ABSTRACT
The causative agent of dengue fever, dengue virus (DENV), is transmitted by mosquitoes, and as distribution of these insects has expanded, so has dengue-related disease. DENV is a member of the Flaviviridae family and has 4 distinct serotypes (DENV-1, -2, -3, and -4). No lasting cross protection is afforded to heterologous serotypes following infection by any one of the individual serotypes. The presence of nonneutralizing antibodies to one serotype can facilitate the occurrence of more-severe dengue hemorrhagic fever through immune enhancement upon infection with a second serotype. For this reason, the development of a safe, tetravalent vaccine to produce a balanced immune response to all four serotypes is critical. We have developed a novel approach to produce safe and effective live-attenuated vaccines for DENV and other insect-borne viruses. Host range (HR) mutants of each DENV serotype were created by truncating transmembrane domain 1 of the E protein and selecting for strains of DENV that replicated well in insect cells but not mammalian cells. These vaccine strains were tested for immunogenicity in African green monkeys (AGMs). No vaccine-related adverse events occurred. The vaccine strains were confirmed to be attenuated in vivo by infectious center assay (ICA). Analysis by 50% plaque reduction neutralization test (PRNT50) established that by day 62 postvaccination, 100% of animals seroconverted to DENV-1, -2, -3, and -4. Additionally, the DENV HR tetravalent vaccine (HR-Tet) showed a tetravalent anamnestic immune response in 100% (16/16) of AGMs after challenge with wild-type (WT) DENV strains.
IMPORTANCE We have generated a live attenuated viral (LAV) vaccine capable of eliciting a strong immune response in African green monkeys (AGMs) in a single dose. This vaccine is delivered by injecting one of four attenuated serotypes into each limb of the animal. 100% of animals given the vaccine generated antibodies against all 4 serotypes, and this response was found to be balanced in nature. This is also one of the first studies of dengue in AGMs, and our study suggests that viremia and antibody response in AGMs may be similar to those seen in DENV infection in humans.
INTRODUCTION
Dengue virus, the etiological agent of dengue fever (DF), is a mosquito-borne virus of the Flaviviridae family (1). DENV is an enveloped, positive-strand RNA virus that is characterized as one of four distinct serotypes (DENV-1, -2, -3, or -4) which can be transmitted to humans by the bite of an aedine mosquito, notably Aedes aegypti and Aedes albopictus (Asian tiger mosquito) (2, 3). DENV infection can lead to a wide spectrum of clinical outcomes ranging from asymptomatic to the classical “breakbone” DF or the much more severe and life-threatening dengue hemorrhagic fever (DHF) and dengue shock syndrome (DSS) (4). The occurrence of DENV follows the distribution of aedine mosquitoes and encompasses tropical and subtropical regions globally, with 3.5 billion people at risk (5).
Dengue is now disseminating out from the tropics; outbreaks have occurred since 1969 in the Caribbean, including Puerto Rico and the U.S. Virgin Islands, and there have been almost 500 confirmed cases on the U.S. mainland, including Florida and Texas (6–8). It is estimated that 100 million cases of dengue fever and 500,000 cases of DHF occur each year, leading to more than 20,000 deaths (1). No efficacious therapeutics currently exists.
There are multiple DENV vaccine candidates currently undergoing clinical trial; however, data from these trials suggest that these vaccines either are incapable of conferring ≥70% tetravalent protection or require multiple injections over months, for up to 1 year, which is impractical and unsafe in rural settings where the disease is endemic and for travelers (6, 9–12).
To be globally successful, it is of critical importance that a dengue vaccine protect against all 4 DENV serotypes. Dengue vaccines that are not effective against all 4 serotypes or that require boosters may leave individuals primed for more-severe and potentially fatal dengue disease such as DHF or DSS should they be exposed to a secondary, heterologous DENV serotype not protected against by the vaccine or if exposed prior to full immunity from secondary or tertiary boosts.
It has recently become evident that the greatest chance of developing an efficacious tetravalent vaccine is through the use of a live, attenuated virus (LAV) vaccine. LAV vaccines are known to produce robust, long-lasting, and broad immune responses and to induce strong humoral and cellular immune responses (13). Recent work has revealed that neutralizing antibodies against DENV are preferentially made against epitopes found only in native, live-virus configurations (14). Live virus is also necessary to induce the appropriate protective T cell response (15, 16). To this end, we have developed a tetravalent LAV vaccine against all four DENV serotypes following a successful monovalent trial using an LAV vaccine against DENV-2 (17). The vaccine approach is based on studies of Sindbis virus (SV) showing that large truncations of the E2 transmembrane domain (TMD) are well tolerated in insect cells but not in mammalian cells. This phenotype is referred to as a host range (HR) mutation because the resulting virus grows successfully in insect cells but is attenuated for growth in mammalian cells (17–19). There are minimal risks associated with a live attenuated vaccine capable of replicating in insect cells. The risk of reversion to the wild-type (WT) phenotype is negligible because the virus is not under any selective pressure in insect cells and the mutations are large deletions that are difficult to “repair.” There is also little to no risk of the virus becoming mosquito borne through human interactions because the levels of viremia present in the serum should be insufficient for contraction by the mosquito (20). These HR viral mutants replicate well in insect cells, which can provide a low-cost system of manufacture while generating stable live virus vaccines that are attenuated in mammalian systems.
This method was used to successfully generate a dengue serotype 2 HR mutant virus with deletions in the first transmembrane domain of the E domain (E-T1) capable of eliciting a protective immune response upon challenge in nonhuman primates (NHPs) (17). We have since developed HR mutant viruses in DENV-1, -3, and -4 and have tested the ability of all four HR mutant viruses as a single-dose LAV vaccine, which we will refer to as HR-Tet in an African green monkey (AGM) model of infection (17, 21, 22). Our results suggest that one dose of HR-Tet is sufficient to induce an anamnestic immune response against all 4 DENV serotypes in NHPs, with no evidence of reactogenicity or reversion of the vaccine. Vaccination resulted in 100% seroconversion against all 4 serotypes of dengue virus with a memory response seen upon heterologous virus challenge. Taken together, these data strongly suggest that the tetravalent HR-Tet is a safe, immunogenic LAV vaccine candidate.
MATERIALS AND METHODS
Virus strains. (i) Host range mutants of dengue 1, 2, 3, and 4 (HR-Tet).
A full-length cDNA clone of dengue serotype 2 (DENV-2; strain 16681, GenBank number U87411) in pGEM3z+ was obtained from R. Putnak of the Walter Reed Army Institute of Research (23). Clones of DENV-1 (strain Western Pacific 74, GenBank number U88536), DENV-3 (partial clone DEN3-YU4N, strain CH53489), and DENV-4 (strain 341750, GenBank number GU289913) were provided by B. Falgout (24–27). A full-length DENV-3 cDNA clone was generated by subcloning the missing piece, nucleotides 434 to 2531 (containing the preM and envelope [E] sequence, plus the first 120 bp of NS1), from WT DENV-3 strain UNC3001 (supplied by A. DeSilva) into pDRIVE. Full-length DENV-3 transcripts were then produced by ligating the two DENV-3 subclones, which were then used as transcription templates (K. M. Smith, B. Falgout, C. J. Spears, C. M. Briggs, M. Quiles, and R. Hernandez, submitted for publication). All the clones produce full-length DENV RNAs when transcribed in vitro with T7 (for DENV-2 and -3) or SP6 (DENV-1 and -4) RNA polymerase, and after transfection into mammalian or insect cells, infectious virions are generated. For the vaccine preparation, all viruses were grown in C6/36 cells and purified on iodixanol gradients.
(ii) Challenge viruses.
Heterologous WT strains of DENV were used to challenge AGMs. DENV-2 strain S16803 (GenBank number GU289914) was provided by R. Putnak (28). DENV-1 Hawaii (HI), DENV-3 H87, and DENV-4 H241 were provided by B. Falgout.
Viremia analysis by RT-PCR.
Viral RNA was extracted from 100 μl of serum collected on days 2 to 7, 9, 11, and 14 postinoculation from the HR-Tet vaccine (monkeys 1095, 1114, 1225, and 1463) or WT DENV-1 to -4 controls (monkeys 1253, 1272, 1407, and 1427) using the e.Z.N.A. viral RNA kit (Omega bio-tek, Norcross, GA). Following elution, RNA was precipitated and resuspended in 10 μl of diethyl pyrocarbonate-double-distilled water (DEPC-dH2O) and used as the template for reverse transcription (RT) (High-Capacity cDNA reverse transcription kit; Applied Biosystems, Carlsbad, CA). PCR was performed using GoTaq Flexi DNA polymerase (Promega, Madison, WI) with a final Mg2+ concentration of 3 mM and 10 μl of cDNA. Nested PCR cycles were as follows: first round, 95°C for 3 min, followed by 45 cycles of 95°C for 45 s, 1 min at 55°C, and 72°C for 1.5 min. Extension was performed for 3 min at 72°C with a 4°C hold. For the second round, 5 μl of the primary PCR was diluted for use under the same conditions with the following alterations: annealing time was 45 s, and extension time was 1 min. Samples were held at 4°C until analysis by gel electrophoresis. Primary PCR primers were designed to amplify the same 1.44-kb region from bp 1151 to 1168 in the E/NS1 domains in all 4 DENV serotypes and vary between serotypes by 2 to 3 nucleotides (nt): DENV-1, sense primer, 5′-GATGTCCAACRCAAGGAG-3′, antisense primer from bp 2578 to 2591, 5′-CGAATTCCACACACACCCTC-3′; DENV-2, sense primer, 5′-GCTGCCCAACACAAGGGG-3′, antisense primer, 5′-CGGATTCCACAAATGCCCTC-3′; DENV-3, sense primer, 5′-GATGTCCTACCCAAGGGG-3′, antisense primer, 5′-CTGATTCCACAGACTCCATT-3′; DENV-4, sense primer, same as DENV-1; antisense primer, 5′-CTAATTCCACAGACCCCATC-3′. The secondary nest primers each amplified a different-size product and were specific to each serotype. Primers were as follows: DENV-1 (product size, 309 bp), sense primer from bp 2220 to 2245, 5′-CACGTCTGTGGGAAAACTGATACACC-3′, antisense primer from bp 2504 to 2529, 5′-GTCGGCCTGGAATTTATATTGCTCTG-3′; DENV-2 (592-bp product), sense primer from bp 1866 to 1888, 5′-GGAAATAGCAGAAACACAACATG-3′, antisense primer from bp 2436 to 2458, 5′-GTTCTTTGTTTTTCCAGCTCACA-3′; DENV-3 (350-bp product), sense primer from bp 2190 to 2209, 5′-CTTTGGATCAGTGGGTGGTG-3′, antisense primer from bp 2518 to 2539, 5′-CCAATCGATTGGGGGAGTCTGC-3′; and DENV-4 (product size, 490 bp), sense primer from 1975 to 1998 bp, 5′-GTGGTTGGGCGTATCATCTCATCC-3′, antisense primer from bp 2441 to 2465, 5′-CACTTCAATTCTTTCCCACTCCATG-3′.
Cell culture and virus culture.
C6/36 (Aedes albopictus, ATCC CRL-1660; American Type Culture Collection, Manassas, VA) and Vero cells (African green monkey kidney, ATCC CCL-81) were maintained as described previously (17). Vero E6 cells (AGM kidney clone 6, ATCC CRL-1586) were maintained in minimal essential medium (MEM) containing Earl's salts supplemented with 10% fetal bovine serum (FBS), 2 mM l-glutamine, 1 mM sodium pyruvate, and 1,500 mg/liter sodium bicarbonate. WT and HR-Tet viruses were grown in C6/36 cells as described previously (17). Confluent T75 flasks of C6/36 cells were inoculated in C6/36 medium with each dengue virus at a multiplicity of infection (MOI) of 0.03 PFU/cell and rocked for 1.5 h at room temperature. Each flask was brought up to 5 ml with C6/36 media, and the flasks were put into the incubator for 7 days at 28°C with 5% CO2. On day 7, supernatants from infected flasks were harvested and clarified by centrifugation.
Primate model and study design.
Thirty-six AGMs (genus Chlorocebus) were employed in this study. All animals were prescreened for the presence of anti-dengue 1 to 4 IgM or IgG by enzyme-linked immunosorbent assay (ELISA) (data not shown), and highly reactive monkeys were excluded. Animals were divided into 9 treatment groups (n = 4) which were as follows: WT tetravalent (WT-Tet), HR-Tet challenged with DENV-1 to -4 (4 groups, n = 4/group; 16 in total), or mock vaccinated (phosphate-buffered saline [PBS]) challenged with DENV-1 to -4 (4 groups, n = 4/group; 16 in total). Blood samples were collected, and clinical observations were made at baseline and on days 2, 3, 5, 7, 9, 11, 14, 30, and 62 after vaccine administration. After blood collection on day 62, animals received 1 of 4 live WT DENV challenge strains (DENV-1 HI, DENV-2 S16803, DENV-3 H87, and DENV-4 H241) before continued blood collection and clinical observations at 63 to 69, 71, 73, and 77 days postvaccination. The WT-Tet group was not challenged. Serum was collected from the blood samples and was subsequently used to measure viremia, dengue-specific IgG, and neutralizing antibody (NAb) response. Vaccine viremia was measured on days 2 to 5 and 7, 9, 11, 14, and 30 postvaccination followed by days 63 to 69 and 71 postchallenge. NAb response was measured on days 0, 14, 30, 62 to 69, 71, 73, and 77. Total DENV-specific IgG levels were also measured for both pre- and postchallenge samples. Clinical observation involved measuring packed cell volume (PCV)/hematocrit and white blood cell and platelet counts. Animals were also observed for any changes in body weight, temperature, respiration, and heart rate twice a day for the duration of the study. Clinical examinations took place daily for the first week of the study, biweekly through day 30, and every month after that out to 74 days (data not shown).
All animal procedures were approved by the Wake Forest University Animal Care and Use Committee at the Wake Forest School of Medicine. Wake Forest is an AAALAC-accredited institution. All research was conducted in compliance with the 8th edition of the Guide for the Care and Use of Laboratory Animals (63).
Viral vaccine preparation and administration.
WT-Tet, HR-Tet, and challenge virus strains were grown as described above. Following clarification of the supernatant, virus was purified and concentrated using iodixanol gradients (OptiPrep Density Gradient Medium; Sigma). Briefly, a layer of 12% iodixanol solution was underlayed with a 35% iodixanol cushion in 38-ml tubes (Becton, Dickinson). Supernatant containing the virus was overlaid on the gradient and then spun overnight at 4°C and 26,000 rpm using a SW28 rotor. Purified virus was visualized by light and harvested. This step also serves to concentrate the virus, increasing titers ∼10-fold. Purified virus was then quantified by infectious center assay (ICA), described below, and diluted for injection using sterile PBS. For vaccination, WT-Tet and HR-Tet serotypes were diluted to 1 × 106 infectious centers (IC)/0.5 ml PBS. Vaccine strains and WT-Tet were administered as four separate injections, one serotype in each limb. Total injected virus was 4 × 106 IC/animal. For challenge, viruses were grown and purified as described above. The titer of purified virus was determined via ICA, and each group (n = 4) was challenged with one serotype of DENV at 1 × 105 IC/0.5 ml PBS.
ICA.
To quantify viremia, an infectious center assay was done as described previously (17). This is a modified plaque assay with cells and virus in soft agar suspension. Titers are calculated and shown as IC/ml.
PRNT.
Prior to the plaque reduction neutralization test (PRNT), monkey sera were heat inactivated for 30 min at 56°C and then diluted serially 1:2 in completed 1× MEM to dilutions ranging from 1:20 to 1:2,560. Twenty-five PFU of WT virus (DENV-1 Western Pacific, DENV-2 New Guinea C [supplied by Vance Vorndam, CDC, Dengue branch], DENV-3 CH53489, and DENV-4 341750) was added and mixed into each dilution, and the mixtures were incubated at 37°C for 35 min. After incubation, subconfluent Vero plates were inoculated with each dilution of mixtures of serum and virus and incubated at 37°C for 1.5 h. Inoculum was then removed from the plates, and 1× MEM containing 1% agarose was added to cover each well. After incubating 7 days at 37°C, plates were stained for 4 h at 37°C with PBS-D containing 1% agarose and 0.06% neutral red stain. After staining, plaques were counted, and serum dilutions at which 50% of the WT virus added was neutralized determined the PRNT50. Seroconversion is defined as a titer of ≥20. Titers of less than 20 are represented by 10 for determination of the geometric mean titer (GMT).
ELISA.
Measurement of dengue-specific IgG was done using an enzyme-linked immunosorbent assay. Briefly, each of the four WT dengue serotypes used for PRNT50 was grown and purified as described above. Following purification, viral protein was quantified using a Micro-bicinchoninic acid (Micro-BCA) protein assay kit (Pierce). Purified virus (100 ng) was added to each well of a 96-well Maxisorp plate (Nunc) in 100 μl carbonate coating buffer, pH 9.5, and incubated overnight at 4°C. Following virus removal, plates were blocked in 300 μl 1× blocking buffer (Sigma) for 1 h at room temperature. Following blocking, 1:2 dilutions of heat-inactivated serum in PBS (1:100 to 1:6,400) were added to the plate. The plate was sealed and incubated at room temperature (25°C) for 1 h. Following incubation, the serum was removed and the plate was washed three times with 300 μl wash buffer containing 0.05% Tween 20. Next, 100 μl of goat anti-monkey IgG horseradish peroxidase (HRP; 1:5,000; Fitzgerald) was added to each well, and the plate was sealed and incubated for 1 h. Following incubation, the plate was washed 5× in PBS–0.05% Tween 20. A 3,3′,5,5′-tetramethylbenzidine dihydrochloride (TMB) substrate was then added, and following incubation in the dark, the reaction was stopped using 2 N H2SO4, and plates were read at 450 nm. Assays were performed in duplicate with positive and negative controls on every plate. The net optical density (OD) was calculated by subtracting the absorbance of test serum from that of a PBS control. Endpoint dilution titers were determined by the dilution at which the OD value was ≥0.10.
Statistical methods.
The statistical significance of previremia, postchallenge viremia, and postchallenge antibody titers was analyzed using the Mann-Whitney U test. Postchallenge antibody titer statistical significance was analyzed using the Kruskal-Wallis test to determine if a significant difference was present between the WT-Tet, HR-Tet, and mock groups. In cases where a significant difference was present, groupwise significance was analyzed using the Nemenyi-Damico-Wolfe-Dunn test. All statistical analysis was performed using the R statistical computing environment.
RESULTS
Production of TMD deletion DENV mutants and HR phenotyping.
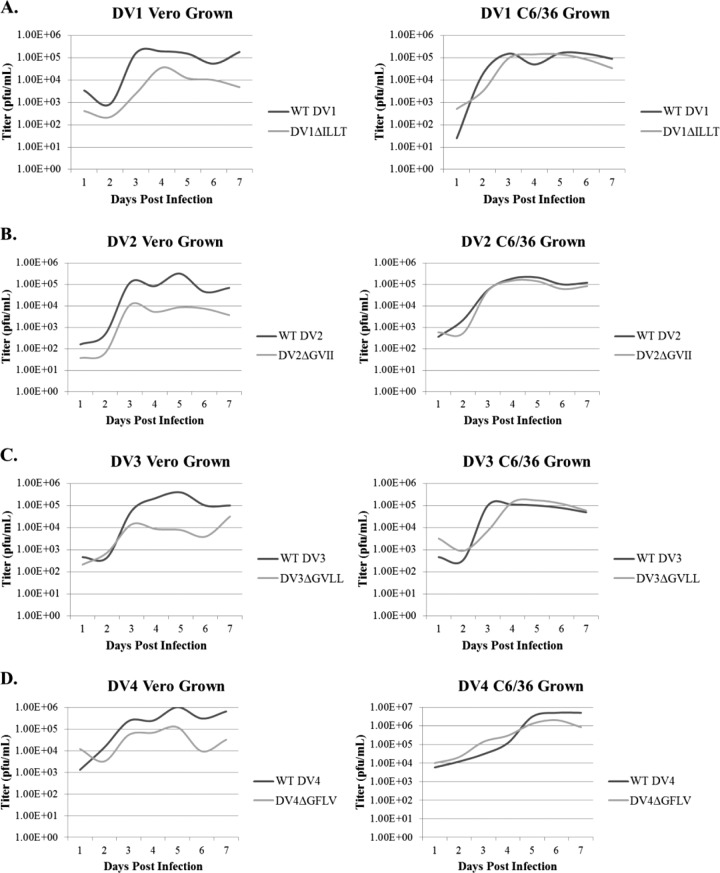
The methodology for producing the DENV-2 E-T1TMD deletion mutant (DENV-2ΔGVII) has been described previously (17, 29). TMD deletion mutants for DENV-1 WP '74, DENV-3 ABV (Smith et al., submitted; patent 61/674,137) and DENV-4 341750 were generated by identifying the consensus sequence in all the E-T1 TMDs and deleting the amino acids analogous to DENV-2 GVII. Sixteen-amino-acid consensus sequences for all four viruses can be seen in Table 1. To generate the mutants, four amino acids were deleted from the E-T1 sequence, leaving 12 amino acids in the TMD. Deletions within the same location in each of the other serotypes were expected to display the same HR phenotype as that previously described for DENV-2ΔGVII (17). These deleted amino acids are shown in boldface in Table 1. The mutant viruses were named according to which amino acids were deleted and are DENV-1ΔILLT, DENV-2ΔGVII, DENV-3ΔGVLL, and DENV-4ΔGFLV. Transcripts from each of the viral clones were transfected into C6/36 cells or Vero cells as described in Materials and Methods to generate live virus for testing the HR phenotype. All viral phenotypes were confirmed to be HR, which we define as >1 log growth reduction in Vero cells compared to C6/36 cells and the inability to plaque in Vero cells.
TABLE 1.
Generation of host range mutants
| Virus | Strain | Mutant | ET-1 TMD sequencea | Virus titer (PFU/ml) |
|
|---|---|---|---|---|---|
| C6/36 | Vero | ||||
| DENV-1 | WP ‘74 | ΔILLT | 452SWTMKIGIGILLTWLG467 | 1.0 × 106 | 1.8 × 103 |
| DENV-2 | 16681 | ΔGVII | 452SWTMKILIGVIITWIG467 | 4.0 × 106 | 1.0 × 104 |
| DENV-3 | ABV | ΔGVLL | 448SWIMKIGIGVLLTWIG465 | 7.0 × 105 | 4.0 × 104 |
| DENV-4 | 341750 | ΔGFLV | 452SWMIRILIGFLVLWIG467 | 1.5 × 106 | 7.5 × 105 |
Consensus sequences for the ET-1 TMD deleted from each virus serotype are shown in boldface.
As shown in Table 1, TMD deletion mutant viruses had titers of 7.0 × 105 PFU/ml (DENV-3ΔGVLL), 1.0 × 106 PFU/ml (DENV-1ΔILLT), 1.5 × 106 PFU/ml (DENV-4ΔGFLV), and 4.0 × 106 PFU/ml (DENV-2ΔGVII, previously reported in reference 17) when grown in C6/36 insect cells. When these TMD deletion viruses were grown in mammalian cells, viral titers dropped by at least 1 log compared to viral titers obtained from insect cells (Table 1). Growth curves for the WT viruses and their mutants in Vero and C6/36 cells can be found in Fig. 1. When these results were combined with the viruses' inability to plaque in Vero cells, we concluded that DENV-1ΔILLT, DENV-3ΔGVLL, and DENV-4ΔGFLV exhibited the same HR phenotype as that observed in DENV-2ΔGVII and were suitable LAV candidates. These vaccine candidates were assayed in a nonhuman primate model to determine their ability to elicit an immune response against dengue virus challenge.
FIG 1.
Growth of HR-Tet vaccine viruses and their parental wild-type viruses in mammalian and insect cells. Each of the four vaccine serotypes and their corresponding parental wild-type viruses were grown in mammalian (Vero) and insect (C6/36) cells for a 7-day time course. Cells were infected with an intial MOI of 0.03 PFU/cell, and infection was allowed to progress for 7 days. Starting at 1 day postinfection, cell supernatant was harvested and a plaque assay done to determine virus titers. As shown, DV1ΔILLT (A), DV2ΔGVII (B), DV3ΔGVLL (C), and DV4ΔGFLV (D) all had ∼1-log reduction in growth in Vero cells compared to their parental WT viruses. In contrast, there was no difference in viral titer between the vaccine viruses and the parental WT viruses when grown in C6/36 cells.
AGM tetravalent DENV study design.
Thirty-six AGMs were selected for use in the trial. The animals were divided into three groups: those tetravalently inoculated with each of the four WT parental viruses (DENV-1 WP '74, DENV-2 16681, DENV-3 ABV, and DENV-4 341750; n = 4), animals tetravalently inoculated with the four TMD deletion mutants (DENV-1ΔILLT, DENV-2ΔGVII, DENV-3ΔGVLL, and DENV-4ΔGFLV; n = 16), and animals inoculated with PBS as a control (n = 16). On day 0, the animals were injected with 1.0 × 106 IC of each of the four purified viruses/0.5 ml PBS, one into each individual limb (i.e., DENV-1 WP'74 or DENV-1ΔILLT, upper left limb; DENV-2 16681 or DENV-2ΔGVII, upper right limb; DENV-3 ABV or DENV-3 ΔGVLL, lower left limb; and DENV-4 341750 or DENV-4ΔGFLV, lower right limb) for a total viral load of 4 × 106 IC/animal. Viruses were administered separately instead of as a mixture to target each individual virus into separate draining lymph nodes. No boost was administered. Animals were monitored for the duration of the trial, and no adverse vaccine reactions were noted for any of the animals by the criteria described in Materials and Methods.
HR-Tet vaccination viremia and antibody response.
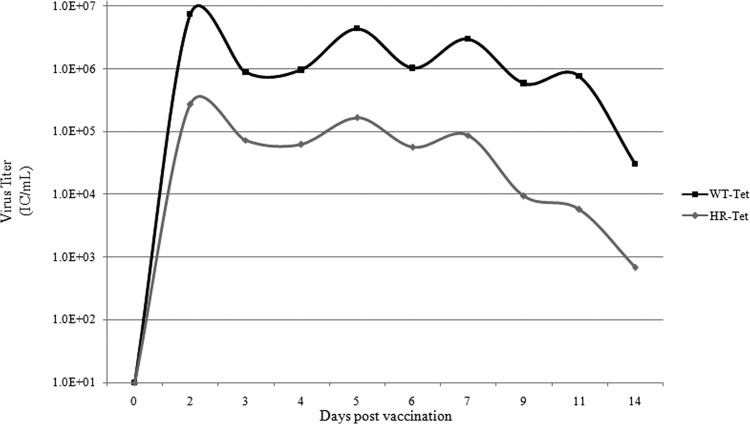
To determine vaccine attenuation in vivo, serum samples were taken on days 1 to 7, and 9, 11, and 14 postvaccination. Levels of viremia in the serum were determined by ICA. As shown in Fig. 2, maximal titers for both WT and HR-Tet were seen about 2 days following vaccination. Animals given 4.0 × 106 IC total virus (1 × 106 each serotype) of the WT dengue viruses (WT-Tet) had an average maximum titer of approximately 7 × 106 IC/ml. In contrast, animals given 4.0 × 106 IC total virus (1 × 106 each serotype) of HR-Tet had an average maximum titer of 2.7 × 105 IC/ml, a difference of more than 1 log (Fig. 2). During the course of viremia, animals given HR-Tet had significantly less virus detected than animals given the WT-Tet viruses on 9 of the 10 days sampled despite both groups being given the same initial dose. These results confirm our in vitro findings that the TMD deletions are HR mutants attenuated for growth in mammalian hosts.
FIG 2.
Viremia following vaccination. The amount of viremia in serum was measured daily out to 14 days postvaccination by ICA. Animals were given either 4 × 106 IC (total) of the 4 parental serotypes (WT-Tet) or 4 × 106 IC (total) of 4 host range mutant serotypes (HR-Tet). Titers from HR-Tet animals were significantly lower than titers from WT-Tet animals on days 2 (P < 0.004), 3, (P < 0.02), 4 (P < 0.03), 5 (P < 0.02), 6 (P < 0.01), 7 (P < 0.007), 9 (P < 0.02), and 14 (P < 0.03) by the Mann-Whitney test.
To ensure that each viral serotype was able to replicate in each animal, RT-PCR was performed on serum samples from all four WT-Tet animals and four randomly chosen HR-Tet animals from eight selected days following vaccination. As shown in Table 2, all four WT-Tet animals had each serotype present at some time during infection, although the four serotypes were not seen simultaneously on any one day tested. In contrast, on day 3 postvaccination, all four HR-Tet animals had simultaneous expression of all four serotypes, and three of the four HR-Tet animals maintained this profile on day 4 postvaccination. Additionally, all remaining 12 HR-Tet serum samples from days 3 and 4 postvaccination were also analyzed by RT-PCR and found to contain all 4 serotypes of DENV (data not shown). This simultaneous expression indicates that each virus was able to replicate in the animal with no viral replication interference, increasing the chances of developing a balanced immune response against all four serotypes.
TABLE 2.
Virus expression in tetravalently vaccinated animals
| Virus | Animal no. | DENV serotype(s) expressed on: |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Day 2 | Day 3 | Day 4 | Day 5 | Day 6 | Day 7 | Day 9 | Day 11 | ||
| WT-Tet | 1253 | 2 | 1, 2, 4 | 1, 2, 3 | 3, 4 | 3 | 4 | 2, 4 | — |
| 1272 | 2, 4 | 1, 2, 4 | 2, 3 | 3, 4 | 3 | 4 | 2, 4 | 4 | |
| 1407 | 2, 4 | 1, 2 | 2, 3 | 3, 4 | 3 | 4 | 2, 4 | 2, 4 | |
| 1427 | 1, 2, 4 | 1, 2, 4 | 2, 3 | 3, 4 | 3 | 4 | 2, 4 | 2, 4 | |
| HR-Tet | 1095 | 1, 2 | 1, 2, 3, 4 | 1, 2, 3, 4 | 2, 4 | 3 | 2, 3, 4 | 2, 4 | 2, 4 |
| 1114 | 1, 2, 4 | 1, 2, 3, 4 | 2 | 2, 3, 4 | 3 | 2, 3, 4 | 2, 4 | 2, 4 | |
| 1225 | 1, 2, 4 | 1, 2, 3, 4 | 1, 2, 3, 4 | 2, 3, 4 | 2, 3 | 2, 3, 4 | 2, 4 | 2, 4 | |
| 1463 | 1, 2 | 1, 2, 3, 4 | 1, 2, 3, 4 | 2, 3, 4 | 2, 3 | 2, 3, 4 | 2, 4 | 2, 4 | |
To determine if HR-Tet elicited the production of DENV-specific antibodies, a dengue-specific ELISA was conducted on serum samples 30 days postvaccination. As shown in Fig. 3, total dengue-specific IgG for each serotype was measured for WT-Tet-, HR-Tet-, and mock-vaccinated animals at 30 and 62 days postvaccination. Ab titers are shown as the average normalized optical density at 450 nm (OD450) for each group obtained. As expected, animals injected with WT-Tet produced IgG against all four of the DENV serotypes at both time points tested. The amount of Ab produced differs significantly from that of mock-vaccinated animals, which had a background level of response. Importantly, animals vaccinated with HR-Tet also elicited a strong IgG Ab response at 30 days postvaccination (Fig. 3A). This response is not significantly different from the WT-Tet response for any of the four serotypes; however, it is significantly different from readings obtained from the mock-vaccinated animals. The same profile was seen on day 62 postvaccination, indicative of a long-lived Ab response (Fig. 3B) and demonstrating that vaccination with HR-Tet initiates a total dengue-specific Ab production similar to that induced by vaccination with WT-Tet.
FIG 3.
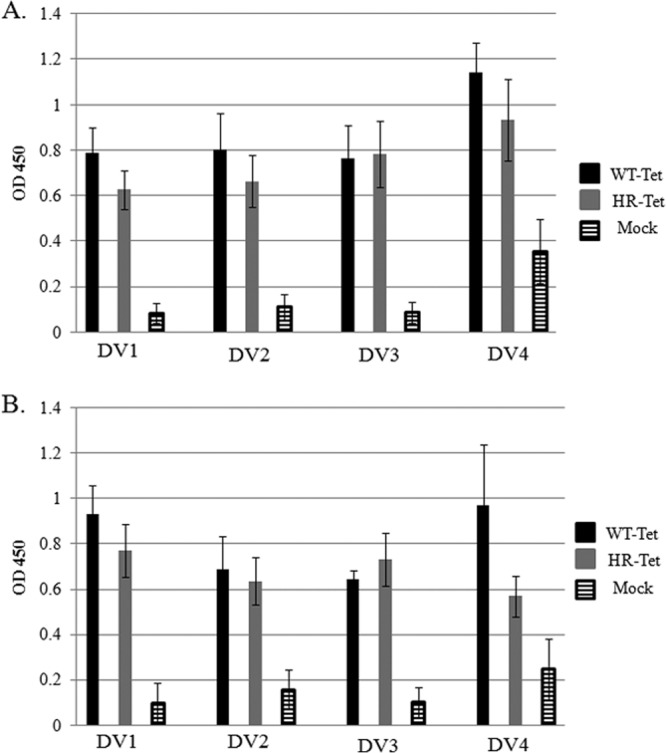
ELISA on postvaccination samples. At 30 (A) and 62 (B) days postvaccination, an ELISA to measure IgG in AGMs was done as described in Materials and Methods. For day 30 (A), WT-Tet and HR-Tet samples are statistically significantly different from mock samples for all DENV serotypes (DV) tested (DV1, WT-Tet versus Mock and HR-Tet versus mock, P < 0.001; DV2, WT-Tet versus mock and HR-Tet versus mock, P < 0.001; DV3, WT-Tet versus mock, P < 0.01, and HR-Tet versus mock, P < 0.001; DV4, WT-Tet versus mock and HR-Tet versus mock, P < 0.001). For day 62 (B), there were significant differences in OD between samples (DV1, WT-Tet versus mock, P < 0.01, and HR-Tet versus mock, P < 0.001; DV2, WT-Tet versus mock, P < 0.01, and HR-Tet versus mock, P < 0.001; DV3, WT-Tet versus mock, not significant, and HR-Tet versus mock, P < 0.001; DV4, WT-Tet versus mock, P < 0.001, and HR-Tet versus mock, P < 0.001). Statistics were performed using the Kruskal-Wallis test with Dunn's posttest.
Once it was ascertained that an Ab response against the vaccine had been elicited, levels of neutralizing antibodies against each of the four DENV serotypes were measured by PRNT50. In Table 3, geometric mean titers (GMTs) for each animal group against each individual serotype are shown along with the GMT for each group. A breakdown of titers seen in each individual animal can be found in Fig. S1 in the supplemental material. The WT-Tet group (n = 4) had 100% seroconversion to DENV-1 by day 14 and 100% seroconversion to DENV-2, -3, and -4 by day 30 postvaccination (see Fig. S1 in the supplemental material). By 62 days postvaccination, 100% of animals (16/16) had tetravalently seroconverted following vaccination with HR-Tet. GMTs from HR-Tet-vaccinated animals differed significantly from those of mock-vaccinated animals, while there was no significant difference in titers between WT-Tet and HR-Tet animals.
TABLE 3.
Geometric mean titers from PRNT50a
| Inoculation group (n) | GMT |
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| DENV-1 |
DENV-2 |
DENV-3 |
DENV-4 |
|||||||||||||
| Day 0 | Day 14 | Day 30 | Day 62 | Day 0 | Day 14 | Day 30 | Day 62 | Day 0 | Day 14 | Day 30 | Day 62 | Day 0 | Day 14 | Day 30 | Day 62 | |
| WT-Tet (4) | 10 | 705 | 1,280 | 640 | 10 | 160 | 538 | 905 | 10 | 269 | 905 | 2,560 | 10 | 18 | 1,810 | 135 |
| HR-Tet (16) | 10 | 104 | 123 | 538 | 10 | 100 | 269 | 129 | 10 | 252 | 123 | 91 | 10 | 21 | 742 | 247 |
| Mock (16) | 10 | 14 | 21 | 16 | 10 | 30 | 10 | 10 | 10 | 11 | 14 | 10 | 10 | 10 | 12 | 10 |
Following execution of plaque reduction neutralization tests (PRNT), the dilution of serum sufficient to reduce viral titer by 50% for each serotype was recorded for each animal. For animals with a titer of <20, the value 10 was used to calculate GMT and report a lack of neutralizing antibody titer. There was no significant difference in antibody titer between the groups on day zero. Beginning on day 14 for all serotypes, statistically significant differences in antibody titer for WT-Tet and HR-Tet when compared to Mock were observed.
Viremia and Ab response in AGM postchallenge.
To test the immunogenicity of HR-Tet, AGMs were given a heterologous viral challenge. On day 62 postvaccination, animals vaccinated with HR-Tet or PBS control (mock) were divided into four groups (n = 4) for challenge. Each group was challenged with 1 × 105 IC of one of the four WT DENV serotypes per animal. The challenge viruses were heterologous strains from those used to generate the vaccine strains: DENV-1 HI, DENV-2 S16803, DENV-3 H87, and DENV-4 H241. Animals given WT-Tet were not challenged. Following challenge, all animals were monitored for any adverse effects. Blood was taken at 10 time points postchallenge (days 62 to 69, 71, 73, and 77 postvaccination) to determine postchallenge viremia and Ab responses.
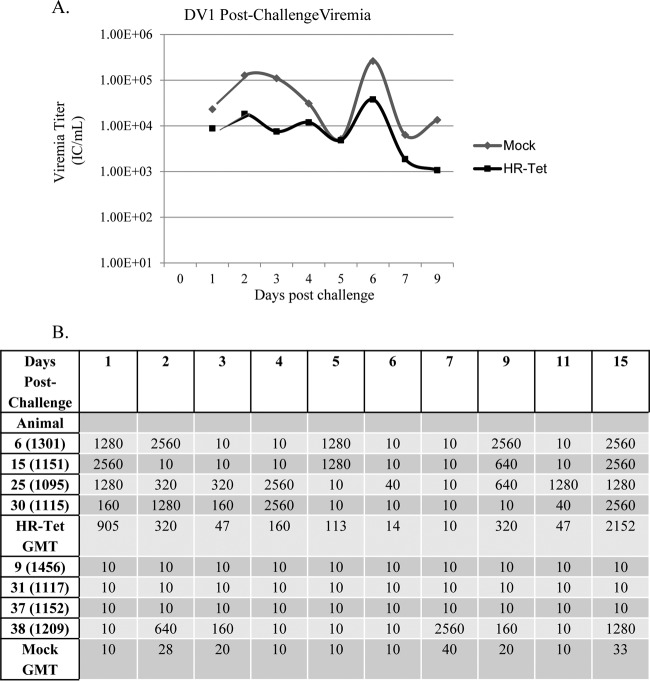
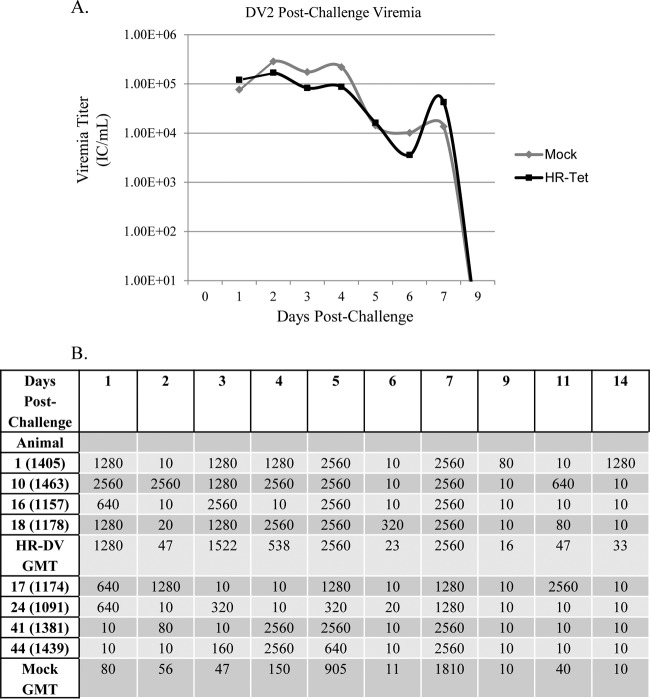
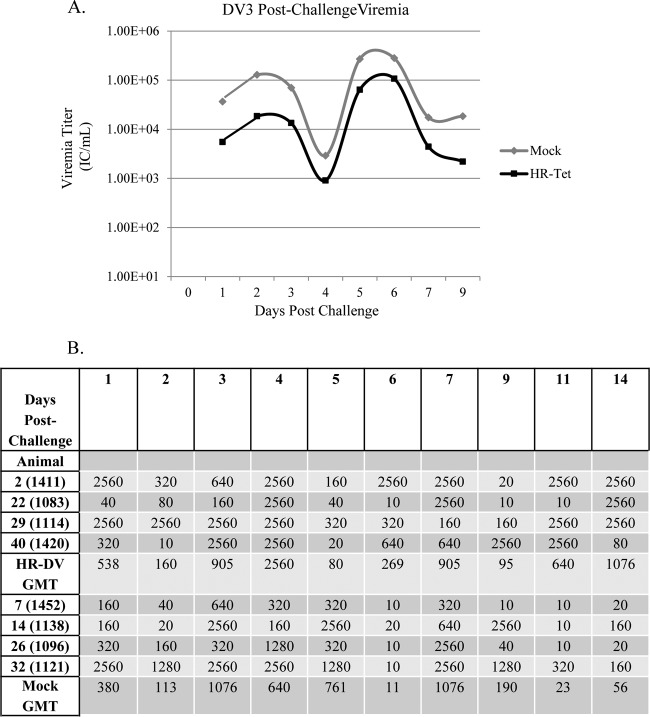
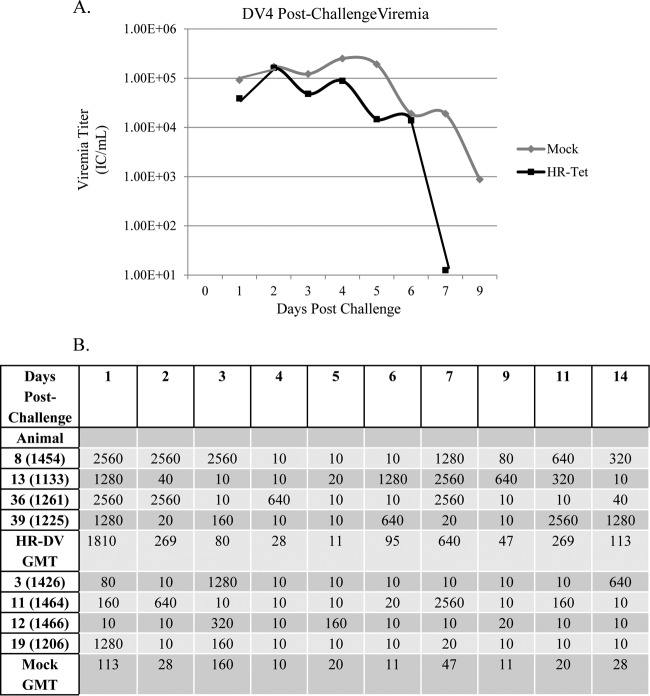
Postchallenge viremia was measured using ICA as described above. Following challenge with DENV-1 HI, mock-vaccinated animals had significantly higher viremia for the duration of infection than did animals vaccinated with HR-Tet (Fig. 4A). By day 71 postvaccination (day 9 postchallenge), there was an almost 10-fold difference in viral titer between vaccinated and nonvaccinated animals. Following challenge with DENV-2 S16803, HR-Tet vaccinated animals displayed a small but measurable decrease in viremia compared to mock-vaccinated animals, although it was not statistically different (Fig. 5A). HR-Tet vaccinated animals challenged with DENV-3 H87 had a 10-fold decrease in viremia compared to mock-vaccinated animals (Fig. 6A). The difference in viremia was most noticeable on days 1 and 5 postchallenge. Lastly, the ability of HR-Tet to protect against challenge from DENV-4 H241 was determined. As shown in Fig. 7A, animals vaccinated with HR-Tet had viremia 10- to 20-fold lower in magnitude on most days and shorter in duration than mock-vaccinated animals. Animals inoculated with the HR-Tet vaccine cleared the viremia by day 9 postchallenge, whereas mock-vaccinated animals still had detectable levels of viremia at this time.
FIG 4.
Viremia and neutralizing antibody titers following challenge with DENV-1 (DV1). (A) HR-Tet animals challenged with DV1 HI had on average 1 log less viremia following challenge compared to mock-vaccinated animals challenged with the same dose of DV1 HI. A significant difference in viremia between the groups was noted on days 2, 3, 6, and 9 (P < 0.02). Graphed titers are averages from four animals challenged with DV1 HI. (B) There was also a large difference in neutralizing antibody titer in mock-vaccinated animals compared to HR-Tet animals following challenge. All animals given HR-TET had high neutralizing antibody titers following challenge with DV1 HI. However, only 25% of mock-vaccinated animals developed neutralizing antibody titers following challenge with DV1 HI.
FIG 5.
Viremia and neutralizing antibody titers following challenge with DENV-2 (DV2). (A) HR-Tet animals challenged with DV2 S16803 had noticeably lower viremia following challenge than did mock-vaccinated animals challenged with the same dose of DV2 S16803. Graphed titers are averages from four animals challenged with DV2 S16803. (B) There was also a large difference in neutralizing antibody titer in mock-vaccinated animals compared to HR-Tet animals following challenge. Animals given HR-Tet prior to challenge had much faster, stronger, and more-sustained neutralizing antibody production than did mock-vaccinated animals challenged with DV2 S16803.
FIG 6.
Viremia and neutralizing antibody titers following challenge with DENV-3 (DV3). (A) HR-Tet animals challenged with DV3 H87 had viremia on average 1 log lower in titer following challenge than mock-vaccinated animals challenged with the same dose of DV3 H87. A significant difference in titer between the two groups was noted on days 2, 3, 5, 6, and 9 (P < 0.02). Graphed titers are averages from four animals challenged with DV3 H87. (B) There was also a difference in neutralizing antibody titer in mock-vaccinated animals compared to HR-Tet animals following challenge. Animals given HR-Tet prior to challenge had much faster, stronger, and more-sustained neutralizing antibody production than did mock-vaccinated animals challenged with DV3 H87.
FIG 7.
Viremia and neutralizing antibody postchallenge with DENV-4 (DV4). (A) HR-Tet animals challenged with DV4 H241 had lower viremia, which was cleared more quickly, than mock-vaccinated animals challenged with the same dose of DV4 H241. A significant difference in titers between the two groups was noted on days 3, 4, and 5 (P < 0.02). Graphed titers are averages from four animals challenged with DV4 H241. (B) There was also a difference in neutralizing antibody titers in mock-vaccinated animals compared to HR-Tet animals following challenge. Animals given HR-Tet prior to challenge had much faster, stronger, and more-sustained neutralizing antibody production than mock-vaccinated animals challenged with DV4 H241.
The presence of NAb against challenge virus was also measured using a PRNT50 as described above. NAb titers were measured daily for 7 days postchallenge and again on days 9, 11, and 15. NAb titers for each individual animal are shown along with the GMT for each group. The assessment for seroconversion was the same as described above, with a value of 10 being used for animals with titers ≤20. For animals challenged with DENV-1 HI, 4 of 4 animals (100%) vaccinated with HR-Tet had NAb titers consistent with seroconversion 1 day postchallenge (Fig. 4B). In contrast, only 1 of 4 naive animals (25%) from the mock-vaccinated group seroconverted to DENV-1 following challenge. That lone animal did not develop antibody against DENV-1 until 2 days postchallenge. There was also a noticeable difference in GMT for each of these groups, with the highest GMT for the HR-Tet animals being seen on day 15 postchallenge (animal 2152, Fig. 4B). The highest GMT achieved by mock-vaccinated animals was 40, seen on day 7. Four of 4 HR-Tet animals (100%) challenged with DENV-2 S16803 retained high levels of neutralizing antibody against DENV-2 as early as 1 day postchallenge (Fig. 5B). While 4 of 4 mock-vaccinated animals (100%) seroconverted to DENV-2, these animals did not fully seroconvert until 3 days postchallenge. HR-Tet vaccination also led to higher NAb titers following DENV-2 S16803 challenge with a maximal GMT of 2,560 observed 5 days postchallenge, whereas the highest GMT seen in the mock-vaccinated animals was 810 and did not occur until 7 days postchallenge (Fig. 5B). When PRNT50 was performed to measure the presence of DENV-3-specific NAb, 100% of animals (4/4) were found to maintain titers of neutralizing antibody following challenge with DENV-3 H87 (Fig. 6B). Animals that received the HR-Tet vaccine also had higher GMT titers of neutralizing antibody with the maximal GMT of 2,560 at 4 days postchallenge compared with mock animals that displayed a maximal GMT of 1,076 on day 3 postchallenge. As shown in Fig. 7B, 4 of 4 (100%) HR-Tet-vaccinated animals retained their seroconverted status following DENV-4 H241 challenge with high titers seen as early as 1 day postchallenge. While 4 of 4 (100%) mock-vaccinated animals did seroconvert against DENV-4, the majority of the animals did not seroconvert until day 3 postchallenge. Furthermore, the maximal GMT elicited by HR-Tet was found to be 1,810 (day 1) compared to 160 for mock-vaccinated animals on day 3 postchallenge.
To determine if the antibody elicited following challenge was indicative of a memory response, sera from these animals were used in an ELISA for serotype-specific DENV IgG on days 9 and 15 postchallenge. These results are presented as fold over control, and an anamnestic response is defined as a titer >4-fold that of the control. As shown in Fig. 8A, animals vaccinated with HR-Tet had a DENV-1-specific Ab response approximately 14-fold greater than that of control animals, whereas mock-vaccinated animals had a <1-fold increase at 9 days postchallenge. This trend continued out to 15 days postchallenge, when HR-Tet-vaccinated animals had an Ab response 8-fold greater than the control samples. However, mock-vaccinated animals were still only 2-fold above the negative control (Fig. 8B). Animals inoculated with the HR-Tet vaccine prior to challenge with DENV-2 S16803 had a >20-fold increase in dengue-specific Ab titer on day 9 following challenge, whereas mock-vaccinated animals had a <1-fold increase at this time point (Fig. 8A). This trend continued out to day 15 postchallenge, when animals vaccinated with HR-Tet exhibited a 5-fold increase in DENV-2-specific IgG compared to mock-vaccinated animals, for which the fold increase was just slightly more than 1 (Fig. 8B). Ab titers from HR-Tet-vaccinated animals challenged with DENV-3 H87 on day 9 postchallenge revealed an 8-fold increase compared to the control, whereas the mock-vaccinated animals had a <1-fold increase (Fig. 8A). This trend continued out to 15 days postchallenge, when HR-Tet-vaccinated animals had a 6-fold increase in DENV-3-specific Ab titer compared to the control and mock-vaccinated animals showed only a 1-fold increase (Fig. 8B). HR-Tet-vaccinated animals challenged with DENV-4 H241 displayed an 8-fold increase in DENV-4-specific IgG on day 9 postchallenge compared to the mock-vaccinated animals, which had a <1-fold increase (Fig. 8A). This trend was sustained out to day 15 postchallenge, when it was established that animals given the HR-Tet vaccine had a 4-fold increase in DENV-4-specific IgG compared to mock-vaccinated animals, which demonstrated only a 1-fold increase (Fig. 8B). As a whole, these data suggest that the HR-Tet vaccine is capable of eliciting a memory immune response following heterologous challenge.
FIG 8.
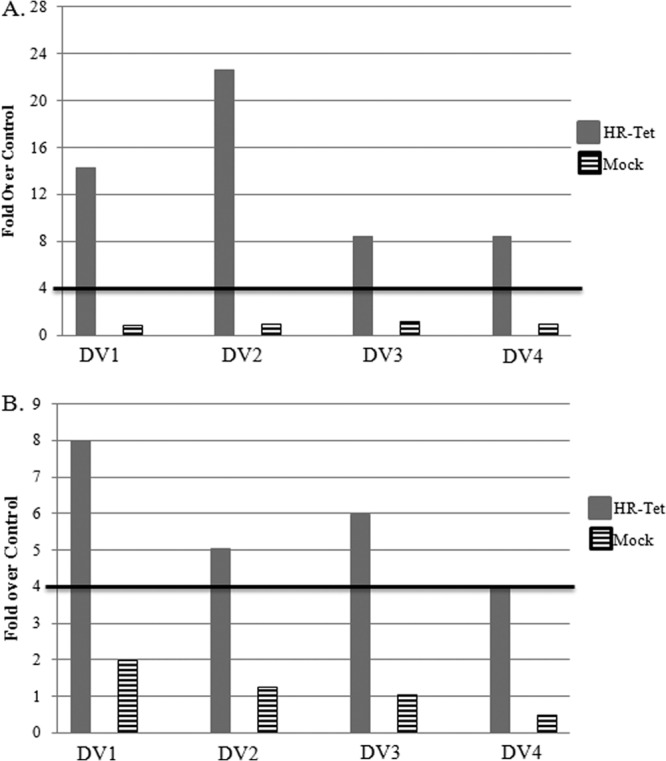
Anamnestic immune response following challenge. On days 71 (A) and 77 (B) postvaccination (days 9 and 15 postchallenge), we looked for evidence of a memory immune response by measuring the fold increase in IgG in animals given HR-Tet prior to challenge compared to IgG in mock-vaccinated animals. An anamnestic response was defined as any response ≥4-fold greater than that of the naive unchallenged control. Animals given HR-Tet prior to challenge with one of the four serotypes of DENV developed a memory immune response against all four serotypes starting at 9 days postchallenge (A, gray bars), and this memory response continued out to 15 days postchallenge (B, gray bars). No evidence of a memory response was seen in mock-vaccinated animals following challenge at either time point (A and B, hatched bars).
DISCUSSION
In light of the recent failure of Sanofi's CDY vaccine (30–32) to protect against all four serotypes, enthusiasm for the success of a live attenuated vaccine formulation to treat dengue fever has waned. However, there is solid evidence that the most effective epitopes are found on the native virion in serotype-specific quaternary conformations (14). This type of epitope may not be present in the CDY vaccine, which is a chimera virus that uses a yellow fever backbone. Additionally, the fundamental importance of the CD8+ T cell response to nonstructural proteins (nsP) during virus replication underscores the importance of presentation of the correct homologous nsP epitopes to the immune system (16). All these antigenic requirements can most successfully be presented in the live attenuated virus. The HR mutants of DENV do not share these deficits. The 4-amino-acid deletions in each mutant are buried within the transmembrane domain. All exposed epitopes available to the host immune system are wild type in sequence.
As shown in the present study, the use of HR mutations for the development of a tetravalent formulation of DENV has met the initial requirements for a safe and immunogenic vaccine in a single dose in an AGM model. The vaccine strains are attenuated for growth in vivo and elicit a strong postvaccination anamnestic memory response that is balanced.
Following vaccination, The WT-Tet-vaccinated animals displayed viremia in a range that is expected for transmission from insect vectors to infected hosts and corresponds with data previously found in naturally acquired human infections (33). Virus titers from the HR-Tet-vaccinated animals are significantly lower than those seen for the WT-Tet-inoculated animals. The viremic period was prolonged past 14 days, but these animals were fighting infection from four different, albeit related, viruses that may allow for more viral replication. We elected to vaccinate in four separate limbs to decrease viral interference during the initial period of infection. As was shown by RT-PCR of the vaccinated animals, all four HR-Tet strains were able to coexist in the animals on day 3 postvaccination. This was repeated on day 4 in 15 of the 16 animals. This result was not seen in the WT-Tet-inoculated animals and may have resulted from the level of virus replication or from the innate ability of the HR-Tet viral mutants to replicate without interference compared to the WT-Tet DENV infections, because all animals were vaccinated in the same manner (one serotype per limb).
One interesting observation was the fact that seroconversion to DENV-2 and DENV-4 came much later for the majority of animals in the study than did seroconversion to DENV-1 and DENV-3 (day 30 versus day 14; see Fig. S1 in the supplemental material). When we look at expression of HR-Tet DENV-2 and DENV-4 following vaccination in AGM serum for all animals tested, DENV-2 was present on 7 of 8 days. Similarly, AGMs were found to have circulating DENV-4 on 6 of the 8 days tested (Table 2). Both DENV-2 and DENV-4, but not DENV-1 or DENV-3, were found in AGM serum at 11 days postvaccination, after which viremia dropped below the limit of detection for RT-PCR. In contrast, AGMs were found to be infected with DENV-1 and DENV-3 on 3 and 5 of the 8 days tested, respectively, with neither serotype present after 7 days postvaccination (Table 2). The extended viremic period seen with DENV-2 and DENV-4 may help explain why seroconversion for these serotypes was delayed compared to DENV-1 and DENV-3, which were cleared much earlier. Indeed, DENV-2 has been shown to be a subdominant dengue virus serotype, which may also be contributing to delayed viral clearance and increased length of time for seroconversion to occur (34).
The accuracy of the IgG response and the tests used to detect it as an appropriate marker of protection has been the subject of much debate (35, 36). Nonetheless, while the magnitude of the response may not correlate with protection in humans, it is indicative of the fact that the immune system has been engaged. It has been suggested that levels of NAb that are protective will differ for each virus serotype. This study found that the serospecific IgG response induced by the HR-Tet vaccine was similar to that seen to be induced by tetravalent WT DENV infection on day 30 postvaccination (Fig. 3A). We suspect that due to the prolonged length of the viremia, this resulted in a change in the normal time frame for IgG, as high levels of IgG were still detected on day 62 postvaccination (Fig. 3B). When the HR-Tet-vaccinated and the mock-vaccinated groups were assayed for protection from challenge, two parameters of protective immunity were measured. First, viremia postchallenge was assayed by ICA. For DENV-4, there was a clear 10- to 20-fold difference in the amount of viremia accompanied by a shorter period of virus replication compared to the control (Fig. 7A). While the duration of viremia was not shortened following challenge with DENV-1 and DENV-3, we did note a difference in the magnitude of viremia (Fig. 3A and 5A). However, this was not the case for DENV-2 (Fig. 5A). It is paradoxical that while DENV-2 displayed the highest NAb titers measured per group/day for overall GMT, this serotype shows the least reduction in viremia after challenge. It is possible that during a tetravalent infection, levels of non- or subneutralizing antibody were still high on day 62 postvaccination. These Abs could hold the virus in immune complexes that are less susceptible to neutralization, creating neutralization interference for a limited time. These findings are confounding, since in contrast to the three other serotypes the tetravalent DENV-2 infection produced the highest levels of NAb. However, sera from DENV-2-challenged HR-Tet-vaccinated animals were tested for their ability to be neutralized by exogenous NAb. This virus population was able to be neutralized completely when reacted exogenously with neutralizing monoclonal antibody (MAb) (data not shown). It is possible that there was also immune interference with the other serotypes at an undetectable level. Neutralization was DENV-2 specific, since serum from DENV-1, -3, and -4 did not show any cross-neutralization (data not shown). It is not surprising that during a tetravalent infection all aspects of interference and possible cross-reactivity seen in monovalent infections would be further complicated (37). In our monovalent study of HR DENV-2, challenge virus was efficiently being cleared on day 6 postchallenge, suggesting that the phenomenon seen here is due to the tetravalent infection.
Recently, it has been reported that DENV-2 grown in insect cells at 28°C can expand in diameter when heated to 37°C (38, 39). This process was not reversible, and the virus retained the original titer; thus, two conformational infectious intermediates were identified. It was concluded that this ability of DENV-2 to expand would explain the ability to identify known neutralizing epitopes on the surface of the larger virus not exposed on the more compacted virus. It was not determined by either group whether this structural difference was a result of a difference in the host environment or a function of the temperature of assembly. Insect host-specific biochemistry, which functions optimally at 28°C, may be responsible for assembly of the compact structure, which can adopt a less compact form at 37°C (40). This temperature-induced expansion phenomenon was not seen in dengue serotype 1 or 4 (39, 41). The ability of DENV-2 to adapt its structure in response to temperature and possibly host environment may explain other aspects of DENV-2 secondary infections and our inability to detect a significant decrease in viremia upon challenge in the presence of significant PRNT50 titers.
The total GMTs of all four serotypes postvaccination tell only part of a complex story. Our prechallenge PRNT50 GMTs from day 14 to 62 HR-Tet serotype 1- to 4-vaccinated animals were not significantly different from those of the homologous WT DENV inoculation. In contrast, the total NAb PRNT50 titer of HR-Tet tetravalent-vaccinated animals postchallenge showed a ranking on day 77 postvaccination in the serodominance order of DENV 4 > 3 > 1 > 2. However, this metric changed when the total NAb GMT titers were compared and showed dominance in the order DENV 2 > 3 > 4 > 1. In theory, the significance of these IgG levels reflects fluctuation in the amount of nonneutralizing antibody, cross-reacting Ab and NAb. In the Sanofi/Mihadol vaccine, DENV-3 16562 PGMK responses predominated (42, 43). In Sanofi's CDY vaccine, DENV-4 1228 responses were serodominant (44). DENV-1 45AZ5 was serodominant in the WRAIR phase II clinical trial and also was antagonistic with the DENV-2 S16803 component (45). These studies are not comparable to the one presented herein due to the differences in the host animal, DENV serotype strain, and method of attenuation.
While no ideal animal model for dengue fever is available, NHP have been shown to be a suitable model for proof of principle. As described in this study and previous work, AGMs are proving to be a useful model for dengue vaccine development (17, 46). Recent studies in humans have shown other dengue vaccines unable to fully protect against challenge from all 4 DENV serotypes despite previous promising studies in rhesus macaques (6, 12, 45, 47–53). Additionally, investigations have shown that AGMs model certain human diseases better than macaques (46, 54–56). Indeed, some macaques are entirely resistant to DENV infection (57). When infected with DENV-2, AGMs showed viremia in the range of 1 to >14 days and a neutralizing antibody response in the range of 100 to 500 PRNT50, which is more similar to what is seen in humans, and showed a longer period of viremia than that observed in related experiments in rhesus and cynomolgus monkeys (17, 58). Our group has previously been successful in using AGMs in DENV vaccine trials (17, 21, 22). Furthermore, Vero cells, which are the prototypic cell for DENV growth and testing, were originally derived from the kidneys of AGMs (59). Thus, we elected to use AGMs as a model for our study. It is also important to note that the amount of LAV vaccine given to humans is normally several orders of magnitude less than what is given to NHP. Future studies will determine whether the responses in AGMs are more representative of the responses in humans.
In summary, the HR-Tet vaccine has several advantages that display merit for development as a tetravalent dengue vaccine candidate. We demonstrate that one dose of HR-Tet is sufficient to tetravalently seroconvert 100% of AGMs within 62 days of vaccination. HR-Tet was found to be attenuated for growth in AGMs compared to wild-type parental strains of dengue virus. These deletions do not change the biochemical structure of the virus or its immunogenic epitopes. Importantly, because HR-Tet was created through sequence deletion, there is virtually no chance of reversion to the parental phenotype. Multiple passages of our DENV-2ΔGVII mutant through insect and AGM cell culture as well as virus isolated from AGM failed to show reversion to the parental sequence (17, 29). Vaccination resulted in increased NAb titers compared to those of nonvaccinated animals, a faster and stronger antibody response following challenge, which is indicative of an anamnestic response. One injection with HR-Tet was sufficient to provide a memory response in terms of antibody production upon challenge with heterologous strains of all four DENV serotypes. The fact that the antibody response to all four serotypes appears to be fairly evenly balanced is consistent with the idea that the vaccine strains are attenuated for growth in mammals and that there is no antigenic competition between the vaccine components. Animals given HR-Tet did not develop any adverse effects due to vaccination, and there was no evidence of reactogenicity at the site of vaccination. Preliminary studies with wild-types strains of DENV indicate that the HR-Tet strains of DENV can be grown to adequate titers in Sf9 cells, which are a clonal cell line of Spodoptera frugiperdia (Fall armyworm) and have been used in the production of the FDA-approved live viral vaccine FluBlok (60–62). Collectively, these data suggest that HR-Tet warrants development as a LAV vaccine for use in humans.
Supplementary Material
ACKNOWLEDGMENTS
Technical support was provided by the Wake Forest Center for Comparative Medicine and Research and a Biological Resources Grant from NIH (grant OD010965; principal investigator, Jay R. Kaplan).
Footnotes
Published ahead of print 2 April 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.00541-14.
REFERENCES
- 1.WHO. 2014. Dengue and severe dengue. Fact sheet no. 17. WHO, Geneva, Switzerland: http://www.who.int/mediacentre/factsheets/fs117/en/ [Google Scholar]
- 2.Halstead SB. 1980. Immunological parameters of Togavirus disease syndromes, p 107–168 In Schlesinger RW. (ed), The togaviruses: biology, structure, replication. Academic Press, New York, NY [Google Scholar]
- 3.Henchal EA, Putnak JR. 1990. The dengue viruses. Clin. Microbiol. Rev. 3:376–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gubler DJ. 2006. Dengue/dengue haemorrhagic fever: history and current status. Novartis Found. Symp. 277:3–16, 16–22. 10.1002/0470058005.ch2 [DOI] [PubMed] [Google Scholar]
- 5.WHO. 2012. Dengue/dengue haemorrhagic fever. WHO, Geneva, Switzerland: http://www.who.int/csr/disease/dengue/en/ [Google Scholar]
- 6.Durbin AP, Kirkpatrick BD, Pierce KK, Elwood D, Larsson CJ, Lindow JC, Tibery C, Sabundayo BP, Shaffer D, Talaat KR, Hynes NA, Wanionek K, Carmolli MP, Luke CJ, Murphy BR, Subbarao K, Whitehead SS. 2013. A single dose of any of four different live attenuated tetravalent dengue vaccines is safe and immunogenic in flavivirus-naive adults: a randomized, double-blind clinical trial. J. Infect. Dis. 207:957–965. 10.1093/infdis/jis936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Morens DM, Fauci AS. 2008. Dengue and hemorrhagic fever: a potential threat to public health in the United States. JAMA 299:214–216. 10.1001/jama.2007.31-a [DOI] [PubMed] [Google Scholar]
- 8.Adalja AA, Sell TK, Bouri N, Franco C. 2012. Lessons learned during dengue outbreaks in the United States, 2001–2011. Emerg. Infect. Dis. 18:608–614. 10.3201/eid1804.110968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sabchareon A, Wallace D, Sirivichayakul C, Limkittikul K, Chanthavanich P, Suvannadabba S, Jiwariyavej V, Dulyachai W, Pengsaa K, Wartel TA, Moureau A, Saville M, Bouckenooghe A, Viviani S, Tornieporth NG, Lang J. 2012. Protective efficacy of the recombinant, live-attenuated, CYD tetravalent dengue vaccine in Thai schoolchildren: a randomised, controlled phase 2b trial. Lancet 380:1559–1567. 10.1016/S0140-6736(12)61428-7 [DOI] [PubMed] [Google Scholar]
- 10.Huang CY, Kinney RM, Livengood JA, Bolling B, Arguello JJ, Luy BE, Silengo SJ, Boroughs KL, Stovall JL, Kalanidhi AP, Brault AC, Osorio JE, Stinchcomb DT. 2013. Genetic and phenotypic characterization of manufacturing seeds for a tetravalent dengue vaccine (DENVax). PLoS Negl. Trop. Dis. 7(5):e2243. 10.1371/journal.pntd.0002243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thomas SJ, Eckels KH, Carletti I, De La Barrera R, Dessy F, Fernandez S, Putnak R, Toussaint JF, Sun W, Bauer K, Gibbons RV, Innis BL. 2013. A phase II, randomized, safety and immunogenicity study of a re-derived, live-attenuated dengue virus vaccine in healthy adults. Am. J. Trop. Med. Hyg. 88:73–88. 10.4269/ajtmh.2012.12-0361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Durbin AP, Whitehead SS. 2013. The dengue human challenge model: has the time come to accept this challenge? J. Infect. Dis. 207:697–699. 10.1093/infdis/jis749 [DOI] [PubMed] [Google Scholar]
- 13.Lauring AS, Jones JO, Andino R. 2010. Rationalizing the development of live attenuated virus vaccines. Nat. Biotechnol. 28:573–579. 10.1038/nbt.1635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Alwis R, Smith SA, Olivarez NP, Messer WB, Huynh JP, Wahala WM, White LJ, Diamond MS, Baric RS, Crowe JE, Jr, de Silva AM. 2012. Identification of human neutralizing antibodies that bind to complex epitopes on dengue virions. Proc. Natl. Acad. Sci. U. S. A. 109:7439–7444. 10.1073/pnas.1200566109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stephens HA, Klaythong R, Sirikong M, Vaughn DW, Green S, Kalayanarooj S, Endy TP, Libraty DH, Nisalak A, Innis BL, Rothman AL, Ennis FA, Chandanayingyong D. 2002. HLA-A and -B allele associations with secondary dengue virus infections correlate with disease severity and the infecting viral serotype in ethnic Thais. Tissue Antigens 60:309–318. 10.1034/j.1399-0039.2002.600405.x [DOI] [PubMed] [Google Scholar]
- 16.Weiskopf D, Angelo MA, de Azeredo EL, Sidney J, Greenbaum JA, Fernando AN, Broadwater A, Kolla RV, De Silva AD, de Silva AM, Mattia KA, Doranz BJ, Grey HM, Shresta S, Peters B, Sette A. 2013. Comprehensive analysis of dengue virus-specific responses supports an HLA-linked protective role for CD8+ T cells. Proc. Natl. Acad. Sci. U. S. A. 110:E2046–E2053. 10.1073/pnas.1305227110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smith KM, Nanda K, Spears CJ, Piper A, Ribeiro M, Quiles M, Briggs CM, Thomas GS, Thomas ME, Brown DT, Hernandez R, McCarl V. 2012. Testing of novel dengue virus 2 vaccines in African green monkeys: safety, immunogenicity, and efficacy. Am. J. Trop. Med. Hyg. 87:743–753. 10.4269/ajtmh.2012.12-0004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hernandez R, Lee H, Nelson C, Brown DT. 2000. A single deletion in the membrane-proximal region of the Sindbis virus glycoprotein E2 endodomain blocks virus assembly. J. Virol. 74:4220–4228. 10.1128/JVI.74.9.4220-4228.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hernandez R, Sinodis C, Horton M, Ferreira D, Yang C, Brown DT. 2003. Deletions in the transmembrane domain of a Sindbis virus glycoprotein alter virus infectivity, stability, and host range. J. Virol. 77:12710–12719. 10.1128/JVI.77.23.12710-12719.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nguyen NM, Thi Hue Kien D, Tuan TV, Quyen NT, Tran CN, Vo Thi L, Thi DL, Nguyen HL, Farrar JJ, Holmes EC, Rabaa MA, Bryant JE, Nguyen TT, Nguyen HT, Nguyen LT, Pham MP, Luong TT, Wills B, Nguyen CV, Wolbers M, Simmons CP. 2013. Host and viral features of human dengue cases shape the population of infected and infectious Aedes aegypti mosquitoes. Proc. Natl. Acad. Sci. U. S. A. 110:9072–9077. 10.1073/pnas.1303395110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bomers MK, Lettinga KD, van Gorp EC, Martina BE, Peerbooms PG, Veenstra J. 2009. Dengue infection with fatal ending. Ned. Tijdschr. Geneeskd. 153:A725 (In Dutch.) [PubMed] [Google Scholar]
- 22.Martin J, Hermida L, Castro J, Romero Y, Cardosa J, Guillen G. 2009. Viremia and the magnitude of the immune response upon infection of green monkeys with dengue virus type 2 are strain-dependent. Curr. Microbiol. 59:579–583. 10.1007/s00284-009-9488-6 [DOI] [PubMed] [Google Scholar]
- 23.Kinney RM, Butrapet S, Chang GJ, Tsuchiya KR, Roehrig JT, Bhamarapravati N, Gubler DJ. 1997. Construction of infectious cDNA clones for dengue 2 virus: strain 16681 and its attenuated vaccine derivative, strain PDK-53. Virology 230:300–308. 10.1006/viro.1997.8500 [DOI] [PubMed] [Google Scholar]
- 24.Puri B, Nelson W, Porter KR, Henchal EA, Hayes CG. 1998. Complete nucleotide sequence analysis of a Western Pacific dengue-1 virus strain. Virus Genes 17:85–88. 10.1023/A:1008009202695 [DOI] [PubMed] [Google Scholar]
- 25.Puri B, Polo S, Hayes CG, Falgout B. 2000. Construction of a full length infectious clone for dengue-1 virus Western Pacific,74 strain. Virus Genes 20:57–63. 10.1023/A:1008160123754 [DOI] [PubMed] [Google Scholar]
- 26.Kelly EP, Puri B, Sun W, Falgout B. 2010. Identification of mutations in a candidate dengue 4 vaccine strain 341750 PDK20 and construction of a full-length cDNA clone of the PDK20 vaccine candidate. Vaccine 28:3030–3037 [DOI] [PubMed] [Google Scholar]
- 27.Puri B, Nelson WM, Henchal EA, Hoke CH, Eckels KH, Dubois DR, Porter KR, Hayes CG. 1997. Molecular analysis of dengue virus attenuation after serial passage in primary dog kidney cells. J. Gen. Virol. 78(Part 9):2287–2291 [DOI] [PubMed] [Google Scholar]
- 28.Kelly EP, Polo S, Sun W, Falgout B. 2011. Evolution of attenuating mutations in dengue-2 strain S16803 PDK50 vaccine and comparison of growth kinetics with parent virus. Virus Genes 43:18–26. 10.1007/s11262-011-0602-z [DOI] [PubMed] [Google Scholar]
- 29.Smith KM, Nanda K, Spears CJ, Ribeiro M, Vancini R, Piper A, Thomas GS, Thomas ME, Brown DT, Hernandez R. 2011. Structural mutants of dengue virus 2 transmembrane domains exhibit host-range phenotype. Virol. J. 8:289. 10.1186/1743-422X-8-289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sabchareon A, Wallace D, Sirivichayakul C, Limkittikul K, Chanthavanich P, Suvannadabba S, Jiwariyavej V, Dulyachai W, Pengsaa K, Wartel TA, Moureau A, Saville M, Bouckenooghe A, Viviani S, Tornieporth NG, Lang J. 2012. Protective efficacy of the recombinant, live-attenuated, CYD tetravalent dengue vaccine in Thai schoolchildren: a randomised, controlled phase 2b trial. Lancet 380:1559–1567. 10.1016/S0140-6736(12)61428-7 [DOI] [PubMed] [Google Scholar]
- 31.Halstead SB. 2012. Dengue vaccine development: a 75% solution? Lancet 380:1535–1536. 10.1016/S0140-6736(12)61510-4 [DOI] [PubMed] [Google Scholar]
- 32.Halstead S. 2013. Identifying protective dengue vaccines: Guide to mastering an empirical process. Vaccine 31:4501–4507. 10.1016/j.vaccine.2013.06.079 [DOI] [PubMed] [Google Scholar]
- 33.Vaughn DW, Green S, Kalayanarooj S, Innis BL, Nimmannitya S, Suntayakorn S, Endy TP, Raengsakulrach B, Rothman AL, Ennis FA, Nisalak A. 2000. Dengue viremia titer, antibody response pattern, and virus serotype correlate with disease severity. J. Infect. Dis. 181:2–9. 10.1086/315215 [DOI] [PubMed] [Google Scholar]
- 34.Sangkawibha N, Rojanasuphot S, Ahandrik S, Viriyapongse S, Jatanasen S, Salitul V, Phanthumachinda B, Halstead SB. 1984. Risk factors in dengue shock syndrome: a prospective epidemiologic study in Rayong, Thailand. I. The 1980 outbreak. Am. J. Epidemiol. 120:653–669 [DOI] [PubMed] [Google Scholar]
- 35.Mahalingam S, Herring BL, Halstead SB. 2013. Call to action for dengue vaccine failure. Emerg. Infect. Dis. 19:1335–1337. 10.3201/eid1908.121864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van Panhuis WG, Gibbons RV, Endy TP, Rothman AL, Srikiatkhachorn A, Nisalak A, Burke DS, Cummings DAT. 2010. Inferring the serotype associated with dengue virus infections on the basis of pre- and postinfection neutralizing antibody titers. J. Infect. Dis. 202:1002–1010. 10.1086/656141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Beltramello M, Williams KL, Simmons CP, Macagno A, Simonelli L, Quyen NT, Sukupolvi-Petty S, Navarro-Sanchez E, Young PR, de Silva AM, Rey FA, Varani L, Whitehead SS, Diamond MS, Harris E, Lanzavecchia A, Sallusto F. 2010. The human immune response to Dengue virus is dominated by highly cross-reactive antibodies endowed with neutralizing and enhancing activity. Cell Host Microbe 8:271–283. 10.1016/j.chom.2010.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang X, Sheng J, Plevka P, Kuhn RJ, Diamond MS, Rossmann MG. 2013. Dengue structure differs at the temperatures of its human and mosquito hosts. Proc. Natl. Acad. Sci. U. S. A. 110:6795–6799. 10.1073/pnas.1304300110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fibriansah G, Ng TS, Kostyuchenko VA, Lee J, Lee S, Wang J, Lok SM. 2013. Structural changes in dengue virus when exposed to a temperature of 37 degrees C. J. Virol. 87:7585–7592. 10.1128/JVI.00757-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.He L, Piper A, Meilleur F, Myles DA, Hernandez R, Brown DT, Heller WT. 2010. The structure of Sindbis virus produced from vertebrate and invertebrate hosts as determined by small-angle neutron scattering. J. Virol. 84:5270–5276. 10.1128/JVI.00044-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kostyuchenko VA, Chew PL, Ng TS, Lok SM. 2014. Near-atomic resolution cryo-electron microscopic structure of dengue serotype 4 virus. J. Virol. 88:477–482. 10.1128/JVI.02641-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sabchareon A, Lang J, Chanthavanich P, Yoksan S, Forrat R, Attanath P, Sirivichayakul C, Pengsaa K, Pojjaroen-Anant C, Chokejindachai W, Jagsudee A, Saluzzo JF, Bhamarapravati N. 2002. Safety and immunogenicity of tetravalent live-attenuated dengue vaccines in Thai adult volunteers: role of serotype concentration, ratio, and multiple doses. Am. J. Trop. Med. Hyg. 66:264–272 [DOI] [PubMed] [Google Scholar]
- 43.Kitchener S, Nissen M, Nasveld P, Forrat R, Yoksan S, Lang J, Saluzzo JF. 2006. Immunogenicity and safety of two live-attenuated tetravalent dengue vaccine formulations in healthy Australian adults. Vaccine 24:1238–1241. 10.1016/j.vaccine.2005.09.029 [DOI] [PubMed] [Google Scholar]
- 44.Morrison D, Legg TJ, Billings CW, Forrat R, Yoksan S, Lang J. 2010. A novel tetravalent dengue vaccine is well tolerated and immunogenic against all 4 serotypes in flavivirus-naive adults. J. Infect. Dis. 201:370–377. 10.1086/649916 [DOI] [PubMed] [Google Scholar]
- 45.Sun W, Cunningham D, Wasserman SS, Perry J, Putnak JR, Eckels KH, Vaughn DW, Thomas SJ, Kanesa-Thasan N, Innis BL, Edelman R. 2009. Phase 2 clinical trial of three formulations of tetravalent live-attenuated dengue vaccine in flavivirus-naive adults. Hum. Vaccin. 5:33–40. 10.4161/hv.5.1.6348 [DOI] [PubMed] [Google Scholar]
- 46.Pandrea I, Apetrei C, Dufour J, Dillon N, Barbercheck J, Metzger M, Jacquelin B, Bohm R, Marx PA, Barre-Sinoussi F, Hirsch VM, Muller-Trutwin MC, Lackner AA, Veazey RS. 2006. Simian immunodeficiency virus SIVagm.sab infection of Caribbean African green monkeys: a new model for the study of SIV pathogenesis in natural hosts. J. Virol. 80:4858–4867. 10.1128/JVI.80.10.4858-4867.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Durbin AP, Schmidt A, Elwood D, Wanionek KA, Lovchik J, Thumar B, Murphy BR, Whitehead SS. 2011. Heterotypic dengue infection with live attenuated monotypic dengue virus vaccines: implications for vaccination of populations in areas where dengue is endemic. J. Infect. Dis. 203:327–334. 10.1093/infdis/jiq059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Durbin AP, Whitehead SS. 2011. Next-generation dengue vaccines: novel strategies currently under development. Viruses 3:1800–1814. 10.3390/v3101800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Barban V, Munoz-Jordan JL, Santiago GA, Mantel N, Girerd Y, Gulia S, Claude JB, Lang J. 2012. Broad neutralization of wild-type dengue virus isolates following immunization in monkeys with a tetravalent dengue vaccine based on chimeric yellow fever 17D/dengue viruses. Virology 429:91–98. 10.1016/j.virol.2012.03.007 [DOI] [PubMed] [Google Scholar]
- 50.Sabchareon A, Wallace D, Sirivichayakul C, Limkittikul K, Chanthavanich P, Suvannadabba S, Jiwariyavej V, Dulyachai W, Pengsaa K, Wartel TA, Moureau A, Saville M, Bouckenooghe A, Viviani S, Tornieporth NG, Lang J. 2012. Protective efficacy of the recombinant, live-attenuated, CYD tetravalent dengue vaccine in Thai schoolchildren: a randomised, controlled phase 2b trial. Lancet 380:1559–1567. 10.1016/S0140-6736(12)61428-7 [DOI] [PubMed] [Google Scholar]
- 51.Sun W, Eckels KH, Putnak JR, Lyons AG, Thomas SJ, Vaughn DW, Gibbons RV, Fernandez S, Gunther VJ, Mammen MP, Jr, Statler JD, Innis BL. 2013. Experimental dengue virus challenge of human subjects previously vaccinated with live attenuated tetravalent dengue vaccines. J. Infect. Dis. 207:700–708 [DOI] [PubMed] [Google Scholar]
- 52.Sun W, Edelman R, Kanesa-Thasan N, Eckels KH, Putnak JR, King AD, Houng HS, Tang D, Scherer JM, Hoke CH, Jr, Innis BL. 2003. Vaccination of human volunteers with monovalent and tetravalent live-attenuated dengue vaccine candidates. Am. J. Trop. Med. Hyg. 69(6 Suppl):24–31 [DOI] [PubMed] [Google Scholar]
- 53.Sun W, Nisalak A, Gettayacamin M, Eckels KH, Putnak JR, Vaughn DW, Innis BL, Thomas SJ, Endy TP. 2006. Protection of Rhesus monkeys against dengue virus challenge after tetravalent live attenuated dengue virus vaccination. J. Infect. Dis. 193:1658–1665. 10.1086/503372 [DOI] [PubMed] [Google Scholar]
- 54.Redmond DE, Jr, Bjugstad KB, Teng YD, Ourednik V, Ourednik J, Wakeman DR, Parsons XH, Gonzalez R, Blanchard BC, Kim SU, Gu Z, Lipton SA, Markakis EA, Roth RH, Elsworth JD, Sladek JR, Jr, Sidman RL, Snyder EY. 2007. Behavioral improvement in a primate Parkinson's model is associated with multiple homeostatic effects of human neural stem cells. Proc. Natl. Acad. Sci. U. S. A. 104:12175–12180. 10.1073/pnas.0704091104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lemere CA, Beierschmitt A, Iglesias M, Spooner ET, Bloom JK, Leverone JF, Zheng JB, Seabrook TJ, Louard D, Li D, Selkoe DJ, Palmour RM, Ervin FR. 2004. Alzheimer's disease abeta vaccine reduces central nervous system abeta levels in a non-human primate, the Caribbean vervet. Am. J. Pathol. 165:283–297. 10.1016/S0002-9440(10)63296-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Goldstein S, Brown CR, Ourmanov I, Pandrea I, Buckler-White A, Erb C, Nandi JS, Foster GJ, Autissier P, Schmitz JE, Hirsch VM. 2006. Comparison of simian immunodeficiency virus SIVagmVer replication and CD4+ T-cell dynamics in vervet and sabaeus African green monkeys. J. Virol. 80:4868–4877. 10.1128/JVI.80.10.4868-4877.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Simmons JS, St John JH, Reynolds FHK. 1931. Experimental studies of dengue. Philippine J. Sci. 44:1–247 [Google Scholar]
- 58.Martin J, Hermida L, Castro J, Lazo L, Martinez R, Gil L, Romero Y, Puente P, Zaragoza S, Cosme K, Guzman MG, Cardosa J, Guillen G. 2009. Viremia and antibody response in green monkeys (Chlorocebus aethiops sabaeus) infected with dengue virus type 2: a potential model for vaccine testing. Microbiol. Immunol. 53:216–223. 10.1111/j.1348-0421.2009.00112.x [DOI] [PubMed] [Google Scholar]
- 59.Ammerman NC, Beier-Sexton M, Azad AF. 2008. Growth and maintenance of Vero cell lines. Curr. Protoc. Microbiol. Appendix:Appendix-4E. 10.1002/9780471729259.mca04es11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Treanor JJ, El Sahly H, King J, Graham I, Izikson R, Kohberger R, Patriarca P, Cox M. 2011. Protective efficacy of a trivalent recombinant hemagglutinin protein vaccine (FluBlok(R)) against influenza in healthy adults: a randomized, placebo-controlled trial. Vaccine 29:7733–7739. 10.1016/j.vaccine.2011.07.128 [DOI] [PubMed] [Google Scholar]
- 61.Cox MM, Hollister JR. 2009. FluBlok, a next generation influenza vaccine manufactured in insect cells. Biologicals 37:182–189. 10.1016/j.biologicals.2009.02.014 [DOI] [PubMed] [Google Scholar]
- 62.Baxter R, Patriarca PA, Ensor K, Izikson R, Goldenthal KL, Cox MM. 2011. Evaluation of the safety, reactogenicity and immunogenicity of FluBlok(R) trivalent recombinant baculovirus-expressed hemagglutinin influenza vaccine administered intramuscularly to healthy adults 50–64 years of age. Vaccine 29:2272–2278. 10.1016/j.vaccine.2011.01.039 [DOI] [PubMed] [Google Scholar]
- 63.National Research Council. 2011. Guide for the care and use of laboratory animals, 8th ed. National Academies Press, Washington, DC [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.