA |
| TH18500 | ΔaraBAD1156::flgM+ |
| TH18527 | ΔaraBAD1156::flgM+ ΔclpX::tetRA |
| TH18528 | ΔaraBAD1156::flgM+ ΔflhDC::FKF |
| TH18549 | ΔaraBAD1156::flgM+ ΔflhDC::FKF ΔclpX::tetRA |
| TH18624 | ΔaraBAD1156::flgM+ ΔfliA5805::tetRA |
| TH18636 | ΔaraBAD1156::flgM+ fliA8088(H14D, R91C, L207P) |
| TH18647 | ΔaraBAD1124::flgM-6His-TEV-δ-SVIE ΔPflhDC8089::(tetR-PtetA) |
| TH18649 | ΔaraBAD1156::flgM+ ΔPflhDC8089::(tetR-PtetA) |
| TH18704 | ΔaraBAD1156::flgM+ ΔinvH-sprB::FKF(ΔSpi1) |
| TH18729 | ΔaraBAD1156::flgM+ ΔfliC7715::tetRA |
| TH18730 | ΔaraBAD1156::flgM+ ΔompT::Km |
| TH18731 | ΔaraBAD1156::flgM+ ΔclpA74::FKF |
| TH18732 | ΔaraBAD1156::flgM+ ΔsseA-ssaU::FKF(ΔSpi2) |
| TH18733 | ΔaraBAD1156::flgM+ ΔclpP::mini-Tn5 |
| TH18737 | ΔaraBAD1156::flgM+ ΔfliA5999(R91C, L207P) |
| TH18739 | ΔaraBAD1156::flgM+ fliA5225(H14D) |
| TH18752 | ΔaraBAD1156::flgM+ ΔydiV251::tetRA |
| TH18769 | ΔaraBAD1124::flgM-6His-TEV-δ-SVIE ΔfliT::Km |
| TH18778 | ΔaraBAD1156::flgM+ fliA5228(V33E) |
| TH18780 | ΔaraBAD1156::flgM+ fliA5223(T138I) |
| TH18782 | ΔaraBAD1156::flgM+ fliA5224(E203D) |
| TH18787 | ΔaraBAD1156::flgM+ ΔflgM5628::FKF |
| TH18788 | ΔaraBAD1156::flgM+ ΔfliT::FKF |
| TH18790 | ΔaraBAD1156::flgM+ ΔflgK7665 |
| TH18793 | ΔaraBAD1156::flgM+ ΔflgL7666 |
| TH18796 | ΔaraBAD1156::flgM+ ΔfliD5630::FRT |
| TH18798 | ΔaraBAD1156::flgM+ ΔdegP::tetRA |
| TH18822 | ΔaraBAD1156::flgM+ ΔcsrA101::Cm |
| TH18823 | ΔaraBAD1156::flgM+ ΔdksA::FKF |
| TH18824 | ΔaraBAD1156::flgM+ ΔrcsB::tetRA |
| TH18830 | ΔaraBAD1124::flgM-6H-TEV-δ-SVIE ΔrcsB::tetRA |
| TH18850 | ΔaraBAD1156::flgM+ ΔlrhA |
| TH18880 | ΔaraBAD1124::flgM-6H-TEV-δ-SVIE PflhDC7793 |
| TH18881 | ΔaraBAD1156::flgM+ PflhDC7793 |
| TH18897 | ΔaraBAD1156::flgM+ fliA5240(L199R) |
| TH18973 | ΔaraBAD1156::flgM+ ΔecnR5::FCF |
| TH19086 | ΔaraBAD1156::flgM+ ΔflgN5626::FKF |
| TH19087 | ΔaraBAD1156::flgM+ ΔfliST5775::FCF |
| TH19099 | ΔaraBAD1156::flgM+ Δhin-fljA7731::tetRA |
| TH19104 | ΔaraBAD1156::flgM+ ΔflgN5626::FKF ΔfliST5775::FCF |
| TH19106 | ΔaraBAD1156::flgM+ ΔfliS5728::FRT |
| TH19113 | ΔaraBAD1156::flgM+ Δhin-fljA7731::tetRA ΔfliC7861::FCF |
| TH19116 | ΔaraBAD1164::flgM-6H-ETK-δ-SVIE |
| TH19118 | ΔaraBAD1164::flgM-6H-ETK-δ-SVIE PflhDC7793 |
| TH19120 | ΔaraBAD1164::flgM-6H-ETK-δ-SVIE ΔlrhA ΔecnR4::FRT ΔfliB-T7771 PflhD7793 ΔydiV252 Δhin-fljA7752 ΔflgMN7753 ΔflgKL7770 |
| TH19145 | ΔaraBAD1164::flgM-6H-ETK-δ-SVIE PflhDC7793 ΔflgM5628::FKF ΔclpX80::tetRA |
| TH19149 | ΔaraBAD1156::flgM+ ΔflgKL5636::FKF |
| TH19320 | ΔaraBAD1156::flgM+ fliA5226(H14N) |
| TH19323 | ΔaraBAD1156::flgM+ ΔclpA74::FKF ΔflhDC8040::tetRA |
| TH19324 | ΔaraBAD1156::flgM+ ΔclpP::mini-Tn5 ΔflhDC8040::tetRA |
| TH19325 | ΔaraBAD1156::flgM+ ΔdegP::tetRA ΔflhDC::FKF |
| TH19326 | ΔaraBAD1156::flgM+ Δhin-fljA7731::tetRA ΔfliC7861::FCF ΔflgKL5739::FKF |
| TH19481 | ΔaraBAD1156::flgM+ ΔfliS8156 (replaced fliST with fliS) |
| TH19673 | ΔaraBAD1124::flgM-6His-TEV-δ-SVIE ΔfliT::Km ΔrcsB::tetRA |
| TH19675 | ΔaraBAD1124::flgM-6H-TEV-δ-SVIE PflhDC7793 ΔrcsB::tetRA |
| TH20042 | ΔaraBAD1156::flgM+ Δhin-5717::FRT |
| TH20043 | ΔaraBAD1156::flgM+ Δhin-5717::FRT ΔfliC7715::tetRA |
| TH20044 | ΔaraBAD1124::flgM-6H-TEV-δ-SVIE PflhDC7793 ΔrcsB::tetRA ΔflgM5628::FKF |
| TH20047 | ΔaraBAD1156::flgM+ fljBenx |
| TH20048 | ΔaraBAD1124::flgM-6His-TEV-δ-SVIE fljBenx vh2 |
| TH20049 | ΔaraBAD1156::flgM+ fljBenx vh2 ΔfliC7715::tetRA |
| TH20050 | ΔaraBAD1124::flgM-6His-TEV-δ-SVIE fljBenx vh2 ΔfliC7715::tetRA |
| TH20053 | ΔaraBAD1156::flgM fljBenx vh2 PflhDC7768::tetRA |
| TH20055 | ΔaraBAD1156::flgM fliA8176 (−18 to +1G replaced by ataaAGGAGGtaaataA) |
| TH20056 | ΔaraBAD1124::flgM-6His-TEV-δ-SVIE fliA8176 |
| TH20057 | ΔaraBAD1156::flgM+ Δhin-5718::FRT |
| TH20058 | ΔaraBAD1156::flgM+ fljBenx vh2 flhD8089::(tetR-PtetA) |
| TH20059 | ΔaraBAD1156::flgM+ Δhin-5718::FRT ΔfliC7715::tetRA |
| TH20060 | ΔaraBAD1156::flgM+ ΔfliC7861::FCF fljBenx vh2 flhD8089::(tetR-PtetA) |
| TH20061 | ΔaraBAD1156::flgM+ fliA8177(H14N, R91C, L207P) |
| TH20062 | ΔaraBAD1156::flgM+ fliA8178(ATG H14N) |
| TH20063 | ΔaraBAD1156::flgM+ fljBenx vh2 flhD8089::(tetR-PtetA) ΔclpX::Tn10dCm |
| TH20064 | ΔaraBAD1156::flgM+ fljBenx vh2 ΔclpX::Tn10dCm |
| TH20065 | ΔaraBAD1156::flgM+ fljBenx vh2 ΔydiV251::tetRA |
| TH20066 | ΔaraBAD1156::flgM+ fljBenx vh2 ΔfliA5805::tetRA |
| TH20067 | ΔaraBAD1156::flgM+ fljBenx vh2 ΔfliB-T7727::tetRA |
| TH20068 | ΔaraBAD1156::flgM+ fljBenx vh2 ΔfliC7715::tetRA ΔclpX::Tn10dCm |
| TH20069 | ΔaraBAD1156::flgM+ fliA8179(GTG:ATG) |
| TH20071 | ΔaraBAD1156::flgM+ fljBenx vh2 fliA8176 ΔfliC7861::FCF |
| TH20072 | ΔaraBAD1156::flgM+ fljBenx vh2 flhD8089::(tetR-PtetA) ΔfliA5805::tetRA |
| TH20073 | ΔaraBAD1156::flgM+ fljBenx vh2 fliA8176 (−18 to +1G replaced by ataaAGGAGGtaaataA) ΔfliC7861::FCF ΔclpX72::FKF |
| TH20074 | ΔaraBAD1156::flgM+ ΔflgMN7753 ΔflgKL7770 Δtrg-7774 ΔycgR7775 ΔfliB-T7771 Δhin-fljA7752 fliA8176(−18 to +1G replaced by ataaAGGAGGtaaataA) |
| TH20075 | ΔaraBAD1124::flgM-6His-TEV-δ-SVIE fljBenx vh2 ΔfliB-T7727::tetRA |
| TH20077 | ΔaraBAD1124::flgM-6His-TEV-δ-SVIE fljBenx vh2 ΔclpX80::tetRA |
| TH20078 | ΔaraBAD1156::flgM+ fliA8176 ΔflgM5794::FCF |
| TH20079 | ΔaraBAD1156::flgM+ fljBenx vh2 flhD8089::(tetR-PtetA) ΔfliA5805::tetRA motA5461::MudJ |
| TH20080 | ΔaraBAD1156::flgM+ fljBenx vh2 flhD8089::(tetR-PtetA) motA5461::MudJ fliA8176 ΔfliC7861::FCF |
| TH20081 | ΔaraBAD1156::flgM+ fliA5226(H14N) ΔfliC7861::FCF |
| TH20082 | ΔaraBAD1156::flgM+ fljBenx vh2 flhD8089::(tetR-PtetA) motA5461::MudJ fliA5226(H14N) ΔfliC7861::FCF |
| TH20083 | ΔaraBAD1124::flgM-6His-TEV-δ-SVIE fljBenx vh2 flhD8089::(tetR-PtetA) ΔclpX::Tn10dCm |
| TH20221 | ΔaraBAD1190::flgM-6His-ETK-NCR247 ΔflgMN7753 ΔflgKL7770 ΔfliB-T7771 Δhin-fljA7752 PflhD7793 fliA5226(H14N) ΔlrhA ΔydiV252 ΔecnR4::FRT |
| TH20222 | ΔaraBAD1191::flgM-6His-ETK-NCR057 ΔflgMN7753 ΔflgKL7770 ΔfliB-T7771 Δhin-fljA7752 PflhD7793 fliA5226(H14N) ΔlrhA ΔydiV252 ΔecnR4::FRT |
| TH20223 | ΔaraBAD1192::flgM-6His-ETK-NCR224 ΔflgMN7753 ΔflgKL7770 ΔfliB-T7771 Δhin-fljA7752 PflhD7793 fliA5226(H14N) ΔlrhA ΔydiV252 ΔecnR4::FRT |
| TH20492 | ΔaraBAD1163::flgM-6His-ETK-MrVIA ΔflgMN7753 ΔflgKL7770 ΔfliB-T7771 Δhin-fljA7752 PflhD7793 fliA5226(H14N) ΔlrhA ΔydiV252 ΔecnR4::FRT |
| TH20685 | ΔaraBAD1195::flgM-6His-ETK-AMA1 ΔflgMN7753 ΔflgKL7770 ΔfliB-T7771 Δhin-fljA7752 PflhD7793 fliA5226(H14N) ΔlrhA ΔydiV252 ΔecnR4::FRT |
All strains were constructed during the course of this study.
FRT, FLP recombination target. Amino acid changes encoded by fliA alleles are shown in parentheses.
SDS-PAGE and Western blotting assays.
SDS-polyacrylamide gel electrophoresis (14%) was carried out using standard procedures using a Bio-Rad system. Levels of secreted, soluble, insoluble, or whole-cell proteins were analyzed by Western blotting. Expressed DnaK, FlgM, and σ28 levels in the whole-cell lysates and culture supernatants were determined. For analysis of strains containing ΔaraBAD::flgM+, equivalents of 50 and 100 optical density (OD) units were loaded for the cellular and supernatant fractions, respectively. In order to analyze strains for FlgM-6His-TEV-δ-SVIE and FlgM-6His-ETK-δ-SVIE secretion, 50 and 300 OD units were loaded for the cellular and supernatant fractions, respectively. For FlgM-6His-ETK-AMA1, FlgM-6His-ETK-NCR peptides, and FlgM-6His-ETK-MrVIA, 10 μl of the supernatant was loaded without the concentration step and detected using anti-His tag antibodies (mouse; Abcam). For NCR and AMA1 constructs, SDS-PAGE followed by Coomassie staining of 20-fold-concentrated supernatant (TCA concentration) was performed.
Anti-DnaK (mouse; Abcam), anti-His tag antibodies (mouse; Abcam), anti-σ28, and anti-FlgM antibodies (rabbit; Hughes lab) were used for detection of Western blots. DnaK was used as a protein standard control for loading concentration and for the presence of protein in the supernatant due to cell lysis. To visualize antigen-antibody complexes, secondary anti-rabbit-IRDye690 and anti-mouse-IRDye800 antibodies (Li-Cor) were used. Densiometric measurements of FlgM, σ28, and DnaK bands were performed using the Li-Cor Odyssey infrared imaging system software. All assays were performed in triplicate.
Motility assay.
Motility assays utilized soft agar tryptone plates (per liter: 10 g Bacto tryptone, 5 g NaCl, and 3 g Bacto agar). A bacterial colony was picked by toothpick and poked through the soft agar, followed by incubation at 37°C for about 5 h. If necessary, either arabinose (0.2%) or anhydrotetracycline (1 μg/ml) was added for ParaBAD or flhDC operon induction, respectively.
RESULTS
FlgM produced from ParaBAD::flgM is secreted.
FlgM is secreted through a completed HBB into the external spent growth medium. It has been shown that fusion of foreign proteins to the C terminus of FlgM can be used for protein purification purposes without the need to lyse cells prior to purification (16). In order to develop the flagellar T3S system for protein purification using FlgM as a secretion signal, we decided to characterize known aspects of FlgM production and secretion to maximize protein production using this system. About 80% of steady-state flgM transcription is from its class 3 promoter. Since FlgM is an anti-σ28 factor, its production via the class 3 promoter is under autoinhibition. For the studies presented here, we removed flgM gene transcription from FlgM autoinhibition by using a construct with the flgM gene transcribed from the arabinose-inducible chromosomal araBAD promoter (ParaBAD). This was accomplished by a targeted deletion of the chromosomal araBAD operon followed by insertion of the flgM+ gene in its place (13). This resulted in the arabinose-dependent induction of FlgM production in strains where the arabinose inducer is not degraded due to deletion of the araBAD structural genes. FlgM transcribed from ParaBAD was produced in the presence of arabinose and secreted at levels higher than those of FlgM produced and secreted from its native promoters (Fig. 1). A fliF deletion strain, which does not form flagellar structures, was unable to secrete ParaBAD-induced FlgM (Fig. 1). These results indicate that FlgM intrinsic-peptide secretion signals are sufficient for high-level FlgM secretion and the flagellar-dependent T3S system is required for FlgM secretion.
FIG 1.
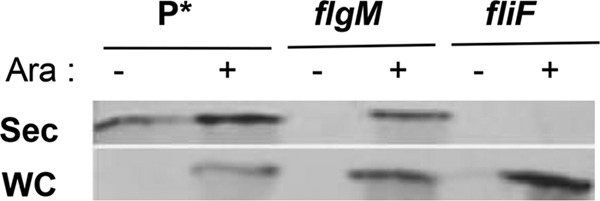
Increased levels of secreted FlgM under ParaBAD overexpression conditions. Western blot analysis of supernatant fractions of strains overexpressing FlgM from the chromosomal ParaBAD promoter. Overnight cultures were diluted 100-fold in LB medium and incubated at 37°C for 2 h, followed by addition of arabinose to 0.2% to induce excess FlgM expression. After 5 h of further incubation at 37°C, cells were separated from the supernatant by centrifugation and TCA was added to the cell supernatant to precipitate secreted proteins (see Materials and Methods). For the secreted proteins, 100 OD units sample was loaded; for the whole-cell protein, 50 OD units sample was loaded. Anti-FlgM antibody was used to determine levels of secreted and whole-cell FlgM (Sec, secreted FlgM; WC, whole-cell FlgM; Ara, 0.2% arabinose). P*, parent strain, TH18500 (ΔaraBAD1156::flgM+).
Mutations affecting FlhD4C2 activity also affect FlgM secretion.
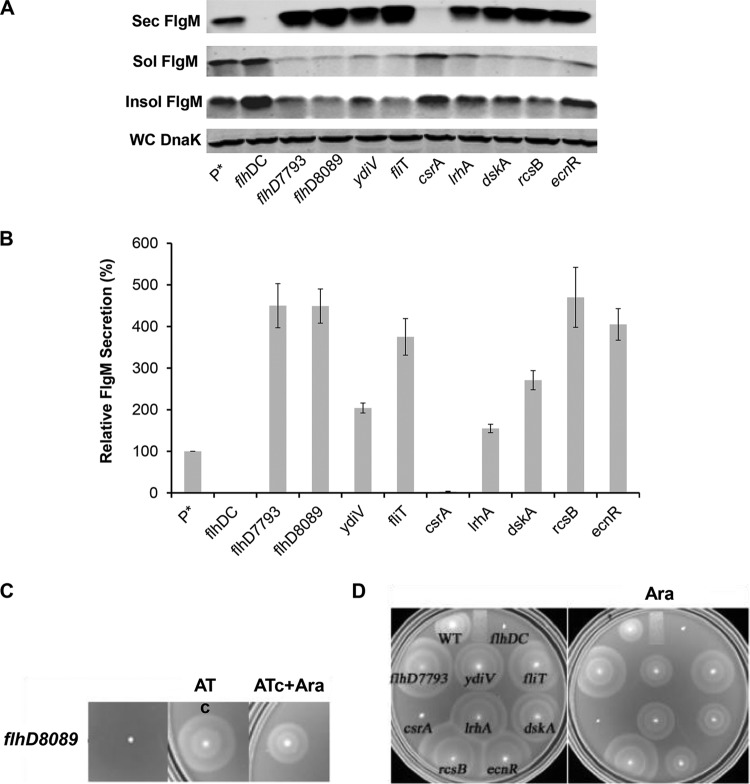
The flhDC operon is at the top of the flagellar transcriptional hierarchy. The regulation of flhDC is complex, and there are six known transcription initiation sites within the flhDC promoter region (20). It was previously reported that changes in the −10 sequences for the P1 and P4 transcription initiation sites within the flhDC promoter region to the canonical TATAAT sequence (the PflhDC7793 allele) resulted in the doubling of the number of HBB structures per cell and increased production and secretion of the flagellar hook protein (21). Other mutations resulting in increased hook protein secretion and presumably increased HBB structures per cell resulted in reduced expression or removal of known inhibitors of flhDC operon transcriptional or posttranscriptional control. We decided to test the effects of flhDC regulatory mutations on the secretion of FlgM transcribed from ParaBAD. The known transcriptional and posttranscriptional inhibitors of flhDC expression included in this study were EcnR, RscB, LrhA, FliT, DskA, and YdiV. EcnR is responsible for FlhDC-mediated autorepression (22). The FlhD4C2 complex directs transcription of ecnR, which in turn results in repression of flhDC transcription in concert with the RcsB protein. RcsB, which regulates capsular polysaccharide synthesis and a number of genes in response to membrane and cell wall damage, is a known repressor of flhDC transcription (23, 24). LrhA is also a known regulator of flhDC that has been shown to bind within the flhDC promoter region to inhibit flhDC operon transcription (25). FliT is transcribed from both class 2 and class 3 flagellar promoters (26). FliT binds to the FlhD4C2 complex and prevents activation of flagellar class 2 transcription (27, 28). FliT is also the secretion chaperone for the flagellar filament capping protein FliD (29). It is thought that secretion of FliD after HBB completion couples inhibition of further class 2 transcription by FlhD4C2 to HBB completion. The DskA protein acts with ppGpp to inhibit flhDC's transcription (30). DskA can also stimulate rpoS translation, which may inhibit flhDC transcription through an RpoS-mediated mechanism (31). YdiV is a posttranscriptional regulator that targets FlhD4C2 to the ClpXP protease for degradation in response to changes in nutrient growth conditions (32, 33). As a control, we included CsrA, which is a positive regulator of flhDC mRNA stability (34). The PflhDC8089 allele was constructed by replacing the flhDC promoter control region with a tetR-PtetA cassette from transposon Tn10. This resulted in the placement of the flhDC operon under the control of the tetA promoter, which can be induced by the addition of the tetracycline analog anhydrotetracycline. We also tested the flhDC promoter changes described previously that increase hook production and secretion (the PflhDC7793 allele) (21). The effects of the flhDC regulatory mutations on levels of secreted FlgM expressed from ParaBAD are shown in Fig. 2.
FIG 2.
Effect of flhDC operon expression on levels of secreted FlgM and cell motility. (A) Secreted levels of FlgM in strains affected in flhDC operon expression were determined by quantitative Western blot analysis using anti-FlgM antibody to detect FlgM in the supernatant of the spent growth medium. (Sec FlgM, secreted FlgM; Sol FlgM, cellular soluble FlgM; Insol FlgM, cellular insoluble FlgM; WC DnaK, whole cellular DnaK). P*, parent strain, TH18500 (ΔaraBAD1156::flgM+). (B) Relative FlgM secreted levels. (C) Swim phenotype on soft agar plates of strain TH18649 (ΔPflhDC8089::tetR-PtetA ΔaraBAD1156::flgM+) with no inducer added, 1 μg/ml anyhdrotetracycline (ATc) added, and both ATc and 0.2% arabinose (Ara) added. (D) Motility assay for various mutant backgrounds depicted with ΔaraBAD1156::flgM+ in the absence and presence of added 0.2% arabinose. WT, wild type.
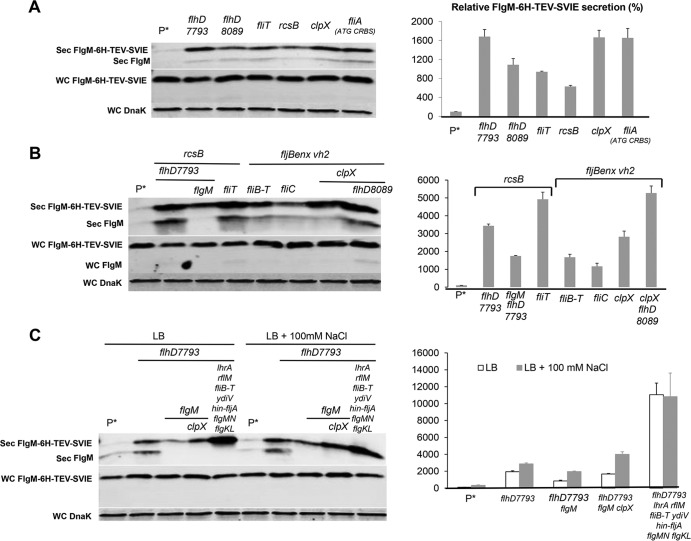
As expected, with deletion of the flhDC structural operon or deletion of the csrA gene, which is required for flhDC mRNA stability, there was no detectable FlgM in the secreted fraction, and FlgM accumulated in the cytoplasm. Mutations defective in known transcriptional and posttranscriptional inhibitors of flhDC expression resulted in increased levels of secreted FlgM, as did the presence of the PflhDC7793 allele. Secreted levels of FlgM in PflhDC7793, PflhDC8089(tetR-PtetA-flhDC), fliT, rcsB, and ecnR mutant strains were 4.5-, 4.5-, 3.8-, 4.7-, and 4-fold higher than that for the wild type, respectively. Secreted levels of FlgM were less in ydiV, lrhA, and dskA mutant strains at 2-, 1.5-, and 2.7-fold compared to that for the wild type, respectively (Fig. 2B). For each strain tested, the accumulated cellular levels of FlgM were inversely proportional to secreted FlgM levels. Significantly, replacement of the flhDC promoter region with the tetA promoter and regulatory region from transposon Tn10 allowed for the production of secreted FlgM at levels meeting or exceeding FlgM secreted levels observed in loss-of-function mutants for the negative regulators of flhDC expression that were tested. Induction of flhDC in the PflhDC8089(tetR-PtetA-flhDC) strain also conferred motility on swim plates (Fig. 2C).
In strains missing negative regulators of flhDC, the swimming phenotypes on soft agar plates indicated increased motility compared to that of wild-type LT2 (Fig. 2D). We were surprised to see substantial levels of motility in the same strains with flgM overexpressed from ParaBAD (Fig. 2D). FlgM inhibits σ28 at a stoichiometry of 1:1. It was expected that induction of FlgM from ParaBAD would prevent all σ28-dependent flagellar class 3 promoter transcription, especially in the wild-type LT2 background. This observation led to the conclusion that overexpressed cellular FlgM aggregated into an inactive form. Thus, the cellular component of FlgM was analyzed from both soluble and insoluble fractions from the cell pellet in the Western blot analysis of secreted and cellular FlgM levels. Although FlgM is a soluble protein, most cellular FlgM produced under overexpression conditions was insoluble (Fig. 2A), suggesting that excess cellular FlgM went into inclusion bodies, and the observed motility under FlgM-inducing conditions suggested that the insoluble form of FlgM was not active.
Effect of the FlgM T3S chaperone, σ28, on FlgM secretion.
As mentioned above, completion of the HBB coincides with a flagellar T3S substrate specificity switch from rod-hook-type secretion substrates to late or filament-type secretion substrates. The late secretion substrates include hook-filament junction proteins (FlgK and FlgL), filament cap protein (FliD), the alternately expressed filament proteins (FliC or FljB), and the anti-σ28 factor FlgM. Efficient secretion of each late secretion substrate requires the aid of a cognate T3S chaperone. These include FlgN (for FlgK and FlgL), FliT (for FliD), FliS (for FliC and FljB), and σ28 (for FlgM) (13, 29, 35). T3S chaperones fall into three classes: (i) those that bind and protect substrates from proteolysis in the system prior to secretion, (ii) those that facilitate substrate secretion, and (iii) those that both stabilize and facilitate secretion. It has been shown that interactions of FlgN, FliS, and FliT flagellar chaperones with the C-terminal cytoplasmic domain of FlhA are required for efficient export of their cognate substrates and that these interactions contribute to coordinating assembly of the flagellar filament (36–38). The σ28 protein falls into the category of T3S chaperones that both stabilize and facilitate secretion; it protects FlgM from proteolysis in the cytoplasm and facilitates the secretion of FlgM, presumably by helping to localize FlgM to the base of the flagellum. A mutant of σ28 with two amino acid substitutions that render it defective in recognition of the −10 and −35 promoter sequences (R91C and L207P) retains its T3S chaperone activity for FlgM secretion (13). This indicated that the T3S chaperone function of σ28 was a separate process from its transcription activity.
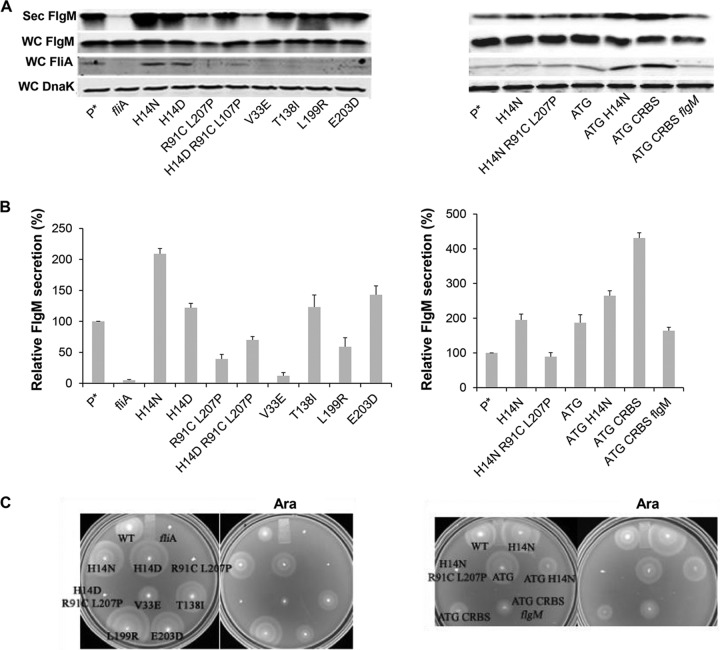
We tested the effects of FlgM bypass mutations in σ28 regions 2, 3, and 4 on secretion of FlgM expressed from ParaBAD (Fig. 3A and B). The region 2.1 mutations included H14D and H14N, which result in increased protein stability, and the V33E allele, whose mutant is defective in binding FlgM. The other FlgM bypass mutations tested included the T138I allele from region 3.1 and L199R from region 4.1 and E203D at the initiation point of region 4.2.
FIG 3.
Effect of fliA (σ28) alleles on levels of secreted FlgM and cell motility. (A) Levels of secreted FlgM were determined for strains carrying different alleles of the σ28 structural gene fliA by quantitative Western blot analysis using anti-FlgM antibody to detect FlgM in the supernatant of the spent growth medium (Sec FlgM, secreted FlgM; WC FlgM, whole cellular FlgM; WC FliA, whole cellular FliA; WC DnaK, whole cellular DnaK). P*, parent strain, TH18500 (ΔaraBAD1156::flgM+). (B) Relative levels of secreted FlgM. (C) Motility assay with various mutant backgrounds, depicted with the ΔaraBAD1156::flgM+ strain (WT), in the absence and presence of added 0.2% arabinose.
As expected from previous studies, the strain with the fliA null allele showed a strong reduction in FlgM secretion, to a level that was 5% of that of the fliA+ strain. The V33E and L199R alleles resulted in FlgM secretion levels that were 12% and 59% of that of the wild type, respectively. Also expected, strains with the H14D and H14N alleles exhibited secreted FlgM levels that were 2.1- and 1.5-fold that seen for the wild type. Unexpectedly, the T138I and E203D alleles, which are defective in binding FlgM, resulted in increased secreted FlgM levels, 1.2- and 1.4-fold that of the wild type, respectively. The promoter binding-defective fliA double mutant (R91C L207P), when also tested and compared to the wild type, showed a 39% level of secreted FlgM. We combined the promoter binding-defective R91C L207P substitutions with the H14D increased-stability substitution and observed secreted FlgM levels in between those observed with either the R91C L207P double mutant or the H14D single mutant.
For wild-type FliA and the H14N, H14D, R91C L207P, and H14D R91C L207P mutants, the FliA level in the cell was consistent with the secreted FlgM level (Fig. 3A). We speculated that the FlgM secretion level was related to the FliA concentration within the cell. Therefore, we introduced other mutations, which would potentially increase the intracellular FliA concentration, to see whether they could affect FlgM secretion. These mutations included an fliA start codon change from GTG to ATG that was combined either with an H14N stabilization substitution or with a change of the fliA ribosome binding sequence (RBS) to a canonical sequence (CRBS). All of these changes resulted in increased FliA intracellular levels and secreted FlgM levels from 2- to 4-fold that of the wild type (Fig. 3A and B). Deletion of the chromosomal flgM gene in the fliA CRBS ATG double mutant background resulted in a reduction of both of intracellular FliA and secreted FlgM. We conclude that the excess σ28 produced in the fliA CRBS ATG double mutant background requires chromosomal flgM expression in addition to ParaBAD-expressed flgM to contribute to σ28 stability, which would also improve FlgM secretion. This is consistent with the results showing that FlgM not only acts as an anti-sigma factor but also protects FliA from being degraded (39).
Motility assays of the fliA mutants under FlgM-induced conditions are showed in Fig. 3C. As expected, the fliA null allele and mutants containing the R91C L207P alleles, which are unable to transcribe flagellar class 3 promoters and therefore unable to produce flagellin, are nonmotile under any condition. The H14N, H14D, and ATG substitution mutants showed increased swarm sizes on motility plates compared to that of the wild type, consistent with previous results (40). V33E, T138I, L199R, and E203D mutants are reported to have flagellar transcriptional activities in the presence of FlgM that are 8.0-, 31-, 45-, and 7-fold higher than that of the wild type, respectively, and their motility phenotypes on swim plates under FlgM-inducing conditions correlate with these levels. The H14N ATG and CRBS ATG double mutants showed decreased motility on swim plates, although they have higher cellular levels of σ28 than the wild-type strain.
Effect of flagellar late T3S substrate competitors on FlgM secretion.
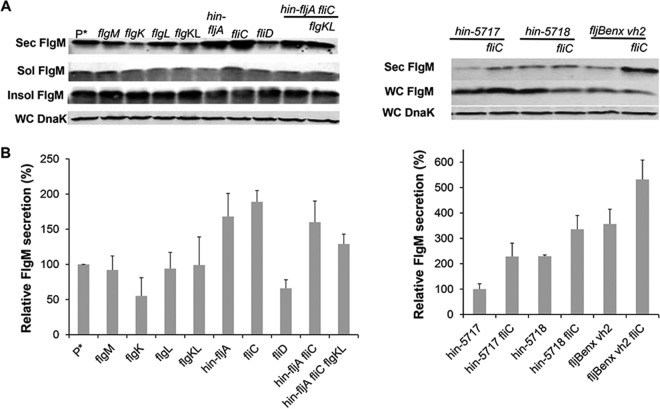
The initial report of FlgM secretion upon HBB completion presented qualitative data suggesting that secreted FlgM levels were higher in strains missing filament protein, a potential secretion competitor for FlgM (9). We tested for the effects of removing potential late secretion substrate competitors on secreted FlgM levels, anticipating that their removal would improve the yield of secreted FlgM. The number of late substrate subunits in the assembled flagellum is about 11 each for the hook-filament junction proteins FlgK and FlgL, 5 for the filament capping protein FliD, and depending on filament length, up to 20,000 for FliC or FljB. The results, presented in Fig. 4A and B, show that removal of FlgK, FlgL, or FliD has little or no effect on secreted FlgM levels, while removal of the filament substrates results in increased levels of secreted FlgM, although only up to 1.9-fold higher than the wild-type level. This result suggests that late T3S substrate levels are not a significant limiting factor in the secretion of FlgM through the flagella.
FIG 4.
Effect of flagellar late substrate deletion, growth phase, and fljBenx vh2 mutations on levels of secreted FlgM. (A) Secreted levels of FlgM were determined for strains carrying different flagellum late substrate gene (flgK, flgL, fliC, fliD, and fljB) deletion mutants, flagellum-specific phase (Δhin-5717 and Δhin-5718) mutants, and fljBenx vh2 mutants by quantitative Western blot analysis using anti-FlgM antibody to detected FlgM in the supernatant of the spent growth medium. Sec FlgM, secreted FlgM; Sol FlgM, cellular soluble FlgM; Insol FlgM, cellular insoluble FlgM; WC DnaK, whole cellular DnaK. P*, parent strain, TH18500 (ΔaraBAD1156::flgM+). (B) Relative secreted FlgM levels.
We also tested the effects of flagellar phase variation on FlgM secretion (Fig. 4A and B). Salmonella enterica alternately expresses one of two flagellin subunit genes, fliC or fljB. Strains carrying the hin-5717 allele are locked in the FliCON FljBOFF flagellin expression mode, while strains carrying the hin-5718 allele are FliCOFF FljBON (41). fljBenx vh2 is a historical relic that is also FliCON FljBOFF and resulted from replacement of the hin-fljBA region from S. enterica strain LT2 with the same region from Salmonella enterica serovar Abortus-equi locked in the FliCON FljBOFF mode (42). The FlgM secretion level in a strain carrying the hin-5717 allele was similar to that of the wild-type strain. Deletion of the fliC gene in the hin-5717 background resulted in a 1.9-fold increase in secreted FlgM compared to that for the wild type. The presence of the hin-5718 allele alone increased secreted FlgM levels by 1.9-fold, and the additional removal of the fliC gene in this background further increased the secreted FlgM level 2.7-fold that of the wild type even though FliC flagellin is not produced in the hin-5718 background. However, fliC mRNA is produced but not translated in the hin-5718 background (41), suggesting that fliC mRNA has a negative effect on FlgM protein secretion. Secreted levels of FlgM in the fljBenx vh2 background were 2.9-fold higher than those in the Δhin-5717 strain, which is also locked for fliCON expression, and further removal of the fliC gene in this background increased secreted FlgM levels about 5-fold of those of the Δhin-5717 strain. We also observed a 2-fold increase in secreted FlgM in the locked fljBON background, which could be accounted for by differences in fliC and fljB promoter strengths. The reason for the difference between the two locked fliCON strain backgrounds (fljBenx vh2 and Δhin-5717) remains to be determined.
The effect of the flagellar late T3S chaperones on FlgM secretion.
Next, we tested the effects of removing the late secretion chaperones FlgN, FliS, and FliT on secreted levels of FlgM expressed from ParaBAD. We reasoned that they might compete with σ28 for delivery of FlgM to the flagellar secretion system for export. Also, T3S chaperones are associated with regulatory functions in the absence of their cognate secretion substrates. FlgN, the T3S chaperone for FlgK and FlgL, is reported to inhibit flgM mRNA translation (43), σ28 is a transcription factor for flagellar class 3 promoters (6), and FliT acts as an anti-FlhD4C2 factor (28). Only FliS is not reported to have a regulatory function in the absence of its cognate secretion substrates, FliC and FljB. However, FliS has recently been shown to bind FlgM with a 1 FliS:1 FlgM stoichiometry, suggesting it does have some regulatory role that has yet to be elucidated (44, 45).
Removal of FlgN had little effect on FlgM secretion (Fig. 5A and B). This was expected, since FlgK and FlgL did not have a significant effect on secreted FlgM levels either (Fig. 4A and B). Removal of FliT, which is also analyzed in Fig. 1, resulted in a 4-fold increase in secreted FlgM levels. We do not have alleles of fliT that separate its anti-FlhD4C2 activity from its chaperone activity. Thus, the increase in secreted FlgM levels is likely due to enhanced FlhD4C2 activity in the absence of FliT. We did observe a 3-fold increase in secreted FlgM levels in the absence of FliS. However, fliS is transcribed in an operon upstream of the fliT gene. Thus, any polar effect of the fliS alleles on fliT would result in increased secreted levels of FlgM due to decreased fliT gene expression. Since we do not have a null allele of fliS resulting from missense mutation, we could not determine an effect of FliS removal on FlgM secretion that was independent of the effects of fliS deletion on FliT expression.
FIG 5.
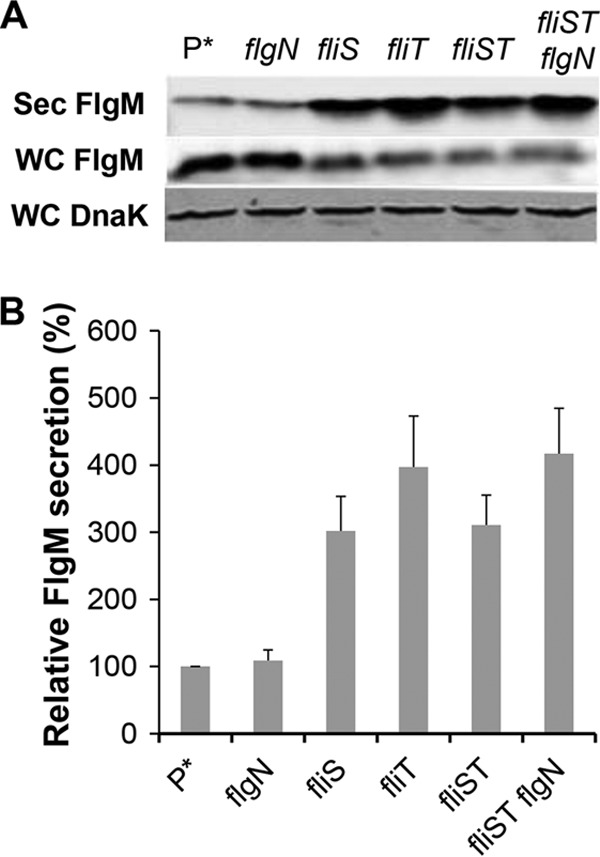
Effects of late flagellum T3S chaperone deletion mutations on levels of secreted FlgM. (A) Secreted levels of FlgM were determined for strains carrying different deletion mutant alleles of the flagellar T3S chaperone genes flgN, fliS, and fliT by quantitative Western blot analysis using anti-FlgM antibody to detect FlgM in the supernatant of the spent growth medium. Sec FlgM, secreted FlgM; WC FlgM, whole cellular FlgM; WC DnaK, whole cellular DnaK. P*, parent strain, TH18500 (ΔaraBAD1156::flgM+). (B) Relative levels of secreted FlgM.
Deletion of Salmonella pathogenicity island 1 (Spi1) results in increased levels of secreted FlgM.
The flhDC operon is the master operon of both the flagellar regulon and the genes of Spi1 (46, 47). The Spi1 regulon encodes genes needed for the structure and assembly of the Spi1 injectisome T3S system. The fliZ gene is transcribed in the fliAZ operon, and FliZ activates the Spi1 transcriptional activator HilD, which in turn activates Spi1 transcription under high salt (200 mM NaCl) microaerobic growth conditions (48). One HilD-activated gene product, RtsB, acts to repress flhDC transcription, providing a feedback loop for the entire flagellar-Spi1 regulon (49). Recent work has demonstrated that HilD activates flhDC transcription late in the cell's growth phase under non-Spi1 induction conditions (50, 51). We tested the effects of deletions of both the Spi1 and Spi2 Salmonella virulence systems on the secreted levels of overexpressed FlgM (from ParaBAD). The loss of Spi1 resulted in decreased FlgM secretion levels, to 55% of that of the wild type under non-Spi1-inducing conditions (aerobic, 86 mM NaCl), whereas loss of Spi2 had no significant effect on secreted levels of FlgM (Fig. 6A and B).
FIG 6.
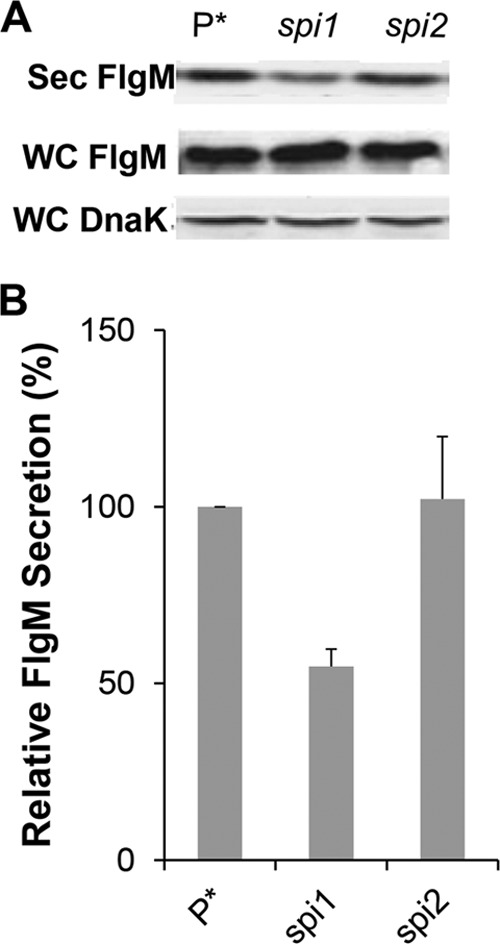
Effects of Spi-1 and Spi-2 deletions on levels of secreted FlgM. (A) Secreted levels of FlgM were determined for strains carrying deletion mutant alleles of the either the Spi1 or Spi2 gene by quantitative Western blot analysis using anti-FlgM antibody to detect FlgM in the supernatant of the spent growth medium. Sec FlgM, secreted FlgM; WC FlgM, whole cellular FlgM; WC DnaK, whole cellular DnaK. P*, parent strain, TH18500 (ΔaraBAD1156::flgM+). (B) Relative levels of secreted FlgM.
Effect of protease removal on FlgM secretion levels.
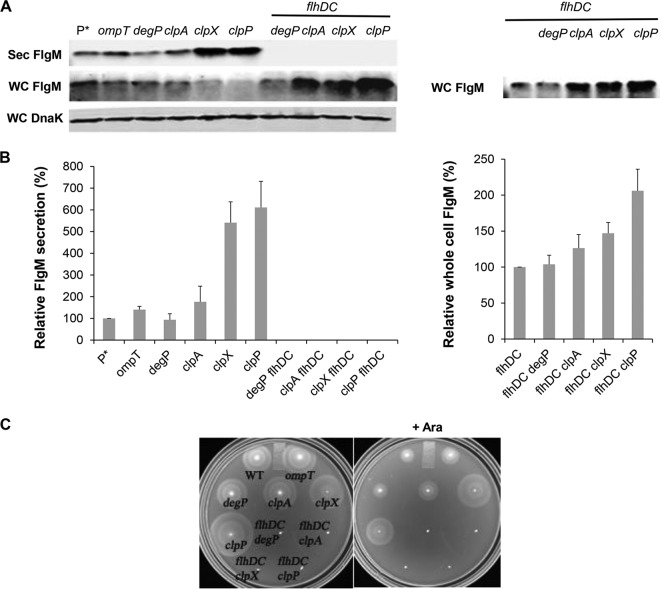
A common technique used to improve protein yield from the cytoplasm is removing cellular proteases. In addition, proteases present in the outer membrane, such as OmpT, can decrease protein yield by degradation after cell lysis (52, 53). The ClpA and ClpX proteins interact with different substrates for delivery to the ClpP serine protease for degradation (54). DegP is a periplasmic protease that exhibits broad substrate specificity. DegP is exclusively directed against unfolded, mislocalized, hybrid and recombinant proteins that are improperly folded from overexpression in the periplasm (55, 56).
The results of protease removal on FlgM secretion levels are shown in Fig. 7. Removal of OmpT resulted in a slight increase in the yield of secreted FlgM, while loss of DegP had little effect. Removal of ClpA, ClpX, and ClpP increased the FlgM secretion yield by 1.7-, 5.4-, and 6.1-fold, respectively, compared to that of the wild type (Fig. 7A and B). This is consistent with the observation that FlhD4C2 is a substrate for YdiV-directed degradation by the ClpXP protease system (32, 33). As a control for cell lysis, deletion of flhDC in the protease mutant backgrounds showed no detectable FlgM in the secreted fraction. In this background, the cellular level of FlgM remained unaffected by loss of DegP mutation, while null alleles of clpA, clpX, and clpP mutation in the flhDC background increased intracellular FlgM accumulation by 1.3-, 1.5-, and 2.1-fold, respectively (Fig. 7B). Our results suggest that ClpA, ClpX, and ClpP are involved in FlgM degradation independent of FlhDC; the increased FlgM secretion in the clpA null strain was due to an increased cellular level of FlgM alone. The effects of the ClpXP protease in the presence of flhD+C+ were likely due to both FlgM stability and FlhD4C2 stability. This is consistent with increased motility observed in the clpP and clpX mutant strains (Fig. 7C).
FIG 7.
Effects of cellular protease mutant alleles on levels of secreted FlgM and cell motility. (A) Secreted levels of FlgM were determined for strains carrying deletion mutant alleles of the ompT, degP, clpA, clpX, or clpP gene with and without a functional flhDC operon by quantitative Western blot analysis using anti-FlgM antibody to detect FlgM in the supernatant of the spent growth medium. Sec FlgM, secreted FlgM; WC FlgM, whole cellular FlgM; WC DnaK, whole cellular DnaK. P*, parent strain, TH18500 (ΔaraBAD1156::flgM+). (B) Relative levels of secreted FlgM in the flhD+C+ and flhDC null backgrounds and relative whole-cell FlgM accumulation level in flhDC null background. (C) Motility assay in various mutant backgrounds, depicted with the ΔaraBAD1156::flgM+ strain (WT) in the absence and presence of added 0.2% arabinose.
Effect of ionic strength on secreted FlgM levels.
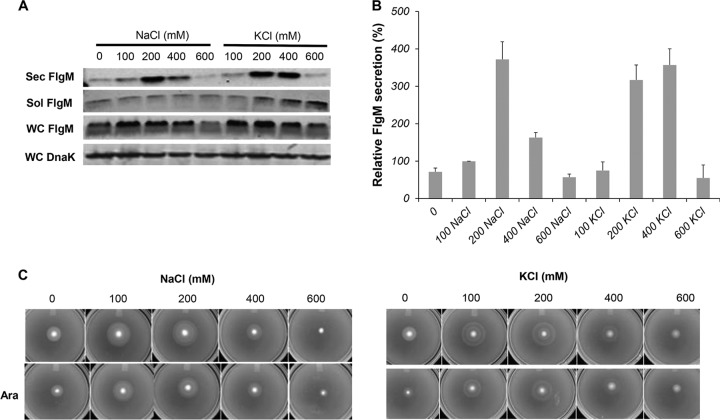
It was reported that high osmolarity increased Spi1 invA gene transcription in S. enterica and that Spi1-dependent type III secretion occurred only in bacteria grown under high-salt conditions (57). We tested the effects of either increased NaCl or KCl on secreted levels of FlgM produced from ParaBAD-flgM+. FlgM secreted levels increased when the NaCl concentration was raised to 200 mM and then dropped at concentrations of 400 and 600 mM. At 200 mM NaCl, the level of secreted FlgM was 3.7-fold higher than the level at 100 mM NaCl (Fig. 8A and B), which is close to the NaCl concentration in our standard LB medium (86 mM). Thus, the NaCl concentration in LB medium is not optimal for FlgM secretion. KCl had a similar effect on secreted FlgM levels; 200 and 400 mM KCl produced the highest levels of secreted FlgM, at 3.2- and 3.6-fold compared to the secreted FlgM level at 100 mM NaCl. This could be due to effects on the solubility of FlgM in the cytoplasm. Increased ionic strength also had a positive effect on motility in soft agar, although this was suppressed under FlgM induction conditions, possibly due to inhibition of σ28-dependent flagellar class 3 transcription (Fig. 8C).
FIG 8.
Effects of NaCl and KCl ionic strengths on levels of secreted FlgM and cell motility. (A) Secreted levels of FlgM were determined with strain P*, the parent strain, TH18500 (ΔaraBAD1156::flgM+), in LB-0.2% arabinose medium with NaCl and KCl added at concentrations depicted by quantitative Western blot analysis, using anti-FlgM antibody to detect FlgM in the supernatant of the spent growth medium. Sec FlgM, secreted FlgM; Sol FlgM, cellular soluble FlgM; Insol FlgM, cellular insoluble FlgM; WC DnaK, whole cellular DnaK. (B) Relative levels of secreted FlgM. (C) Motility assay using various concentrations of NaCl and KCl, depicted with strain TH18500, in the absence and presence of added 0.2% arabinose (Ara).
Effect of multiple conditions on FlgM secretion.
Individual results described above that enhanced the yield of secreted FlgM were combined in order to obtain a strain and conditions that maximized this yield under conditions of FlgM overexpression from ParaBAD::flgM+ (Fig. 9A and B). Some of these strains, such as the fljBenx vh2 strain, strains containing clpX mutations, and strains containing flhD8089, provided the highest FlgM secretion levels. All of them produced secreted FlgM levels at about 5-fold that of the wild type. In these strains, only a trace amount of FlgM accumulated within the cell, suggesting that expression and stability of cellular FlgM were limiting.
FIG 9.
Effects of different combined mutations on levels of secreted FlgM. (A) Secreted levels of FlgM were determined for strains carrying different combined mutations, depicted by quantitative Western blot analysis using anti-FlgM antibody to detect FlgM in the supernatant of the spent growth medium (Sec FlgM, secreted FlgM; WC FlgM, whole cellular FlgM; WC DnaK, whole cellular DnaK). P*, parent strain, TH18500 (ΔaraBAD1156::flgM+). (B) Relative levels of secreted FlgM.
Use of FlgM as a T3S signal to secrete δ-SVIE, MrVIA, AMA1, and NCR peptides.
We tested the use of FlgM as a secretion signal to secrete and purify peptides and proteins that are otherwise difficult to purify. For this purpose, we asked collaborators to provide DNA clones of proteins they were unable to purify by standard methods. These include a malarial surface protein, AMA-1 (apical membrane antigen 1), synthesized by Plasmodium falciparum, in collaboration with Ray Norton, the defensin-like peptides from Sinorhizobium meliloti, NCR (nodule-specific cysteine-rich) peptides, in collaboration with Graham Walker, and the neuroactive conopeptides from Conus sp., δ-SVIE and MrVIA, in collaboration with Baldomero Olivera. The δ-SVIE protein is a small peptide produced by a venomous marine cone snail and inhibits sodium channels in vertebrate neuromuscular systems. This 31-amino-acid peptide (DGCSSGGTFCGIHPGLCCSEFCFLWCITFID) is hydrophobic and contains 6 cysteine residues that form 3 pairs of intramolecular disulfide bonds. The hydrophobic nature of the peptide and requirement for multiple disulfide bond formation to produce an active conformation of δ-SVIE are an impediment to proper folding when this peptide is overexpressed in E. coli. We wanted to test whether δ-SVIE could be produced in an active form via FlgM-mediated secretion. We reasoned that because secretion from the cell initiates with the N terminus of FlgM, δ-SVIE sequence of the FlgM-δ-SVIE fusion would exit the cell as it exits the ribosome: from the N terminus to the C terminus. This might facilitate proper folding and disulfide formation of δ-SVIE into an active conformation. C-terminal fusions of δ-SVIE to hexa-His-tagged (6His) FlgM with either TEV (tobacco etch virus) or enterokinase (ETK) protease cleavage sites engineered between the two proteins were expressed from the chromosomal ParaBAD expression locus. The 6His tag facilitates purification using a Ni-agarose affinity column, and the TEV and ETK cleavage sites are recognized by TEV protease and enterokinase, respectively, allowing the secreted δ-SVIE to be separated from its FlgM secretion signal.
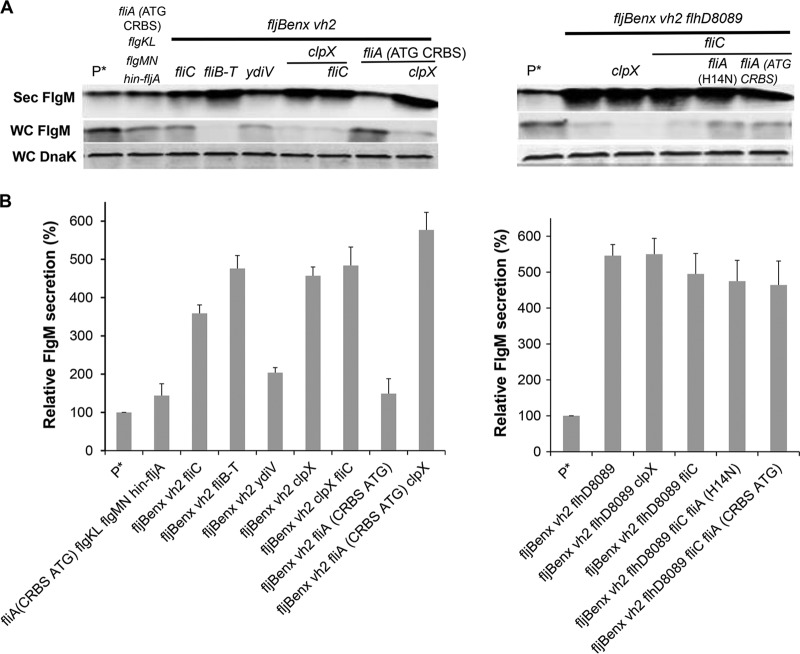
All of the selected single mutants increased FlgM-6His-TEV-δ-SVIE secretion (see Fig. 10A and B). As expected, the secreted levels of ParaBAD-expressed fusion protein levels were enhanced in mutant backgrounds that produced higher secreted FlgM levels. Secreted levels of FlgM-6His-TEV-δ-SVIE in the flhD7793 (TH18880), flhD8089 (TH18647), fliT (TH18769), rcsB (TH18830), clpX (TH18353), and fliA (CRBS ATG) (TH20056) strains were 16.8-, 10.9-, 9.4-, 6.3-, 16.7-, and 16.6-fold that of the wild type (TH17831). We also combined different mutations together to test FlgM-6His-TEV-δ-SVIE secretion levels. In the multiple mutant background flhD7793 rcsB (TH19675), flhD7793 rcsB flgM (TH20044), rcsB fliT (TH19673), fljBenx vh2 fliB-fliT (TH20075), fljBenx vh2 fliC (TH20050), fljBenx vh2 clpX (TH20077), and flhD8089 fljBenx vh2 clpX (TH20044) strains, levels were 34-, 17-, 49-, 16.7-, 11.7-, 28.2-, and 52.7-fold of that of the wild type. When 6His-ETK was inserted between FlgM and SVIE, the flhD7793 (TH19118), flhD7793 flgM (TH19122), flhD7793 flgM clpX (TH19145), and flhD7793 lrhA fliB-fliT ydiV hin-fljA flgMN flgKL (TH19120) strains increased FlgM-6His-ETK-δ-SVIE secretion levels up to 19-, 8.5-, 17-, and 110-fold of that of wild type. Except for the flhD7793 lrhA fliB-fliT ydiV hin-fljA flgMN flgKL strain (TH19120), addition of 100 mM NaCl resulted in increased secretion levels of the fusion protein.
FIG 10.
Effects of different combined mutations and added salt concentrations on levels of secreted FlgM-6His-TEV-δ-SVIE and FlgM-6His-ETK-δ-SVIE. (A, B, and C) Levels of secreted FlgM-6His-TEV-δ-SVIE and FlgM-6His-ETK-δ-SVIE were determined for strains carrying different combined mutations, depicted by quantitative Western blot analysis using anti-FlgM antibody to detected FlgM-6His-TEV-δ-SVIE and FlgM-6His-ETK-δ-SVIE in the supernatant of the spent growth medium (Sec FlgM-6His-TEV-SVIE, secreted FlgM-6His-TEV-δ-SVIE; Sec FlgM-6H-ETK-δ-SVIE, secreted FlgM-6His-ETK-δ-SVIE; Sec FlgM, secreted FlgM; WC DnaK, whole cellular DnaK). Relative levels of secreted FlgM-6His-TEV-δ-SVIE and FlgM-6His-ETK-δ-SVIE are presented at the right of the gels. P*, parent strain, TH17831 (ΔaraBAD1124::flgM-6His-TEV-δ-SVIE).
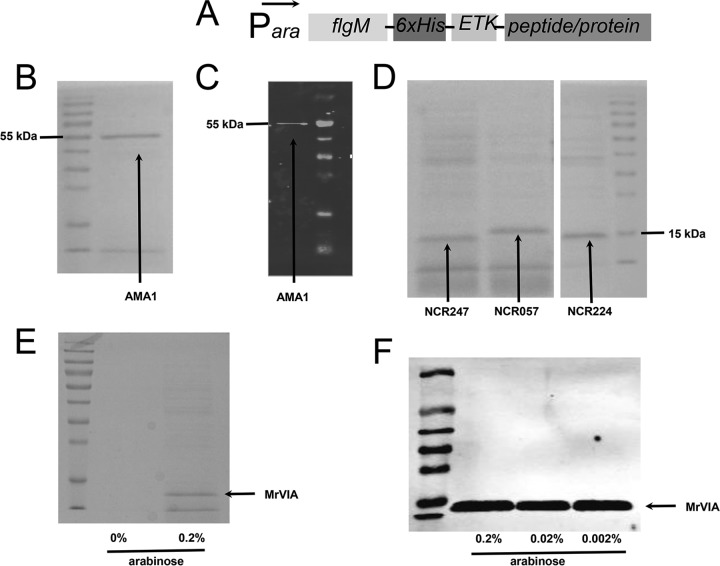
Figure 11 shows the secretion of ParaBAD-expressed FlgM-6His-ETK-AMA1, FlgM-6His-ETK-NCR, and FlgM-6His-ETK-MrVIA constructs in the flhDC7793, lrhA, ydiV fliB-fliT hin-fljBA, flgMN, and flgKL mutant backgrounds. The culture supernatant was concentrated 20-fold by TCA precipitation and run on SDS-PAGE gels. Coomassie staining revealed the FlgM fusion constructs to be the prominent proteins present in the cell supernatant.
FIG 11.
Secretion of AMA1, NCR, and MrVIA peptides fused to a His-tagged FlgM secretion signal. (A) Schematic diagram of the constructs tested. (B) Coomassie-stained SDS-PAGE gel of the supernatant from a culture expressing FlgM-His6-ETK-AMA1 in an optimized secretion strain (TH20685). The supernatant was concentrated 20-fold by TCA precipitation. (C) Western blot against 20 μl of nonconcentrated supernatant from panel B with anti-His antibody. (D) Coomassie-stained SDS-PAGE gel of the supernatant from cultures expressing three different NCR peptides fused to a His-tagged FlgM secretion signal in an optimized secretion strain background (NCR247, TH20221; NCR057, TH20222; NCR224, TH20223). The supernatants were concentrated 20-fold by TCA precipitation. (E) Coomassie-stained SDS-PAGE gel of the supernatant from a culture expressing conopeptide MrVIA fused to a His-tagged FlgM secretion signal in an optimized secretion strain background (TH20492). The supernatants were concentrated 20-fold by TCA precipitation. (F) Western blot against 20 μl of nonconcentrated supernatant from strain TH20492 (FlgM-His-ETK-MrVIA) with anti-His antibody grown under different arabinose induction concentrations.
DISCUSSION
The type III secretion (T3S) systems of the flagellum and injectisome provide a conduit for the production and purification of recombinant proteins that are fused to the T3S signal. Although the nature of the T3S signal is not fully understood, there appears to be a universal requirement for an N-terminal disordered peptide signal and a coupling to the proton motive force as the secretion fuel (58, 59). Many but not all T3S substrates require cognate T3S chaperones for their stability in the cytoplasm and/or as secretion pilots for targeting the T3S substrate to the secretion apparatus in the cytoplasmic membrane. Another feature of T3S systems is the ability to undergo a secretion specificity switch from early to late secretion substrate specificity. In the flagellar system, this occurs upon completion of extracellular hook growth that is more than 40 nm in length, resulting in the transition to filament-type substrate secretion and assembly (60, 61). The FlgM protein is an integral part of the transition from hook completion to filament assembly. FlgM is a late, filament-type flagellar secretion substrate and an anti-σ28 factor. FlgM is a small, 97-amino-acid protein. The N-terminal half of FlgM includes the T3S signal, while the C-terminal half includes the anti-σ28 interaction domain. It was initially determined that either domain of FlgM could serve as a late flagellar secretion substrate. Later it was found that σ28 was the T3S chaperone for FlgM, which accounted for the ability for low-level C-terminal FlgM secretion to occur in the absence of the N-terminal secretion signal. It has been shown that fusion of recombinant proteins to the C terminus of FlgM would allow for their secretion and purification without a requirement to lyse cells prior to purification (16). In this study, we examined conditions that might facilitate FlgM production and secretion. These conditions were then applied to produce the recombination proteins FlgM-6His-TEV-δ-SVIE, FlgM-6His-ETK-δ-SVIE, FlgM-6His-ETK-MrVIA, FlgM-6His-ETK-AMA1, and FlgM-6His-ETK-NCR peptides to produce and purify these proteins without cell lysis using FlgM as a vector to direct the secretion of the fused peptide/protein from the cell via the flagellar T3S system.
FlgM and FlgM fusion constructs were overexpressed from the chromosomal ParaBAD promoter by replacing the araBAD coding region with the flgM or FlgM fusion coding regions. By removing araBAD, we ensured that an arabinose inducer would not be consumed as a carbon source. This inducing system proved sufficient to produce FlgM in quantities in excess of what the cell could secrete (Fig. 2). We then manipulated the cells by introducing mutations that might increase FlgM stability and secretion in order to increase production of secreted FlgM.
Initially we examined the effect of mutations that were known to increase the number of flagellar T3S systems per cell on secreted FlgM levels. These included null mutations in known negative regulators of flhDC transcription (ecnR, rcsB, lrhA, and dskA) and flhDC promoter up alleles, PflhDC7793 and PflhDC8089. We also tested null alleles of fliT and ydiV that inhibit FlhD4C2 function at a posttranscriptional level. Such mutations had been shown to increase the production and secretion of the flagellar hook protein into the periplasm. All mutant backgrounds tested resulted in increased levels of secreted FlgM. After determining the effects of different mutations on secreted levels of FlgM, we tried to combine our observations to construct an optimized secretion strain to maximize the amount of FlgM secreted from the cell. We found that all of the strains containing fliB-fliT, clpX, or flhD8089 increased FlgM secretion by about 5-fold that of the wild-type strain, but the combination did not increase FlgM secretion further. This suggests that there is a limit on the number of flagellar secretion portals (hook-basal bodies) that can be achieved by increased FlhD4C2-dependent transcription and the maximal effect was achieved with the individual mutations. Under conditions of increased FlhD4C2-dependent transcription, it is possible that the cellular levels of FlgM became a limiting factor in FlgM secretion.
We explored the effects of various alleles of the FlgM T3S chaperone σ28, encoded by the fliA gene. Any allele that resulted in increased cellular levels of σ28 resulted in a corresponding increase in FlgM secretion, especially under FlgM overexpression conditions. These results are consistent with the role of σ28 as the T3S chaperone for FlgM.
Removal of late secretion competitors of FlgM secretion or their cognate chaperones had mixed results. Of the 4 secretion competitors FlgK, FlgL, FliD, and FliC/FljB, only removal of the filament late secretion substrates FliC/FljB had a significant effect on FlgM secretion. This is not surprising, since the amount of filament subunits in the flagellum is about 1,000 times that of the other three components.
Removal of the filament T3S chaperone protein, FliS, resulted in a 3-fold increase in secreted FlgM levels, while removal of the FlgK and FlgL T3S chaperone FlgN had no effect. Recently, FliS was shown to bind FlgM, but FliS was not able to dislodge FlgM in complex with σ28 (44, 45). Since fliS and fliT are cotranscribed in the same operon (fliS is upstream of fliT), any deletion mutation of fliS may affect fliT gene expression. In this work, we could not conclude how removal of FliS might affect FlgM secretion. Unexpectedly, a flagellar phase variation mutant allele, fljBenx vh2, showed a significant increase in secreted FlgM levels compared to results with the hin-5717 allele.
We observed decreased secreted FlgM levels in a Spi1 deletion strain. This is in agreement with recent results demonstrating a positive effect of the Spi1 transcription factor HilD on flhDC transcription under Spi1-noninducing conditions (50, 51).
Removal of cellular proteases also produced mixed results on FlgM secretion. The ClpXP protease was known to regulate the number of flagella per cell by degradation of the FlhD4C2 complex, which is directed by the YdiV protein. YdiV is produced during poor nutrient growth conditions. YdiV binds the FlhD component of FlhD4C2, which prevents further interaction between FlhD4C2 and DNA. YdiV then targets FlhD4C2 to the ClpXP protease for degradation (32, 33). As expected, removal of either ClpX or ClpP resulted in increased FlgM secreted levels. Removal of the DegP or OmpT protease was also tested for effects on FlgM secretion, and no effect was observed.
The last variable tested for secreted levels of overexpressed FlgM was ionic strength. It was reported that type III secretion was induced by higher osmolarity (57). We tested the effect of NaCl and KCl concentrations on FlgM secretion and observed that addition of NaCl to 200 mM or KCl to 200 to 400 mM resulted in an about 4-fold increase in secreted FlgM levels compared to results with 100 mM NaCl, which is close to the concentration of NaCl in LB (0.5%, or 86 mM). It was possible that the NaCl effect was due to increased potential of the proton motive force. However, the same effect was observed with the addition of KCl, suggesting that it is ionic strength that controls secreted FlgM levels. It is possible that ionic strength simply results in increased stability of FlhD4C2, but this remains to be determined.
In summary, while many factors could be altered to increase FlgM secretion, combinations of these factors were not additive, and the rate of FlgM secretion appeared limited. The apparent rate of FlgM secretion was enhanced by increasing σ28 levels to facilitate FlgM delivery to hook-basal body (HBB) secretion conduits but again to a limited extent, since intracellular FlgM still accumulated. The effect of increased FlhD4C2-dependent transcription on the number of HBB structures did enhance secreted FlgM levels but was also limited. This is consistent with previous results showing that increased FlhD4C2-dependent transcription produced at most a 2- to 3-fold increase in the number of HBB structures per cell (21). Increased ionic strength improved secreted FlgM levels but not when combined with other factors, suggesting that ionic strength contributed to other factors tested, such as enhanced FlhDC or σ28 levels or facilitation of σ28-mediated FlgM secretion or FlhD4C2-dependent transcription.
Finally, we tested the practicality of the optimized FlgM secretion system to purify peptides which are otherwise difficult to produce by using standard bacterial expression systems. These included a cysteine-rich, hydrophobic peptide, contoxin δ-SVIE, nodule-specific, cysteine-rich (NCR) antimicrobial peptides produced by the root nodules of certain legumes, and a malaria surface antigen domain of apical membrane antigen AMA-1. Figure 10 shows the effects of different conditions on secretion of the ParaBAD-expressed FlgM-6His-ETK-δ-SVIE construct. As reported earlier, the flhDC7793 promoter up allele resulted in a substantial increased in the level of secreted FlgM-6His-ETK-δ-SVIE. Removal of FlgM reduced the positive effect of the flhDC7793 allele, probably due to increased expression of secretion competitors, which are upregulated in the absence of FlgM. A strain that combines the flhDC7793 allele with mutations resulting in increased FlhDC stability (lrhA, ydiV, and fliT [within the fliBCDST deletion]) and mutations that remove secretion competitors (fliC [within the fliBCDST deletion], fljB [within the hin-fljBA deletion], flgMN, and flgKL) resulted in more than a 100-fold-increased secreted-protein level compare to that of the wild-type strain. For the FlgM-6His-ETK-MrVIA, FlgM-6His-ETK-AMA1, and FlgM-6His-ETK-NCR constructs, Coomassie-stained supernatants from cell cultures revealed the secreted FlgM-peptide fusions to be the prominent secreted proteins.
This work utilized the Salmonella Typhimurium strain LT2, which is attenuated for virulence. For commercial protein production, a similar strain background could be constructed and tested for E. coli K-12, or the Spi2 region of LT2 could simply be deleted to eliminate any virulence potential of Salmonella protein production strains using FlgM-mediated secretion.
ACKNOWLEDGMENTS
This work was supported by PHS grant GM056141 (to K.T.H.) from the National Institutes of Health and by a Technology Commercialization Project grant from the University of Utah.
Footnotes
Published ahead of print 4 April 2014
REFERENCES
- 1.Sourjik V, Wingreen NS. 2012. Responding to chemical gradients: bacterial chemotaxis. Curr. Opin. Cell Biol. 24:262–268. 10.1016/j.ceb.2011.11.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chevance FF, Hughes KT. 2008. Coordinating assembly of a bacterial macromolecular machine. Nat. Rev. Microbiol. 6:455–465. 10.1038/nrmicro1887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang S, Fleming RT, Westbrook EM, Matsumura P, McKay DB. 2006. Structure of the Escherichia coli FlhDC complex, a prokaryotic heteromeric regulator of transcription. J. Mol. Biol. 355:798–808. 10.1016/j.jmb.2005.11.020 [DOI] [PubMed] [Google Scholar]
- 4.Macnab RM. 2003. How bacteria assemble flagella. Annu. Rev. Microbiol. 57:77–100. 10.1146/annurev.micro.57.030502.090832 [DOI] [PubMed] [Google Scholar]
- 5.Macnab RM. 2004. Type III flagellar protein export and flagellar assembly. Biochim. Biophys. Acta 1694:207–217. 10.1016/j.bbamcr.2004.04.005 [DOI] [PubMed] [Google Scholar]
- 6.Ohnishi K, Kutsukake K, Suzuki H, Iino T. 1990. Gene fliA encodes an alternative sigma factor specific for flagellar operons in Salmonella typhimurium. Mol. Gen. Genet. 221:139–147 [DOI] [PubMed] [Google Scholar]
- 7.Berg HC, Anderson RA. 1973. Bacteria swim by rotating their flagellar filaments. Nature 245:380–382. 10.1038/245380a0 [DOI] [PubMed] [Google Scholar]
- 8.Ohnishi K, Kutsukake K, Suzuki H, Lino T. 1992. A novel transcriptional regulation mechanism in the flagellar regulon of Salmonella typhimurium: an antisigma factor inhibits the activity of the flagellum-specific sigma factor, sigma F. Mol. Microbiol. 6:3149–3157. 10.1111/j.1365-2958.1992.tb01771.x [DOI] [PubMed] [Google Scholar]
- 9.Hughes KT, Gillen KL, Semon MJ, Karlinsey JE. 1993. Sensing structural intermediates in bacterial flagellar assembly by export of a negative regulator. Science 262:1277–1280. 10.1126/science.8235660 [DOI] [PubMed] [Google Scholar]
- 10.Kutsukake K. 1994. Excretion of the anti-sigma factor through a flagellar substructure couples flagellar gene expression with flagellar assembly in Salmonella typhimurium. Mol. Gen. Genet. 243:605–612 [DOI] [PubMed] [Google Scholar]
- 11.Namba K. 2001. Roles of partly unfolded conformations in macromolecular self-assembly. Genes Cells 6:1–12. 10.1046/j.1365-2443.2001.00384.x [DOI] [PubMed] [Google Scholar]
- 12.Fattori J, Prando A, Martins AM, Rodrigues FH, Tasic L. 2011. Bacterial secretion chaperones. Protein Pept. Lett. 18:158–166. 10.2174/092986611794475048 [DOI] [PubMed] [Google Scholar]
- 13.Aldridge PD, Karlinsey JE, Aldridge C, Birchall C, Thompson D, Yagasaki J, Hughes KT. 2006. The flagellar-specific transcription factor, sigma28, is the type III secretion chaperone for the flagellar-specific anti-sigma28 factor FlgM. Genes Dev. 20:2315–2326. 10.1101/gad.380406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Daughdrill GW, Chadsey MS, Karlinsey JE, Hughes KT, Dahlquist FW. 1997. The C-terminal half of the anti-sigma factor, FlgM, becomes structured when bound to its target, sigma 28. Nat. Struct. Biol. 4:285–291. 10.1038/nsb0497-285 [DOI] [PubMed] [Google Scholar]
- 15.Gillen KL, Hughes KT. 1991. Molecular characterization of flgM, a gene encoding a negative regulator of flagellin synthesis in Salmonella typhimurium. J. Bacteriol. 173:6453–6459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Singer HM, Erhardt M, Steiner AM, Zhang MM, Yoshikami D, Bulaj G, Olivera BM, Hughes KT. 2012. Selective purification of recombinant neuroactive peptides using the flagellar type III secretion system. mBio 3(3):e00115–12. 10.1128/mBio.00115-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640–6645. 10.1073/pnas.120163297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Davis RW, Botstein D, Roth JR. 1980. Advanced bacterial genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 19.Karlinsey JE. 2007. lambda-Red genetic engineering in Salmonella enterica serovar Typhimurium. Methods Enzymol. 421:199–209. 10.1016/S0076-6879(06)21016-4 [DOI] [PubMed] [Google Scholar]
- 20.Yanagihara S, Iyoda S, Ohnishi K, Iino T, Kutsukake K. 1999. Structure and transcriptional control of the flagellar master operon of Salmonella typhimurium. Genes Genet. Syst. 74:105–111. 10.1266/ggs.74.105 [DOI] [PubMed] [Google Scholar]
- 21.Erhardt M, Hughes KT. 2010. C-ring requirement in flagellar type III secretion is bypassed by FlhDC upregulation. Mol. Microbiol. 75:376–393. 10.1111/j.1365-2958.2009.06973.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wozniak CE, Lee C, Hughes KT. 2009. T-POP array identifies EcnR and PefI-SrgD as novel regulators of flagellar gene expression. J. Bacteriol. 191:1498–1508. 10.1128/JB.01177-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Francez-Charlot A, Laugel B, Van Gemert A, Dubarry N, Wiorowski F, Castanie-Cornet MP, Gutierrez C, Cam K. 2004. RcsCDB His-Asp phosphorelay system negatively regulates the flhDC operon in Escherichia coli. Mol. Microbiol. 49:823–832. 10.1046/j.1365-2958.2003.03601.x [DOI] [PubMed] [Google Scholar]
- 24.Wang Q, Zhao Y, McClelland M, Harshey RM. 2007. The RcsCDB signaling system and swarming motility in Salmonella enterica serovar Typhimurium: dual regulation of flagellar and SPI-2 virulence genes. J. Bacteriol. 189:8447–8457. 10.1128/JB.01198-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lehnen D, Blumer C, Polen T, Wackwitz B, Wendisch VF, Unden G. 2002. LrhA as a new transcriptional key regulator of flagella, motility and chemotaxis genes in Escherichia coli. Mol. Microbiol. 45:521–532. 10.1046/j.1365-2958.2002.03032.x [DOI] [PubMed] [Google Scholar]
- 26.Yokoseki T, Kutsukake K, Ohnishi K, Iino T. 1995. Functional analysis of the flagellar genes in the fliD operon of Salmonella typhimurium. Microbiology 141:1715–1722. 10.1099/13500872-141-7-1715 [DOI] [PubMed] [Google Scholar]
- 27.Aldridge C, Poonchareon K, Saini S, Ewen T, Soloyva A, Rao CV, Imada K, Minamino T, Aldridge PD. 2010. The interaction dynamics of a negative feedback loop regulates flagellar number in Salmonella enterica serovar Typhimurium. Mol. Microbiol. 78:1416–1430. 10.1111/j.1365-2958.2010.07415.x [DOI] [PubMed] [Google Scholar]
- 28.Yamamoto S, Kutsukake K. 2006. FliT acts as an anti-FlhD2C2 factor in the transcriptional control of the flagellar regulon in Salmonella enterica serovar Typhimurium. J. Bacteriol. 188:6703–6708. 10.1128/JB.00799-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fraser GM, Bennett JC, Hughes C. 1999. Substrate-specific binding of hook-associated proteins by FlgN and FliT, putative chaperones for flagellum assembly. Mol. Microbiol. 32:569–580. 10.1046/j.1365-2958.1999.01372.x [DOI] [PubMed] [Google Scholar]
- 30.Lemke JJ, Durfee T, Gourse RL. 2009. DksA and ppGpp directly regulate transcription of the Escherichia coli flagellar cascade. Mol. Microbiol. 74:1368–1379. 10.1111/j.1365-2958.2009.06939.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Osterberg S, del Peso-Santos T, Shingler V. 2011. Regulation of alternative sigma factor use. Annu. Rev. Microbiol. 65:37–55. 10.1146/annurev.micro.112408.134219 [DOI] [PubMed] [Google Scholar]
- 32.Takaya A, Erhardt M, Karata K, Winterberg K, Yamamoto T, Hughes KT. 2012. YdiV: a dual function protein that targets FlhDC for ClpXP-dependent degradation by promoting release of DNA-bound FlhDC complex. Mol. Microbiol. 83:1268–1284. 10.1111/j.1365-2958.2012.08007.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wada T, Morizane T, Abo T, Tominaga A, Inoue-Tanaka K, Kutsukake K. 2011. EAL domain protein YdiV acts as an anti-FlhD4C2 factor responsible for nutritional control of the flagellar regulon in Salmonella enterica serovar Typhimurium. J. Bacteriol. 193:1600–1611. 10.1128/JB.01494-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wei BL, Brun-Zinkernagel AM, Simecka JW, Pruss BM, Babitzke P, Romeo T. 2001. Positive regulation of motility and flhDC expression by the RNA-binding protein CsrA of Escherichia coli. Mol. Microbiol. 40:245–256. 10.1046/j.1365-2958.2001.02380.x [DOI] [PubMed] [Google Scholar]
- 35.Auvray F, Thomas J, Fraser GM, Hughes C. 2001. Flagellin polymerisation control by a cytosolic export chaperone. J. Mol. Biol. 308:221–229. 10.1006/jmbi.2001.4597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bange G, Kummerer N, Engel C, Bozkurt G, Wild K, Sinning I. 2010. FlhA provides the adaptor for coordinated delivery of late flagella building blocks to the type III secretion system. Proc. Natl. Acad. Sci. U. S. A. 107:11295–11300. 10.1073/pnas.1001383107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kinoshita M, Hara N, Imada K, Namba K, Minamino T. 2013. Interactions of bacterial flagellar chaperone-substrate complexes with FlhA contribute to co-ordinating assembly of the flagellar filament. Mol. Microbiol. 90:1249–1261. 10.1111/mmi.12430 [DOI] [PubMed] [Google Scholar]
- 38.Minamino T, Kinoshita M, Hara N, Takeuchi S, Hida A, Koya S, Glenwright H, Imada K, Aldridge PD, Namba K. 2012. Interaction of a bacterial flagellar chaperone FlgN with FlhA is required for efficient export of its cognate substrates. Mol. Microbiol. 83:775–788. 10.1111/j.1365-2958.2011.07964.x [DOI] [PubMed] [Google Scholar]
- 39.Barembruch C, Hengge R. 2007. Cellular levels and activity of the flagellar sigma factor FliA of Escherichia coli are controlled by FlgM-modulated proteolysis. Mol. Microbiol. 65:76–89. 10.1111/j.1365-2958.2007.05770.x [DOI] [PubMed] [Google Scholar]
- 40.Tomoyasu T, Ohkishi T, Ukyo Y, Tokumitsu A, Takaya A, Suzuki M, Sekiya K, Matsui H, Kutsukake K, Yamamoto T. 2002. The ClpXP ATP-dependent protease regulates flagellum synthesis in Salmonella enterica serovar Typhimurium. J. Bacteriol. 184:645–653. 10.1128/JB.184.3.645-653.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bonifield HR, Hughes KT. 2003. Flagellar phase variation in Salmonella enterica serovar Typhimurium is mediated by a posttranscriptional control mechanism. J. Bacteriol. 185:3567–3574. 10.1128/JB.185.12.3567-3574.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Enomoto M, Stocker BA. 1975. Integration, at hag or elsewhere, of H2 (phase-2 flagellin) genes transduced from Salmonella to Escherichia coli. Genetics 81:595–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Karlinsey JE, Lonner J, Brown KL, Hughes KT. 2000. Translation/secretion coupling by type III secretion systems. Cell 102:487–497. 10.1016/S0092-8674(00)00053-2 [DOI] [PubMed] [Google Scholar]
- 44.Galeva A, Moroz N, Yoon YH, Hughes KT, Samatey FA, Kostyukova AS. 2014. Bacterial flagellin-specific chaperone FliS interacts with anti-sigma factor FlgM. J. Bacteriol. 196:1215–1221. 10.1128/JB.01278-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xu S, Peng Z, Cui B, Wang T, Song Y, Zhang L, Wei G, Wang Y, Shen X. 2013. FliS modulates FlgM activity by acting as a non-canonical chaperone to control late flagellar gene expression, motility and biofilm formation in Yersinia pseudotuberculosis. Environ. Microbiol. 10.1111/1462-2920.12222 [DOI] [PubMed] [Google Scholar]
- 46.Iyoda S, Kamidoi T, Hirose K, Kutsukake K, Watanabe H. 2001. A flagellar gene fliZ regulates the expression of invasion genes and virulence phenotype in Salmonella enterica serovar Typhimurium. Microb. Pathog. 30:81-90. 10.1006/mpat.2000.0409 [DOI] [PubMed] [Google Scholar]
- 47.Lucas RL, Lostroh CP, DiRusso CC, Spector MP, Wanner BL, Lee CA. 2000. Multiple factors independently regulate hilA and invasion gene expression in Salmonella enterica serovar Typhimurium. J. Bacteriol. 182:1872–1882. 10.1128/JB.182.7.1872-1882.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chubiz JE, Golubeva YA, Lin D, Miller LD, Slauch JM. 2010. FliZ regulates expression of the Salmonella pathogenicity island 1 invasion locus by controlling HilD protein activity in Salmonella enterica serovar Typhimurium. J. Bacteriol. 192:6261–6270. 10.1128/JB.00635-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ellermeier CD, Slauch JM. 2003. RtsA and RtsB coordinately regulate expression of the invasion and flagellar genes in Salmonella enterica serovar Typhimurium. J. Bacteriol. 185:5096–5108. 10.1128/JB.185.17.5096-5108.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mouslim C, Hughes KT. 2014. The effect of cell growth phase on the regulatory cross-talk between flagellar and Spi1 virulence gene expression. PLoS Pathog. 10:e1003987. 10.1371/journal.ppat.1003987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Singer HM, Kuhne C, Deditius JA, Hughes KT, Erhardt M. 2014. The Salmonella Spi1 virulence regulatory protein HilD directly activates transcription of the flagellar master operon flhDC. J. Bacteriol. 196:1448–1457. 10.1128/JB.01438-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Baneyx F, Georgiuo G. 1990. In vivo degradation of secreted fusion proteins by the Escherichia coli outer membrane protease OmpT. J. Bacteriol. 172:491–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hughes KT, Dessen A, Gray JP, Grubmeyer C. 1993. The Salmonella typhimurium nadC gene: sequence determination by use of Mud-P22 and purification of quinolinate phosphoribosyltransferase. J. Bacteriol. 175:479–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Flynn JM, Neher SB, Kim YI, Sauer RT, Baker TA. 2003. Proteomic discovery of cellular substrates of the ClpXP protease reveals five classes of ClpX-recognition signals. Mol. Cell 11:671–683. 10.1016/S1097-2765(03)00060-1 [DOI] [PubMed] [Google Scholar]
- 55.Dorel C, Lejeune P, Rodrigue A. 2006. The Cpx system of Escherichia coli, a strategic signaling pathway for confronting adverse conditions and for settling biofilm communities? Res. Microbiol. 157:306–314. 10.1016/j.resmic.2005.12.003 [DOI] [PubMed] [Google Scholar]
- 56.Merdanovic M, Clausen T, Kaiser M, Huber R, Ehrmann M. 2011. Protein quality control in the bacterial periplasm. Annu. Rev. Microbiol. 65:149–168. 10.1146/annurev-micro-090110-102925 [DOI] [PubMed] [Google Scholar]
- 57.Galán JE, Curtiss R., III 1990. Expression of Salmonella typhimurium genes required for invasion is regulated by changes in DNA supercoiling. Infect. Immun. 58:1879–1885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Minamino T, Namba K. 2008. Distinct roles of the ATPase and proton motive force in bacterial flagellar protein export. Nature 451:485–488. 10.1038/nature06449 [DOI] [PubMed] [Google Scholar]
- 59.Paul K, Erhardt M, Hirano T, Blair DF, Hughes KT. 2008. Energy source of flagellar type III secretion. Nature 451:489–492. 10.1038/nature06497 [DOI] [PubMed] [Google Scholar]
- 60.Erhardt M, Singer HM, Wee DH, Keener JP, Hughes KT. 2011. An infrequent molecular ruler controls flagellar hook length in Salmonella enterica. EMBO J. 30:2948–2961. 10.1038/emboj.2011.185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Uchida K, Aizawa SI. 21 February 2014. The flagellar soluble protein FliK determines the minimal length of the hook in Salmonella enterica serovar Typhimurium. J. Bacteriol. 10.1128/JB.00050-14 [DOI] [PMC free article] [PubMed] [Google Scholar]