Abstract
The transcription factor STAT1 is essential for interferon (IFN)-mediated immunity in humans and mice. STAT1 function is tightly regulated, and both loss- and gain-of-function mutations result in severe immune diseases. The two alternatively spliced isoforms, STAT1α and STAT1β, differ with regard to a C-terminal transactivation domain, which is absent in STAT1β. STAT1β is considered to be transcriptionally inactive and to be a competitive inhibitor of STAT1α. To investigate the functions of the STAT1 isoforms in vivo, we generated mice deficient for either STAT1α or STAT1β. As expected, the functions of STAT1α and STAT1β in IFN-α/β- and IFN-λ-dependent antiviral activity are largely redundant. In contrast to the current dogma, however, we found that STAT1β is transcriptionally active in response to IFN-γ. In the absence of STAT1α, STAT1β shows more prolonged IFN-γ-induced phosphorylation and promoter binding. Both isoforms mediate protective, IFN-γ-dependent immunity against the bacterium Listeria monocytogenes, although with remarkably different efficiencies. Our data shed new light on the potential contributions of the individual STAT1 isoforms to STAT1-dependent immune responses. Knowledge of STAT1β's function will help fine-tune diagnostic approaches and help design more specific strategies to interfere with STAT1 activity.
INTRODUCTION
Signal transducer and activator of transcription 1 (STAT1) regulates a wide variety of cellular activities, such as differentiation, proliferation, apoptosis, and homeostasis. STAT1 is a constituent of the Janus kinase (JAK)/STAT pathway through which many cytokines, including interferons (IFNs), transmit their biological effects. Ligand binding to cognate receptors leads to auto- and/or trans phosphorylation of JAKs, a family of nonreceptor tyrosine kinases. Subsequent phosphorylation events on the intracellular receptor domains and on STAT proteins result in activation of STAT homo- and/or heterodimers, which translocate to the nucleus and induce gene expression. In response to type I IFNs (IFN-α subtypes and IFN-β) and type III IFNs (IFN-λ subtypes), STAT1/2 heterodimers and IFN regulatory factor 9 (IRF9) form IFN-stimulated gene factor 3 (ISGF3), which binds to IFN-stimulated response elements (ISREs) in the regulatory regions of target genes. Type II IFN (IFN-γ) mainly activates STAT1 homodimers, which bind to IFN-γ activation sites (GAS) (1). IFN-induced activation of STAT1 occurs by phosphorylation on two amino acid residues. Phosphorylation of tyrosine 701 (Tyr701) is essential for the translocation of STAT1 dimers into the nucleus, whereas phosphorylation of serine 727 (Ser727), which is located in the C-terminal transactivation domain (TAD), has both positive and negative effects on gene transcription (1–5). The absence of STAT1 leads in mice and humans to higher susceptibility to viral and bacterial pathogens as a result of impaired IFN signaling (6–8).
Two STAT1 isoforms are generated by alternative splicing in most, if not all, cells and tissues: full-length STAT1α (91 kDa) and C-terminally truncated STAT1β (84 kDa). The latter lacks 38 amino acids, including the Ser727 site and most of the TAD. It is currently believed that STAT1β homodimers are unable to induce transcription due to their inability to recruit essential cofactors, such as the histone acetyltransferases CBP/p300 (9–11). However, both isoforms can be phosphorylated on Tyr701, translocate to the nucleus, and bind equally well to GAS sites (10–13). Both STAT1 isoforms are considered to be fully functional within an ISGF3 complex, as the essential TAD is provided by STAT2 (9, 14, 15). As STAT1β overexpression can inhibit STAT1α function (16) and as infection with certain pathogens increases the ratio of STAT1β to STAT1α in murine and human cells (17–19), it has been postulated that STAT1β acts as a dominant-negative regulator in IFN-γ signaling. However, it is notable that all data on STAT1 isoform functions have been obtained from studies with transfected cell lines and/or in vitro assays. The functions of the two isoforms have not yet been investigated in vivo.
We now report the generation of mice deficient for either the STAT1α or the STAT1β isoform. Gene expression profiling in primary cells revealed that STAT1β is transcriptionally active and induces responses that are different from those induced by STAT1α. In the absence of STAT1α, STAT1β shows prolonged Tyr701 phosphorylation and promoter binding activity but does not induce a generally enhanced transcriptional response. Importantly, STAT1β alone is capable of eliciting IFN-γ-dependent protective immunity against infections in vivo, consistent with the conclusion that STAT1β contributes uniquely to host immunity and is not merely an inactive or dominant-negative factor.
MATERIALS AND METHODS
Generation of STAT1α- and STAT1β-deficient mice.
We used gene-targeting vectors containing the terminal portions of the cDNA for either Stat1α or Stat1β fused in frame into exon 19 of the genomic Stat1 gene (accession no. AF349678). A human β-globin splice and a floxed neomycin cassette were inserted downstream of the cDNAs and placed between two regions of homology. Targeting constructs were linearized and transfected into embryonic day 14 (E14) embryonic stem (ES) cells (129P2) (20). Clones with homologously integrated targeting constructs and C57BL/6N blastocysts were used to generate chimeric mice. Offspring carrying the targeted alleles were crossed to Tg(CMV-Cre) mice (21) to remove the neomycin cassette and subsequently intercrossed to obtain Stat1α/α and Stat1β/β homozygous mice. The mice were backcrossed to the C57BL/6N background by speed congenics (22).
ES cell screening and Southern blotting.
To confirm correct integration of the targeting constructs in transfected ES cells after positive PCR screening (forward primer, 5′-ACGAGATCAGCAGCCTCTGT-3′; reverse primer, 5′-CTCTAGCCCAAACTTCTCTAAAAGC-3′; fragment size, 1,420 bp), Southern blot analysis was performed as described previously (20). Homologously integrated constructs were screened by detection of a 7.4-kb (wild-type [WT]), a 5.7-kb (Stat1α), and a 5.6-kb (Stat1β) long fragment after digestion with BsrGI by using a 5′ [α-32P]ATP-labeled probe (Random labeling kit; Invitrogen) obtained by PCR with the following primers: forward, 5′-ACAAAGCCACCATTCGTAGG-3′; reverse, 5′-GTGCATCTCTGAGCAAAACC-3′. Isolated DNAs from ES cell clones were used as templates.
Genotyping.
Genetic screening of WT, Stat1α/α, and Stat1β/β mice was performed by a duplex PCR with primers (purchased from Sigma-Aldrich) using DNA from mouse tail biopsy specimens. The primers were Tα/β wt fwd (5′-CAAAGCGTCTCCATTCATCTC-3′), Tα/β wt rev (5′-CATTTCCACAACACGTTTCC-3′), and huβ rev (5′-AGAAAACATCAAGGGTCCCATA-3′) and resulted in the following fragment sizes: WT, 367 bp; Stat1β, 493 bp; and Stat1α, 603 bp. Standard PCRs were performed in a final volume of 20 μl containing 1× PCR buffer (20 mM Tris-HCl, pH 8.8, 50 mM KCl), 1.5 mM MgCl2, 0.2 mM deoxynucleoside triphosphates (dNTPs), 1 μM primer for Tα/β wt fwd and 0.5 μM for Tα/β wt rev and huβ rev, and 1 U Biotaq (Agrobiogen). The cycling conditions consisted of 5 min denaturation at 95°C, followed by 30 cycles of 30 s denaturation at 95°C, 40 s annealing at 62°C, and 40 s elongation at 72°C, and a single step of 5 min at 72°C. Twenty to 40 ng genomic DNA was used as the template.
Mice and ethics statement.
C57BL/6N (WT) mice were purchased from Charles River Laboratories. Stat1α/α, Stat1β/β, and Stat1−/− mice (7) were housed under specific-pathogen-free conditions according to Federation of European Laboratory Animal Science Associations (FELASA) guidelines. All animal experiments were discussed and approved by the Ethics and Animal Welfare Committee of the University of Veterinary Medicine, Vienna, Austria, and conform to the guidelines of the national authority (Austrian Federal Ministry of Science and Research; section 8ff of the Animal Science and Experiments Act, Tierversuchsgesetz [TVG], BMWF-68.205/0204-C/GT/2007, BMWF-68.205/0210-II/10b/2009, and BMWF-68.205/0243-II/3b/2011).
Cell culture and cytokines.
Bone marrow-derived macrophages (BMMϕ) and E14 ES cells were cultivated as previously described (23). Recombinant mouse IFN-β and IFN-γ were purchased from Calbiochem (Merck Biosciences, Germany) and used at concentrations of 100 U/ml.
Whole-cell extracts and Western blotting.
Proteins from cultivated cells or organs were isolated and used for Western blot (WB) analysis as described previously (24). Anti-phospho-Tyr701 STAT1 and anti-STAT1 (N terminal) were purchased from Cell Signaling Technology; anti-phospho-Ser727 STAT1 was kindly provided by Pavel Kovarik from the Max F. Perutz Laboratories (MFPL), University of Vienna, Vienna, Austria (25). Anti-pan-ERK was purchased from BD Transduction Laboratories (the antibody detects p42, p44, p56, and p85; the p42 or the p85 band is depicted in our experiments). Peroxidase-conjugated secondary antibodies (mouse and rabbit) were purchased from GE Healthcare.
Electrophoretic mobility shift assay (EMSA).
BMMϕ were stimulated with IFN-γ (100 U/ml) for 1 h or left untreated, and whole-cell extracts were prepared as described above. The extracts (10 μg or 15 μl) were mixed with 2 μl of poly(dI-dC) (15.6 U/ml; Sigma) and incubated for 5 min at room temperature. Eight microliters of probe mixture containing 2.5 μl of 10× Licor binding buffer (LBB) (100 mM Tris-HCl, pH 7.5, 500 mM KCl, 10 mM dithiothreitol [DTT]), 2.5 μl of DTT-Tween (25 mM DTT, 2,5% Tween 20), 2 μl H2O, and 1 μl of 5′-IRDye 700/800-labeled oligonucleotides with a GAS binding site (5′-GTCGACATTTCCCGTAAATC-3′; purchased from Invitrogen) was added and incubated for 20 min at room temperature; 2.5 μl of 10× orange loading dye (Licor) was added, samples were separated on a native polyacrylamide gel, and subsequently, the gel was scanned with the Odyssey infrared imager from Licor.
Immunofluorescence.
BMMϕ (4 × 104 cells) were plated on glass slides and stimulated with IFN-γ (100 U/ml) for 1 h, 8 h, or 24 h or left untreated. The cells were fixed with 4% Histofix (purchased from Roth) and then quenched with glycine (100 mM) for 15 min. The cells were permeabilized by methanol treatment at −20°C for 5 min. Slides were blocked with 1% bovine serum albumin (BSA) (purchased from Roth) in Dulbecco's phosphate-buffered saline (PBS) (purchased from PAA) at room temperature for 1 h. Anti-phospho-Tyr701 STAT1 was used as a primary antibody (1:100 in 1% BSA-PBS solution at 4°C overnight), and a fluorescently labeled anti-rabbit secondary antibody (Alexa Fluor 488) was used for detection (1:200 in 1% BSA-PBS solution at room temperature for 1 h). Both antibodies were purchased from Cell Signaling Technology. For counterstaining of nuclei, diamidino-2-phenylindole (DAPI) (100 ng/ml; Sigma) was used at room temperature for 5 min. For detection of fluorescence signals, a Leica DM5500B microscope was used.
Nuclear extracts.
BMMϕ were stimulated with IFN-γ (100 U/ml) for 1 h, 24 h, or 48 h or left untreated. The cells were washed with Dulbecco's PBS, resuspended in buffer A (10 mM HEPES, pH 7.9, 10 mM KCl, 0.1 mM [each] EDTA and EGTA), and left on ice for 15 min. Igepal CA-630 (NP-40; Sigma) was added at a final concentration of 0.6%, vortexed vigorously for 10 s, and centrifuged for 1 min (13,000 × g at 4°C). The supernatant, containing cytoplasmic proteins, was collected. The nuclear pellets were washed with Dulbecco's PBS, resuspended in buffer B (20 mM HEPES, pH 7.9, 25% glycerol, 400 mM NaCl, 1 mM [each] EDTA and EGTA), and incubated for 40 min on a shaker at 4°C. Samples were centrifuged for 5 min (13,000 × g at 4°C), and the supernatants, containing nuclear proteins, were collected. Both buffers contain 2 mM DTT, 0.4 mM Na3VO4, 25 mM NaF, 1 mM phenylmethylsulfonyl fluoride (PMSF), 20 μg/ml leupeptin, 20 U/ml aprotinin, 2 μg/ml pepstatin A. Proteins were used for WB analysis as described above. Anti-specificity protein 1 (anti-SP1) was purchased from Santa Cruz Biotechnology and anti-GAPDH (anti-glyceraldehyde-3-phosphate dehydrogenase) from Cell Signaling Technology.
ChIP assays and RT-qPCR.
The chromatin immunoprecipitation (ChIP) protocol was adapted from Nissen and Yamamoto (26), and quantitative PCRs (qPCRs) were performed with the following primers (purchased from Sigma-Aldrich): Gbp2 dist fwd (5′-TGATTTCCCAGCATTTGACA-3′) and rev (5′-AGGGTGAAAAGGGTGTGGTT-3′) (4); Irf1 fwd (5′-GGAGCACAGCTGCCTTGTACTT-3′) and rev (5′-CCCACTCGGCCTCATCATT-3′) (NCBI reference sequence NC000077.5). For reverse transcription (RT)-qPCR, 2 μl of DNA was used in a total volume of 25 μl containing 2.5 mM MgCl2, 300 nM fwd/rev primer, 0.2× EvaGreen dye (Biotium), 1 unit HotFire DNA polymerase (Solis Biodyne), 1× HotFire buffer B (Solis Biodyne), and 200 μM dNTP mixture (MBI Fermentas). The cycling conditions were as follows: 15 min at 95°C, followed by 40 cycles of 20 s at 95°C and 1 min at 60°C, with a subsequent melting curve for quality control. RT-qPCR was performed in duplex on a Stratagene MX3000 machine. Values were normalized to the input control and calculated relative to unstimulated WT cells. Statistical analysis (analysis of variance [ANOVA]) was performed with SPSS software.
RT-PCR and RT-qPCR.
Total RNA was isolated from cells or organs using TRIzol reagent (Invitrogen) according to the manufacturer's instructions. cDNA synthesis and statistical analysis of RT-qPCR data were performed as described previously (27). RT-PCRs were performed with the following primers specific for the detection of Stat1α and Stat1β: Stat1α/β fwd (5′-ATGATGGGTGCATTA-3′), Stat1α rev (5′-GTGCTCATCATACTGTCAA-3′), and Stat1β rev (5′-TTACACTTCAGACACA-3′). RT-PCR was done as described above using the following PCR program: 95°C for 5 min, followed by 32 cycles of 95°C for 30 s, 55°C for 30 s, and 72°C for 45 s. Cyclophilin was used as the endogenous control with the primers Cyclophilin-fwd (5′-GACGCCACTGTCGCTTTTCG-3′) and -rev (5′-CAGGACATTGCGAGCAGATGG-3′) (PCR program: 95°C for 5 min, followed by 25 cycles of 95°C for 30 s, 64°C for 30 s, and 72°C for 30 s). All primers were purchased from Sigma-Aldrich or Qiagen and used at a final concentration of 1 μM. The primers used for RT-qPCR are accessible via the Real-Time Primer and Probe Database (http://medgen.ugent.be/rtprimerdb/): Ube2d2 (ID 3377), myxovirus resistance 1 gene (Mx1) (ID 3887), C-X-C motif chemokine 10 gene (Cxcl10) (ID 3890), Irf1 (ID 3848), and nitric oxide synthase 2 gene (Nos2) (ID 3483). qPCR for the guanylate-binding protein 2 gene (Gbp2) using EvaGreen Dye (Biotium) was performed as described previously (28). All other primers for qPCRs were purchased from Qiagen: nitric oxide synthase 2 gene (Nos2) (QT01059268), class II transactivator gene (CIIta) (QT00153398), Cnn3 (QT00164283), lymphocyte antigen 6 complex locus I gene (Ly6i) (QT02328882), Sell (QT00101164), placenta-specific gene 8 (Plac8) (QT00126917), G protein-coupled receptor 33 gene (Gpr33) (QT00248353), Isg20 (QT01055803), and Upp1 (QT00108850). QPCRs were performed in duplicate on a Stratagene MX3000 machine.
Infections of mice.
Infections with encephalomyocarditis virus (EMCV), vesicular stomatitis virus (VSV), and salivary gland-derived murine cytomegalovirus (MCMV) were performed as previously described (24, 29). Listeria monocytogenes strain EGD was intraperitoneally (i.p.) injected. Age-matched (8 to 12 weeks) and sex-matched (males for L. monocytogenes and VSV; mixed sexes for all others) mice were used. Murine rotavirus (strain EDIM) was applied orally in 20 μl of a 10-fold-diluted virus stock prepared as described previously (30). At the indicated times, two fresh stool pellets were collected from each animal and homogenized, and the rotavirus antigen concentration was determined with the rotavirus ELISA kit (R-Biopharm) according to the manufacturer's instructions. Mice were anesthetized by i.p. injection of a mixture of ketamine and xylazine before intranasal infection with influenza A virus strain SC35M (H7N7) in 40 μl of PBS-0.3% BSA. Statistical analysis of survival curves and rotavirus loads was carried out with Graph Pad Prism (log-rank Mantel-Cox and ANOVA).
Determination of bacterial loads in organs.
Livers and spleens were isolated on day 3 or 5 postinfection and homogenized in 5 ml and 2 ml endotoxin-free PBS (Sigma-Aldrich), respectively. The homogenates were serially diluted 1:10 in PBS and plated in triplicate on Listeria selective agar plates (Oxford Listeria selective agar and supplement; Merck). After 2 days of incubation at 37°C, colonies were counted. Statistical analysis (ANOVA) was performed with SPSS software.
Microarray analysis.
Isolation of total RNA was performed as described above. Sample quality was checked using a Bioanalyzer (Agilent). Only RNA samples with RNA integrity numbers (RIN factor) of >9 were used for further processing. Affymetrix GeneChip Mouse Gene 1.0 ST array analysis was performed by the Center of Medical Research (ZMF), Division Core Facility Molecular Biology, Medical University Graz, Graz, Austria. Affymetrix GeneChip Mouse Gene 1.0 ST Arrays (Affymetrix) were used to determine the transcriptional profile of tissue samples (in biological triplicates) using the Ambion WT Expression kit (Life Technologies), the GeneChip WT Terminal Label and Controls kit (Affymetrix), and the GeneChip Hybridization, Wash and Stain kit (Affymetrix) according to the manufacturers' protocols. Briefly, 100 ng of total RNA was used for reverse transcription, cDNA synthesis, and purification (Ambion). Fragmentation and labeling were performed using the GeneChip WT terminal label and controls kit (Affymetrix) according to the manufacturer's instructions. Labeled samples were loaded and hybridized to GeneChip Mouse Gene 1.0 ST arrays for 16 h overnight at 45°C with rotation in a hybridization oven. The arrays were washed and stained in an Affymetrix GeneChip fluidics station 450, according to the Affymetrix protocols. The arrays were scanned with the GeneChip Scanner GCS3000. For generation of probe set expression values, CEL files containing probe level data were normalized using the robust multichip average (RMA) algorithm (including background correction, quantile normalization across all arrays, and median polished summarization based on log-transformed expression values) implemented in Partek Software v.6.5 (Partek Inc.). Subsequently, the data were log2 transformed. Differences among genotypes within time points and within genotypes among time points were tested with ANOVA or t tests using the appropriate contrasts. As criteria for filtering strongly regulated transcripts, a minimum difference in the log mean of 2-fold up- or downregulation and significance at a P value of ≤0.05 were used throughout.
Microarray data accession number.
The microarray data have been deposited in the Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE48970.
RESULTS
Both STAT1α and STAT1β maintain their basal expression levels.
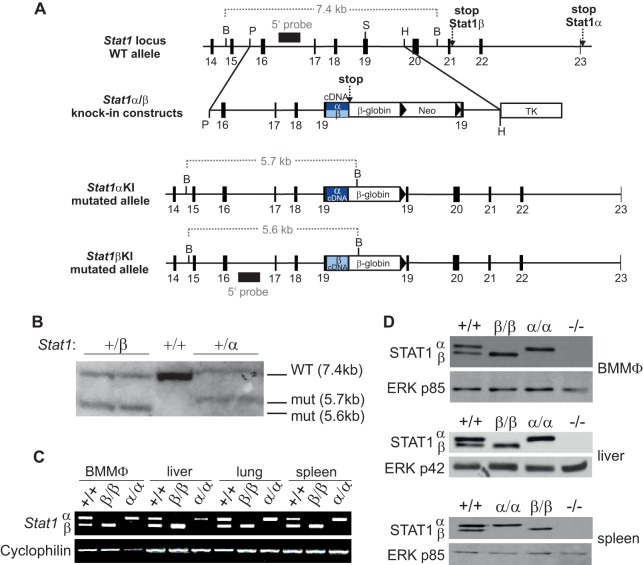
Mice deficient for either STAT1α or STAT1β were generated by a strategy similar to that previously used for STAT3 isoform-deficient mice (31) (Fig. 1A). Homologous integration of targeting constructs in transfected ES cells was verified by PCR (data not shown) and Southern blot analysis (Fig. 1B). B6.129P2-Stat1β/βtmBiat mice (expressing only the STAT1β isoform and referred to here as Stat1β/β mice) and B6.129P2-Stat1α/αtmBiat mice (expressing only the STAT1α isoform and referred to here as Stat1α/α mice) were viable and were born at Mendelian ratios. Similarly to mice deficient for both STAT1 isoforms (7, 8), they showed no gross differences in body size and weight from WT mice. The mRNAs and proteins of both isoforms were detectable in WT cells and organs, whereas Stat1β/β cells/organs expressed only the Stat1β and Stat1α/α cells/organs and only the Stat1α mRNA and protein (Fig. 1C and D). The levels of the single STAT1 isoforms were comparable to the sum of both isoforms in WT cells (Fig. 1D). As STAT1 protein levels are maintained by constitutive low-level type I IFN signaling (32), the data are consistent with the notion that STAT1 isoforms are functionally redundant within an ISGF3 complex.
FIG 1.
Generation of STAT1α and STAT1β isoform-specific knock-in mice. (A) Schematic illustration of the genomic organization of the Stat1 locus from exon 14 to exon 23 (black boxes). Knock-in (KI) constructs and the structure of recombined alleles after insertion are shown below. Restriction endonuclease sites used for cloning and Southern blot analysis and the resulting fragment sizes are indicated. B, BamHI; P, PmlI; S, SmaI; H, HindIII; β-globin, human β-globin splice; Neo, neomycin cassette flanked by loxP sites (triangles); TK, herpes simplex virus thymidine kinase cassette; α, end of Stat1α cDNA (exons 19 to 23); β, end of Stat1β cDNA (exons 19 to 21); stop, stop codon. (B) Southern blot analysis after digestion with BsrGI of either two Stat1+/β or two Stat1+/α heterozygous ES cell clones and a Stat1+/+ control. Restriction sites and fragment sizes are depicted in panel A. WT, WT allele; mut, mutated allele. (C) Total RNA was extracted from BMMϕ and the indicated organs from Stat1+/+, Stat1β/β, and Stat1α/α mice and subjected to RT-PCR. Cyclophilin was used as the endogenous control. (D) Total-protein extracts of BMMϕ, liver, and spleen from WT (+/+), Stat1β/β (β/β), Stat1α/α (α/α), and Stat1−/− (−/−) mice were used for Western blot analysis. Protein expression of STAT1α and STAT1β was detected with a STAT1 antibody against the N-terminal region of the protein. A pan-ERK antibody was used as a loading control (p85 is depicted).
In the absence of STAT1α, STAT1β shows prolonged Tyr701 phosphorylation.
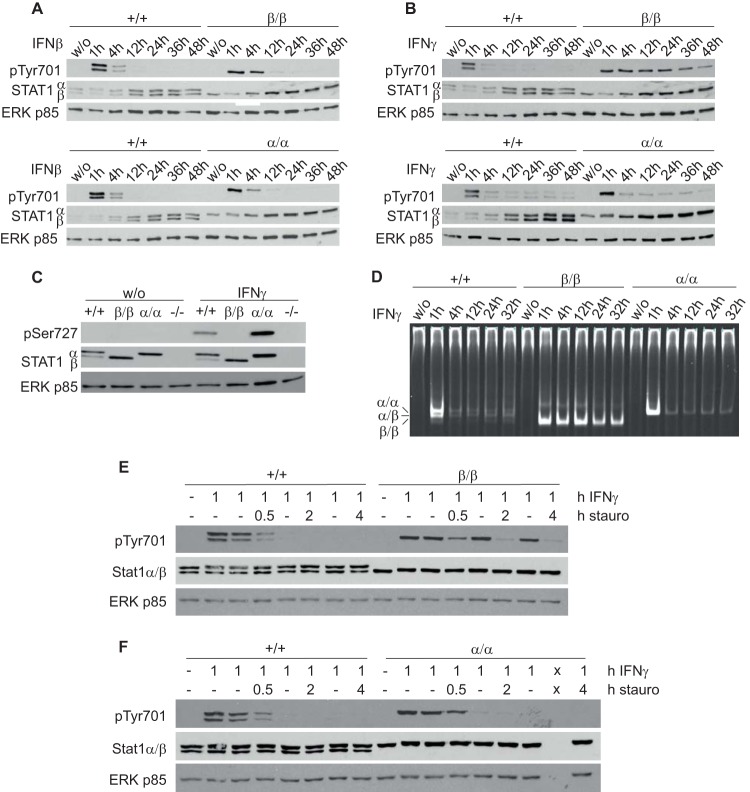
IFN-β induced similar tyrosine 701 phosphorylation (pTyr701) of STAT1 in WT, Stat1β/β, and Stat1α/α BMMϕ (Fig. 2A). In contrast, IFN-γ induced prolonged pTyr701 STAT1 in Stat1β/β compared to WT and Stat1α/α cells (Fig. 2B). Consistent with the autoregulatory function of STAT1 (9, 33, 34), we found upregulation of STAT1 protein at later time points after IFN-β and IFN-γ treatment (Fig. 2A and B). STAT1 protein was also upregulated by IFN-γ in cells expressing only the STAT1β isoform, indicating that STAT1β retains an autoregulatory capacity (Fig. 2B). As expected, phosphorylation of serine 727 (pSer727) could not be detected in cells expressing only the STAT1β isoform (Fig. 2C). In line with the tyrosine phosphorylation data, we found IFN-γ-induced DNA-binding activity of STAT1β homodimers for up to 32 h (Fig. 2D), while in WT- and STAT1α-expressing cells, DNA-binding activity decreased sharply at 4 h. Notably, dimers formed in WT macrophages were mainly STAT1α-STAT1α and STAT1α-STAT1β dimers, both of which showed similar DNA-binding kinetics.
FIG 2.
STAT1β shows prolonged tyrosine 701 phosphorylation. (A to D) BMMϕ derived from WT (+/+), Stat1β/β (β/β), and Stat1α/α (α/α) mice were stimulated with IFN-β (A) or IFN-γ (B to D) for the times indicated or left untreated (w/o). (A to C) Total-protein extracts were used for detection of Tyr701-phosphorylated STAT1 (A and B) or Ser727-phosphorylated STAT1 (C) by Western blotting. (A to C) Membranes were reprobed with N-terminus-specific STAT1 and pan-ERK antibodies. (D) Total cell extracts were used for EMSA using a GAS consensus sequence-containing oligonucleotide (IRDye 700 labeled). α/α, STAT1α-STAT1α; α/β, STAT1α-STAT1β; β/β, STAT1β-STAT1β. (A to D) Results are representative of three independent experiments. (E and F) BMMϕ isolated from WT (+/+) and Stat1β/β (β/β) (E) and WT (+/+) and Stat1α/α (α/α) (F) mice were stimulated with IFN-γ for 1 h or left untreated (−), followed by incubation with or without (−) staurosporine (stauro) (500 nM) for the times indicated. Total-protein extracts were used for detection of Tyr701-phosphorylated STAT1 by Western blotting. The data are representative of four independent experiments. x, empty lane.
To investigate the cause of prolonged Tyr701 phosphorylation of STAT1β, cells were treated with IFN-γ for 1 h before addition of the kinase inhibitor staurosporine to inhibit ongoing STAT1 phosphorylation. Addition of staurosporine led to a rapid decrease of STAT1α and STAT1β phosphorylation in WT cells (Fig. 2E). Exposure of Stat1β/β cells to staurosporine for 0.5 h resulted in a clear reduction in STAT1β phosphorylation, indicating that STAT1β is not resistant to phosphatases. As expected, the rate of dephosphorylation of STAT1α in Stat1α/α cells was similar to that observed in WT cells (Fig. 2F).
In the absence of STAT1α, STAT1β shows prolonged nuclear localization and prolonged binding to endogenous promoter elements.
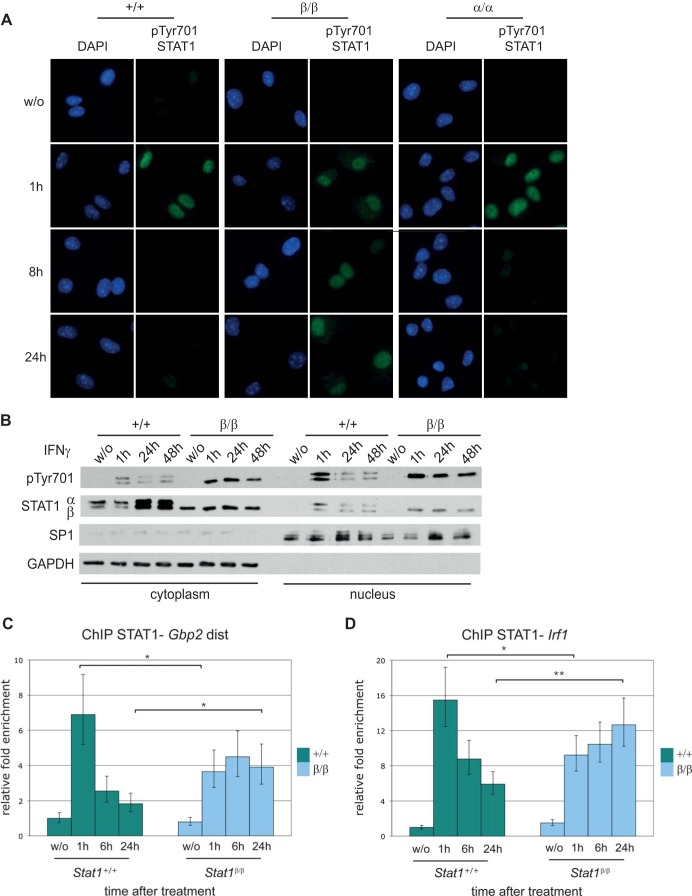
We next addressed whether prolonged pTyr701 of STAT1β affects the protein's intracellular localization. Nuclear pTyr701 STAT1 was detectable by immunofluorescence for up to 24 h after IFN-γ stimulation in Stat1β/β cells, whereas nuclear levels of pTyr701 STAT1 decreased markedly between 1 h and 8 h after treatment in both WT and Stat1α/α cells (Fig. 3A). Similar results were obtained in cellular fractionation experiments: we found persistent nuclear localization of pTyr701 STAT1β for up to 48 h after IFN-γ treatment in Stat1β/β cells, whereas levels of nuclear STAT1α and STAT1β decreased sharply between 1 h and 24 h after IFN-γ treatment in WT cells (Fig. 3B).
FIG 3.
STAT1β shows prolonged nuclear localization and prolonged promoter binding after IFN-γ treatment compared to STAT1α. (A) BMMϕ derived from WT (+/+), Stat1β/β (β/β), and Stat1α/α (α/α) mice were stimulated with IFN-γ (100 U/ml) for the times indicated or left untreated (w/o). Phosphorylation of Tyr701 STAT1 was detected with a specific phospho-STAT1 primary antibody and an IRDye 800-labeled secondary antibody (green). DAPI (100 ng/ml) was used for nuclear staining (blue). Fluorescence signals were analyzed with a Leica DM5500B microscope. (B) BMMϕ derived from WT (+/+) and Stat1β/β (β/β) mice were stimulated with IFN-γ as described for panel A. Cytoplasmic and nuclear proteins were isolated and used for Western blotting. Membranes were probed with an anti-phosphotyrosine 701 STAT1 antibody (pTyr701) and an antibody against the STAT1 N-terminal region (STAT1α/β). The purity of fractions was determined by detection of GAPDH (cytoplasm specific) and SP1 (nucleus specific). (C and D) STAT1 binding to the endogenous distal (dist) Gbp2 (C) or Irf1 (D) promoter region was examined by ChIP analysis using an antibody against the N-terminal portion of STAT1 and qPCR. The values were normalized to the input control and calculated relative to untreated WT cells. Mean values ± standard errors (SE) from at least three independent experiments are shown; *, P ≤ 0.05; **, P ≤ 0.01.
To test whether binding of STAT1β to endogenous promoters is also prolonged, we performed site-directed ChIP experiments. WT and Stat1β/β BMMϕ were stimulated with IFN-γ for 1 h, 6 h, and 24 h, and STAT1 binding to guanylate-binding protein 2 gene (Gbp2) and Irf1 promoters was analyzed. These promoters are strongly dependent on STAT1 (35). In WT cells, STAT1 bound strongly to the Gbp2 promoter 1 h after IFN-γ treatment, and binding was clearly lower after 6 h and 24 h (Fig. 3C). The level of STAT1 binding 1 h after treatment was lower in Stat1β/β than in WT cells. However, STAT1 binding was more persistent in Stat1β/β cells than in WT cells, as it did not decrease between 1 h and 24 h after IFN-γ treatment. A consistent pattern was found for STAT1 in the promoter region of Irf1, although binding in WT cells was less transient than at the Gbp2 promoter (Fig. 3D). Again, STAT1 binding persisted for up to 24 h after IFN-γ treatment in Stat1β/β cells.
In response to IFN-γ, STAT1β is transcriptionally active but induces IFN-stimulated genes (ISGs) with a delay or at lower levels than STAT1α.
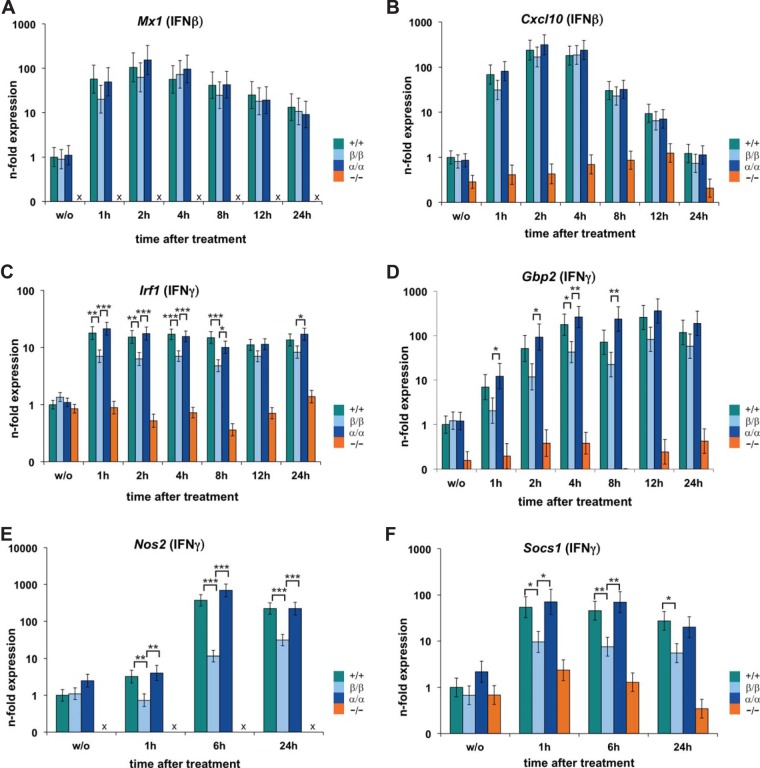
Consistent with redundant roles of STAT1 isoforms within an ISGF3 complex, we found similar induction of Mx1 and Cxcl10 in Stat1β/β, Stat1α/α, and WT cells in response to IFN-β (Fig. 4A and B).
FIG 4.
STAT1β is transcriptionally active in response to IFN-β and IFN-γ. BMMϕ isolated from WT (+/+), Stat1β/β (β/β), Stat1α/α (α/α), and Stat1−/− (−/−) mice were stimulated with IFN-β (A and B) or IFN-γ (C to F) for the times indicated or left untreated (w/o). Total RNA was extracted and used for RT-qPCR analysis for the genes indicated. Ube2d2 was used for normalization, and expression levels were calculated relative to untreated WT cells. Mean values ± SE are given (log scale), and data from at least three independent experiments are shown; x, not detectable; *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001.
Surprisingly, cells expressing only the STAT1β isoform showed clear upregulation of ISGs in response to IFN-γ (Fig. 4C to F). Induction of Irf1 and Gbp2 was around 3- to 10-fold lower in Stat1β/β than in Stat1α/α and WT cells at 1 h to 8 h after stimulation but reached levels similar to those in Stat1α/α and WT cells at later time points (Fig. 4C and D). Similar results were obtained for Cxcl10 (data not shown), whereas the expression of Nos2 and Socs1 was persistently reduced in Stat1β/β compared to Stat1α/α and WT cells (Fig. 4E and F). We did not observe any differences in the induction of ISGs between Stat1α/α and WT cells (Fig. 4C to F and data not shown). The data clearly show that STAT1β is transcriptionally active, albeit at a generally lower level than STAT1α. The transcriptional activity of STAT1β was not specific for macrophages: we found induction of ISGs (i.e., Irf1, Cxcl10, Gbp2, and Stat1) and antiviral activity in IFN-γ-treated Stat1β/β primary embryonic fibroblasts (data not shown).
STAT1α and STAT1β affect the transcription of distinct sets of genes.
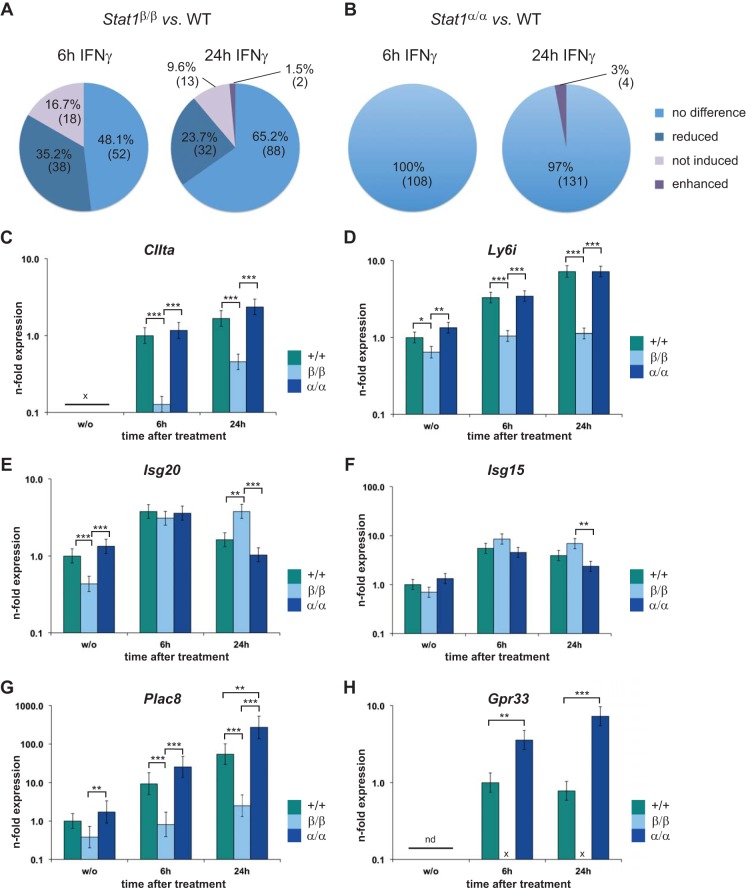
Microarray analysis of WT, Stat1β/β, Stat1α/α, and Stat1−/− BMMϕ before and after 6 h or 24 h of IFN-γ treatment was used to dissect STAT1 isoform-specific transcriptional activities (Fig. 5A and B). Expression data were filtered for at least 4-fold induction by IFN-γ in WT cells and for dependence on STAT1. Unexpectedly, between 48% and 65% of the transcripts were expressed similarly in WT and Stat1β/β BMMϕ (Fig. 5A; see Table S1 in the supplemental material). They included many of the classical ISGs regulated by GAS (e.g., gamma interferon-inducible protein 47 gene [Ifi47]), ISRE (e.g., Mx1), or both consensus sites (e.g., Gbp4). Around 52% and 35% of transcripts were differentially expressed between WT and Stat1β/β cells at 6 h and 24 h after treatment (Fig. 5A; see Table S2 in the supplemental material). Consistent with the RT-qPCR data (Fig. 4E and F), these transcripts included Nos2 and Socs1. The majority of the differentially expressed transcripts were still induced by STAT1β, but a subset (16.7% and 9.6%) were very weakly or not at all induced in Stat1β/β cells, indicating an absolute requirement for STAT1α for the regulation of some IFN-γ-responsive genes (Fig. 5A; see Table S2 in the supplemental material). This pattern was confirmed by RT-qPCR validation experiments for a set of genes (Fig. 5C and D and data not shown). Promoter analysis showed no correlation between the presence of GAS or ISRE consensus sites in gene promoters and the requirement for the presence of STAT1α for their induction (data not shown). The levels of some transcripts were enhanced in Stat1β/β cells at 24 h after treatment (Fig. 5A): this was the case for two transcripts that were at least 4-fold induced in WT cells (see Table S2 in the supplemental material) and for 14 transcripts that the microarray analysis indicated to be induced by less than 4-fold in WT cells, including the classical ISGs Isg20 and Isg15 (Fig. 5E and F and data not shown).
FIG 5.
Transcriptional activities of STAT1α and STAT1β overlap but are nonredundant. BMMϕ isolated from WT (+/+), Stat1β/β (β/β), Stat1α/α (α/α), and Stat1−/− (−/−) mice were stimulated with IFN-γ for the times indicated or left untreated (w/o). Total RNA was extracted and subjected to microarray (A and B) or RT-qPCR (C to H) analysis. (A and B) Summary of data derived from three independent experiments. Transcripts at least 4-fold (significantly) induced in WT cells were selected for each time point. To specifically analyze STAT1-dependent genes, only transcripts that were differentially expressed between WT and Stat1−/− cells were included in the analysis. Expression of the transcripts (108 at 6 h and 135 at 24 h of treatment) was compared between Stat1β/β and WT (A) and Stat1α/α and WT (B) cells, and the percentages and numbers of transcripts that showed common expression patterns are depicted. No difference, <2-fold difference and/or P > 0.05; reduced, lower expression than in WT cells but still induced by treatment; not induced, lower expression than in WT cells and not induced by treatment; differential expression was defined as at least 2-fold difference and a P value of ≤0.05. (C to H) RT-qPCR validation of expression patterns was performed as described in the legend to Fig. 4 for CIIta (C), Ly6i (D), Isg20 (E), Isg15 (F), Plac8 (G), and probable Gpr33 (H). Expression levels of genes that were not detectable in untreated WT cells were calculated relative to 6-h-treated WT cells. Mean values ± SE are given (log scale), and data from at least three independent experiments (different from those used for microarray analysis) are shown; x or nd, not detectable; *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001. See Tables S1 and S2 in the supplemental material for lists of differentially expressed genes.
The absence of STAT1β had a comparatively small effect. None of the genes regulated by STAT1 required STAT1β for upregulation by IFN-γ (see Table S3 in the supplemental material). In contrast, a few transcripts showed increased levels of expression in Stat1α/α compared to WT cells at 24 h after treatment (Fig. 5B; see Table S4 in the supplemental material). Furthermore, three genes, including Plac8 and Gpr33, which the microarray analysis indicated to be less than 4-fold induced in WT cells, showed increased expression in Stat1α/α cells (Fig. 5G and H; see Table S4 in the supplemental material). Induction of these two genes was strictly dependent on the presence of STAT1α.
STAT1α and STAT1β make distinct contributions to host defense against infections.
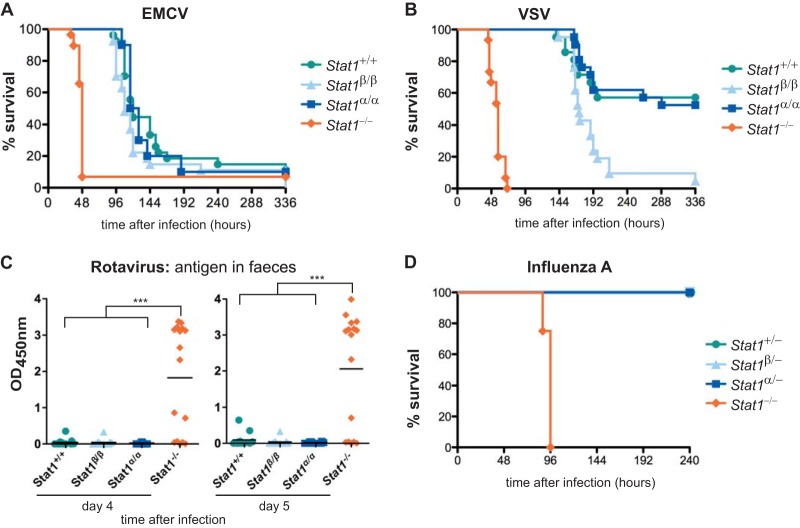
To test the contributions of the two STAT1 isoforms to type I IFN-dependent immunity in vivo, we infected mice with EMCV and VSV, two viruses that are mainly controlled by type I IFNs (36–38). C57BL/6N mice are sensitive to EMCV infection, and 90% of mice died within 8 days of infection with 50 PFU. WT, Stat1α/α, and Stat1β/β mice showed no differences in survival, but survival was clearly extended compared to Stat1−/− mice (Fig. 6A). Stat1β/β mice were more sensitive than WT and Stat1α/α mice to intranasal infection with VSV (Fig. 6B). However, Stat1β/β mice showed strongly increased survival over Stat1−/− mice, confirming that STAT1β significantly contributes to host immunity in vivo. Lack of STAT1β had no impact on survival after challenge with EMCV and VSV, indicating that STAT1α alone is sufficient to establish full antiviral immunity.
FIG 6.
STAT1α and STAT1β can mediate type I and type III IFN-dependent antiviral immunity in vivo. (A) EMCV (50 PFU/mouse) was administered i.p. to WT (Stat1+/+), Stat1β/β, Stat1α/α, and Stat1−/− mice, and survival was monitored for 14 days. The data are derived from two independent experiments; n = 20 (Stat1+/+), n = 18 (Stat1β/β), n = 10 (Stat1α/α), and n = 18 (Stat1−/−). (B) VSV (104 PFU/mouse) was administered intranasally to mice of the genotypes indicated, and survival was monitored for 14 days. The data are from two independent experiments; n = 21 (Stat1+/+, Stat1β/β, and Stat1α/α) and n = 15 (Stat1−/−). Significances were as follows: Stat1−/− versus all others, P < 0.001; Stat1β/β versus WT, P < 0.01; Stat1β/β versus Stat1α/α, P < 0.001. (C) Rotavirus was administered orally to adult mice, and virus shedding in feces on days 4 and 5 postinfection was determined by enzyme-linked immunosorbent assay (ELISA). Pooled data from two independent experiments with 16 or 17 animals per genotype are shown. ***, P ≤ 0.001. (D) Influenza A virus strain SC35M (104 PFU/mouse) was administered intranasally to mice carrying one functional allele of the IFN-regulated Mx1 gene, and survival was monitored for 10 days. n = 6 (Stat1+/−), n = 7 (Stat1β/−), n = 8 (Stat1α/−), and n = 4 (Stat1−/−). Significance, Stat1−/− versus all others, P < 0.001.
Control of rotavirus growth in gut epithelial cells is strongly dependent on a functional IFN-λ system (30). We orally infected WT and mutant mice with murine rotavirus and measured virus antigen in feces. Antigen levels in feces of Stat1α/α and Stat1β/β mice were as low as in those of WT mice but were very high in Stat1−/− mice (Fig. 6C), indicating that each STAT1 isoform is capable of transmitting activating signals from the IFN-λ receptor. The antiviral immune response to influenza A virus in the respiratory tract depends on the IFN-α/β system and, to a lesser extent, on the IFN-λ system (39, 40). As the IFN-mediated protection against influenza A virus is pronounced only if mice carry at least one functional allele of the IFN-regulated Mx1 gene (41), we crossed Stat1α/α and Stat1β/β females to Mx1+/+ Stat1−/− males and challenged the Mx1-heterozygous F1 offspring with mouse-adapted influenza A virus strain SC35M (42). The 50% lethal dose of SC35M for Stat1−/− mice is below 10 PFU, whereas it is approximately 105 PFU for Stat1+/+ mice with functional Mx1 alleles (reference 42 and unpublished results). Stat1α/α and Stat1β/β mice showed only minimal transient weight loss when challenged with 104 PFU of SC35M, and all survived the infection, as did the Stat1+/− control animals (Fig. 6D and data not shown).
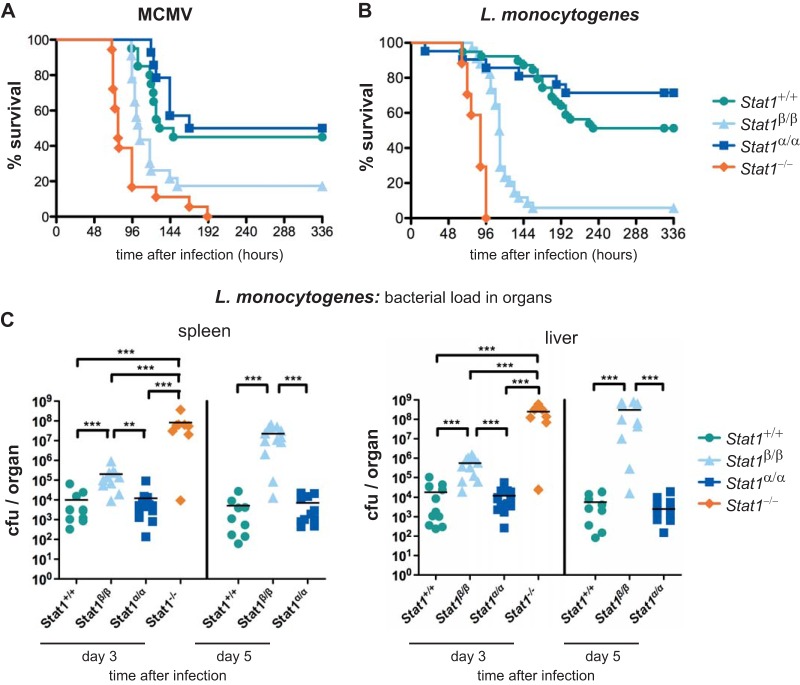
To assess the ability of STAT1β to mediate IFN-γ-dependent antiviral and antibacterial immunity, we challenged mice with MCMV and the intracellular bacterium L. monocytogenes. Host survival upon MCMV infection relies on STAT1 and requires functional IFN-α/β and IFN-γ signaling (43). WT and Stat1α/α mice showed similar survival rates, but Stat1β/β mice were clearly more susceptible to infection (Fig. 7A). Nevertheless, Stat1β/β mice survived significantly longer than Stat1−/− mice. Protective innate immunity against L. monocytogenes requires IFN-γ/STAT1-dependent macrophage activation (44, 45). Stat1β/β mice were more susceptible to infection than WT and Stat1α/α mice but were clearly more resistant than Stat1−/− mice (Fig. 7B). The slightly increased rate of survival of Stat1α/α over WT mice was not statistically significant. Consistent with the survival rates, bacterial loads in organs from Stat1β/β mice were significantly lower than in those from Stat1−/− mice (Fig. 7C). In contrast to its activity against MCMV, STAT1β's anti-Listeria activity cannot be attributed to its effect on IFN-α/β signaling, as this is deleterious rather than protective in the infection model used (46–48). No difference in bacterial load between WT and Stat1α/α mice was found at 3 and 5 days postinfection (Fig. 7C). Although STAT1α is of major importance for immunity against MCMV and L. monocytogenes, STAT1β can induce antimicrobial immunity, albeit with reduced effectiveness compared to the full-length isoform.
FIG 7.
STAT1α and STAT1β show differential efficiencies in immune defense against MCMV and L. monocytogenes infections. (A) WT (Stat1+/+), Stat1β/β, Stat1α/α, and Stat1−/− mice were infected i.p. with MCMV (4 × 105 PFU/mouse), and survival was monitored for 14 days. The data are derived from two independent experiments; n = 20 (Stat1+/+), n = 23 (Stat1β/β), n = 14 (Stat1α/α), and n = 18 (Stat1−/−). Significances were as follows: Stat1β/β versus all others, P < 0.01; Stat1−/− versus WT and Stat1α/α, P < 0.001. (B) WT (Stat1+/+), Stat1β/β, Stat1α/α, and Stat1−/− mice were infected i.p. with L. monocytogenes (2 × 105 CFU/mouse), and survival was monitored for 14 days. The data are derived from four independent experiments. n = 39 (Stat1+/+), n = 34 (Stat1β/β), n = 21 (Stat1α/α), and n = 17 (Stat1−/−). Significances were as follows: Stat1β/β versus all others, P < 0.0001; Stat1−/− versus all others, P < 0.0001. (C) Mice were infected as for panel B and killed 3 or 5 days postinfection. Whole spleens and livers were removed and homogenized, and bacterial loads were determined with standard CFU assays. The data are from two independent experiments. Days 3 and 5, n = 11 and 9 (Stat1+/+), n = 11 and 12 (Stat1β/β), n = 13 and 11 (Stat1α/α), and n = 8 and 0 (Stat1−/−); because of their early lethality after infection, Stat1−/− mice had to be excluded from the analysis at day 5. **, P ≤ 0.01; ***, P ≤ 0.001.
DISCUSSION
It was previously believed that STAT1β is transcriptionally inactive and thus a dominant-negative regulator of STAT1α homodimers. We present two novel gene-targeted mice that allow us to assess specific functions of the STAT1 isoforms in primary cells and in vivo. Our data show for the first time that STAT1β is not merely a transcriptionally inactive STAT1 isoform. More than half of the IFN-γ-responsive genes can be induced by STAT1β, although some of them at reduced levels compared to STAT1α (Fig. 4C to F and 5A and C; see Table S1 in the supplemental material). Despite the large overlap in the genes regulated, the two isoforms display distinct transcriptional profiles. Regulation of some genes strictly depends on the presence of STAT1α (Fig. 5D to F; see Table S1 in the supplemental material), whereas STAT1β is required to limit the expression of a small number of genes, particularly at later time points (Fig. 5G and H; see Table S2 in the supplemental material). Importantly, STAT1β's transcriptional activity is associated with functional biological responses, i.e., IFN-γ-induced antiviral activity in vitro (data not shown) and IFN-γ-dependent antimicrobial immunity in vivo (Fig. 7).
We have no evidence for functional differences between STAT1α and STAT1β in IFN-α/β- and IFN-λ-mediated immune responses. This finding is in line with previous reports that STAT2 provides the activating TAD within an ISGF3 complex (15, 49–52). Stat1β/β, Stat1α/α, and WT mice survive at similar rates after challenge with EMCV (Fig. 6A), a virus controlled by type I IFNs in vivo (36, 38). STAT1β also contributes to the immune defense against VSV (Fig. 6B), which is cleared as a result of type I IFN signaling (37, 53), but surprisingly, less efficiently than STAT1α. The reason for the discrepancy is unclear but may be related to a contribution of IFN-γ to anti-VSV defense in the central nervous system (54). The STAT1 isoforms were also equally effective in controlling rotavirus infections (Fig. 6C), which are controlled by IFN-λ-mediated antiviral immunity (30). Further support for the notion that STAT1 isoforms are functionally redundant within an ISGF3 complex, irrespective of whether activated by IFN-α/β or IFN-λ, comes from our finding that Stat1β/β and Stat1α/α mice show similar resistance against influenza A virus infection (Fig. 6D) (39, 40).
STAT3 and STAT4 also express alternatively spliced β-isoforms. As for STAT1, the β-isoforms were originally considered to be transcriptionally inactive, but this belief has been challenged by the analysis of Stat3 isoform-specific knock-in (31, 55) and Stat4 isoform-transgenic mice (56–58). Both STAT3β and STAT4β are known to be transcriptionally active and to exert functions that overlap, but are not identical to, those of the α-isoforms. It is unknown how the STAT1, STAT3, and STAT4 β-isoforms induce transcription despite lacking a C-terminal TAD. One possibility is that they interact with other transcription factors that provide a functional TAD. This has been shown to be the case for STAT3β and Jun/Fos family members at the α-macroglobulin promoter (59, 60). Alternatively, essential coactivators could be recruited through regions other than the C-terminal TAD. For example, the N-terminal domain of STAT1 has been reported to interact with CBP (61). The interaction may occur with lower affinity but might nevertheless suffice for transcriptional activation. Other proteins could direct coactivator complexes to regions other than the C-terminal TAD or may provide an alternative recruitment interface: N-myc interactor (Nmi) interacts with the coiled-coil domain of all STATs except STAT2 and enhances recruitment of CBP (62).
STAT1β is able to drive the expression of more than half of the STAT1-dependent IFN-γ target genes. ISRE-driven genes may be induced through IFN-γ-activated ISGF3 or STAT1/IRF9 complexes (63–65). The ability of STAT1β to induce Irf1 may account for ISRE-regulated gene induction during secondary responses. It is unclear how STAT1β induces Irf1, but the mechanism may involve a recently identified ISRE around 7 kb upstream of the transcription start site of the Irf1 gene (66). However, STAT1β is not able to perform all the functions of STAT1α, and our data reveal that over 16% and 9% of genes are induced only by STAT1α. Induction was weak or nonexistent in Stat1β/β cells at 6 h and 24 h after IFN-γ treatment, whereas it was unimpaired in Stat1α/α cells (Fig. 5A and B; see Tables S2 and S3 in the supplemental material). Differential responsiveness to the STAT1 isoforms does not obviously correlate with the presence of GAS, ISREs, or both (data not shown), arguing against the possibility that STAT1β acts solely in conjunction with IRF9 and/or STAT2 or through the induction of IRF1. It thus seems as though the requirement for the STAT1 C-terminal TAD depends on the precise structure of the target gene promoter and its ability to recruit other gene-specific transcription factors and/or coregulators. A few genes showed enhanced expression in cells expressing STAT1β alone (Fig. 5B, E, and F; see Table S2 in the supplemental material). This may be due to the prolonged activation of STAT1β and increased formation of ISGF3-II, which contains unphosphorylated STAT2 and regulates ISGF3-driven genes, in particular, at late time points after IFN-γ treatment (67).
The importance of the C-terminal TAD of STAT1 for the regulation of IFN-γ responses is consistent with previous results suggesting that CDK8-mediated Ser727 phosphorylation of STAT1α is required for full induction of IFN-γ-responsive genes (2, 4). However, in some cases, phosphorylation of STAT1 on Ser727 inhibits gene expression (2). Although the genes whose expression was reduced in Stat1β/β cells were similar to those reduced in Stat1S727A cells (e.g., Irf1, Gbp2, and Nos2), we found fewer genes with enhanced expression. Future work will dissect which aspects of STAT1-dependent transcriptional control rely on the presence of the C-terminal region and which on its serine phosphorylation.
The prolonged activation, nuclear localization, and chromatin association of STAT1β in cells lacking STAT1α may also play a part in the protein's function (Fig. 2B and D and 3). Prolonged activation of STAT1β is not due to resistance against phosphatases (Fig. 2E), although we cannot exclude the possibility that there are subtle differences in the dephosphorylation of the two isoforms. The prolonged activation of STAT1β most likely stems from the reduced induction of Socs1 (Fig. 4F). SOCS1 is a negative-feedback regulator that can bind to and inhibit all four members of the JAK family (68). Absence of SOCS1 results in hyperresponsiveness to IFN-γ and sustained STAT1 phosphorylation (68–70). STAT1β's more persistent activation in the absence of STAT1α may compensate for its weaker transactivation capacity. This explanation would imply that the lower induction of a single target gene, Socs1, affects the overall transcriptional activity of STAT1β. Thus, STAT1β's function in the absence of STAT1α may not be a true reflection of its function in WT cells. Further analysis will be required to determine whether and to what extent STAT1β's functions depend on its prolonged activity.
Our transcriptional data are clearly of physiological relevance. STAT1β alone can mediate IFN-γ-dependent inhibition of bacterial growth and increase host survival after infection with L. monocytogenes (Fig. 7B and C). However, despite the large overlap between STAT1α- and STAT1β-regulated genes, STAT1β is much less potent than STAT1α against MCMV, VSV, and L. monocytogenes infection. This suggests that IFN-γ-dependent innate immunity depends on an appropriately dosed response or on genes selectively regulated by STAT1α. An alternative explanation is that macrophage transcriptional responses may not be representative of the complex immune responses in vivo and that STAT1 isoforms have cell-type-specific transcriptional activities depending on other interacting transcription factors and/or coregulators. Additional experiments will be required to address this possibility.
Another intriguing finding of our study is that mice lacking STAT1β are phenotypically indistinguishable from WT mice with regard to their susceptibility to viral and bacterial infections (Fig. 6 and 7). Furthermore, IFN-γ-mediated transcriptional responses are very similar in cells lacking STAT1β and in WT cells (Fig. 5B; see Table S3 in the supplemental material). Thus, although STAT1β represses some genes (Fig. 5B, G, and H), it is not an inhibitor of STAT1α during host defense against microbial infections. This may be related to its relatively low level in WT cells. STAT1β may have to be present in large excess to block STAT1α activity, an explanation that would be consistent with the fact that inhibitory functions have been reported only when STAT1β is overexpressed (16, 17). In most cells, STAT1α is more abundant than STAT1β, but there has not yet been a thorough analysis of the ratios of STAT1 isoforms in distinct cell types and throughout the course of immune challenges. This raises the possibility that STAT1β functions only in certain cell types and/or during particular immune responses.
More detailed experimentation will be required to understand the distinct functions of the STAT1 isoforms in vivo. It is becoming increasingly evident that the availability of STAT1 determines biological responses (6, 32, 43, 71, 72). The finding that the STAT1 isoforms show distinct functional properties underlines the importance of differentiating between them. Many functions have been ascribed to STAT1, and dividing them into those regulated by STAT1α, STAT1β, or both may help in the design of specific strategies to interfere with STAT1 function and to fine-tune diagnostic approaches.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by Austrian Science Fund (FWF) grants FWF-P25642-B22 to B.S. and SFB-F28 to B.S., M.M., and T.D. T.M. was supported in part by the Excellence Initiative of the German Research Foundation (GSC-4, Spemann Graduate School). M.P. participated in the FWF-funded DK-IAI (W1212).
We declare that we have no conflicts of interest.
We thank Ingeborg Klymiuk for help with the microarray analysis. We are grateful to Pavel Kovarik for fruitful discussions throughout the project and to Graham Tebb for critically reading and editing the manuscript.
Footnotes
Published ahead of print 7 April 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.00295-14.
REFERENCES
- 1.Levy DE, Darnell JE., Jr 2002. Stats: transcriptional control and biological impact. Nat. Rev. Mol. Cell Biol. 3:651–662. 10.1038/nrm909 [DOI] [PubMed] [Google Scholar]
- 2.Bancerek J, Poss ZC, Steinparzer I, Sedlyarov V, Pfaffenwimmer T, Mikulic I, Dolken L, Strobl B, Muller M, Taatjes DJ, Kovarik P. 2013. CDK8 kinase phosphorylates transcription factor STAT1 to selectively regulate the interferon response. Immunity 38:250–262. 10.1016/j.immuni.2012.10.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sadzak I, Schiff M, Gattermeier I, Glinitzer R, Sauer I, Saalmuller A, Yang E, Schaljo B, Kovarik P. 2008. Recruitment of Stat1 to chromatin is required for interferon-induced serine phosphorylation of Stat1 transactivation domain. Proc. Natl. Acad. Sci. U. S. A. 105:8944–8949. 10.1073/pnas.0801794105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Varinou L, Ramsauer K, Karaghiosoff M, Kolbe T, Pfeffer K, Muller M, Decker T. 2003. Phosphorylation of the Stat1 transactivation domain is required for full-fledged IFN-gamma-dependent innate immunity. Immunity 19:793–802. 10.1016/S1074-7613(03)00322-4 [DOI] [PubMed] [Google Scholar]
- 5.Wen Z, Zhong Z, Darnell JE., Jr 1995. Maximal activation of transcription by Stat1 and Stat3 requires both tyrosine and serine phosphorylation. Cell 82:241–250. 10.1016/0092-8674(95)90311-9 [DOI] [PubMed] [Google Scholar]
- 6.Boisson-Dupuis S, Kong XF, Okada S, Cypowyj S, Puel A, Abel L, Casanova JL. 2012. Inborn errors of human STAT1: allelic heterogeneity governs the diversity of immunological and infectious phenotypes. Curr. Opin. Immunol. 24:364–378. 10.1016/j.coi.2012.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Durbin JE, Hackenmiller R, Simon MC, Levy DE. 1996. Targeted disruption of the mouse Stat1 gene results in compromised innate immunity to viral disease. Cell 84:443–450. 10.1016/S0092-8674(00)81289-1 [DOI] [PubMed] [Google Scholar]
- 8.Meraz MA, White JM, Sheehan KC, Bach EA, Rodig SJ, Dighe AS, Kaplan DH, Riley JK, Greenlund AC, Campbell D, Carver-Moore K, DuBois RN, Clark R, Aguet M, Schreiber RD. 1996. Targeted disruption of the Stat1 gene in mice reveals unexpected physiologic specificity in the JAK-STAT signaling pathway. Cell 84:431–442. 10.1016/S0092-8674(00)81288-X [DOI] [PubMed] [Google Scholar]
- 9.Muller M, Laxton C, Briscoe J, Schindler C, Improta T, Darnell JE, Jr, Stark GR, Kerr IM. 1993. Complementation of a mutant cell line: central role of the 91 kDa polypeptide of ISGF3 in the interferon-alpha and -gamma signal transduction pathways. EMBO J. 12:4221–4228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shuai K, Stark GR, Kerr IM, Darnell JE., Jr 1993. A single phosphotyrosine residue of Stat91 required for gene activation by interferon-gamma. Science 261:1744–1746. 10.1126/science.7690989 [DOI] [PubMed] [Google Scholar]
- 11.Zakharova N, Lymar ES, Yang E, Malik S, Zhang JJ, Roeder RG, Darnell JE., Jr 2003. Distinct transcriptional activation functions of STAT1alpha and STAT1beta on DNA and chromatin templates. J. Biol. Chem. 278:43067–43073. 10.1074/jbc.M308166200 [DOI] [PubMed] [Google Scholar]
- 12.Schindler C, Darnell JE., Jr 1995. Transcriptional responses to polypeptide ligands: the JAK-STAT pathway. Annu. Rev. Biochem. 64:621–651. 10.1146/annurev.bi.64.070195.003201 [DOI] [PubMed] [Google Scholar]
- 13.Vinkemeier U, Cohen SL, Moarefi I, Chait BT, Kuriyan J, Darnell JE., Jr 1996. DNA binding of in vitro activated Stat1 alpha, Stat1 beta and truncated Stat1: interaction between NH2-terminal domains stabilizes binding of two dimers to tandem DNA sites. EMBO J. 15:5616–5626 [PMC free article] [PubMed] [Google Scholar]
- 14.Bhattacharya S, Eckner R, Grossman S, Oldread E, Arany Z, D'Andrea A, Livingston DM. 1996. Cooperation of Stat2 and p300/CBP in signalling induced by interferon-alpha. Nature 383:344–347. 10.1038/383344a0 [DOI] [PubMed] [Google Scholar]
- 15.Qureshi SA, Leung S, Kerr IM, Stark GR, Darnell JE., Jr 1996. Function of Stat2 protein in transcriptional activation by alpha interferon. Mol. Cell. Biol. 16:288–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baran-Marszak F, Feuillard J, Najjar I, Le Clorennec C, Bechet JM, Dusanter-Fourt I, Bornkamm GW, Raphael M, Fagard R. 2004. Differential roles of STAT1alpha and STAT1beta in fludarabine-induced cell cycle arrest and apoptosis in human B cells. Blood 104:2475–2483. 10.1182/blood-2003-10-3508 [DOI] [PubMed] [Google Scholar]
- 17.Alvarez GR, Zwilling BS, Lafuse WP. 2003. Mycobacterium avium inhibition of IFN-gamma signaling in mouse macrophages: Toll-like receptor 2 stimulation increases expression of dominant-negative STAT1 beta by mRNA stabilization. J. Immunol. 171:6766–6773 [DOI] [PubMed] [Google Scholar]
- 18.Bhardwaj N, Rosas LE, Lafuse WP, Satoskar AR. 2005. Leishmania inhibits STAT1-mediated IFN-gamma signaling in macrophages: increased tyrosine phosphorylation of dominant negative STAT1beta by Leishmania mexicana. Int. J. Parasitol. 35:75–82. 10.1016/j.ijpara.2004.10.018 [DOI] [PubMed] [Google Scholar]
- 19.Ruvolo V, Navarro L, Sample CE, David M, Sung S, Swaminathan S. 2003. The Epstein-Barr virus SM protein induces STAT1 and interferon-stimulated gene expression. J. Virol. 77:3690–3701. 10.1128/JVI.77.6.3690-3701.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mamo S, Kobolak J, Borbiro I, Biro T, Bock I, Dinnyes A. 2010. Gene targeting and calcium handling efficiencies in mouse embryonic stem cell lines. World J. Stem Cells 2:127–140. 10.4252/wjsc.v2.i6.127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schwenk F, Baron U, Rajewsky K. 1995. A cre-transgenic mouse strain for the ubiquitous deletion of loxP-flanked gene segments including deletion in germ cells. Nucleic Acids Res. 23:5080–5081. 10.1093/nar/23.24.5080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Teppner I, Aigner B, Schreiner E, Muller M, Windisch M. 2004. Polymorphic microsatellite markers in the outbred CFW and ICR stocks for the generation of speed congenic mice on C57BL/6 background. Lab Anim. 38:406–412. 10.1258/0023677041958882 [DOI] [PubMed] [Google Scholar]
- 23.Karaghiosoff M, Neubauer H, Lassnig C, Kovarik P, Schindler H, Pircher H, McCoy B, Bogdan C, Decker T, Brem G, Pfeffer K, Muller M. 2000. Partial impairment of cytokine responses in Tyk2-deficient mice. Immunity 13:549–560. 10.1016/S1074-7613(00)00054-6 [DOI] [PubMed] [Google Scholar]
- 24.Strobl B, Bubic I, Bruns U, Steinborn R, Lajko R, Kolbe T, Karaghiosoff M, Kalinke U, Jonjic S, Muller M. 2005. Novel functions of tyrosine kinase 2 in the antiviral defense against murine cytomegalovirus. J. Immunol. 175:4000–4008 [DOI] [PubMed] [Google Scholar]
- 25.Kovarik P, Stoiber D, Novy M, Decker T. 1998. Stat1 combines signals derived from IFN-gamma and LPS receptors during macrophage activation. EMBO J. 17:3660–3668. 10.1093/emboj/17.13.3660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nissen RM, Yamamoto KR. 2000. The glucocorticoid receptor inhibits NFkappaB by interfering with serine-2 phosphorylation of the RNA polymerase II carboxy-terminal domain. Genes Dev. 14:2314–2329. 10.1101/gad.827900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leitner NR, Strobl B, Bokor M, Painz R, Kolbe T, Rulicke T, Muller M, Karaghiosoff M. 2006. A time- and dose-dependent STAT1 expression system. BMC Biotechnol. 6:48. 10.1186/1472-6750-6-48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stockinger S, Reutterer B, Schaljo B, Schellack C, Brunner S, Materna T, Yamamoto M, Akira S, Taniguchi T, Murray PJ, Muller M, Decker T. 2004. IFN regulatory factor 3-dependent induction of type I IFNs by intracellular bacteria is mediated by a TLR- and Nod2-independent mechanism. J. Immunol. 173:7416–7425 [DOI] [PubMed] [Google Scholar]
- 29.Prchal-Murphy M, Semper C, Lassnig C, Wallner B, Gausterer C, Teppner-Klymiuk I, Kobolak J, Muller S, Kolbe T, Karaghiosoff M, Dinnyes A, Rulicke T, Leitner NR, Strobl B, Muller M. 2012. TYK2 kinase activity is required for functional type I interferon responses in vivo. PLoS One 7:e39141. 10.1371/journal.pone.0039141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pott J, Mahlakoiv T, Mordstein M, Duerr CU, Michiels T, Stockinger S, Staeheli P, Hornef MW. 2011. IFN-lambda determines the intestinal epithelial antiviral host defense. Proc. Natl. Acad. Sci. U. S. A. 108:7944–7949. 10.1073/pnas.1100552108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maritano D, Sugrue ML, Tininini S, Dewilde S, Strobl B, Fu X, Murray-Tait V, Chiarle R, Poli V. 2004. The STAT3 isoforms alpha and beta have unique and specific functions. Nat. Immunol. 5:401–409. 10.1038/ni1052 [DOI] [PubMed] [Google Scholar]
- 32.Gough DJ, Messina NL, Hii L, Gould JA, Sabapathy K, Robertson AP, Trapani JA, Levy DE, Hertzog PJ, Clarke CJ, Johnstone RW. 2010. Functional crosstalk between type I and II interferon through the regulated expression of STAT1. PLoS Biol. 8:e1000361. 10.1371/journal.pbio.1000361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rusinova I, Forster S, Yu S, Kannan A, Masse M, Cumming H, Chapman R, Hertzog PJ. 2013. Interferome v2.0: an updated database of annotated interferon-regulated genes. Nucleic Acids Res. 41:D1040–D1046. 10.1093/nar/gks1215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wong LH, Sim H, Chatterjee-Kishore M, Hatzinisiriou I, Devenish RJ, Stark G, Ralph SJ. 2002. Isolation and characterization of a human STAT1 gene regulatory element. Inducibility by interferon (IFN) types I and II and role of IFN regulatory factor-1. J. Biol. Chem. 277:19408–19417. 10.1074/jbc.M111302200 [DOI] [PubMed] [Google Scholar]
- 35.Ramsauer K, Farlik M, Zupkovitz G, Seiser C, Kroger A, Hauser H, Decker T. 2007. Distinct modes of action applied by transcription factors STAT1 and IRF1 to initiate transcription of the IFN-gamma-inducible gbp2 gene. Proc. Natl. Acad. Sci. U. S. A. 104:2849–2854. 10.1073/pnas.0610944104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen LS, Wei PC, Liu T, Kao CH, Pai LM, Lee CK. 2009. STAT2 hypomorphic mutant mice display impaired dendritic cell development and antiviral response. J. Biomed. Sci. 16:22. 10.1186/1423-0127-16-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Detje CN, Meyer T, Schmidt H, Kreuz D, Rose JK, Bechmann I, Prinz M, Kalinke U. 2009. Local type I IFN receptor signaling protects against virus spread within the central nervous system. J. Immunol. 182:2297–2304. 10.4049/jimmunol.0800596 [DOI] [PubMed] [Google Scholar]
- 38.Fensterl V, Wetzel JL, Ramachandran S, Ogino T, Stohlman SA, Bergmann CC, Diamond MS, Virgin HW, Sen GC. 2012. Interferon-induced Ifit2/ISG54 protects mice from lethal VSV neuropathogenesis. PLoS Pathog. 8:e1002712. 10.1371/journal.ppat.1002712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mordstein M, Kochs G, Dumoutier L, Renauld JC, Paludan SR, Klucher K, Staeheli P. 2008. Interferon-lambda contributes to innate immunity of mice against influenza A virus but not against hepatotropic viruses. PLoS Pathog. 4:e1000151. 10.1371/journal.ppat.1000151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mordstein M, Neugebauer E, Ditt V, Jessen B, Rieger T, Falcone V, Sorgeloos F, Ehl S, Mayer D, Kochs G, Schwemmle M, Gunther S, Drosten C, Michiels T, Staeheli P. 2010. Lambda interferon renders epithelial cells of the respiratory and gastrointestinal tracts resistant to viral infections. J. Virol. 84:5670–5677. 10.1128/JVI.00272-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Grimm D, Staeheli P, Hufbauer M, Koerner I, Martinez-Sobrido L, Solorzano A, Garcia-Sastre A, Haller O, Kochs G. 2007. Replication fitness determines high virulence of influenza A virus in mice carrying functional Mx1 resistance gene. Proc. Natl. Acad. Sci. U. S. A. 104:6806–6811. 10.1073/pnas.0701849104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kochs G, Koerner I, Thiel L, Kothlow S, Kaspers B, Ruggli N, Summerfield A, Pavlovic J, Stech J, Staeheli P. 2007. Properties of H7N7 influenza A virus strain SC35M lacking interferon antagonist NS1 in mice and chickens. J. Gen. Virol. 88:1403–1409. 10.1099/vir.0.82764-0 [DOI] [PubMed] [Google Scholar]
- 43.Gil MP, Salomon R, Louten J, Biron CA. 2006. Modulation of STAT1 protein levels: a mechanism shaping CD8 T-cell responses in vivo. Blood 107:987–993. 10.1182/blood-2005-07-2834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kernbauer E, Maier V, Stoiber D, Strobl B, Schneckenleithner C, Sexl V, Reichart U, Reizis B, Kalinke U, Jamieson A, Muller M, Decker T. 2012. Conditional Stat1 ablation reveals the importance of interferon signaling for immunity to Listeria monocytogenes infection. PLoS Pathog. 8:e1002763. 10.1371/journal.ppat.1002763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lu B, Ebensperger C, Dembic Z, Wang Y, Kvatyuk M, Lu T, Coffman RL, Pestka S, Rothman PB. 1998. Targeted disruption of the interferon-gamma receptor 2 gene results in severe immune defects in mice. Proc. Natl. Acad. Sci. U. S. A. 95:8233–8238. 10.1073/pnas.95.14.8233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Auerbuch V, Brockstedt DG, Meyer-Morse N, O'Riordan M, Portnoy DA. 2004. Mice lacking the type I interferon receptor are resistant to Listeria monocytogenes. J. Exp. Med. 200:527–533. 10.1084/jem.20040976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Carrero JA, Calderon B, Unanue ER. 2004. Type I interferon sensitizes lymphocytes to apoptosis and reduces resistance to Listeria infection. J. Exp. Med. 200:535–540. 10.1084/jem.20040769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.O'Connell RM, Saha SK, Vaidya SA, Bruhn KW, Miranda GA, Zarnegar B, Perry AK, Nguyen BO, Lane TF, Taniguchi T, Miller JF, Cheng G. 2004. Type I interferon production enhances susceptibility to Listeria monocytogenes infection. J. Exp. Med. 200:437–445. 10.1084/jem.20040712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Horvath CM, Stark GR, Kerr IM, Darnell JE., Jr 1996. Interactions between STAT and non-STAT proteins in the interferon-stimulated gene factor 3 transcription complex. Mol. Cell. Biol. 16:6957–6964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Park C, Lecomte MJ, Schindler C. 1999. Murine Stat2 is uncharacteristically divergent. Nucleic Acids Res. 27:4191–4199. 10.1093/nar/27.21.4191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Paulson M, Pisharody S, Pan L, Guadagno S, Mui AL, Levy DE. 1999. Stat protein transactivation domains recruit p300/CBP through widely divergent sequences. J. Biol. Chem. 274:25343–25349. 10.1074/jbc.274.36.25343 [DOI] [PubMed] [Google Scholar]
- 52.Paulson M, Press C, Smith E, Tanese N, Levy DE. 2002. IFN-stimulated transcription through a TBP-free acetyltransferase complex escapes viral shutoff. Nat. Cell Biol. 4:140–147. 10.1038/ncb747 [DOI] [PubMed] [Google Scholar]
- 53.Muller U, Steinhoff U, Reis LF, Hemmi S, Pavlovic J, Zinkernagel RM, Aguet M. 1994. Functional role of type I and type II interferons in antiviral defense. Science 264:1918–1921. 10.1126/science.8009221 [DOI] [PubMed] [Google Scholar]
- 54.Kundig TM, Hengartner H, Zinkernagel RM. 1993. T cell-dependent IFN-gamma exerts an antiviral effect in the central nervous system but not in peripheral solid organs. J. Immunol. 150:2316–2321 [PubMed] [Google Scholar]
- 55.Yoo JY, Huso DL, Nathans D, Desiderio S. 2002. Specific ablation of Stat3beta distorts the pattern of Stat3-responsive gene expression and impairs recovery from endotoxic shock. Cell 108:331–344. 10.1016/S0092-8674(02)00636-0 [DOI] [PubMed] [Google Scholar]
- 56.Hoey T, Zhang S, Schmidt N, Yu Q, Ramchandani S, Xu X, Naeger LK, Sun YL, Kaplan MH. 2003. Distinct requirements for the naturally occurring splice forms Stat4alpha and Stat4beta in IL-12 responses. EMBO J. 22:4237–4248. 10.1093/emboj/cdg393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mo C, Chearwae W, O'Malley JT, Adams SM, Kanakasabai S, Walline CC, Stritesky GL, Good SR, Perumal NB, Kaplan MH, Bright JJ. 2008. Stat4 isoforms differentially regulate inflammation and demyelination in experimental allergic encephalomyelitis. J. Immunol. 181:5681–5690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.O'Malley JT, Eri RD, Stritesky GL, Mathur AN, Chang HC, Hogenesch H, Srinivasan M, Kaplan MH. 2008. STAT4 isoforms differentially regulate Th1 cytokine production and the severity of inflammatory bowel disease. J. Immunol. 181:5062–5070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang X, Darnell JE., Jr 2001. Functional importance of Stat3 tetramerization in activation of the alpha 2-macroglobulin gene. J. Biol. Chem. 276:33576–33581. 10.1074/jbc.M104978200 [DOI] [PubMed] [Google Scholar]
- 60.Zhang X, Wrzeszczynska MH, Horvath CM, Darnell JE., Jr 1999. Interacting regions in Stat3 and c-Jun that participate in cooperative transcriptional activation. Mol. Cell. Biol. 19:7138–7146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang JJ, Vinkemeier U, Gu W, Chakravarti D, Horvath CM, Darnell JE., Jr 1996. Two contact regions between Stat1 and CBP/p300 in interferon gamma signaling. Proc. Natl. Acad. Sci. U. S. A. 93:15092–15096. 10.1073/pnas.93.26.15092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhu M, John S, Berg M, Leonard WJ. 1999. Functional association of Nmi with Stat5 and Stat1 in IL-2- and IFNgamma-mediated signaling. Cell 96:121–130. 10.1016/S0092-8674(00)80965-4 [DOI] [PubMed] [Google Scholar]
- 63.Kimura T, Kadokawa Y, Harada H, Matsumoto M, Sato M, Kashiwazaki Y, Tarutani M, Tan RS, Takasugi T, Matsuyama T, Mak TW, Noguchi S, Taniguchi T. 1996. Essential and non-redundant roles of p48 (ISGF3 gamma) and IRF-1 in both type I and type II interferon responses, as revealed by gene targeting studies. Genes Cells 1:115–124. 10.1046/j.1365-2443.1996.08008.x [DOI] [PubMed] [Google Scholar]
- 64.Zimmermann A, Trilling M, Wagner M, Wilborn M, Bubic I, Jonjic S, Koszinowski U, Hengel H. 2005. A cytomegaloviral protein reveals a dual role for STAT2 in IFN-{gamma} signaling and antiviral responses. J. Exp. Med. 201:1543–1553. 10.1084/jem.20041401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bluyssen HA, Muzaffar R, Vlieststra RJ, van der Made AC, Leung S, Stark GR, Kerr IM, Trapman J, Levy DE. 1995. Combinatorial association and abundance of components of interferon-stimulated gene factor 3 dictate the selectivity of interferon responses. Proc. Natl. Acad. Sci. U. S. A. 92:5645–5649. 10.1073/pnas.92.12.5645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Begitt A, Droescher M, Meyer T, Schmid CD, Baker M, Antunes F, Owen MR, Naumann R, Decker T, Vinkemeier U. 2014. STAT1-cooperative DNA binding distinguishes type 1 from type 2 interferon signaling. Nat. Immunol. 15:168–176. 10.1038/ni.2794 [DOI] [PubMed] [Google Scholar]
- 67.Morrow AN, Schmeisser H, Tsuno T, Zoon KC. 2011. A novel role for IFN-stimulated gene factor 3II in IFN-gamma signaling and induction of antiviral activity in human cells. J. Immunol. 186:1685–1693. 10.4049/jimmunol.1001359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ilangumaran S, Rottapel R. 2003. Regulation of cytokine receptor signaling by SOCS1. Immunol. Rev. 192:196–211. 10.1034/j.1600-065X.2003.00020.x [DOI] [PubMed] [Google Scholar]
- 69.Bullen DV, Darwiche R, Metcalf D, Handman E, Alexander WS. 2001. Neutralization of interferon-gamma in neonatal SOCS1−/− mice prevents fatty degeneration of the liver but not subsequent fatal inflammatory disease. Immunology 104:92–98. 10.1046/j.1365-2567.2001.01294.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Alexander WS, Starr R, Fenner JE, Scott CL, Handman E, Sprigg NS, Corbin JE, Cornish AL, Darwiche R, Owczarek CM, Kay TW, Nicola NA, Hertzog PJ, Metcalf D, Hilton DJ. 1999. SOCS1 is a critical inhibitor of interferon gamma signaling and prevents the potentially fatal neonatal actions of this cytokine. Cell 98:597–608. 10.1016/S0092-8674(00)80047-1 [DOI] [PubMed] [Google Scholar]
- 71.Miyagi T, Gil MP, Wang X, Louten J, Chu WM, Biron CA. 2007. High basal STAT4 balanced by STAT1 induction to control type 1 interferon effects in natural killer cells. J. Exp. Med. 204:2383–2396. 10.1084/jem.20070401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Regis G, Pensa S, Boselli D, Novelli F, Poli V. 2008. Ups and downs: the STAT1:STAT3 seesaw of interferon and gp130 receptor signalling. Semin. Cell Dev. Biol. 19:351–359. 10.1016/j.semcdb.2008.06.004 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.