Abstract
Dryvax (Wyeth Laboratories, Inc., Marietta, PA) is representative of the vaccinia virus preparations that were previously used for preventing smallpox. While Dryvax was highly effective, the national supply stocks were depleted, and there were manufacturing concerns regarding sterility and the clonal heterogeneity of the vaccine. ACAM2000 (Acambis, Inc./Sanofi-Pasteur Biologics Co., Cambridge, MA), a single-plaque-purified vaccinia virus derivative of Dryvax, recently replaced the polyclonal smallpox vaccine for use in the United States. A substantial amount of sequence heterogeneity exists within the polyclonal proteome of Dryvax, including proteins that are missing from ACAM2000. Reasoning that a detailed comparison of antibody responses to the polyclonal and monoclonal vaccines may be useful for identifying unique properties of each antibody response, we utilized a protein microarray comprised of approximately 94% of the vaccinia poxvirus proteome (245 proteins) to measure protein-specific antibody responses of 71 individuals receiving a single vaccination with ACAM2000 or Dryvax. We observed robust antibody responses to 21 poxvirus proteins in vaccinated individuals, including 11 proteins that distinguished Dryvax responses from ACAM2000. Analysis of protein sequences from Dryvax clones revealed amino acid level differences in these 11 antigenic proteins and suggested that sequence variation and clonal heterogeneity may contribute to the observed differences between Dryvax and ACAM2000 antibody responses.
INTRODUCTION
The eradication of smallpox in the 1980s was a historical milestone that marked the first successful vaccination campaign to conquer a global infectious disease. Shortly after natural infections were declared eradicated, the commercial production of smallpox vaccines was discontinued. However, the United States reinstituted the smallpox vaccine stockpile program based on concerns that an act of biological terrorism could result in reemergence of smallpox due to the cessation of routine vaccination (1–4). The standard smallpox vaccine Dryvax, used predominantly throughout the United States, was derived from lymphatic fluid collected from the skin of live animals after scarification with replicating vaccinia virus (VACV; New York City Board of Health [NYCBOH] strain). The Dryvax product consists of a heterogeneous pool of VACV clones that could potentially become contaminated by bovine pathogens or other adventitious material during processing, as well as having an increased risk for selection of more-virulent strains of VACV (5–9). Dryvax vaccination has significant limitations, including risks to pregnant and immunocompromised individuals, serious and occasional lethal adverse events such as myopericarditis, and the potential for transmission of VACV to others who are at risk for adverse events (5, 10–20). Further, there is significant variability in human antibody responses to traditional polyclonal VACV vaccines (21–23). The levels of expression of antigens that are important for protective immunity or that may influence adverse reactions are difficult to control because Dryvax and similar vaccine preparations are comprised of heterogeneous VACV clones, some more virulent than others (6–9). This type of molecular and biological diversity within Dryvax vaccine preparations was demonstrated through examination of individual VACV clones isolated from the pooled vaccine (6–9).
An improved vaccine production method was needed in order to address the shortcomings of polyclonal smallpox vaccines and their manufacturing process. One approach to improving the vaccine was initiated through the isolation of a single clone from polyclonal VACV preparations (9, 24). In some cases, immune responses to plaque-purified VACV preparations were altered significantly by large genomic deletions that accompanied clone attenuation (21). Ultimately, ACAM2000 (Acambis, Inc./Sanofi-Pasteur Biologics Co., Cambridge, MA), a cell culture product of a single VACV clone, was approved in 2007 by the U.S. Food and Drug Administration (FDA) as a replacement for Dryvax (6, 25). This new vaccine clone, which was isolated from multiple doses of the original polyclonal Dryvax, maintains monoclonality and was shown to be free of adventitious bacterial, fungal, or viral pathogenic contaminants (6, 9). ACAM2000 is similar to the polyclonal vaccine in terms of cutaneous vaccination lesions, viral shedding, and general humoral or cell-mediated immune responses, while myopericarditis and other adverse side effects are also comparable to those of Dryvax (6, 9, 26, 27). Clinical trials comparing Dryvax and ACAM2000 showed similar vaccine efficacy at the highest dose. Efficacy of Dryvax was maintained with vaccine dilution, whereas dilution of ACAM2000 resulted in decreased efficacy (26, 28).
In 2007, Osborne et al. (7) compared the genomes of ACAM2000 and the neurovirulent Dryvax clone CL3, which was isolated during ACAM2000 production. There are 625 nucleotide substitutions within the coding sequence of ACAM2000 compared to CL3, consisting of 572 single nucleotide polymorphisms that result in 290 amino acid changes, as well as insertions or deletions (indels) of various sizes (7). While most proteins are conserved, there are substantial differences between the two clones for a subset of open reading frames (ORFs). For example, the full-length alpha/beta interferon (IFN-α/β) receptor, ankyrin-like protein, and tumor necrosis factor alpha receptor are missing from ACAM2000 (7, 8). The potential impact of these missing or variant proteins on long-term immunity to smallpox is unknown, and a detailed analysis of immune responses to the ACAM2000 proteome may be useful for identifying unique properties of this vaccine. In a previously reported study (23), we developed a microarray of the vaccinia virus proteome that was used to identify antigens comprising the human antibody response to Dryvax vaccination. Expansion of the vaccinia virus protein microarray to include the proteome of the monkeypox virus allowed us to distinguish antibody responses to smallpox vaccination from infection by monkeypox virus (29). On the basis of these previous results, we reasoned that it should be possible to compare human antibody responses to the monoclonal ACAM2000 vaccine and Dryvax by using a microarray consisting of 94% coverage of the VACV proteome. We report the results of the proteome-wide analysis of viral antigens identified by this study.
MATERIALS AND METHODS
Vaccinations.
Sera were collected prior to and 28 days after primary vaccination with ACAM2000 (Acambis, Inc./Sanofi-Pasteur Biologics Co., Cambridge, MA) from volunteers who gave consent (n = 61). In addition, sera were collected from 10 individuals who gave consent 28 days following primary vaccination with Dryvax (Wyeth Laboratories, Inc., Marietta, PA) (derived from NYCBOH), as previously described (23). Peripheral venous blood from each healthy donor was collected for the preparation of serum, following written informed consent and in accordance with the protocols approved by the U.S. Army Medical Research Institute of Infectious Diseases (USAMRIID) Institutional Review Board (IRB). The ACAM2000 and Dryvax studies used the same inclusion/exclusion criteria (listed in the package insert), and only vaccinia virus-naive subjects were eligible to participate in these protocols. ACAM2000 subjects were divided into two groups on day 7 of the study to examine the spread of virus from the vaccination site (unpublished data): subjects in the treatment group had povidone iodine applied to the vaccination site starting on day 7; subjects in the control group did not have povidone iodine applied and the vaccination site was monitored as usual. Vaccination site treatments with povidone iodine do not impact antibody responses (30); therefore, data from all ACAM2000 vaccinations were processed as a single group. After the subjects gave informed consent, blood was drawn before and following scarification. Serum was separated and stored frozen using standard procedures. ACAM2000, smallpox (vaccinia virus [VACV]) vaccine, live, was derived from Dryvax (Wyeth Laboratories, Marietta, PA; calf lymph vaccine, NYCBOH) using plaque purification techniques and grown in African green monkey kidney (Vero) cells. The freeze-dried vaccine was reconstituted using the provided diluent per package insert instructions (25). Each reconstituted vaccine vial contained approximately 100 doses of 0.0025 ml of live VACV, containing 2.5 × 105 to 12.5 × 105 PFU/dose. A sterile bifurcated needle (provided with the vaccine) was used to remove vaccine from the vial and subsequently used to administer ACAM2000 percutaneously using 15 jabs. The vaccination site was kept covered using a semipermeable bandage, with the bandage changed every 1 to 3 days until the scab fell off. Vaccination was deemed successful if the vaccinee developed a major cutaneous reaction such as a vesicular or pustular lesion, an area of palpable induration, or congestion surrounding the vaccination site.
Vaccinia virus proteome microarray.
Proteins encoded by VACV (Copenhagen; GenBank accession no. M35027.1) were produced as described previously (23, 29). Briefly, 273 pENTR221 entry clones that were fully sequenced and characterized were recombined into the pDEST20 glutathione S-transferase (GST) expression vector using Gateway cloning methods (Invitrogen, CA). All GST-tagged recombinant vaccinia virus proteins were expressed in Sf9 insect cells using Gateway baculovirus expression (Invitrogen) and purified using glutathione-based affinity purification. The 245 vaccinia virus proteins that passed quality control criteria, as well as several control proteins, were printed in a microarray on thin-film nitrocellulose PATH slides (GenTel Biosciences, WI). The protein microarrays were stored at −20°C until use.
All incubations and microarray manipulations were automated by using a HS 400 Pro hybridization station (Tecan Group Ltd., NC), set at 22°C, using previously described methods (23, 29). Briefly, arrays were blocked for 1 h in blocking buffer consisting of 50 mM HEPES (pH 7.5), 200 mM NaCl, 0.08% Triton X-100, 25% glycerol, 20 mM reduced glutathione, 1% bovine serum albumin (BSA), and 1 mM dithiothreitol (DTT). Protein microarrays were rinsed with wash buffer (1× phosphate-buffered saline [PBS] [pH 7.4], 0.2% Tween 20, 1% BSA) and probed (1 h) with 2 μl of serum diluted 1:150 in probe buffer (1× PBS [pH 7.4], 0.1% Tween 20, 1% BSA). Protein microarrays were rinsed (wash buffer), and antibody binding was detected by incubation (1 h) with 1:2,000 dilution of goat-anti human IgG (H+L) Alexa Fluor 647.
Microarray data analysis.
Dried microarray slides were scanned by a confocal laser scanner (GenePix 4000B; Molecular Devices, CA), using a wavelength of 635 nm. Raw pixel counts were generated by imaging the microarrays using a power setting of 100% and the highest photomultiplier tube (PMT) gain that did not produce saturated signals. Data acquired from GenePix software were analyzed using ProtoArray Prospector v5.1 (Invitrogen, CA) in Immune Response Profiling mode. Quantile normalization was performed on raw pixel counts from nonvaccinated and vaccinated (Dryvax and ACAM2000) groups of individuals separately. Following normalization, an M-statistics algorithm (IRBP Toolbox v5.1; Invitrogen) was used to calculate statistical significance, implementing a minimal signal of 500 relative fluorescence units (RFU) with a minimal signal gap of 200 RFU. A Bonferroni's correction was performed for comparisons of nonvaccinated and vaccinated groups. Outliers among the data replicates were identified by using a modified Z-score (median absolute deviation of >3.5) and removed from further analysis. The data were log2 transformed for hierarchical clustering analysis with the MeV v4.4.1 TM4 Microarray Software Suite (31), using Euclidean distance as the dissimilarity metric.
Bioinformatics.
Amino acid sequences encoded by ACAM2000 (GenBank accession no. AY313847) and 14 other Dryvax clonal sequences obtained from the Viral Bioinformatics Resource Center (32, 33) were examined. The 14 Dryvax sequences consisted of 11 different plaque-purified clones (GenBank accession no. JN654977 to JN654986) isolated from a stored vial of Dryvax vaccine (lot 1556-14), two plaque-purified clones (VACV-3737 [GenBank accession no.DQ377945]; and VACV-DUKE [GenBank accession no. DQ439815]) harvested from a Dryvax vaccination site as well as the virulent plaque-purified clone (ACAM3/CL3 [GenBank accession no.AY313848]) that was isolated during ACAM2000 production. Multiple sequence alignments were performed in MegAlign (DNASTAR Lasergene software suite v.8), using the ClustalW alignment program and the Gonnet 250 protein weight matrix to score each alignment. The multiple alignment parameters consisted of the following: gap penalty, 10; gap length penalty, 0.2; delay divergent sequences, 30%.
Percent length identity was calculated to compare the sequence length of proteins in the Dryvax vaccine to those in ACAM2000. In order to take into account protein heterogeneity among the different Dryvax clones, percent length identity was calculated by taking the number of Dryvax clones that expressed VACV proteins of equal length to that of ACAM2000 divided by the total number of Dryvax clones used for sequence analysis (n = 14).
Relative entropy was also used as a measure of protein sequence variability, using the following formula:
where mi is the ith column of alignment m, cia is the count of character a in column i, and pia is the probability of character a in column i, given a 21-letter alphabet.
A 21-letter alphabet (20 possible amino acids, plus a dash for gaps) was used in the calculation of entropy for each amino acid residue in the alignment. Protein sequence alignments were evaluated (Gblocks v0.91b [34]) with gaps allowed within final blocks to eliminate poorly aligned columns or regions that may skew relative entropy calculations. Because the B19R gene product is not produced by one of the Dryvax strains and is truncated in most other strains (265 amino acids in length in the ACAM2000 reference sequence and nine other Dryvax sequences), a conserved block consisting of the first 259 residues was used for B19R entropy calculation. The C21/B27R protein is truncated in two strains (79 amino acids instead of 113 residues) and also exhibits a high degree of variability in the two truncated proteins. For this reason, a conserved block consisting of the first 49 residues was used for C21/B27R entropy calculation.
For each protein, a delta entropy value per residue was calculated as follows: ΔH = Hobserved − Hconserved where Hobserved is the summation of entropies for individual proteins divided by the total number of amino acid residues in that protein and Hconserved is the entropy value of the same alignment if all residues were completely conserved.
RESULTS
Smallpox vaccinations and protein microarrays.
Seventy-one vaccinia virus-naive subjects participated in independent Dryvax (n = 10) and ACAM2000 (n = 61) studies. Among the subjects that completed the Dryvax study, their ages ranged from 23 to 46 years old, 6% were male, 10% were African American, and the remaining 90% were Caucasian. In the ACAM2000 study, 68% of subjects were male, 78.3% were Caucasian, 20.0% were African-American, 1.7% were Asian, with a mean age of 25 years and a range of 19 to 39 years. All vaccinated individuals in both completed studies, had a classic “take” reaction or major cutaneous reaction following scarification. There were no serious adverse events after vaccination.
Blood for serum isolation was collected from individuals in each study before and 28 days following primary administration of either Dryvax or ACAM2000 vaccine. We used microarrays covering 94% (245 recombinant proteins) of the VACV proteome to examine antigen-specific antibody responses of the 71 primary Dryvax- or ACAM2000-vaccinated individuals. Each protein included in the microarray was derived from a fully sequenced plasmid clone, and the products expressed in eukaryotic cells were affinity purified for inclusion in the final platform.
Similarities in antibody responses to Dryvax and ACAM2000 vaccination.
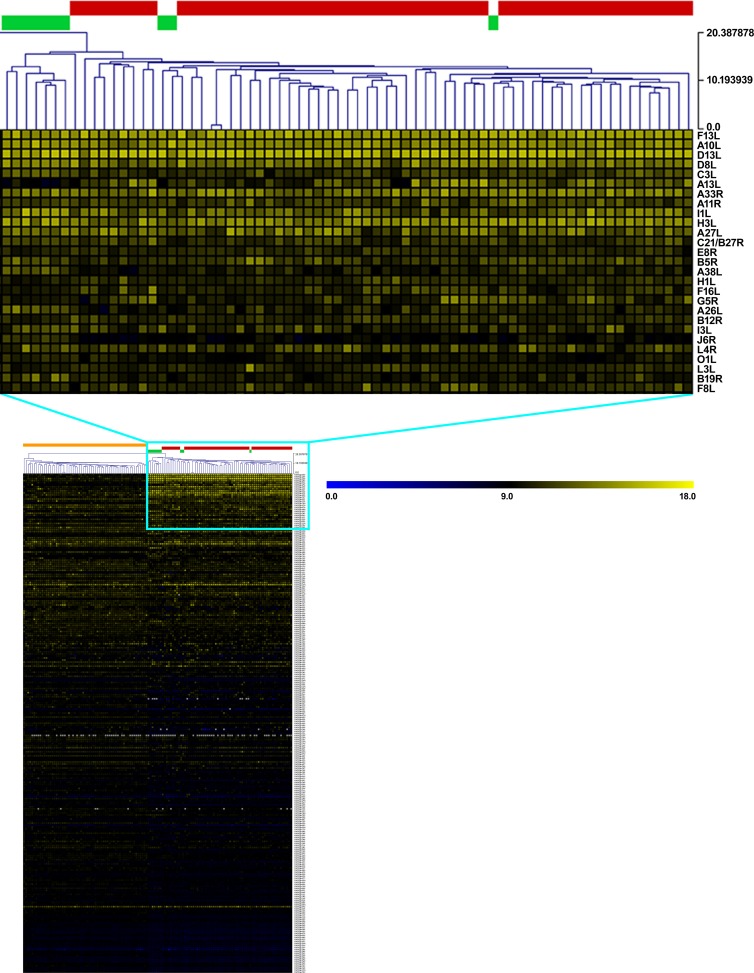
Dilutions of sera collected from control or vaccinated subjects were incubated separately on the proteome microarray in order to measure antibody binding to specific VACV antigens (Fig. 1). Hierarchical clustering of serum antibody binding results indicated that all vaccinated individuals grouped together, while sera from nonvaccinated controls formed a separate cluster (Fig. 1). Further, we noted that seven out of 10 Dryvax-vaccinated individuals clustered independently of the ACAM2000-vaccinated subjects. While these results suggested that antibody responses of individuals receiving either vaccine were similar, subtle differences in antibody responses between polyclonal and monoclonal smallpox vaccines were evident. Upon further analysis, 21 viral proteins were observed to elicit a robust antibody response in Dryvax- and ACAM2000-vaccinated individuals in comparison to nonvaccinated controls (P ≤ 0.0002) (Table 1). In addition to the 21 VACV proteins, the late transcription factor H5R was recognized by human antibody from vaccinated individuals (∼74%; P = 0.0012), but these results did not meet the Bonferroni's corrected P value cutoff (P ≤ 0.0002). As shown in Table 1, the 21 antigens from vaccinia virus that were targeted by antibody responses represented proteins that were secreted, intracellular, or associated directly with the extracellular enveloped virion (EEV) or intracellular mature virion (IMV). Notably, antibody responses in over 90% of smallpox-vaccinated individuals involved the EEV membrane protein F13L, IMV membrane proteins D13L and A27L, and the IMV core protein A10L.
FIG 1.
Hierarchical clustering of vaccinia virus proteins recognized by antibody from vaccinia virus-naive and Dryvax- and ACAM2000-vaccinated individuals. Protein microarrays were probed with sera from 61 vaccinia virus-naive individuals and 71 primary vaccinated individuals (10 individuals vaccinated with Dryvax and 61 vaccinated with ACAM2000). Antibody binding to the vaccinia virus proteins was measured with Alexa Fluor 647-conjugated anti-human IgG (H+L). Hierarchical clustering, using Euclidean distance average linkage method with normalized and log2-transformed data, was used to visualize protein microarray results. Naive and vaccinated sera are listed in columns while vaccinia virus protein-antibody interactions (in rows) are sorted by increasing P value. The color key at the top right of the heat map at the bottom of the figure (main heat map) shows the scale of log2-transformed signal intensity (0 to 18), and the nodal scale at the top right of the magnified section of the heat map, also corresponding to minimum and maximum signal intensity units, uses a true branch length structure. The color blocks at the top of the main heat map represent the subject groups: orange before primary vaccination (naive); red primary ACAM2000 vaccination; green, primary Dryvax vaccination. Only significant protein-antibody binding interactions (P < 0.0002; blue box) among vaccinated individuals are magnified (top) for visualization.
TABLE 1.
Proteins of vaccinia virus recognized by antibody from vaccinia virus-naive or Dryvax- and ACAM2000-vaccinated individuals
| Protein | Descriptiona | Location of viral antigenb | Nonvaccinated prevalence (%) | Vaccinated prevalence (%) | P value | References |
|---|---|---|---|---|---|---|
| F13L | IEV membrane wrapping palmytilated protein | EEV membrane | 2 | 99 | 3.86E−39 | 52–55 |
| A10L | Core protein P4a, assembly of nucleoprotein complex | IMV core | 3 | 97 | 1.67E−35 | 54–58 |
| D13L | Rifampin resistance protein, spicule coat formation of IV | IMV membrane | 2 | 93 | 2.61E−33 | 54, 56, 59, 60 |
| D8L | Cell surface chondroitin sulfate binding protein | IMV membrane | 2 | 69 | 2.67E−19 | 54–56, 58, 61 |
| C3L | Complement regulatory protein | EC/EEV membrane | 8 | 79 | 4.93E−19 | 48, 62, 63 |
| A13L | IV to IMV assembly phosphoprotein | IMV membrane | 6 | 77 | 8.13E−19 | 54–56, 58, 64 |
| A33R | EEV glycoprotein, actin tail formation with A36R | EEV membrane | 3 | 68 | 9.88E−18 | 55, 65, 66 |
| A11R | IMV membrane assembly protein | IC, viral factory | 13 | 84 | 1.39E−16 | 54, 67 |
| I1L | Intermediate-class gene promoter | IMV core | 3 | 62 | 4.08E−15 | 54–56, 68 |
| H3L | IMV heparan sulfate binding surface protein, IMV assembly | IMV membrane | 10 | 70 | 5.57E−14 | 54–56, 58, 69, 70 |
| A27L | Heparan sulfate binding surface protein, IMV microtubule-dependent transport | IMV membrane | 32 | 92 | 1.32E−12 | 54–56, 58, 71, 72 |
| C21/B27R | Ankyrin-like protein | Unknown | 11 | 77 | 1.61E−11 | |
| B5R | Plaque size, host range protein precursor, EEV formation and dissemination | EEV membrane | 11 | 63 | 4.27E−10 | 55, 73–75 |
| A38L | Integrin/CD47-associated protein, Ca2+ influx | Host cell membrane | 2 | 40 | 2.26E−08 | 76, 77 |
| G5R | Putative FEN1-like nuclease | IMV corec | 10 | 45 | 1.33E−06 | 78, 79 |
| A26L | IMV A-type inclusion, laminin binding protein | IMV membrane | 14 | 60 | 1.56E−06 | 54, 56, 80 |
| I3L | Single-stranded DNA-binding phosphoprotein | IMV core | 6 | 34 | 2.21E−05 | 54–56, 81, 82 |
| J6R | DNA-directed RNA polymerase 147-kDa subunit | IMV core | 3 | 40 | 2.28E−05 | 54–56, 83, 84 |
| L4R | Single- and double-stranded DNA and ssRNA-binding virion core protein vp8 | IMV core | 10 | 41 | 3.95E−05 | 54–56, 58, 85, 86 |
| O1L | ERK1/2 signaling modulator | ICd | 5 | 33 | 4.37E−05 | 49 |
| B19R | IFN-α/β receptor | EC, host cell membrane | 2 | 25 | 9.65E−05 | 51, 87, 88 |
IEV, intracellular enveloped virion; IV, immature virion; IMV, intracellular mature virus; EEV, extracellular enveloped virus; ssRNA, single-stranded RNA.
EEV, extracellular enveloped virus; IMV, intracellular mature virus; EC, extracellular; IC, intracellular.
It should be noted that while da Fonseca et al. (78) found G5R to be located within the IMV, other studies (54–56) did not.
The study of Manes et al. (89) in 2008 suggests that O1L may also be detected within VACV virion particles.
Antibody responses that differentiate Dryvax from ACAM2000 vaccination.
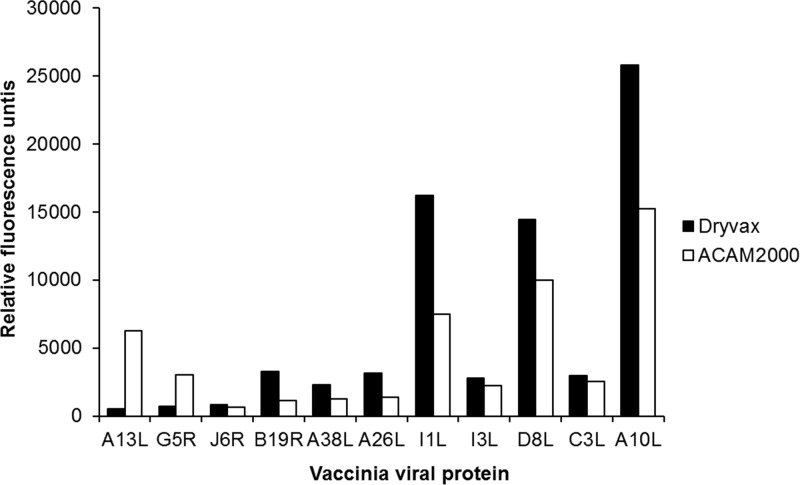
Upon further analysis of the 21 vaccinia virus antigens that elicited a specific VACV antibody response in all vaccinated individuals, we noted that antibody responses to 11 antigenic proteins were statistically different between Dryvax- and ACAM2000-vaccinated individuals (P ≤ 0.05; Fig. 2). Antibody binding to two vaccinia virus proteins, the IMV structural phosphoprotein A13L and IMV putative nuclease protein G5R, were higher in the ACAM2000-vaccinated group, while antibody binding to the remaining nine vaccinia virus proteins (J6R, B19R, A38L, A26L, I1L, I3L, D8L, C3L, and A10L) were found to be higher in the Dryvax-vaccinated group (Fig. 2). The EEV membrane protein F13L exhibited a marginal difference in antibody responses (50% prevalence in Dryvax versus 79% in ACAM2000; data not shown), but it was excluded from further analysis because it did not meet our significance criteria (P ≤ 0.052).
FIG 2.
Antigens that differentiate Dryvax from the monoclonal ACAM2000 vaccine. Serum antibody responses from 71 individuals receiving a primary vaccination with Dryvax (n = 10) or ACAM2000 (n = 61) were measured by protein microarray. Data shown represent a significant difference (P < 0.05) in antibody response between vaccinated groups of individuals.
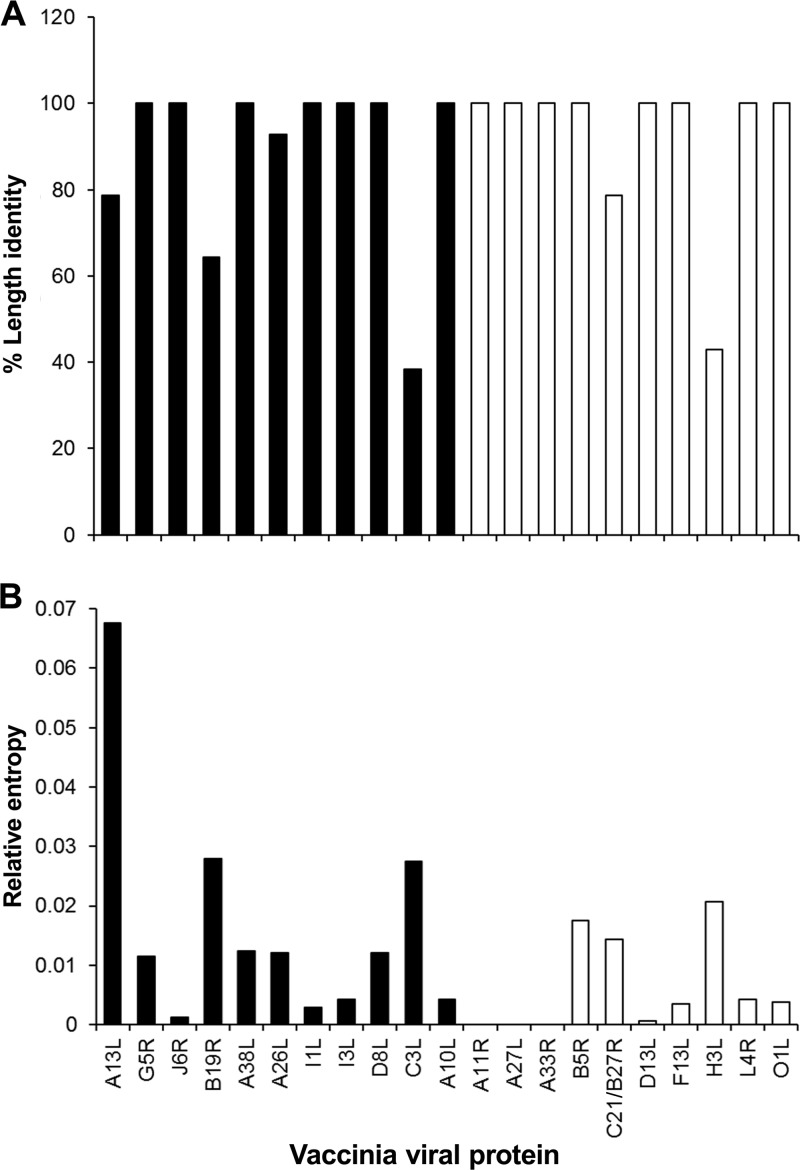
Due to the clonal heterogeneity of Dryvax ORFs, variations in proteins were anticipated to lead to changes in antibody epitopes. Therefore, we examined the sequences of the 11 viral proteins that presented significant differences between antibody responses to ACAM2000 and Dryvax vaccine and compared these sequence results to those of the 10 viral proteins that had similar levels of antibody binding in both vaccinated groups. Multiple sequence alignments were generated for the proteins present in ACAM2000 and 14 Dryvax clones (see Fig. S1 in the supplemental material). Among the sequences that were examined, 11 were from Dryvax clones that were plaque purified (8) from a specific lot of Dryvax vaccine (Dryvax clones designated by DPP prefix before a number), two were from a vaccinia pustule following Dryvax vaccination (VACV-3737 and VACV-DUKE) (35; VACV-3737 genome directly submitted to the Genome Sequencing Center of the Washington University School of Medicine), and the last, slightly more virulent clone (CL3/ACAM3) was isolated from pooled vials of Dryvax vaccine during ACAM2000 (7) vaccine production (32, 33). Four antigens (A13L, B19R, A26L, and C3L) differed in sequence length among the 11 proteins that distinguished antibody responses between vaccinated groups, while only two (C21/B27R and H3L) of the 10 VACV antigens common to both vaccinated groups varied in sequence length (Fig. 3A). Vaccinia virus IFN-α/β receptor protein B19R is present in all VACV clones except for clone 17 (DPP17), which has an 11.7-kbp deletion that results in the loss of this protein (8). For clones that express the B19R gene, the protein is usually truncated, with the exception of three VACV strains (CL3, DPP13, and DPP21) that express a full-length protein (Fig. S1). Further, three (DPP13, DPP15, and DUKE) of the Dryvax clones have in-frame amino acid deletions in A13L, while nine clones have in-frame amino acid deletions in C3L. In VACV3737, A26L is elongated by 2 amino acid residues. Furthermore, all 11 of the vaccinia virus proteins that distinguished Dryvax from ACAM2000 vaccine varied by one or more amino acid residues, while 7 of the 10 that were common to both vaccinated groups varied. A11R, A27L, and A33R were 100% conserved among all Dryvax clones, D13L and L4R proteins varied by only one amino acid residue in one to four Dryvax clones, and the remaining five proteins (B5R, C21/B27R, D13L, H3L, and O1L) varied by more than one amino acid residue (Fig. S1). Relative entropy scores of the sequence alignment were used to visualize overall sequence variations for each protein. Entropy scores varied independently of the differences observed in protein length (Fig. 3B). As shown in Fig. 3B, none of the 11 proteins that differentiated Dryvax from ACAM2000 antibody responses had an entropy score of zero, indicating that no sequence was completely conserved, whereas 3 of the 10 VACV antigens (A11R, A27L, and A33R) that showed similar antibody binding in both vaccinated groups had 100% sequence conservation. Collectively, these observations suggested that variability within VACV antigens as well as clonal heterogeneity contributed to the observed differences between Dryvax and ACAM2000 antibody responses.
FIG 3.
Analysis of vaccinia virus proteins that differentiate Dryvax from ACAM2000 antibody responses. The 11 vaccinia virus proteins that distinguished Dryvax from ACAM2000 antibody responses are indicated by black bars, and the antigens that were common among both vaccinated groups are indicated by white bars. (A) Percent protein length identity. (B) Relative sequence entropy. Fifteen Dryvax clones, including ACAM2000, were used for sequence analysis (20, 21).
DISCUSSION
We identified 21 VACV proteins that collectively comprised the human antibody response to both Dryvax and ACAM2000 vaccines. We also noted subtle differences in vaccine responses, as antibody results from most (70%) Dryvax-vaccinated individuals grouped independently from antibody results from the ACAM2000-vaccinated individuals. The VACV proteins A13L and G5R appeared to elicit a higher antibody response in ACAM2000-vaccinated individuals, whereas antibody responses to J6R, B19R, A38L, A26L, I1L, I3L, D8L, C3L, and A10L were higher in Dryvax-vaccinated individuals. Because Dryvax consists of a heterogeneous mixture of VACV clones, we examined the possibility that the differences in antibody recognition may be due to protein variations between vaccine strains. We noted that the 11 proteins that distinguished antibody responses between vaccines were less conserved than antigens that were common to both ACAM2000 and Dryvax. For example, A13L, A26L, and C3L differed in sequence length, the IFN-α/β receptor protein B19R is missing or truncated in several VACV clones, and amino acid sequences for all 11 proteins varied by 1 to 88 amino acid residues. Surprisingly, the longest protein, J6R (1,276 residues), was highly conserved (ΔH = 0.001) compared to the shortest protein, A13L (68 to 70 residues; ΔH = 0.068). Perhaps mutations are less tolerated for the essential enzyme J6R, a DNA-directed RNA polymerase subunit, whereas mutations may be more advantageous for the IMV surface protein A13L. It should also be noted that an antibody neutralization epitope was mapped to amino acid residues 59 to 69 of A13L, a region that is conserved in ACAM2000 and Dryvax (36). Although the precise relationship between antibody responses to the VACV proteins we identified and smallpox immunity will require extensive study, our results suggest that variations in protein sequences may contribute to differences in antibody responses to the smallpox vaccine strains.
In general, the efficacy of smallpox vaccines relies on immune responses to multiple VACV proteins, rather than a single immunodominant target (37). The VACV antigens that we identified included viral surface proteins (IMV, EEV), as well as secreted and intracellular proteins. In animal studies, a combination of antigens from both IMV and EEV infectious virion forms were required for complete protection following VACV challenge (38, 39). Six of the VACV antigens (D8L, A13L, A33R, H3L, A27L, and B5R) are IMV or EEV surface proteins that we identified as common components of the antibody response to Dryvax and ACAM2000. These six antigens were previously shown to be targets of neutralizing antibodies or were critical for protective immunity against a VACV infection (36, 38–44). Further, for VACV antigens such as B5R and A33R, antibody interactions required complement activation to neutralize viral infection (45). In contrast to IMV and EEV surface proteins, secreted and intracellular VACV proteins can have an indirect role in protective immunity. For the case of C3L, this secreted VACV protein was shown to enhance pathogenesis of VACV and monkeypoxvirus infections by affecting complement activation (46–48). As an example of an intracellular antigen recognized by vaccine antibody responses observed in our study, the poxvirus protein O1L enhances virulence by continuous activation of the extracellular signal-regulated kinase (ERK) pathway, which may promote viral replication and dissemination (49).
While the VACV antigen-antibody interactions that we identified were common to the study population as a group, immune responses will vary from person to person. The prevalence of significant antibody responses to any given antigen ranged from 25 to 99% in our study. We found that human antibody responses to VACV involved <10% of the total viral proteome in our previous study (23) and that eight individual vaccinia virus proteins were useful biomarkers of smallpox immunity following vaccination with Dryvax. Using an expanded protein microarray to obtain the results presented here, we confirmed that <10% (21 of 261 predicted proteins and 245 tested proteins) of the total viral proteome is recognized by human antibodies following vaccination, while reporting antibody recognition of additional VACV antigens (Table 1). Another published report (50) included four additional proteins (WR148, A17L, A4L, and WR169) as antigens recognized by polyclonal smallpox vaccine and ACAM2000 vaccine antibody responses. It should be noted that the purified proteins we used were expressed in eukaryotic cells, whereas the previous report (50) used nonpurified proteins produced in an Escherichia coli expression system. The VACV Western Reserve (WR) antigens were not included on our protein microarray. Further, antibody recognition of A17L was found to be marginally significant in our study (53% in all vaccinated individuals; P = 0.015) but below our significance criteria (P ≤ 0.0002), and we observed antibody binding to A4L in sera from both nonvaccinated and vaccinated individuals.
By measuring common deletion alleles of Dryvax clones isolated from a specific vaccine lot, Qin et al. (8) determined that ACAM2000-like viruses were the dominant form (∼60%), approximately 40% were VACV-DUKE-like viruses, and less than 1% were similar to clones CL3 and DPP17 (includes a large 11.7-kbp deletion, including B19R). Therefore, in contrast to ACAM2000, selective replication of VACV clones from the polyclonal Dryvax vaccine at the site of skin inoculation may be a source of variability in individual antibody responses due to differences in protein sequences or protein abundance (29). For example, a Dryvax variant (VACV-DUKE) was isolated from a vaccinated patient who developed vaccinia necrosum (35). VACV-DUKE was similar to the slightly more virulent CL3 Dryvax clone in that it had a nearly full-length (351 residues) IFN-α/β receptor gene (B19R in COP; B18R in WR), whereas this protein is truncated by ∼90 amino acid residues in the ACAM2000 strain due to a 4-kb DNA deletion (7, 8). A C-terminal deletion of B19R, as is the case in ACAM2000, caused a decrease in IFN-α/β binding affinity due to the elimination of the third immunoglobulin domain, while complete deletion of B19R in the VACV-WR strain resulted in severe attenuation of viral infection from intranasal challenge in mice (51). It is likely that in vivo selection of clones that alter innate immunity by activities of B19R will also impact antibody responses.
The results of our study with a comprehensive VACV protein microarray (245 recombinant proteins) confirmed previously reported vaccine antigens and also identified novel antibody-binding proteins that could be important biomarkers of vaccinia immunity. We identified 21 VACV proteins with significant antibody binding from sera of all vaccinated individuals, while 11 proteins distinguished vaccination with Dryvax from the monoclonal replacement ACAM2000. The precise relationship between smallpox immunity and the VACV proteins we identified as targets of antibody responses needs to be established. The antibody response biomarkers described here may provide useful information for improving smallpox vaccination, especially for those individuals with contraindications to vaccination with live, replicating VACV.
Supplementary Material
ACKNOWLEDGMENTS
This research was supported in part by an appointment of C.P. and S.K. to the Student Research Participation Program at the U.S. Army Medical Research Institute of Infectious Disease administered by Oak Ridge Institute for Science and Education through an interagency agreement between the U.S. Department of Energy and USAMRMC. This clinical study was sponsored by the Military Vaccine Agency, Office of the Surgeon General, United States Army.
Opinions, interpretations, conclusions, and recommendations are those of the authors and are not necessarily endorsed by the U.S. Army or the U.S. government.
Footnotes
Published ahead of print 23 April 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/CVI.00035-14.
REFERENCES
- 1.Alibek K. 2004. Smallpox: a disease and a weapon. Int. J. Infect. Dis. 8(Suppl 2):S3–S8 [DOI] [PubMed] [Google Scholar]
- 2.Arita I. 2005. Smallpox vaccine and its stockpile in 2005. Lancet Infect. Dis. 5:647–652. 10.1016/S1473-3099(05)70242-5 [DOI] [PubMed] [Google Scholar]
- 3.Henderson DA, Inglesby TV, Bartlett JG, Ascher MS, Eitzen E, Jahrling PB, Hauer J, Layton M, McDade J, Osterholm MT, O'Toole T, Parker G, Perl T, Russell PK, Tonat K. 1999. Smallpox as a biological weapon: medical and public health management. Working Group on Civilian Biodefense. JAMA 281:2127–2137 [DOI] [PubMed] [Google Scholar]
- 4.Whitley RJ. 2003. Smallpox: a potential agent of bioterrorism. Antiviral Res. 57:7–12. 10.1016/S0166-3542(02)00195-X [DOI] [PubMed] [Google Scholar]
- 5.Fenner F, Henderson DA, Arita I, Jezek Z, Ladnyi ID. 1988. Smallpox and its eradicationM, p 277–314 World Health Organization, Geneva, Switzerland: http://www2a.cdc.gov/nip/isd/spoxclincian/contents/references/sp_eradication_toc.pdf [Google Scholar]
- 6.Monath TP, Caldwell JR, Mundt W, Fusco J, Johnson CS, Buller M, Liu J, Gardner B, Downing G, Blum PS, Kemp T, Nichols R, Weltzin R. 2004. ACAM2000 clonal Vero cell culture vaccinia virus (New York City Board of Health strain)–a second-generation smallpox vaccine for biological defense. Int. J. Infect. Dis. 8(Suppl 2):S31–S44. 10.1016/j.ijid.2004.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Osborne JD, Da Silva M, Frace AM, Sammons SA, Olsen-Rasmussen M, Upton C, Buller RM, Chen N, Feng Z, Roper RL, Liu J, Pougatcheva S, Chen W, Wohlhueter RM, Esposito JJ. 2007. Genomic differences of vaccinia virus clones from Dryvax smallpox vaccine: the Dryvax-like ACAM2000 and the mouse neurovirulent clone-3. Vaccine 25:8807–8832. 10.1016/j.vaccine.2007.10.040 [DOI] [PubMed] [Google Scholar]
- 8.Qin L, Upton C, Hazes B, Evans DH. 2011. Genomic analysis of the vaccinia virus strain variants found in Dryvax vaccine. J. Virol. 85:13049–13060. 10.1128/JVI.05779-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weltzin R, Liu J, Pugachev KV, Myers GA, Coughlin B, Blum PS, Nichols R, Johnson C, Cruz J, Kennedy JS, Ennis FA, Monath TP. 2003. Clonal vaccinia virus grown in cell culture as a new smallpox vaccine. Nat. Med. 9:1125–1130. 10.1038/nm916 [DOI] [PubMed] [Google Scholar]
- 10.Aragon TJ, Ulrich S, Fernyak S, Rutherford GW. 2003. Risks of serious complications and death from smallpox vaccination: a systematic review of the United States experience, 1963–1968. BMC Public Health 3:26. 10.1186/1471-2458-3-26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Centers for Disease Control and Prevention. 2003. Smallpox (vaccinia) vaccine contraindications. Centers for Disease Control and Prevention, Atlanta, GA: http://www.bt.cdc.gov/agent/smallpox/vaccination/contraindications-clinic.asp [Google Scholar]
- 12.Copeman PW, Wallace HJ. 1964. Eczema vaccinatum. Br. Med. J. 2:906–908. 10.1136/bmj.2.5414.906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Engler RJ, Kenner J, Leung DY. 2002. Smallpox vaccination: risk considerations for patients with atopic dermatitis. J. Allergy Clin. Immunol. 110:357–365. 10.1067/mai.2002.128052 [DOI] [PubMed] [Google Scholar]
- 14.Entwistle DM, Bray PT, Laurence KM. 1962. Prenatal infection with vaccinia virus: report of a case. Br. Med. J. 2:238–239. 10.1136/bmj.2.5299.238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Freed ER, Duma RJ, Escobar MR. 1972. Vaccinia necrosum and its relationship to impaired immunologic responsiveness. Am. J. Med. 52:411–420. 10.1016/0002-9343(72)90031-9 [DOI] [PubMed] [Google Scholar]
- 16.Greenberg M. 1948. Complications of vaccination against smallpox. Am. J. Dis. Child. 76:492–502 [DOI] [PubMed] [Google Scholar]
- 17.Halsell JS, Riddle JR, Atwood JE, Gardner P, Shope R, Poland GA, Gray GC, Ostroff S, Eckart RE, Hospenthal DR, Gibson RL, Grabenstein JD, Arness MK, Tornberg DN, Department of Defense Smallpox Vaccination Clinical Evaluation Team 2003. Myopericarditis following smallpox vaccination among vaccinia-naive US military personnel. JAMA 289:3283–3289. 10.1001/jama.289.24.3283 [DOI] [PubMed] [Google Scholar]
- 18.Lane JM, Ruben FL, Neff JM, Millar JD. 1969. Complications of smallpox vaccination, 1968. N. Engl. J. Med. 281:1201–1208. 10.1056/NEJM196911272812201 [DOI] [PubMed] [Google Scholar]
- 19.Lane JM, Ruben FL, Neff JM, Millar JD. 1970. Complications of smallpox vaccination, 1968: results of ten statewide surveys. J. Infect. Dis. 122:303–309. 10.1093/infdis/122.4.303 [DOI] [PubMed] [Google Scholar]
- 20.Levine MM. 1974. Live-virus vaccines in pregnancy. Risks and recommendations. Lancet ii:34–38 [DOI] [PubMed] [Google Scholar]
- 21.Midgley CM, Putz MM, Weber JN, Smith GL. 2008. Vaccinia virus strain NYVAC induces substantially lower and qualitatively different human antibody responses compared with strains Lister and Dryvax. J. Gen. Virol. 89:2992–2997. 10.1099/vir.0.2008/004440-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Putz MM, Midgley CM, Law M, Smith GL. 2006. Quantification of antibody responses against multiple antigens of the two infectious forms of vaccinia virus provides a benchmark for smallpox vaccination. Nat. Med. 12:1310–1315. 10.1038/nm1457 [DOI] [PubMed] [Google Scholar]
- 23.Schmid K, Keasey SL, Pittman P, Emerson GL, Meegan J, Tikhonov AP, Chen G, Schweitzer B, Ulrich RG. 2008. Analysis of the human immune response to vaccinia by use of a novel protein microarray suggests that antibodies recognize less than 10% of the total viral proteome. Proteomics Clin. Appl. 2:1528–1538. 10.1002/prca.200780113 [DOI] [PubMed] [Google Scholar]
- 24.Tartaglia J, Perkus ME, Taylor J, Norton EK, Audonnet JC, Cox WI, Davis SW, van der Hoeven J, Meignier B, Riviere M, Languet B, Paoletti E. 1992. NYVAC: a highly attenuated strain of vaccinia virus. Virology 188:217–232. 10.1016/0042-6822(92)90752-B [DOI] [PubMed] [Google Scholar]
- 25.U.S. Food and Drug Administration. 2009. ACAM2000 package insert. U.S. Food and Drug Administration, Rockville, MD: http://www.fda.gov/BiologicsBloodVaccines/Vaccines/ApprovedProducts/ucm180810.htm [Google Scholar]
- 26.Artenstein AW, Johnson C, Marbury TC, Morrison D, Blum PS, Kemp T, Nichols R, Balser JP, Currie M, Monath TP. 2005. A novel, cell culture-derived smallpox vaccine in vaccinia-naive adults. Vaccine 23:3301–3309. 10.1016/j.vaccine.2005.01.079 [DOI] [PubMed] [Google Scholar]
- 27.Frey SE, Newman FK, Kennedy JS, Ennis F, Abate G, Hoft DF, Monath TP. 2009. Comparison of the safety and immunogenicity of ACAM1000, ACAM2000 and Dryvax in healthy vaccinia-naive adults. Vaccine 27:1637–1644. 10.1016/j.vaccine.2008.11.079 [DOI] [PubMed] [Google Scholar]
- 28.Frey SE, Couch RB, Tacket CO, Treanor JJ, Wolff M, Newman FK, Atmar RL, Edelman R, Nolan CM, Belshe RB, National Institute of Allergy and Infectious Diseases Smallpox Vaccine Study Group 2002. Clinical responses to undiluted and diluted smallpox vaccine. N. Engl. J. Med. 346:1265–1274. 10.1056/NEJMoa020534 [DOI] [PubMed] [Google Scholar]
- 29.Keasey S, Pugh C, Tikhonov A, Chen G, Schweitzer B, Nalca A, Ulrich RG. 2010. Proteomic basis of the antibody response to monkeypox virus infection examined in cynomolgus macaques and a comparison to human smallpox vaccination. PLoS One 5:e15547. 10.1371/journal.pone.0015547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hammarlund E, Lewis MW, Hanifin JM, Simpson EL, Carlson NE, Slifka MK. 2008. Traditional smallpox vaccination with reduced risk of inadvertent contact spread by administration of povidone iodine ointment. Vaccine 26:430–439. 10.1016/j.vaccine.2007.10.070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, Braisted J, Klapa M, Currier T, Thiagarajan M, Sturn A, Snuffin M, Rezantsev A, Popov D, Ryltsov A, Kostukovich E, Borisovsky I, Liu Z, Vinsavich A, Trush V, Quackenbush J. 2003. TM4: a free, open-source system for microarray data management and analysis. Biotechniques 34:374–378 [DOI] [PubMed] [Google Scholar]
- 32.Ehlers A, Osborne J, Slack S, Roper RL, Upton C. 2002. Poxvirus orthologous clusters (POCs). Bioinformatics 18:1544–1545. 10.1093/bioinformatics/18.11.1544 [DOI] [PubMed] [Google Scholar]
- 33.Upton C, Slack S, Hunter AL, Ehlers A, Roper RL. 2003. Poxvirus orthologous clusters: toward defining the minimum essential poxvirus genome. J. Virol. 77:7590–7600. 10.1128/JVI.77.13.7590-7600.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Castresana J. 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 17:540–552. 10.1093/oxfordjournals.molbev.a026334 [DOI] [PubMed] [Google Scholar]
- 35.Li G, Chen N, Feng Z, Buller RM, Osborne J, Harms T, Damon I, Upton C, Esteban DJ. 2006. Genomic sequence and analysis of a vaccinia virus isolate from a patient with a smallpox vaccine-related complication. Virol. J. 3:88. 10.1186/1743-422X-3-88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu C, Meng X, Yan B, Crotty S, Deng J, Xiang Y. 2011. An epitope conserved in orthopoxvirus A13 envelope protein is the target of neutralizing and protective antibodies. Virology 418:67–73. 10.1016/j.virol.2011.06.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kennedy RB, Ovsyannikova IG, Jacobson RM, Poland GA. 2009. The immunology of smallpox vaccines. Curr. Opin. Immunol. 21:314–320. 10.1016/j.coi.2009.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fogg C, Lustig S, Whitbeck JC, Eisenberg RJ, Cohen GH, Moss B. 2004. Protective immunity to vaccinia virus induced by vaccination with multiple recombinant outer membrane proteins of intracellular and extracellular virions. J. Virol. 78:10230–10237. 10.1128/JVI.78.19.10230-10237.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lustig S, Fogg C, Whitbeck JC, Eisenberg RJ, Cohen GH, Moss B. 2005. Combinations of polyclonal or monoclonal antibodies to proteins of the outer membranes of the two infectious forms of vaccinia virus protect mice against a lethal respiratory challenge. J. Virol. 79:13454–13462. 10.1128/JVI.79.21.13454-13462.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Benhnia MR, McCausland MM, Laudenslager J, Granger SW, Rickert S, Koriazova L, Tahara T, Kubo RT, Kato S, Crotty S. 2009. Heavily isotype-dependent protective activities of human antibodies against vaccinia virus extracellular virion antigen B5. J. Virol. 83:12355–12367. 10.1128/JVI.01593-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Benhnia MR, McCausland MM, Su HP, Singh K, Hoffmann J, Davies DH, Felgner PL, Head S, Sette A, Garboczi DN, Crotty S. 2008. Redundancy and plasticity of neutralizing antibody responses are cornerstone attributes of the human immune response to the smallpox vaccine. J. Virol. 82:3751–3768. 10.1128/JVI.02244-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Davies DH, McCausland MM, Valdez C, Huynh D, Hernandez JE, Mu Y, Hirst S, Villarreal L, Felgner PL, Crotty S. 2005. Vaccinia virus H3L envelope protein is a major target of neutralizing antibodies in humans and elicits protection against lethal challenge in mice. J. Virol. 79:11724–11733. 10.1128/JVI.79.18.11724-11733.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moss B. 2011. Smallpox vaccines: targets of protective immunity. Immunol. Rev. 239:8–26. 10.1111/j.1600-065X.2010.00975.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rudraraju R, Ramsay AJ. 2010. Single-shot immunization with recombinant adenovirus encoding vaccinia virus glycoprotein A27L is protective against a virulent respiratory poxvirus infection. Vaccine 28:4997–5004. 10.1016/j.vaccine.2010.05.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Benhnia MR, Maybeno M, Blum D, Aguilar-Sino R, Matho M, Meng X, Head S, Felgner PL, Zajonc DM, Koriazova L, Kato S, Burton DR, Xiang Y, Crowe JE, Jr, Peters B, Crotty S. 2013. Unusual features of vaccinia virus extracellular virion form neutralization resistance revealed in human antibody responses to the smallpox vaccine. J. Virol. 87:1569–1585. 10.1128/JVI.02152-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Girgis NM, Dehaven BC, Xiao Y, Alexander E, Viner KM, Isaacs SN. 2011. The vaccinia virus complement control protein modulates adaptive immune responses during infection. J. Virol. 85:2547–2556. 10.1128/JVI.01474-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hudson PN, Self J, Weiss S, Braden Z, Xiao Y, Girgis NM, Emerson G, Hughes C, Sammons SA, Isaacs SN, Damon IK, Olson VA. 2012. Elucidating the role of the complement control protein in monkeypox pathogenicity. PLoS One 7:e35086. 10.1371/journal.pone.0035086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kotwal GJ, Isaacs SN, McKenzie R, Frank MM, Moss B. 1990. Inhibition of the complement cascade by the major secretory protein of vaccinia virus. Science 250:827–830. 10.1126/science.2237434 [DOI] [PubMed] [Google Scholar]
- 49.Schweneker M, Lukassen S, Spath M, Wolferstatter M, Babel E, Brinkmann K, Wielert U, Chaplin P, Suter M, Hausmann J. 2012. The vaccinia virus O1 protein is required for sustained activation of extracellular signal-regulated kinase 1/2 and promotes viral virulence. J. Virol. 86:2323–2336. 10.1128/JVI.06166-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Townsend MB, Keckler MS, Patel N, Davies DH, Felgner P, Damon IK, Karem KL. 2013. Humoral immunity to smallpox vaccines and monkeypox virus challenge: proteomic assessment and clinical correlations. J. Virol. 87:900–911. 10.1128/JVI.02089-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Alcami A, Symons JA, Smith GL. 2000. The vaccinia virus soluble alpha/beta interferon (IFN) receptor binds to the cell surface and protects cells from the antiviral effects of IFN. J. Virol. 74:11230–11239. 10.1128/JVI.74.23.11230-11239.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Blasco R, Moss B. 1991. Extracellular vaccinia virus formation and cell-to-cell virus transmission are prevented by deletion of the gene encoding the 37,000-dalton outer envelope protein. J. Virol. 65:5910–5920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Husain M, Moss B. 2002. Similarities in the induction of post-Golgi vesicles by the vaccinia virus F13L protein and phospholipase D. J. Virol. 76:7777–7789. 10.1128/JVI.76.15.7777-7789.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Resch W, Hixson KK, Moore RJ, Lipton MS, Moss B. 2007. Protein composition of the vaccinia virus mature virion. Virology 358:233–247. 10.1016/j.virol.2006.08.025 [DOI] [PubMed] [Google Scholar]
- 55.Yoder JD, Chen TS, Gagnier CR, Vemulapalli S, Maier CS, Hruby DE. 2006. Pox proteomics: mass spectrometry analysis and identification of vaccinia virion proteins. Virol. J. 3:10. 10.1186/1743-422X-3-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chung CS, Chen CH, Ho MY, Huang CY, Liao CL, Chang W. 2006. Vaccinia virus proteome: identification of proteins in vaccinia virus intracellular mature virion particles. J. Virol. 80:2127–2140. 10.1128/JVI.80.5.2127-2140.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Heljasvaara R, Rodriguez D, Risco C, Carrascosa JL, Esteban M, Rodriguez JR. 2001. The major core protein P4a (A10L gene) of vaccinia virus is essential for correct assembly of viral DNA into the nucleoprotein complex to form immature viral particles. J. Virol. 75:5778–5795. 10.1128/JVI.75.13.5778-5795.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jensen ON, Houthaeve T, Shevchenko A, Cudmore S, Ashford T, Mann M, Griffiths G, Krijnse Locker J. 1996. Identification of the major membrane and core proteins of vaccinia virus by two-dimensional electrophoresis. J. Virol. 70:7485–7497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mohandas AR, Dales S. 1995. Involvement of spicules in the formation of vaccinia virus envelopes elucidated by a conditional lethal mutant. Virology 214:494–502. 10.1006/viro.1995.0060 [DOI] [PubMed] [Google Scholar]
- 60.Zhang Y, Moss B. 1992. Immature viral envelope formation is interrupted at the same stage by lac operator-mediated repression of the vaccinia virus D13L gene and by the drug rifampicin. Virology 187:643–653. 10.1016/0042-6822(92)90467-4 [DOI] [PubMed] [Google Scholar]
- 61.Hsiao JC, Chung CS, Chang W. 1999. Vaccinia virus envelope D8L protein binds to cell surface chondroitin sulfate and mediates the adsorption of intracellular mature virions to cells. J. Virol. 73:8750–8761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.DeHaven BC, Girgis NM, Xiao Y, Hudson PN, Olson VA, Damon IK, Isaacs SN. 2010. Poxvirus complement control proteins are expressed on the cell surface through an intermolecular disulfide bridge with the viral A56 protein. J. Virol. 84:11245–11254. 10.1128/JVI.00372-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.McKenzie R, Kotwal GJ, Moss B, Hammer CH, Frank MM. 1992. Regulation of complement activity by vaccinia virus complement-control protein. J. Infect. Dis. 166:1245–1250. 10.1093/infdis/166.6.1245 [DOI] [PubMed] [Google Scholar]
- 64.Unger B, Traktman P. 2004. Vaccinia virus morphogenesis: A13 phosphoprotein is required for assembly of mature virions. J. Virol. 78:8885–8901. 10.1128/JVI.78.16.8885-8901.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Roper RL, Payne LG, Moss B. 1996. Extracellular vaccinia virus envelope glycoprotein encoded by the A33R gene. J. Virol. 70:3753–3762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wolffe EJ, Weisberg AS, Moss B. 2001. The vaccinia virus A33R protein provides a chaperone function for viral membrane localization and tyrosine phosphorylation of the A36R protein. J. Virol. 75:303–310. 10.1128/JVI.75.1.303-310.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Resch W, Weisberg AS, Moss B. 2005. Vaccinia virus nonstructural protein encoded by the A11R gene is required for formation of the virion membrane. J. Virol. 79:6598–6609. 10.1128/JVI.79.11.6598-6609.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Knutson BA, Liu X, Oh J, Broyles SS. 2006. Vaccinia virus intermediate and late promoter elements are targeted by the TATA-binding protein. J. Virol. 80:6784–6793. 10.1128/JVI.02705-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.da Fonseca FG, Wolffe EJ, Weisberg A, Moss B. 2000. Effects of deletion or stringent repression of the H3L envelope gene on vaccinia virus replication. J. Virol. 74:7518–7528. 10.1128/JVI.74.16.7518-7528.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lin CL, Chung CS, Heine HG, Chang W. 2000. Vaccinia virus envelope H3L protein binds to cell surface heparan sulfate and is important for intracellular mature virion morphogenesis and virus infection in vitro and in vivo. J. Virol. 74:3353–3365. 10.1128/JVI.74.7.3353-3365.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chung CS, Hsiao JC, Chang YS, Chang W. 1998. A27L protein mediates vaccinia virus interaction with cell surface heparan sulfate. J. Virol. 72:1577–1585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sanderson CM, Hollinshead M, Smith GL. 2000. The vaccinia virus A27L protein is needed for the microtubule-dependent transport of intracellular mature virus particles. J. Gen. Virol. 81:47–58 [DOI] [PubMed] [Google Scholar]
- 73.Engelstad M, Howard ST, Smith GL. 1992. A constitutively expressed vaccinia gene encodes a 42-kDa glycoprotein related to complement control factors that forms part of the extracellular virus envelope. Virology 188:801–810. 10.1016/0042-6822(92)90535-W [DOI] [PubMed] [Google Scholar]
- 74.Herrera E, Lorenzo MM, Blasco R, Isaacs SN. 1998. Functional analysis of vaccinia virus B5R protein: essential role in virus envelopment is independent of a large portion of the extracellular domain. J. Virol. 72:294–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mathew E, Sanderson CM, Hollinshead M, Smith GL. 1998. The extracellular domain of vaccinia virus protein B5R affects plaque phenotype, extracellular enveloped virus release, and intracellular actin tail formation. J. Virol. 72:2429–2438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Parkinson JE, Sanderson CM, Smith GL. 1995. The vaccinia virus A38L gene product is a 33-kDa integral membrane glycoprotein. Virology 214:177–188. 10.1006/viro.1995.9942 [DOI] [PubMed] [Google Scholar]
- 77.Sanderson CM, Parkinson JE, Hollinshead M, Smith GL. 1996. Overexpression of the vaccinia virus A38L integral membrane protein promotes Ca2+ influx into infected cells. J. Virol. 70:905–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.da Fonseca FG, Weisberg AS, Caeiro MF, Moss B. 2004. Vaccinia virus mutants with alanine substitutions in the conserved G5R gene fail to initiate morphogenesis at the nonpermissive temperature. J. Virol. 78:10238–10248. 10.1128/JVI.78.19.10238-10248.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Senkevich TG, Koonin EV, Moss B. 2009. Predicted poxvirus FEN1-like nuclease required for homologous recombination, double-strand break repair and full-size genome formation. Proc. Natl. Acad. Sci. U. S. A. 106:17921–17926. 10.1073/pnas.0909529106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chiu WL, Lin CL, Yang MH, Tzou DL, Chang W. 2007. Vaccinia virus 4c (A26L) protein on intracellular mature virus binds to the extracellular cellular matrix laminin. J. Virol. 81:2149–2157. 10.1128/JVI.02302-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Rochester SC, Traktman P. 1998. Characterization of the single-stranded DNA binding protein encoded by the vaccinia virus I3 gene. J. Virol. 72:2917–2926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Welsch S, Doglio L, Schleich S, Krijnse Locker J. 2003. The vaccinia virus I3L gene product is localized to a complex endoplasmic reticulum-associated structure that contains the viral parental DNA. J. Virol. 77:6014–6028. 10.1128/JVI.77.10.6014-6028.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Broyles SS, Moss B. 1986. Homology between RNA polymerases of poxviruses, prokaryotes, and eukaryotes: nucleotide sequence and transcriptional analysis of vaccinia virus genes encoding 147-kDa and 22-kDa subunits. Proc. Natl. Acad. Sci. U. S. A. 83:3141–3145. 10.1073/pnas.83.10.3141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hooda-Dhingra U, Thompson CL, Condit RC. 1989. Detailed phenotypic characterization of five temperature-sensitive mutants in the 22- and 147-kilodalton subunits of vaccinia virus DNA-dependent RNA polymerase. J. Virol. 63:714–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bayliss CD, Smith GL. 1997. Vaccinia virion protein VP8, the 25 kDa product of the L4R gene, binds single-stranded DNA and RNA with similar affinity. Nucleic Acids Res. 25:3984–3990. 10.1093/nar/25.20.3984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yang WP, Bauer WR. 1988. Purification and characterization of vaccinia virus structural protein VP8. Virology 167:578–584. 10.1016/0042-6822(88)90120-1 [DOI] [PubMed] [Google Scholar]
- 87.Colamonici OR, Domanski P, Sweitzer SM, Larner A, Buller RM. 1995. Vaccinia virus B18R gene encodes a type I interferon-binding protein that blocks interferon alpha transmembrane signaling. J. Biol. Chem. 270:15974–15978. 10.1074/jbc.270.27.15974 [DOI] [PubMed] [Google Scholar]
- 88.Ueda Y, Morikawa S, Matsuura Y. 1990. Identification and nucleotide sequence of the gene encoding a surface antigen induced by vaccinia virus. Virology 177:588–594. 10.1016/0042-6822(90)90524-U [DOI] [PubMed] [Google Scholar]
- 89.Manes NP, Estep RD, Mottaz HM, Moore RJ, Clauss TR, Monroe ME, Du X, Adkins JN, Wong SW, Smith RD. 2008. Comparative proteomics of human monkeypox and vaccinia intracellular mature and extracellular enveloped virions. J. Proteome Res. 7:960–968. 10.1021/pr070432+ [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.