Abstract
In this study, we demonstrate that constitutive activation of Raf-1 oncogenic signaling induces stabilization and accumulation of Aurora-A mitotic kinase that ultimately drives the transition from an epithelial to a highly invasive mesenchymal phenotype in estrogen receptor α-positive (ERα+) breast cancer cells. The transition from an epithelial- to a mesenchymal-like phenotype was characterized by reduced expression of ERα, HER-2/Neu overexpression and loss of CD24 surface receptor (CD24 –/low). Importantly, expression of key epithelial-to-mesenchymal transition (EMT) markers and upregulation of the stemness gene SOX2 was linked to acquisition of stem cell-like properties such as the ability to form mammospheres in vitro and tumor self-renewal in vivo. Moreover, aberrant Aurora-A kinase activity induced phosphorylation and nuclear translocation of SMAD5, indicating a novel interplay between Aurora-A and SMAD5 signaling pathways in the development of EMT, stemness and ultimately tumor progression. Importantly, pharmacological and molecular inhibition of Aurora-A kinase activity restored a CD24+ epithelial phenotype that was coupled to ERα expression, downregulation of HER-2/Neu, inhibition of EMT and impaired self-renewal ability, resulting in the suppression of distant metastases. Taken together, our findings show for the first time the causal role of Aurora-A kinase in the activation of EMT pathway responsible for the development of distant metastases in ERα+ breast cancer cells. Moreover, this study has important translational implications because it highlights the mitotic kinase Aurora-A as a novel promising therapeutic target to selectively eliminate highly invasive cancer cells and improve the disease-free and overall survival of ERα+ breast cancer patients resistant to conventional endocrine therapy.
Keywords: breast cancer, stemness, metastases
INTRODUCTION
Breast cancer is one of the most common malignancies affecting women in western countries. Prognosis of breast cancer is strictly dependent on tumor recurrence and development of distant metastases.1 One of the major problems in eradicating metastatic cancer cells is their ability to self-renewal and to gain resistance to conventional anticancer drugs.2 Invasive cancer cells undergo an epithelial-to-mesenchymal transition (EMT) process and acquire a more aggressive phenotype with increased capacity to develop distant metastases.3–5 Specifically, EMT is a physiological process essential during embryonic development during which epithelial cells convert to a mesenchymal cell phenotype characterized by a loss of cell polarity and cell–cell adhesion, and gain active cell motility.6 The increased motility and invasive behavior of cancer cells undergoing EMT is largely due to the loss of adhesion molecules such as E-cadherin and claudin and gain of mesenchymal proteins such as N-cadherin and vimentin.7,8Importantly, it has been demonstrated that breast cancer cells that display EMT and invasive properties also lose the CD24 surface receptor and acquire a CD44+/CD24 –/low cancer stem cell-like phenotype implicated in tumor self-renewal, acquired chemoresistance and tumor progression.9–11 Translational studies have demonstrated a relationship between a higher fraction of CD44+/CD24–/low breast cancer cells within the tumor cell population and shorter disease-free and ovERαll survival and with greater incidence of distant metastases.12
Importantly, CD44+/CD24–/low cancer cells isolated from tumors that were initiated by these cells were able to form tumors when implanted at low density as secondary xenografts, demonstrating their capacity of self-renewal.13 Studies on the molecular characteristics of breast cancer stem cells have demonstrated that CD44+/CD24–/low cells lack differentiated cell lineage markers, are generally estrogen receptor α negative (ERα–) and have a 10- to 50-fold increased ability to form tumors in non-obese diabetic/severe combined immunodeficiency mice when compared with the bulk of tumor cells.14,15 Notably, molecular analysis of the global genomic signature of CD44+ and CD24+ cells (normal and malignant) has shown how CD44+/CD24–/low breast cancer cells show similar gene expression profiles to stem cells.16 Moreover, tumors rich in CD44+/CD24–/low breast cancer cells were characterized by the amplification of transforming growth factor-β signaling pathway, indicating the key role of transforming growth factor-β in the development of EMT, stemness and tumor progression.17,18
Constitutive activation of mitogen-activated protein kinase (MAPK) signaling pathway has also been implicated in the progression of breast cancer through the induction of chemoresistance and development of distant metastases.19–22 Deregulation of MAPK signaling pathway alone is usually not sufficient to induce a fully invasive phenotype, suggesting that additional ‘hits’ are required to promote the generation of metastatic cancer cells.23,24 Notably, cooperation between transforming growth factor-β and sustained MAPK signaling pathways induces EMT and an invasive phenotype in cultured mammary epithelial cells.25 HER-2/Neu is one of the major growth factor receptors overexpressed in approximately 25% of breast tumors that activate the MAPK signaling pathway and is strongly associated with poor patient survival.26 Recent evidence indicates that the constitutive activation of HER-2/Neu pathway induces EMT and has an important role in the maintenance of cancer stem cells.27–32 Although these studies have identified an important role for the MAPK signaling pathway in the development of EMT and clonal expansion of cancer stem cells, the molecular mechanisms linking constitutive MAPK activity with development of EMT, acquisition of a stem cell-like phenotype and tumor progression have not been fully elucidated. Here we identify a novel non-canonical cross-talk between MAPK and Aurora-A oncogenic signalings responsible for the development of distant metastases through the activation of EMT pathway in ERα+ breast cancer cells.
RESULTS
Aberrant activation of MAPK signaling pathway induces the development of metastatic breast cancer xenografts
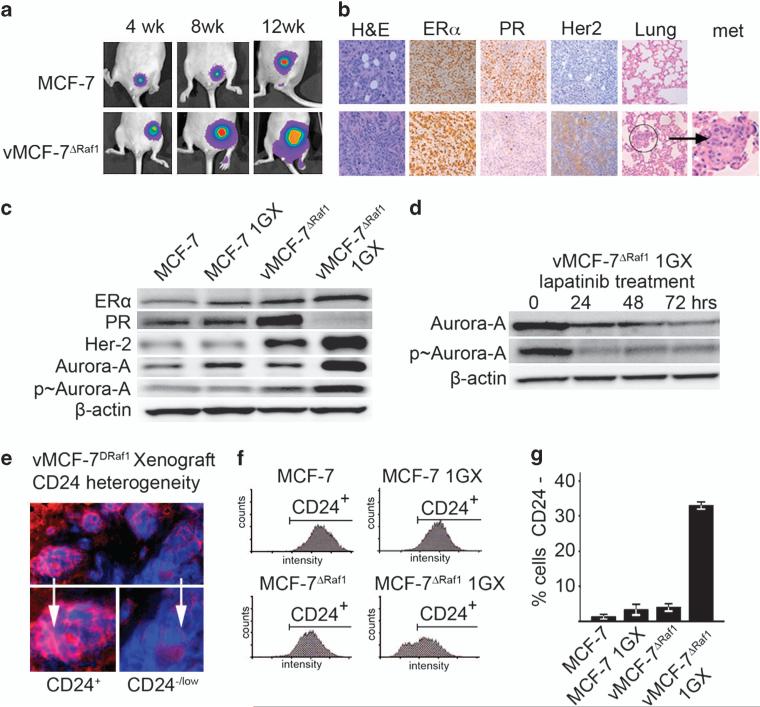
To identify the molecular mechanisms by which aberrant activation of MAPK signaling pathway induces breast cancer progression, we employed a variant MCF-7 cell line overexpressing a constitutive active Raf-1 oncoprotein (vMCF-7ΔRaf-1). vMCF-7ΔRaf-1 cells displayed constitutive phosphorylation of MAPK, retained positivity for ERα and progesterone receptor and showed increased expression of HER-2/Neu compared with parental MCF-7 cells (Supplementary Figure S1A). Immunofluorescence demonstrated the maintenance of an epithelial phenotype of vMCF-7ΔRaf-1 cells characterized by positivity for epithelial markers E-cadherin and β-catenin and lack of expression of the mesenchymal protein vimentin (Supplementary Figure S1B). Next, we developed breast cancer xenograft models employing vMCF-7ΔRaf-1 and parental MCF-7 cells as control to assess the extent to which constitutive activation of MAPK signaling pathway in vivo will induce the development of distant metastases. In contrast to MCF-7 parental xenografts, vMCF-7ΔRaf-1 tumors were ERα+, PR– and HER-2/Neu+, displayed a poorly differentiated phenotype characterized by anaplastic cancer cells with high nuclear pleiomorphism and showed a higher rate of tumor growth, and also gave rise to spontaneous lung metastases by 90 days of tumor growth (Figures 1a and b and Supplementary Figure S1C). vMCF-7ΔRaf-1 breast tumors also exhibited centrosome amplification that was not observed in parental xenografts, indicating deregulation of cell cycle control during tumor progression in breast cancer cells harboring constitutive activation of MAPK pathway (Supplementary Figure S1D).
Figure 1.
Establishment of MCF-7 and vMCF-7ΔRaf-1 breast cancer xenografts. (a) Tumor xenografts imaging in live animals of MCF-7 (upper row) and vMCF-7ΔRaf-1 (lower row) expressing the firefly luciferase reporter lentivector at 4, 8 and 12 weeks after mammary fat pad injection. (b) Paraffin sections of xenograft tumors (12 weeks) showing: hematoxylin and eosin (H&E) staining of low-grade tubular tumors for MCF-7 (upper row) and high-grade vMCF-7ΔRaf-1 tumors (lower row); expression of ER in both xenografts; loss of progesterone receptor (PR) and HER-2/Neu expression in vMCF-7ΔRaf-1 xenografts; and H&E staining of lungs showing development of metastases in vMCF-7Raf-1 xenografts. (c) Immunoblot analysis of parental and cancer cells re-cultured from tumor xenografts (1GX) showing that vMCF-7ΔRaf-1 1GX cells retain the expression of ER, lack expression of progesterone receptor and overexpress HER-2/Neu and Aurora-A. (d) Immunoblot analysis of vMCF-7ΔRaf-1 1GX cells treated with 1 μM lapatinib showing reduced expression of total and p-Aurora-A. (e) Immunofluorescence analysis showing tumor cell heterogeneity for the luminal marker CD24 in vMCF-7ΔRaf-1 xenografts. CD24 receptor was labeled in red and DNA was labeled in blue with Hoechst dye. (f) FACS analysis showing that only vMCF-7ΔRaf-1 1GX cells developed a subpopulation of CD24–/low cells (~30%), while MCF-7, MCF-7 1GX and vMCF-7ΔRaf-1 displayed a CD24+ phenotype. (g) Graph showing the percentage of MCF-7 and variant cells displaying a CD24–/low phenotype from three independent experiments (±s.d.).
Invasive breast cancer cells display Aurora-A kinase overexpression and loss of CD24 epithelial marker
Immunoblotting of re-cultured cells from tumor xenografts (referred to as first generation derived from xenografts, 1GX) confirmed the immunohistochemistry results on breast cancer xenografts (Figure 1c). Because vMCF-7ΔRaf-1 tumor xenografts developed centrosome amplification, we characterized the expression of the mitotic kinase Aurora-A, which has an important role in the development of centrosome abnormalities, genomic instability and tumor progression. Aurora-A expression and phosphorylation was greatly increased in vMCF-7ΔRaf-1 1GX compared with parental cells (Figure 1c). To establish whether HER-2/Neu and Aurora-A overexpression in vMCF-7ΔRaf-1 1GX cells was the result of gene amplification, we performed a fluorescence in situ hybridization analysis employing probes for chromosomes 17, 20 and specific probes for HER-2/Neu and Aurora-A genes and found no indication of amplification of HER-2/Neu and Aurora-A genes in vMCF-7ΔRaf-1 xenografts (Table 1). To determine the extent to which HER-2/Neu and Aurora-A overexpression was due to increased transcriptional activity, we performed quantitative real-time reverse transcription–polymerase chain reaction. While HER-2/Neu mRNAs were overexpressed only in vMCF-7ΔRaf-1 1GX cells, Aurora-A mRNA expression displayed similar levels in all cell lines (Supplementary Figure S1E). These results indicate that HER-2/Neu overexpression is linked to increased transcriptional activity, whereas post-translational mechanisms likely regulate Aurora-A overexpression. To investigate whether Aurora-A overexpression was dependent on HER-2/Neu signaling, vMCF-7ΔRaf-1 1GX cells were treated with lapatinib, a small-molecule inhibitor of HER-2/Neu tyrosine kinase activity (Figure 1d). Evidence of HER-2/Neu inhibition following treatment with 1 μM lapatinib is provided in Supplementary Figure S5A. Following treatment with 1 μM lapatinib, Aurora-A expression and phosphorylation was decreased significantly over time (Figure 1d). Importantly, treatment of vMCF-7ΔRaf-1 1GX cells with lapatinib did not affect the level of expression of Aurora-A mRNA (Supplementary Figure S1F), demonstrating that abnormal accumulation of Aurora-A depends on activation of HER-2/Neu signaling pathway leading to phosphorylation and consequent stabilization of Aurora-A kinase.
Table 1.
FISH analysis of MCF-7 and vMCF-7ΔRaf-1 xenografts
| Summary of the percentage of
cells with HER-2 and CEP 17 FISH probe signals |
Results and
interpretations |
|||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 HER-2 | 2 HER-2 | 3 HER-2 | 4 HER-2 | 5 HER-2 | 6 HER-2 | >7 HER-2 | 1 CEP 17 | 2 CEP 17 | 3 CEP 17 | 4 CEP 17 | 5 CEP 17 | 6 CEP 17 | + HER-2 | + CEP 17 | – HER-2 | – CEP 17 | Interpretation | |
| MCF-7 | 29 | 68 | 2 | 0 | 0 | 0 | 0 | 16 | 40 | 41 | 2 | 0 | 0 | 0 | 0 | 99 | 98 | No gene amplification |
| vMCF-7ΔRaf-1 | 38 | 60 | 1 | 1 | 0 | 0 | 0 | 21 | 40 | 37 | 1 | 0 | 0 | 1 | 1 | 98 | 61 | No gene amplification |
| Summary of the percentage of
cells with Aurora and CEP 20 FISH probe signals |
Results and
interpretations |
|||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 Aurora | 2 Aurora | 3 Aurora | 4 Aurora | 5 Aurora | 6 Aurora | >7 Aurora | 1 CEP 20 | 2 CEP 20 | 3 CEP 20 | 4 CEP 20 | 5 CEP 20 | 6 CEP 20 | + Aurora | + CEP 20 | – Aurora (gr) | – CEP 20 (or) | Interpretation | |
| MCF-7 | 0 | 0 | 9 | 16 | 17 | 20 | 37 | 3 | 16 | 35 | 27 | 13 | 4 | 75 | 18 | 9 | 54 | No gene amplification |
| vMCF-7ΔRaf-1 | 0 | 11 | 25 | 30 | 19 | 6 | 7 | 6 | 23 | 46 | 18 | 5 | 0 | 61 | 22 | 10 | 31 | No gene amplification |
Abbreviations: FISH, fluorescence in situ hybridization. FISH analysis employing probes for chromosomes 17, 20 and specific probes for HER-2 and Aurora-A genes showing that amplification of HER-2/Neu and Aurora-A genes was not present in breast tumor xenografts.
To investigate whether in vivo growth of vMCF-7ΔRaf-1 cells leads to transciptome changes that are linked to their metastatic phenotype, we employed a microarray analysis using human Affymetrix chips to assess global genome expression in MCF-7 and variant cancer cells. From the comparison between breast cancer cells, we identified a differentially expressed invasive gene signature of 10 genes involved in EMT and tumor progression (Supplementary Figures S2A and B). Next, we investigated the status of CD24 epithelial marker to determine whether loss of CD24 receptor was coupled to the development of distant metastases. Significantly, vMCF-7ΔRaf-1 tumor xenografts developed tumor cell heterogeneity for the CD24 receptor (Figure 1e). Reduced expression of CD24 was confirmed in re-cultured cancer cells from vMCF-7ΔRaf-1 xenografts compared with parental cells (Figures 1f and g).
Aurora-A kinase overexpression is functionally linked to EMT and breast cancer stemness
To establish the relationship between loss of CD24 expression and breast cancer progression, we injected vMCF-7ΔRaf-1 and parental MCF-7 cells that express endogenous CD24 receptor into the tail vein of nude mice. The highly invasive basal-like MDA-MB 231 cell line that lacks CD24 expression was used as a positive control. Notably, only MDA-MB 231 cells consistently established lung metastases, whereas mice injected with MCF-7 and vMCF-7ΔRaf-1 cells did not display any evidence of secondary lesions (Figure 2a). Next, we investigated the extent to which loss of CD24 expression in invasive vMCF-7ΔRaf-1 xenografts was linked to a metastatic transcriptome signature. We employed human Affymetrix chips to assess global genome expression in CD24+ and CD24–/low subpopulations isolated from vMCF-7ΔRaf-1 1GX cancer cells by fluorescence-activated cell sorting (FACS) (Figure 2b). Comparison between CD24+ and CD24–/low subpopulations identified an invasive transcriptome signature of 11 genes involved in EMT, stemness and distant metastases (SMAD5, TIMP2, PCDH8, THBS1, CLDN1, SOX2, PAI-RBP1, SKY, NME3, HPSE and CHFR) (Figure 2b). We than determined the expression and localization of important proteins involved in EMT, tumor progression and stemness in CD24+ and CD24–/low cancer cells. The breast cancer EMT marker CD44 and the tyrosine kinase receptor HER-2/Neu were overexpressed in the CD24–/low subpopulation compared with CD24+ cells (Figure 2c). CD24–/low cancer cells showed a mesenchymal-like morphology characterized by the loss of ERα, epithelial markers E-cadherin and β-catenin, and gain of the mesenchymal marker vimentin (Figures 2c and d). Moreover, they overexpressed proteins promoting EMT and stemness (SMAD5 and SOX2) and downregulation of tumor suppressors involved in epithelial cell adhesion (PCDH8), Aurora-A kinase degradation (CHFR), suppression of metastasis (TIMP2) and angiogenesis (THBS1) that were identified earlier in the transcriptome analysis. Importantly, we also detected an increase in phosphorylation of SMAD5 (p-SMAD5) that was linked to its nuclear localization only in the CD24–/low subpopulation (Figures 2c and d) indicative of induction of SMAD5 transcriptional activity during the epithelial–mesenchymal phenotypic switch of breast cancer cells. Notably, the mitotic kinase Aurora-A was overexpressed and displayed a robust phosphorylation only in CD24–/low cancer cells that also showed significant centrosome amplification (Supplementary Figures S3A and B), indicating that Aurora-A kinase activity may promote the genesis of CD24–/low cancer cells (Figure 2c). To establish whether mesenchymal-like CD24–/low cancer cells also displayed stem cell-like characteristics, we tested the ability of CD24+ and CD24–/low cancer cells to grow under non-adherent conditions and to generate mammospheres (an in vitro surrogate assay to determine self-renewal ability) from single-cell suspensions. Notably, only the CD24–/low subpopulation successfully developed mammospheres from single-cell suspensions after three passages (Figure 2e). To validate in vitro the self-renewal ability of the CD24–/low subpopulation, we performed a fluorescence-activated cell sorting (FACS) analysis of CD24+ and CD24–/low cancer cells. Following sorting isolation, CD24+ and CD24–/low subpopulations were cultured for 10 days and FACS analysis was performed to determine CD24 expression. Only the CD24–/low subpopulation reconstituted the extent of CD24 heterogeneity characteristic of bulk vMCF-7ΔRaf-11GX cells, indicative of self-renewal ability (Figure 2f). Finally, we investigated in vivo the tumorigenic activity of CD24+ and CD24–/low subpopulations. CD24–/low cancer cells showed a 20-fold increased ability to form tumors in nude mice compared with CD24+ cancer cells (Figure 2g). Taken together, these results demonstrate that cancer cells harboring a CD24–/low mesenchymal phenotype arise from CD24+ cancer cells in ERα+ breast tumors and display Aurora-A overexpression, activation of EMT pathway and acquisition of stemness properties.
Figure 2.
Molecular characterization of CD24+ and CD24–/low breast cancer cells. (a) Lung metastases imaging in live animals of MDA-MB 231, MCF-7 and vMCF-7ΔRaf-1 cells expressing the firefly luciferase reporter lentivector 1 week after tail vein injection. (b) Heat map representing the unsupervised cluster analysis of global gene expression in CD24+ and CD24–/low subpopulations derived from vMCF-7ΔRaf-1 1GX cells and identification of an invasive transcriptome signature. (c) Immunoblot analysis showing that CD24–/low cells overexpress HER-2/Neu, Aurora-A, EMT and cancer stem cell markers. (d) Immunofluorescence analysis showing activation of EMT in CD24–/low cells characterized by loss of E-cadherin and β-catenin, expression of vimentin and nuclear localization of p-SMAD5. (e) Light microscopy analysis showing development of mammospheres from CD24–/low cells grown under non-adherent conditions at 0, 2, 8 and 24 days (three passages). (f) FACS analysis showing the percentage of CD24+ cells in CD24+ and CD24–/low subpopulations at day 0 and after 10 days following sorting analysis from three independent experiments (±s.d). (g) Tumor xenografts imaging in live animals of CD24+ and CD24–/low cancer cells expressing the firefly luciferase reporter lentivector at 4 weeks after mammary fat pad injection. CD24+ and CD24–/low cells were injected into the mammary fat pad of nude mice and only CD24– cells showed tumorigenic activity at a concentration of 100 000 cells.
Aberrant Aurora-A kinase activity induces EMT and development of CD24low/– cancer cells
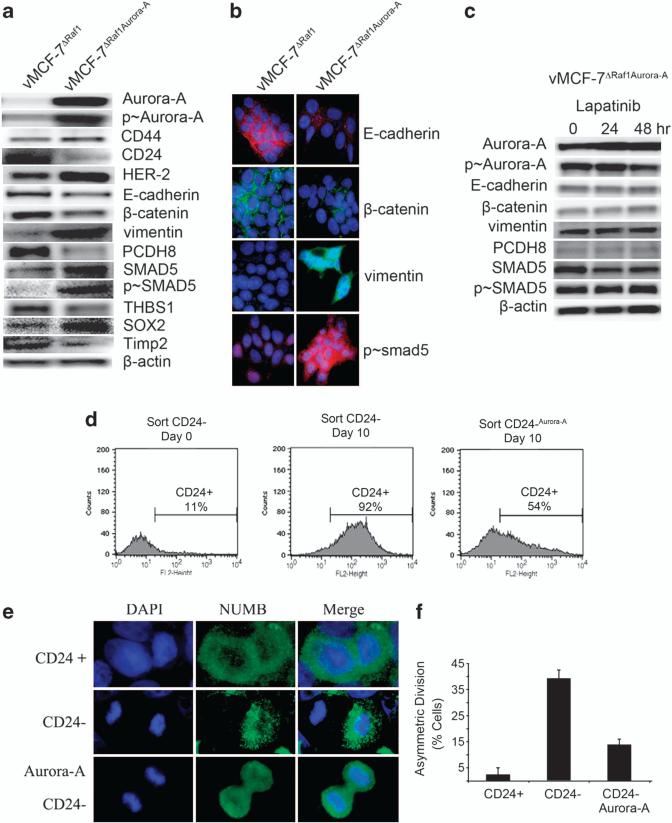
To investigate the causal role of Aurora-A kinase activity in the activation of EMT pathway, we overexpressed Aurora-A kinase to generate vMCF-7Aurora-A and vMCF-7ΔRaf-1/Aurora-A variants. Notably, Aurora-A overexpression in vMCF-7ΔRaf-1 cells induced a similar expression profile as observed in vMCF-7ΔRaf-1 1GX cancer cells. Overexpression of Aurora-A in parental MCF-7 cells resulted in a similar phenotype, although vMCF-7Raf-1/Aurora-A cells displayed a stronger induction of EMT, suggesting a synergism between MAPK and Aurora-A signalings in the activation of EMT pathway (data not shown). Specifically, vMCF-7ΔRaf-1/Aurora-A cells displayed Aurora-A hyperphosphorylation, CD44 and HER-2/Neu overexpression and loss of CD24 leading to the development of EMT and a mesenchymal-like cell phenotype (Figures 3a and b). Importantly, vMCF-7ΔRaf-1/Aurora-A cells showed overexpression of proteins promoting EMT and stemness (SMAD5 and SOX2) and downregulation of tumor suppressors involved in the Aurora-A kinase degradation (CHFR), suppression of metastasis (TIMP2) and angiogenesis (THBS1) previously identified in CD24–/low cancer cells (Figure 3a). We also detected an increase in the phosphorylation status of SMAD5 that was linked to its nuclear localization, demonstrating that Aurora-A kinase activity induced p-SMAD5 and its nuclear activation (Figures 3a and b). Next, we investigated whether activation of EMT pathway in vMCF-7ΔRaf-1/Aurora-A cells was directly dependent on the deregulation of Aurora-A kinase activity or conversely was the result of HER-2/Neu overexpression. Evidence of HER-2/Neu inhibition following treatment with 1 μM lapatinib is provided in Supplementary Figure S5B. As these cells overexpressed the Aurora-A lentivector construct, we did not expect a reduced Aurora-A kinase activity following inhibition of HER-2/Neu signaling as observed in vMCF-7ΔRaf-1 1GX cells (Figure 1d). As expected, treatment with lapatinib did not reduce Aurora-A expression. Importantly, inhibition of HER-2/Neu signaling in vMCF-7DRAF/Aurora cells leads to a reduction of phosphorylation of Aurora-A kinase, demonstrating that phosphorylation/stabilization of Aurora-A kinase remains HER-2-dependent (Figure 3c). Lack of suppression of Aurora-A kinase activity was linked to the maintenance of an EMT phenotype characterized by low expression of E-cadherin and β-catenin, and expression of vimentin and p-SMAD5. In addition, to investigate the causal role of Aurora-A kinase in the induction of stemness, we employed ER+ and HER-2+ BT-474 breast cancer cells that overexpressed Aurora-A kinase (data not shown). BT-474 cells generate mammospheres at high efficiency from single-cell suspensions (Supplementary Figures S6A and B). Treatment of mammospheres with 0.1 μM Alisertib for 72 h greatly reduced the number and size of mammospheres, demonstrating that inhibition of Aurora-A kinase activity impairs the self-renewal capacity of breast cancer cells (Supplementary Figures S6A and B).
Figure 3.
Overexpression of Aurora-A induces EMT and a cancer stem cell-like phenotype. (a) Immunoblot analysis of vMCF-7ΔRaf-1 and vMCF-7ΔRaf-1 cells engineered to overexpress Aurora-A (vMCF-7ΔRaf-1/Aurora-A) showing that vMCF-7ΔRaf-1/Aurora-A cells overexpress CD44, HER-2/Neu and Aurora-A, CD24 downregulation, and expression of EMT and cancer stem cell markers. (b) Immunofluorescence analysis of vMCF-7ΔRaf-1 and vMCF-7ΔRaf-1/Aurora-A cells showing activation of EMT in vMCF-7ΔRaf-1/Aurora-A cells characterized by loss of E-cadherin and β-catenin, expression of vimentin and nuclear localization of p-SMAD5. (c) Immunoblot analysis of vMCF-7ΔRaf-1/Aurora-A cells treated with 1 μM lapatinib showing that constitutive activation of Aurora-A kinase activity is essential for the development of EMT regardless inhibition of HER-2/Neu signaling. (d) FACS analysis showing that sorted CD24–/low cells infected with a lentivector overexpressing Aurora-A maintain a higher percentage of cells carrying a CD24–/low phenotype after 10 days in culture compared with control CD24–/low cells that give rise to a CD24+ subpopulation. (e) Asymmetric division in CD24–/low cells, and symmetric division in CD24+ and CD24–/low cells overexpressing Aurora-A (CD24–/low/Aurora-A). The marker of asymmetric mitotic divisions NUMB was labeled in green and DNA was labeled in blue with Hoechst dye. (f) Graph showing the percentage of cells with asymmetric divisions in CD24+, CD24–/low and CD24–/low/Aurora-A cells from three independent experiments (±s.d.).
Deregulated Aurora-A kinase activity induces the maintenance of CD24–/low cancer cells through impairment of asymmetric cell divisions
To establish at the mechanistic level the causal role of constitutive activation of Aurora-A kinase in the maintenance of a CD24–/low mesenchymal-like phenotype, we performed FACS to isolate the CD24–/low subpopulation from vMCF-7ΔRaf-11GX cells (Figure 3d). CD24–/low cells were grown under culture conditions for 10 days and FACS analysis was performed to determine CD24 expression. A separate group of CD24–/low cells was infected with a lentivector-expressing Aurora-A and cultured for 10 days as the control groups. While 92% of initially CD24–/low cells became CD24+ after 10 days, only 54% of CD24–/low/Aurora-A cells gained positivity for the CD24 receptor, demonstrating that deregulated Aurora-A kinase activity induces the maintenance of a CD24–/low phenotype (Figure 3d). To investigate whether the mechanism by which Aurora-A increased the CD24–/low subpopulation occurs through impairment of asymmetric divisions, we isolated CD24+ and CD24low/– cancer cells from vMCF-7ΔRaf-11GX cancer cells by FACS cell sorting and performed an immunofluorescence assay to label cells with NUMB protein, a well-characterized marker of asymmetric divisions (Figure 3e). While only 3% of CD24+ cells displayed asymmetric partition of NUMB into only one daughter cell, 39% of the CD24–/low cells showed asymmetric partition of NUMB during cell division (Figures 3e and f). Significantly, when CD24–/low cells were infected with a lentivector-expressing Aurora-A, their asymmetric divisions were significantly suppressed (14%), resulting in the maintenance of a CD24–/low phenotype in the majority of cells (Figures 3e and f). In view of the fact that asymmetric divisions of stem cells are compromised in the presence of centrosome amplification,33 we analyzed the centrosome phenotype in vMCF-7ΔRaf-1 cells overexpressing Aurora-A. vMCF-7ΔRaf-1/Aurora-A cells displayed centrosome amplification compared with parental vMCF-7ΔRaf-1 cells (Supplementary Figures S3C and D), indicative of maintenance of CD24–/low cells through the development of centrosome amplification and impairment of asymmetric divisions.
Pharmacological inhibition of Aurora-A kinase activity in vitro reverses EMT and impairs the self-renewal ability of CD24– cancer cells
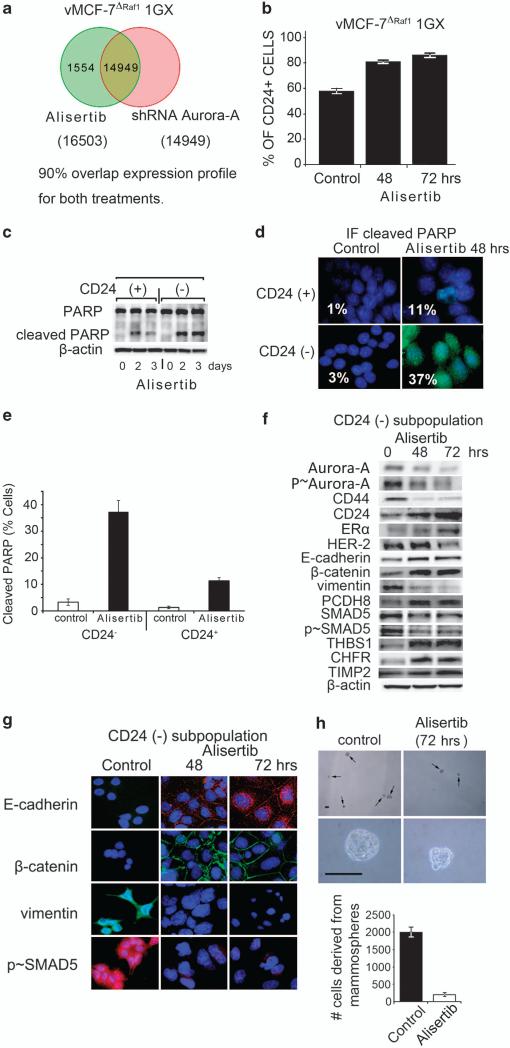
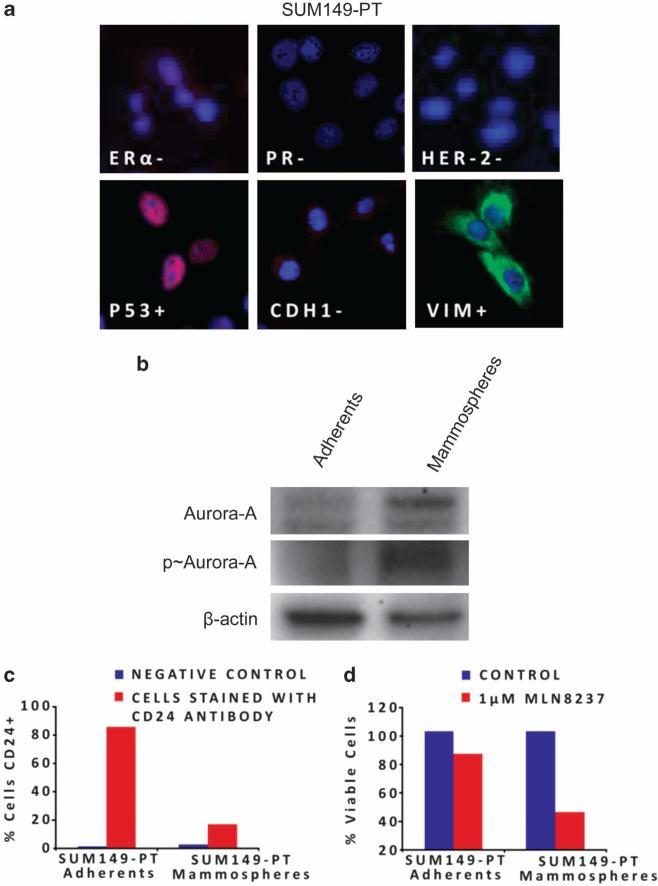
To investigate in vitro the causal role of Aurora-A kinase activity in the activation of EMT pathway and stemness, breast cancer cells were treated with a novel small-molecule inhibitor of Aurora-A, Alisertib (Millennium, Cambridge, MA, USA). First, we established the selectivity of Alisertib in the inhibition of Aurora-A kinase activity through global genome expression in vMCF-7ΔRaf-1 1GX cells that express high levels of endogenous Aurora-A (Figure 1c). vMCF-7ΔRaf-11GX cells were treated with 1 μM Alisertib or with an expression vector carrying short hairpin RNA (shRNA) Aurora-A (Supplementary Figure S4A). The resulting transcriptome analysis showed that treatment with Alisertib or shRNA Aurora-A compared with control cells resulted in 90% overlap gene expression profiles (Figure 4a). To determine the effect of inhibition of Aurora-A kinase activity on cell cycle progression, breast cancer cells were treated with 1 μM Alisertib for 24 h and cells were arrested in the G2/M phase of the cell cycle (Supplementary Figure S4B). Immunoblot analysis of breast cancer cells showed that treatment with 1 μM Alisertib suppressed Aurora-A kinase activity, reduced PLK1 expression and activated the p53-mediated G1/S cell cycle checkpoint. Importantly, activation of apoptosis was detected by cleaved poly-ADP ribose polymerase (PARP) only in vMCF-7ΔRaf-1 1GX cells that express high levels of Aurora-A (Supplementary Figure S4C). These results demonstrate that vMCF-7DRAF1 1GX cells overexpressing displaying Aurora-A are more chemosensitive to molecular inhibition of Aurora-A kinase activity by Alisertib. Treatment of cancer cells with Alisertib also induced a multinucleated phenotype in vMCF-7ΔRaf-1 1GX cells (Supplementary Figure S4D). Next, we wanted to determine whether Alisertib restored a CD24+ epithelial phenotype through selective activation of apoptosis in CD24–/low cells. Treatment of vMCF-7ΔRaf-1 1GX cells with 1 μM Alisertib significantly increased the percentage of cells expressing the CD24 receptor (Figure 4b). Importantly, we determined that the genesis of a CD24+ phenotype following treatment with Alisertib was linked to a higher cytotoxic effect in the CD24–/low subpopulation that overexpressed Aurora-A (Figure 4c). Additionally, at the cellular level, ~37% of CD24–/low cells displayed cleaved PARP following Alisertib treatment in contrast to CD24+ cells where only ~11% showed activation of apoptosis (Figures 4d and e). These results demonstrate that CD24–/low cancer cells were more sensitive to the cytotoxic effects of Alisertib. To establish whether Alisertib also induced a CD24+ epithelial phenotype through reversion of EMT, we isolated the CD24–/low subpopulation from bulk vMCF-7ΔRaf-1 1GX cells by FACS sorting and CD24–/low cells were treated with 1 μM Alisertib (Figure 4f). At the molecular level, inhibition of Aurora-A kinase activity increased the expression of ERα and CD24, while CD44 and HER-2/Neu expressions were significantly reduced (Figure 4f). Reversion of EMT by Alisertib treatment was characterized by increased expression of epithelial markers E-cadherin, β-catenin and PCDH8 and loss of the mesenchymal marker vimentin, demonstrating that Aurora-A kinase activity has an important role in the development of EMT and the genesis of CD24–/low mesenchymal-like cancer cells (Figures 4f and g). Treatment with Alisertib was also coupled to decreased p-SMAD5, demonstrating a mechanistic linkage between Aurora-A kinase activity and nuclear activation of SMAD5 in the development of EMT (Figures 4f and g). Moreover, inhibition of Aurora-A kinase activity induced expression of THBS1, CHFR and TIMP2 tumor suppressors (Figures 4f and g). To investigate the extent to which inhibition of Aurora-A kinase activity also interferes with self-renewal capacity and stemness of CD24–/low cells, we treated cell suspensions generated from the CD24–/low subpopulation with 0.1 μM Alisertib for 72 h. Treatment with Alisertib greatly reduced the number and size of mammospheres, demonstrating that inhibition of Aurora-A kinase activity impairs the self-renewal ability of CD24–/low cells (Figure 4h). Moreover, to validate the chemosensitivity of CD24–/low cells to Alisertib in a different breast cancer cell model, we employed the triple-negative SUM149-PT cells (Figure 5a). Mammospheres derived from SUM149-PT cells displayed an increased Aurora-A kinase activity compared with adherent cells (Figure 5b). Importantly, mammo-spheres developed a CD24–/low phenotype compared with adherent cells and were more sensitive to the cytotoxic effects of Alisertib (Figures 5c and d). Taken together, these results demonstrate for the first time the causal role of Aurora-A kinase activity in the activation of EMT pathway and the genesis of CD24–/low mesenchymal-like cancer cells.
Figure 4.
Molecular inhibition of Aurora-A kinase activity in vitro reverses EMT and suppresses self-renewal ability of CD24–/low breast cancer cells. (a) Venn diagram of the unsupervised cluster analysis of global gene expression between vMCF-7ΔRaf-1 1GX cells treated with Alisertib or with an shRNA Aurora-A vector showing 90% overlap expression profile. (b) Graph showing FACS analysis of vMCF-7ΔRaf-1 1GX cells treated with 1 μM Alisertib for 48 and 72 h displaying a CD24+ phenotype from three independent experiments (±s.d). (c) Immunoblot analysis of CD24+ and CD24–/low cells showing that treatment with 1 μM Alisertib induced cleaved PARP mainly in the CD24–/low subpopulation. (d) Immunofluorescence analysis of CD24+ and CD24–/low cells treated with 1 μM Alisertib for 48 h showing activation of apoptosis mainly in CD24–/low cells. Cleaved PARP was labeled in green and DNA was labeled in blue with Hoechst dye. (e) Graph showing the percentage of CD24+ and CD24–/low cells displaying cleaved PARP before and after treatment with Alisertib. Experiments were performed in triplicate (±s.d.). (f) Immunoblot analysis of CD24–/low cells treated with Alisertib for 48 and 72 h showing reversion of EMT and restoration of an epithelial CD44–/CD24+ phenotype. (g) Immunofluorescence analysis of CD24–/low cells treated with1 μM MLN8237 for 48 and 72 h showing reversion of EMT. E-cadherin and p-SMAD5 were labeled in red, β-catenin and vimentin were labeled in green, and DNA was labeled in blue with Hoechst dye. (h) Light microscopy analysis showing inhibition of mammospheres growth derived from CD24–/low cells after treatment with 0.1 μM Alisertib for 72 h. Upper panels: low magnification; lower panels: high magnification. Graph showing the percentage of cells derived from mammospheres before and after treatment with Alisertib. Experiments were performed in triplicate (±s.d.).
Figure 5.
Chemosensitivity of SUM149-PT breast cancer cells to Alisertib. (a) Immunofluorescence showing a triple-negative and fibroblastoid-like phenotype of SUM149-PT cells. ER, PR, P53, CDH1 were labeled in red, HER-2 and Vimentin were labeled in green and DNA was labeled in blue with Hoechst dye. (b) Immublotting assay showing increased expression and activity of Aurora-A kinase in SUM149-PT mammospheres. (c) FACS analysis showing loss of CD24 receptor in SUM149-PT mammospheres. (d) 3-[4,5-Dimethylthiazol-2-yl]-2,5-diphenyl tetrazolium bromide (MTT) assay showing chemosensitivity of SUM149-PT mammospheres to Alisertib.
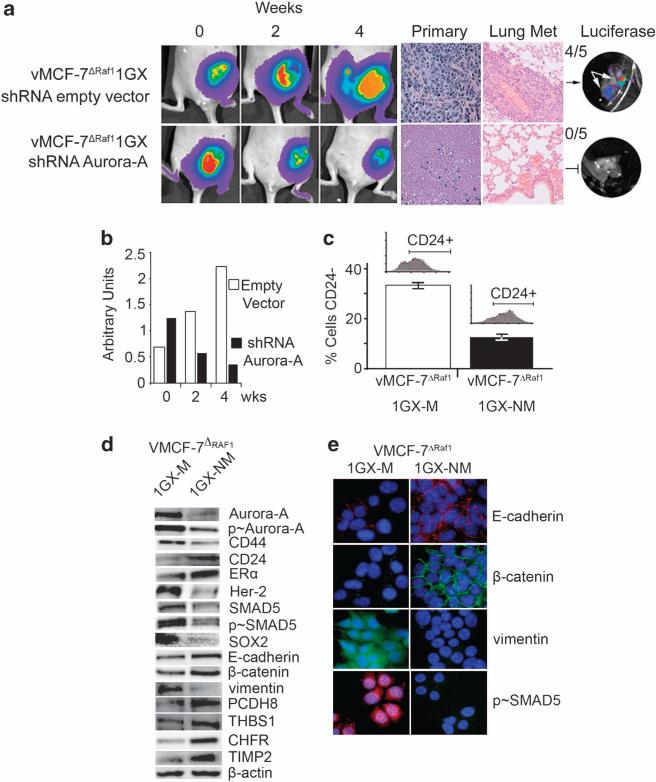
Molecular targeting of Aurora-A kinase activity in vivo restores a CD24+ epithelial phenotype and inhibits the development of distant metastases
We employed the shRNA Aurora-A vector (Supplementary Figure S4A) to inhibit in vivo Aurora-A kinase activity and suppress the development of breast cancer metastases. First, we established vMCF-7ΔRaf-1 xenograft orthotopic tumors expressing lucifERαse to monitor tumor growth in living animals over a period of 8 weeks (Figure 6a). Following 8 weeks of tumor growth, primary tumors were injected with vectors expressing Aurora-A-targeted shRNAs (lower row), while a separate group of animals were injected with ‘empty’ vectors and used as control groups (upper row). Three intratumoral sites were injected for each animal and repeated three times a week for up to 4 weeks, the animals were than euthanized, primary breast tumors and lungs were removed, prepared for histology and assayed to determine the presence of metastatic dissemination. vMCF-7ΔRaf-1 xenografts that did not receive treatment with shRNA Aurora-A developed a poorly differentiated phenotype that was associated with the development of lung metastases (Figure 6a). Remarkably, the Aurora-A-targeted shRNA treatment resulted in significant tumor shrinkage, extensive necrotic tumor tissue and failure to give rise to lung metastases (Figures 6a and b). To determine whether the mechanism responsible for the suppression of lung metastases was linked to the decrease of CD24–/low mesenchymal subpopulation, we re-cultured cancer cells from metastatic and nonmetastatic xenografts (vMCF-7ΔRaf-1 1GX-M and vMCF-7ΔRaf-1 1GX-NM, respectively). Importantly, FACS analysis showed that the percentage of CD24–/low cancer cells was significantly decreased in vMCF-7ΔRaf-1 1GX-NM cells (~15%) compared with the invasive vMCF-7ΔRaf-1 1GX-M cells (~32%) (Figure 6c). At the molecular level, vMCF-7ΔRaf-1 1GX-NM cells displayed suppression of EMT pathway linked to a CD24+ epithelial phenotype, gain of ERα expression, loss of Aurora-A phosphorylation and reduced CD44 and HER-2/Neu expression in contrast to vMCF-7ΔRaf-1 1GX-M cells (Figure 6d). Suppression of EMT was linked to reduced expression and phosphorylation levels of SMAD5, reduced expression of the stemness gene SOX2, re-expression of epithelial markers E-cadherin, β-catenin and PCDH8 and loss of the mesenchymal marker vimentin (Figures 6d and e). Moreover, the lack of development of distant metastases in vMCF-7ΔRaf-1 1GX-NM cells was linked to increased expression of inhibitors of Aurora-A kinase activity (CHFR), invasion (TIMP2) and angiogenesis (THBS1) (Figure 6d). Taken together, these results demonstrate that deregulated Aurora-A kinase activity induces EMT in initially CD24+ breast cancer cells, conferring the ability to develop a CD24–/low mesenchymal-like phenotype and metastasize to distant organs. Importantly, they also validate in vivo the causal role of Aurora-A kinase activity in the induction of EMT responsible for the genesis of CD24–/low mesenchymal-like cancer cells and ultimately distant metastases.
Figure 6.
Molecular targeting of Aurora-A kinase activity in vivo restores an epithelial phenotype and suppresses breast cancer metastases. (a) Tumor imaging in live animals of vMCF-7ΔRaf-1 xenografts expressing the firefly luciferase reporter lentivector. Following 8 weeks growth, tumor xenografts were treated with an empty shRNA vector (upper row) and an shRNA targeting Aurora-A (lower row) and imaged at 0, 2 and 4 weeks after treatment. Paraffin sections of xenograft tumors showing hematoxylin and eosin (H&E) staining of primary tumors and lung tissue. (b) Graph showing the area of tumor xenografts growth using NIH Image J program from three independent experiments. (c) Graph showing that nonmetastatic cells (vMCF-7ΔRaf-1 1GX-NM) derived from tumor xenografts treated with shRNA Aurora-A decreased the percentage of CD24–/low cells compared with control metastatic cells (vMCF-7ΔRaf-1 1GX-M). The percentage of CD24–/low cells was detected by FACS analysis from three independent experiments (±s.d). (d) Immunoblot analysis of vMCF-7ΔRaf-1 1GX-M and vMCF-7ΔRaf-1 1GX-NM cells showing suppression of EMT and restoration of an epithelial phenotype in vMCF-7ΔRaf-1 1GX-NM cells. (e) Immunofluorescence analysis showing expression of epithelial markers E-cadherin and β-catenin, lack of vimentin and inhibition of nuclear SMAD5 phosphorylation in vMCF-7ΔRaf-1 1GX-NM cells. E-cadherin and p-SMAD5 were labeled in red, β-catenin and vimentin were labeled in green and DNA was labeled in blue with Hoechst dye.
DISCUSSION
In this study, we established a novel oncogenic cross-talk between Raf/MAPK and Aurora-A signaling pathways in the development of EMT, stemness and tumor progression in ERα+ breast cancer cells. Earlier, it was demonstrated that constitutive activation of Raf/MAPK signaling pathway drives EMT, stem-like properties and metastases.34,35 Moreover, recent findings also demonstrate that Raf-1 gene amplification activates p-ERK-β-catenin signaling to promote the expansion of breast cancer stem cells.36 Overexpression of HER-2/Neu also induces EMT, chemoresistance and breast cancer metastases and is found in approximately 25% of invasive breast carcinomas and is strongly associated with poor patient survival.37,38 However, the presence of HER-2/Neu overexpression and increased MAPK activity in 50–60% ductal carcinoma in situ raises the question of whether other oncogenic signaling pathways are necessary during tumor growth to induce EMT, stemness and the transition from a non-invasive to a fully invasive breast cancer phenotype.39
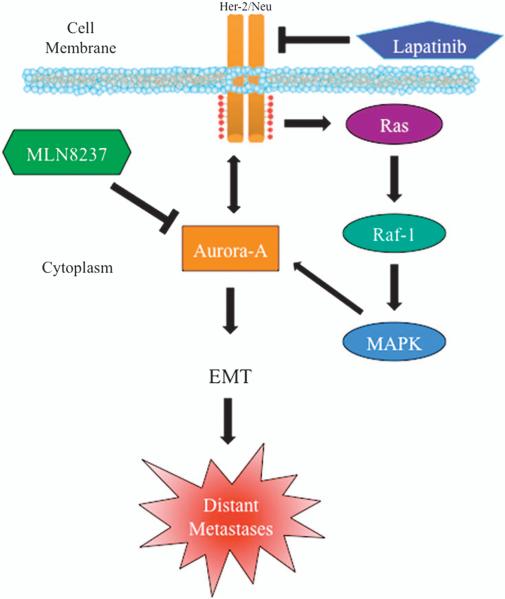
Here we show that constitutive activation of the MAPK alone is not sufficient to confer EMT in cultured ERα+ vMCF-7ΔRaf-1 breast cancer cells. However, during in vivo growth vMCF-7ΔRaf-1 tumor cell xenografts acquired tumor cell heterogeneity for CD24 epithelial marker, overexpression of HER-2/Neu receptor and gave rise to distant metastases. At the molecular level, HER-2/Neu overexpression induced accumulation of the mitotic kinase Aurora-A, which was associated with a CD24–/low mesenchymal phenotype and cancer stem-like cell properties, including activation of SMAD5, self-renewal, the ability to form mammospheres from single-cell suspensions and high tumorigenic activity in vivo. Importantly, another alternative mechanism to induce Aurora-A overexpression is linked to the loss of CHFR, a tumor suppressor involved in Aurora-A degradation.40 We also identified the mechanism by which Aurora-A maintains the CD24–/low mesenchymal phenotype through the development of centrosome amplification and disruption of asymmetric divisions. Overexpression of Aurora-A induced the upregulation of HER-2/Neu, demonstrating a positive feedback loop between these oncogenic signaling pathways, yet inhibition of HER-2/Neu signaling did not reverse EMT, thereby demonstrating that the Aurora-A kinase activity is downstream to HER-2/Neu signaling. Molecular inhibition of Aurora-A kinase activity in vitro and in vivo resulted in the suppression of HER-2/Neu expression, reduced p-SMAD5, gain of a CD24+ epithelial phenotype and suppression of distant metastases. Taken together, these experiments demonstrate for the first time a non-mitotic role for Aurora-A kinase in the development of EMT, stemness and tumor progression (Figure 7). Finally, this study highlights the translational relevance of Aurora-A kinase as a promising therapeutic target to suppress EMT and tumor progression in ERα+ breast cancer patients.
Figure 7.
Induction of EMT and breast cancer progression through aberrant activation of Aurora-A kinase activity. Constitutive activation of HER-2/MAPK signaling pathway during tumor growth leads to stabilization and accumulation of Aurora-A kinase. Aurora-A kinase activation in turn increases the expression levels of HER-2/Neu receptor, creating a ‘positive feedback loop’ between HER-2/Neu and Aurora-A tumorigenic activity. Aberrant Aurora-A kinase activity induces EMT and the development of a mesenchymal phenotype responsible for distant metastases. Targeting Aurora-A kinase signaling pathway can be effective for eliminating highly invasive breast cancer cells and suppress tumor progression.
MATERIALS AND METHODS
Human breast cancer cell lines
The human breast cancer cell lines MCF-7, MDA-MB 231, SUM149-PT and BT-474 were obtained from ATCC (Manassas, VA, USA). The MCF-7 cells overexpressing the Raf-1 oncoprotein were generated as described previously.41
Cell cycle profile, indirect immunofluorescence and immunoblotting
For FACS, indirect immunofluorescence and protein expression analyses, breast cancer cells were treated as described previously.42,43,44
Antibodies
Antibodies employed in this study were as follows: tMAPK, p-MAPK, tHER-2/Neu, p-HER-2/Neu, Aurora-A, p-Aurora-A, SMAD5, p-SMAD5, SOX2, E-cadherin, CHFR (Cell Signaling Technology, Boston, MA, USA); CD44, CD24 (Abcam, Cambridge Science Park, UK); ERα, PR, β-catenin, vimentin, PCDH8 (Santa Cruz Biotechnology, Delaware Avenue Santa Cruz, CA, USA); THBS1 (LSBio, Seattle, WA, USA) and β-actin (Sigma, Northbrook, IL, USA).
Human breast cancer xenografts
Procedures established by the Institutional Animal Care and Use Committee based on US NIH guidelines for the care and use of laboratory animals were followed for all experiments. A 4-week-old non-ovariectomized female NCR/Nu/Nu nude mice were anesthetized by exposure to 3% isoflurane and injected subcutaneously with 2 × 106 cells suspended in 50 ml of 50% Matrigel (BD Bioscience, Bedford, MA, USA). Tumor localization and growth was monitored using the IVS imaging system from the ventral view 10 min after luciferin injection. After 12 weeks, mice were killed and xenograft tumors were processed for histology, immunohistochemistry and immunofluorescence analyses as described previously.43,44 To re-establish cultures from 1GX explants, tumors were excised from killed animals, minced using sterile scissors, transferred to complete culture medium and fibroblast-free tumor cell lines were established by serial passages in culture. A separated group of nude mice (five in each group) received intratumoral injection (three times a week) of 100 μg of PSSH1 empty shRNA (control) or PSSH1 Aurora-A shRNA plasmids diluted in 100 μl of FuGENE 6 (Roche, Indianapolis, IN, USA) for a total of 4 weeks.
Isolation of CD24+ and CD24–/low cancer cells
CD24+ and CD24–/low subpopulations were isolated from 20 × 106 vMCF-7Raf-1 1GX cells by FACS analysis employing a CD24-FITC antibody (BD Pharmingen, San Jose, CA, USA) according to the manufacturer's protocol.
Mammospheres formation
CD24+ and CD24–/low cells were plated in ultra-low attachment 96-well culture dishes in 100 μl of MammoCult medium (STEMCELL Technologies, Vancouver, BC, Canada), 25 μl of medium was added every 2 days for a maximum of 8 days. Mammospheres’ formation was recorded every 2 days through a digital camera (Nikon, Melville, NY, USA).
Genomic analysis
Gene microarray and cytogenetic analyses were performed as described previously.5 The raw data regarding the transcriptome analysis can be accessed at: http://www.ncbi.nlm.nih.gov/geo/info/linking.html.
Quantitative real-time reverse transcription–polymerase chain reaction
Quantitative real-time reverse transcription–polymerase chain reaction was performed by employing the LightCycler 480 Real-Time PCR System (Roche) according to the manufacturer's protocol. The probes for HER-2/Neu (left primer: 5′-GGGAAACCTGGAACTCACCT-3′; right primer: 5′-AGCGATGAGCACGTAGCC-3′) and Aurora-A (left primer: 5′-TGTTTCTT TTTATTTGTCTTTGTCTGA-3′; right primer: 5′-TGAAATTAGCACACTGTCCA CAA-5′) genes were selected by searching the Universal Probe Library for Human (Roche).
Construction of shRNA vectors
The PSSH1 shRNA suppression plasmid was described previously. Briefly, it contains the H1 RNA polymerase III-dependent promoter for the generation of shRNA molecules. shRNA oligos directed against the 39 untranslated region of Aurora A (5′-TAGGGATTTGCTTGGGATA-3′) were annealed and cloned into the Bg/II/HindIII cloning site at the 3′ end of the RNA polymerase III-dependent H1 RNA promoter-driven vector. Clones containing the insert were identified and sequenced to ensure fidelity.
Supplementary Material
ACKNOWLEDGEMENTS
This study was supported by NCI CA72836 to JLS, USAMRMC BC022276 and Intramural RECDA Award to ABD, the Italian Association for Cancer Research (AIRC) to AA, the Mayo Clinic Breast Cancer Specialized Program of Research Excellence NIH CA116201 to JI and the Mayo Clinic School of Medicine. This study was also supported by the Atwater Foundation. We also wish to acknowledge the Cytogenetic Shared Resource and TACMA core facilities of the Mayo Clinic Comprehensive Cancer Center for performing SKY, routine cytogenetic and immunohistochemistry analysis and assisting us with the interpretation of the results.
Footnotes
CONFLICT OF INTEREST
The authors declare no conflict of interest.
REFERENCES
- 1.Basaran G, Devrim C, Caglar HB, Gulluoglu B, Kaya H, Seber S, et al. Clinical outcome of breast cancer patients with N3a (>/ = 10 positive lymph nodes) disease: has it changed over years? Med Oncol. 2010;28:726–732. doi: 10.1007/s12032-010-9516-1. [DOI] [PubMed] [Google Scholar]
- 2.Chang H, Rha SY, Jeung HC, Im CK, Ahn JB, Kwon WS, et al. Association of the ABCB1 gene polymorphisms 2677G>T/A and 3435C>T with clinical outcomes of paclitaxel monotherapy in metastatic breast cancer patients. Ann Oncol. 2009;20:272–277. doi: 10.1093/annonc/mdn624. [DOI] [PubMed] [Google Scholar]
- 3.Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY, et al. The epithelial–mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hwang-Verslues WW, Kuo WH, Chang PH, Pan CC, Wang HH, Tsai ST, et al. Multiple lineages of human breast cancer stem/progenitor cells identified by profiling with stem cell markers. PLoS One. 2009;4:e8377. doi: 10.1371/journal.pone.0008377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.D'Assoro AB, Leontovich A, Amato A, Ayers-Ringler JR, Quatraro C, Hafner K, et al. Abrogation of p53 function leads to metastatic transcriptome networks that typify tumor progression in human breast cancer xenografts. Int J Oncol. 2010;37:1167–1176. doi: 10.3892/ijo_00000768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guarino M, Rubino B, Ballabio G. The role of epithelial–mesenchymal transition in cancer pathology. Pathology. 2007;39:305–318. doi: 10.1080/00313020701329914. [DOI] [PubMed] [Google Scholar]
- 7.Beavon IR. The E-cadherin–catenin complex in tumour metastasis: structure, function and regulation. Eur J Cancer. 2000;36:1607–1620. doi: 10.1016/s0959-8049(00)00158-1. [DOI] [PubMed] [Google Scholar]
- 8.Strathdee G. Epigenetic versus genetic alterations in the inactivation of E-cadherin. Semin Cancer Biol. 2002;12:373–379. doi: 10.1016/s1044-579x(02)00057-3. [DOI] [PubMed] [Google Scholar]
- 9.Dean M, Fojo T, Bates S. Tumour stem cells and drug resistance. Nat Rev Cancer. 2005;5:275–284. doi: 10.1038/nrc1590. [DOI] [PubMed] [Google Scholar]
- 10.Turley EA, Veiseh M, Radisky DC, Bissell MJ. Mechanisms of disease: epithelial–mesenchymal transition—does cellular plasticity fuel neoplastic progression? Nat Clin Pract Oncol. 2008;5:280–290. doi: 10.1038/ncponc1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wicha MS, Liu S, Dontu G. Cancer stem cells: an old idea—a paradigm shift. Cancer Res. 2006;66:1883–1890. doi: 10.1158/0008-5472.CAN-05-3153. discussion 95–96. [DOI] [PubMed] [Google Scholar]
- 12.Abraham BK, Fritz P, McClellan M, Hauptvogel P, Athelogou M, Brauch H. Prevalence of CD44 +/CD24−/low cells in breast cancer may not be associated with clinical outcome but may favor distant metastasis. Clin Cancer Res. 2005;11:1154–1159. [PubMed] [Google Scholar]
- 13.Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci USA. 2003;100:3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Honeth G, Bendahl PO, Ringner M, Saal LH, Gruvberger-Saal SK, Lovgren K, et al. The CD44+/CD24− phenotype is enriched in basal-like breast tumors. Breast Cancer Res. 2008;10:R53. doi: 10.1186/bcr2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Giatromanolaki A, Sivridis E, Fiska A, Koukourakis MI. The CD44 /CD24 phenotype relates to ‘triple-negative’ state and unfavorable prognosis in breast cancer patients. Med Oncol. 2011;28:745–752. doi: 10.1007/s12032-010-9530-3. [DOI] [PubMed] [Google Scholar]
- 16.Shipitsin M, Campbell LL, Argani P, Weremowicz S, Bloushtain-Qimron N, Yao J, et al. Molecular definition of breast tumor heterogeneity. Cancer Cell. 2007;11:259–273. doi: 10.1016/j.ccr.2007.01.013. [DOI] [PubMed] [Google Scholar]
- 17.Wendt MK, Smith JA, Schiemann WP. Transforming growth factor-beta-induced epithelial–mesenchymal transition facilitates epidermal growth factor-dependent breast cancer progression. Oncogene. 2010;29:6485–6498. doi: 10.1038/onc.2010.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Murohashi M, Hinohara K, Kuroda M, Isagawa T, Tsuji S, Kobayashi S, et al. Gene set enrichment analysis provides insight into novel signalling pathways in breast cancer stem cells. Br J Cancer. 2009;102:206–212. doi: 10.1038/sj.bjc.6605468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eralp Y, Derin D, Ozluk Y, Yavuz E, Guney N, Saip P, et al. MAPK overexpression is associated with anthracycline resistance and increased risk for recurrence in patients with triple-negative breast cancer. Ann Oncol. 2008;19:669–674. doi: 10.1093/annonc/mdm522. [DOI] [PubMed] [Google Scholar]
- 20.Jin W, Wu L, Liang K, Liu B, Lu Y, Fan Z. Roles of the PI-3K and MEK pathways in Ras-mediated chemoresistance in breast cancer cells. Br J Cancer. 2003;89:185–191. doi: 10.1038/sj.bjc.6601048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ghayad SE, Vendrell JA, Larbi SB, Dumontet C, Bieche I, Cohen PA. Endocrine resistance associated with activated ErbB system in breast cancer cells is reversed by inhibiting MAPK or PI3K/Akt signaling pathways. Int J Cancer. 2010;126:545–562. doi: 10.1002/ijc.24750. [DOI] [PubMed] [Google Scholar]
- 22.Whyte J, Bergin O, Bianchi A, McNally S, Martin F. Key signalling nodes in mammary gland development and cancer. Mitogen-activated protein kinase signalling in experimental models of breast cancer progression and in mammary gland development. Breast Cancer Res. 2009;11:209. doi: 10.1186/bcr2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang F, Weaver VM, Petersen OW, Larabell CA, Dedhar S, Briand P, et al. Reciprocal interactions between beta1-integrin and epidermal growth factor receptor in three-dimensional basement membrane breast cultures: a different perspective in epithelial biology. Proc Natl Acad Sci USA. 1998;95:14821–14826. doi: 10.1073/pnas.95.25.14821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marampon F, Ciccarelli C, Zani BM. Down-regulation of c-Myc following MEK/ERK inhibition halts the expression of malignant phenotype in rhabdomyosarcoma and in non muscle-derived human tumors. Mol Cancer. 2006;5:31. doi: 10.1186/1476-4598-5-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim ES, Kim MS, Moon A. Transforming growth factor (TGF)-beta in conjunction with H-ras activation promotes malignant progression of MCF10A breast epithelial cells. Cytokine. 2005;29:84–91. doi: 10.1016/j.cyto.2004.10.001. [DOI] [PubMed] [Google Scholar]
- 26.Slamon DJ, Godolphin W, Jones LA, Holt JA, Wong SG, Keith DE, et al. Studies of the HER-2/neu proto-oncogene in human breast and ovarian cancer. Science. 1989;244:707–712. doi: 10.1126/science.2470152. [DOI] [PubMed] [Google Scholar]
- 27.Muthuswamy SK, Li D, Lelievre S, Bissell MJ, Brugge JS. ErbB2, but not ErbB1, reinitiates proliferation and induces luminal repopulation in epithelial acini. Nat Cell Biol. 2001;3:785–792. doi: 10.1038/ncb0901-785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aranda V, Haire T, Nolan ME, Calarco JP, Rosenberg AZ, Fawcett JP, et al. Par6-aPKC uncouples ErbB2 induced disruption of polarized epithelial organization from proliferation control. Nat Cell Biol. 2006;8:1235–1245. doi: 10.1038/ncb1485. [DOI] [PubMed] [Google Scholar]
- 29.Kim IY, Yong HY, Kang KW, Moon A. Overexpression of ErbB2 induces invasion of MCF10A human breast epithelial cells via MMP-9. Cancer Lett. 2009;275:227–233. doi: 10.1016/j.canlet.2008.10.013. [DOI] [PubMed] [Google Scholar]
- 30.Korkaya H, Paulson A, Iovino F, Wicha MS. HER2 regulates the mammary stem/ progenitor cell population driving tumorigenesis and invasion. Oncogene. 2008;27:6120–6130. doi: 10.1038/onc.2008.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roesler R, Cornelio DB, Abujamra AL, Schwartsmann G. HER2 as a cancer stem-cell target. Lancet Oncol. 2010;11:225–226. doi: 10.1016/S1470-2045(09)70404-8. [DOI] [PubMed] [Google Scholar]
- 32.Korkaya H, Wicha MS. Cancer stem cells: nature versus nurture. Nat Cell Biol. 2010;12:419–421. doi: 10.1038/ncb0510-419. [DOI] [PubMed] [Google Scholar]
- 33.Basto R, Brunk K, Vinadogrova T, Peel N, Franz A, Khodjakov A, et al. Centrosome amplification can initiate tumorigenesis in flies. Cell. 2008;133:1032–1042. doi: 10.1016/j.cell.2008.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial–mesenchymal transitions in development and disease. Cell. 2009;139:871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 35.Kerkhoff E, Fedorov LM, Siefken R, Walter AO, Papadopoulos T, Rapp UR. Lung-targeted expression of the c-Raf-1 kinase in transgenic mice exposes a novel oncogenic character of the wild-type protein. Cell Growth Differ. 2000;11:185–190. [PubMed] [Google Scholar]
- 36.Chang CJ, Yang JY, Xia W, Chen CT, Xie X, Chao CH, et al. EZH2 promotes expansion of breast tumor initiating cells through activation of RAF1-beta-catenin signaling. Cancer Cell. 2011;19:86–100. doi: 10.1016/j.ccr.2010.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yu D, Hung MC. Overexpression of ErbB2 in cancer and ErbB2-targeting strategies. Oncogene. 2000;19:6115–6121. doi: 10.1038/sj.onc.1203972. [DOI] [PubMed] [Google Scholar]
- 38.Yu D, Hung MC. Role of erbB2 in breast cancer chemosensitivity. BioEssays. 2000;22:673–680. doi: 10.1002/1521-1878(200007)22:7<673::AID-BIES10>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 39.Lu J, Guo H, Treekitkarnmongkol W, Li P, Zhang J, Shi B, et al. 14-3-3zeta Cooperates with ErbB2 to promote ductal carcinoma in situ progression to invasive breast cancer by inducing epithelial–mesenchymal transition. Cancer Cell. 2009;16:195–207. doi: 10.1016/j.ccr.2009.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yu X, Minter-Dykhouse K, Malureanu L, Zhao WM, Zhang D, Merkle CJ, et al. Chfr is required for tumor suppression and Aurora A regulation. Nat Genet. 2005;37:401–406. doi: 10.1038/ng1538. [DOI] [PubMed] [Google Scholar]
- 41.Davis JM, Navolanic PM, Weinstein-Oppenheimer CR, Steelman LS, Hu W, Konopleva M, et al. Raf-1 and Bcl-2 induce distinct and common pathways that contribute to breast cancer drug resistance. Clin Cancer Res. 2003;9:1161–1170. [PubMed] [Google Scholar]
- 42.D'Assoro AB, Barrett SL, Folk C, Negron VC, Boeneman K, Busby R, et al. Amplified centrosomes in breast cancer: a potential indicator of tumor aggressiveness. Breast Cancer Res Treat. 2002;75:25–34. doi: 10.1023/a:1016550619925. [DOI] [PubMed] [Google Scholar]
- 43.D'Assoro AB, Busby R, Suino K, Delva E, Almodovar-Mercado GJ, Johnson H, et al. Genotoxic stress leads to centrosome amplification in breast cancer cell lines that have an inactive G1/S cell cycle checkpoint. Oncogene. 2004;23:4068–4075. doi: 10.1038/sj.onc.1207568. [DOI] [PubMed] [Google Scholar]
- 44.D'Assoro AB, Busby R, Acu ID, Quatraro C, Reinholz MM, Farrugia DJ, et al. Impaired p53 function leads to centrosome amplification, acquired ERalpha phenotypic heterogeneity and distant metastases in breast cancer MCF-7 xenografts. Oncogene. 2008;27:3901–3911. doi: 10.1038/onc.2008.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.