Abstract
Appropriate remediation targets or universal guidelines for polar regions do not currently exist, and a comprehensive understanding of the effects of diesel fuel on the natural microbial populations in polar and subpolar soils is lacking. Our aim was to investigate the response of the bacterial community to diesel fuel and to evaluate if these responses have the potential to be used as indicators of soil toxicity thresholds. We set up short- and long-exposure tests across a soil organic carbon gradient. Utilizing broad and targeted community indices, as well as functional genes involved in the nitrogen cycle, we investigated the bacterial community structure and its potential functioning in response to special Antarctic blend (SAB) diesel fuel. We found the primary effect of diesel fuel toxicity was a reduction in species richness, evenness, and phylogenetic diversity, with the resulting community heavily dominated by a few species, principally Pseudomonas. The decline in richness and phylogenetic diversity was linked to disruption of the nitrogen cycle, with species and functional genes involved in nitrification significantly reduced. Of the 11 targets we evaluated, we found the bacterial amoA gene indicative of potential ammonium oxidation, the most suitable indicator of toxicity. Dose-response modeling for this target generated an average effective concentration responsible for 20% change (EC20) of 155 mg kg−1, which is consistent with previous Macquarie Island ecotoxicology assays. Unlike traditional single-species tolerance testing, bacterial targets allowed us to simultaneously evaluate more than 1,700 species from 39 phyla, inclusive of rare, sensitive, and functionally relevant portions of the community.
INTRODUCTION
Petroleum hydrocarbon contamination in polar regions occurs commonly as a result of resource extraction, human habitation, and subsequent activities (1). Toxicity information relating to the effects of petroleum hydrocarbon contamination on terrestrial ecosystems in polar and subpolar regions is limited (2), yet available evidence suggests that the effects of oil spills are more damaging than in temperate regions due to low temperatures and slower ecosystem recovery (1). Currently, available remediation trigger concentrations in polar and subpolar soils are largely based upon countries' domestic guidelines for diesel fuel. The subsequent values are highly variable, ranging between 100 mg kg−1 and 2,000 mg kg−1, with no evidence-based guidance for site-specific modifications (3). It has been recommended that a weight-of-evidence approach integrating site-specific ecotoxicological, chemical, and ecological assessments is needed to develop universally accepted guidelines for petroleum hydrocarbon contamination in cold regions (4).
Single-species tolerance testing, used commonly for ecotoxicological assessments, is resource- and time-intensive. The confidence surrounding five to 10 model organisms accurately representing the sensitivity of an ecosystem is also questionable (5, 6). Further, the selection criterion of model organisms has resulted in a bias toward temperate and Northern Hemisphere species, to the exclusion of rare and often sensitive species (6). In the Antarctic and subantarctic regions, traditional toxicology indicator species, such as earthworms and large invertebrates, are sparse or nonexistent, further compounding the uncertainty surrounding the application of traditional tolerance testing “model” organisms. In 2008, Hickey et al. suggested that new, rapid tolerance testing approaches that target a suite of organisms across many taxa were required to increase the confidence in the resulting toxicity estimates. Rapid tolerance testing has been broadly defined as toxicity testing that aims to reduce the time and resources per species, by reducing replication, the number of treatments, and testing species concurrently. Rather than small numbers of precise estimates, rapid tests are expected to promote greater numbers of species assessed (more approximately), with increased relevance to the ecosystem, thereby delivering more accurate estimates of community sensitivity overall.
Microorganisms are important to soil health due to their integral role in biogeochemical cycles and ecosystem sustainability (7–9). With developments in sequencing technologies, the phylogenetic and functional understanding of environmental microbial communities is rapidly expanding and in turn promoting the development of molecular microbial indicators as integrated measures of ecosystem health (10, 11). To date, the majority of studies assessing the impact of petroleum hydrocarbons on polar soil microbial populations have focused on the stimulated portion of the bacterial community, with the aim of establishing the natural attenuation capacity of a soil or the effects of active bioremediation. These studies have identified a loss of diversity, combined with a selection toward heterotrophic species with known petroleum hydrocarbon-degrading potential (12–15). By comparison, little information exists on the species sensitive to petroleum hydrocarbon toxicity. Of those studies available, assessments are confined to monitoring predetermined microbial functional groups, noninclusive of species with unknown sensitivities or potential ecosystem functional services (16, 17).
A key challenge for developing microbial indicators is the high variability of microbial communities observed naturally across temporal and spatial scales. Many studies have linked this microbial variation to changes in the physical and chemical properties of soil, with factors such as carbon content and pH particularly important (18, 19). For the subantarctic region, bacterial and alkane-degrading diversity has already been correlated with organic carbon and total nitrogen availability (20). However, it is unclear how the variability in soil types and resulting microbial community composition will affect the resilience of soil bacterial communities when exposed to petroleum hydrocarbons. Another key challenge for implementing microbial monitoring specifically for petroleum hydrocarbons is the concurrent stimulation and inhibition effect of petroleum hydrocarbons on bacteria, which complicates toxicity assessments. To counter this, key nutrient-cycling parameters can be targeted that are sensitive to petroleum hydrocarbon toxicity but not stimulated by increases in available carbon (9). For example, ammonia oxidation as a chemolithic process is not affected by increases in organic carbon, yet it is inhibited by hydrocarbons through the general mechanism of biochemical toxicity (nonpolar narcosis), as well as specifically inhibiting ammonium monooxygenase (21, 22). As bacteria and archaea are primarily responsible for the conversion of ammonia to nitrite, toxicity or inhibition suggests overall ecosystem impairment. Similarly, functional genes, including nifH and nosZ, that are responsible for key enzymatic steps within the nitrogen cycle have been targeted to assess functional aspects of soil microbial communities (23, 24).
Macquarie Island, a world heritage-listed Australian subantarctic territory, is located approximately 1,500 km southeast of Tasmania (54°37′53′′S, 158°52′15′′E). The island has been the location of a permanently occupied research station since 1949. Historical and recent activity at the research station has led to three contaminated sites exhibiting medium to high levels of petroleum hydrocarbons (25, 26). All three sites are contaminated with hydrocarbons in the diesel fuel range (C9 to C28), with heavier hydrocarbons also present, originating from relic oil dumping and burn pits in the C29 to C36 range. However, the majority of the contamination originates from the widespread use of special Antarctic blend (SAB) diesel fuel (C9 to C18), a lighter aromatic diesel fuel suitable for use in cold regions. Following a site-specific bioassessment of the site, bioremediation combining nutrient addition and aeration in situ began in 2009 (25–27). As yet, no remediation trigger values or site-specific remediation target concentrations exist for this site.
Our aim was to identify the microbial indicators of fresh diesel fuel toxicity in Macquarie Island soils. The effects of SAB on the bacterial community were evaluated with broad and targeted community indices, as well as the abundances of functional genes encoding key enzymes within the nitrogen cycle. We utilized the results in dose-response curves to further test the utility of microbial targets as toxicity indicators. We hypothesized that the bacterial community would change with increasing fuel contamination, with a loss of diversity and a selection toward species capable of hydrocarbon degradation. We further hypothesized that the targeted portions of the bacterial community would provide more sensitive indicators than communitywide diversity estimates.
MATERIALS AND METHODS
Sampling sites and diesel fuel spiking.
Macquarie Island's climate is heavily influenced by its oceanic position. Average air temperatures range from 3.3°C in the winter to 7.0°C in the summer. Precipitation events occur approximately 312 days per year, and strong northwest to westerly winds, often gale force (>55.6 km h−1), predominate (Australian Bureau of Meteorology). For this experiment, four uncontaminated bulk soil plots consisting of sandy to peaty soil were selected from the isthmus, approximately 100 to 150 m from the major contaminated site (54°37′53′′S, 158°52′15′′E). Approximately 500 g of soil was collected from each plot (a total of 2 kg) to a depth of 30 cm, to target the most active microbial zone and to correspond with related invertebrate studies (28). Each of the four bulk soils were homogenized by mixing, separated into 10 individual subsamples of 50 g, and placed in 100-ml amber jars. One control sample for each of the four bulk soils was left unamended. The remaining soils were spiked with SAB diesel fuel to three target concentration ranges: low (0 to 400 mg kg−1; n = 3), medium (401 to 5,000 mg kg−1; n = 3), and high (5,000 to 20,000 mg kg−1; n = 3) (Table 1). Bulk soils were chosen to reduce the variability of soil properties within plots and to have control over the range of concentrations. All soils were incubated aerobically, in the dark. Three of the bulk soils (P1 to P3) were incubated for 21 days (short exposure), while the remaining bulk soil was incubated for 18 months (long exposure; P4). While no comment can be made on the short-term response of P4, the extended exposure was set up to determine if microbial community responses to the diesel fuel were observed between both short- and long-term samples (29). For the extended exposure, the lids of the amber jars were opened every 4 weeks to maintain aerobic conditions. Although not highly aerobic, these conditions will provide aerobic and anaerobic pockets within the jars, consistent with the soil matrix in situ.
TABLE 1.
Summary of measured physicochemical parameters
| Soil property | Concn (mg kg−1)a |
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P1 (low C) |
P2 (medium C) |
P3 (high C) |
P4 (long exposure) |
|||||||||||||
| Control | Low (n = 3)d | Medium (n = 3) | High (n = 3) | Control | Low (n = 3) | Medium (n = 3) | High (n = 3) | Control | Low (n = 3) | Medium (n = 3)d | High (n = 3) | Control | Low (n = 3) | Medium (n = 3) | High (n = 3)d | |
| TPH (avg) | <DL | 157 | 1,146 | 10,329 | <DL | 41 | 1,185 | 10,894 | <DL | 206 | 1,442 | 17,845 | <DL | 47 | 279 | 11,447 |
| SD | 84 | 815 | 6,583 | 54 | 1,055 | 6,582 | 173 | 1,117 | 12,407 | 18 | 39 | 6,449 | ||||
| Carbonb | 5 | 5 | 5 | 5 | 8 | 8 | 8 | 8 | 36 | 36 | 36 | 36 | 6.7 | 6.7 | 6.7 | 6.7 |
| Dry matter fraction | 0.8 | 0.83 | 0.85 | 0.83 | 0.75 | 0.77 | 0.70 | 0.70 | 0.33 | 0.30 | 0.30 | 0.30 | 0.42 | 0.43 | 0.50 | 0.30 |
| SD | 0.03 | 0.05 | 0.03 | 0.03 | 0.06 | 0.00 | 0.00 | 0.00 | 0.00 | 0.03 | 0.00 | 0.00 | ||||
| Conductivityc | 102 | 103 | 95 | 101 | 271 | 290 | 308 | 275 | 670 | 633 | 631 | 633 | 776 | 1,333 | 915 | 980 |
| SD | 9 | 3 | 2 | 4 | 6 | 27 | 8 | 1 | 3 | 286 | 404 | 343 | ||||
| pH | 5.7 | 5.7 | 5.7 | 5.6 | 6.6 | 6.6 | 6.5 | 6.7 | 5.3 | 5.3 | 5.4 | 5.2 | 4.8 | 4.7 | 5.2 | 5.1 |
| SD | 0.0 | 0.1 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.1 | 0.0 | 0.0 | 0.2 | 0.2 | ||||
| Nitrite | <0.15 | 0.7 | 0.6 | 1.1 | 5.3 | 4.3 | 4.4 | 2.2 | 96.9 | 104.7 | 63.4 | 43.3 | <0.15 | <0.15 | <0.15 | <0.15 |
| SD | 0.6 | 0.4 | 0.5 | 0.2 | 0.7 | 1.1 | 8.1 | 4.1 | 1.7 | 0 | 0 | 0 | ||||
| Nitrate | 65 | 66 | 51 | 75 | 302 | 353 | 380 | 254 | 1,415 | 1,457 | 1,329 | 1,403 | 20 | 21 | 20 | 8 |
| SD | 5 | 7 | 3 | 8 | 27 | 50 | 34 | 16 | 22 | 11 | 4 | 6 | ||||
| Ammonium | 3.1 | 1.1 | 0.8 | 1.4 | 2.9 | 3.9 | 5.3 | 4.9 | 52.8 | 56.1 | 51.8 | 32.9 | 7.8 | 8.1 | 4.8 | 5.6 |
| SD | 0.3 | 0.2 | 0.1 | 0.3 | 0.4 | 0.8 | 1.7 | 3.5 | 2.8 | 0.7 | 1.0 | 1.8 | ||||
| Phosphate | 12.5 | 10.5 | 9.8 | 8.3 | 32.7 | 31.6 | 34.9 | 34.8 | 379.8 | 416.9 | 442.6 | 427.0 | 9.0 | 9.2 | 10.2 | 7.7 |
| SD | 0.7 | 0.8 | 0.4 | 0.6 | 1.3 | 0.5 | 18.9 | 2.5 | 8.5 | 2.7 | 1.4 | 0.7 | ||||
| Sulfate | 30.8 | 34.0 | 31.4 | 34.2 | 65.1 | 71.3 | 77.1 | 69.8 | 84.4 | 83.6 | 80.7 | 77.6 | 70 | 89 | 63 | 80 |
| SD | 0.7 | 1.8 | 0.5 | 0.8 | 1.7 | 6.4 | 1.2 | 1.3 | 0.8 | 44 | 31 | 19 | ||||
Controls are unamended. Low, low TPH concentration (range of detection limit [DL] to 400 mg kg−1); medium, medium TPH concentration (range of 401 to 5,000 mg kg−1); high, high TPH concentration (>5,000 mg kg−1). All values are in mg kg−1 unless otherwise stated.
Values are % LOI (loss on ignition).
Values are in μS/cm.
One sample in this group had substantially less sequencing reads than the other samples; as such, they were excluded from further phylogenetic analysis.
Nutrients and soil parameters were analyzed after the incubation period according to the Australasian Standard Soil protocols (30) (Table 1). Briefly, total carbon was determined by the loss-on-ignition (LOI) method and expressed as a percentage. The concentrations of anions and cations in the soil were measured on a water extract (1 g/5 ml water) and expressed as mg kg−1. Conductivity and pH were also measured on the same water extract. A 10-g subsample of soil from each sample was extracted with hexane and assessed by gas chromatography to determine the total petroleum hydrocarbon (TPH) concentration (9). The detection limit (DL) for TPH concentrations was 20 mg kg−1. TPH concentrations below the detection limit were estimated with a substitution method based on half the detection limit, i.e., estimated TPH concentrations of 10 mg kg−1 (Table 1). Average measured TPH concentrations were calculated for each of the fuel categories within the four plots (Fig. 1A, Table 1). Dry matter fraction was determined gravimetrically using the same samples analyzed for TPH. We have reported all nutrient and TPH concentrations on a dry matter basis unless stated otherwise.
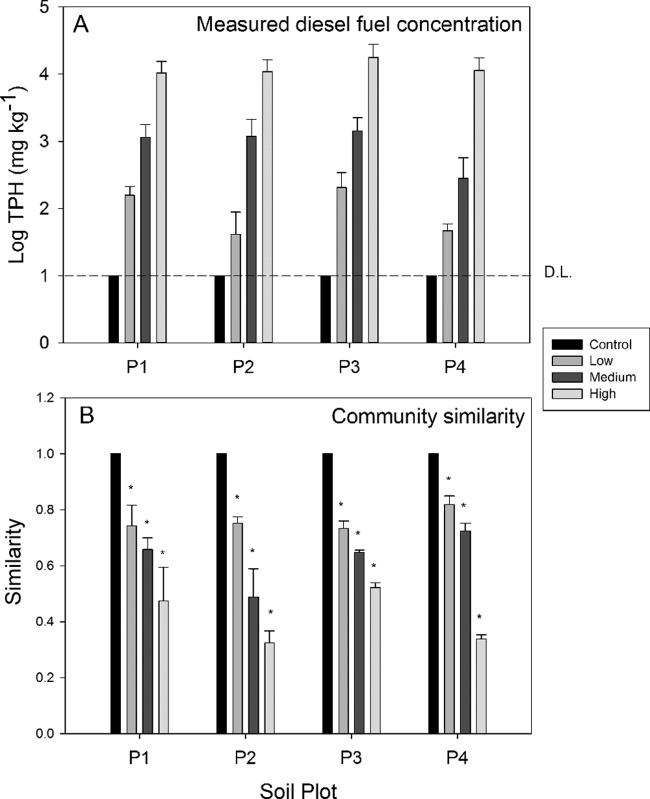
FIG 1.
The average measured total petroleum hydrocarbon (TPH) concentration and bacterial community similarity of samples across SAB spiking fuel concentration ranges and soil plots. (A) The average measured TPH log concentration for the spiking fuel ranges within each soil plot. The dashed line indicates the TPH detection limit (DL). (B) The average community similarity within each soil plot based on the weighted UniFrac distance. Error bars account for the standard deviation between biological replicates. *, significant decline in community similarity was found (P < 0.05).
Barcoded amplicon pyrosequencing targeting the 16S rRNA gene.
After the short (21 days) and long (18 months) incubation periods, total soil genomic DNA was extracted from 50-mg subsamples of each soil sample in triplicate with the FastDNA SPIN kit for soil (MP Biomedicals, NSW, Australia). In total, 120 DNA extracts (40 samples in triplicate) were titrated to a standard working concentration range of 5 to 10 ng μl−1. The technical replicates were analyzed with automated ribosomal intergenic spacer analysis (ARISA) to evaluate inter/intrasample similarity according to van Dorst et al. (31). Intersample replication was evaluated with analysis of similarity (ANOSIM) in PRIMER v6. After intersample similarity was confirmed (see Fig. S1 and Table S1 in the supplemental material), the extracted genomic DNA from a randomly selected replicate was used as a template for barcoded tag pyrosequencing. The V1, V2, and V3 hypervariable regions of the small-subunit (SSU) ribosomal gene were targeted using the 27F and 519R universal primers (32). The PCR amplification and barcoded sequencing were performed at the Research and Testing Laboratory (Lubbock, TX, USA). Although systematic errors remain with barcoding sequencing technologies, stochastic replication (3×) was performed to limit some of the PCR bias. Samples were run on the Roche 454 FLX Titanium platform. Data were provided from the sequencing facility in the form of standard flowgram format (sff) files (33).
Processing amplicon pyrosequencing data.
The sequence data were processed using the MOTHUR software package (34). This involved extraction of the sequence information and flowgrams from the sff files, demultiplexing and removal of short reads (<200 bp), truncation of long reads (>500 bp), and sequences with homopolymers (>8-bp repeats) and ambiguous bases. Remaining reads were screened for quality by taking an average length phred read score of ≥25. Reads were aligned against the SILVA 16S rRNA database file provided with the MOTHUR package (35). Chimera checking with the UCHIME algorithm (36) allowed removal of any chimeric artifacts, and sequences were preclustered at 1% to negate instrument error rate. The reads were then clustered into operational taxonomic units (OTUs) based on 96% sequence similarity for species-level OTUs (37). Taxonomic assignment was determined against the Greengenes database provided within the MOTHUR package (38). An OTU abundance-by-sample matrix was generated from the resulting output, and various diversity indices were calculated. A neighbor-joining tree was created through MOTHUR (34) with the Clearcut program addition (39). The neighbor-joining tree was used to run the weighted UniFrac algorithm (40).
Data analysis.
Multivariate data analysis was conducted with the software packages PRIMER v6 and Permanova+ (41). To account for the log-normal distribution of the data, the abundance-by-sample matrix was square root transformed. After transformation, the skewness and kurtosis was reduced closer to 0. For sample comparison, the abundance-by-sample matrix was then standardized to express the OTU abundances as relative abundances. Resemblance matrices were calculated with both the Bray-Curtis similarity coefficient and the weighted UniFrac measurements. To test the null hypothesis of no differences between plots, ANOSIM was performed on the resemblance matrices with 999 permutations. To test the null hypothesis of no differences between diesel fuel categories, a one-way analysis of variance (ANOVA) was used to test for significant differences in the microbial communities between fuel categories within each plot.
Results from the chemical and physical analysis of the soils were used to evaluate the influence of environmental variables. The results were normalized, and a similarity matrix was calculated with the Euclidean distance metric. The relative relationship of the environmental variables to the biological distribution was analyzed using a distance-based linear model (DistLM). The selection criterion for the model was adjusted R2. The selection procedure was stepwise, with 999 permutations. The DistLM was based on the null hypothesis of no relationship between the environmental variables and the biological resemblance matrix. All statistical tests were considered significant at a P value of <0.05.
Individual OTUs were evaluated across the SAB spiking range to determine if they were significantly inhibited or stimulated with increasing TPH concentration. After OTUs present in only one sample were removed, the log-transformed abundance of individual OTUs was plotted against the log-transformed TPH data. The resulting dose responses of each OTU were then fitted to a linear equation in R (http://www.R-project.org/). The number of OTUs significantly stimulated or inhibited with TPH was determined based on the quality of fit (P < 0.05) and slope of the line. The percentage abundance of each OTU was aggregated into genera to create a heatmap within R. Representative OTUs that were unable to be reliably classified to the genus level were listed with the closest classification level. The OTUs were then aggregated into phylum-level phylogeny. All phyla were analyzed for positive or negative correlations to increasing fuel concentrations. The Acidobacteria/Proteobacteria ratio, considered representative of the oligotrophs/copitrophs ratio in the environment, was calculated and used as an additional community index.
Targeting the nitrogen cycle with quantitative PCR.
Quantitative PCR (qPCR) was used to measure the abundance of the nifH, amoA, and nosZ genes present. The same DNA extracts used in the barcoded pyrosequencing analysis were utilized for the qPCR analysis, and each of the selected genomic DNA extracts was analyzed by qPCR in triplicate. Samples were analyzed using the QuantiTect Fast SYBR green PCR master mix real-time PCR kit (Qiagen, Doncaster, VIC, Australia) and run on the ABI 7500 real-time PCR machine (Applied Biosystems). Each 20-μl reaction mixture contained 12.5 μl of master mix, 1.25 μl of template DNA, and 1 μM the forward and reverse primers. The thermal cycling program consisted of 94°C for 5 min, 45 cycles of 94°C for 20 s, 54°C for 50 s, followed by a melt-curve step from 50°C to 95°C. The quantitative fluorescence data were collected during the 54°C step. To test for the potential presence of PCR inhibitors, a four- to five-point curve (in duplicate) of different DNA concentrations for each soil sample was analyzed according to Ma et al. (42). At the DNA concentrations used, no inhibition was detected. Quantification data were collected only when there was no detected PCR inhibition, no amplification in the “no-template control,” a single peak in the melt curve consistent with specific amplification, and a reaction efficiency of 100% ± 10%.
Subunits of the nitrogenase enzyme are encoded by the genes nifH, nifD, and nifK. The nifH gene is the most widely sequenced and utilized marker for nitrogen fixation. Here, we used the primers IGK3 and DVV (43) to target the nifH gene as a proxy for potential nitrogen fixation activity (Table 2). A standard curve was generated with the IGK3/DVV-amplified PCR product from a control subantarctic soil. The number of copies was determined spectrophotometrically. The standard curves were linear over six orders of magnitude, with amplification efficiencies of 93.8 to 95.1% and an R2 value of 0.997.
TABLE 2.
Details of primers used in qPCR to target functional genes within the nitrogen cycle
| Process | Target gene | Primers | Reference | Primer sequence | Positive control |
|---|---|---|---|---|---|
| Nitrogen fixation | nifH | IGK3 | 43 | GCIWTHTAYGGIAARGGIATHGGIA | gDNA from soil |
| DVV | ATIGCRAAICCICCRCAIACIACRTC | ||||
| Ammonium oxidation | B.amoAa | amoA1F | 44 | GGGGTTTCTACTGGTGGT | gDNA from soil |
| amoA2R | CCCCTCKGSAAAGCCTTCTTC | ||||
| A.amoAb | A.amoAF | 45 | STAATGGTCTGGCTTAGACG | gDNA from soil | |
| A.amoAR | GCGGCCATCCATCTGTATGT | ||||
| Denitrification | nosZ | nosZ2F | 46 | CGCRACGGCAASAAGGTSMSSGT | Pseudomonas stutzeri |
| nosZ2R | CAKRTGCAKSGCRTGGCAGAA |
Bacterial amoA.
Archaeal amoA.
Nitrification refers to the oxidation of ammonia into nitrite and the subsequent oxidation of nitrite into nitrate. As PCR primers targeting the entire nitrite oxidation functional group are not currently available, the ammonium oxidizing step was used as a proxy for potential nitrifying activity. The ammonium oxidizing step can be performed by ammonium oxidizing bacteria (AOB) and ammonium oxidizing archaea (AOA). We evaluated both groups with primer sets targeting the AOB (amoA1F/amoA2R) (44) and AOA (Arch.amoAF/Arch.amoAR) (45) amoA genes (Table 2). Standard curves were generated with the amplified PCR products from a control subantarctic soil, with the number of copies determined spectrophotometrically. For bacterial amoA, the standard curve was linear over seven orders of magnitude with amplification efficiencies of 91.6 to 92.0%, R2 = 0.998. The archaeal amoA standard curve was linear over six orders of magnitude, with amplification efficiencies of 90.6 to 94.8% (R2 = 0.974).
Classified genes known to be present in denitrifying microorganisms include nir (nitrate reductase) and nos (nitrous oxide reductase). We chose to target the nosZ gene as a proxy for potential denitrification with the primer set nosZ2-F/nosZ2-R (46). These primers target primarily Proteobacteria, excluding denitrifiers within Firmicutes. Although not present in all denitrifying species, the nosZ gene has been widely evaluated and applied in polar soils due to its relevance in nitrous oxide production (a potent greenhouse gas). Further, in Siciliano et al., the applicability of quantifying nosZ abundance from mixed templates was specifically tested with positive results (47). In another study, nosZ was more sensitive to environmental changes than nirS and nirK (48). To determine the copy numbers of the nosZ gene, a standard curve was generated using the nosZ2-F/nosZ2-R (46)-amplified PCR product from Pseudomonas stutzeri, a well-known denitrifier species. The number of copies of the nosZ gene was determined spectrophotometrically (Table 2). The standard curve was linear over five orders of magnitude; the amplification efficiency determined from the slope of the standard curve was 99.8% (R2 = 0.96).
Dose-response modeling.
For dose-response modeling, a large number of data points are recommended over high replication to best capture possible shifts in the curve (49, 50). Hence, we used individual samples and their TPH concentrations instead of the concentration ranges to maximize the confidence surrounding the dose-response curves. The dose-response measurements were calculated as a percentage of the control to enable comparison between soil types (P1 to P4). The dose response curves were generated within the drc R package (51). This included plotting the data and selecting the most suitable model as evidenced by Akaike's information criterion (AIC) and ANOVA comparisons of models. Effective concentration values (ECx) (including standard error and confidence intervals) were then calculated from the curve generated by the most suitable model for each data set. The resulting effective concentration responsible for 20% change (EC20) values calculated from the dose-response curves were used to compare the relative sensitivities of soil types and community measures.
RESULTS
Bacterial community composition.
A total of 127,053 quality-checked sequence reads were obtained, averaging 3,304 (±1,626) sequence reads per sample. For further comparative analysis, the sequences were subsampled at 1,450 sequences, resulting in approximately 14,000 per plot and 4,350 sequences per SAB treatment. Three samples (within P1-low, P3-med, and P4-high) had exceptionally low numbers of sequence reads at <800 and were excluded from further analysis. The distribution of the bacterial communities was used to create two similarity matrices, based on the Bray-Curtis correlation coefficient and UniFrac distance. Utilizing the weighted UniFrac similarity matrix, a one-way ANOVA confirmed significant differences between bacterial communities according to their TPH concentration ranges: control, low (10 to 400 mg kg−1), medium (401 to 5,000 mg kg−1), and high (>5,000 mg kg−1). Significant dissimilarity between the control and the treatments was observed in all four soil plots (Fig. 1B, Table 3). The soil plots, P1 to P4, were also found to be significantly different in a one-way ANOSIM, global R value = 0.73 (P < 0.001) (Table 3).
TABLE 3.
ANOVA and ANOSIM results testing the amplicon pyrosequencing community UniFrac and Bray-Curtis dissimilarities between fuel categories and soil plots
| Differences and testb | Pairwise test result | t/R statistic | P valuec |
|---|---|---|---|
| P1 differences between fuel categories (1-way ANOVA) | Global | 17.4 | 0.006 |
| Control, high | 8.0 | 0.009 | |
| Control, med | 5.5 | 0.026 | |
| Control, low | 4.2 | 0.032 | |
| Low, high | 5.5 | 0.027 | |
| Low, med | 1.9 | 0.241 | |
| Med, high | 3.8 | 0.044 | |
| P2 differences between fuel categories (1-way ANOVA) | Global | 57.9 | 0.001 |
| Control, high | 12.8 | <0.001 | |
| Control, med | 9.7 | 0.001 | |
| Control, low | 4.7 | 0.016 | |
| Low, high | 11.4 | <0.001 | |
| Low, med | 7.1 | 0.003 | |
| Med, high | 4.4 | 0.021 | |
| P3 differences between fuel categories (1-way ANOVA) | Global | 213.6 | <0.001 |
| Control, high | 28 | <0.001 | |
| Control, med | 19.4 | <0.001 | |
| Control, low | 15.6 | <0.001 | |
| Low, high | 17.4 | <0.001 | |
| Low, med | 6.2 | 0.007 | |
| Med, high | 9.4 | 0.001 | |
| P4 differences between fuel categories (1-way ANOVA) | Global | 278.5 | <0.001 |
| Control, high | 28.6 | <0.001 | |
| Control, med | 13.1 | <0.001 | |
| Control, low | 8.3 | <0.001 | |
| Low, high | 28 | <0.001 | |
| Low, med | 6.6 | 0.005 | |
| Med, high | 23.6 | <0.001 | |
| Differences between plotsa (1-way ANOSIM) | Global | 0.729 | <0.001 |
| P1, P2 | 0.722 | <0.001 | |
| P1, P3 | 0.807 | <0.001 | |
| P1, P4 | 0.765 | <0.001 | |
| P2, P3 | 0.781 | <0.001 | |
| P2, P4 | 0.781 | <0.001 | |
| P3, P4 | 0.767 | 0.002 |
Plots include P1 (low-carbon soil), P2 (medium-carbon soil), P3 (high-carbon soil), and P4 (aged soil).
Fuel categories refer to TPH concentrations. Unamended control, low TPH concentration (range of DL to 400 mg kg−1), medium TPH concentration (range of 401 to 5,000 mg kg−1), and high TPH concentration (>5,000 mg kg−1).
Significant differences are in bold. P values were considered significant at <0.05.
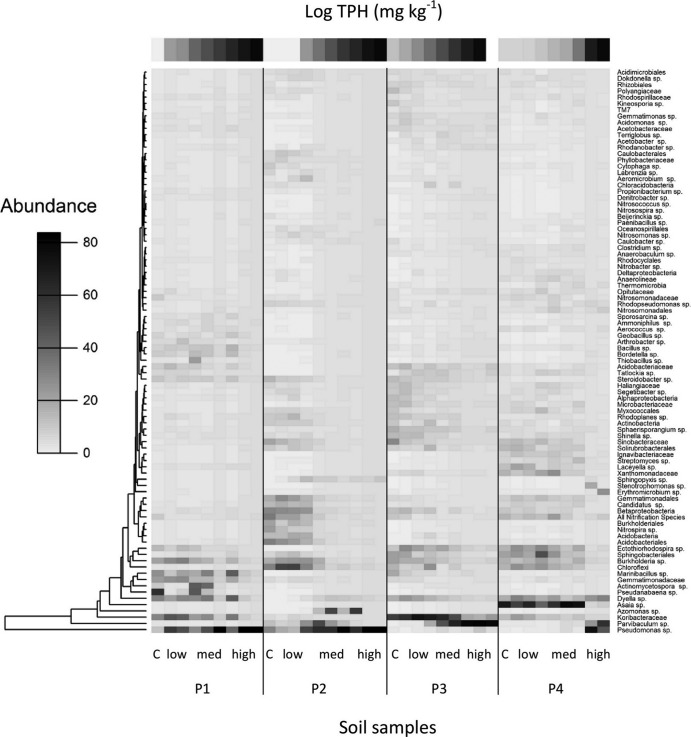
Diesel fuel substantially reduced the similarity of communities from the controls largely by stimulating or reducing the relative abundances of key lineages rather than removing entire lineages of bacteria (Fig. 2). The communities shifted from large numbers of species in relatively low abundance to communities heavily dominated by only a few species. We found that the OTUs consisting of 26% to 79% of the relative abundance were significantly inhibited across the SAB spiking range, while only a small proportion of OTUs, 0.4% to 7.8%, were stimulated (Table 4). The genus most stimulated was Pseudomonas, contributing a maximum of 5.5% relative abundance in the control soils compared to >60% relative abundance in the highest-concentration samples for the low (P1)- and medium (P2)-carbon soils and >20% in the long-exposure spiked soils (P4). By comparison, the high-organic-carbon soil (P3) exhibited a lower relative abundance of Pseudomonas (<1%) and maintained a relative abundance between 0.06% and 4% across the entire spiking range. Instead, the genus Parvibaculum, which contributed 0.2% of the total abundance in the control soils, increased substantially, contributing to 43% of the final relative abundance (Fig. 2). Bacteria involved in the nitrification process, specifically ammonium oxidation or the oxidation of nitrite to nitrate, were significantly inhibited following the addition of diesel fuel. A functional group for the nitrification species, “All nitrification species,” was consolidated, consisting of Nitrosospira species, Nitrosococcus species, Nitrobacter species, Nitrosomonas species, Nitrospira species, and unclassified species from the family Nitrosomonadaceae and the order Nitrosomonadales. The group was significantly inhibited across the diesel fuel concentration range for each soil plot (P < 0.05) (Fig. 2).
FIG 2.
Relative abundances of genera present in soil samples from low to high special Antarctic blend (SAB) diesel fuel concentration. Samples are sorted according to SAB fuel concentration within each soil plot and are labeled according to fuel category (control, low, medium, and high). The log SAB fuel concentration is on a scale of 0 to 4.5, with 4.5 ∼ 31,000 mg kg−1. Representative operational taxonomic units (OTUs) that were unable to be reliably classified to the genus level were listed with the closest classification level.
TABLE 4.
OTUs significantlya stimulated and inhibited across the SAB diesel fuel spiking range
| Soil plot | Total no. of unique OTUs | Inhibited OTUs |
Stimulated OTUs |
||||
|---|---|---|---|---|---|---|---|
| No. | Relative abundance in control soils | Relative abundance in high soils (SE) | No. | Relative abundance in control soils | Relative abundance in high soils (SE) | ||
| P1 | 386 | 28 | 39 | 6.5 (1.95) | 7 | 0.4 | 68.0 (10.4) |
| P2 | 723 | 178 | 56 | 5.3 (0.8) | 8 | 7.8 | 73.8 (7.7) |
| P3 | 712 | 77 | 26 | 2.5 (3.1) | 11 | 0.6 | 51.5 (1.3) |
| P4 | 715 | 119 | 79 | 15.9 (0.3) | 14 | 0.8 | 52.5 (1.4) |
P < 0.05.
Environmental predictors.
Carbon was a significant predictor variable (F = 5.13, P = 0.001) when analyzed with a marginal DistLM model (Table 5). However, in sequential DistLM tests, organic carbon contributed only 4.1% to the total biological variation observed between soil samples. The environmental variables of pH, nitrate, phosphate, and TPH all had a greater influence on the biological distribution than organic carbon. The soil pH was the greatest predictor value, accounting for 18% of the total variation between all samples, as calculated with Pearson's correlation (P < 0.001) (Table 5).
TABLE 5.
DistLM results indicating strength of environmental variable as a predictor of the biological distribution and patterns of bacterial communities across all samples
| Variable | Value (mg kg−1)a |
||||||
|---|---|---|---|---|---|---|---|
| Marginal tests |
Sequential tests |
||||||
| Pseudo-F | P | Prop | Pseudo-F | P | Prop | Cumul | |
| pH | 7.28 | <0.001 | 0.18 | 7.28 | <0.001 | 0.18 | 0.18 |
| Nitrate | 5.09 | <0.001 | 0.13 | 5.73 | <0.001 | 0.12 | 0.30 |
| Phosphate | 4.30 | <0.001 | 0.12 | 6.45 | <0.001 | 0.12 | 0.42 |
| TPH | 2.28 | 0.019 | 0.06 | 3.99 | <0.001 | 0.07 | 0.49 |
| Carbonb | 5.13 | <0.001 | 0.13 | 2.58 | 0.006 | 0.04 | 0.53 |
| Nitrite | 4.44 | <0.001 | 0.12 | 2.22 | 0.013 | 0.03 | 0.57 |
| Bromine | 2.43 | 0.008 | 0.06 | 1.52 | 0.116 | 0.02 | 0.59 |
| Sulfate | 2.77 | 0.005 | 0.08 | 1.69 | 0.079 | 0.02 | 0.62 |
| Conductivityc | 4.06 | 0.002 | 0.11 | 1.58 | 0.110 | 0.02 | 0.64 |
| Chlorine | 5.33 | <0.001 | 0.14 | 1.46 | 0.140 | 0.02 | 0.66 |
| Ammonium | 1.30 | 0.137 | 0.04 | 1.18 | 0.289 | 0.02 | 0.68 |
| Dry matter fraction | 5.40 | <0.001 | 0.14 | 1.04 | 0.377 | 0.02 | 0.69 |
Marginal test results are based on individual variables. Sequential test results are based on the relative proportion of influence when all variables are considered. Significant differences are in bold; P values were considered significant at <0.05. Pseudo-F, a multivariate version of Fisher's F statistic utilized in PRIMER E; Prop, the proportion of variance predicted with environmental variable; Cumul, the cumulative proportion of variance explained. Values are in mg kg−1 unless otherwise specified.
Values are % LOI.
Values are in μS/cm.
Community indices.
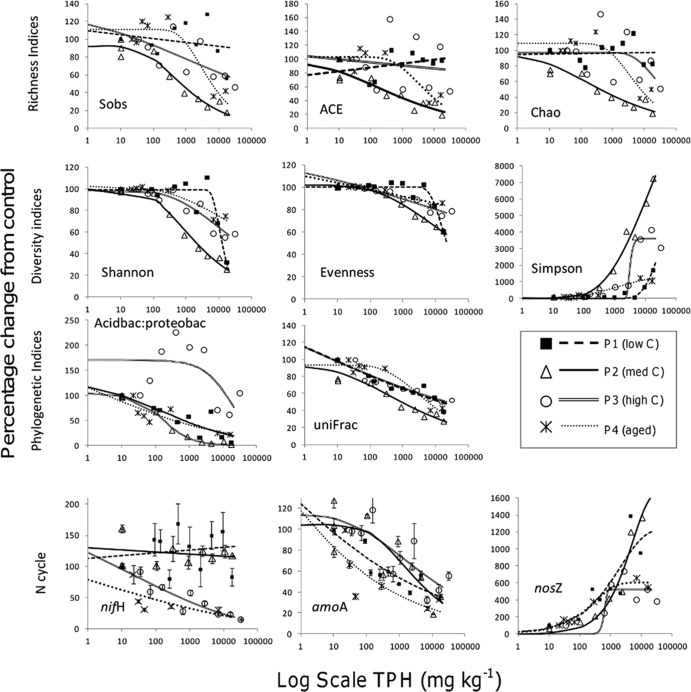
With increasing diesel fuel concentration, the total species richness, species diversity, species evenness, similarity indices, and UniFrac measurements across most soils declined from the controls (Fig. 3). The control communities within the low-carbon soils (P1) consisted of substantially lower species numbers, diversity, and evenness than the higher-carbon soils, and as a result the effect of the diesel fuel on the communities was less severe. The species richness within the medium-carbon soils (P2) was sensitive to increasing diesel fuel, with abundance-based coverage estimation (ACE), Chao1, and species observed (Sobs) values decreasing by up to 20%. The Sobs value in the higher-carbon soils (P3) declined almost 50% from the control, while ACE and Chao1 values were variable across all fuel concentrations. In the long-exposure soils (P4), the loss of species richness from the control occurred at higher concentrations than in the acute soils for Sobs, ACE, and Chao1, with a total decrease of 60% observed.
FIG 3.
The effect of increasing SAB fuel concentrations on richness, diversity, and phylogenetic indices and key functional genes within the nitrogen cycle (nifH, nitrogen fixation; amoA, ammonium oxidation; nosZ, denitrification). Values are expressed as percentage change from the control. The best-fitting dose response curve as determined by AIC was fitted to each data set in the drc R package. The selected dose-response regression model was used to calculate EC20 values.
The Shannon diversity index (H′), Simpson diversity index, and Pielou's evenness index (J′) in the low-carbon soils (P1) were not significantly impacted by SAB concentrations below 5,000 mg kg−1. In the medium carbon (P2) soils, a gradual decline in the Shannon diversity index was observed from 100 mg kg−1, resulting in a 70% decrease from the control. This decrease was less severe in the higher-carbon and aged soils (P3 and P4, respectively), with a 40% decrease from the control observed. Pielou's evenness index in high-carbon soils (P3) decreased by approximately 20% from the control, with the decline detectable at concentrations below 100 mg kg−1. The P4 soil exposed to the contaminant for 18 months exhibited different sensitivity compared with that of the short-exposure soils, with the evenness index particularly inhibited.
The UniFrac dissimilarity measurement provided a sensitive target for all soils (Fig. 3). The phylogenetic distance measurement changed between 40% and 60% from the control soils for all four samples. The long-exposure soils (P4) exhibited a higher level of similarity to the control soil and to soils spiked with <1,000 mg kg−1 fuel, suggesting a higher tolerance or recovery in the genetic potential of the community than short-exposure soils. The ratio of Acidobacteria to Proteobacteria, which is indicative of the oligotrophic/copiotrophic species ratio, was reduced, suggesting a shift toward faster opportunistic species within gamma- and betaproteobacteria. The low- and medium-carbon soils and long-exposure soils were sensitive to this ratio, with 75% decreases observed compared to the control soils. The high-carbon soils were not sensitive to this measurement, with very high EC20 values generated (Table 6).
TABLE 6.
Generated EC20s across a range of community indices
| Community indexa | Modelb | Plot | Treatment |
EC20 |
Model parameterse |
||||
|---|---|---|---|---|---|---|---|---|---|
| % C | Exposure | Concnc | SEd | b | d | e | |||
| Sobs | W2.3 | P1 | 5 | Short | NAf | NA | −0.03 | 171.0 | 1.9 |
| SE (25.32) | P2 | 8 | Short | 140 | 80 | −0.44 | 91.9 | 403.4 | |
| df = 23 | P3 | 36 | Short | 30 | 340 | −0.15 | 124.8 | 871.3 | |
| P4 | 6.7 | Long | 1,300 | 1,000 | −0.66 | 111.4 | 2,763.7 | ||
| ACE | W2.3 | P1 | 5 | Short | NA | NA | 0.04 | 154.0 | 4,867.1 |
| SE (26.17) | P2 | 8 | Short | 20 | 220 | −0.26 | 94.9 | 149.1 | |
| df = 23 | P3 | 36 | Short | NA | NA | −0.03 | 153.2 | 51.5 | |
| P4 | 6.7 | Long | 1,300 | 2,000 | −0.53 | 103.2 | 3,118.4 | ||
| Chao1 | W2.3 | P1 | 5 | Short | NA | NA | 0.00 | 157.3 | 0.00001 |
| SE (20.5) | P2 | 8 | Short | 30 | 16 | −0.28 | 93.9 | 146.6 | |
| df = 23 | P3 | 36 | Short | 11,000 | 6,400 | −0.61 | 97.9 | 23,398 | |
| P4 | 6.7 | Long | 1,500 | 620 | −0.59 | 109.1 | 3,334 | ||
| Shannon (H′) | W2.3 | P1 | 5 | Short | 8,100 | 850 | −1.91 | 98.8 | 10,384 |
| SE (7.14) | P2 | 8 | Short | 250 | 110 | −0.40 | 99.9 | 816.4 | |
| df = 23 | P3 | 36 | Short | 2,200 | 1,900 | −0.28 | 98.5 | 11,865 | |
| P4 | 6.7 | Long | 3,700 | 5,300 | −0.21 | 102.3 | 36,297 | ||
| Pielou's evenness (J′) | W2.3 | P1 | 5 | Short | 10,400 | 830 | −1.13 | 100.5 | 15,863 |
| SE (3.30) | P2 | 8 | Short | 140 | 470 | −0.24 | 102.0 | 9,992.5 | |
| df = 23 | P3 | 36 | Short | 50 | 390 | −0.09 | 125.4 | 11,809 | |
| P4 | 6.7 | Long | NA | NA | 50,273 | 145.6 | 882.4 | ||
| Weighted UniFrac | W2.3 | P1 | 5 | Short | 10 | 1.28 | −0.13 | 159.4 | 6.42 |
| SE (7.41) | P2 | 8 | Short | 60 | 110 | −0.26 | 91.9 | 348.4 | |
| df = 23 | P3 | 36 | Short | 10 | 0.16 | −0.12 | 172.9 | 1.98 | |
| P4 | 6.7 | Long | 940 | 750 | −0.41 | 93.4 | 3,027.9 | ||
| Acidobacteria/Proteobacteria ratio | LL3 | P1 | 5 | Short | 10 | 56 | 0.38 | 1.3 | 184.1 |
| SE (3.4) | P2 | 8 | Short | 40 | 110 | 1.03 | 1.04 | 159.5 | |
| P3 | 36 | Short | 4,500 | 2,700 | 1.05 | 1.7 | 17,195 | ||
| P4 | 6.7 | Long | 10 | 0.14 | 0.24 | 2.06 | 2.7 | ||
| nifH | W1.3 | P1 | 5 | Short | NA | NA | −0.02 | 282.5 | 15,863 |
| SE(25.39) | P2 | 8 | Short | NA | NA | 0.01 | 320.4 | 9,992.5 | |
| df = 90 | P3 | 36 | Short | 10 | 0.1 | 0.13 | 244.0 | 11,809 | |
| P4 | 6.7 | Long | 10 | 0.1 | 0.09 | 219 | 882.4 | ||
| amoA | W2.3 | P1 | 5 | Short | 10 | 0.06 | −0.16 | 214. | 0.4 |
| SE (17.0) | P2 | 8 | Short | 410 | 240 | −0.40 | 104.2 | 1,356.6 | |
| df = 90 | P3 | 36 | Short | 200 | 490 | −0.25 | 113.7 | 1,337.6 | |
| P4 | 6.7 | Long | 10 | 0.06 | −0.20 | 228.4 | 0.2 | ||
| nosZ | W2.3 | P1 | 5 | Short | 130 | 110 | 0.54 | 1,244.0 | 2,066.2 |
| SE (17.37) | P2 | 8 | Short | 740 | 540 | 0.78 | 1,702.2 | 5,119.1 | |
| df = 90 | P3 | 36 | Short | 520 | 830 | 5.84 | 526.2 | 671.5 | |
| P4 | 6.7 | Long | 30 | 41 | 0.66 | 605.6 | 306.8 | ||
Sobs, species observed; ACE, abundance-based coverage estimation; nifH, amoA, and nosZ, the abundance of functional genes within the nitrogen cycle as determined through qPCR.
The best-fitting dose response curve for the data as determined by AIC. W2.3, Weibull 2 curve/3 parameters; W1.3, Weibull 1 curve/3 parameters; LL3, log logistic curve/3 parameters.
The concentration that corresponds to a 20% effective change on the community.
The associated standard error of the EC20 estimate only.
Model parameters are defined as b = relative slope, d = upper limit, and e = point of inflection.
NA, no reliable dose response curve could be fitted.
Abundance of functional genes.
Of the functional genes evaluated, bacterial amoA (indicative of nitrification) was the most sensitive indicator of hydrocarbon toxicity in soil (Fig. 3). The low-carbon and aged soils (P1 and P4, respectively) were most sensitive to the bacterial amoA measurement, with a significant decline in copy numbers observed at very low concentrations. A decline of bacterial amoA copy numbers in the medium- and high-carbon soils (P2 and P3, respectively) was also observed for SAB concentrations >100 mg kg−1. The archaeal amoA gene was present in low to nondetectable levels in all but one of the soils (P2; see Fig. S2 in the supplemental material). Within P2, the abundance of archaeal amoA was an order of magnitude lower than the bacterial amoA gene, ranging from 5.02 × 102 to 4.07 × 101, with abundances declining with increasing SAB concentrations. As such, only the bacterial amoA was pursued as an indicator, and archaeal amoA was excluded from further analysis. The abundance of the nifH gene (representative of nitrogen fixation), was variable across all soils and diesel fuel concentrations analyzed; thus, no significant or measurable impact was observed. The nosZ gene abundance (indicative of denitrification) was stimulated in all soils spiked with diesel fuel. While the greatest overall increases were observed in the low- and medium-carbon soils, similar increases in the nosZ gene copy numbers occurred at low TPH concentrations for all soils, and the low-carbon and long-term exposure soils generated similar sensitive dose-response curves between 100 and 1,000 mg kg−1.
Dose-response modeling.
Within the low-carbon soil (P1), the EC20 values, based on percentage change from the control, generated high values outside the experimental spiking range for the Simpson index, and no significant response was measurable for ACE, Chao1, and Sobs (Table 6). Overall, the calculations of EC20s across the traditional community diversity measures, including Sobs, ACE, Chao1, Shannon, Simpson, and Pielou's evenness (J′), generated results with high associated errors and large variability between soil types (Fig. 4, Table 6). In contrast, the abundances of the bacterial amoA and nosZ genes along with the community UniFrac measurements generated sensitive EC20 estimates with less associated error and a response that was consistent across all soil types (Fig. 4, Table 6). For bacterial amoA, EC20 concentrations ranged from 10 mg kg−1 to 400 mg kg−1, with an average EC20 of 155 mg kg−1 (standard deviation [SD], 195 mg kg−1). For the nosZ gene and UniFrac measurements, EC20 concentrations ranged between 250 mg kg−1 and 700 mg kg−1, with an average of 355 mg kg−1 (SD, 330 mg kg−1) for nosZ and 250 mg kg−1 (SD, 460 mg kg−1) for UniFrac (Table 6). For bacterial amoA and nosZ and the Acidobacteria/Proteobacteria ratio, the soil plots with the lowest carbon content (P1 and P4) generated the most sensitive EC20 values. For the UniFrac measurement, the most sensitive EC20 values were found in the low (P1)- and high (P3)-carbon soils (Table 6).
FIG 4.
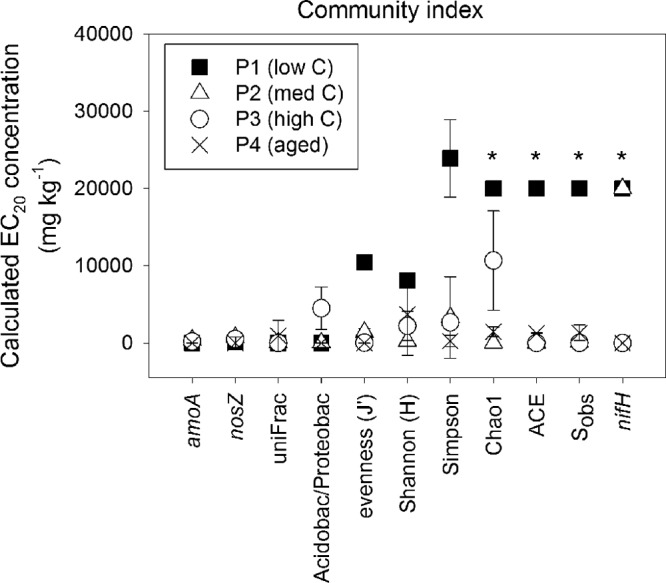
Comparison of EC20 values derived from community measures across variable carbon contents. *, no reliable dose-response model could be fitted.
DISCUSSION
The primary observation of diesel fuel impacts on subantarctic soil microbial communities was a reduction in evenness and richness, resulting in a bacterial community that was heavily dominated by a few specific genera. Our results suggest that the decline in diversity and richness observed after diesel fuel contamination was linked to the disruption of the nitrogen cycle and affected niche-specific species. The sensitivity, low associated estimate errors, and sustained inhibition of the bacterial amoA gene across variable soil types suggest inhibition of potential nitrification activity is likely to be the best microbial indicator of soil sensitivity to diesel fuel for subantarctic soils. The nosZ gene abundance, Acidobacteria/Proteobacteria ratio, and UniFrac similarity are also potentially valuable microbial targets. The average EC20 value (155 mg kg−1 [standard error, 95 mg kg−1]), generated from the abundance of the bacterial amoA gene, was within our range of low diesel fuel concentration (<400 mg kg−1). This concentration is consistent with previous ecotoxicology investigations at Macquarie Island, where an EC20 of 190 mg kg−1 was generated from an acute potential nitrification enzyme assay and a protective concentration between 50 and 200 mg kg−1 was recommended based on avoidance, survival, and reproduction tests of the endemic Macquarie Island earthworm (9, 28). Within wetland assessments, community indices that relate to ecosystem-specific functionality, such as the AOB/AOA ratio, have likewise been identified as promising microbial indicators (10).
In animal and plant ecosystems, a loss of biodiversity is often linked to a decline in community functionality and a decreased resilience to disturbance (52). In the past, microbial ecosystems have been thought to be more resilient to disturbance than plant and animal ecosystems, due to their high functional redundancy (53). However, uncertainty on the extent of this functional redundancy in microbial communities is growing. For example, Allison and Martiny in 2008 reviewed over 110 studies investigating the effects of heavy metals, hydrocarbons, and fertilizer amendments and found that microbial communities are functionally variable (54) and therefore not as resistant or resilient to disturbances as previously thought. More recently, Cravo-Laureau et al. reported that a moderate loss of microbial diversity resulted in a significant decline in functional phenanthrene degradation capacity, highlighting that a small reduction in diversity can result in significant impacts on microbial ecosystem functionality (55).
High-throughput sequencing and qPCR targeting functional genes involved in the nitrogen cycle were used here to simultaneously identify the stimulated and inhibited portions of the bacterial community. This combined approach enabled the vulnerable taxa within the bacterial community to be identified. Across the four subantarctic soil types, the overall phylogenetic diversity, the potential nitrification species, and the Acidobacteria phylum were found to be significantly inhibited in response to SAB diesel fuel. This was especially important in the low-carbon soils where the broad community diversity estimates of Sobs, ACE, and Chao1 were unable to detect a significant response to the SAB, due to preexisting low species richness. While the limitations of widely used diversity estimates have been previously reported (56, 57), it is important to note that without the targeted and functionally important indices used here, the establishment of sensitive species would have been missed due to the limited response in the low-carbon soils or masked by the dominance of the few stimulated species.
One of the novel potential microbial indicators found to be sensitive to SAB contamination was the Acidobacteria/Proteobacteria ratio (Fig. 3). In 2007, Fierer et al. suggested ecological partitioning within the total bacterial diversity based on the r and K selection criteria (58). Although encompassing high levels of phylogenetic and physiological diversity, it was found certain phyla exhibit oligotrophic (slow-growing, K selection criteria) or copiotrophic (rapid growth, r selection criteria) life strategies. In Fierer et al., high numbers of Acidobacteria were found to correlate with low-nutrient environments with slow-growing K-selected species occupying specific environmental niches. Conversely, Proteobacteria, in particular Betaproteobacteria, correlated with copiotrophic conditions and species capable of rapid r-selected growth (58). The ratio of Acidobacteria to Proteobacteria observed here was consistent with the ecological partitioning trend and generated sensitive EC20 values for P1, P2, and P4 between 10 and 110 mg kg−1, all within our low fuel concentration range (<400 mg kg−1). The low EC20 values suggest that low-nutrient subantarctic soils were particularly susceptible to reductions in niche-specific species. The high nutrient soils, with preexisting high ratios of rapidly growing opportunistic species, had a much higher sensitivity threshold at 4,500 mg kg−1.
Pseudomonas, the most stimulated genus found here, has well-known hydrocarbon-degrading capabilities, and its presence has been widely reported in petroleum hydrocarbon-contaminated sites in Antarctica and Macquarie Island (13, 14, 59). The other genus most stimulated in response to high diesel fuel concentrations was Parvibaculum, which has also been reported to have the genetic potential for hydrocarbon degradation (60). While the rapid stimulation of potential hydrocarbon degraders that we observed has been well documented, the full extent of species inhibited with diesel fuel (contributing up to 80% of the total abundance in control communities) has not previously been highlighted (Table 4, Fig. 2). Of those species most inhibited by diesel fuel, nitrifying bacteria were notably vulnerable, with significant declines in both the relative abundance of nitrification species and bacterial amoA gene copy numbers (Fig. 2, 3, and 4).
Ammonium oxidation to nitrite and the subsequent oxidation of nitrite into nitrate are crucial processes in soil. The inhibition of nitrification species and bacterial amoA copy numbers is consistent with previous reports, identifying nitrification as a sensitive measurement for microbial communities following contamination with petroleum hydrocarbons (9, 61), heavy metals (62), and solvents (63). The specific toxicity of petroleum hydrocarbons on nitrification potential has been attributed previously to competitive inhibition with the ammonia monooxygenase enzyme by small aliphatic compounds competing directly with ammonia for the active binding site (22), as well as noncompetitive inhibition from compounds with larger molecular weight through binding to a hydrophobic region of the ammonia monooxygenase (AMO) enzyme and influencing enzyme turnover (22, 64). Doubt has been raised concerning the use of ammonia oxidizers to assess toxicity, because these organisms are thought to recover from hydrocarbon pollution (65). We observed here that unlike the control, the soils chronically exposed to SAB in P4 for 18 months had low to nondetectable levels of the genera involved in nitrification, as well as the bacterial amoA copy numbers. It is important to note that in many soils, the AOA are more abundant than AOB, and the role of archaea in ammonium oxidation should be considered in these soils. Additionally, the bacterial and archaea amoA gene and the nifH and nosZ genes were targeted with only one primer set each in this study. Many other primers capable of targeting these genes exist, and while we are confident the trends would be consistent, different primers may generate slightly different results.
The amount of organic carbon in soils is thought to mediate toxicity in terrestrial systems by binding toxic substrates, thereby limiting their bioavailability in the environment. Carbon present in a system may also provide an alternative carbon source for microorganisms unable to utilize petroleum hydrocarbons. Previously on Macquarie Island, organic carbon has been correlated to the microbial distribution and hydrocarbon-degrading capacity of soils (20). Here, we found the sensitivity to potential nitrification inhibition, denitrification stimulation, and decreases in the Acidobacteria/Proteobacteria ratio were greatest in the soils with the lowest organic carbon content (Fig. 3, Table 6). However, we found the carbon content did not mediate sensitivity of soils to all indices; for example, the UniFrac distance measurement was least sensitive in the chronically exposed soil, despite relatively low soil carbon content (Table 6).
While carbon was correlated with the biological distribution, pH, soil moisture, and available nutrient levels had more significant effects on the resulting bacterial communities (Table 5). Furthermore, the soil characteristics within each soil plot had more influence on the bacterial distribution than the diesel fuel concentration alone. In a previous study assessing the influence of soil properties on alkane-degrading communities at Macquarie Island, the soil parameters of 48 samples from two chronically contaminated sites and nine reference sites were measured (20). The soil plots of P1 and P4 had carbon content similar to that of the two contaminated sites but also shared similar pH, nitrate, and phosphate levels. The P2 and P4 soil plots were closer in resemblance to the reference soils of the island, with substantially higher levels of carbon, nitrate, and phosphate and, for P2, less acidic soil. Given the observed inhibition of potential nitrification and stimulation of potential denitrification, the decline in nitrate observed in the highest fuel concentration samples from P4 was of particular interest. The extended exposure of high diesel fuel concentrations shared low nitrate levels similar to those of the chronically contaminated sites reported by Powell et al. (20). It is difficult to infer if the low levels of nitrite and nitrate were driven by nutrient limitations in the low-carbon soils or were a result of higher toxicity levels resulting from lower levels of organic matter available for binding. We can conclude that the response of the bacterial population was different across soil types, with toxicity more likely to be greatest in low-carbon soils. The mode of action and extent to which other environmental characteristics mediate sensitivity or resilience, especially pH, moisture, and nutrients, need to be further explored.
With increasing numbers of new and untested chemical compounds reaching the environment, there is an urgent need for more rapid ecotoxicology approaches than the traditional single-species tolerance tests (5, 6). The application of high-throughput sequencing and qPCR technologies used in this study evaluated the sensitivity of more than 1,700 species, across 266 families, from 39 separate phyla. Consistent with rapid tolerance testing approaches, we believe the species coverage and confidence in the ecosystem relevance are high. However, it must be noted that the lack of replication in our control samples may have introduced bias into the final analysis, and increased replication, particularly for the control soils, would improve confidence in the individual toxicity values obtained here. Given their integral roles in terrestrial ecosystem function and the capacity for high-throughput analysis, we believe microbial populations have a lot to offer as biological indicators. The large number of species in low abundance found in the control soils also provided an indication of what a nonimpacted bacterial community should resemble. Given the proposed reduction in functional capacity with disturbance to this community structure, an emerging challenge for remediation technologies is also highlighted: that is, how to restore a balanced microbial community capable of sustainable biogeochemical cycling.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by UNSW Australia internal grant funding schemes and the Australian Antarctic Division.
We thank the researchers and technical officers: Tom Mooney, Sharyn Gaskin, Susan Ferguson, Tim Spedding, Ben Raymond, Jim Walworth, Claire Wooldridge, Brett Quinton, Dan Wilkins, Greg Hince, Anne Palmer, Lauren Wise, and Jane Wasley. These people took part in the 2009 and 2010 Macquarie Island summer field seasons or provided support from Kingston, Tasmania, Australia.
Footnotes
Published ahead of print 25 April 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03939-13.
REFERENCES
- 1.AMAP. 1998. AMAP assessment report: Arctic pollution issues, p 859. Arctic Monitoring and Assessment Programme, Oslo, Norway [Google Scholar]
- 2.Wall DH, Virginia RA. 1999. Controls on soil biodiversity: insights from extreme environments. Appl. Soil Ecol. 13:137–150. 10.1016/S0929-1393(99)00029-3 [DOI] [Google Scholar]
- 3.Snape I, Acomb L, Barnes DL, Bainbridge S, Eno R, Filler DM, Plato N, JS P, Raymond T, Rayner J, Riddle M, Rike AG, Rutter A, Schafer A, Siciliano SD, Walworth J. 2008. Contamination, regulation, and remediation: an introduction to bioremediation of petroleum hydrocarbons in cold regions, p 1–37. Cambridge University Press, Cambridge, United Kingdom [Google Scholar]
- 4.Dagnino A, Sforzini S, Dondero F, Fenoglio S, Bona E, Jensen J, Viarengo A. 2008. A weight-of-evidence approach for the integration of environmental “triad” data to assess ecological risk and biological vulnerability. Integr. Environ. Assess. Manag. 4:314–326. 10.1897/IEAM_2007-067.1 [DOI] [PubMed] [Google Scholar]
- 5.Hickey GL, Kefford BJ, Dunlop JE, Craig PS. 2008. Making species salinity sensitivity distributions reflective of naturally occurring communities: using rapid testing and Bayesian statistics. Environ. Toxicol. Chem. 27:2403–2411. 10.1897/08-079.1 [DOI] [PubMed] [Google Scholar]
- 6.Kefford BJ, Palmer CG, Jooste S, Warne MSJ, Nugegoda D. 2005. What is meant by “95% of species”? An argument for the inclusion of rapid tolerance testing. Human Ecol. Risk Assess. 11:1025–1046. 10.1080/10807030500257770 [DOI] [Google Scholar]
- 7.Neilson MN, Winding A. 2002. Microorganisms as indicators of soil health, p 14–16 In NERI (ed), NERI technical report no. 338. Institute NER, Roskilde, Denmark [Google Scholar]
- 8.Avidano L, Gamalero E, Cossa G, Carraro E. 2005. Characterization of soil health in an Italian polluted site by using microorganisms as bioindicators. Appl. Soil Ecol. 30:21–33. 10.1016/j.apsoil.2005.01.003 [DOI] [Google Scholar]
- 9.Schafer AN, Snape I, Siciliano SD. 2007. Soil biogeochemical toxicity end points for sub-Antarctic islands contaminated with petroleum hydrocarbons. Environ. Toxicol. Chem. 26:890–897. 10.1897/06-420R.1 [DOI] [PubMed] [Google Scholar]
- 10.Sims A, Zhang Y, Gajaraj S, Brown PB, Hu Z. 2013. Toward the development of microbial indicators for wetland assessment. Water Res. 47:1711–1725. 10.1016/j.watres.2013.01.023 [DOI] [PubMed] [Google Scholar]
- 11.Nazaries L, Pan Y, Bodrossy L, Baggs EM, Millard P, Murrell JC, Singh BK. 2013. Evidence of microbial regulation of biogeochemical cycles from a study on methane flux and land use change. Appl. Environ. Microbiol. 79:4031–4040. 10.1128/AEM.00095-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ruberto L, Dias R, Lo Balbo A, Vazquez SC, Hernandez EA, MacCormack WP. 2009. Influence of nutrients addition and bioaugmentation on the hydrocarbon biodegradation of a chronically contaminated Antarctic soil. J. Appl. Microbiol. 106:1101–1110. 10.1111/j.1365-2672.2008.04073.x [DOI] [PubMed] [Google Scholar]
- 13.Aislabie JM, Balks MR, Foght JM, Waterhouse EJ. 2004. Hydrocarbon spills on Antarctic soils: effects and management. Environ. Sci. Technol. 38:1265–1274. 10.1021/es0305149 [DOI] [PubMed] [Google Scholar]
- 14.Saul DJ, Aislabie JM, Brown CE, Harris L, Foght JM. 2005. Hydrocarbon contamination changes the bacterial diversity of soil from around Scott Base, Antarctica. FEMS Microbiol. Ecol. 53:141–155. 10.1016/j.femsec.2004.11.007 [DOI] [PubMed] [Google Scholar]
- 15.Hamamura N, Olson SH, Ward DM, Inskeep WP. 2006. Microbial population dynamics associated with crude-oil biodegradation in diverse soils. Appl. Environ. Microbiol. 72:6316–6324. 10.1128/AEM.01015-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guo GX, Deng H, Qiao M, Mu YJ, Zhu YG. 2011. Effect of pyrene on denitrification activity and abundance and composition of denitrifying community in an agricultural soil. Environ. Poll. 159:1886–1895. 10.1016/j.envpol.2011.03.035 [DOI] [PubMed] [Google Scholar]
- 17.Deng H, Guo GX, Zhu YG. 2011. Pyrene effects on methanotroph community and methane oxidation rate, tested by dose-response experiment and resistance and resilience experiment. J. Soils Sedim. 11:312–321. 10.1007/s11368-010-0306-3 [DOI] [Google Scholar]
- 18.Lauber CL, Hamady M, Knight R, Fierer N. 2009. Pyrosequencing-based assessment of soil pH as a predictor of soil bacterial community structure at the continental scale. Appl. Environ. Microbiol. 75:5111–5120. 10.1128/AEM.00335-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fierer N, Jackson RB. 2006. The diversity and biogeography of soil bacterial communities. Proc. Natl. Acad. Sci. U. S. A. 103:626–631. 10.1073/pnas.0507535103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Powell SM, Bowman JP, Ferguson SH, Snape I. 2010. The importance of soil characteristics to the structure of alkane-degrading bacterial communities on sub-Antarctic Macquarie Island. Soil Biol. Biochem. 42:2012–2021. 10.1016/j.soilbio.2010.07.027 [DOI] [Google Scholar]
- 21.Hyman MR, Murton IB, Arp DJ. 1988. Interaction of ammonia monooxygenase from Nitrosomonas europaea with alkanes, alkenes, and alkynes. Appl. Environ. Microbiol. 54:3187–3190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Keener WK, Arp DJ. 1993. Kinetic studies of ammonia moonooxygenase inhibition in Nitrosomonas europaea by hydrocarbons and halogenated hydrocarbons in an optimized whole-cell assay. Appl. Environ. Microbiol. 59:2501–2510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bissett A, Richardson AE, Baker G, Thrall PH. 2011. Long-term land use effects on soil microbial community structure and function. Appl. Soil Ecol. 51:66–78. 10.1016/j.apsoil.2011.08.010 [DOI] [Google Scholar]
- 24.Colloff MJ, Wakelin SA, Gomez D, Rogers SL. 2008. Detection of nitrogen cycle genes in soils for measuring the effects of changes in land use and management. Soil Biol. Biochem. 40:1637–1645. 10.1016/j.soilbio.2008.01.019 [DOI] [Google Scholar]
- 25.Deprez P, Arens M, Locher H. 1994. Identification and preliminary assessment of contaminated sites in the Australian Antarctic Territory. Australian Antarctic Division, Hobart, Australia [Google Scholar]
- 26.Rayner JL, Snape I, Walworth JL, Harvey PM, Ferguson SH. 2007. Petroleum-hydrocarbon contamination and remediation by microbioventing at sub-Antarctic Macquarie Island. Cold Reg. Sci. Technol. 48:139–153. 10.1016/j.coldregions.2006.11.001 [DOI] [Google Scholar]
- 27.Walworth J, Pond A, Snape I, Rayner J, Ferguson S, Harvey P. 2007. Nitrogen requirements for maximizing petroleum bioremediation in a sub-Antarctic soil. Cold Reg. Sci. Technol. 48:84–91. 10.1016/j.coldregions.2006.07.001 [DOI] [Google Scholar]
- 28.Mooney TJ, King CK, Wasley J, Andrew NR. 2013. Toxicity of diesel contaminated soils to the subantarctic earthworm Microscolex macquariensis. Environ. Toxicol. Chem. 32:370–377. 10.1002/etc.2060 [DOI] [PubMed] [Google Scholar]
- 29.Li H, Zhang Y, Kravchenko I, Xu H, Zhang C-G. 2007. Dynamic changes in microbial activity and community structure during biodegradation of petroleum compounds: a laboratory experiment. J. Environ. Sci. 19:1003–1013. 10.1016/S1001-0742(07)60163-6 [DOI] [PubMed] [Google Scholar]
- 30.Rayment G, Lyons D. 2011. Soil chemical methods—Australasia. CSIRO Publishing, Collingwood, VIC, Australia [Google Scholar]
- 31.van Dorst J, Bissett A, Palmer AS, Brown M, Snape I, Stark JS, Raymond B, McKinlay J, Ji M, Winsley T, Ferrari BC. 21 March 2014. Community fingerprinting in a sequencing world. FEMS Microbiol. Ecol. 10.1111/1574-6941.12308 [DOI] [PubMed] [Google Scholar]
- 32.Lane DJ. 1991. 16S/23S rRNA sequencing, p 115–175 In Stackebrandt E, Goodfellow M. (ed), Nucleic acid techniques in bacterial systematics. John Wiley & Sons, Chichester, United Kingdom [Google Scholar]
- 33.Dowd SE, Sun Y, Wolcott RD, Domingo A, Carroll JA. 2008. Bacterial tag-encoded FLX amplicon pyrosequencing (bTEFAP) for microbiome studies: bacterial diversity in the ileum of newly weaned Salmonella-infected pigs. Foodborne Pathog. Dis. 5:459–472. 10.1089/fpd.2008.0107 [DOI] [PubMed] [Google Scholar]
- 34.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF. 2009. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75:7537–7541. 10.1128/AEM.01541-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J, Glöckner FO. 2007. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 35:7188–7196. 10.1093/nar/gkm864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. 2011. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27:2194–2200. 10.1093/bioinformatics/btr381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim M, Morrison M, Yu Z. 2011. Evaluation of different partial 16S rRNA gene sequence regions for phylogenetic analysis of microbiomes. J. Microbiol. Meth. 84:81–87. 10.1016/j.mimet.2010.10.020 [DOI] [PubMed] [Google Scholar]
- 38.Labbé D, Margesin R, Schinner F, Whyte LG, Greer CW. 2007. Comparative phylogenetic analysis of microbial communities in pristine and hydrocarbon-contaminated Alpine soils. FEMS Microbiol. Ecol. 59:466–475. 10.1111/j.1574-6941.2006.00250.x [DOI] [PubMed] [Google Scholar]
- 39.Sheneman L, Evans J, Foster JA. 2006. Clearcut: a fast implementation of relaxed neighbor joining. Bioinformatics 22:2823–2824. 10.1093/bioinformatics/btl478 [DOI] [PubMed] [Google Scholar]
- 40.Lozupone C, Hamady M, Knight R. 2006. UniFrac—an online tool for comparing microbial community diversity in a phylogenetic context. BMC Bioinformatics 7:371. 10.1186/1471-2105-7-371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Clarke KR, Warwick RM. 2001. Changes in marine communities: an approach to statistical analysis and interpretation. PRIMER-E, Plymouth, United Kingdom [Google Scholar]
- 42.Ma WK, Bedard-Haughn A, Siciliano SD, Farrell RE. 2008. Relationship between nitrifier and denitrifier community composition and abundance in predicting nitrous oxide emissions from ephemeral wetland soils. Soil Biol. Biochem. 40:1114–1123. 10.1016/j.soilbio.2007.12.004 [DOI] [Google Scholar]
- 43.Gaby JC, Buckley DH. 2012. A comprehensive evaluation of PCR primers to amplify the nifH gene of nitrogenase. PLoS One 7:e42149. 10.1371/journal.pone.0042149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rotthauwe JH, Witzel KP, Liesack W. 1997. The ammonia monooxygenase structural gene amoA as a functional marker: molecular fine-scale analysis of natural ammonia-oxidizing populations. Appl. Environ. Microbiol. 63:4704–4712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Francis CA, Roberts KJ, Beman JM, Santoro AE, Oakley BB. 2005. Ubiquity and diversity of ammonia-oxidizing archaea in water columns and sediments of the ocean. Proc. Natl. Acad. Sci. U. S. A. 102:14683–14688. 10.1073/pnas.0506625102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Henry S, Bru D, Stres B, Hallet S, Philippot L. 2006. Quantitative detection of the nosZ gene, encoding nitrous oxide reductase, and comparison of the abundances of 16S rRNA, narG, nirK, and nosZ genes in soils. Appl. Environ. Microbiol. 72:5181–5189. 10.1128/AEM.00231-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Siciliano SD, Ma W, Powell S. 2007. Evaluation of quantitative polymerase chain reaction to assess nosZ gene prevalence in mixed microbial communities. Can. J. Microbiol. 53:636–642. 10.1139/W07-014 [DOI] [PubMed] [Google Scholar]
- 48.Zhang X, Liu W, Schloter M, Zhang G, Chen Q, Huang J, Li L, Elser JJ, Han X. 2013. Response of the abundance of key soil microbial nitrogen-cycling genes to multi-factorial global changes. PLoS One 8(10):e76500. 10.1371/journal.pone.0076500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cottingham KL, Lennon JT, Brown BL. 2005. Knowing when to draw the line: designing more informative ecological experiments. Front. Ecol. Environ. 3:145–152. 10.1890/1540-9295(2005)003[0145:KWTDTL]2.0.CO;2 [DOI] [Google Scholar]
- 50.Knezevic SZ, Streibig JC, Ritz C. 2007. Utilizing R software package for dose-response studies: the concept and data analysis. Weed Technol. 21:840–848. 10.1614/WT-06-161.1 [DOI] [Google Scholar]
- 51.Ritz C, Streibig JC. 2005. Bioassay analysis using R. J. Stat. Software 12:1–22 http://www.jstatsoft.org/v12/i05/paper [Google Scholar]
- 52.Tilman D, Wedin D, Knops J. 1996. Productivity and sustainability influenced by biodiversity in grassland ecosystems. Nature 379:718–720. 10.1038/379718a0 [DOI] [Google Scholar]
- 53.Yachi S, Loreau M. 1999. Biodiversity and ecosystem productivity in a fluctuating environment: the insurance hypothesis. Proc. Natl. Acad. Sci. U. S. A. 96:1463–1468. 10.1073/pnas.96.4.1463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Allison SD, Martiny JBH. 2008. Resistance, resilience, and redundancy in microbial communities. Proc. Natl. Acad. Sci. U. S. A. 105:11512–11519. 10.1073/pnas.0801925105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cravo-Laureau C, Hernandez-Raquet G, Vitte I, Jézéquel R, Bellet V, Godon J-J, Caumette P, Balaguer P, Duran R. 2011. Role of environmental fluctuations and microbial diversity in degradation of hydrocarbons in contaminated sludge. Res. Microbiol. 162:888–895. 10.1016/j.resmic.2011.04.011 [DOI] [PubMed] [Google Scholar]
- 56.Curtis TP, Sloan WT. 2004. Prokaryotic diversity and its limits: microbial community structure in nature and implications for microbial ecology. Curr. Opin. Microbiol. 7:221–226. 10.1016/j.mib.2004.04.010 [DOI] [PubMed] [Google Scholar]
- 57.Bent SJ, Forney LJ. 2008. The tragedy of the uncommon: understanding limitations in the analysis of microbial diversity. ISME J. 2:689–695. 10.1038/ismej.2008.44 [DOI] [PubMed] [Google Scholar]
- 58.Fierer N, Bradford MA, Jackson RB. 2007. Toward an ecological classification of soil bacteria. Ecology 88:1354–1364. 10.1890/05-1839 [DOI] [PubMed] [Google Scholar]
- 59.Powell SM, Ma WK, Siciliano SD. 2006. Isolation of denitrifying bacteria from hydrocarbon-contaminated Antarctic soil. Polar Biol. 30:69–74. 10.1007/s00300-006-0161-2 [DOI] [Google Scholar]
- 60.Flocco CG, Gomes NCM, Mac Cormack W, Smalla K. 2009. Occurrence and diversity of naphthalene dioxygenase genes in soil microbial communities from the maritime Antarctic. Environ. Microbiol. 11:700–714. 10.1111/j.1462-2920.2008.01858.x [DOI] [PubMed] [Google Scholar]
- 61.Sverdrup LE, Ekelund F, Krogh PH, Nielsen T, Johnsen K. 2002. Soil microbial toxicity of eight polycyclic aromatic compounds: effects on nitrification, the genetic diversity of bacteria, and the total number of protozoans. Environ. Toxicol. Chem. 21:1644–1650. 10.1002/etc.5620210815 [DOI] [PubMed] [Google Scholar]
- 62.Pereira R, Sousa J, Ribeiro R, Gonçalves F. 2006. Microbial indicators in mine soils (S. Domingos mine, Portugal). Soil Sedim. Contam. 15:147–167. 10.1080/15320380500506813 [DOI] [Google Scholar]
- 63.Miller JL, Sardo MA, Thompson TL, Miller RM. 1997. Effect of application solvents on heterotrophic and nitrifying populations in soil microcosms. Environ. Toxicol. Chem. 16:447–451. 10.1002/etc.5620160309 [DOI] [Google Scholar]
- 64.Chang SW, Hyman MR, Williamson KJ. 2002. Cooxidation of napthalene and other polycyclic aromatic hydrocarbons by the nitrifying bacterium, Nitrosomonas europaea. Biodegradation 13:373–381. 10.1023/A:1022811430030 [DOI] [PubMed] [Google Scholar]
- 65.Deni J, Penninckx MJ. 2004. Influence of long-term diesel fuel pollution on nitrite-oxidising activity and population size of nitrobacter spp. in soil. Microbiol. Res. 159:323–329. 10.1016/j.micres.2004.06.004 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.