Abstract
Alkylation and oxidation constitute major routes of DNA damage induced by endogenous and exogenous genotoxic agents. Understanding the biological consequences of DNA lesions often necessitates the availability of oligodeoxyribonucleotide (ODN) substrates harboring these lesions, and sensitive and robust methods for validating the identities of these ODNs. Tandem mass spectrometry is well suited for meeting these latter analytical needs. In the present study, we evaluated how the incorporation of an ethyl group to different positions (i.e., O2, N3 and O4) of thymine and the oxidation of its 5-methyl carbon impact collisionally activated dissociation (CAD) pathways of electrospray-produced deprotonated ions of ODNs harboring these thymine modifications. Unlike an unmodified thymine, which often manifests poor cleavage of the C3′-O3′ bond, the incorporation of an alkyl group to the O2 position and, to a much lesser extent, the O4 position, but not the N3 position of thymine, led to facile cleavage of the C3′-O3′ bond on the 3′ side of the modified thymine. Similar efficient chain cleavage was observed when thymine was oxidized to 5-formyluracil or 5-carboxyluracil, but not 5-hydroxymethyluracil. Additionally, with the support of computational modeling, we revealed that proton affinity and acidity of the modified nucleobases govern the fragmentation of ODNs containing the alkylated and oxidized thymidine derivatives, respectively. These results provided important insights into the effects of thymine modifications on ODN fragmentation.
Introduction
The integrity and stability of DNA are challenged by both exogenous and endogenous agents, which generate a diverse array of modifications in DNA. These include, but are not limited to, abasic sites, single-nucleobase lesions, DNA-DNA crosslinks, and strand breaks [1, 2]. Owing to their widespread presence and important biological consequences, alkylated and oxidized nucleobases are two heavily investigated groups of DNA lesions [3-6].
DNA possesses multiple nucleophilic sites [7], and alkylating agents are ubiquitously present in the environment [8, 9] and within the living cells [10]. Hence, DNA alkylation is generally unavoidable. Through SN1/SN2 reaction, alkylating agents can react with the nitrogen or oxygen atoms of nucleobases to generate a variety of covalent adducts [11].
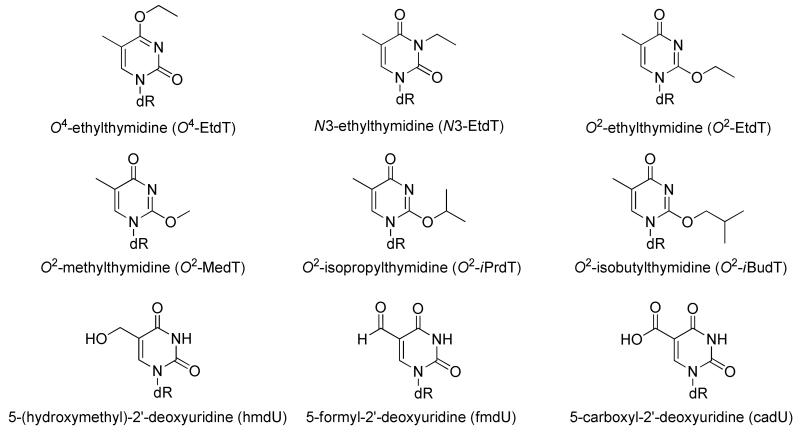
Reactive oxygen species (ROS), which are generated inside cells by various exogenous and endogenous agents and can attack DNA, may play a key role in the development of cancer and other human diseases [12, 13]. In this context, the 5-(2′-deoxyuridinyl)methyl radical can form on thymidine via either hydroxyl radical-mediated hydrogen atom abstraction from the 5-methyl group [14, 15] or deprotonation of the radical cation intermediate of thymidine produced by one-electron oxidation [16]. The 5-methyl radical is subsequently trapped by oxygen and eventually generates 5-(hydroxymethyl)-2′-deoxyuridine (hmdU) and 5-formyl-2′-deoxyuridine (fmdU) [17]. 5-Carboxyl-2′-deoxyuridine (cadU) can be generated from γ-irradiation or 2-methyl-1,4-naphthoquinone-sensitized photooxidation of aqueous solution of thymidine under aerobic conditions [18]. In addition, cadU can be generated in DNA upon iterative oxidation of thymidine by thymine hydroxylase [19].
Mass spectrometry (MS) and tandem MS (MS/MS) have evolved as important tools for the structure elucidation of oligodeoxyribonucleotides (ODNs) carrying normal and/or modified nucleobases [20-23]. To facilitate the application of MS for ODN analysis, it is important to fully understand the behaviors and mechanisms of ODN fragmentation. Although many factors can influence the fragmentation of ODNs, it is now accepted that proton transfer from the phosphate backbone to the nucleobase initiates the nucleobase loss, which forges the subsequent cleavage of the adjacent 3′ C-O bond and the formation of wn and its complementary [an-Base] ions [24-26]. As a result, proton affinities of nucleobases play an important role in the fragmentation patterns of ODNs. In this context, thymidine has the lowest proton affinity among the four canonical nucleosides, which is consistent with the observations that strand cleavages at the 3′ side of dA, dC, and dG are more facile than that at the 3′ side of dT upon the collisional activation of deprotonated ions of ODNs [27, 28]. Currently, there is no systematic evaluation about how incorporation of alkylated and oxidized thymidine derivatives into ODNs affects their fragmentation. In this study, we examined the fragmentation behavior of ODNs harboring these two types of modified thymidines (Scheme 1), with the goal to establishing a foundation for the future structural elucidation of ODNs housing these and other modified thymidines by MS/MS.
Scheme 1.
Experimental
Synthesis and Purification of Modified Oligodeoxyribonucleotides
ODNs were synthesized on controlled pore glass (CPG) solid support resin using a Beckman Oligo 1000S DNA synthesizer at 1 μmole scale (Fullerton, CA). The phosphoramidite building blocks of alkylated and oxidized thymidines were independently synthesized following established literature procedures [23, 29-34]. All modified phosphoramidites were dissolved in anhydrous acetonitrile at a concentration of 0.067 M. Commercially available phosphoramidites (ultramild) were used for the incorporation of the unmodified nucleotides (Glen Research Inc., Sterling, VA) following the standard ODN assembly protocol.
All ODNs, 5′-TTXTT-3′ (‘X’ represents dA, dT, dC, dG and alkylated thymidines) and 5′-ATGGCGXGCTAT-3′ (‘X’ designates unmodified, alkylated and oxidized thymidines) were cleaved from the solid support with concentrated ammonium hydroxide at room temperature for 1 h. The 5-mer ODNs were used directly for ESI-MS and MS/MS experiments, whereas the 12-mer ODNs were purified by HPLC prior to mass spectrometric analyses.
HPLC separation of synthetic ODNs was performed on an Agilent 1100 HPLC system with a Kinetex XB-C18 column (250 × 4.60 mm, 5 μm in particle size and 100 Å in pore size; Phenomenex Inc., Torrance, CA). For the purification of ODNs, a triethylammonium acetate (TEAA) buffer (50 mM, pH 6.8, Solution A) and a mixture of solution A and acetonitrile (70/30, v/v, Solution B) were employed as mobile phases. The flow rate was 0.8 mL/min, and the gradient profile in terms of solution B was 5-25% in 5 min followed by 25-65% in 60 min.
Electrospray Ionization Mass Spectrometry (ESI-MS)
ESI-MS and MS/MS experiments were performed on an LCQ Deca XP ion-trap mass spectrometer (ThermoFinnigan, San Jose, CA). The ODNs were prepared in a 10-μM solution containing water and methanol (50:50, v/v). These solutions were infused directly into the mass spectrometer with a syringe pump at a flow rate of 10.0 μL/min. The mass spectrometer was operated in the negative-ion mode. The spray voltage was 4.0 kV, and the temperature for the ion transport tube was maintained at 300°C. Tandem mass spectra were collected by selecting the deprotonated ions for collisional activation, and the mass width for precursor ion selection was set at 3 m/z units for all ODNs. The normalized collision energy, which represented the resonance excitation voltage, was reported in the present study as the percentage value of the maximum available voltage, which was 5 V peak-to-peak. The activation Q and activation time were 0.250 and 30 ms, respectively. Each spectrum was an average of approximately 60 scans, and the acquisition time for each scan was 1000 ms. The fraction of a given ion was calculated based on its peak intensity.
Computational Methods
All calculations were carried out using density functional theory (DFT) methods included in the Gaussian 09W package [35]. Geometry optimizations and frequency calculations (without scaling) were performed at the B3LYP/6-31+G(d) level of theory in the gas phase. Single-point energies (SPE) were calculated using B3LYP/6-311+G(2d,p) on B3LYP/6-31+G(d) optimized structures [. The corrected energy (Ecorrected) for a given molecule is equivalent to the sum of the SPE [at 6-311+G(2d,p)] and the zero point correction energy [at 6-31+G(d)].
Our choice of basis sets stemmed from earlier studies showing that 6-31G(d) geometries yielded final relative energies in excellent agreement with geometries obtained using the larger basis set, 6-31+G(d,p) [36, 37], as well as its successful application in our previous computational work [38]. These studies also showed that the C3′ and C5′ hydroxyl groups of the 2′-deoxyribose interact with nucleobase, which cannot occur in duplex DNA [36, 37]. As such, all C3′ and C5′ hydroxyl groups in the nucleosides and 2′-deoxyribose structures modeled herein were replaced with methoxyl groups for more accurate calculations. Similarly, for proton affinity calculations, the hydrogen atom on the N1 position of the modified thymine bases was replaced with a methyl group. Although modified, all structures are referred in the text by their common name.
Proton affinity (PA; kcal/mol) at O2, N3, and O4 positions was calculated as follows, −PA = ΔH298K = [Hcorrected(M+H+) − Hcorrected(M)] − 5/2RT, where Hcorrected designates the zero point-corrected enthalpy, (M+H+) is the protonated ion, M is the neutral nucleobase, R = 1.987×10−3 kcal•K−1•mol−1, and T = 298 K. For the nucleobases protonated at O2 and O4, the PA was calculated for conformers with the proton orientated on either side of the oxygen. Similarly, PA for fmU and caU were determined with the carbonyl positioned toward O4 or C6. Acidity (kcal/mol) was calculated in terms of the change in enthalpy (ΔH) at the N1 position of uracil, 5-X-uracil (X = hm, fm, and ca), thymine, and O2-, N3-, and O4-alkylthymine derivatives [alkyl = methyl (Me), ethyl (Et), isopropyl (iPr), and isobutyl (iBu)] at the B3LYP/6-31+G(d) level of theory as follows, ΔH = [Hcorrected(H+) + Hcorrected(Base−)N1] − Hcorrected(BaseH)N1, where H+, Base−, and BaseH represent proton, deprotonated (N1 position) nucleobase, and neutral nucleobase, respectively. We also calculated the bond dissociation energy (ΔE; kcal/mol) for the nucleoside using SPE as follows, ΔE = Ecorrected(Base−) + Ecorrected(dR+)] − Ecorrected(nucleoside), where Ecorrected represents the zero point-corrected electronic energy, and Base−, dR+, and nucleoside refer to the deprotonated (N1 position) nucleobase, cationic 2′-deoxyribose, and neutral nucleoside, respectively. The lowest energy conformers are represented herein for PA, acidity, and BDE.
Results and Discussion
Fragmentation of 5′-TTXTT-3′
We first sought to assess how the fragmentation behaviors of ODNs are altered by the nature of the alkyl modifications on thymidine. Previously, it has been demonstrated that proton transfer from the backbone phosphate to nucleobase drives the nucleobase loss and subsequent strand cleavage, which is supported, in part, by the observations that the lowest proton affinity of thymidine is associated with a less facile strand cleavage at the 3′ side of this nucleoside [27, 28].
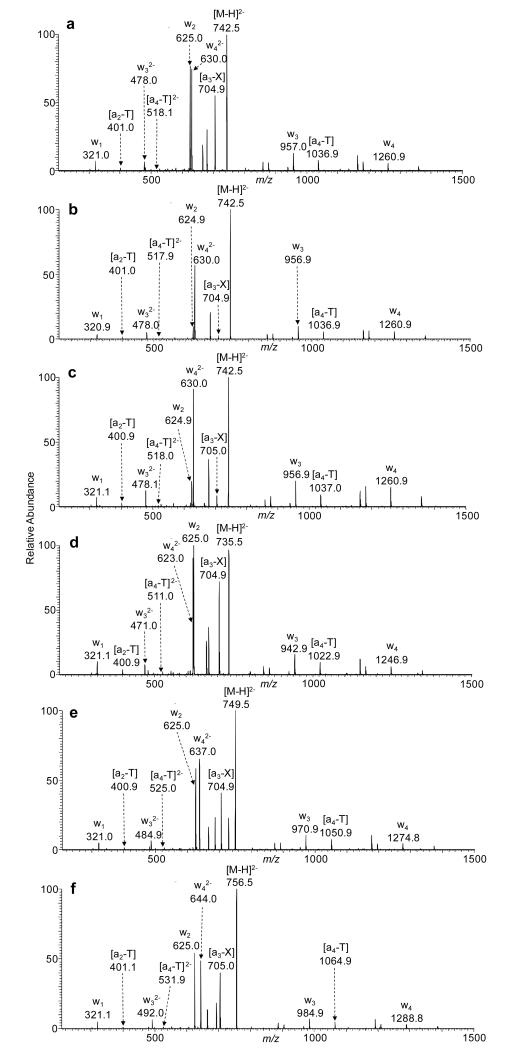
The MS/MS of the ESI-produced [M-2H]2− ions of 5′-TTXTT-3′ (‘X’ = alkylated dT) and 5′-TTNTT-3′ (‘N’ = dA, dT, dG and dC) are shown in Figure 1 and Figure S1, respectively. Our results revealed that the alkylation at thymidine, particularly at the O2 position of thymidine, results in a facile cleavage of the C-O bond at the 3′ of the modified thymidine, generating the w2 and [a3-X] ions. In this respect, we found that [a3-X] ion is more abundant than any other [an-Base] ions for the O2-EtdT-containing ODN (Figure 1), though an abundant w42− ion was also observed for all the T-rich pentameric ODNs studied here (Figure 1 and Figure S1).
Figure 1.
The product-ion spectra of the ESI-produced [M-2H]2− ions of d(TTXTT), where X = O2-EtdT (a), N3-EtdT (b), O4-EtdT (c), O2-MedT (d), O2-iPrdT (e), and O2-iBudT (f). The spectra were acquired at a normalized collision energy of 23%.
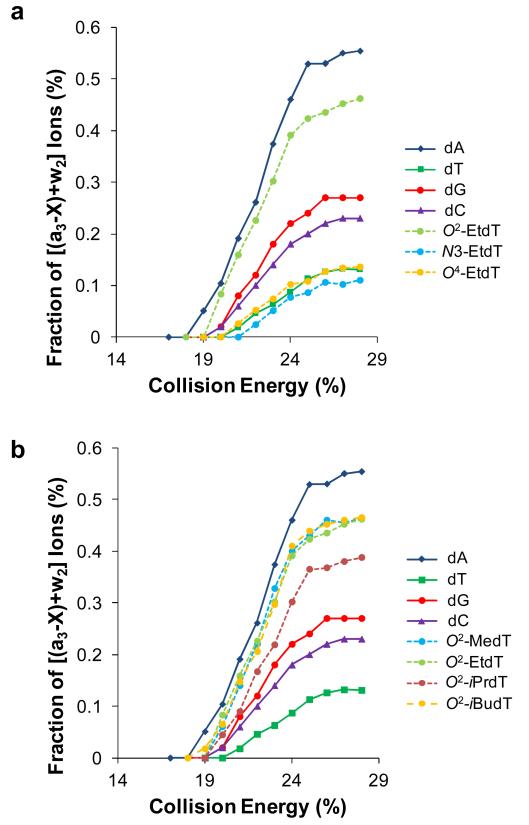
We next acquired MS/MS at different collisional energies to compare the fragmentation efficiencies of the 3′ C-O bond of these alkylated thymidines. Here, we use the relative abundances of a3-X and w2 ions, which both arise from the cleavage of C-O bond on the 3′ side of the modified thymidine, to directly compare the impact of alkylation on strand cleavage on the 3′ side of the modified thymine (Figure 2). Our results demonstrated that the strand cleavage at the 3′ side of the O2-EtdT occurs much more efficiently than that at the 3′ side of N3-EtdT or O4-EtdT. Facile chain cleavage was also observed at the 3′-side of other O2-alkylated thymidine derivatives, where the fragmentation patterns of ODNs containing O2-alkylthymidines are very similar to that of the corresponding dA-containing ODN (Figure 2). By contrast, the fragmentation behaviors of ODNs carrying O4- or N3-EtdT resemble more closely the respective dT-containing ODN (Figure 2). Additionally, we observe that this cleavage of unmodified thymidine is the least facile among all four canonical nucleosides (Figure 2), which is consistent with previous findings [25-28].
Figure 2.
Fraction of [(a3-X)+w2] ions in the MS/MS of the ESI-produced [M-2H]2− ions of d(TTXTT), where ‘X’ = dA, dT, dC, dG, O2-EtdT, N3-EtdT, O4-EtdT (a), and dA, dT, dC, dG, O2-MedT, O2-iPrdT and O2-iBudT (b).
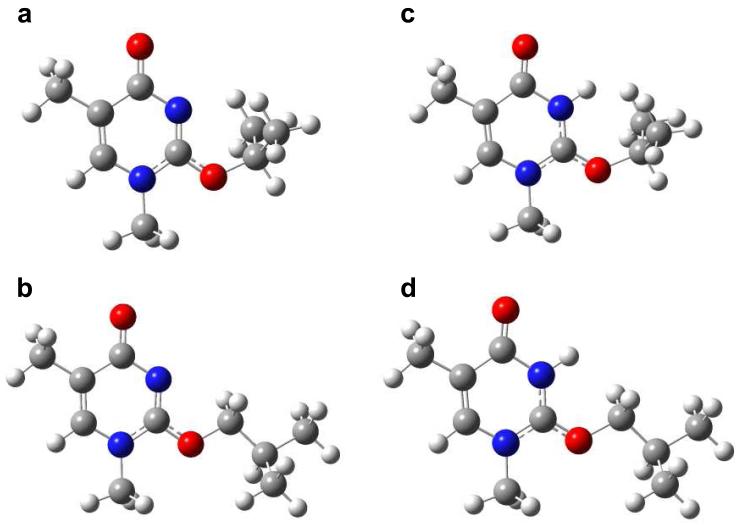
One interesting feature is that nearly all O2 alkylations result in similar patterns of collisionally activated dissociation (CAD) spectra, with the exception of O2-iPrdT (Figure 2b). The lower level of cleavage on the 3′ side of O2-iPrdT perhaps can be attributed to the inhibition of proton transfer from the phosphate to this modified nucleobase, as a consequence of steric hindrance imposed by the iPr group. In this vein, it is worth noting that the B3LYP/6-31+G(d)-optimized geometries revealed the steric hindrance conferred by the iPr moiety in O2-isopropylthymine, but not by the iBu group in O2-isobutylthymine, for the proton transfer to the highest proton affinity site in these modified nucleobases (i.e., the N3 position, Figure 3).
Figure 3.
B3LYP/6-31+G(d)-optimized geometries of N1-methyl-O2-isopropylthymine (a) and N1-methyl-O2-isobutylthymine (b). The corresponding optimized geometries for the [M+H]+ ions of these two modified thymine derivatives with protonation at the site with the highest proton affinity (i.e., N3 position) are shown in (c) and (d), respectively.
Fragmentation of 5′-ATGGCGXGCTAT-3′
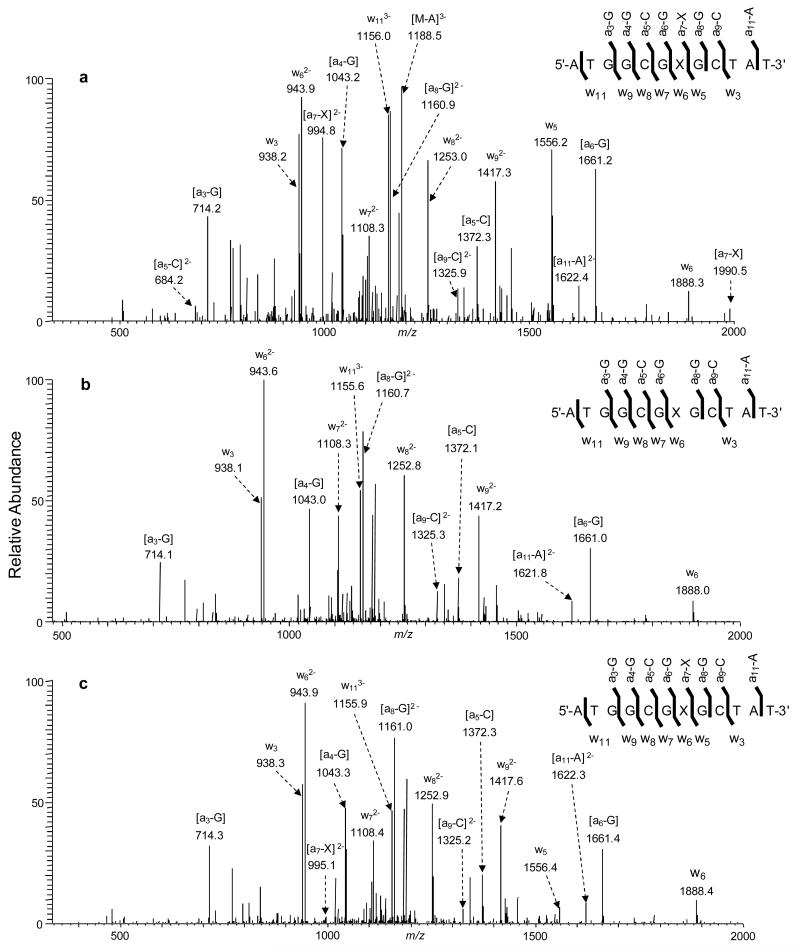
Having examined the effect of alkylation at different positions of thymine on the fragmentation of a T-rich ODN, we next explored whether the findings are general by using a 12-mer ODN with a diverse sequence context, i.e. 5′-ATGGCGXGCTAT-3′ (‘X’= O2-EtdT, N3-EtdT, and O4-EtdT). The MS/MS of the ESI-produced [M-3H]3− ions of these ODNs (Figure 4) showed that the [a7-X] and w5 ions are abundant in the MS/MS for the O2-EtdT-containing ODN; however, neither the [a7-X] nor w5 ion was observed for the corresponding N3-EtdT-containing ODN. This phenomenon is in good agreement with the findings made for the 5-mer T-rich ODNs (Figure 1). In addition, it is of note that the [a7-X] and w5 ions were observed readily in the MS/MS for the [M-2H]2− and [M-4H]4- ions of the O2-EtdT-containing 12-mer ODN (Figure S2), suggesting that the charge states of the precursor ion do not affect appreciably the facile cleavage on the 3′ side of the O2-alkylated thymidine derivatives.
Figure 4.
The product-ion spectra of the ESI-produced [M-3H]3− ions of d(ATGGCGXGCTAT), (a) X = O2-EtdT, (b) X = N3-EtdT, and (c) X = O4-EtdT. The spectra were acquired at a normalized collision energy of 30%.
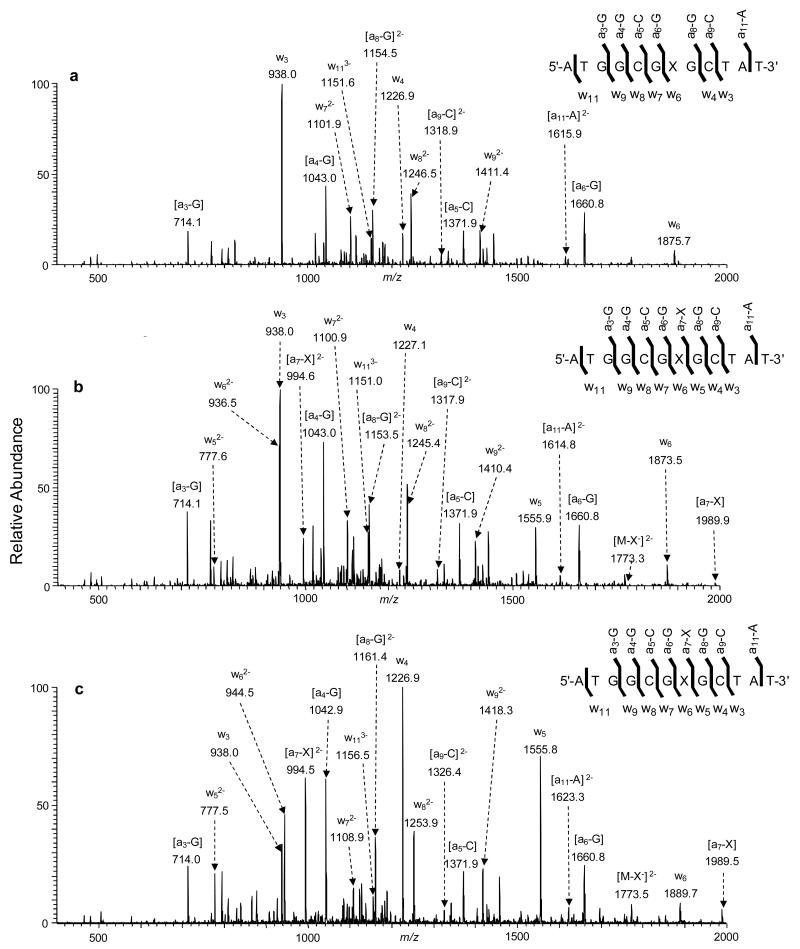
To further investigate the impact of thymidine modifications on strand cleavage, we extended our study to include three important oxidized thymidines. The MS/MS of the ESI-produced [M-3H]3− ions of the 12-mer 5′-ATGGCGXGCTAT-3′ ODNs (‘X’ = hmdU, fmdU and cadU) are displayed in Figure 5. It is important to note that neither the [a7-X] nor the w5 ion was observed for the 12-mer hmdU-containing ODNs, which is similar to what we found for the corresponding thymidine-bearing ODN. This result indicates that, similar to N3-alkylation, the oxidation of the 5-methyl functionality to a hydroxymethyl group exerts a minimal effect on ODN fragmentation.
Figure 5.
The product-ion spectra of the ESI-produced [M-3H]3− ions of d(ATGGCGXGCTAT), (a) X = hmdU, (b) X = fmdU, and (c) X = cadU. The spectra were acquired at a normalized collision energy of 30%.
Interestingly, a new [M-X−]2− ion was observed in the ESI-MS/MS of ODNs containing fmdU and cadU (Figure 5b and 5c), which is absent in all the MS/MS for the ODNs containing alkylated thymidines (Figure 4) or hmdU (Figure 5a). This finding aligns well with previous reports showing that the formation of 5-formyluracil weakens the N-glycosidic bond, and the modified nucleobase can be selectively released from DNA as a deprotonated ion [21, 39]. Along this line, results from a recent DFT calculation predict that the oxidation of the 5-methyl group to a formyl or carboxyl group, but not a hydroxymethyl functionality, results in more facile cleavage of the N-glycosidic bond via a hydroxide ion-mediated nucleophilic reaction [38]. It is also important to note that, at a relatively low collisional energy (16%), the [M-X−]2− ion is more abundant (Figure S3), and the MS/MS of the cadU-containing ODN displays more abundant [M-X−]2− ion than the corresponding fmdU-containing ODN. Furthermore, the [M-X−]2− ion was still not detectable for the hmdU-containing ODN at this collisional energy.
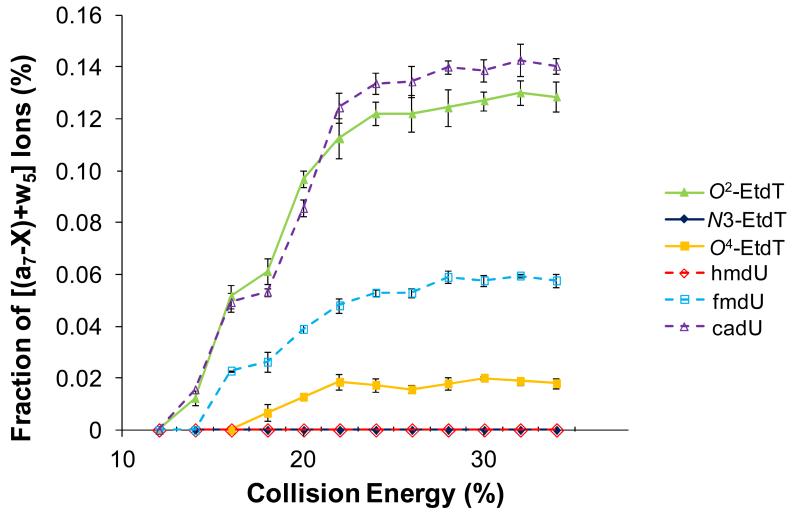
We next obtained the CAD spectra at different collisional energies for ODNs bearing both types of modified thymidine-containing ODNs (Figure 6). It is obvious that alkylation at the O2 and O4 positions, and the oxidation of the 5-methyl group to a formyl or carboxyl group, have a significant impact on ODN fragmentation. However, neither the alkylation at the N3 of thymidine nor the oxidation of the 5-methyl functionality to a hydroxymethyl group exerts substantial effect on the fragmentation behavior. With these results, we can determine the order of cleavage efficiency for the three regiomeric alkylated thymidine to be O2 > O4 > N3. Furthermore, the above results indicate that these two types of modified thymidines undergo different fragmentation pathways.
Figure 6.
Fraction of [(a7-X)+w5] ions in the MS/MS of the ESI-produced [M-3H]3− ions of d(ATGGCGXGCTAT) (‘X’ = O2-EtdT, N3-EtdT, O4-EtdT, hmdU, fmdU and cadU). Both the singly and doubly charged (a7-X) and w5 ions are used in fraction calculation.
Computational Studies
We next performed computational modeling to determine the major characteristics that impact the fragmentation pathways of ODNs carrying the modified thymidine derivatives (Table 1).
Table 1.
Gas-phase proton affinities (PA), acidity and bond dissociation energy (BDE) of alkylated and oxidized thymine bases
| Base | PA (kcal/mol) | Acidity (kcal/mol) | BDE (kcal/mol) |
|---|---|---|---|
| T | 208.19 | 332.22 | 133.92 |
| O2-MeT | 233.01 | 611.32 | 410.18 |
| O2-EtT | 234.54 | 609.18 | 407.95 |
| O2-iPrT | 235.47 | 607.52 | 406.19 |
| O2-iBuT | 235.28 | 608.08 | 406.77 |
| O4-EtT | 227.04 | 615.08 | 416.14 |
| N3-EtT | 213.33 | 617.01 | 416.41 |
| hmU | 205.99 | 332.81 | 134.93 |
| fmU | 213.61 | 315.73 | 118.54 |
| caU | 216.23 | 318.45 | 119.02 |
As indicated in Table 1, the two types of modified bases, i.e. alkylated and oxidized thymines, exhibit different proton affinities, gas-phase acidities and bond dissociation energies of the N-glycosidic bond. Despite the fact that both modifications result in elevated proton affinities of the nucleobases, the alkyl modifications introduce greater changes. The degree of increase in PA follows the order of O2-MedT ~ O2-EtdT ~ O2-iPrdT ~ O2-iBudT > O4-EtdT > N3-EtdT (Table 1), which parallels the preference of chain cleavage observed on the 3′ side of the modified thymidines (Figures 1-2 & 4-6). Whereas alkylation gives rise to elevated acidity of the N1 nitrogen and bond dissociation energy (BDE) of the N-glycosidic bond, the oxidation of the 5-methyl group in thymine to a formyl or carboxyl group decreases the acidity and BDE (Table 1), rendering the ODN more susceptible to lose the modified nucleobase as an anion. In contrast, oxidation of the 5-methyl carbon in thymine to a hydroxymethyl group introduces little change to the acidity or BDE (Table 1). These results are in keeping with the observation of the [M-X−]2− ion in the MS/MS of the ESI-produced [M-3H]3− ions of ODNs carrying fmdU and cadU, but not hmU (Figure 5 and Figure S3). Taken together, our results support that the proton affinity plays an important role in the fragmentation of ODNs containing O2-alkylated thymidines, whereas acidity and BDE appear to be the driving force for the fragmentation of ODNs containing fmdU or cadU.
Conclusions
We prepared ODNs containing alkylated and oxidized thymidine derivatives and examined the effects of these modified thymidines on the cleavage efficiencies on their 3′ C-O bond induced by low-energy collisional activation on an ion trap mass spectrometer. Additionally, we performed computational studies to rationalize the observed fragmentation patterns. The systematic CAD studies of deprotonated ions of the modified thymidine-containing ODNs revealed that alkylation at the O2 position of thymidine renders the ODNs more prone to fragmentation at the 3′ side of the modified base compared to the alkylation at N3 and O4 positions, with the alkylation at the N3 position displaying the lowest fragmentation efficiency. In this context, it is worth noting that the O2-alkylated thymidine derivatives are known to be resistant to DNA repair and persist in mammalian tissues [40, 41].
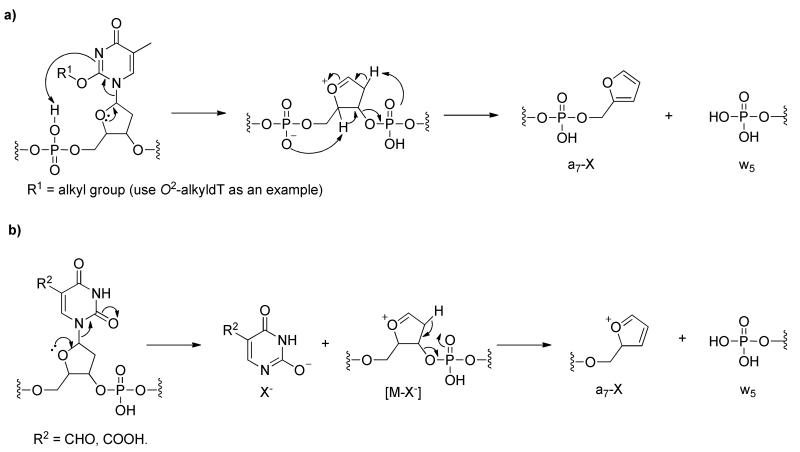
The specific alkylation and oxidation were observed to directly influence ODN fragmentation. The alkylation promotes fragmentation by increasing the proton affinity of the modified nucleobase, whereas oxidation decreases the stability of N-glycosidic bond, facilitating the fragmentation of lesion-containing ODNs. Furthermore, our computational studies provided insights underlying the experimental findings, revealing two different mechanisms contributing to the observed facile cleavages on the 3′ side of some alkylated and oxidized thymidine derivatives (Scheme 2, where fragmentations of the 12-mer ODNs were used as examples here). Together, the results from the present study built a solid foundation for understanding the fragmentations of modified thymidine-carrying ODNs.
Scheme 2.
Supplementary Material
Acknowledgements
This work was supported by the National Institutes of Health (R01 CA101864).
References
- 1.Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993;362:709–715. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- 2.Sander M, Cadet J, Casciano DA, Galloway SM, Marnett LJ, Novak RF, Pettit SD, Preston RJ, Skare JA, Williams GM, Van Houten B, Gollapudi BB. Proceedings of a workshop on DNA adducts: biological significance and applications to risk assessment Washington, DC, April 13-14, 2004. Toxicol. Appl. Pharmacol. 2005;208:1–20. doi: 10.1016/j.taap.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 3.Fu D, Calvo JA, Samson LD. Balancing repair and tolerance of DNA damage caused by alkylating agents. Nat. Rev. Cancer. 2012;12:104–120. doi: 10.1038/nrc3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Feng S, Jacobsen SE, Reik W. Epigenetic reprogramming in plant and animal development. Science. 2010;330:622–627. doi: 10.1126/science.1190614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kryston TB, Georgiev AB, Pissis P, Georgakilas AG. Role of oxidative stress and DNA damage in human carcinogenesis. Mutat. Res. 2011;711:193–201. doi: 10.1016/j.mrfmmm.2010.12.016. [DOI] [PubMed] [Google Scholar]
- 6.Rai P, Onder TT, Young JJ, McFaline JL, Pang B, Dedon PC, Weinberg RA. Continuous elimination of oxidized nucleotides is necessary to prevent rapid onset of cellular senescence. Proc. Natl. Acad. Sci. U. S. A. 2009;106:169–174. doi: 10.1073/pnas.0809834106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shrivastav N, Li D, Essigmann JM. Chemical biology of mutagenesis and DNA repair: cellular responses to DNA alkylation. Carcinogenesis. 2010;31:59–70. doi: 10.1093/carcin/bgp262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hecht SS. DNA adduct formation from tobacco-specific N-nitrosamines. Mutat. Res. 1999;424:127–142. doi: 10.1016/s0027-5107(99)00014-7. [DOI] [PubMed] [Google Scholar]
- 9.Hamilton JTG, McRoberts WC, Keppler F, Kalin RM, Harper DB. Chloride methylation by plant pectin: An efficient environmentally significant process. Science. 2003;301:206–209. doi: 10.1126/science.1085036. [DOI] [PubMed] [Google Scholar]
- 10.De Bont R, van Larebeke N. Endogenous DNA damage in humans: a review of quantitative data. Mutagenesis. 2004;19:169–185. doi: 10.1093/mutage/geh025. [DOI] [PubMed] [Google Scholar]
- 11.Drablos F, Feyzi E, Aas PA, Vaagbo CB, Kavli B, Bratlie MS, Pena-Diaz J, Otterlei M, Slupphaug G, Krokan HE. Alkylation damage in DNA and RNA - repair mechanisms and medical significance. DNA Repair. 2004;3:1389–1407. doi: 10.1016/j.dnarep.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 12.Wiseman H, Halliwell B. Damage to DNA by reactive oxygen and nitrogen species: role in inflammatory disease and progression to cancer. Biochem. J. 1996;313:17–29. doi: 10.1042/bj3130017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cadet J, Teoule R. Comparative-study of oxidation of nucleic-acid components by hydroxyl radicals, singlet oxygen and superoxide anion radicals. Photochem. Photobiol. 1978;28:661–667. doi: 10.1111/j.1751-1097.1978.tb06991.x. [DOI] [PubMed] [Google Scholar]
- 14.Romieu A, Bellon S, Gasparutto D, Cadet J. Synthesis and UV photolysis of oligodeoxynucleotides that contain 5-(phenylthiomethyl)-2′-deoxyuridine: a specific photolabile precursor of 5-(2′-deoxyuridilyl)methyl radical. Org. Lett. 2000;2:1085–1088. doi: 10.1021/ol005643y. [DOI] [PubMed] [Google Scholar]
- 15.Box HC, Dawidzik JB, Budzinski EE. Free radical-induced double lesions in DNA. Free Radic. Biol. Med. 2001;31:856–868. doi: 10.1016/s0891-5849(01)00653-0. [DOI] [PubMed] [Google Scholar]
- 16.Delatour T, Douki T, D’Ham C, Cadet J. Photosensitization of thymine nucleobase by benzophenone through energy transfer, hydrogen abstraction and one-electron oxidation. J. Photochem. Photobiol. B. 1998;44:191–198. [Google Scholar]
- 17.Lin G, Li L. Oxidation and reduction of the 5-(2-deoxyuridinyl)methyl radical. Angew. Chem. Int. Ed. Engl. 2013;52:5594–5598. doi: 10.1002/anie.201209454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Berthod T, Cadet J, Molko D. 5-carboxy-2′-deoxyuridine, a new photooxidation product of thymidine. J. Photochem. Photobiol. A Chem. 1997;104:97–104. [Google Scholar]
- 19.Thornburg LD, Lai MT, Wishnok JS, Stubbe J. A nonheme iron protein with heme tendencies - an Iinvestigation of the substrate-specificity of thymine hydroxylase. Biochemistry. 1993;32:14023–14033. doi: 10.1021/bi00213a036. [DOI] [PubMed] [Google Scholar]
- 20.Marzilli LA, Wang D, Kobertz WR, Essigmann JM, Vouros P. Mass spectral identification and positional mapping of aflatoxin B-1-guanine adducts in oligonucleotides. J. Am. Soc. Mass Spectrom. 1998;9:676–682. doi: 10.1016/S1044-0305(98)00039-7. [DOI] [PubMed] [Google Scholar]
- 21.Wang YS, Men LJ, Vivekananda S. Fragmentation of deprotonated ions of oligodeoxynucleotides carrying a 5-formyluracil or 2-aminoimidazolone. J. Am. Soc. Mass Spectrom. 2002;13:1190–1194. doi: 10.1016/S1044-0305(02)00452-X. [DOI] [PubMed] [Google Scholar]
- 22.Liu H, Yoo HJ, Hakansson K. Characterization of phosphate-containing metabolites by calcium adduction and electron capture dissociation. J. Am. Soc. Mass Spectrom. 2008;19:799–808. doi: 10.1016/j.jasms.2008.03.004. [DOI] [PubMed] [Google Scholar]
- 23.Andersen N, Wang P, Wang Y. Replication across regioisomeric ethylated thymidine lesions by purified DNA polymerases. Chem. Res. Toxicol. 2013;26:1730–8. doi: 10.1021/tx4002995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McLuckey SA, Vanberkel GJ, Glish GL. Tandem mass-spectrometry of small, multiply charged oligonucleotides. J. Am. Soc. Mass Spectrom. 1992;3:60–70. doi: 10.1016/1044-0305(92)85019-G. [DOI] [PubMed] [Google Scholar]
- 25.Wan KX, Gross J, Hillenkamp F, Gross ML. Fragmentation mechanisms of oligodeoxynucleotides studied by H/D exchange and electrospray ionization tandem mass spectrometry. J. Am. Soc. Mass Spectrom. 2001;12:193–205. doi: 10.1016/S1044-0305(00)00208-7. [DOI] [PubMed] [Google Scholar]
- 26.Wan KX, Gross ML. Fragmentation mechanisms of oligodeoxynucleotides: effects of replacing phosphates with methylphosphonates and thymines with other bases in T-rich sequences. J. Am. Soc. Mass Spectrom. 2001;12:580–589. doi: 10.1016/S1044-0305(01)00223-9. [DOI] [PubMed] [Google Scholar]
- 27.Liguori A, Napoli A, Sindona G. Survey of the proton affinities of adenine, cytosine, thymine and uracil dideoxyribonucleosides, deoxyribonucleosides and ribonucleosides. J. Mass Spectrom. 2000;35:139–144. doi: 10.1002/(SICI)1096-9888(200002)35:2<139::AID-JMS921>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 28.Green-Church KB, Limbach PA. Mononucleotide gas-phase proton affinities as determined by the kinetic method. J. Am. Soc. Mass Spectrom. 2000;11:24–32. doi: 10.1016/S1044-0305(99)00116-6. [DOI] [PubMed] [Google Scholar]
- 29.Xu YZ, Swann PF. A simple method for the solid-phase synthesis of oligodeoxynucleotides containing O4-alkylthymine. Nucleic Acids Res. 1990;18:4061–4065. doi: 10.1093/nar/18.14.4061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu YZ, Swann PF. Oligodeoxynucleotides containing O2-alkylthymine - synthesis and characterizations. Tetrahedron Lett. 1994;35:303–306. [Google Scholar]
- 31.Dai Q, He C. Syntheses of 5-formyl- and 5-carboxyl-dC containing DNA oligos as potential oxidation products of 5-hydroxymethylcytosine in DNA. Org. Lett. 2011;13:3446–3449. doi: 10.1021/ol201189n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shiau GT, Schinazi RF, Chen MS, Prusoff WH. Synthesis and biological-activities of 5-(hydroxymethyl, azidomethyl, or zminoethyl)-2′-deoxyuridine and related 5′-substituted analogs. J. Med. Chem. 1980;23:127–133. doi: 10.1021/jm00176a005. [DOI] [PubMed] [Google Scholar]
- 33.TardyPlanechaud S, Fujimoto J, Lin SS, Sowers LC. Solid phase synthesis and restriction endonuclease cleavage of oligodeoxynucleotides containing 5-(hydroxymethyl)-cytosine. Nucleic Acids Res. 1997;25:553–558. doi: 10.1093/nar/25.3.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Berthod T, Petillot Y, Guy A, Cadet J, Molko D. Synthesis of oligonucleotides containing 5-carboxy-2′-deoxyuridine at defined sites. J. Org. Chem. 1996;61:6075–6078. [Google Scholar]
- 35.Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, Scalmani G, Barone V, Mennucci B, Petersson GA, Nakatsuji H, Caricato M, Li X, Hratchian HP, Izmaylov AF, Bloino J, Zheng G, Sonnenberg JL, Hada M, Ehara M, Toyota K, Fukuda R, Hasegawa J, Ishida M, Nakajima T, Honda Y, Kitao O, Nakai H, Vreven T, Montgomery J, J A, Peralta JE, Ogliaro F, Bearpark M, Heyd JJ, Brothers E, Kudin KN, Staroverov VN, Keith T, Kobayashi R, Normand J, Raghavachari K, Rendell A, Burant JC, Iyengar SS, Tomasi J, Cossi M, Rega N, Millam JM, Klene M, Knox JE, Cross JB, Bakken V, Adamo C, Jaramillo J, Gomperts R, Stratmann RE, Yazyev O, Austin AJ, Cammi R, Pomelli C, Ochterski JW, Martin RL, Morokuma K, Zakrzewski VG, Voth GA, Salvador P, Dannenberg JJ, Dapprich S, Daniels AD, Farkas O, Foresman JB, Ortiz JV, Cioslowski J, Fox DJ. Gaussian 09, Revision B.01. Gaussian, Inc.; Wallingford CT: 2010. [Google Scholar]
- 36.Millen AL, Archibald LAB, Hunter KC, Wetmore SD. A kinetic and thermodynamic study of the glycosidic bond cleavage in deoxyuridine. J. Phys. Chem. B. 2007;111:3800–3812. doi: 10.1021/jp063841m. [DOI] [PubMed] [Google Scholar]
- 37.Millen AL, Wetmore SD. Glycosidic bond cleavage in deoxynucleotides — a density functional study1. Can. J. Chem. 2009;87:850–863. [Google Scholar]
- 38.Williams RT, Wang Y. A density functional theory study on the kinetics and thermodynamics of N-glycosidic bond cleavage in 5-substituted 2′-deoxycytidines. Biochemistry. 2012;51:6458–6462. doi: 10.1021/bi300797q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bjelland S, Eide L, Time RW, Stote R, Eftedal I, Volden G, Seeberg E. Oxidation of thymine to 5-formyluracil in DNA - mechanisms of formation, structural implications, and base excision by human cell-free-extracts. Biochemistry. 1995;34:14758–14764. doi: 10.1021/bi00045a017. [DOI] [PubMed] [Google Scholar]
- 40.Brent TP, Dolan ME, Fraenkelconrat H, Hall J, Karran P, Laval F, Margison GP, Montesano R, Pegg AE, Potter PM, Singer B, Swenberg JA, Yarosh DB. Repair of O4-alkylpyrimidines in mammalian-cells - a present consensus. Proc. Natl. Acad. Sci. U. S. A. 1988;85:1759–1762. doi: 10.1073/pnas.85.6.1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Upadhyaya P, Kalscheuer S, Hochalter JB, Villalta PW, Hecht SS. Quantitation of pyridylhydroxybutyl-DNA adducts in liver and lung of F-344 rats treated with 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone and enantiomers of its metabolite 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanol. Chem. Res. Toxicol. 2008;21:1468–1476. doi: 10.1021/tx8001109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.