ABSTRACT
Oncolytic viruses (OVs) are attractive avenues of cancer therapy due to the absence of toxic side effects often seen with current treatment modalities. Bovine herpesvirus 1 (BHV-1) is a species-specific virus that does not induce cytotoxicity in normal primary human cells but can infect and kill various human immortalized and transformed cell lines. To gain a better understanding of the oncolytic breadth of BHV-1, the NCI panel of established human tumor cell lines was screened for sensitivity to the virus. Overall, 72% of the panel is permissive to BHV-1 infection, with corresponding decreases in cellular viability. This sensitivity is in comparison to a sensitivity of only 32% for a herpes simplex virus 1 (HSV-1)-based oncolytic vector. Strikingly, while 35% of the panel supports minimal or no BHV-1 replication, significant decreases in cellular viability still occur. These data suggest that BHV-1 is an OV with tropism for multiple tumor types and is able to induce cytotoxicity independent of significant virus replication. In contrast to other species-specific OVs, cellular sensitivity to BHV-1 does not correlate with type I interferon (IFN) signaling; however, mutations in KRAS were found to correlate with high levels of virus replication. The knockdown or overexpression of KRAS in human tumor cell lines yields modest changes in viral titers; however, overexpression of KRAS in normal primary cells elicits permissivity to BHV-1 infection. Together, these data suggest that BHV-1 is a broad-spectrum OV with a distinct mechanism of tumor targeting.
IMPORTANCE Cancer remains a significant health issue, and novel treatments are required, particularly for tumors that are refractory to conventional therapies. Oncolytic viruses are a novel platform given their ability to specifically target tumor cells while leaving healthy cells intact. For this strategy to be successful, a fundamental understanding of virus-host interactions is required. We previously identified bovine herpesvirus 1 as a novel oncolytic virus with many unique and clinically relevant features. Here, we show that BHV-1 can target a wide range of human cancer types, most potently lung cancer. In addition, we show that enhanced KRAS activity, a hallmark of many cancers, is one of the factors that increases BHV-1 oncolytic capacity. These findings hold potential for future treatments, particularly in the context of lung cancer, where KRAS mutations are a negative predictor of treatment efficacy.
INTRODUCTION
Oncolytic virotherapy (OVT) is based on the observation that viruses, either through genetic engineering or by an inherent mechanism, preferentially replicate in and kill cancer cells while having minimal detrimental effects on normal cells (1). Oncolytic viruses (OVs) elicit the destruction of cancer cells as a direct result of viral replication and the induction of tumor-specific immune responses (2). The safety of OVs and their ability to induce antitumor activity in patients have been demonstrated in phase I and II clinical trials (reviewed in reference 3). Wild-type (wt) OVs, such as reovirus, Newcastle disease virus (NDV), vesicular stomatitis virus (VSV), and bovine herpesvirus 1 (BHV-1), do not require mutations to render them oncotropic. Alternatively, OVs that require genetic modification for selective oncolysis include herpes simplex virus 1 (HSV-1) and adenovirus (1). The collection of gain- or loss-of-function mutations within a tumor type dictates permissivity to OVs. A common aberration in cancer cells involves loss-of-function mutations within the interferon (IFN) signaling pathway (4).
HSV-1 was the first virus used to show that gene deletion can render a virus oncolytic (5). The oncolytic HSV-1 vector KM100 (ICP0n212VP16in1814 [6]) possesses lesions in infected cell protein 0 (ICP0), an immediate early (IE) protein that acts as a transcriptional activator during infection and counteracts host cellular responses to viral infection mediated by IFN (6–8). Accordingly, KM100 is unable to infect and replicate in nontransformed, nonimmortalized fibroblasts because it cannot effectively block the IFN-induced antiviral state (9, 10) but effectively replicates in human and murine transformed cells with deficiencies in IFN responsiveness (10). Moreover, unlike small RNA viruses such as VSV, the ability of HSV-1 OVs to elicit a viral burst and induce cytotoxicity in vitro does not correlate with efficacy in vivo (11–13).
BHV-1 is a member of the Herpesviridae family, in the Alphaherpesviridae subfamily. BHV-1 is a species-specific, neurotropic virus that initiates bovine respiratory disease in cattle through transient immunosuppression (14). It establishes lifelong latency in neurons, with reactivation occurring due to stress (14–16). The structure of BHV-1 is similar to that of HSV-1. BHV-1 binds attachment and entry receptors used by HSV-1, such as heparan-sulfate and nectin-1 (17). However, it is unable to bind nectin-2 but binds CD155 instead (17–19). Genes expressed by BHV-1 are generally named after the coinciding HSV-1 genes, which often have similar functions (8, 20, 21). While BHV-1 is unable to productively infect normal human cells (14, 22), human immortalized, transformed, and breast cancer-initiating cells are permissive to infection (22, 23). Interestingly, the ability of BHV-1 to kill human breast tumor cells and breast cancer-initiating cells is not contingent upon virus replication or the production of a viral burst (23). Furthermore, in contrast to other species-specific viruses, sensitivity to BHV-1 does not correlate with type I IFN signaling (22). Thus, the determinants of permissivity for BHV-1 in human cells are unknown.
Ras is a superfamily of plasma membrane-associated proteins whose members, particularly HRAS, KRAS, and NRAS, can be found in almost every human cell type (24, 25). Ras proteins and their downstream signaling effectors are pleiotropic, with roles in cell growth, migration, adhesion, cytoskeletal integrity, survival, and differentiation (25–27). Ras is a GTPase that is active in its GTP-bound form and inactive in its GDP-bound form. In addition, Ras is able to mediate cellular responses to virus infection by mediating the activity of antiviral pathways (28–32).
Mutations in Ras, the most common involving constitutive activation through mutation of the GTP binding cleft (33), are associated with particular tumor types, with approximately 20% of all tumors having an activating Ras mutation (25, 34). KRAS mutations have been associated with the progression of mammary tumors, acute myelogenous leukemia, lung cancer, and gastrointestinal cancer (24, 35, 36).
Many cancer cells are resistant to apoptosis due to mutations in the Ras pathway, which functions to inhibit double-stranded RNA (dsRNA)-activated protein kinase R (PKR), an IFN-stimulated gene (ISG) (28). Viruses that are sensitive to the effects of PKR, such as reovirus, can infect and replicate in cells with a Ras-activating mutation, such as cancer cells, while remaining inhibited in normal cells (37). Additionally, HSV-1 ICP34.5-null mutants preferentially replicate in cancer cells with Ras gain-of-function mutations (38, 39).
Here, we report that BHV-1 is able to infect and kill human tumor cell lines from a variety of histological origins and that the oncolytic activity of BHV-1 does not correlate with the extent of virus replication. Of interest, high levels of virus replication were observed in lung, colon, and prostate tumor cell lines, which have been associated with mutations in KRAS. While the knockdown or overexpression of oncogenic KRAS in human tumor cell lines yielded a modest effect, overexpression in normal primary cells conferred permissivity to BHV-1 infection. Together, these data indicate that BHV-1 holds promise as a broad-spectrum oncolytic agent with the ability to infect and kill a wide variety of human tumor cell types, particularly those expressing oncogenic KRAS. These studies also shed light on aspects of the cellular environment within human cells that determine permissivity to this species-specific virus.
MATERIALS AND METHODS
Cell lines.
All cell types were maintained at 37°C with 5% CO2 in medium supplemented with 2 mM l-glutamine. Cell lines with KRAS knockdown or overexpression were maintained in the appropriate medium supplemented with 2 mM penicillin-streptomycin. Madin-Darby bovine kidney (MDBK) cells were obtained from Vikram Misra (University of Saskatchewan) and were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 5% horse serum. Human osteosarcoma U2OS cells and human embryonic lung (HEL) cells were maintained in DMEM with 10% fetal bovine serum (FBS). 293 T cells were grown in DMEM with 10% FBS. Human lung carcinoma cells (A549; ATCC) were grown in α-minimal essential medium (α-MEM) with 10% FBS. The human immortalized fibroblast cell lines MSU-1 and MSU-1.1 were maintained in Eagle's MEM with 10% FBS supplemented with sodium pyruvate (1 mM), l-serine (0.25 mM; Sigma), and l-aspartic acid (0.15 mM; Sigma). Cells within the National Cancer Institute (NCI) panel of 60 established human cancer cell lines were maintained in Roswell Park Memorial Institute (RPMI) medium supplemented with 10% FBS.
Viruses.
BHV-1 constructs expressing green fluorescent protein (GFP) under the control of an endogenous immediate early (IE) promoter were a kind gift from Guenther Keil (Friedrich Loeffler Institut, Germany). Viral stocks were propagated and titrated on MDBK cells, followed by sucrose cushion purification (23). The oncolytic HSV-1 vector KM100 (ICP0n212VP16in1814), expressing GFP under the control of an IE cytomegalovirus promoter, was propagated and titrated on U2OS cells. Stocks of this virus were also prepared by sucrose cushion purification (23).
Measurement of virus replication and cytotoxicity.
Cells were seeded into 96-well plates, and 1 day later, 90 to 95% confluent cell monolayers were infected with BHV-1 or KM100 at multiplicities of infection (MOIs) of 10, 5, 2.5, 1, and 0.5 on each half of the plate, respectively. Infection was carried out for 1 h at 37°C, after which a maintenance overlay of RPMI medium plus 1% FBS was applied. At 1 and 2 days postinfection (p.i.), plates were scanned on a Typhoon BioAnalyzer (GE Healthcare, Piscataway, NJ), and virus replication was quantified as a function of GFP fluorescence and expressed as a fold change over the background fluorescence. Minimum (1.00) and maximum (22.06) fold change values were set at dark blue and red, respectively. White represents median values. Virus replication fold change values of <1 were set to 1 for the heat map analysis. Cytotoxicity, in terms of decreases in cellular metabolism, was assessed at 2 days p.i. by using alamarBlue (5% [vol/vol]; Biosource, Carlsbad, CA). Cells were incubated for 30 min at 37°C, after which fluorescence was read by using a Safire fluorescence plate reader (Tecan, Mannedorf, Switzerland). Data were analyzed relative to data from uninfected controls, corrected for background fluorescence, and used to generate a heat map. Minimum and maximum levels of toxicity are represented by dark blue and red, respectively. White represents median values. At least three independent experiments were performed for each cell line.
Graphical analysis of NCI panel data.
Comparisons of virus replication and cellular viability data were used to assess whether cell death of a particular cancer type correlates with high or low levels of viral replication. Partek Genomics Suite software (Partek Inc., St. Louis, MO) was used to generate box-and-whisker plots and to perform principal component analysis (PCA). Box-and-whisker plots were used to indicate the amount and distribution of variability in panel data. Data were grouped according to tissue origin and analyzed at an MOI of 0.5. PCA was used to visualize the correlation between virus replication, cellular metabolism, and membrane integrity. Each point represents the x, y, and z coordinates of each cell line within the multivariate data space. Data were analyzed at an MOI of 0.5.
Western blot analysis.
Cells were mock infected or infected with BHV-1, and whole-cell lysates were collected at various times postinfection in whole-cell extract buffer (20 mM HEPES [pH 7.4], 100 mM NaCl, 10 mM β-glycerophosphate, 0.2% Triton X-100, 1 mM sodium orthovanadate, 50 mM sodium fluoride, 1 mM phenylmethylsulfonyl fluoride [PMSF], 2 mM dithiothreitol [DTT], 1× protease inhibitor cocktail [Sigma, St. Louis, MO]) and lysed on ice for 30 min. Lysates were centrifuged at 1,000 rpm for 10 min at 4°C, and the supernatants were collected. Protein was quantified by using a Bradford assay kit (Bio-Rad Laboratories, Mississauga, ON, Canada). Whole-cell extracts were boiled in sample buffer containing sodium dodecyl sulfate (SDS) and β-mercaptoethanol and run on a 10% polyacrylamide gel for bovine ICP0 (bICP0), a 12% polyacrylamide gel for E2F1, and a 15% polyacrylamide gel for KRAS knockdown/overexpression confirmation. Gels were transferred onto polyvinylidene difluoride (PVDF) membranes (Millipore, Billerica, MA) with a wet transfer apparatus at 100 V for 1 h. All blots were blocked in 5% nonfat milk in Tris-buffered saline (TBS) at room temperature for 2 h. Blots were probed with primary antibodies specific for bICP0 (1:500; courtesy of Clinton Jones, University of Nebraska), E2F1 (1:500; Sigma, St. Louis, MO), or KRAS (1:1,000; Sigma) diluted in TBS-Tween (0.1%) overnight at 4°C. Blots were probed with an anti-rabbit (bICP0) or anti-mouse (E2F1 and KRAS) secondary antibody conjugated to horseradish peroxidase (HRP; Sigma), diluted 1:2,000 in 5% nonfat milk in 0.1% TBS–Tween. Blots were visualized by chemiluminescence.
Factors that dictate cellular sensitivity to BHV-1.
NCI panel cell lines were divided into groups based on low (0 to 5,000 relative fluorescence units [RFU]), medium (5,000 to 20,000 RFU), and high (>20,000 RFU) levels of BHV-1 replication. The Sanger Institute COSMIC database was used to determine the mutations present in each cell line. The number of cell lines possessing each mutation in low-, medium-, and high-replication groups was graphed.
Overexpression of oncogenic KRAS in human tumor and normal primary cells.
The human tumor cell lines COLO-205, NCI-H226, and BT-549 as well as the normal primary cell line HEL were included in KRAS overexpression studies. Activated KRAS effector mutant constructs were a generous gift from Patrick Lee (Dalhousie University). All overexpression plasmids are based on the pBABE-puro retrovirus backbone with activated KRAS generated through the introduction of an activating G12V mutation (40). Cells were transduced with retroviral constructs to make stable cell lines for use in subsequent experiments. Specifically, 293 T cells were transfected with 2 μg pVSV-G, 12 μg pHITgag/pol, and 10.5 μg pBABE vectors in Opti-MEM for 16 h. Retrovirus-containing supernatants were collected at 24 and 48 h posttransfection. Cell debris was pelleted by centrifugation at 1,000 rpm for 10 min, and supernatants were collected and subsequently filtered through a 0.45-μm filter. Virus was flash-frozen and stored at −80°C until use.
Cell lines were infected with the retrovirus for 3 h at 37°C, after which a maintenance overlay of RPMI medium plus 5% FBS was applied. At 2 days p.i., cells were subjected to puromycin selection at 1.0 μg/ml (COLO-205), 1.0 μg/ml (NCI-H226), 1.5 μg/ml (BT-549), and 1.0 μg/ml (HEL). Western blot analysis was performed to verify KRAS overexpression.
Knockdown of KRAS in human tumor and HELKRAS cells.
To achieve knockdown of KRAS, a short hairpin RNA (shRNA) was made with the sequence 5′-CGATACAGCTAATTCAGAATC-3′ (sense) (33). The sense sequence was used to design the following forward and reverse oligonucleotides, respectively: 5′-CCGGCGATACAGCTAATTCAGAATCCTCGAGGATTCTGAATTAGCTGTATCGTTTTTG-3′ (forward) and 5′-AATTCAAAAACGATACAGCTAATTCAGAATCCCGAGGATTCTGAATTAGCTGTATCG-3′ (reverse).
Forward and reverse oligonucleotides were annealed by using 1× annealing buffer (100 mM potassium acetate, 30 mM HEPES [pH 7.4], 2 mM magnesium acetate) and heated at 95°C for 5 min, followed by 30 min of incubation at 70°C. Oligonucleotides were phosphorylated by using T4 polynucleotide kinase and then purified via phenol chloroform extraction. The lentiviral vector pLKO-puro was digested with the EcoRI and AgeI restriction enzymes and ligated with the insert using T4 ligase. The plasmid was amplified on 2× low-salt lysogeny broth (LB) agar plates containing 100 μg/ml ampicillin. The following day, select colonies were inoculated into 2× low-salt LB broth and incubated overnight at 37°C with shaking. Plasmid DNA was extracted by using the QiaPrep Miniprep Spin kit (Qiagen, Hilden, Germany). Clones were verified by sequencing.
293 T cells were cotransfected with 16 μg of pLKO-shLUC, 8 μg of pCMV.DR.8.91, 4 μg of pVSV-G, and 48 μl of Lipofectamine 2000 (Life Technologies, Carlsbad, CA) in Opti-MEM for 4 h, after which medium was changed to DMEM plus 10% FBS. The following day, medium was changed to DMEM plus 30% FBS, the supernatant virus was harvested 24 h later, and medium was replaced with DMEM plus 30% FBS for a second round of supernatant harvesting 24 h later. Cell debris present within the supernatant fraction was pelleted by centrifugation at 1,500 rpm for 5 min and subsequently filtered through a 0.45-μm filter and a sterile syringe. Virus was pelleted by centrifugation at 25,000 rpm for 1.5 h, after which pellets were resuspended in serum-free DMEM.
Cell lines in the KRAS knockdown panel include HCT116, which has a G13D amino acid mutation, and A549, which has a G12S amino acid mutation (Sanger Institute, 2010). KRAS was also knocked down in HELKRAS cells as a control. Cells were seeded into 6-well plates, and a day later, 60% confluent monolayers were infected with a lentivirus expressing shKRAS or control shLUC for 1 h at 37°C, after which a maintenance overlay of DMEM plus 5% FBS was applied. Medium was changed at 24 h p.i., followed by puromycin (Sigma) selection at 48 h p.i. using 2 μg/μl (A549), 1.5 μg/μl (HCT116), or 1.0 μg/μl (HELKRAS). Cells were maintained and split 2 days later to expand the population of survivor cells carrying the shKRAS or shLUC construct. Western blot analysis was used to confirm KRAS knockdown.
Viral burst.
Cells were infected with BHV-1 at MOIs of 1, 3, 5, and 10. Viral supernatants and infected cells were collected at 1 and 2 days p.i. Samples were freeze-thawed three times prior to centrifugation at 1,000 rpm for 10 min at 4°C. The supernatant was collected and titrated by serial dilution in serum-free DMEM. Dilutions were applied onto 90 to 95% confluent monolayers of MDBK cells for 1 h at 37°C. MDBK monolayers were maintained in DMEM supplemented with 0.5% horse serum. At 2 days p.i., cells were scanned on a Typhoon BioAnalyzer (GE Healthcare), and PFU were counted.
Cytopathic effect assays.
HEL, HELempty, HELKRAS, and HELKRASshKRAS cells were mock infected or infected with BHV-1 at the indicated MOIs for 1 h at 37°C. Following viral adsorption, cells were maintained in DMEM plus 5% FBS. At 2 days p.i., cells were fixed with methanol and stained by using Giemsa to visualize cytopathic effects (CPEs).
RESULTS
BHV-1 replicates and reduces cytotoxicity in a wide range of human tumor cell lines.
To fully appreciate the oncolytic capacity of BHV-1, particularly in light of its unique properties (22, 23), we screened the NCI panel of 59 established human tumor cell lines, comparing BHV-1 to our prototypic HSV-1 vector KM100. The capacities of BHV-1 and KM100 to initiate replication in and decrease the viability of the NCI panel were assessed.
Initiation of virus replication, as a function of GFP fluorescence, was analyzed as the fold change over background fluorescence and used to construct a heat map (Fig. 1). GFP expression is not a measure of productive virus infection but was used to assess viral entry and the initiation of replication. For simplicity, we refer to the initiation of virus replication, as indicated by GFP fluorescence, as virus replication. Cellular toxicity, in terms of a reduction in cellular viability, was evaluated at 2 days p.i. and used to generate a heat map (Fig. 1).
FIG 1.
Heat maps showing BHV-1 (a) and KM100 (b) replication and effects on cellular viability of NCI panel cell lines. Cells were infected at MOIs of 0.5, 1.0, 2.5, 5.0, and 10.0 for 1 h at 37°C. Virus replication as a function of GFP fluorescence was quantified at 2 days p.i. by using a Typhoon BioAnalyzer and is represented as the fold change over background fluorescence. Minimum (1.00) and maximum (22.06) fold change values were set at dark blue and red, respectively. White represents median values. Cellular toxicity, in terms of decreases in cellular metabolism, was quantified at 2 days p.i., as measured by using alamarBlue. Data were analyzed as the fold change over background values, and minimum and maximum fold change values were set at dark blue and red, respectively. White represents median values. Replication and cellular toxicity are arranged in order of increasing MOIs. At least three independent trials were completed for each cell line. CNS, central nervous system.
In general, NCI panel cell lines fall into five different categories: high levels of virus replication with significant decreases in cellular viability (category 1), low levels of virus replication with significant decreases in cellular viability (category 2), minimal to no virus replication with significant decreases in cellular viability (category 3), virus replication with no significant decrease in cellular viability (category 4), and no significant virus replication or decrease in cellular viability (category 5) (Tables 1 and 2).
TABLE 1.
Summary of NCI panel categories based on effects of BHV-1 infectiona

NCI panel cell lines were divided into five different categories: high-level virus replication (20,000+ RFU) with a significant decrease in cellular viability (minimum decrease of 20% at an MOI of 10) (category 1), low-level virus replication (5 to 20,000 RFU) with a significant decrease in cellular viability (category 2), minimal or no virus replication (0 to 5,000 RFU) with a significant decrease in cellular viability (category 3), virus replication with no significant decrease in cellular viability (category 4), and no significant virus replication or decrease in cellular viability (category 5).
Overall, 95% of the panel cell lines supported some extent of BHV-1 replication (Fig. 1a), with 72% having a corresponding increase in cytotoxicity, defined as a decrease in cellular viability assays of at least 20% at an MOI of 10 (Fig. 1a). In reference, a decrease in cellular viability of 80% (MOI of 10) was observed for the U2OS cell line, a human osteosarcoma cell line which acts as a prototypic cell line in our screen, as it is highly permissive to BHV-1 infection (data not shown). Taken together, 22% (13/60) of the cell lines showed high-level BHV-1 replication and a corresponding decrease in cellular viability, 15% (9/60) showed low-level BHV-1 replication and a decrease in cellular viability, 35% (21/60) showed minimal or no replication and a decrease in cellular viability, 23% (14/60) showed replication but no effect on cellular viability, and, finally, 5% (3/60) showed no replication and no effect on cellular viability (Table 1).
In contrast, the ability of KM100 to replicate and induce cytotoxicity in panel cell lines was markedly decreased in comparison to that of BHV-1 (Fig. 1b). Only 3% (2/60) of cell lines showed high-level KM100 replication and a corresponding decrease in cellular viability, 15% (9/60) showed low-level KM100 replication and a decrease in cellular viability, 13% (8/60) showed minimal or no replication and a decrease in cellular viability, 63% (38/60) showed replication but no effect on cellular viability, and, finally, 5% (3/60) showed no replication and no effect on cellular viability (Fig. 1b and Table 2).
TABLE 2.
Summary of NCI panel categories based on effects of KM100 infectiona
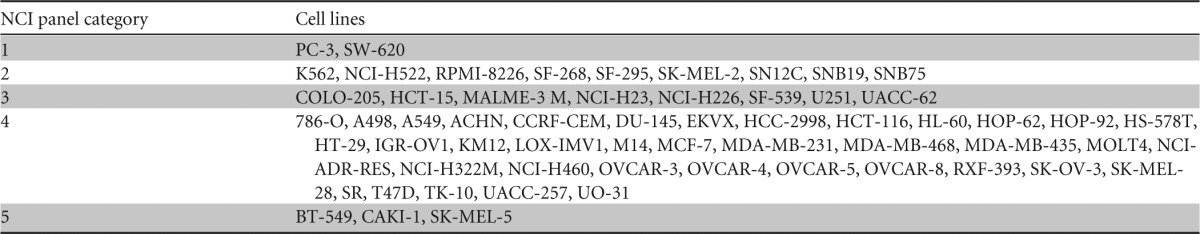
NCI panel cell lines were divided into five different categories: high-level virus replication (20,000+ RFU) with a significant decrease in cellular viability (minimum decrease of 20% at an MOI of 10) (category 1), low-level virus replication (5 to 20,000 RFU) with a significant decrease in cellular viability (category 2), minimal or no virus replication (0 to 5,000 RFU) with a significant decrease in cellular viability (category 3), virus replication with no significant decrease in cellular viability (category 4), and no significant virus replication or decrease in cellular viability (category 5).
To ensure that GFP fluorescence is an accurate measure of the initiation of virus replication, SF-268 (category 1), T47D (category 3), and UACC-257 (category 5) cells were selected to study the expression of bICP0, an IE/early (E) gene that is required for efficient BHV-1 infection of bovine cells by acting as a transcriptional activator throughout productive infection (41, 42). Expression of bICP0 was not detected in T47D and UACC-257 cells at any of the time points examined but was apparent in SF-268 cells, similar to the control permissive cell lines MDBK and U2OS (Fig. 2). For category 5 cells, such as UACC-257 cells, it is possible that BHV-1 fails to bind and enter, or this process is inefficient. However, wild-type HSV-1, which uses similar receptors for viral entry as BHV-1, is able to initiate replication in this cell line (data not shown), suggesting that the blockade to replication may be downstream of entry.
FIG 2.
bICP0 expression in NCI panel cell lines. Cells were infected with BHV-1 at an MOI of 3 for 1 h at 37°C. Whole-cell extracts were harvested at 4, 8, 12, 24, and 48 h p.i. for Western blot analysis. MDBK cells served as a positive control, as they are highly permissive to the virus. The human osteosarcoma cell line U2OS was also used as a positive control, as it is a human tumor cell line which has been shown to be highly permissive to BHV-1. Asterisks indicate a nonspecific band.
Box-and-whisker plots were used to compare the distribution, variability, and median values of BHV-1 and KM100 data sets within each tissue type relative to virus replication and cytotoxicity. Box-and-whisker plots showed a large range of variability within leukemic cell lines with regard to BHV-1 and KM100 replication, while lung and colon cell lines possessed a large range in variability in cytotoxicity for both viruses (Fig. 3). The median values for BHV-1 replication and cytotoxicity were higher than those for KM100. This indicates that BHV-1 is able to initiate replication and induce cytotoxicity at lower MOIs than KM100. PCA validated the box-and-whisker plot analysis (data not shown).
FIG 3.
Box-and-whisker plots of NCI panel data with BHV-1 and KM100. Box-and-whisker plots show BHV-1 (left) and KM100 (right) replication and cellular toxicity at an MOI of 0.5 for the NCI panel. Virus replication, as a function of GFP fluorescence, was quantified at 2 days p.i. by using a Typhoon BioAnalyzer and is represented as relative fluorescence units. Cellular toxicity, in terms of decreases in cellular metabolism, was quantified at 2 days p.i. by using alamarBlue. Box-and-whisker plots were generated by using Partek Genomics Suite software.
Knockdown of mutant KRAS decreases BHV-1 titers.
Knowledge of pathways that dictate the permissivity of human tumor cells to OVs allows more efficient targeting and aids in increased antitumor efficacy due to an improved understanding of host-virus interactions. Data from our group indicate that BHV-1 oncolysis does not correlate with defects in type I IFN signaling (22). Furthermore, BHV-1 is able to distinguish between normal and immortalized cell types (22), suggesting that immortalization and cellular changes occurring during this process may confer sensitivity to BHV-1. However, mutations in TP53 and retinoblastoma (RB1) protein, which are commonly deregulated during immortalization, do not correlate with permissivity to BHV-1 (Fig. 4).
FIG 4.
Correlation of gene mutations with BHV-1 replication in NCI panel cell lines. NCI panel cell lines were divided into groups based on low (0 to 5,000 RFU), medium (5,000 to 20,000 RFU), and high (>20,000 RFU) levels of replication. The mutation status of major oncogenes and tumor suppressors was determined by using the Sanger Institute NCI panel database. APC, adenomatous polyposis coli.
NCI panel cell lines were divided into groups based on low, medium, and high levels of BHV-1 replication. The mutations present within each cell line were determined by using the Sanger Institute COSMIC database (http://cancer.sanger.ac.uk/cancergenome/projects/cosmic/). The incidence of each mutation in low-, medium-, and high-replication groups was quantified to determine which genes are differentially expressed between low-replication and high-replication cohorts. Only genes for which a large difference was observed between low- and high-replication groups were explored. The results indicate that mutations in KRAS correlate with high levels of virus replication (Fig. 4). Moreover, cancer types most commonly associated with KRAS mutations (e.g., lung and colon cancers) were highly permissive to BHV-1 (Fig. 1). Ras proteins are the principal activators of multicomponent signaling cascades, with implications for cellular differentiation, proliferation, and apoptosis. Given the cellular processes controlled by Ras signaling cascades, it is not surprising that they are commonly mutated during immortalization and transformation (34).
The impact of KRAS knockdown on BHV-1 replication in A549 (lung) (G12S) and HCT116 (colon) (G13D) cells, which express a constitutively active form of the protein (http://cancer.sanger.ac.uk/), was evaluated by using a lentiviral shRNA system. These cell types were selected as representatives of lung and colon cancer cells, tissue types which are commonly associated with KRAS mutations and are also highly permissive to BHV-1. Stable cell lines were generated by puromycin selection and maintained for subsequent experiments. KRAS knockdown was confirmed by Western blotting (Fig. 5a). Assessment of the effects of KRAS knockdown on BHV-1 replication (Fig. 5b) and cellular viability (data not shown) did not delineate differences between control and KRAS knockdown cell lines. However, when viral titers were examined, a decrease of 1 order of magnitude was observed at 1 day p.i. at MOIs of 1 and 3 between A549shLUC and A549shKRAS cells (Fig. 5c). This effect was not present at MOIs of 5 and 10. Conversely, a decrease of approximately half an order of magnitude was observed at 2 days p.i. at each MOI (Fig. 5c). Similar results were observed for HCT116 cells (data not shown).
FIG 5.

KRAS knockdown decreases sensitivity of A549 cells to BHV-1 infection. (a) Confirmation of KRAS knockdown in A549shKRAS cells. Whole-cell extracts were harvested for Western blot analysis to confirm the knockdown of KRAS. (b) BHV-1 replication in A549shLUC and A549shKRAS cell lines. Virus replication, as a function of GFP fluorescence, was quantified at 2 days p.i. by using a Typhoon BioAnalyzer and is represented as absolute fluorescence units. (c) BHV-1 titers on A549shLUC and A549shKRAS cell lines. Cells were infected with BHV-1 at the indicated MOIs for 1 h at 37°C. Triplicate samples of viral supernatants and cell-associated virus particles were collected at 1 and 2 days p.i. and titrated on naive MDBK monolayers. Error bars represent means and standard errors of the means (n = 3).
The overexpression of oncogenic KRAS in COLO205, NCI-H226, and BT-549 cells, which express wild-type (wt) KRAS (http://cancer.sanger.ac.uk/), was achieved by retroviral transduction with stable cell lines created through puromycin selection. However, differences in BHV-1 replication and cellular viability between control and KRAS overexpression cell lines were not detected (data not shown). This observation is not surprising, as these cell lines contain a plethora of mutations (http://cancer.sanger.ac.uk/) that likely make the addition of active KRAS redundant and its individual effect difficult to ascertain.
KRAS overexpression sensitizes normal primary cells to BHV-1 infection.
Studies of the effects of oncogenic KRAS within the context of a normal primary cell type will allow determination of the effects of this single mutant protein on BHV-1 infection. To better address whether active KRAS confers enhanced sensitivity to BHV-1 infection, the nonimmortalized, nontransformed cell line HEL was transduced with a control retrovirus (empty) or a retrovirus expressing constitutively activated KRAS (G12V). Western blot analysis revealed that retrovirus transduction alone increases cellular levels of KRAS (Fig. 6a). HEL, HELempty, and HELKRAS cells were subsequently infected with BHV-1 (Fig. 6b). In comparison to wt HEL, for which CPE was observed at an MOI of 10 at 3 days p.i., almost complete destruction of HELKRAS monolayers was present at an MOI of 2.5 at the same time point, with CPE being noticeable at an MOI of 0.5. Intermediate levels of CPE were apparent in the control cell line HELempty, suggesting that the process of transduction itself or the resultant increase in endogenous KRAS expression levels predisposes these cells to BHV-1 infection. To confirm that the increase in sensitivity of HEL cells to BHV-1 infection is due to the overexpression of functionally active KRAS, KRAS was knocked down in HELKRAS cells. The results showed that CPE was apparent at an MOI of 10 in HELKRASshKRAS cells, at levels similar to those seen in wt HEL cells. To evaluate whether KRAS overexpression increases permissivity of HEL cells to BHV-1, viral titers were determined at 1 and 2 days p.i. and compared between wt HEL, HELempty, HELKRAS, and HELKRASshKRAS cells. At 1 day p.i., BHV-1 titers were 1 to 2 logs higher in HELempty and HELKRAS cells than in wt HEL cells, with little difference being observed between HELempty and HELKRAS cells (Fig. 6c). However, at 2 days p.i., BHV-1 titers were significantly higher in HELKRAS cells than in both wt HEL and HELempty cells. Knockdown of KRAS in HELKRASshKRAS cells significantly lowered BHV-1 titers. Control HELKRASshLUC cells expressed the same level of KRAS as HELKRAS cells and behaved in a similar fashion, as expected (data not shown). Together, these data suggest that overexpression of endogenous or mutant KRAS in HEL cells increases their permissivity to BHV-1 infection.
FIG 6.
KRAS overexpression increases susceptibility of HEL cells to BHV-1. (a) Confirmation of KRAS overexpression and knockdown in HELKRAS and HELKRASshKRAS cells, respectively. Whole-cell extracts were harvested for Western blot analysis to confirm the overexpression or knockdown of KRAS. −ve indicates the negative control 293 T whole-cell extract. (b) Permissiveness of wt HEL, HELempty, HELKRAS, and HELKRASshKRAS cells to BHV-1. Cells were infected at the indicated MOIs for 1 h at 37°C. At 2 days p.i., cells were Giemsa stained to visualize CPE. (c) BHV-1 titers on wt HEL, HELempty, HELKRAS, and HELKRASshKRAS cell lines. Cells were infected with BHV-1 at the indicated MOIs for 1 h at 37°C. Triplicate samples of viral supernatants and cell-associated virus particles were collected at 1 and 2 days p.i. and titrated on naive MDBK monolayers. Error bars represent means and standard errors of the means (n = 3).
MSU-1.0 and MSU-1.1 are cell lines derived by sequential clonal selection of human foreskin fibroblast cells following the introduction of the v-myc oncogene, which is sufficient for immortalization but not transformation (43). The MSU-1.1 cell line additionally possesses mutations in HRAS and KRAS (43). However, neither cell line is able to form tumors in athymic mice (43). Therefore, these cell lines allow us to examine BHV-1 infection at early stages in the transformation process. An increase in permissivity to BHV-1 was observed for MSU-1.1 cells in comparison to MSU-1.0 cells, suggesting that cells that have incurred additional mutations at late stages of the immortalization process have an increased sensitivity to BHV-1 infection (data not shown).
KRAS overexpression increases the expression levels of E2F1 in normal primary cells.
The BHV-1 IE/E protein bICP0 stimulates productive infection by inducing viral gene synthesis (42, 44). Infection of MDBK cells with a bICP0-null virus results in a reduction in titers of approximately 100-fold (44). The E2F family of transcriptional regulators has roles in cell proliferation, cell cycle progression, the activity of tumor suppressor proteins, and p53-dependent/independent apoptosis. A member of the E2F family, E2F1, binds and activates the bICP0 E promoter 100-fold in transient-transfection assays (45). BHV-1 has also been shown to increase E2F1 protein levels during productive infection (46). Conversely, knockdown of E2F1 has been shown to significantly reduce the efficacy of infection (46). Mutations in KRAS have been shown to impact many cellular processes, including cell cycle progression (25–27, 47, 48). To determine whether overexpression of activated KRAS elicits an increase in the E2F1 expression level, thereby enhancing BHV-1 infection, we examined basal E2F1 levels in the HEL series of cell lines. Western blot analysis showed a correlation between levels of KRAS (Fig. 6a) and those of E2F1 (Fig. 7). Furthermore, small interfering RNA (siRNA)-mediated knockdown of E2F1 in the MCF7 cell line, a breast cancer cell line that is highly permissive to BHV-1 infection, reduced viral titers by approximately 3 orders of magnitude at all MOIs examined (data not shown). These data suggest a potential mechanism by which oncogenic KRAS enhances BHV-1 infection in human transformed and HELKRAS cells.
FIG 7.
Overexpression of KRAS increases expression levels of basal E2F1 in normal primary cells. Whole-cell extracts were harvested for Western blot analysis to examine basal E2F1 expression in wt HEL, HELempty, HELKRAS, and HELKRASshKRAS cell lines. A 293 T cell lysate was used as a positive control.
DISCUSSION
The use of OVs to target and lyse cancer cells is a novel approach to cancer therapy that lacks the toxic side effects of many current cancer treatments. HSV-1 was the first virus used to demonstrate that a genetic mutation can render a virus oncolytic (5, 49, 50). In fact, HSV-1 has been studied extensively as an OV. The safety of oncolytic HSV-1 at current maximum feasible doses has been demonstrated in phase I and II clinical trials (reviewed in reference 3). Although cross-priming and amplification of antitumor immunity have been demonstrated following intratracheal (i.t.) administration of oncolytic HSV-1 (51, 52), systemic delivery will be required for the treatment of metastatic and minimal residual disease. However, the high incidence of preexisting immunity to HSV-1 may limit systemic delivery of the virus. To date, clinical trials have failed to demonstrate whether direct tumor lysis is required for patient responses. In fact, evidence suggests that direct tumor lysis may be linked to dose (53), which could restrict the use of certain OVs due to manufacturing difficulties. These obstacles warrant the development of nonhuman viruses for OVT. Furthermore, the use of wt nonhuman viruses circumvents safety concerns regarding the risk of unexpected toxicities due to the use of genetically manipulated viruses (54).
The results presented here indicate that BHV-1 is an OV with the ability to infect a large range of human tumor cell types and induce cytotoxicity at low MOIs in comparison to KM100, which has been well characterized for its in vivo and in vitro oncolytic capacity (10, 12). While the majority of panel cell lines support both BHV-1 and KM100 replication, BHV-1 displays a more significant increase in virus replication over background levels at lower MOIs than does KM100, with a corresponding decrease in cellular viability. In the majority of cases where low-level/no BHV-1 replication was observed, decreases in cellular viability still occurred. Overwhelmingly, while 72% of panel cell lines screened with BHV-1 showed a decrease in cellular viability, only 32% showed a decrease with KM100 infection. Furthermore, 35% of the panel supported minimal/no BHV-1 replication; however, a decrease in cellular viability occurred. Together, these data suggest that BHV-1 holds potential as an OV possessing tropism for multiple cancer types and is able to induce cytotoxicity independent of significant virus replication.
We have previously reported that in the majority of human breast tumor cells studied, cellular death occurs in the absence of a viral burst (23). Thus, although we did not measure viral burst on NCI panel cell lines, previous studies indicate that virus replication data are not predictive of cytotoxicity and cellular death from BHV-1 infection (23). While these data suggest that a soluble cytotoxic factor may be responsible for cellular death, we have shown that supernatants from breast cancer cells infected with BHV-1 are unable to decrease the cellular viability of MDBK cells (23). The mechanism by which BHV-1 elicits cellular death remains unknown; however, possible methods include epigenetic alterations and microRNA (miRNA) production (55–59; discussed in reference 23). Future studies will investigate these possibilities.
The ability of BHV-1 to infect and kill a large range of human tumor cell types suggests that a ubiquitous factor(s), pathway, or process is responsible for dictating cellular sensitivity to BHV-1. Panel screen data implicate a broader mechanism for the restriction of BHV-1 replication other than sensitivity to type I IFN signaling, unlike other species-specific viruses such as myxoma virus (MV), NDV, and VSV. Furthermore, the ability of BHV-1 to infect and kill human immortalized cells is exciting, as it suggests that the virus may be able to infect preneoplastic cells and therefore target developing lesions (22).
The Ras family of proteins has pleiotropic roles in the cell, including mediation of PKR activity. Activated Ras inhibits autophosphorylation of PKR, blocking its downstream effects, including inhibition of viral protein synthesis by phosphorylating the α subunit of eukaryotic initiation factor 2 (eIF2α) (28). Some viruses, such as HSV-1, have developed mechanisms to counteract the effects of PKR and thus productively infect cells without the aid of activated Ras. Other viruses, such as reovirus, rely on activated Ras in order to counteract the effects of PKR. BHV-1 does not encode a homologue of ICP34.5 and, like reovirus, may rely on oncogenic Ras to establish productive infection. Our data suggest that KRAS plays a role in dictating sensitivity to BHV-1 infection; however, it is not the sole factor in this process. While the effects of KRAS knockdown and overexpression on the sensitivity of human tumor cell lines to BHV-1 are variable, this observation is not surprising, as these cell lines contain a plethora of mutations, which may make the addition or knockdown of oncogenic KRAS insignificant. Most striking are the changes in permissivity of normal primary HEL cells to BHV-1 infection from KRAS overexpression. Unexpectedly, control retroviral transduction increased the sensitivity of HEL cells to BHV-1, likely due to a corresponding increase in endogenous KRAS levels. In fact, HELempty cells were intermediate in terms of permissivity to BHV-1 relative to parental HEL and HELKRAS cells in all assays performed. However, the HELKRAS cell line was found to have an altered phenotype in comparison to wt HEL and HELempty cells, adopting a spindle-like and elongated appearance (data not shown). Although activated KRAS is insufficient for transformation (43), its ability to enhance BHV-1 replication is consistent with our previous observations that immortalized, but nontransformed, cells are permissive for infection (22).
The intricacies of Ras signaling, including its role in multiple cell processes and multitude of downstream effectors, make comprehensive interrogation of this pathway difficult. Although, to our knowledge, a direct link between KRAS activity and E2F1 has not been established, it is not surprising that we observed increased E2F1 levels in HELempty and HELKRAS cells, as E2F1 levels correlate with the mitotic index of many cancer cells (60, 61). Although E2F1 directly stimulates BHV-1 replication due to E2F1 binding sites within the bICP0 E promoter, it is likely that additional pathways and factors, activated directly or indirectly by KRAS, work in concert with E2F1 to render HELempty and HELKRAS cells permissive to infection by BHV-1. Further studies are required to unravel the relationship between KRAS, E2F1, immortalization, and cellular permissivity to BHV-1. Although KRAS is not the only factor that dictates permissivity to BHV-1, an understanding of the relationship between KRAS and BHV-1 activity has clinical relevance, particularly in lung cancer, given that KRAS mutations are predictive of poor response rates to lung cancer therapy (62–65). By understanding the factors that govern permissivity (or lack thereof) of human cells to nonhuman viruses, important insights into the evolution of host antiviral mechanisms are gained, which can ultimately be exploited for the development of novel therapeutics.
ACKNOWLEDGMENTS
B.P.C. holds a fellowship from the Canadian Breast Cancer Foundation. This work was sponsored by operating grants from the Cancer Research Society and the Canadian Cancer Society Research Institute (formerly the Canadian Breast Cancer Research Alliance).
We acknowledge that there are no financial conflicts of interest related to this research.
We thank Vikram Misra (University of Saskatchewan, Canada), Günther Keil (Friedrich Loeffler Institut, Germany), Patrick Lee (Dalhousie University, Canada), and Clinton Jones (University of Nebraska) for reagents and Nicole Vidinu and Michael Herman for their help in screening the NCI panel.
Footnotes
Published ahead of print 2 April 2014
REFERENCES
- 1.Cervantes-Garcia D, Ortiz-Lopez R, Mayek-Perez N, Rojas-Martinez A. 2008. Oncolytic virotherapy. Ann. Hepatol. 7:34–45 http://www.medigraphic.com/pdfs/hepato/ah-2008/ah081e.pdf [PubMed] [Google Scholar]
- 2.Vaha-Koskela MJ, Heikkila JE, Hinkkanen AE. 2007. Oncolytic viruses in cancer therapy. Cancer Lett. 254:178–216. 10.1016/j.canlet.2007.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Russell SJ, Peng KW, Bell JC. 2012. Oncolytic virotherapy. Nat. Biotechnol. 30:658–670. 10.1038/nbt.2287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shmulevitz M, Marcato P, Lee PW. 2005. Unshackling the links between reovirus oncolysis, Ras signaling, translational control and cancer. Oncogene 24:7720–7728. 10.1038/sj.onc.1209041 [DOI] [PubMed] [Google Scholar]
- 5.Martuza RL, Malick A, Markert JM, Ruffner KL, Coen DM. 1991. Experimental therapy of human glioma by means of a genetically engineered virus mutant. Science 252:854–856. 10.1126/science.1851332 [DOI] [PubMed] [Google Scholar]
- 6.Mossman KL, Saffran HA, Smiley JR. 2000. Herpes simplex virus ICP0 mutants are hypersensitive to interferon. J. Virol. 74:2052–2056. 10.1128/JVI.74.4.2052-2056.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mossman KL, Smiley JR. 2002. Herpes simplex virus ICP0 and ICP34.5 counteract distinct interferon-induced barriers to virus replication. J. Virol. 76:1995–1998. 10.1128/JVI.76.4.1995-1998.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Everett RD, Boutell C, McNair C, Grant L, Orr A. 2010. Comparison of the biological and biochemical activities of several members of the alphaherpesvirus ICP0 family of proteins. J. Virol. 84:3476–3487. 10.1128/JVI.02544-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mossman KL, Smiley JR. 1999. Truncation of the C-terminal acidic transcriptional activation domain of herpes simplex virus VP16 renders expression of the immediate-early genes almost entirely dependent on ICP0. J. Virol. 73:9726–9733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hummel JL, Safroneeva E, Mossman KL. 2005. The role of ICP0-null HSV-1 and interferon signaling defects in the effective treatment of breast adenocarcinoma. Mol. Ther. 12:1101–1110. 10.1016/j.ymthe.2005.07.533 [DOI] [PubMed] [Google Scholar]
- 11.Workenhe ST, Pol JG, Lichty BD, Cummings DT, Mossman KL. Combining oncolytic HSV-1 with immunogenic cell death-inducing drug mitoxantrone breaks cancer immune tolerance and improves therapeutic efficacy. Cancer Immunol. Res. 5:1–11. 10.1158/2326-6066.CIR-13-0059-T [DOI] [PubMed] [Google Scholar]
- 12.Sobol PT, Boudreau JE, Stephenson K, Wan Y, Lichty BD, Mossman KL. 2011. Adaptive antiviral immunity is a determinant of the therapeutic success of oncolytic virotherapy. Mol. Ther. 19:335–344. 10.1038/mt.2010.264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Workenhe ST, Simmons G, Pol JG, Lichty BD, Halford WP, Mossman KL. 2014. Immunogenic HSV mediated oncolysis shapes the antitumor immune response and contributes to therapeutic efficacy. Mol. Ther. 22:123–131. 10.1038/mt.2013.238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hushur O, Takashima Y, Matsumoto Y, Otsuka H. 2004. Restriction of bovine herpesvirus 1 (BHV-1) growth in non-permissive cells beyond the expression of immediate early genes. J. Vet. Med. Sci. 66:453–455. 10.1292/jvms.66.453 [DOI] [PubMed] [Google Scholar]
- 15.Turin L, Russo S, Poli G. 1999. BHV-1: new molecular approaches to control a common and widespread infection. Mol. Med. 5:261–284 [PMC free article] [PubMed] [Google Scholar]
- 16.Jones C, Chowdhury S. 2007. A review of the biology of bovine herpesvirus type 1 (BHV-1), its role as a cofactor in the bovine respiratory disease complex and development of improved vaccines. Anim. Health Res. Rev. 8:187–205. 10.1017/S146625230700134X [DOI] [PubMed] [Google Scholar]
- 17.Campadelli-Fiume G. 2000. Virus receptor arrays, CD46 and human herpesvirus 6. Trends Microbiol. 8:436–438. 10.1016/S0966-842X(00)01804-7 [DOI] [PubMed] [Google Scholar]
- 18.Merrill MK, Bernhardt G, Sampson JH, Wikstrand CJ, Bigner DD, Gromeier M. 2004. Poliovirus receptor CD155-targeted oncolysis of glioma. Neuro Oncol. 6:208–217. 10.1215/S1152851703000577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pende D, Spaggiari GM, Marcenaro S, Martini S, Rivera P, Capobianco A, Falco M, Lanino E, Pierri I, Zambello R, Bacigalupo A, Mingari MC, Moretta A, Moretta L. 2005. Analysis of the receptor-ligand interactions in the natural killer-mediated lysis of freshly isolated myeloid or lymphoblastic leukemias: evidence for the involvement of the poliovirus receptor (CD155) and Nectin-2 (CD112). Blood 105:2066–2073. 10.1182/blood-2004-09-3548 [DOI] [PubMed] [Google Scholar]
- 20.Henderson G, Zhang Y, Jones C. 2005. The bovine herpesvirus 1 gene encoding infected cell protein 0 (bICP0) can inhibit interferon-dependent transcription in the absence of other viral genes. J. Gen. Virol. 86:2697–2702. 10.1099/vir.0.81109-0 [DOI] [PubMed] [Google Scholar]
- 21.Saira K, Zhou Y, Jones C. 2007. The infected cell protein 0 encoded by bovine herpesvirus 1 (bICP0) induces degradation of interferon response factor 3 and, consequently, inhibits beta interferon promoter activity. J. Virol. 81:3077–3086. 10.1128/JVI.02064-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rodrigues R, Cuddington B, Mossman K. 2010. Bovine herpesvirus type 1 as a novel oncolytic virus. Cancer Gene Ther. 17:344–355. 10.1038/cgt.2009.77 [DOI] [PubMed] [Google Scholar]
- 23.Cuddington BP, Dyer AL, Workenhe ST, Mossman KL. 2013. Oncolytic bovine herpesvirus type 1 infects and kills breast tumor cells and breast cancer-initiating cells irrespective of tumor subtype. Cancer Gene Ther. 20:282–289. 10.1038/cgt.2013.18 [DOI] [PubMed] [Google Scholar]
- 24.Ayllon V, Rebollo A. 2000. Ras-induced cellular events. Mol. Membr. Biol. 17:65–73. 10.1080/09687680050117093 [DOI] [PubMed] [Google Scholar]
- 25.Downward J. 2003. Targeting RAS signalling pathways in cancer therapy. Nat. Rev. Cancer 3:11–22. 10.1038/nrc969 [DOI] [PubMed] [Google Scholar]
- 26.Rodriguez-Viciana P, Tetsu O, Oda K, Okada J, Rauen K, McCormick F. 2005. Cancer targets in the Ras pathway. Cold Spring Harb. Symp. Quant. Biol. 70:461–467. 10.1101/sqb.2005.70.044 [DOI] [PubMed] [Google Scholar]
- 27.Ferro E, Trabalzini L. 2010. RalGDS family members couple Ras to Ral signalling and that's not all. Cell. Signal. 22:1804–1810. 10.1016/j.cellsig.2010.05.010 [DOI] [PubMed] [Google Scholar]
- 28.Everts B, van der Poel HG. 2005. Replication-selective oncolytic viruses in the treatment of cancer. Cancer Gene Ther. 12:141–161. 10.1038/sj.cgt.7700771 [DOI] [PubMed] [Google Scholar]
- 29.Christian SL, Zu D, Licursi M, Komatsu Y, Pongnopparat T, Codner DA, Hirasawa K. 2012. Suppression of IFN-induced transcription underlies IFN defects generated by activated Ras/MEK in human cancer cells. PLoS One 7:e44267. 10.1371/journal.pone.0044267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Noser JA, Mael AA, Sakuma R, Ohmine S, Marcato P, Lee PW, Ikeda Y. 2007. The RAS/Raf1/MEK/ERK signaling pathway facilitates VSV-mediated oncolysis: implication for the defective interferon response in cancer cells. Mol. Ther. 15:1531–1536. 10.1038/sj.mt.6300193 [DOI] [PubMed] [Google Scholar]
- 31.Battcock SM, Collier TW, Zu D, Hirasawa K. 2006. Negative regulation of the alpha interferon-induced antiviral response by the Ras/Raf/MEK pathway. J. Virol. 80:4422–4430. 10.1128/JVI.80.9.4422-4430.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shmulevitz M, Pan LZ, Garant K, Pan D, Lee PW. 2010. Oncogenic Ras promotes reovirus spread by suppressing IFN-beta production through negative regulation of RIG-I signaling. Cancer Res. 70:4912–4921. 10.1158/0008-5472.CAN-09-4676 [DOI] [PubMed] [Google Scholar]
- 33.Wee S, Jagani Z, Xiang KX, Loo A, Dorsch M, Yao YM, Sellers WR, Lengauer C, Stegmeier F. 2009. PI3K pathway activation mediates resistance to MEK inhibitors in KRAS mutant cancers. Cancer Res. 69:4286–4293. 10.1158/0008-5472.CAN-08-4765 [DOI] [PubMed] [Google Scholar]
- 34.Bos JL. 1989. ras oncogenes in human cancer: a review. Cancer Res. 49:4682–4689 [PubMed] [Google Scholar]
- 35.Zhao S, Wang Y, Cao L, Ouellette MM, Freeman JW. 2010. Expression of oncogenic K-ras and loss of Smad4 cooperate to induce the expression of EGFR and to promote invasion of immortalized human pancreas ductal cells. Int. J. Cancer 127:2076–2087. 10.1002/ijc.25412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Leslie K, Gao SP, Berishaj M, Podsypanina K, Ho H, Ivashkiv L, Bromberg J. 2010. Differential interleukin-6/Stat3 signaling as a function of cellular context mediates Ras-induced transformation. Breast Cancer Res. 12:R80. 10.1186/bcr2725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Strong JE, Coffey MC, Tang D, Sabinin P, Lee PW. 1998. The molecular basis of viral oncolysis: usurpation of the Ras signaling pathway by reovirus. EMBO J. 17:3351–3362. 10.1093/emboj/17.12.3351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Davis JJ, Fang B. 2005. Oncolytic virotherapy for cancer treatment: challenges and solutions. J. Gene Med. 7:1380–1389. 10.1002/jgm.800 [DOI] [PubMed] [Google Scholar]
- 39.Farassati F, Yang AD, Lee PW. 2001. Oncogenes in Ras signalling pathway dictate host-cell permissiveness to herpes simplex virus 1. Nat. Cell Biol. 3:745–750. 10.1038/35087061 [DOI] [PubMed] [Google Scholar]
- 40.Norman KL, Hirasawa K, Yang AD, Shields MA, Lee PW. 2004. Reovirus oncolysis: the Ras/RalGEF/p38 pathway dictates host cell permissiveness to reovirus infection. Proc. Natl. Acad. Sci. U. S. A. 101:11099–11104. 10.1073/pnas.0404310101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fraefel C, Zeng J, Choffat Y, Engels M, Schwyzer M, Ackermann M. 1994. Identification and zinc dependence of the bovine herpesvirus 1 transactivator protein BICP0. J. Virol. 68:3154–3162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Saira K, Chowdhury S, Gaudreault N, da Silva L, Henderson G, Doster A, Jones C. 2008. The zinc RING finger of bovine herpesvirus 1-encoded bICP0 protein is crucial for viral replication and virulence. J. Virol. 82:12060–12068. 10.1128/JVI.01348-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Morgan TL, Yang DJ, Fry DG, Hurlin PJ, Kohler SK, Maher VM, McCormick JJ. 1991. Characteristics of an infinite life span diploid human fibroblast cell strain and a near-diploid strain arising from a clone of cells expressing a transfected v-myc oncogene. Exp. Cell Res. 197:125–136. 10.1016/0014-4827(91)90489-H [DOI] [PubMed] [Google Scholar]
- 44.Geiser V, Zhang Y, Jones C. 2005. Analysis of a bovine herpesvirus 1 recombinant virus that does not express the bICP0 protein. J. Gen. Virol. 86:1987–1996. 10.1099/vir.0.80921-0 [DOI] [PubMed] [Google Scholar]
- 45.Workman A, Jones C. 2010. Productive infection and bICP0 early promoter activity of bovine herpesvirus 1 are stimulated by E2F1. J. Virol. 84:6308–6317. 10.1128/JVI.00321-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Workman A, Jones C. 2011. Analysis of the cell cycle regulatory protein (E2F1) after infection of cultured cells with bovine herpesvirus 1 (BHV-1) or herpes simplex virus type 1 (HSV-1). Virus Res. 160:66–73. 10.1016/j.virusres.2011.05.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fan J, Bertino JR. 1997. K-ras modulates the cell cycle via both positive and negative regulatory pathways. Oncogene 14:2595–2607. 10.1038/sj.onc.1201105 [DOI] [PubMed] [Google Scholar]
- 48.Monticone M, Biollo E, Maffei M, Donadini A, Romeo F, Storlazzi CT, Giaretti W, Castagnola P. 2008. Gene expression deregulation by KRAS G12D and G12V in a BRAF V600E context. Mol. Cancer 7:92. 10.1186/1476-4598-7-92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jia W, Zhou Q. 2005. Viral vectors for cancer gene therapy: viral dissemination and tumor targeting. Curr. Gene Ther. 5:133–142. 10.2174/1566523052997460 [DOI] [PubMed] [Google Scholar]
- 50.Shen Y, Nemunaitis J. 2006. Herpes simplex virus 1 (HSV-1) for cancer treatment. Cancer Gene Ther. 13:975–992. 10.1038/sj.cgt.7700946 [DOI] [PubMed] [Google Scholar]
- 51.Kaufman HL, Kim DW, DeRaffele G, Mitcham J, Coffin RS, Kim-Schulze S. 2010. Local and distant immunity induced by intralesional vaccination with an oncolytic herpes virus encoding GM-CSF in patients with stage IIIc and IV melanoma. Ann. Surg. Oncol. 17:718–730. 10.1245/s10434-009-0809-6 [DOI] [PubMed] [Google Scholar]
- 52.Senzer NN, Kaufman HL, Amatruda T, Nemunaitis M, Reid T, Daniels G, Gonzalez R, Glaspy J, Whitman E, Harrington K, Goldsweig H, Marshall T, Love C, Coffin R, Nemunaitis JJ. 2009. Phase II clinical trial of a granulocyte-macrophage colony-stimulating factor-encoding, second-generation oncolytic herpesvirus in patients with unresectable metastatic melanoma. J. Clin. Oncol. 27:5763–5771. 10.1200/JCO.2009.24.3675 [DOI] [PubMed] [Google Scholar]
- 53.Breitbach CJ, Burke J, Jonker D, Stephenson J, Haas AR, Chow LQ, Nieva J, Hwang TH, Moon A, Patt R, Pelusio A, Le Boeuf F, Burns J, Evgin L, De Silva N, Cvancic S, Robertson T, Je JE, Lee YS, Parato K, Diallo JS, Fenster A, Daneshmand M, Bell JC, Kirn DH. 2011. Intravenous delivery of a multi-mechanistic cancer-targeted oncolytic poxvirus in humans. Nature 477:99–102. 10.1038/nature10358 [DOI] [PubMed] [Google Scholar]
- 54.Chen N, Bellone CJ, Schriewer J, Owens G, Fredrickson T, Parker S, Buller RM. 2011. Poxvirus interleukin-4 expression overcomes inherent resistance and vaccine-induced immunity: pathogenesis, prophylaxis, and antiviral therapy. Virology 409:328–337. 10.1016/j.virol.2010.10.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Niller HH, Wolf H, Minarovits J. 2009. Epigenetic dysregulation of the host cell genome in Epstein-Barr virus-associated neoplasia. Semin. Cancer Biol. 19:158–164. 10.1016/j.semcancer.2009.02.012 [DOI] [PubMed] [Google Scholar]
- 56.Adhya D, Basu A. 2010. Epigenetic modulation of host: new insights into immune evasion by viruses. J. Biosci. 35:647–663. 10.1007/s12038-010-0072-9 [DOI] [PubMed] [Google Scholar]
- 57.Paschos K, Allday MJ. 2010. Epigenetic reprogramming of host genes in viral and microbial pathogenesis. Trends Microbiol. 18:439–447. 10.1016/j.tim.2010.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cullen BR. 2011. Viruses and microRNAs: RISCy interactions with serious consequences. Genes Dev. 25:1881–1894. 10.1101/gad.17352611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lu F, Stedman W, Yousef M, Renne R, Lieberman PM. 2010. Epigenetic regulation of Kaposi's sarcoma-associated herpesvirus latency by virus-encoded microRNAs that target Rta and the cellular Rbl2-DNMT pathway. J. Virol. 84:2697–2706. 10.1128/JVI.01997-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.DeGregori J, Kowalik T, Nevins JR. 1995. Cellular targets for activation by the E2F1 transcription factor include DNA synthesis- and G1/S-regulatory genes. Mol. Cell. Biol. 15:4215–4224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Harbour JW, Dean DC. 2000. The Rb/E2F pathway: expanding roles and emerging paradigms. Genes Dev. 14:2393–2409. 10.1101/gad.813200 [DOI] [PubMed] [Google Scholar]
- 62.Webb JD, Simon MC. 2010. Novel insights into the molecular origins and treatment of lung cancer. Cell Cycle 9:4098–4105. 10.4161/cc.9.20.13588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Okudela K, Woo T, Kitamura H. 2010. KRAS gene mutations in lung cancer: particulars established and issues unresolved. Pathol. Int. 60:651–660. 10.1111/j.1440-1827.2010.02580.x [DOI] [PubMed] [Google Scholar]
- 64.Singh N, Bal A, Aggarwal AN, Das A, Behera D. 2010. Clinical outcomes in non-small-cell lung cancer in relation to expression of predictive and prognostic biomarkers. Future Oncol. 6:741–767. 10.2217/fon.10.30 [DOI] [PubMed] [Google Scholar]
- 65.Rossi A, Galetta D, Gridelli C. 2009. Biological prognostic and predictive factors in lung cancer. Oncology 77(Suppl 1):90–96. 10.1159/000258500 [DOI] [PubMed] [Google Scholar]








