Abstract
The relative positions that genes occupy on their respective chromosomes can play a critical role in determining how they are regulated at the transcriptional level. For example, a significant fraction of the genes from a variety of coregulated gene sets, including the ribosomal protein (RP) and the rRNA and ribosome biogenesis (RRB) regulons, exist as immediate, adjacent gene pairs. These gene pairs occur in all possible divergent, tandem, and convergent orientations. Adjacent-gene pairing in these regulons is associated with a tighter transcriptional coregulation than is observed for nonpaired genes of the same regulons. In order to define the cis and trans factors that regulate adjacent-gene coregulation (AGC), we conducted a mutational analysis of the convergently oriented RRB gene pair MPP10-YJR003C. We observed that coupled corepression of the gene pair under heat shock was abrogated when the two genes were separated by an actively expressed RNA polymerase (Pol) II transcription unit (the LEU2 gene) but not when the inserted LEU2 gene was repressed. In contrast, the insertion of an RNA Pol III-transcribed tRNA (Thr) gene did not disrupt the coregulated repression of MPP10 and YJR003C. A targeted screen of mutants defective in regulating chromosome architecture revealed that the Spt20, Snf2, and Chd1 proteins were required for coupling the repression of YJR003C to that of MPP10. Nucleosome occupancy assays performed across the MPP10 and YJR003C promoter regions revealed that the mechanism of corepression of the gene pair was not related to the repositioning of nucleosomes across the respective gene promoters.
INTRODUCTION
One of the essential regulatory challenges that all cells face is the need to maintain an internal homeostasis as well as to react appropriately to changing external environments. Cells monitor their surroundings for a wide range of factors, including nutritional levels, temperature, and osmolarity changes, and they respond to changing conditions by regulating the activity of appropriate metabolic pathways. Given that these metabolic pathways can encompass hundreds of gene products, cells require mechanisms to quickly, effectively shift their use of large classes of genes simultaneously. Often this level of control is affected at the level of transcription, and depending on the particular environmental cue, a cell may either activate or repress hundreds to thousands of genes simultaneously (1, 2). Transcriptional reprogramming can be accomplished on many levels, including through the regulation of transcription factors, through nucleosome modifications, through chromatin remodeling, and even through changing the subnuclear localization of specific genes (3, 4, 5, 6, 7).
Among the more complex, important, and tightly regulated metabolic pathways are those that are involved in the production of ribosomes. Ribosome biogenesis can represent a substantial fraction of the total cellular economy, and it depends on the coordinated activity of hundreds of genes (8). For example, in budding yeast, the proper synthesis and assembly of the 79 ribosomal proteins (RPs) and the 4 highly processed rRNAs that constitute a ribosome involves 137 RP genes, some 150 rRNA gene repeats, as well as over 200 rRNA and ribosome biogenesis (RRB, or ribi) genes (9, 10). All three of these large sets of genes are tightly coregulated under changing conditions, including activation by nutrient replenishment and repression by temperature or osmolarity shocks. Since an actively dividing yeast cell needs to produce roughly 2,000 of the 3.6-MDa ribosomes per minute, it is an important and nontrivial task to effectively regulate the overall synthesis and relative stoichiometry of the numerous ribosome biogenesis factors (8).
Genome-wide expression analysis in S. cerevisiae has revealed that the expression profiles of hundreds of ribosome biogenesis-related genes are rapidly altered in response to stressors or nutrient availability (1, 11). This response is mediated in part by highly conserved signal transduction pathways that ultimately control the expression levels of the RRB, RP, and rRNA gene sets. The TORC1 and ras/PKA/cyclic AMP pathways converge on the Sch9p kinase and Sfp1 that targets the rRNA and ribosome biosynthesis (RRB) transcriptional repressors Stb3p, Dot6p, and Tod6p (12, 13, 14, 15). These proteins in turn recruit the Rpd3L histone deacetylase complex to the promoters of RRB genes and mediate the budding yeast stress response (16). TOR kinase signaling also regulates the activity of the RP promoter binding transcription factor Fhl1 through controlling the subcellular location of the corepressor Crf1 (17). Furthermore, TOR also regulates the transcription of the rRNA genes by RNA polymerase (Pol) I through the displacement of the Rrn3 transcription factor from ribosomal DNA (rDNA) promoters (18, 19). Altogether, these pathways ultimately target a large fraction of the genome (20, 21).
We previously reported that there is an additional level of transcriptional control for the members of the RP and RRB regulons that depends on the relative positions of the genes on the chromosomes. A significant fraction of the RP and RRB genes exist as immediately adjacent gene pairs, and this arrangement results in a tighter transcriptional coordination than those of genes within the same regulons that are not paired (10, 22). This phenomenon of adjacent-gene pairing extends to other large coregulated gene sets, including those related to DNA damage response, carbohydrate metabolism, nitrogen metabolism, and heat shock response (22). Importantly, more than half of the adjacent gene pairs are found in tandem or convergent orientations, suggesting that their coregulation is not simply a consequence of bidirectional promoters, like that described for the GAL1-GAL10 genes (23). Whereas it has long been appreciated that adjacent genes are more closely coregulated than nonadjacent genes, the precise mechanisms by which it is achieved had not been elucidated (24). The coupled regulation of adjacent genes has been proposed to be significant on a genomic scale, including in the so-called neighboring gene effect, where deletions in one gene yield phenotypes associated with the disruption of its neighbor (25).
In this study, we have taken a mutational approach to identify both cis and trans factors that regulate the coordinated repression of the convergently oriented MMP10-YJR003C RRB gene pair following a heat shock. We have found that the coupled coregulation of the MPP10 and YJR003C genes depends less on their relative positions than it does on whether or not they are separated by an actively expressed RNA Pol II transcription unit. Furthermore, the coupled repression of YJR003C depends upon the activity of Chd1, the SWI/SNF complex member Snf2, and the SAGA complex member Spt20. While these trans factors include chromatin remodelers, we did not observe a correlation between transcriptional repression and changes in the nucleosome occupancy profiles at the MPP10 or YJR003C promoter.
MATERIALS AND METHODS
Yeast strains.
A complete list of all strains used in this study, as well as their relevant genotypes, is included in Table 1. Strain YMM13 (MATa leu2Δ1trp1Δ63 ura3-52) was used as a wild type and is the parent strain used to generate the various mutants. The insertions in the intergenic region of MPP10 and YJR003C were generated using the two-step, delitto perfetto method (28), targeting the integration of the LEU2 gene in either orientation between MPP10 and YJR003C. A complete list of the oligonucleotide primers used in this study is provided in Table 2. The primers are named according to their targeted gene, the strand and position that they anneal to (W or C), and whether they were used for mRNA expression studies (quantitative reverse transcription [qRT]) or nucleosome-scanning assay (NSA).
TABLE 1.
Yeast strains used in this study
| Strain | Genotype | Source or reference |
|---|---|---|
| YMM13 | MATa leu2Δ1 trp1Δ63 ura3-52 | 10, 26 |
| YMM514 | MATa leu2Δ1 trp1Δ63 ura3-52 (ΔPACΔRRPE)MPP10 | 27 |
| YMM554 | MATa leu2Δ1 trp1Δ63 ura3-52 MPP10.LEU2 | This study |
| YMM555 | MATa leu2Δ1 trp1Δ63 ura3-52 MPP10.tRNA-Thr | This study |
| YMM556 | MATa leu2Δ1 trp1Δ63 ura3-5 MPP10.Ty1.tRNA-Thr | This study |
| YMM559 | MATa leu2Δ1 trp1Δ63 ura-52 MPP10.LEU2 | This study |
| YMM557 | MATa leu2Δ0 met15Δ0 ura3Δ0 yjr003cΔ::KANr | Open Biosystems |
| YMM593 | MATa leu2Δ0 met15Δ0 ura3Δ0 snf2Δ::KANr | Open Biosystems |
| YMM565 | MATa leu2Δ0 met15Δ0 ura3Δ0 chd1Δ::KANr | Open Biosystems |
| YMM594 | MATa leu2Δ0 met15Δ0 ura3Δ0 isw1Δ::KANr | Open Biosystems |
| YMM566 | MATa leu2Δ0 met15Δ0 ura3Δ0 isw2Δ::KANr | Open Biosystems |
| YMM595 | MATa leu2Δ0 met15Δ0 ura3Δ0 asf1Δ::KANr | Open Biosystems |
| YMM596 | MATa leu2Δ0 met15Δ0 ura3Δ0 swr1Δ::KANr | Open Biosystems |
| YMM562 | MATa leu2Δ0 met15Δ0 ura3Δ0 spt20Δ::KANr | Open Biosystems |
TABLE 2.
Oligonucleotides used in this study
| Name | Forward primer (5′–3′) | Annealing site | Reverse primer (5′–3′) | Annealing site | Use |
|---|---|---|---|---|---|
| ACT1qRT | ATCGTTATGTCCGGTGGTACC | 1196 | TGGAAGATGGAGCCAAAGC | 1281 | qPCR |
| KANqRT | CACCGGATTCAGTCGTCACTCATGG | 559 | GGCAAGATCCTGGTATCGGT CTGCGATTCC | 684 | qPCR |
| EBP2qRT | AACGCTACCTTACAGAAACG | 957 | TCCGTTAGGCCTGCCTCTATCGAA | 1122 | qPCR |
| MPP10qRT | CGAGGAGGAGGAGGCTATTTAT | 674 | CTTCCTCATCCGCAAATAAGTC | 844 | qPCR |
| YJR003CqRT | ACCACCATTGACCCATACTCTC | 147 | GACCACTTCCATCAGTTCATCA | 447 | qPCR |
| SAG1-NSA-1 | GGTTACTTTGAGCACACGGCTTTG | −634 | GATACCGGACAATTGGCTTCCTG | −475 | NSA |
| SAG1-NSA-2 | CTCATCTCAGGGAACGAAAATTG | −566 | CATAGGTTGAAATCATGAGAGAAG | −32 | NSA |
| SAG1-NSA-3 | CTCAGCTGAGCTCGGTTCGATC | −410 | GAAGAAAAAAAGAGCCAGGATG | −272 | NSA |
| YJR003C-NSA-1 | GTACAGTTAGTACATTGAGTC | −346 | GACGGAAAAGGATAAGAACTAG | −205 | NSA |
| YJR003C-NSA-2 | CATCCTGGCTCTTTTTTTCTTC | −294 | CTATGATAAAATCTGCGGTG | −182 | NSA |
| YJR003C-NSA-3 | CTAGTTCTTATCCTTTTCCGTC | −224 | GAAACAGCCTTCGGGTAATG | −105 | NSA |
| YJR003C-NSA-5 | CATTACCCGAAGGCTGTTTC | −124 | GAATGGCGGTTAGCTGTTAAG | −5 | NSA |
| YJR003C-NSA-6 | CACCATGAAAGAGTTCGATGAG | −64 | GAGAGAGTAAAACCTCTTGTTAG | 57 | NSA |
| YJR003C-NSA-7 | CTTAACAGCTAACCGCCATTC | −26 | GCTTTTTGATTATGTTCTTTC | 92 | NSA |
| YJR003C-NSA-8 | CATTCTGCGGCTACGTTATCTAAC | 15 | GATGACGAATTGGATCGAAAG | 129 | NSA |
| YJR003C-NSA-9 | ATAGAAAGAACATAATCAAAAAGC | 68 | CATTCTGCGATAGAGAGTATG | 181 | NSA |
| YJR003C-NSA-10 | CTTTCGATCCAATTCGTCATC | 108 | CATATTGTGTACCATGGCCGCATC | 243 | NSA |
| YJR003C-NSA-11 | CATACTCTCTATCGCAGAATG | 160 | CAACGCCGTAGTCAAGATCAC | 280 | NSA |
| YJR003C-NSA-12 | GATGCGGCCATGGTACACAATATG | 219 | GTTTGATGGCGGGAAGTGAAG | 343 | NSA |
| YJR003C-NSA-13 | GTGATCTTGACTACGGCGTTG | 259 | CAAGAGCTTTGTACTCTTCCTG | 383 | NSA |
| MPP10-NSA-1 | TTATGACTATCATTCCTATCGCAAAG | −640 | GAAGGCCTTTCGCAGCTCTTC | −506 | NSA |
| MPP10-NSA-2 | GTATTGGACGTTCTGATGAATGTG | −584 | GCTATCAAAACGAAGACAAC | −460 | NSA |
| MPP10-NSA-3 | GAAGAGCTGCGAAAGGCCTTC | −526 | GTAAAACACAAACCGGCCCCCAG | −400 | NSA |
| MPP10-NSA-4 | GTTGTCTTCGTTTTGATAGC | −479 | GTTACGTGACAAGCCACTCTCTC | −313 | NSA |
| MPP10-NSA-5 | CTGGGGGCCGGTTTGTGTTTTAC | −422 | CACCAACACCTAATGTGGACAAC | −272 | NSA |
| MPP10-NSA-6 | GAGAGAGTGGCTTGTCACGTAAC | −335 | CAGAAGTACAGAGCTAATG | −197 | NSA |
| MPP10-NSA-7 | GTTGTCCACATTAGGTGTTGGTG | −294 | GAAAAGGCGGTGAATATTTTATG | −124 | NSA |
| MPP10-NSA-8 | CATTAGCTCTGTACTTCTG | −215 | CGTGCACACATCATTTATTCATAAC | −24 | NSA |
| MPP10-NSA-9 | CGTAATATACATATTTTCGTGTAG | −179 | GACATTATACGACTTTCCTTGGGTC | 5 | NSA |
| MPP10-NSA-10 | CTGGCCGCCGGCATGCGAG | −119 | CAATACTCCAAAGAGTTCTGAC | 24 | NSA |
| MPP10-NSA-10.5 | GAAACAGTGTTTTGTTATGAATAAATG | −61 | GCTTTAACATCTTTAGAAGTGGC | 86 | NSA |
| MPP10-NSA-11 | GTGCACGAAAGACCCAAGGAAAG | −30 | CTTACAGGTATTGATAACTGAATC | 117 | NSA |
| MPP10-NSA-12 | GTCAGAACTCTTTGGAGTATTG | 3 | CATCAACAGTGATTTCGTCCAG | 166 | NSA |
| MPP10-NSA-13 | GATTCAGTTATCAATACCTGTAAG | 94 | GATCACCATCAATACTATCAAGAA | 229 | NSA |
| MPP10-NSA-14 | CTGGACGAAATCACTGTTGATG | 145 | GGCGTGACAACATCTTTGAGTTCTTG | 269 | NSA |
| MPP10-NSA-15 | TTCTTGATAGTATTGATGGTGATC | 206 | CTTTCACTTGATCTCCTCCGTC | 340 | NSA |
| HSP104-NSA-1 | GAAACCGTGGATGTTCAGGAC | −700 | CGCCTTTGAATCGATGACAAT | −588 | NSA |
| HSP104-NSA-2 | CACCGAGCCGGGGAAATTCG | −657 | GTCGTCGATCCAGTCCATTTC | −546 | NSA |
| HSP104-NSA-3 | ATTGTCATCGATTCAAAGGCG | −608 | GCCCTTGGAGTTTGGATTCTTG | −501 | NSA |
| HSP104-NSA-5 | CAAGAATCCAAACTCCAAGGGC | −522 | GAATAAATAAGTGAATAGGTAG | −413 | NSA |
| HSP104-NSA-6 | GGTTTAAAAACCTTCTGCACCA | −474 | CGATGGAGGGTTCAATGTTAAT | −358 | NSA |
| HSP104-NSA-7 | CTACCTATTCACTTATTTATTC | −434 | TAACCCTTCTAGAAAATTCTGG | −279 | NSA |
| HSP104-NSA-8 | ATTAACATTGAACCCTCCATCG | −379 | CTTTGAGATGGGCCCCCTGTTG | −203 | NSA |
| HSP104-NSA-9 | CCAGAATTTTCTAGAAGGGTTA | −300 | GTTTGCGCCCCTTTGCCTTTTAC | −161 | NSA |
| HSP104-NSA-11 | GTAAAAGGCAAAGGGGCGCAAAC | −183 | TTGCTGATTCGATTCAAGGG | −52 | NSA |
| HSP104-NSA-12 | GGCATTGTAATCTTGCCTCAATTCC | −132 | CTGTATATTTTATGGTACGTGTAG | −5 | NSA |
| HSP104-NSA-13 | CCCTTGAATCGAATCAGCAA | −71 | GCCAACGTCAAAATCGTTAGAGCCC | 53 | NSA |
| HSP104-NSA-14 | CTACACGTACCATAAAATATACAG | −29 | GTATAGGTTGTAATTGTGGATG | 100 | NSA |
| HSP104-NSA-15 | GGGCTCTAACGATTTTGACGTTGGC | 29 | CTGATCCATCTTCTGGCGTTTC | 142 | NSA |
| HSP104-NSA-16 | CATCCACAATTACAACCTATAC | 79 | GATCATAGTCGTAACGGCCC | 190 | NSA |
| HSP104-NSA-17 | GAAACGCCAGAAGATGGATCAG | 121 | GCAGGTTGCTGTTGAGGAATTC | 245 | NSA |
| HSP104-NSA-18 | GGGCCGTTACGACTATGATC | 171 | CCCCAAAGCATAACTTGGAG | 279 | NSA |
| HSP104-NSA-19 | GAATTCCTCAACAGCAACCTGC | 224 | GCGCTATAAATGAGTCCTTCTG | 337 | NSA |
Updating of RRB gene annotations.
The lists of the predicted membership of the RRB regulon (10, 26) were tabulated and updated for gene function annotations according to the Saccharomyces Genome Database (SGD; http://www.yeastgenome.org/) as of 15 November 2013.
Identification of putative trans regulators of RRB and RP gene expression.
The data set from the 165-gene deletion chromatin interaction study (29) was analyzed to identify mutants that preferentially disrupted RRB and RP gene expression. The number of genes disrupted (P < 0.05) in the RRB, RP, paired RRB, and paired RP gene sets were determined, and the significance of the disruption was calculated by a hypergeometric probability density function:
where P is the probability, K is the total number of genes disrupted, k is the number of genes in the subset disrupted, n is the number of genes in the subset, and N is the total number of genes with measured P values in the original experiment.
Culture conditions for heat shock response.
Strains were grown in YPD (1% yeast extract, 2% peptone, 2% dextrose) medium to early to mid-log phase (optical density at 600 nm of 0.40 to 0.90). A heat shock time course was induced by growing cultures at 30°C and transferring cells to 37°C medium (1).
RNA preparation and expression analysis.
Aliquots of yeast were obtained across a time course and washed at 4°C to remove the medium, and RNA was obtained by a hot acid phenol extraction (30) with the following modifications. Samples were extracted twice with phenol and once with chloroform and then ethanol precipitated prior to resuspension in diethyl pyrocarbonate (DEPC) water. Ten micrograms of RNA was cleared of genomic contaminants by treatment with DNase I according to the manufacturer's instructions (DNA-free; Ambion) and were checked by PCR using primers directed to the ACT1 coding region. cDNA was generated with oligo(dT) primers using the Retro-script kit according to the manufacturer's instructions (Ambion). Linear conditions were identified by the titration of cDNA template for PCR, followed by native PAGE. Quantitative PCR (qPCR) was then performed across the time course, and the products were analyzed by native PAGE stained with Sybr Gold (Invitrogen). Images were obtained on either a Typhoon or a Storm phosphorimager scanner (Molecular Dynamics) and quantified using the manufacturer's ImageQuant software.
Mapping of nucleosome positions.
Nucleosome positions were mapped using the nucleosome-scanning assay as described in reference 31. Aliquots of 100 ml of cells were fixed with 2% formaldehyde for 30 min, and then the reaction was quenched with 125 mM glycine for 10 min. The cells were then washed once in Tris-buffered saline (TBS) buffer and spheroplasted with Zymolyase 20T for 40 min (until approximately 85% of cells had spheroplasted). Spheroplasts were then washed twice and resuspended in 1.7 ml of MNase digestion buffer. Aliquots (360 μl) were then digested with limiting concentrations of MNase I (New England BioLabs) for 40 min. The digestion reaction was stopped by the addition of Tris-EDTA (TE)-SDS buffer, and cross-links were reversed by incubating the samples overnight at 65°C in the presence of proteinase K (New England BioLabs). DNA was recovered by phenol-chloroform/isoamyl alcohol extraction and ethanol precipitation. RNA was removed from the sample by treating the samples with RNase A for 60 min, at which point the DNA was again extracted by phenol-chloroform/isoamyl alcohol extraction and ethanol precipitation. The DNA samples were air dried and then resuspended in TE buffer.
The digestion reactions were then visualized on a 1.0% agarose gel, and the sample that resulted in the generation of mononucleosome-sized fragments was subsequently analyzed by real-time PCR. Real-time PCR was performed on an Applied Biosystems 7300 instrument utilizing Sybr green chemistry (Life Biosystems) and analyzed using the manufacturer's software. Twenty-μl reactions from each chromatin preparation were run in triplicate, and outliers were removed based on the manufacturer's criteria prior to analysis. The ratio of nucleosome-protected to nucleosome-depleted regions within the GAL locus was used as a control, and nucleosome positioning data were determined as previously described (32).
RESULTS
The RRB regulon membership predictions were accurate.
The RRB regulon was originally defined as a set of 65 transcriptionally coregulated genes that were enriched for the PAC and RRPE promoter motifs and whose products were suggested to play a role in the rRNA and ribosome biogenesis pathways (26). A full 30 of the original set of 65 RRB genes were uncharacterized at the time, and they were known only as unannotated open reading frames. By analyzing genome-wide expression profiles of yeast cells progressing through multiple changing environmental conditions, as well as through the analysis of gene promoter sequences, we expanded the predicted membership of the RRB regulon to include some 188 genes (10). This expansion indicated that the RRB regulon is at least as important in overall ribosome biogenesis as the 137-member ribosomal protein (RP) regulon and the large, 150-member tandem array of rRNA gene repeats. The expanded RRB gene set also included as-yet uncharacterized genes, as well as genes for which other non-RRB functions had been ascribed. To investigate the degree to which the RRB regulon membership assignments accurately predicted activities in the rRNA and ribosome biogenesis pathways, we reevaluated the known functions and annotations of the gene set (Table 3). We found that of the predicted 38 RRB genes for which there were previously no known functions, 34 (89%) have subsequently been shown to play a role in rRNA or ribosome biogenesis. Furthermore, of the 70 predicted RRB genes that had reported functions in pathways other than that of rRNA and ribosome biogenesis, 53 (76%) have subsequently been shown to exhibit additional RRB-consistent activities (Table 4). Therefore, the predictions of the RRB regulon membership were accurate, indicating that a similar approach could be useful in the identification and characterization of other coregulated gene sets in other metabolic pathways and species.
TABLE 3.
Previously uncharacterized RRB gene members with annotated RRB functions
| Systematic name | Standard name | Function and or pathway per SGD | Systematic name | Standard name | Function and/or pathway per SGD |
|---|---|---|---|---|---|
| YAL036C | RBG1 | Ribosome-associated protein | YJL109C | UTP10 | Processing of the 18S rRNA |
| YBL028C | Nucleolar protein | YJL122W | ALB1 | Production of the 60S ribosomal subunit | |
| YBL054W | TOD6 | Ribosome biogenesis transcription factor | YKL082C | RRP14 | Production of the 60S ribosomal subunit |
| YBR247C | ENP1 | Production of the 40S ribosomal subunit | YKL099C | UTP11 | 18S rRNA single-subunit processome |
| YCR072C | RSA4 | Production of the 60S ribosomal subunit | YKL143W | LTV1 | Production of the 40S ribosomal subunit |
| YDL063C | SYO1 | Production of the 60S ribosomal subunit | YLR002C | NOC3 | Production of the 60S ribosomal subunit |
| YDR101C | ARX1 | Production of the 60S ribosomal subunit | YLR276C | DBP9 | Helicase, 27S rRNA processing |
| YDR365C | ESF1 | Processing of the 18S rRNA | YLR401C | DUS3 | Dihydrouridine synthase, RNA modification |
| YER049W | TPA1 | Translation factor | YML093W | UTP14 | 18S rRNA single-subunit processome |
| YGL099W | LSG1 | Production of the 60S ribosomal subunit | YNL110C | NOP15 | Production of the 60S ribosomal subunit |
| YGR103W | NOP7 | Production of the 60S ribosomal subunit | YNL132W | KRE33 | Production of the 40S ribosomal subunit |
| YGR145W | ENP2 | Production of the 40S ribosomal subunit | YNR053C | NOG2 | Production of the 60S ribosomal subunit |
| YGR283C | Ribosome-associated methyltransferase | YOL124C | TRM11 | Guanosine methyltransferase | |
| YHL039W | EFM1 | Elongation factor methyltransferase | YOR091W | TMA46 | Ribosome-associated protein |
| YHR196W | UTP9 | 18S rRNA single-subunit processome | YPL012W | RRP12 | Ribosomal subunit export factor |
| YIL096C | Production of the 60S ribosomal subunit | YPL093W | NOG1 | 60S ribosomal subunit biogenesis | |
| YIL127C | RRT14 | rRNA biogenesis factor | YPR143W | RRP15 | Production of the 60S ribosomal subunit |
TABLE 4.
RRB gene members with newly annotated RRB functions
| Systematic name | Standard name | Function and/or pathway per SGD | Systematic name | Standard name | Function and/or pathway per SGD |
|---|---|---|---|---|---|
| YAL025C | MAK16 | Production of the 60S ribosomal subunit | YKL021C | MAK11 | Production of the 60S ribosomal subunit |
| YAL036C | RBG1 | Ribosome-associated protein | YKL191W | DPH2 | Modifies histidine residues in translation elongation factor 2 |
| YBL024W | NCL1 | tRNA methyltransferase | YKR056W | TRM2 | tRNA methyltransferase |
| YBR034C | HMT1 | Methyltransferase of ribosomal protein Rps2p | YKR092C | SRP40 | Pre-ribosomal assembly and transport |
| YBR267W | REI1 | Production of the 60S ribosomal subunit | YLR009W | RLP24 | Production of the 60S ribosomal subunit |
| YCL037C | SRO9 | Ribosome-associated protein | YLR249W | YEF3 | Translational elongation factor eEF1B subunit |
| YCR055C | PWP2 | 35S pre-rRNA processing | YLR401C | DUS3 | Dihydrouridine synthase |
| YDL201W | TRM8 | tRNA methyltransferase | YMR014W | BUD22 | Production of the 40S ribosomal subunit |
| YDR060W | MAK21 | Production of the 60S ribosomal subunit | YMR131C | RRB1 | Production of the 60S ribosomal subunit |
| YDR101C | ARX1 | Production of the 60S ribosomal subunit | YMR309C | NIP1 | Subunit of eukaryotic translation initiation factor 3 |
| YDR120C | TRM1 | tRNA methyltransferase | YNL062C | GCD10 | tRNA methyltransferase |
| YDR165W | TRM82 | tRNA methyltransferase | YNL110C | NOP15 | Production of the 60S ribosomal subunit |
| YDR299W | BFR2 | Component of 90S preribosomes | YNL119W | NCS2 | tRNA uridine modification |
| YDR465C | RMT2 | Ribosomal protein Rpl12 methyltransferase | YNL175C | NOP13 | Preribosomal complex nucleolar protein |
| YDR496C | PUF6 | Production of the 60S ribosomal subunit | YNL247W | Cysteinyl-tRNA synthetase | |
| YER126C | NSA2 | Production of the 60S ribosomal subunit | YNL308C | KRI1 | Production of the 40S ribosomal subunit |
| YGL099W | LSG1 | Production of the 60S ribosomal subunit | YNR053C | NOG2 | Production of the 60S ribosomal subunit |
| YGL111W | NSA1 | Production of the 60S ribosomal subunit | YNR054C | ESF2 | Involved in pre-18S rRNA processing |
| YGR162W | TIF4631 | Production of the 60S ribosomal subunit | YOL124C | TRM11 | tRNA methyltransferase |
| YGR245C | SDA1 | Production of the 60S ribosomal subunit | YOR206W | NOC2 | Production of the 60S ribosomal subunit |
| YHR052W | CIC1 | Production of the 60S ribosomal subunit | YOR243C | PUS7 | 5S rRNA pseudouridine synthase |
| YHR070W | TRM5 | tRNA methyltransferase | YOR272W | YTM1 | Production of the 60S ribosomal subunit |
| YHR170W | NMD3 | Production of the 60S ribosomal subunit | YPL093W | NOG1 | Production of the 60S ribosomal subunit |
| YIR012W | SQT1 | Production of the 60S ribosomal subunit | YPL146C | NOP53 | Production of the 60S ribosomal subunit |
| YIR026C | YVH1 | Production of the 60S ribosomal subunit | YPL212C | PUS1 | tRNA export protein |
| YJL125C | GCD14 | tRNA methyltransferase | YPL226W | NEW1 | Production of the 40S ribosomal subunit |
| YJR041C | URB2 | Ribosome biogenesis protein |
Defining the cis elements required for adjacent-gene coregulation (AGC).
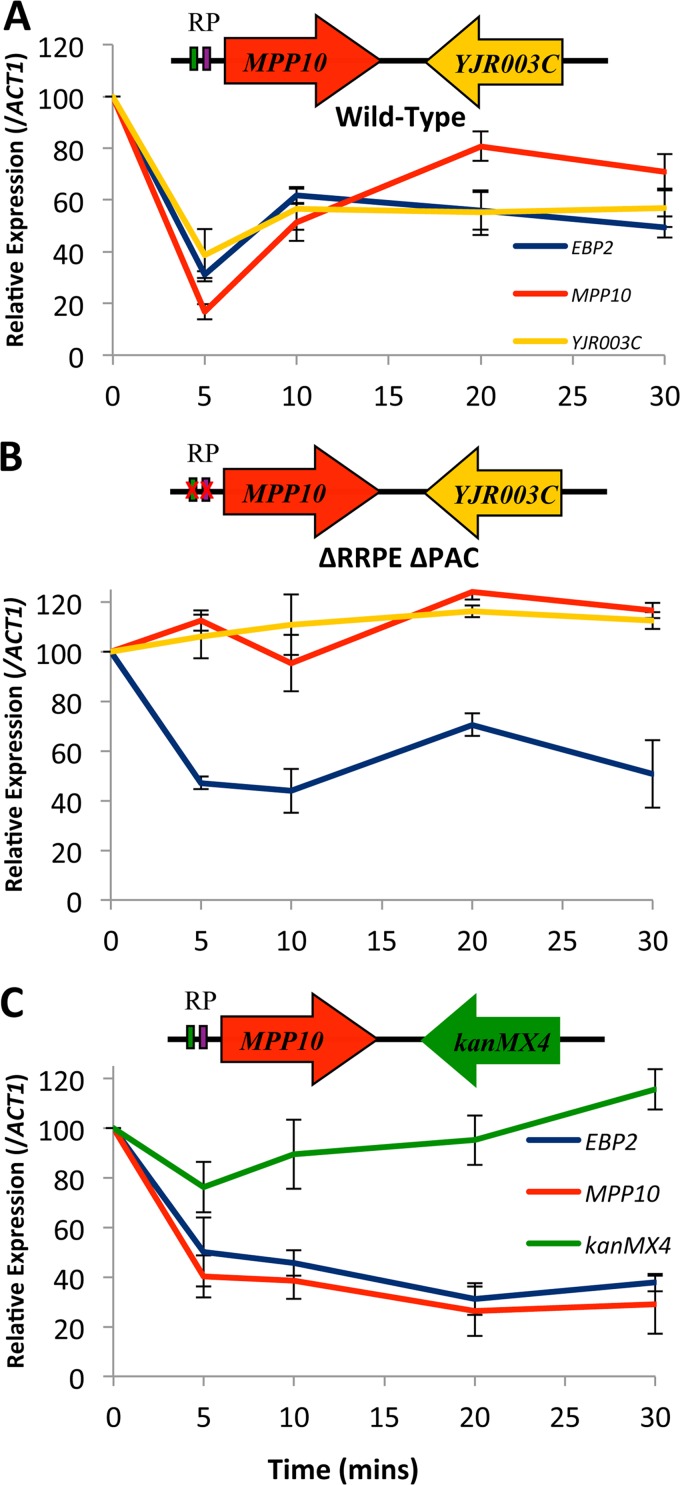
When we first identified and characterized the membership of the RRB regulon, we noticed that a significant fraction (roughly 15%) of the RRB genes were located on the chromosomes as immediate, adjacent gene pairs. Significant levels of adjacent-gene pairing were subsequently found in other coregulated gene sets in budding yeast and in the RRB and RP regulons across divergent eukaryotes (22). Additionally, the sets of paired genes were found to be more tightly coregulated across multiple changing growth conditions than were those of the unpaired genes of the same regulons. In order to define the cis elements that control the coregulated expression of the adjacent gene pairs, we initiated a mutational analysis of the convergently transcribed gene pair MPP10-YJR003C (26). We created a mutant yeast strain that contained discrete substitutions within the PAC and RRPE promoter motifs of the MPP10 gene, and we monitored the gene expression profiles following heat shock (27). We found that the MPP10 promoter substitutions were sufficient to disrupt the regulated expression of not only MPP10 but also of the adjacent YJR003C gene, even though the promoter for YJR003C lies some 3.5 kbp away and is oriented in the opposite direction (Fig. 1). We also altered the relative positions of the two genes through the insertion of a URA3 kanMX4 pCORE cassette that separated the two genes by an additional 3.8 kbp. In this case, the heat shock-induced repression of the MPP10 gene remained intact, but it did not extend to the separated YJR003C gene (27).
FIG 1.
Relative expression profiles of RRB genes following heat shock. Strains were grown in YPD media to early log phase, subjected to a 37°C heat shock, and monitored for their expression profiles at the EBP2, MPP10, YJR003C, and kanMX4 genes by RT-PCR. (A) YMM13 (WT); (B) YMM514 Δ RPPE/ΔPAC; (C) YMM557 yjr003cΔ::kanMX4.
In order to further define the DNA sequence elements that play a role in mediating adjacent gene coregulation, we created and tested new sets of mutants that altered the positions and relationships of the MPP10 and YJR003C genes. Given that sequences from the promoter of the MPP10 gene were able to direct the expression pattern of the adjacent YJR003C gene, we tested whether the MPP10 promoter could similarly exert a regulatory influence on an exogenous gene that replaced YJR003C. To do this, we took advantage of a genome-wide deletion library in which nonessential genes were deleted and replaced with a kanMX4 marker that is driven from the TEF promoter from Ashbya gosypii. The yjr003cΔ::kanMX4 deletion mutant was subjected to a 37°C heat shock and monitored by reverse transcription-PCR (RT-PCR) for the expression levels of MPP10, kanMX4, EBP2, and ACT1 as an internal reference control (Fig. 1). The MPP10 gene exhibited the classic heat shock repression response, as did the RRB control gene EBP2. However, the exogenous kanMX4 gene was not repressed during the heat shock, indicating that it was not subject to repression via the adjacent MPP10 promoter.
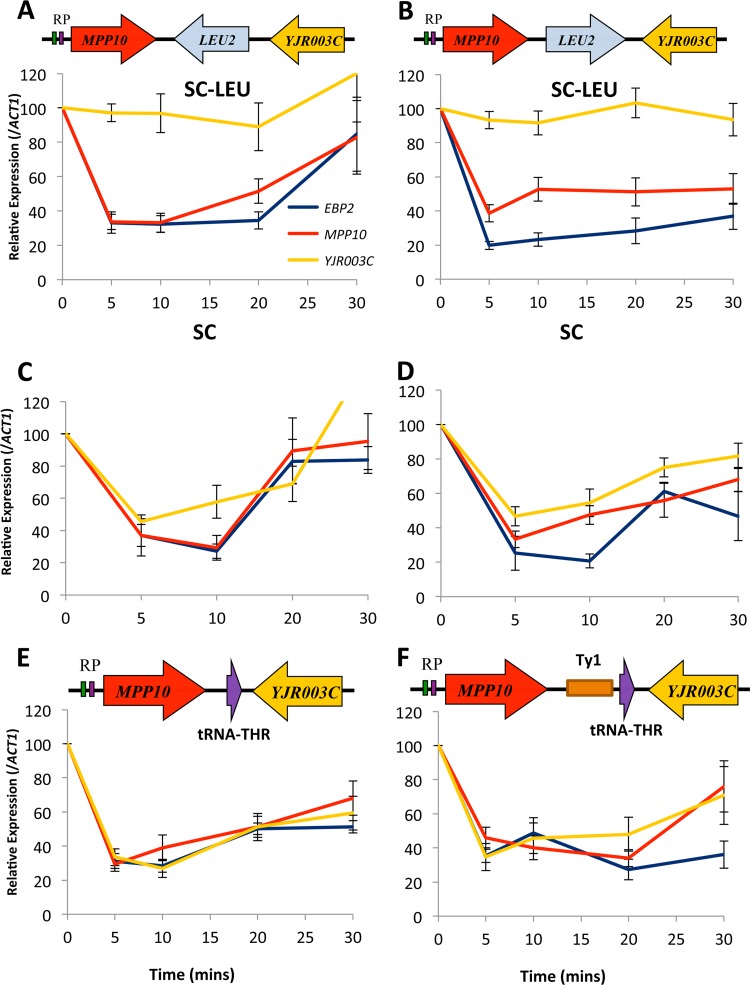
The creation of the pCORE- and kanMX4-associated mutants described above involved the use of exogenous, constitutively expressed genes and promoters that are not native to budding yeast: the kanMX4 gene is derived from bacteria, and the URA3 gene comes from K. lactis. To further characterize the sequence elements that play a role in adjacent gene coregulation, we engineered mutants that involved more native and potentially regulatable gene insertions. We used the pCORE-based delitto perfetto approach to engineer the native LEU2 gene and promoter from budding yeast between MPP10 and YJR003C (28). The advantage of the LEU2 insertion construct is that the LEU2 gene could be repressed or induced by growing the cells in media containing or lacking leucine, respectively (33). We created the LEU2 insertion strain, subjected it to heat shock, and monitored the transcript levels. When the strain was grown in synthetic complete (SC)-leucine media (i.e., under conditions where the LEU2 gene was expressed), we again observed that the YJR003C gene was no longer subjected to heat shock repression. However, when the strain was grown in SC or YPD medium containing leucine, the regulated repression of YJR003C remained intact (Fig. 2A and B; also see Fig. S1 in the supplemental material). We monitored the expression levels of the LEU2 gene under the two conditions and observed that LEU2 expression levels were consistent across the time course and were 4.5-fold lower in YPD than they were in SC-leucine (see Fig. S2). To test whether these findings were related to the orientation of the LEU2 insertion, we created another strain harboring the LEU2 insertion in the opposite direction. We observed the same effect, namely, that the coregulated repression of YJR003C after heat shock remained intact under conditions where the intervening LEU2 gene was not expressed but not when the LEU2 gene was expressed (Fig. 2C and D).
FIG 2.
Insertions of active RNA Pol II-transcribed genes can abrogate AGC. Strains were grown in either SC-leucine (A and B), SC media (C and D), or YPD (E and F), subjected to a 37°C heat shock, and monitored for their expression profiles at the EBP2, MPP10, and YJR003C genes by RT-PCR. The profiles of the leftward-oriented MPP10.LEU2 insert (YMM554) are represented in panels A and C, and the profiles of the rightward-oriented MPP10.LEU2 insert (YMM559) are presented in panels B and D. (E) YMM555 MPP10. tRNA (Thr); (F) YMMM556 MPP10.Ty tRNA (Thr).
Given that the insertion of an active, native RNA Pol II-transcribed gene was able to uncouple the regulated corepression of the MPP10 and YJR003C genes, we sought to determine whether this phenomenon extended to genes that are expressed by a different RNA polymerase. We integrated an RNA Pol III-transcribed tRNA (Thr) gene between MPP10 and YJR003C by the delitto perfetto method and monitored the expression of the genes following heat shock (Fig. 2E and F). We observed that the integration of this 0.7-kbp insert did not abrogate the regulated repression of MPP10 or YJR003C. We also integrated a larger tRNA (Thr) construct between MPP10 and YJR003C which contained an associated Ty element, a construct that was previously found to exhibit nucleosome boundary activity (34, 35). Again, the tRNA(Thr) Ty insert did not uncouple the regulated repression of YJR003C from MPP10. Thus, it appears that the adjacent coregulation of the MPP10 and YJR003C genes depends less upon the distance that separates them than it does on whether or not they are separated by an active, RNA polymerase II-transcribed gene.
Trans-acting factors related to chromatin remodeling mediate AGC.
We reasoned that in addition to depending on cis-acting DNA sequence elements, AGC will depend on the activity of trans-acting factors to couple the coregulated gene expression. In order to identify factors that may play a role in mediating AGC, we used a bioinformatics approach to survey a large gene expression data set that targeted 165 nonessential genes that have been implicated to function in regulating gene expression and chromatin architecture (29). The members of this extensive gene deletion study included factors that function in nucleosome remodeling (SWI/SNF, RSC, and INO80), histone assembly (FACT and CAF-1), histone modification (COMPASS, Rpd3L/S, NuA4, and SAGA), and transcription factors and transcriptional coactivators (Mediator). We screened the 165-gene knockout data set for mutants that preferentially disrupted the expression patterns of RRB and RP genes, as these factors would represent likely candidates for controlling their regulation (Table 5). For each knockout strain, we determined whether the expression profile of each gene in the genome deviated significantly (P < 0.05) from that in the wild-type strain. We then analyzed the sets of disrupted genes to determine whether they were significantly enriched for members of the RRB or RP regulons. We found that a substantial fraction of the 165 mutants did indeed preferentially disrupt the expression of the RRB genes over the other genes in the genome (P < 0.005), including mutants associated with the SWI/SNF, SAGA, RSC, NuA4, and Mediator complexes. Other mutants that were not associated with larger complexes also preferentially disrupted the RRB gene expression, including, as would be expected, the RRB promoter-associated PAC motif binding factor Tod6 (Table 5). The wide range of mutants that were found to affect RRB gene expression may be related to the fact that the expression of the RRB gene set is tightly controlled under a wide range of changing conditions, and that it may be subjected to multiple forms of regulation. We also identified a smaller set of mutants that exhibit preferential (P < 0.05) disruptions in RP gene expression, including members of the SAGA, CAF-1, and SET3 complexes. To determine whether any of the 165 mutants specifically play a role in AGC, we identified those mutants that preferentially (P < 0.05) disrupted the expression of any of the RRB or RP genes that were members of an adjacent pair. This analysis identified components of the same transcriptional regulators, including members of the SWI/SNF, SAGA, COMPASS, and Mediator complexes. Overall, this analysis suggests that the coordinated control of the RRB and RB genes involves multiple classes of transcriptional regulators, and that the same classes of transcriptional regulators control the expression of both the paired and nonpaired gene sets.
TABLE 5.
List of chromatin architecture-related deletion mutants that preferentially disrupt the expression of the indicated gene sets
| Disrupted gene(s) | |||||||
|---|---|---|---|---|---|---|---|
| RRB (P < 0.005) |
RP (P < 0.05) |
RRB gene pairs (P < 0.05) |
RP gene pairs (P < 0.05) |
||||
| Mutant | Complex | Mutant | Complex | Mutant | Complex | Mutant | Complex |
| SPT20, ADA2, GCN5, HFI1 | SAGA/ADA | SPT7, GCN5 | SAGA/ADA | SPT20 | SAGA | MED2, MED15 | MEDIATOR |
| MED2, MED9, MED15, MED16 | MEDIATOR | MSI1, CAC1 | CAF-1 | SWD3, SDC1 | COMPASS | SNF6 | SWI/SNF |
| NPL6, RSC1, RSC2, RSC30 | RSC | SET3 | SET3 | IES3 | INO80 | ||
| SNF2, SNF5, SNF6, SWI3, SNF12 | SWI/SNF | RRT109 | NOT3 | CCR4/NOT | |||
| CCR4, NOT4, CAF130, CAF40 | CCR4/NOT | ||||||
| EAF1, EAF6, EAF7 | NuA4 | ||||||
| TOD6, LEO1, SAS4, HIR1, JHD2, RTT109, SSN6, TUP1, CPR1, HST1 | |||||||
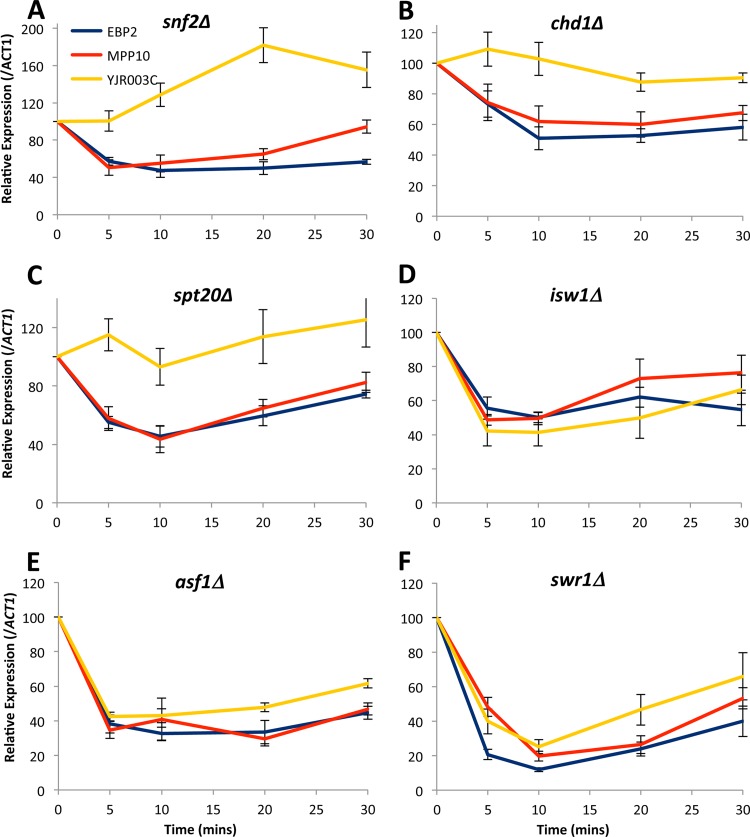
In order to test directly whether the candidate chromatin modifier complexes identified above function in mediating AGC, we screened a select panel of mutants for potential defects in the coregulation of the MPP10 and YJR003C genes after a heat shock. We reasoned that mutations in putative coordinating trans factors mimic the phenotype that we observed in the cis mutants, namely, that the regulation of MPP10 gene would proceed normally but the regulated repression of YJR003C would be compromised (Fig. 3). We chose mutants from the SAGA complex (spt20Δ), from the SWI/SNF complex (snf2Δ), and from other regulators, including chromatin remodelers (chd1Δ, isw2Δ, and swr1Δ) and a nucleosome assembly factor (asf1Δ). Each of the mutants was subjected to a heat shock, and the relative expression levels of the MPP10, YJR003C, EBP2, and ACT1 genes were determined by RT-PCR. Interestingly, we did observe an uncoupling defect in the spt20Δ, chd1Δ, and snf2Δ mutants, since in these strains the regulated repression of MPP10 remained intact but that of the YJR003C gene was lost. However, we did not see the same effect in other mutants, indicating that the activities of the Isw1, Swr2, and Asf1 proteins are not required for mediating this case of AGC. Thus, the coordinated repression of the MPP10 and YJR003C genes appears to depend on the activity of the trans factors Snf2, Chd1, and Spt20.
FIG 3.
Mutations in trans factors abrogate AGC. The indicated yeast strains were grown in YPD media to early log phase, subjected to a 37°C heat shock, and monitored for their expression profiles by RT-PCR. (A) YMM593 snf2Δ; (B) YMM565 chd1Δ; (C) YMM562 spt20Δ; (D) YMM566 isw1Δ; (E) YMM595 asf1Δ; (F) YMM596 swr1Δ.
Adjacent-gene coregulation does not appear to be mediated by nucleosome repositioning.
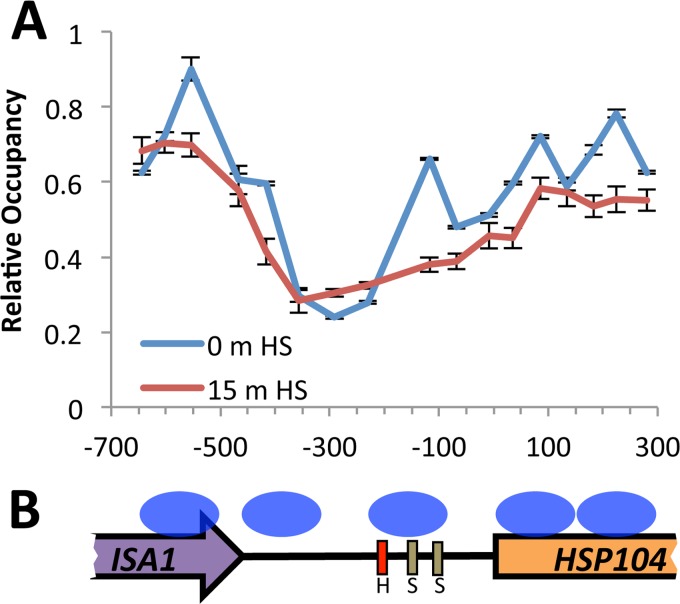
Given that the Snf2 and Chd1 proteins are components of chromatin remodeler complexes, one possible mechanism whereby AGC could be mediated is through the repositioning of nucleosomes within the respective gene promoters. Typically, the promoters of actively transcribed genes contain nucleosome-depleted regions (NDRs) that favor the association of RNA polymerase II and the initiation of transcription, and the dynamic repositioning of nucleosomes in NDRs can play an important role in regulating gene expression. To investigate whether this aspect of chromatin management plays a role in the regulated repression of the MPP10 and YJR003C genes, we monitored the positions of the nucleosomes in the respective gene promoters before and after heat shock by a micrococcal nuclease sensitivity assay. As a control, we monitored the nucleosome occupancy of the HSP104 promoter, since it has been shown previously that a temperature shift-induced activation of the gene is associated with the displacement of a particular nucleosome in its promoter (Fig. 4). We compared our nucleosome occupancy profiles with those that have been previously reported for the HSP104 gene (36), and we did observe the displacement of a nucleosome located approximately 150 bp upstream of the HSP104 translational start site. The position of this nucleosome overlaps the positions of the heat shock response (HSE) and stress response promoter elements (STRE), which are the binding sites for the Hsf1 and Msn4/Msn2 transcriptional activators, respectively. The clearing of this nucleosome allows for the binding of these transcriptional regulators and the induction of transcription after heat shock.
FIG 4.
Stress response-associated nucleosome displacement occurs at the HSP104 promoter. (A) Nucleosome positions were determined across the HSP104 promoter by a nucleosome-scanning assay in the wild-type background before and after a 15-min heat shock at 39°C. (B) Previously published nucleosome positions at the HSP104 promoter as determined by ChIP are represented as blue ovals (36). H, heat shock response element; S, stress response elements.
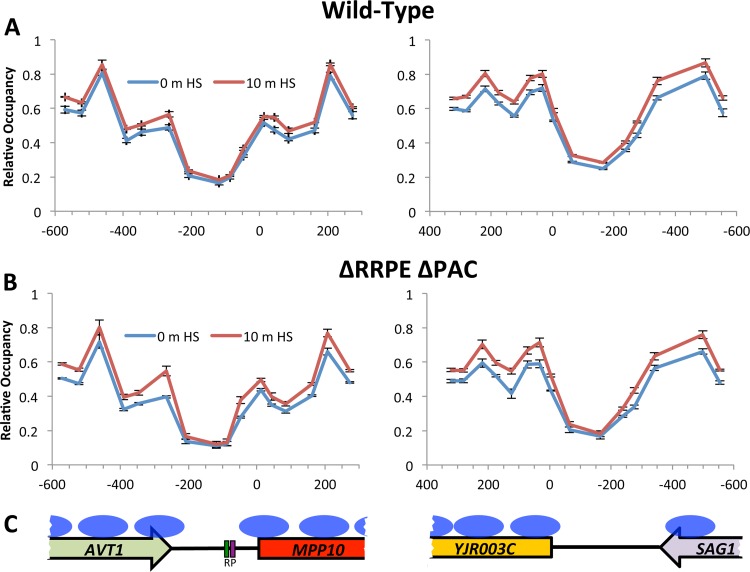
Nucleosome occupancy levels were also monitored across the MPP10 and YJR003C promoters, and in each case we could detect an NDR region that corresponded well with the nucleosome occupancy pattern detected previously (4). However, when the cells were subjected to a heat shock, we could detect no significant changes in the nucleosome occupancy profiles, as neither the MPP10 nor the YJR003C promoter exhibited a significant alteration in nucleosome position (Fig. 5). We also tested the MPP10 ΔPAC-ΔRRPE promoter mutant by this assay before and after heat shock and likewise found that this cis mutant showed no significant changes in the positions of its MPP10 and YJR003C promoter-associated nucleosomes. As a positive control, we monitored the nucleosome occupancy profiles in the snf2Δ mutant, and as predicted, they were disrupted across the MPP10 and YJR003C promoters (see Fig. S3 in the supplemental material). Thus, while factors associated with protein remodeler complexes are required for the coordinated repression of the adjacent MPP10-YJR003C gene pair, they do not appear to be affecting their control at the level of nucleosome repositioning.
FIG 5.
Nucleosome mapping at the MPP10 and YJR003C promoter regions. Nucleosome positions were determined across both promoters by a nucleosome-scanning assay in the wild-type background (A) and in the YMM514 ΔRRPE ΔPAC background (B) before and after a 10-min heat shock at 37°C. (C) Previously published nucleosome positions at the MPP10 and YJR003C promoters as determined by ChIP are represented as blue ovals (35).
DISCUSSION
The discovery and characterization of the RRB regulon (also known as the ribi regulon) considerably expanded our understanding concerning the classes and numbers of genes that contribute to ribosome biogenesis (10, 26, 37). Previously, it had been recognized that the expression levels of the set of 137 RP and 150 rRNA genes was subjected to tight regulatory control, albeit through different RNA polymerases (RNA Pol II for the RPs, RNA pols I and III for the rRNAs) (37, 38). The addition of the genes of the RRB regulon reveals that the overall ribosome biogenesis pathway is dependent on the coordinated expression of some 500 genes or more. This is a significant fraction of the entire yeast genome, and given that all cells must make their own ribosomes, it is reasonable to assume that all organisms will likewise contain similarly large classes of genes. Defining these gene sets will represent an important component of the gene annotation projects that are arising from the rapidly accumulating DNA sequence data sets of newly described species. Because ribosomes are highly conserved, it is relatively easy to identify rRNA and RP homologs in new species (39, 40). The validation of the approach that we used originally to predict the membership of the RRB regulon suggests that a similar approach could be successfully applied to identify RRB genes in new species, many of which may have limited other bases for gene annotations. Furthermore, the approach that we used is not limited to identifying genes associated with rRNA and ribosome biogenesis pathways, since the regulon membership was based solely on classifying genes through common promoter motifs and common transcriptional responses to changing conditions (10).
The observation that the coupled repression of the MPP10 and YJR003C genes can be maintained even after they have been separated by the insertion of an exogenous 1.5-kbp DNA fragment indicates that the mechanism of coregulation is not strictly distance limited. It also argues against a model in which the two genes are coordinately regulated via colliding RNA polymerases or the interactions between overlapping 3′ untranslated region (UTR) transcripts, since it would be unlikely for the MPP10 and YJR003C transcripts to extend across the additional intervening DNA. Furthermore, the finding that the coordinated repression of YJR003C was consistently abrogated when it was separated from the MPP10 gene by an actively expressed RNA Pol II transcription unit (LEU2 URA3 kanMX) but not by an RNA Pol III transcription unit [tRNA (Thr)] indicates that disrupting the mechanism of coupling between the two genes is RNA polymerase promoter specific. This finding is consistent with the native positions of the nonpaired and paired RRB and RP genes; they were found as either isolated single genes or immediately adjacent pairs. We did not observe cases in which two RRB or two RP genes were separated by a single gene from outside the respective regulons. Again, since the relative activity, but not the relative orientation of the intervening LEU2 transcription unit, was the determining factor in abrogating corepression, the disruption of AGC was unlikely to be mediated through the interactions of mRNA transcripts. That the inserted LEU2 or kanMX4 gene from the yjr003cΔ::kanMX4 deletion strain did not fall under the repressive influence of the MPP10 promoter indicates that the YJR003C promoter sequences are specifically receptive to repression. Defining those sequences should be as straightforward as it was to identify the relevant motifs in the MPP10 promoter.
The identification of relevant trans factors also yields insight as to how AGC is mediated. The Spt20 protein is a structural component of the SAGA complex, a multisubunit histone acetyltransferase that interacts with the TATA-binding protein TBP and promotes the formation of the preinitiation complex (41, 42). Snf2 is a catalytic subunit of the SWI/SNF chromatin remodeling ATPase that can regulate gene expression by altering the positions of nucleosomes on DNA, including those that have been modified by SAGA (43, 44, 45). Chd1 is an ATP-dependent chromatin remodeling enzyme that regulates various aspects of transcription (44). It contains an Snf2/Swi2-type helicase domain and a C-terminal nucleosome-binding domain. All of these factors are known to be important transcriptional regulators, and they may contribute to AGC directly or through their association with or recruitment of other transcriptional regulators, including components of the basal transcription machinery.
One model for mediating AGC across the gene pairs is through the formation of short DNA loops that can form transiently in a transcription-dependent manner (46). Gene loops can physically bridge distant segments of DNA and, through the interactions of associated factors and complexes, bring the promoter and terminator regions of genes into close contact (46, 47, 48). Such interactions have the potential for impacting levels of gene expression, including gene silencing. The HMR-E and HMR-I silencers are separated by several kilobase pairs of DNA, yet they can be seen to physically and functionally interact in vivo (49). In the tandemly arranged SNA3-INO1 gene pair, regulated inositol-induced repression of SNA3 was mediated through Ino2/Ino4 binding proteins that recognize E-box consensus sequences not from within the SNA3 promoter but from within the downstream (intergenic region) INO1 promoter (50). Furthermore, a short, stable DNA loop linking the promoter and terminator regions of the INO1 gene could be seen during activated transcription (51), and it was suggested to be formed through interactions between transcriptional activators and TFIIB. Interestingly, gene looping has even been shown to be important in the regulated expression of divergently transcribed genes, as in the case of the establishment of transcriptional memory at the GAL1-GAL10 locus (46).
Therefore, one possibility is that the heat shock-induced corepression of YJR003C is mediated through a DNA loop that juxtaposes its promoter next to the promoter of MPP10. This physical association could transmit a repressive signal that is mediated through the recognition of the MPP10 PAC and RRPE promoter motifs. Our analysis of the trans mutants suggest that the putative DNA looping arrangement between the MPP10 and YJR003C promoters depends on the activity of the Snf2, Chd1, and Spt20 proteins, and our analysis of the cis mutants suggests that it cannot extend past another active RNA Pol II promoter. Together, the identification of relevant cis and trans elements that regulate AGC provides important insights and direction for further investigations as to how it is achieved at the molecular level.
Supplementary Material
Footnotes
Published ahead of print 4 April 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/EC.00317-13.
REFERENCES
- 1.Gasch AP, Spellman PT, Kao CM, Carmel-Harel O, Eisen MB, Storz G, Botstein D, Brown PO. 2000. Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell 11:4241–4257. 10.1091/mbc.11.12.4241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.de Nadal E, Ammerer G, Posas F. 2011. Controlling gene expression in response to stress. Nat. Rev. Genet. 12:833–845. 10.1038/nrg3055 [DOI] [PubMed] [Google Scholar]
- 3.Fraser P, Bickmore W. 2007. Nuclear organization of the genome and the potential for gene regulation. Nature 447:413–417. 10.1038/nature05916 [DOI] [PubMed] [Google Scholar]
- 4.Lee W, Tillo D, Bray N, Morse RH, Davis RW, Hughes TR, Nislow C. 2007. A high-resolution atlas of nucleosome occupancy in yeast. Nat. Genet. 39:1235–1244. 10.1038/ng2117 [DOI] [PubMed] [Google Scholar]
- 5.Karlic R, Chung HR, Lasserre J, Vlahovicek K, Vingron M. 2010. Histone modification levels are predictive for gene expression. Proc. Natl. Acad. Sci. U. S. A. 107:2926–2931. 10.1073/pnas.0909344107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weake VM, Workman JL. 2010. Inducible gene expression: diverse regulatory mechanisms. Nat. Rev. Genet. 11:426–437. 10.1038/nrg2781 [DOI] [PubMed] [Google Scholar]
- 7.Lelli KM, Slattery M, Mann RS. 2012. Disentangling the many layers of eukaryotic transcriptional regulation. Annu. Rev. Genet. 46:43–68. 10.1146/annurev-genet-110711-155437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Warner JR. 1999. The economics of ribosome biosynthesis in yeast. Trends Biochem. Sci. 24:437–440. 10.1016/S0968-0004(99)01460-7 [DOI] [PubMed] [Google Scholar]
- 9.Fatica A, Tollervey D. 2002. Making ribosomes. Curr. Opin. Cell Biol. 14:313–318. 10.1016/S0955-0674(02)00336-8 [DOI] [PubMed] [Google Scholar]
- 10.Wade CH, Umbarger MA, McAlear MA. 2006. The budding yeast rRNA and ribosome biosynthesis (RRB) regulon contains over 200 genes. Yeast 23:293–306. 10.1002/yea.1353 [DOI] [PubMed] [Google Scholar]
- 11.Zaman S, Lippman SI, Zhao X, Broach JR. 2008. How Saccharomyces responds to nutrients. Annu. Rev. Genet. 42:27–81. 10.1146/annurev.genet.41.110306.130206 [DOI] [PubMed] [Google Scholar]
- 12.Cipollina C, van den Brink J, Daran-Lapujade P, Pronk JT, Vai M, de Winde JH. 2008. Revisiting the role of yeast Sfp1 in ribosome biogenesis and cell size control: a chemostat study. Microbiology 154:337–346. 10.1099/mic.0.2007/011767-0 [DOI] [PubMed] [Google Scholar]
- 13.Fingerman I, Nagaraj V, Norris D, Vershon AK. 2003. Sfp1 plays a key role in yeast ribosome biogenesis. Eukaryot. Cell 2:1061–1068. 10.1128/EC.2.5.1061-1068.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Urban J, Soulard A, Huber A, Lippman S, Mukhopadhyay D, Deloche O, Wanke V, Anrather D, Ammerer G, Riezman H, Broach JR, De Virgilio C, Hall MN, Loewith R. 2007. Sch9 is a major target of TORC1 in Saccharomyces cerevisiae. Mol. Cell 26:663–674. 10.1016/j.molcel.2007.04.020 [DOI] [PubMed] [Google Scholar]
- 15.Bosio MC, Negri R, Dieci G. 2011. Promoter architectures in the yeast ribosomal expression program. Transcription 2:71–77. 10.4161/trns.2.2.14486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alejandro-Osorio AL, Huebert DJ, Porcaro DT, Sonntag ME, Nillasithanukroh S, Will JL, Gasch AP. 2009. The histone deacetylase Rpd3p is required for transient changes in genomic expression in response to stress. Genome Biol. 10:R57. 10.1186/gb-2009-10-5-r57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martin DE, Soulard A, Hall MN. 2004. TOR regulates ribosomal protein gene expression via PKA and the Forkhead transcription factor FHL1. Cell 119:969–979. 10.1016/j.cell.2004.11.047 [DOI] [PubMed] [Google Scholar]
- 18.Laferte A, Favry E, Sentenac A, Riva M, Carles C, Chedin S. 2006. The transcriptional activity of RNA polymerase I is a key determinant for the level of all ribosome components. Genes Dev. 20:2030–2040. 10.1101/gad.386106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Claypool J, French S, Johzuka K, Eliason K, Vu L, Dodd J, Beyer A, Nomura M. 2003. Tor pathway regulates Rrn3p-dependent recruitment of yeast RNA polymerase I to the promoter but does not participate in alteration of the number of active genes. Mol. Biol. Cell 15:946–956. 10.1091/mbc.E03-08-0594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liko D, Slattery MG, Heideman W. 2007. Stb3 binds to ribosomal RNA processing element motifs that control transcriptional responses to growth in Saccharomyces cerevisiae. J. Biol. Chem. 282:26623–26628. 10.1074/jbc.M704762200 [DOI] [PubMed] [Google Scholar]
- 21.Wang Y, Pierce M, Schneper L, Guldal CG, Zhang X, Tavazoie S, Broach JR. 2004. Ras and Gpa2 mediate one branch of a redundant glucose signaling pathway in yeast. PLoS Biol. 2:E128. 10.1371/journal.pbio.0020128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Arnone JT, Robbins-Pianka A, Arace JR, Kass-Gergi S, McAlear MA. 2012. The adjacent positioning of coregulated gene pairs is widely conserved across eukaryotes. BMC Genomics 13:546. 10.1186/1471-2164-13-546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.West RW, Jr, Yocum RR, Ptashne M. 1984. Saccharomyces cerevisiae GAL1-GAL10 divergent promoter region: location and function of the upstream activating sequence UASG. Mol. Cell. Biol. 4:2467–2478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cohen BA, Mitra RD, Hughes JD, Church GM. 2000. A computational analysis of whole-genome expression data reveals chromosomal domains of gene expression. Nat. Genet. 26:183–186. 10.1038/79896 [DOI] [PubMed] [Google Scholar]
- 25.Ben-Shitrit T, Yosef N, Shemesh K, Sharan R, Ruppin E, Kupiec M. 2012. Systematic identification of gene annotation errors in the widely used yeast mutation collections. Nat. Methods 9:373–378 [DOI] [PubMed] [Google Scholar]
- 26.Wade C, Shea KA, Jensen RV, McAlear MA. 2001. EBP2 is a member of the yeast RRB regulon, a transcriptionally coregulated set of genes that are required for ribosome and rRNA biosynthesis. Mol. Cell. Biol. 21:8638–8650. 10.1128/MCB.21.24.8638-8650.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Arnone JT, McAlear MA. 2011. Adjacent gene pairing plays a role in the coordinated expression of ribosome biogenesis genes MPP10 and YJR003C in Saccharomyces cerevisiae. Eukaryot. Cell 10:43–53. 10.1128/EC.00257-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Storici F, Resnick MA. 2006. The delitto perfetto approach to in vivo site-directed mutagenesis and chromosome rearrangements with synthetic oligonucleotides in yeast. Methods Enzymol. 409:329–345. 10.1016/S0076-6879(05)09019-1 [DOI] [PubMed] [Google Scholar]
- 29.Lenstra TL, Benschop JJ, Kim T, Schulze JM, Brabers NA, Margaritis T, van de Pasch LA, van Heesch SA, Brok MO, Groot Koerkamp MJ, Ko CW, van Leenen D, Sameith K, van Hooff SR, Lijnzaad P, Kemmeren P, Hentrich T, Kobor MS, Buratowski S, Holstege FC. 2011. The specificity and topology of chromatin interaction pathways in yeast. Mol. Cell 42:536–549. 10.1016/j.molcel.2011.03.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. 1993. Current protocols in molecular biology. Wiley Interscience, New York, NY [Google Scholar]
- 31.Hainer SJ, Pruneski JA, Mitchell RD, Monteverde RM, Martens JA. 2011. Intergenic transcription causes repression by directing nucleosome assembly. Genes Dev. 25:29–40. 10.1101/gad.1975011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mavrich TN, Ioshikhes IP, Venters BJ, Jiang C, Tomsho LP, Qi J, Schuster SC, Albert I, Pugh BF. 2008. A barrier nucleosome model for statistical positioning of nucleosomes throughout the yeast genome. Genome Res. 18:1073–1083. 10.1101/gr.078261.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Andreadis A, Hsu YP, Hermodson M, Kohlhaw G, Schimmel P. 1984. Yeast LEU2. Repression of mRNA levels by leucine and primary structure of the gene product. J. Biol. Chem. 259:8059–8062 [PubMed] [Google Scholar]
- 34.Bi X, Broach JR. 2001. Chromosomal boundaries in S. cerevisiae. Curr. Opin. Genet. Dev. 11:199–204. 10.1016/S0959-437X(00)00179-9 [DOI] [PubMed] [Google Scholar]
- 35.Bi X, Yu Q, Sandmeier JJ, Zou Y. 2004. Formation of boundaries of transcriptionally silent chromatin by nucleosome-excluding structures. Mol. Cell. Biol. 24:2118–2131. 10.1128/MCB.24.5.2118-2131.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Uffenbeck SR, Krebs JE. 2006. The role of chromatin structure in regulating stress-induced transcription in Saccharomyces cerevisiae. Biochem. Cell Biol. 84:477–489. 10.1139/o06-079 [DOI] [PubMed] [Google Scholar]
- 37.Jorgensen P, Rupes I, Sharom JR, Schneper L, Broach JR, Tyers M. 2004. A dynamic transcriptional network communicates growth potential to ribosome synthesis and critical cell size. Genes Dev. 18:2491–2505. 10.1101/gad.1228804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fromont-Racine M, Senger B, Saveanu C, Fasiolo F. 2003. Ribosome assembly in eukaryotes. Gene 313:17–42. 10.1016/S0378-1119(03)00629-2 [DOI] [PubMed] [Google Scholar]
- 39.Granneman S, Baserga SJ. 2004. Ribosome biogenesis: of knobs and RNA processing. Exp. Cell Res. 296:43–50. 10.1016/j.yexcr.2004.03.016 [DOI] [PubMed] [Google Scholar]
- 40.Li L, Stoeckert CJ, Jr, Roos DS. 2003. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 13:2178–2189. 10.1101/gr.1224503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lagesen K, Hallin P, Rodland EA, Staerfeldt HH, Rognes T, Ussery DW. 2007. RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 35:3100–3108. 10.1093/nar/gkm160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sterner DE, Grant PA, Roberts SM, Duggan LJ, Belotserkovskaya R, Pacella LA, Winston F, Workman JL, Berger SL. 1999. Functional organization of the yeast SAGA complex: distinct components involved in structural integrity, nucleosome acetylation, and TATA-binding protein interaction. Mol. Cell. Biol. 19:86–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huisinga KL, Pugh BF. 2004. A genome-wide housekeeping role for TFIID and a highly regulated stress-related role for SAGA in Saccharomyces cerevisiae. Mol. Cell 13:573–585. 10.1016/S1097-2765(04)00087-5 [DOI] [PubMed] [Google Scholar]
- 44.Clapier CR, Cairns BR. 2009. The biology of chromatin remodeling complexes. Annu. Rev. Biochem. 78:273–304. 10.1146/annurev.biochem.77.062706.153223 [DOI] [PubMed] [Google Scholar]
- 45.Chandy M, Gutierrez JL, Prochasson P, Workman JL. 2006. SWI/SNF displaces SAGA-acetylated nucleosomes. Eukaryot. Cell 5:1738–1747. 10.1128/EC.00165-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hampsey M, Singh BN, Ansari A, Laine JP, Krishnamurthy S. 2011. Control of eukaryotic gene expression: gene loops and transcriptional memory. Adv. Enzyme Regul. 51:118–125. 10.1016/j.advenzreg.2010.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Laine JP, Singh BN, Krishnamurthy S, Hampsey M. 2009. A physiological role for gene loops in yeast. Genes Dev. 23:2604–2609. 10.1101/gad.1823609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Singh BN, Ansari A, Hampsey M. 2009. Detection of gene loops by 3C in yeast. Methods 48:361–367. 10.1016/j.ymeth.2009.02.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Valenzuela L, Dhillon N, Dubey RN, Gartenberg MR, Kamakaka RT. 2008. Long-range communication between the silencers of HMR. Mol. Cell. Biol. 28:1924–1935. 10.1128/MCB.01647-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shetty A, Swaminathan A, Lopes JM. 2013. Transcription regulation of a yeast gene from a downstream location. J. Mol. Biol. 425:457–465. 10.1016/j.jmb.2012.11.018 [DOI] [PubMed] [Google Scholar]
- 51.El Kaderi B, Medler S, Raghunayakula S, Ansari A. 2009. Gene looping is conferred by activator-dependent interaction of transcription initiation and termination machineries. J. Biol. Chem. 284:25015–25025. 10.1074/jbc.M109.007948 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.