Abstract
Protein localization has been traditionally explored in unicellular organisms, whose ease of genetic manipulation facilitates molecular characterization. The two rod-shaped bacterial models Escherichia coli and Bacillus subtilis have been prominently used for this purpose and have displaced other bacteria whose challenges for genetic manipulation have complicated any study of cell biology. Among these bacteria is the spherical pathogenic bacterium Staphylococcus aureus. In this report, we present a new molecular toolbox that facilitates gene deletion in staphylococci in a 1-step recombination process and additional vectors that facilitate the insertion of diverse reporter fusions into newly identified neutral loci of the S. aureus chromosome. Insertion of the reporters does not add any antibiotic resistance genes to the chromosomes of the resultant strains, thereby making them amenable for further genetic manipulations. We used this toolbox to reconstitute the expression of mreB in S. aureus, a gene that encodes an actin-like cytoskeletal protein which is absent in coccal cells and is presumably lost during the course of speciation. We observed that in S. aureus, MreB is organized in discrete structures in association with the membrane, leading to an unusual redistribution of the cell wall material. The production of MreB also caused cell enlargement, but it did not revert staphylococcal shape. We present interactions of MreB with key staphylococcal cell wall-related proteins. This work facilitates the use S. aureus as a model system in exploring diverse aspects of cellular microbiology.
INTRODUCTION
A significant number of studies of protein localization has been traditionally performed by using bacterial models, which are relatively simple and tractable haploid organisms (1–3). However, only a few bacterial species, such as the model organisms Escherichia coli and Bacillus subtilis, are generally considered for these studies, mostly because of the availability of a large number of molecular tools. In an attempt to have a broader and more general idea of the fundamental processes within the microbial world, there is growing interest in exploring other bacterial species that differ in shape, developmental properties, or infective potential from conventional bacterial models (4–6). Among these species is the spherical bacterium Staphylococcus aureus. It is an opportunistic pathogen that causes hard-to-treat acute and chronic infections in humans (7). It is indeed the study of its pathogenic attributes that makes this bacterium very attractive. Besides the interest in S. aureus as a major human pathogen, it is also an appealing model to explore questions related to cell shape, cell division, and cell proliferation, because it propagates very efficiently under laboratory conditions and is morphologically different from the conventional models E. coli and B. subtilis.
In order to genetically manipulate S. aureus in the laboratory, several batteries of plasmids have been constructed over the past years (8–12), yet performance of physiologically relevant experiments is often required, where the copy number of the expressed gene is equivalent to that of the chromosome, which is usually not the case when using replicative plasmids. For this reason, further approaches have been developed to integrate entire plasmids into specific loci within the staphylococcal chromosome, such as the phage attachment site attP (10, 13–17). Although this approach allowed the expression of genes in chromosome-equivalent copy numbers, the insertion of large DNA fragments into the staphylococcal chromosome negatively influences numerous physiological aspects of S. aureus, including the expression of the reporter itself (18–21). In this report, we present a new set of vectors to fulfill different molecular purposes related to the manipulation of the staphylococcal chromosome. Our new set of plasmids is constituted by a plasmid (pSceI) that allows gene deletion in an improved one-step recombination process and an additional set of vectors that allows the integration of reporter systems into several neutral loci of the staphylococcal chromosome. These tools permit the generation of single- and double-labeled strains by inserting reporters into neutral chromosomal loci without requiring the use of antibiotic selection markers. This leaves the resulting strain more amenable to further genomic manipulations. Labeled strains can express an implemented set of stable fluorophore proteins necessary to perform experiments related to cellular, molecular, and infection biology.
In this study, we have used this toolbox to explore a question related to the shape of S. aureus. Specifically, it is postulated that its spherical shape is irreversible and is derived from rod ancestors (22–24) that lost mreB and, thus, cell wall elongation (25–33) during the course of speciation. Indeed, mreB is absent in most spherical species such as S. aureus. However, the chromosome of S. aureus still presents the reminiscent mreCD operon and all mreB-associated genes that are required for bacteria to maintain a rod shape (34). Consequently, we generated an artificial mreB-expressing strain to investigate the effects of MreB production on S. aureus cells using the tools presented in this study.
MATERIALS AND METHODS
Strains, media, and culture conditions.
The strain used in this study was S. aureus Newman (35). S. aureus RN4220 (36) and Escherichia coli DH5α (37) were used for cloning purposes. A complete strain list is shown in Table S2 in the supplemental material. S. aureus cells were normally propagated in tryptic soy broth (TSB) medium or TSB agar. When needed, antibiotics were used at the following final concentrations: 100 μg/ml ampicillin, 50 μg/ml kanamycin, 5 μg/ml chloramphenicol, and 2 μg/ml erythromycin. When required, the culture medium was supplemented with a final concentration of 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG), 10 mM xylose, or 40 μg/ml 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal). For growth of S. aureus biofilms, a culture grown overnight was diluted 1:100 in a solution containing TSB, 0.5% (wt/vol) glucose, and 3% (wt/vol) NaCl (38); dispensed into polystyrene well plates; and incubated overnight statically at 37°C. Biofilms formed by S. aureus were stained with crystal violet (1% [wt/vol]) for better visualization, according to methods described previously (39). Secretion of hemolytic toxins was monitored by spotting 3 μl of a culture grown overnight onto TSB plates containing 5% sheep blood and checking the size of the halo after 48 h of incubation at 37°C. To quantify the hemolytic capacity, supernatants of liquid cultures of the different staphylococcal strains were filter sterilized and used to lyse a 2% solution of erythrocytes in phosphate-buffered saline (PBS) buffer. After 30 min of incubation and further removal of the unlysed erythrocytes, the hemolytic activity was quantified by monitoring the absorbance at 405 nm (40).
pSceI, pAmy, and pLac construction and cargo insertion.
In order to construct pSceI, the I-SceI gene was amplified from pSW-I (41) by using primer pair AYG147-AYG148 and cloned into plasmid pDR111 (see Table S3 in the supplemental material) (42, 43) to generate a cassette in which the expression of I-SceI is under the control of an IPTG-inducible promoter. A nested PCR using primer pairs AYG149-AYG151N and AYG150-AYG151N served to add an I-SceI restriction sequence upstream of the expression cassette. The whole fragment was further subcloned into the pMAD vector (44).
To construct pAmy and pLac, the respective upstream and downstream DNA regions of the amy and lac loci were PCR amplified (using primer pairs AYG63-AYG104 and AYG105-AYG66 to amplify the upstream and downstream regions of amy and primer pairs AYG59B-AYG60B and AYG61B-AYG62 to amplify the upstream and downstream regions of lac). The two PCR products generated for each locus were joined in a second PCR. A multicloning site (MCS) was introduced artificially with the designated primers. The resultant joining PCR rendered new amy and lac regions with an MCS positioned in the middle of the sequence. These cassettes were subsequently cloned into the pMAD vector, creating plasmids pAmy and pLac.
In order to improve the tractability of S. aureus, a collection of plasmids harboring different cargo was designed as follows. The PHP-lacI inducible promoter was obtained from pDR111 (XhoI/BamHI) and cloned into pAmy and pLac. The Pxyl inducible promoter was obtained from pSG1154 (45) and cloned into pAmy and pLac (BamHI/NheI). The yellow fluorescent protein (YFP)-encoding gene was amplified from pKM003 (a gift from David Rudner [Harvard University, USA]) and cloned into pAmy (SalI/BamHI) and pLac (SalI/NheI). The red fluorescent protein Mars-encoding gene was amplified from pmRFPmars (46) and cloned into pAmy (SalI/BamHI) and pLac (SalI/NheI), respectively. The SNAP-encoding region was amplified from psnapF (New England BioLabs) and cloned into pAmy (SalI/BamHI) and pLac (SalI/NheI). The His6 tag derivatives were constructed by amplifying amy and lac loci by using primer pairs AYG63-GK146 and GK144-AYG66 and primer pairs AYG59B-GK145 GK144-AYG62, respectively, to further join them and clone them into pMAD.
Generation of deletion mutants by using pSceI.
amy::Cm and lac::Cm deletion cassettes were generated by using long-flanking-homology PCR (47) (primers are listed in Table S4 in the supplemental material) and cloned into pSceI. The resultant plasmid was inserted into S. aureus RN4220 by electroporation and propagated in TSB medium at 30°C. Liquid cultures were set at 30°C with shaking, and 1 mM IPTG was added to the cultures at an optical density at 540 nm (OD540) of approximately 0.5. After 4 h of incubation, the cultures were shifted for two more hours to 42°C and then plated onto TSB–chloramphenicol–X-Gal. After 48 h of incubation at 42°C, the blue colonies still carrying the plasmid were discarded. White colonies resistant to chloramphenicol were checked by PCR to confirm that the deletion cassette was integrated. Constructs were transferred from strain RN4220 to strain Newman by Phi11 phage transduction (48).
Generation of labeled strains using pAmy and pLac.
Reporters and overexpressing cassettes were subcloned into plasmids pAmy and pLac and propagated in strain RN4220. The plasmids were integrated into the neutral amy and lac loci via a first recombination by growing the strains for 6 h at 42°C. Subsequently, the cultures were plated onto selective medium (erythromycin plus X-Gal). The genetic constructs obtained from the first recombination process were transferred to strain Newman by Phi11 phage transduction. To allow a second recombination in strain Newman as the genetic background and, hence, the exclusive integration of the construct, cultures of blue colonies were incubated for 6 h at 30°C, then shifted for 3 h to 42°C, and subsequently plated onto TSB plus X-Gal. After 48 h of incubation at 42°C, the blue colonies still carrying the plasmid were discarded. PCR was used to validate whether the white colonies carried the corresponding insertion in the neutral loci. This second recombination process rendered a markerless labeled strain.
Image capture and analysis.
Unless otherwise specified in the figure legends, samples were fixed in 4% paraformaldehyde prior to single-cell analysis and resuspended in PBS buffer. For membrane staining, samples were incubated for 10 min with Nile red (Sigma) at a final concentration of 0.5 μg/ml. For DNA staining, samples were incubated for 10 min with Hoechst (Invitrogen) at a final concentration of 1 μg/ml. Samples were repeatedly washed prior to single-cell analysis. Images were processed by using Leica Application Suite V3.7 software. Microscopy images were taken on a Leica DMI6000B microscope equipped with a Leica CRT6000 illumination system and a Leica DFC630FX color camera. Image processing (Z-stack analysis) was done by using Leica Application Suite Advance Fluorescence software. Deconvolution of the fluorescence signal was done by using AutoQuant Deconvolution Algorithms software (Media Cibernetics). Signals were detected by using the following filters (Leica). The green fluorescent protein (GFP) signal was detected by using a BP480/40 excitation filter and a BP527/30 emission filter. The red fluorescent protein (RFP) signal was detected by using a BP546/40 excitation filter and a BP600/40 emission filter. Hoechst and Nile red were detected by using BP360/40 and BP546/40 excitation filters and BP470/40 and BP600/40 emission filters, respectively.
Flow cytometry.
Cultures were fixed with 4% paraformaldehyde, washed, and resuspended in PBS buffer. Dilution of samples (1:100) was necessary prior to flow cytometry analyses. Further sonication treatment consisting of three consecutive series of 12 pulses (power output of 50% with 0.7-s cycles) was required to separate single cells. Flow cytometry analysis was carried out with a MACS Quant flow cytometer. The photomultiplier voltage was set between 400 and 500 V. No gates were required during the analysis of the samples. Every sample was analyzed by measuring 50,000 events using FACS Diva software (BD Biosciences) to capture the data. Further data analysis was performed with FlowJo 9.6 (FlowJo Software).
Cell fractionation and MreB purification.
The cell pellet was harvested, and cells were lysed in SMM buffer (1 M sucrose, 0.04 M maleic acid, 0.04 M MgCl2 [pH 6.5]) by treating them with lysostaphin (10 μg/ml), followed by French press disruption. Cell debris was eliminated by centrifugation (13,000 rpm for 10 min). To separate the membrane fraction from the cytoplasmic fraction, the supernatant was subjected to ultracentrifugation (100,000 × g for 1 h). The supernatant constituted the cytoplasmic fraction, and the cell pellet represented the membrane fraction. The membrane fraction was solubilized in Tris buffer (20 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1% DDM [n-dodecyl β-d-maltoside], 1 mM phenylmethylsulfonyl fluoride [PMSF]). Mars-MreB purification was done by using RFP-Trapbeads (Chromotek) according to the manufacturer's instructions.
Transmission electron microscopy.
Bacteria were grown in TSB for 6 h at 30°C and harvested by centrifugation at 6,000 rpm. The pellets were fixed for 2 h at 4°C with 0.1 M sodium cacodylate (pH 7.2) containing 2.5% glutaraldehyde and 2% formaldehyde. Specimens were washed 3 times each for 5 min with 50 mM sodium cacodylate (pH 7.2) and then fixed for 2 h at 4°C with 2% osmium tetroxide in 50 mM sodium cacodylate (pH 7.2). Samples washed with distilled water were stained overnight at 4°C with 0.5% aqueous uranyl acetate, dehydrated, and embedded in Epon 812. Stained ultrathin sections were inspected with an EM900 electron microscope (Zeiss). Negatives were digitalized by scanning and processed with Adobe Photoshop.
Immunoblotting.
Immunoblotting was carried out as previously described (49), using a polyclonal antibody against MreB of B. subtilis, kindly provided by Peter Graumann (University of Marburg, Germany), and anti-mCherry (BioVision). When specified, the protein content was adjusted to 25 μg of total protein per lane by using a Nanodrop ND-1000 spectrophotometer to quantify the protein concentrations of the samples.
Bacterial two-hybrid analysis.
The bacterial adenylate cyclase-based two-hybrid (BACTH) system (EuroMedex) was used to measure interactions between MreB and cell wall- or cell division-related proteins. Therefore, the coding sequences of mreB, pbp2, pbp3, mreC, mreD, ftsZ, ftsA, and ezrA were PCR amplified and cloned in frame into bacterial hybrid expression vectors (pKT25, pKNT25, pUT18, and pUT18C) (a complete list of strains can be found in the supplemental material). This leads to the generation of C- or N-terminal fusions to the T25 or T18 catalytic domain of the adenylate cyclase from Bordetella pertussis. E. coli strain BTH101 was cotransformed with all combinations of plasmid pairs in regard to mreB. This strain harbors a lacZ gene, the expression of which is under the control of a cyclic AMP (cAMP)-inducible promoter. If an interaction occurs, the T25 and T18 catalytic domains of the adenylate cyclase reconstitute the enzyme, and cAMP is produced. This in turn triggers the expression of the reporter and leads cells to turn blue in the presence of X-Gal. BACTH assays were performed on LB plates containing ampicillin, kanamycin, and X-Gal according to a protocol described previously by Karimova et al. (50). Plates were incubated for 48 h at 30°C. Efficiencies of interactions were quantified by assessing β-galactosidase activities as described previously (51). Therefore, cells were grown overnight in LB with IPTG and appropriate antibiotics. The OD540 was recorded prior to cell permeabilization using SDS and chloroform.
Additional software used in this study.
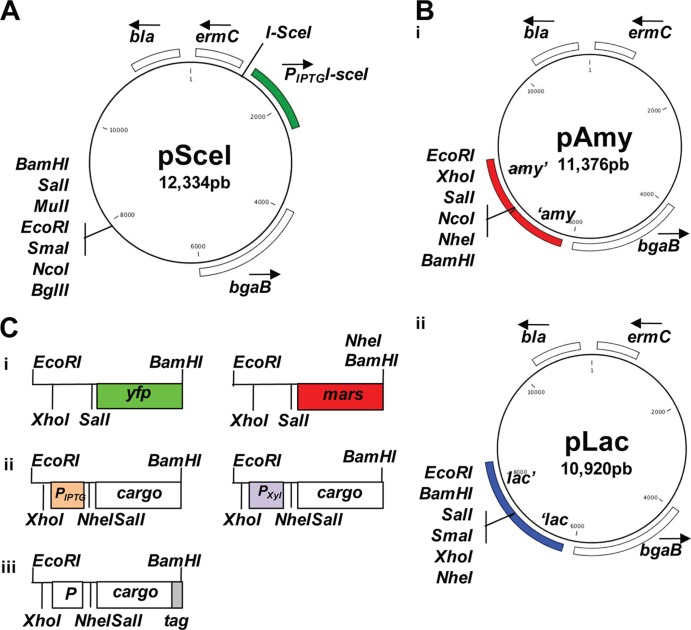
In addition to the software that was used in association with specific equipment (e.g., AutoQuant for the deconvolution of the fluorescence images), the following collection of supplementary software was considered for data processing. Unless specified otherwise, the bioinformatic analysis was performed by using SMART (http://smart.embl-heidelberg.de/) and STRING (http://string-db.org/). Fluorescence images were processed by using Fiji (http://fiji.sc/). The schematic representations of the plasmids presented in Fig. 1 were created by using CLC Workbench (CLCbio).
FIG 1.
Molecular toolbox for genomic manipulation of S. aureus. (A) Scheme of plasmid pSceI. The I-SceI recognition sequence is adjacent to the I-SceI gene. The bla gene encodes a β-lactamase (ampicillin resistance), and the ermC gene encodes an rRNA methylase (erythromycin resistance). The bgaB gene encodes a β-galactosidase. (B) Scheme of plasmids pAmy (i) and pLac (ii). (C) Representation of the diverse possibilities of cargoes that are inserted into plasmids pAmy and pLac. (i) Transcriptional fusions to the fluorescent proteins GFP and Mars (green and red fluorescence, respectively). (ii) Promoters inducible by IPTG and xylose for protein expression. (iii) His6 and SNAP tags for protein detection.
RESULTS
Novel toolbox for boosting the tractability of S. aureus.
To contribute to improving genetic manipulations of S. aureus, we have designed a molecular toolbox based on the tools that are currently available for B. subtilis. The tractability of B. subtilis relies on its ability to incorporate exogenous DNA into its genome (which permits gene deletion in a one-step process of double recombination) (52, 53) and the possibility of integrating single-copy genetic constructs into two neutral loci in the chromosome (54). The neutral loci are the amyE and lacA genes, which encode an extracellular α-amylase and β-galactosidase, respectively, and are nonessential proteins for the propagation of B. subtilis under laboratory conditions. Bioinformatic analysis identified two loci in the genome of S. aureus (strain Newman) that displayed a high level of similarity to the neutral amyE and lacA loci in B. subtilis. These loci are the NWMN2354 and NWMN2098 genes, which are currently annotated as glutamyl-aminopeptidase- and β-galactosidase-encoding genes, respectively (here referred to as amy and lac loci) (55) (see Table S1 in the supplemental material). In order to determine whether amy and lac can be used as neutral loci under laboratory conditions, we first tested if the disruption of these genes affected key physiological features that are traditionally tested in laboratory studies. Therefore, the deletion cassettes were cloned into our newly synthesized replicative plasmid pSceI (Fig. 1A), which allows the expression of the I-SceI restriction enzyme under the control of an IPTG-inducible promoter. I-SceI recognizes an 18-bp-long palindromic DNA region that is present exclusively in mitochondrial DNA of Saccharomyces cerevisiae (56) and artificially inserted into the backbone of pSceI. Upon the addition of IPTG to S. aureus cultures carrying the plasmid, the I-SceI enzyme is expressed, and the plasmid is linearized by single cutting, preventing its replication and favoring the insertion of the construct by double recombination (57–60).
The amy and lac deletion cassettes were cloned into the pMAD and pSceI vectors and transformed into S. aureus cells. Cells were allowed to grow in the presence of IPTG until they reached stationary phase and were then plated onto TSB medium containing X-Gal and the appropriate antibiotics (see Fig. S1 in the supplemental material). After 2 days of incubation, white colonies were selected, and the deletion of the amy or lac gene was verified by PCR analysis and Sanger sequencing. The results showed that the procedure of gene deletion in a 1-step recombination process using the pSceI vector increased the efficiency of double recombination in S. aureus up to 5-fold compared to that of its parental backbone (plasmid pMAD) (data not shown).
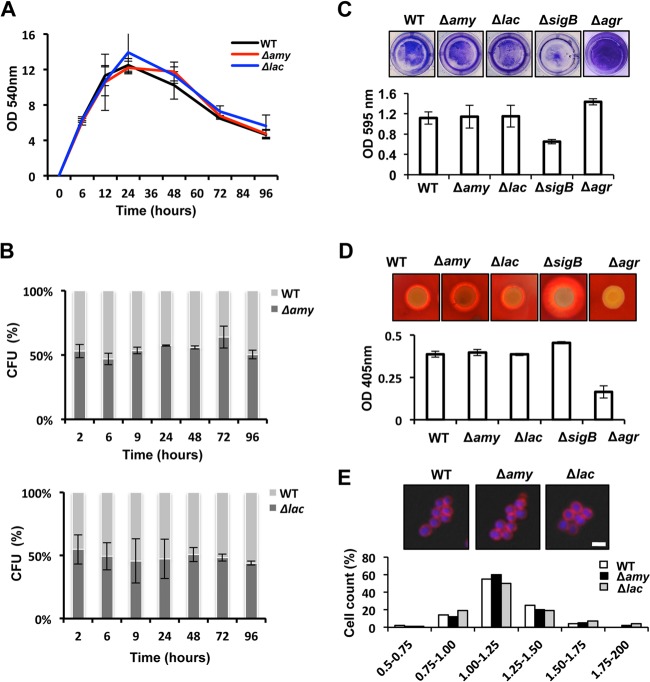
The resulting Δamy and Δlac strains were analyzed for their phenotypic properties. In TSB medium, they displayed growth rates comparable to that of the wild-type strain (Fig. 2A), suggesting that these mutants can be efficiently propagated under laboratory conditions without any physiological disadvantage. To determine if the absence of the amy or lac gene negatively affects the cellular fitness of S. aureus, we performed competition experiments with the Δamy or the Δlac mutant strain and the parental strain in mixed communities. Therefore, plasmids pAmy and pLac were used to insert a constitutive fluorescence reporter into the amy or lac locus. These fluorescently labeled strains with truncated amy and lac genes, respectively, were mixed with the wild-type strain in a 1:1 ratio at an initial OD540 of 0.05. The mixture was incubated in TSB medium at 37°C for 96 h. Progression of the strains was analyzed by quantifying the number of CFU per ml of culture at different time points during incubation. Wild-type colonies and Δamy or Δlac mutant colonies were distinguished based on the emission of fluorescence of the labeled amy- or lac-defective mutant (Fig. 2B). Analysis of CFU/ml showed no changes during the time course of incubation, suggesting that no alterations in cellular fitness occurred between the wild type and the Δamy and Δlac strains.
FIG 2.
Physiological assays of the wild-type strain and the amy- and lac-defective mutants. (A) Growth analysis of wild-type (WT) strain Newman and the Δamy and Δlac strains. The OD540 was monitored for 96 h in TSB cultures grown at 37°C. (B) Competition assays of mixed populations (1:1 ratio) of WT and Δamy::YFP strains (Δamy) and of WT and Δlac::YFP strains (Δlac). Mixed communities were incubated at an initial OD540 of 0.05. Samples were taken over 96 h, and CFU were counted by using a dissection scope equipped with a fluorescence excitation detection system. (C) Biofilm formation assay of cultures grown in 24-well titer plates. Biofilms were stained with crystal violet (1%) and quantified by spectrophotometry analysis (n = 3) (error bars represent standard deviations). (D) Hemolytic activity of the different strains. Cultures grown overnight were spotted onto TSB agar containing 5% sheep blood and incubated at 37°C during 48 h. Quantification of hemolytic activity was performed by measuring the OD405 of a solution of 2% erythrocytes previously incubated with the strains' supernatants (n = 3) (error bars represent standard deviations). (E) Fluorescence microscopy pictures used to monitor the cell sizes of the different strains. Cultures were grown until they reached the exponential phase. The membrane of the cell was stained with Nile red (false colored in red), and the DNA was stained with Hoechst 3342 (false colored in blue). Quantification of the cell diameter was performed with Leica Application software (see Materials and Methods).
Developmental assays were performed by monitoring the ability of the mutants to form surface-associated biofilms under laboratory conditions, which entails growing the cells in standing cultures in 24-well polystyrene plates and further staining the cells attached to the bottom of the well with crystal violet (39). This assay provided evidence for the ability of the Δamy and Δlac mutants to form biofilms. Their biofilm formation ability was comparable to that of the wild type but differed from those of the biofilm-defective ΔsigB and Δagr mutants, which present higher levels of biofilm formation. These strains were used as controls in this experiment (Fig. 2C) (61). Furthermore, the ability of the mutants to secrete hemolytic toxins was monitored by culturing S. aureus on 5% sheep blood agar (62) (Fig. 2D). After 48 h of incubation at 37°C, the colonies produced a hemolysis halo as a consequence of the secretion of hemolytic toxins. The diameter sizes of the halos of the Δamy and Δlac strains were similar to that of the wild-type strain. In contrast, the ΔsigB mutant developed a hemolysis halo with a larger diameter, as it is known that this strain produces larger amounts of toxins. In contrast, the Δagr mutant is defective in the secretion of hemolytic toxins, and consequently, it showed poor hemolytic activity (63, 64). A quantitative hemolytic assay was performed according to a protocol described previously (40). Altogether, the Δamy and Δlac strains had hemolytic activities similar to that of the wild-type strain.
Finally, the cellular dimensions of Δamy, Δlac, and wild-type cells were determined by quantifying the diameters of approximately 500 cells of each strain. To do this, liquid cultures of Δamy, Δlac, and wild-type strains were grown in TSB medium at 37°C until they reached stationary phase. Cells were stained for further microscopy analysis using Nile red and Hoechst 3342 dyes. Nile red is a lipophilic, fluorescent dye that allows membrane staining and, thus, determination of cellular boundaries (Fig. 2E) (65). In contrast, Hoechst 3342 is a blue fluorescent dye used to stain DNA and allows the differentiation of single cells in microscopy samples (Fig. 2E) (66). No significant variation in cell diameter was found between the wild type and the Δamy and Δlac mutants.
Since the disruption of the amy and lac loci did not perturb any of the physiological parameters tested so far, it is likely that these genes do not play an important physiological role under the tested conditions and that the loci can be used for further genetic manipulation. Consequently, we constructed a collection of plasmids to enable the single-copy insertion of cargo within these neutral loci of S. aureus (Fig. 1B). In order to construct these plasmids, upstream and downstream DNA regions of the amy and lac loci were introduced into temperature-sensitive plasmid pMAD (44). The counterscreening process described previously for the excision of the pMAD vector leads to the integration of the construct into the staphylococcal chromosome with a recombination rate of approximately 0.7 to 0.8%. We found this rate to be highly dependent on the extension of the flanking homology regions, showing an optimal integration rate for flanking regions of between 0.7 and 1.0 kb. When flanking homology regions with a length of <0.7 kb were used, the recombination rate decreased to 0.3%. A detailed protocol for plasmid integration is presented in Fig. S2 in the supplemental material.
We generated different versions of the plasmids that include distinct cargo possibilities to integrate into the amy and/or lac locus. The different cargo possibilities are presented in Fig. 1C, which include stable fluorescence proteins that are useful for the generation of transcriptional and translational reporter fusions (Fig. 1Ci). We also constructed diverse gene expression cassettes whose induction is under the control of IPTG- and xylose-inducible promoters (Fig. 1Cii). These inducible promoters are conventionally used for a wide range of Gram-positive bacteria, including S. aureus (12, 67, 68). This toolbox includes versions of plasmids containing His6 and SNAP C-terminal tags for the expression of recombinant proteins. This allows the detection of the expressed proteins by immunoblotting or the purification of tagged proteins (Fig. 1Ciii). Finally, this collection of plasmids includes vectors harboring a multicloning site that can be used for any type of cloning. The use of these vectors allows the generation of labeled strains that do not harbor any antibiotic resistance cassette in the chromosome, thus making them more amenable to further genomic manipulations. An example of resultant strains that can be generated by using this collection of vectors is shown in Fig. 2B.
Heterologous expression of mreB in S. aureus.
We have used this toolbox to explore a fundamental question related to the spherical shape of S. aureus cells. Most spherical species do not have mreB in their chromosome, suggesting that round species lost mreB during the course of speciation. Accordingly, phylogenetic studies together with biochemical evidences suggest that spherical bacteria are derived from ancestors that were initially rods (22–24). However, the staphylococcal chromosome still preserves a two-gene operon with mreC and mreD, which is probably residual from the mreBCD ancestral operon (26, 27, 69). In addition to this, the genome of S. aureus presents all the genes encoding MreB-related proteins (e.g., RodA-, MraY-, and MurG-like proteins). Thus, we wanted to investigate the effects of cell shape on the expression of mreB in S. aureus.
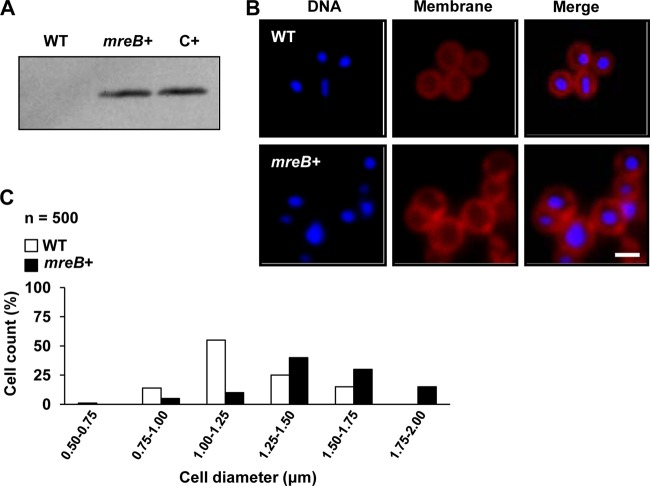
We generated a synthetic strain of S. aureus that expressed the native mreB gene from B. subtilis, as this is the taxonomically closest rod-shaped microorganism to S. aureus. Thus, mreB was cloned into plasmid pAmy under the control of an IPTG-inducible promoter and subsequently inserted into the amy locus (see Fig. S2 in the supplemental material). The resulting strain was grown in TSB medium in the presence of IPTG at 37°C for 3 h. MreB accumulation in S. aureus cells was detected by Western blot analysis using polyclonal antibodies against MreB of B. subtilis (70) (Fig. 3A). Next, we constructed a strain producing Mars-tagged MreB (red fluorescence protein [46]) under the expression control of a xylose-inducible promoter. This strain was used to purify Mars-MreB from cell extracts by using RFP-Trapbeads, which selectivity bind red fluorescent proteins. The eluted protein was resolved by SDS-PAGE and identified by mass spectrometry (see Fig. S3C in the supplemental material). Altogether, these results were consistent with the idea that the synthetic strain of S. aureus expressed recombinant mreB.
FIG 3.
S. aureus cells expressing mreB of B. subtilis. (A) Western blot analysis detecting the presence of the MreB protein in cell extracts by using polyclonal antibodies against MreB of B. subtilis. WT is wild-type strain Newman. mreB+ is a Newman strain expressing mreB of B. subtilis under the control of an IPTG-inducible promoter. The positive control (C+) is B. subtilis strain PY79. (B) Comparison of cell sizes of wild-type and mreB+ strains. Cultures were grown until exponential phase was reached, and the cell size was monitored by fluorescence microscopy. The cellular membrane was stained with Nile red (false colored red), and the DNA was stained with Hoechst 3342 (false colored blue). Bar, 1 μm. (C) Quantification of the diameters of 500 cells selected from wild-type and mreB+ microscopic fields.
Next, we wanted to evaluate the effect of MreB on the cell shape of S. aureus. Interestingly, upon MreB production in S. aureus, a significant enlargement of the cells compared to size of wild-type cells was observed, as was apparent by the increase in cell diameter (Fig. 3B). Variations in cell size were analyzed for 500 randomly selected cells from the fluorescence microscopy images (Fig. 3C). The MreB-producing cells showed a mean diameter of 1.46 μm ± 0.19 μm, compared to the wild-type cells, which had a mean diameter of 1.08 μm ± 0.12 μm. The Newman mreB+ strain showed an increase in cell diameter of approximately 35% compared to that of the wild-type strain. In contrast, control strains that overproduced a cytoplasmic GFP (amy::PIPTGgfp) or a GFP-fused membrane protein (amy::PIPTGsa1402-gfp) (see Fig. S5A in the supplemental material) showed cell sizes similar to those of the wild type, suggesting that the increase in cell size observed for the mreB+ strain specifically resulted from the production of MreB. We also used flow cytometry analysis to determine cell size variations of a subpopulation of 50,000 cells expressing mreB in comparison to wild-type cells (see Fig. S4 in the supplemental material). Both techniques provided evidence for the enlargement of MreB-producing cells in comparison to wild-type cells, which was unlikely to be due to a defective cell division process because similar growth rates were found for wild-type and mreB-expressing cells (see Fig. S5B in the supplemental material).
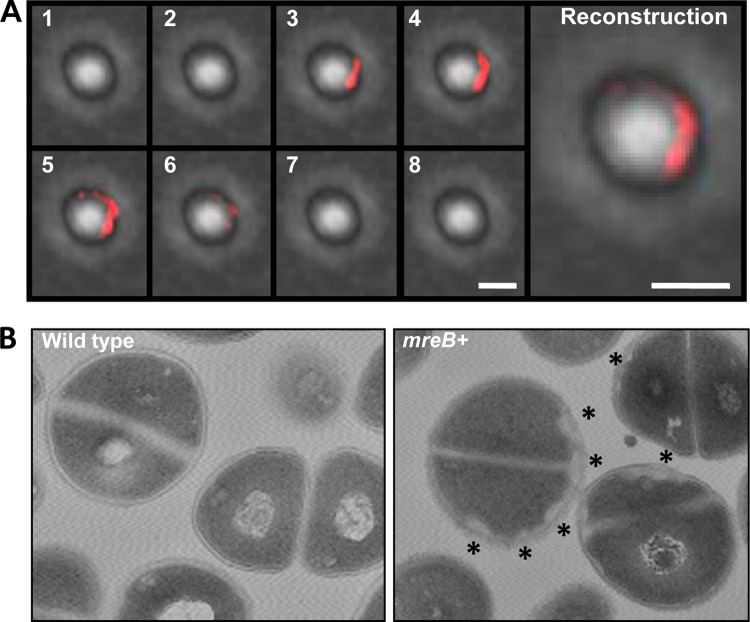
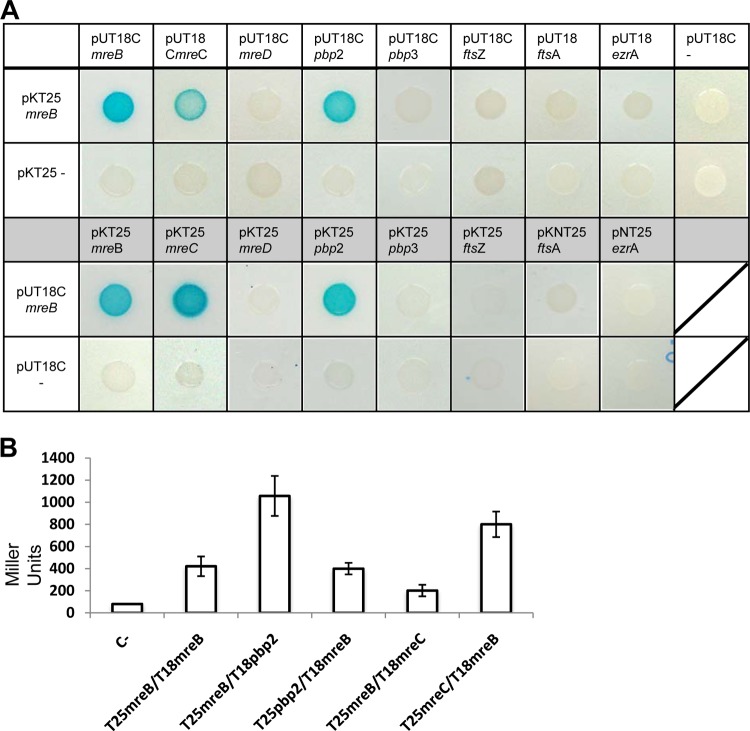
We used Mars-MreB to explore the subcellular organization of MreB in S. aureus cells by using fluorescence microscopy. Cells showed a red fluorescence signal distributed in discrete regions throughout the cellular membrane (Fig. 4A; see also Fig. S6 in the supplemental material). Generally, the red fluorescence signal defined small curved structures associated with the membrane. To gain more insight in this observation, cell extracts were separated into cytosolic fractions and membrane fractions and used to detect Mars-MreB (see Fig. S3A in the supplemental material). The immunoblot analysis showed a signal associated with the membrane fraction, despite the fact that MreB is a cytoplasmic protein, similar to what was previously reported (69). This subcellular organization of MreB together with the cell enlargement found prompted us to investigate any alteration in the usual morphology of the staphylococcal cell wall. Electron microscopy analysis was performed on S. aureus mreB-expressing cells and wild-type cells (Fig. 4B). The cells were sectioned at different planes, which prevented a comparative analysis of cell size. However, S. aureus mreB+ cells consistently showed an increase in the thickness of the cell wall, which was restricted to discrete regions of the cellular membrane compared to wild-type cells. To further explore the hypothetical influence of MreB on cell wall-related proteins, we used bacterial two-hybrid assays to monitor possible interactions between MreB and key components of cell wall turnover, such as Pbp2 and Pbp3, and also staphylococcal MreC and MreD homologs (Fig. 5A). In parallel, we also tested also the interaction between MreB and key components of the cell division protein machinery, such as FtsZ, FtsA, and EzrA. The bacterial two-hybrid assay evidenced a strong interaction between MreB and the staphylococcal penicillin-binding protein Pbp2, as was described previously for B. subtilis (31). An additional interaction was detected between MreB and the staphylococcal protein MreC, similar to what was reported previously for other bacterial models (26, 33, 71). In contrast, no interactions between MreB and any of the protein components of the cell division machinery were detected. This corroborates our previous results indicating that the expression of mreB does not interfere with the cell division protein machinery but leads to cell enlargement and an increase in cell wall thickness, likely due to the interaction between MreB and cell wall-related proteins.
FIG 4.
Subcellular distribution of MreB in S. aureus. (A) Fluorescent pictures of S. aureus cells expressing the Mars-MreB translational fusion (false colored red). Shown are Z-stack image series of a single cell expressing mreB. The pictures are sequentially numbered according to their position in the Z-stack. The fluorescence signal was deconvoluted and false-colored red. Bar, 1 μm. A reconstruction of the Z-stack series is presented at the right. In this panel, the fluorescence signals from the 8 planes of the Z-stack are combined into one single plane. Bar, 1 μm. (B) MreB-expressing cells show an altered disposition of cell wall material. Shown are transmission electron microscopy micrographs of sectioned cells from wild-type and MreB-expressing (mreB+) cultures. The unusual reorganization of the cell wall material is observed in discrete regions of the bacterial membrane in MreB-expressing cells, which are labeled with asterisks.
FIG 5.
Analysis of protein-protein interactions between MreB and cell wall-related proteins. (A) Bacterial two-hybrid analysis showing interactions between MreB and the protein components of the staphylococcal cell wall elongation machinery (Pbp2, Pbp3, MreC, and MreD) or the cell division protein machinery (FtsZ, FtsA, and EzrA). The MreB-MreB interaction was used as a positive control (pKT25-mreB/pUT18C-mreB). (B) Quantification of positive interactions using β-galactosidase activity. Cells were grown at 30°C for 24 h. Values are the mean results from three independent cultures; error bars represent standard deviations. The Zip/Zip control reached 3,000 Miller units (results not shown). C−, empty vectors.
DISCUSSION
The new toolbox presented in this report can be used to improve several aspects of the current methodology of genetic manipulation of staphylococci. First, we developed plasmid pSceI, which facilitates gene deletion in a 1-step recombination event and expedites the often tedious process of generating mutants in S. aureus. This system replaces the gene of interest by double recombination using an antibiotic resistance cassette, which allows the selection of mutants in medium supplemented with the antibiotic. Using this approach, the resultant allele can be transferred from one strain to another via phage transduction (72, 73). However, this technique is not suitable for the generation of markerless deletion mutants, as it offers no possibility for the selection of mutants in selective growth medium. The generation of markerless gene deletion strains still relies on the traditional 2-step recombination process, which involves the use of pMAD (44).
Second, the pAmy and pLac vector systems allow the integration of different reporters into the bacterial chromosome. These systems can generate single- or double-labeled strains without any antibiotic resistance cassettes, thus making them more amenable to further genetic manipulation. In our opinion, the most important advantage presented by the pAmy and pLac vector systems is the possibility of generating labeled strains that contain a stable copy number of the reporter of interest that is equivalent to that of the chromosome. This will allow us to be more precise in the study of factors involved in the virulence traits of S. aureus. In addition, the stable integration of DNA into the chromosome will also allow the use of strains of interest in infection models, since plasmids are usually less stable during the progression of infection. This will also contribute to explorations of trending concepts in microbiology, such as, for instance, the study of specialized cell types within populations of genetically identical bacteria (74, 75). We expect that these new protocols and tools will expand the already existing tools to make S. aureus a more appealing bacterial model not only for molecular pathogenesis but also in the field of cellular and synthetic biology.
Finally, we have made use of our toolbox to investigate the effects of mreB expression on S. aureus cells. MreB is organized in discrete structures positioned beneath the cellular membrane that rotate around the long axis of E. coli and B. subtilis cells (76–78). Indeed, we were also able to detect MreB in the membrane fraction of S. aureus, being organized mainly in discrete regions of the membrane that were visible by using fluorescence microscopy. Together with the increase in cell size that was observed, we concluded that MreB was actively expressed in S. aureus. The fact that we could not obtain a rod-shaped bacterium upon mreB expression was not totally surprising, since other rod-shape-related proteins (e.g., Pbp2, RodA, MreC, and MreD) could have evolved differently in spherical bacteria and have just partially conserved function, being important for cell wall synthesis but not necessarily related to cylindrical growth. We found that interactions between MreB and the staphylococcal cell wall-related proteins were similar to those naturally existing in rod-shaped bacteria (25–28, 33, 79–82). These interactions are consistent with the increase in cell wall thickness that we observed in the electron micrographs.
It is generally assumed that spherical morphology is irreversible in bacteria, since no genetic manipulation has proven otherwise thus far. Thus, it is plausible that the mreB gene was selectively eliminated from S. aureus during an adaptive process that could benefit S. aureus in an as-yet-unknown fashion. One possibility, considering our data, is that a hypothetical reduction of cell size may have benefited S. aureus in dispersing through media of higher viscosity, a particularly important feature of pathogens like S. aureus that intrude fluids without the assistance of any flagella.
Supplementary Material
ACKNOWLEDGMENTS
We thank all members of the Institute of Molecular Infection Biology (IMIB), especially Isa Westedt for technical assistance and Stan Gorski for help in editing the manuscript. We thank Victor de Lorenzo (CNB-CSIC, Spain) for kindly providing the collection of pSEVA vectors including the I-SceI enzyme cassette. We thank Peter Graumann (University of Marburg, Germany) for kindly providing the polyclonal antibody against MreB of B. subtilis. We thank Knut Ohlsen for kindly providing the antibody against PknB of S. aureus and Martin Fraunholz (University of Würzburg, Germany) for kindly providing the Mars fluorescence expression vector.
This work was funded by the Young Investigator Program of the Research Center of Infectious Diseases (ZINF) of the University of Würzburg, Germany, and by grant LO 1804/2-1 from The German Research Foundation (DFG). G.K. is a recipient of a Schrödinger fellowship (FWF). J.-C.G.-B. is a recipient of a Ph.D. fellowship from the Graduate School of Life Sciences (GSLS) of the University of Würzburg.
Footnotes
Published ahead of print 18 April 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.00759-14.
REFERENCES
- 1.Henning UL. 1975. Determination of cell shape in bacteria. Annu. Rev. Microbiol. 29:45–60. 10.1146/annurev.mi.29.100175.000401 [DOI] [PubMed] [Google Scholar]
- 2.Young KD. 2010. Bacterial shape: two-dimensional questions and possibilities. Annu. Rev. Microbiol. 64:223–240. 10.1146/annurev.micro.112408.134102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Errington J, Daniel RA, Scheffers DJ. 2003. Cytokinesis in bacteria. Microbiol. Mol. Biol. Rev. 67:52–65. 10.1128/MMBR.67.1.52-65.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pereira PM, Filipe SR, Tomasz A, Pinho MG. 2007. Fluorescence ratio imaging microscopy shows decreased access of vancomycin to cell wall synthetic sites in vancomycin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 51:3627–3633. 10.1128/AAC.00431-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pereira SF, Henriques AO, Pinho MG, de Lencastre H, Tomasz A. 2007. Role of PBP1 in cell division of Staphylococcus aureus. J. Bacteriol. 189:3525–3531. 10.1128/JB.00044-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Turner RD, Ratcliffe EC, Wheeler R, Golestanian R, Hobbs JK, Foster SJ. 2010. Peptidoglycan architecture can specify division planes in Staphylococcus aureus. Nat. Commun. 1:26. 10.1038/ncomms1025 [DOI] [PubMed] [Google Scholar]
- 7.Otto M. 2012. MRSA virulence and spread. Cell. Microbiol. 14:1513–1521. 10.1111/j.1462-5822.2012.01832.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brzoska AJ, Firth N. 2013. Two-plasmid vector system for independently controlled expression of green and red fluorescent fusion proteins in Staphylococcus aureus. Appl. Environ. Microbiol. 79:3133–3136. 10.1128/AEM.00144-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Corrigan RM, Foster TJ. 2009. An improved tetracycline-inducible expression vector for Staphylococcus aureus. Plasmid 61:126–129. 10.1016/j.plasmid.2008.10.001 [DOI] [PubMed] [Google Scholar]
- 10.Lei MG, Cue D, Alba J, Junecko J, Graham JW, Lee CY. 2012. A single copy integration vector that integrates at an engineered site on the Staphylococcus aureus chromosome. BMC Res. Notes 5:5. 10.1186/1756-0500-5-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lofdahl S, Sjostrom JE, Philipson L. 1978. A vector for recombinant DNA in Staphylococcus aureus. Gene 3:161–172. 10.1016/0378-1119(78)90059-8 [DOI] [PubMed] [Google Scholar]
- 12.Pereira PM, Veiga H, Jorge AM, Pinho MG. 2010. Fluorescent reporters for studies of cellular localization of proteins in Staphylococcus aureus. Appl. Environ. Microbiol. 76:4346–4353. 10.1128/AEM.00359-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Luong TT, Lee CY. 2007. Improved single-copy integration vectors for Staphylococcus aureus. J. Microbiol. Methods 70:186–190. 10.1016/j.mimet.2007.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sohail M, Dyke KG. 1995. Sites for co-integration of large staphylococcal plasmids. Gene 162:63–68. 10.1016/0378-1119(95)00324-Y [DOI] [PubMed] [Google Scholar]
- 15.Lee CY, Buranen SL, Ye ZH. 1991. Construction of single-copy integration vectors for Staphylococcus aureus. Gene 103:101–105. 10.1016/0378-1119(91)90399-V [DOI] [PubMed] [Google Scholar]
- 16.Saunders CW, Schmidt BJ, Mirot MS, Thompson LD, Guyer MS. 1984. Use of chromosomal integration in the establishment and expression of blaZ, a Staphylococcus aureus beta-lactamase gene, in Bacillus subtilis. J. Bacteriol. 157:718–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schwesinger MD, Novick RP. 1975. Prophage-dependent plasmid integration in Staphylococcus aureus. J. Bacteriol. 123:724–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee CY, Iandolo JJ. 1986. Integration of staphylococcal phage L54a occurs by site-specific recombination: structural analysis of the attachment sites. Proc. Natl. Acad. Sci. U. S. A. 83:5474–5478. 10.1073/pnas.83.15.5474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee CY, Iandolo JJ. 1986. Lysogenic conversion of staphylococcal lipase is caused by insertion of the bacteriophage L54a genome into the lipase structural gene. J. Bacteriol. 166:385–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mainiero M, Goerke C, Geiger T, Gonser C, Herbert S, Wolz C. 2010. Differential target gene activation by the Staphylococcus aureus two-component system saeRS. J. Bacteriol. 192:613–623. 10.1128/JB.01242-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Prax M, Lee CY, Bertram R. 2013. An update on the molecular genetics toolbox for staphylococci. Microbiology 159:421–435. 10.1099/mic.0.061705-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gupta RS. 2000. The phylogeny of proteobacteria: relationships to other eubacterial phyla and eukaryotes. FEMS Microbiol. Rev. 24:367–402. 10.1111/j.1574-6976.2000.tb00547.x [DOI] [PubMed] [Google Scholar]
- 23.Siefert JL, Fox GE. 1998. Phylogenetic mapping of bacterial morphology. Microbiology 144(Part 10):2803–2808 [DOI] [PubMed] [Google Scholar]
- 24.Tamames J, Gonzalez-Moreno M, Mingorance J, Valencia A, Vicente M. 2001. Bringing gene order into bacterial shape. Trends Genet. 17:124–126. 10.1016/S0168-9525(00)02212-5 [DOI] [PubMed] [Google Scholar]
- 25.Divakaruni AV, Loo RR, Xie Y, Loo JA, Gober JW. 2005. The cell-shape protein MreC interacts with extracytoplasmic proteins including cell wall assembly complexes in Caulobacter crescentus. Proc. Natl. Acad. Sci. U. S. A. 102:18602–18607. 10.1073/pnas.0507937102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kruse T, Bork-Jensen J, Gerdes K. 2005. The morphogenetic MreBCD proteins of Escherichia coli form an essential membrane-bound complex. Mol. Microbiol. 55:78–89. 10.1111/j.1365-2958.2004.04367.x [DOI] [PubMed] [Google Scholar]
- 27.Leaver M, Errington J. 2005. Roles for MreC and MreD proteins in helical growth of the cylindrical cell wall in Bacillus subtilis. Mol. Microbiol. 57:1196–1209. 10.1111/j.1365-2958.2005.04736.x [DOI] [PubMed] [Google Scholar]
- 28.Divakaruni AV, Baida C, White CL, Gober JW. 2007. The cell shape proteins MreB and MreC control cell morphogenesis by positioning cell wall synthetic complexes. Mol. Microbiol. 66:174–188. 10.1111/j.1365-2958.2007.05910.x [DOI] [PubMed] [Google Scholar]
- 29.Alyahya SA, Alexander R, Costa T, Henriques AO, Emonet T, Jacobs-Wagner C. 2009. RodZ, a component of the bacterial core morphogenic apparatus. Proc. Natl. Acad. Sci. U. S. A. 106:1239–1244. 10.1073/pnas.0810794106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bendezu FO, Hale CA, Bernhardt TG, de Boer PA. 2009. RodZ (YfgA) is required for proper assembly of the MreB actin cytoskeleton and cell shape in E. coli. EMBO J. 28:193–204. 10.1038/emboj.2008.264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kawai Y, Daniel RA, Errington J. 2009. Regulation of cell wall morphogenesis in Bacillus subtilis by recruitment of PBP1 to the MreB helix. Mol. Microbiol. 71:1131–1144. 10.1111/j.1365-2958.2009.06601.x [DOI] [PubMed] [Google Scholar]
- 32.Takacs CN, Poggio S, Charbon G, Pucheault M, Vollmer W, Jacobs-Wagner C. 2010. MreB drives de novo rod morphogenesis in Caulobacter crescentus via remodeling of the cell wall. J. Bacteriol. 192:1671–1684. 10.1128/JB.01311-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.White CL, Kitich A, Gober JW. 2010. Positioning cell wall synthetic complexes by the bacterial morphogenetic proteins MreB and MreD. Mol. Microbiol. 76:616–633. 10.1111/j.1365-2958.2010.07108.x [DOI] [PubMed] [Google Scholar]
- 34.Margolin W. 2009. Sculpting the bacterial cell. Curr. Biol. 19:R812–R822. 10.1016/j.cub.2009.06.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lipinski B, Hawiger J, Jeljaszewicz J. 1967. Staphylococcal clumping with soluble fibrin monomer complexes. J. Exp. Med. 126:979–988. 10.1084/jem.126.5.979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kornblum J, Kreiswirth B, Projan Ross SJH, Novick RP. 1990. Agr: a polycistronic locus regulating exoprotein synthesis in Staphylococcus aureus, p 373–402 In Novick RP. (ed), Molecular biology of the staphylococci. VCH Publishers, New York, NY [Google Scholar]
- 37.Reusch RN, Hiske TW, Sadoff HL. 1986. Poly-beta-hydroxybutyrate membrane structure and its relationship to genetic transformability in Escherichia coli. J. Bacteriol. 168:553–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Blevins JS, Elasri MO, Allmendinger SD, Beenken KE, Skinner RA, Thomas JR, Smeltzer MS. 2003. Role of sarA in the pathogenesis of Staphylococcus aureus musculoskeletal infection. Infect. Immun. 71:516–523. 10.1128/IAI.71.1.516-523.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.O'Toole GA, Kolter R. 1998. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol. Microbiol. 28:449–461. 10.1046/j.1365-2958.1998.00797.x [DOI] [PubMed] [Google Scholar]
- 40.Morrison JM, Miller EW, Benson MA, Alonzo F, III, Yoong P, Torres VJ, Hinrichs SH, Dunman PM. 2012. Characterization of SSR42, a novel virulence factor regulatory RNA that contributes to the pathogenesis of a Staphylococcus aureus USA300 representative. J. Bacteriol. 194:2924–2938. 10.1128/JB.06708-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Silva-Rocha R, Martinez-Garcia E, Calles B, Chavarria M, Arce-Rodriguez A, de Las Heras A, Paez-Espino AD, Durante-Rodriguez G, Kim J, Nikel PI, Platero R, de Lorenzo V. 2013. The Standard European Vector Architecture (SEVA): a coherent platform for the analysis and deployment of complex prokaryotic phenotypes. Nucleic Acids Res. 41:D666–D675. 10.1093/nar/gks1119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Britton RA, Eichenberger P, Gonzalez-Pastor JE, Fawcett P, Monson R, Losick R, Grossman AD. 2002. Genome-wide analysis of the stationary-phase sigma factor (sigma-H) regulon of Bacillus subtilis. J. Bacteriol. 184:4881–4890. 10.1128/JB.184.17.4881-4890.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nakano S, Kuster-Schock E, Grossman AD, Zuber P. 2003. Spx-dependent global transcriptional control is induced by thiol-specific oxidative stress in Bacillus subtilis. Proc. Natl. Acad. Sci. U. S. A. 100:13603–13608. 10.1073/pnas.2235180100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Arnaud M, Chastanet A, Debarbouille M. 2004. New vector for efficient allelic replacement in naturally nontransformable, low-GC-content, gram-positive bacteria. Appl. Environ. Microbiol. 70:6887–6891. 10.1128/AEM.70.11.6887-6891.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim L, Mogk A, Schumann W. 1996. A xylose-inducible Bacillus subtilis integration vector and its application. Gene 181:71–76. 10.1016/S0378-1119(96)00466-0 [DOI] [PubMed] [Google Scholar]
- 46.Paprotka K, Giese B, Fraunholz MJ. 2010. Codon-improved fluorescent proteins in investigation of Staphylococcus aureus host pathogen interactions. J. Microbiol. Methods 83:82–86. 10.1016/j.mimet.2010.07.022 [DOI] [PubMed] [Google Scholar]
- 47.Wach A. 1996. PCR-synthesis of marker cassettes with long flanking homology regions for gene disruptions in S. cerevisiae. Yeast 12:259–265. [DOI] [PubMed] [Google Scholar]
- 48.Rudin L, Sjostrom JE, Lindberg M, Philipson L. 1974. Factors affecting competence for transformation in Staphylococcus aureus. J. Bacteriol. 118:155–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lopez D, Fischbach MA, Chu F, Losick R, Kolter R. 2009. Structurally diverse natural products that cause potassium leakage trigger multicellularity in Bacillus subtilis. Proc. Natl. Acad. Sci. U. S. A. 106:280–285. 10.1073/pnas.0810940106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Karimova G, Pidoux J, Ullmann A, Ladant D. 1998. A bacterial two-hybrid system based on a reconstituted signal transduction pathway. Proc. Natl. Acad. Sci. U. S. A. 95:5752–5756. 10.1073/pnas.95.10.5752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Miller JH. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 52.Dubnau D. 1991. Genetic competence in Bacillus subtilis. Microbiol. Rev. 55:395–424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dubnau D. 1999. DNA uptake in bacteria. Annu. Rev. Microbiol. 53:217–244. 10.1146/annurev.micro.53.1.217 [DOI] [PubMed] [Google Scholar]
- 54.Nagami Y, Kimura M, Teranishi Y, Tanaka T. 1988. Construction of a new shuttle expression vector for Bacillus subtilis and Escherichia coli by using a polycistronic system. Gene 69:59–69. 10.1016/0378-1119(88)90378-2 [DOI] [PubMed] [Google Scholar]
- 55.Lechat P, Hummel L, Rousseau S, Moszer I. 2008. GenoList: an integrated environment for comparative analysis of microbial genomes. Nucleic Acids Res. 36:D469–D474. 10.1093/nar/gkm1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Plessis A, Perrin A, Haber JE, Dujon B. 1992. Site-specific recombination determined by I-SceI, a mitochondrial group I intron-encoded endonuclease expressed in the yeast nucleus. Genetics 130:451–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Viret JF. 1993. Meganuclease I-SceI as a tool for the easy subcloning of large DNA fragments devoid of selection marker. Biotechniques 14:325–326 [PubMed] [Google Scholar]
- 58.Schweizer HP, Hoang TT. 1995. An improved system for gene replacement and xylE fusion analysis in Pseudomonas aeruginosa. Gene 158:15–22. 10.1016/0378-1119(95)00055-B [DOI] [PubMed] [Google Scholar]
- 59.Cohen-Tannoudji M, Robine S, Choulika A, Pinto D, El Marjou F, Babinet C, Louvard D, Jaisser F. 1998. I-SceI-induced gene replacement at a natural locus in embryonic stem cells. Mol. Cell. Biol. 18:1444–1448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Martinez-Garcia E, de Lorenzo V. 2011. Engineering multiple genomic deletions in Gram-negative bacteria: analysis of the multi-resistant antibiotic profile of Pseudomonas putida KT2440. Environ. Microbiol. 13:2702–2716. 10.1111/j.1462-2920.2011.02538.x [DOI] [PubMed] [Google Scholar]
- 61.Lauderdale KJ, Boles BR, Cheung AL, Horswill AR. 2009. Interconnections between sigma B, agr, and proteolytic activity in Staphylococcus aureus biofilm maturation. Infect. Immun. 77:1623–1635. 10.1128/IAI.01036-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Herbert S, Ziebandt AK, Ohlsen K, Schafer T, Hecker M, Albrecht D, Novick R, Gotz F. 2010. Repair of global regulators in Staphylococcus aureus 8325 and comparative analysis with other clinical isolates. Infect. Immun. 78:2877–2889. 10.1128/IAI.00088-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Balaban N, Novick RP. 1995. Autocrine regulation of toxin synthesis by Staphylococcus aureus. Proc. Natl. Acad. Sci. U. S. A. 92:1619–1623. 10.1073/pnas.92.5.1619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cheung AL, Chien YT, Bayer AS. 1999. Hyperproduction of alpha-hemolysin in a sigB mutant is associated with elevated SarA expression in Staphylococcus aureus. Infect. Immun. 67:1331–1337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Greenspan P, Mayer EP, Fowler SD. 1985. Nile red: a selective fluorescent stain for intracellular lipid droplets. J. Cell Biol. 100:965–973. 10.1083/jcb.100.3.965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Latt SA, Stetten G, Juergens LA, Willard HF, Scher CD. 1975. Recent developments in the detection of deoxyribonucleic acid synthesis by 33258 Hoechst fluorescence. J. Histochem. Cytochem. 23:493–505. 10.1177/23.7.1095650 [DOI] [PubMed] [Google Scholar]
- 67.Liew ATF, Theis T, Jensen SO, Garcia-Lara J, Foster SJ, Firth N, Lewis PJ, Harry EJ. 2011. A simple plasmid-based system that allows rapid generation of tightly controlled gene expression in Staphylococcus aureus. Microbiology 157:666–676. 10.1099/mic.0.045146-0 [DOI] [PubMed] [Google Scholar]
- 68.Pinho MG, Errington J. 2005. Recruitment of penicillin-binding protein PBP2 to the division site of Staphylococcus aureus is dependent on its transpeptidation substrates. Mol. Microbiol. 55:799–807. 10.1111/j.1365-2958.2004.04420.x [DOI] [PubMed] [Google Scholar]
- 69.Jones LJ, Carballido-Lopez R, Errington J. 2001. Control of cell shape in bacteria: helical, actin-like filaments in Bacillus subtilis. Cell 104:913–922. 10.1016/S0092-8674(01)00287-2 [DOI] [PubMed] [Google Scholar]
- 70.Defeu Soufo HJ, Graumann PL. 2006. Dynamic localization and interaction with other Bacillus subtilis actin-like proteins are important for the function of MreB. Mol. Microbiol. 62:1340–1356. 10.1111/j.1365-2958.2006.05457.x [DOI] [PubMed] [Google Scholar]
- 71.Vats P, Shih YL, Rothfield L. 2009. Assembly of the MreB-associated cytoskeletal ring of Escherichia coli. Mol. Microbiol. 72:170–182. 10.1111/j.1365-2958.2009.06632.x [DOI] [PubMed] [Google Scholar]
- 72.Novick RP. 1991. Genetic systems in staphylococci. Methods Enzymol. 204:587–636. 10.1016/0076-6879(91)04029-N [DOI] [PubMed] [Google Scholar]
- 73.Prax M, Lee CY, Bertram R. 2013. An update on the molecular genetics toolbox for staphylococci. Microbiology 159:421–435. 10.1099/mic.0.061705-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lopez D, Vlamakis H, Kolter R. 2009. Generation of multiple cell types in Bacillus subtilis. FEMS Microbiol. Rev. 33:152–163. 10.1111/j.1574-6976.2008.00148.x [DOI] [PubMed] [Google Scholar]
- 75.Lopez D, Kolter R. 2010. Extracellular signals that define distinct and coexisting cell fates in Bacillus subtilis. FEMS Microbiol. Rev. 34:134–149. 10.1111/j.1574-6976.2009.00199.x [DOI] [PubMed] [Google Scholar]
- 76.Dominguez-Escobar J, Chastanet A, Crevenna AH, Fromion V, Wedlich-Soldner R, Carballido-Lopez R. 2011. Processive movement of MreB-associated cell wall biosynthetic complexes in bacteria. Science 333:225–228. 10.1126/science.1203466 [DOI] [PubMed] [Google Scholar]
- 77.Garner EC, Bernard R, Wang W, Zhuang X, Rudner DZ, Mitchison T. 2011. Coupled, circumferential motions of the cell wall synthesis machinery and MreB filaments in B. subtilis. Science 333:222–225. 10.1126/science.1203285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.van Teeffelen S, Wang S, Furchtgott L, Huang KC, Wingreen NS, Shaevitz JW, Gitai Z. 2011. The bacterial actin MreB rotates, and rotation depends on cell-wall assembly. Proc. Natl. Acad. Sci. U. S. A. 108:15822–15827. 10.1073/pnas.1108999108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Begg KJ, Spratt BG, Donachie WD. 1986. Interaction between membrane proteins PBP3 and rodA is required for normal cell shape and division in Escherichia coli. J. Bacteriol. 167:1004–1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Henriques AO, Glaser P, Piggot PJ, Moran CP., Jr 1998. Control of cell shape and elongation by the rodA gene in Bacillus subtilis. Mol. Microbiol. 28:235–247. 10.1046/j.1365-2958.1998.00766.x [DOI] [PubMed] [Google Scholar]
- 81.Lee JC, Stewart GC. 2003. Essential nature of the mreC determinant of Bacillus subtilis. J. Bacteriol. 185:4490–4498. 10.1128/JB.185.15.4490-4498.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Land AD, Winkler ME. 2011. The requirement for pneumococcal MreC and MreD is relieved by inactivation of the gene encoding PBP1a. J. Bacteriol. 193:4166–4179. 10.1128/JB.05245-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.