Abstract
Protein methylation is a post-translational modification (PTM) which modulates cellular and biological processes including transcription, RNA processing, protein interactions and protein dynamics. Methylation, catalyzed by highly specific methyltransferase enzymes, occurs on several amino acids including arginine, lysine, histidine and dicarboxylic amino acids like glutamate. Mass spectrometry (MS) based techniques continue to be the methods of choice for the study of protein PTMs. These approaches are powerful and sensitive tools that have been used to identify, quantify and characterize protein methylation. In addition, metabolic labeling strategies can be coupled to MS detection in order to measure dynamic and differential in vivo protein methylation rates. In this review, different applications of mass spectrometry technologies and methods to study protein methylation are discussed.
Introduction
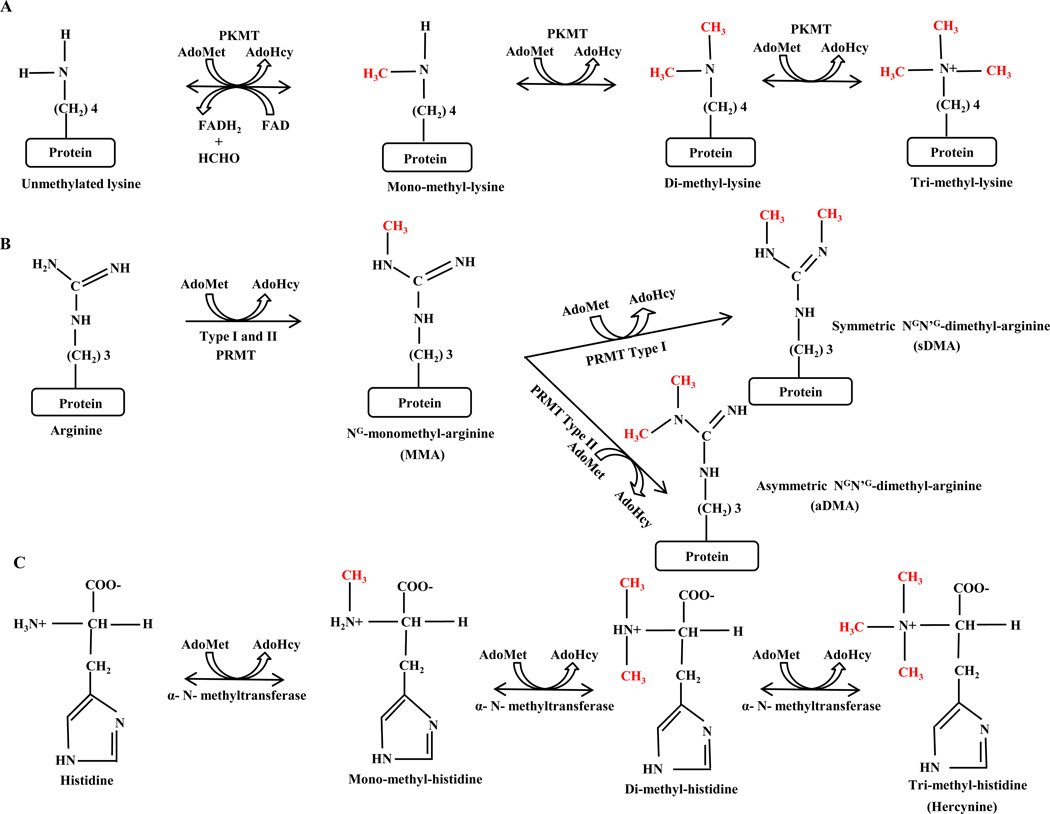
Post-translational modifications (PTM)s play crucial roles in modulating protein activity, turnover, and protein-protein interactions. Protein methylation is a fairly common type of protein PTM and has been implicated in several biological processes such as transcriptional regulation, RNA processing, metabolism and signal transduction [1]. Although methylation has been most commonly observed on lysine and arginine residues, methylation of other amino acids including histidine (H), cysteine (C), aspartic acid (D), glutamic acid (E), serine (S) and threonine (T) has been reported [2, 3]. Lysine-methylation occurs by transferring one to three methyl groups from S-Adenosyl-Methionine (SAM) to the lysine ε -amine side chain, which leads to monomethylated (me1), dimethylated (me2) or trimethylated (me3) lysines (Figure 1). In the case of arginine, one or two methyl groups are added to its guanidine group which leads to mono- or di-methylation [2] (Figure 1). Methylation specific enzymes (methyltransferases) can read specific protein sequence/motifs and further propagate existing methylation marks [2]. For example, arginine methyltransferase enzymes often target proteins sequence including an RGG- RNA binding motif [4]. Furthermore, methylation has been shown to depend on a protein’s existing methylation state and to be a dynamic modification. For instance, methylated lysines within histones have been shown to have measurable differential turnover rates [5].
Figure 1.
Biochemical mechanism of lysine, arginine and histidine methylation. (A) Lysine methylation. Formation of mono-, di- and tri-methylated lysine by adding methyl-group to ε amine of lysine residue. Conversion of S-adenosyl-L-methionine (AdoMet) to S-adenosyl-Lhomocysteine (AdoHcy) leads to methyl-group transfer to a protein. The methylation reaction is catalyzed by Protein Lysine-Methyl-Transferase (PKMT). The reversibility of the methylation reaction in presence of Fe (II) and α-ketoglutarate has been proved by the discovery of a demethylase. (B) Arginine methylation. Addition of methyl groups to guanidine nitrogens of arginine forms NG -monomethyl-, NGN’G -dimethyl- symmetric (sDMA) and asymmetric (aDMA) - arginines. Type-I and –II protein Arginine methyltransferase (PRMT) are the catalytic enzyme for Arginine methylation. (C) Histidine methylation. A methyl-group will be added to the α-amino nitrogen atom of Histidine and result in mono-, di- and trimethyl- histidine (Hercynine). Histidine methylation is catalyzed by a single enzyme, Histidine-α-N-methyltransferase [3].
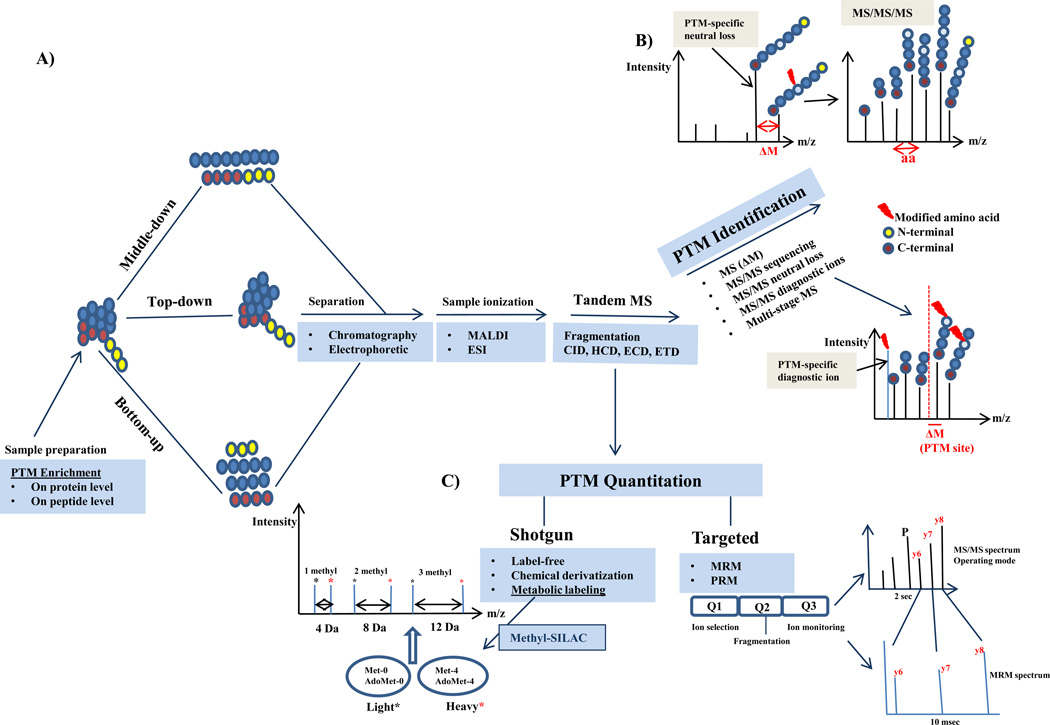
PTMs, including protein methylation, have been traditionally identified by Edman degradation, amino acid analysis, radio isotope labeling or antibody based methods including peptide and protein arrays. These methods suffer from being unspecific, low throughput, and having a low dynamic range for quantitative measurements. In addition, they fail to identify specific modification sites, cannot distinguish methylation state, and they often rely on prior knowledge of the modification. For instance, protein methylation has been detected by radioactive methods (review in [6]) including using tritiated methyltransferase cofactor S-Adenosyl methionine (SAM) as a methyl-donor. The weakness of this radioactive method is that radioisotopes of carbon and hydrogen are weak radio emitters and it is difficult to detect modified peptides efficiently. Another issue with current technologies is the small size of the methyl group which makes it challenging to develop high quality methylation specific antibodies. Protein methylation substrates can be identified by protein and peptide arrays, however, any hit needs to be validated with purified endogenous proteins by mass spectrometry. In recent years, MS based methodology has proven to be superior for the analysis of PTMs including methylation due to improvements in the accuracy and sensitivity of MS instrumentation. MS methods have been developed to identify proteins carrying PTMs, to map (novel) PTM sites, to quantify the changes in PTM abundance at individual sites, and to characterize the cooperativity between interrelated PTMs at several sites on proteins [6–9] (Figure 2).
Figure 2.
Workflow for PTM analysis including methylation-specific mass spectrometry (MS). A) Protein samples are prefractionated, purified and eventually enriched for methylation either on the protein or peptide level. The samples will be subjected to mass spectrometric analysis based on one of these following strategies: Bottom-up, Middle-down and Top-down methods. B) The methylated peptides are identified by tandem mass spectrometry using different fragmentation techniques. Different PTM-specific MS/MS strategies are used to identify the methylation, localize the methylation sites and characterize the subtype of methylation including di-methylation types (aDMA, sDMA). C) Methylation is quantified by mass spectrometry based on two main quantitative approaches: shotgun (discovery) and targeted MS. Shotgun approaches which have been used for methylation are mainly labelfree, SILAC (methyl-SILAC) as well as chemical derivatization. In the methyl-SILAC approach, the cells metabolically convert [13CD3] methionine to [13CD3] S-adenosyl methionine. Heavy methyl-groups are fully incorporated into methylation sites in vivo. This method allows confident identification and relative quantitation for proteins with methylation sites. The specific mass shifts allow differentiation between mono-, di- and tri-methylated species. Targeted quantitative (absolute and relative) strategies including SRM and most recently PRM can be used to study methylation. Label-free and labeling strategies can be applied to targeted approach as well.
Mass Spectrometry based technology for PTM analysis: application to protein methylation
Post-translational modifications are functional groups including chemical species (phosphate, carbohydrate or methyl-group) and functional polypeptides (ubiquitin and SUMO). PTMs can be added or removed from an amino acid side chain or protein termini or created by the cleavage of signal peptides from proteins or by covalent cross linking between separate protein domains [10]. These chemical changes on modified amino acids form a mass shift that can be measured by mass spectrometry (MS). Tandem mass spectrometry (MS/MS) provides valuable information about modified peptides. However, during MS/MS sequencing, it can be challenging to assign the mass shifts because the identified mass shifts may represent isobaric modifications or the sum of a few modifications. For example, the mass difference between tri-methylation (+42.05 Da) and acetylation (+42.01 Da) is very small (0.0364 Da) and can only be discriminated within <30 ppm mass accuracy on sensitive instruments such as the Fourier-Transform Ion Cyclotron MS (FT-ICRMS) or Orbitrap systems. An additional way to discriminate between tri-methylation and acetylation is by the presence of diagnostic marker ions and neutral loss in MS/MS spectra [11].
1-Top-down, Bottom-up and Middle-down mass spectrometry approach: application to histone-methylation
MS-based analysis of PTMs, can globally be classified into three groups based on whether the fragmentation is carried out on: intact protein ions (Top-down) [12–14], short peptide ions post in vitro protein digest (Bottom-up) [12, 15], or more recently on large polypeptides (3–20 kDa) (Middle-down [12, 16–18]). These technologies differ in their capacity for shotgun or large-scale discovery and differ in MS instrumentation. The Top-down approach normally requires high mass accuracy instruments and has been used mostly with FT-ICR-MS, but this method has now been extended to other MS instruments [19]. Top-down is suitable for combinatorial PTM analysis, but less suitable for a shotgun sequencing based identification of low stoichiometric modification sites. For Bottom-up experiments, trap-based instruments with high speed MS/MS scanning, which have the capacity for MS/MS sequencing and for shotgun proteomics are mostly in use. The Middle-down method was introduced after the development of recent fragmentation methods including Electron Capture Dissociation (ECD) [20] and Electron Transfer Dissociation (ETD) [21] which allows efficient ionization of large peptides. The Middle-down technique has been developed to analyze larger peptides, in addition to labile and backbone modifications. While Middle-down has a comparable quantitative capacity to Bottom-up, it also has the capacity to characterize combinatorial PTMs like Top-down.
These methodologies have been extensively applied to study histone modifications, revealing new methylation, phosphorylation, ubiquitination and acetylation sites (review in [22]). With a Bottom-up approach, mono-, di- and tri-methylation at lysine 4, lysine 27 and lysine 36 in H3 were identified. Lysine 27 has been reported to be predominantly mono-methylated while lysine 36 was mostly tri-methylated [23]. A Top-down approach using ECD fragmentation revealed several acetylation, phosphorylation and methylation sites on different H3 forms. Lysine 4 mono-methylation was observed in 5% and lysine 9 dimethylation in about 50% of histone H3.1 in Hela cells [13]. Garcia et al., developed a Middle-down approach using ECD fragmentation to identify and characterize H3 variants and the site occupancy of the most abundantly modified H3 residues: lysine K4, K9, K23, K27 and K36 in ten different rat tissues [24]. Young et al., reported methods to detect histone modification combinations in a single experiment by a Middle-down strategy using a novel saltless pH gradient for weak-cation exchange-hydrophilic interaction chromatography (WCX-HILIC). Using ETD fragmentation, they were able to characterize over 200 modified histone H3.2 forms and 70 histone H4 forms [17].
2- Methylation-specific mass spectrometry
Since protein modifications are often transient and labile with a low stoichiometry (< femtomole), a number of complementary methods such as PTM-targeted MS/MS and selective enrichment of modified peptides prior to MS/MS analysis are needed to improve the sensitivity and quality of mass spectrometric analysis. Although different enrichment strategies are available for the efficient enrichment of various modified peptides, only relatively weak affinity based enrichment methods are available for methylation (review in [7]). Most of these methylation enrichment strategies utilize either methyl-lysine [24, 25] or methyl-arginine [26, 27, 28] specific antibodies, for purification of methylated peptides by immunoprecipitation (IP) (Table 1). Ong et al. [25], used anti-methyl lysine and -arginine antibodies to enrich for methylated peptides following methyl-specific labeling of proteins using heavy methionine SILAC labeling. The authors were able to use SILAC in combination with enrichment to identify 59 methylation sites, corresponding to 58 methylated peptides, in HeLa cells. These methylation sites were identified in 33 different proteins (Table 1). More recently, Uhlmann et al., improved the enrichment by combining different separation methods including Strong-Cation-Exchange (SCX), Isoelectric-Focusing (IEF) and HILIC prior to immunoprecipitation to increase the enrichment efficiency and reduce the sample complexity. Individual comparisons of different separation methods with antibody enrichment demonstrated that HILIC identifies 3–5 times more methylation sites compared to other methods. In total, Uhlmann et al., identified 249 arginine methylation sites in 131 proteins. They identified 190 new methylation sites and 93 proteins which were not previously described to be methylated as well [28] (Table 1).
Table 1.
Examples of large-scale mass spectrometry analysis of protein methylation
| Enrichment | Type of Mass spectrometer |
Separation method |
Number of identified methylated peptides/proteins |
Labeling-method | Reference |
|---|---|---|---|---|---|
| Methyl-arginine (R)-specific antibody | Q-exactive | HILIC1 | Methyl-R sites: 215 Novel methyl-R sites:171 |
Methyl-SILAC | [28] |
| SCX2 | Methyl-R sites: 39 Novel Meth-R sites: 25 |
||||
| IEF3 | Methyl-R sites: 66 Novel methyl-R sites: 34 |
||||
| Total number of methyl-R: 249 sites (190 novel sites) Methylated proteins: 131 (93 not previously described as methylated proteins) |
|||||
| Methyl-arginine (R)- specific antibody | Q-TOF | Gel-based | Methylated sites: 59 Methylated proteins: 33 |
Methyl-SILAC | [25] |
| LTQ-FT | |||||
| Methyl-lysine (K)- specific antibody | |||||
| Methyl-arginine (R)-specific antibody | Q-Star | Gel-based | Methylated proteins: 200 | Label-free | [27] |
HILIC Hydrophilic Interaction Chromatography,
SCX Strong Cation Exchange,
IEF Isoelectric Focusing
Identification and site localization of methylation by Tandem Mass spectrometry
Tandem mass spectra generated by different fragmentation techniques can provide valuable information for peptide sequencing, PTM identification, the PTM subtype and PTM site localization. Collision Induced Dissociation (CID) [25, 29–31, 35] as well as ETD [29–33] spectra of methylated peptides contain methylation specific ion signals in the low mass range. These low mass ions resulting from neutral losses and other peptide backbone fragments can be used to identify, validate and localize the methylation sites (Table 2). The methylation related losses in CID spectra are mostly water losses [33], but other abundant low-mass losses including methylamine, methylguanidine, or methylcarbodiimide have been identified [25, 29, 30]. ETD fragmentation has been applied to study lysine [32] and arginine [31–33] methylation. Methylarginine-associated neutral losses from charge reduced precursor ions during ETD fragmentation [32] result in highly-abundant low-mass product ions which allow for the reliable discrimination of symmetric and asymmetric dimethylarginine [29, 30] (Table 2). In contrast, lysine methylated peptides do not produce significant losses during ETD fragmentation. In addition to neutral loss and immonium ions identification, there are other PTM-specific tandem MS based strategies including precursor ion scanning [34, 36] and multistage MS/MS [37]. Couttas et al. [36], applied immonium ion scanning to discover new histone methylation sites and this method improved the discovery rate of modified peptides 4 fold in comparison to control experiments.
Table 2.
Examples for immonium ions, diagnostic ions and neutral losses of methylated peptides using CID and ETD fragmentation by MS
| Modification | Abbreviation | Mass shift (Δm) (Da) |
Instrument | Neutral loss (Da) | Immonium ions (m/z) | References |
|---|---|---|---|---|---|---|
| Methyl-arginine | ||||||
| Arginine mono-methylation | MMA | 14.0156 | LTQ-FT QSTAR Triple-quadrupole LTQ-Orbitrap-velos Quadrupole-ion trap HCT-ultra ion trap |
31.04221,* 73.064 2,* |
143 (not unique for arginine) | [25, 30–33, 36] |
| Arginine asymmetric di-methylation | aDMA | 28.0312 | Quadrupole- ion trap LTQ-FT LTQ/Orbitrap LTQ-Orbitrap velos HCT-ultra ion trap |
45.05783,* 87.0874,* |
71.06 | [25, 30–34] |
| Arginine symmetric di-methylation | sDMA | 28.0312 | LTQ-FT LTQ/Orbitrap |
31.04221 87.087 |
71.06 | [25, 30] |
| Methyl-lysine | ||||||
| Lysine mono-methylation | 4.0156 | Triple-quadrupole MALDI-TOF |
98.096 84.081 (C-terminal lysine) 101 (N-terminal lysine) |
[35, 36] | ||
| Lysine di-methylation | 28.0312 | Triple-quadrupole | 98.096 112.1 84.081 |
[35, 36] | ||
| Lysine tri-methylation | 42.0470 | LTQ-FT QSTAR MALDI-TOF |
59.0735 | [25, 35] | ||
MMA (mono-methylamine),
MMG (mono-methylguanidine),
DMA (Di-methylamine),
DMG (Di-methylguanidine),
ETD (Electron-transfer-dissociation), The numbers in bold represent the higher ion intensity in mass spectra comparing to unbold
3- Quantitative Mass spectrometry to study PTM: Application to methylation
Quantitative MS-based proteomics techniques [38] are classified into two main groups: shotgun (discovery, large scale) and targeted MS (review in [39]). Shotgun MS [38] is based on Data-Dependent Acquisition (DDA) and intensity-based product ion scanning. Here, there is no need for information on predefined peptides. In targeted methods, the mass spectrometer- often a triple quadrupole instrument - identifies specific and predefined peptide/fragment ion pairs called transitions over time in a Data-Independent Acquisition (DIA) mode. The most common targeted method is “selected reaction monitoring” (SRM) [39], which has the advantages of a better detection limit and extended dynamic range compared to shotgun approaches. Most recently, the Coon group, proposed a new targeted approach “Parallel Reaction Monitoring”(PRM) on a Q-Exactive instrument (Hybrid Quadrupole-Orbitrap) [40]. The advantages of PRM over traditional SRM are a wider dynamic range, high specificity, parallel detection of all target product ion in one concerted high resolution mass analysis rather than 3–5 transitions to validate the peptide identity, minimum upfront method development, and higher tolerance for co-isolated background peptides/species [40].
Label-free and labeling based quantitative MS methods have been applied to study and quantify histone modifications (Review in [42]). In a Middle-down label free ETD based approach, 74 discrete combinatorial modification codes on the tail of histone H4 in differentiating human embryonic stem cells (ES) were identified and quantified. Significant changes in the methylation and acetylation patterns of histone H4 isoforms during differentiation were observed, thus describing a context-specific PTM pattern. For example, H4R3 methylation was found only in the presence of H4K20 dimethylation [43]. Chemical derivatization in combination with stable isotope labeling is another quantitative method which has been widely used to study histone modification. Smith et al [44], used stable isotope labeling (deuterated acetic anhydride) to quantify Histone H4 lysine acetylation. In 2005, Garcia et al., introduced a double derivatization technique to quantify histone PTMs [45]. The first derivatization was applied to the free amino group in the N-terminus and unmodified or mono-methylated internal lysine so that similar sized fragments were formed from reproducible cleavage of histone protein by trypsin C-terminal to Arginine mimicking an ArgC-digest. They modified carboxylic acid groups with a normal (d0-methanol) or stableisotope labeled reagent d4-methanol (esterification reaction on peptide level) for relative quantitation. Further optimization of this method was done using d0- or d10-propionic anhydride to label the newly formed free N-terminal amino group during the second round of derivatization after digestion to overcome some limitations of labeled methanol [46]. With this method histone methylation can be directly quantified by comparing peak pairs separated by a +5 Da mass shift.
Another quantitative method to study methylation, is MS quantitation using isotopic reductive methylation (MassSQUIRM) [47]. This method differentiates between lysine methylation states by reductive methylation with heavy formaldehyde, causing the addition of up to two methyl-groups to lysine residues. All the peptides (mono-, di- and unmethylated) are converted to the same chemical species to have the same ionization efficiency. The dimethylated peptide has 28 Da (m/z), with one heavy methyl (30 Da) and with two-methyl groups (32 Da) in the unmodified form. MassSQUIRM can measure demethylase dynamics and their capacity to remove mono- and dimethyl marks from lysine residues. As briefly mentioned earlier, in a variation of SILAC (Stable Isotope Labeling by Amino acids in Cell culture), cells can be cultured in media with 13CD3-methionine instead of heavy leucine, lysine, or arginine as in regular SILAC approaches [25]. The heavy methionine can be converted to 13CD3-adenosyl methionine, a biological methyl-donor (Figure 1), which, in turn can be used by methyl transferases to label with heavy amino acids methyl groups. The relative peak intensity of methyl-modified peptide pairs are used to identify and quantify methylated species (Figure 2). Zee et al, reported the steady state kinetics of global methylation of histones on a residue-specific basis using such heavy methyl-SILAC labeling [5]. Their work showed a progressively slower rate of formation of mono-, di-, and trimethylated residues and different methylation rates associated with either active or silent genes.
4- Identification of methylation cross talk with other modifications by quantitative MS
Identification of multiple PTMs in peptide sequences may be a hint at the synergistic or antagonistic interaction of these modifications. Methylation cross talk with other modifications such as acetylation, phosphorylation and ubiquitination has been described [6, 48]. Lysine methylation increases protein half-life by blocking ubiquitination (review in [6]). Darwanto et al., quantified (absolute and relative) 20 histone modification sites on H2A, H2B, H3 and H4 including acetylation, propionylation, methylation and ubiquitination by a MRM approach. The authors discovered an inverse correlation between Histone H2B ubiquitination and H3-Lysine 79 methylation [48].
5- Conclusion
Taken together, the advancements in the mass spectrometry field including new developments in instrumentation, technologies and methodology have been applied to improve research into protein methylation. Besides lysine and arginine methylation, there are scarce publications on other methylated amino acids [49] and there are insufficient validation strategies able to discriminate between real modification sites and artifacts [50], technical methylation and functional methylation. Therefore, there is room for the improvement of methylation detection strategies that will undoubtedly advance these fields.
Highlights.
-
-
Mass spectrometry based technologies and methodologies are powerful tools to confidently identify and characterize methylated amino-acids.
-
-
They also enable to distinguish methylation subtypes, and to localize modification sites.
-
-
Mass spectrometry can quantitatively measure protein methylation states and their dynamics.
Acknowledgements
BAG acknowledges funding from a National Science Foundation (NSF) Early Faculty CAREER award and NSF CBET-091143 grant, and the NIH Innovator grant (DP2OD007447) from the Office of the Director, National Institutes of Health. Funding from Eli Lilly is also gratefully acknowledged.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bedford MT, Clarke SG. Protein arginine methylation in mammals: who, what, and why. Mol Cell. 2009;33:1–13. doi: 10.1016/j.molcel.2008.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Paik WK, Paik DC, Kim S. Historical review: the field of protein methylation. Trends Biochem Sci. 2007;32:146–152. doi: 10.1016/j.tibs.2007.01.006. **This review is an introduction to protein methylation, history of research and the functional impact of protein methylation.
- 3.Ishikawa Y, Melville DB. The enzymatic alpha-N-methylation of histidine. J Biol Chem. 1970;245:5967–5973. [PubMed] [Google Scholar]
- 4.Lee YH, Stallcup MR. Minireview: Protein arginine methylation of nonhistone proteins in transcriptional regulation. Mol Endocrinol. 2009;23:425–433. doi: 10.1210/me.2008-0380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zee BM, Levin RS, Xu B, LeRoy G, Wingreen NS, Garcia BA. In vivo residuespecific histone methylation dynamics. J Biol Chem. 2010;285:3341–3350. doi: 10.1074/jbc.M109.063784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Erce MA, Pang CN, Hart-Smith G, Wilkins MR. The methylproteome and the intracellular methylation network. Proteomics. 2012;12:564–586. doi: 10.1002/pmic.201100397. *In this review, analytical techniques to detect and localize methylation sites are discussed. In addition, methylation, interactome, cross-talks are reviewed.
- 7.Zhao Y, Jensen ON. Modification-specific proteomics: strategies for characterization of post-translational modifications using enrichment techniques. Proteomics. 2009;9:4632–4641. doi: 10.1002/pmic.200900398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choudhary C, Mann M. Decoding signaling networks by mass spectrometrybased proteomics. Nat Rev Mol Cell Biol. 2010;11:427–439. doi: 10.1038/nrm2900. [DOI] [PubMed] [Google Scholar]
- 9. Jensen ON. Interpreting the protein language using proteomics. Nat Rev Mol Cell Biol. 2006;7:391–403. doi: 10.1038/nrm1939. **This review discuss challenges in PTM analysis, PTM analysis by mass spectrometry and modification-specific proteomics including enrichment strategies. Further, new technologies and integrative computational stretegies are reviewed.
- 10. Walsh CT, Garneau-Tsodikova S, Gatto GJ Jr. Protein posttranslational modifications: the chemistry of proteome diversifications. Angew Chem Int Ed Engl. 2005;44:7342–7372. doi: 10.1002/anie.200501023. * This review is about the chemical mechanism of protein post-translational modification, covering almost all types of modifications.
- 11.Zhang K, Yau PM, Chandrasekhar B, New R, Kondrat R, Imai BS, Bradbury ME. Differentiation between peptides containing acetylated or tri-methylated lysines by mass spectrometry: an application for determining lysine 9 acetylation and methylation of histone H3. Proteomics. 2004;4:1–10. doi: 10.1002/pmic.200300503. [DOI] [PubMed] [Google Scholar]
- 12. Garcia BA. What does the future hold for Top Down mass spectrometry? J Am Soc Mass Spectrom. 2010;21:193–202. doi: 10.1016/j.jasms.2009.10.014. **A comprehensive technical review on Top-down mass spectrometry. Instrumentation, data analysis software and separation methods for Top-down are discussed. A comparison to other methods, including middle-down and bottom-up, is made.
- 13.Thomas CE, Kelleher NL, Mizzen CA. Mass spectrometric characterization of human histone H3: a bird's eye view. J Proteome Res. 2006;5:240–247. doi: 10.1021/pr050266a. [DOI] [PubMed] [Google Scholar]
- 14.Tran JC, Zamdborg L, Ahlf DR, Lee JE, Catherman AD, Durbin KR, Tipton JD, Vellaichamy A, Kellie JF, Li M, Wu C, Sweet SM, Early BP, Siuti N, LeDuc RD, Compton PD, Thomas PM, Kelleher NL. Mapping intact protein isoforms in discovery mode using top-down proteomics. Nature. 2011;480:254–258. doi: 10.1038/nature10575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Plazas-Mayorca MD, Bloom JS, Zeissler U, Leroy G, Young NL, DiMaggio PA, Krugylak L, Schneider R, Garcia BA. Quantitative proteomics reveals direct and indirect alterations in the histone code following methyltransferase knockdown. Mol Biosyst. 2010;6:1719–1729. doi: 10.1039/c003307c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cannon J, Lohnes K, Wynne C, Wang Y, Edwards N, Fenselau C. High-throughput middle-down analysis using an orbitrap. J Proteome Res. 2010;9:3886–3890. doi: 10.1021/pr1000994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Young NL, DiMaggio PA, Plazas-Mayorca MD, Baliban RC, Floudas CA, Garcia BA. High throughput characterization of combinatorial histone codes. Mol Cell Proteomics. 2009;8:2266–2284. doi: 10.1074/mcp.M900238-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garcia BA, Pesavento JJ, Mizzen CA, Kelleher NL. Pervasive combinatorial modification of histone H3 in human cells. Nat Methods. 2007;4:487–489. doi: 10.1038/nmeth1052. [DOI] [PubMed] [Google Scholar]
- 19.Macek B, Waanders LF, Olsen JV, Mann M. Top-down protein sequencing and MS3 on a hybrid linear quadrupole ion trap-orbitrap mass spectrometer. Mol Cell Proteomics. 2006;5:949–958. doi: 10.1074/mcp.T500042-MCP200. [DOI] [PubMed] [Google Scholar]
- 20.Kelleher NL, Zubarev RA, Bush K, Furie B, Furie BC, McLafferty FW, Walsh CT. Localization of labile posttranslational modifications by electron capture dissociation: the case of gamma-carboxyglutamic acid. Anal Chem. 1999;71:4250–4253. doi: 10.1021/ac990684x. [DOI] [PubMed] [Google Scholar]
- 21. Syka JE, Coon JJ, Schroeder MJ, Shabanowitz J, Hunt DF. Peptide and protein sequence analysis by electron transfer dissociation mass spectrometry. Proc Natl Acad Sci U S A. 2004;101:9528–9533. doi: 10.1073/pnas.0402700101. ** First paper on ETD fragmentation mechanism and development. They used gas-phase ion-ion chemistry and MS/MS to analyze peptide sequence and to induce backbone fragmentation. This fragmentation (ETD) is done by transfer of singly charged anthracene anions to multiply protonated peptides. They used ETD to analyse phospho-peptides by MS/MS using a quadrupole-linear-trap.
- 22. Sidoli S, Cheng L, Jensen ON. Proteomics in chromatin biology and epigenetics: Elucidation of post-translational modifications of histone proteins by mass spectrometry. J Proteomics. 2012;75:3419–3433. doi: 10.1016/j.jprot.2011.12.029. * This review focuses on histone PTMs. The separation of histone variants, enrichment methods, and mass spectrometry analysis for histone analysis as well as quantitation are discussed. Further, Chromatin-IP and Mass spectrometry analysis as well as data handling are reviewed.
- 23.Wu T, Yuan T, Tsai SN, Wang C, Sun SM, Lam HM, Ngai SM. Mass spectrometry analysis of the variants of histone H3 and H4 of soybean and their posttranslational modifications. BMC Plant Biol. 2009;9:98. doi: 10.1186/1471-2229-9-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Garcia BA, Thomas CE, Kelleher NL, Mizzen CA. Tissue-specific expression and post-translational modification of histone H3 variants. J Proteome Res. 2008;7:4225–4236. doi: 10.1021/pr800044q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ong SE, Mittler G, Mann M. Identifying and quantifying in vivo methylation sites by heavy methyl SILAC. Nat Methods. 2004;1:119–126. doi: 10.1038/nmeth715. ** This study describes the methyl-SILAC method, which allows for studying invivo methylation sites with direct methyl-group labeling. This method allows confident identification and relative quantitation of protein methylation by mass spectrometry.
- 26.Iwabata H, Yoshida M, Komatsu Y. Proteomic analysis of organ-specific post-translational lysine-acetylation and -methylation in mice by use of anti-acetyllysine and -methyllysine mouse monoclonal antibodies. Proteomics. 2005;5:4653–4664. doi: 10.1002/pmic.200500042. [DOI] [PubMed] [Google Scholar]
- 27.Boisvert FM, Lam YW, Lamont D, Lamond AI. A quantitative proteomics analysis of subcellular proteome localization and changes induced by DNA damage. Mol Cell Proteomics. 2010;9:457–470. doi: 10.1074/mcp.M900429-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Uhlmann T, Geoghegan VL, Thomas B, Ridlova G, Trudgian DC, Acuto O. A method for large-scale identification of protein arginine methylation. Mol Cell Proteomics. 2012 doi: 10.1074/mcp.M112.020743. * They used multi-dimensional chromatography in combination with a methylarginine antibody to enrich methylated arginines from a complex mixture. Methylation sites were identified using HCD fragmentation on a Q-Exactive instrument.
- 29.Gehrig PM, Hunziker PE, Zahariev S, Pongor S. Fragmentation pathways of N(G)- methylated and unmodified arginine residues in peptides studied by ESIMS/ MS and MALDI-MS. J Am Soc Mass Spectrom. 2004;15:142–149. doi: 10.1016/j.jasms.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 30.Brame CJ, Moran MF, McBroom-Cerajewski LD. A mass spectrometry based method for distinguishing between symmetrically and asymmetrically dimethylated arginine residues. Rapid Commun Mass Spectrom. 2004;18:877–881. doi: 10.1002/rcm.1421. [DOI] [PubMed] [Google Scholar]
- 31. Wang H, Straubinger RM, Aletta JM, Cao J, Duan X, Yu H, Qu J. Accurate localization and relative quantification of arginine methylation using nanoflow liquid chromatography coupled to electron transfer dissociation and orbitrap mass spectrometry. J Am Soc Mass Spectrom. 2009;20:507–519. doi: 10.1016/j.jasms.2008.11.008. ** They used an alternating CID/ETD method to localize and characterize arginine methylation in proteins with arginine-rich motifs.
- 32. Snijders AP, Hung ML, Wilson SA, Dickman MJ. Analysis of arginine and lysine methylation utilizing peptide separations at neutral pH and electron transfer dissociation mass spectrometry. J Am Soc Mass Spectrom. 2010;21:88–96. doi: 10.1016/j.jasms.2009.09.010. ** An ETD fragmentation method was used to identify and characterize methylated peptides. The researchers identified diagnostic ions in ETD spectra for methylated arginine.
- 33. Hart-Smith G, Low JK, Erce MA, Wilkins MR. Enhanced methylarginine characterization by post-translational modification-specific targeted data acquisition and electron-transfer dissociation mass spectrometry. J Am Soc Mass Spectrom. 2012;23:1376–1389. doi: 10.1007/s13361-012-0417-8. ** The authors used targeted data acquisition to study methyl-arginine by ETD fragmentation. They determined that ETD is the preferred fragmentation method for the characterization of methyl-arginine.
- 34.Rappsilber J, Friesen WJ, Paushkin S, Dreyfuss G, Mann M. Detection of arginine dimethylated peptides by parallel precursor ion scanning mass spectrometry in positive ion mode. Anal Chem. 2003;75:3107–3114. doi: 10.1021/ac026283q. [DOI] [PubMed] [Google Scholar]
- 35.Hirota J, Satomi Y, Yoshikawa K, Takao T. Epsilon -N,N,N-trimethyllysine-specific ions in matrix-assisted laser desorption/ionization-tandem mass spectrometry. Rapid Commun Mass Spectrom. 2003;17:371–376. doi: 10.1002/rcm.924. [DOI] [PubMed] [Google Scholar]
- 36.Couttas TA, Raftery MJ, Bernardini G, Wilkins MR. Immonium ion scanning for the discovery of post-translational modifications and its application to histones. J Proteome Res. 2008;7:2632–1641. doi: 10.1021/pr700644t. [DOI] [PubMed] [Google Scholar]
- 37.Schroeder MJ, Shabanowitz J, Schwartz JC, Hunt DF, Coon JJ. A neutral loss activation method for improved phosphopeptide sequence analysis by quadrupole ion trap mass spectrometry. Anal Chem. 2004;76:3590–3598. doi: 10.1021/ac0497104. [DOI] [PubMed] [Google Scholar]
- 38. Cox J, Mann M. Quantitative, high-resolution proteomics for data-driven systems biology. Annu Rev Biochem. 2011;80:273–299. doi: 10.1146/annurev-biochem-061308-093216. ** A comprehensive review on large-scale protein analysis and quantitative proteomics.
- 39. Picotti P, Aebersold R. Selected reaction monitoring-based proteomics: workflows, potential, pitfalls and future directions. Nat Methods. 2012;9:555–566. doi: 10.1038/nmeth.2015. ** In this review selected reaction monitoring method development and other quantitation techniques are discussed. In addition, SRM’s application to proteomics, to PTMs and clinical applications are reviewed. Further, the pitfalls and outlooks for SRM approaches are discussed.
- 40. Peterson AC, Russell JD, Bailey DJ, Westphall MS, Coon JJ. Parallel reaction monitoring for high resolution and high mass accuracy quantitative, targeted proteomics. Mol Cell Proteomics. 2012 doi: 10.1074/mcp.O112.020131. ** The authors introduced the new quantitative targeted method:"PRM”, an adaption of the SRM method for the Q-exactive instrument with applications for proteomics.
- 41.Garcia BA. Mass spectrometric analysis of histone variants and post-translational modifications. Front Biosci. 2009;1:142–153. doi: 10.2741/S14. [DOI] [PubMed] [Google Scholar]
- 42.Pesavento JJ, Bullock CR, LeDuc RD, Mizzen CA, Kelleher NL. Combinatorial modification of human histone H4 quantitated by two-dimensional liquid chromatography coupled with top down mass spectrometry. J Biol Chem. 2008;283:14927–14937. doi: 10.1074/jbc.M709796200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Phanstiel D, Brumbaugh J, Berggren WT, Conard K, Feng X, Levenstein ME,McAlister GC, Thomson JA, Coon JJ. Mass spectrometry identifies and quantifies 74 unique histone H4 isoforms in differentiating human embryonic stem cells. Proc Natl Acad Sci U S A. 2008;105:4093–4098. doi: 10.1073/pnas.0710515105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Smith CM, Gafken PR, Zhang Z, Gottschling DE, Smith JB, Smith DL. Massspectrometric quantification of acetylation at specific lysines within the amino-terminal tail of histone H4. Anal Biochem. 2003;316:23–33. doi: 10.1016/s0003-2697(03)00032-0. [DOI] [PubMed] [Google Scholar]
- 45. Garcia BA, Mollah S, Ueberheide BM, Busby SA, Muratore TL, Shabanowitz J, Hunt DF. Chemical derivatization of histones for facilitated analysis by mass spectrometry. Nat Protoc. 2007;2:933–938. doi: 10.1038/nprot.2007.106. ** A method paper on double-chemical derivatization using propionic anhydride to mimic reproducible Arg-C digests. They combined the method with stable isotope labeling to characterize and quantify histone modifications.
- 46.Plazas-Mayorca MD, Zee BM, Young NL, Finger man IM, LeRoy G, Briggs SD, Garcia BA. One-pot shotgun quantitative mass spectrometry characterization of histones. J Proteome Res. 2009;8:5367–5374. doi: 10.1021/pr900777e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Blair LP, Avert NL, Huang R, Cole PA, Tavern SD, Tackett AJ. MassSQUIRM: An assay for quantitative measurement of lysine demethylase activity. Epigenetics. 2011;6:490–499. doi: 10.4161/epi.6.4.14531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Darwanto A, Curtis MP, Schrage M, Kirsch W, Liu P, Xu G, Neigh JW, Zhang K. A modified "cross-talk" between histone H2B Lys-120 ubiquitination and H3 Lys-79 methylation. J Biol Chem. 2010;285:21868–21876. doi: 10.1074/jbc.M110.126813. * One of the very few MRM studies on histone modifications and cross-talk events. Twenty different modified sites in histones were quantified by MS. The authors showed inverse correlation of H2B ubiquitination and H3 Lys-79 methylation.
- 49.Savitski MM, Nielsen ML, Zubarev RA. ModifiComb, a new proteomic tool for mapping sub-stoichiometric post-translational modifications, finding novel types of modifications, and fingerprinting complex protein mixtures. Mol Cell Proteomics. 2006;5:935–948. doi: 10.1074/mcp.T500034-MCP200. [DOI] [PubMed] [Google Scholar]
- 50. Jung SY, Li Y, Wang Y, Chen Y, Zhao Y, Qin J. Complications in the assignment of 14 and 28 Da mass shift detected by mass spectrometry as in vivo methylation from endogenous proteins. Anal Chem. 2008;80:1721–1729. doi: 10.1021/ac7021025. * The assignment of 14 or 28 Da mass shifts identified by MS as a result of methylation, is complicated. A large number of peptides can be modified on lysine, arginine, histidine, and glutamic acid residues with a mass increase of 14 or 18. Using Methyl-SILAC method, they described that technical methylation (gel-based method) can cause complications in correctly of endogenous methylation.