Abstract
In a search for genes and pathways implicated in T-cell lymphoblastic lymphoma (T-LBL) development, we used a murine lymphoma model, where mice of the NMRI-inbred strain were inoculated with murine leukemia virus mutants. The resulting tumors were analyzed by integration analysis and global gene expression profiling to determine the effect of the retroviral integrations on the nearby genes, and the deregulated pathways in the tumors. Gene expression profiling identified increased expression of genes involved in the minichromosome maintenance and origin of recognition pathway as well as downregulation in negative regulators of G1/S transition, indicating increased S-phase initiation in murine T-LBLs.
Keywords: T-cell lymphoblastic lymphoma, T-LBL, Mouse model, Insertional mutagenesis, MLV, Gene expression Profiling, SL3-3
1. Introduction
Human T-cell lymphoblastic lymphomas (T-LBLs) are neoplasms of immature T-cells and constitute a group of rare, heterogeneous and clinically very aggressive tumors [1–3]. The molecular pathogenesis that contributes to T-LBL development is not fully elucidated. However, since murine T-LBLs are histopatho-logically and phenotypically comparable to human T-LBLs [4], mouse models of T-LBL are ideal to obtain additional insight into the mechanism of T-LBL development in humans.
When injected into newborn NMRI inbred mice, the SL3-3 murine leukemia virus (MLV) induces hematological malignancies, including T-LBLs [5–8]. The SL3-3 enhancers in the U3 regions of the LTRs, consist of 2.5 tandem copies of a 72 bp sequence with binding sites for Runx, c-Myb, Ets, and Nf1 transcription factors [5,6,8–10]. Within these tandem repeats, there are three identical glucocorticoid response element (GRE)-overlapping E-boxes (Egre) which contain binding sites for the glucocorticoid receptor (GR) and basic helix-loop-helix (bHLH) factors [7,11]. Mutations in the SL3-3 enhancer at the binding site for GR have no effect on the pathogenicity of SL3-3, and induce T-LBLs with a similar latency period as the wild type virus [7]. On the other hand, mutations in the Egre binding sites increase SL3-3 latency significantly by up to several months, and broaden disease from strictly T-LBL to various hematopoietic malignancies, including B-cell lymphomas and leukaemias of myeloid and erythroid origin. The oncogenic effects of SL3-3 MLV are caused by integration of the viral genome into the host cell DNA through multiple rounds of infection, and deregulation of nearby cellular genes – a process defined as insertional mutagenesis [12,13]. If the integration occurs near or within a gene of importance for cancer development, the cell in which the virus has integrated, may gain a growth advantage, leading to malignant transformation and tumor development. Screening the end-stage tumors for resulting integration sites, is therefore an efficient method for identifying genes involved in murine and potentially also human T-cell lymphomagenic processes. To gain more insight into the mechanisms by which murine T-LBLs develop, we performed integration analysis and gene expression profiling on thymus and spleen T-LBL tumors. Our data shows that genes involved in G1/S-phase transition are frequently targeted by retroviral integrations in the murine T-LBLs. Additional identification of increased expression of genes from the minichromosome maintenance (Mcm)–origin of recognition (Orc) pathway, as well as downregulation of G1/S-phase inhibitors, implicate S-phase initiating genes in the development of murine T-LBLs.
2. Results
2.1. Retroviral integrations in SL3-3 3mGR and SL3-3 3mEgre induced tumors
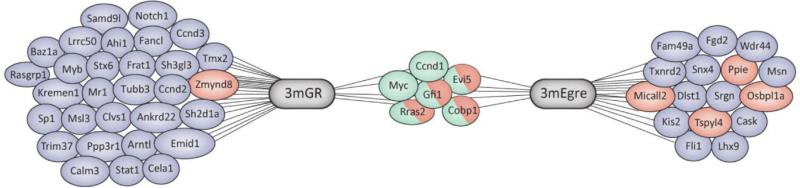
Ninety-five integrations were identified in 10 SL3-3 3mGR and 9 SL3-3 3mEgre induced tumors, of which 78 were located within 10 kb distance of a RefSeq gene (Fig. 1 and Table 1). Gfi1, Evi5, Myc, Rras2, and D-type cyclins were common integration sites (CISs) for both the 3mGR and 3mEgre virus mutants and targeted multiple times in some of the tumors (Table 1). In the 3mGR induced tumors, 03/127 had two integrations in Zmynd8 and Ccnd3 was targeted in both the 03/12 and the 03/210 tumor, whereas two integrations in each of the genes Ppie, Osbpl1a, Tpsy14 and Micall2 were identified in the 3mEgre induced tumors 04/524, 04/525, and 04/1260, respectively. All tumors were previously identified as T-LBLs by immunohistopathological examination of liver, spleen and lymph nodes [7].
Fig. 1.
Retroviral integrations in SL3-3 3mGR and SL3-3 3mEgre induced tumors. Of 95 retroviral integrations 78 were located within a 10 kb distance of a RefSeq gene representing a total of 52 different genes. Blue illustrating single hits, red illustrating multiple hits in the same tumor at different locations and green illustrating common integrations in both SL3-3 3mGR and SL3-3 3mEgre induced tumors. Integrations identified in the genomic region carrying Gfi1 and Evi5 are registered so that integrations downstream of Evi5 are counted as Gfi1 integrations and only integrations within the Evi5 gene itself are registered as Evi5 integrations. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of the article.)
Table 1.
Overview of tumors and integrations. Neighboring genes within 10 kb of the retroviral integrations are summarized in the table. T, thymus; S, spleen; LN, lymph node.
| Tumor | Tissue of integration | Tissue for array | Virus | Latency (days) | Integration |
|
|---|---|---|---|---|---|---|
| Gene name (symbol) | Hitsa | |||||
| 04/478 | LN | S | SL3-3 3mEgre | 69 | Cyclin D1 (Ccnd1) | 1 |
| Moesin (Msn) | 1 | |||||
| Thioredoxin reductase 2 (Txnrd2) | 1 | |||||
| Growth factor independent 1 (Gfi1) | 1 | |||||
| 04/512 | LN | S | SL3-3 3mEgre | 76 | Ecotropic viral integration site 5 (Evi5) | 1 |
| Family with sequence similarity 49, member A (Fam49a) | 1 | |||||
| 04/524 | S | S | SL3-3 3mEgre | 82 | Ecotropic viral integration site 5 (Evi5) | 1 |
| Peptidylprolyl isomerase E (Ppie) | 2 | |||||
| 04/525 | S | T | SL3-3 3mEgre | 82 | Growth factor independent 1 (Gfi1) | 1 |
| Oxysterol binding protein-like 1A (Osbpl1a) | 2 | |||||
| Testis-specific Y-encoded-like protein 4 (Tspyl4) | 2 | |||||
| 04/712 | LN | S | SL3-3 3mEgre | 98 | Related RAS viral (r-ras) oncogene Homolog 2 (Rras2)/coatomer protein complex, subunit beta 1 (Cobp1) | 2 |
| FYVE, RhoGEF and PH domain containing 2 (Fgd2) | 1 | |||||
| 04/1123 | T | T, S | SL3-3 3mEgre | 122 | Growth factor independent 1 (Gfi1) | 1 |
| Kaplan integration site 2 (Kis2) | 1 | |||||
| Myelocytomatosis oncogene (Myc) | 1 | |||||
| Sorting nexin 4 (Snx4) | 1 | |||||
| 04/1260 | S | T | SL3-3 3mEgre | 146 | MICAL-like 2 (Micall2) | 2 |
| 04/1328 | T | T | SL3-3 3mEgre | 157 | Calcium/calmodulin-dependent serine protein kinase (MAGUK family) (Cask)/G protein-coupled receptor 34 (Gpr34) | 1 |
| Serglycin (Srgn) | 1 | |||||
| Dihydrolipoamide S-succinyltransferase (E2 component of 2-oxo-glutarate complex) (Dlst) | 1 | |||||
| 04/1345 | T | T | SL3-3 3mEgre | 163 | WD repeat domain 44 (Wdr44) | 1 |
| LIM homeobox protein 9 (Lhx9) | 1 | |||||
| Cyclin D1 (Ccnd1) | 1 | |||||
| Friend leukemia integration 1 (Fli1) | 1 | |||||
| 03/12 | LN | S | SL3-3 3mGr | 44 | Major histocompatibility complex, class I-related (Mr1)/syntaxin 6 (Stx6) | 1 |
| Abelson helper integration site 1 (Ahi1)/myeloblastosis oncogene (Myb) | 1 | |||||
| Cyclin D3 (Ccnd3) | 1 | |||||
| Growth factor independent 1 (Gfi1) | 2 | |||||
| 03/21 | LN | S | SL3-3 3mGr | 48 | Oxysterol binding protein-like 1A (Osbpl1a) | 1 |
| Frequently rearranged in advanced T cell lymphomas (Frat1) | 1 | |||||
| Thioredoxin-related transmembrane protein 2 (Tmx2) | 1 | |||||
| Ecotropic viral integration site 5 (Evi5) | 2 | |||||
| SH3-domain GRB2-like 3 (Sh3g13) | 1 | |||||
| 03/66 | S | S | SL3-3 3mGr | 51 | Aryl hydrocarbon receptor nuclear translocator-like (Arntl) | 1 |
| Sterile alpha motif domain containing 9-like (Samd9l) | 1 | |||||
| Growth factor independent 1 (Gfi1) | 2 | |||||
| RAS guanyl releasing protein 1 (RasGrp1) | 1 | |||||
| Fanconi anemia, complementation group L (Fancl) | 1 | |||||
| Tripartite motif-containing 37 (Trim37) | 1 | |||||
| 03/94 | T | T, S | SL3-3 3mGr | 55 | Male-specific lethal 3 homolog (Drosophila) (Msl3) | 1 |
| Dynein, axonemal assembly factor 1 (Dnaaf1) | 1 | |||||
| Notch 1 (Notch1) | 1 | |||||
| RAS guanyl releasing protein 1 (RasGrp1) | 2 | |||||
| 03/97 | T | S, T | SL3-3 3mGr | 55 | Tubulin, beta 3 class III (Tubb3) | 1 |
| Cyclin D1 (Ccnd1) | 1 | |||||
| Myelocytomatosis oncogene (Myc) | 1 | |||||
| 03/127 | S | T | SL3-3 3mGr | 58 | RAS guanyl releasing protein 1 (RasGrp1) | 1 |
| Zinc finger, MYND-type containing 8 (Zmynd8) | 2 | |||||
| Calmodulin 3 (Calm3) | 1 | |||||
| 03/130 | LN | T | SL3-3 3mGr | 58 | SH2 domain protein 1A (Sh2d1a) | 1 |
| Growth factor independent 1 (Gfi1) | 2 | |||||
| Kringle containing transmembrane protein 1 | 1 | |||||
| (Kremen1)/EMI domain containing 1 (Emid1) Myeloblastosis oncogene (Myb) | 1 | |||||
| 03/131 | T | T | SL3-3 3mGr | 58 | Related RAS viral (r-ras) oncogene homolog 2 (Rras2)/coatomer protein complex, subunit beta 1 (Cobp1) | 1 |
| RAS guanyl releasing protein 1 (RasGrp1) | 2 | |||||
| Chymotrypsin-like elastase family, member 1 (Cela1) | 1 | |||||
| Trans-acting transcription factor 1 (Sp1) | 1 | |||||
| Signal transducer and activator of transcription 1 (Stat1) | 1 | |||||
| 03/196 | LN | T | SL3-3 3mGr | 65 | Cyclin D2 (Ccnd2) | 1 |
| Ankyrin repeat domain 22 (Ankrd22) | 1 | |||||
| 03/210 | T | S | SL3-3 3mGr | 68 | Related RAS viral (r-ras) oncogene homolog 2 (Rras2)/coatomer protein complex, subunit beta 1 (Cobp1) | 1 |
| Clavesin 1 (Clvs1) | 1 | |||||
| RAS guanyl releasing protein 1 (RasGrp1) | 1 | |||||
| Cyclin D3 (Ccnd3) | 1 | |||||
| Protein phosphatase 3, regulatory subunit B, alpha isoform (calcineurin B, type I) (Ppp3r1) | 1 | |||||
Number of integrations identified in or near the specific gene.
2.2. Global mRNA expression in T-LBLs vs. normal tissue
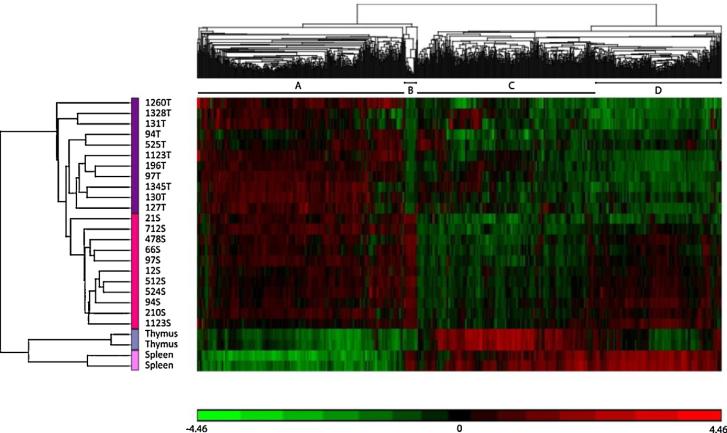
Gobal microarray analysis was performed on 22 tumors induced by the SL3-3 MLV bearing mutations in 3mGR (6 spleen tumors and 6 thymus tumors) and 3mEgre (5 spleen tumors and 5 thymus tumors) and compared to healthy murine spleen and thymus tissue. Initial analysis was performed using unsupervised hierarchical clustering with Pearson's dissimilarity and Ward's method, which indicated that the tumors cluster, first according to disease and second according to tissue (Fig. 2). The three-paired thymus and spleen samples included in our study did not group together, but followed the tissue specific clustering. Overall, we observed four main clusters (A–D). Clusters A and C represent genes that are either upregulated or downregulated, respectively, in tumor tissue, while clusters B and D represent mainly tissue specific genes or genes that are not affected in the tumors compared to control tissue.
Fig. 2.
Unsupervised hierarchical clustering of tumor and normal samples for genes that passed the criteria of ±2-fold change and p ≤ 0.05. Red: upregulated, green: downregulated. (A) Upregulated in tumors, (B and D): upregulated in spleen tumors, downregulated in thymus tumors, and (C) downregulated in tumors. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of the article.)
2.3. Gene expression in SL3-3 3mGR vs. SL3-3 3mEgre induced tumors
To determine which genes differ between tumors induced by the SL3-3 3mGR and SL3-3 3mEgre virus mutants, tumors induced by the two virus mutants were compared within the tissue groups. Surprisingly, there was no difference in gene expression between tumors induced by the two virus mutants (set by the criteria ±1.5-fold difference and p ≤ 0.05), indicating that although they induce tumors with mean latencies of 56 and 110 days for SL3-3 3mGR and SL3-3 3mEgre, respectively (Table 1), the alterations that lead to tumor development are indistinguishable at the gene expression level between the two. Therefore, the influence of the virus mutants was not included in following analyses.
2.4. The effect of the retroviral integrations on expression of nearby genes
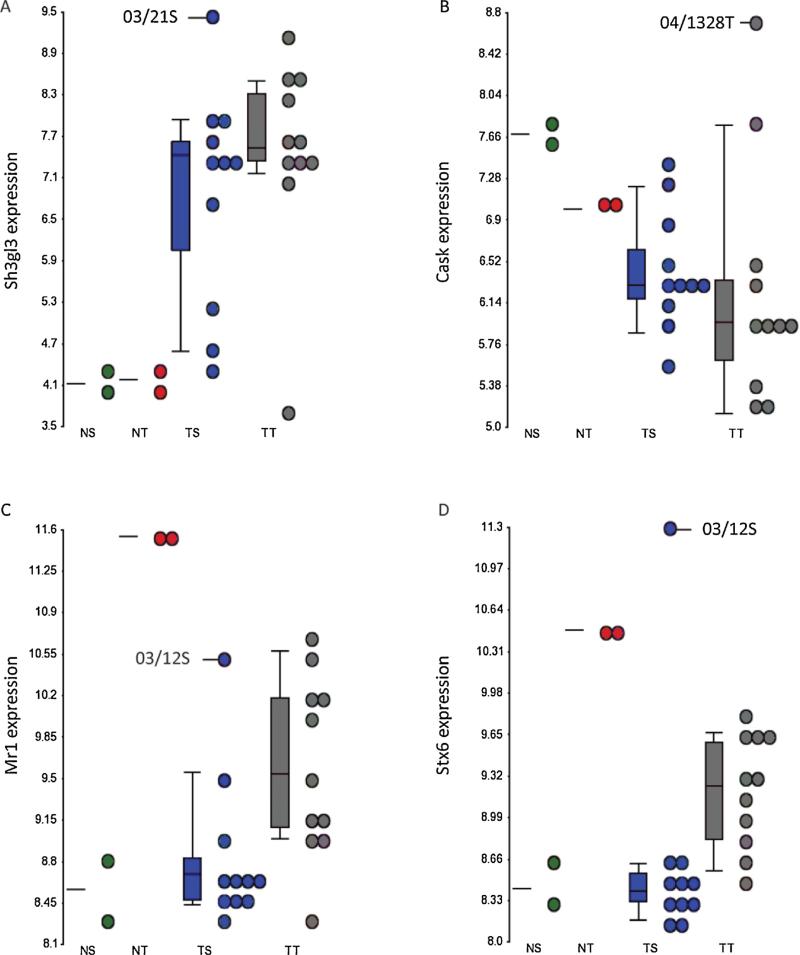
In order to determine if the effect of the retroviral integrations in the individual tumors could be seen from the overall gene expression profile, thymus and spleen tumors were compared to healthy controls within the two tissue groups. Genes with a ±1.5-fold change in tumor relative to normal sample and a p-value ≤0.05 were considered differentially expressed with statistical significance. Based on these criteria, 3725 and 4841 genes were found differentially expressed in thymus and spleen tumors compared to tissue controls. We determined which of the genes, within 10 kbs of a viral integration, were commonly deregulated in thymus and spleen tumors. Here we found Gfi1, and Ppie upregulated in both tissue groups, while Ankrd22, Fam49a, and Arntl were downregulated (Table 2). No other genes within 10 kbs of the retroviral integration sites were affected in the tumors. Several of the genes that were identified as single hits were affected in either the thymus or the spleen tumor group. Sh2d1a, Sh3gl3, Myb, and Trim37 were upregulated in spleen tumors, whereas Fgd2, Cask, Fli1 and Evi5 were generally downregulated. None of these eight genes were significantly affected in the thymus tumors. This tumor group, however, showed upregulation of Txnrd2 and downregulation of Mr1, Stx6, Kremen1, Stat1 and Micall2. To determine if the retroviral integrations had an effect on transcript expression in the tumors they were identified in, we looked at gene expression dot-plots where individual tumors could be visualized. Here, Sh3gl3, Cask, Mr1 and Stx6 (Fig. 3) had the highest expression in the tumors, where they were identified as integration sites, indicating an activating effect of the integrated retrovirus on these genes. There was no similar correlation between the remaining integrations and the expression of the nearby genes (data not shown), indicating that, although some of the genes in vicinity of the retroviral integrations seem to be affected by insertional mutagenesis, the majority of the insertions have little or no effect on expression levels of the genes within 10 kbs.
Table 2.
Overview of deregulated of genes within 10 kbs of a retroviral integration. Tumor (T) vs. normal (N) fold change is given.
| Gene | Description | Thymus tumors (fold change T vs. N) | Spleen tumors (fold change T vs. N) |
|---|---|---|---|
| Gfi1 | Growth factor independent 1 | 3.2* | 10.8** |
| Ppie | Peptidylprolyl isomerase E | 2.0** | 1.7** |
| Ankrd22 | Ankyrin repeat domain 22 | –1.7* | –3.8** |
| Fam49a | Family with sequence similarity 49, member A | –4.7** | –2.8* |
| Arntl | Aryl hydrocarbon receptor nuclear translocator-like | –3.2* | –2.0** |
| Sh2d1a | SH2 domain protein 1A | 6.0** | |
| Sh3gl3 | SH3-domain GRB2-like 3 | 5.0* | |
| Myb | Myeloblastosis oncogene | 2.6** | |
| Trim37 | Tripartite motif-containing 37 | 1.7** | |
| Fgd2 | FYVE, RhoGEF and PH domain containing 2 | –2.3* | |
| Cask | Calcium/calmodulin-dependent serine protein kinase | –2.0* | |
| Fli1 | Friend leukemia integration 1 | –1.7* | |
| Evi5 | Ecotropic viral integration site 5 | –1.7* | |
| Txnrd2 | Thioredoxin reductase 2 | 2.2** | |
| Mr1 | Major histocompatibility complex, class I-related | –5.8** | |
| Kremen1 | Kringle containing transmembrane protein 1 | –5.3* | |
| Stx6 | Syntaxin 6 | –3.1** | |
| Stat1 | Signal transducer and activator of transcription 1 | –2.0* | |
| Micall2 | MICAL-like 2 | –1.8* |
p ≤ 0.05.
p ≤ 0.005.
Fig. 3.
Dot plots showing expression of (A) Sh3gl3, (B) Cask, (C) Mr1, and (D) Stx6 for the individual tumors. NS: normal spleen; NT, normal thymus; TS, tumor spleen; TT, tumor thymus. Tumor ID for tumors with integrations in the specific genes, as well as mean values and standard deviations are indicated.
2.5. Genes associated with S-phase initiation are deregulated in murine T-LBL
We continued the analysis to identify new genes potentially associated with the development of murine T-LBLs. Tumor samples were compared to healthy controls within the two tissue groups, where genes with a ±1.5-fold change in tumor relative to normal sample and p ≤ 0.05 were considered differentially expressed. The main affected pathways in both tissue groups included downregulation of JAK-STAT pathway, MAPK pathway, p53 pathway and genes involved in cytokine receptor interactions (data not shown), whereas cell-cycle associated genes were generally upregulated in both thymus and spleen tumors (Supplementary Table 1). There was an overall increased expression of genes involved in the initiation of replication, including genes that constitute the origin recognition (Orc) and minichromosome maintenance (Mcm) complexes, as well as E2f, Cdt1, Ccne2, Ccna2, Cdc6, Cdc7, Cdc45, CDk2, Dbf4, Rpa and DNA polymerase genes. This, together with the simultaneous downregulation of negative regulators of G1/S-phase (Ccng2 and Cdkn1b), suggested a general increase in cell cycle progression in murine T-LBLs. Surprisingly, we also observed increased expression of genes commonly known to induce apoptosis (Bak, Bax, and Pdcd1) and cell cycle check point repressors (p21, p16 and p18) as well as downregulation of antiapoptotic genes (Bcl2 and Bcl2l1).
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.leukres.2013.04.012.
3. Discussion
Insertional mutagenesis in MLV infected mice has been widely used to study lymphoma biology, and several genes, that have since been associated with the development of human lymphomas, including Myc [15], Myb [16], D-type cyclines [17] and RasGrp1 [18] have been identified using these models. In this study, we aimed to identify new genes and pathways deregulated in murine T-LBL by performing integration analysis and gene expression profiling on tumors induced by the SL3-3 MLV. A total of 95 integration sites were detected in 19 SL3-3 3mEgre and SL3-3 3mGR induced tumors targeting 52 different genes. For 9 different genes more than one retroviral integration site was observed pr gene in the same tumor (Table 1). The exact genomic nucleotide positions of the integrations were not similar illustrating that two independent integration events into the same gene had occurred within the same tumor. Since integration sites were determined on pools of tumor cells and not individual cell it was not possible to decipher whether the original integration event occurred in one or two different cells. Due to limited material the fraction or clonality of cells harboring specific integrations were not evaluated but in former studies [14] insertions have been documented to be mono or oligo clonal thereby representing relative high fractions of tumor cells
Of the 52 different genes 19 of could be assigned a function in cell cycle progression or apoptosis, and 15 were directly involved in G1/S-phase transition (Supplementary Table 2). We found recurring integrations in Gfi1, Evi5, Myc, RasGrp1, Rras2, and Ccnd1, which are well known CISs from other murine lymphoma models [19–21] and have a direct or indirect effect on G1/S-phase transition [22–24]. Previously studies have shown that retroviral integrations in many of these loci correlate with high levels of mRNA [21,26,27]. By performing global gene expression profiling on the T-LBL tumors, we could determine both the effect of the integrated retrovirus on the nearby genes and the pathways that were deregulated in the tumors compared to normal tissue. In the experimental setting, we included two groups of tumors induced by SL3-3 with mutations in the 3mGR and 3mEgre binding sites, respectively. Both mutants give rise to T-cell lymphoblastic lymphomas, but with significantly different latencies [7]. Based on a minimum of ±1.5-fold changes, there was no difference in the gene expression profile when comparing tumors induced by the two virus variants. This may be due to the low number of samples in each group, but may also be due to smaller differences in gene expression that are still significant, but not detected due to the cut-off at ±1.5-fold change. Alternatively, the difference in latency of the two viruses may be caused by changes in expression of genes that do not differ on the mRNA level, but have varying protein expression. We also speculate that the latency periods reflect the speed of tumor development. If the only difference between the 3mGR and 3mEgre induced tumors is that the 3mGR tumor cells are pushed faster through the G1 checkpoint than the 3mEgre tumor cells, and the cell-cycle related gene products are subsequently degraded, this may not be reflected in the overall gene expression profile. Based on these observations, the remaining gene expression analyses were performed with no regard to the virus variant.
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.leukres. 2013.04.012.
Although some genes (Gfi1, Ppie, Ankrd, Fam49a, and Arntl) were generally up- or downregulated in both thymus and spleen tumors, many were specifically deregulated in either spleen (Sh2d1a, Sh3gl3, Myb, Trim37, Fgd2, Cask, Fli1, and Evi5) or thymus (Txnrd2, Mr1, Kremen1, Stx6, Stat1, and Micall2). Sh2d1a, Cask, Mr1, and Stx6 showed the highest upregulation in the tumors where they were found as integrations, indicating an activating effect of the integrated retrovirus on these genes. The expression of Notch1, Myc, Rasgrp1, Rras2 and the D-type cyclines, which are major common integration target in many MLV induced lymphomas [20,22,28,30], remained unchanged in both overall gene expression and in the individual tumors where they were found as integrations. Although our results indicate that the presence of a virus in the vicinity of a gene does not always alter its expression, we cannot exclude whether or not there is an oncogenic effect of the integrations on protein stability or alternative splicing, as we only analyzed the summarized gene expression.
Gene expression profiling identified additional upregulation of the Mcm–Orc pathway (Cdc6, Cdc7, Dbf4, Cdc45, Rpa1-3, Cdk2, Cdt1, Ccne2, Ccna2, E2f, Mcm, Orc, and DNA polymerase genes) and simultaneous down-regulation of cell cycle inhibitors (Ccng2 and Cdkn1b) [30,31], which were not found as retroviral targets by the integration analysis, but have well-established functions in G1–S-phase transition [32–34]. Although this might suggest that the murine T-LBLs have a general increased S-phase initiation, we further observed an increase in genes commonly known to induce apoptosis [35] or inhibit cell cycle progression (Bax, Bak, p21, p16 and p18) and decrease in anti-apoptotic genes (Bcl2 and Bcl2l1). While this may reflect increased apoptosis in the tumors, there was no upregulation in the expression of downstream effectors, such as caspases (data not shown), although there may exist a caspase-independent activation of apoptosis through calpains, cathepsins, granzymes, and the proteasome complex, which all have been implicated in promoting cell death [36–38]. There is some evidence that regulators of the cell death programme are also able to affect cell cycle progression [39–42]. It has been shown that both Bcl2 and Bcl2l1 are able to inhibit cell growth at the transition from the G0 stage into S-phase, and that this function can be reduced by co-expression of Bax [39]. A decrease in cell proliferation has been observed in Bcl2 overexpressing Jurkat cells [42], supporting a function for Bcl2 in the inhibition of cellular proliferation. Furthermore, upregulation of pro-apoptotic Bcl2 family members, as Baxα, can facilitate the S-phase entry in T cells, whereas Bcl2 delays this entry via modulation of p27Kip1 [41]. Whether or not our observations of increased expression in Bax and Bak and decreased expression of Bcl2 reflect increased apoptosis or increased S-phase entry is still unclear.
We further found upregulation of several cyclin-dependent kinase (Cdk) inhibitors (p21, p16 and p18) which are known to bind and antagonize the activity of Cyclin/Cdk complexes, at the G1/S-phase in particular. An alternative role as negative regulators of apoptosis has been demonstrated for some members of the INK family, as p19 [43], indicating that their overexpression might contribute to progression through the G1 checkpoint. We speculate whether the upregulation in Bax, Bak, p16, p18, p21 and the Mcm–Orc pathway together with a downregulation in Bcl2 and Bcl2l1, reflect increased S-phase entry in the murine T-LBL cells.
Moreover, Pdcd1/Pd1, which is associated with mechanisms of tumor immune evasion [44–47], was upregulated 14-fold in the spleen tumors. Activation of the Pdcd1 pathway contributes to tumor progression by negatively regulating TCR signaling and thus, effector T-cell responses, such as cytokine secretion [48,49], which also correlates well with our observations that the vast majority of the genes involved in cytokine signaling were significantly down-regulated (data not shown).
The chromosomal abnormalities that lead to development of human T-LBLs are poorly characterized. The few studies that address the T-cell lymphoma biology show that TAL1, HOX11, LYL1, LMO1 and LMO2 are frequently overexpressed in patients with TLBL [51,52]. However, we did not find any upregulation in these genes in our T-LBL model.
In conclusion, both the integration analysis and patterns of mRNA expression identified by gene expression profiling strongly indicate that genes involved in G1/S-phase transition and/or S-phase initiation are deregulated in murine T-LBLs.
4. Materials and methods
4.1. Identification of integration sites
Newborn mice of the NMRI inbred strain were inoculated with the SL3-3 MLV bearing mutations in Gr (SL3-3 3mGR) and Egre (SL3-3 3mEgre) binding sites as described previously [7]. On sickness, mice were sacrificed and splenic and thymic tumors were removed. Genomic DNA was isolated using the DNeasy Tissue kit (Qiagen) and retroviral integration sites were identified by a splinklerette-based PCR and sequencing as described previously [50]. One tumor was analyzed from each mouse and an overview is presented in Table 1.
4.2. Tumors and tissue
The tumors were characterized by an experienced pathologist as T-cell lymphoblastic lymphomas based on immunohistopathological examination of liver, spleen and lymph nodes [7]. For tumors induced by SL3-3 3mEgre, clonality was analyzed by Southern blotting where clonal TCRß but not Ig(κ) light chain gene rearrangements were found in all investigated tumors, consistent with T-cell lymphoma [7]. Normal thymus, spleen and lymph node tissue was dissected from six 4 weeks old NMRI inbred mice and pooled in two groups of three samples.
4.3. RNA extraction and array hybridization
Total RNA was purified using the TRIzol® reagent (Invitrogen) and mirVana™ miRNA isolation kit (Ambion). 10–20 mg of snap-frozen tissue was cut on dry ice and homogenized in 1 mL TRIzol®. Samples were added 200 μL chloroform and centrifuged at 12,000 × g at 4 °C where after the aqueous phase was dissolved in 1.25 times its volume in 100% ethanol and transferred to a mirVana™ filter cartridge. Samples were centrifuged, washed and eluted. RNA concentration was measured on a Nanodrop where an OD260/OD280 ratio of 1.9–2.2 was considered to indicate pure RNA. RNA quality and integrity were measured on a Bioanalyzer where a 28S/18S ratio of 1.7–2.3 and RIN values of 9.4–10.0 indicated that intact RNA. cDNA was synthetized according to the Ambion WT Expression Kit (Ambion). Samples were labeled and hybridized to the Mouse Exon 1.0 ST Arrays (Affymetrix) according to the GeneChip® WT Terminal Labeling and Hybridization manual (Ambion). Arrays were scanned using the Affymetrix Scanner and digital images were processed by the GeneChip® Operating Software v1.4 to generate .CEL intensity files.
4.4. Data analysis
Data from the exon arrays were processed using Partek Genomic Suite 6.4 (Partek Inc., St. Louis, MO). The July 2007 mouse (Mus musculus) genome Build 37 assembly by NCBI and the Mouse Genome Sequencing Consortium were used as reference genome. Exons were summarized to genes to generate an overall expression signal value of the single represented gene. Differentially expressed genes were identified using Analysis of Variance (ANOVA). A tumor-related list of significantly differentially expressed genes with p ≤ 0.05 and ±2-fold variations was obtained. Unsupervised hierarchical clustering was performed using Pearson's dissimilarity and Ward's method.
Supplementary Material
Acknowledgements
We are very grateful for the technical assistance of Lone Højgaard Nielsen on RNA purification and Maria Jensen on gene expression profiling. This work was supported by the PhD School for Genetic Medicine, The Danish Cancer Research Foundation, The Danish Cancer Society, and The Danish Research Council for Health and Disease.
Funding
This work was supported by the PhD School for Genetic Medicine, The Danish Cancer Research Foundation, The Danish Cancer Society, and The Danish Research Council for Health and Disease.
Footnotes
Contributions: F.S.P. and K.D. contributed equally to this work. M.J.D., K.D., F.S.P. and H.E.J. participated in the design and coordination of the study. M.J.D. and K.D. analyzed the results. M.J.D. drafted the manuscript. D.E.M. performed the pathogenicity study. L.B.L. dissected the murine control tissue. M.W. performed the integration analysis. All authors read and approved the manuscript.
Conflict of interests
All authors declare no competing interests.
References
- 1.Burkhardt B, Woessmann W, Zimmermann M, Kontny U, Vormoor J, Doerffel W, et al. Impact of cranial radiotherapy on central nervous system prophylaxis in children and adolescents with central nervous system-negative stage III or IV lymphoblastic lymphoma. J Clin Oncol. 2006;24:491–9. doi: 10.1200/JCO.2005.02.2707. [DOI] [PubMed] [Google Scholar]
- 2.Reiter A, Schrappe M, Ludwig W-D, Tiemann M, Parwaresch R, Schirg E, et al. Intensive ALL-type therapy without local radiotherapy provides a 90% event-free survival for children with T-cell lymphoblastic lymphoma: a BFM Group report. Hematology. 2012:416–21. [PubMed] [Google Scholar]
- 3.Abromowitch M, Sposto R, Perkins S, Zwick D, Siegel S, Finlay J, et al. Shortened intensified multi-agent chemotherapy and non-cross resistant maintenance therapy for advanced lymphoblastic lymphoma in children and adolescents: report from the Children's Oncology Group. Br J Haematol. 2008;143:261–7. doi: 10.1111/j.1365-2141.2008.07320.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Morse HC, III, Anver MR, Fredrickson TN, Haines DC, Harris AW, Harris NL, et al. Hematopathology subcommittee of the Mouse Models of Human Cancers Consortium. Bethesda proposals for classification of lymphoid neoplasms in mice. Blood. 2002;100:246–58. doi: 10.1182/blood.v100.1.246. [DOI] [PubMed] [Google Scholar]
- 5.Hallberg B, Schmidt J, Luz A, Pedersen FS, Grundstrom T. SL3-3 enhancer factor 1 transcriptional activators are required for tumor formation by SL3-3 murine leukemia virus. J Virol. 1991;65:4177–81. doi: 10.1128/jvi.65.8.4177-4181.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ethelberg S, Hallberg B, Lovmand J, Schmidt J, Luz A, Grundström T, et al. Second-site proviral enhancer alterations in lymphomas induced by enhancer mutants of SL3-3 murine leukemia virus: negative effect of nuclear factor 1 binding site. Microbiology. 1997;71:1196–206. doi: 10.1128/jvi.71.2.1196-1206.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ejegod D, Sørensen KD, Mossbrugger I, Quintanilla-Martinez L, Schmidt J, Pedersen F. Control of pathogenicity and disease specificity of a T-lymphomagenic gammaretrovirus by E-box motifs but not by an overlapping glucocorticoid response element. J Virol. 2009;83:336–46. doi: 10.1128/JVI.01368-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sørensen KD, Quintanilla-martinez L, Kunder S, Schmidt J, Pedersen F. Mutation of all Runx (AML1/Core) sites in the enhancer of T-lymphomagenic SL3-3 murine leukemia virus unmasks a significant potential for myeloid leukemia induction and favors enhancer evolution toward induction of other disease patterns mutation of A. J Virol. 2004;78:13216–31. doi: 10.1128/JVI.78.23.13216-13231.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nieves A, Levy LS, Lenz J. Importance of a c-Myb binding site for lymphomagenesis by the retrovirus SL3-3. J Virol. 1997;71:1213–9. doi: 10.1128/jvi.71.2.1213-1219.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zaiman Al, Nieves A, Lenz J. CBF, Myb, and Ets binding sites are important for activity of the core I element of the murine retrovirus SL3-3 in T lymphocytes. J Virol. 1998;72:3129–37. doi: 10.1128/jvi.72.4.3129-3137.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Golemis EA, Speck NA, Hopkins N. Alignment of U3 region sequences of mammalian type C viruses: identification of highly conserved motifs and implications for enhancer design. J Virol. 1990;64:534–42. doi: 10.1128/jvi.64.2.534-542.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fan H, Johnson C. Insertional oncogenesis by non-acute retroviruses: implications for gene therapy. Viruses. 2011;3:398–422. doi: 10.3390/v3040398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pedersen FS, Sørensen AB. Pathogenesis of oncoviral infections. In: Kurth Reinhard, Bannert Norbert., editors. Retroviruses: Molecular Microbiology and Genomics. Horizon Press; Norfolk, UK: 2010. pp. 237–68. [Google Scholar]
- 14.Nielsen AA, Kjartansdóttir KR, Rasmussen MH, Sørensen AB, Wang B, Wabl M, et al. Activation of the brain-specific neurogranin gene in murine T-cell lymphomas by proviral insertional mutagenesis. Gene. 2009;442(1–2):55–62. doi: 10.1016/j.gene.2009.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kadam P, Steele P, Li YQ, Preisler H. Analysis of the c-myc and N-ras genes in acute myelogenous leukemia cells which manifest the constitutive expression of the c-myc gene. Anticancer Res. 1993;13:747–51. [PubMed] [Google Scholar]
- 16.Lahortiga I, De Keersmaecker K, Van Vlierberghe P, Graux C, Cauwelier B, Lambert F, et al. Duplication of the MYB oncogene in T cell acute lymphoblastic leukemia. Nat Genet. 2007;39:593–5. doi: 10.1038/ng2025. [DOI] [PubMed] [Google Scholar]
- 17.Cao X, Fan L, Fang C, Zhu D-X, Dong H-J, Wang D-M, et al. The expression of SOX11, cyclin D1, cyclin D2, and cyclin D3 in B-cell lymphocytic proliferative diseases. Med Oncol. 2011 doi: 10.1007/s12032-011-9937-5. [Epub ahead] [DOI] [PubMed] [Google Scholar]
- 18.Oki T, Kitaura J, Watanabe-Okochi N, Nishimura K, Maehara A, Uchida T, et al. Aberrant expression of RasGRP1 cooperates with gain-of-function NOTCH1 mutations in T-cell leukemogenesis. Leukemia. Epub ahead. 2011:1–8. doi: 10.1038/leu.2011.328. [DOI] [PubMed] [Google Scholar]
- 19.de Ridder J, Uren A, Kool J, Reinders M, Wessels L. Detecting statistically significant common insertion sites in retroviral insertional mutagenesis screens. PLoS Comput Biol. 2006;2:1530–42. doi: 10.1371/journal.pcbi.0020166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dabrowska MJ, Dybkaer K, Johnsen HE, Wang B, Wabl M, Pedersen F. Loss of MicroRNA targets in the 3′ untranslated region as a mechanism of retroviral insertional activation of growth factor independence 1. J Virol. 2009;83:8051–61. doi: 10.1128/JVI.00427-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Uren AG, Kool J, Matentzoglu K, Ridder JD, Uitert MV, Lagcher W, et al. Largescale mutagenesis in p19ARF- and p53-deficient mice identifies cancer genes and their collaborative networks. Cell. 2008;133:727–41. doi: 10.1016/j.cell.2008.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Korotayev K, Chaussepied M, Ginsberg D. ERK activation is regulated by E2F1 and is essential for E2F1-induced S phase entry. Cell Signal. 2008;20:1221–6. doi: 10.1016/j.cellsig.2008.02.012. [DOI] [PubMed] [Google Scholar]
- 23.Berns K, Martins C, Dannenberg J-H, Berns A, Riele H, Bernards  . p27 kip1 independent cell cycle regulation by MYC. Oncogene. 2000;19:4822–7. doi: 10.1038/sj.onc.1203879. [DOI] [PubMed] [Google Scholar]
- 24.Eldridge AG, Loktev AV, Hansen DV, Verschuren EW, Reimann JD, Jackson P. The evi5 oncogene regulates cyclin accumulation by stabilizing the anaphase-promoting complex inhibitor emi1. Cell. 2006;124:367–80. doi: 10.1016/j.cell.2005.10.038. [DOI] [PubMed] [Google Scholar]
- 26.Kim R, Trubetskoy A, Suzuki T, Jenkins NA, Copeland NG, Lenz J. Genome-based identification of cancer genes by proviral tagging in mouse retrovirus-induced T-cell lymphomas. J Virol. 2003;77:2056–62. doi: 10.1128/JVI.77.3.2056-2062.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Suzuki T, Shen H, Akagi K, Morse HC, Malley JD, Naiman DQ, et al. New genes involved in cancer identified by retroviral tagging. Nat Genet. 2002;32:166–74. doi: 10.1038/ng949. [DOI] [PubMed] [Google Scholar]
- 28.Dumortier A, Jeannet R, Kirstetter P, Kleinmann E, Sellars M, dos Santos NR, et al. Notch activation is an early and critical event during T-cell leukemogenesis in Ikaros-deficient mice. Mol Cell Biol. 2006;26:209–20. doi: 10.1128/MCB.26.1.209-220.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Toyoshima H, Hunter T. p27, a novel inhibitor of G1 cyclin-Cdk protein kinase activity, is related to p21. Cell. 1994;78:67–74. doi: 10.1016/0092-8674(94)90573-8. [DOI] [PubMed] [Google Scholar]
- 31.Bennin DA, Don ASA, Brake T, McKenzie JL, Rosenbaum H, Ortiz L, et al. Cyclin G2 associates with protein phosphatase 2A catalytic and regulatory B′ subunits in active complexes and induces nuclear aberrations and a G1/S phase cell cycle arrest. J Biol Chem. 2002;277:27449–67. doi: 10.1074/jbc.M111693200. [DOI] [PubMed] [Google Scholar]
- 32.Ohtani K, Degregori J, Nevins J. Regulation of the cyclin E gene by transcription factor E2F1. Proc Natl Acad Sci U S A. 1995;92:12146–50. doi: 10.1073/pnas.92.26.12146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sclafani R. Cdc7p-Dbf4p becomes famous in the cell cycle. J Cell Sci. 2000;113:2111–7. doi: 10.1242/jcs.113.12.2111. [DOI] [PubMed] [Google Scholar]
- 34.Schaarschmidt D. Human Mcm proteins at a replication origin during the G1 to S phase transition. Nucleic Acids Res. 2002;30:4176–85. doi: 10.1093/nar/gkf532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dewson G, Kluck R. Mechanisms by which Bak and Bax permeabilise mitochondria during apoptosis. J Cell Sci. 2009;16:2801–9. doi: 10.1242/jcs.038166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guicciardi ME, Deussing J, Miyoshi H, Bronk SF, Svingen PA, Peters C, et al. contributes to TNF-alpha-mediated hepatocyte apoptosis by promoting mitochondrial release of cytochrome c. J Clin Investig. 2000;106:1127–37. doi: 10.1172/JCI9914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stoka V, Turk B, Schendel SL, Kim TH, Cirman T, Snipas SJ, et al. Lysosomal protease pathways to apoptosis. Cleavage of bid, not pro-caspases, is the most likely route. J Biol Chem. 2001;276:3149–57. doi: 10.1074/jbc.M008944200. [DOI] [PubMed] [Google Scholar]
- 38.Bröker LE, Kruyt FAE, Giaccone G. Cell death independent of caspases: a review. Clin Cancer Res. 2005;11:3155–62. doi: 10.1158/1078-0432.CCR-04-2223. [DOI] [PubMed] [Google Scholar]
- 39.Reilly LAO, Huang DCS, Strasser A. The cell death inhibitor Bcl-2 and its influence control of cell cycle entry. EMBO J. 1996;15:6979–90. [PMC free article] [PubMed] [Google Scholar]
- 40.Zinkel S, Gross A, Yang E. BCL2 family in DNA damage and cell cycle control. Cell Death Differ. 2006;13:1351–9. doi: 10.1038/sj.cdd.4401987. [DOI] [PubMed] [Google Scholar]
- 41.Brady HJM, Gil-gomez G, Kirberg J, Berns A. Baxa perturbs T cell development and affects cell cycle entry of T cells. EMBO J. 1996;15:6991–7001. [PMC free article] [PubMed] [Google Scholar]
- 42.Johnson VL, Cooper IR, Jenkins JR, Chow S. Effects of differential overexpression of Bcl-2 on apoptosis, proliferation, and telomerase activity in Jurkat T cells. Exp Cell Res. 1999;251:175–84. doi: 10.1006/excr.1999.4557. [DOI] [PubMed] [Google Scholar]
- 43.Scassa ME, Ceruti JM, Marazita MC, Carcagno AL, Ca ET, Sirkin PF, et al. Critical review INK4 proteins, a family of mammalian CDK inhibitors with novel biological functions. EMBO J. 2007;59:419–26. doi: 10.1080/15216540701488358. [DOI] [PubMed] [Google Scholar]
- 44.Barber DL, Wherry E, Masopust D, Zhu B, Allison JP, Sharpe AH, et al. Restoring function in exhausted CD8 T cells during chronic viral infection. Nature. 2006;439:682–7. doi: 10.1038/nature04444. [DOI] [PubMed] [Google Scholar]
- 45.Trautmann L, Janbazian L, Chomont N, Said EA, Gimmig S, Bessette B, et al. Upregulation of PD-1 expression on HIV-specific CD8+ T cells leads to reversible immune dysfunction. Nat Med. 2006;12:1198–202. doi: 10.1038/nm1482. [DOI] [PubMed] [Google Scholar]
- 46.Petrovas C, Casazza JP, Brenchley JM, Price DA, Gostick E, Adams WC, et al. PD-1 is a regulator of virus-specific CD8+ T cell survival in HIV infection. J Exp Med. 2006;203:2281–92. doi: 10.1084/jem.20061496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mumprecht S, Schürch C, Schwaller J, Solenthaler M, Ochsenbein A. Programmed death 1 signaling on chronic myeloid leukemia-specific T cells results in T-cell exhaustion and disease progression. Blood. 2009;114:1528–36. doi: 10.1182/blood-2008-09-179697. [DOI] [PubMed] [Google Scholar]
- 48.Iwai Y, Ishida M, Tanaka Y, Okazaki T, Honjo T, Minato N. Involvement of PD-L1 on tumor cells in the escape from host immune system and tumor immunotherapy by PD-L1 blockade. Proc Natl Acad Sci. 2002;99:12293–7. doi: 10.1073/pnas.192461099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Blank C, Brown I, Peterson AC, Spiotto M, Iwai Y, Honjo T, et al. PD-L1/B7H-1 inhibits the effector phase of tumor rejection by T cell receptor (TCR) transgenic CD8+ T cells. Cancer Res. 2004;64:1140–5. doi: 10.1158/0008-5472.can-03-3259. [DOI] [PubMed] [Google Scholar]
- 50.Mikkers H, Allen J, Knipscheer P, Romeijn L, Hart A, Vink E, et al. High-throughput retroviral tagging to identify components of specific signaling pathways in cancer. Nat Genet. 2002;32:153–9. doi: 10.1038/ng950. [DOI] [PubMed] [Google Scholar]
- 51.Kaneko Y, Frizzera G, Maseki N, Sakurai M, Komada Y, Sakurai M, et al. A novel translocation, t(9;17)(q34;q23), in aggressive childhood lymphoblastic lymphoma. Leukemia. 1988;2:745–8. [PubMed] [Google Scholar]
- 52.Lones MA, Heerema NA, Le Beau MM, Sposto R, Perkins SL, Kadin ME, et al. Chromosome abnormalities in advanced stage lymphoblastic lymphoma of children and adolescents: a report from CCG-E08. Cancer Genet Cytogenet. 2007;172:1–11. doi: 10.1016/j.cancergencyto.2006.07.011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.