Abstract
Recent metagenomic studies on saltern ponds with intermediate salinities have determined that their microbial communities are dominated by both Euryarchaeota and halophilic bacteria, with a gammaproteobacterium closely related to the genera Alkalilimnicola and Arhodomonas being one of the most predominant microorganisms, making up to 15% of the total prokaryotic population. Here we used several strategies and culture media in order to isolate this organism in pure culture. We report the isolation and taxonomic characterization of this new, never before cultured microorganism, designated M19-40T, isolated from a saltern located in Isla Cristina, Spain, using a medium with a mixture of 15% salts, yeast extract, and pyruvic acid as the carbon source. Morphologically small curved cells (young cultures) with a tendency to form long spiral cells in older cultures were observed in pure cultures. The organism is a Gram-negative, nonmotile bacterium that is strictly aerobic, non-endospore forming, heterotrophic, and moderately halophilic, and it is able to grow at 10 to 25% (wt/vol) NaCl, with optimal growth occurring at 15% (wt/vol) NaCl. Phylogenetic analysis based on 16S rRNA gene sequence comparison showed that strain M19-40T has a low similarity with other previously described bacteria and shows the closest phylogenetic similarity with species of the genera Alkalilimnicola (94.9 to 94.5%), Alkalispirillum (94.3%), and Arhodomonas (93.9%) within the family Ectothiorhodospiraceae. The phenotypic, genotypic, and chemotaxonomic features of this new bacterium showed that it constitutes a new genus and species, for which the name Spiribacter salinus gen. nov., sp. nov., is proposed, with strain M19-40T (= CECT 8282T = IBRC-M 10768T = LMG 27464T) being the type strain.
INTRODUCTION
Hypersaline environments are widely distributed habitats represented by a variety of aquatic and terrestrial systems (1). Most microbiological studies in these extreme environments have been carried out on aquatic habitats, especially saline lakes and marine salterns, which are used for the commercial production of salt by evaporation of seawater (1, 2). One of the salterns that has been most extensively studied during the last 30 years is located in Santa Pola, Alicante, Spain; these studies have been based on culture-dependent (3–5) as well as culture-independent (6–11) techniques. The studies carried out in this hypersaline habitat have permitted the in-depth determination of the microbial diversity of the ponds of the saltern, especially that of the crystallizers (ponds in which the NaCl is precipitated). The crystallizers are dominated by the square-shaped haloarchaeal species Haloquadratum walsbyi, the Gram-negative rod Salinibacter ruber, a member of the phylum Bacteroidetes, and the recently described Nanohaloarchaeota (6, 7, 12–16).
In contrast to the most concentrated ponds, few studies have been carried out on the ponds of intermediate salinity designated concentrators (4, 5, 10, 17, 18). Early studies based on culture-dependent techniques indicated that the most frequently isolated halophilic bacteria were members of the genera Pseudomonas-Alteromonas-Alcaligenes (probably the current members of the Halomonadaceae) and Salinivibrio, as well as haloarchaeal members of the genera Haloferax and Haloarcula (4). However, later molecular studies based on PCR amplification of the 16S rRNA gene and denaturing gradient gel electrophoresis showed that these were not the dominant groups (10, 18).
Very recently, we have carried out metagenomic studies on three intermediate-salinity ponds (with 13, 19, and 33% total salts, respectively) and a crystallizer pond (with 37% salts) by deep shotgun sequencing of environmental DNA. These studies have permitted the determination of the microbial diversity of these ponds with intermediate salinities in comparison with that of the crystallizer pond (15, 19). Comparison of the 16S rRNA reads with sequences in databases indicated that the major groups at the intermediate-salinity ponds were the Euryarchaeota (which was the predominant group at the crystallizer pond), as well as the phyla Proteobacteria and Bacteroidetes. At the genus level, several haloarchaea (members of the genera Halorubrum, Haloquadratum, and Natronomonas) were still very abundant in ponds with 19 or 33% salts, but a large diversity composed of members of many different bacterial genera was also evident (with representatives related to the genera Salinibacter, Alkalilimnicola, Arhodomonas, Roseovarius, and Oceanicola) (15, 19).
The assembly of the metagenomic data sets corresponding to the intermediate-salinity saltern ponds permitted us to determine that several organisms that had not been previously isolated or characterized were abundant in these habitats, including two groups consisting of Euryarchaeota with high G+C contents and Euryarchaeota with low G+C contents (different from Haloquadratum walsbyi), a group of Actinobacteria with low G+C contents, and interestingly, a group belonging to the class Gammaproteobacteria which may account for up to 15% of the total prokaryotic population of the intermediate-salinity ponds. This abundant group of microbes was phylogenetically distantly related to the genera Alkalilimnicola, Arhodomonas, and Nitrococcus. The pI of the predicted proteins was very similar to that of Alkalilimnicola ehrlichii and indicated that this microorganism is a salt-out strategist (15, 19).
In this paper, we describe the isolation and characterization of the most abundant bacterium, according to the previous metagenomic studies. This bacterium is a member of the Ectothiorhodospiraceae, with Alkalilimnicola and Arhodomonas being the most closely related genera. We have characterized this new isolate taxonomically and describe it as a new genus and species.
MATERIALS AND METHODS
Isolation and culture conditions.
Strain M19-40T was isolated from a water sample obtained in October 2011 from a pond of a marine saltern located in Isla Cristina, Huelva, southwest Spain. The isolation medium used was designed on the basis of the nutritional requirements described for Alkalilimnicola ehrlichii (20), after incubation at 37°C for 6 weeks. The composition of the isolation medium was (wt/vol) 11.7% NaCl, 1.95% MgCl2·6H2O, 3.05% MgSO4·7H2O, 0.05% CaCl2, 0.3% KCl, 0.01% NaHCO3, 0.035% NaBr, 0.1% yeast extract (Difco), 0.11% pyruvic acid, and 1.8% agar. For routine growth, the strain was cultivated in SM15 medium with 15% (wt/vol) total salts. The composition of SM15 medium was (wt/vol) 11.7% NaCl, 1.95% MgCl2·6H2O, 3.05% MgSO4·7H2O, 0.05% CaCl2, 0.3% KCl, 0.01% NaHCO3, 0.035% NaBr, 0.5% casein digest, 0.25% yeast extract (Difco), 0.11% pyruvic acid, and 0.1% glucose. The pH was adjusted to 7.5 with 1 M KOH. When necessary, solid media were prepared by adding 1.8% (wt/vol) agar. The culture was maintained at −80°C in SM15 medium containing 50% (vol/vol) glycerol.
Phylogenetic analysis.
Genomic DNA was prepared using the method described by Marmur (21). The 16S rRNA gene was amplified by PCR with the forward primer 16F27 and the reverse primer 16R1488 (22). Direct determination of the sequence of the PCR-amplified DNA was carried out using an automated DNA sequencer (model ABI 3130XL; Applied Biosystems). The identification of phylogenetic relatives and calculation of pairwise 16S rRNA gene sequence similarities were achieved using the EzTaxon-e server (23). The 16S rRNA gene sequence analysis was performed with the ARB software package (24). The 16S rRNA gene sequences were aligned with the published sequences of the closely related bacteria, and the alignment was confirmed and checked against both the primary and secondary structures of the 16S rRNA molecule using the alignment tool of the ARB software package. Phylogenetic trees were constructed using three different methods, the maximum likelihood (25), maximum parsimony (26), and neighbor-joining (27) methods, and the algorithms integrated in the ARB software for phylogenetic inference. A bootstrap test (28) was performed by calculating 1,000 replicate trees in order to assess the robustness of the topology. The 16S rRNA gene sequences used for phylogenetic comparisons were obtained from the GenBank database, and the strain designations and accession numbers are shown in Fig. 1.
FIG 1.
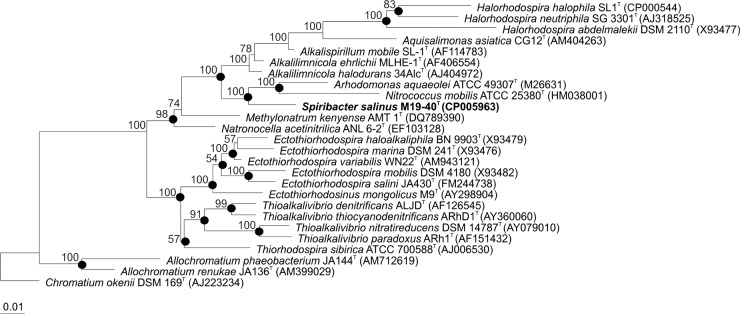
Phylogenetic tree of 16S rRNA gene sequences based on the maximum parsimony algorithm showing the position of Spiribacter salinus M19-40T and closely related species. GenBank sequence accession numbers are shown in parentheses. Bootstrap values higher than 50% are indicated at branch points. Chromatium okenii DSM 169T was used as the outgroup. Filled circles indicate that the corresponding nodes were also obtained in the trees generated with the maximum likelihood and neighbor-joining algorithms. Bar, 0.01 substitution per nucleotide position.
Genome recruitment.
The recruitment of the complete genome of strain M19-40T (29) from metagenomic data sets recently obtained from hypersaline habitats with different salt concentrations (15, 30, 31) was carried out via BLASTN analysis. A restrictive cutoff of 95% identity in at least 50 bp was established to guarantee that only similarities at the level of nearly identical microbes were counted.
Morphological, physiological, and biochemical characteristics.
For the determination of cellular morphology and motility, a culture from SM15 liquid medium was examined under a phase-contrast microscope. The morphology of the colonies and their size and pigmentation were observed on SM15 solid medium after 10 days of incubation at 37°C. Optimal conditions for growth were determined by growing the strain in SM liquid medium in 0.5, 3.0, 5, 7.5, 10, 12.5, 15, 20, 25, and 30% (wt/vol) NaCl and at temperatures of 15, 20, 28, 30, 37, 40, and 45°C. Growth rates were determined by monitoring the increase in the optical density at 600 nm. The pH range for the isolate was tested in SM15 medium adjusted to pH 5.0, 6.0, 7.0, 7.5, 8.0, 9.0, or 10.0 with the addition of the appropriate buffering capacity to each medium (32). All biochemical tests were carried out with 15% salts and 37°C, unless it is stated otherwise. Growth under anaerobic conditions was determined by incubating the strain in an anaerobic chamber in SM15 agar medium. Catalase activity was determined by adding a 1% (wt/vol) H2O2 solution to colonies on SM15 agar medium. The oxidase test was performed using a DrySlide assay (Difco). The following tests were conducted as described by Cowan and Steel (33) with the addition of 15% total salts and pyruvic acid to the medium (5, 34): tests for hydrolysis of esculin, casein, gelatin, Tweens 20, 60, and 80, tyrosine, and starch; the urease, methyl red, and Vogues-Proskauer tests; and tests for lysine, arginine, or ornithine decarboxylases and nitrate reduction. Citrate utilization was determined on Simmons citrate medium supplemented with 15% salts. For determining the range of substrates used as carbon and energy sources, the classical medium of Koser (35) as modified by Ventosa et al. (5) was used. Substrates were added as filter-sterilized solutions to give a final concentration of 1 g liter−1, except for carbohydrates, which were used at 2 g liter−1. Acid production from carbohydrates was performed as recommended by Ventosa et al. (5).
Electron microscopy.
Exponential- to stationary-phase cells grown in SM15 liquid medium incubated at 37°C on a shaking incubator at 200 rpm were collected by centrifugation and fixed in 1.6% glutaraldehyde buffered with 0.1 M cacodylate (pH 7.4) for 1 h at room temperature and 24 h at 4°C, and they were then postfixed in a 1% (wt/vol) osmium tetroxide solution for 1 h at 4°C. The dehydration was performed in an increasing series of acetone (50 to 100%), and the samples were embedded in Spurr resin and stained with 2% (wt/vol) aqueous uranyl acetate for 2 h. Blocks were obtained by polymerization at 70°C for 8 h. After hardening, ultrathin sections were cut with a diamond knife and then counterstained with uranyl acetate and lead citrate using an automatic counterstaining machine (Leica AC20). The thin sections were observed using a Philips CM-10 transmission electron microscope operating at 80 kV.
Chemotaxonomic analysis.
Fatty acid analysis was performed using a MIDI microbial identification system (36). Cells of strain M19-40T were cultured on SM15 agar medium at 37°C for 4 days. The extraction and analysis of fatty acids were performed according to the recommendations for the MIDI system. This analysis was carried out by CECT (Spanish Collection of Type Cultures, Valencia, Spain) using gas chromatography (Agilent 6850) and a standardized protocol for the MIDI Sherlock system (37).
The analysis of respiratory quinones and polar lipids of strain M19-40T was carried out by the Identification Service of DSMZ (Braunschweig, Germany). The quinones were determined according to the method of Collins et al. (38). The polar lipids were analyzed using the method of Minnikin et al. (39) and Collins and Jones (40).
Determination of DNA G+C content.
The G+C content of the genomic DNA of strain M19-40T was determined from the midpoint temperature (Tm) of the thermal denaturation profile (41), using the equation of Owen and Hill (42), obtained with a Perkin-Elmer UV-visible Lambda 20 spectrophotometer at 260 nm equipped with a PTP-1 Peltier system programmed with an increase in temperature of 1°C/min.
RESULTS AND DISCUSSION
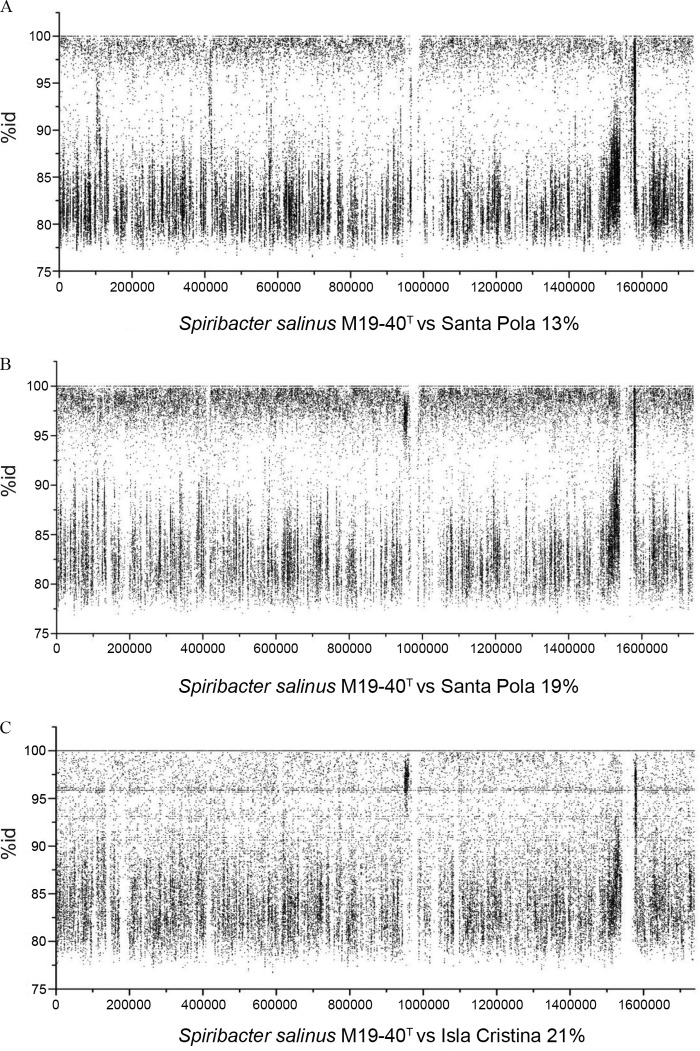
In order to isolate the gammaproteobacterium phylogenetically related to the genera Alkalilimnicola and Arhodomonas identified by metagenomic approaches (15), we sampled several saltern ponds during different periods from 2011 to 2012 using many growth media and conditions. The salterns sampled were located in Santa Pola, Alicante (where the metagenomic studies were previously carried out), and Huelva and Isla Cristina, Huelva, Spain. We used culture media with different salt concentrations ranging from 7.5 to 25% to which yeast extract (at concentrations ranging from 0.01 to 0.1%) and different carbon sources were added; the carbon source was either 10 mM glycerol, pyruvic acid, or sodium acetate. The pH of the media was adjusted to 7.0 to 9.0, and agar or agarose was used as the solidifying agent. Besides, aerobic, microaerophilic, or anaerobic conditions were used. More than 2,000 colonies randomly selected from these media were analyzed by PCR amplification and sequencing of the 16S rRNA gene. Several colonies were phylogenetically related to the genera Arhodomonas or Alkalilimnicola, but they were not able to grow after subculturing. One tiny colony of >0.5 mm showing pink pigmentation was subcultured on the same medium, although its growth was very slow and limited. In an effort to optimize the growth of this strain under laboratory conditions, different media and growth conditions were tested. The medium that showed optimal growth was based on a medium previously used for the isolation of the species Arhodomonas aquaeolei (43). For the isolation of the new bacterium, we took into consideration the data obtained from the previous metagenomic studies and the growth media already used for the cultivation of the most closely related microorganisms, members of the genera Alkalilimnicola and Arhodomonas. The isolate, designated strain M19-40T, was phylogenetically related to the genus Alkalilimnicola, with a 16S rRNA sequence similarity of 94.9%, a percentage similar to that reported previously on the basis of metagenomic studies for the contigs that represented one of the prokaryotic organisms abundant in intermediate-salinity ponds (15, 19). In order to determine if this new isolate corresponded to the previously reported abundant gammaproteobacterium, we sequenced the complete genome of strain M19-40T (29, 44). Comparison of the complete genome sequence with the metagenomic data sets previously obtained from several saltern ponds located in Santa Pola (with salinities of 13, 19, 33, and 37% total salts) and Isla Cristina (with 21% total salts) showed that the M19-40T genome sequence recruited best from the sequences of the metagenome data sets of the ponds with 13 to 21% salts (Fig. 2) but not with those of the other data sets, indicating that strain M19-40T is quite abundant in the water of ponds with intermediate salinities but is practically absent from ponds with salinities of from 33 to 37% salts (see Fig. S1 in the supplemental material). Besides the intermediate-salinity ponds of the Santa Pola saltern, sequences related to strain M19-40T (analyzed by recruitment of its genome from the available metagenomes of hypersaline habitats or by comparison of 16S rRNA gene sequences from the RDP database) have been observed in ponds of a saltern in San Diego, CA, with 12 to 14% salinity and several other habitats in Asia and South America with intermediate salinities (44).
FIG 2.
Recruitment of Spiribacter salinus M19-40T genome (1739487 bp) against metagenomes of hypersaline aquatic habitats of different salinities. (A) Metagenome from a Santa Pola saltern (13% salinity pond); (B) metagenome from a Santa Pola saltern (19% salinity pond); (C) metagenome from an Isla Cristina saltern (21% salinity pond). A restrictive cutoff of 95% identity in at least 50 bp of the length of the metagenomic reads was used.
The identification of phylogenetic neighbors and calculation of pairwise 16S rRNA gene sequence similarity were achieved using the EzTaxon-e server (23) and the ARB software package (24). For these analyses, we used both the 16S rRNA gene sequence obtained in this study (1,418 bp) and the sequence obtained from the complete sequence of the genome of strain M19-40T (both sequences were identical). The phylogenetic analysis, based on the maximum-parsimony algorithm, revealed that strain M19-40T constituted a monophyletic branch clearly separated from the most closely related species of the genera Alkalilimnicola, Arhodomonas, Alkalispirillum, and Nitrococcus, all of which are members of the family Ectothiorhodospiraceae (Fig. 1). Neighbor-joining and maximum likelihood methods resulted in highly similar tree topologies (Fig. 1). The closest phylogenetic similarity of strain M19-40T with other bacterial species was with the type strains of Alkalilimnicola ehrlichii (MLHE-1T; 94.7% 16S rRNA gene sequence similarity), Alkalilimnicola halodurans (34AlcT; 94.3%), Alkalispirillum mobile (SL-1T; 94.2%), Arhodomonas aquaeolei (ATCC 49307T; 93.6%), and Nitrococcus mobilis (ATCC 25380T; 93.1%). These low percentages clearly support the placement of the new isolate within a new genus and species separate from the previously described bacterial taxa. Besides, the phylogenetic position of strain M19-40T was confirmed when we constructed the phylogenetic tree using a concatenated sequence of 277 conserved proteins in all genomes available for members of the family Ectothiorhodospiraceae (44).
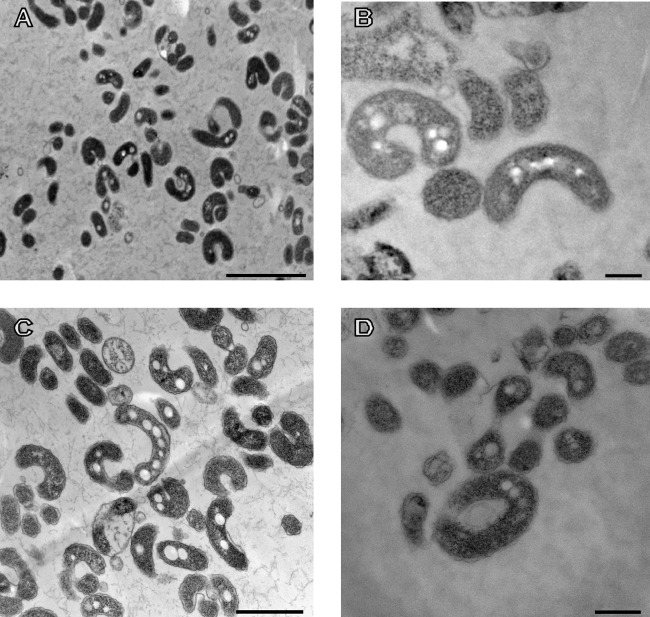
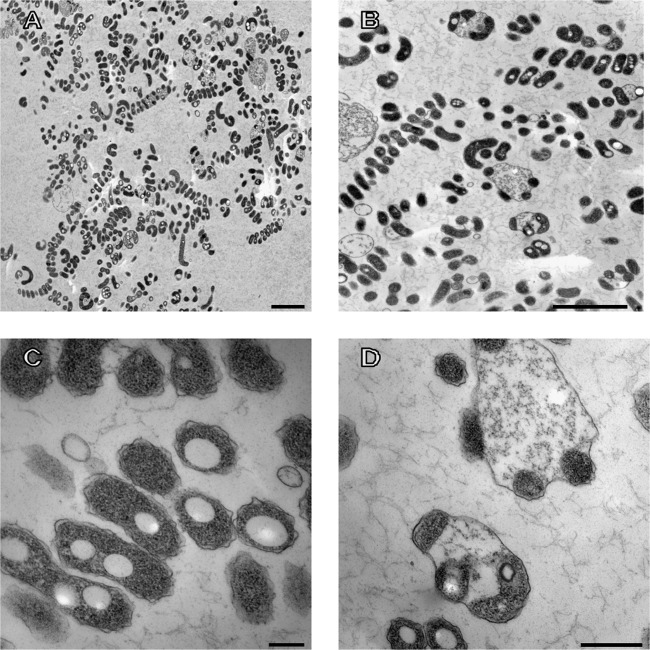
Strain M19-40T was Gram stain negative, nonmotile, and strictly aerobic. It was able to grow in medium containing 10 to 25% (wt/vol) NaCl and optimally in a medium containing 15% (wt/vol) NaCl (see Fig. S2 in the supplemental material). No growth was observed in the absence of NaCl. Thus, this bacterium can be considered a moderately halophilic microorganism (45). The optimal temperature and pH were 37°C and pH 7.5. When grown on SM15 liquid medium at 37°C, the cells were short and thin curved rods on young cultures, but long spiral cells were produced at the stationary stage (see Fig. S3 in the supplemental material). These spiral cells were not so abundant when strain M19-40T was grown at 28°C. By electron microscopy, the cells of strain M19-40T at early exponential stage were curved slender rods with a size of 0.3 μm (width) by 0.8 to 1.8 μm (length) (Fig. 3A and B). Later, the cells elongated and long curved cells with a tendency to produce spirals were produced (Fig. 3C and D). At the late stationary phase of growth, long spiral cells were more evident (Fig. 4A and B), showing large polyalkanoate inclusion bodies (Fig. 4C). Besides, a Gram-negative bacterium-type cell wall structure was observed (Fig. 4C). Some of the spirals were surrounded by an external envelope, as observed in Fig. 4B and D. These resembled the rotund bodies often observed in Vibrio marinus (46), Oceanithermus (47, 48), and some species of the genus Thermus, such as Thermus aquaticus (49), and in a regulation-defective S-layer mutant of Thermus thermophilus, in which round multicellular bodies surrounded by a common envelope were observed as the culture approached the stationary phase (50, 51). However, in strain M19-40T, it seems that only a single spiral cell is included within such an envelope. This bacterium showed catalase and oxidase activity; however, it was not able to reduce nitrate to nitrite. Other phenotypic features of strain M19-40T are detailed below in the new species description.
FIG 3.
Transmission electron microphotographs (A to D) of thin sections of cells of Spiribacter salinus M19-40T grown on SM15 liquid medium at early exponential phase. Bars, 2 μm (A), 300 nm (B), 1 μm (C), and 500 nm (D).
FIG 4.
Transmission electron microphotographs (A to D) of thin sections of cells of Spiribacter salinus M19-40T grown on SM15 liquid medium at late stationary phase. Bars, 2 μm (A, B), 200 nm (C), and 500 nm (D).
The G+C content of the DNA of strain M19-40T was 60.0 mol% (determined by the Tm method). This value is similar to the genomic G+C content calculated from the complete genome sequence (62.7%) (44). This value is within the range described for members of the family Ectothiorhodospiraceae (50.5 to 69.9 mol%) (52) but different from the values reported for species of the genera Alkalilimnicola (65.6 to 67.5 mol%) (20, 53), Alkalispirillum (66.2 mol%) (54), and Arhodomonas (67.0 mol%) (43).
The cellular fatty acid profile of strain M19-40T was characterized by the fatty acids C18:1ω6c/C18:1ω7c (60.6%), C16:0 (13.4%), 3-OH C10:0 (6.4%), and C12:0 (5.7%) as the major fatty acids, with minor amounts of C16:1ω7c/C16:1ω6c (3.6%), C19:0ω8c cyc (2.2%), 3-OH C14:0 (2.0%), C18:0 (1.6%), C10:0 (1.2%), and 3-OH C12:0 (1.2%) being detected. This fatty acid profile is quite different from that reported for Alkalilimnicola, for which the major fatty acids are C18:1ω7c/C18:1ω9c but in which C18:1ω6c (the major fatty acid determined in strain M19-40T) is absent; besides, the other major fatty acids present in strain M19-40T are not found in species of Alkalilimnicola (53). Similarly, the major fatty acids present in strain M19-40T are also absent in species of the genera Arhodomonas and Alkalispirillum, for which the major fatty acids are C18:1 cys11 and C18:1, respectively (43, 54), both of which are absent in strain M19-40T. The respiratory isoprenoid quinone found in strain M19-40T was ubiquinone Q8, similar to that reported for species of the family Ectothiorhodospiraceae (52). The polar lipids present in strain M19-40T are phosphatidylglycerol, phosphatidylethanolamine, a phosphoglycolipid, a phosphoaminoglycolipid, and three phospholipids (see Fig. S4 in the supplemental material). The presence of phosphatidylglycerol and phosphatidylethanolamine but not the other lipids was also reported for Alkalilimnicola halodurans, with M19-40T thus showing a polar lipid profile different from that of this related bacterium (53).
We have recently sequenced the complete genome of strain M19-40T and assembled it into a single contig (29, 44). The genome has a size of 1.7 Mbp, being the smallest genome described within the Ectothiorhodospiraceae. It is composed of a single circular replicon with only one rRNA operon and 1,706 protein-coding genes. The genome is very streamlined with a median intergenic spacer of only 14 to 19 nucleotides, also the smallest intergenic spacer of any species of the Ectothiorhodospiraceae (44). The analysis of this genome showed that the new bacterium has a simplified metabolic versatility, missing the chemolithotrophic and carbon fixation pathways. It was also determined to have a small number of insertion sequence and other mobile genetic elements and the absence of a CRISPR system and flagellum. These features are frequently observed in oligotrophic microorganisms with streamlined genomes that reach high population densities in aquatic habitats, as has been reported for “Candidatus Pelagibacter ubique” in the ocean (44). Metabolically, it is a heterotrophic microorganism, but the presence of xanthorhodopsin-coding genes might indicate an additional energy source when light is available. The presence of transporter genes and a complete ectoine biosynthesis gene cluster suggests a salt-out strategy for osmoregulation (44). The widespread distribution of strain M19-40T in habitats with intermediate salinities (13 to 25%) indicates that it can be considered an ecologically moderate halophile, in contrast to the previously defined moderately halophilic bacteria, on the basis of growth under laboratory conditions (44, 45).
The comparison of strain M19-40T with the most closely related species for which complete genome sequences are available permitted the calculation of the average nucleotide sequence identity (ANI) according to the proposal of Konstantinidis and Tiedje (55). The ANI of strain M19-40T with respect to the sequence of Alkalilimnicola ehrlichii MLHE-1T is 68.0%, a value low enough to justify the placement of the new isolate in a separate genus and species (55).
The difficulty with growing this bacterium in laboratory culture medium and under laboratory conditions has hampered the phenotypic characterization of the bacterium following the classical methods. The available genomic sequence data have been of considerable importance for the phenotypic characterization of this new bacterium. For example, the presence in the genome of genes related to the production of urease has permitted us to modify the test for determination of urease production; we originally used the classical Christensen method (56), and the test was negative, but we were able to show a positive result when we tested it carefully by the most sensitive modified method described by Cowan and Steel (33). Gas vesicles have not been observed in cells of this bacterium, and this finding was confirmed by the absence of genes related to gas vesicle biosynthesis.
In conclusion, the polyphasic taxonomic study, based on a combination of phylogenetic, genotypic, chemotaxonomic, and phenotypic data, supports the classification of strain M19-40T within a separate genus and species, for which we propose the new name Spiribacter salinus gen. nov., sp. nov. The differentiation of the new genus from other related genera of the family Ectothiorhodospiraceae can be accomplished by use of a combination of phylogenetic analysis based on 16S rRNA gene sequence comparison and genotypic and chemotaxonomic features. The predominant fatty acids or polar lipids and the genomic G+C content of the new genus Spiribacter are different from those of the related genera Alkalilimnicola, Arhodomonas, and Alkalispirillum, as discussed above.
Description of Spiribacter gen. nov.
Spiribacter (Spi.ri.bac′ter. L. n. spira, a spiral; N.L. masc. n. bacter, a rod; N.L. masc. n. Spiribacter, spiral rod).
Cells are Gram negative, nonmotile, and curved to spiral shaped. Non-endospore forming. Strictly aerobic. Heterotrophic. Moderately halophilic. Not able to grow in the absence of NaCl. Catalase and oxidase positive. The major cellular fatty acids are C18:1ω6c/C18:1ω7c, C16:0, and 3-OH C10:0. The respiratory isoprenoid quinone is ubiquinone Q8. The polar lipids are phosphatidylglycerol, phosphatidylethanolamine, a phosphoglycolipid, a phosphoaminoglycolipid, and three phospholipids. The genus Spiribacter belongs to the family Ectothiorhodospiraceae and is distantly related to the genera Alkalilimnicola, Alkalispirillum, and Arhodomonas (≤94.9% similarity on the basis of 16S rRNA gene sequence analysis). The type species is Spiribacter salinus.
Description of Spiribacter salinus sp. nov.
Spiribacter salinus (sa.li′nus. N.L. masc. adj. salinus, salted, salty).
The species shows the following characteristics, in addition to the aforementioned features of the genus. When grown on SM15 liquid medium at 37°C, curved cells of 0.3 by 0.8 to 1.8 μm are observed in young cultures, while in stationary phase, most cells are long spiral shaped and may reach 30 to 35 μm. The spiral cells are not so abundant when grown at 28°C. Cells are nonmotile. Colonies are circular, entire, smooth, convex, pink to dark pink pigmented, and 0.5 to 1.5 mm in diameter on SM15 agar medium after 10 days of incubation at 37°C. Moderately halophilic, growing at 10 to 25% (wt/vol) NaCl, with optimal growth at 15% (wt/vol) NaCl. No growth occurs in the absence of NaCl. Growth at 15 to 40°C, showing optimal growth at 37°C, and at pH values over the range of 6.0 to 9.0, with optimal growth occurring at pH 7.5 to 8.0. Gelatin, casein, starch, Tweens 20, 60, and 80, tyrosine, and esculin are not hydrolyzed. Nitrate is not reduced to nitrite. Urease and H2S are produced. Simmons citrate and phosphatase are negative. Methyl red and Voges-Proskauer tests are negative. It is able to utilize pyruvate or glycerol as carbon and energy sources; however, the following compounds are not utilized as sole sources of carbon and energy when assayed by the conventional methods: d-arabinose, fructose, d-galactose, d-glucose, maltose, d-melizitose, l-raffinose, sucrose, d-trehalose, ethionine, d-mannitol, d-sorbitol, citrate, formate, fumarate, malate, propionate, succinate, tartrate, l-cysteine, l-threonine, phenylalanine, or serine. Acid is not produced from d-glucose and other carbohydrates. Lysine, arginine, and ornithine decarboxylases are negative.
The type strain is M19-40T (= CECT 8282T = IBRC-M 10768T = LMG 27464T), which was isolated from a saltern in Isla Cristina, Huelva, Spain. The DNA G+C content of the type strain is 60.0 mol% (determined by the Tm method) or 62.7% (determined from the genome sequence).
Supplementary Material
ACKNOWLEDGMENTS
This research was supported by grants from the Spanish Ministry of Science and Innovation (CGL2010-19303 and BIO2011-12879-E), the National Science Foundation (grant DEB-0919290), the Junta de Andalucía (P10-CVI-6226), and the Generalitat Valenciana (ACOMP/2009/155). FEDER funds and the Plan Andaluz de Investigación also supported this project. Maria José León and Ana B. Fernández were recipients of postgraduate and postdoctoral fellowships, respectively, from the Junta de Andalucía. Rohit Ghai was supported by a Juan de la Cierva scholarship from the Spanish Ministry of Science and Innovation.
We thank Jean Euzéby for his help with the etymology of the novel taxa and Juan Luis Ribas and Asunción Fernández from the Microscopy Service of CITIUS (General Research Services, Universidad de Sevilla, Seville, Spain) for technical assistance with the electron microscopy study.
Footnotes
Published ahead of print 18 April 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.00430-14.
REFERENCES
- 1.Ventosa A. 2006. Unusual micro-organisms from unusual habitats: hypersaline environments, p 223–253 In Logan NA, Lappin-Scott HM, Oyston PCF. (ed), Prokaryotic diversity: mechanism and significance. Cambridge University Press, Cambridge, United Kingdom [Google Scholar]
- 2.Oren A. 2013. Life at high salt concentrations, intracellular KCl concentrations, and acidic proteomes. Front. Microbiol. 4:315. 10.3389/fmicb.2013.00315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rodriguez-Valera F, Ruiz-Berraquero F, Ramos-Cormenzana A. 1981. Characteristics of the heterotrophic bacterial populations in hypersaline environments of different salt concentrations. Microb. Ecol. 7:235–243. 10.1007/BF02010306 [DOI] [PubMed] [Google Scholar]
- 4.Rodriguez-Valera F, Ventosa A, Juez G, Imhoff JF. 1985. Variation of environmental features and microbial populations with salt concentrations in a multi-pond saltern. Microb. Ecol. 11:107–115. 10.1007/BF02010483 [DOI] [PubMed] [Google Scholar]
- 5.Ventosa A, Quesada E, Rodriguez-Valera F, Ruiz-Berraquero F, Ramos-Cormenzana A. 1982. Numerical taxonomy of moderately halophilic Gram-negative rods. J. Gen. Microbiol. 128:1959–1968 [Google Scholar]
- 6.Antón J, Llobet-Brossa E, Rodríguez-Valera F, Amann R. 1999. Fluorescence in situ hybridization analysis of the prokaryotic community inhabiting crystallizer ponds. Environ. Microbiol. 1:517–523. 10.1046/j.1462-2920.1999.00065.x [DOI] [PubMed] [Google Scholar]
- 7.Antón J, Rosselló-Mora R, Rodríguez-Valera F, Amann R. 2000. Extremely halophilic bacteria in crystallizer ponds from solar salterns. Appl. Environ. Microbiol. 66:3052–3057. 10.1128/AEM.66.7.3052-3057.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Benlloch S, Martínez-Murcia A, Rodriguez-Valera F. 1995. Sequencing of bacterial and archaeal 16S rDNA genes directly amplified from a hypersaline environment. Syst. Appl. Microbiol. 18:574–581. 10.1016/S0723-2020(11)80418-2 [DOI] [Google Scholar]
- 9.Benlloch S, Acinas SG, Antón J, López-López A, Luz SP, Rodríguez-Valera F. 2001. Archaeal biodiversity in crystallizer ponds from a solar saltern: culture versus PCR. Microb. Ecol. 41:12–19. 10.1007/s002480000069 [DOI] [PubMed] [Google Scholar]
- 10.Benlloch S, López-López A, Casamayor EO, Øvreås L, Goddard V, Daae FL, Smerdon G, Massana R, Joint I, Thingstad F, Pedrós-Alió C, Rodríguez-Valera F. 2002. Prokaryotic genetic diversity throughout the salinity gradient of a coastal solar saltern. Environ. Microbiol. 4:349–360. 10.1046/j.1462-2920.2002.00306.x [DOI] [PubMed] [Google Scholar]
- 11.Legault BA, Lopez-Lopez A, Alba-Casado JC, Doolittle WF, Bolhuis H, Rodriguez-Valera F, Papke RT. 2006. Environmental genomics of “Haloquadratum walsbyi” in a saltern crystallizer indicates a large pool of accessory genes in an otherwise coherent species. BMC Genomics 7:171. 10.1186/1471-2164-7-171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Antón J, Oren A, Benlloch S, Rodríguez-Valera F, Amann R, Rosselló-Mora R. 2002. Salinibacter ruber gen. nov., sp. nov., a novel, extremely halophilic member of the Bacteria from saltern crystallizer ponds. Int. J. Syst. Evol. Microbiol. 52:485–491. 10.1099/ijs.0.01913-0 [DOI] [PubMed] [Google Scholar]
- 13.Bolhuis H, Poele EM, Rodríguez-Valera F. 2004. Isolation and cultivation of Walsby's square archaeon. Environ. Microbiol. 6:349–360. 10.1111/j.1462-2920.2004.00692.x [DOI] [PubMed] [Google Scholar]
- 14.Burns DG, Janssen PH, Itoh T, Kamekura M, Li Z, Jensen G, Rodríguez-Valera F, Bolhuis H, Dyall-Smith ML. 2007. Haloquadratum walsbyi gen. nov., sp. nov., the square haloarchaeon of Walsby, isolated from saltern crystallizers in Australia and Spain. Int. J. Syst. Evol. Microbiol. 57:387–392. 10.1099/ijs.0.64690-0 [DOI] [PubMed] [Google Scholar]
- 15.Ghai R, Pašić L, Fernández AB, Martin-Cuadrado AB, Mizuno CM, McMahon KD, Papke RT, Stepanauskas R, Rodriguez-Brito B, Rohwer F, Sánchez-Porro C, Ventosa A, Rodríguez-Valera F. 2011. New abundant microbial groups in aquatic hypersaline environments. Sci. Rep. 1:135. 10.1038/srep00135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Narasingarao P, Podell S, Ugalde JA, Brochier Armanet C, Emerson JB, Brocks JJ, Heidelberg KB, Banfield JF, Allen EE. 2012. De novo metagenomic assembly reveals abundant novel major lineage of Archaea in hypersaline microbial communities. ISME J. 6:81–93. 10.1038/ismej.2011.78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pedrós-Alió C, Calderón-Paz JI, MacLean MH, Medina G, Marrasé C, Gasol JM, Guixa-Boixereu N. 2000. The microbial food web along salinity gradients. FEMS Microbiol. Ecol. 32:143–155. 10.1111/j.1574-6941.2000.tb00708.x [DOI] [PubMed] [Google Scholar]
- 18.Casamayor EO, Massana R, Benlloch S, Øvreås L, Díez B, Goddard VJ, Gasol JM, Joint I, Rodríguez-Valera F, Pedrós-Alió C. 2002. Changes in archaeal, bacterial and eukaryal assemblages along a salinity gradient by comparison of genetic fingerprinting methods in a multipond solar saltern. Environ. Microbiol. 4:338–348. 10.1046/j.1462-2920.2002.00297.x [DOI] [PubMed] [Google Scholar]
- 19.Fernández AB, Ghai R, Martin-Cuadrado A-B, Sánchez-Porro C, Rodriguez-Valera F, Ventosa A. Prokaryotic taxonomic and metabolic diversity of an intermediate salinity hypersaline habitat assessed by metagenomics. FEMS Microbiol. Ecol., in press. 10.1111/1574-6941.12329 [DOI] [PubMed] [Google Scholar]
- 20.Hoeft SE, Blum JS, Stolz JF, Tabita FR, Witte B, King GM, Santini JM, Oremland RS. 2007. Alkalilimnicola ehrlichii sp. nov., a novel, arsenite-oxidizing haloalkaliphilic gammaproteobacterium capable of chemoautotrophic or heterotrophic growth with nitrate or oxygen as the electron acceptor. Int. J. Syst. Evol. Microbiol. 57:504–512. 10.1099/ijs.0.64576-0 [DOI] [PubMed] [Google Scholar]
- 21.Marmur J. 1961. A procedure for the isolation of deoxyribonucleic acid from micro-organisms. J. Mol. Biol. 3:208–218. 10.1016/S0022-2836(61)80047-8 [DOI] [Google Scholar]
- 22.Márquez MC, Carrasco IJ, Xue Y, Ma Y, Cowan DA, Jones BE, Grant WD, Ventosa A. 2008. Aquisalibacillus elongatus gen. nov., sp. nov., a moderately halophilic bacterium of the family Bacillaceae isolated from a saline lake. Int. J. Syst. Evol. Microbiol. 58:1922–1926. 10.1099/ijs.0.65813-0 [DOI] [PubMed] [Google Scholar]
- 23.Kim OS, Cho YJ, Lee K, Yoon SH, Kim M, Na H, Park SC, Jeon YS, Lee JH, Yi H, Won S, Chun J. 2012. Introducing EzTaxon-e: a prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species. Int. J. Syst. Evol. Microbiol. 62:716–721. 10.1099/ijs.0.038075-0 [DOI] [PubMed] [Google Scholar]
- 24.Ludwig W, Strunk O, Westram R, Richter L, Meier H, Yadhukumar , Buchner A, Lai T, Steppi S, Jobb G, Förster W, Brettske I, Gerber S, Ginhart AW, Gross O, Grumann S, Hermann S, Jost R, König A, Liss T, Lüssmann R, May M, Nonhoff B, Reichel B, Strehlow R, Stamatakis A, Stuckmann N, Vilbig A, Lenke M, Ludwig T, Bode A, Schleifer KH. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363–1371. 10.1093/nar/gkh293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Felsenstein J. 1981. Evolutionary trees from DNA sequences: a maximum likelihood approach. J. Mol. Evol. 17:368–376. 10.1007/BF01734359 [DOI] [PubMed] [Google Scholar]
- 26.Fitch WM. 1971. Toward defining the course of evolution: minimum change for a specific tree topology. Syst. Zool. 20:406–416. 10.2307/2412116 [DOI] [Google Scholar]
- 27.Saitou N, Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406–425 [DOI] [PubMed] [Google Scholar]
- 28.Felsenstein J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791. 10.2307/2408678 [DOI] [PubMed] [Google Scholar]
- 29.Leon MJ, Ghai R, Fernandez AB, Sanchez-Porro C, Rodriguez-Valera F, Ventosa A. 2013. Draft genome of Spiribacter salinus M19-40, an abundant gammaproteobacterium in aquatic hypersaline environments. Genome Announc. 1(1):e00179-12. 10.1128/genomeA.00179-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fernandez AB, Ghai R, Martin-Cuadrado AB, Sanchez-Porro C, Rodriguez-Valera F, Ventosa A. 2013. Metagenome sequencing of prokaryotic microbiota from two hypersaline ponds of a marine saltern in Santa Pola, Spain. Genome Announc. 1(6):e00933-13. 10.1128/genomeA.00933-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fernández AB, León MJ, Vera B, Sanchez-Porro C, Ventosa A. 2014. Metagenomic sequence of prokaryotic microbiota from an intermediate-salinity pond of a saltern in Isla Cristina, Spain. Genome Announc. 2(1):e00045-14. 10.1128/genomeA.00045-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sánchez-Porro C, de la Haba RR, Soto-Ramírez N, Márquez MC, Montalvo-Rodríguez R, Ventosa A. 2009. Description of Kushneria aurantia gen. nov., sp. nov., a novel member of the family Halomonadaceae, and a proposal for reclassification of Halomonas marisflavi as Kushneria marisflavi comb. nov., of Halomonas indalinina as Kushneria indalinina comb. nov. and of Halomonas avicenniae as Kushneria avicenniae comb. nov. Int. J. Syst. Evol. Microbiol. 59:397–405. 10.1099/ijs.0.001461-0 [DOI] [PubMed] [Google Scholar]
- 33.Cowan ST, Steel KJ. 1977. Manual for the identification of medical bacteria. Cambridge University Press, London, United Kingdom [Google Scholar]
- 34.Quesada E, Ventosa A, Ruiz-Berraquero F, Ramos-Cormenzana A. 1984. Deleya halophila, a new species of moderately halophilic bacteria. Int. J. Syst. Bacteriol. 34:287–292. 10.1099/00207713-34-3-287 [DOI] [Google Scholar]
- 35.Koser SA. 1923. Utilization of the salts of organic acids by the colon-aerogenes group. J. Bacteriol. 8:493–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sasser M. 1990. Identification of bacteria by gas chromatography of cellular fatty acids, MIDI technical note 101. MIDI Inc, Newark, DE [Google Scholar]
- 37.MIDI Inc. 2008. Sherlock microbial identification system operating manual, version 6.1. MIDI Inc, Newark, DE [Google Scholar]
- 38.Collins MD, Pirouz T, Goodfellow M, Minnikin DE. 1977. Distribution of menaquinones in actinomycetes and corynebacteria. J. Gen. Microbiol. 11:221–230 [DOI] [PubMed] [Google Scholar]
- 39.Minnikin DE, Collins MD, Goodfellow M. 1979. Fatty acid and polar lipid composition in the classification of Cellulomonas, Oerskovia and related taxa. J. Appl. Bacteriol. 47:87–95. 10.1111/j.1365-2672.1979.tb01172.x [DOI] [PubMed] [Google Scholar]
- 40.Collins MD, Jones D. 1980. Lipids in the classification and identification of coryneform bacteria containing peptidoglycans based on 2,4-diaminobutyric acid. J. Appl. Microbiol. 48:459–470 [Google Scholar]
- 41.Marmur J, Doty P. 1962. Determination of the base composition of deoxyribonucleic acid from its thermal denaturation temperature. J. Mol. Biol. 5:109–118. 10.1016/S0022-2836(62)80066-7 [DOI] [PubMed] [Google Scholar]
- 42.Owen RJ, Hill LR. 1979. The estimation of base compositions, base pairing and genome size of bacterial deoxyribonucleic acids, p 217–296 In Skinner FA, Lovelock DW. (ed), Identification methods for microbiologists, 2nd ed. Academic Press, London, United Kingdom [Google Scholar]
- 43.Adkins JP, Madigan MT, Mandelco L, Woese CR, Tanner RS. 1993. Arhodomonas aquaeolei gen. nov., sp. nov., an aerobic, halophilic bacterium isolated from a subterranean brine. Int. J. Syst. Bacteriol. 43:514–520. 10.1099/00207713-43-3-514 [DOI] [PubMed] [Google Scholar]
- 44.López-Pérez M, Ghai R, Leon MJ, Rodríguez-Olmos A, Copa-Patiño JL, Soliveri J, Sanchez-Porro C, Ventosa A, Rodriguez-Valera F. 2013. Genomes of “Spiribacter,” a streamlined, successful halophilic bacterium. BMC Genomics 14:787. 10.1186/1471-2164-14-787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ventosa A, Nieto JJ, Oren A. 1998. Biology of moderately halophilic aerobic bacteria. Microbiol. Mol. Biol. Rev. 62:504–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Felter RA, Colwell RR, Chapman GB. 1969. Morphology and round body formation in Vibrio marinus. J. Bacteriol. 99:326–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Miroshnichenko ML, L'Haridon S, Jeanthon C, Antipov AN, Kostrikina NA, Tindall BJ, Schumann P, Spring S, Stackebrandt E, Bonch-Osmolovskaya EA. 2003. Oceanithermus profundus gen. nov., sp. nov., a thermophilic, microaerophilic, facultatively chemolithoheterotrophic bacterium from a deep-sea hydrothermal vent. Int. J. Syst. Evol. Microbiol. 53:747–752. 10.1099/ijs.0.02367-0 [DOI] [PubMed] [Google Scholar]
- 48.Mori K, Kakegawa T, Higashi Y, Nakamura K, Maruyama A, Hanada S. 2004. Oceanithermus desulfurans sp. nov., a novel thermophilic, sulfur-reducing bacterium isolated from a sulfide chimney in Suiyo Seamount. Int. J. Syst. Evol. Microbiol. 54:1561–1566. 10.1099/ijs.0.02962-0 [DOI] [PubMed] [Google Scholar]
- 49.Brock TD, Edwards MR. 1970. Fine structure of Thermus aquaticus, an extreme thermophile. J. Bacteriol. 104:509–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Castán P, de Pedro MA, Risco C, Vallés C, Fernández LA, Schwarz H, Berenguer J. 2001. Multiple regulatory mechanisms act on the 5′ untranslated region of the S-layer gene from Thermus thermophilus HB8. J. Bacteriol. 183:1491–1494. 10.1128/JB.183.4.1491-1494.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Castán P, Zafra O, Moreno R, de Pedro MA, Vallés C, Cava F, Caro E, Schwarz H, Berenguer J. 2002. The periplasmic space in Thermus thermophilus: evidence from a regulation-defective S-layer mutant overexpressing an alkaline phosphatase. Extremophiles 6:225–232. 10.1007/s00792-001-0246-3 [DOI] [PubMed] [Google Scholar]
- 52.Imhoff JF. 1984. Reassignment of the genus Ectothiorhodospira Pelsh 1936 to a new family, Ectothiorhodospiraceae fam. nov., and emended description of the Chromatiaceae Bavendamm 1924. Int. J. Syst. Bacteriol. 34:338–339. 10.1099/00207713-34-3-338 [DOI] [Google Scholar]
- 53.Yakimov MM, Giuliano L, Chernikova TN, Gentile G, Abraham WR, Lünsdorf H, Timmis KN, Golyshin PN. 2001. Alcalilimnicola halodurans gen. nov., sp. nov., an alkaliphilic, moderately halophilic and extremely halotolerant bacterium, isolated from sediments of soda-depositing Lake Natron, East Africa Rift Valley. Int. J. Syst. Evol. Microbiol. 51:2133–2143. 10.1099/00207713-51-6-2133 [DOI] [PubMed] [Google Scholar]
- 54.Rijkenberg MJ, Kort R, Hellingwerf KJ. 2001. Alkalispirillum mobile gen. nov., spec. nov., an alkaliphilic non-phototrophic member of the Ectothiorhodospiraceae. Arch. Microbiol. 175:369–375. 10.1007/s002030100274 [DOI] [PubMed] [Google Scholar]
- 55.Konstantinidis KT, Tiedje JM. 2005. Genomic insights that advance the species definition for prokaryotes. Proc. Natl. Acad. Sci. U. S. A. 102:2567–2572. 10.1073/pnas.0409727102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Christensen WB. 1946. Urea decomposition as a means of differentiating Proteus and paracolon cultures from each other and from Salmonella and Shigella types. J. Bacteriol. 52:461–466 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.